Abstract
We established a reverse genetics system for the nonstructural (NS) gene segment of influenza A virus. This system is based on the use of the temperature-sensitive (ts) reassortant virus 25A-1. The 25A-1 virus contains the NS gene from influenza A/Leningrad/134/57 virus and the remaining gene segments from A/Puerto Rico (PR)/8/34 virus. This particular gene constellation was found to be responsible for the ts phenotype. For reverse genetics of the NS gene, a plasmid-derived NS gene from influenza A/PR/8/34 virus was ribonucleoprotein transfected into cells that were previously infected with the 25A-1 virus. Two subsequent passages of the transfection supernatant at 40°C selected viruses containing the transfected NS gene derived from A/PR/8/34 virus. The high efficiency of the selection process permitted the rescue of transfectant viruses with large deletions of the C-terminal part of the NS1 protein. Viable transfectant viruses containing the N-terminal 124, 80, or 38 amino acids of the NS1 protein were obtained. Whereas all deletion mutants grew to high titers in Vero cells, growth on Madin-Darby canine kidney (MDCK) cells and replication in mice decreased with increasing length of the deletions. In Vero cells expression levels of viral proteins of the deletion mutants were similar to those of the wild type. In contrast, in MDCK cells the level of the M1 protein was significantly reduced for the deletion mutants.
The influenza A virus genome contains eight segments of single-stranded RNA of negative polarity, coding for nine structural proteins and one nonstructural protein (NS1). The NS1 protein is abundant in influenza virus-infected cells but has not been detected in virions. NS1 is a phosphoprotein found in the nucleus early during infection and also in the cytoplasm at later times of the viral cycle. Moreover, NS1 has been found in complexes with polysomes (2, 14–16, 27). Studies of temperature-sensitive (ts) influenza virus mutants carrying lesions in the NS gene suggested that the NS1 protein is a transcriptional and posttranscriptional regulator of mechanisms by which the virus is able to inhibit host cell gene expression and to stimulate its own protein synthesis (11, 12, 19). Like many other proteins that regulate posttranscriptional processes, the NS1 protein interacts with specific RNA sequences and structures. The NS1 protein has been reported to bind to different RNA species, including viral RNA, poly(A) RNA, U6 snRNA, 5′ untranslated regions of viral mRNAs, and double-stranded RNA (dsRNA) (11, 13, 26, 28, 29). Expression of the NS1 protein from cDNA in transfected cells has been associated with several effects: inhibition of nucleocytoplasmic transport of mRNA (9), inhibition of pre-mRNA splicing (17), and stimulation of translation of viral mRNA (2, 5). It was also shown that binding of NS1 protein to dsRNA may prevent activation of the dsRNA-activated protein kinase in infected cells, and it is postulated that by this mechanism the virus counteracts the interferon-mediated inhibition of translation of viral proteins (18). In addition to possessing RNA binding activities, the influenza virus NS1 protein interacts with cellular proteins such as factor NS1-I (33) and a 230-amino-acid cellular protein which specifically binds to the effector domain of the NS1 protein (23).
Several functional domains of the influenza A virus NS1 protein were mapped. These studies were based on the evaluation of the properties of mutated NS1 proteins expressed from cDNA. For example, within the NS1 protein two nuclear localization signals were mapped (10). Qiu and Krug (28) suggested the presence of two other functional domains in the influenza A virus NS1 protein. The domain near the amino end corresponding to amino acids 19 to 38 was shown to be the RNA binding domain. The domain corresponding to amino acids 134 to 161 at the carboxyl half of the molecule was presumed to be the effector domain that interacts with host nuclear proteins to prevent the export of RNA from the nucleus (28). However, it was shown that influenza A (and influenza B) viruses can tolerate carboxy-terminal deletions of the NS1 protein, which suggests that the effector domain is not essential for viral replication (24). Experiments with truncated forms of the NS1 protein expressed from cDNAs showed that the N-terminal 81 amino acids of the NS1 protein of influenza A virus are sufficient to display RNA binding activity but that the 113 N-terminal amino acids are required for a full set of activities typical for normal-size NS1 protein of 237 amino acid residues (21, 22). Although the properties of the NS1 protein were extensively characterized in vitro by expression of its mutants, the lack of an efficient rescue system for the NS gene segment made it difficult to analyze the biological properties of influenza viruses bearing modified NS1 proteins. Rescue of NS1 genes coding for the wild-type NS1 protein was possible (7, 8, 30).
