Abstract
The ventromedial prefrontal cortex (vmPFC) is a part of the limbic system engaged in the regulation of social, emotional, and cognitive states, which are characteristically impaired in disorders of the brain such as schizophrenia and depression. Here, we show that intrinsically photosensitive retinal ganglion cells (ipRGCs) modulate, through light, the integrity, activity, and function of the vmPFC. This regulatory role, which is independent of circadian and mood alterations, is mediated by an ipRGC-thalamic-corticolimbic pathway. Lack of ipRGC signaling in mice causes dendritic degeneration, dysregulation of genes involved in synaptic plasticity, and depressed neuronal activity in the vmPFC. These alterations primarily undermine the ability of the vmPFC to regulate emotions. Our discovery provides a potential light-dependent mechanism for certain PFC-centric disorders in humans.
An input from retina to the prefrontal cortex has major effects on its molecular, morphological, and functional characteristics.
INTRODUCTION
In mammals, non–image-forming visual functions such as circadian photoentrainment, pupillary light response, and mood regulation by light exposure are dependent on a subpopulation of retinal ganglion cells (RGCs) made intrinsically photosensitive (thus ipRGCs) by their expression of the photopigment melanopsin (Opn4) (1–5). These ipRGCs are diverse, comprising at least six subtypes (M1 to M6) (6–8) distinguished by size, dendritic stratification, and electrophysiological properties. Their central projections also differ, reflecting the diverse biological functions that ipRGCs regulate (9). Among the brain regions ipRGCs innervate is the perihabenula (PHb) (10), a nucleus of the dorsal thalamus that mediates the direct effects of light on mood (10, 11). The PHb densely innervates the ventral prelimbic (PL) and infralimbic (IL) region of the prefrontal cortex (PFC) in a nonspecific manner (10) and thus relays retinal input directly to this region.
The PFC organizes goal-directed behaviors, its main domains being planning, decision-making, executive attention, and emotional regulation (12, 13). A distinction is drawn between the dorsolateral PFC, implicated in purely cognitive functions, and a paralimbic PFC, comprising the orbital and ventromedial areas, involved in evaluating the emotional significance of events and regulating emotions for the purposes of goal-directed behavior (12–14). Although functional dissociations of the mouse PFC are not clear cut, the ventromedial PFC (vmPFC) groups together the ventral PL and IL cortex both of which are heavily interconnected (13). It is part of a broader network interconnecting limbic and associative areas such as the basomedial amygdala and the ventral striatum, the hypothalamus, and the brainstem (15, 16) and thereby contributes to a wide range of functions. Lesion studies in mice implicate the vmPFC in social cognition (17), the regulation of fear and anxiety (18), and the cognitive control of actions. These behavioral deficits are a core feature of many clinical populations including those suffering from depression and schizophrenia, who are known to have a compromised vmPFC (17, 19). Despite its clinical importance, and the fact that it is interconnected with many areas directly or indirectly receiving photic input, to date, there is no indication that this region may be affected by light exposure, an important environmental variable. Here, we show that light exposure regulates the integrity, activity, and function of the vmPFC through an ipRGC-thalamic-prefrontal pathway that is independent of the visual cortex.
RESULTS
Dark-reared mice or mice lacking ipRGC exhibit reduced apical dendritic arborization in the vmPFC
Within the vmPFC, PHb innervation encompasses layers I and III/IV of the IL and PL cortex. Because mice and other rodents lack a well-defined granular layer IV, we nominally refer to this PHb-recipient area as layer III/IV. Previous studies showed that chronic exposure of animals to constant darkness (known as a dark-dark or DD) delays the normal development of the primary visual cortex (20, 21). To address whether light exposure affects the vmPFC, we prepared and analyzed Golgi-stained coronal sections along the anterior-posterior extent of the IL and PL cortex. One group of control mice were maintained on a standard 12-hour light–12-hour dark cycle (12:12 LD), and another group of experimental mice were kept in DD before birth and up to 4 months after birth. We then reconstructed the apical dendritic arbors of pyramidal layer III/IV neurons in the vmPFC because this population is well known for their plasticity in response to environmental stimuli (22) (fig. S1, A and B, and Fig. 1A). We observed significant differences between light regimes with mice in the DD group displaying severe dendritic atrophy, with a reduction in total apical dendritic length of ~36% relative to LD controls, and a similar (~40%) significant reduction in the number of branch points (Fig. 1, B and C). This shows that light is required for the normal development of the vmPFC.
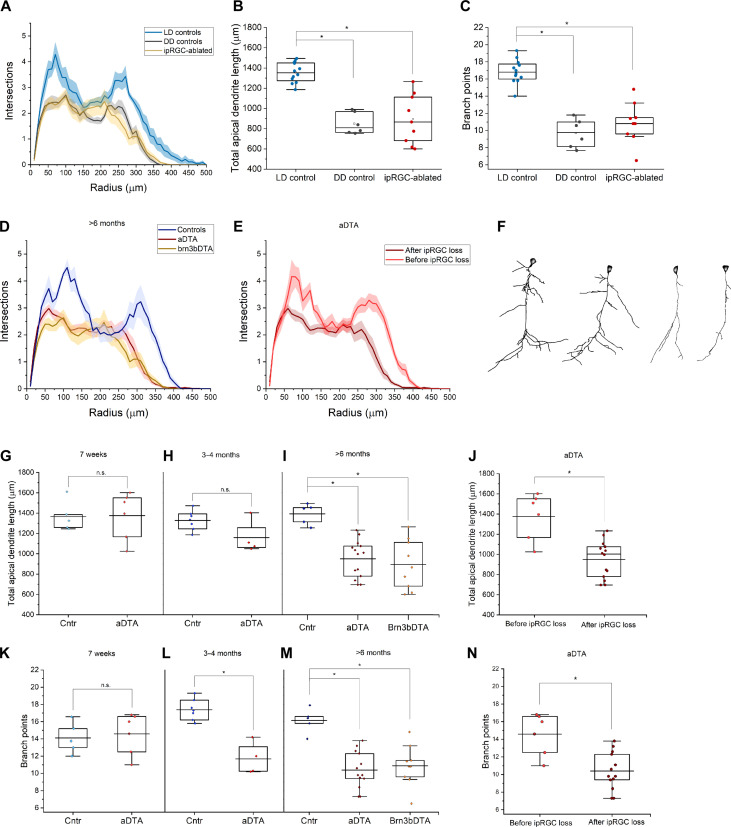
Fig. 1. Loss of ipRGCs leads to apical dendritic degeneration in the vmPFC.
(A) Sholl analysis revealed a significant degeneration of both proximal branches and distal tufts of vmPFC layer III/IV neurons in animals conceived and reared in constant darkness (DD) and ipRGC-ablated animals (specifically, Brn3bDTA) [P < 0.001, repeated-measures analysis of variance (ANOVA)]. (B) Total apical dendritic length is significantly (P < 0.001) reduced in DD mice (849.4 ± 43.1) relative to LD controls (1327.9 ± 36.2). (C) The number of branch nodes was also significantly reduced (P < 0.001) in DD mice (9.7 ± 0.7) relative to LD controls (17.4 ± 0.5). (D) Sholl analysis revealed a significant dendritic degeneration of vmPFC layer III/IV neurons in aDTA and Brn3bDTA animals relative to LD littermate controls (P < 0.001, repeated-measures ANOVA). (E) In aDTA mice, apical dendrites had a wild-type morphology at 7 weeks of age (light red), significantly different than at 6 months [dark red, replotted from (C)] (P < 0.001, repeated-measures ANOVA). (F) Marked examples of apical dendritic shrinkage; compare neurons from wild-type (leftmost) and young (7 weeks) aDTA mice (center-left) with affected neurons in old (>6 months) aDTA (center-right) and brn3bDTA mice (rightmost). (G) Total apical dendritic length was not significantly different between controls (1360.1 ± 85.2) and aDTAs (1374.8 ± 94.3) at 7 weeks of age; (H) it is still not significantly different at 3 to 4 months of age (controls 1327 ± 36.2 versus aDTAs 1196.3 ± 104.3); (I) it becomes significantly different (P < 0.0001) at older than 6 months (controls 1358.6 ± 31.3 versus aDTAs 950.1 ± 48.6); the same significant difference is observed in age-matched Brn3bDTAs (894.1 ± 81.8). (J) Data replotted from aDTAs pre- and post-ipRGC degeneration for between-ages comparison (P < 0.001, t test). (K) The number of branch points was not significantly different between controls and aDTAs at 7 weeks of age (14.2 ± 1.0 versus 14.6 ± 1.0 respectively); (L) an incipient significant difference was observed at 3 to 4 months (17.4 ± 0.5 versus 11.7 ± 0.9, P < 0.001), (M) which increased at older ages than 6 months (16.1 ± 0.6 versus 10.4 ± 0.6, P < 0.001). (N) There is a significant difference within aDTA genotype between 7 weeks and over 6 months of age (P < 0.001, t test). All values are means ± SEM. In the plots, each dot represents one animal.
