Abstract
Ferroptosis is a form of iron-dependent, lipid peroxidation–driven regulatory cell death that has been implicated in the pathogenesis of multiple diseases, including organ injury, ischemia/reperfusion, and neurodegenerative diseases. However, inhibitors that directly and specifically target ferroptosis are not yet available. Here, we identify the compound AS-252424 (AS) as a potent ferroptosis inhibitor through kinase inhibitor library screening. Our results show that AS effectively inhibits lipid peroxidation and ferroptosis in both human and mouse cells. Mechanistically, AS directly binds to the glutamine 464 of ACSL4 to inhibit its enzymatic activity, resulting in the suppression of lipid peroxidation and ferroptosis. By using nanoparticle-based delivery systems, treatment with AS-loaded nanoparticles effectively alleviate ferroptosis-mediated organ injury in mouse models, including kidney ischemia/reperfusion injury and acute liver injury (ALI). Thus, our results identify that AS is a specific and targeted inhibitor of ACSL4 with remarkable antiferroptosis function, providing a potential therapeutic for ferroptosis-related diseases.
AS-252424 ameliorates ferroptosis-related diseases by directly targeting inhibition of ACSL4.
INTRODUCTION
Ferroptosis is a form of regulated cell death driven by the excessive accumulation of lipid peroxidation on cell membranes in an iron-dependent manner (1, 2). Recent advances in ferroptosis have showed that it plays a pivotal role in tumor suppression (3). The tumor suppressors p53 and breast cancer 1-associated protein 1 have been found to promote ferroptosis by inhibiting the expression of solute carrier family 7 member 11 (SLC7A11) and thus exerting its tumor-suppressive function (4, 5). Moreover, CD8+ T cells promote tumor cell lipid peroxidation and ferroptosis by releasing interferon-γ (IFN-γ) and down-regulating the expression of SLC3A2 and SLC7A11 during immunotherapy and radiotherapy (6, 7). Arachidonic acid (AA) in combination with IFN-γ synergistically induces tumor cell ferroptosis in an acyl–coenzyme A (CoA) synthetase long-chain family member 4 (ACSL4)–dependent manner, and AA supplementation promotes immunotherapy (8). Although ferroptosis contributes to tumor suppression and immunotherapy, excessive ferroptosis can also lead to the pathogenesis of multiple diseases, including organ injury, ischemia/reperfusion (I/R), and neurodegenerative diseases (9, 10), suggesting that ferroptosis might be a potential target for the treatment of these diseases.
Now, with the mechanism of ferroptosis continues to be explored, several proteins have been found to defend against ferroptosis by limiting the production of lipid peroxidation, including glutathione peroxidase 4 (GPX4), ferroptosis suppressor protein 1 (FSP1), and guanosine triphosphate cyclohydrolase-1 (GCH1) (11–14). GPX4 is a glutathione (GSH)–dependent selenoenzyme that plays an essential role in inhibiting ferroptosis by converting toxic lipid hydroperoxides (L-OOH) to nontoxic lipid alcohols (L-OH) (15, 16). FSP1 acts as a ferroptosis suppressor in parallel with GPX4, which inhibits lipid peroxidation by reducing the lipophilic radical-trapping antioxidants (RTAs) ubiquinone to ubiquinol in a GSH-independent manner (17, 18). GCH1 causes lipid remodeling by promoting the synthesis of its metabolic derivatives tetrahydrobiopterin/dihydrobiopterin (BH4/BH2), which also trap lipid peroxidation to inhibit ferroptosis independently of the GPX4/GSH system (11). In addition to these ferroptosis suppressors that promote lipid peroxide clearance, ACSL4, a lipid metabolism enzyme, has been reported to enhance lipid peroxide generation (19, 20). ACSL4 contributes to ferroptosis by promoting the esterification of polyunsaturated fatty acids (PUFAs) to acyl-CoA, especially adrenic acid (C22:4) and AA (C20:4) (21). Therefore, ACSL4 is an important pharmacological target for ferroptosis in the treatment of ferroptosis-related diseases. However, compounds that directly target ACSL4 are not yet available.
AS-252424 (AS) is a furan-2-ylmethylene thiazolidinedione that acts as a selective small-molecule PI 3-kinase γ (PI3Kγ) inhibitor with potent anti-PI3Kγ kinase activity (22). AS has been found to be associated with inhibition of reactive oxide species production (23), but its role in ferroptosis is still unclear. Moreover, AS blocks neutrophil infiltration by markedly reducing chemotaxis to monocyte chemoattractant protein 1 in a PI3Kγ-independent manner (22), suggesting the existence of other targets for AS. In this study, we identified AS as a targeted ferroptosis inhibitor through kinase inhibitor library screening, which directly binds to the glutamine 464 of ACSL4 to inhibit lipid peroxidation and ferroptosis, contributing to the treatment of kidney I/R injury and acute liver injury (ALI). Together, our findings reveal the potential role and molecular mechanisms of AS in controlling ferroptosis and provide a promising therapeutic approach for the treatment of ferroptosis-related diseases.
RESULTS
Identification of AS inhibits RSL3-induced lipid peroxidation and ferroptosis
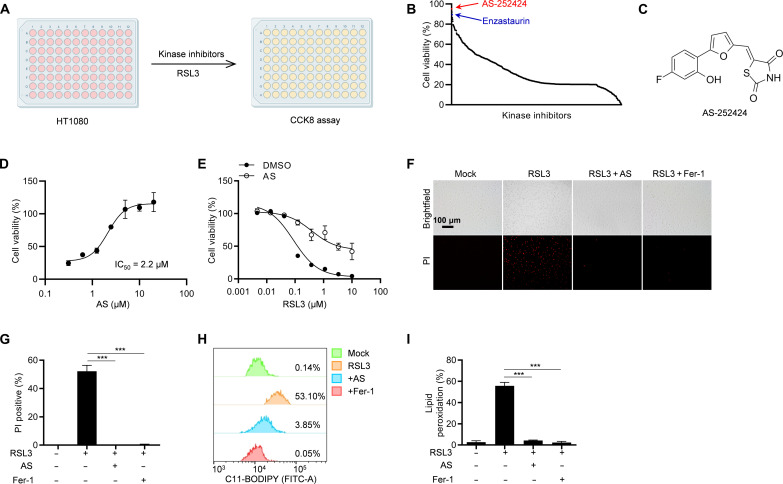
To identify small molecular compounds that inhibit ferroptosis, we performed a kinase inhibitor library screening in HT-1080 cells (Fig. 1A). The results showed that enzastaurin, an inhibitor of ferroptosis regulator protein kinase C (24), was identified, which confirmed the robustness of our screening system for ferroptosis inhibitors (Fig. 1B and table S1). Encouragingly, among these previously uncharacterized ferroptosis inhibitors, AS greatly inhibited RSL3-induced ferroptosis (Fig. 1, B and C). Moreover, the half-maximal inhibitory concentration (IC50) of AS for RSL3-induced ferroptosis was approximately 2.2 μM (Fig. 1D). To further determine the inhibitory effect of AS on ferroptosis, we first pretreated HT-1080 cells with AS and then stimulated with various concentrations of RSL3, showing that AS substantially rescued cells from ferroptosis (Fig. 1E). Similarly, AS almost completely blocked RSL3-induced cell death indicated by staining with propidium iodide (PI) (Fig. 1, F and G). Since iron-dependent lipid peroxidation is a key hallmark of ferroptosis, the levels of lipid peroxidation were detected in AS-treated HT-1080 cells by staining with BODIPY-C11 581/591. We found that AS was as effective as ferrostatin-1 (Fer-1) in eliminating RSL3-induced lipid peroxidation (Fig. 1, H and I). Therefore, these results suggested that AS efficiently suppressed RSL3-induced lipid peroxidation and ferroptosis.
Fig. 1. AS inhibits RSL3-induced lipid peroxidation and ferroptosis.
(A) Screening scheme of ferroptosis inhibitors. HT-1080 cells were pretreated with DMSO or 10 μM inhibitors for 1 hour and then stimulated with 1 μM RSL3 for 8 hours, and cell viability was detected by CCK-8 assay. (B) Results of ferroptosis inhibitor screening. (C) Chemical structure of AS. (D) Cell viability analysis of HT-1080 cells pretreated with different concentrations of AS for 1 hour and then stimulated with 1 μM RSL3 for 8 hours. (E) Cell viability analysis of HT-1080 cells pretreated with or without AS for 1 hour and then stimulated with different concentrations of RSL3 for 8 hours. (F) Images of HT-1080 cells pretreated with AS or ferroptosis inhibitor Fer-1 for 1 hour and then stimulated with 1 μM RSL3 for 8 hours in the presence of PI. (G) The percentage of PI-positive HT-1080 cells pretreated with AS or Fer-1 for 1 hour and then stimulated with 1 μM RSL3 for 8 hours in the presence of PI. (H) Lipid peroxidation was analyzed by flow cytometry in HT-1080 cells pretreated with AS or Fer-1 for 1 hour and then stimulated with 1 μM RSL3 for 8 hours in the presence of C11-BODIPY. (I) The percentage of C11-BODIPY–positive HT-1080 cells pretreated with AS or Fer-1 for 1 hour and then stimulated with 1 μM RSL3 for 8 hours in the presence of C11-BODIPY. Data are the mean ± SEM, n = 3 biologically independent experiments [(G) and (I)]. Statistical analysis was performed using an unpaired two-tailed Student’s t test. ***P < 0.001.
