Abstract
Genetic load refers to the accumulated and potentially life-threatening deleterious mutations in populations. Understanding the mechanisms underlying genetic load variation of transposable element (TE) insertion, a major large-effect mutation, during range expansion is an intriguing question in biology. Here, we used 1,115 global natural accessions of Arabidopsis (Arabidopsis thaliana) to study the driving forces of TE load variation during its range expansion. TE load increased with range expansion, especially in the recently established Yangtze River basin population. Effective population size, which explains 62.0% of the variance in TE load, high transposition rate, and selective sweeps contributed to TE accumulation in the expanded populations. We genetically mapped and identified multiple candidate causal genes and TEs, and revealed the genetic architecture of TE load variation. Overall, this study reveals the variation in TE genetic load during Arabidopsis expansion and highlights the causes of TE load variation from the perspectives of both population genetics and quantitative genetics.
The genetic load of transposable elements accumulated during Arabidopsis range expansion and effective population size could explain 62.0% of the variance in genetic load of transposable elements.
IN A NUTSHELL.
Background: Genetic load refers to accumulated deleterious mutations that could reduce organism fitness. Range expansion promotes adaptation but increases genetic load. Transposable elements (TEs) are a type of repetitive DNA sequence mobilizing across the genome; this mobilization could rapidly produce large-effect mutations. Understanding genetic load and its drivers during range expansion has important implications for human health, crop breeding, and conservation biology. However, the genetic load of TEs during range expansion remains unclear.
Question: How did the genetic load of TEs vary during Arabidopsis (Arabidopsis thaliana) range expansion? What are the driving forces of TE load variation?
Findings: By analyzing the genomes of 1,115 worldwide Arabidopsis accessions, we determined that the deleterious effect of TEs is between that of deleterious missense mutations and loss-of-function mutations. TE load accumulated along the expansion axis, particularly in the recently established Yangtze River basin population. Effective population size explained 62.0% of the variance in TE load. High transposition rates and selective sweeps also contributed to TE accumulation in the expanded populations. In addition, we genetically mapped the candidate causal genes or TEs and revealed the genetic architecture of TE load variation among natural populations.
Next steps: It is important to incorporate multiple genome assemblies or long-read sequencing data to capture the full landscape of TE variation. We must also clarify the relative contribution of each driving force to TE load variation and perform experimental validation of candidate genes or TEs associated with TE load variation in natural populations.
Introduction
Range expansion is a common process of adaptive evolution in which enhanced genetic drift usually increases the frequency of deleterious mutations on expanding wave fronts and incurs expansion load, reducing fitness and affecting the persistence of newly colonizing populations (Peischl et al. 2013). Understanding the genetic load and its causes during range expansion is crucial for understanding species adaptation to not only the local conditions but also climate change. Expansion load has been extensively studied in diverse species (Wang et al. 2017; Takou et al. 2021), especially in humans (Lohmueller et al. 2008; Henn et al. 2016). However, most previous studies mainly focused on nonsynonymous or loss-of-function (LoF) mutations (Bertorelle et al. 2022).
The genetic load of transposable elements (TEs), one of the main contributors to large-effect mutations (Lisch 2013), remains largely unknown. TEs are repetitive DNA fragments that can transpose across the genome and constitute a large portion of the genome in many organisms (Wells and Feschotte 2020). TE gain or loss is fast, the transposition rate of TEs is higher than that of single nucleotide substitution and small indel mutations (Quadrana et al. 2019; Ho et al. 2021). Transposition of TEs can affect gene function and genome stability, promote phenotypic divergence, and contribute to speciation and adaptation (Van't Hof et al. 2016; Wei and Cao 2016; Niu et al. 2019).
TE insertions are generally regarded as deleterious and can reduce fitness via 3 routes: gene or gene expression disruption, ectopic TE recombination, and deleterious action of TE transcript and protein products (Barron et al. 2014). Accordingly, in natural populations, most TEs are rare insertions and are depleted in genic regions (Barron et al. 2014; Quadrana et al. 2016; Stuart et al. 2016; Baduel et al. 2021a). In particular, a single TE or TE family can compromise host fitness (Hill et al. 2016). Therefore, clarifying the genetic load of TEs (TE load) during range expansion is important for revealing the mechanism of invasion and environmental adaptation. However, the high turnover, nonconstant transposition rate (Bergman and Bensasson 2007), and insertion preference (Quadrana et al. 2019) hinder the systematic study of deleterious effects of TE insertions at genome level.
Consistent with other mutations, the evolutionary dynamics of TEs are shaped by forces acting on the processes of mutation generation (transposition and excision) and mutation maintenance (selection and genetic drift) (Charlesworth and Charlesworth 1983; Tenaillon et al. 2010). The transposition rate and deletion rate differ among TE families and host genotypes (Adrion et al. 2017; Baduel et al. 2021a; Ho et al. 2021), and can affect the TE load. In particular, features of TEs and hosts can affect the transposition rate, such as the transposition capacity (Chen et al. 2020) and robustness of the TE-silencing system (He et al. 2022). In addition, environmental factors can also affect the transposition rate of TEs (Baduel et al. 2021a).
In the TE mutation maintenance process, the strength of purifying selection and genetic drift is correlated with effective population size (Ne); the smaller the Ne value, the more relaxed the purifying selection. Therefore, TE load is associated with Ne (Lynch and Conery 2003; Lockton et al. 2008). Demographic processes (such as range expansion and founder effects) that reduce the Ne could lead to the accumulation of TEs. For example, the TE number in the invasive populations of spotted-wing Drosophila (Drosophila suzukii) is considerably higher than that in its native populations (Merel et al. 2021). Similarly, Capsella rubella, a close relative of Arabidopsis (Arabidopsis thaliana), originated through an extreme bottleneck (Foxe et al. 2009; Guo et al. 2009), which strongly reduced its Ne and exhibits a much higher TE load than its sister species (Niu et al. 2019).
Arabidopsis is a selfing species globally distributed and has over 1,000 resequenced genomes (Cao et al. 2011; Long et al. 2013; 1001 Genomes Consortium 2016; Durvasula et al. 2017; Zou et al. 2017). Arabidopsis underwent a postglacial spread of a human commensal nonrelict group, which originated near the Balkans, expanded mainly along the east-west axis, and comprised 95% of the natural populations (Lee et al. 2017). Therefore, Arabidopsis is a great model for understanding the TE load variation during range expansion.
In Arabidopsis, previous studies mainly focused on European accessions and revealed the contribution of TE silencing systems and environmental factors to transposition modulation (Quadrana et al. 2016; Baduel et al. 2021a). Here, we explored 1,115 globally distributed natural Arabidopsis accessions, including 204 accessions (117 sequenced in this study) from the eastern edge of the species. Given that (i) the dissection of the deleterious effect of TE insertions is a fundamental question, (ii) the spectrum of TE load in natural populations is important for understanding the dynamics of TE load, (iii) the demographic history and genetic features potentially affect the TE load, and (iv) the determinant loci of TE load are crucial for understanding the TE load variation, we comprehensively investigated these questions in Arabidopsis. Overall, we elucidate the variation in TE load during Arabidopsis expansion and highlight the causes of TE load variation.
Results
The mutational landscapes of TEs in Arabidopsis natural populations
To study the genetic load of TEs during Arabidopsis range expansion, the short-read sequencing data of 1,114 globally distributed Arabidopsis nonreference accessions were utilized (Fig. 1A). While 986 Arabidopsis accessions were sequenced in previous studies (Supplemental Data Set 1) (1001 Genomes Consortium 2016; Durvasula et al. 2017; Zou et al. 2017), those of 128 accessions from northwestern China and the Yangtze River basin were sequenced in this study (Supplemental Data Set 2). The 1,114 nonreference accessions were grouped into 1 relict, 10 nonrelict populations, and an admixed group (see Materials and Methods).
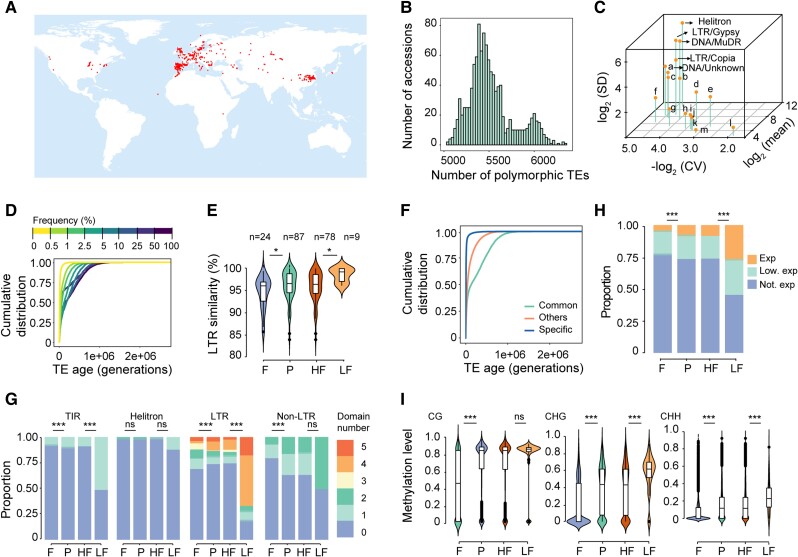
Figure 1.
Transposable elements identification and characterization in natural Arabidopsis populations. A) Geographical distribution of 1,114 nonreference accessions. Dots indicate sample locations. B) Polymorphic TE numbers per accession. C) Copy number variation of 18 TE superfamilies in 1,115 natural accessions. X-axis represents copy number variation in each TE superfamily across accessions, evaluated as coefficient of variation (CV); y-axis represents the average number of TEs in each superfamily; z-axis represents the overall contribution of each superfamily to the variation in total TE number, evaluated as standard deviation (SD). a, LINE/L1; b, DNA/En-Spm; c, DNA/HAT; d, DNA/Harbinger; e, RathE1_cons; f, DNA/Pogo; g, DNA/Mariner; h, short interspersed nuclear elements (SINE); i, Unassigned; j, DNA/Tc1; k, RathE3_cons; l, LINE? ; m, RathE2_cons. D) Age distribution of TEs in different frequency bins. E) Long terminal repeat (LTR) similarity of LTR TEs in 4 TE categories: fixed (F; TEs with read coverage in all accessions), polymorphic (P), high frequency (HF; frequency ≥5%), and low frequency (LF; frequency <5%). Mann–Whitney U test was used for the significance test. ***P < 0.001; **P < 0.01; *P < 0.05; ns, not significant. F) Age distribution of TEs with different geographical distribution patterns. Common, TEs present in all populations; Others, TEs present in at least 2, but not all populations; Specific, population-specific TEs. G) Proportion of TEs containing transposase domains in the 4 TE categories. TIR, terminal inverted repeat. Fisher's exact test was used for the significance test. H) Proportion of TEs in 4 categories, based on expression potential (the potential to express in TE-activated mutant): Exp, expressed and annotated; Low. exp, low expressed; Not. exp, not expressed. Fisher's exact test was used for the significance test. I) DNA methylation levels of TEs in the 4 TE categories. Mann–Whitney U test was used for the significance test.
A total of 31,189 TEs belonging to 320 families and 18 superfamilies were annotated in the Col-0 genome (TAIR10). The transposable element polymorphism identification (TEPID) software (Stuart et al. 2016), which combines evidence regarding split reads and discordant reads, was used to determine the presence-and-absence variation (PAV) of TEs in Arabidopsis, with Col-0 as the reference accession. Compared with Col-0, these accessions contained, on average, 441 TE presences and 1,257 TE absences (Supplemental Fig. S1A). Comparison between Col-0 and genome assemblies from 8 regional accessions suggested that the precision rate of TE PAV detection ranged from 0.54 to 0.71, and on average 0.66, which was comparable to most TE polymorphism detection tools (Kosugi et al. 2019; Vendrell-Mir et al. 2019) (Supplemental Data Set 3). Among different TE superfamilies, most of them have high precision rate (>0.6), except for DNA/En-Spm and long-terminal repeat (LTR)/Gypsy superfamilies (Supplemental Fig. S1B). In addition, the precision rate of intergenic TEs was lower than genic TEs, probably resulting from the poor read mapping in these regions (Supplemental Fig. S1C). In the Col-0 genome, 20.4% of TEs (6,351/31,189) were polymorphic (present in at least 1 accession but not in all accessions), and in terms of different TEs, the highest fraction (26.7%) of polymorphic TEs were retrotransposons (Supplemental Fig. S1D).
The total number of TEs in each accession ranged from 29,979 to 31,201, and a total of 67,429 TE loci were identified among the 1,115 accessions, of which 42,756 were polymorphic. The number of polymorphic TEs per accession varied from 5,306 to 6,528 (Fig. 1B). All analyses described below were conducted on polymorphic TEs unless stated otherwise. Based on the standard variation of TE numbers in diverse superfamilies across all 1,115 accessions, we estimated the contribution of each TE superfamily to the variation of TE number. The results revealed RC/Helitron, LTR/Gypsy, DNA/MuDR, LTR/Copia, and DNA/Unknown superfamilies as the top 5 major contributors to the variation in the total TE number (Fig. 1C).
To estimate the age of polymorphic TEs, we used the Genealogical Estimation of Variant Age (GEVA), which relies on the sequence divergence of regions around TEs (Albers and McVean 2020). The results indicated that most of the polymorphic TEs transposed after the divergence of Arabidopsis from its sister species Arabidopsis lyrata, approximately 10 million years ago (Hu et al. 2011) (Fig. 1D). In addition, only 5,022 TEs (16.1% of the Col-0 TEs) are shared between Arabidopsis and A. lyrata (MN47), which implies the rapid evolution and fast turnover of TEs. TE frequency was commonly used to reflect TE age, of which low-frequency TEs are much younger (Quadrana et al. 2016; Baduel et al. 2019). Accordingly, except for the 25% to 50% bin, here the TE frequency corresponds with TE age (Fig. 1D), as reported in previous study (Baduel et al. 2021a). The age estimation of LTR TEs in Col-0, based on the diversification of 2 LTR sequences on either end of each intact LTR TE, also indicated that polymorphic TEs and low-frequency TEs (frequency < 5%) were younger than fixed TEs and high-frequency TEs (frequency ≥ 5%), respectively (Fig. 1E). Additionally, in terms of the geographical distribution, regional distributed TEs were much younger than the globally distributed TEs (Fig. 1F). Taken together, these results indicated that TE gain and loss is fast and most polymorphic TEs were transposed recently after speciation.