Dark rearing causes depression-like mood alterations (23) and lengthens the period of circadian rhythms in rodents (24). Depression is associated with metabolic changes in the vmPFC (25). This raised the possibility that our DD results might have been secondary to these alterations. To address this, we used two mouse lines that detect light but lack a subset of ipRGCs (5, 26). We analyzed the vmPFC pyramidal neurons of a transgenic mouse line that expresses an attenuated form of diphtheria toxin A (“aDTA”) under the control of the melanopsin promoter (Opn4aDTA/aDTA). In aDTA mice, M1 ipRGCs develop and function normally until about 8 weeks of age, when they start degenerating; the loss progresses linearly with complete die-off by 6 months (5). Basal mood, as assessed by forced swim and sucrose preference tests, is normal in these animals, their visual system develops normally, and they have intact circadian rhythms (5, 11). In addition, ipRGC degeneration in aDTA mice occur without traumatic or stressful procedures known to affect PFC neuronal morphology (22). In mice older than 6 months, we observed major differences between genotypes, with reconstructed neurons from aDTAs displaying severe dendritic atrophy (Fig. 1, D to F), similar to what was observed in wild-type mice reared in DD. The reduced number of intersections in Scholl analysis after ipRGC loss (Fig. 1D) confirmed a reduction in total apical dendritic length by ~30% in aDTA mice relative to controls (Fig. 1I), with a ~36% significant reduction in the number of branch points (Fig. 1M).
The time course of ipRGC loss in aDTAs afforded an internal control. We repeated the morphometric analysis in aDTA mice at 7 weeks old, before the onset of cell degeneration, and at 3 to 4 months old, when ~50% of ipRGCs had degenerated. At 3 to 4 months of age, aDTA mice displayed an early form of the atrophic phenotype showing ~10% reduction in total apical dendritic length and ~33% reduction in number of branch points compared age matched control mice (Fig. 1, H and L). In contrast, the atrophic phenotype was not observed in 7-week-old aDTA mice, when degeneration of ipRGCs had not started (Fig. 1, E, G, K, J, and N). Thus, the severity of apical dendritic atrophy seen in this neuronal population was proportional to the extent of ipRGC loss and confirms an essential role for light in the dendritic development of the vmPFC.
To investigate if the atrophic phenotype we observed is specific to regions of the cortex that receive ipRGC input, we reconstructed neurons in the motor cortex as a control region. No differences between the wild-type and aDTA mice in the motor cortex were observed (fig. S1C), indicating that the atrophic phenotype was specific to the vmPFC.
Chronic dark rearing and the aDTA mice lack the ability to adjust their day/night activity to the 24-hour day in what is known as circadian photoentrainment (5, 24). The lack of circadian photoentrainment could underlie the atrophic phenotype we observed in the vmPFC. Therefore, we repeated our analysis in a transgenic line, Opn4Cre/+;Brn3bzDTA/+ (Brn3bDTAs) mice that have normal circadian photoentrainment but lack ipRGC input to the PHb-vmPFC (26). These mice express a strong variant of diphtheria toxin A under the control of the Brn3b promoter in a Cre-dependent manner, and Cre itself under the control of the melanopsin promoter. Brn3b is a transcription factor expressed in all ipRGCs except a subset of M1 cells that project to the suprachiasmatic nucleus (SCN) and are sufficient for circadian photoentrainment (26). So, Brn3bDTA mice can synchronize their circadian rhythms to the environmental light cycle but lose in developmental stages non–SCN-projecting ipRGCs. In this mouse model, we found vmPFC abnormalities identical to those present in dark reared and aDTA mice showing an ~33% average reduction in total apical dendritic length and ~37% fewer branch points versus littermate controls (Fig. 1, D, I, and M). We conclude that lack of light-driven ipRGC input leads to apical dendritic degeneration in the vmPFC and that this effect is not secondary to any SCN-based circadian disruptions, mood alterations, or developmental effects.
vmPFC response to a stressful stimulus is reduced in ipRGC-ablated mice
We then measured the activity of the vmPFC in ipRGC-ablated mice by photometry recording in freely moving mice. We transduced vmPFC neurons of Brn3bDTA mice and wild-type littermates with the Ca2+ sensor, GCaMP6f, under the control of the synapsin promoter. An optical fiber was implanted above the vmPFC region (Fig. 2A) and patched to a photometry apparatus capable of exciting GCaMP and measuring emitted fluorescence without restricting the movement of animals. We recorded the activity of the vmPFC at different circadian times, because the vmPFC is known to display strong circadian rhythms (21). Contrary to the presence of strong molecular circadian clock in the vmPFC, the activity of the vmPFC did not show robust daily variations. However, the vmPFC of ipRGC-ablated animals did indeed exhibit lower intrinsic activity, specifically by showing fewer endogenous activity peaks compared to controls (Fig. 2, B to D). These data suggest that ipRGC input to the PHb maintain vmPFC integrity and function.
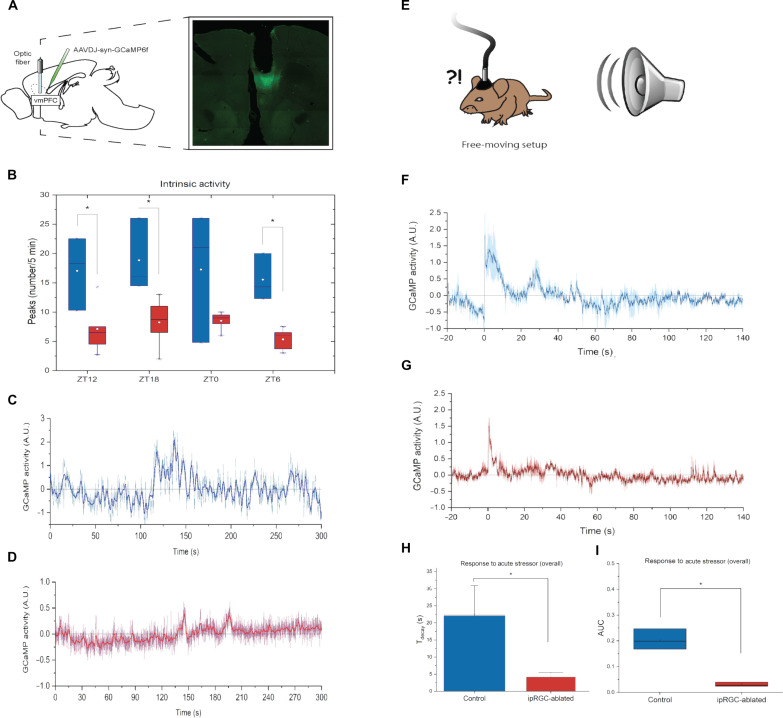
Fig. 2. vmPFC activity in response to a stressful stimulus is depressed in ipRGC-ablated mice.
(A) Neurons were transduced with GCaMP6f; an optical fiber was implanted above the area. (B) We recorded in vivo neuronal activity from the vmPFC. Recordings of endogenous activity showed a significantly higher number of peaks in controls relative to Brn3bDTAs at ZT12 (P = 0.037), ZT18 (P = 0.028), and ZT6 (P = 0.002). A trend in the same direction is present at ZT0 (P = 0.116). All values are means ± SEM. (C and D) Examples to highlight high intrinsic activity recording from a control mouse (C) versus a low intrinsic activity trace from a Brn3bDTA (D). Traces are recordings of individual animals; the dark/thicker line is a rolling time average. (E) Mice were exposed to a loud sound—a nonvisual fear-evoking stimulus. (F and G) Normalized Ca2+-dependent fluorescence in response to the noise (delivered at time = 0) in controls (F) and Brn3bDTA mice (G). Traces are means (±SEM, hued area) of recordings from three animals, each averaged over multiple trials. (H) Fluorescence decayed to baseline more slowly in controls than in ipRGC-ablated mutants (22.1 ± 5.9 s versus 4.1 ± 0.9 s) (P = 0.039). (I) The area under the curve of vmPFC activity in response to this stimulus was significantly higher in controls (0.204 ± 0.0230) versus ipRGC-ablated mice (0.030 ± 0.0048) (P = 0.002).
