Abstract
Plants have unique responses to fluctuating light conditions. One such response involves chloroplast photorelocation movement, which optimizes photosynthesis under weak light by the accumulation of chloroplasts along the periclinal side of the cell, which prevents photodamage under strong light by avoiding chloroplast positioning toward the anticlinal side of the cell. This light-responsive chloroplast movement relies on the reorganization of chloroplast actin (cp-actin) filaments. Previous studies have suggested that CHLOROPLAST UNUSUAL POSITIONING 1 (CHUP1) is essential for chloroplast photorelocation movement as a regulator of cp-actin filaments. In this study, we conducted comprehensive analyses to understand CHUP1 function. Functional, fluorescently tagged CHUP1 colocalized with and was coordinately reorganized with cp-actin filaments on the chloroplast outer envelope during chloroplast movement in Arabidopsis thaliana. CHUP1 distribution was reversibly regulated in a blue light- and phototropin-dependent manner. X-ray crystallography revealed that the CHUP1-C-terminal domain shares structural homology with the formin homology 2 (FH2) domain, despite lacking sequence similarity. Furthermore, the CHUP1-C-terminal domain promoted actin polymerization in the presence of profilin in vitro. Taken together, our findings indicate that CHUP1 is a plant-specific actin polymerization factor that has convergently evolved to assemble cp-actin filaments and enables chloroplast photorelocation movement.
CHLOROPLAST UNUSUAL POSITIONING 1 is an actin polymerization factor that is unique to plants and essential for chloroplast movement in response to high-light stress.
IN A NUTSHELL.
Background: Chloroplasts dynamically adjust their intracellular positioning in response to the intensity and direction of light, mediating the accumulation response, which optimizes photosynthesis under weak light conditions, and the avoidance response, which mitigates photodamage under strong light conditions. The blue light photoreceptors, phototropin1 and 2, redundantly mediate these responses—both phot1 and phot2 contribute to the accumulation response, while phot2 alone governs the avoidance response. Specifically, short actin filaments associated with chloroplasts, referred to as cp-actin filaments, facilitate chloroplast movement through asymmetrical reorganizations at the front side of moving chloroplasts, specifically in the space between the chloroplast and plasma membrane. CHLOROPLAST UNUSUAL POSITIONING 1 (CHUP1) regulates the cp-actin filaments.
Question: How do phototropins regulate cp-actin filaments, and what role does CHUP1 play in the regulation of cp-actin filaments?
Findings: Using a comprehensive approach that includes cell biology, X-ray crystallography, biochemistry, electron microscopy, and plant physiology, our study reveals CHUP1 as a type of actin nucleator controlling chloroplast movement. Noteworthy discoveries include CHUP1's pivotal role in polymerizing cp-actin filaments under weak light and depolymerizing them under strong light, in a blue light- and phototropin-dependent manner. Notably, the C-terminal half of CHUP1 (CHUP1-C) exhibits structural similarity to the formin FH-2 domain, despite lacking amino acid sequence homology. Furthermore, CHUP1-C's ability to promote actin polymerization when complexed with profilin in vitro implies that CHUP1 evolved independently as a plant-specific actin nucleator for chloroplast movement.
Next steps: Our present model relies on actin polymerization as the motive force for chloroplast movements. The coordinated rearrangement of cp-actin filaments at the leading edge of moving chloroplasts is governed by the asymmetric distribution of CHUP1. However, the mechanism by which CHUP1 and cp-actin filaments are aligned in one direction and their collective contribution to the motive force of chloroplast relocation remains to be elucidated.
Introduction
Plants have evolved a variety of systems that allow them to quickly acclimate to fluctuations in their natural environments, such as changes in light intensity. Light is essential for photosynthesis but harmful when too bright. The chloroplast is a specialized organelle in plants and algae that converts solar energy into chemical energy through photosynthesis. In fluctuating light conditions, chloroplasts dynamically change their positions within the cell according to the quality, intensity, and direction of incident light (for review, see Wada et al. 2003). Chloroplasts move toward weak light to capture as much energy as possible for photosynthesis (the so-called accumulation response; Zurzycki 1955; Gotoh et al. 2018) but they rapidly move away from strong light to limit light absorption (the avoidance response) (Zurzycki 1957; Kasahara et al. 2002). Chloroplasts can move in any direction without rotating or rolling (Tsuboi et al. 2009; Tsuboi and Wada 2011). When a part of a prothalial cell of the Southern maidenhair fern (Adiantum capillus-veneris) is irradiated with a microbeam of 30 W m−2 of red light for 1 min, the chloroplasts in the cell move to the irradiated area (Kagawa and Wada 1994). By contrast, when part of a chloroplast is irradiated directly with strong blue light, the chloroplast escapes from the light (Tsuboi and Wada 2011).
Light-induced chloroplast movements are mediated by blue light receptors, specifically phototropins (phot1 and phot2 in seed plants) (Jarillo et al. 2001; Kagawa et al. 2001; Sakai et al. 2001). In Arabidopsis (Arabidopsis thaliana), phot1 functions only in the accumulation response (Sakai et al. 2001), whereas phot2 functions in both the accumulation and avoidance responses (Jarillo et al. 2001; Kagawa et al. 2001; Sakai et al. 2001). phot2 mutants that are defective in the avoidance response are more susceptible than wild-type (WT) and phot1 mutant plants to strong light-induced photodamage (Kasahara et al. 2002; Sztatelman et al. 2010; Cazzaniga et al. 2013). Thus, the avoidance response is essential for plant survival, especially under fluctuating light conditions (Wada et al. 2003). To control these movements, chloroplasts continuously monitor signals released from the photoreceptors (Tsuboi and Wada 2013).
Actin filaments are essential components of the plant cell cytoskeleton, contributing to numerous cellular programs that are vital for plant growth, development, and acclimation to the environment. The spatial arrangement of actin filaments in plant cells is highly dynamic and responsive to a range of development and environmental cues. Various actin-binding proteins regulate the organization and dynamics of actin filaments, enabling the cell to adjust to different physiological demands and stimuli (for reviews, see the studies by Staiger and Blanchoin 2006; Higaki et al. 2007; Li et al. 2015). For instance, plant cells contain high concentrations of profilin, which binds to monomeric actin and prevents spontaneous nucleation for polymerization. The formin family of nucleation factors binds to the profilin:actin complexes, nucleates, and catalyzes the elongation of linear actin filaments. Chloroplast movement is mediated by the dynamic reorganization of short actin filaments, called chloroplast actin (cp-actin) filaments, that are located at the interspace between each chloroplast and the plasma membrane (Kadota et al. 2009). It has thus been speculated that a formin or a functionally similar nucleation factor mediates the assembly of cp-actin filaments. In response to strong blue light, cp-actin filaments are rapidly disrupted and disappear in a phot2-dependent manner and then reappear on the leading edge of moving chloroplasts (Kadota et al. 2009; Kong et al. 2013a). For this rapid disruption of cp-actin filaments, the involvement of a severing protein, such as a member of the actin depolymerization factor/cofilin family or a functionally similar protein, is assumed. The distribution of cp-actin filaments around the chloroplast determines the speed and direction of chloroplast movement; the larger the asymmetry in filament distribution, the faster the chloroplast will move (Kadota et al. 2009; Kong et al. 2013a) (reviews; Kong and Wada 2011; Suetsugu and Wada 2016).
In Arabidopsis, CHLOROPLAST UNUSUAL POSITIONING 1 (CHUP1) functions in chloroplast positioning and movement (Kasahara et al. 2002; Oikawa et al. 2003). CHUP1 has multiple functional domains in its N-terminal half, including a hydrophobic region, a coiled-coil region, and an actin-binding domain (Oikawa et al. 2003, 2008). The N-terminal hydrophobic domain attaches CHUP1 to the chloroplast outer envelope (Oikawa et al. 2003, 2008). The actin-binding domain in the N-terminal half binds to F-actin (Oikawa et al. 2003). By contrast, little is known about the functional domains in the C-terminal half of CHUP1, except that there is a proline-rich region (Oikawa et al. 2003) that binds to profilin (Schmidt von Braun and Schleiff 2008). The domain structure of CHUP1, its intracellular localization, and the chup1 mutant phenotype suggest that CHUP1 functions in the regulation of cp-actin filaments. Indeed, cp-actin filaments were not detected in chup1 mutant cells, even though cytoplasmic actin filaments were clearly observed (Kadota et al. 2009; Kong et al. 2013a). CHUP1 is highly conserved across a broad range of plant species. The protein is found in all plant lineages above the green algae (Charophytes) that show chloroplast photorelocation movement (Suetsugu and Wada 2016). The conserved functional roles of CHUP1 as the main player necessary for chloroplast positioning and movement have been also confirmed in various plant species including A. capillus-veneris (Suetsugu et al. 2012), the moss Physcomitrium patens (Usami et al. 2012), and Nicotiana benthamiana (Angel et al. 2013).
In this study, to obtain insights into CHUP1 as a plant-specific class of actin-binding protein that promotes actin polymerization functioning in chloroplast photorelocation movement, we performed multiple analyses ranging from cell biology, X-ray crystallography, biochemistry, electron microscopy, total internal reflection fluorescence (TIRF) microscopy, and plant physiology. We demonstrated that CHUP1 is dynamically reorganized on the chloroplast outer membrane in a blue light- and phototropin-dependent manner. Furthermore, we revealed that the CHUP1-C-terminal domain is structurally similar to the formin homology 2 (FH2) domain, although they share little amino acid sequence similarity. In addition, we evaluated the function of the CHUP1-C-terminal domain to promote actin polymerization in the presence of profilin in vitro.
Results
CHUP1-YFP and CHUP1-tdTomato fusion proteins are fully functional in transgenic Arabidopsis plants
To examine CHUP1 function in the regulation of cp-actin filaments, we transformed a CHUP1-deficient Arabidopsis mutant (chup1-3) with constructs encoding CHUP1-fluorescent fusion proteins [CHUP1-YFP (yellow fluorescent protein) and CHUP1-tdTomato] driven by the native CHUP1 promoter to produce CHUP1pro:CHUP1-YFP (C1Y) and CHUP1pro:CHUP1-tdTomato (C1tdT) transgenic plants (Supplementary Fig. S1A). C1Y and C1tdT transgenic plants accumulated CHUP1-YFP and CHUP1-tdTomato to levels comparable to those of endogenous CHUP1 in WT (Supplementary Fig. S1B). The expression of CHUP1-YFP and CHUP1-tdTomato successfully complemented the chloroplast positioning and photorelocation movement defects of the chup1-3 mutant (Supplementary Fig. S1, C and D). Under weak blue light (3.2 µmol m−2 s−1), chloroplasts accumulated on the periclinal wall in both C1Y and C1tdT transgenic plants as in WT plants but not in chup1-3 mutant plants. Under strong blue light conditions (21 and 48 µmol m−2 s−1), we observed a chloroplast avoidance response in both C1Y and C1tdT transgenic plants as in WT plants but not in chup1-3 mutant plants. The chloroplasts of chup1-3 mutant plants were at the bottom of the mesophyll cells under all light conditions, as previously described (Oikawa et al. 2003). Together, these data show that CHUP1-YFP and CHUP1-tdTomato in the transgenic plants are as functional as endogenous CHUP1 in WT plants.
CHUP1 is reorganized on the chloroplast outer membrane in a blue light-dependent manner
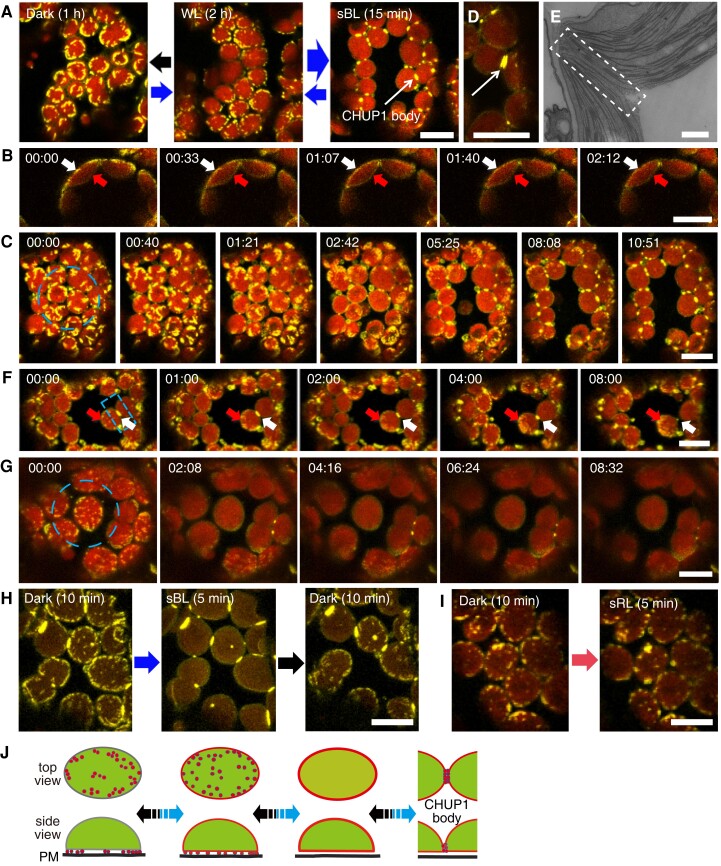
We examined the subcellular localization of CHUP1 under different light conditions by confocal microscopy of palisade cells in the rosette leaves of C1Y plants. When white light (20 µmol m−2 s−1) was applied for 2 h, we detected CHUP1-YFP fluorescence as small dots along the periphery of the plasma membrane side of chloroplasts (Fig. 1A middle panel and B 00:00). After dark adaptation for 1 h, the dots aggregated to form longer and larger fluorescent areas, mainly along the chloroplast periphery (Fig. 1A left panel and C 00:00). When a part of a cell in a dark-adapted leaf was illuminated with strong blue light (458-nm laser) for several minutes, the dots of CHUP1-YFP fluorescence disappeared and were replaced by fluorescence distributed over the entire chloroplast envelope, including on the vacuole side (Fig. 1, B and C).
Figure 1.