To characterize the transposition activity of polymorphic TEs, which are most probably active, we analyzed their structural integrity, transcription potential, and DNA methylation level. Given that data on these genetic features are mostly abundant in Col-0, all comparative analyses were based on Col-0. First, to characterize the transposition potential of TEs, we searched the total number of transposition-related domains for each TE. Higher fraction of polymorphic TEs had transposition-related domains than that of fixed TEs, except Helitrons (whose transposition-related domains were similar between polymorphic and fixed TEs) and LTR TEs (Fig. 1G). Second, given that most TEs are repressed by the host genome, to evaluate the transcription potential of TEs, we utilized the published long-read TE transcriptome data of Col-0 triple mutants (ddm1 rdr6 pol V), which lack multiple layers of TE repression and could potentially reflect the transcription potential of TEs (Panda and Slotkin 2020). Compared with fixed TEs, a much higher proportion of polymorphic TEs were expressed in the TE-activated mutants (Fig. 1H). Third, to clarify the extent of DNA methylation-induced TE repression, we calculated the DNA methylation levels of polymorphic and fixed TEs. The results showed that cytosines in all contexts were methylated to a much higher degree in polymorphic TEs than in fixed TEs (Fig. 1I).
Given that low-frequency TEs are a more accurate indicator of recent mobilization than high-frequency TEs, we compared the above 3 features between low-frequency (frequency < 5%) and high-frequency (frequency ≥ 5%) TEs. Low-frequency TEs possessed more transposition-related domains, especially for terminal inverted repeat (TIR) and LTR TEs (Fig. 1G), and showed higher transcription potential (Fig. 1H) and higher CHG and CHH methylation levels than high-frequency TEs (Fig. 1I). Taken together, these results suggest that polymorphic TEs, particularly low-frequency TEs, contain more transposition-related domains and are more likely to be transcribed, which help themselves or other nonautonomous members to transpose. However, at the same time, these recently active TEs tend to be silenced by DNA methylation, suggesting that hosts could identify and silence potentially active TEs.
Deleterious effects of TEs vary between deleterious nonsynonymous mutations and loss-of-function mutations, and synergistic epistasis is present among TEs with large fitness effects
TE insertions have been demonstrated to often have deleterious effects (Pasyukova et al. 2004; Barron et al. 2014; Hill et al. 2016). At genome level, the site frequency spectrum (SFS) of TE insertions could be used to evaluate the deleterious effect of TEs. The more deleterious mutations are more skewed toward low frequency because of purifying selection (Williamson et al. 2004). Comparison of the SFS of TE insertions to 4-fold degenerate sites (which are putatively neutral) indicates that TE insertions are under purifying selection, as previously demonstrated (Baduel et al. 2021a), and in particular, here we found that the deleterious effect of TE insertions varies between that of tolerated nonsynonymous single nucleotide polymorphisms (tnSNPs) and deleterious nonsynonymous SNPs (dnSNPs) predicted by Provean (Choi et al. 2012) (Fig. 2A).
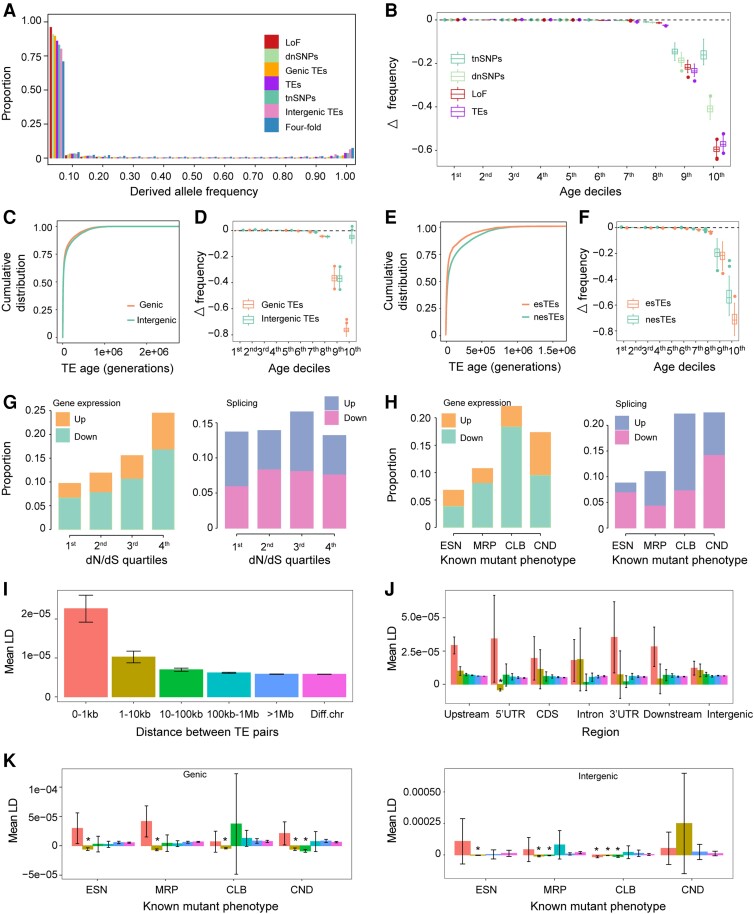
Figure 2.
Evaluation of the deleterious effects of TE insertions. A) Derived allele frequency of TE insertions and other variants. Four-fold, 4-fold degenerate sites; dnSNPs, deleterious nonsynonymous SNPs; tnSNPs, tolerated nonsynonymous SNPs; LoF, loss-of-function variants; Genic TEs, TEs in the gene body and flanking region (2 kb upstream and 1 kb downstream); Intergenic TEs, TEs outside genic regions. B) Correlation of Δ frequency (TEs/dnSNPs/tnSNPs/LoF frequency–neutral site frequency) and age deciles. Age deciles increased from 1st to 10th. C) Age distribution of genic and intergenic TEs. D) Correlation of Δ frequency (genic/intergenic TEs frequency–neutral site frequency) and age deciles. Age deciles increased from 1st to 10th. E) Age distribution of TE insertions capable of affecting gene expression or splicing (esTEs) and not capable of affecting gene expression and splicing (nesTEs). F) Correlation of Δ frequency (esTEs/nesTEs frequency–neutral site frequency) and age deciles. Age deciles increased from 1st to 10th. G and H) Proportion of TE insertions associated with at least 2-fold change in expression level or PSI value compared with the nearest genes grouped into 4 conservation categories based on their ratio of nonsynonymous to synonymous substitutions (dN/dS) quartiles (G) or mutant phenotypes (H). In (G), the first quartile (1st) was the most constrained, while the last quartile (4th) was the least constrained. In (H), the categories defined according to known mutant phenotypes were as follows: ESN, essential; MRP, morphological; CLB, cellular-biochemical; CND, conditional. Up, upregulated gene expression level or PSI value in accessions with TEs compared with accessions without TEs. Down, downregulated gene expression level or PSI value in accessions with TEs compared with accessions without TEs. I) Mean LD of all TE pairs with different physical distances. LD, linkage disequilibrium; Diff. chr, TE pairs located on different chromosomes. Error bar indicates 95% confidence interval. J) Mean LD of TE pairs located in different genomic regions. Asterisk (*) denotes significant negative LD with a completely negative 95% confidence interval. Bar color denotes the physical distance between TEs of a pair, as shown in I. K) Mean LD of TE pairs located within genic or intergenic regions of genes in different functional categories.
However, the excess of low-frequency TEs could also come from recent TE burst. To control the confounding effect of variable transposition rate, we leveraged the age distribution of TEs and performed age-adjusted SFS comparison between TE insertions and neutral sites, which was robust to TE burst (Horvath et al. 2022). If TE insertions are deleterious, purifying selection would prevent the accumulation of TEs to high frequencies based on the deviation of Δ frequency (TE frequency–neutral site frequency) from 0, especially in older age bins, and thus create a negative correlation between Δ frequency and age. The negative correlation (Spearman's rho = −0.98) between Δ frequency and age deciles confirmed that TE insertions are under purifying selection and are more deleterious than dnSNPs (Spearman's rho = −0.94) but not LoF (Spearman's rho = −0.99) (Fig. 2B).
To further characterize the deleterious effect of different types of TE insertions, we adopted 3 approaches that rely on the relationship of TEs with their inserted or adjacent genes. The first approach is based on the location of TE insertion. As previously reported (Quadrana et al. 2016; Stuart et al. 2016; Baduel et al. 2021a), TEs are enriched in pericentromeric regions, where genes are strongly depleted (Supplemental Fig. S2A). Similarly, TEs are enriched in intergenic regions as well (Supplemental Fig. S2B), which implies that TE insertions in genic regions are more deleterious, or TEs are more likely to insert into intergenic regions. Accordingly, the SFS of genic TE insertions is more skewed than that of total TEs, tnSNPs, and intergenic TEs, but less skewed than that of dnSNPs and LoF mutations (Fig. 2A), which implies that the deleterious effect of genic TE insertions varies between that of total TEs and dnSNPs. Similarly, based on the SFS, the potential fitness effect of intergenic TE insertions varies between that of 4-fold degenerate sites and tnSNPs (Fig. 2A), and intergenic TE insertions are less deleterious than genic TEs. The potential higher false positive rate of intergenic TE calls would lead to an excess of rare intergenic TEs rather than the observed excess of common intergenic TEs, thus would be less likely to bias our conclusion.
Consistent with previous studies about SNPs, which showed that deleterious mutations are younger than neutral or benign mutations (Kiezun et al. 2013; Albers and McVean 2020), we found that the more deleterious genic TE insertions were younger than the less deleterious intergenic TE insertions (Fig. 2C, P = 2.4e-06, Mann–Whitney U test). To control the confounding effect of insertion preference, we also compared the age-adjusted SFS of genic TE insertions and intergenic TE insertions. The more negative correlation (Spearman's rho = −0.97) between Δ frequency (TE frequency–neutral site frequency) and age deciles of genic TEs suggested that genic TE insertions are more deleterious than intergenic TE insertions (Spearman's rho = −0.90) (Fig. 2D).
The second approach is based on the ability of TE insertions to affect gene expression and splicing. We used the RNA-seq data of 413 accessions (Kawakatsu et al. 2016) to evaluate the effects of TE insertions on gene transcription and splicing. The results showed that 23.1% and 14.2% of the TE insertions were associated with at least 2-fold change in the expression level and percent spliced in (PSI) value of the inserted or adjacent genes, respectively. Accordingly, these TE insertions affecting gene expression or splicing were regarded as esTEs, and the others were regarded as nesTEs. Similar to the reported transcriptional effects of TEs (Baduel et al. 2021a), in general, TEs in the coding sequence or intronic regions were more likely to regulate gene expression and splicing than other regions, and mainly downregulated gene expression and inhibited splicing (Supplemental Fig. S2, C and D).
Given that genic TE insertions were more deleterious (Fig. 2, A and D), and that stabilizing selection constrains the variation in gene expression (Hill et al. 2021), TE insertions affecting gene expression or splicing were most probably deleterious. To validate this assumption, we compared the SFS of esTEs with that of nesTEs. As expected, the SFS of esTEs was more skewed toward rare variations than that of nesTEs, which implies that esTEs are more deleterious than nesTEs (Supplemental Fig. S2E). Consistent with our result that the more deleterious genic TEs were younger than the less deleterious intergenic TEs (Fig. 2C), the more deleterious esTEs were much younger than the less deleterious nesTEs (Fig. 2E, P < 2.2e-16, Mann–Whitney U test). The age-adjusted SFS also supports that esTEs have a more negative correlation (Spearman's rho = −0.99) between Δ frequency (TE frequency–neutral site frequency) and age deciles than nesTEs (Spearman's rho = −0.94) (Fig. 2F). Thus, TE insertions altering gene expression or splicing are more deleterious.
The 3rd approach relies on the functional importance of genes adjacent to or contained TEs. The observed TEs would be away from functional important genes due to either purifying selection or insertion preference (Quadrana et al. 2019). However, TE insertions affecting function of important genes would potentially be more deleterious than those that do not affect gene function and thus would be selected against. To verify this assumption, we categorized TEs based on the conservation of the nearest genes and tested if genes with more essential functions were depleted of TE insertions that affect gene function. The conservation categories were based on the dN/dS ratio; the smaller the dN/dS ratio, the more conserved and functionally essential the gene (Larracuente et al. 2008; Waterhouse et al. 2011). Accordingly, more constrained genes were depleted of TE insertions that could affect the gene expression level (Fig. 2G). We also categorized the functional essentiality of genes based on their known mutant phenotypes (Lloyd and Meinke 2012). The functionally important genes were depleted of TE insertions that could affect the gene expression level and splicing (Fig. 2H). These results suggest that TE insertions located close to functionally important genes are more deleterious than TE insertions near less important genes.
The classic theoretical framework predicts that purifying selection acting on the synergistic epistasis of deleterious TE insertions (whereby an additional TE copy exacerbates the reduction of fitness) is predominantly responsible for TE abundance maintenance (Charlesworth and Charlesworth 1983; Charlesworth 1991). To explore the possible effect of the synergistic epistasis of TEs, we utilized the repulsion linkage disequilibrium (LD) of TE pairs generated by purifying selection and computed the mean LD of TE pairs with different physical distances. Despite the noise of LD calculation with rare variants, we used TEs with frequency <1% to maximize the deleterious effect of TE insertions and thus improve the likelihood of the identification of the synergistic epistasis of TEs. In particular, we also included SNPs with similar frequencies and at similar locations for comparison.
