Abstract
The gynoecium is critical for the reproduction of flowering plants as it contains the ovules and the tissues that foster pollen germination, growth, and guidance. These tissues, known as the reproductive tract (ReT), comprise the stigma, style, and transmitting tract (TT). The ReT and ovules originate from the carpel margin meristem (CMM) within the pistil. SHOOT MERISTEMLESS (STM) is a key transcription factor for meristem formation and maintenance. In all above-ground meristems, including the CMM, local STM downregulation is required for organ formation. However, how this downregulation is achieved in the CMM is unknown. Here, we have studied the role of HISTONE DEACETYLASE 19 (HDA19) in Arabidopsis (Arabidopsis thaliana) during ovule and ReT differentiation based on the observation that the hda19-3 mutant displays a reduced ovule number and fails to differentiate the TT properly. Fluorescence-activated cell sorting coupled with RNA-sequencing revealed that in the CMM of hda19-3 mutants, genes promoting organ development are downregulated while meristematic markers, including STM, are upregulated. HDA19 was essential to downregulate STM in the CMM, thereby allowing ovule formation and TT differentiation. STM is ectopically expressed in hda19-3 at intermediate stages of pistil development, and its downregulation by RNA interference alleviated the hda19-3 phenotype. Chromatin immunoprecipitation assays indicated that STM is a direct target of HDA19 during pistil development and that the transcription factor SEEDSTICK is also required to regulate STM via histone acetylation. Thus, we identified factors required for the downregulation of STM in the CMM, which is necessary for organogenesis and tissue differentiation.
HISTONE DEACETYLASE19 regulates ovules and reproductive tract formation by repressing SHOOT MERISTEMLESS expression.
Introduction
The number of seeds produced by a plant is important for the reproductive success of a species and, in the case of crops, for food security. Seed yield depends on several characteristics, like flower number, the number of ovules per pistil, as well as the capacity of the gynoecium to foster their fertilization, among others.
The gynoecium or pistil contains the ovules and the tissues required for pollen germination, growth, and guidance. These tissues are known as the reproductive tract (ReT) and comprise the stigma, style, and transmitting tract (TT). Both structures emerge from the carpel margin meristem (CMM) (Reyes-Olalde et al. 2013; Chávez Montes et al. 2015), a determinate meristem that appears as a ridge in the medial part of the gynoecium at stage 7 of floral development (Fig. 1A; Smyth et al. 1990). As pistil development progresses, the CMM produces the ovules and the different structures composing the ReT (Fig. 1A). During stages 8 to 9, it produces the placenta and ovule primordia (OP), and the medial ridges fuse, forming the septum (Fig. 1A). In stages 10 to 12, all the organs complete their developmental program, and the gynoecium becomes competent for fertilization (Bowman et al. 1999; Roeder and Yanofsky 2006; Reyes-Olalde et al. 2013; Chávez Montes et al. 2015; Herrera-Ubaldo et al. 2019).
Figure 1.
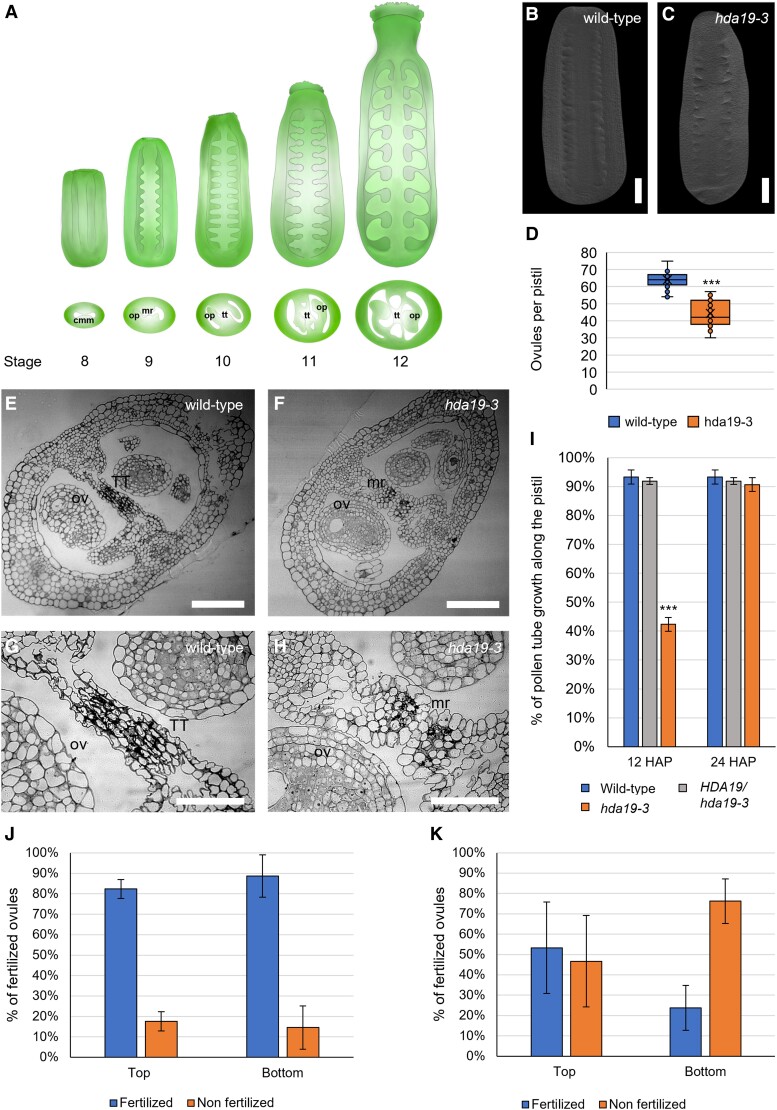
Ovule density and TT are abnormal in the hda19-3 mutant. A) Scheme of the development of the ReT from the CMM from stages 8 to 12 of flower development according to Smyth et al. (1990). Abbreviations: CMM, carpel margin meristem; OP, ovule primordia; mr, medial ridge; TT, transmitting tract. B, C) Cleared pistils of the wild type (B) and hda19-3(C) at stage 9, showing differences in ovule density. Images were digitally extracted for comparison. Scale bar = 50 µm. D) Ovule number of wild-type (n = 70 pistils) and hda19-3 (n = 85 pistils) plants. Center line = median; X = average; box limits = upper and lower quartiles; whiskers = 1.5× interquartile range; points = single measures and outliers. ***Student's t-test with P-value < 0.001. E to H) Transversal section of wild-type (E, G) and hda19-3(F, H) pistils at stage 12. hda19-3 shows unfused septum and defective TT due to the lack of degeneration of septum cells. Images G and H are close-ups of a portion of images E and F, respectively. Abbreviations: TT, transmitting tract; OV, ovule; mr, medial ridge. Scale bar = 50 µm. I) Ratio of pollen tube length/pistil length at 12 and 24 HAP in wild-type (n = 8), heterozygous (n = 8), and homozygous (n = 12) hda19-3 pistils, hand-pollinated with wild-type pollen. Bars represent the mean ± SE value. ***Student's t-test with P-value < 0.001, comparing wild type with HDA19/hda19-3 or hda19-3. J, K) Seed set in the top half and bottom half of wild-type (J) and hda19-3(K) siliques, hand-pollinated with wild-type pollen, showing that the bottom half of hda19-3 siliques shows higher levels of unfertilized ovules than the top half. Bars represent the mean ± SE value of the percentage of unfertilized ovules in 10 wild-type and 10 hda19-3 siliques.
Ovule formation encompasses two steps: (1) boundary formation and primordium outgrowth and (2) identity determination. The first step is controlled by general modulators of lateral organ formation, such as CUP-SHAPED COTYLEDON 1/2/3 (CUC1/2/3), BLADE-ON-PETIOLE (BOP), PIN-FORMED 1 (PIN1), and AINTEGUMENTA (ANT) (Ishida et al. 2000; Mizukami and Fischer 2000; Vernoux et al. 2000; Norberg et al. 2005). Particularly, OP emerge from the placenta asynchronously. Indeed, ovules start to emerge from the placenta at stage 9a, while the final ovule number is reached only at stage 9c. This results in the presence of ovules of different sizes and developmental stages within the same pistil (Yu et al. 2020).
The second step is controlled by organ-specific genes. Ovule identity is redundantly determined by three MADS-domain transcription factors, SEEDSTICK (STK) and SHATTERPROOF1/2 (SHP1/2). In the respective triple mutant, ovules are replaced by carpelloid structures (Favaro et al. 2003; Pinyopich et al. 2003).
Regarding ReT tissues, the TT forms in the center of the septum from the top to the base of the pistil (Fig. 1A) (Crawford and Yanofsky 2008) and supports the growth of pollen tubes to reach the ovules. In Arabidopsis (Arabidopsis thaliana), pollen tubes grow through the intercellular spaces between TT cells, aided by the production of a growth-facilitating cell wall and the death of the TT cells. This process is developmentally regulated and starts before anthesis (Crawford and Yanofsky 2008; Pereira et al. 2021). The first stage of TT formation depends on transcription factors controlling the formation of the septum, such as ETTIN (ETT), SPATULA (SPT), SEUSS (SEU), ANT, and INDEHISCENT (IND) (Sessions and Zambryski 1995; Alvarez and Smyth 2002; Azhakanandam et al. 2008; Kay et al. 2013; Pereira et al. 2021). Later, genes like NO TRANSMITTING TRACT (NTT) and HECATE1/2/3 (HEC1/2/3) control the maturation of TT cells (Gremski et al. 2007, Marsch-Martínez et al. 2014; Herrera-Ubaldo et al. 2019). Interestingly, STK and SHP1/2 are also required for TT formation in cooperation with NTT (Marsch-Martínez et al. 2014; Herrera-Ubaldo et al. 2019) and CESTA/HALF-FILLED (CES/HAF) (Di Marzo et al. 2020a, 2020b).
Genetic and transcriptomic analyses have revealed similarities between the CMM and undetermined meristems like the shoot apical meristem (SAM) (Wynn et al. 2011; Heidstra and Sabatini 2014; Gaillochet and Lohmann 2015; Villarino et al. 2016; Reyes-Olalde and de Folter 2019; Zúñiga-Mayo et al. 2019). However, how the activity of the CMM is controlled, leading to the consumption of the stem cells once organ formation is completed, is not known (Alvarez and Smyth 2002; Reyes-Olalde and de Folter 2019).
HISTONE DEACETYLASE 19 (HDA19) is a major regulator of the maintenance of the SAM (Long et al. 2006; Gorham et al. 2018; Chung et al. 2019), the flower meristem (Krogan et al. 2012; Bollier et al. 2018), and the root apical meristem (Pi et al. 2015). HDA19 is an epigenetic modifier that removes acetyl groups from histones, repressing the transcription of the associated genes (Kumar et al. 2021). HDA19 target specificity relies on the interaction with sequence-specific transcription factors and other cofactors (Kumar et al. 2021). For instance, in the inflorescence meristem (IM), HDA19 interacts with the auxin response factors ETT and AUXIN RESPONSE FACTOR 4 (ARF4) and the YABBY protein FILAMENTOUS FLOWERS (FIL) to downregulate the KNOTTED-1-like homeobox (KNOX) transcription factor gene SHOOT MERISTEMLESS (STM), allowing flower primordia specification (Chung et al. 2019). Interestingly, it was previously suggested that STM's role is similar in the SAM and CMM (Scofield et al. 2007; Durbak and Tax 2011; Poulios and Vlachonasios 2018): STM has to be locally downregulated to allow the formation of organ primordia from the SAM (Long et al. 1996; Kanrar et al. 2006; Landrein et al. 2015). Likewise, in pistils, STM overexpression interferes with ovule identity determination and septum formation (Scofield et al. 2007).