The changes in the basal activity and morphology of the vmPFC prompted us to investigate the intra-vmPFC Ca2+ dynamics in response to a stressful stimulus. A sudden loud sound was chosen (Fig. 2E) because it is independent of the visual system ensuring that any difference between ipRGC-ablated mice and controls could not be due directly to loss of the visual sensory input. Neuronal activity in controls and Brn3bDTAs surges after the auditory stimulation (Fig. 2, F and G), reaching a peak ~500 ms on average after the sound is emitted, with no significant difference between genotypes on time to peak. The amplitude of the peak response was also not significantly different between genotypes. However, the time course of the decay differed significantly (Fig. 2H) in that fluorescence decayed to baseline (Tdecay) in 4.13 s in Brn3bDTAs versus 22.08 s in controls. This longer decay means that the cumulative activity, quantified by area under the curve, is significantly higher in controls (Fig. 2I). Thus, the absence of ipRGC input causes deficits in the vmPFC that are manifested at the morphological and neuronal activity levels and its response to stressful stimulations.
Lack of ipRGC input leads to severe gene-expression dysregulation in the vmPFC
The profound dendritic atrophy and weakened activity in the vmPFC may reflect a more generalized network-level dysfunction that would leave a characteristic transcriptomic signature. To investigate this, we performed high-throughput single-cell RNA sequencing (scRNA-seq) on cortical tissue from Opn4Cre/+;Brn3bzDTA/+ (Brn3bDTA) and littermate Opn4+/+;Brn3bzDTA/+ (control) mice (fig. S2A). This line was chosen over aDTAs because it was possible to harvest tissues at a younger age that is more amenable to tissue dissociation. We generated high-quality single-cell transcriptome profiles for 24,340 cells (11,800 control, 12,540 Brn3bDTA) and used unsupervised learning approaches to identify more than 20 transcriptionally distinct cell types within the vmPFC (fig. S2, B and C). We first identified, on the basis of the expression of canonical marker genes, major cell types including glutamatergic neurons (Slc17a7 a.k.a. Vglut1), GABAergic neurons (Gad2), astrocytes (Gja1), oligodendrocytes (Olig1), endothelial cells (Flt1), mural cells (Rgs5), microglia (Selplg), perivascular macrophages (F13a1), and fibroblasts (Dcn) (27, 28) (fig. S2, D and E). Consistently with the prior literature (29), no brn3b transcripts were detected in any cluster. We then extracted the identified neuronal cells and performed an additional round of unsupervised clustering to resolve 16 discrete neuron types within the vmPFC (6914 control, 7278 Brn3bDTA) (Fig. 3, A and B).
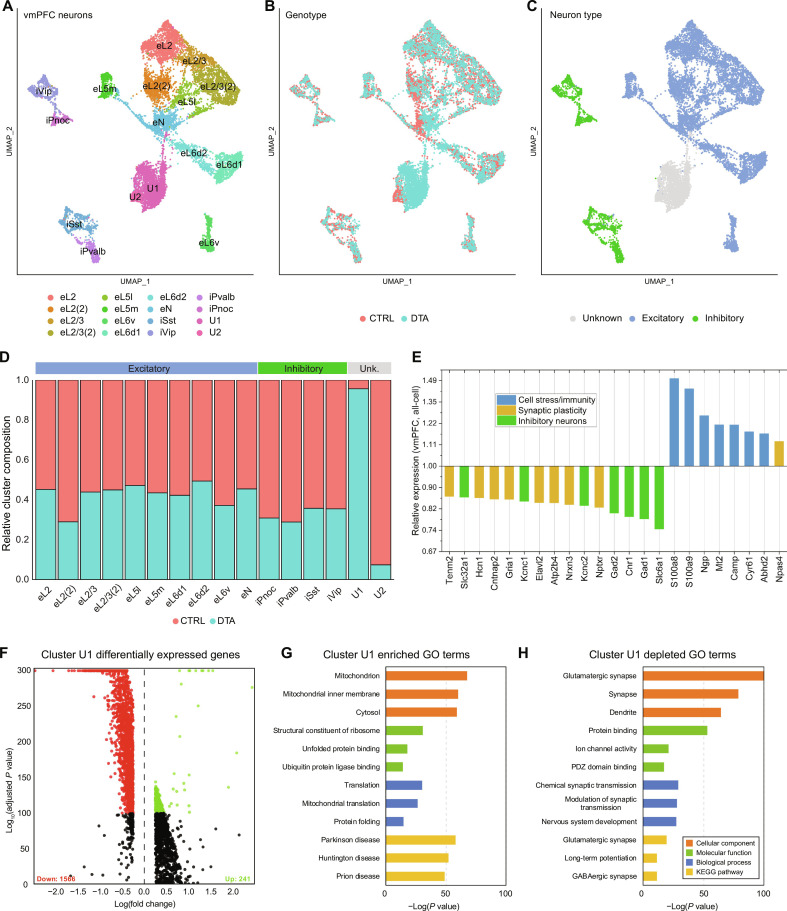
Fig. 3. Genes associated with neurotransmission and synaptic plasticity are severely dysregulated in the vmPFC of ipRGC-ablated mice.
(A) Uniform manifold approximation and projection (UMAP) plot of all neurons identified in the vmPFC. (B) Same as in (A) overlaid with sample genotype. (C) Same as in (A) overlaid with major neuron type assignment. (D) Relative neuron cluster composition by genotype (red, control; blue, DTA). (E) Relative expression of differentially expressed genes in DTA samples averaged across all major cell types of the vmPFC. (F) Volcano plot of differentially expressed genes between DTA and control samples within unspecified neuronal cluster U1. (G and H) Gene ontology (GO) terms significantly enriched in the up-regulated (G) or down-regulated (H) genes from cluster U1.
To correlate each neuronal cluster to a specific anatomical layer of the cortex, we identified marker genes for each cluster and examined their expression in the Allen Brain Atlas ISH database (fig. S3, A to D). For 14 of these clusters, we were able to identify their origin layer on the basis of patterns of gene expression; 10 were excitatory (Slc17a7 positive) and 4 were inhibitory (Slc32a1 positive) (Fig. 3C). Each contained cells originating from both control and Brn3bDTA tissue, though in most cases there were fewer Brn3bDTA neurons, despite their outnumbering control neurons overall (Fig. 3D). The remaining two neuronal clusters, on the other hand, exhibited extreme divergence (Fig. 3D). These clusters did not contain any strongly specific marker genes, appearing instead to be weakly Slc17a7-positive glutamatergic neurons markedly depleted of RNA content. We first ranked the most significantly differentially expressed genes across all cell clusters, to identify those subject to the largest dysregulation in the Brn3bDTA vmPFC (Fig. 3E). We found that genes associated with neurotransmission and synaptic plasticity are severely dysregulated in the vmPFC of ipRGC-ablated mice, whereas genes associated with cell stress and immune responses are up-regulated (Fig. 3E).
We then examined the two unknown clusters, U1 and U2. The largest unspecified cluster, U1, contained a notable 2727 Brn3bDTA but only 101 control neurons. When we examined the differentially expressed genes between the U1 and the remaining clusters, we found that it was substantially depleted of hundreds of transcripts (>900 genes, P < 10−100) and enriched for relatively few (Fig. 3F). To see what genetic programs were depleted in these cells, we performed gene ontology analysis on all significantly up-regulated or down-regulated genes (Fig. 3, G and H). Down-regulated genes were involved in pathways including ion channel activity, ion transport, nervous system development, and many other canonical neuronal functions. Conversely, within the up-regulated set of genes in U1, we detected signatures of cellular stress and/or degeneration including unfolded protein binding and pathways implicated in Huntington’s and Parkinson’s disease. Thus, it appears that cluster U1 neurons, overwhelmingly from Brn3bDTA animals, broadly down-regulate normal neuronal processes in a shift to stress-related responses. The smaller unknown cluster, U2, containing predominantly control neurons (8 Brn3bDTA, 297 control), shared a similar set of genes as measured by gene ontology (fig. S3G) but down-regulated to a much less significant degree (fig. S3, E and F). Together, our analyses showed large changes in genes involved in synaptic plasticity and signaling specifically in ipRGC-ablated animals.
To determine whether the effect of ipRGC loss was specific to the vmPFC, we collected tissue from the nearby motor cortex in both control and Brn3bDTA animals and repeated the scRNA-seq analysis. Again, we could identify all major cell types from both control and Brn3bDTA tissue in the motor cortex. Among neurons, we saw an even overlap and distribution of neuronal cell types, with no large cluster containing degenerate neurons or an enrichment of Brn3bDTA cells (fig. S4, A and B). Thus, we can confirm that the detrimental effect of ipRGC loss was indeed specific to the vmPFC.
