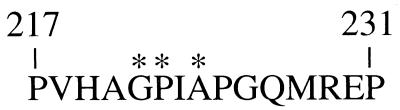Abstract
Human immunodeficiency virus type 1 (HIV-1) Gag and the cellular protein cyclophilin A form an essential complex in the virion core: virions produced by proviruses encoding Gag mutants with decreased cyclophilin A affinity exhibit attenuated infectivity, as do virions produced in the presence of the competitive inhibitor cyclosporine. The A224E Gag mutant has no effect on cyclophilin A affinity but renders HIV-1 replication cyclosporine resistant in Jurkat T cells. In contrast, A224E mutant virus is dead in H9 T cells, although replication is rescued by cyclosporine or by expression in cis of a Gag mutant that decreases cyclophilin A-affinity. The observation that disruption of the Gag-cyclophilin A interaction rescues A224E mutant replication in H9 cells prompted experiments which revealed that, relative to Jurkat cells, H9 cells express greater quantities of cyclophilin A. The resulting larger quantity of cyclophilin A shown to be packaged into virions produced by H9 cells is presumably disruptive to the A224E mutant virion core. Further evidence that increased cyclophilin A expression in H9 cells is of functional relevance was provided by the finding that Gag mutants with decreased cyclophilin A affinity are dead in Jurkat cells but capable of replication in H9 cells. Similarly, cyclosporine concentrations which inhibit wild-type HIV-1 replication in Jurkat cells stimulate HIV-1 replication in H9 cells. These results suggest that HIV-1 virion infectivity imposes narrow constraints upon cyclophilin A stoichiometry in virions and that infectivity is finely tuned by host cyclophilin A expression levels.
One property of human immunodeficiency virus type 1 (HIV-1) that makes it unique among retroviruses is that the cytoplasmic host protein cyclophilin A is required for its replication (27). The capsid domain of the HIV-1 Gag polyprotein contains a proline-rich loop which forms a stable complex with cyclophilin A (7, 14, 17). The amino acid sequence of the proline-rich loop is shown in Fig. 1. Three residues located at the apex of the proline-rich loop, A220, G221, and P222, make intimate contact with the hydrophobic pocket of cyclophilin A (5, 9, 17, 45) and are required for binding to cyclophilin A in vitro, for packaging of cyclophilin A into virions, and for the production of infectious virions (6, 7, 14, 43). Mutant phenotypes relevant to this paper are summarized in Table 1.
FIG. 1.
Primary structure of the HIV-1 Gag polyprotein proline-rich loop that binds cyclophilin A and confers cyclophilin A dependence on HIV-1 replication. The numbers refer to amino acid residues with respect to the amino terminus of the Gag polyprotein. The asterisks indicate the positions of G221, P222, and A224, residues important to this study.
TABLE 1.
Phenotypes of HIV-1 Gag mutantsa
| Gag mutation(s) | CyPA binding and packaging | Inhibition of CyPA binding and packaging by CsAb | Replication in:
|
|||
|---|---|---|---|---|---|---|
| Jurkat cells
|
H9 cells
|
|||||
| Without CsA | With CsA | Without CsA | With CsA | |||
| None (wild type) | + | + | + | − | + | + |
| G221A | − | ND | − | ND | + | ND |
| P222A | − | ND | − | ND | + | ND |
| A224E | + | + | + | + | − | + |
| P222A/A224E | − | ND | + | + | + | ND |
CyPA, cyclophilin A; CsA, cyclosporine; ND, not done.
These results have been reported previously. See text for references.
The normal cellular function of cyclophilin A in vivo is not known. In vitro studies have demonstrated that cyclophilin A catalyzes the isomerization of peptidyl-prolyl bonds and that it also exhibits classic chaperone activity, increasing the yield of properly folded protein substrate (11, 15, 36). Structural studies of an uncomplexed HIV-1 Gag fragment and of Gag fragments in complex with cyclophilin A indicate that the peptidyl-prolyl bond connecting Gag residues G221 and P222 does not undergo isomerization as a result of binding to cyclophilin A (17, 18, 45). These data, together with the observation that HIV-1 replication requires the formation of a stable complex between Gag and cyclophilin A, suggest that cyclophilin A functions as a Gag chaperone rather than as a Gag isomerase (27).
Cyclophilin A was first discovered because of its high affinity for cyclosporine, a drug commonly used to suppress allograft rejection (19). Like Gag, cyclosporine also binds to the hydrophobic pocket of cyclophilin A (23, 29, 38). As a competitive inhibitor of the HIV-1 Gag-cyclophilin A interaction (28), cyclosporine disrupts cyclophilin A incorporation into virions and, by doing so, attenuates virion infectivity (3, 13, 14, 22, 32, 37, 40).
Two Gag mutations, A224E and G226D, alter the sensitivity of HIV-1 replication to cyclosporine (1). The location of these mutations indicates not only that Gag binds to cyclophilin A but also that a gag-encoded function requires cyclophilin A. Also, since these mutations have no effect on Gag’s affinity for cyclophilin A (4), they demonstrate that Gag’s primary sequence requirements for binding to cyclophilin A can be distinguished from Gag’s primary sequence requirements for dependence upon cyclophilin A (Table 1).