Here, we show that mutations affecting HDA19 lead to ovule number reduction and TT defects. We identified targets of HDA19 during ovule formation and TT differentiation through transcriptomic analysis of cells isolated by fluorescence-activated cell sorting (FACS), using a marker line active in TT, placenta, and ovules. We observed that STM was over- and ectopically expressed in TT and ovules in hda19-3 compared to the wild type and that the STM locus showed more histone acetylation in hda19-3 flowers. Our work also shows that the stk mutant has similar defects as hda19-3 regarding ovule number and TT differentiation, and that STK binds the STM locus and regulates its acetylation state. Overall, our results indicate that the correct development of ovules and TT requires repression of STM by HDA19 and that STK is also necessary for histone acetylation-mediated downregulation of STM in the CMM.
Results
Development of the ReT requires HDA19
Several histone deacetylases (HDAs) are important for plant development (Kumar et al. 2021). HDA6, HDA9, and HDA19 are partially redundant for reproductive traits like flowering time (Kang et al. 2015; Ning et al. 2019), but HDA19 mutants are the only ones that show a reduced seed set (Kang et al. 2015; Ning et al. 2019). Although this phenotype was previously reported (Zhou et al. 2013; Kuhn et al. 2020), its molecular basis has not been investigated. Here, we focus on the role of HDA19 during ovule number determination and TT differentiation as prefertilization characters contributing to the final seed number.
For that, we analyzed ovule number in wild type and hda19-3 flowers. We observed that, although hda19-3 pistils are similar in size (Fig. 1, B and C), they contain significantly fewer ovules (51 ± 3 ovules) than the wild type (67 ± 4 ovules) (Fig. 1D), indicating a lower ovule density. Next, we analyzed pistil radial organization, observing that, in hda19-3 flowers, the medial ridges remain unfused at stage 12 (Fig. 1, E to H). Also, the degeneration of the subepidermal cells required for TT maturation is drastically reduced (Fig. 1, F and H) in comparison to the wild type (Fig. 1, E and G). These defects impair pollen tube growth inside the pistil (Fig. 1I; Supplemental Fig. S1) and reduce seed set predominantly in the bottom half of the silique (Fig. 1, J and K). In hand-pollination experiments using wild-type pollen, the percentage of unfertilized ovules in wild-type siliques at 16 h after pollination (HAP) was low and similar in the top and bottom halves of the siliques (Fig. 1J). In hda19-3 flowers, fertility is overall lower compared to the wild type (Fig. 1, J and K), as noted before (Zhou et al. 2023). However, the percentage of unfertilized ovules in the bottom half of the siliques was notably higher with respect to the top half (Fig. 1K). Altogether, we conclude that HDA19 is required for proper OP specification and TT development.
Transcriptomic analysis of hda19-3 sorted cells supports the role of HDA19 in ReT development and ovule number determination
HDA19 is highly expressed in most tissues of the inflorescence (Zhou et al. 2005). As we wanted to focus on its role during ovule number determination and TT differentiation, we designed a strategy for the isolation of these specific cell types for transcriptomic analysis.
We crossed hda19-3 with a line containing the genomic region containing STK fused to the gene encoding the Green Fluorescent Protein (pSTK::STK-GFP, Mizzotti et al. 2014), driving the expression of STK-GFP in the placenta, ovules, and TT from stage 8 to 12 (Mizzotti et al. 2014; Herrera-Ubaldo et al. 2019). After verifying that the spatial expression pattern of STK-GFP was equivalent in both genetic backgrounds (Supplemental Fig. S2), we collected whole inflorescences, except open flowers, directly in a solution (Villarino et al. 2016) to produce protoplast. GFP-positive protoplasts were then collected by FACS, and RNA was extracted to identify differentially expressed genes by RNA-seq.
Enrichment of STK-GFP in sorted protoplasts was validated by RT-qPCR (Supplemental Fig. S3) before library preparation. Three and four samples of RNA from GFP-positive cells (80 to 100,000 cells/replicate) from wild-type and hda19-3 plants, respectively, were used for library preparation and RNA Illumina sequencing (HiSeq2500).
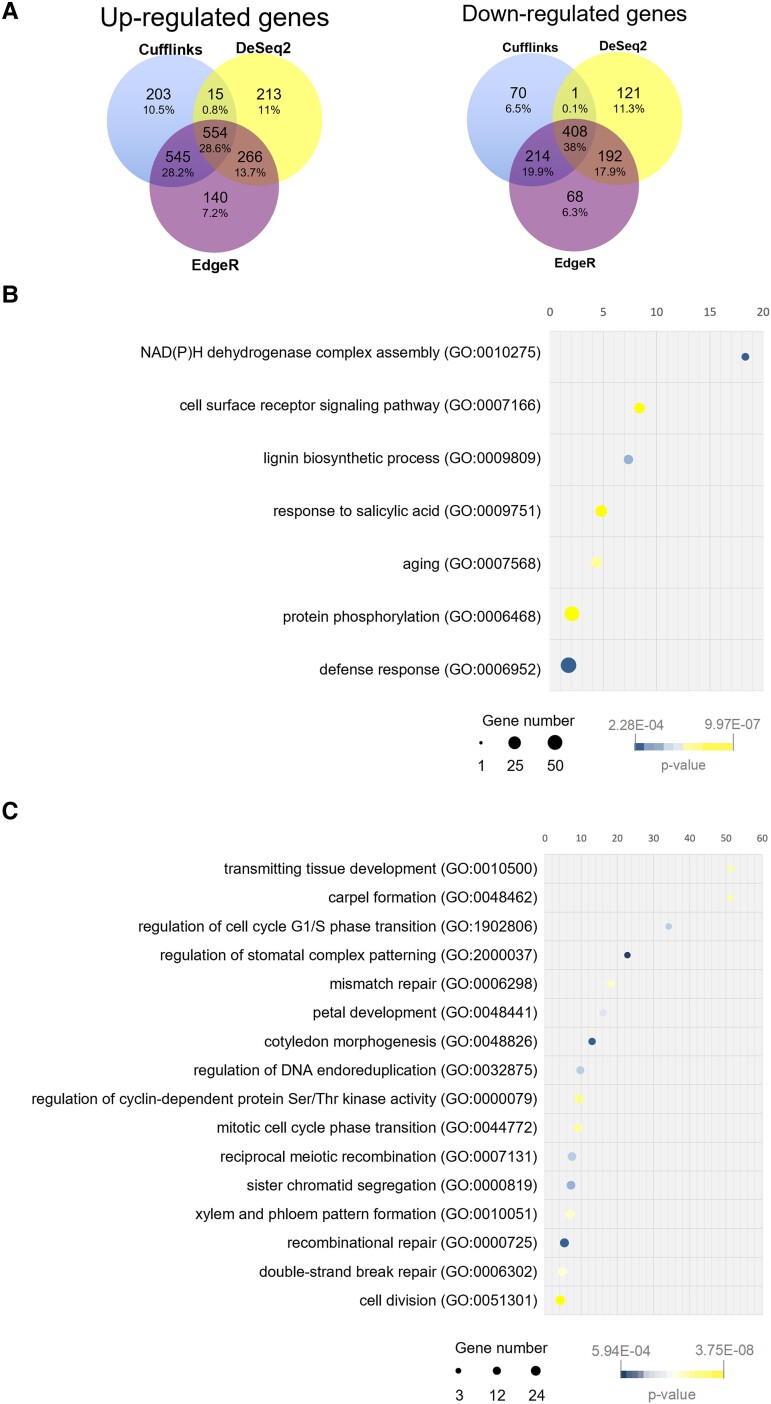
Since HDA19 is an epigenetic regulator, we used a stringent approach to classify differentially expressed genes (DEGs). We analyzed the RNA-seq data using Cufflinks (Trapnell et al. 2012), DeSeq2 (Love et al. 2014), and EdgeR (Robinson et al. 2009) (Supplemental Data Set 1 to S3). Then, we compared the DEGs and selected only those common in all three analyses (Supplemental Data Set 4). This approach identified 554 up- and 408 downregulated genes (Fig. 2A).
Figure 2.
Reproductive tract transcriptomic comparison of wild-type and hda19-3 flowers. A) Venn diagrams showing the overlap of up- and downregulated genes (FDR < 0.001 and logFC > |1.5|) detected by Cufflinks, EdgeR, and DeSeq2. B) Enrichment analysis of “Biological Process” GO terms associated with upregulated genes (logFC > |1.5|) detected by all three programs. C) Enrichment analysis of “Biological Process” GO terms associated with downregulated genes (logFC > |1.5|) detected by all three programs (fold enrichment > 4). Graphs show overrepresented child-most (tip) GO terms. Parent GO terms (root terms) have not been plotted for simplicity. Enrichment analysis was performed using Panther (Mi et al. 2018, 2019) with the default parameters set by the program.
To survey the roles of DEGs, we performed a gene ontology (GO) enrichment analysis of “Biological Process’ terms associated with the up- and downregulated genes, respectively (Fig. 2, B and C; Supplemental Data Set 5 and 6), using Panther (Mi et al. 2018, 2019). Among the upregulated genes, we found the enriched GO terms “response to salicylic acid”, “defense response”, and “aging”, which reflect previously described roles of had19 (Zhou et al. 2005; Kim et al. 2008; Tanaka et al. 2008; Zhu et al. 2010; Choi et al. 2012) (Fig. 2B; Supplemental Data Set 5). Likewise, “NAD(P)H dehydrogenase complex assembly” relates to the role ofHDA19 in regulating the light response (Benhamed et al. 2006; Guo et al. 2008; Jan et al. 2011).
Among the downregulated genes, the most enriched GO terms (Fig. 2C) were “transmitting tissue development” and “carpel formation”, with a fold enrichment of 51.28 (Fig. 2C; Supplemental Data Set 6).
Overall, the enrichment analysis of GO terms showed that genes associated with known roles of HDA19 were present among the DEGs. Moreover, genes necessary for the normal progression of carpel and TT development were downregulated in hda19-3 sorted cells, supporting the validity of our dataset and sampling strategy.
Meristematic markers and genes involved in ovule and TT development are deregulated in the had19-3 mutant
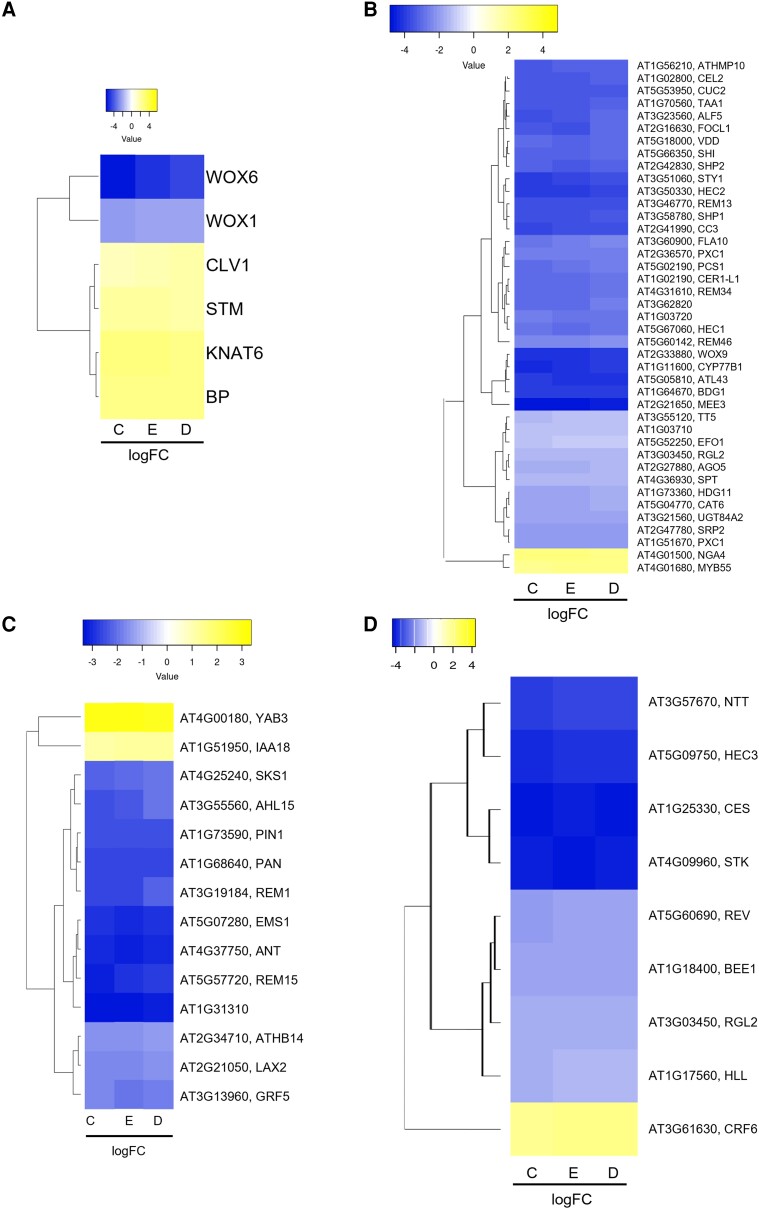
HDA19 regulates the maintenance of stem cell populations in several meristems (Long et al. 2006; Krogan et al. 2012; Pi et al. 2015; Bollier et al. 2018). Therefore, we checked for deregulated meristematic markers in our dataset, and we found that CLAVATA1 (CLV1), as well as the KNOX genes STM, KNAT1/BREVIPEDICELLUS (BP) and KNAT6, showed higher expression in hda19-3 compared to wild-type sorted cells (Fig. 3A).