To validate the scRNA-seq gene expression results, we performed RNAscope experiments with positive results. We selected genes that showed strong differences in DTA neurons, including both up-regulated and down-regulated transcripts with established neuronal function. RNAscope confirmed that DTA neurons exhibit significantly decreased overall expression of Map2, Nlgn2, and Gls and significantly higher levels of Ubiquitin (Ubb) as compared to controls (fig. S5).
ipRGC-ablated mice show impaired regulation of aversive emotions
ipRGC-ablated mice have normal mood, as assessed by forced swim and sucrose preference tests, and their hippocampal-dependent memory is unimpaired (10). Prefrontal injuries in humans show changes in general cognition and social demeanor, and in both primates and rodents, PFC lesions result in anomalous social behavior (30–32). We opted, therefore, to examine the behavior of ipRGC-ablated mice in a social context. The ipRGC-ablated and littermate control animals were housed in standard colony conditions with their same-sex littermates for at least 6 months and no exposure to females after weaning. Male mice were tested in a free social interaction test (Fig. 4A) in which the mouse was exposed for 5 min in an unfamiliar setting to a strange conspecific of the same sex. The interactions between the two mice were tracked and scored for many different parameters of social behavior, including total time interacting together. Moreover, because we have noticed in our laboratory that socially isolated aDTA mice become hyper-reactive relative to wild types, and social isolation is a known stressor to rodents capable of altering PFC myelination patterns (33), we tested mice before social isolation, after a short 4-day period of social isolation, and again after long-term 23-day isolation (Fig. 4A).
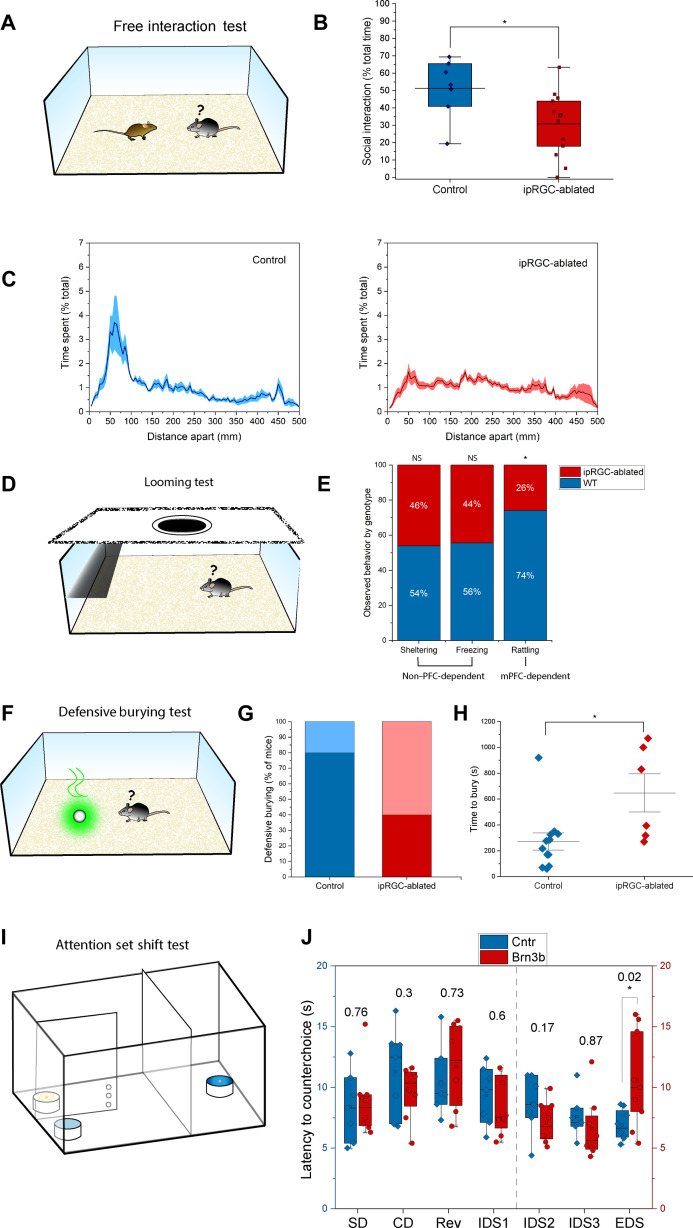
Fig. 4. ipRGC-ablated mice show impaired regulation of emotion and attention.
(A) In a free-interaction test, a test mouse is exposed to a strange conspecific in an unfamiliar arena. (B) ipRGC-ablated mice were less likely to interact socially, spending 30.9 ± 5.1% of their time in this activity versus 51.4 ± 6.4% for controls (P = 0.025). (C) Percent time spent with conspecific in control and ipRGC-ablated mice. (D) In the looming test, a mouse can react to a simulated predator by running under a shelter, freezing, and/or rattling his tail. (E) A similar proportion of ipRGC-ablated mice ran under the shelter and froze relative to controls (sheltering: 73.3% versus 85.7%; freezing: 40% versus 50%), but they were less likely to rattle their tails (20% versus 57.1%, P = 0.027) (normal approximation proportion test). Tail rattling is a known mPFC-dependent behavior. The plot shows what proportion of specified behavior was displayed by control versus ipRGC-ablated mice (normalized). NS, not significant; WT, wild-type. (F) In defensive burying test, mice bury a fox urine–soaked Q-tip if they can overcome their fear of the scent. (G) ipRGC-ablated mice were less likely to bury the noxious object: 40% did (dark red), versus 80% of littermate controls (dark blue), P = 0.025) (normal approximation proportion test). (H) For those that buried the object, a difference was evident in the latency to enact this behavior (647 ± 147.4 s for ipRGC-ablated mice versus 271.1 ± 66.2 s for controls, P = 0.015). (I) Attention set shift test is a task in which cognitive flexibility and attention interact with emotional drives. (J) We recorded the latency necessary for a mouse to correct its choice after a reward-association rule was learned. For each stage of the test, the P value of the performance difference between genotypes is reported over the bars. ipRGC-ablated mice took longer to enact corrective behavior in the EDS (10.6 ± 1.3 s versus 6.9 ± 0.4 s, P = 0.023). Each dot represents one animal.
After long-term isolation, ipRGC-ablated mice substantially reduced social interactions with their conspecific (Fig. 4B) by avoiding the conspecific when approached and were averse to interacting closely by spending less time engaging in social behaviors such as nose touching and sniffing (Fig. 4, B and C, and movie S1). As these mice were never exposed to females, there was little immobility and severe fighting was uncommon (fig. S6A). Thus, the decreased social interactions (Fig. 4C) were not due to aggressive behavior. Without prior social isolation, there was no significant difference between genotypes in social contact (fig. S6B).
To assess social preference further, we used the three-chamber test, where a mouse is given a choice between a conspecific and an object (fig. S7A). The ipRGC-ablated mice were significantly less likely to spend time in the conspecific-mouse compartment during the first 2 min of the trial—consistent with heightened fear toward unfamiliar conspecifics displayed in the free-interaction assay (fig. S7, B and C, and movies S2 and S3). The reduced social contact exhibited by the ipRGC-ablated mice suggested to us an increased state of generalized anxiety. We first used the marble burying test and measured defensive burying of 20 marbles perceived as a potential threat. The number of marbles buried by ipRGC-ablated mice was not different to littermate controls (fig. S6C) confirming that ipRGC ablation does not heighten fear toward potentially threatening stimuli.
We then subjected the ipRGC-ablated mice to the looming test, where fear is induced by stimulating a predator attack. In this test, mice typically react to an expanding dark disc used to mimic a predator approaching or looming from above by escaping to the shelter, freezing, or rattling their tails (Fig. 4D and movie S4). Tail rattling during the looming test is directly mediated by the medial PFC (34) and has been used as a proxy for social fear associated with PFC dysregulation, whereas the shelter escape and the freezing behaviors are dependent on image-forming vision. Consistent with previous data, the ipRGC-ablated mice showed normal image functions as they escaped to the shelter and froze in a similar proportion to their littermate controls (Fig. 4E). The ipRGC-ablated mice were significantly less prone to rattling their tails (only 20.0% of ipRGC-ablated mice did versus 57.1% of controls; Fig. 4E), indicating a dysfunction in the medial PFC.
We then used the defensive burying assay using a noxious odor stimulus, a behavior that is independent of image-forming functions but still depends on the vmPFC (35). In this test (Fig. 4F), a Q-tip dipped in concentrated fox-urine odorant [trimethylthiazoline (TMT)] was placed in the cage (fig. S6D). As expected, the mice buried the Q-tip as a defensive strategy except that only 40% of the ipRGC-ablated mice buried within the 20-min limit of the test relative to 80% of the wild types (Fig. 4G). The ipRGC-ablated mice also took significantly longer to bury the Q-tip relative to controls (650 s versus 270 s) (Fig. 4H). Moreover, these mice typically distanced themselves from the Q-tip, often confining themselves to the opposite end of the arena (movie S5). Thus, an olfactory stimulus elicited behavioral abnormalities just like visual stimuli did. Together, these data indicate that the behavioral deficits observed in ipRGC-ablated mice are not due to sensory dysfunction but instead are a manifestation of emotional dysregulation due to an impaired vmPFC.