Reorganization of CHUP1-YFP on the chloroplast envelope in response to blue light. A) Reversible changes in the distribution pattern of CHUP1-YFP under the indicated light conditions. CHUP1-YFP in the palisade cells of C1Y transgenic Arabidopsis plants was observed following incubation in darkness for 1 h (black arrow), weak white light (20 μmol m−2 s−1) for 2 h (thin blue arrow), or strong blue light (sBL; 458-nm laser scan with an output power of 2.8 µW) for 15 min (thick blue arrow). The images were captured at a resolution of 512 × 512 pixels using a 4× digital zoom and are presented with false-color indicating YFP (yellow) and chlorophyll (red) fluorescence. The time (min:s) of image acquisition is shown in the upper left corner of each image. B) Redistribution of CHUP1-YFP from the interface between the plasma membrane and the chloroplast envelope (white arrows) to the entire chloroplast envelope, including the vacuole side (red arrows). The whole cell was irradiated with strong blue light. Time-lapse images were captured at 33-s intervals. The image acquisition and presentation are the same as in A). C) Blue light-dependent reorganization of CHUP1-YFP during a strong light-induced avoidance response followed by dark adaptation for 20 min. The dashed blue circle indicates the area that was irradiated. A time-lapse movie of this response can be seen in Supplementary Movie S1. D) A CHUP1 body appears as 2 lines along the interface between 2 chloroplasts after 5 min of strong blue light irradiation. E) Transmission electron microscopy image of the region of contact (at the center of the rectangle) between 2 chloroplasts. F) Redistribution of a CHUP1 body (white arrows) to small dots (red arrows) at the leading edge of a moving chloroplast. G) Redistribution of CHUP1-YFP on an isolated irradiated chloroplast. The dotted CHUP1-YFP signal rapidly disappeared but no CHUP1 body formed. A time-lapse movie of this response can be seen in Supplementary Movie S2. The area irradiated with strong blue light is indicated as a dashed blue circle (20 µm in diameter) in C and G), and a dashed blue rectangle (10 μm × 5 μm) in F) superimposed on the first image (00:00) of each series. Strong blue light was provided by 458-nm laser scans (an output power of 2.8 μW). Time-lapse images were collected at 40-, 33-, 60-, and 30-s intervals in C, F, and G), respectively. The image acquisition and presentation are the same as in A). H and I) Blue light-specific reorganization of CHUP1-YFP. H) CHUP1-YFP was sequentially observed in the same palisade cell of C1Y transgenic Arabidopsis plants following incubation in the dark for 10 min (Dark 10 min), irradiation with strong blue light (sBL; 458-nm laser scans) for 5 min (sBL 5 min), and further incubation in the dark for 10 min (Dark 10 min). I) CHUP1-YFP was sequentially observed in a palisade mesophyll cell following incubation in the dark for 10 min and then irradiated with strong red-light (sRL) by 633-nm laser scans for 5 min. The image acquisition and presentation are the same as in A). Scale bar, 500 nm in E), 10 μm in all other panels. J) Diagram illustrating the reorganization of CHUP1 on the chloroplast envelope. CHUP1 (red) localizes as dots at the interface between the plasma membrane and a chloroplast in darkness. In continuous strong light, CHUP1 disperses to the entire chloroplast envelope, first as dots at the interface and then diffusely throughout the chloroplast outer envelope. Finally, CHUP1 accumulates as a CHUP1 body where 2 chloroplasts are in contact. These steps are reversibly regulated in a blue light-dependent manner. The lower panel shows side views of a chloroplast (side view) and the upper panel shows the chloroplast surface that faces the plasma membrane (top view). Blue arrows indicate phototropin-dependent responses under blue light and black arrows indicate the reverse steps in the dark.
We assessed the avoidance response of C1Y plants by illuminating a region of interest (ROI) with a microbeam of strong blue light (458-nm laser). After 15 min, we clearly observed chloroplast avoidance from the microbeam, and the dispersed CHUP1-YFP accumulated at contact sites between chloroplasts, forming a specific structure named the “CHUP1 body” (Fig. 1A right panel and C and Supplementary Movie S1). Sometimes, a CHUP1 body appeared as 2 discrete parallel lines of the same length, each belonging to one of the 2 adjacent chloroplasts (Fig. 1D). Presumably, CHUP1 on the outer membrane of each chloroplast accumulated along the contact area and formed a CHUP1 body. We attempted to observe CHUP1 bodies in WT plants by electron microscopy. However, we detected no notable structures where there was close contact between the outer membranes of 2 chloroplasts (Fig. 1E). When a CHUP1 body in a rectangular ROI was illuminated with strong blue light (458-nm laser), CHUP1-YFP fluorescence gradually decreased and small dots of fluorescence appeared at the leading edge of the moving chloroplast, suggesting that CHUP1 becomes redistributed on the chloroplast envelope (Fig. 1F). If a chloroplast did not come into contact with other chloroplasts, we observed no CHUP1 body (Fig. 1G and Supplementary Movie S2). The above-mentioned reorganization patterns of CHUP1 were specifically induced by blue light but not by red light (Fig. 1, H and I). These data collectively suggest that the position of CHUP1 is reversibly reorganized on the chloroplast outer membrane in a blue light-dependent manner (Fig. 1J).
Reorganization of CHUP1 is regulated in a phototropin-dependent manner
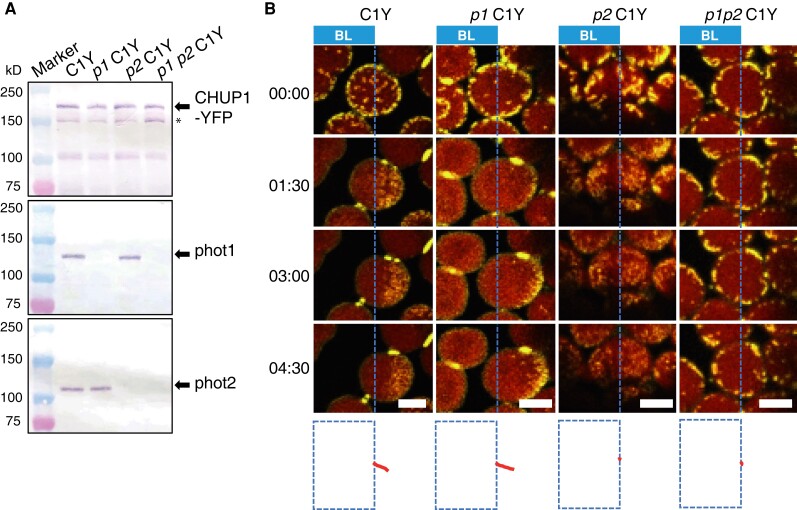
Blue light induces not only chloroplast movements but also the reorganization of CHUP1 on the chloroplast surface. We examined the involvement of phototropin in the regulation of CHUP1 localization. To this end, we crossed the C1Y line into phot1 chup1, phot2 chup1, and phot1 phot2 chup1 mutant backgrounds to produce phot1 C1Y, phot2 C1Y, and phot1 phot2 C1Y lines, respectively. The abundance of CHUP1-YFP was similar in all lines (Fig. 2A). When the avoidance response was induced in the WT line (C1Y) and in phot1 C1Y by partial-cell irradiation with strong blue light (458-nm laser), the chloroplasts moved out of the strong light-irradiated area, which was accompanied by an asymmetric distribution of CHUP1-YFP fluorescence: higher intensity on the leading end and lower intensity on the rear end of the moving chloroplast (Fig. 2B and Supplementary Movies S3 and S4). However, CHUP1-YFP was only partially reorganized in the phot2 C1Y line (Fig. 2B and Supplementary Movie S5). The dots of CHUP1-YFP fluorescence became smaller but never disappeared, and no CHUP1 body formed (Fig. 2B and Supplementary Movie S5). We observed no such reorganization for CHUP1-YFP in the phot1 phot2 C1Y line (Fig. 2B and Supplementary Movie S6). By contrast, the dots of CHUP1-YFP fluorescence disappeared normally in the phot1 C1Y line, indicating that phot2 alone is sufficient for CHUP1 reorganization and that phot1 also participates in the regulation of CHUP1 reorganization. Furthermore, the trajectories of chloroplast movement were well aligned with the asymmetric distribution of CHUP1-YFP on moving chloroplasts (Fig. 2B). Therefore, CHUP1 localization is dynamically regulated in a blue light- and phototropin-dependent manner.
Figure 2.
Identification of the main photoreceptor regulating CHUP1 localization. A) Immunoblot analysis of CHUP1-YFP abundance in transgenic plants in the WT, phot1, phot2, and phot1 phot2 mutant backgrounds (C1Y, p1 C1Y, p2 C1Y, p1 p2 C1Y). Protein samples (50 μg) were separated on 7.5% SDS-PAGE gels and probed with anti-CHUP1 (top panel), anti-PHOT1 (middle), and anti-PHOT2 (bottom) polyclonal antibodies. Arrows from top to bottom blots indicate CHUP1-YFP, phot1, and phot2, respectively. The band indicated by * most likely represents truncated CHUP1-YFP generated by endogenous proteases, since this band is absent in cells lacking CHUP1-YFP. B) Series of images at the indicated time points (min:s) showing the reorganization of CHUP1-YFP in strong blue light in WT and phot mutant cells. In each image, the region to the left of the blue dotted line was irradiated with strong blue light by 458-nm laser scans with an output power of 2.8 μW at 30-s intervals to induce the avoidance response. The images were captured at a resolution of 512 × 256 pixels using a 4× digital zoom and are presented with false-color indicating YFP (yellow) and chlorophyll (red) fluorescence. The time (min:s) of image acquisition is shown to the left. The paths of individual chloroplasts are indicated in red at the bottom of each image. The centers of each chloroplast were traced. Time-lapse movies of these responses can be seen in Supplementary Movies S3 to S6. Scale bars, 5 μm.
CHUP1 is reorganized coordinately with cp-actin filaments
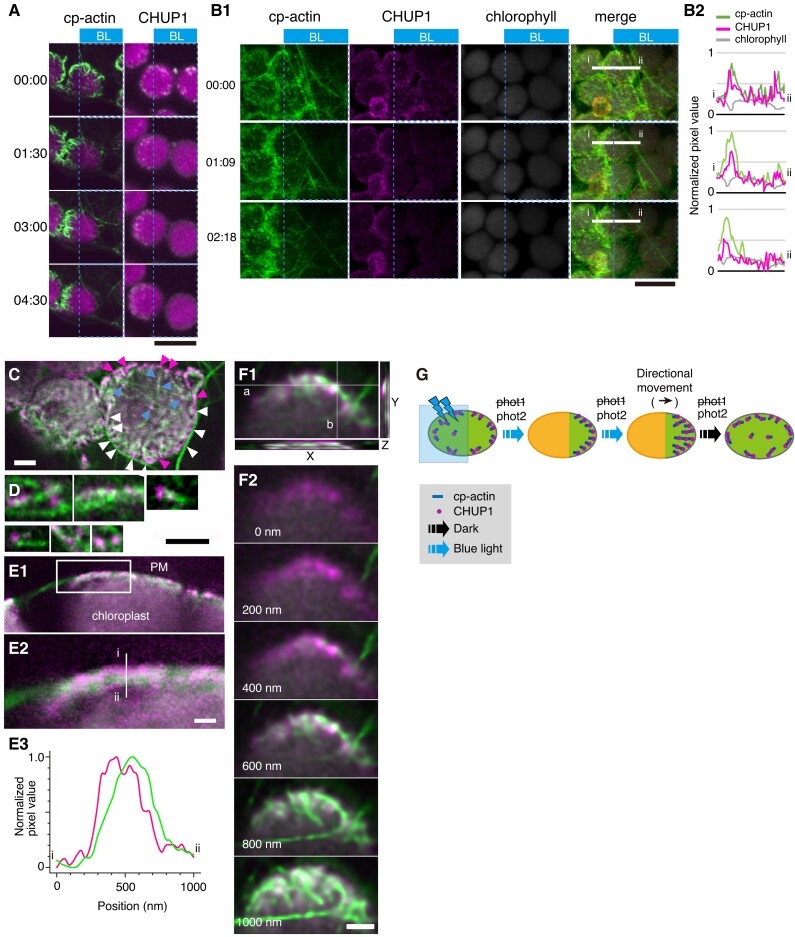
We visualized cp-actin filaments in Arabidopsis transgenic plants harboring a construct encoding GFP fused to the actin-binding domain of mouse Talin (GFP-mTalin). We observed asymmetric distributions for cp-actin filaments between the plasma membrane and moving chloroplasts (Kadota et al. 2009; Kong et al. 2013a). The distribution pattern and dynamics of CHUP1-YFP were similar to those of cp-actin filaments on moving chloroplasts (Fig. 3A; Kong et al. 2013a). Therefore, we studied the relationship between cp-actin filaments and CHUP1 during chloroplast movements in a transgenic line expressing both CHUP1-tdTomato and GFP-mTalin (CHUP1-tdTomato GFP-mTalin line). CHUP1 and cp-actin filaments closely colocalized (Fig. 3, B to D) with a Mander's colocalization coefficient of 0.965 calculated from Fig. 3C. Both CHUP1 and cp-actin filaments disappeared rapidly in response to strong blue light and reappeared at the front ends of moving chloroplasts (Fig. 3, A and B and Supplementary Movie S7), as previously observed for cp-actin filaments (Kadota et al. 2009; Kong et al. 2013a).
Figure 3.