AS is a common inhibitor of ferroptosis
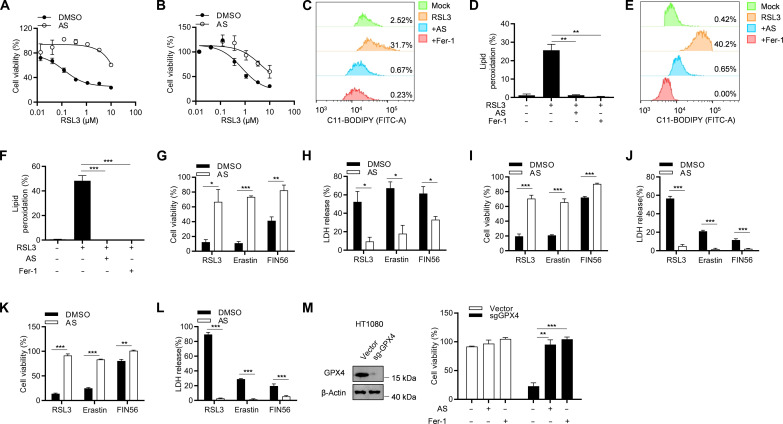
To further verify the inhibitory effect of AS on ferroptosis, we tested the antiferroptotic effect of AS on another eight different human and murine cell lines, including MDA-MB-231, Hela, A549, L-02, HK-2, HT22, MCA205, and B16-F10. Similar to HT-1080 cells, AS also inhibit RSL3-induced ferroptosis in these cell lines (Fig. 2, A and B, and fig. S1, A to F), suggesting that AS has a universal antiferroptotic activity in a cell-independent effect. Moreover, AS effectively blocked lipid peroxidation in MDA-MB-231 and HT22 cells (Fig. 2, C to F). In addition to RSL3, other compounds, including erastin (2) and FIN56 (25), have also been reported to induce ferroptosis. To explore whether AS has an inhibitory effect on erastin- and FIN56-induced ferroptosis, we stimulated AS-pretreated HT-1080 cells with erastin or FIN56 and found that AS greatly improved cell viability and rescue cell death (Fig. 2, G and H). The similar results were also shown in MDA-MB-231 and HT22 cells (Fig. 2, I to L). Moreover, AS could also inhibit GPX4 knockout–induced ferroptosis as Fer-1 (Fig. 2M), indicating that AS is a common inhibitor of ferroptosis.
Fig. 2. AS is a common inhibitor of ferroptosis.
(A and B) Cell viability analysis of MDA-MB-231 cells (A) or HT22 cells (B) pretreated with or without AS for 1 hour and then stimulated with different concentrations of RSL3 for 8 hours. (C and E) Lipid peroxidation was analyzed by flow cytometry in MDA-MB-231 cells (C) or HT22 cells (E) pretreated with AS or Fer-1 for 1 hour and then stimulated with 1 μM RSL3 for 8 hours in the presence of C11-BODIPY. (D and F) The percentage of C11-BODIPY–positive MDA-MB-231 cells (D) or HT22 cells (F) pretreated with AS or Fer-1 for 1 hour and then stimulated with 1 μM RSL3 for 8 hours in the presence of C11-BODIPY. (G and H) Cell viability (G) or LDH release (H) assay of HT-1080 cells pretreated with or without AS for 1 h and then stimulated with 1 μM RSL3, 20 μM erastin, or 40 μM FIN56. (I and J) Cell viability (I) or LDH release (J) assay of MDA-MB-231 cells pretreated with or without AS for 1 hour and then stimulated with 1 μM RSL3, 20 μM erastin, or 40 μM FIN56. (K and L) Cell viability (K) or LDH release (L) assay of MDA-MB-231 cells pretreated with or without AS for 1 hour and then stimulated with 1 μM RSL3, 20 μM erastin, or 40 μM FIN56. (M) CRISPR-Cas9–generated GPX4-knockout HT-1080 cells were treated with or without AS or Fer-1. The expression of GPX4 was detected by immunoblotting (left), and the cell viability was detected by CCK-8 assay (right). Data are the mean ± SEM, n = 3 biologically independent experiments [(D) and (F) to (M)]. Statistical analysis was performed using an unpaired two-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001.
AS directly targets ACSL4 to inhibit ferroptosis
AS has long been known as a potent and selective inhibitor of PI3Kγ and is involved in a variety of pathophysiological events by inhibiting the kinase activity of PI3Kγ (22, 26). To understand whether AS rescues ferroptosis by inhibiting PI3Kγ, we first examined the effect of several other inhibitors of PI3Kγ, including taselisib, CAY10505 (CAY), AS-605240, IITZ-01, and CZC24832, on ferroptosis. The results showed that these inhibitors had no effect on RSL3-induced ferroptosis in HT-1080 and MDA-MB-231 cells (fig. S2, A and B), implying that AS may inhibit ferroptosis in a PI3Kγ-independent manner. To further confirm this result, we knocked out PI3Kγ in HT-1080 and MDA-MB-231 cells with CRISPR-Cas9 and found that PI3Kγ deficiency neither rescued cell death nor affected the inhibitory effect of AS on ferroptosis (fig. S2, C and D). Moreover, AKT, a well-known downstream target of PI3K, has previously been found to be inhibited by AS (27). We treated cells with several other AKT inhibitors, including Afuresertib, PHT-427, Ipatasertib, and AKT inhibitor VIII, and found that these inhibitors did not affect RSL3-induced ferroptosis (fig. S2, E and F). Thus, these results suggest that AS inhibits ferroptosis independent of the PI3K-AKT pathway.
Since ferroptosis is an iron-dependent cell death caused primarily by unrestricted lipid peroxidation, we next explored whether AS inhibits ferroptosis by regulating iron metabolism and oxidative stress. We first examined the iron level in the cells by staining with FerroOrange, a Fe2+-selective fluorescent probe, and the results showed that AS had no effect on RSL3-induced iron accumulation (fig. S3A). Furthermore, we used ultraviolet (UV)−vis (visible) spectroscopy to analyze the iron chelation ability of AS and found that desferrioxamine (DFO) could chelate iron as previously reported (28), but AS could not chelate iron directly (fig. S3, B and C). We next explored the antioxidant activity of AS by using 2,2-diphenyl-1-picrylhydrazyl (DPPH) analyze, and the results showed that Fer-1, a classical RTA, sharply reduced the levels of free radicals, but AS had no free radical scavenging activity (fig. S3D). These results indicate that AS cannot act as iron chelator or RTA to inhibit ferroptosis.
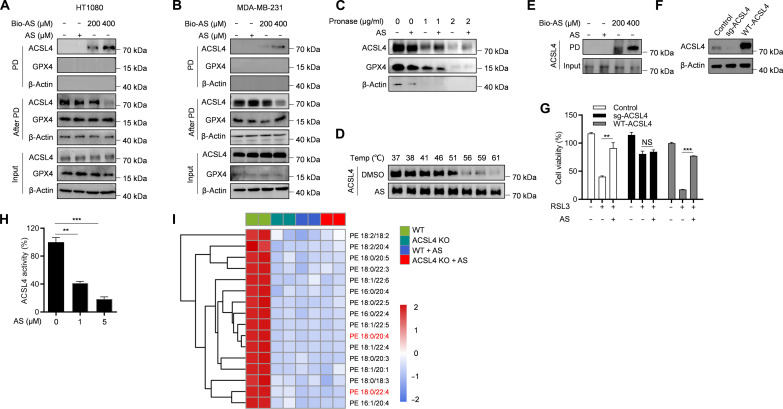
To investigate the specific molecular mechanism by which AS inhibits ferroptosis, we screened for the potential biological target of AS. A biotin-tagged AS (Bio-AS) was designed and synthesized (fig. S4A). We first tested the effect of Bio-AS on RSL3-induced ferroptosis in HT-1080 and MDA-MB-231 cells, and the results showed that Bio-AS had a favorable antiferroptotic activity as free AS (fig. S4, B and C), indicating that the addition of the biotin tag had no effect on the function of AS. Next, Bio-AS was used as an affinity probe and incubated with cell lysates of HT-1080 cells. Bio-AS was pulled down with streptavidin beads, and the Bio-AS–binding proteins were affinity-purified and further analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) (fig. S4D). Unexpectedly, the LC-MS/MS analysis indicated that the protein intensity of ACSL4 was the highest (fig. S4E and table S2). To determine the interaction between AS and ACSL4, a biotin-streptavidin pull-down assay was performed with Bio-AS, and the results showed that ACSL4, but not GPX4, was pulled down by Bio-AS both in HT-1080 and MDA-MB-231 cells in a dose-dependent manner (Fig. 3, A and B). Furthermore, the in situ binding of AS to ACSL4 was verified by the drug affinity–responsive target stability (DARTS) assay, a method for identifying label-free small-molecule target based on the reduction in protease sensitivity of target protein upon drug binding. The results showed that AS protected ACSL4 from the degradation by pronase in HT-1080 and MDA-MB-231 cells but had no effect on other ferroptosis-related proteins, such as GPX4 (Fig. 3C and fig. S4F). In addition, the engagement between AS and ACSL4 was further characterized by cell thermal shift assay (CETSA), and the data showed that AS sharply improved the thermostability of ACSL4 compared with the control (Fig. 3D and fig. S4G). To determine whether AS interacts directly with ACSL4, Bio-AS was incubated with recombinant ACSL4 protein, and we found that recombinant ACSL4 protein could be pulled down by Bio-AS (Fig. 3E and fig. S4H), indicating that AS directly binds to ACSL4. Then, to verify whether AS alleviated ferroptosis by targeting ACSL4, we knocked out ACSL4 in HT-1080 cells. The results showed that ACSL4 knockout (ACSL4-KO) substantially abolished the antiferroptotic function of AS, whereas reexpression of wild type (WT)–ACSL4 (ACSL4 isoform 1, UniProt accession no. O60488-1) restored the inhibitory effect of AS on ferroptosis (Fig. 3, F and G). ACSL4 is a lipid metabolism enzyme that has been reported to mediate ferroptosis by promoting the esterification of PUFAs to acyl-CoA (19). We found that AS treatment hugely inhibited the enzymatic activity of ACSL4 (Fig. 3H). Moreover, we further performed lipidomic analysis of WT and ACSL4-KO HT-1080 cells with or without AS treatment and found that AS treatment of WT HT-1080 cells phenocopied the lipidomic changes of different long-chain PUFA-containing phospholipids species triggered by ACSL4-KO (Fig. 3I). Moreover, AS treatment could not further reduce the level of these long-chain PUFA-containing phospholipids species in ACSL4-KO HT-1080 cells (Fig. 3I). Collectively, these results suggest that AS prevents ferroptosis via directly targeting ACSL4 and inhibiting its enzymatic activity.