We also tested mice for high-order cognitive processing of focused attention and flexibility using a classic vmPFC test, the IntraDimensional/ExtraDimensional (ID/ED) set shifting task (36). Briefly, mice were trained to dig into ramekins for a buried pellet of food. The ramekins were filled with different digging media, and during the test (Fig. 4I), the mouse performed a series of discriminations that differed along two dimensions (i.e., odor and texture). Two measures of performance were recorded: (i) the number of trials to reach a criterion level of performance as a measure of accuracy and (ii) the latency to correct their choice following an error (counter choice). In the Simple Discrimination (SD) stage, the mouse learnt that a specific digging medium was associated with reward. In the subsequent Compound Discrimination (CD) phase, an additional dimension was introduced; now, the ramekins differed by both digging medium and scent, but the mouse must focus on the digging medium as the relevant cue. In the Reversal (Rev) stage, the reward contingency was reversed such that the previously unrewarding scent was now rewarded. In all these stages of learning, (SD, CD, and Rev) the ipRGC-ablated mice and the controls did not differ in their performance (Fig. 4J and fig. S7D) indicating normal learning capabilities in ipRGC-ablated mice.
We then subjected the mice to the IntraDimensional Shift (IDS) during which the mice were required to discriminate three new exemplars of digging media/odors (IDS1, IDS2, and IDS3). In all cases, the reward was associated with the digging media and not the odor. For the most part, the animals performed normally on the IDS discriminations with no major changes between the ipRGC-ablated mice and the controls in terms of accuracy and the latency to counter-choice (Fig. 4J and fig. S7D). We observed an unexpected deficit in IDS2 rule acquisition because the animals were trained in this stage on the following day (fig. S7D). We think this deficit reflects a transient mnemonic impairment for two reasons: (i) The trial number to criterion for IDS1 and IDS3 were in the normal range for which there was no memory requirement, and (ii) the speed at which they corrected their errors for each intradimensional shift was not impaired (Fig. 4J). The main difficulty came during the ExtraDimensional Shift (EDS) test when for the first time in the course of testing, odor become the rewarding stimulus and the digging medium had to be ignored. Now, the ipRGC-ablated mice were markedly impaired, evidenced by long latencies to make their response to the new rewarding dimension (Fig. 4J). Performance in this complex cognitive task is linked to specific prefrontal areas: The Rev. stage recruits an orbitofrontal network, while the EDS relies on the medial portions of the PFC. Therefore, our data indicate that cognitive networks in the orbitofrontal and dorsomedial PFC are normal, and importantly, the deficits in the vmPFC we observed are not due to a generalized prefrontal dysfunction.
DISCUSSION
We report, using two genetic lines and constant dark rearing, that light via ipRGCs affects the activity and function of vmPFC in mice, independent of circadian, developmental, and mood alterations. Using a genetic line (Brn3bDTA mice), which has normal circadian photoentrainment because only non–SCN-projecting ipRGCs are ablated at early stages, we uncover that the effects of ipRGCs on vmPFC are not a corollary of circadian disruptions. In addition, using a genetic line (aDTA mice) where ipRGCs are ablated only at adult stages starting at 2 months and completed by ~6 months (5), we demonstrate similar deficits to those of Brn3DTA mice, indicating that there is no developmental critical period for the effects of ipRGCs on vmPFC. Last, both genetic lines show normal basal mood as assessed by classical depression tests (10, 11), highlighting that the effects of ipRGCs on vmPFC are not a consequence of mood alterations. Even in the presence of ipRGCs, the absence of light (dark rearing) leads to similar deficits in the vmPFC dendritic structures as those observed in the two ipRGC-ablated genetic lines.
We decided to investigate the vmPFC because it receives input from the PHb that is innervated directly by ipRGCs (10). In this study, we refer to the PHb-recipient area of the PFC, broadly encompassing the IL and PL cortex, as vmPFC (37). Previously, it was shown that the PFC dendritic structure of pyramidal neurons are sensitive to circadian disruptions and stress (21), with stress inducing the largest deficits. The deficits in the dendritic structures we observe in this study, using the two genetic lines and dark rearing, are greater than those observed with stress. Simply ablating ipRGCs or just depriving a nocturnal animal from light led to changes in the vmPFC that are more substantial than stress or circadian disruptions. We also investigated the time course of the changes in the dendritic structures and correlated them to ipRGC loss in aDTA mice. We observed that deficits in the dendritic structures of pyramidal neurons directly correspond with the extent of ipRGC loss, showing normal morphology at ages below 2 months and then showing progressive disruptions starting at 3 to 4 months. The morphological changes we observed are correlated with physiological deficits in the response of the vmPFC to acute stressors such a loud sound. The use of a noxious auditory stimulus avoids the complications of using a retina-dependent stressor. We used photometry to show that vmPFC of ipRGC-ablated animals respond initially to the sound but show faster decay and smaller area under the curve in response to the acute stressor. Our studies indicate that ipRGCs of the retina and their light reception have a profound influence on the morphology and the physiology of the vmPFC.
Neurons within the vmPFC are relatively plastic and respond to a variety of stressors and behavioral manipulations. For our single-cell sequencing analysis from both control and ipRGC-ablated animals, most neurons are healthy and layer specific. However, we observed two unspecified clusters (U1 and U2), with most cells in U1 hailing from ipRGC-ablated animals. Both the U1 and U2 clusters represent a regressed neuronal state, rather than a neuron type, that normally exists within the vmPFC in adult animals. In ipRGC-ablated animals, the numbers of cells with regressed neuronal state were substantially higher with thousands of them appearing in the U1 cluster. Within scRNA-seq experiments, there are often large number of reads generated from ambient RNA binding to transcriptome capture beads, which when sequenced create the illusion of real cells with low gene content. These low-quality cells often contain very few genes that are of mixed origin between major cell types. In our case, the U1 and U2 all contain a strong complement of genes (>500 each) and are not positive for non-neuronal cell type markers. For this reason, we have categorized these cells as unhealthy neurons. Specifically, for U1 cluster, the general decrease in neurons with layer- or type-specific genetic signatures was accompanied by a notable increase in gene-poor neurons that likely represent degenerate cells. This observation is in strong agreement with histological data taken from the vmPFC of ipRGC-ablated animals that show a substantial retraction of pyramidal apical dendrites compared to wild-type littermate controls. This dendritic retraction nicely matches the genetic retraction that we see in U1, which are significantly depleted for more than 1000 genes. These down-regulated genes even show expected signatures of dendritic atrophy including gene ontology terms associated with synapses, cell membrane, and cell adhesion. For example, inhibitory interneuron markers (Gat1, Gad1, and Gad2, Cnr1, Kcnc1, and Kcnc2, Vgat) were the most down-regulated genes, followed by regulators of synaptic transmission, synaptogenesis and dendritic outgrowth, and neuronal excitability (Neurexins, Hcn1, GluA1, Tenm2, Elavl2, and Cntnap2); the most relatively up-regulated genes were signatures of cell stress, inflammation, or immune system reactivity, with the exception of Npas4, a transcription factor that regulates the number and distribution of inhibitory synapses on dendritic arbors (38).
In rodents, the PFC is important for social behaviors. We have conducted several behavioral tests (free interaction and three-chamber) to examine the social behavior of the animals lacking input to the PHb, and a selected set of behaviors (looming, defensive burying, and attention set shift tests) that are known to be associated with the medial PFC. Animals lacking input to the PHb showed severe social dysregulation. This emotional dysregulation was not due to an increased state of generalized anxiety (marble burying test), in agreement with previous studies (11). The looming, defensive burying, and attention set shift tests measure mPFC function directly. The looming is particularly informative because it measures two aspects, vision, by the freezing and sheltering of the mouse and mPFC function by the tail rattling. Animals lacking input to the PHb showed deficits only in tail rattling, indicating cortical vision to be intact. Defensive burying test, a nonvisual behavior for the mPFC function, further confirmed that the deficits in the mPFC are not due to cortical visual effects. Last, the most comprehensive test was the attention shift set, which measure seven aspects that involves memory and discrimination, and the animals lacking input to the PHb only showed deficits in the EDS stage, which is highly dependent on the medial portions of the PFC but not the Rev. stage, which is dependent on the orbitofrontal region that does not receive input from ipRGCs. The light-modulated circuit appears to be mainly involved in the top-down regulation of negative emotions, possibly via an attentional effect, as revealed by the performance of ipRGC-ablated mice in the EDS stage of the attention set shift test. On the basis of the impaired rule-acquisition of ipRGC-ablated animals in the IDS2 stage of the same test, a deficit in the retention of procedural memory also cannot be ruled out; it has been suggested that the PFC may mediate “episodic control,” i.e. the storage of a rule in memory and its use to adapt behavior (39). These results show that the morphological, physiological, and molecular deficits in the vmPFC lead to medial PFC–specific behavioral deficits.