Figure 3.
CMM-specific genes are deregulated in the hda19-3 mutant. A) Meristem marker genes deregulated in the hda19-3 mutant. B) CMM-specific genes from stages 6 to 8 of pistil development according to Villarino et al. (2016) that are deregulated in the hda19-3 mutant. C) CMM-specific genes from stages 8 to 10 of pistil development according to Wynn et al. (2011) that are deregulated in hda19-3.D) Notable ovule number and TT regulators are deregulated in hda19-3. Genes plotted correspond to genes with an FDR < 0.001 and logFC > |1.2| in the RNA-Seq data according to Cufflinks (C), EdgeR (E), and DeSeq2 (D). Genes were hierarchically clustered by average clustering using Euclidean distances. Heatmaps were obtained using Heatmapper (http://heatmapper.ca/). The scales are relative to the maximum and minimum values of each table and represents the logFC (wild-type vs hda19-3) as detected by each program.
STM, BP, and KNAT6 are partially redundant for SAM maintenance (Byrne et al. 2002), and their local downregulation at the boundaries of the meristem is necessary for organ initiation (Belles-Boix et al. 2006; Ragni et al. 2008; Zhao et al. 2015). Therefore, their upregulation in hda19-3 plants could hinder the formation of CMM-derived structures.
Furthermore, to identify CMM-specific genes associated with ReT differentiation and ovule initiation, we cross-referenced our dataset with two pre-existing sets of CMM-specific genes (Wynn et al. 2011; Villarino et al. 2016). We found that several genes involved in TT and style development like HEC1, HEC2, SPT, and STYLISH (STY1) (Sohlberg et al. 2006; Gremski et al. 2007; Nahar et al. 2012; Kuhn et al. 2020) were downregulated in hda19-3 plants (Fig. 3B), possibly explaining the defects in the TT (Fig. 1, E to H) and style (Kuhn et al. 2020). Additionally, several well-known players of ovule initiation, such as PHABULOSA (PHB), ANT, PERIANTHIA (PAN) and PIN1, were downregulated in the hda19-3 mutant (Fig. 3C), consistent with their reduced number of ovules (Fig. 1D).
Only four CMM-specific genes were upregulated in hda19-3: NGATHA4 (NGA4) and MYB55 (Villarino et al. 2016) (Fig. 3B); and YABBY3 (YAB3) and INDOLE-3-ACETIC ACID INDUCIBLE 18 (IAA18) (Wynn et al. 2011) (Fig. 3C). NGA4 and YAB3 are negative regulators of lateral organ formation (Alvarez et al. 2009; González-Reig et al. 2012; Lee et al. 2015), consistent with the ReT phenotype of hda19-3 plants. The role of IAA18 and MYB55 in the pistil is not known, but iaa18 mutants have reduced fertility (Uehara et al. 2008), and MYB55 is a direct target of BRASSINAZOLE-RESISTANT 1 (BZR1) (He 2005), participating in organ boundary establishment in the SAM (Bell et al. 2012; Gendron et al. 2012). Therefore, the upregulation of these genes might also interfere with ReT development.
Finally, we checked the expression of some additional regulators of TT and ovule number determination that were not included in these two datasets (Wynn et al. 2011; Villarino et al. 2016) due to their experimental conditions. For example, HEC3 and STK itself were downregulated (Fig. 3D). It is worth noting that, although STK shows a log fold-change (logFC) close to −4 in hda19-3 mutants, its spatial expression pattern was unchanged with respect to the wild type (Supplemental Fig. S2).
Overall, analysis of CMM-specific genes revealed that several positive regulators of TT differentiation and ovule initiation were downregulated (PIN 1, ANT), while KNOX genes involved in meristem maintenance (STM, BP, KNAT6) and negative regulators of lateral organ development (NGA4, YAB3) were upregulated in hda19-3 sorted cells. As HDA19 is a transcriptional repressor, their upregulation suggests that HDA19 might directly regulate meristematic genes and/or genes involved in inhibiting the formation and differentiation of CMM-derived structures.
STM is ectopically expressed and over-acetylated in the ReT of the hda19-3 mutant
Among the possible candidates contributing to hda19-3's defects in ovule formation and TT differentiation (Fig. 1), we focused our attention on STM, which was upregulated in hda19-3 mutants compared to the wild type (Fig. 3A). HDA19 downregulates STM in the IM to allow organ development from this meristem (Chung et al. 2019). Therefore, we hypothesized that a similar situation could occur in the CMM: as STM is required for pistil development (Scofield et al. 2007), HDA19 may also regulate it there.
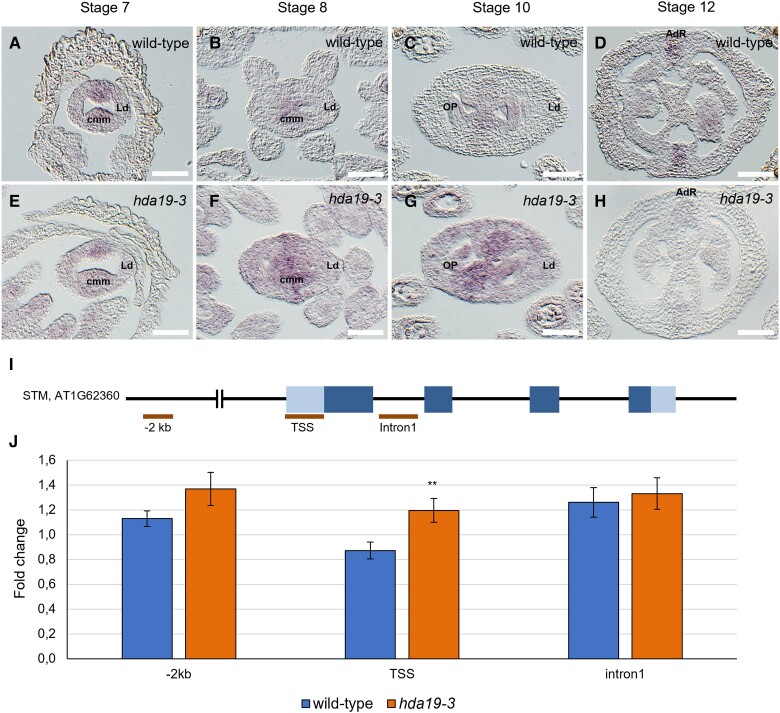
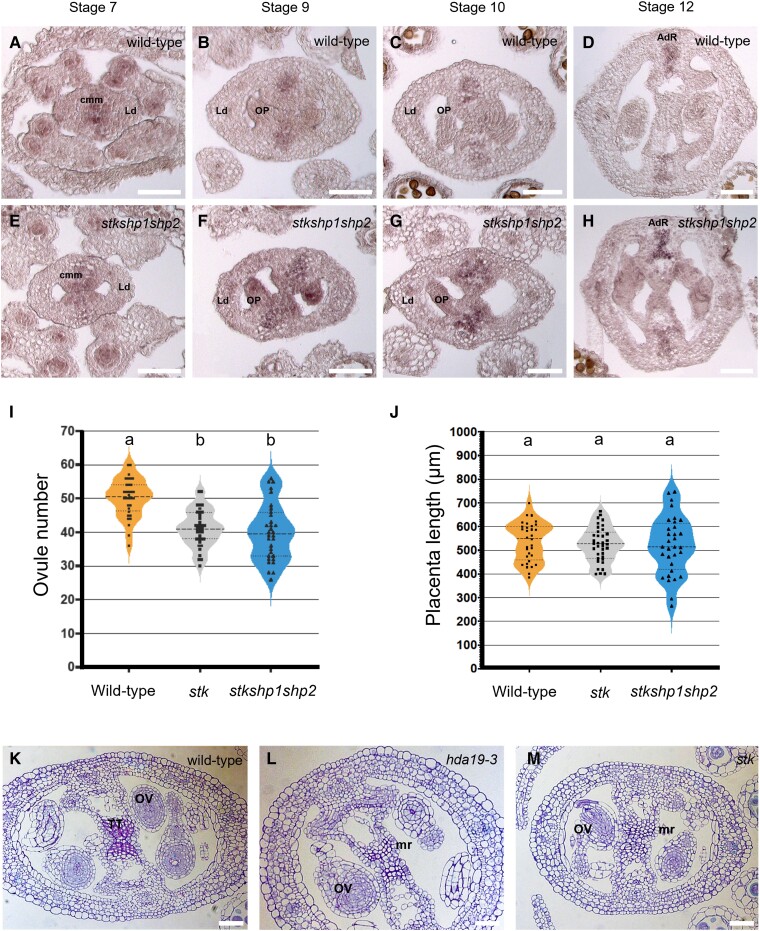
We investigated the spatial expression pattern of STM in wild-type and hda19-3 pistils by in situ hybridization (ISH) using an STM-specific antisense probe (Fig. 4, A to H). At stage 7, the expression pattern of STM was similar in both genotypes (Fig. 4, A and E). However, from stages 8 to 10, the expression of STM in the medial ridge increased in hda19-3 and expanded to the placenta, OP, and even the lateral domain (Fig. 4, B and C, F and G). After stage 10, STM expression became restricted to the adaxial replum zone in both genotypes (Fig. 4, D and H). STM promotes meristematic fate; therefore, ectopic expression in the placenta, OP, and TT from stages 8 to 10 might interfere with ovule initiation and TT development, causing the phenotypes observed in the hda19-3 mutant.
Figure 4.
STM is ectopically expressed and highly acetylated in hda19-3 pistils. A to H) Detection of STM mRNA by ISH in wild-type (A to D) and hda19-3(E to H) pistils. At stage 6, STM mRNA is specifically expressed in the CMM of wild-type pistils (A, B). Then expression decreases during the following stages until it is restricted to the adaxial replum zone (C, D). Instead, in the hda19-3 mutant during stages 7 to 10 when ovule primordia arise, the expression of STM increases and expands outside the CMM (F, G), to finally become restricted to the replum later on (H). Abbreviations: CMM, carpel margin meristem; LD, lateral domain; OP, ovule primordia; AdR, adaxial replum. Scale bar = 50 µm. I) Regions of the STM locus tested for changes in H3K9/14ac are indicated by bars. J) H3K9/14ac ChIP-qPCR of STM locus in wild-type and hda19-3 flowers. Graph shows the mean ± SD result of STM regions enrichment in the hda19-3 background with respect to the wild type, among three independent biological replicates of the H3K9/K14ac ChIP-qPCR experiment performed on wild-type, hda19-3, stk, and stkshp1shp2 flowers. **Student's t-test with P-value < 0.01.