Association of CHUP1 with cp-actin filaments. A) Asymmetric distribution of cp-actin filaments (green color, left panels) and CHUP1 (white color right panels) on moving chloroplasts. Time-lapse images were collected at 30-s intervals for 4 min 30 s under strong blue light irradiation at the blue rectangle ROI using 458-nm laser scans (output power of 2.8 μW) in the intervals between image acquisitions. Image acquisition time stamps (min:s) are shown. Fluorescence images were captured at a resolution of 512 × 512 pixels using a 4× digital zoom in GFP-mTalin and CHUP1-YFP transgenic plants, respectively. The fluorescent images show cp-actin (green) or CHUP1 (white) merged with chlorophyll (magenta). The time (min:s) of image acquisition is shown on the left. Scale bar, 10 μm. B) Reorganization of CHUP1 and cp-actin filaments on moving chloroplasts in a CHUP1-tdTomato GFP-mTalin line. Time-lapse images were collected at the indicated time points (min:s). The other details are the same as in A) except that images were acquired at 34-s intervals for 3 min 27 s. Scale bar, 10 µm. A time-lapse movie of this response is shown in Supplementary Movie S7. B1) fluorescence images show cp-actin filaments (cp-actin, green), CHUP1-tdTomato (CHUP1, magenta), chloroplast (chlorophyll, gray), and the merged image of cp-actin filaments, CHUP1, and chlorophyll (merge). B2) fluorescence intensity profiles of CHUP1-tdTomato (magenta) and cp-actin filaments (green) along the white line (i and ii) in B1) (merge). C) Top view of chloroplasts facing the plasma membrane of the periclinal wall of palisade cells. Cp-actin filaments, CHUP1, and chloroplasts are shown with fluorescent images obtained from GFP-mTalin (green), CHUP1-tdTomato (magenta), and chlorophyll (gray), respectively, in a CHUP1-tdTomato GFP-mTalin line. Notably, CHUP1 was predominantly found in the peripheral region of chloroplast (magenta arrowheads). White arrowheads indicate CHUP1 attached to long cytoplasmic actin filaments. Blue arrowheads indicate CHUP1 localized at the tips or along the sides of cp-actin filaments. D) Magnified views of CHUP1 localized close to cp-actin filaments. E) Distribution of CHUP1 and cp-actin filaments within the region between the plasma membrane and the chloroplast envelope. CHUP1 localized to the plasma membrane side and cp-actin filaments along the chloroplast side. E1) An optical slice of a side view of a chloroplast next to the plasma membrane along an anticlinal wall. E2) An enlarged image of the area within the rectangle in E1). E3) Fluorescence intensity profiles of CHUP1-tdTomato (magenta) and cp-actin filaments (green) along the white line (i and ii) in E2) from the plasma membrane side (i, top of the line) to the chloroplast side (ii, bottom of the line). F) Separate distributions of CHUP1 at the plasma membrane side and cp-actin filaments on the chloroplast side. Top views of part of a chloroplast are shown. F1) A Z-series of 6 confocal images of fluorescence of CHUP1-tdTomato (magenta) and GFP-mTalin (green) taken with 200-nm steps from the plasma membrane side to the chloroplast side and superimposed on an X–Y plane using the maximum intensity projection method. In the X–Z plane at white line a and the Y–Z plane at white line b, the CHUP1-tdTomato and GFP-mTalin fluorescence for the entire Z-series was compiled, confirming that CHUP1 is on the plasma membrane side and cp-actin filaments are on the chloroplast side. The individual images are shown in F2). Scale bars, 1 μm in C, D, and F), and 500 nm in E). G) Diagram illustrating the coordinated phototropin-dependent dynamics of CHUP1 and cp-actin filaments. Clusters of CHUP1 (magenta dots) closely localize with cp-actin filaments. Asymmetric redistribution of CHUP1 and cp-actin filaments is coordinately regulated by phototropin in response to irradiation of part of a chloroplast with strong blue light (blue rectangle). phot2 is the main photoreceptor regulating the strong light-induced avoidance response.
CHUP1-tdTomato fluorescence appeared as dots mainly around the periphery of the plasma membrane side of chloroplasts (Fig. 3, B and C). In many cases, the CHUP1 dots appeared to be connected to one end or the side of cp-actin filaments (Fig. 3, C and D). Optical sectioning demonstrated that CHUP1-tdTomato is observed close to the plasma membrane, but not to the chloroplasts (Fig. 3, E and F). We confirmed this observation by fluorescence intensity analysis of the interface between the plasma membrane and a chloroplast (Fig. 3E). We frequently observed long cytoplasmic actin filaments running along the chloroplast periphery and attaching to the CHUP1 dots there (Fig. 3C). Therefore, these data collectively suggest that CHUP1 closely localizes and is reorganized coordinately with cp-actin filaments in a blue light- and phototropin-dependent manner (Fig. 3G).
We further investigated the effects of the actin-depolymerizing reagent latrunculin B (LatB) on the dynamic reorganization of CHUP1-YFP. As described previously (Figs. 2B, 3, A and B), the asymmetric distribution of CHUP1-YFP was normal on moving chloroplasts in the mock-treated rosette leaf cells with DMSO only. Notably, although LatB treatment severely reduced chloroplast movement, as previously described (Kadota et al. 2009; Kong et al. 2013a), we still observed the asymmetric distribution of CHUP1-YFP following microbeam irradiation with strong blue light (Supplementary Fig. S2A). Furthermore, we clearly observed the reversible distribution of CHUP1-YFP in the presence of LatB during dark incubation (Supplementary Fig. S2B), suggesting that actin polymerization is not necessary for CHUP1 movement on the chloroplast outer membrane.
The CHUP1-C-terminal fragment share structural homology with formin homology-2 domains
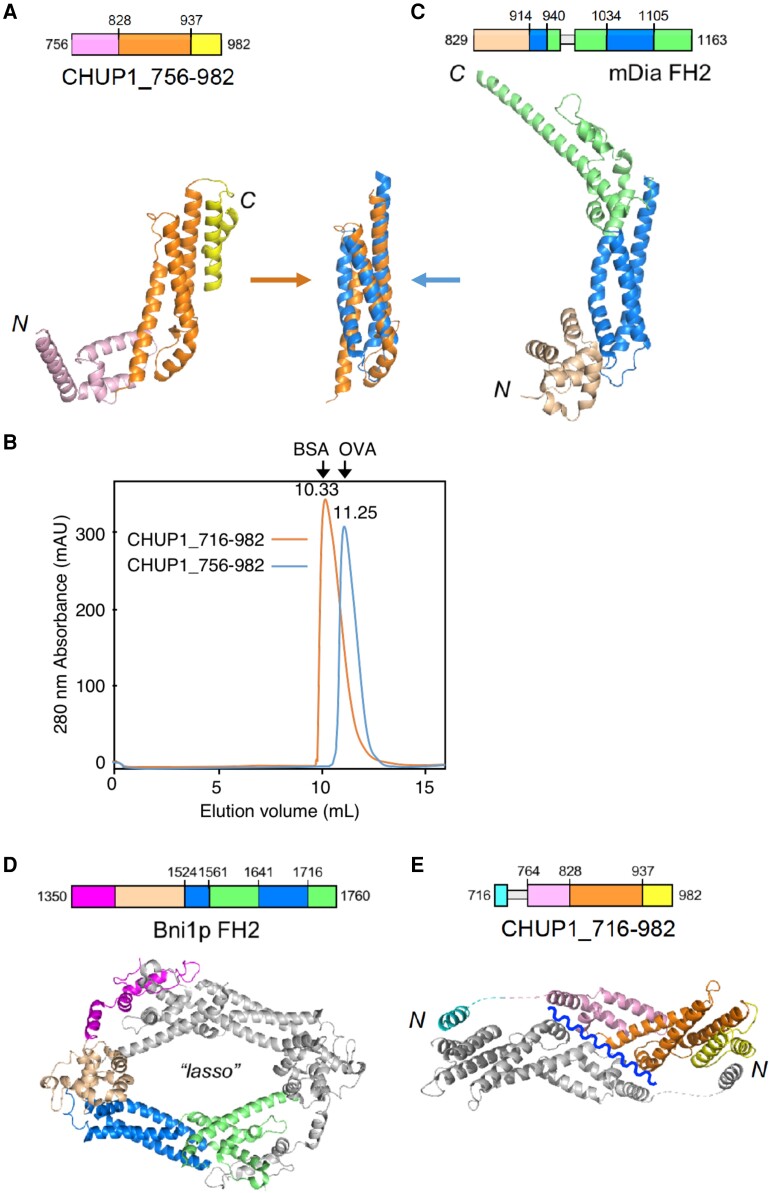
To gain insight into the CHUP1 function, we determined the crystal structure of the CHUP1-C-terminal fragment, CHUP1_756–982 (amino acids 756 to 982) (Fig. 4A; Table 1). The crystal contained a single molecule in the asymmetric unit. Size-exclusion chromatography suggested that CHUP1_756–982 prefers to exist as a monomer in solution (Fig. 4B). The overall structure of CHUP1_756–982 takes on an elongated shape consisting entirely of α-helices. Intriguingly, the 3-helix bundle located at the center of the C-terminal domain was reminiscent of the FH2 domains found in all well-characterized actin nucleators (Fig. 4, A and C; Shimada et al. 2004; Xu et al. 2004). Although there is no obvious amino acid sequence similarity between the 2 domains and the corresponding region of the FH2 sequence is non-contiguous, the 2 structures were superimposable with a root-mean-square deviation of 1.82 Å over the 52 Cα atoms.
Figure 4.
Three-dimensional structures of the C-terminal fragments of CHUP1. A) Crystal structure of CHUP1_756–982. B) The elution profiles of CHUP1_756–982 and CHUP1_716–982 were obtained by gel filtration chromatography using a Superdex 75 10/300 column. The elution volumes of BSA (67 kD) and ovalbumin (OVA, 43 kD) used for calibration are presented. The molecular weights of the recombinant proteins were calculated as 42 kD for CHUP1_756–982 and 64 kD for CHUP1_716–982. C) Crystal structure of the mDia1 FH2 domain (PDB 1V9D, Shimada et al. 2004). The central 3-helix bundles of CHUP1_756–982 and FH2 are superimposed in the middle between panels A and C). D) Functional dimer form of Bni1p FH2 (PDB 1UX5, Xu et al. 2004). E) Crystal structure of CHUP1_716–982. This fragment adopts a closed dimeric structure in crystal, which might open along the wavy line to become a functional dimer. In C and E), loops that are missing from the crystal structure due to poor electron density are indicated by thin gray boxes in the amino acid sequences.
Table 1.
Data collection and refinement statistics
| Statistics | CHUP1_756–982/C864S (SeMet) | CHUP1_716–982 (Native) |
|---|---|---|
| Data collection | ||
| Space group | P6222 | I213 |
| Cell dimensions | ||
| a, b, c (Å) | 123.3, 123.3, 160.8 | 180.7, 180.7, 180.7 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0 to 2.8 (2.85 to 2.80) |
50.0 to 3.0 (3.05 to 3.00) |
| Rmerge | 0.201 (>1) | 0.114 (>1) |
| I/σI | 27.2 (2.2) | 30.6 (2.2) |
| Unique reflections | 18,407 (882) | 19,727 (973) |
| Completeness (%) | 99.9 (100.0) | 99.9 (100.0) |
| Redundancy | 41.9 (43.8) | 21.7 (22.8) |
| Refinement | ||
| Resolution (Å) | 50.0 to 2.8 | 50.0 to 3.0 |
| No. reflections | 18,407 | 19,727 |
| Rwork/Rfree | 0.202/0.228 | 0.174/0.224 |
| No. atoms | ||
| Protein | 1,815 | 3,791 |
| Water | 7 | 0 |
| B-factors (Å2) | ||
| Protein | 72.0 | 94.8 |
| Water | 57.5 | |
| Rms deviations | ||
| Bond lengths (Å) | 0.009 | 0.010 |
| Bond angles (°) | 1.09 | 1.29 |
| PDB ID | 8WAF | 8WAG |
Values in parentheses are for highest-resolution shell. The Rfree value was calculated for the R factor using a test set (5%) of reflections not used in the refinement.
Rms, root-mean-square.
The functional form of the FH2 domain is a homodimer, in which the 2 subunits interact at both ends of their elongated structures (Fig. 4D; Xu et al. 2004). The CHUP1-C-terminal fragment used in the crystallization did not contain the sequence corresponding to the N-terminal interacting site of the FH2 domain. Therefore, we extended the original CHUP1 sequence by 40 residues in the direction of the N terminus and determined the crystal structure of CHUP1_716–982 (Table 1). The asymmetric unit contained 2 molecules. A dimeric structure in the crystal (Fig. 4E) agrees with the dimeric state in the solution suggested by size-exclusion chromatography (Fig. 4B). The newly added N-terminal portion formed a short α-helix and connected with another CHUP1 molecule in the crystal (Fig. 4E). The 30-residue segment that connected with the short, N-terminal α-helix and CHUP1_756–982 had no clear electron density (broken lines in Fig. 4E). We speculate that this segment serves as a flexible tether, facilitating the formation of a functional homodimer in a manner that resembles the FH2 domain–actin complex. Because the dimeric structure in the asymmetric unit is a closed form, it is necessary to open along the wavy blue line to become a functional dimer (Fig. 4E). For reference, the structures assembled according to the crystallographic symmetry do not match a functional homodimer structure like that of formin. Thus, the crystallographic results do not directly suggest any biological form but did provide a useful hint for the design of the loss-of-function mutations (see below).
CHUP1-C interacts with the barbed ends and sides of actin filaments in vitro
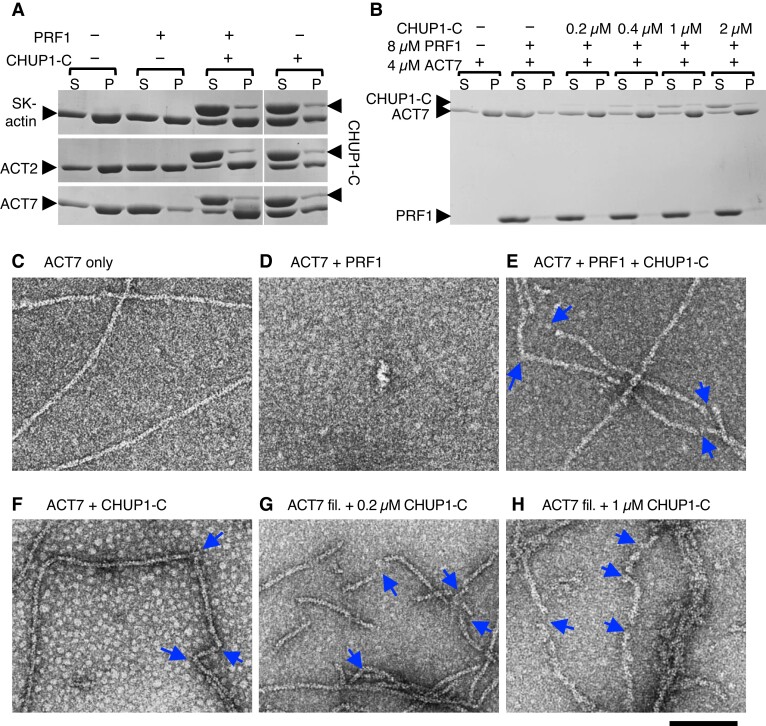
To examine how CHUP1 interacts with actin filaments, we performed a preliminary cosedimentation assay using CHUP1_611–1004, hereafter called CHUP1-C, and rabbit skeletal muscle actin (SK-actin) filaments. SK-actin was chosen because it is relatively stable and available in large quantities and has been used to characterize various plant-derived actin-binding proteins (e.g. Ito et al. 2003; Michelot et al. 2005; Yi et al. 2005). These properties of SK-actin were considered appropriate for the initial screening of the biochemical activities of CHUP1-C.
When 1 µM CHUP1-C was allowed to interact with 10 µM SK-actin filaments, a fraction of CHUP1-C co-sedimented with the actin filaments following ultracentrifugation. When the actin filaments were sheared by vigorous pipetting to increase the number of filament ends prior to the addition of CHUP1-C, the co-sedimented fraction of CHUP1-C increased from 4.7% to 10.4%, suggesting that CHUP1-C interacts with the ends of actin filaments. By contrast, fragmentation of the actin filaments with gelsolin did not increase the amount of co-sedimented CHUP1-C (Supplementary Fig. S3, A and B and Table S1). Because gelsolin remains bound to the barbed ends of actin filaments after severing (Sun et al. 1999), this result suggested that CHUP1-C binds to the barbed ends, but not to the pointed ends, of actin filaments. Nonetheless, the fact that some CHUP1-C bound to gelsolin-treated actin filaments suggested that CHUP1-C can bind along the sides of actin filaments as well, which we confirmed by electron microscopy, as described in the next section. The above experiments were performed in the presence of Ca2+, which was required to activate gelsolin. However, interactions of CHUP1-C with actin filaments did not depend on Ca2+, since chelating Ca2+ with excess EGTA did not change the amount of CHUP1-C that co-sedimented with control actin filaments (Supplementary Fig. S3A and Table S1).