Interestingly, the phenotype of viruses bearing either the A224E mutation or the G226D mutation depends on the host cell in which viral replication is assayed. Mutant virus stocks bearing these mutations were originally selected by serial passage of HIV-1 in CD4+ HeLa cells cultured in the continuous presence of a cyclosporine analog (1). In these cells the mutant viruses are cyclosporine dependent: they will not replicate in the absence of the drug. In Jurkat T cells, in contrast, these mutations confer cyclosporine resistance but not drug dependence (4): the mutant viruses are capable of replicating in the presence or in the absence of the drug.
After noting that A224E virus replication is drug dependent in H9 cells, we demonstrated that H9 cells express higher levels of cyclophilin A than do Jurkat cells. We compared the replication of a panel of Gag mutants in Jurkat cells and in H9 cells (Table 1), and the observed differences between the two cell lines suggest that HIV-1 replication is exquisitely sensitive to cyclophilin A expression levels.
MATERIALS AND METHODS
Plasmid DNAs and viruses.
Proviral DNAs were propagated in Escherichia coli JM109 clone 3226 (Life Technologies, Inc., Gaithersburg, Md.) at 30°C by standard methods (33). Supercoiled plasmids for use in transfection experiments were purified by using Plasmid Maxi kit (Qiagen, Chatsworth, Calif.).
pNL4-3 is a plasmid containing a complete infectious clone of HIV-1 (2). The retrovirus sequence is numbered with respect to the 5′ edge of the 5′ long terminal repeat of the DNA provirus. Amino acid numbering is with respect to the amino-terminal residue of the Gag polyprotein. The engineering of the G221A, P222A, A224E, and P222A/A224E mutants has been described previously (4, 6). pNL4-3ΔVif contains a deletion in vif coding sequences (21) and was a gift from Klaus Strebel. pNL4-3ΔVif plasmids expressing each of the P222A, A224E, and P222A/A224E Gag mutants were engineered by standard methods (33). Each mutant virus is otherwise isogenic to the parent virus pNL4-3.
Cell culture, transfections, and infections.
The human lymphocyte lines, Jurkat (41) and H9 (31), were obtained from the National Institutes of Health AIDS Research and Reference Program and maintained in RPMI 1640 supplemented with 10% fetal calf serum. Human fibroblast 293T cells were maintained in Dulbecco modified Eagle medium-F12 (1:1) supplemented with 10% fetal bovine serum.
Viral infections were initiated in 107 Jurkat or H9 cells by using 2 μg of proviral DNA and 250 μg of DEAE-dextran (Pharmacia Biotech Inc., Piscataway, N.J.) per ml in 1 ml of serum-free RPMI medium for 20 min at room temperature. Cells were washed in serum-free medium, resuspended in 3 ml of conditioned medium with 6 ml of fresh medium, and split into 3-ml cultures with cyclosporine added at various concentrations. Every 2 days supernatant was harvested and frozen, and cells were passaged. At the conclusion of the experiment, the frozen samples were thawed and analyzed for reverse transcriptase activity as described below.
For experiments in which infection was initiated by infection with exogenous virus, viral stocks were prepared by calcium phosphate transfection of 10 μg of supercoiled proviral DNA into 293T cells by using a Mammalian Cell Transfection Kit (Specialty Media, Lavellette, N.J.). All stocks utilized in a given experiment were normalized for virion content by reverse transcriptase activity. Jurkat or H9 cells (107) were pelleted and resuspended in 1 ml of RPMI containing 103 infectious units of virus (as determined by limiting dilution). At 1 h the cells were washed, expanded to 9 ml, and split three ways.
Cyclosporine.
Cyclosporine was obtained from the former Sandoz Pharmaceuticals Corporation (East Hanover, N.J.). Prior to addition to tissue culture medium, the drug powder was dissolved in ethanol to make stock solutions of less than 2.5 mM. All samples in a given experiment in which cyclosporine was used received the same volume of ethanol, including the no-drug controls.
Exogenous reverse transcriptase assay.
Ten microliters of cell culture supernatant (for viral replication assays) or 10 μl of precleared, filtered supernatant (for transient transfections of proviral DNA) was added to 50 μl of reverse transcriptase cocktail [60 mM Tris-HCl (pH 8.0), 180 mM KCl, 6 mM MgCl2, 6 mM dithiothreitol, 0.6 mM EGTA, 0.12% Triton-X 100, 6 μg of oligo(dT) per ml, 12 μg of poly(rA) per ml, 0.05 mM [α-32P]dTTP (800 Ci/mmol)] and left for 1 h at 37°C. Ten microliters of each sample was spotted onto DE-81 paper. Unincorporated nucleotides were washed from the filter, and a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) was used to quantitate the radioactivity incorporated, as an indication of relative reverse transcriptase activity.
Virion purification.
Supernatant collected from infected Jurkat cells or H9 cells at the peak of reverse transcriptase activity was centrifuged at 1,000 rpm for 5 min and passed through a 0.45-μm-pore-size filter to remove cellular debris. The filtrate was gently layered on top of a single-step gradient consisting of 2 ml of 25% sucrose over 2 ml of 45% sucrose in TNE (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, and 1 mM EDTA) and subjected to centrifugation at 80,000 × g for 2 h in a Beckman SW41 rotor. The virion-containing interface was harvested and brought up to 6 ml with TNE to dilute the sucrose. The virions were pelleted again at 80,000 × g for 2 h over 2 ml of 25% sucrose in an SW41 rotor. The pellet was resuspended in 50 μl of 2× sodium dodecyl sulfate (SDS) sample buffer for Western blot analysis as described below.