Our results allow for a simple model. Light through ipRGCs that innervate the PHb activates the vmPFC, and this activation is important for the integrity of the morphological, physiological, and molecular components of the vmPFC. Our discovery of modulation by light of the vmPFC raises an intriguing possibility. The PFC is the most evolutionarily recent area of the neocortex, and its most primordial core is the vmPFC. Its modulation by ipRGCs, simple photodetectors found in all vertebrates, invites the speculation that its evolutionary origin and ancestral function may lie in the regulation of light-driven daily activity patterns.
A notable aspect of this study is the fact that dark rearing led to major deficits in the vmPFC. Dark rearing has been shown to affect the primary visual cortex. However, this is not unexpected considering the massive input from the retina to the primary visual cortex. However, are the deficits in dark rearing occur secondarily through the primary visual cortex? Animals that lack only ipRGCs and have normal input to the primary visual cortex still exhibit deficits in the vmPFC, even though the PFC receives input broadly from sensory areas, including visual ones (40). Therefore, the absence of an ancient photoreceptive system that is subconscious in nature is sufficient to cause deficits in the vmPFC, even in the context of functional cortical vision.
In humans, the PFC expands during development eventually adding up to 29% of the overall volume of the cortex (41), a large proportion to total brain size, which sets a record for the entire animal kingdom (42). It is well established in the literature that the PFC is one of the first brain region to undergo age-dependent degeneration, a phenomenon characterized by morphological changes (dendritic atrophy) notably similar to those observed in this study after light deprivation. Aging causes a progressive yellowing of the eye lens, an effect that reduces the intensity of blue-shifted light reaching the retina. It was shown that the transmission of 480-nm light decreased by 72% at age 80 relative to age 10 (43). It is precisely to these short-wavelength photons that ipRGCs are most responsive. The outcome is an age-dependent, progressive loss of ipRGC signaling. This was thought only to result in the characteristic circadian and sleep disturbances seen in the elderly. We may now have reason to suspect a role of ipRGC signaling in PFC age-dependent degeneration.
A marked trend in recent decades has been the proliferation of artificial blue-light sources, and human nighttime exposure to them, an unprecedented behavior in the natural history of our species known to disrupt physiological ipRGC signaling. The emotion-regulatory function of the vmPFC is specifically affected in many disorders, from schizophrenia to autism to syndromes characterized by fear generalization (37). Some of the most dysregulated genes in the PFC of mice lacking ipRGC signaling (e.g., cntnap2) are mutated in well-established mouse models of these disorders (44). More generally, in this study we show that light, via a retinal cell population, profoundly affects the PFC across multiple domains—neuronal morphology, gene expression, network activity, and behavior—with effect sizes so considerable as to suggest an ecologically important regulation. This unexpected feature, if confirmed in humans, may prove to be of considerable interest.
MATERIALS AND METHODS
Animals
Opn4aDTA/aDTA and Opn4Cre/+;Brn3bzDTA/+ mouse lines were used in this study, housed under a 12-hour light: 12-hour dark (T24) cycle at a temperature of 22° to 25°C with food and water ad libitum unless otherwise noted. In Brn3bDTAs, ~200 ipRGCs survive per retina, which are the only ipRGCs that do not express the transcription factor Brn3b (POU4F2); these cells exclusively innervate the SCN. Brn3bDTA mice were obtained by mating mice that express the Cre-dependent diphtheria toxin under the control of the Brn3b promoter (Brn3bzDTA/+), with Opn4Cre/Cre mice. Opn4Cre/+ littermates were used as controls. All animals were handled in accordance with guidelines of the Animal Care and Use Committees of Johns Hopkins University and the National Institutes of Health.
Golgi staining and cell morphometry
Animals were socially housed with littermates in standard colony conditions until the indicated age; rapidly anesthetized with 5% isoflurane, their brain was extracted without any preliminary fixation, rinsed with phosphate-buffered saline (PBS), and processed with FD Neurotechnologies’ Rapid Golgi staining kit, in accordance with the kit protocol. The resulting 200-mm coronal sections were imaged at ×40 to ×100 magnification. We reconstructed the apical dendritic trees of pyramidal neurons in layer III/IV, throughout the anterio-posterior extension of the IL and PL areas (+1.9 to +1.3 mm relative to bregma), from Golgi-stained coronal sections. The dendritic morphology of neurons in the Golgi-stained sections was reconstructed using the Neurolucida tracing system (MicroBrightField). The neurons to be traced were selected according to the following criteria: (i) Position in the PFC area shown by previous tracing to receive PHb afferents (PL and IL cortices). (ii) Pyramidal type, with a roughly triangular soma, a clear single apical dendrite extending from the soma toward the pial surface, and two or more dendritic trees extending from the base of the soma. (iii) Distance from the soma to the pial surface <390 and >280 μm (~layer 3). (iv) Shape of the apical dendritic tree not suggestive of a layer 5 or short-shaft layer 2/3 neuron. (v) Apical dendritic tree mainly unfolding within the middle third of the section, well isolated and not obscured by neighbors. Six to 10 cells typically qualified to be analyzed per animal (with little discretion for the experimenter). The cells were traced and reconstructed by two independent investigators, one of whom was blind to genotype. Analysis of the variables of interest was performed automatically by Neurolucida Explorer.
Single-cell RNA sequencing
Generation of single-cell suspensions from vmPFC tissue
Brains from 1-month-old Brn3bzDTA animals and littermate controls were processed in tandem d for scRNA-seq experiments to minimize potential batch effects. After rapid anesthesia induction with 5% isoflurane, the brain was extracted the vmPFC was manually excised. Subsequently, cells were dissociated following a published protocol (45). Cell viability was then measured with Trypan blue and resuspended in ice-cold PBS to desired concentrations for 10X Genomics single-cell capture.
Data analysis
Single cells were loaded into a 10X Genomics Chromium controller and processed with the 3′ single-cell gene expression solution V2 (10X Genomics) according to manufacturer instructions. Up to three libraries were multiplexed at once and sequenced on an Illumina NextSeq 500 platform. Reads were demultiplexed and aligned and digital gene expression matrices were generated with the CellRanger pipeline from 10X Genomics. Read demultiplexing and alignment was performed on the Maryland Advanced Research Computing Center, and data analysis was performed on local Mac/Linux devices using the Seurat package in R.
Digital gene expression matrices for each replicate were loaded into Seurat, retaining only cells that expressed at least 500 unique genes. Samples from both genotypes were scaled, normalized, and integrated for comparative analysis in Seurat (46). We then performed principal components analysis with uniform manifold approximation and projection, and clustering by k-nearest neighbor method to reveal distinct clusters. Cluster identities were determined by differential expression of distinct cell type–specific markers. For detailed analysis of individual cell types, cells of interest were extracted and reprocessed following the same procedure.
Gene ontology analysis
Gene ontology analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (47).
Bulk gene dysregulation ranking
A subset of data from high-quality, non–RNA-depleted cells only was used for this analysis.
RNAscope validation and analysis
Brain section of two control and three DTA mice (4 to 5 weeks) were cut at 20-μm thickness and affixed onto microscope slides. For in situ hybridization, the sections were initially fixed with 4% paraformaldehyde for 60 min at room temperature. This was followed by two consecutive 1-min washes with PBS before undergoing a dehydration process involving 50% ethanol (1 × 5 min), 70% ethanol (1 × 5 min), and two rounds of 100% ethanol (2 × 5 min each) and then placed in 100% ethanol at −20°C overnight. On the following day, the slides were allowed to air-dry at room temperature for 10 min. A hydrophobic pen was used to delineate a barrier around the sections, and the sections were allowed to dry for an additional 10 to 15 min. The sections were then treated with Protease Pretreat-4 solution for 30 min at room temperature. The slides were then washed twice with ddH2O (2 × 1 min each) before being incubated with the appropriate probes.