CHUP1 is a type of actin-binding protein that promotes actin polymerization in the presence of profilin
Since CHUP1-C binds to the barbed ends of actin filaments and includes the formin homology-1 (FH1)-like polyproline sequences (Supplementary Fig. S3A and Table S1; Oikawa et al. 2003) followed by the segment that forms the 3-helix bundle sharing spatial arrangement with the core of FH2 (Fig. 4), we tested whether CHUP1-C promotes polymerization of actin in the presence or absence of profilin. To assess the polymerization-promoting activities of CHUP1-C, we used ACT2 and ACT7, 2 major isoforms of Arabidopsis actin in vegetative tissues, in addition to SK-actin. Both ACT2 and ACT7 are localized on the surface of chloroplasts (Kijima et al. 2018). As reported previously (Kijima et al. 2016), ultracentrifugation assays demonstrated that the prior addition of excess Arabidopsis PROFILIN1 (PRF1) strongly inhibits salt-induced spontaneous polymerization of ACT7, while polymerization of ACT2 or SK-actin was only partially inhibited by PRF1 (Fig. 5A). This inhibition was fully reversed by the addition of 4 µM CHUP1-C in the case of ACT2 and ACT7, but only partially reversed in the case of SK-actin. These results suggest that CHUP1-C has a potent ability to polymerize autologous actin complexed with autologous profilin, but has a weaker activity with heterologous SK-actin (Fig. 5A). When CHUP1-C was added to G-actin in the absence of PRF1, however, polymerization was partially inhibited (Fig. 5A and Supplementary Figs. S3C and S4A).
Figure 5.
Interaction of CHUP1-C with actin filaments in vitro. A) Ultracentrifugation assays of the effects of profilin and CHUP1-C on actin polymerization. Rabbit skeletal muscle actin (SK-actin) or Arabidopsis ACT2 or ACT7 was allowed to polymerize in F-buffer 1 in the presence or absence of 8 μM PRF1 and 4 μM CHUP1-C. After ultracentrifugation, the supernatant (S) and pellet (P) fractions were separately run on SDS-PAGE. B) Similar to A), except that only ACT7 was used and the concentration of CHUP1-C varied from 0 to 2 μM. C to F) Electron micrographs of actin polymerized in the presence or absence of profilin and CHUP1-C for 20 min. In C to F), 4 μM ACT7 was allowed to polymerize in F-buffer 1 in the absence of PRF1 and CHUP1-C C), in the presence of 4 μM PRF1 D), in the presence of 4 μM PRF1 and 0.4 μM CHUP1-C E), and in the presence of 0.4 μM CHUP1-C F). Actin solutions were diluted to 0.7 µM ACT7 in EM buffer, and then fixed and negatively stained with uranyl acetate. For particles observed in F), see Supplementary Fig. S7C. G and H) ACT7 filaments polymerized in the absence of PRF1 and CHUP1-C were diluted to 0.7 µM and applied onto an EM grid. After ∼30 s, 0.2 µM G) or 1 µM H) CHUP1-C was added, and the sample was negatively stained within several seconds. Blue arrows indicate the positions of sharp bent or gap of actin filaments. Scale bar, 100 nm.
We further examined the ability of CHUP1-C to promote polymerization of actin complexed with PRF1 using ACT7. Polymerization-promoting activity was evident when 0.2 µM CHUP1-C was added to 4 µM ACT7 and 8 µM PRF1, and the activity was saturated at 0.4 µM (Fig. 5B and Supplementary Fig. S4B). We confirmed the ability of CHUP1-C to promote the polymerization of ACT7 in the presence of PRF1 by kinetic experiments (Supplementary Fig. S5) and electron microscopy observations after negative staining with uranyl acetate (Fig. 5, C to E). At higher magnifications, we sometimes observed blob-like structures at the end or along the side of ACT7 filaments polymerized in the presence of CHUP1-C (arrows in Supplementary Fig. S6A). These blobs were not detected when ACT7 alone was polymerized (Fig. 5C), indicating that they are CHUP1-C-containing structures. The blobs were larger than the particles observed when CHUP1-C alone was negatively stained (Supplementary Fig. S6B), but were similar in shape and size to the particles observed in the ACT7–CHUP1-C mixture (various, but typically round or oval with ≥12 nm in diameter) (Supplementary Fig. S6C). Thus, we reasoned that they are CHUP1-C molecules complexed with ACT7. By contrast, the typical particles observed in the CHUP1-C alone sample were ellipses of ∼9 nm × 7 nm (Supplementary Fig. S6B), resembling the dimer structure of the FH2 domain of formin Bud Neck Involved 1 (Bni1p) from budding yeast (Saccharomyces cerevisiae; Fig. 4D). These results are consistent with the idea that CHUP1-C forms a dimeric ring structure that resembles an FH2 dimeric ring and binds to 2 actin monomers.
We noted in the electron micrographs that, unlike control ACT7 filaments (Fig. 5C), ACT7 filaments that polymerized in the presence of PRF1 and CHUP1-C had frequent sharp bends, and some filaments showed many gaps along otherwise continuous filaments (arrows in Fig. 5E). These bends and gaps were probably caused by weakened interactions between actin molecules in the filaments. Similarly, we observed contorted filaments with frequent gaps when CHUP1-C was added to filaments of ACT7 (Fig. 5, G and H) or when ACT7 was allowed to polymerize in the presence of CHUP1-C but in the absence of PRF1 (Fig. 5F). These results were consistently and reproducibly observed when ACT7 filaments, as well as SK-actin filaments (Supplementary Fig. S7), were allowed to interact with CHUP1-C either during or after polymerization, but never in control actin filaments in the absence of CHUP1-C. We sometimes observed side-bound, blob-like structures close to the gaps along the filament (red arrows in Supplementary Figs. S6E, S7, E and F). These results suggest that side-binding of CHUP1-C destabilizes the filament structure.
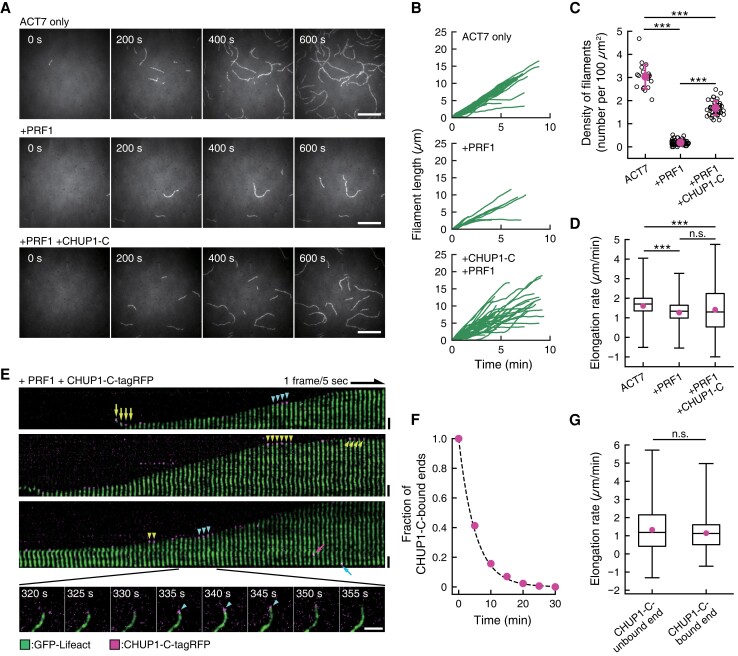
We next followed CHUP1-C-mediated ACT7 polymerization by TIRF microscopy (Fig. 6 and Supplementary Fig. S8). Accordingly, we visualized ACT7 filaments by labeling them with GFP-Lifeact (Belyy et al. 2020), since it was technically difficult to label labile ACT7 molecules with a fluorescent dye, and ACT7 filaments are not labeled with fluorescent phalloidin in vitro (Kijima et al. 2016). When 1.5 µM monomeric ACT7 was allowed to polymerize in F-buffer 1, a number of filaments appeared and elongated on the lipid surface in a flow chamber within the time course of 10 min. Prior incubation of ACT7 with 3 µM PRF1 greatly decreased the number of filaments (Fig. 6, A and C). Addition of 0.25 µM (final concentration) CHUP1-C-tagRFP before induction of polymerization partially but significantly reversed the inhibitory effect of PRF1 on the number of ACT7 filaments (Fig. 6, A and C). This increase in filament number must be due to either filament severing or enhanced de novo nucleation and assembly of actin filaments. Under the experimental condition for TIRF microscopy, however, we observed no filament severing during the time-lapse window of observation. We thus concluded that the larger number of filaments in the presence of PRF1 and CHUP1-C-tagRFP compared with in the presence of PRF1 only is due to enhanced de novo nucleation and assembly of actin filaments. This idea is consistent with the results of the bulk kinetic assays (Supplementary Fig. S5).
Figure 6.
CHUP1-C-dependent polymerization of ACT7 filaments from ACT7–profilin complexes. A) Nucleation and elongation of ACT7 filaments. Monomeric ACT7 at 1.5 μM was allowed to polymerize in F-buffer 2 containing 0.3% (w/v) methylcellulose, 2 mM Trolox, and 0.25 μM GFP-Lifeact (E17K) in a chamber. Prior incubation of ACT7 with a 2-fold molar excess of profilin PRF1 strongly inhibited spontaneous nucleation (middle row). However, the addition of 0.5 µM CHUP1-C-tagRFP partially reversed the effects of PRF1 and allowed nucleation and polymerization of a large number of filaments (bottom row). Scale bars, 10 µm. B) Polymerization kinetics of individual filaments under the 3 conditions in A). C) Quantification of nucleation activities of ACT7 under the 3 conditions in A). Numbers of ACT7 filaments were counted at about 10 min after the induction of polymerization. The means of the densities (the numbers of filaments per 100 µm2) were 3.04 ± 0.54/100 µm2 for ACT7 only (21 ROIs from 2 chambers), 0.19 ± 0.11/100 µm2 for +PRF1 (68 ROIs from 3 chambers) and 1.68 ± 0.31/100 µm2 for +PRF1 +CHUP1-C (40 ROIs from 3 chambers). ***P < 0.001 (Mann–Whitney U-test). D) Boxplot showing the elongation rates during 10-s intervals. The box represents the 25 to 75th percentiles, and the median is indicated by the black line. The whiskers show the complete range from minimum to maximum values. Means (represented by red circle) ± standard deviation (Sd) and the numbers of samples were 1.61 ± 0.61 µm/min for ACT7 only (1,235 intervals of 44 filaments from 2 chambers), 1.27 ± 0.62 µm/min for +PRF1 (347 intervals of 15 filaments from 3 chambers) and 1.41 ± 1.08 µm/min for +PRF1 +CHUP1-C (1,482 intervals of 47 filaments from 3 chambers). ***P < 0.001 (Mann–Whitney U-test). E) Kymographic representation of polymerization of an ACT7 filament in the presence of PRF1 and CHUP1-C-tagRFP. Green shows fluorescence of GFP-Lifeact (E17K), and magenta shows that of CHUP1-C-tagRFP. Raw fluorescence images of a polymerizing ACT7 filament (shown in the bottom row) were straightened and aligned to assemble a kymograph after the background was subtracted using ImageJ software. Spots of CHUP1-C-tagRFP were observed both during the growing phases (cyan arrowheads) and the stationary phases (yellow arrowheads) at the ACT7 filament ends. CHUP1-C-tagRFP spots were also sometimes observed at the end of short ACT7 filaments (yellow arrows), which presumably represent complexes of CHUP1-C-tagRFP and actin filaments at the initial phase of polymerization. Binding to the filament side (magenta arrow) and the less active end (cyan arrow) was also observed, but these frequencies were much lower than that of active-end binding (Supplementary Fig. S7, B and C). Scale bars, 2 µm. F) Dwell time of CHUP1-C-tagRFP at active ends of the filaments. Time-lapse images taken at 5-s intervals were analyzed, and the minimum lifetime of each fluorescent spot were cumulated and plotted (red circles). The dashed line is the exponential fit yielding the dissociation rate of CHUP1-C-tagRFP from the elongating end koff = 0.19 s−1. G) Comparison of the elongation rates between CHUP1-C-unbound ends and CHUP1-C-bound ends. Boxplot shows the elongation rates during 5-s interval of 36 filaments to which CHUP1-C-tagRFP was transiently bound (from 3 chambers), analyzed separately for CHUP1-C-tagRFP-bound phases and unbound phases. The box represents the 25 to 75th percentiles, and the median is indicated by the black line. The whiskers show the complete range from minimum to maximum values. Means (represented by red circle) ± Sd and the numbers of samples were 1.32 ± 1.14 µm/min for unbound ends (2,437 intervals) and 1.14 ± 0.85 µm/min (274 intervals). P = 0.053 (Mann–Whitney U-test).
In all observed cases, one of the 2 filament ends grew preferentially. CHUP1-C-tagRFP was often detected at the growing ends of ACT7 filaments, but was rarely observed at the less active ends (Fig. 6E and Supplementary Fig. S8). These results are consistent with the idea that CHUP1-C binds to the barbed ends of the filaments (Supplementary Fig. S3A and Table S1).
The association of CHUP1-C-tagRFP with the growing ends was transient, and many filament ends grew without bound CHUP1-C-tagRFP in the presence of PRF1 (Fig. 6, E and F). Interestingly, the distribution of the elongation rates in the presence of PRF1 and CHUP1-C-tagRFP was wider than in the absence of the 2 proteins or the presence of PRF1 only, although the average elongation rates were not very different among the 3 conditions (Fig. 6D). Thus, CHUP1-C somehow affects the elongation rates, although the association of CHUP1-C at the end did not correlate with faster or slower elongation rates (Fig. 6G).