In this study, we established an efficient reverse genetics system to rescue synthetic NS genes of different lengths into influenza A viruses. A plasmid-derived NS gene of influenza A/Puerto Rico/8/34 (PR8) virus was ribonucleoprotein (RNP) transfected into Vero cells. These had previously been infected with a ts reassortant influenza A virus whose NS gene was found to be responsible for the ts phenotype (4). Passages in Vero cells at 40°C selected viruses containing the transfected NS gene derived from PR8 virus. This transfection system permitted us to obtain functional transfectant influenza viruses carrying truncated NS1 proteins. Depending on the size of the NS1 protein, transfectant viruses showed different growth patterns in Vero cells, Madin-Darby canine kidney (MDCK) cells, and embryonated eggs and were attenuated in mice.
MATERIALS AND METHODS
Viruses and cells.
Influenza A virus 25A-1 is a reassortant virus containing the NS gene segment from the cold-adapted strain A/Leningrad (Len)/134/47/57 and the remaining genes from the PR8 virus (4). The 25A-1 virus is ts in mammalian cells and was used previously as a helper virus for rescuing the wild-type NS gene of the PR8 virus into infectious particles (30). For this study, the 25A-1 virus was adapted to Vero cells (ATCC CCL-81) by 20 sequential passages at 34°C. The maximum titer of the Vero-adapted 25A-1 virus was 108 PFU/ml at 34°C, whereas at 40°C the maximum titer achieved was 103.8 PFU/ml. Vero cells were used for transfection experiments, selection and plaque purification of the rescued transfectant viruses, and virus titrations. The Vero cells were cultivated in serum-free medium (Baxter-Immuno, Orth Donau, Austria). In addition, MDCK cells and 10-day-old embryonated chicken eggs were used for virus titrations. MDCK cells were cultivated in Dulbecco modified Eagle medium containing 2% fetal calf serum.
Construction of plasmids.
Viral RNA from the PR8 virus was extracted by using Ultraspec RNA purification reagent (Biotecx Laboratories) and served as the template for subsequent amplification of the viral NS gene by reverse transcription-PCR (RT-PCR). Sense (5′-ACTACTTCTAGAGAAGACAAAGCAAAAGCAGGGTGACA-3′) and antisense (5′-ACTACTCTGCAGATTAACCC TCACTAAAAGTAGAAACAAG-3′) primers used for the reactions were selected according to the PR8 NS gene sequence published by Baez et al. (1). The sense primer also contains the restriction sites XbaI for cloning and BpuAI for plasmid linearization. The antisense primer contains a PstI restriction site for cloning and a T3 promoter sequence, allowing in vitro transcription from cloned NS cDNA. Resulting amplification products were XbaI/PstI digested and cloned into the plasmid vector pUC19 (New England Biolabs, Inc., Beverly, Mass.). The resulting construct was called pPUC19-T3/NS PR8. Starting from this plasmid, three constructs were prepared.
(i) NS1/124.
dTTP was introduced between NS PR8 nucleotide positions 400 and 401 by inverse PCR (25), using the back-to-back primers 3′NS-400 (5′-TCCATGATCGCCTGGTCCA-3′) and 5′NS-T-401 (5′-TTAAGAACATCATACTGAAAGCGAAC-3′) (CODON Genetic Systems, Weiden, Austria) in order to create a stop codon (TAA) and a Tru9I restriction site (T/TAA). Following phosphorylation and Klenow enzyme treatment, amplified DNA was self-ligated and propagated in Escherichia coli TG1. The resulting construct was called NS1/124. Digestion with Tru9I confirmed the presence of the introduced nucleotide.