Fig. 3. AS directly targets ACSL4 to inhibit ferroptosis.
(A and B) The cell lysates of HT-1080 (A) or MDA-MB-231 (B) were incubated with different concentrations of Bio-AS for 2 hours and then pulled down by using streptavidin beads. The total proteins (input), bound proteins (PD), and remained proteins (after PD) were immunoblotted as indicated. (C) The cell lysates of HT-1080 were incubated with or without AS for 2 hours and then treated with different concentrations of pronase. The expression of ACSL4 and GPX4 was detected by immunoblotting. (D) The cell lysates of HT-1080 were incubated with or without AS for 2 hours and then treated with increasing melting temperature (37° to 61°C). The expression of ACSL4 was detected by immunoblotting. (E) The recombinant ACSL4 protein was incubated with different concentrations of Bio-AS for 2 hours and then pull-down by using streptavidin beads. The expression of ACSL4 was detected by immunoblotting. (F and G) CRISPR-Cas9–generated ACSL4-knockout HT-1080 cells were transduced with or without human ACSL4 and then treated with AS for 1 hour and stimulated with RSL3. The expression of ACSL4 was detected by immunoblotting (F), and the cell viability was detected by CCK-8 assay (G). (H) Recombinant ACSL4 protein was incubated with different concentrations of AS for 10 min at 37°C. The enzymatic activity of ACSL4 was analyzed by measuring the formation of AA-CoA from AA following the protocol described in Material and Methods. (I) Heatmap of major PE species in WT and ACSL4 KO HT-1080 cells treated with or without AS and then stimulated with RSL3 for 6 hours. Each PE/PC species was normalized to the corresponding mean value. Data are the mean ± SEM, n = 3 biologically independent experiments [(G) and (H)]. Statistical analysis was performed using an unpaired two-tailed Student’s t test. NS, P > 0.05, **P < 0.01, ***P < 0.001. PE, phosphatidylethanolamine; PC, phosphatidyl cholines.
AS binds to the glutamine 464 of ACSL4
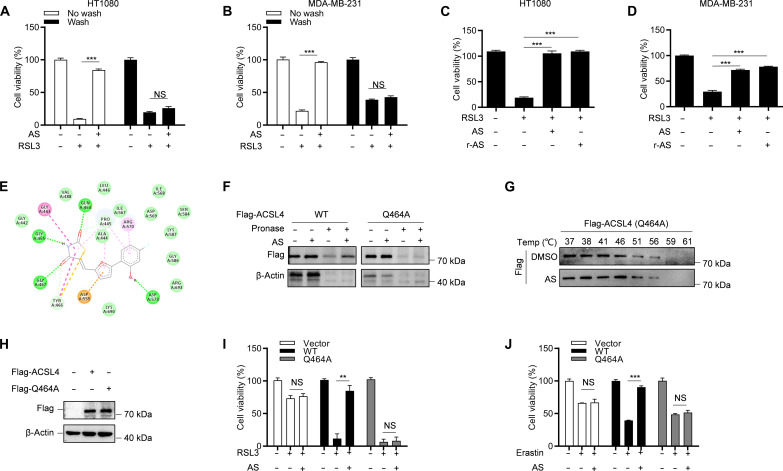
Next, we investigated in detail how AS binds to ACSL4. Since AS structure has an acrylamide group, which is an electrophile that usually covalently and irreversibly bind to the cysteine in target protein through a Michael addition reaction (29), we next explored whether AS has an irreversible inhibitory effect on ferroptosis. After treating HT-1080 and MDA-MB-231 cells with AS for 1 hour, we washed the cells three times to remove the free compound and then stimulated the cells with RSL3. The results showed that the antiferroptotic effect of AS disappeared after washing (Fig. 4, A and B), indicating that the inhibition of ferroptosis by AS was reversible. To investigate the contribution of acrylamide group to the antiferroptotic effect of AS, we synthesized reduced AS (r-AS) by a reduction reaction to destroy the acrylamide group of AS (fig. S5A). Similar to AS, r-AS still inhibited RSL3-induced ferroptosis (Fig. 4, C and D). These results indicate that AS may bind to ACSL4 through a noncovalent bond and reversibly inhibits ferroptosis.
Fig. 4. AS binds to the Gln464 residue of ACSL4 to inhibit its activity.
(A and B) Cell viability analysis of HT-1080 cells (A) or MDA-MB-231 cells (B) pretreated with AS for 30 min and washed three times and then stimulated with 1 μM RSL3 for 8 hours. (C and D) Cell viability analysis of HT-1080 cells (C) or MDA-MB-231 cells (D) pretreated with AS or r-AS for 1 hour and then stimulated with 1 μM RSL3 for 8 hours. (E) Three-dimensional binding mode diagrams between ACSL4 and AS. (F) The cell lysates of HEK293T overexpressing WT or Q464A mutant Flag-ACSL4 were incubated with or without AS for 2 hours and then treated with different concentrations of pronase. The expression of ACSL4 was detected by immunoblotting. (G) The cell lysates of HEK293T overexpressing Flag-ACSL4 Q464A were incubated with or without AS for 2 hours and then treated with increasing melting temperature (37° to 61°C). The expression of ACSL4 was detected by immunoblotting. (H to J) CRISPR-Cas9–generated ACSL4-knockout HT-1080 cells were transduced with WT or Q464A mutant Flag-ACSL4 and then treated with AS for 1 hour and stimulated with RSL3 or erastin. The expression of ACSL4 was detected by immunoblotting (H), and the cell viability was analyzed by CCK-8 [(I) and (J)]. Data are the mean ± SEM, n = 3 biologically independent experiments [(A) to (D) and (I) and (J)]. Statistical analysis was performed using an unpaired two-tailed Student’s t test. NS, P > 0.05, **P < 0.01, ***P < 0.001.
We further identify the possible binding sites of AS and ACSL4. Since the crystal structure of ACSL4 is not yet available, we used the AlphaFold-based three-dimensional structure of ACSL4 for molecular interaction docking to predict the binding site of AS. The results showed that AS was readily docked into the pocket of ACSL4 with a binding energy of −6.37 kcal/mol (fig. S5B). Furthermore, AS may bind to ACSL4 through hydrogen bonding at four residues, including Q464, G465, G467, and D573 (Fig. 4E). To decipher which residue in ACSL4 binds to AS, DARTS was performed in human embryonic kidney (HEK) 293T cells overexpressing either WT or mutant plasmids of ACSL4. We found that only the Q464A mutation in ACSL4 abolished its binding to AS (Fig. 4F and fig. S5C), suggesting that AS may bind to Q464 of ACSL4. To further clarify this result, CETSA analysis was performed, and we found that AS had no effect on ACSL4 thermostability compared to control upon Q464A mutation (Fig. 4G and fig. S5D). Furthermore, we found that AS inhibited RSL3- and erastin-induced ferroptosis in ACSL4-KO HT-1080 cells reconstituted with WT ACSL4 but not in ACSL4-KO HT-1080 cells reconstituted with mutant ACSL4 (Q464A) (Fig. 4, H to J). Collectively, these results suggest that AS binds directly to Q464 of ACSL4 to inhibit ferroptosis.
Next, we investigate the specificity of ACSL4 binding and inhibition by AS. The mammalian ACSL family contains five members, namely, ACSL1, ACSL3, ACSL4, ACSL5, and ACSL6. We analyze the conservation of Q464 in these ACSL proteins and found that Q464 was present only in ACSL3 and ACSL4, whereas it was absent in ACSL1, ACSL5, and ACSL6 (fig. S6A). Since Q464 is conserved in ACSL3 and ACSL4, we explored whether AS could also bind to ACSL3. By using the DARTS analysis, we found that AS does not bind to ACSL3 (fig. S6B). To explore the effect of AS on the enzymatic activities of ACSL1 and ACSL3, we purified these two proteins (fig. S6C). By using the enzyme activity assays in vitro, we found that AS had no effect on the activities of ACSL1 and ACSL3 (fig. S6, D and E). Thus, these results indicate that AS specifically binds to ACSL4 and inhibits its enzymatic activity.
Since several compounds, including triacsin C, PRGL493, rosiglitazone (ROSI), pioglitazone (PIO), and troglitazone (TRO), have recently been reported to be ACSL4 inhibitors (30), we thus compared the inhibitory efficacy and specificity of AS and these compounds on ACSL4 and ferroptosis. The IC50 of these compounds on RSL3-induced ferroptosis were tested, and we found that AS was more effective in inhibiting ferroptosis than other ACSL4 inhibitors except TRO (fig. S7, A to D). Unexpectedly, one of the inhibitors of ACSL4, the natural product triacsin C, failed to inhibit ferroptosis (fig. S7E). One possible reason is that triacsin C may be more inclined to induce apoptosis and necrosis (31). Since TRO has a better inhibitory effect on ferroptosis, we next explore whether it specifically targets ACSL4 like AS. DARTS analysis was performed in HT-1080 cells, and we found that TRO did not bind to ACSL4 (fig. S7F). Previous studies reported that TRO has antioxidant activity (32), and our results also showed that TRO was able to scavenger free radicals by DPPH analysis (fig. S7G), implying that TRO may have a broad antioxidant effect rather than being a specific inhibitor of ferroptosis. In addition, since AS, ROSI, PIO, and TRO belong to the thiazolidinediones compounds and share the same compound scaffold, we thus investigate whether AS targets ACSL4 via the thiazolidinediones motif. The results showed that thiazolidinedione could not bind to ACSL4 (fig. S7H). Together, these results indicate that AS is a potent, specific, and targeted inhibitor of ACSL4.