Experimental support for a hypothetical model of the CHUP1_716–982-actin complex
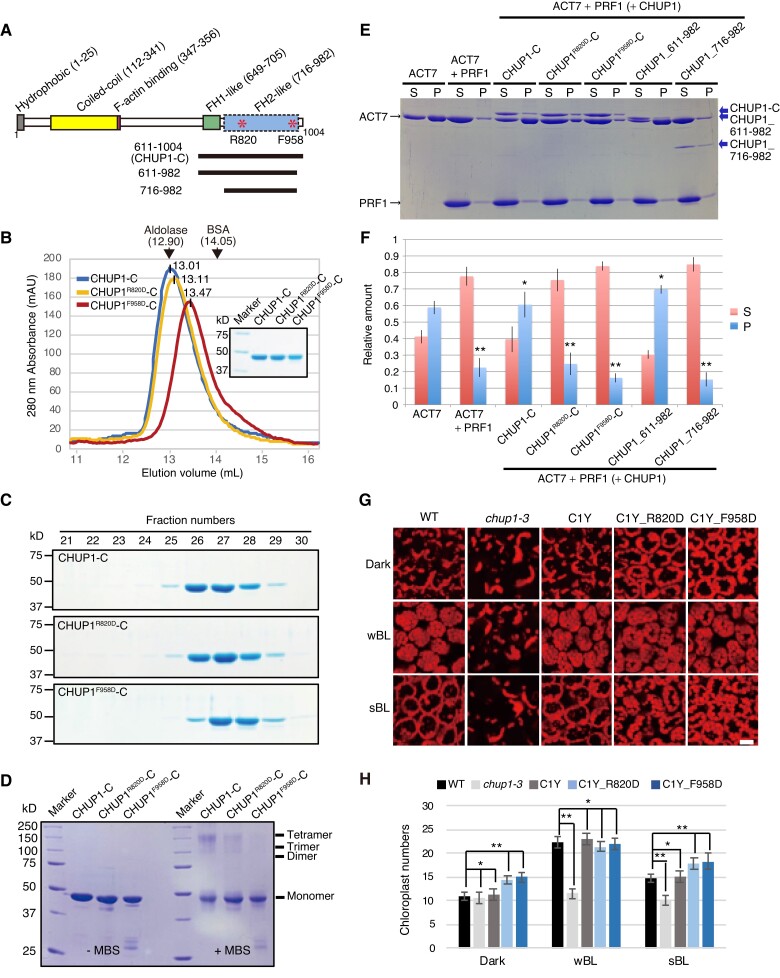
We constructed a hypothetical model of the CHUP1_716–982-actin complex with the assumption that CHUP1_716–982 forms a dimer that is structurally and functionally similar to FH2 (Supplementary Fig. S9, A and B). By reference to the model, we selected 2 amino acid positions for mutagenesis (Fig. 7A and Supplementary Fig. S9A). The R820 residue is positioned close to the contact sites with the actin molecules, so that the substitution of this positively charged residue may decrease affinity for actin, which is negatively charged. The F958 residue is located near the C-terminal end of the CHUP1_716–982 fragment and interacts with the short α-helix in the N-terminally extended region of the other subunit in the dimer, and we expected that its substitution may disrupt the formation of a functional dimer.
Figure 7.
Effects of point mutations at R820 or F958 of CHUP1-C on the structure and activity of CHUP1. A) Diagram of CHUP1 showing its functional domains: the hydrophobic region, the coiled-coil domain, the F-actin-binding domain, the proline-rich FH1-like domain, and the FH2-like domain. Asterisks indicate R820, which is involved in the association with actin, and F958, which is involved in the dimerization of the FH2-like domain. The positions of the 611 to 1,004 fragment (CHUP1-C) and the 611 to 982 and 716 to 982 fragments used for crystallization are indicated by lines below the structure. B and C) Effect of point mutations R820D and F958D on CHUP1-C dimer structure. B) The elution profiles of CHUP1-C, CHUP1R820D-C, and CHUP1F958D-C recombinant proteins were obtained by gel filtration chromatography using a Superdex 200 10/300 column. The elution volumes of aldolase (158 kD) and BSA (67 kD) used for calibration are presented. The molecular weights of the CHUP1-C recombinant proteins were calculated as 151.5 kD for CHUP1-C, 144.7 kD for CHUP1R820D-C, and 122.5 kD for CHUP1F958D-C. C) The protein profiles were confirmed using 10% SDS-PAGE gels. D) Effect of the point mutations R820D and F958D on the multimeric structure of CHUP1-C. Two µM of CHUP1-C, CHUP1R820D-C, and CHUP1F958D-C recombinant proteins was incubated without (left panel) or with (right panel) 20 µM MBS (m-maleimidobenzoyl-N-hydroxysuccinimide ester) for 1 h, and their multimer formations were analyzed using 10% SDS-PAGE. E and F) Effect of deletion and point mutations of CHUP1-C on actin polymerization. E) Ultracentrifugation assay of actin polymerization. ACT7 (4 µM) was allowed to polymerize in F-buffer 1 containing 8 µM PRF1 and 0.4 µM intact or mutant CHUP1-C, and a representative set of supernatant (S) and pellet (P) fractions after ultracentrifugation were analyzed by SDS-PAGE. F) Quantitative analysis of actin polymerization showing the fractions of ACT7 in the pellet and the supernatant under each condition as shown in E). The data are presented as means ± Sd (n = 3). Asterisks indicate statistically significant differences between ACT7 and each line of pellet detected by Student's t-test (* not significant P > 0.05; ** significant P < 0.0001). G) Effect of point mutations at R820D and F958D of CHUP1 on chloroplast positioning. The 4th rosette leaves of WT, chup1-3, C1Y, C1Y_R820D and C1Y_F958D plants were detached after dark adaptation (dark) for 14 h and then were further irradiated with a weak blue light of 2 µmol m−2 s−1 or a strong blue light (sBL) of 50 µmol m−2 s−1 for 2 h, respectively. Chloroplasts were shown with chlorophyll autofluorescence that was captured using confocal microscopy at a resolution of 512 × 512 pixels in a depth of 2 µm. Scale bar, 20 µm. H) Average number of chloroplasts observed on the periclinal side of palisade cells of WT, chup1-3, C1Y, C1Y_R820D, and C1Y_F958D plants. The average numbers of chloroplasts shown in G) were counted from 20 palisade cells from 3 rosette leaves. The data are presented as means ± SE (n = 20). Asterisks indicate statistical differences detected by Student's t-test (* not significant P > 0.05; ** significant P < 0.0001).
We used size-exclusion chromatography and crosslinking treatment to measure the ability of mutagenized CHUP1-C domains to form complexes. CHUP1-C with a F958D point mutation (CHUP1F958D-C) eluted more slowly than CHUP1-C and CHUP1-C with a R820D point mutation (CHUP1R820D-C_) (Fig. 7, B and C). In agreement, following crosslinking with a protein crosslinker, MBS (m-maleimidobenzoyl-N-hydroxysuccinimide ester), some fractions of CHUP1-C and CHUP1R820D-C were found in a multimeric state around 150 kD with the majority in a monomeric state (∼42.7 kD) on SDS-PAGE. This suggests a configuration of either a trimer (∼128.1 kD) or a tetramer (∼170.8 kD). Notably, CHUP1F958D-C did not exhibit such a multimeric form (Fig. 7D). These data were collectively consistent with our prediction that the F958 residue is involved in multimer formation. If the multimer formation is essential for CHUP1-C activity, as is the case for formins, the CHUP1F958D-C mutant should be unable to promote the polymerization of ACT7 in the presence of PRF1. We confirmed this prediction by sedimentation experiments (Fig. 7, E and F). Likewise, CHUP1R820D-C lacked the ability to promote the polymerization of ACT7 in the presence of PRF1 (Fig. 7, E and F).
To further confirm the physiological activity of the R820 and F958 residues, we generated transgenic plants expressing CHUP1R820D-YFP (C1Y_R820D) or CHUP1F958D-YFP (C1Y_F958D) in the chup1-3 background and analyzed them for chloroplast movement (Fig. 7, G and H and Supplementary Fig. S9, C and D). When chloroplast movements in the 2 transgenic lines were examined by a red-light transmittance assay, we detected neither a normal accumulation response nor a normal avoidance response (Supplementary Fig. S9D). The defect in blue light-dependent chloroplast repositioning seen in the chup1 mutant was partially rescued in the C1Y_R820D and C1Y_F958D transgenic plants. However, the number of chloroplasts observed at the cell surface in the dark and strong blue light was significantly higher in C1Y_R820D and C1Y_F958D transgenic plants than in WT and C1Y transgenic plants (Figs. 7, G and H). Thus, the chloroplast movement phenotype of the chup1-3 mutant is not completely rescued in the 2 transgenic plants, suggesting that both R820 and F958 are essential for normal CHUP1 function.
The region between amino acid residues 611 and 716 includes polyproline sequences (Supplementary File 1 and Fig. S10), which correspond to the FH1 domain implicated in profilin binding (Schmidt von Braun and Schleiff 2008). Removal of this polyproline region, together with a C-terminal truncation at residue 982, completely inhibited the ability of CHUP1 to promote the polymerization of ACT7 in the presence of PRF1 (Fig. 7, E and F). However, the C-terminal truncation at residue 982 on its own had no noticeable effect on the actin-polymerizing activity (Fig. 7, E and F). Thus, the polyproline sequence is also important for CHUP1-mediated polymerization of actin bound to profilin. Based on our results, we propose that CHUP1-C promotes actin polymerization in the presence of profilin in a manner dependent on the FH1-like polyproline sequence and the dimeric FH2 -like structure.
Discussion
CHUP1 assembles actin filaments in a blue-light-regulated manner
The distribution of CHUP1 on the chloroplast envelope varies, with different patterns, including spots, a diffuse distribution, and CHUP1 bodies, under different light conditions (Fig. 1, A and J). The dynamic reorganization of CHUP1 is reversibly regulated—mainly by phot2 but with contribution from phot1—in response to blue light (Figs. 2B and 3G). During chloroplast photorelocation movements, cp-actin filaments are generated at the leading edge of each chloroplast, specifically at the interface between the plasma membrane and the chloroplast (Kadota et al. 2009; Kong et al. 2013a). This asymmetric distribution of cp-actin filaments coordinately changed with the asymmetric distribution of CHUP1 on the front and rear ends of moving chloroplasts (Fig. 3, A, B, and G). The reorganization of CHUP1 from dots into a diffuse pattern in the rear part of a moving chloroplast is probably related to cp-actin depolymerization by strong light. By contrast, small CHUP1 dots at the front of moving chloroplasts are probably involved in cp-actin polymerization. Therefore, CHUP1 determines the sites of cp-actin nucleation to polymerize cp-actin filaments responding to the intensity and position of incident blue light (Fig. 3G).
CHUP1 is a class of plant-specific, actin-binding protein that promotes actin polymerization in the presence of profilin
Our study further shows that CHUP1 promotes actin polymerization in the presence of profilin (Figs. 5 and 6 and Supplementary Fig. S8). Although the crystal structure of CHUP1_756–982 has some structural homology to the FH2 domain of formin (Fig. 4A), there is no detectable similarity between the 2 protein sequences. Nonetheless, based on the structural similarity with FH2, we were able to construct a structural model of the CHUP1_716–982-actin complex, which allowed us to predict that the F958 residue is important for oligomerization of CHUP1 and that the R820 residue is part of a potential actin-binding site. We confirmed that these residues are essential for actin polymerization in vitro and for chloroplast movement in vivo (Fig. 7 and Supplementary Fig. S9). These results demonstrate that the structural similarity between the CHUP1 C-terminal domain (CHUP1_716–982) and the FH2 domain of formin has functional significance.
Formins are a major group of well-characterized actin nucleators that form tethered dimer structures (Xu et al. 2004) and are present in plants as well as other organisms (van Gisbergen and Bezanilla 2013). The proline-rich FH1 domain of formin mediates interactions with actin–profilin complexes, and the FH2 domain mediates actin polymerization by incorporating G-actin liberated from the actin–profilin complexes. The lasso domain of FH2 is important for its ability to form a dimer. CHUP1-C forms multimers in solution (Fig. 7, B to D) and catalyzes actin polymerization in the presence of profilin (Fig. 5). There are 2 aspects regarding structural similarities between formins and CHUP1-C. First, both proteins have polyproline sequences, which are called FH1 and are implicated in binding profilin–actin complex in formins. The polyproline sequence is followed by a domain that contains a similar 3-helix bundle in both proteins. In the case of formins, this domain is called FH2 and has been shown to form an open dimer that accommodates 2 actin molecules inside and is critical for the nucleating activity. Our crystallography analysis demonstrated that CHUP1_716–982 forms a closed dimer. Negative stain electron microscopy showed that CHUP1-C in the absence of actin is mostly visible as small ellipses, but in the presence of monomeric actin forms ovoid structures that are similar in size to an FH2 dimer–actin dimer complex. Moreover, gel filtration chromatography demonstrated that CHUP1-C forms multimers, and that the multimer formation was inhibited by a point mutation (F958D) predicted to disrupt dimerization when FH2-like dimerization of CHUP1-C is assumed. Taken together, our data are consistent with the hypothesis that the polyproline sequence in CHUP1-C captures an actin–profilin complex, which is then fed to the nucleation activity of the FH2-like dimeric structure, as shown as a model in Supplementary Fig. S9A.
However, unlike formin, which promotes the polymerization of dilute actin solution in the absence of profilin (Pruyne et al. 2002), CHUP1-C promotes actin polymerization only in the presence of profilin and inhibits actin polymerization in the absence of profilin (Fig. 5, A and B and Supplementary Figs. S3C, S4, and S5). CHUP1-C-mediated actin polymerization in the presence of PRF1 is less efficient than the pure actin solution, and again, this is distinct from formin, which nucleates actin polymerization both in the presence and in the absence of profilin more efficiently than pure actin (Sagot et al. 2002). Moreover, unlike well-characterized vertebrate and yeast formins that remain bound to the fast-growing barbed ends of actin filaments and protect them from capping proteins (Paul and Pollard 2009), association of CHUP1-C with growing ends of ACT7 filaments was transient, and the association did not accelerate the elongation rates (Supplementary Fig. S8). However, certain Arabidopsis formins do not stay bound to (FH1; Michelot et al. 2006) or only transiently associate with (FH14; Zhang et al. 2016) filament ends, and therefore, the lack of stable association of CHUP1-C with the growing ends cannot be considered as a clear distinction of CHUP1-C from formins. Nonetheless, despite the potential structural similarity, functional similarities between CHUP1-C and formin FH1-FH2 in stimulating actin polymerization in vitro are rather limited.
It should be noted, however, that a large number of actin-binding proteins regulate polymerization of actin in vivo (reviewed by Paavilainen et al. 2004). Thus, kinetic comparison of CHUP1-C-mediated polymerization of ACT7 in the presence of PRF1 with spontaneous polymerization of pure actin is physiologically irrelevant. What is relevant is that CHUP1-C reverses the inhibition of de novo formation of actin filaments by profilin. Our TIRF observation relies on the excluded volume effect of methylcellulose to press actin filaments to the glass surface and maintain them within the evanescent field and is incapable of visualizing nascent actin oligomers immediately after formation in solution outside the evanescent field. Rigorous demonstration that CHUP1-C is an actin nucleator awaits visualization that the nascent actin oligomers that form in the presence of profilin have bound CHUP1-C-tagRFP.