(ii) NS1/80.
A frameshift at NS PR8 nucleotide position 263 was generated as follows. Plasmid pUC19-T3/NS PR8 was digested with StyI at NS PR8 position 265, and single-stranded overhangs produced by the restriction enzyme were filled by treatment with Klenow enzyme (Boehringer, Mannheim, Germany). Then DNA was self-ligated, propagated in E. coli TG1, and purified. The resulting construct was called NS1/80. The anticipated frameshift due to the removal of four nucleotides was confirmed by sequencing.
(iii) NS1/38.
A cassette of stop codons (TGAATAACTAGCTGA) was introduced at NS PR8 nucleotide position 140 by inverse PCR using the back-to-back primers 3′NS140-stop (5′-TTATTCATCGGCGAAGCCGATCAAGG-3′) and 5′NS141-stop (5′-CTAGCTGATCAGAAATCCCTAAGAGG-3′) (CODON Genetic Systems). Following phosphorylation and Klenow treatment, amplified DNA was self-ligated, propagated in E. coli TG1, and purified. The resulting construct was called NS1/38 and confirmed by sequencing.
Generation of transfectant viruses.
Generation of NS transfectant viruses was performed according to the standard DEAE-dextran transfection protocol described by Luytjes et al. (20), with several modifications. Briefly, six-well plastic plates containing approximately 106 Vero cells/well were infected with the 25A-1 virus at a multiplicity of infection (MOI) of 1. After incubation for 30 min, the inoculum was removed and cells were treated with a DEAE-dextran-dimethyl sulfoxide solution at room temperature. After aspiration of this solution, cells were transfected with reconstituted RNP complexes. The RNPs were formed by T3 RNA polymerase transcription from NS plasmids linearized with BpuAI in the presence of purified influenza A virus 25A-1 polymerase preparations (6, 8). Transfected cells were incubated for 18 h at 37°C. Subsequently the transfection supernatant was passaged twice at 40°C. Rescued transfectant viruses were plaque purified on Vero cells three times at 37°C. The isolated viruses were analyzed by RT-PCR with the sense primer 5′-AGCAAAAGCAGGGTGACAAAG-3′ (corresponding to nucleotide positions 1 to 21 of the NS gene of influenza A/Len/134/47 virus) and the antisense primers 5′-CTCTTGCTCCACTTCAAGC-3′ and 5′-CTCTTGTTCCACTTCAAAT-3′ (corresponding to nucleotide positions 834 to 816 of the NS genes of PR8 and A/Len/134/47 viruses, respectively). The antisense primers permit one to distinguish whether the NS genes are derived from the 25A-1 helper virus or from the transfected PR8-derived NS gene.
Growth in tissue culture and embryonated eggs.
Confluent monolayers of MDCK or Vero cells on six-well plates were infected with viruses at an MOI of 0.05, overlaid with RPMI medium containing 5 μg of trypsin (Sigma), and incubated at 37°C. At different time points, supernatants were assayed for infectious virus particles in plaque assays on Vero cells. Embryonated chicken eggs were infected with each virus at 105 PFU/egg; after 48 h of incubation at 34°C, allantoic fluids were harvested and titrated for infectivity by plaquing on Vero cells.
Viral replication in mice lungs.
To determine viral replication in lungs, mice were infected intranasally with each virus at 105.2 PFU/animal under slight ether narcosis. At days 2, 4, and 6, lungs were aseptically removed from four animals of each group, and 10% tissue extracts were prepared by grinding the tissue samples with a porcelain homogenizer containing glass sand in phosphate-buffered saline containing antibiotics. The suspensions were centrifuged (3,000 × g, 5 min), and the supernatants were assayed for infectious virus particles in plaque assays on Vero cells.
Analysis of viral protein synthesis.