Western blots and antibodies.
For analysis of cyclophilin A content, soluble proteins were prepared from Jurkat cells and H9 cells grown to a cell density of 106/ml. Cells were lysed with radioimmunoprecipitation assay buffer and normalized by cell number or for total protein content by using a bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.). Samples were then processed by SDS-polyacrylamide gel electrophoresis under standard conditions (20). Virion-associated proteins normalized for reverse transcriptase activity were also processed by SDS-polyacrylamide gel electrophoresis. Size-separated cellular proteins or virion-associated proteins were transferred to polyvinylidene difluoride membranes with a Bio-Rad (Hercules, Calif.) mini-blotting apparatus and probed with a rabbit anti-cyclophilin A antibody (Affinity BioReagents, Heshanic Station, N.J.) or with a murine monoclonal anti-HIV-1 capsid antibody (Dupont NEN). Binding of primary antibodies was detected with the appropriate peroxidase-coupled secondary antibody and the Renaissance chemiluminescence kit (Dupont).
RESULTS
The A224E mutation renders HIV-1 replication cyclosporine resistant in Jurkat cells and cyclosporine dependent in H9 cells.
We compared the replication of A224E virus and of other mutant viruses in two human T-cell leukemia lines, Jurkat and H9. Each of these previously described mutants (Fig. 1 and Table 1) is otherwise isogenic to the parent virus pNL4-3. Since it was our intention to compare viral replication phenotypes in these two T-cell lines, we wanted to minimize effects on viral replication that might be attributable to peculiarities of producer cell lines. Therefore, infection was initiated in the Jurkat and H9 cells by direct transfection of proviral DNAs. Nonetheless, results similar to those reported here were obtained when infection was initiated exogenously with viral stocks produced in 293T cells (data not shown). Once DNA transfection was complete, the accumulation of reverse transcriptase activity in the culture supernatant was monitored as an indication of virus replication in the T-cell cultures.
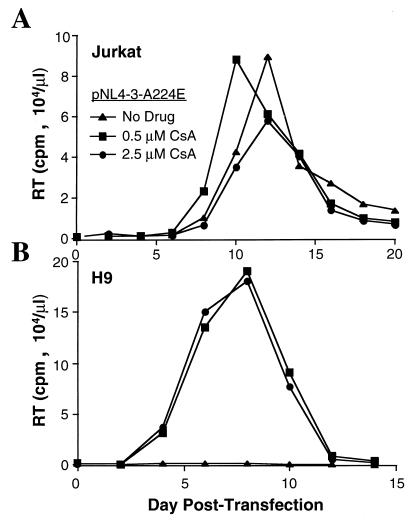
A224E virus was capable of replicating in Jurkat cells in either the presence or the absence of cyclosporine (Fig. 2A); it exhibited a slight stimulation of replication in the presence of 0.5 μM cyclosporine and only minimal inhibition at 2.5 μM. The ability of this mutant to replicate in the presence of cyclosporine is not due to increased affinity for cyclophilin A compared to the wild type but is due to the ability of the virus to replicate in the absence of cyclophilin A packaging (4).
FIG. 2.
HIV-1 A224E mutant virus is cyclosporine resistant in Jurkat cells and cyclosporine dependent in H9 cells. Infection of the Jurkat T-cell line (A) or of the H9 T-cell line (B) was initiated by transfection of proviral DNA pNL4-3 encoding the A224E amino acid mutation. Cyclosporine (CsA) was maintained in the cell culture medium at the indicated concentrations. The accumulation of extracellular virions in the culture supernatant resulting from viral spread through the culture is indicated by the reverse transcriptase (RT) activity in the supernatant at the indicated times posttransfection.
In contrast to the results obtained with Jurkat cells, in the absence of cyclosporine, no replication was detected with the A224E mutant virus in H9 cells (Fig. 2B). When either 0.5 or 2.5 μM cyclosporine was added to the culture medium, there was a dramatic rescue of A224E virus replication in H9 cells (Fig. 2B), such that the virus replicated at least as well as the wild-type virus (for example, compare these data with the wild-type replication kinetics shown in Fig. 3B). The A224E virus replication kinetics and magnitude of reverse transcriptase activity in H9 cells were identical at the two cyclosporine concentrations, indicating that there was no inhibition of replication at the higher dose relative to the lower one.
FIG. 3.
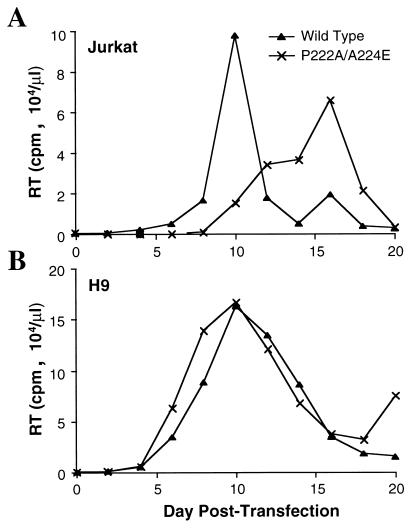
P222A, a Gag mutation that confers reduced cyclophilin A affinity to the Gag polyprotein, rescues the replication of HIV-1 mutant A224E in H9 cells when the two mutations are expressed in cis. Jurkat T cells (A) or H9 T cells (B) were transfected with the wild-type proviral DNA pNL4-3 or with pNL4-3 encoding the P222A/A224E double mutation. RT, reverse transcriptase.