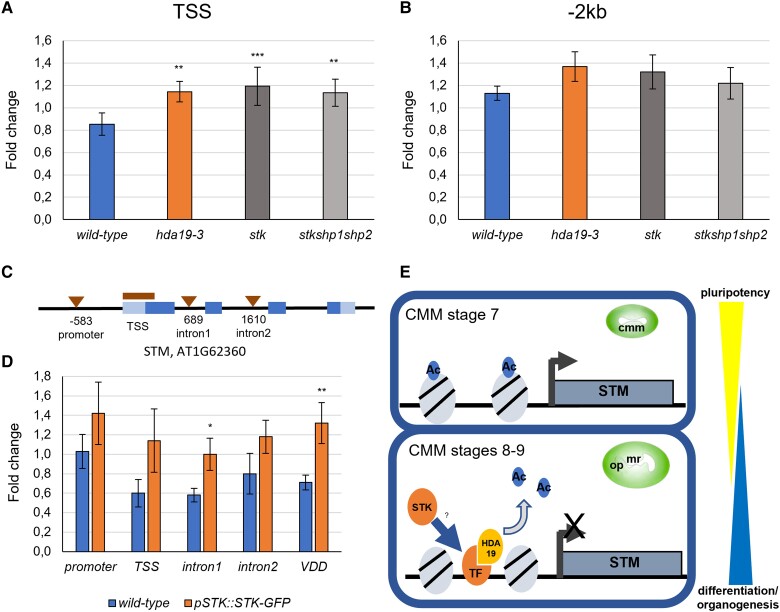
To analyze whether the changes in STM's expression pattern could be caused by variations in histone acetylation levels resulting from the absence of HDA19, we collected the inflorescence apex (including flowers up to stage 12 but excluding the IM) from wild-type and hda19-3 plants and performed ChIP-qPCR. ChIP-qPCR is a widely used technique to reveal differences in the levels of specific histone modifications at a specific locus (Saleh et al. 2008; Petrella et al. 2020). We used an antibody against HISTONE 3 (H3) acetylated at lysine 9 and 14 (K9/14) (H3K9/14ac) (Fig. 4, I and J) to capture DNA fragments associated with acetylated histone H3. Since no other histone de/acetylases are substantially deregulated in hda19-3 sorted cells (Supplemental Fig. S4), the changes in STM histone acetylation observed with this antibody should be caused mostly by the direct action of HDA19.
We tested the acetylation levels in several regions of the STM locus: (1) the distal promoter (−2 kb) (Fig. 4I) because it was reported that HDA19 can deacetylate these regions of the STM locus (Chung et al. 2019); (2) the transcription start-site (TSS), as its acetylation level generally is associated with active gene expression (Wang et al. 2017; Kim et al. 2021; Kumar et al. 2021); and (3) the first intron. We observed a significant increase in acetylation at the STM TSS in hda19-3 and a moderate increase at the promoter region, while no notable increment in acetylation was found in the first intron (Fig. 4J). In conclusion, our results suggest that, at the stages tested, HDA19 is required for the deacetylation of the STM locus. Moreover, this acetylation pattern differs from that of Chung et al. (2019), suggesting that HDA19 acts on different regions in each meristem.
Downregulation of STM alleviates ReT defects of the hda19-3 mutant in an expression level-dependent manner
To verify if the ectopic expression of STM was involved in the reduction of ovule number and TT defects in the hda19-3 mutant, we generated an RNA interference (RNAi) construct to reduce STM levels in the hda19-3 background. We used the pFRH vector as, in our experience, it causes a moderate downregulation of the target gene (Fornara et al. 2004) that is compatible with the objective of our experiment.
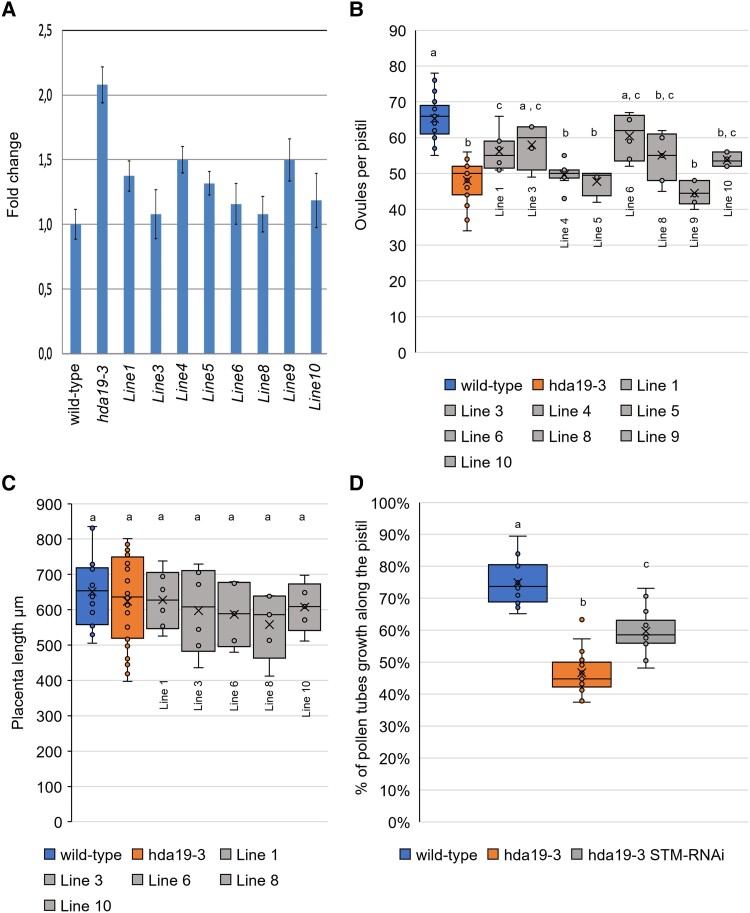
We analyzed eight independent hda19-3 lines transformed with p35S::STM-RNAi::pFRH (Fig. 5; Supplemental Fig. S5). We characterized the T1 generation to avoid random silencing of the construct in subsequent generations. We measured STM transcript levels in wild-type, hda19-3, and hda19-3 p35S::STM-RNAi::pFRH (hda19-3 STMRNAi hereafter) inflorescences by RT-qPCR. The levels of STM transcript in the hda19-3 mutant were twice those in the wild type (Fig. 5A), and all the hda19-3 STMRNAi transformants showed STM mRNA levels in between those of hda19-3 and the wild type (Fig. 5A).
Figure 5.
Downregulation of STM alleviates hda19-3 ReT phenotypes. A) Level of STM mRNA in inflorescences of wild-type, hda19-3, and eight independent hda19-3 STMRNAi lines. Error bars represent the mean ± SE among three replicates. B) Ovule number per pistil of wild-type (n = 25), hda19-3 (n = 27) and hda19-3 STMRNAi lines (in order from Line 1 to Line 10: n = 8, 4, 10, 4, 6, 7, 6, 6). Center line = median; X = average; box limits = upper and lower quartiles; whiskers = 1.5× interquartile range; points = single measures and outliers. The statistical significance of differences in ovule number was determined by a one-way ANOVA and a Bonferroni post hoc test for multiple comparisons. Letters over boxes denote significantly different averages (P < 0.05), comparing wild type, hda19-3 and hda19-3 STMRNAi lines. C) Placenta length of wild-type, hda19-3, and hda19-3 STMRNAi lines that showed significant changes in ovule number (Lines 1, 3, 6, 8, 10). Center line = median; X = average; box limits = upper and lower quartiles; whiskers = 1.5× interquartile range; points = single measures and outliers. The statistical significance of differences in placenta length (P < 0.05) was determined by a one-way ANOVA with a Bonferroni post hoc test for multiple comparisons among wild-type, hda19-3 and hda19-3 STMRNAi lines. D) Pollen tube growth measured as pollen tube length/total length of the pistil. Pollen tubes were allowed to grow for 14 HAP of emasculated flowers. Growth was visualized by aniline-blue staining and the lengths of pollen tubes and pistils were measured using ImageJ. Wild-type (n = 17 pistils); hda19-3 (n = 17 pistils); hda19-3 STMRNAi (pool of Lines 1 and 10) (n = 18 pistils). Center line = median; X = average; box limits = upper and lower quartiles; whiskers = 1.5× interquartile range; points = single measures and outliers Letters over boxes indicate statistical differences as determined by a one-way ANOVA (P < 0.05) and a Bonferroni post hoc test for multiple comparisons.
Ovule number and density increased in the hda19-3 STMRNAi plants according to the level of STM downregulation (Fig. 5B), suggesting that the STM expression level in the gynoecium is related to ovule number determination. Lines 4 and 9 showed the mildest reduction of STM levels (ca. 75% of hda19-3 levels) and, in accordance, did not show significant changes in ovule number (Fig. 5B). Line 5 was an exception, as it showed a reduction in STM levels similar to line 1 (ca. 35% reduction) (Fig. 5A), but while Line 1 showed an increase in ovule number, Line 5 did not (Fig. 5B). As expected, placenta length remained unchanged in all inspected lines (Fig. 5C). Therefore, the increase in ovule number represents an increase in ovule density (Fig. 5, B and C).
We selected Lines 1 and 10 for the inspection of TT phenotypes as they had comparable STM expression levels and ovule density, and flowers were still available after the experiments shown in Fig. 5, A and B.
Although the septum of Lines 1 and 10 was still partially unfused (Supplemental Fig. S5, A to C), the ridges were contacting more closely (Supplemental Fig. S5, B and C), and the subepidermal cells exhibited a wild-type-like phenotype (Supplemental Fig. S5, A to C). In addition, hand-pollination experiments revealed an improved pollen tube growth rate in the transgenic lines compared to the hda19-3 mutant (Fig. 5D; Supplemental Fig. S5, D to F). We hand-pollinated wild-type, hda19-3, and hda19-3 STMRNAi pistils (Lines 1 and 10) with wild-type pollen and stained them with aniline blue 14 HAP (Fig. 5D; Supplemental Fig. S5, D to F). In a preliminary experiment, we determined that pollen tubes reach the bottom of wild-type pistils 16 HAP. Therefore, at 14 HAP, we would be able to precisely quantify the differences between each genotype. At 14 HAP, pollen tubes grew through 70% of wild-type pistils and 60% of hda19-3 STMRNAi pistils, a significant increase with respect to hda19-3, where pollen tubes barely grew 40% of the pistil length (Fig. 5D; Supplemental Fig. S5, D to F). Overall, even if the anatomical changes in the TT were mild (Supplemental Fig. S5, A to C), they improved pollen tube growth in comparison to the hda19-3 mutant.
In conclusion, our results strongly suggest that ectopic expression of STM (Fig. 4, A to H) contributes to the ReT phenotypes present in the hda19-3 mutant (Fig. 1) because reducing STM expression in hda19-3 to wild-type levels led to a recovery of ovule number and density (Fig. 5, B and C) and of TT function (Fig. 5D; Supplemental Fig. S5, D to F).
Stk mutants show defects in the ReT and increased histone acetylation at the STM locus
HDA19 does not directly bind DNA; therefore, it has to be recruited to DNA by DNA-binding proteins to regulate its targets. In the IM, HDA19 interacts with ETT, ARF4, and FIL (Chung et al. 2019) to downregulate STM. However, in the pistil, ETT, ARF4, and FIL are mainly expressed in the lateral domains at stages 8 and 9 (Sessions et al. 1997; Pekker et al. 2005; González-Reig et al. 2012). Consequently, the action of HDA19 in the CMM is probably mediated by a different complex.
Among the transcription factors that are co-expressed with HDA19 in the CMM, we highlight STK as a possible candidate for several reasons. STK controls several aspects of ovule development (in redundancy with SHP1/2) (Favaro et al. 2003; Pinyopich et al. 2003) and is required for septum and TT development (Colombo et al. 2010; Herrera-Ubaldo et al. 2019; Di Marzo et al. 2020a, 2020b) Moreover, STK shows repressor activity associated with histone deacetylation of its direct targets (Mizzotti et al 2014), suggesting it might act in conjunction with HDAs.