Confluent monolayers of MDCK or Vero cells in six-well plates were infected with transfectant viruses at an MOI of 5. After 30 min, the inoculum was removed and RPMI medium was added. After 6 h of incubation at 37°C, the RPMI medium was replaced with 0.5 ml of cysteine-methionine-free minimal essential medium supplemented with [35S]methionine-[35S]cysteine (50 μCi/ml; Amersham) and incubated for 30 min. Then cells were washed two times with phosphate-buffered saline and lysed directly in the dishes by adding 200 μl of electrophoresis sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, 10% glycerol). Proteins were then analyzed by SDS-gel electrophoresis on 13% polyacrylamide gels containing 5 M urea. After electrophoresis for 13 h at 100 V, protein gels were fixed in a solution of 20% methanol and 5% acetic acid, rinsed with water, and dried for autoradiography.
RESULTS
Rescue of NS transfectant influenza viruses.
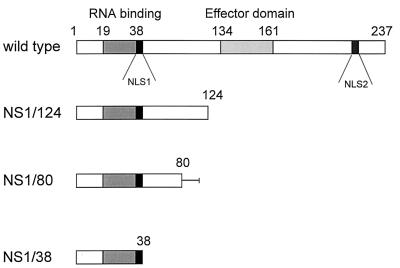
Plasmid-derived NS RNA from wild-type PR8 virus as well as NS RNA containing stop codons at nucleotide positions 400, 298, and 140 in the NS1 open reading frame were successfully rescued into viral particles. A schematic drawing of these constructs is shown in Fig. 1. The corresponding transfectant viruses were designated NS1/124, NS1/80, and NS1/38, containing the N-terminal NS1-specific 124, 80, and 38 amino acids, respectively. For analyses of the origin of the NS genes in the viral progeny after selection, RT-PCR with PR8-specific primers was performed. The stability of the introduced stop codons was analyzed by sequencing the NS1 gene segments of the transfectant viruses after five passages in Vero cells. No revertants were found (data not shown).
FIG. 1.
Rescued transfectant viruses. The bars represent the NS proteins of the generated transfectant viruses; amino acid positions are indicated. The RNA binding domain and the effector domain are represented by shaded bars (28). The two nuclear localization signals (NLS1 and NLS2) described by Greenspan et al. (10) are represented by black bars. The line after amino acid position 80 of NS1/80 corresponds to the amino acids HGLCTCVALPN which resulted from the frameshift at nucleotide position 263. Generation of transfectant viruses is described in Materials and Methods.
Growth of NS transfectants in tissue culture.
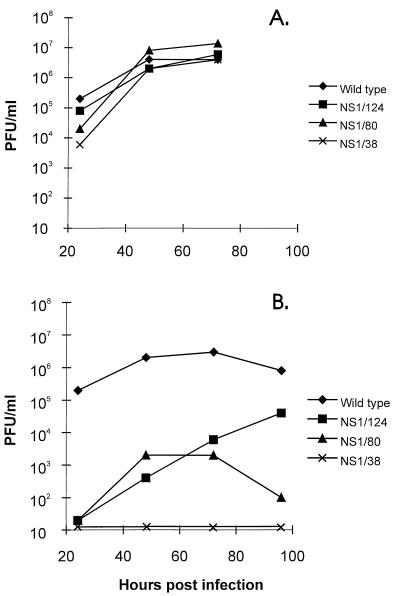
We next evaluated the potential of the rescued transfectant viruses to replicate in Vero and MDCK cells (Fig. 2). Cells were infected at an MOI of 0.05, and the viral yields in the supernatant were titrated on Vero cells at different time points. All transfectants showed similar patterns of growth in Vero cells. The maximum titer was reached at 72 h postinfection and was in the range of 106.5 to 107 PFU/ml (Fig. 2A). The hemagglutination titer achieved was 64 (data not shown). In MDCK cells, in contrast to Vero cells, viruses containing truncated forms of NS1 protein were less productive than the wild-type transfectant virus. NS1/124 virus reached its maximum titer of 104.5 PFU/ml 96 h after infection. The maximum titer of NS1/80 was only 103.2 PFU/ml. Transfectant NS1/38 virus was fully restricted in its growth in MDCK cells (Fig. 2B). In embryonated eggs, all transfectant viruses except NS1/38 were able to grow as well as the PR8 wild-type transfectant virus, achieving titers of 108.5 PFU/ml.