At the genome level, the mean LD and 95% confidence interval of all TE pairs with different physical distances were positive (Fig. 2I), which resulted from the demographic history as pointed previously (Sandler et al. 2021). Because purifying selection is expected to generate more negative LD among the highly deleterious TE pairs, we focused on the mean LD of TE pairs with more deleterious effects. Among the potentially more deleterious genic TEs, the LD of TE pairs located 1 to 10 kb apart and in 5′ untranslated regions (5'UTRs) was significantly negative (Fig. 2J). We also analyzed the LD of TE pairs inserted or adjacent to functionally important genes. When genes were categorized according to their conservation score, we observed significantly negative LD values of TE pairs located in the intergenic regions (1 to 10 kb bin) of the most constrained genes (Supplemental Fig. S2F). When genes were categorized according to their mutant phenotype, the LD values of TE pairs located in genic regions were significantly negative for all phenotypic categories (1 to 10 kb bin) and conditional categories (10 to 100 kb bin) (Fig. 2K). Interestingly, we also observed significantly negative LD for TE pairs located in intergenic regions when the mutant phenotype of the closest gene was categorized as essential (1 to 10 kb bin), morphological (1 to 10 and 10 to 100 kb bins), and cellular-biochemical (0 to 1, 1 to 10, and 10 to 100 kb bins) (Fig. 2K).
However, unlike in Drosophila (Drosophila melanogaster) (Lee 2022), we found significantly negative LD only between the more deleterious TE pairs but not at genome level, which might be ascribed to the weakly deleterious effects of TE insertions in Arabidopsis (the SFS of LoF mutations was more skewed to low frequency than that of TEs) compared with the strongly deleterious effects of TE insertions in Drosophila (the SFS of TE insertions is more skewed to low frequency than that of LoF mutations). Accordingly, among SNPs with varying degrees of deleterious impact, only the most deleterious LoF mutation pairs located in conserved genes or functionally important genes displayed significantly negative LD (Supplemental Fig. S3). Overall, these results suggest that the synergistic epistasis of TEs is present in Arabidopsis and tends to be prevalent among TE pairs with more deleterious effects.
TE load increased with the distance from the origin and was negatively correlated with Ne
To study the TE load variation during the range expansion of Arabidopsis, we focused on 10 nonrelict populations. The Balkans population was regarded as the origin of nonrelicts as it is located near the predicted origin of nonrelicts (Fig. 3A) (1001 Genomes Consortium 2016; Lee et al. 2017). Consistent with the expansion from Balkans, the genetic diversity of natural populations decreased with expansion from the origin (Fig. 3B). All expanded populations, except the Spain population, showed significantly lower genetic diversity than the Balkans population (Fig. 3B, P < 0.01, Mann–Whitney U test); the higher genetic diversity of the Spain population could have resulted from the introgression of Iberian relicts (Lee et al. 2017). Populations located on the margin of expansion, especially the Yangtze River basin population, had much lower genetic diversity (reduced by 60.7% relative to the Balkans) (Fig. 3B).
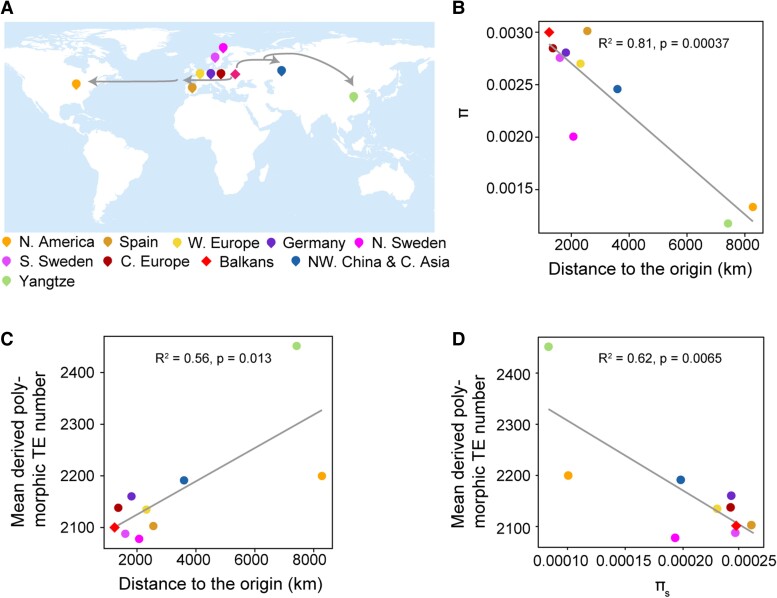
Figure 3.
Transposable element load variation during Arabidopsis range expansion. A) Geographical representation of the range expansion of nonrelicts. Locations of 10 nonrelict populations are indicated on the map. The putative expansion trace is indicated with arrows. B) Pearson correlation between genetic diversity (π; calculated in nonoverlapping 10 kb windows) and the distance to the putative origin. C) Pearson correlation between TE load and the distance to the putative origin. D) Pearson correlation between the mean derived TE number and effective population size, used 4-fold degenerate sites diversity (πs) as proxy. Genetic diversity was calculated in nonoverlapping 10 kb windows.
To estimate TE load variation during range expansion, the derived polymorphic TE number per individual (TEs present in Arabidopsis but absent in A. lyrata and C. rubella) was used as a load proxy. Consistent with the theoretical prediction of expansion load (Peischl et al. 2013), TE load increased with the distance from the origin. Most expanded populations showed higher TE load than the origin Balkans population (P < 0.01, Mann–Whitney U test), and populations on the expansion wave front, especially the recently established Yangtze River basin population, exhibited the largest TE load (increased by 16.7% relative to the Balkans) (Fig. 3C).
Ne, reflected by nucleotide diversity at 4-fold degenerate sites, was directly correlated with the effectiveness of purifying selection (Lynch and Conery 2003; Charlesworth 2009). The linear regression between TE load and nucleotide diversity at 4-fold degenerate sites in nonrelict populations suggested that Ne alone explained 62.0% of the TE load variation among the natural populations (Fig. 3D). Accordingly, populations on the expansion front had much lower Ne and accumulated much higher TE load than the origin Balkans population, such as Yangtze River basin population, northwestern China and Central Asia and North America population (Fig. 3D). The exception of the Spain population might be explained by the introgression of Iberian relicts, which increased the genetic diversity of this population (Lee et al. 2017). However, the reason for the exception of the 2 Swedish populations remains unclear and awaits further study. It is noteworthy that TE load increase was weaker in North America population relative to the drastic decrease of its Ne. This pattern may result from the recent introduction of diverse European lineages and the admixture among introduced lineages, which could increase genetic diversity and reduce genetic load to some extent (Shirsekar et al. 2021).
Given TE number detection was sensitive to sequencing coverage (Stritt et al. 2018), especially TE insertions (Supplemental Fig. S4, A and B), we leveraged 398 accessions with very high coverage (≥25×) to check the consistency of TE load variation between these 2 datasets. The results suggested that TE load accumulation during range expansion was robust to sequencing coverage (Supplemental Fig. S4C). In addition, we also used the genic TEs which have higher precision rates in TE calls to check the consistency of TE load variation between the overall TE sets and the high-quality TE sets. The results were largely reproduced (Supplemental Fig. S4D).
In addition, reference bias was another confounding factor that might bias our conclusion. To evaluate the effect of reference bias, we used 9 genome assemblies from 9 populations as a reference to identify polymorphic TEs. The results suggested that TE load accumulation along expansion axes was robust to reference bias, since the TE load pattern was consistent in nearly all cases (except the Spain reference genome) when using reference genomes of different populations (Supplemental Fig. S5). The exception of Spain as reference genome could be resulted from the introgression of Iberian relicts (Lee et al. 2017).
To explore if expansion fronts also accumulated deleterious SNPs (dnSNPs predicted by Provean), we focused on 3 east-west expanded populations with the smallest Ne. Compared to the origin population, the genetic load of Yangtze River basin population, northwestern China and C. Asia population and North America population, are 1.09-fold, 1.02-fold, and 1.004-fold, respectively. The increase indicated that both TEs and deleterious SNPs accumulated at the expansion fronts. However, TE accumulation was much higher (1.17-fold, 1.04-fold, and 1.05-fold TE load in Yangtze River basin population, northwestern China and Central Asia population, and North America population, compared to the origin, respectively).
Nonrelicts were demonstrated to spread mainly along the east-west axis (Lee et al. 2017). Intriguingly, TE load differed between the western and eastern expansion fronts (Fig. 3C). On the western expansion fronts, the reduction in Ne was mild, and the TE load was only slightly higher in the expanded populations. In contrast to the western expansion fronts, the eastern expansion fronts showed a clear expansion trace (Zou et al. 2017; Hsu et al. 2019). Ne decreased with the distance to Balkans, and TE load increased along the expansion axis, reaching the highest at the most eastern expansion front. In subsequent analyses, we focused on gaining in-depth insights into the dynamics and causes of the TE load on the eastern expansion fronts, especially the Yangtze River basin population.
High transposition rate and selective sweeps contribute to high TE load in the Yangtze River basin population
To gain further insight into the TE load variation during range expansion, we focused on the Yangtze River basin population. The load of TEs with different deleterious effects, as predicted in Fig. 2, was compared between the closely related northwestern China and Central Asia populations. Between the 2 populations, the load of TEs with different deleterious effects was much higher in the Yangtze River basin population than in the northwestern China and Central Asia population (Fig. 4A and Supplemental Fig. S6A, P < 0.01, Mann–Whitney U test).
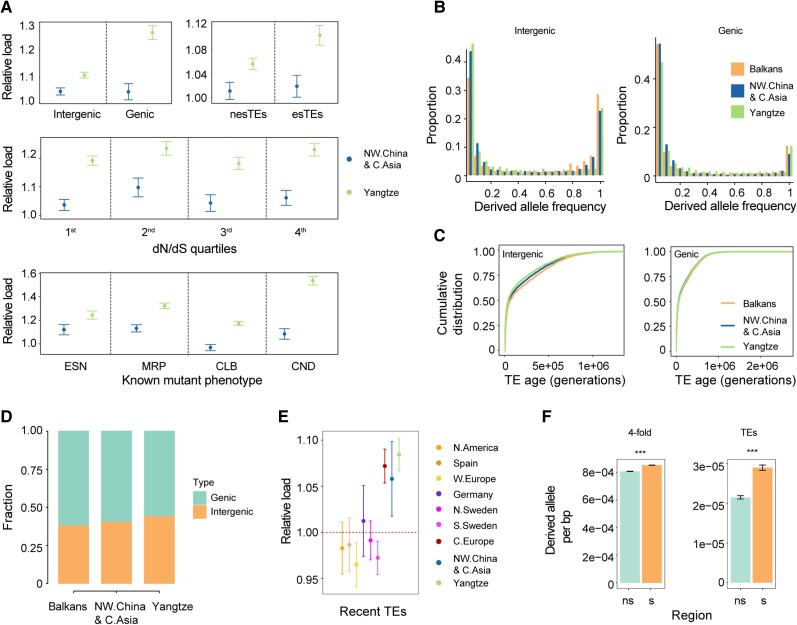
Figure 4.
Causes of high TE load in the Yangtze River basin population. A) Load of TEs with different deleterious effects as classified in Fig. 2. The mean number of derived alleles in each variant category in the Balkans population was used as the standard for calculating the relative load. esTEs, TE insertions can affect gene expression or splicing. nesTEs, TE insertions can’t affect gene expression and splicing. ESN, essential; MRP, morphological; CLB, cellular-biochemical; CND, conditional. Error bars represent 95% confidence intervals. B) Comparison of the derived allele frequency of different TEs among 3 populations. C) Age distribution of TEs with different deleterious effects in the 3 populations. D) Proportion of genic and intergenic TEs in different populations. E) Comparison of recent TE load (top 5% youngest TEs) among nonrelict populations. The mean number of recent TEs in the Balkans population was used as a standard for calculating the relative load. Error bars represent 95% confidence intervals. F) Derived allele counts per base pair in selective sweep regions (s) and nonselective sweep regions (ns) of the Yangtze River basin population. Error bars represent 95% confidence intervals. Mann–Whitney U test was used for significance test. ***P < 0.001.
Range expansion is assumed to cause genetic surfing, a phenomenon where allele frequency increases because of strong genetic drift (Klopfstein et al. 2006). To test the effect of genetic surfing on the accumulation of TEs in the Yangtze River basin population, we compared the SFS of TEs in the Yangtze River basin population with that in the northwestern China and Central Asia and Balkans populations. The Yangtze River basin population showed fewer low-frequency TEs and an excess of high-frequency and fixed TEs in all TE classes, except intergenic TEs, which supported the effect of genetic surfing on the accumulation of TEs in the Yangtze River basin population (Fig. 4B). As theoretically expected (Peischl et al. 2016), genetic surfing was most likely caused by the lowest effective population size of the Yangtze River basin population (Fig. 3D).
The exception that the SFS of intergenic TEs did not show a high-frequency shift compared with the Balkans population and its sister populations implied that other factors might contribute to the higher intergenic TE load in the Yangtze River basin population as well. Given the less efficient purifying selection in the Yangtze River basin population, the excess of rare intergenic TEs in this population suggested that these rare TEs were possibly recent transpositions, indicating increased transposition rate. Intergenic TEs in the Yangtze River basin population were much younger than those in the northwestern China and Central Asia and Balkans populations (Fig. 4C, P < 0.01, Mann–Whitney U test), while genic TEs in the Yangtze River basin population showed a similar age distribution as the northwestern China and Central Asia and Balkans populations (Fig. 4C, P > 0.05, Kolmogorov–Smirnov test). The relative proportion of genic TEs and intergenic TEs also suggested that the Yangtze River basin population had more intergenic TEs than each of the other 2 populations (Fig. 4D, P < 0.01, Chi-square test).