AS-loaded nanoparticles inhibit ferroptosis in vitro
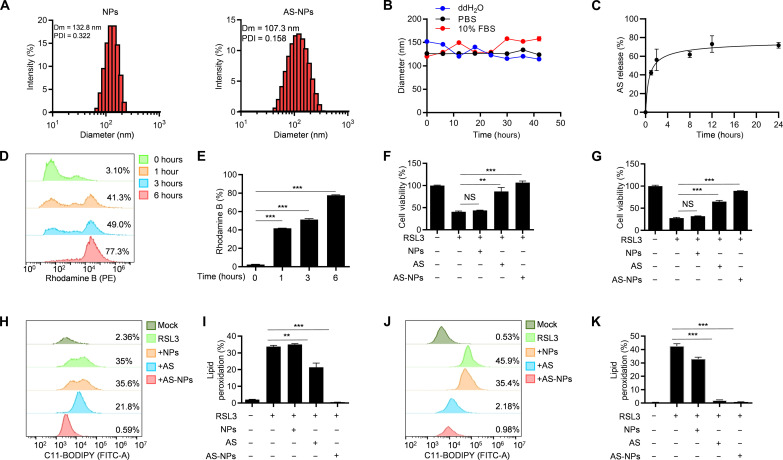
Since AS is a hard-to-dissolve compound with a short half-life (t1/2 = 1 hour) and a high clearance rate (2.25 liters/kg per hour) (22), we developed a methoxy poly(ethylene glycol)-poly (ε-caprolactone) (mPEG-PCL) copolymer–based nanoparticle for AS delivery (fig. S8A). Dynamic light scattering analysis revealed that the size of AS-loaded nanoparticles (AS-NPs) was approximately 107.3 nm (Fig. 5A), and transmission electron microscopy images also showed that AS-NPs were spherical with an average size of 100 nm (fig. S8B). The average ζ-potentials of these AS-NPs was around −1.0 mV (fig. S8C), and AS-NPs had superior stability in purified water, phosphate-buffered saline (PBS), or PBS containing 10% fetal bovine serum (FBS) (Fig. 5B). Moreover, AS-NPs have a good drug release ability, which can release the encapsulated AS rapidly and efficiently (Fig. 5C). To investigate the cellular uptake of AS-NPs, HT-1080 cells were incubated with rhodamine B loaded nanoparticles for various time periods. We found that the percentage of rhodamine B–positive cells reach up to 85% at 6 hours (Fig. 5, D and E), suggesting that AS-NPs could easily deliver AS into the cells.
Fig. 5. AS-NPs inhibit ferroptosis.
(A) The diameter of the NPs and AS-NPs. (B) Stability of the NPs and AS-NPs in different media (ddH2O, PBS, 10% and FBS). Dm, diameter; PDI, polydispersity index. (C) Percentage of AS-NPs released at different time points. (D) The uptake of nanoparticle was analyzed by flow cytometry in HT-1080 cells. (E) The percentage of rhodamine B–positive HT-1080 cells. (F and G) Cell viability analysis of HT-1080 cells (F) or MDA-MB-231 cells (G) pretreated with AS or AS-NPs for 3 hours and then stimulated with 1 μM RSL3 for 8 hours. (H and J) Lipid peroxidation was analyzed by flow cytometry in HT-1080 cells (H) or MDA-MB-231 cells (J) pretreated with AS or AS-NPs for 3 hours and then stimulated with 1 μM RSL3 for 8 hours in the presence of C11-BODIPY. (I and K) The percentage of C11-BODIPY–positive HT-1080 cells (I) or MDA-MB-231 cells (K) pretreated with AS or AS-NPs for 3 hours and then stimulated with 1 μM RSL3 for 8 hours in the presence of C11-BODIPY. Data are the mean ± SEM, n = 3 biologically independent experiments [(E) to (G), (I), and (K)]. Statistical analysis was performed using an unpaired two-tailed Student’s t test. NS, P > 0.05, **P < 0.01, ***P < 0.001.
Next, to investigate whether AS-NPs could efficiently inhibit ferroptosis in vitro, we pretreated HT-1080 and MDA-MB-231 cells with free AS or AS-NPs and then stimulated with RSL3. The results showed that AS-NPs rescued cells from ferroptosis as useful as free AS (Fig. 5, F and G) and also blocked RSL3-induced cell death indicated by PI staining (fig. S9, A to D). Moreover, erastin- or FIN56-induced cell death was also inhibited by AS-NPs in both HT-1080 and MDA-MB-231 cells (fig. S9, E to H). Lipid peroxidation as a biomarker of ferroptosis was also examined, and we found that AS-NPs could effectively inhibit lipid peroxidation (Fig. 5, H to K). In sum, these data suggest that AS-NPs have a notable antiferroptotic effect in vitro.
AS-NPs protect against I/R–induced acute kidney injury
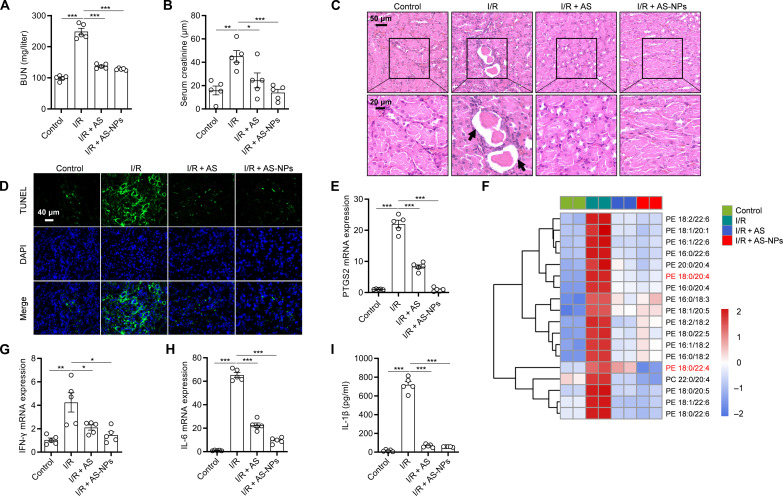
On the basis of the favorable antiferroptotic effect of AS-NPs in vitro, we further investigated the protective effect of AS-NPs against ferroptosis-related diseases in vivo. Previous studies have reported that ferroptosis has a dominant role in I/R-induced acute kidney injury (AKI) (33), suggesting that ferroptosis is a potential target for the treatment of kidney injury. To examine the effect of AS-NPs on AKI in mice, AS-NPs (20 mg/kg) were intraperitoneally injected at 1 hour before renal I/R. We found a markedly reduction in blood urea nitrogen (BUN) and serum creatinine levels after AS-NP treatment (Fig. 6, A and B). Kidney injury was evaluated by hematoxylin and eosin (H&E) staining and terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) staining, and the results showed that AS-NPs markedly reduced the widespread degeneration of tubular architecture, renal tubular cell swelling, and damage (Fig. 6, C and D). We next investigated the effect of AS-NPs on kidney inflammation and ferroptosis. Prostaglandin-endoperoxide synthase 2 (PTGS2) is a key enzyme in prostaglandin biosynthesis, which was recently identified as a biomarker of ferroptosis (14). The result showed that the expression of PTGS2 was markedly decreased after AS-NP treatment (Fig. 6E). Furthermore, we performed lipidomic analysis of kidney and found that AS-NP treatment hugely reduced the level of different long-chain PUFA-containing phospholipids species (Fig. 6F). In addition, the expression of proinflammatory cytokines IFN-γ and interleukin-6 (IL-6) in kidney and the level of IL-1β in serum were also inhibited by AS-NPs (Fig. 6, G to I). All these results suggest that AS-NPs inhibit renal inflammation and ferroptosis and alleviate AKI after I/R.
Fig. 6. AS-NPs protect against I/R-induced AKI.
(A) The levels of BUN. (B) The levels of serum creatinine. (C) Representative images of kidney sections with H&E staining. Black arrows indicate renal tubular injury during renal IRI. (D) Representative images of kidney sections with TUNEL staining. (E) The mRNA levels of PTGS2. (F) Heatmap of major PE/PC species in kidney of mice pretreated with AS or AS-NPs for 1 hour before kidney I/R. Each PE/PC species was normalized to the corresponding mean value. (G and H) IFN-γ (G) and IL-6 (H) in kidney were evaluated by real-time polymerase chain reaction (PCR). (I) Enzyme-linked immunosorbent assay (ELISA) analysis of IL-1β in serum of C57BL/6J mice after kidney I/R injury and pretreated with free AS or AS-NPs (20 mg/kg). Data are the mean ± SEM, n = 5 biologically independent mice [(A), (B), (E), and (G) to (I)]. Statistical analysis was performed using an unpaired two-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001.
Since AS is involved in a variety of pathophysiological events by acting on the known target PI3Kγ (24), we therefore investigated whether the remission of AKI by AS-NPs was dependent on PI3Kγ. We first tested the expression level and activity of PI3Kγ in the kidney tissues, and the results showed that AS-NPs inhibited PI3Kγ expression and activation (fig. S10A). Furthermore, we examined the effect of CAY, another specific inhibitor of PI3Kγ, on AKI in mice. The results showed that CAY did not affect the production of BUN and creatinine (fig. S10, B and C), and the pathological injury, inflammatory cytokine expression, and ferroptosis level of the kidney were also not changed after CAY treatment (fig. S10, D to H). Together, these results suggest that AS-NPs alleviate AKI by acting on ACSL4 but not PI3Kγ.