FIG. 2.
Growth of transfectant viruses in Vero (A) and MDCK (B) cells. Confluent monolayers of Vero or MDCK cells were infected with viruses at an MOI of 0.05 and incubated at 37°C. At different time points, supernatants were assayed for infectious virus particles in plaque assays on Vero cells as described in Materials and Methods.
Replication in mice lungs.
The restricted replication pattern of the viruses in MDCK cells and eggs suggested that the transfectants bearing the truncated forms of the NS1 protein might be attenuated in vivo. BALB/c mice were infected intranasally with 105.2 PFU of each transfectant virus. The wild-type transfectant virus was highly pathogenic, causing lethal pneumonia as a result of viral replication. As shown in Fig. 3, 106.8 PFU of the wild type per g of lung tissue was detected at day 2. Transfectant viruses were attenuated to different extents. Although the titers of the NS1/124 virus were only slightly reduced in mouse lungs and pulmonary lesions were visible at day 6, the NS1/124 virus did not kill the animals at the given dose. The peak titer of the NS1/80 transfectant was 104.3 PFU/g at day 4. This transfectant was cleared at day 6, and no lung lesions were detected. Attempts to reisolate transfectant virus NS1/38 from lungs and nasal turbinates failed at all time points.
FIG. 3.
Viral titers in lung tissue of mice. BALB/c mice were infected intranasally with 105.2 PFU of transfectant viruses. On days 2, 4, and 6 following virus administration, mice were sacrificed and their lungs were removed for virus quantitation. Lungs of four mice were pooled, homogenized, and assayed for infectious virus particles in plaque assays on Vero cells.
Synthesis of viral polypeptides.
Viral polypeptide synthesis in the virus-infected Vero and MDCK cells was analyzed at 6 h postinfection. In Vero cells, the level of viral protein synthesis by the deletion mutants was comparable to that of the wild type; in contrast, in MDCK cells the level of the M1 protein was significantly reduced for the transfectant viruses NS1/124, NS1/80, and NS1/38 compared to that for the wild-type transfectant (Fig. 4). The bands were quantified by an optical density scanner (Phoretix 1D version 2.01; Nonlinear Dynamics). The calculated NP/M1 ratios for the different viruses are as follows: wild type, 0.53 in Vero cells and 0.78 in MDCK cells; NS1/124, 1.00 in Vero and 8.17 in MDCK; NS1/80, 1.07 in Vero and 5.88 in MDCK; NS1/38, 0.47 in Vero and 6.81 in MDCK. The lower amount of M1 protein expressed in MDCK cells was also reflected by a lower amount of M1 polypeptide incorporated into viral particles (data not shown).
FIG. 4.
Synthesis of viral proteins. Confluent monolayers of Vero (lanes 1 to 5) or MDCK (lanes 6 to 10) cells were infected with transfectant viruses at an MOI of 5. Five hours postinfection, cells were labeled with [35S]methionine and [35S]cysteine for 30 min and lysed. Cell extracts were analyzed by SDS-gel electrophoresis on 13% polyacrylamide gels containing 5 M urea. Lane 1, uninfected Vero cell extract; lanes 2 and 6, wild-type-infected cell extracts; lanes 3 and 7, transfectant NS1/124-infected cell extracts; lanes 4 and 8, transfectant NS1/80-infected cell extracts; lanes 5 and 9, transfectant NS1/38-infected cell extracts; line 10, uninfected MDCK cell extract. The positions of the viral proteins are indicated on the left. The faint band migrating between the M1 and NS1 proteins corresponds to the HA2 subunit. For transfectant NS1/38, the NS1 protein is not visible due to the small size and lower number of methionine and cysteine residues.