To investigate if STK has a role in the regulation of STM in the CMM, we performed ISH using an STM-specific antisense probe (Fig. 6, A to H). To account for the possible redundancy with SHP1/2, we probed stk shp1 shp2 triple mutant pistils. STM was ectopically and overexpressed in stk shp1 shp2 (Fig. 6, E to H), in a similar fashion to what we had previously observed in the hda19-3 mutant (Fig. 4, A to H). This observation suggests that STK or SHP1/2 could be involved in STM regulation during ReT development.
Figure 6.
STK regulates STM expression in the CMM. A to H) Detection of STM mRNA in the wild type (A to D) and the stk shp1 shp2 triple mutant (E to H) by ISH. STM is over- and ectopically expressed in the stk shp1 shp2 triple mutant at stages 7 to 10 of pistil development (F, G) as compared to the wild type (B, C). Abbreviations: CMM, carpel margin meristem; LD, lateral domain; OP, ovule primordia; AdR, adaxial replum. Scale bar, 50 µm. I, J) Ovule number (I) and placenta length (J) in wild type (n = 31), stk (n = 35), and stk shp1 shp2 (n = 31). Center dashed line = median; Upper and lower dotted lines = upper and lower quartiles; Violin ends = 1.5× interquartile range; Squares or triangles within the violins = single measures and outliers Letters over violin-plots indicate statistical differences as determined by a one-way ANOVA (P < 0.05) and a Bonferroni post hoc test. K–M) Transversal sections of stages 10 to 11 wild-type (K), hda19-3(L), and stk pistils (M). hda19-3 and stk pistils show defective subepidermal cells in the TT. Abbreviations: TT, transmitting tract; OV, ovule; mr, medial ridge. Scale bar, 50 µm.
Then, we measured ovule number and placenta length: the stk mutant showed a reduced ovule number (Fig. 6I) without changes in placenta length (Fig. 6J), similar to hda19-3 mutants (Fig. 1). The stk shp1 shp2 triple mutant did not show statistically significant additive effects (Fig. 6, I and J), suggesting that SHP1 and SHP2 are not involved in ovule number reduction.
Finally, we evaluated the TT morphology in stk pistils. While ridges were fused as previously described (Di Marzo et al. 2020a, 2020b), we found that the subepidermal cells looked similar to those in hda19-3 mutants (Fig. 6, K to M).
STK directly binds the STM locus and is involved in its deacetylation
To explore if STK was required to regulate the acetylation state of the STM locus in the CMM, we performed ChIP-qPCR using an anti H3K9/14ac antibody on inflorescences (including flowers up to stage 12 but excluding the IM) of wild-type, hda19-3, stk, and stk shp1 shp2 plants (Fig. 7, A and B), analyzing the same regions tested in hda19-3 mutants (Fig. 4I). Similar to what we had observed in hda19-3, stk and stk shp1 shp2 mutants showed an increase in H3K9/14ac at the level of the TSS, and a moderate increase at the promoter (Fig. 7, A and B). This observation supports the hypothesis that HDA19 and STK might cooperatively regulate the acetylation state of these regions.
Figure 7.
STK binds to the STM locus. A, B) H3K9/14ac ChIP-qPCR of STM TSS (A) and of a distal promoter region (−2 kb) (B) in the wild type, hda19-3, stk, and stk shp1 shp2 mutants. The pattern of acetylation at the STM locus is similar among the different mutant genotypes. Graphs show the mean ± SD value of enrichment among three independent biological replicates of the H3K9/14ac ChIP-qPCR experiment performed on wild-type, hda19-3, stk, and stk shp1 shp2 flowers. Student's t-test, **P-value < 0.01. ***P-value < 0.001. C) Regions of the STM locus tested by anti-GFP ChIP-qPCR in wild-type and pSTK::STK-GFP flowers. Triangles mark CaRG boxes detected by AthaMap (Steffens et al. 2004) while the bar marks the TSS region of STM. D) ChIP-qPCR to test binding of STK (pSTK::STK-GFP) to the promoter, TSS, first intron, and second intron of STM. VDD is a known direct target of STK, so it was used as positive control. Graph shows the mean ± SD enrichment value among four independent biological replicates of the anti-GFP ChIP-qPCR experiment. Student's t-test, *P-value < 0.05. **P-value < 0.01. E) Model of the regulation of STM expression in the CMM involving HDA19 and STK. Until stage 7, the expression of STM in the CMM is high due to the presence of acetylated histones in association with the STM locus. At stages 8 to 9, when the CMM starts producing the ReT organs, HDA19 deacetylates the histones associated with the STM locus, leading to its downregulation and thereby allowing the formation of OP and TT. Direct binding of STK to the STM locus facilitates the action of HDA19, potentially by generating multimeric complexes involved in HDA19 binding and stabilization to the STM locus. Abbreviations and symbols: CMM, carpel margin meristem; mr, medial ridge; OP, Ovule primordia; TF, transcriptional factor; Ac, acetyl group.
To provide further evidence, we verified whether STK could directly bind to the STM genomic region by performing ChIP-qPCR on inflorescences (including flowers up to stage 12 but excluding the IM) of wild-type and pSTK::STK-GFP plants using an anti-GFP antibody. We found one MADS-domain DNA-binding motif (a CaRG box) in the proximal promoter area and two in the first and second introns of the STM locus (Fig. 7C). In addition to those, we also tested the region around the TSS of STM that was enriched in H3K9/14ac (Fig. 7A). As a positive control, we used VERDANDI (VDD), a direct target of STK (Matias-Hernandez et al. 2010) (Fig. 7D). The first STM intron showed the strongest enrichment, followed by the TSS. In contrast, the second intron and the promoter showed no enrichment (Fig. 7D). This result indicates that STK can directly bind the STM locus and that its action is required to regulate its expression level (Fig. 6, A to H).
Taken together, our phenotypic analysis, expression studies, and ChIP-qPCR experiments show that HDA19 and STK activity have similar effects on the STM expression domain and transcript level. Their activity is required to regulate CMM functionality and to allow ReT development and ovule number determination, both essential traits for plant reproductive fitness (Fig. 7E).
Discussion
Here, we show that HDA19 is necessary for ovule initiation and TT differentiation (Fig. 1; Supplemental Fig. S1). To pinpoint the role of HDA19 in these tissues, we performed an RNA-seq of pSTK::STK-GFP-expressing cells isolated by FACS from wild-type and hda19-3 backgrounds. We found that several genes involved in meristem maintenance were upregulated in hda19-3 mutant ReTs (Fig. 3). We focused on studying STM because of its well-known role in meristematic activity (Barton and Poethig 1993; Clark et al. 1997) and the recent report that HDA19 is necessary to downregulate STM in the IM (Chung et al. 2019). Other upregulated KNOX genes like BP (Byrne et al. 2002) were less attractive as, in the pistil, BP is associated with the differentiation of the replum (Alonso-Cantabrana et al. 2007; González-Reig et al. 2012), rather than the ovules and TT. CLV1 was another potential target of HDA19, but the simultaneous upregulation of CLV1 and STM suggested that the upregulation of CLV1 could be a consequence of the spatial and temporal expansion of the meristematic activity of hda19-3 CMMs (Fig. 3), rather than its cause, as STM is downregulated in clv1 mutants (Durbak and Tax 2011). Nevertheless, it is still possible that these genes play a role together with STM in mediating the CMM phenotypes of the hda19-3 mutant, and it will be interesting to study their contribution in the future. Instead, genes involved in organogenesis like SPT, CUC1/2, ANT, PAN, PIN1, HEC1 or NTT were downregulated in sorted cells of hda19-3 plants (Fig. 3), suggesting that they are not directly regulated by HDA19.
Loss-of-function stm mutants lack a functional SAM and, consequently, fail to produce flowers. Therefore, to assess whether STM deregulation was involved in the ReT phenotypes of hda19-3, we reduced STM levels in hda19-3 plants by RNAi using a weak constitutive promoter. The downregulation of STM in the hda19-3 background partially restored ovule number and density and pollen tube growth in the TT (Fig. 5; Supplemental Fig. S5). None of the transgenic lines displayed a complete rescue of the hda19-3 phenotype. However, this is not surprising given that HDA19 regulates many genes, so it is unlikely that ectopic expression of STM is the only cause of all of hda19-3's ReT defects. So, after all, downregulation of KNAT6 and BP, partially redundant with STM in the SAM (Byrne et al. 2002), might be necessary for a complete rescue.
Previously, Scofield and collaborators observed that overexpressing STM through an inducible system led to the transformation of ovules into carpelloid structures (Scofield et al. 2007). This aligns with our observations but represents a more extreme phenotype, probably because the expression levels of STM achieved by their strategy were much higher than those observed in hda19-3, leading to a strong enough downregulation of STK and SHP1/2 to cause the conversion of ovules into carpelloid structures (Scofield et al. 2007).
Our results support the idea that the CMM functions in a similar way as its indeterminate counterparts like the SAM or IM (Long et al. 2006; Chung et al. 2019) and provide insights into how its activity is terminated. However, this aspect requires further research, as ISH showed that, from stage 10, STM expression in the hda19-3 mutant is similar to that in the wild type (Fig. 4, A to H). This finding suggests that, in addition to HDA19, other factors contribute to STM silencing in the CMM since lack of HDA19 delays STM silencing but does not prevent it completely.
Then, as HDA19 is unable to bind DNA on its own, we searched for partners that might mediate its interaction with DNA. We turned our attention to STK, based on its expression in the TT, placenta, and OP (Supplemental Fig. S2), its known roles in ovule and TT development (Pinyopich et al. 2003, Herrera-Ubaldo et al. 2019, Di Marzo et al. 2020a, 2020b), and that fact that the repression of some of its targets is associated with changes in histone acetylation (Mizzotti et al. 2014). The existence of similar defects in CMM-derived organs (Figs. 1 and 6, I to M), and a similar histone acetylation pattern at the STM locus in hda19-3 and stk mutants (Figs. 4, I, J and 7, A, B), supports STK as a player required for STM regulation by HDA19 in the CMM. Nevertheless, it should be noted that, while the stk mutant shows a reduction of ovule number and density similar to hda19-3, the TT defects are milder in stk than in hda19-3 plants. The role of STK in the development of the TT was already described (Marsch-Martínez et al. 2014; Herrera-Ubaldo et al. 2019, Di Marzo et al. 2020a, 2020b), but single stk mutants showed very mild defects that became more severe when stk was combined with the ntt and ces mutants. Overall, this suggests that, in the TT, HDA19 might act in cooperation with additional proteins to regulate STM.
Mutant analysis indicated that SHP1/2 are not involved in this process. Although STK was first described as a member of the ovule identity complex redundant with SHP1/2 (Favaro et al. 2003; Pinyopich et al. 2003), they also have nonredundant functions during reproductive development (Liljegren et al. 2000; Pinyopich et al. 2003; Mizzotti et al. 2012, 2014; Ezquer et al. 2016; Di Marzo et al. 2020a, 2020b). Our results suggest that the function in the CMM is another instance of the specific roles of the three paralogs.
Finally, we found that STK binds the STM locus at the level of the TSS and the first intron (Fig. 7, C and D). MADS-domain proteins form tetramers that are able to bind two DNA sequences simultaneously, creating DNA loops (Smaczniak et al. 2012). Based on this and the fact that the TSS shows a higher acetylation level in both mutants, a plausible model is that a MADS-domain complex containing STK and HDA19 binds to the STM locus, forming a loop between the TSS and the first intron. This would facilitate HDA19's deacetylation of the TSS region of STM, thereby reducing its expression and allowing organ initiation from the CMM (Fig. 7E).