Expression of the P222A mutation in cis rescues A224E virus replication in H9 cells.
We suspected that the rescue of A224E replication in H9 cells by cyclosporine was related to the drug’s ability to function as a competitive inhibitor of the Gag-cyclophilin A interaction. If this is correct, expression in cis of a mutation that lowers Gag affinity for cyclophilin A would also rescue A224E virus replication in H9 cells. The A224E mutation was combined in cis with the mutation P222A. The latter mutation disrupts cyclophilin A affinity and virion packaging roughly fourfold (6), as does the double mutation P222A/A224E (4).
The P222A/A224E virus was able to replicate in Jurkat cells, although with delayed kinetics with respect to those of the wild type (Fig. 3A). In H9 cells, the P222A/A224E double mutant virus replicated at least as well as the wild-type virus (Fig. 3B). Thus, reduction of cyclophilin A incorporation by pharmacologic means with cyclosporine, or by genetic means with the P222A mutation expressed in cis, restores infectivity to the A224E mutant virus in H9 cells.
Cyclophilin A expression levels in Jurkat and H9 cells.
The A224E mutation has no effect on cyclophilin A affinity or on the efficiency of cyclophilin A packaging into virions (4). The fact that the A224E virus replicates well in Jurkat cells but does not replicate detectably in H9 cells unless rescued by factors which inhibit cyclophilin A packaging into virions suggested that the two cell lines might differ with respect to cyclophilin A expression levels. If H9 cells expressed higher levels of cyclophilin A than Jurkat cells and the packaging of relatively large quantities of cyclophilin A into virions was not tolerated by the A224E mutant virion core, cyclosporine or the P222A mutation would restore A224E infectivity in H9 cells by decreasing the levels of cyclophilin A in the virions.
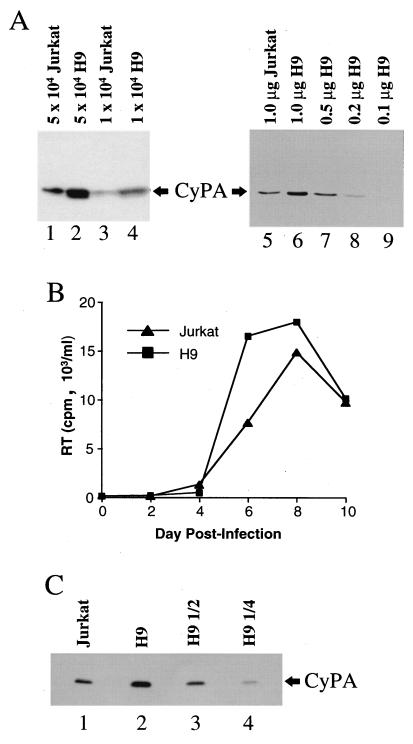
To determine if H9 cells express higher levels of cyclophilin A than do Jurkat cells, total soluble protein from the two cell lines was probed in immunoblots with a rabbit anti-cyclophilin A antibody. Samples from Jurkat cells (Fig. 4A, lanes 1 and 3) and H9 cells (Fig. 4A, lanes 2 and 4) were normalized by cell number. Samples from Jurkat cells and H9 cells were also normalized for total protein content (Fig. 4A, lanes 5 to 9). Whether samples were normalized for cell number or for total cellular protein, it was evident that H9 cells express higher levels of cyclophilin A than do Jurkat cells. Similar results were obtained when cyclophilin A expression was examined by using cells grown at the lower density of 2 × 105/ml (data not shown). To compare relative protein levels, the H9 sample was serially diluted (Fig. 4A, lanes 6 to 9), and these semiquantitative immunoblots indicated that H9 cells express roughly threefold more cyclophilin A than do Jurkat cells.
FIG. 4.
H9 cells express more cyclophilin A (CyPA) than do Jurkat cells. (A) Jurkat cells (lanes 1, 3, and 5) and H9 cells (lanes 2, 4, and 6 to 9) were grown to a density of 106/ml, lysed in radioimmunoprecipitation assay buffer, and processed for Western blotting with a primary anti-cyclophilin A antibody. Lanes 1 to 4, samples normalized by cell number. The quantity of soluble cell lysate loaded per lane, as measured by number of cell equivalents, is indicated above the lanes. Lanes 5 to 9, samples normalized by quantity of total protein in the soluble cell lysate. The numbers above the lanes indicate the amount of total protein loaded per well. (B) Replication of wild-type HIV-1 in Jurkat or H9 cells after initiation of infection with a common stock of virus produced by transfection of 293T cells with pNL4-3. RT, reverse transcriptase. (C) Virions were purified from the supernatants of the cultures used for panel B on day 8 postinfection. Virion-associated protein was probed in a Western blot with a primary anti-cyclophilin A antibody. Lanes 1 and 2, samples from Jurkat and H9 cell cultures, respectively, normalized for reverse transcriptase activity and capsid protein. Lanes 3 and 4, serial twofold dilutions of the H9 sample, as indicated.