DISCUSSION
We established a reverse genetics method which permitted us to rescue influenza viruses containing in vitro-mutagenized NS gene segments. We used this technique to obtain transfectant influenza viruses with deletions in the NS1 protein. The growth characteristics in different hosts and expression levels of viral proteins of the transfectant viruses were dependent on the lengths of the deletions.
Replication of NS1/124, a transfectant virus whose NS1 segment codes for the N-terminal 124 amino acids, was not affected in Vero cells and embryonated chicken eggs and was only slightly diminished on MDCK cells. Moreover, this virus was only slightly attenuated in mice, as indicated by its growth in mouse lungs. The expression levels of the viral proteins in infected cells were similar to those of the wild type except for the M1 protein, which was reduced in MDCK cells. This latter finding is in accordance with data of Enami et al. (5), who reported the stimulation by NS1 protein of the translation of a chimeric chloramphenicol acetyltransferase mRNA containing 5′ sequences derived from the M gene. In another study, de la Luna et al. (2) showed that coexpression of NS1 protein led to increases in the translation of M1 mRNA. This effect was also shown to be dependent on 5′-terminal extracistronic sequences of the M1 gene.
Recently, it was shown that plasmid-driven expression of the N-terminal 113 amino acids of the NS1 protein in COS-1 cells was sufficient to retain functions of the wild-type NS1 protein such as nuclear retention of mRNA and stimulation of viral mRNA translation and binding to dsRNA. However, the truncated form of the NS1 protein containing the N-terminal 81 amino acids was sufficient only for dsRNA binding, not for nuclear retention and stimulation of translation of mRNA (22).
In this regard, it is surprising that the growth characteristics and expression of viral proteins of NS1/80, an NS1 transfectant virus that contains 80 amino acids of the N terminus of the NS1 protein, were not affected in Vero cells and embryonated chicken eggs and only slightly diminished on MDCK cells. However, in contrast to the NS1/124 transfectant, this virus was significantly attenuated in mice.
Since the NS1/80 mutant was viable, we transfected an NS gene segment coding for the N-terminal 38 amino acids of the NS1 protein. Surprisingly, transfection of this segment also yielded viable transfectant viruses. This virus grew to high titers in Vero cells but did not grow in MDCK cells and embryonated chicken eggs. Moreover, this virus did not replicate in mouse lungs and nasal turbinates.
The transfectants’ differential ability to grow in Vero cells and other host cells could be due to the fact that Vero cells are deficient in the expression of functional interferon (3). Interferon induces the dsRNA-activated protein kinase (PKR). In the presence of dsRNA, PKR becomes activated and phosphorylates the alpha subunit of the eukaryotic translation initiation factor 2 (eIF2). As a result, protein synthesis within the cell is blocked. Thus, PKR activation and phosphorylation of eIF-2 can represent major effectors of the interferon antiviral response at the level of protein synthesis. The NS1 binds to dsRNA and subsequently blocks the activation of dsRNA-activated PKR in vitro. For this reason it was suggested that one of the mechanisms employed by the influenza virus to evade the antiviral effects of interferon might involve the NS1 protein. This hypothesis might explain why transfectant influenza viruses containing large deletions in the NS1 protein are capable of replicating efficiently in the interferon-deficient Vero cells but not in normal host cells. However, there might be other host cell factors that inhibit viral replication which are absent in Vero cells but present in MDCK cells and mice. Another possibility is that Vero cells have a host factor, lacking in MDCK cells and mice, that compensates for the NS1 deletions. We are currently investigating if the host cell tropism of the mutant viruses containing deletions in the NS1 gene is related to the interferon-mediated antiviral response.