To further support this result, we compared the load of the most recent TEs (top 5% of the age distribution, less than 700 generations), which likely reflect recent mobilization and are less likely under selection, among multiple nonrelict populations. The most recent TE load, both in genic and intergenic regions, in the Yangtze River basin population was also higher than in Balkans and northwestern China and Central Asia population (Fig. 4E and Supplemental Fig. S6B, P < 0. 01, Mann–Whitney U test). Since genic region TE calls have much higher accuracy (Supplemental Fig. S1C) and TE calls in all populations consist of false positives, we conclude that high transposition rate rather than the confounding effect of false positives underlies the high TE load of Yangtze River basin population. The inconsistency between genic TEs and intergenic TEs in SFS and age distribution might result from the stronger purifying selection of genic TEs than intergenic TEs, which could erode recent burst signal.
Hitch-hiking through positive selection has been demonstrated to contribute to the accumulation of deleterious mutations (Hartfield and Otto 2011; Marsden et al. 2016). To test if positive selection contributes to TE accumulation in the Yangtze River basin population, we compared the derived allele count per base-pair in selective sweep regions with that in regions not under selective sweep. Although both the 4-fold degenerate sites and TEs were significantly enriched in selective sweep regions, the enrichment was stronger for TEs (1.35-fold enrichment) than for 4-fold degenerate sites (1.06-fold enrichment) (Fig. 4F). In contrast, in the northwestern China and Central Asia population, at the syntenic region of the selective sweep in the Yangtze River basin population, both the 4-fold degenerate sites and TEs were not enriched (Supplemental Fig. S6C), thus we could rule out the confounding effect of recombination. Because TEs could be highly adaptive to new environments (Niu et al. 2019), it is likely that the enrichment of TEs in selective sweep regions was caused not only by the hitch-hiking effect but also by the TEs themselves as targets of positive selection. Thus, either positive selection or hitch-hiking effect could have contributed to TE accumulation in the Yangtze River basin population.
Genetic architecture of TE expression variation
To dissect the genetic architecture of natural variation in TE load, we focused on 2 stages of the TE transposition process (Fig. 5A): the initiation stage of transcription and the final stage of copy number variation. We first aimed to disentangle the genetic factors responsible for TE expression.
Figure 5.
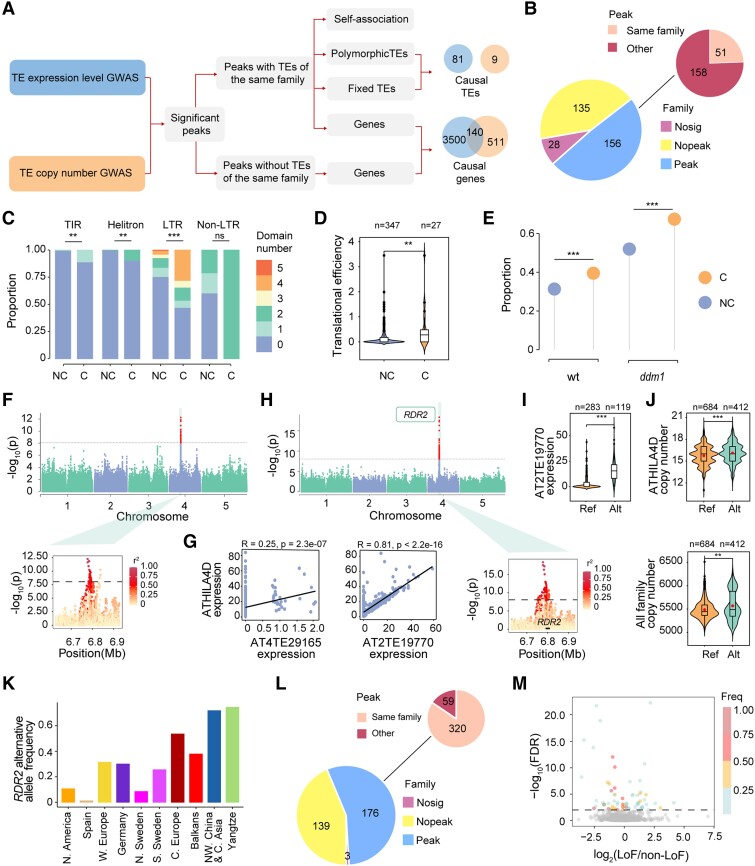
Genetic factors associated with TE family expression and copy number variation. A) Pipeline and strategy used to identify causal loci associated with TE load variation. GWAS, genome-wide association studies. B) Summary of the results of TE family expression level GWAS. Nosig, TE families without significant SNPs; Nopeak, TE families without significant peaks; Peak, TE families with significant peaks; Same family, peaks with TEs of the same family; Other, peaks without TEs of the same family. C) Proportion of TEs containing the transposase domain. TEs were divided into 2 categories: noncausal (NC; TEs of the same family but not in the candidate interval) and causal (C). TIR, terminal inverted repeat; LTR, long terminal repeat. n = 2,252 (TIR, NC), 26 (TIR, C), 3,030 (Helitron, NC), 10 (Helitron, C), 1,056 (LTR, NC), 43 (LTR, C), 286 (non-LTR, NC), 3 (non-LTR, C). Fisher's exact test was used for significance test. ***P < 0.001; **P < 0.01; ns, not significant. D) Translational efficiency of causal TEs. Mann–Whitney U test was used for significance test. E) Proportion of LTR TEs capable of producing transposition intermediates in 2 genotypes: wild type (wt) and ddm1 mutant (ddm1). n = 331 (wt, NC), 17 (wt, C), 549 (ddm1, NC), and 29 (ddm1, C). Fisher's exact test was used for significance test. F) Manhattan plot and local Manhattan plot of GWAS of the ATHILA4D family expression level. The dots above dashed line were significant SNPs. The significant threshold was set based on Bonferroni correction (0.01/number of passed SNPs). G) Pearson correlation between the ATHILA4D family expression level and the expression level of 2 members of this family. H) Manhattan plot and local Manhattan plot of the GWAS of AT2TE19770 expression level. The dots above dashed line were significant SNPs. The significant threshold was set based on Bonferroni correction (0.01/number of passed SNPs). I) Expression levels of the 2 alleles of AT2TE19770. Alleles were grouped based on the lead SNP since the deleterious missense SNPs in RDR2 were in strong LD with the lead SNP (r2 = 0.88). Ref, reference allele; Alt, alternative allele. Mann–Whitney U test was used for significance test. J) Copy number variation of the ATHILA4D family and polymorphic TEs between accessions with reference and nonreference RDR2 alleles. Dots in the box indicate mean copy number. Mann–Whitney U test was used for significance test. K) Frequency of the RDR2 alternative allele in 10 nonrelict populations. L) Summary of TE family copy number GWAS results. M) Differences in TE load related phenotypes (TE family expression level and copy number) between accessions with loss-of-function (LoF) mutations and those without LoF mutations in candidate genes identified by GWAS. Horizontal line indicates 1% FDR. LoF/non-LoF indicates the ratio of TE load related phenotypes in accessions with LoF mutations to those in accessions without LoF mutations in candidate genes. Freq indicates the frequency of LoF alleles in the Yangtze River basin population.
Based on the published high-coverage RNA-seq data of 414 Arabidopsis accessions (including Col-0) (Kawakatsu et al. 2016), we measured TE expression at both the family and locus levels. At the superfamily level, by taking the TE superfamily size into account, the Unassigned superfamilies showed the highest expression level, followed by LTR/Copia, LINE, SINE, and DNA/Harbinger superfamilies (Supplemental Fig. S7A). At the locus level, among the 31,189 TEs in the Col-0 (reference) genome, 16,974 (53.8%) were expressed in at least 1 accession (Supplemental Fig. S7B). A total of 13 TEs were expressed in all 414 accessions. Most TEs (81.1%) were expressed in only a few accessions (frequency < 0.05) (Supplemental Fig. S7C). Analysis of the percentage of expressed TEs of each type revealed that more retrotransposons were transcribed than DNA transposons (including TIR and Helitron) (Supplemental Fig. S7D).
The quantification of TE expression is much more accurate at the family level than at the single locus level because of sequence similarity among TE members within the same family. Therefore, expression level variation among TE families, instead of among the different TE loci, was utilized to identify the causal loci in the subsequent genome-wide association study (GWAS). Among the 319 TE families expressed in at least 1 accession, 291 families showed significant association signals (22,167 significant SNPs) (Fig. 5B). Here, we focused on 209 peaks from the GWAS of 156 TE families, which are usually regarded as strong GWAS signals (Schaid et al. 2018) (Supplemental Data Set 4). Among the 209 peaks, 51 peaks encompassed at least 1 TE belonging to the family under investigation (Fig. 5B, Supplemental Data Set 4). In total, we identified 40 fixed and 41 polymorphic candidate causal TEs in these 51 peaks (Supplemental Data Set 5).
Among the 81 candidate causal TEs, the expression levels of 23 causal TEs were strongly correlated with those of their corresponding families (Pearson's r > 0.75), indicating that these causal TEs were high-confidence causal candidates for the expression variation of their families (Supplemental Data Set 6). The strong expression-level correlation between candidate TEs and their families suggested that most members of these families, except candidate TEs, were nearly not expressed. The association signal might result from TE sequence variation or TE PAV, and in the case of VANDAL families, the anti-silencing factor encoded by TE themselves would be one of the causes (Fu et al. 2013). However, among all VANDAL families, only VANDALNX2 family has members whose expression level are strongly correlated with their family expression level, indicating the scarcity of trans demethylation in our GWAS.
In terms of fixed TEs as causal loci, AT4TE52315, the causal locus of ATCOPIA10 family expression variation, is an example (Supplemental Fig. S8A). The expression level of ATCOPIA10 family (or AT4TE52315) was significantly lower in accessions with the reference allele (alleles were grouped based on the lead SNP) (Supplemental Fig. S8, B and C). By contrast, in cases where polymorphic TEs acted as the causal loci, for some TE families, such as the ATCOPIA70 family, both the TE PAV and sequence variation contributed to its expression variation. We identified AT2TE29450 as the causal locus in the association analysis of ATCOPIA70 family expression level (Supplemental Fig. S9A); accessions with AT2TE29450 showed much higher ATCOPIA70 expression levels than accession without AT2TE29450 (Supplemental Fig. S9B). Additionally, AT2TE29450 was identified again in the association analysis of the ATCOPIA70 family expression level, which was performed using only 348 accessions with AT2TE29450 (Supplemental Fig. S9C). Accessions with the nonreference allele showed higher ATCOPIA70 family and AT2TE29450 expression levels than those with the reference allele (Supplemental Fig. S9D).
To characterize the transposition potential of candidate causal TEs, we compared the transposition potential of causal TEs with that of other TEs belonging to the same family. We divided the TEs into 2 categories: noncausal (NC) (TEs belonging to the same family but not in the peak) and causal. Compared with NC TEs, a higher proportion of causal TEs, especially TIR TEs, LTR TEs, and Helitrons, contained transposition-related domains (Fig. 5C). Furthermore, the translational efficiency of causal TE genes (TEs with gene structure) was significantly higher than that of NC TE genes (Fig. 5D). In addition, we measured the transposition potential of causal TEs using the DNA-seq data of virus-like particles (VLPs), which reflect the capacity of LTR TEs to produce transposition intermediates (Lee et al. 2020). The results showed that a much higher proportion of causal LTR TEs was able to produce transposition intermediates (Fig. 5E). Together, these results suggest that causal TEs have higher transposition potential than other TEs of the same family.
Despite 51 peaks containing TEs of the same family, it is possible that causal loci within some peaks are genes rather than TEs. In these 51 peaks, there are a total of 1,373 genes (Supplemental Data Set 7). For example, in the association analysis of the ATHILA4D family, RNA-DEPENDENT RNA POLYMERASE 2 (RDR2), which functions in RNA-directed DNA methylation (RdDM) pathway by converting single-stranded RNA into double-stranded RNA (Matzke and Mosher 2014), was identified at a peak on chromosome 4 (chr4: 6,777,205) (Fig. 5F). This peak also contained a member of the ATHILA4D family, AT4TE29165; however, AT4TE29165 was silenced in most accessions, and its expression level was weakly correlated with that of the ATHILA4D family (Fig. 5G). Instead, another member of the ATHILA4D family on chromosome 2, AT2TE19770, was highly expressed, and its expression level was strongly correlated with that of the ATHILA4D family (Fig. 5G). We further conducted association analysis of the expression level of AT2TE19770 (unique mapping rate = 0.97) and identified the same peak (chr4: 6,777,205) in the GWAS of the family expression level (Fig. 5H). These results indicate that RDR2, rather than AT4TE29165, is the candidate causal locus of this peak.
The expression level of RDR2 was similar between the 2 alleles grouped by lead SNP. Additionally, 5 significant SNPs were identified in the RDR2 gene, 2 of which were missense mutations, and the missense mutation (chr4: 6,784,172) that occurred in the functionally important C-terminal head domain (Fukudome et al. 2021) was deleterious as predicted by Provean. Accessions with the nonreference allele exhibited a higher expression level of AT2TE19770 (Fig. 5I). In addition, the copy number of the ATHILA4D family and all TE families at the genome level was significantly higher in accessions with the nonreference allele (Fig. 5J). Interestingly, the nonreference allele of RDR2 occurred at high frequency in the Yangtze River basin population but at low frequency in North America population, and potentially contributed to the higher TE expansion in Yangtze River basin population (Fig. 5K). Except for ATHILA4D family, we did not identify RDR2 in the association analysis of other families, unlike the general contribution of CHROMOMETHYLASE 2 (CMT2) and NUCLEAR RNA POLYMERASE D1B (NRPE1) to DNA methylation variation (Sasaki et al. 2019), which only used fixed TEs that will rule out the cis effect. In contrast, here we focused on TE expression, which could be affected by either cis or trans regulation, therefore it is likely that the power of TE expression GWAS to detect trans association is not consistent among different TE families. Nevertheless, the confirmation of causality of RDR2 would benefit from further experimental validation.