AS-NP treatment prevents ConA-induced ALI
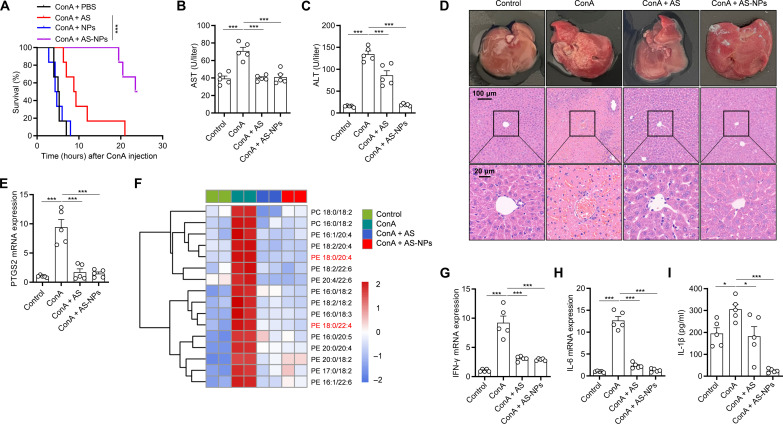
In addition to contributing to AKI, ferroptosis was also found to be involved in concanavalin A (ConA)–induced ALI (34). To investigate whether AS-NPs can alleviate ConA-induced ALI, the C57BL/6 mice were pretreated with AS-NPs before challenge with a lethal dose of ConA (30 mg/kg). The results showed that AS-NPs greatly improved the survival rate of mice compared with free AS and control (Fig. 7A). In addition, the function of liver was analyzed, and we found that AS-NP treatment inhibits serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels (Fig. 7, B and C). Furthermore, the pathological injury of the liver was also greatly improved by H&E staining analysis (Fig. 7D). To investigate the inhibitory effect of AS-NPs on ferroptosis in the liver, we examined the expression of PTGS2 and found that it was steeply inhibited by AS-NPs (Fig. 7E). We further performed lipidomic analysis of liver and found that AS-NP treatment sharply reduced the level of different long-chain PUFA-containing phospholipids species (Fig. 7F). In addition, by examining the levels of liver inflammation, we found that AS-NPs markedly inhibited IFN-γ and IL-6 expression in liver and IL-1β production in serum (Fig. 7, G to I). Together, these results suggest that AS-NPs have a highly potent inhibitory effect on liver inflammation and ferroptosis in mice of ConA-induced ALI.
Fig. 7. AS-NP treatment prevents ConA-induced ALI.
(A) Survival analysis of C57BL/6J mice pretreated with free AS or AS-NPs (20 mg/kg) and then intraperitoneally injected with ConA (30 mg/kg). n = 6 per group. (B to I) C57BL/6J mice pretreated with free AS or AS-NPs (20 mg/kg) and then intraperitoneally injected with ConA (15 mg/kg) for 24 hours. n = 5 biologically independent mice. Serum levels of AST (B) and ALT (C). Images of H&E staining in liver cross sections (D). The mRNA expression of PTGS2 was evaluated by real-time PCR (E). Heatmap of all major PE/PC species in liver with hierarchical clustering of control, ConA, ConA + AS, and ConA + AS-NPs. Each PE/PC species was normalized to the corresponding mean value (F). The mRNA expression of IFN-γ (G) and IL-6 (H) in liver was evaluated by real-time PCR. IL-1β (I) in serum was analyzed by ELISA. Data are the mean ± SEM. Statistical analysis was performed using an unpaired two-tailed Student’s t test [(B), (C), (E), and (G) to (I)] or generalized Wilcoxon test (A). *P < 0.05, ***P < 0.001.
We next explored whether the effect of AS-NPs on preventing liver injury was dependent on PI3Kγ. The results showed that AS-NPs inhibited PI3Kγ expression and activation in the liver (fig. S11A). Furthermore, we tested the effect of CAY on ALI in mice and found that CAY had no effect on the production of AST and ALT (fig. S11, B and C). Moreover, the pathological injury, inflammatory cytokine expression, and ferroptosis level of the liver were also not changed after CAY treatment (fig. S11, D to H). In summary, these results suggest that AS-NPs attenuate ConA-induced AKI independent of PI3Kγ.
DISCUSSION
Ferroptosis is a type of programmed cell death driven by iron-dependent lipid peroxidation that has been implicated in the pathogenesis of multiple diseases, including organ injury, I/R, and neurodegenerative diseases (9, 10). Therefore, the development of targeted inhibitors of ferroptosis may provide candidate strategies for the treatment of these diseases. In this study, we identified that AS can directly bind to the glutamine 464 of ACSL4 and thus act as a specific and targeted ferroptosis inhibitor. Furthermore, AS has a notable therapeutic effect on ferroptosis-related diseases, including AKI and ALI. These findings not only reveal the mechanism by which AS inhibits ferroptosis but also provide a potential therapeutic approach for the treatment of ferroptosis-related diseases.
Now, a few small-molecule compounds have shown potential inhibitory effects on ferroptosis. Deferoxamine as an iron chelator has been found to inhibit ferroptosis and has neuroprotective effects on primary cortical neurons (34). However, iron is an essential element for many fundamental physiological activities, and chelation iron by deferoxamine may increase the risk of iron imbalance (35, 36). Vitamin K, serotonin (5-hydroxytryptamine), 3-hydroxyanthranilic acid, Fer-1, and liproxstatin-1 were found to inhibit ferroptosis by acting as potent RTAs (37–39), suggesting that these compounds may have a wide range of antioxidant effects. In addition, itaconate, quercetin, and resveratrol have been found to inhibit ferroptosis via defense against oxidative stress (40, 41). Despite the existence of different ferroptosis inhibitors, the efficacy of their clinical application is still unclear. Our results demonstrate that AS is a specific ferroptosis inhibitor by directly targeting the Q464 residue of ACSL4, which provides a potential candidate drug for the clinical treatment of ferroptosis-related diseases.
AS is a potent PI3Kγ inhibitor that prevents tumor cell growth and enhances tumor cell therapy (42, 43). However, its role in ferroptosis is now unknown. Our data identified that AS is a potent inhibitor of ferroptosis through kinase inhibitor library screening. AS inhibited ferroptosis with an IC50 value of 2.2 μM, indicating that it was more effective than most other ferroptosis inhibitors. Furthermore, we found that AS was able to inhibit ferroptosis induced by various activators in a variety of human and murine cell lines, suggesting that AS is a common inhibitor of ferroptosis. Unexpectedly, we found that AS inhibited lipid peroxidation and ferroptosis in an PI3Kγ-independent manner, suggesting that the antiferroptotic effect of AS may be dependent on other targets. Further identification by mass spectrometry showed that AS directly binds to Q464 of ACSL4 to inhibit its enzymatic activity. However, the Q464A mutation of ACSL4 has no effect on ferroptosis, but it inhibits the antiferroptotic effect of AS. One possible mechanism is that AS might trigger a conformational change in ACSL4 and thereby inhibit its enzymatic activity.
Since I/R-induced AKI and ConA-induced ALI are two classical animal models of ferroptosis (32, 33), we evaluated the efficacy of AS in vivo by using these two animal models. To overcome the low water solubility and short half-life of AS, we developed an AS-NP. Our study demonstrates that AS-NPs notably inhibited ferroptosis in vivo and in vitro. Furthermore, AS-NP treatment notably reduced the inflammatory response and improved the pathogenesis of AKI and ALI. Considering that ferroptosis is not only implicated in AKI and ALI, it has also been reported to contribute to the pathogenesis of other ferroptosis-related diseases, such as neurodegenerative diseases, pulmonary fibrosis, and autoimmune diseases (10, 44, 45). Therefore, AS may have great potential in the treatment of these ferroptosis-related diseases.
MATERIALS AND METHODS
Chemicals
AS (T6208), RSL3 (T3646), erastin (T1765), taselisib (T1999), rosiglitazone (T0334), troglitazone (T3170), pioglitazone (T0214), PRGL493 (T35666), CAY (T6433), AS-605240 (T2248), IITZ-01 (T6799), CZC24832 (T1949), afuresertib (T1911), PHT-427 (T2420), ipatasertib (T6252), and AKT inhibitor VIII (T3346) were purchased from Topscience; FIN56 (HY-103087) and Fer-1 (HY-100579) were purchased from MedChemExpress; triacsin C (T139793) was purchased from Aladdin; and 2,4-thiazolidinedione (T819408) was purchased from Macklin.
Cells lines
HT-1080, MDA-MB-231, HT22, L-02, A549, Hela, B16-F10, MCA205, HK-2, and HEK293T cells were cultured in Dulbecco’s minimum essential medium (DMEM) supplemented with 10% FBS and 1% penicillin/streptomycin (P/S) at 37°C in an incubator with a humidified atmosphere of 20% O2 and 5% CO2. All cell lines in our laboratory were routinely tested for mycoplasma contamination, and the cells used in this study were negative for mycoplasma.
Animals
C57BL/6 male mice aged 8 to 10 weeks were obtained from GemPharmatech. All mice were kept under specific pathogen–free conditions in the Animal Facility of the Jiangnan University Animal Center. All procedures involving mice and experimental protocols were approved by the Ethics Committee of Jiangnan University Animal Center.
Assessment of lipid peroxidation
Cells were seeded in six-well plates at a density of 5 × 104 cells per well. On the next day, the cells were treated with the indicated compounds and then collected, stained with 5 μM BODIPY 581/591 C11 (Invitrogen, D3861) at 37°C for 30 min, and analyzed by flow cytometry. At least 5000 cells were analyzed in each group, and all experiments were repeated at least three times.