It would have been interesting to study the mode of interaction between STK and HDA19. However, the fact that only repressed direct targets of STK are regulated through histone deacetylation (Mizzotti et al. 2014) suggests that STK must be able to form different multimeric complexes comprising different proteins. In other words, not every complex containing STK contains HDA19. This suggests that the mechanism of interaction between STK and HDA19 is either indirect (e.g. through other MADS-domain proteins) or regulated by post-translational modifications or alternative splicing.
SUMOylation is an interesting candidate modification to mediate the interaction of HDA19 and STK. HDA19 and other proteins of its core complex, like TOPLESS, are SUMOylated (Miller et al. 2010, 2013), while STK has predicted SUMO-interacting motifs (Zhao et al. 2014). When a protein is SUMOylated, usually, only a small fraction of the existing protein (5% to 10%) is modified (Xiao et al. 2015). Therefore, at a given time, only the HDA19-SUMOylated fraction would interact with STK, explaining the simultaneous action of STK as an activator (non-HDA19 interacting) and as a repressor (interacting with SUMOylated HDA19). Another possibility is alternative splicing. STK has six different splicing forms that differ in the presence/absence of several N-terminal exons. Therefore, it is possible that not all proteins derived from alternative splicing are able to interact with HDA19. Overall, this prompted us to discard the idea of testing the interaction between STK and HDA19 in the current work, as it would represent a project on its own.
An interesting hypothesis derived from our work is that STK expression at stage 8 (Pinyopich et al. 2003), before OP start to form, might be necessary to reduce STM levels (Fig. 7C) to allow OP initiation and septum fusion. This is supported by the observation that STM levels are normal in hda19-3 mutants before stage 8 (Fig. 4, A to H), suggesting that HDA19 regulates STM during stages 8 to 10 when STK starts to accumulate.
Finally, we performed FACS-coupled transcriptomics of gynoecium cells without inducible systems, avoiding variability and artifacts reported previously (Villarino et al. 2016). In addition, our dataset containing TT, placenta, and sporophytic ovule cells will be highly useful for future studies of pistil and ovule development. Indeed, we already uncovered possible roles of genes like YAB3 or MYB55. For example, YAB3, which represses medial factors (González-Reig et al. 2012) and mutations thereof affect CMM development, was proposed to act noncell autonomously as ISH did not reveal expression in the CMM (Wynn et al. 2011). As pSTK::STK-GFP is not expressed in the lateral domain, our data indicates that, in the wild-type, YAB3 is indeed expressed in the middle domain at low levels that might be undetectable by ISH.
In conclusion, our results show that the tight control of STM levels achieved through the action of HDA19 and STK is essential to maximize the reproductive fitness of the plant, as even moderate changes in its expression lead to defects in the ReT that reduce seed set.
Materials and methods
Plant materials and growth conditions
Plants for phenotypic analyses were grown in a greenhouse under long-day conditions (16 h light, 8 h dark) at 22 to 24 °C. For cell sorting experiments, plants were grown in a walk-in growth chamber under continuous light at 22 °C.
Arabidopsis (A. thaliana) wild-type Col-0 seeds were available in the laboratory. hda19-3 (SALK_139445) was provided by the Nottingham Arabidopsis Stock Center (NASC). The pSTK::STK-GFP line was available in the laboratory (Mizzotti et al. 2014).
stk and STK/stk shp1 shp2 seeds (used to obtain the stk shp1 shp2 triple mutant by segregation) were previously described and available in the laboratory (Favaro et al. 2003; Pinyopich et al. 2003).
Ovule number and placenta length measurement
Inflorescences were fixed overnight in ethanol:acetic acid 9:1 and rehydrated in an ethanol series. Pistils (stages 7 to 10) were dissected and mounted in chloral hydrate:glycerol:water (8:1:3, w/v/v) and immediately observed under a Zeiss Axiophot D1 microscope (Carl Zeiss MicroImaging) equipped with DIC optics and an Axiocam MRc5 camera (Zeiss) with Axiovision software (version 4.1). Ovules were counted manually. Placenta length was measured using the Axiovision software. For statistical analysis in Fig. 1 wild type and hda19-3 were confronted by Student's t-test (two-tailed distribution, homoscedastic). For experiments in Figs. 5 and 6 the statistical significance of differences in ovule number and placenta length was determined by a one-way ANOVA and a Bonferroni post hoc test for multiple comparisons (P < 0.05).
Pistil histology and imaging
Individual wild type, hda19-3, p35SRNAi:STM, and stk flowers were fixed in 2% (w/v) paraformaldehyde and 2.5% (w/v) glutaraldehyde in PIPES buffer [0.025 M, pH 7, 0.001% (v/v) Tween-80], placed under vacuum for 1 h, and then stored at 4 °C overnight. The material was dehydrated in an ethanol series and embedded in LR White resin. Thick sections (500 µm) were obtained with a Leica EM UC7 Ultramicrotome, stained with 1% (w/v) toluidine blue (Sigma-Aldrich, St Louis, MO, USA), and mounted with DPX (Sigma). Slides were observed under a Zeiss AxioImager AZ microscope equipped with a Zeiss Axiocam MRc3 camera using Zen Imaging software (Zen 2011 SP1).
Aniline blue staining for pollen tube growth analysis
Flowers were emasculated and pollinated with wild-type pollen. Pollen tubes were allowed to grow for different times depending on the experiment (12, 14, 16 or 24 HAP). Then, pistils were fixed overnight in 9:1 ethanol:acetic acid, washed three times with water, incubated in 1 M NaOH overnight, washed three times in water, and incubated in 0.1% aniline blue (w/v). Pistils were imaged using a Zeiss AxioPhot D1 microscope equipped with fluorescence filters. Images were recorded with an Axiocam MRc5 camera (Zeiss) with Axiovision software (version 4.1). Pollen tube length was measured using ImageJ (Schneider et al. 2012). For experiments reported in Fig. 1 and Supplemental Fig. S1, statistical significance was determined by Student's t-test (two-tailed distribution, homoscedastic), comparing wild type with HDA19/hda19-3 or hda19-3. For the experiment in Fig. 5 and Supplemental Fig. S5, statistical significance was determined by a one-way ANOVA and a Bonferroni post hoc test for multiple comparisons (P < 0.05).
Protoplast recovery and fluorescence activated cell sorting
The pSTK::STK-GFP construct was introgressed into the hda19-3 background and then used for cell sorting, along with the parental pSTK::STK-GFP line. pSTK::STK-GFP is expressed specifically in the TT, placenta, and ovules during stages 8 to 12 (Mizzotti et al. 2014). To be able to isolate sufficient numbers of cells expressing STK-GFP in a short time, we collected whole inflorescences, except for open flowers, directly in the solution used for protoplasting (Villarino et al. 2016).
Protoplasting and sorting were performed as described (Villarino et al. 2016), with some modifications. Namely, cells were sorted into RLT buffer from the RNAeasy Microkit (Qiagen) supplemented with 1% β-mercaptoethanol (v/v), keeping a ratio of 3 volumes of RLT buffer per 1 volume of sorted cells (in our sorting conditions, 50,000 cells in 200 µL). Cell sorting was performed using a Moflo XDP (Beckman Coulter Inc.). Each experiment yielded between 80,000 and 100,000 STK-GFP-positive cells.
RNA extraction and expression analysis
RNA was extracted immediately after sorting using an RNAeasy Microkit (Qiagen) with a few modifications. We used one column for every 25,000 cells. Columns were eluted twice, using the first eluate for the second elution. Finally, all eluates from the same sample were pooled, yielding 70 to 100 ng of total RNA. For RT-qPCR validation, the columns were eluted a third time, using 14 µL of nuclease-free water that was sequentially used to elute all the columns corresponding to the same sample. This yielded around 5 ng of total RNA that was retro-transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen/Life Technologies). Then, the expression of STK-GFP was measured by RT-qPCR using a Thermal Cycler from Applied Biosystems and a QuantiTect SYBR Green PCR Kit (Qiagen). MON1 (AT2G28390) was used as a reference gene for normalization (Czechowski et al. 2005).
For evaluation of STM levels in RNAi lines, RNA was extracted using Trizol (Invitrogen), and 500 ng of RNA was retrotranscribed using the iScript cDNA synthesis kit (BioRad). RT-qPCRs were performed using iTaq Universal SYBR Green Supermix (BioRad) in a Bio-Rad iCycler iQ thermal cycler. The ACT8 gene (AT1G49240) was used as the reference gene for normalization because, according to our RNA-seq data, its RNA levels did not change in hda19-3 plants.
All primers used in RT-qPCR experiments showed an amplification efficiency close to 100%, so the 2−ΔΔCT method was used for the calculation of expression changes. All primers used are listed in Supplemental Table S1.
RNA sequencing and analysis
Three samples of RNA from wild-type GFP-positive cells and four samples from hda19-3 GFP-positive cells were used for library preparation. Libraries were sequenced in one lane of the HiSeq2500 Illumina platform with a yield of 250 million reads and an average of 35 million reads per sample. Around 10 million reads were filtered out. Of the 240 million reads remaining, 230 million were mapped uniquely (96% of the reads after filtering) and were used for analysis.
Library preparation, RNA sequencing, and bioinformatic analysis were essentially performed as described (Villarino et al. 2016). Briefly, we used Cufflinks (Trapnell et al. 2012), DeSeq2 (Love et al. 2014), and EdgeR (Robinson et al. 2009) to analyze the RNA-seq data (Supplemental Data Set 1 to S3). Then, we crossed the data obtained by the three software packages and selected the DEGs detected by all three programs, with an FDR < 0.001 and a logFC > |1.2| (Supplemental Data Set 4). Genes whose expression value was zero in either wild-type or hda19-3 samples were filtered out.
Chromatin immunoprecipitation and qPCR analysis
Chromatin immunoprecipitation was performed as described (Mizzotti et al. 2014). We used a rabbit anti-H3K9/K14ac antibody (Upstate 07-352, Sigma-Aldrich) for the immunoprecipitation of DNA fragments associated with acetylated histones and a mouse monoclonal anti-GFP antibody (Roche, #11814460001) for the immunoprecipitation of DNA fragments associated with STK-GFP. We collected the whole inflorescences, leaving the IM out by cutting the flowers (up to stage 12) just below their base with tweezers. qPCR was performed using iTaq Universal SYBR Green Supermix (BioRad) in a Bio-Rad iCycler iQ thermal cycler.
Enrichment of tested areas was calculated as fold-change against a control, nonenriched region (GAPDH locus) as previously described (Matias-Hernandez et al. 2010). Primers used for ChIP-qPCR are listed in Supplemental Table S1. For the anti-H3K9/K14ac antibody ChIP-qPCR experiment, statistical significance was evaluated by Student's t-test (two-tailed distribution, homoscedastic) comparing wild type with hda19-3, stk or stkshp1shp2 among three independent biological replicates. For the anti-GFP antibody ChIP-qPCR, wild type and pSTK::STK-GFP enrichment values were compared with Student's t-test (two-tailed distribution, homoscedastic) among four independent biological replicates.
In situ hybridization
Sectioning and ISH were performed as described (Dreni et al. 2007). The STM probe was obtained as described (Simonini and Kater 2014).
Generation of RNAi lines
For the p35S::STM-RNAi construct, a fragment of 352 bp of STM coding sequence was amplified (see primer list in Supplemental Table S3), cloned into pDONR207, and then subcloned into pFRH (derived from pFGC5941; NCBI accession number AY310901) using Gateway technology (Invitrogen).
The construct was transformed into Agrobacterium tumefaciens strain EHA105, and hda19-3 plants were transformed by the floral dip method (Clough and Bent 1998). Transformants were selected on MS media (Murashige and Skoog 1962) supplemented with 20 mg/L of hygromycin. The presence of the construct was confirmed by genotyping (see primers in Supplemental Table S1).
Clustering analysis
Heatmaps were obtained using Heatmapper (Babicki et al. 2016). Genes were hierarchically clustered by Euclidean distances using the average linkage method.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number PRJNA1031145.