We demonstrated by electron microscopy (Fig. 5, E to G and Supplementary Figs. S6E, S7, B, C, E and S8F) that both SK-actin and ACT7 filaments treated with CHUP1-C were sharply bent or even fragmented on the carbon-coated grids, suggesting that side-binding of CHUP1-C destabilizes the filament structure. The destabilization of filament structures led to frequent filament fragmentation on carbon-coated grids, but no severing events were detected during TIRF observation of ACT7 filaments in the presence of CHUP1-C-tagRFP. Further studies are thus needed to investigate specific physiological conditions that might promote efficient severing of ACT7 filaments in a CHUP1-C-dependent manner.
It is intriguing that plants evolved to have CHUP1 as a separate type of actin polymerization factor, even though they also have their own formin family proteins. Interestingly, we identified sequences similar to the CHUP1 C-terminal half (named CHUC) in some non-Charophyte single-celled algae, including Coccomyxa subellipsoidea (Chlorophyta), Micromonas commoda (Chlorophyta), and Guillardia theta (Cryptophyta), as well as in land plants (Supplementary Fig. S11). These algae swim and do not show intracellular chloroplast movement. Thus, CHUC might not be involved in chloroplast movement in these algae. Consequently, the origin of CHUP1 and the physiological function of CHUC are unresolved.
Together, our structural and biochemical analyses of the conserved CHUP1 C-terminal domain revealed that CHUP1 is a previously unknown class of plant-specific actin-binding protein that promotes actin polymerization in the presence of profilin. We suggest that the structural similarity between CHUP1-C and formins FH1 and FH2 is the consequence of convergent evolution, judging from the lack of sequence similarity, as well as the limited functional similarities, between the 2 actin-binding protein families.
Does cp-actin polymerization generate the motive force for chloroplast movement?
Based on the results of the LatB experiment (Supplementary Fig. S2), cp-actin filaments appeared to have no effect on the blue light-induced localization changes of CHUP1-YFP. This result suggests that the mechanisms driving the asymmetric distribution of CHUP1-YFP might be influenced by factors beyond simple cytoskeletal distribution, since LatB likely binds to G-actin, inducing actin depolymerization (Kandasamy and Meagher 1999). These observations raise intriguing questions about the interplay between the actin cytoskeleton, chloroplast movement, and the localization of CHUP1-YFP. Further research is required to understand the molecular basis of these observations and to uncover the underlying mechanisms.
Several studies have indicated that the acto-myosin system was involved in chloroplast movement; that is, the chloroplasts move along a long cytoplasmic actin filament, driven by myosins. For example, anti-myosin antibody responsive protein(s) and a myosin XI tail domain were identified on the chloroplast envelope (Krzeszowiec and Gabryś 2007; Sattarzadeh et al. 2009), and the myosin inhibitors 2,3-butanedione monoxime (BDM) and N-ethylmaleimide (NEM) effectively inhibited chloroplast movement (Paves and Truve 2007; Yamada et al. 2011; Kong et al. 2013a). However, quadruple knockout lines of the major myosin XIs in mesophyll cells of Arabidopsis leaves showed normal chloroplast movements, even though the movements of other organelles were defective (Suetsugu et al. 2010).
Our alternative model relies on actin polymerization for generating motive force during chloroplast movements (Wada and Kong 2018). In response to blue light, CHUP1 is reorganized and becomes asymmetrically redistributed to the front end of the chloroplast (Fig. 3G). Once this redistribution occurs, cp-actin filaments are polymerized at the interface between the plasma membrane and the chloroplast. Polymerized cp-actin filaments are bundled and anchored to the plasma membrane by a plasma membrane–localized protein, THRUMIN1 (Whippo et al. 2011; Kong et al. 2013a). If the polymerized cp-actin filaments are anchored to the plasma membrane, the cp-actin filaments that are newly polymerized by CHUP1 may push CHUP1 itself forward, thereby pushing the chloroplast forward. This proposed mechanism is reminiscent of the comet-tail movement of the bacterium Listeria in animal cells (Lambrechts et al. 2008). In the case of Listeria, actin polymerization is structurally restricted to one end of the rod-shaped bacterial cell, ensuring that the force vector of individual polymerizing actin filaments is oriented more or less parallel to the long axis of the cell. It remains to be elucidated how cp-actin filaments are aligned in one direction and collectively generate the motive force of chloroplast photorelocation movement.
Materials and methods
Plant materials
Arabidopsis (A. thaliana) accession Columbia (Col-0) was used for all experiments. The chup1-3 mutant line (Oikawa et al. 2003) was crossed to the phot1-5, phot2-1, and phot1-5 phot2-1 mutants (Suetsugu et al. 2013) to produce the mutants chup1-3 phot1-5, chup1-3 phot2-1, and chup1-3 phot1-5 phot2-1. Transgenic Arabidopsis lines expressing CHUP1-fluorescent fusion constructs (CHUP1-YFP and CHUP1-tdTomato) from the native CHUP1 promoter were produced via Agrobacterium (Agrobacterium tumefaciens strain GV3101)-mediated transformation of the chup1-3 mutant (Supplementary Fig. S1A). The CHUP1-YFP 10-5 transgenic line (C1Y) was further crossed to the chup1-3 phot1-5, chup1-3 phot2-1, and chup1-3 phot1-5 phot2-1 mutants to produce phot1 CHUP1pro:CHUP1-YFP (phot1 C1Y), phot2 CHUP1pro:CHUP1-YFP (phot2 C1Y), and phot1 phot2 CHUP1pro:CHUP1-YFP (phot1 phot2 C1Y) transgenic plants, respectively. The GFP-mTalin 2-28 transgenic line (Kadota et al. 2009; Kong et al. 2013a) was crossed to the CHUP1-tdTomato 8-1 transgenic line. EGFP-mouse Talin was originally developed by Kost et al. (1998) and simply referred to as GFP-mTalin in this study.
Immunoblot analysis
Arabidopsis plants were cultured on half-strength Murashige and Skoog (MS) medium (Nihon Pharmaceutical, Japan) supplemented with 1× MS vitamin (Sigma, USA), 1% (w/v) sucrose, and 0.8% (w/v) agar in a growth chamber set to a 16-h light/8-h dark photoperiod at 23 °C. One hundred milligrams of rosette leaves frozen in liquid nitrogen was homogenized on ice using a glass homogenizer in a 1:3 (w/v) ratio with extraction buffer [50 mM Tris–HCl, pH 7.5, 10% (v/v) glycerol, 5 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol, and complete proteinase inhibitors (Roche, Germany)]. The homogenates were clarified by centrifugation at 8,000 × g for 15 min at 4 °C. Fifty micrograms of protein extracts was separated on 7.5% (w/v) SDS-PAGE gels, transferred onto a nitrocellulose membrane (Amersham, Switzerland), and probed with anti-CHUP1 (Oikawa et al. 2008), anti-phot1, or anti-phot2 (Kong et al. 2013b) polyclonal antibodies at 1:2,000 dilutions.
Analysis of chloroplast photorelocation movement
Chloroplast movement was analyzed by measuring changes in red-light transmittance or chloroplast positioning, as previously described (Kong and Wada 2011). For these assays, the 4th rosette leaf was detached from 3-wk-old Arabidopsis plants cultured on half-strength MS medium. The detailed light conditions are described in Fig. 7G and Supplementary Fig. S1, B and C.
Confocal microscopy
GFP, YFP, tdTomato, and chlorophyll fluorescence were detected using a Leica SP5 confocal microscope as previously described (Kong et al. 2013a). The excitation wavelengths were 488 nm for GFP, 518 nm for YFP, and 561 nm for tdTomato. The emission ranges were 500 to 530 nm for GFP, 530 to 560 nm for YFP, and 575 to 630 nm for tdTomato. The fluorescence images were captured with an objective lens (63×/1.20 W, Leica) at resolutions of 256 × 256, 512 × 256, or 512 × 512 pixels using 4× digital zooming for the indicated cycles. Chloroplast avoidance response was induced by strong blue light (458-nm laser with 2.8-µW output power through a 63× objective lens) at the appropriate circular or rectangular ROI during intervals for the indicated times (see figure legends), with an output power of 2.8 µW
The fluorescence images of GFP, tdTomato, and chlorophyll shown in Fig. 3 were obtained using a Nikon A1 confocal microscope (Nikon, Tokyo) with a 63× water immersion objective. The excitation lasers were 488 nm for GFP and 561 nm for tdTomato. Mander's colocalization coefficient between the green and red channels was calculated using Coloc2 Fiji plug-in after background removal in images using the rolling ball method (20 pixels radius) (Manders et al. 1993).
Thin section electron microscopy
The detached 4th rosette leaves of 4-wk-old WT plants were irradiated on agar plates with strong blue light (50 µmol m−2 s−1) for 30 min. The leaves were cut into about 2 mm × 2 mm pieces and were fixed in 2.5% (v/v) glutaraldehyde, postfixed in 1% (w/v) osmium tetroxide, dehydrated in acetone, and embedded in Spurr's resin. Thin sections were stained with 4% (w/v) uranyl acetate and 0.4% (w/v) lead citrate and examined with a transmission electron microscope (model H-7600; Hitachi).
Image processing
All microscopy data were validated through at least 3 independent experiments. Representative images are presented after reorganization using image processing software, such as Adobe Photoshop CS6 (Adobe Systems, San Jose, CA, USA) or ImageJ (https://imageJ.nih.gov/ij/).
Plasmid construction
The sequences encoding the C-terminal fragments of CHUP1 including CHUP1_611–1004 (CHUP1-C), CHUP1_611–982, CHUP1_716–982, and CHUP1_756–982 were amplified from the cloned CHUP1 cDNA (AGI: At3g25690; Oikawa et al. 2008) using PCR with the specific primer pairs (Supplementary Table S2). The PCR fragments were cloned into the BamHI and SalI restriction sites of the expression vector pGEX-6P-1 (GE Healthcare) to produce pGEX-CHUP1_611–1004, pGEX-CHUP1_611–982, pGEX-CHUP1_716–982, and pGEX-CHUP1_756–982, respectively. The backbone of CHUP1_611–1004-tagRFP (for CHUP1-C-tRFP) was also amplified from the CHUP1 cDNA and tagRFP vector (evrogen, Russia) using PCR with the specific primer pairs (Supplementary Table S2). The PCR fragments were cloned into the BamHI and XhoI restriction sites of the vector pGEX-6P-1 (GE Healthcare) to produce pGEX-CHUP1_611–1004-tagRFP.
The plasmid to produce His-tagged EGFP-lifeact (E17K) was constructed by PCR-based methods, using pColdI vector (Takara, Japan) with an engineered TEV cleavage site between the His tag and the multicloning site (Ngo et al. 2015). The amino acid sequence of the linker and Lifeact (E17K) is as follows: …LYKGPGKDPQMGVADLIKKFESISKEK, where the first 3 residues (LYK) are the C terminus of EGFP, GPGKDPQ is the linker, and MGVADLIKKFESISKEK is Lifeact with the E17K mutation.
Genomic DNA encompassing the promoter and the coding region of CHUP1 was amplified from genomic DNA prepared from Arabidopsis leaf tissues using PCR with the specific primer pair (Supplementary Table S2). The PCR fragment was cloned into the BamHI and SalI restriction sites of pPZP211/35S-nosT to produce CHUP1pro:CHUP1-YFP (or tdTomato) in the pPZP211 backbone.
Protein preparations for X-ray crystallography
Selenomethionine (SeMet)-substituted CHUP1_756–982 with a C864S mutation was produced as a glutathione S-transferase (GST)-fusion protein using the pGEX-6P-1 vector (GE Healthcare). The transformed Escherichia coli strain B834(DE3) cells (Novagen) were grown at 37 °C, until the OD620 reached 0.4 in the SeMet core medium (Wako), containing 25 mg L−1 L-SeMet (Nacalai Tesque). After overnight induction with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 20 °C, the cells were harvested by centrifugation at 6,000 × g at 4 °C for 15 min and resuspended in 50 mM Tris–HCl, pH 7.5, 500 mM NaCl, 2 mM 1,4-dithiothreithol (DTT), 2 mM EDTA, and 1% (v/v) glycerol (buffer A), supplemented with one tablet of complete EDTA-free protease inhibitor (Roche) per 50-mL solution. After disruption by sonication and centrifugation at 18,000 × g at 4 °C for 30 min, the supernatant was loaded onto a glutathione Sepharose 4B (GE Healthcare) column equilibrated with buffer A. The column was then washed with buffer A. After cleavage with PreScission protease (GE Healthcare) on the column overnight, the protein was eluted with buffer A. The protein was further purified by gel filtration on a Superdex 200 10/300 GL column (GE Healthcare) in 50 mM Tris–HCl, pH 7.5, 200 mM NaCl, and 2 mM DTT (buffer B). The purified protein was concentrated to 10 mg mL−1 for crystallization with an Amicon Ultra-15 centrifugal concentrator (Merck Millipore), and the buffer exchanged with 20 mM HEPES-NaOH, pH 7.0, containing 3 mM Tris(2-carboxyethyl)phosphine [TCEP (buffer C)].
The longer C-terminal fragment, CHUP1_716–982, was produced as a GST-fusion protein using the pGEX-6P-1 vector. The transformed E. coli BL21-Gold [DE3 (Agilent Technologies)] cells were grown at 37 °C, until the OD600 reached 0.8. After overnight induction with 0.2 mM IPTG at 20 °C, the cells were harvested by centrifugation at 6,000 × g at 4 °C for 15 min and resuspended in 50 mM Tris–HCl, pH 8.0, 50 mM NaCl, 5 mM DTT, 2 mM MgCl2, and 10% (v/v) glycerol (buffer D), supplemented with one tablet of cOmplete EDTA-free protease inhibitor (Roche) and 5 µL Benzonase Nuclease (Merck Millipore) per 50-mL solution. After disruption by sonication and centrifugation at 18,000 × g at 4 °C for 30 min, the supernatant was loaded on a glutathione Sepharose 4B column equilibrated with buffer D. The column was sequentially washed with 50 mM Tris–HCl, pH 8.0, 400 mM NaCl, 5 mM DTT, and 10% (v/v) glycerol; 50 mM Tris–HCl, pH 8.0, 100 mM KCl, 10 mM MgCl2, 5 mM DTT, 0.25 mM ATP, and 5% (v/v) glycerol; and 50 mM Tris–HCl, pH 6.8, 150 mM NaCl, 1 mM EDTA (adjusted to pH 8.0), and 1 mM DTT (buffer E). After cleavage with PreScission protease on the column overnight, the protein was eluted with buffer E. The protein was further purified by gel filtration on a Superdex 200 10/300 GL column in 50 mM Tris–HCl, pH 8.0, 50 mM NaCl, 5 mM DTT, and 5% (v/v) glycerol (buffer F). The purified protein was concentrated to 20 mg mL−1 with a Vivaspin Turbo 15 centrifugal concentrator (Sartorius), without changing the buffer composition.