Targeting essential functional sites within the NS1 protein might be a promising strategy to obtain stable attenuated influenza virus vaccine strains. It remains to be established whether functions such as the regulation of nuclear export of mRNA, inhibition of splicing, and inhibition of host cell mRNA polyadenylation are associated with the attenuation phenotype of the generated NS1 deletion mutants. Since the size of the deletion correlates with the degree of attenuation, we should be able to obtain a tailor-made virus with the desired balance between attenuation and immunogenicity. The possibility of deleting large parts of the protein should reduce the probability of repairing the deletion or generating second-site suppressor mutations. It should, however, be considered that even deletion mutants might phenotypically revert by acquiring intragenic or extragenic suppressor mutations (31, 32).
ACKNOWLEDGMENTS
This work was supported in part by Austrian Science Fund project 11366-MOB (T.M.). T.M. was supported by the Austrian Programme for Advanced Research and Technology of the Austrian Academy of Sciences.
REFERENCES
- 1.Baez M, Taussig R, Zazra J J, Young J F, Palese P, Reisfeld A, Skalka A M. Complete nucleotide sequence of the influenza A/PR/8/34 virus NS gene and comparison with the NS genes of the A/Udorn/72 and A/FPV/Rostock/34 strains. Nucleic Acids Res. 1980;8:5845–5858. doi: 10.1093/nar/8.23.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de la Luna S, Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaz M O, Ziemin S, Le Beau M M, Pitha P, Smith S D, Chilcote R R, Rowley J D. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc Natl Acad Sci USA. 1988;85:5259–5263. doi: 10.1073/pnas.85.14.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egorov A, Garmashova L M, Lukashok I V, Nevedomskaia G N, Aleksandrova G I, Klimov A I. The NS gene—a possible determinant of apathogenicity of a cold-adapted donor of attenuation A/Leningrad/134/47/57 and its reassortants. Vopr Virusol. 1994;39:201–205. . (In Russian.) [PubMed] [Google Scholar]
- 5.Enami K, Sato T A, Nakada S, Enami M. Influenza virus NS1 protein stimulates translation of the M1 protein. J Virol. 1994;68:1432–1437. doi: 10.1128/jvi.68.3.1432-1437.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enami M, Luytjes W, Krystal M, Palese P. Introduction of site-specific mutations into the genome of influenza virus. Proc Natl Acad Sci USA. 1990;87:3802–3805. doi: 10.1073/pnas.87.10.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enami M, Palese P. High-efficiency formation of influenza virus transfectants. J Virol. 1991;65:2711–2713. doi: 10.1128/jvi.65.5.2711-2713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enami M, Sharma G, Benham C, Palese P. An influenza virus containing nine different RNA segments. Virology. 1991;185:291–298. doi: 10.1016/0042-6822(91)90776-8. . (Erratum, 186:798, 1992.) [DOI] [PubMed] [Google Scholar]
- 9.Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenspan D, Palese P, Krystal M. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J Virol. 1988;62:3020–3026. doi: 10.1128/jvi.62.8.3020-3026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatada E, Fukuda R. Binding of influenza A virus NS1 protein to dsRNA in vitro. J Gen Virol. 1992;73:3325–3329. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- 12.Hatada E, Hasegawa M, Shimizu K, Hatanaka M, Fukuda R. Analysis of influenza A virus temperature-sensitive mutants with mutations in RNA segment 8. J Gen Virol. 1990;71:1283–1292. doi: 10.1099/0022-1317-71-6-1283. [DOI] [PubMed] [Google Scholar]
- 13.Hatada E, Saito S, Okishio N, Fukuda R. Binding of the influenza virus NS1 protein to model genome RNAs. J Gen Virol. 1997;78:1059–1063. doi: 10.1099/0022-1317-78-5-1059. [DOI] [PubMed] [Google Scholar]
- 14.Krug R M, Etkind P R. Cytoplasmic and nuclear virus-specific proteins in influenza virus-infected MDCK cells. Virology. 1973;56:334–348. doi: 10.1016/0042-6822(73)90310-3. [DOI] [PubMed] [Google Scholar]