LDH release assay
Cell death assays were performed by measuring the release of LDH from cells into the culture medium by the LDH Cytotoxicity Assay Kit (Beyotime, C0016) according to the manufacturer’s instructions. The fluorescence intensity was measured at a wavelength of 490 nm and was determined using a microplate reader (BioTek Epoch).
Cell viability assay
Cell viability was measured using a Cell Counting Kit-8 (CCK-8, Topscience) according to the manufacturer’s instructions. Briefly, cells were seeded in a 96-well plate at a density of 5000 cells per well. The next day, after the treatment of different compounds for a certain time, each well was added with 10 μl of CCK-8 and cultured for 2 hours (37°C, 5% CO2), and the absorbance was detected by using a microplate reader (BioTek Epoch) at 450 nm.
PI staining
Cells were seeded in a 12-well plate at a density of 2.5 × 105 cells per well. On the second day, the cells treated with the corresponding compounds for a certain time were stained with PI (5 μg/ml) and photographed by a Nikon microscope (NiKon Ti-U). All experiments were repeated at least three times.
Western blot
Cells were washed in cold PBS and lysed in NP-40 lysis buffer (Beyotime, P0013F). Lysates were incubated on ice for 30 min and cleared by centrifugation at 12,000g for 10 min. Protein concentration was quantified using a BCA Protein Assay Kit (Beyotime, P0012). Total protein was mixed with sample buffer and denatured at 100°C for 10 min. Sample was separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane (Millipore, IPVH00010). Membranes were blocked with 5% (w/v) nonfat dry milk and incubated with primary antibodies β-actin rabbit monoclonal antibody (mAb; Abmart, P30002), PI3Kγ rabbit mAb (Zenbio, R25369), p-AKT (Ser473) rabbit mAb (Cell Signaling, #9271), GPX4 rabbit mAb (ABclonal, A1933), ACSL3 rabbit mAb (Zenbio, 161226), and ACSL4 rabbit mAb (Zenbio, R24265), overnight at 4°C followed by washing in 0.1% Tween/PBS. Membranes were incubated with corresponding secondary antibodies at 25°C for 1 hour and then washed three times before signal detection.
Real-time PCR
Total RNA was prepared using the TRIzol Reagent (Vazyme, R401-01) according to the manufacturer’s instructions. cDNA was synthesized using HiScript II Q RT SuperMix for quantitative polymerase chain reaction (qPCR; Vazemy, R223-01). qPCR was performed on cDNA using Hieff qPCR SYBR Green Master Mix (Yeasen, 11201ES08) on a StepOnePlus Real-Time PCR System. Gene expression was quantified using the following primers: human β-actin forward: GCTCCTCCTGAGCGCAAGTAC; human β-actin reverse: GGACTCGTCATACTCCTGCTTGC; human PTGS2 forward: CGGTGAAACTCTGGCTAGACAG; human PTGS2 reverse: GCAAACCGTAGATGCTCAGGGA; mouse PTGS2 forward: CGGACTGGATTCTATGGTGAAA; mouse PTGS2 reverse: CTTGAAGTGGGTCAGGATGTAG; mouse IFN-γ forward: AGGTCAACAACCCACAGGTC; mouse IFN-γ reverse: ATCAGCAGCGACTCCTTTTC; mouse IL-6 forward: CCGGAGAGGAGACTTCACAG; mouse IL-6 reverse: CAGAATTGCCATTGCACAAC. The expression levels of target genes were normalized by subtracting the corresponding β-actin threshold cycle value. All fold changes were expressed normalized to the untreated control.
Drug affinity–responsive target stability
The cells were collected and the total proteins were separated by the use of lysis buffers. The cell lysates were centrifuged at 12,000g at 4°C for 10 min. The supernatant was treated with dimethyl sulfoxide (DMSO) or AS. After incubation at 4°C for 2 hours, pronase (Roche, 10165921001) was added to the lysate at room temperature and incubated for another 2 hours. The reaction was terminated by adding a protease inhibitor mixture and SDS-PAGE sample buffer, followed by Western blotting.
Cell thermal shift assay
Cells were collected, and the pelleted cells were washed with PBS, followed by lysis with NP-40 (containing phosphatase inhibitors) for 30 min at 4°C. Subsequently, the samples were centrifuged at 12,000g for 10 min at 4°C. The protein supernatant after centrifugation was treated with DMSO or AS. After being coincubated with drugs for 2 hours, the cells in each group were heated at different temperatures (37° to 61°C) for 5 min and then centrifuged at 4°C for 10 min, and the supernatants were analyzed by immunoblotting.
Chemical synthesis
Structure the HS-PEG-H-AS (r-AS)
AS (163.9 μl, 30.5 mg/ml) was mixed with HS-PEG-OH (0.5 μl, 78.8 mg/ml) and 2,2-dimethoxy-2-phenylacetophenone (1 ml, 10.0 mg/ml), and the reaction was stirred overnight under UV (350 nm) at room temperature. Then, the vacuum freeze dryer was used for the mixture solution to dry and to give the desired product r-AS. The bonding efficiency was measured by nuclear magnetic resonance (NMR; 30.1 mg, 80%) as a yellow solid.
Structure the Bio-AS
d-Biotin (2.61 mg) was mixed with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (1.3 mg) and 4-dimethylaminopyridine (1.65 mg) in 100 μl of DMSO. Then, the freezing dryer was used for the mixture solution to dry and to give the desired product Bio-AS. The conjugation efficacy was measured by 1H-NMR (10.0 mg, 70%) as a yellow solid.
DPPH assay
Substances (Fer-1, AS, and DMSO) were diluted in methanol and pipetted in 96-well plates in triplicate. DPPH solution (final concentration, 200 μM) was added to start the reaction. After the equilibrium settled (generally after 0.5 hours), the fluorescence intensity was measured at a wavelength of 517 nm and was determined using a microplate reader (BioTek Epoch) instrument. All values were normalized to DMSO.
Iron chelating studies
FeSO4 or FeCl3 (1.5 mM) solution was added into the solution containing DFO or AS (1.5 mM) at the concentration of 50 μM in Hepes buffer (20 mM, pH = 7.4) with 150 mM NaCl. The UV-vis spectra in the range of 290 to 600 nm for DFO and 290 to 600 nm for AS were recorded by a microplate reader (Synergy H4).
Detection of intracellular Fe2+
Cells were seeded in a 12-well plate at a density of 2.5 × 105 cells per well. On the second day, the cells treated with the corresponding compounds for a certain time were stained with FerroOrange (1 μM). Nuclei were costained with antifade mounting medium with 4′,6-diamidino-2-phenylindole (DAPI; Beyotime, P0131), and images were captured under a microscope (ZEISS, Axio Imager Z2).
Histology and immunostaining
The samples were collected and immediately fixed in 4% paraformaldehyde fix solution (Beyotime, P0099) overnight. After being washed once with PBS, samples were transferred into 70% ethanol and subjected to embedding, and sections were prepared and stained with H&E. Slides were examined under a Nikon ECL IPSE Ci biological microscope, and images were acquired with a Nikon DS-U3 color digital camera.
TUNEL staining
TUNEL staining was used to evaluate sample apoptosis. The samples were fixed in 4% paraformaldehyde fix solution (Beyotime, P0099) overnight at room temperature and then washed with PBS, and the samples were treated with the CoraLite Plus 488 TUNEL Assay Apoptosis Detection Kit (Proteintech, PF00006) according to the manufacturer’s instructions. Nuclei were costained with antifade mounting medium with DAPI (Beyotime, P0131), and images were captured under a microscope (ZEISS, Axio Imager Z2).
Plasmid construction and lentivirus infection
We used CRISPR-Cas9 technology to knock out PI3Kγ, GPX4, and ACSL4. Each single guide RNA (sgRNA) was cloned into the empty backbone of lenti-CRISPR v2. The following sgRNA sequences were used: sgPI3Kγ, 5′-TGGGCAGCACGAACTCGATG-3′ and 3′-CATCGAGTTCGTGCTGCCCA-5′; sgGPX4: 5′-CGTGTGCATCGTCACCAACG-3′ and 3′-CGTTGGTGACGATGCACACG-5′; sgACSL4, 5′-GACATTGAACGAATGTATGG-3′ and 3′-CCATACATTCGTTCAATGTC-5′. LentiGuide clones were transfected into HEK293T cells with a psPAX2 packaging plasmid and pMD2.G expressing plasmid. Cells were infected with lentivirus and selected with puromycin (Biosharp, BS111) for 7 days, and then single cells were sorted into 96-well plates. Single cells were maintained in DMEM with 10% FBS and 1% P/S at 37°C in an incubator with 20% O2 and 5% CO2 for 3 to 4, weeks and each colony was verified by Western blot to confirm target gene deletion.
ACSL4 activity assay
ACSL4 activity was measured following a previously described methodology (45). Briefly, recombinant ACSL4 protein was coincubated with different concentrations of AS for 10 min at 37°C in a 100-μl reaction system containing 175 mM tris (pH 7.4), 10 mM adenosine triphosphate, 8 mM MgCl2, 5 mM dithiothreitol, 250 μM CoA, and 50 μM [3H]-AA. After terminating the reaction with 1 ml of ethyl acetate, the upper layer was removed, and the lower layer aqueous phase was further extracted twice. Then, the radioactivity of the extracted aqueous phase was evaluated by the liquid scintillation counter. The result was expressed as a percentage of the total enzymatic activity.
Mass spectrometry analysis
Lipids were extracted from approximately 30 mg of tissues, or 5 × 106 cells using the lipids were extracted using the Bligh and Dyer method. The organic phase was dried and dissolved in 50 μl of mobile phase A [trichloromethane/isopropanol 2:1 (v/v)] for vortex. Then, the phospholipids were mixed with mobile phase B [acetonitrile/isopropanol/water 7:9:4 (v/v/v) and 10 mM ammonium acetate], and MS and MS/MS analysis of PLs was performed on a Q-Exactive hybrid quadrupole-orbitrap mass spectrometer (Thermo Fisher Scientific).