Virions produced in H9 cells contain more cyclophilin A than do virions produced in Jurkat cells.
Since H9 cells express higher levels of cyclophilin A than do Jurkat cells, the cyclophilin A contents of virions produced by these two cell lines were compared. To obtain sufficient quantities of virions for biochemical analysis, infection was initiated in Jurkat and H9 cells by using a single virus stock produced by transient transfection of 293T cells with pNL4-3. Following infection, virion accumulations in the supernatants of the two cell lines were roughly comparable, although the reverse transcriptase activity peaked slightly earlier and achieved higher magnitudes in the H9 cell cultures (Fig. 4B).
Virions were purified from supernatants taken at day 8 from the two cultures (Fig. 4B) by centrifugation through sucrose gradients as described in Materials and Methods. After normalization for capsid protein content and for reverse transcriptase activity, samples were probed in Western blots with anti-cyclophilin A antibodies. Virions produced in H9 cells were repeatedly found to contain greater quantities of cyclophilin A than virions produced in Jurkat cells. Results from a representative experiment are shown in Fig. 4C. Serial dilution of the sample produced in H9 cells indicated that these virions contained between two- and threefold more cyclophilin A than the virions produced by Jurkat cells (Fig. 4C).
HIV-1 mutants with decreased cyclophilin A affinity replicate in H9 cells.
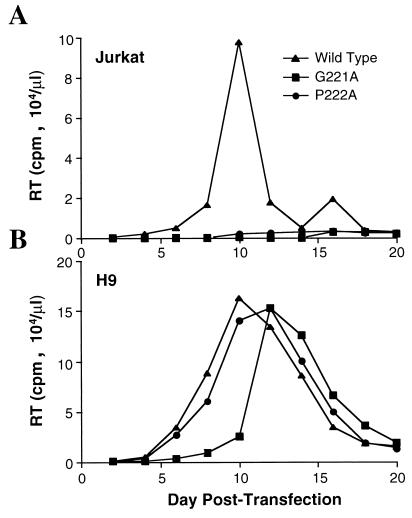
If the higher level of cyclophilin A expression in H9 cells than in Jurkat cells is of functional relevance to HIV-1 replication, mutations which decrease Gag polyprotein affinity for cyclophilin A would have more drastic effects on viral replication in Jurkat cells than in H9 cells. Therefore, the effect on viral replication of two mutations which decrease cyclophilin A affinity, G221A and P222A (6), was examined. The presence of either mutation was sufficient to inhibit the accumulation of extracellular virions in Jurkat T-cell cultures to levels below the limit of detection by assays for reverse transcriptase activity; no significant accumulation of virions was seen with either mutant G221A or P222A for up to 3 weeks of culture (Fig. 5A). Under identical conditions, the wild-type virus peaked at 10 days posttransfection (Fig. 5A).
FIG. 5.
HIV-1 proviruses expressing Gag mutants with attenuated cyclophilin A affinity cannot promote a spreading infection in Jurkat cells (A) but do replicate in H9 cells (B). Infection was initiated by direct transfection of wild-type or mutant proviral DNA pNL4-3 into the T-cell lines. RT, reverse transcriptase.
In contrast to what was observed following transfection of the Jurkat cells, the G221A and P222A mutants were both capable of replicating quite well in H9 cells. Both mutant viruses peaked by 12 days posttransfection, in comparison with the replication peak at 10 days in these cells for the wild-type virus (Fig. 5B). Although the mutant viruses were able to replicate in H9 cells, there was a slight inhibition of replication, the magnitude of which correlated with the previously reported effect of these mutations on cyclophilin A binding and packaging: G221A has a larger effect than P222A on cyclophilin A affinity (6), and it produced a more significant reduction in replication in H9 cells.
Replication of wild-type HIV-1 is cyclosporine resistant in H9 cells.
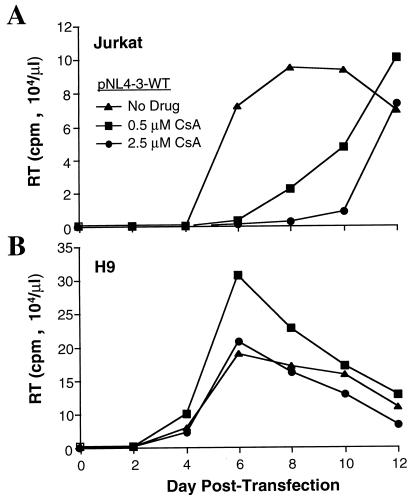
Additional evidence that higher-level cyclophilin A expression in H9 cells is of functional relevance was provided by comparing the effects of cyclosporine on replication of the wild-type virus in the two cell lines. Since cyclosporine competes with Gag for binding to cyclophilin A, a given concentration of the drug would be expected to disrupt HIV-1 replication more effectively in Jurkat cells than in H9 cells. Infection was initiated by transfection with wild-type pNL4-3, and cyclosporine was maintained continuously in the culture medium at 0, 0.5, or 2.5 μM. In Jurkat cells, viral replication was inhibited by cyclosporine in a titratable fashion (Fig. 6A). When the identical experiment was conducted with H9 cells, no inhibition of viral replication was observed (Fig. 6B). In fact, in the presence of 0.5 μM cyclosporine, the peak of reverse transcriptase activity was 75% higher than that produced without the drug. Even with 2.5 μM cyclosporine, viral replication kinetics and magnitude were as robust as those produced without the drug.