Accession numbers of the major Arabidopsis genes mentioned in this work are as follows: HDA19 = AT4G38130; STM = AT1G62360; STK = AT4G09960; SHP1 = AT3G58780; SHP2 = AT2G42830.
Supplementary Material
Acknowledgments
Thanks to Sarah Schuett (NCSU) and Claudia Bazzini (UniMi) for technical assistance with FACS and the Genomic Sciences Laboratory Research Facility (NCSU) for library preparation and Illumina sequencing.
Contributor Information
Silvia Manrique, Dipartimento di Bioscienze, Università degli Studi di Milano, Via Giovanni Celoria 26, Milan 20133, Italy.
Alex Cavalleri, Dipartimento di Bioscienze, Università degli Studi di Milano, Via Giovanni Celoria 26, Milan 20133, Italy.
Andrea Guazzotti, Dipartimento di Bioscienze, Università degli Studi di Milano, Via Giovanni Celoria 26, Milan 20133, Italy.
Gonzalo H Villarino, Department of Plant and Microbial Biology, North Carolina State University, Raleigh, NC 27606, USA.
Sara Simonini, Department of Plant and Microbial Biology & Zurich-Basel Plant Science Center, University of Zurich, Zollikerstrasse 107, Zurich CH-8008, Switzerland.
Aureliano Bombarely, Dipartimento di Bioscienze, Università degli Studi di Milano, Via Giovanni Celoria 26, Milan 20133, Italy.
Tetsuya Higashiyama, Institute of Transformative Bio-Molecules (ITbM), Nagoya University, Furo-cho, Chikusa-ku, Nagoya, Aichi 464-8601, Japan.
Ueli Grossniklaus, Department of Plant and Microbial Biology & Zurich-Basel Plant Science Center, University of Zurich, Zollikerstrasse 107, Zurich CH-8008, Switzerland.
Chiara Mizzotti, Dipartimento di Bioscienze, Università degli Studi di Milano, Via Giovanni Celoria 26, Milan 20133, Italy.
Ana Marta Pereira, Faculdade de Ciências da Universidade do Porto, Departamento de Biologia, Universidade do Porto, rua do Campo Alegre, Porto 4169-007, Portugal; LAQV Requimte, Sustainable Chemistry, Universidade do Porto, Porto 4169-007, Portugal.
Silvia Coimbra, Faculdade de Ciências da Universidade do Porto, Departamento de Biologia, Universidade do Porto, rua do Campo Alegre, Porto 4169-007, Portugal; LAQV Requimte, Sustainable Chemistry, Universidade do Porto, Porto 4169-007, Portugal.
Subramanian Sankaranarayanan, Department of Biological Sciences and Engineering, Indian Institute of Technology Gandhinagar, Palaj, Gujarat 382355, India.
Elisabetta Onelli, Dipartimento di Bioscienze, Università degli Studi di Milano, Via Giovanni Celoria 26, Milan 20133, Italy.
Simona Masiero, Dipartimento di Bioscienze, Università degli Studi di Milano, Via Giovanni Celoria 26, Milan 20133, Italy.
Robert G Franks, Department of Plant and Microbial Biology, North Carolina State University, Raleigh, NC 27606, USA.
Lucia Colombo, Dipartimento di Bioscienze, Università degli Studi di Milano, Via Giovanni Celoria 26, Milan 20133, Italy.
Author contributions
S.Man., L.C., A.C., A.G., and R.G.F. conceived and design the research. S.Man., A.C., A.G., G.V., E.O., A.M.P., S.C., S.Sim., C.M., and S.Sans. performed the experiments. L.C., R.G.F., T.H., U.G., and S.Mas. contributed the reagents/materials/analysis tools. S.Man., A.C., G.V., and A.B. analyzed the data. S.Man., S.Mas., A.C., A.G., C.M., S.C., U.G., L.C. raised funds. S.Man., A.C., L.C., and A.G. wrote the manuscript with input from all authors.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Growth of pollen tubes in pistils of wild-type, HDA19/hda19-3, and hda19-3 plants.
Supplemental Figure S2. STK-GFP expression in the pSTK::STK-GFP marker line.
Supplemental Figure S3. Enrichment of the STK-GFP transcript.
Supplemental Figure S4. Expression levels of histone acetylases and deacetylases in STK-GFP expressing hda19-3 cells as compared to the wild type in RNA-seq data.
Supplemental Figure S5. Transmitting tract phenotypes of wild-type, had19-3, and hda19-3 STMRNAi plants.
Supplemental Table S1. Primers used in this study.
Supplemental Data Set 1. Results of RNA-seq analysis with Cufflinks.
Supplemental Data Set 2. Results of RNA-seq analysis with DeSeq2.
Supplemental Data Set 3. Results of RNA-seq analysis with EdgeR.
Supplemental Data Set 4. Common DEGs among Cufflinks, DeSeq2, and EdgeR.
Supplemental Data Set 5. Enrichment analysis of “Biological Process’ GO terms associated with the upregulated genes.
Supplemental Data Set 6. Enrichment analysis of “Biological Process’ GO terms associated with downregulated genes.
Funding
S.Man., A.C., and L.C. were funded by MIUR (PRIN2017). A.G. and L.C. were funded by H2020 Excellent Science (SEXSEED ID: 690946). S.Man. was funded by an EMBO short-term fellowship (2015). S.Man., S.Mas., and C.M. were funded by FP7 People: Marie-Curie Actions “International Research Staff Exchange Scheme” (FRUITLOOK ID: 612640). S.C. was funded by Fundação para a Ciência e a Tecnologia (Project—POCI-01-0145-FEDER-027839). U.G. was funded by the Swiss National Science Foundation (grant 31003A_179553).
Data availability
RNA-seq data are available under the accession number PRJNA1031145 (www.ncbi.nlm.nih.gov/bioproject/PRJNA1031145).
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Alonso-Cantabrana H, Ripoll JJ, Ochando I, Vera A, Ferrándiz C, Martínez-Laborda A. Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development. 2007:134(14):2663–2671. 10.1242/dev.02864 [DOI] [PubMed] [Google Scholar]
- Alvarez J, Smyth DR. CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int J Plant Sci. 2002:163(1):17–41. 10.1086/324178 [DOI] [Google Scholar]
- Alvarez JP, Goldshmidt A, Efroni I, Bowman JL, Eshed Y. The NGATHA distal organ development genes are essential for style specification in Arabidopsis. The Plant Cell. 2009:21(5):1373–1393. 10.1105/tpc.109.065482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhakanandam S, Nole-Wilson S, Bao F, Franks RG. SEUSS and AINTEGUMENTA mediate patterning and ovule initiation during gynoecium medial domain development. Plant Physiol. 2008:146(3):1165–1181. 10.1104/pp.107.114751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016:44(W1):W147–W153. 10.1093/nar/gkw419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK, Poethig RS. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development. 1993:119(3):823–831. 10.1242/dev.119.3.823 [DOI] [Google Scholar]
- Bell EM, Lin WC, Husbands AY, Yu L, Jaganatha V, Jablonska B, Mangeon A, Neff MM, Girke T, Springer PS. Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc Natl Acad Sci U S A. 2012:109(51):21146–21151. 10.1073/pnas.1210789109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belles-Boix E, Hamant O, Witiak SM, Morin H, Traas J, Pautot V. KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell. 2006:18(8):1900–1907. 10.1105/tpc.106.041988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M, Bertrand C, Servet C, Zhou DX. Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell. 2006:18(11):2893–2903. 10.1105/tpc.106.043489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollier N, Sicard A, Leblond J, Latrasse D, Gonzalez N, Gévaudant F, Benhamed M, Raynaud C, Lenhard M, Chevalier C, et al. At-MINI ZINC FINGER2 and SL-INHIBITOR of MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in arabidopsis and tomato. Plant Cell. 2018:30(1):83–100. 10.1105/tpc.17.00653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Baum SF, Eshed Y, Putterill J, Alvarez J. Molecular genetics of gynoecium development in Arabidopsis. Curr Top Dev Biol. 1999:45:155–161. 10.1016/s0070-2153(08)60316-6 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. ASYMMETRIC LEAVES1 reveals know gene redundancy in Arabidopsis. Development. 2002:129(8):1957–1965. 10.1242/dev.129.8.1957 [DOI] [PubMed] [Google Scholar]
- Chávez Montes RA, Herrera-Ubaldo H, Serwatowska J, de Folter S. Towards a comprehensive and dynamic gynoecium gene regulatory network. Curr Plant Biol. 2015:3–4:3–12. 10.1016/j.cpb.2015.08.002 [DOI] [Google Scholar]
- Choi S-M, Song H-R, Han S-K, Han M, Kim C-Y, Park J, Lee Y-H, Jeon J-S, Noh Y-S, Noh B. HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 2012:71(1):135–146. 10.1111/j.1365-313X.2012.04977.x [DOI] [PubMed] [Google Scholar]
- Chung Y, Zhu Y, Wu MF, Simonini S, Kuhn A, Armenta-Medina A, Jin R, Østergaard L, Gillmor SC, Wagner D. Auxin response factors promote organogenesis by chromatin-mediated repression of the pluripotency gene SHOOTMERISTEMLESS. Nat Commun. 2019:10(1):886. 10.1038/s41467-019-08861-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1Gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997:89(4):575–585. 10.1016/S0092-8674(00)80239-1 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998:16(6):735–743. 10.1046/j.1365-313X.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Colombo M, Brambilla V, Marcheselli R, Caporali E, Kater MM, Colombo L. A new role for the SHATTERPROOF genes during Arabidopsis gynoecium development. Dev Biol. 2010:337(2):294–302. 10.1016/j.ydbio.2009.10.043 [DOI] [PubMed] [Google Scholar]
- Crawford BCW, Yanofsky MF. The formation and function of the female reproductive tract in flowering plants. Curr Biol. 2008:18(20):R972–R978. 10.1016/j.cub.2008.08.010 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005:139(1):5–17. 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marzo M, Herrera-Ubaldo H, Caporali E, Novák O, Strnad M, Balanzà V, Ezquer I, Mendes MA, de Folter S, Colombo L. SEEDSTICK controls Arabidopsis fruit size by regulating Cytokinin levels and FRUITFULL. Cell Rep. 2020a:30(8):2846–2857.e3. 10.1016/j.celrep.2020.01.101 [DOI] [PubMed] [Google Scholar]
- Di Marzo M, Roig-Villanova I, Zanchetti E, Caselli F, Gregis V, Bardetti P, Chiara M, Guazzotti A, Caporali E, Mendes MA, et al. MADS-box and bHLH transcription factors coordinate transmitting tract development in Arabidopsis thaliana. Front Plant Sci. 2020b:11:526. 10.3389/fpls.2020.00526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PBF, An G, Colombo L, Kater MM. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 2007:52(4):690–699. 10.1111/j.1365-313X.2007.03272.x [DOI] [PubMed] [Google Scholar]
- Durbak AR, Tax FE. CLAVATA signaling pathway receptors of arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems. Genetics. 2011:189(1):177–194. 10.1534/genetics.111.130930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezquer I, Mizzotti C, Nguema-Ona E, Gotté M, Beauzamy L, Viana VE, Dubrulle N, Costa de Oliveira A, Caporali E, Koroney A-S. The developmental regulator SEEDSTICK controls structural and mechanical properties of the Arabidopsis seed coat. Plant Cell. 2016:28(10):2478–2492. 10.1105/tpc.16.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky MF, Kater MM, Colombo L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell. 2003:15(11):2603–2611. 10.1105/tpc.015123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Pařenicová L, Falasca G, Pelucchi N, Masiero S, Ciannamea S, Lopez-Dee Z, Altamura MM, Colombo L, Kater MM. Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol. 2004:135(4):2207–2219. 10.1104/pp.104.045039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillochet C, Lohmann JU. The never-ending story: from pluripotency to plant developmental plasticity. Development. 2015:142(13):2237–2249. 10.1242/dev.117614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Liu JS, Fan M, Bai MY, Wenkel S, Springer PS, Barton KM, Wang Z-Y. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc Natl Acad Sci U S A. 2012:109(51):21152–21157. 10.1073/pnas.1210799110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Reig S, Ripoll JJ, Vera A, Yanofsky MF, Martínez-Laborda A. Antagonistic gene activities determine the formation of pattern elements along the mediolateral axis of the Arabidopsis fruit. PLoS Genet. 2012:8(11):e1003020. 10.1371/journal.pgen.1003020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorham SR, Weiner AI, Yamadi M, Krogan NT. HISTONE DEACETYLASE 19 and the flowering time gene FD maintain reproductive meristem identity in an age-dependent manner. J Exp Bot. 2018:69(20):4757–4771. 10.1093/jxb/ery239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development. 2007:134(20):3593–3601. 10.1242/dev.011510 [DOI] [PubMed] [Google Scholar]