The probes were from Advanced Cell Diagnostics and included Mm-Nlgn2 (T2; 406681), Mm-Ubb (T8; 591071), Mm-Hspa8 (T7; 1137391), Mm-Pfn2 (T6; 559691), Mm-Gls-O1 (T4; 535651), Mm-Map2 (T3; 431151), Mm-Gad2 (T5; 439371), and Mm-Cntnap2 (T1; 449381). These probes were hybridized to the sections for a period of 2 hours at 40°C, followed by washing and hybridization with target-binding amplifiers to facilitate signal amplification for the detection of single RNA transcripts. In the initial round of hybridization, the first 3 of the 12 target genes (T1 to T3) were labeled with three distinct fluorophores. Subsequently, the sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) for 30 s and mounted with ProLong Gold Antifade Mountant (Jackson ImmunoResearch, catalog no. 017–000-121).
Image acquisition was carried out using an ECLIPSE Ti2 inverted microscope from Nikon. After imaging the sections, the coverslips were removed in 4× SSC buffer (G. Biosciences, catalog no. 786–023), and the fluorophores were cleaved using the cleaving solution provided in the kit. This process was repeated iteratively, targeting sets of three genes at a time (T4 to T6, T7 to T9), until all eight target genes were successfully imaged and analyzed.
The procedure for image analysis involved several steps. First, each confocal image file was imported into FIJI ImageJ. Subsequently, a maximum intensity z-projection was generated. Following this, each channel within the image was separated into distinct image files, and each newly created image was saved in the TIFF file format. To align and combine images obtained from multiple rounds of tissue imaging, we used the ACD Biosciences Image Registration Software, following the manufacturer’s specified protocol as outlined in ACD Biosciences Document No. 30065UM. In a concise summary of the process, one of the DAPI images from the various rounds of imaging served as the reference image. All other DAPI images from subsequent imaging rounds were registered to this reference image. This registration process produced a transformation matrix that contained coordinate conversions, which were subsequently applied to all the other gene-specific images and images from each round of imaging. This step effectively established a unified coordinate system for all images from all imaging rounds. Within the same software tool, we also cropped nonoverlapping regions present around the edges of the images. Following cropping, each registered image was saved separately as a TIFF file.
To quantify the data, we standardized our approach by selecting a uniform region of interest (ROI) for each section. These ROIs were chosen to cover an equivalent area encompassing layers I to III of the vmPFC, as illustrated in fig. S1. Subsequently, in ImageJ, a manual threshold was applied to each image, leading to the creation of binary images. The count of positively stained pixels within these binary images served as our measure of the average signal, which was then subjected to statistical analysis.
Fiber photometry
Mice were maintained under 1 to 1.5% isoflurane anesthesia and stereotactically injected with 120 nl of AAVDJ-syn-GCaMP6f, titer 1012 gc/ml, over approximately 10 min. The empirically optimized injection coordinates were Medial-Lateral (ML) 0.15, Anterior-Posterior (AP) 1.72, Dorsal-Ventral (DV) 2.84–2.64, relative to bregma; the DV coordinate was adjusted for each animal (the goal being to inject 2 ± 0.1 mm from the dorsal surface of the brain). An optical fiber ⌀400 mm with a conical tip (Doric Lenses) was implanted over the injection site, in the PL cortex (DV ~2.5) (see fig. S4A). After 5 weeks, the animals were transferred into a special cage and connected to a photometry apparatus (Doric Lenses), where all experimental manipulations were conducted. Ca2+-dependent and isosbestic fluorescence wavelengths were recorded, the latter as an internal control. Every trace was rectified and corrected by fitting first for exponential decay and secondly linearly using the isosbestic fluorescence as the regressor. A classic problem with photometry is inter-individual comparisons, as many hard-to-control-for variables affect even normalized fluorescence amplitude (e.g. transfection efficiency, distance of transfected cells from fiber tip, differences sometimes in light-emitting diode optical power output). To get around this hurdle, we took advantage of the “background noise” produced, in each recording, by uncorrelated single-cell activity at rest. We calculated the median absolute deviation (MAD) of stretches of such minimal activity for every trace and equalized them across animals. The possibility of differential GCaMP expression between groups, which would result in different amplitude of this activity, was thus accounted for. MAD normalization affects only measures of fluorescence amplitude, not time- or frequency-based variables.
Recordings from mice in which the transfection site was subsequently found to be outside the ILC were excluded (these recordings typically did not show sound- and/or light-evoked responses). All the mice used in this experiment had completed the behavioral tests described below and were 11 to 13 months old (age-matched between genotypes).
Behavioral tests
Tests were performed during the mice’s active phase under dim red light. In every case, the animals were habituated by keeping them at least 1 hour in the test room before the test was performed. All test sessions, except for IDED, were videotaped, and every single video is available for the reader’s viewing pleasure at the link provided in the next section. Particularly salient examples are enclosed here as the Supplementary Materials. IDED sessions require 5 to 10 hours’ total time per animal and were therefore not recorded, except for the representative snippets in the Supplementary Materials. All tests were performed with the experimenter blind to genotype.
Free interaction test
The test mouse was placed in an unfamiliar cage with new bedding, acclimatized for 20 min, and exposed for 5 min to a strange wild-type conspecific of the same sex of approximately 7 to 12 weeks of age. Neither participant was a breeder mouse (to reduce aggression). Interactions between mice were analyzed by TopScan (CleverSys Inc.).
Three-chamber test
The test mouse was placed in the middle chamber and acclimatized for 10 min, and then partitions were removed allowing the animal to explore the other two chambers, for 10 min. Again, movements were analyzed with TopScan.
Marble burying test
The mouse was dropped into a square-footprint arena with 10-cm-thick corncob bedding and a regular array of 20 1-cm black marbles. After 10 min, he was extracted from the cage and the number of completely or partially buried marbles counted.
Looming test
As previously described, the test mouse was placed in a custom-built arena and allowed to acclimatize for 10 min. A maximum of 10 iterations of the expanding disc animations were played (the series was cut short if the mouse displayed all the three behaviors of interest).
Defensive burying test
Mice were acclimatized to the unfamiliar test cage, with a 5-cm-thick layer of new bedding, for 10 min. A Q-tip soaked in 4 ml of TMT was dropped into the cage in the second quadrant, and the mouse’s reactions videotaped for 20 min.
IDED test
Every mouse was preliminarily handled by the experiment for 7 days and weighed to determine a body weight baseline. Food restriction followed, for 4 days, with 1 to 2 g of food dispensed every day in the ramekins subsequently used for the test. Then, the mice were acclimated to the test arena for 2 days and trained to dig into bedding-filled ramekins for food for 1 or 2 days depending on performance. Body weight typically dropped to 80% of baseline by the beginning of the test sessions.
Statistical analysis
Statistical analysis of results was made by a Student’s t test or a two-way analysis of variance (ANOVA), unless otherwise stated.
Acknowledgments
We thank D. Pine and J. N. Crawley for very insightful suggestions on the behavioral phenotypes. We also thank D. Lowery and D. Adamovich for support.
Funding: This work was supported by NIH grants GM076430 and EY027202 (to H.Z.), the Intramural Research Program of the National Institute of Mental Health (ZIA MH002964 to S.H. and ZIA MH002951 to Y.C.), and the Rodent Behavioral Core (ZIC MH002952 to Y.C.). LLO was self-funded during part of the study.
Author contributions: L.L.O. designed the experiments, performed and analyzed the fiber photometry experiments, and performed the behavioral tests. L.L.O., F.M., and J.J.Z. performed the morphometric analysis. H.W. provided the animals with the correct littermate controls. J.J.Z. and L.L.O. performed the scRNA-seq experiments. M.B.T. and J.J.Z. analyzed the scRNA-seq data. R.K. and Q.T. performed the post RNA-seq validation with RNAscope. L.L.O., J.J.Z., J.d.H., and K.C. analyzed the behavioral tests. L.L.O., J.J.Z., M.B.T., Y.C., H.Z., and S.H. wrote the manuscript. Y.C., H.Z. and S.H. discussed and supervised research.
Competing interests: The authors declare they have no competing interests.