A total of 158 peaks did not contain TEs belonging to the same family (Supplemental Data Set 4), and most of the casual loci in these peaks were probably genes. These 158 peaks contained 2,518 genes (Supplemental Data Set 7), including genes involved in epigenetic regulation, such as the DNA demethylation gene DEMETER-LIKE 2 (DML2) (Supplemental Fig. S10). In summary, we identified 81 candidate TEs and 3,640 candidate genes, including 22 well-known TE regulatory genes (Supplemental Data Set 7), from the 209 peaks of GWAS. Overall, these TEs and genes represent the candidate causal loci responsible for natural variation in the TE family expression level.
Genetic architecture of TE copy number variation
To further investigate the mechanism of natural variation in TE load, we performed association analysis and determined the genetic architecture of TE family copy number variation among the 1,115 natural accessions. To exclude the confounding effects of sequencing coverage, we also performed association analysis utilizing 398 accessions with coverage at least 25×. The largely overlapping (70% signals found in 398 accessions were also identified in 1,115 accessions) and additional association signals in GWAS with 1,115 accessions (283 signals) prompted us to use 1,115 accessions to perform GWAS to increase detection power. Among the 318 TE families with copy number variation, 315 showed significant association signals (156,995 significant SNPs) (Fig. 5L). To identify the causal loci, we focused on 176 TE families with a total of 379 significant peaks (Supplemental Data Set 8).
Out of 379 significant peaks, 320 peaks encompassed at least 1 TE belonging to the same TE family as that being studied (Fig. 5L, Supplemental Data Set 8). This association signal might have resulted from differences in the PAV or sequence variation of TEs or from the linkage between TE and the adjacent causal SNP. To identify candidate causal TEs (Fig. 5A), we first excluded 315 peaks with polymorphic TEs belonging to the same family; this strategy was different from that adopted in the association study of TE family expression level. In the TE expression level GWAS, the PAV of actively transcribed TEs could affect the fate of the TE family through transcription and further transposition. However, the association in TE copy number GWAS probably originated from the PAV of TEs (self-associated, the presence of a TE is associated with a single copy number gain) rather than from the real contribution of active TEs (the presence of a TE is associated with multiple copy number gain). The remaining 5 out of 320 peaks contained fixed TEs, which were likely causal TEs (Supplemental Data Set 9). For example, in the GWAS of the ATCOPIA68 family copy number, significant SNPs were detected in AT1TE62960, a member of this family (Supplemental Fig. S11A). Sequence variation of AT1TE62960 potentially contributed to its family copy number variation (Supplemental Fig. S11B). However, these causal TEs did not overlap with causal TEs identified in the family expression level GWAS, mostly probably because we ruled out the self-associated TEs in the copy number analyses, leaving behind only a few candidate causal TEs (Fig. 5A).
Although the 320 peaks contained TEs belonging to the same family, it is possible that the causal elements in some peaks were genes rather than TEs, especially in peaks without candidate causal TEs. There are 88 candidate genes in the 5 peaks that contained fixed TEs of the same family (Supplemental Data Set 10). In the 59 out of 379 peaks that did not contain any TEs belonging to the same family, genes were most likely the causal loci. These 59 peaks harbored a total of 590 genes (Supplemental Data Set 10), 2 of which (DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 [DRD1] and NEEDED FOR RDR2-INDEPENDENT DNA METHYLATION [NERD]) were previously reported to repress TEs (Kanno et al. 2004; Pontier et al. 2012).
In summary, among the 379 peaks of GWAS of TE family copy number variation, 315 peaks were potentially self-associated, and 651 genes (Supplemental Data Set 10) and 9 TEs (Supplemental Data Set 9) were identified in the remaining 64 peaks as candidate causal loci of TE family copy number variation. Intriguingly, 140 candidate genes identified at the copy number variation stage overlapped with those identified at the TE expression stage and were enriched in the salicylic acid signaling pathway (Fig. 5A, Supplemental Data Set 11). The observed number of overlapping genes was significantly higher than the expected number (P < 0.01), implying that some genes or pathways were involved in both TE expression and copy number variation.
LoF mutations in candidate genes are associated with TE family expression and copy number variation
To verify the causality of candidate genes identified by the GWAS of TE expression and copy number, we utilized the natural LoF mutations and determined if the candidate genes contributed to TE load variation. Furthermore, we tested if accessions with LoF mutations in candidate genes differed from those without LoF mutations in TE family expression level or copy number.
In the 4,151 genes identified from the GWAS peaks, 2,363 harbored LoF mutations, of which 864 possessed LoF mutations with minor allele frequency (MAF) > 5%. The accessions with LoF and without LoF alleles for these 864 genes possessed LoF mutations with MAF > 5%, providing the possibility to test the causality of these genes. In total, 61 genes exhibited a significant difference (false discovery rate [FDR] < 1%) in TE family expression level or copy number between accessions with LoF and non-LoF alleles. Among these 61 genes, 30 exhibited elevated TE expression level or copy number; 30 displayed diminished TE expression level or copy number; and 1 showed dual roles, depending on the associated TE family (Fig. 5M, Supplemental Data Set 12).
Discussion
Understanding the genetic load as well as its causes during range expansion are long-standing questions, with important implications for human health (Lynch 2016), crop breeding (Liu et al. 2017; Ramu et al. 2017; Wang et al. 2017), and conservation biology (Kleinman-Ruiz et al. 2022). Although TEs are a major component of the genomes of diverse species, the genetic load of TEs at the genome level in different natural populations remains largely unexplored. Moreover, the types of demographic factors and molecular processes or mechanisms that determine the TE load remain largely unknown.
The present study revealed the TE load variation among natural populations and pointed out the increased TE load at the expanding wave fronts. Although previous studies have demonstrated the general deleterious effect of TE insertions (Baduel et al. 2021a), nonconstant transposition rate and insertion preferences could mimic the effect of purifying selection on shape of SFS. Here we leveraged TE age distribution to control these confounding factors, and confirmed the deleterious effects of TE insertions. More importantly, we further revealed that the deleterious effect of TE insertions varies between that of dnSNPs and LoF mutations. In addition, we adopted 3 approaches for inferring the deleterious effect of different TE insertions, according to their insertion position and capacity of altering gene function and the functional importance of inserted or adjacent genes. These 3 methods enabled us to study the deleterious effects of TE insertions at high resolution in natural populations. Nevertheless, evaluating the fitness effect of each TE insertion is still a big challenge. Methods based on the evolutionary conservation across multispecies alignment, such as genomic evolutionary rate profiling (Davydov et al. 2010), have been frequently used to classify the deleterious extent of base substitutions, which could not be used to evaluate the deleterious extent of TE insertions since TEs evolve fast and lack conservation among species.
More importantly, we revealed several factors that could affect TE load variation. First, Ne was identified as a major contributor of the TE load variation and explained 62.0% of the variation in TE load along western and eastern expansion routes of Arabidopsis populations. Accordingly, in spotted wing Drosophila, TE load is correlated with Ne (R2 = 0.90) (Merel et al. 2021). Apparently, to maintain a relatively large population size, which could reduce the deleterious mutations in natural populations, especially those of endangered species, would be an effective strategy in conservation biology. Second, consistent with a theoretical study, which pointed out that transposition rate also contributes to TE number variation (Charlesworth and Charlesworth 1983), we found that higher transposition rate also contributed to higher TE load in the expanded populations. To infer the mutation rate of TEs in natural populations is difficult because of the purging of deleterious TEs. Here, we used intergenic TEs as well as youngest TEs, which are less likely to undergo selection-mediated purging. This approach revealed that the Yangtze River basin population has a higher transposition rate. Nevertheless, mutation accumulation lines (several generations of manipulated populations) could be used to estimate the mutation rate more accurately in the future (Adrion et al. 2017; Ho et al. 2021). Third, positive selection or hitch-hiking effect could contribute to the accumulation of TEs in the expanded populations. TEs have been demonstrated to be adaptive in diverse species (González et al. 2008; Barron et al. 2014; Chuong et al. 2017; Li et al. 2018; Rech et al. 2019). It will be highly interesting to determine which TEs are under positive selection and which phenotypic traits are affected by these TEs. Nevertheless, the relative contribution of each factor remains to be studied in-depth.
The genetic architecture of TE load has been investigated in both Arabidopsis and Drosophila, based on TE copy number variation in TE families or whole genome (Quadrana et al. 2016; Baduel et al. 2021a; Merel et al. 2021). Here, to investigate the genetic architecture of natural transposition variation, besides considering the result of transposition as in previous studies, we studied both the initiation stage (TE expression) and final stage (TE copy number variation) of the TE transposition process. Transcription marks the initiation stage of TEs, particularly retrotransposons and autonomous transposons. It is crucial to identify determinants of TE expression, which are largely unknown. By contrast, TE copy number variation represents the final result of the TE transposition process. Thus, we addressed TE load variation from 2 different angles and identified candidate TE-regulating genes and active TEs that might affect TE gain and loss rates. We demonstrated that RDR2, identified in the GWAS of TE expression level, was correlated with the total TE number at the genome level in natural populations, and the nonreference allele of RDR2 occurred at high frequency in the Yangtze River basin population and potentially contributed to TE expansion. Particularly, we identified 140 candidate genes at both stages and these genes were enriched in the salicylic acid signaling pathway, indicating the potential role of this pathway in TE regulation.
However, detailed functional analyses are needed to reveal the causal mutations responsible for TE load and epigenetic regulation, which are essential for understanding the mechanism of TE load variation. Besides TE expression level and copy number variation, approaches that focus on the DNA methylation variation of TEs could also identify potential TE load regulators. Based on the DNA methylation level of TEs, as determined in the 1001 Genomes Project of Arabidopsis, the causal genes of DNA methylation variation of TEs have been identified via GWAS (Sasaki et al. 2019, 2022). Nevertheless, the results of our present study and those of previous studies suggest that TE load is associated with many diverse molecular pathways. Similar to more than 7,000 human height-associated genomic segments discovered recently in the GWAS (Yengo et al. 2022), the genomes of natural Arabidopsis populations potentially contain numerous TE load-associated variants.
Nevertheless, there are some issues that should be taken into account in further studies. First, although we performed quality evaluation of short-read sequencing using genome assembly, and excluded the confounding effects of identification quality in different regions; given the repetitive nature of TEs (Baduel et al. 2021b; Rech et al. 2022), similar studies performed using long-read sequencing methods will be beneficial for the study of TE load variation mechanisms. Second, reference bias occurred when using 1 reference genome to map the PAV of TEs. The power to detect TEs in different accessions probably varies with genetic distance or demographic history to the reference genome, such as when Spain accession was used as reference in the present study. Apparently, although we adopted multiple approaches to support our conclusion, similar analysis using genome assemblies of multiple accessions would provide a full landscape of TE load variation. Third, the transposition activity analysis was only based on the Arabidopsis reference genome and its mutants, as these datasets were only available for the reference genome. In-depth comparison of mutants of multiple accessions would benefit such study.
Materials and methods
Plant materials and high-throughput DNA sequencing
The paired-end resequencing data of 1,114 globally distributed nonreference Arabidopsis (Arabidopsis thaliana) accessions were obtained from 4 sources. The 810 accessions were obtained from the 1001 Genomes Project (1001 Genomes Consortium 2016), and those 60 accessions were retrieved from the published data of Africa (Durvasula et al. 2017). Of the 244 accessions collected from China, 116 were sequenced by our laboratory previously (Zou et al. 2017) (Supplemental Data Set 1), and 128 collected from the Yangtze River basin and northwestern China were sequenced in this study (Supplemental Data Set 2). Coverage for each accession was over 10×.
DNA of the 128 Arabidopsis natural accessions sequenced in this study was extracted from leaves with the cetyltrimethylammonium bromide method (Doyle 1987). Paired-end sequencing libraries, with an insert size of approximately 350 bp, were constructed and sequenced on the Illumina HiSeq X Ten platform to generate 150 bp paired-end reads.
The natural accession (5–15) of Yangtze River basin population was sequenced with Pacbio Sequel. Genomic DNA for long-read sequencing was extracted from 3-wk-old rosette leaves using the QIAGEN DNA Midi Kit (Cat. No. 13343).
Genome assembling and scaffolding
Canu (v1.4) (Koren et al. 2017) was used to assemble the genome of 5–15 with default parameters, and genome size was set to 140 Mb. Pilon (Walker et al. 2014) was used to polish the assembly with short reads. RagTag (v.2.0.1) (Alonge et al. 2022) was used to scaffold contigs into chromosomes, using TAIR10 as the reference genome.
Population structure
According to a previous study, the worldwide Arabidopsis includes relicts (accessions that are distantly related to others) and nonrelicts (1001 Genomes Consortium 2016). African accessions that are at least as divergent as relicts defined in the 1001 Genomes Project (Durvasula et al. 2017) were also treated as relicts. Two accessions from southwestern China, which are more divergent from nonrelicts than those of relicts defined in the 1001 Genomes Project, were also treated as relicts (Supplemental Fig. S12A).
Nonrelict accessions from the 1001 Genomes Project were classified into 8 populations and an admixed group, as described previously (1001 Genomes Consortium 2016). Because accessions from North America represent a newly colonized population (Exposito-Alonso et al. 2018), we designated these accessions as the North America population. Additionally, 38 accessions (including 11 accessions sequenced in this study) from northwestern China clustered with the Central Asia population of the 1001 Genomes Project (Supplemental Fig. S12B); together, they were grouped as the northwestern China and Central Asia population. A total of 204 accessions from the Yangtze River basin (117 sequenced in this study) formed a cluster, which was designated as the Yangtze River basin population (Supplemental Fig. S12B). In total, all these 1,114 nonreference accessions were grouped into 1 relict, 10 nonrelict populations, and an admixed group.
TE identification and validation
TEPID (Stuart et al. 2016) was employed for the detection of polymorphic TEs in 1,114 nonreference Arabidopsis accessions. Using the TE annotation of Col-0 (TAIR10) as a reference, the TE presence/absence calls for each accession were determined with tepid-map and tepid-discover algorithm, and the tepid-refine algorithm was further used to reduce false negative calls.