In situ pull-down experiment and target validation
HT-1080 cells were grown in culture medium until 70 to 80% confluence. The cells were lysed by NP-40 lysis buffer (Beyotime, P0013F) for 30 min at 4°C. Next, the lysis buffer was subsequently collected by centrifugation at 12,000g for 10 min at 4°C. The supernatants were incubated with Bio-AS for 2 hours at 4°C, and then the mixture was incubated with streptavidin beads (Thermo Fisher Scientific, 20359). The beads were washed with PBS. The enriched proteins were suspended by loading buffer and boiled for 15 min and then separated by SDS-PAGE for target validation and analyzed by LC-MS/MS.
Preparation and characterization of AS-NPs
One milligram of mPEG-PCL (20.0 mg/ml) mixed with 50 μg of AS (30.5 mg/ml) was stirred in 700 μl of polyvinylpyrrolidone water (1 mg/ml) for 3 to 5 min at 60°C, the dialyzed solution was then centrifuged at 4°C at 21,000g for 1 hour, the supernatant was removed, and 1 ml of distillation-distillation H2O (ddH2O) was added to resuspend. The NanoSight NS300 was used to measure particle size and zeta potential.
AS-NP stability
The dispersion solution of 300 μl of nanoparticles was placed in a glass bottle at 37°C for 0, 6, 12, 18, 24, 30, 36, and 42 hours to observe the state of the dispersion solution, and the particle size of AS-NPs was measured.
In vitro drug release studies
A certain amount of the prepared drug-carrying nanoparticles was loaded into the dialysis bag and then sealed and put into 20 ml of ddH2O (pH 7.4) to investigate the drug release behavior in vitro. At a specific point in time, 1 ml of ddH2O was taken out to determine the content of AS, so as to obtain the release curve in vitro.
ConA-induced acute liver injury mouse models
C57BL/6 male mice were housed in a dedicated pathogen-free animal facility. For survival studies, the dose of ConA (Sigma-Aldrich, 11028-71-0) used was 30 mg/kg. In ALI studies, the ConA dose was reduced to 15 mg/kg. ConA was injected into the tail vein of mice. For measurement of proinflammatory cytokines, blood was collected from the eyeball of mice 3 hours after ConA injection. Mice were euthanized 24 hours after injection. At this time point, liver and blood samples were collected for H&E staining and measurement of AST/ALT (Nanjing Jiancheng Bioengineering Institute) according to the instructions in the manual.
IRI-AKI mouse experiment
C57BL/6 male mice were randomly divided into four groups (n = 5). Intraperitoneal injection of AS and AS-NPs 1 hour before molding and then IRI-AKI was induced by clamping the renal pedicles for 25 min. The mice in the control group received operations in which the kidneys were exposed but not clamped, and they were treated with sterile PBS. After reperfusion for 24 hours, the mice were euthanized, and the blood and kidneys were obtained for analysis. Kidney and blood samples were collected for H&E staining and BUN/creatinine (Nanjing Jiancheng Bioengineering Institute) according to the instructions in the manual.
Statistical analysis
The data were presented based on at least three independent experiments as mean ± SEM. The comparison of indicated two groups was performed by Student’s t test (two-tailed, unpaired) using GraphPad Prism 8: *P < 0.05, **P < 0.01, ***P < 0.001, and not significant (NS). All of the statistical details can be found in the figure legends.
Acknowledgments
We thank R. Zhou (University of Science and Technology of China) for helpful and insightful comments on the article.
Funding: This work was supported by the National Natural Science Foundation of China (82202038), the Natural Science Foundation of Jiangsu Province (BK20221085), the Wuxi Science and Technology Development Fund (K20231057), and the General Program of Wuxi Medical Center, Nanjing Medical University (WMCG202308).
Author contributions: Conceptualization: Q.H., Y.R., F.G., and Y.H. Formal analysis: Q.H., Y.R., and D.L. Investigation: Q.H., Y.R., X.L., X.Z., M.L., and Y.H. Validation: Q.H., Y.R., X.L., X.Z., M.L., and Y.H. Methodology: Q.H., Y.R., X.L., Y.M., M.L., F.G., and Y.H. Project administration: Q.H., Y.R., X.Z., W.X., and Y.H. Funding acquisition: W.X., F.G., and Y.H. Data curation: Q.H., Y.R., and Y.H. Software and data visualizations: Q.H., Y.R., X.L., M.L., W.X., and Y.H. Resources: Y.R., Y.L., X.L., X.Z., and M.L. Supervision: M.L., W.X., and Y.H. Writing—original draft: Q.H. and Y.H. Writing—review and editing: Q.H., Y.R., and Y.H. All authors approved the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Proteomic data have been deposited in the ProteomeXchange Consortium via the iProX partner repository under accession code PXD046602.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Legends for tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.Stockwell B. R., Friedmann Angeli J. P., Bayir H., Bush A. I., Conrad M., Dixon S. J., Fulda S., Gascon S., Hatzios S. K., Kagan V. E., Noel K., Jiang X., Linkermann A., Murphy M. E., Overholtzer M., Oyagi A., Pagnussat G. C., Park J., Ran Q., Rosenfeld C. S., Salnikow K., Tang D., Torti F. M., Torti S. V., Toyokuni S., Woerpel K. A., Zhang D. D., Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dixon S. J., Lemberg K. M., Lamprecht M. R., Skouta R., Zaitsev E. M., Gleason C. E., Patel D. N., Bauer A. J., Cantley A. M., Yang W. S., Morrison B. III, Stockwell B. R., Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lei G., Zhuang L., Gan B., Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer 22, 381–396 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., Shi J., Liu X., Feng L., Gong Z., Koppula P., Sirohi K., Li X., Wei Y., Lee H., Zhuang L., Chen G., Xiao Z. D., Hung M. C., Chen J., Huang P., Li W., Gan B., BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat. Cell Biol. 20, 1181–1192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang L., Kon N., Li T., Wang S. J., Su T., Hibshoosh H., Baer R., Gu W., Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang W., Green M., Choi J. E., Gijon M., Kennedy P. D., Johnson J. K., Liao P., Lang X., Kryczek I., Sell A., Xia H., Zhou J., Li G., Li J., Li W., Wei S., Vatan L., Zhang H., Szeliga W., Gu W., Liu R., Lawrence T. S., Lamb C., Tanno Y., Cieslik M., Stone E., Georgiou G., Chan T. A., Chinnaiyan A., Zou W., CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 569, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lang X., Green M. D., Wang W., Yu J., Choi J. E., Jiang L., Liao P., Zhou J., Zhang Q., Dow A., Saripalli A. L., Kryczek I., Wei S., Szeliga W., Vatan L., Stone E. M., Georgiou G., Cieslik M., Wahl D. R., Morgan M. A., Chinnaiyan A. M., Lawrence T. S., Zou W., Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Discov. 9, 1673–1685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao P., Wang W., Wang W., Kryczek I., Li X., Bian Y., Sell A., Wei S., Grove S., Johnson J. K., Kennedy P. D., Gijon M., Shah Y. M., Zou W., CD8(+) T cells and fatty acids orchestrate tumor ferroptosis and immunity via ACSL4. Cancer Cell 40, 365–378.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y., Feng D., Wang Z., Zhao Y., Sun R., Tian D., Liu D., Zhang F., Ning S., Yao J., Tian X., Ischemia-induced ACSL4 activation contributes to ferroptosis-mediated tissue injury in intestinal ischemia/reperfusion. Cell Death Differ. 26, 2284–2299 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang X., Stockwell B. R., Conrad M., Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 22, 266–282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraft V. A. N., Bezjian C. T., Pfeiffer S., Ringelstetter L., Muller C., Zandkarimi F., Merl-Pham J., Bao X., Anastasov N., Kossl J., Brandner S., Daniels J. D., Schmitt-Kopplin P., Hauck S. M., Stockwell B. R., Hadian K., Schick J. A., GTP Cyclohydrolase 1/Tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent. Sci. 6, 41–53 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bersuker K., Hendricks J. M., Li Z., Magtanong L., Ford B., Tang P. H., Roberts M. A., Tong B., Maimone T. J., Zoncu R., Bassik M. C., Nomura D. K., Dixon S. J., Olzmann J. A., The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doll S., Freitas F. P., Shah R., Aldrovandi M., da Silva M. C., Ingold I., Goya Grocin A., Xavier da Silva T. N., Panzilius E., Scheel C. H., Mourao A., Buday K., Sato M., Wanninger J., Vignane T., Mohana V., Rehberg M., Flatley A., Schepers A., Kurz A., White D., Sauer M., Sattler M., Tate E. W., Schmitz W., Schulze A., O’Donnell V., Proneth B., Popowicz G. M., Pratt D. A., Angeli J. P. F., Conrad M., FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Yang W. S., SriRamaratnam R., Welsch M. E., Shimada K., Skouta R., Viswanathan V. S., Cheah J. H., Clemons P. A., Shamji A. F., Clish C. B., Brown L. M., Girotti A. W., Cornish V. W., Schreiber S. L., Stockwell B. R., Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alim I., Caulfield J. T., Chen Y., Swarup V., Geschwind D. H., Ivanova E., Seravalli J., Ai Y., Sansing L. H., Ste Marie E. J., Hondal R. J., Mukherjee S., Cave J. W., Sagdullaev B. T., Karuppagounder S. S., Ratan R. R., Selenium drives a transcriptional adaptive program to block ferroptosis and treat stroke. Cell 177, 1262–1279.e25 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Ingold I., Berndt C., Schmitt S., Doll S., Poschmann G., Buday K., Roveri A., Peng X., Porto Freitas F., Seibt T., Mehr L., Aichler M., Walch A., Lamp D., Jastroch M., Miyamoto S., Wurst W., Ursini F., Arner E. S. J., Fradejas-Villar N., Schweizer U., Zischka H., Friedmann Angeli J. P., Conrad M., Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell 172, 409–422.e21 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Frei B., Kim M. C., Ames B. N., Ubiquinol-10 is an effective lipid-soluble antioxidant at physiological concentrations. Proc. Natl. Acad. Sci. U.S.A. 87, 4879–4883 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Littarru G. P., Tiano L., Bioenergetic and antioxidant properties of coenzyme Q10: Recent developments. Mol. Biotechnol. 