Data and materials availability: All videos of recorded behavioral tests, as well as all original databases, are freely and publicly available at https://livejohnshopkins-my.sharepoint.com/:f:/g/personal/llazzer1_jh_edu/ElJEwX1ivCxBgmIAeyz27D4BLs4iGfZFEnHA8_WJjOOlFg?e=QIHj7D. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Legends for movies S1 to S5
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S5
REFERENCES AND NOTES
- 1.Warthen D. M., Provencio I., The role of intrinsically photosensitive retinal ganglion cells in nonimage-forming responses to light. Eye Brain 4, 43–48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hattar S., Liao H. W., Takao M., Berson D. M., Yau K. W., Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science 295, 1065–1070 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berson D. M., Dunn F. A., Takao M., Phototransduction by retinal ganglion cells that set the circadian clock. Science 295, 1070–1073 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Provencio I., Rodriguez I. R., Jiang G., Hayes W. P., Moreira E. F., Rollag M. D., A novel human opsin in the inner retina. J. Neurosci. 20, 600–605 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guler A. D., Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature 453, 102–105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ecker J. L., Dumitrescu O. N., Wong K. Y., Alam N. M., Chen S. K., LeGates T., Renna J. M., Prusky G. T., Berson D. M., Hattar S., Melanopsin-expressing retinal ganglion-cell photoreceptors: cellular diversity and role in pattern vision. Neuron 67, 49–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quattrochi L. E., Stabio M. E., Kim I., Ilardi M. C., Michelle Fogerson P., Leyrer M. L., Berson D. M., The M6 cell: A small-field bistratified photosensitive retinal ganglion cell. J. Comp. Neurol. 527, 297–311 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt T. M., Alam N. M., Chen S., Kofuji P., Li W., Prusky G. T., Hattar S., A role for melanopsin in alpha retinal ganglion cells and contrast detection. Neuron 82, 781–788 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazzerini Ospri L., Prusky G., Hattar S., Mood, the Circadian System, and Melanopsin Retinal Ganglion Cells. Annu. Rev. Neurosci. 40, 539–556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez D. C., Fogerson P. M., Lazzerini Ospri L., Thomsen M. B., Layne R. M., Severin D., Zhan J., Singer J. H., Kirkwood A., Zhao H., Berson D. M., Hattar S., Light Affects Mood and Learning through Distinct Retina-Brain Pathways. Cell 175, 71–84.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeGates T. A., Altimus C. M., Wang H., Lee H. K., Yang S., Zhao H., Kirkwood A., Weber E. T., Hattar S., Aberrant light directly impairs mood and learning through melanopsin-expressing neurons. Nature 491, 594–598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuster J. M., The Prefrontal Cortex—An Update. Neuron 30, 319–333 (2001). [DOI] [PubMed] [Google Scholar]
- 13.K. B. J. Franklin, Y. Chudasama, in The Mouse Nervous System, G. Paxinos, L. Puelles, C. Watson, Eds. (Elsevier Inc., 2012), pp. 727–735. [Google Scholar]
- 14.Morgan M. A., Romanski L. M., LeDoux J. E., Extinction of emotional learning: contribution of medial prefrontal cortex. Neurosci. Lett. 163, 109–113 (1993). [DOI] [PubMed] [Google Scholar]
- 15.Adhikari A., Lerner T. N., Finkelstein J., Pak S., Jennings J. H., Davidson T. J., Ferenczi E., Gunaydin L. A., Mirzabekov J. J., Ye L., Kim S. Y., Lei A., Deisseroth K., Basomedial amygdala mediates top-down control of anxiety and fear. Nature 527, 179–185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vertes R. P., Differential projections of the infralimbic and prelimbic cortex in the rat. Synapse 51, 32–58 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Bicks L. K., Koike H., Akbarian S., Morishita H., Prefrontal Cortex and Social Cognition in Mouse and Man. Front. Psychol. 6, 1805 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sotres-Bayon F., Quirk G. J., Prefrontal control of fear: more than just extinction. Curr. Opin. Neurobiol. 20, 231–235 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chudasama Y., Robbins T. W., Functions of frontostriatal systems in cognition: comparative neuropsychopharmacological studies in rats, monkeys and humans. Biol. Psychol. 73, 19–38 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Gianfranceschi L., Siciliano R., Walls J., Morales B., Kirkwood A., Huang Z. J., Tonegawa S., Maffei L., Visual cortex is rescued from the effects of dark rearing by overexpression of BDNF. Proc. Natl. Acad. Sci. U.S.A. 100, 12486–12491 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Cruz C., Simon M., Flugge G., Fuchs E., Czeh B., Diurnal rhythm and stress regulate dendritic architecture and spine density of pyramidal neurons in the rat infralimbic cortex. Behav. Brain Res. 205, 406–413 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Izquierdo A., Wellman C. L., Holmes A., Brief uncontrollable stress causes dendritic retraction in infralimbic cortex and resistance to fear extinction in mice. J. Neurosci. 26, 5733–5738 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y., Zhang H. K., Liu F., Lei G., Liu P., Jiao T., Dang Y. H., Altered Light Conditions Contribute to Abnormalities in Emotion and Cognition Through HINT1 Dysfunction in C57BL/6 Mice. Front. Behav. Neurosci. 12, 110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chew K. S., Renna J. M., McNeill D. S., Fernandez D. C., Keenan W. T., Thomsen M. B., Ecker J. L., Loevinsohn G. S., VanDunk C., Vicarel D. C., Tufford A., Weng S., Gray P. A., Cayouette M., Herzog E. D., Zhao H., Berson D. M., Hattar S., A subset of ipRGCs regulates both maturation of the circadian clock and segregation of retinogeniculate projections in mice. eLife 6, e22861 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drevets W. C., Videen T. O., Price J. L., Preskorn S. H., Carmichael S. T., Raichle M. E., A functional anatomical study of unipolar depression. J. Neurosci. 12, 3628–3641 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S. K., Badea T. C., Hattar S., Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature 476, 92–95 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saunders A., Macosko E. Z., Wysoker A., Goldman M., Krienen F. M., de Rivera H., Bien E., Baum M., Bortolin L., Wang S., Goeva A., Nemesh J., Kamitaki N., Brumbaugh S., Kulp D., McCarroll S. A., Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L. E., la Manno G., Codeluppi S., Furlan A., Lee K., Skene N., Harris K. D., Hjerling-Leffler J., Arenas E., Ernfors P., Marklund U., Linnarsson S., Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badea T. C., Cahill H., Ecker J., Hattar S., Nathans J., Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron 61, 852–864 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brody E. B., Roxvold H. E., Influence of prefrontal lobotomy on social interaction in a monkey group. Psychosom. Med. 14, 406–415 (1952). [DOI] [PubMed] [Google Scholar]
- 31.Sato M., Prefrontal cortex and emotional behaviors. Folia Psychiatr. Neurol. Jpn. 25, 69–78 (1971). [DOI] [PubMed] [Google Scholar]
- 32.Franzen E. A., Myers R. E., Neural control of social behavior: prefrontal and anterior temporal cortex. Neuropsychologia 11, 141–157 (1973). [DOI] [PubMed] [Google Scholar]
- 33.Liu J., Dietz K., DeLoyht J. M., Pedre X., Kelkar D., Kaur J., Vialou V., Lobo M. K., Dietz D. M., Nestler E. J., Dupree J., Casaccia P., Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nat. Neurosci. 15, 1621–1623 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salay L. D., Ishiko N., Huberman A. D., A midline thalamic circuit determines reactions to visual threat. Nature 557, 183–189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Boer S. F., Koolhaas J. M., Defensive burying in rodents: ethology, neurobiology and psychopharmacology. Eur. J. Pharmacol. 463, 145–161 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Heisler J. M., Morales J., Donegan J. J., Jett J. D., Redus L., O’Connor J. C., The attentional set shifting task: a measure of cognitive flexibility in mice. J. Vis. Exp., 51944 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiser J., Koenigs M., The Multifaceted Role of the Ventromedial Prefrontal Cortex in Emotion, Decision Making, Social Cognition, and Psychopathology. Biol. Psychiatry 83, 638–647 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bloodgood B. L., Sharma N., Browne H. A., Trepman A. Z., Greenberg M. E., The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature 503, 121–125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koechlin E., Ody C., Kouneiher F., The architecture of cognitive control in the human prefrontal cortex. Science 302, 1181–1185 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Noudoost B., Clark K. L., Moore T., Working memory gates visual input to primate prefrontal neurons. eLife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brodmann K., Neue Ergebnisse über die vergleichende histologische Lokalisation der Grosshirnrinde mit besonderer Berücksichtigung. Anat. Anzeiger 41, 1 (1912). [Google Scholar]
- 42.Uylings H. B., van Eden C. G., Qualitative and quantitative comparison of the prefrontal cortex in rat and in primates, including humans. Prog. Brain Res. 85, 31–62 (1990). [DOI] [PubMed] [Google Scholar]
- 43.Kessel L., Lundeman J. H., Herbst K., Andersen T. V., Larsen M., Age-related changes in the transmission properties of the human lens and their relevance to circadian entrainment. J Cataract Refract Surg 36, 308–312 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Ellegood J., Crawley J. N., Behavioral and Neuroanatomical Phenotypes in Mouse Models of Autism. Neurotherapeutics 12, 521–533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saxena A., Wagatsuma A., Noro Y., Kuji T., Asaka-Oba A., Watahiki A., Gurnot C., Fagiolini M., Hensch T. K., Carninci P., Trehalose-enhanced isolation of neuronal sub-types from adult mouse brain. Biotechniques 52, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.da W. H., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legends for movies S1 to S5
Movies S1 to S5