Crystallization and structure determination
CHUP1_756–982 and CHUP1_716–982 crystals were grown at 20 °C using the hanging-drop vapor-diffusion method. The reservoir solution for CHUP1_756–982 was 50 mM CHES, pH 9.0, 0.2 M Li2SO4, and 36% (v/v) PEG400, whereas that for CHUP1_716–982 was 1.1 M sodium malonate, 100 mM HEPES-NaOH, pH 7.4, 0.5% (v/v) Jeffamine ED-2001, pH 7.0 (Hampton Research). The crystals were briefly dipped into cryoprotectant solution containing 40% (v/v) PEG400 for CHUP1_756–982 or 16% (v/v) glycerol for CHUP1_716–982 and then flash-cooled in liquid nitrogen.
Data sets were collected at the SPring-8 beamline BL44XU (Hyogo, Japan) and were processed with HKL2000 (Otwinowski and Minor 1997). The structure of SeMet-substituted CHUP1_756–982 was solved by the SAD method, using the program PHENIX (Adams et al. 2010). The structure of the native CHUP1_716–982 was solved by the molecular replacement method with PHENIX, using the structure of the SeMet-substituted CHUP1_756–982 as a search model. Model building was refined using PHENIX. Figures were generated using PyMOL.
Recombinant protein production for actin polymerization assay and EM observation
The proteins were produced in E. coli strain Rosetta gami B [DE3 (Novagen)] by 0.5 mM IPTG induction at 20 °C after the E. coli cells were cultured to the late log phase (OD600 = 0.8 to 1.0). The cells were harvested by centrifugation at 6,500 × g at 4 °C for 20 min and homogenized in extraction buffer [50 mM Tris–HCl, pH 7.5 at 23 °C, 150 mM NaCl, 5% (v/v) glycerol, 5 mM DTT, 1% (v/v) Triton X-100, 1 mM EDTA, and 0.2 mM PMSF]. The GST proteins were trapped using a glutathione Sepharose 4B matrix (GE Healthcare), and the CHUP1 C-terminal fragments were eluted by on-column cleavage using PreScission protease. The eluted CHUP1-C was further purified in buffer (10 mM HEPES pH 7.4, 100 mM NaCl, 2 mM MgCl2, and 1 mM DTT) by size-exclusion chromatography using a Superdex 200 10/300 column (GE Healthcare). The purified fractions were concentrated using a centrifugal filter unit (Amicon Ultra filter 30K, Millipore).
Arabidopsis profilin1 [PRF1 (AGI: At2g19760)] was produced in E. coli strain Rosetta (DE3) (Novagen) and purified as described previously (Kijima et al. 2016). Arabidopsis vegetative actins ACT2 and ACT7 were produced as fusion proteins with thymosin-His in Sf9 insect cells or Dictyostelium discoideum, and purified as described previously (Kijima et al. 2016). Rabbit skeletal muscle actin was prepared according to a previously published method (Spudich and Watt 1971). Each purified protein was snap-frozen in liquid N2 and stored at −80 °C.
His-TEV-EGFP-Lifeact (E17K) was produced in E. coli strain Rosetta (DE3) and purified using Ni2+-affinity chromatography as instructed by the manufacturer and released from the His tag using His-tagged TEV protease. The resulting protein is referred to as GFP-Lifeact (E7K).
Actin polymerization assay
Actin (4 µM) was allowed to polymerize in F-buffer 1 (10 mM HEPES-NaOH pH 7.4, 50 mM KCl, 2 mM MgCl2, 0.2 mM ATP, and 1 mM DTT) in the presence of various concentrations of PRF1 and CHUP1-C. After 30 min of incubation at 22 °C, the solution was ultracentrifuged at 250,000 × g at 22 °C for 10 min. The supernatant and pellet fractions were separately subjected to SDS-PAGE. The relative amounts of actin were estimated by analyzing intensities of Coomassie-stained bands using ImageJ (https://imagej.nih.gov/ij/).
Negative stain electron microscopy
Actin filaments diluted in EM buffer (10 mM potassium phosphate buffer pH 7.4, 25 mM KCl, 2.5 mM MgCl2, 0.2 mM ATP, and 0.5 mM DTT) were placed on carbon-coated copper grids, stained with 1% (w/v) uranyl acetate, and observed using an FEl Tecnai F20 electron microscope. The images were recorded with a Gatan ORIUS SC600 CCD camera and processed with Adobe Photoshop to adjust the contrast and decrease noise.
TIRF observation
A 10-µL observation chamber was assembled with a KOH-washed 22 × 32 mm2 coverslip (thickness No. 1, Matsunami), a 18 × 18 mm2 coverslip (thickness No. 1, Matsunami), and double-sided tape (thickness ∼100 µm). To passivate the surface of the coverslips against nonspecific adhesion of protein, the chamber was filled with DPPC vesicle solution (1,2-dipamitoyl-sn-glycero-3-phosphocholine, Avanti Polar Lipids, USA) prepared as described previously (Yamamoto et al. 2010). After 1 h of incubation at 37 °C in a humid environment, the chamber was washed with 40 µL of F-buffer 2 (10 mM HEPES-KOH pH 7.4, 50 mM KCl, 2 mM MgCl2, 0.2 mM ATP, 1 mM EGTA, 10 mM DTT) to remove excess vesicles and lipid bilayer. The chamber was then rinsed with 10 mg mL−1 bovine serum albumin (BSA) in F-buffer 2, incubated for 10 min, and mounted on the microscope stage.
After mixing 2.5 µL of 6 µM ACT7 and 2.5 µL of 12 µM PRF1 in G-buffer (2 mM Tris–HCl pH 8.0, 0.2 mM CaCl2, 10 mM DTT, 0.2 mM ATP), polymerization was initiated by the addition of 5 µL of 2X F-buffer 2 [20 mM HEPES-KOH pH 7.4, 100 mM KCl, 4 mM MgCl2, 0.4 mM ATP, 2 mM EGTA, 20 mM DTT, 0.6% (w/v) 4,000 cP methylcellulose, 4 mM Trolox, 0.5 µM GFP-Lifeact (E17K)] alone or with 0.5 µM CHUP1-C-tagRFP. Soon after mixing by gently pipetting up and down 5 times, the solution was introduced into the observation chamber. For 2-color TIRF microscopy, images were acquired at 5-s intervals for a total of 10 min using an inverted microscope (IX-71, Olympus) equipped with a 100× objective lens (UPlanApo NA 1.40 oil, Olympus) and an EMCCD camera (iXon3, Andor Technology). The excitation lasers were 488 nm (OBIS 488LS, COHERENT) for GFP-Lifeact (E17K), and 532 nm (Compass 315M, COHERENT) for CHUP1-C-tagRFP. Shutters and the camera were controlled by Micro-Manager (Edelstein et al. 2010).
Statistical analysis
Statistical analyses were performed as described in each figure legend. Statistical data are provided in Supplementary Data Set 1.
Data and materials availability
The coordinates and structure factors have been deposited in the Protein Data Bank, with the entries for CHUP1_756–982 with the C864S mutation and for CHUP1_716–982 being 8WAF and 8WAG, respectively.
Accession numbers
Sequence data from this article can be found at The Arabidopsis Information Resource (https://www.arabidopsis.org) under accession numbers: ACT7 (AT5G09810), CHUP1 (AT3G25690); PHOT1 (AT3G45780), PHOT2 (AT5G58140), PRF1 (AT2G19760).
Supplementary Material
Acknowledgments
We thank Takachika Tanaka, Yukiko Eda, and Hee-Jung Chung, for excellent technical assistance, and Dr Makito Miyazaki of RIKEN for the support. We thank the beamline staff at the SPring-8 beamlines—BL26B1, BL26B2, BL44XU, and BL32XU (Harima, Japan)—and the Photon Factory beamlines—BL-1A and BL-5A (Tsukuba, Japan)—for technical support. The experiments at the Photon Factory were performed with the approval of the Photon Factory Program Advisory Committee, as Proposal 2017G009, and those at SPring-8 BL44XU were performed under the Cooperative Research Program of the Institute for Protein Research, Osaka University, Osaka, Japan, as Proposals 20166617, 20176718, and 20186815.
Contributor Information
Sam-Geun Kong, Department of Biological Sciences, College of Natural Sciences, Kongju National University, Chungnam 32588, Korea; Department of Biology, Faculty of Sciences, Kyushu University, Fukuoka 812-8581, Japan.
Yosuke Yamazaki, Department of Physics, Faculty of Science and Engineering, Waseda University, Tokyo 169-8555, Japan; Department of Physics, Graduate School of Science, Kyoto University, Kyoto 606-8502, Japan.
Atsushi Shimada, Division of Structural Biology, Medical Institute of Bioregulation, Kyushu University, Fukuoka 812-8582, Japan.
Saku T Kijima, Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology, Ibaraki 305-8562, Japan.
Keiko Hirose, Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology, Ibaraki 305-8562, Japan.
Kaoru Katoh, Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology, Ibaraki 305-8562, Japan.
Jeongsu Ahn, Department of Biological Sciences, College of Natural Sciences, Kongju National University, Chungnam 32588, Korea.
Hyun-Geun Song, Department of Biological Sciences, College of Natural Sciences, Kongju National University, Chungnam 32588, Korea.
Jae-Woo Han, Department of Biological Sciences, College of Natural Sciences, Kongju National University, Chungnam 32588, Korea.
Takeshi Higa, Institute for Protein Research, Osaka University, Osaka 565-0871, Japan.
Akira Takano, Division of Structural Biology, Medical Institute of Bioregulation, Kyushu University, Fukuoka 812-8582, Japan.
Yuki Nakamura, Division of Structural Biology, Medical Institute of Bioregulation, Kyushu University, Fukuoka 812-8582, Japan.
Noriyuki Suetsugu, Department of Biology, Faculty of Sciences, Kyushu University, Fukuoka 812-8581, Japan.
Daisuke Kohda, Division of Structural Biology, Medical Institute of Bioregulation, Kyushu University, Fukuoka 812-8582, Japan.
Taro Q P Uyeda, Department of Physics, Faculty of Science and Engineering, Waseda University, Tokyo 169-8555, Japan; Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology, Ibaraki 305-8562, Japan.
Masamitsu Wada, Department of Biology, Faculty of Sciences, Kyushu University, Fukuoka 812-8581, Japan; Graduate School of Science, Tokyo Metropolitan University, Tokyo 192-0397, Japan.
Author contributions
S.-G.K., D.K., T.Q.P.U., and M.W. conceived this project. S.-G.K., Y.Y., A.S., S.T.K., K.H., D.K., T.Q.P.U., and M.W. planned the experiments and analyzed the data. S.-G.K., Y.Y., A.S., S.T.K., K.H., K.K., J.A., H.-G.S., J.-W.H., T.H., A.T., Y.N., N.S., and T.Q.P.U. carried out the experiments: S.-G.K., K.K., J.A., H.-G.S., J.-W.H., T.H., N.S., and M.W. were involved in the plant physiological and cell biological experiments; A.S., A.T., Y.N., and D.K. were involved in the crystallography studies; and S.-G.K., Y.Y., S.T.K., K.H., and T.Q.P.U. were involved in the actin polymerization assays, TIRF and EM observations. S.-G.K., A.S., D.K., T.Q.P.U., and M.W. wrote the manuscript.
Supplementary data
The following materials are available in the online version of this article.
Supplementary Figure S1. Rescue of chloroplast positioning and movements in C1Y transgenic plants.
Supplementary Figure S2. The effects of actin-depolymerizing reagent, latrunculin B (LatB), on dynamic reorganizations of CHUP1-YFP.
Supplementary Figure S3. Interaction of CHUP1-C with skeletal muscle actin filaments.
Supplementary Figure S4. Quantification of the effects of CHUP1-C on polymerization of ACT7.
Supplementary Figure S5. Kinetic analysis of ACT7 polymerization in the presence of PRF1 and CHUP1-C.
Supplementary Figure S6. Electron micrographs of negatively stained ACT7 filaments.
Supplementary Figure S7. Electron micrographs of skeletal muscle actin filaments incubated with CHUP1-C.
Supplementary Figure S8. TIRF microscopy observation of growing ACT7 filaments in the presence of PRF1 and CHUP1-C-tagRFP.
Supplementary Figure S9. Model of the CHUP1-C–actin complex and its functional analysis in transgenic plants.
Supplementary Figure S10. Protein sequence alignment of the C-terminal region of CHUP1.
Supplementary Figure S11. Organismal lineages and distributions of CHUP1 and CHUC genes.
Supplementary Table S1. Binding of CHUP1-C to skeletal muscle actin filaments.
Supplementary Table S2. List of primers used in this study.
Supplementary Data Set 1. Statistical data.
Supplementary File 1. FASTA File for the alignment shown in Supplementary Fig. S10.
Supplementary Movie S1. Reorganization of CHUP1-YFP in a WT palisade cell during the blue light-induced chloroplast avoidance response.
Supplementary Movie S2. Reorganization of CHUP1-YFP in a WT palisade cell during the blue light-induced chloroplast avoidance response.
Supplementary Movie S3. Reorganization of CHUP1-Y in a WT palisade cell during the blue light-induced chloroplast avoidance response.
Supplementary Movie S4. Reorganization of CHUP1-YFP in a phot1 mutant palisade cell during the blue light-induced chloroplast avoidance response.
Supplementary Movie S5. Reorganization of CHUP1-YFP in a phot2 mutant palisade cell during the blue light-induced chloroplast avoidance response.
Supplementary Movie S6. Reorganization of CHUP1-YFP in a phot1 phot2 mutant palisade cell during the blue light-induced chloroplast avoidance response.
Supplementary Movie S7. Reorganization of CHUP1 and cp-actin filaments in a CHUP1-tdTomato × GFP-mTalin palisade cell during the blue light-induced chloroplast avoidance response.