To evaluate the identification accuracy of TE polymorphisms, we utilized the genome assembly of 8 accessions from 8 populations (Supplemental Data Set 3) (Jaegle et al. 2023; Wlodzimierz et al. 2023). Whole-genome alignment between Col-0 (TAIR10) and the 8 assemblies was performed with AnchorWave (Song et al. 2022) and used to check if the identified TE PAV are real PAV.
TE feature analyses
GEVA, which relies on sequence divergence around a TE site, was used to estimate the age of polymorphic TEs with the parameters “–Ne 300000” and “–mut 7e-9” (Albers and McVean 2020), and the mean of the composite posterior distribution under joint model was used as the age estimates for a given TE site. Full-length LTR TEs were annotated using LTRpred (Drost 2020) in Col-0, and LTR similarity was calculated by LTRpred. Conserved domain search (CD-Search) (Lu et al. 2020) was used to search for TE transposition-related domains; a transposase for TIR transposons; replication protein A and helicase for Helitrons; reverse transcriptase and endonuclease for LINEs; and GAG protein, aspartic proteinase, reverse transcriptase (RT), RNaseH, and integrase for LTR TEs (Wicker et al. 2007). The expression potential of TEs was measured using the published TE-transcript annotation data of Col-0 (TAIR10) (Panda and Slotkin 2020), which was produced using TE-activated mutants. In the TE-transcript annotation, TEs were grouped into 3 categories (“Expressed and Annotated”, “Low expressed”, “.”) based on transcript abundance. The “Expressed and Annotated” and “Low expressed” categories were supported by reads, and reads were more abundant in “Expressed and Annotated” category than in the “Low expressed” category, while the “.” category had no read support. DNA methylation data were obtained from a previous study (Kawakatsu et al. 2016), and the weighted methylation level of TEs was calculated as described previously (Schultz et al. 2012).
Evaluation of the deleterious effect of TE insertions
To determine the derived allele frequency spectrum of 4-fold degenerate sites and deleterious mutations, only SNP sites with missing rate < 10% were used. SNPs and indels were called using the genome analysis toolkit (GATK) pipeline (GATK v2.1.8) (DePristo et al. 2011) and annotated with SnpEff (version 4.3t) (Cingolani et al. 2012). Provean (Choi et al. 2012) was used to predict the deleterious effect of nSNPs against the NCBI nonredundant protein database. The nSNPs with score of Provean analysis ≤ −2.5 were defined as deleterious (dnSNPs), and those with score > −2.5 were defined as tolerated (tnSNPs). The LoF mutations (including stop-gain, splice site, and frameshift) were identified and filtered as described previously (Xu et al. 2019). However, 3 or more frameshift mutations found in the same gene of an accession were not excluded in the filtering step; only the frameshift mutations that restored the reading frame were filtered out. Ancestral state inference was based on the genome sequence alignment of Col-0 and its 2 close outgroups, Arabidopsis lyrata (MN47) and Capsella rubella (MTE), using LASTZ (Harris 2007). Alleles that matched the 2 outgroups were defined as ancestral alleles. Alleles different from the alleles of 2 outgroups, which were identical, were defined as derived alleles.
To determine the derived allele frequency spectrum of TEs, only polymorphic TE sites with missing rate < 10% were used. The ancestral state inference of TEs was based on the genome sequence alignment of Col-0, A. lyrata, and C. rubella using AnchorWave (Song et al. 2022), which is more sensitive in making TE presence/absence calls than LASTZ. TEs absent in the 2 outgroups but present in Arabidopsis were defined as derived.
To control the variable transposition rate of TEs, TEs were grouped into 10 equally sized bins according to their age. The SFS of TEs relative to 4-fold degenerate sites were plotted, as previously described (Horvath et al. 2022). GEVA was used to estimate the age of SNPs with the parameters “–Ne 300000” and “–mut 7e-9” (Albers and McVean 2020), and the mean of the composite posterior distribution under joint model was used as the age estimates for a given SNP site.
To study the effect of TE insertions on gene expression, the normalized transcriptome data of 413 accessions were obtained from a previous study (Kawakatsu et al. 2016). To calculate fold-change in the expression level of each gene with polymorphic TEs in or nearby, the gene expression level in accessions with TEs was normalized relative to that in accessions without TEs.
To study the effect of TE insertions on alternative splicing (AS), the transcript model of Col-0 was downloaded from TAIR (Araport11). SUPPA2 (Trincado et al. 2018) was used to identify 7 types of AS events (skipping exon [SE], alternative 5′ splice sites [A5], alternative 3′ splice sites [A3], mutually exclusive exons [MX], retained introns [RI], alternative first exons [AF], alternative last exons [AL]). The transcripts per million (TPM) value of each transcript was measured with TopHat (Trapnell et al. 2009) and Salmon (Patro et al. 2017), and the RNA-seq data obtained from a previous study (Kawakatsu et al. 2016) were used to calculate the TPM value of each transcript in 413 accessions. The PSI value of each AS event was calculated with SUPPA2 based on the TPM value of each accession. The PSI value indicates splicing efficacy; the larger the PSI value, the lower the splicing efficacy. To calculate fold-change in the PSI value of each gene with polymorphic TEs in or nearby, the PSI value in accessions with TEs was normalized relative to that in accessions without TEs.
The importance of gene function was measured based on sequence conservation and known mutant phenotypes. To evaluate gene sequence conservation, orthologous genes between Arabidopsis and A. lyrata were identified using MCScanX (Wang et al. 2012). The dN/dS ratios of the orthologous genes of Arabidopsis and A. lyrata were calculated with KaKs_calculator 3.0 (Zhang 2022). Genes were categorized into 4 bins, according to the dN/dS quartiles; the first quartile was the most constrained, and the 4th quartile was the least conserved. The mutant phenotypes of 2,400 genes were grouped into 4 categories (essential, morphological, cellular-biochemical, and conditional) according to their effect (Lloyd and Meinke 2012).
Evaluation of the synergistic epistasis of TEs and SNPs
To evaluate the synergistic epistasis of deleterious TE insertions, only TEs with frequency lower than 1%, which are more likely to be deleterious, were used. TEs in pericentromeric regions, which were determined based on genome-wide DNA accessibility analysis using DNase I sensitivity assays (Shu et al. 2012), were ruled out because pericentromeric regions have extensive LD and would likely induce a bias. PLINK (v1.90b4) (Purcell et al. 2007) was used to calculate the correlation coefficient (r) of each TE pair, and the raw value of LD was back-calculated using the equation below:
where pi and pj are the frequency of TEs i and j, respectively.
Each TE pair was categorized according to its physical distance (on the same chromosome or on different chromosomes) and deleterious effect, and the mean LD of TE pairs in each category was calculated. At least 5 TE pairs were required to be included in each category.
Similar to the LD analysis of TEs, LD analysis was performed using rare SNPs (frequency < 1%) with different deleterious effects (4-fold degenerate sites, tnSNPs, dnSNPs, and LoF). Each SNP pair was categorized according to its physical distance and deleterious effect, and the mean LD of SNP pairs in each category was calculated. At least 5 SNP pairs were required to be included in each category.
Geographical distance, genetic diversity, and TE load calculation
The Haversine distance of each accession to the putative nonrelict origin predicted previously was calculated using the “geosphere” package of R, and the mean distance of all accessions within a population was used as the distance of this population to the origin. Genetic diversity (π) was calculated using VCFtools (Danecek et al. 2011) in nonoverlapping 10 kb windows for each population. To estimate the TE load, only polymorphic TE sites with missing rate <10% were used. Derived polymorphic TE counts were used as load proxies and compared among 10 nonrelict populations. Derived allele counts of dnSNPs predicted by Provean were used as load proxies of the genetic load of SNPs.
To evaluate the influence of sequencing coverage on TE PAV detection, we randomly selected 20 accessions from different populations and with different coverages. For each accession, we down-sampled the coverage at a 10× step, as previously described (Stritt et al. 2018), and detected TE presence and absence at each step. TE load of accessions with enough coverage (no less than 25×) was then calculated to evaluate the influence of coverage on TE load.
To evaluate the influence of reference bias on TE PAV detection, 9 accessions from 9 populations, with assembled genomes (Supplemental Data Set 3, including accession Sha (Jiao and Schneeberger 2020) from Central Asia population), were separately used as the reference genome to detect TE load in each population. For each population, 2 accessions with similar coverage (30 to 35×) were randomly selected and mapped to each of the 9 genomes. EDTA (Ou et al. 2019) was used to annotate TEs in each of the 9 genomes. TE PAV detection was performed with TEPID (Stuart et al. 2016).
Selective sweep region identification
OmegaPlus (version 3.0.3) (Alachiotis et al. 2012) was used to identify selective sweep regions. OmegaPlus is based on LD, and the ω statistic was computed at 10 kb intervals with the parameters “-minwin 10000” and “-maxwin 100000”. The top 5% regions with high ω values were defined as selective sweep regions. To test the enrichment of TEs in selective sweep regions, the derived allele counts per base-pair of 4-fold degenerate sites and TE sites were compared between selective sweep and nonsweep regions. Pericentromeric regions were removed from sweep and nonsweep regions when the derived allele count per base-pair was calculated since these regions exhibit high TE density and low gene density, which could bias the results.
TE expression analysis
Because TEs are highly repetitive in nature, exhibit polymorphic insertions, and display cotranscription with genes, the quantification of TE expression using short-read data has been a challenging task for a long time. Recently, many tools have been developed to quantify TE expression using RNA-seq data, at both the family and locus levels (Lanciano and Cristofari 2020). However, accurate counting at the locus level is still challenging because of ambiguous mapping, especially for young TEs. Therefore, we focused on quantifying TE family expression using a modified version of TEtranscripts pipeline (Jin et al. 2015; Chung et al. 2019), which largely excludes the transcripts that cotranscribed with genes.
High-coverage RNA-seq data of 414 natural Arabidopsis accessions (including Col-0) were obtained from a previous study (Kawakatsu et al. 2016). RNA-seq reads were mapped to TAIR10 using Spliced Transcripts Alignment to a Reference (STAR) software (Dobin et al. 2013) with the parameters “–winAnchorMultimapNmax 100 –outFilterMultimapNmax 100”. A modified version of TEtranscripts (Jin et al. 2015; Chung et al. 2019) was used to quantify TE expression level. Two output files (multiple mapping and unique mapping) for each TE family and individual TEs were generated with the parameters “–mode multi” and “–mode uniq”, respectively. In the “uniq” mode, only reads uniquely mapped to TEs were counted, and in the “multi” mode, all reads mapped to TEs were counted. The multiple mapping file was used to measure the transcription level of each TE family, and the unique mapping file was used to quantify the transcripts of each individual TE. The scaling factor for each sample was calculated using edgeR (Robinson et al. 2010) with the relative log expression method and was then used to normalize the expression data of each TE family and individual TE. To compare the expression of 18 TE superfamilies, the expression level of each superfamily was further normalized relative to the corresponding superfamily size. The unique mapping rate of each TE was calculated based on raw read counts and defined as the percentage of uniquely mapped reads relative to all mapped reads.
GWAS analysis
Biallelic SNPs with MAF > 5% and missing rate < 10% were used in GWAS analysis. In the TE family expression GWAS analysis, expression levels of 319 expressed TE families were used as phenotypes. In the TE family copy number GWAS analysis, the copy number of 318 TE families with polymorphic TEs was used as phenotypes. GWAS was performed with efficient mixed-model association expedited (EMMAX) software (linear mixed model) for each phenotype using principal components and kinship matrix to control for population structure (Kang et al. 2010). Significant thresholds were set based on Bonferroni correction (0.01/number of passed SNPs); candidate interval was determined based on the lead SNP of a peak and the SNPs linked to the lead SNP (r2 > 0.2), and pairwise LD was calculated using PLINK (v1.90b4) (Purcell et al. 2007). The candidate intervals were then intersected with gene annotation (Araport11), reference TE annotation (TAIR10), and polymorphic TEs to identify the causal element. Gene ontology enrichment analysis was conducted with agriGO (Tian et al. 2017).
Analysis of the transposition potential of candidate causal TEs
To estimate the capacity of TEs to produce transposition intermediates, the Oxford Nanopore Technology (ONT) long-read VLP DNA-seq data of Col-0 were retrieved from a previous study (Lee et al. 2020). The long-read data of 2 genotypes (wild-type of Col-0 and ddm1 mutant) were mapped to the reference genome (TAIR10) using Minimap2 (Li 2018). The number of reads mapped to annotated TEs (TAIR10) was calculated using featureCounts (Liao et al. 2014). LTR TEs with at least 1 read were regarded as LTR TEs capable of producing transposition intermediates. To calculate the translational efficiency of TE genes, polysomal RNA-seq and RNA-seq data of the ddm1 mutant (Col-0 background) were retrieved from a previous study (Lee et al. 2020). The translational efficiency of TE genes (Araport 11) was calculated as the ratio of the fragments per kilobase per million mapped fragments value of polysomal RNA to that of total RNA.
Analysis of LoF mutations in candidate genes
To validate the causality of candidate genes identified by GWAS, we utilized the natural LoF mutations of these genes and determined if their respective phenotypes (expression level or copy number) differed with the PAV of LoF allele. Among the LoF alleles with MAF > 5%, Mann–Whitney U test was used to test the difference in TE expression level or copy number between LoF and non-LoF alleles. After multiple test correction, genes with FDR < 0.01 were defined as significant.
Statistical analyses
All statistical analyses were performed in R (http://www.r-project.org/) (Supplemental Data Set 13).
Accession numbers
The raw sequence data and genome assembly reported in this paper have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation (Genome Sequence Archive: CRA008569, GWHDRII00000000) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
The accession numbers of published data used in the study are listed below. Resequencing data of published accessions were obtained from NCBI SRP056687, European Nucleotide Archive PRJEB19780 and NCBI SRP062811. DNA methylation data were retrieved from NCBI GSE43857. RNA-seq data of 414 accessions were retrieved from NCBI GSE80744. VLP DNA-seq data, polysomal RNA-seq and RNA-seq data of Col-0 and mutant were obtained from NCBI GSE128932.