FIG. 6.
HIV-1 replication in H9 cells is resistant to inhibition by cyclosporine. Infection of Jurkat T cells (A) or of H9 T cells (B) was initiated by transfection of wild-type (WT) proviral DNA pNL4-3. Cyclosporine (CsA) was maintained in the cell culture medium for the duration of the experiment at the indicated concentrations. RT, reverse transcriptase.
Gag mutants with altered cyclophilin A affinity or cyclophilin A dependence do not rescue the replication of Vif-defective HIV-1 in H9 cells.
With respect to HIV-1 replication, cyclophilin A shares several features with the HIV-1 Vif protein (8, 12, 16, 21, 26, 34, 39). Both proteins are incorporated into virions, and both are required for reverse transcription after virions fuse membranes with susceptible target cells. Interestingly, as with the A224E mutant, H9 cells are nonpermissive for the replication of Vif-defective HIV-1. Vif is not required for HIV-1 replication in other cell lines, such as CEM-SS or, to a lesser extent, Jurkat cells.
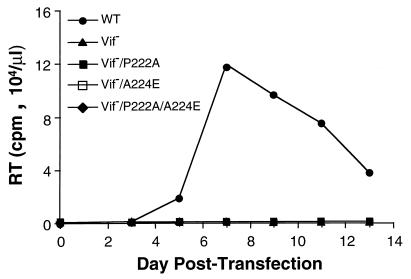
To test the possibility that cyclophilin A and Vif perform related functions in HIV-1 replication, Vif-defective proviruses bearing various gag mutations that affect interactions with cyclophilin A were constructed. As expected, the Vif-defective virus was unable to replicate in H9 cells (Fig. 7). This virus replicated weakly in Jurkat cells but as well as the wild type in CEM-SS cells (data not shown). Since the P222A mutant was able to rescue A224E replication in H9 cells, the effect of the P222A, A224E, or P222A/A224E mutant on replication of the Vif-defective virus in H9 cells was tested. None of the Gag mutants were found to rescue Vif-defective viral replication in H9 cells (Fig. 7).
FIG. 7.
HIV-1 Gag mutants expressed in cis do not rescue the replication of Vif-defective HIV-1 in H9 cells. Infection of H9 T cells was initiated by transfection of wild-type (WT) or mutant proviral DNA pNL4-3. RT, reverse transcriptase. The Vif− virus is pNL4-3 with a deletion in the vif open reading frame (21).
DISCUSSION
By examining the effect of pharmacologic and genetic factors that alter the interaction between HIV-1 Gag and cyclophilin A in two T-cell lines that differ in cyclophilin A expression levels, we have obtained evidence that HIV-1 replication is modulated by levels of cyclophilin A expression in host cells. The A224E mutant replicates well in Jurkat cells but is dead in H9 cells (Fig. 2). The latter cell line expresses relatively high levels of cyclophilin A (Fig. 4), and by disrupting the interaction between Gag and cyclophilin A with the competitive inhibitor cyclosporine (Fig. 2) or by expressing the A224E mutation in cis with another Gag mutation that reduces cyclophilin A affinity (Fig. 3), the replication of the A224E virus in H9 cells was rescued. Conversely, Gag mutants that decrease cyclophilin A affinity and completely block viral replication in Jurkat cells (Fig. 5A) are, in effect, rescued by the higher concentrations of cyclophilin A expressed in H9 cells (Fig. 5B). Similarly, cyclosporine concentrations which effectively block HIV-1 replication in Jurkat cells have no effect on HIV-1 replication in the presence of the higher quantities of cyclophilin A expressed by H9 cells (Fig. 6B).
We believe that the differences in virus replication in Jurkat cells and H9 cells are explained by the higher levels of cyclophilin A expressed in H9 cells (Fig. 4A) and the resulting greater quantity of cyclophilin A packaged into the virions that these cells produce (Fig. 4C). Cyclophilin A is required for an early event in the infection cycle, after membrane fusion but before initiation of reverse transcription (6). Based on genetic and structural data, current models propose that cyclophilin A interferes with contacts between capsid monomers in the virion core (17, 27). At a ratio of 1:10 with respect to capsid (14, 37), cyclophilin A weakens the core structure to an extent required for virion uncoating. The linear decrease in infectivity observed with linear decreases in cyclophilin A packaging (6) is consistent with this model and suggests that the capsid/cyclophilin A ratio must be finely tuned for infectivity. When there is too much cyclophilin A, the core is unstable; when there is too little cyclophilin A, the core is too stiff for uncoating. Subtle weakening of capsid-capsid interactions by the A224E mutation would destabilize the core and permit virion uncoating to proceed in Jurkat cells in the absence of cyclophilin A. Since cyclophilin A affinity is not attenuated by the A224E mutation (4), a slight increase in cyclophilin A expression, as observed with H9 cells, would weaken capsid-capsid interactions further, rendering the virion core unstable. Consistent with this model is the observation that A224E virus replication in H9 cells is rescued by factors that decrease cyclophilin A packaging.