- 15.Krug R M, Soeiro R. Studies on the intranuclear localization of influenza virus-specific proteins. Virology. 1975;64:378–387. doi: 10.1016/0042-6822(75)90114-2. [DOI] [PubMed] [Google Scholar]
- 16.Lazarowitz S G, Compans R W, Choppin P W. Influenza virus structural and nonstructural proteins in infected cells and their plasma membranes. Virology. 1971;46:830–843. doi: 10.1016/0042-6822(71)90084-5. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y, Qian X Y, Krug R M. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 1994;8:1817–1828. doi: 10.1101/gad.8.15.1817. [DOI] [PubMed] [Google Scholar]
- 18.Lu Y, Wambach M, Katze M G, Krug R M. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 19.Ludwig S, Vogel U, Scholtissek C. Amino acid replacements leading to temperature-sensitive defects of the NS1 protein of influenza A virus. Arch Virol. 1995;140:945–950. doi: 10.1007/BF01314970. [DOI] [PubMed] [Google Scholar]
- 20.Luytjes W, Krystal M, Enami M, Pavin J D, Palese P. Amplification, expression, and packaging of foreign gene by influenza virus. Cell. 1989;59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 21.Marion R M, Aragon T, Beloso A, Nieto A, Ortin J. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 1997;25:4271–4277. doi: 10.1093/nar/25.21.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marion R M, Zurcher T, de la Luna S, Ortin J. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J Gen Virol. 1997;78:2447–2451. doi: 10.1099/0022-1317-78-10-2447. [DOI] [PubMed] [Google Scholar]
- 23.Nemeroff M E, Wang W, Chen Z, Li Y, Chien C, Montelione G, Liu J, Lynch P, Berman H, Krug R M. Abstracts of the 10th International Conference on Negative Strand Viruses, Dublin, Ireland. 1997. Unique interactions of the influenza virus NS1 protein with host cell nuclear functions, abstr. 229; p. 164. [Google Scholar]
- 24.Norton G P, Tanaka T, Tobita K, Nakada S, Buonagurio D A, Greenspan D, Krystal M, Palese P. Infectious influenza A and B virus variants with long carboxyl terminal deletions in the NS1 polypeptides. Virology. 1987;156:204–213. doi: 10.1016/0042-6822(87)90399-0. [DOI] [PubMed] [Google Scholar]
- 25.Ochman H, Gerber A S, Hartl D L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park Y W, Katze M G. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J Biol Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 27.Privalsky M L, Penhoet E E. The structure and synthesis of influenza virus phosphoproteins. J Biol Chem. 1981;256:5368–5376. [PubMed] [Google Scholar]
- 28.Qiu Y, Krug R M. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A) J Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu Y, Nemeroff M, Krug R M. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA. 1995;1:304–316. [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw M W, Kiseleva I V, Egorov A Y, Hemphill M L, Xu X. Nucleocapsid protein alone is sufficient for the generation of influenza transfectants. In: Brown L E, Hampson A W, Webster R G, editors. Options for the control of influenza III. Amsterdam, The Netherlands: Elsevier Science B.V.; 1996. pp. 433–436. [Google Scholar]
- 31.Snyder M H, London W T, Maassab H F, Chanock R M, Murphy B R. A 36 nucleotide deletion mutation in the coding region of the NS1 gene of an influenza A virus RNA segment 8 specifies a temperature-dependent host range phenotype. Virus Res. 1990;15:69–83. doi: 10.1016/0168-1702(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 32.Treanor J J, Buja R, Murphy B R. Intragenic suppression of a deletion mutation of the nonstructural gene of an influenza A virus. J Virol. 1991;65:4204–4210. doi: 10.1128/jvi.65.8.4204-4210.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolff T, O’Neill R E, Palese P. Interaction cloning of NS1-I, a human protein that binds to the nonstructural NS1 proteins of influenza A and B viruses. J Virol. 1996;70:5363–5372. doi: 10.1128/jvi.70.8.5363-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]