Funding
This work was supported in part by Grants-in-Aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT)/the Japan Society for the Promotion of Science (JSPS) KAKENHI grant numbers 20227001, 23120523, 25120721, 25251033, and 16K14758 (to M.W.); 21770050 (to S.-G.K.); 20687006, 24687014, and 25121726 (to A.S.); 26119002 (to D.K.); and 24117008 (to T.Q.P.U.). Other support was provided by the Basic Science Research Program through the National Research Foundation of Korea (NRF), Ministry of Education (2021R1I1A3059996 to S.-G.K.), the Next-Generation Biogreen 21 Program grant, Rural Development Administration (RDA) (Project No. PJ01366901 to S.-G.K.), “Regional Innovation Mega Project” Program through the Korea Innovation Foundation funded by Ministry of Science and ICT (Project No. 2023-DD-UP-007 to S.-G.K.), and by the Ohsumi Frontier Science Foundation (to M.W.).
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010:66(2):213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel CA, Lutz L, Yang X, Rodriguez A, Adair A, Zhang Y, Leisner SM, Nelson RS, Schoelz JE. The P6 protein of Cauliflower mosaic virus interacts with CHUP1, a plant protein which moves chloroplasts on actin microfilaments. Virology. 2013:443(2):363–374. 10.1016/j.virol.2013.05.028 [DOI] [PubMed] [Google Scholar]
- Belyy A, Merino F, Sitsel O, Raunser S. Structure of the Lifeact–F-actin complex. PLoS Biol. 2020:18(11):e3000925. 10.1371/journal.pbio.3000925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzaniga S, Dall’ Osto L, Kong S-G, Wada M, Bassi R. Interaction between avoidance of photon absorption, excess energy dissipation and zeaxanthin synthesis against photooxidative stress in Arabidopsis. Plant J. 2013:76(4):568–579. 10.1111/tpj.12314 [DOI] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using µManager. Curr Protoc Mol Biol. 2010:92(1):14.20.11–14.20.17. 10.1002/0471142727.mb1420s92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh E, Suetsugu N, Yamori W, Ishishita K, Kiyabu R, Fukuda M, Higa T, Shirouchi B, Wada M. Chloroplast accumulation response enhances leaf photosynthesis and plant biomass production. Plant Physiol. 2018:178(3):1358–1369. 10.1104/pp.18.00484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T, Sano T, Hasezawa S. Actin microfilament dynamics and actin side-binding proteins in plants. Curr Opin Plant Biol. 2007:10(6):549–556. 10.1016/j.pbi.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Ito K, Kashiyama T, Shimada K, Yamaguchi A, Awata J, Hachikubo Y, Manstein DJ, Yamamoto K. Recombinant motor domain constructs of Chara corallina myosin display fast motility and high ATPase activity. Biochem Biophys Res Commun. 2003:312(4):958–964. 10.1016/j.bbrc.2003.10.202 [DOI] [PubMed] [Google Scholar]
- Jarillo JA, Gabrys H, Capel J, Alonso JM, Ecker JR, Cashmore AR. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature. 2001:410(6831):952–954. 10.1038/35073622 [DOI] [PubMed] [Google Scholar]
- Kadota A, Yamada N, Suetsugu N, Hirose M, Saito C, Shoda K, Ichikawa S, Kagawa T, Nakano A, Wada M. Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc Natl Acad Sci U S A. 2009:106(31):13106–13111. 10.1073/pnas.0906250106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M. Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science. 2001:291(5511):2138–2141. 10.1126/science.291.5511.2138 [DOI] [PubMed] [Google Scholar]
- Kagawa T, Wada M. Brief irradiation with red or blue light induces orientational movement of chloroplasts in dark-adapted prothallial cells of the fern Adiantum. J Plant Res. 1994:107(4):389–398. 10.1007/BF02344062 [DOI] [Google Scholar]
- Kandasamy MK, Meagher RB. Actin-organelle interaction: association with chloroplast in Arabidopsis leaf mesophyll cells. Cell Motil Cytoskeleton. 1999:44(2):110–118. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Kagawa T, Oikawa K, Suetsugu N, Miyao M, Wada M. Chloroplast avoidance movement reduces photodamage in plants. Nature. 2002:420(6917):829–832. 10.1038/nature01213 [DOI] [PubMed] [Google Scholar]
- Kijima ST, Hirose K, Kong S-G, Wada M, Uyeda TQP. Distinct biochemical properties of Arabidopsis thaliana actin isoforms. Plant Cell Physiol. 2016:57(1):46–56. 10.1093/pcp/pcv176 [DOI] [PubMed] [Google Scholar]
- Kijima ST, Staiger CJ, Katoh K, Nagasaki A, Ito K, Uyeda TQP. Arabidopsis vegetative actin isoforms, AtACT2 and AtACT7, generate distinct filament arrays in living plant cells. Sci Rep. 2018:8(1):4381. 10.1038/s41598-018-22707-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong S-G, Arai Y, Suetsugu N, Yanagida T, Wada M. Rapid severing and motility of chloroplast-actin filaments are required for the chloroplast avoidance response in Arabidopsis. Plant Cell. 2013a:25(2):572–590. 10.1105/tpc.113.109694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong S-G, Suetsugu N, Kikuchi S, Nakai M, Nagatani A, Wada M. Both phototropin 1 and 2 localize on the chloroplast outer membrane with distinct localization activity. Plant Cell Physiol. 2013b:54(1):80–92. 10.1093/pcp/pcs151 [DOI] [PubMed] [Google Scholar]
- Kong S-G, Wada M. New insights into dynamic actin-based chloroplast photorelocation movement. Mol Plant. 2011:4(5):771–781. 10.1093/mp/ssr061 [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH. A GFP-mouse talin fusion labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 1998:16(3):393–401. 10.1046/j.1365-313x.1998.00304.x [DOI] [PubMed] [Google Scholar]
- Krzeszowiec W, Gabryś H. Phototropin mediated relocation of myosins in Arabidopsis thaliana. Plant Signal Behav. 2007:2(5):333–336. 10.4161/psb.2.5.4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts A, Gevaert K, Cossart P, Vandekerckhove J, Van Troys M. Listeria comet tails: the actin-based motility machinery at work. Trends Cell Biol. 2008:18(5):220–227. 10.1016/j.tcb.2008.03.001 [DOI] [PubMed] [Google Scholar]
- Li J, Blanchoin L, Staiger CJ. Signaling to actin stochastic dynamics. Annu Rev Plant Biol. 2015:66(1):415–440. 10.1146/annurev-arplant-050213-040327 [DOI] [PubMed] [Google Scholar]
- Manders EMM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dual-colour confocal images. J Microsc. 1993:169(3):375–382. 10.1111/j.1365-2818.1993.tb03313.x [DOI] [PubMed] [Google Scholar]
- Michelot A, Derivery E, Paterski-Boujemaa R, Guérin C, Huang S, Parcy F, Staiger CJ, Blanchoin L. A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr Biol. 2006:16(19):1924–1930. 10.1016/j.cub.2006.07.054 [DOI] [PubMed] [Google Scholar]
- Michelot A, Guérin C, Huang S, Ingouff M, Richard S, Rodiuc N, Staiger CJ, Blanchoin L. The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell. 2005:17(8):2296–2313. 10.1105/tpc.105.030908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo KX, Kodera N, Katayama E, Ando T, Uyeda TQP. Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. eLife. 2015:4:e04806. 10.7554/eLife.04806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K, Kasahara M, Kiyosue T, Kagawa T, Suetsugu N, Takahashi F, Kanegae T, Niwa Y, Kadota A, Wada M. CHLOROPLAST UNUSUAL POSITIONING1 is essential for proper chloroplast positioning. Plant Cell. 2003:15(12):2805–2815. 10.1105/tpc.016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K, Yamasato A, Kong S-G, Kasahara M, Nakai M, Takahashi F, Ogura Y, Kagawa T, Wada M. Chloroplast outer envelope protein CHUP1 is essential for chloroplast anchorage to the plasma membrane and chloroplast movement. Plant Physiol. 2008:148(2):829–842. 10.1104/pp.108.123075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997:276:307–326. 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Paavilainen VO, Bertling E, Falck S, Lappalainen P. Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol. 2004:14(7):386–394. 10.1016/j.tcb.2004.05.002 [DOI] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton. 2009:66(8):606–617. 10.1002/cm.20379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paves H, Truve E. Myosin inhibitors block accumulation movement of chloroplasts in Arabidopsis thaliana leaf cells. Protoplasma. 2007:230(3-4):165–169. 10.1007/s00709-006-0230-y [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmong S, Bretcher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002:297(5581):612–615. 10.1126/science.1072309 [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002:4(8):626–631. 10.1038/ncb834 [DOI] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci U S A. 2001:98(12):6969–6974. 10.1073/pnas.101137598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattarzadeh A, Krahmer J, Germain AD, Hanson MR. A myosin XI tail domain homologous to the yeast myosin vacuole-binding domain interacts with plastids and stromules in Nicotiana benthamiana. Mol Plant. 2009:2(6):1351–1358. 10.1093/mp/ssp094 [DOI] [PubMed] [Google Scholar]
- Schmidt von Braun S, Schleiff E. The chloroplast outer membrane protein CHUP1 interacts with actin and profilin. Planta. 2008:227(5):1151–1159. 10.1007/s00425-007-0688-7 [DOI] [PubMed] [Google Scholar]
- Shimada A, Nyitrai M, Vetter IR, Kuhlmann D, Bugyi B, Narumiya S, Geeves MA, Wittinghofer A. The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Mol Cell. 2004:13(4):511–522. 10.1016/S1097-2765(04)00059-0 [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971:246(15):4866–4871. 10.1016/S0021-9258(18)62016-2 [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Blanchoin L. Actin dynamics: old friends with new stories. Curr Opin Plant Biol. 2006:9(6):554–562. 10.1016/j.pbi.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Suetsugu N, Dolja VV, Wada M. Why have chloroplasts developed a unique motility system? Plant Signal Behav. 2010:5(10):1190–1196. 10.4161/psb.5.10.12802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N, Kong S-G, Kasahara M, Wada M. Both LOV1 and LOV2 domains of phototropin2 function as the photosensory domain for hypocotyl phototropic responses in Arabidopsis thaliana (Brassicaceae). Am J Bot. 2013:100(1):60–69. 10.3732/ajb.1200308 [DOI] [PubMed] [Google Scholar]
- Suetsugu N, Sato Y, Tsuboi H, Kasahara M, Imaizumi T, Kagawa T, Hiwatashi Y, Hasebe M, Wada M. The KAC family of kinesin-like proteins is essential for the association of chloroplasts with the plasma membrane in land plants. Plant Cell Physiol. 2012:53(11):1854–1865. 10.1093/pcp/pcs133 [DOI] [PubMed] [Google Scholar]
- Suetsugu N, Wada M. Evolution of the cp-actin-based motility system of chloroplasts in green plants. Front Plant Sci. 2016:7:561. 10.3389/fpls.2016.00561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HQ, Yamamoto M, Mejillano M, Yin HL. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem. 1999:274(47):33179–33182. 10.1074/jbc.274.47.33179 [DOI] [PubMed] [Google Scholar]
- Sztatelman O, Waloszek A, Banaś AK, Gabryś H. Photoprotective function of chloroplast avoidance movement: in vivo chlorophyll fluorescence study. J Plant Physiol. 2010:167(9):709–716. 10.1016/j.jplph.2009.12.015 [DOI] [PubMed] [Google Scholar]
- Tsuboi H, Wada M. Chloroplasts can move in any direction to avoid strong light. J Plant Res. 2011:124(1):201–210. 10.1007/s10265-010-0364-z [DOI] [PubMed] [Google Scholar]
- Tsuboi H, Wada M. Chloroplasts continuously monitor photoreceptor signals during accumulation movement. J Plant Res. 2013:126(4):557–566. 10.1007/s10265-012-0542-2 [DOI] [PubMed] [Google Scholar]
- Tsuboi H, Yamashita H, Wada M. Chloroplasts do not have a polarity for light-induced accumulation movement. J Plant Res. 2009:122(1):131–140. 10.1007/s10265-008-0199-z [DOI] [PubMed] [Google Scholar]
- Usami H, Maeda T, Fujii Y, Oikawa K, Takahashi F, Kagawa T, Wada M, Kasahara M. CHUP1 mediates actin-based light-induced chloroplast avoidance movement in the moss Physcomitrella patens. Planta. 2012:236(6):1889–1897. 10.1007/s00425-012-1735-6 [DOI] [PubMed] [Google Scholar]
- van Gisbergen PA, Bezanilla M. Plant formins: membrane anchors for actin polymerization. Trends Cell Biol. 2013:23(5):227–233. 10.1016/j.tcb.2012.12.001 [DOI] [PubMed] [Google Scholar]
- Wada M, Kagawa T, Sato Y. Chloroplast movement. Annu Rev Plant Biol. 2003:54(1):455–468. 10.1146/annurev.arplant.54.031902.135023 [DOI] [PubMed] [Google Scholar]
- Wada M, Kong S-G. Actin-mediated movement of chloroplasts. J Cell Sci. 2018:131(2):jcs210310. 10.1242/jcs.210310 [DOI] [PubMed] [Google Scholar]
- Whippo CW, Khurana P, Davis PA, DeBlasio SL, DeSloover D, Staiger CJ, Hangarter RP. THRUMIN1 is a light-regulated actin-bundling protein involved in chloroplast motility. Curr Biol. 2011:21(1):59–64. 10.1016/j.cub.2010.11.059 [DOI] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004:116(5):711–723. 10.1016/S0092-8674(04)00210-7 [DOI] [PubMed] [Google Scholar]
- Yamada N, Suetsugu N, Wada M, Kadota A. Phototropin-dependent biased relocalization of cp-actin filaments can be induced even when chloroplast movement is inhibited. Plant Signal Behav. 2011:6(11):1651–1653. 10.4161/psb.6.11.17767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto D, Uchihashi T, Kodera N, Yamashita H, Nishikori S, Ogura T, Shibata M, Ando T. High-speed atomic force microscopy techniques for observing dynamic biomolecular processes. Methods Enzymol. 2010:475:541–564. 10.1016/S0076-6879(10)75020-5 [DOI] [PubMed] [Google Scholar]
- Yi K, Guo C, Chen D, Zhao B, Yang B, Ren H. Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis. Plant Physiol. 2005:138(2):1071–1082. 10.1104/pp.104.055665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Liu C, Wang J, Ren Z, Staiger CJ, Ren H. A processive Arabidopsis formin modulates actin filament dynamics in association with profilin. Mol Plant. 2016:9(6):900–910. 10.1016/j.molp.2016.03.006 [DOI] [PubMed] [Google Scholar]
- Zurzycki J. Chloroplast arrangement as a factor in photosynthesis. Acta Soc Bot Pol. 1955:24(1):27–63. 10.5586/asbp.1955.003 [DOI] [Google Scholar]
- Zurzycki J. The destructive effect of light on the photosynthetic apparatus. Acta Soc Bot Pol. 1957:26(1):157–175. 10.5586/asbp.1957.008 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.