It remains a possibility that cellular properties other than cyclophilin A expression levels are responsible for the differences in viral phenotypes in Jurkat and H9 cells. It seems unlikely that differences in cyclophilin A primary structure could be responsible for the observations reported here, since there is absolute cyclophilin A amino acid conservation among four different primate species (42). Alternatively, Jurkat and H9 cells might differ with respect to the expression levels of an unknown factor that regulates the Gag-cyclophilin A interaction.
Although cyclophilin A accumulates to reasonably high levels in all cell lines and tissue types examined (25), threefold increases in cyclophilin A expression following heat shock in yeast (35) and following concanavalin A stimulation of mouse splenocytes (25) have been reported. The experiments presented here suggest that a threefold change in cyclophilin A expression could have profound effects on HIV-1 replication. Several steps of the HIV-1 life cycle, for example, reverse transcription (44) and proviral transcription (10, 24, 30), are regulated by the activation state of the cell. Perhaps increases in cyclophilin A expression are yet another cellular activation response that HIV-1 exploits to maximize viral production.
Disruption of HIV-1 vif leads to a cell-type-dependent phenotype similar to that of cyclophilin A-deficient virions (8, 12, 16, 21, 26, 34, 39). Although H9 cells are nonpermissive for replication of Vif-defective HIV-1 and for the A224E mutant, factors which rescue A224E replication do not rescue that of Vif-defective virus. These results suggest that the role of Vif in the early events of HIV-1 replication is distinct from the role of cyclophilin A.
ACKNOWLEDGMENTS
We thank Klaus Strebel for the vif mutant HIV-1 provirus and William McVicker of the former Sandoz Pharmaceuticals, Inc., for providing cyclosporine. Jurkat and H9 cells were obtained from the National Institutes of Health AIDS Research and Reference Program.
This work was supported by grant AI36199 from the National Institute of Allergy and Infectious Diseases. J.L. is an Irma T. Hirschl Scholar.
REFERENCES
- 1.Aberham C, Weber S, Phares W. J. Virol. 70:3536–3544. 1996. Spontaneous mutations in the human immunodeficiency virus type 1 gag gene that affect viral replication in the presence of cyclosporins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartz S R, Hohenwalter E, Hu M-K, Rich D H, Malkovsky M. Inhibition of human immunodeficiency virus replication by nonimmunosuppressive analogs of cyclosporin A. Proc Natl Acad Sci USA. 1995;92:5381–5385. doi: 10.1073/pnas.92.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braaten D, Aberham C, Franke E K, Yin L, Phares W, Luban J. Cyclosporin A-resistant human immunodeficiency virus type 1 mutants demonstrate that gag encodes the functional target of cyclophilin A. J Virol. 1996;70:5170–5176. doi: 10.1128/jvi.70.8.5170-5176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braaten D, Ansari H, Luban J. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 1997;71:2107–2113. doi: 10.1128/jvi.71.3.2107-2113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braaten D, Franke E K, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 prior to the initiation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukovsky A A, Weimann A, Accola M A, Göttlinger H G. Transfer of the HIV-1 cyclophilin-binding site to simian immunodeficiency virus from Macaca mulatta can confer both cyclosporin sensitivity and cyclosporin dependence. Proc Natl Acad Sci USA. 1997;94:10943–10948. doi: 10.1073/pnas.94.20.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camaur D, Trono D. Characterization of human immunodeficiency virus type 1 Vif particle incorporation. J Virol. 1996;70:6106–6111. doi: 10.1128/jvi.70.9.6106-6111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorfman T, Weimann A, Borsetti A, Walsh C T, Göttlinger H G. Active-site residues of cyclophilin A are crucial for its incorporation into human immunodeficiency virus type 1 virions. J Virol. 1997;71:7110–7113. doi: 10.1128/jvi.71.9.7110-7113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinberg M B, Greene W C. Molecular insights into human immunodeficiency virus type 1 pathogenesis. Curr Opin Immunol. 1992;4:466–474. doi: 10.1016/s0952-7915(06)80041-5. [DOI] [PubMed] [Google Scholar]
- 11.Fischer G, Wittmann-Liebold B, Lang K, Kiefhaber T, Schmid F X. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature. 1989;337:476–478. doi: 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]
- 12.Fouchier R A, Simon J H, Jaffe A B, Malim M H. Human immunodeficiency virus type 1 Vif does not influence expression or virion incorporation of gag-, pol-, and env-encoded proteins. J Virol. 1996;70:8263–8269. doi: 10.1128/jvi.70.12.8263-8269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke E K, Luban J. Inhibition of HIV-1 replication by cyclosporine A or related compounds correlates with the ability to disrupt the Gag-cyclophilin A interaction. Virology. 1996;222:279–282. doi: 10.1006/viro.1996.0421. [DOI] [PubMed] [Google Scholar]
- 14.Franke E K, Yuan H E H, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 15.Freskgard P-O, Bergenhem N, Jonsson B-H, Svensson M, Carlsson U. Isomerase and chaperone activity of prolyl isomerase in the folding of carbonic anhydrase. Science. 1992;258:466–468. doi: 10.1126/science.1357751. [DOI] [PubMed] [Google Scholar]