Supplementary Material
Acknowledgments
We would like to thank Magnus Nordborg (Gregor Mendel Institute of Molecular Plant Biology), Fei Lu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences), Jinfeng Chen (Institute of Zoology, Chinese Academy of Sciences) and Fu-Min Zhang (Institute of Botany, Chinese Academy of Sciences) for helpful suggestions about the study. Especially, we thank the anonymous reviewers for their valuable comments that improves our manuscript a lot.
Contributor Information
Juan Jiang, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Yong-Chao Xu, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China.
Zhi-Qin Zhang, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Jia-Fu Chen, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Xiao-Min Niu, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China.
Xing-Hui Hou, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China.
Xin-Tong Li, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Li Wang, Agricultural Synthetic Biology Center, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen 518000, China.
Yong E Zhang, College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China; State Key Laboratory of Integrated Management of Pest Insects and Rodents & Key Laboratory of the Zoological Systematics and Evolution, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China.
Song Ge, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Ya-Long Guo, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Author contributions
Y.-L.G. conceived the study; X.-H.H. and X.-M.N. performed the experiments; J.J., Y.-C.X., Z.-Q.Z., J.-F.C., X.-T.L., L.W., Y.E.Z., S.G., and Y.-L.G. analyzed and interpreted the data; J.J. and Y.-L.G. wrote the paper with contribution from all authors.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of polymorphic transposable elements (TEs).
Supplemental Figure S2. Classifying the deleterious extent of transposable element (TE) insertions.
Supplemental Figure S3. Mean linkage disequilibrium (LD) of SNP pairs with different physical distances and located in genes of different functional categories.
Supplemental Figure S4. The influence of sequencing coverage and precision rate on transposable element (TE) load.
Supplemental Figure S5. The influence of reference bias on detected transposable element (TE) number.
Supplemental Figure S6. Causes of transposable element (TE) load accumulation in Yangtze River basin population.
Supplemental Figure S7. Transposable element (TE) expression in different accessions at family level and locus level.
Supplemental Figure S8. Genome-wide association study (GWAS) of ATCOPIA10 family expression level.
Supplemental Figure S9. Genome-wide association study (GWAS) on the expression level of ATCOPIA70 family.
Supplemental Figure S10. Genome-wide association study (GWAS) results of TA12 family expression level.
Supplemental Figure S11. Genome-wide association study (GWAS) results of ATCOPIA68 copy number.
Supplemental Figure S12. Population structure of 1,114 nonreference accessions.
Supplemental Data Set 1. Summary of the published natural accessions used in this study.
Supplemental Data Set 2. Summary of the newly sequenced 128 natural accessions in this study.
Supplemental Data Set 3. Summary of the parameters of transposable element (TE) polymorphism identification accuracy.
Supplemental Data Set 4. Summary of significant peaks in transposable element (TE) family expression level genome-wide association study (GWAS) analysis.
Supplemental Data Set 5. Summary of candidate causal transposable element (TE) associated with TE family expression level variation.
Supplemental Data Set 6. The correlation coefficients of candidate causal transposable element (TE) expression levels and their family expression levels.
Supplemental Data Set 7. Summary of candidate genes associated with transposable element (TE) family expression level variation.
Supplemental Data Set 8. Summary of significant peaks in transposable element (TE) family copy number genome-wide association study (GWAS) analysis.
Supplemental Data Set 9. Summary of causal transposable element (TE) associated with TE family copy number variation.
Supplemental Data Set 10. Summary of candidate genes in the genome-wide association study (GWAS) analysis of transposable element (TE) family copy number.
Supplemental Data Set 11. Summary of the overlapping candidate genes in the two-stage genome-wide association study (GWAS) analysis.
Supplemental Data Set 12. Summary of the candidate genes whose loss-of-function (LoF) mutation is associated with transposable element (TE) expression level or copy number variation.
Supplemental Data Set 13. Summary of the statistic tests used.
Funding
This work was supported by the National Natural Science Foundation of China (31925004) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27010305).
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- 1001 Genomes Consortium . 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016:166(2):481–491. 10.1016/j.cell.2016.05.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrion JR, Song MJ, Schrider DR, Hahn MW, Schaack S. Genome-wide estimates of transposable element insertion and deletion rates in Drosophila melanogaster. Genome Biol Evol. 2017:9(5):1329–1340. 10.1093/gbe/evx050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alachiotis N, Stamatakis A, Pavlidis P. OmegaPlus: a scalable tool for rapid detection of selective sweeps in whole-genome datasets. Bioinformatics. 2012:28(17):2274–2275. 10.1093/bioinformatics/bts419 [DOI] [PubMed] [Google Scholar]
- Albers PK, McVean G. Dating genomic variants and shared ancestry in population-scale sequencing data. PLoS Biol. 2020:18(1):e3000586. 10.1371/journal.pbio.3000586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, Lebeigle L, Kirsche M, Jenike K, Ou S, Aganezov S, Wang X, Lippman ZB, Schatz MC, Soyk S. Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing. Genome Biol. 2022:23(1):258. 10.1186/s13059-022-02823-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baduel P, Leduque B, Ignace A, Gy I, Gil J Jr, Loudet O, Colot V, Quadrana L. Genetic and environmental modulation of transposition shapes the evolutionary potential of Arabidopsis thaliana. Genome Biol. 2021a:22(1):138. 10.1186/s13059-021-02348-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baduel P, Quadrana L, Colot V. Efficient detection of transposable element insertion polymorphisms between genomes using short-read sequencing data. Methods Mol Biol. 2021b:2250:157–169. 10.1007/978-1-0716-1134-0_15 [DOI] [PubMed] [Google Scholar]
- Baduel P, Quadrana L, Hunter B, Bomblies K, Colot V. Relaxed purifying selection in autopolyploids drives transposable element over-accumulation which provides variants for local adaptation. Nat Commun. 2019:10(1):5818. 10.1038/s41467-019-13730-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron MG, Fiston-Lavier A-S, Petrov DA, Gonzalez J. Population genomics of transposable elements in Drosophila. Annu Rev Genet. 2014:48(1):561–581. 10.1146/annurev-genet-120213-092359 [DOI] [PubMed] [Google Scholar]
- Bergman CM, Bensasson D. Recent LTR retrotransposon insertion contrasts with waves of non-LTR insertion since speciation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007:104(27):11340–11345. 10.1073/pnas.0702552104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertorelle G, Raffini F, Bosse M, Bortoluzzi C, Iannucci A, Trucchi E, Morales HE, van Oosterhout C. Genetic load: genomic estimates and applications in non-model animals. Nat Rev Genet. 2022:23(8):492–503. 10.1038/s41576-022-00448-x [DOI] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011:43(10):956–963. 10.1038/ng.911 [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Transposable elements in natural populations with a mixture of selected and neutral insertion sites. Genet Res. 1991:57(2):127–134. 10.1017/S0016672300029190 [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009:10(3):195–205. 10.1038/nrg2526 [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The population dynamics of transposable elements. Genet Res. 1983:42(1):1–27. 10.1017/S0016672300021455 [DOI] [Google Scholar]
- Chen J, Lu L, Robb SMC, Collin M, Okumoto Y, Stajich JE, Wessler SR. Genomic diversity generated by a transposable element burst in a rice recombinant inbred population. Proc Natl Acad Sci U S A. 2020:117(42):26288–26297. 10.1073/pnas.2015736117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012:7(10):e46688. 10.1371/journal.pone.0046688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung N, Jonaid GM, Quinton S, Ross A, Sexton CE, Alberto A, Clymer C, Churchill D, Navarro Leija O, Han MV. Transcriptome analyses of tumor-adjacent somatic tissues reveal genes co-expressed with transposable elements. Mob DNA. 2019:10(1):39. 10.1186/s13100-019-0180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 2017:18(2):71–86. 10.1038/nrg.2016.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012:6(2):80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011:27(15):2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010:6(12):e1001025. 10.1371/journal.pcbi.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011:43(5):491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013:29(1):15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987:19:11–15. [Google Scholar]
- Drost H-G. LTRpred: de novo annotation of intact retrotransposons. The Journal of Open Source Software. 2020:5(50):2170. 10.21105/joss.02170 [DOI] [Google Scholar]
- Durvasula A, Fulgione A, Gutaker RM, Alacakaptan SI, Flood PJ, Neto C, Tsuchimatsu T, Burbano HA, Pico FX, Alonso-Blanco C, et al. African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2017:114(20):5213–5218. 10.1073/pnas.1616736114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Alonso M, Becker C, Schuenemann VJ, Reiter E, Setzer C, Slovak R, Brachi B, Hagmann J, Grimm DG, Chen J, et al. The rate and potential relevance of new mutations in a colonizing plant lineage. PLoS Genet. 2018:14(2):e1007155. 10.1371/journal.pgen.1007155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxe JP, Slotte T, Stahl EA, Neuffer B, Hurka H, Wright SI. Recent speciation associated with the evolution of selfing in Capsella. Proc Natl Acad Sci U S A. 2009:106(13):5241–5245. 10.1073/pnas.0807679106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Kawabe A, Etcheverry M, Ito T, Toyoda A, Fujiyama A, Colot V, Tarutani Y, Kakutani T. Mobilization of a plant transposon by expression of the transposon-encoded anti-silencing factor. EMBO J. 2013:32(17):2407–2417. 10.1038/emboj.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukudome A, Singh J, Mishra V, Reddem E, Martinez-Marquez F, Wenzel S, Yan R, Shiozaki M, Yu Z, Wang JC-Y, et al. Structure and RNA template requirements of Arabidopsis RNA-DEPENDENT RNA POLYMERASE 2. Proc Natl Acad Sci U S A. 2021:118(51):e2115899118. 10.1073/pnas.2115899118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J, Lenkov K, Lipatov M, Macpherson JM, Petrov DA. High rate of recent transposable element-induced adaptation in Drosophila melanogaster. PLoS Biol. 2008:6(10):e251. 10.1371/journal.pbio.0060251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y-L, Bechsgaard JS, Slotte T, Neuffer B, Lascoux M, Weigel D, Schierup MH. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc Natl Acad Sci U S A. 2009:106(13):5246–5251. 10.1073/pnas.0808012106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS. Improved pairwise alignment of genomic DNA [PhD thesis]. The Pennsylvania State University; 2007.