37, 31–37 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Doll S., Proneth B., Tyurina Y. Y., Panzilius E., Kobayashi S., Ingold I., Irmler M., Beckers J., Aichler M., Walch A., Prokisch H., Trumbach D., Mao G., Qu F., Bayir H., Fullekrug J., Scheel C. H., Wurst W., Schick J. A., Kagan V. E., Angeli J. P., Conrad M., ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 13, 91–98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan H., Li X., Zhang X., Kang R., Tang D., Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem. Biophys. Res. Commun. 478, 1338–1343 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Kagan V. E., Mao G., Qu F., Angeli J. P., Doll S., Croix C. S., Dar H. H., Liu B., Tyurin V. A., Ritov V. B., Kapralov A. A., Amoscato A. A., Jiang J., Anthonymuthu T., Mohammadyani D., Yang Q., Proneth B., Klein-Seetharaman J., Watkins S., Bahar I., Greenberger J., Mallampalli R. K., Stockwell B. R., Tyurina Y. Y., Conrad M., Bayir H., Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 13, 81–90 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pomel V., Klicic J., Covini D., Church D. D., Shaw J. P., Roulin K., Burgat-Charvillon F., Valognes D., Camps M., Chabert C., Gillieron C., Francon B., Perrin D., Leroy D., Gretener D., Nichols A., Vitte P. A., Carboni S., Rommel C., Schwarz M. K., Ruckle T., Furan-2-ylmethylene thiazolidinediones as novel, potent, and selective inhibitors of phosphoinositide 3-kinase gamma. J. Med. Chem. 49, 3857–3871 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Prasad A., Jia Y., Chakraborty A., Li Y., Jain S. K., Zhong J., Roy S. G., Loison F., Mondal S., Sakai J., Blanchard C., Snyder S. H., Luo H. R., Inositol hexakisphosphate kinase 1 regulates neutrophil function in innate immunity by inhibiting phosphatidylinositol-(3,4,5)-trisphosphate signaling. Nat. Immunol. 12, 752–760 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H.-L., Hu B.-X., Li Z.-L., Du T., Shan J.-L., Ye Z.-P., Peng X.-D., Li X., Huang Y., Zhu X.-Y., Chen Y.-H., Feng G.-K., Yang D., Deng R., Zhu X.-F., PKCβII phosphorylates ACSL4 to amplify lipid peroxidation to induce ferroptosis. Nat. Cell Biol. 24, 88–98 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Shimada K., Skouta R., Kaplan A., Yang W. S., Hayano M., Dixon S. J., Brown L. M., Valenzuela C. A., Wolpaw A. J., Stockwell B. R., Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat. Chem. Biol. 12, 497–503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin M., Zhou Q., Lee E., Dan S., Duan H. Q., Kong D., AS252424, a PI3Kγ inhibitor, downregulates inflammatory responsiveness in mouse bone marrow-derived mast cells. Inflammation 37, 1254–1260 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Graves B. M., Simerly T., Li C., Williams D. L., Wondergem R., Phosphoinositide-3-kinase/akt - dependent signaling is required for maintenance of [Ca(2+)](i), I(Ca), and Ca(2+) transients in HL-1 cardiomyocytes. J. Biomed. Sci. 19, 59 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Z., Lin J., Sun K., Guo J., Yao X., Wang G., Hou L., Xu J., Guo J., Guo F., Deferoxamine alleviates osteoarthritis by inhibiting chondrocyte ferroptosis and activating the Nrf2 pathway. Front. Pharmacol. 13, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boike L., Henning N. J., Nomura D. K., Advances in covalent drug discovery. Nat. Rev. Drug Discov. 21, 881–898 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castillo A. F., Orlando U. D., Maloberti P. M., Prada J. G., Dattilo M. A., Solano A. R., Bigi M. M., Ríos Medrano M. A., Torres M. T., Indo S., Caroca G., Contreras H. R., Marelli B. E., Salinas F. J., Salvetti N. R., Ortega H. H., Lorenzano Menna P., Szajnman S., Gomez D. E., Rodríguez J. B., Podesta E. J., New inhibitor targeting Acyl-CoA synthetase 4 reduces breast and prostate tumor growth, therapeutic resistance and steroidogenesis. Cell. Mol. Life Sci. 78, 2893–2910 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dechandt C. R. P., Zuccolotto-dos-Reis F. H., Teodoro B. G., Fernandes A. M. A. P., Eberlin M. N., Kettelhut I. C., Curti C., Alberici L. C., Triacsin C reduces lipid droplet formation and induces mitochondrial biogenesis in primary rat hepatocytes. J. Bioenerg. Biomembr. 49, 399–411 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Linkermann A., Skouta R., Himmerkus N., Mulay S. R., Dewitz C., De Zen F., Prokai A., Zuchtriegel G., Krombach F., Welz P. S., Weinlich R., Vanden Berghe T., Vandenabeele P., Pasparakis M., Bleich M., Weinberg J. M., Reichel C. A., Brasen J. H., Kunzendorf U., Anders H. J., Stockwell B. R., Green D. R., Krautwald S., Synchronized renal tubular cell death involves ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 111, 16836–16841 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Li X., Ge C., Min J., Wang F., The multifaceted role of ferroptosis in liver disease. Cell Death Differ. 29, 467–480 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Fan B. Y., Pang Y. L., Shen W. Y., Wang X., Zhao C. X., Li W. X., Liu C., Kong X. H., Ning G. Z., Feng S. Q., Yao X., Neuroprotective effect of deferoxamine on erastininduced ferroptosis in primary cortical neurons. Neural Regen. Res. 15, 1539–1545 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dev S., Babitt J. L., Overview of iron metabolism in health and disease. Hemodial. Int. 21, S6–S20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bajbouj K., Shafarin J., Hamad M., High-dose deferoxamine treatment disrupts intracellular iron homeostasis, reduces growth, and induces apoptosis in metastatic and nonmetastatic breast cancer cell lines. Technol. Cancer Res. Treat. 17, 1533033818764470 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishima E., Ito J., Wu Z., Nakamura T., Wahida A., Doll S., Tonnus W., Nepachalovich P., Eggenhofer E., Aldrovandi M., Henkelmann B., Yamada K. I., Wanninger J., Zilka O., Sato E., Feederle R., Hass D., Maida A., Mourao A. S. D., Linkermann A., Geissler E. K., Nakagawa K., Abe T., Fedorova M., Proneth B., Pratt D. A., Conrad M., A non-canonical vitamin K cycle is a potent ferroptosis suppressor. Nature 608, 778–783 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu D., Liang C. H., Huang B., Zhuang X., Cui W., Yang L., Yang Y., Zhang Y., Fu X., Zhang X., Du L., Gu W., Wang X., Yin C., Chai R., Chu B., Tryptophan metabolism acts as a new anti-ferroptotic pathway to mediate tumor growth. Adv. Sci. 10, e2204006 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zilka O., Shah R., Li B., Friedmann Angeli J. P., Griesser M., Conrad M., Pratt D. A., On the Mechanism of Cytoprotection by Ferrostatin-1 and Liproxstatin-1 and the Role of Lipid Peroxidation in Ferroptotic Cell Death. ACS Cent. Sci. 3, 232–243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato K., Takahashi M., Oh-Hashi K., Ando K., Hirata Y., Quercetin and resveratrol inhibit ferroptosis independently of Nrf2-ARE activation in mouse hippocampal HT22 cells. Food Chem. Toxicol. 172, 113586 (2023). [DOI] [PubMed] [Google Scholar]
- 41.He R., Liu B., Xiong R., Geng B., Meng H., Lin W., Hao B., Zhang L., Wang W., Jiang W., Li N., Geng Q., Itaconate inhibits ferroptosis of macrophage via Nrf2 pathways against sepsis-induced acute lung injury. Cell Death Discov. 8, 43 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cintas C., Douche T., Dantes Z., Mouton-Barbosa E., Bousquet M. P., Cayron C., Therville N., Pont F., Ramos-Delgado F., Guyon C., Garmy-Susini B., Cappello P., Burlet-Schiltz O., Hirsch E., Gomez-Brouchet A., Thibault B., Reichert M., Guillermet-Guibert J., Phosphoproteomics identifies PI3K inhibitor-selective adaptive responses in pancreatic cancer cell therapy and resistance. Mol. Cancer Ther. 20, 2433–2445 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Sweetlove M., Wrightson E., Kolekar S., Rewcastle G. W., Baguley B. C., Shepherd P. R., Jamieson S. M., Inhibitors of pan-PI3K signaling synergize with BRAF or MEK inhibitors to prevent BRAF-mutant melanoma cell growth. Front. Oncol. 5, 135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu C. L., Nydes M., Shanley K. L., Morales Pantoja I. E., Howard T. A., Bizzozero O. A., Reduced expression of the ferroptosis inhibitor glutathione peroxidase-4 in multiple sclerosis and experimental autoimmune encephalomyelitis. J. Neurochem. 148, 426–439 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z., Yao Z., Wang L., Ding H., Shao J., Chen A., Zhang F., Zheng S., Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy 14, 2083–2103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Legends for tables S1 and S2
Tables S1 and S2