- 16.Gabuzda D H, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine W A, Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamble T R, Vajdos F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 18.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 19.Handschumacher R, Harding M, Rice J, Drugge R. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 21.Karczewski M K, Strebel K. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J Virol. 1996;70:494–507. doi: 10.1128/jvi.70.1.494-507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karpas A, Lowdell M, Jacobson S, Hill F. Inhibition of human immunodeficiency virus and growth of infected T cells by the immunosuppressive drugs cyclosporin A and FK506. Proc Natl Acad Sci USA. 1992;89:8351–8355. doi: 10.1073/pnas.89.17.8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ke H, Mayrose D, Belshaw P J, Alberg D G, Schreiber S L, Chang Z Y, Etzkorn F A, Ho S, Walsh C T. Crystal structures of cyclophilin A complexed with cyclosporin A and N-methyl-4-[(E)-2-butenyl]-4,4-dimethylthreonine cyclosporin A. Structure. 1994;2:33–44. doi: 10.1016/s0969-2126(00)00006-x. [DOI] [PubMed] [Google Scholar]
- 24.Kinoshita S, Su L, Amano M, Timmerman L A, Kaneshima H, Nolan G P. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity. 1997;6:235–244. doi: 10.1016/s1074-7613(00)80326-x. [DOI] [PubMed] [Google Scholar]
- 25.Koletsky A J, Harding M W, Handschumacher R E. Cyclophilin: distribution and variant properties in normal and neoplastic tissues. J Immunol. 1986;137:1054–1059. [PubMed] [Google Scholar]
- 26.Liu H W X, Newman M, Shaw G M, Hahn B H, Kappes J C. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J Virol. 1995;69:7630–7638. doi: 10.1128/jvi.69.12.7630-7638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luban J. Absconding with the chaperone: essential cyclophilin-Gag interaction in HIV-1 virions. Cell. 1996;87:1157–1159. doi: 10.1016/s0092-8674(00)81811-5. [DOI] [PubMed] [Google Scholar]
- 28.Luban J, Bossolt K A, Franke E K, Kalpana G V, Goff S P. Human immunodeficiency virus type 1 gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 29.Pflugl G, Kallen J, Schirmer T, Jansonius J, Zurini M, Walkinshaw M. X-ray structure of a decameric cyclophilin-cyclosporin crystal complex. Nature. 1993;361:91–94. doi: 10.1038/361091a0. [DOI] [PubMed] [Google Scholar]
- 30.Poli G, Fauci A S. The effect of cytokines and pharmacologic agents on chronic HIV infection. AIDS Res Hum Retroviruses. 1992;8:191–197. doi: 10.1089/aid.1992.8.191. [DOI] [PubMed] [Google Scholar]
- 31.Popovic M, Sarngadharan M G, Read E, Gallo R C. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 32.Rosenwirth B, Billich A, Datema R, Donatsch P, Hammerschmid F, Harrison R, Hiestand P, Jaksche H, Mayer P, Peichl P, Quesniaux V, Schatz F, Schuurman H-J, Traber R, Wenger R, Wolff B, Zenke G, Zurini M. Inhibition of human immunodeficiency virus type 1 replication by SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog. Antimicrob Agents Chemother. 1994;38:1763–1772. doi: 10.1128/aac.38.8.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Sova P, Volsky D J. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with Vif-negative human immunodeficiency virus type 1. J Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sykes K, Gething M-J, Sambrook J. Proline isomerases function during heat shock. Proc Natl Acad Sci USA. 1993;90:5853–5857. doi: 10.1073/pnas.90.12.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi N, Hayano T, Suzuki M. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature. 1989;337:473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- 37.Thali M, Bukovsky A A, Kondo E, Rosenwirth B, Walsh C T, Sodroski J, Göttlinger H G. Specific association of cyclophilin A with human immunodeficiency virus type 1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 38.Theriault Y, Logan T M, Meadows R, Yu L, Olejniczak E T, Holzman T F, Simmer R L, Fesik S W. Solution structure of the cyclosporin A/cyclophilin complex by NMR. Nature. 1993;361:88–91. doi: 10.1038/361088a0. [DOI] [PubMed] [Google Scholar]
- 39.von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wainberg M, Dascal A, Blain N, Fitz-Gibbon L, Boulerice F, Numazaki K, Tremblay M. The effect of cyclosporine A on infection of susceptible cells by human immunodeficiency virus type 1. Blood. 1988;72:1904–1910. [PubMed] [Google Scholar]
- 41.Weiss A, Wiskocil R, Stobo J. The role of T3 surface molecules in the activation of human T cells: a two stimulus requirement for IL-2 production reflects events occurring at a pretranslational level. J Immunol. 1984;133:123–128. [PubMed] [Google Scholar]
- 42.Yin L, Luban J. The HIV-1 replication block in non-human primates is not explained by differences in Cyclophilin A primary structure. AIDS Res Hum Retroviruses. 1998;14:95–97. doi: 10.1089/aid.1998.14.95. [DOI] [PubMed] [Google Scholar]
- 43.Yoo S, Myszka D G, Yeh C, McMurray M, Hill C P, Sundquist W I. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J Mol Biol. 1997;269:780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]
- 44.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Chen Y, Schutkowski M, Fischer G, Ke H. Cyclophilin A complexed with a fragment of HIV-1 gag protein: insights into HIV-1 infectious activity. Structure. 1997;5:139–146. doi: 10.1016/s0969-2126(97)00172-x. [DOI] [PubMed] [Google Scholar]