- Hartfield M, Otto SP. Recombination and hitchhiking of deleterious alleles. Evolution. 2011:65(9):2421–2434. 10.1111/j.1558-5646.2011.01311.x [DOI] [PubMed] [Google Scholar]
- He L, Huang H, Bradai M, Zhao C, You Y, Ma J, Zhao L, Lozano-Duran R, Zhu J-K. DNA methylation-free Arabidopsis reveals crucial roles of DNA methylation in regulating gene expression and development. Nat Commun. 2022:13(1):1335. 10.1038/s41467-022-28940-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Botigue LR, Peischl S, Dupanloup I, Lipatov M, Maples BK, Martin AR, Musharoff S, Cann H, Snyder MP, et al. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci U S A. 2016:113(4):E440–E449. 10.1073/pnas.1510805112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MS, Vande Zande P, Wittkopp PJ. Molecular and evolutionary processes generating variation in gene expression. Nat Rev Genet. 2021:22(4):203–215. 10.1038/s41576-020-00304-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T, Schlotterer C, Betancourt AJ. Hybrid dysgenesis in Drosophila simulans associated with a rapid invasion of the P-element. PLoS Genet. 2016:12(3):e1005920. 10.1371/journal.pgen.1005920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho EKH, Bellis ES, Calkins J, Adrion JR, Latta LC IV, Schaack S. Engines of change: transposable element mutation rates are high and variable within Daphnia magna. PLoS Genet. 2021:17(11):e1009827. 10.1371/journal.pgen.1009827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath R, Menon M, Stitzer M, Ross-Ibarra J. Controlling for variable transposition rate with an age-adjusted site frequency spectrum. Genome Biol Evol. 2022:14(2):evac016. 10.1093/gbe/evac016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C-W, Lo C-Y, Lee C-R. On the postglacial spread of human commensal Arabidopsis thaliana: journey to the east. New Phytol. 2019:222(3):1447–1457. 10.1111/nph.15682 [DOI] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng J-F, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011:43(5):476–481. 10.1038/ng.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaegle B, Pisupati R, Soto-Jimenez LM, Burns R, Rabanal FA, Nordborg M. Extensive sequence duplication in Arabidopsis revealed by pseudo-heterozygosity. Genome Biol. 2023:24(1):44. 10.1186/s13059-023-02875-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao WB, Schneeberger K. Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat Commun. 2020:11(1):989. 10.1038/s41467-020-14779-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Tam OH, Paniagua E, Hammell M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics. 2015:31(22):3593–3599. 10.1093/bioinformatics/btv422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010:42(4):348–354. 10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJM. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004:14(9):801–805. 10.1016/j.cub.2004.04.037 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Huang S-C, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y, et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell. 2016:166(2):492–505. 10.1016/j.cell.2016.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A, Pulit SL, Francioli LC, van Dijk F, Swertz M, Boomsma DI, van Duijn CM, Slagboom PE, van Ommen GJ, Wijmenga C, et al. Deleterious alleles in the human genome are on average younger than neutral alleles of the same frequency. PLoS Genet. 2013:9(2):e1003301. 10.1371/journal.pgen.1003301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman-Ruiz D, Lucena-Perez M, Villanueva B, Fernandez J, Saveljev AP, Ratkiewicz M, Schmidt K, Galtier N, Garcia-Dorado A, Godoy JA. Purging of deleterious burden in the endangered Iberian lynx. Proc Natl Acad Sci U S A. 2022:119(11):e2110614119. 10.1073/pnas.2110614119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfstein S, Currat M, Excoffier L. The fate of mutations surfing on the wave of a range expansion. Mol Biol Evol. 2006:23(3):482–490. 10.1093/molbev/msj057 [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017:27(5):722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Momozawa Y, Liu X, Terao C, Kubo M, Kamatani Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019:20(1):117. 10.1186/s13059-019-1720-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciano S, Cristofari G. Measuring and interpreting transposable element expression. Nat Rev Genet. 2020:21(12):721–736. 10.1038/s41576-020-0251-y [DOI] [PubMed] [Google Scholar]
- Larracuente AM, Sackton TB, Greenberg AJ, Wong A, Singh ND, Sturgill D, Zhang Y, Oliver B, Clark AG. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008:24(3):114–123. 10.1016/j.tig.2007.12.001 [DOI] [PubMed] [Google Scholar]
- Lee CR, Svardal H, Farlow A, Exposito-Alonso M, Ding W, Novikova P, Alonso-Blanco C, Weigel D, Nordborg M. On the post-glacial spread of human commensal Arabidopsis thaliana. Nat Commun. 2017:8(1):14458. 10.1038/ncomms14458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Ernst E, Berube B, Borges F, Parent J-S, Ledon P, Schorn A, Martienssen RA. Arabidopsis retrotransposon virus-like particles and their regulation by epigenetically activated small RNA. Genome Res. 2020:30(4):576–588. 10.1101/gr.259044.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YCG. Synergistic epistasis of the deleterious effects of transposable elements. Genetics. 2022:220(2):iyab211. 10.1093/genetics/iyab211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018:34(18):3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z-W, Hou X-H, Chen J-F, Xu Y-C, Wu Q, González J, Guo Y-L. Transposable elements contribute to the adaptation of Arabidopsis thaliana. Genome Biol Evol. 2018:10(8):2140–2150. 10.1093/gbe/evy171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014:30(7):923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lisch D. How important are transposons for plant evolution? Nat Rev Genet. 2013:14(1):49–61. 10.1038/nrg3374 [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhou Y, Morrell PL, Gaut BS. Deleterious variants in Asian rice and the potential cost of domestication. Mol Biol Evol. 2017:34(4):908–924. 10.1093/molbev/msw296 [DOI] [PubMed] [Google Scholar]
- Lloyd J, Meinke D. A comprehensive dataset of genes with a loss-of-function mutant phenotype in Arabidopsis. Plant Physiol. 2012:158(3):1115–1129. 10.1104/pp.111.192393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockton S, Ross-Ibarra J, Gaut BS. Demography and weak selection drive patterns of transposable element diversity in natural populations of Arabidopsis lyrata. Proc Natl Acad Sci U S A. 2008:105(37):13965–13970. 10.1073/pnas.0804671105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, Hubisz MJ, Sninsky JJ, White TJ, Sunyaev SR, Nielsen R, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008:451(7181):994–997. 10.1038/nature06611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, Platzer A, Zhang Q, Vilhjalmsson BJ, Korte A, Nizhynska V, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 2013:45(8):884–890. 10.1038/ng.2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020:48(D1):D265–D268. 10.1093/nar/gkz991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Mutation and human exceptionalism: our future genetic load. Genetics. 2016:202(3):869–875. 10.1534/genetics.115.180471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003:302(5649):1401–1404. 10.1126/science.1089370 [DOI] [PubMed] [Google Scholar]
- Marsden CD, Ortega-Del Vecchyo D, O'Brien DP, Taylor JF, Ramirez O, Vila C, Marques-Bonet T, Schnabel RD, Wayne RK, Lohmueller KE. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc Natl Acad Sci U S A. 2016:113(1):152–157. 10.1073/pnas.1512501113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014:15(6):394–408. 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- Merel V, Gibert P, Buch I, Rodriguez Rada V, Estoup A, Gautier M, Fablet M, Boulesteix M, Vieira C. The worldwide invasion of Drosophila suzukii is accompanied by a large increase of transposable element load and a small number of putatively adaptive insertions. Mol Biol Evol. 2021:38(10):4252–4267. 10.1093/molbev/msab155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X-M, Xu Y-C, Li Z-W, Bian Y-T, Hou X-H, Chen J-F, Zou Y-P, Jiang J, Wu Q, Ge S, et al. Transposable elements drive rapid phenotypic variation in Capsella rubella. Proc Natl Acad Sci U S A. 2019:116(14):6908–6913. 10.1073/pnas.1811498116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, Lugo CSB, Elliott TA, Ware D, Peterson T, et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 2019:20(1):275. 10.1186/s13059-019-1905-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda K, Slotkin RK. Long-read cDNA sequencing enables a “gene-like” transcript annotation of transposable elements. Plant Cell. 2020:32(9):2687–2698. 10.1105/tpc.20.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasyukova EG, Nuzhdin SV, Morozova TV, Mackay TFC. Accumulation of transposable elements in the genome of Drosophila melanogaster is associated with a decrease in fitness. J Hered. 2004:95(4):284–290. 10.1093/jhered/esh050 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017:14(4):417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peischl S, Dupanloup I, Bosshard L, Excoffier L. Genetic surfing in human populations: from genes to genomes. Curr Opin Genet Dev. 2016:41:53–61. 10.1016/j.gde.2016.08.003 [DOI] [PubMed] [Google Scholar]
- Peischl S, Dupanloup I, Kirkpatrick M, Excoffier L. On the accumulation of deleterious mutations during range expansions. Mol Ecol. 2013:22(24):5972–5982. 10.1111/mec.12524 [DOI] [PubMed] [Google Scholar]
- Pontier D, Picart C, Roudier F, Garcia D, Lahmy S, Azevedo J, Alart E, Laudie M, Karlowski WM, Cooke R, et al. NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in Arabidopsis. Mol Cell. 2012:48(1):121–132. 10.1016/j.molcel.2012.07.027 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007:81(3):559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Bortolini Silveira A, Mayhew GF, LeBlanc C, Martienssen RA, Jeddeloh JA, Colot V. The Arabidopsis thaliana mobilome and its impact at the species level. Elife. 2016:5:15716. 10.7554/eLife.15716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Etcheverry M, Gilly A, Caillieux E, Madoui MA, Guy J, Bortolini Silveira A, Engelen S, Baillet V, Wincker P, et al. Transposition favors the generation of large effect mutations that may facilitate rapid adaption. Nat Commun. 2019:10(1):3421. 10.1038/s41467-019-11385-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu P, Esuma W, Kawuki R, Rabbi IY, Egesi C, Bredeson JV, Bart RS, Verma J, Buckler ES, Lu F. Cassava haplotype map highlights fixation of deleterious mutations during clonal propagation. Nat Genet. 2017:49(6):959–963. 10.1038/ng.3845 [DOI] [PubMed] [Google Scholar]
- Rech GE, Bogaerts-Marquez M, Barron MG, Merenciano M, Villanueva-Canas JL, Horvath V, Fiston-Lavier AS, Luyten I, Venkataram S, Quesneville H, et al. Stress response, behavior, and development are shaped by transposable element-induced mutations in Drosophila. PLoS Genet. 2019:15(2):e1007900. 10.1371/journal.pgen.1007900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech GE, Radio S, Guirao-Rico S, Aguilera L, Horvath V, Green L, Lindstadt H, Jamilloux V, Quesneville H, Gonzalez J. Population-scale long-read sequencing uncovers transposable elements associated with gene expression variation and adaptive signatures in Drosophila. Nat Commun. 2022:13(1):1948. 10.1038/s41467-022-29518-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010:26(1):139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler G, Wright SI, Agrawal AF. Patterns and causes of signed linkage disequilibria in flies and plants. Mol Biol Evol. 2021:38(10):4310–4321. 10.1093/molbev/msab169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Gunis J, Reichardt-Gomez I, Nizhynska V, Nordborg M. Conditional GWAS of non-CG transposon methylation in Arabidopsis thaliana reveals major polymorphisms in five genes. PLoS Genet. 2022:18(9):e1010345. 10.1371/journal.pgen.1010345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Kawakatsu T, Ecker JR, Nordborg M. Common alleles of CMT2 and NRPE1 are major determinants of CHH methylation variation in Arabidopsis thaliana. PLoS Genet. 2019:15(12):e1008492. 10.1371/journal.pgen.1008492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, Chen W, Larson NB. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet. 2018:19(8):491–504. 10.1038/s41576-018-0016-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MD, Schmitz RJ, Ecker JR. ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 2012:28(12):583–585. 10.1016/j.tig.2012.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirsekar G, Devos J, Latorre SM, Blaha A, Queiroz Dias M, Gonzalez Hernando A, Lundberg DS, Burbano HA, Fenster CB, Weigel D. Multiple sources of introduction of North American Arabidopsis thaliana from across Eurasia. Mol Biol Evol. 2021:38(12):5328–5344. 10.1093/molbev/msab268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu H, Wildhaber T, Siretskiy A, Gruissem W, Hennig L. Distinct modes of DNA accessibility in plant chromatin. Nat Commun. 2012:3:1281. 10.1038/ncomms2259 [DOI] [PubMed] [Google Scholar]
- Song B, Marco-Sola S, Moreto M, Johnson L, Buckler ES, Stitzer MC. AnchorWave: sensitive alignment of genomes with high sequence diversity, extensive structural polymorphism, and whole-genome duplication. Proc Natl Acad Sci U S A. 2022:119(1):e2113075119. 10.1073/pnas.2113075119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritt C, Gordon SP, Wicker T, Vogel JP, Roulin AC. Recent activity in expanding populations and purifying selection have shaped transposable element landscapes across natural accessions of the Mediterranean grass Brachypodium distachyon. Genome Biol Evol. 2018:10(1):304–318. 10.1093/gbe/evx276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Eichten SR, Cahn J, Karpievitch YV, Borevitz JO, Lister R. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. Elife. 2016:5:e20777. 10.7554/eLife.20777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takou M, Hamala T, Koch EM, Steige KA, Dittberner H, Yant L, Genete M, Sunyaev S, Castric V, Vekemans X, et al. Maintenance of adaptive dynamics and no detectable load in a range-edge outcrossing plant population. Mol Biol Evol. 2021:38(5):1820–1836. 10.1093/molbev/msaa322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Hollister JD, Gaut BS. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 2010:15(8):471–478. 10.1016/j.tplants.2010.05.003 [DOI] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017:45(W1):W122–W129. 10.1093/nar/gkx382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009:25(9):1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trincado JL, Entizne JC, Hysenaj G, Singh B, Skalic M, Elliott DJ, Eyras E. SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018:19(1):40. 10.1186/s13059-018-1417-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van't Hof AE, Campagne P, Rigden DJ, Yung CJ, Lingley J, Quail MA, Hall N, Darby AC, Saccheri IJ. The industrial melanism mutation in British peppered moths is a transposable element. Nature. 2016:534(7605):102–105. 10.1038/nature17951 [DOI] [PubMed] [Google Scholar]
- Vendrell-Mir P, Barteri F, Merenciano M, Gonzalez J, Casacuberta JM, Castanera R. A benchmark of transposon insertion detection tools using real data. Mob DNA. 2019:10(1):53. 10.1186/s13100-019-0197-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014:9(11):e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Beissinger TM, Lorant A, Ross-Ibarra C, Ross-Ibarra J, Hufford MB. The interplay of demography and selection during maize domestication and expansion. Genome Biol. 2017:18(1):215. 10.1186/s13059-017-1346-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YP, Tang HB, DeBarry JD, Tan X, Li JP, Wang XY, Lee T-H, Jin HZ, Marler B, Guo H, et al. MCScanx: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012:40(7):e49. 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Zdobnov EM, Kriventseva EV. Correlating traits of gene retention, sequence divergence, duplicability and essentiality in vertebrates, arthropods, and fungi. Genome Biol Evol. 2011:3:75–86. 10.1093/gbe/evq083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Cao X. The effect of transposable elements on phenotypic variation: insights from plants to humans. Sci China Life Sci. 2016:59(1):24–37. 10.1007/s11427-015-4993-2 [DOI] [PubMed] [Google Scholar]
- Wells JN, Feschotte C. A field guide to eukaryotic transposable elements. Annu Rev Genet. 2020:54(1):539–561. 10.1146/annurev-genet-040620-022145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007:8(12):973–982. 10.1038/nrg2165 [DOI] [PubMed] [Google Scholar]
- Williamson S, Fledel-Alon A, Bustamante CD. Population genetics of polymorphism and divergence for diploid selection models with arbitrary dominance. Genetics. 2004:168(1):463–475. 10.1534/genetics.103.024745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodzimierz P, Rabanal FA, Burns R, Naish M, Primetis E, Scott A, Mandáková T, Gorringe N, Tock AJ, Holland D, et al. Cycles of satellite and transposon evolution in Arabidopsis centromeres. Nature. 2023:618(7965):557–565. 10.1038/s41586-023-06062-z [DOI] [PubMed] [Google Scholar]
- Xu Y-C, Niu X-M, Li X-X, He WR, Chen J-F, Zou Y-P, Wu Q, Zhang YE, Busch W, Guo Y-L. Adaptation and phenotypic diversification in Arabidopsis through loss-of-function mutations in protein-coding genes. Plant Cell. 2019:31(5):1012–1025. 10.1105/tpc.18.00791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo L, Vedantam S, Marouli E, Sidorenko J, Bartell E, Sakaue S, Graff M, Eliasen AU, Jiang Y, Raghavan S, et al. A saturated map of common genetic variants associated with human height. Nature. 2022:610(7933):704–712. 10.1038/s41586-022-05275-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. KaKs_calculator 3.0: calculating selective pressure on coding and non-coding sequences. Genomics Proteomics Bioinformatics. 2022:20(3):536–540. 10.1016/j.gpb.2021.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y-P, Hou X-H, Wu Q, Chen J-F, Li Z-W, Han T-S, Niu X-M, Yang L, Xu Y-C, Zhang J, et al. Adaptation of Arabidopsis thaliana to the Yangtze River basin. Genome Biol. 2017:18(1):239. 10.1186/s13059-017-1378-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.