- Guo L, Zhou J, Elling AA, Charron JBF, Xing WD. Histone modifications and expression of light-regulated genes in arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol. 2008:147(4):2070–2083. 10.1104/pp.108.122929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J-X. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005:307:1634–1638. 10.1126/science.1107580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Sabatini S. Plant and animal stem cells: similar yet different. Nat Rev Mol Cell Biol. 2014:15(5):301–312. 10.1038/nrm3790 [DOI] [PubMed] [Google Scholar]
- Herrera-Ubaldo H, Lozano-Sotomayor P, Ezquer I, di Marzo M, Montes RAC, Goḿez-Felipe A, Pablo-Villa J, Diaz-Ramirez D, Ballester P, Ferrándiz C, et al. New roles of NO TRANSMITTING TRACT and SEEDSTICK during medial domain development in arabidopsis fruits. Development (Cambridge). 2019:146(1):dev172395. 10.1242/dev.172395 [DOI] [PubMed] [Google Scholar]
- Ishida T, Aida M, Takada S, Tasaka M. Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol. 2000:41(1):60–67. 10.1093/pcp/41.1.60 [DOI] [PubMed] [Google Scholar]
- Jan IC, Chun PJ, Hemmes H, Jung C, Chua NH. Rapid and reversible light-mediated chromatin modifications of arabidopsis phytochrome A locus. Plant Cell. 2011:23(2):459–470. 10.1105/tpc.110.080481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M-J, Jin H-S, Noh Y-S, Noh B. Repression of flowering under a noninductive photoperiod by the Hda 9 - Agl 19 - Ft module in Arabidopsis. New Phytologist. 2015:206(1):281–294. 10.1111/nph.2015.206.issue-1 [DOI] [PubMed] [Google Scholar]
- Kanrar S, Onguka O, Smith HMS. Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta. 2006:224(5):1163–1173. 10.1007/s00425-006-0298-9 [DOI] [PubMed] [Google Scholar]
- Kay P, Groszmann M, Ross JJ, Parish RW, Swain SM. Modifications of a conserved regulatory network involving INDEHISCENT controls multiple aspects of reproductive tissue development in Arabidopsis. New Phytol. 2013:197(1):73–87. 10.1111/j.1469-8137.2012.04373.x [DOI] [PubMed] [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008:20(9):2357–2371. 10.1105/tpc.107.055566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Piquerez SJM, Ramirez-Prado JS, Mastorakis E, Veluchamy A, Latrasse D, Manza-Mianza D, Brik-Chaouche R, Huang Y, Rodriguez-Granados NY, et al. GCN5 modulates salicylic acid homeostasis by regulating H3K14ac levels at the 5′ and 3′ ends of its target genes. Nucleic Acids Res. 2021:48(11):5953–5966. 10.1093/NAR/GKAA369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Hogan K, Long JA. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development. 2012:139(22):4180–4190. 10.1242/dev.085407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Harborough SR, McLaughlin HM, Natarajan B, Verstraeten I, Friml J, Kepinski S, Østergaard L. Direct ETTIN-auxin interaction controls chromatin states in gynoecium development. Elife. 2020;9:e51787. 10.7554/eLife.51787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Thakur JK, Prasad M. Histone acetylation dynamics regulating plant development and stress responses. Cell Mol Life Sci. 2021:78(10):4467–4486. 10.1007/s00018-021-03794-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B, Kiss A, Sassi M, Chauvet A, Das P, Cortizo M, Laufs P, Takeda S, Aida M, Traas J, et al. Mechanical stress contributes to the expression of the STM homeobox gene in Arabidopsis shoot meristems. Elife. 2015;4:e07811. 10.7554/eLife.07811.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Kwon SH, Lee SJ, Park SK, Song JT, Lee S, Lee MM, Hwang Y-H, Kim JH. The Arabidopsis thaliana NGATHA transcription factors negatively regulate cell proliferation of lateral organs. Plant Mol Biol. 2015:89(4–5):529–538. 10.1007/s11103-015-0386-y [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowmant JL, Yanofsky MF. SHATTERPROOF MADS-box genes control dispersal in Arabidopsis. Nature. 2000:404(6779):766–770. 10.1038/35008089 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996:379(6560):66–69. 10.1038/379066a0 [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006:312(5779):1520–1523. 10.1126/science.1123841 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsch-Martínez N, Zúñiga-Mayo VM, Herrera-Ubaldo H, Ouwerkerk PBF, Pablo-Villa J, Lozano-Sotomayor P, Greco R, Ballester P, Balanzá V, Kuijt SJH, et al. The NTT transcription factor promotes replum development in Arabidopsis fruits. Plant J. 2014:80(1):69–81. 10.1111/tpj.12617 [DOI] [PubMed] [Google Scholar]
- Matias-Hernandez L, Battaglia R, Galbiati F, Rubes M, Eichenberger C, Grossniklaus U, Kater MM, Colombo L. VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis. Plant Cell. 2010:22(6):1702–1715. 10.1105/tpc.109.068627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2018:47(D1):D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, Thomas PD. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat Protoc. 2019:14(3):703–721. 10.1038/s41596-019-0128-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci U S A. 2010:107(38):16512–16517. 10.1073/pnas.1004181107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Scalf M, Rytz TC, Hubler SL, Smith LM, Vierstra RD. Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in arabidopsis. Mol Cell Proteomics. 2013:12(2):449–463. 10.1074/mcp.M112.025056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci U S A. 2000:97(2):942–947. 10.1073/pnas.97.2.942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzotti C, Ezquer I, Paolo D, Rueda-Romero P, Guerra RF, Battaglia R, Rogachev I, Aharoni A, Kater MM, Caporali E, et al. SEEDSTICK is a master regulator of development and metabolism in the Arabidopsis seed coat. PLoS Genet. 2014:10(12):e1004856. 10.1371/journal.pgen.1004856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzotti C, Mendes MA, Caporali E, Schnittger A, Kater MM, Battaglia R, Colombo L. The MADS box genes SEEDSTICK and ARABIDOPSIS Bsister play a maternal role in fertilization and seed development. Plant J. 2012:70(3):409–420. 10.1111/j.1365-313X.2011.04878.x [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962:15(3):473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Nahar Most. A-U, Ishida T, Smyth DR, Tasaka M, Aida M. Interactions of CUP-SHAPED COTYLEDON and SPATULA genes control carpel margin development in Arabidopsis thaliana. Plant Cell Physiol. 2012:53(6):1134–1143. 10.1093/pcp/pcs057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y-Q, Chen Q, Lin R-N, Li Y-Q, Li L, Chen S, He X-J. The Hda 19 histone deacetylase complex is involved in the regulation of flowering time in a photoperiod-dependent manner. Plant J. 2019:98(3):448–464. 10.1111/tpj.2019.98.issue-3 [DOI] [PubMed] [Google Scholar]
- Norberg M, Holmlund M, Nilsson O. The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development. 2005:132(9):2203–2213. 10.1242/dev.01815 [DOI] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005:17(11):2899–2910. 10.1105/tpc.105.034876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira AM, Moreira D, Coimbra S, Masiero S. Paving the way for fertilization: the role of the transmitting tract. Int J Mol Sci. 2021:22(5):2603. 10.3390/ijms22052603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella R, Caselli F, Roig-Villanova I, Vignati V, Chiara M, Ezquer I, Tadini L, Kater MM, Gregis V. BPC transcription factors and a polycomb group protein confine the expression of the ovule identity gene SEEDSTICK in Arabidopsis. Plant J. 2020:102(3):582–599. 10.1111/tpj.14673 [DOI] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev Cell. 2015:33(5):576–588. 10.1016/j.devcel.2015.04.024 [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003:424(6944):85–88. 10.1038/nature01741 [DOI] [PubMed] [Google Scholar]
- Poulios S, Vlachonasios KE. Synergistic action of GCN5 and CLAVATA1 in the regulation of gynoecium development in Arabidopsis thaliana. New Phytol. 2018:220(2):593–608. 10.1111/nph.15303 [DOI] [PubMed] [Google Scholar]
- Ragni L, Belles-Boix E, Günl M, Pautot V. Interaction of KNAT6 and KNAT2 with brevipedicellus and pennywise in Arabidopsis inflorescences. Plant Cell. 2008:20(4):888–900. 10.1105/tpc.108.058230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Olalde JI, de Folter S. Control of stem cell activity in the carpel margin meristem (CMM) in Arabidopsis. Plant Reprod. 2019:32(2):123–136. 10.1007/s00497-018-00359-0 [DOI] [PubMed] [Google Scholar]
- Reyes-Olalde JI, Zuñiga-Mayo VM, Chávez Montes RA, Marsch-Martínez N, de Folter S. Inside the gynoecium: at the carpel margin. Trends Plant Sci. 2013:18(11):644–655. 10.1016/j.tplants.2013.08.002 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009:26(1):139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AHK, Yanofsky MF. Fruit development in Arabidopsis. Arabidopsis Book. 2006:4:e0075. 10.1199/tab.0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc. 2008:3(6):1018–1025. 10.1038/nprot.2008.66 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012:9(7):671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S, Dewitte W, Murray JAH. The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J. 2007:50(5):767–781. 10.1111/j.1365-313X.2007.03095.x [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997:124(22):4481–4491. 10.1242/dev.124.22.4481 [DOI] [PubMed] [Google Scholar]
- Sessions RA, Zambryski PC. Arabidopsis gynoecium structure in the wild type and in ettin mutants. Development. 1995:121(5):1519–1532. 10.1242/dev.121.5.1519 [DOI] [PubMed] [Google Scholar]
- Simonini S, Kater MM. Class I BASIC PENTACYSTEINE factors regulate HOMEOBOX genes involved in meristem size maintenance. J Exp Bot. 2014:65(6):1455–1465. 10.1093/jxb/eru003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Angenent GC, Kaufmann K. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development. 2012:139(17):3081–3098. 10.1242/dev.074674 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990:2(8):755–767. 10.1105/tpc.2.8.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlberg JJ, Myrenås M, Kuusk S, Lagercrantz U, Kowalczyk M, Sandberg G, Sundberg E. STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 2006:47(1):112–123. 10.1111/j.1365-313X.2006.02775.x [DOI] [PubMed] [Google Scholar]
- Steffens NO. AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res. 2004:32(90001):368D–372. 10.1093/nar/gkh017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kikuchi A, Kamada H. The arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2008:146(1):149–161. 10.1104/pp.107.111674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SJ, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-Seq experiments with TopHat and Cufflinks. Nat Protoc. 2012:7(3):562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H. Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol. 2008:49(7):1025–1038. 10.1093/pcp/pcn079 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Kronenberger J, Grandjean O, Laufs P, Traas J. PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development. 2000:127(23):5157–5165. 10.1242/dev.127.23.5157 [DOI] [PubMed] [Google Scholar]
- Villarino GH, Hu Q, Manrique S, Flores-Vergara M, Sehra B, Robles L, Brumos J, Stepanova AN, Colombo L, Sundberg E. Transcriptomic signature of the SHATTERPROOF2 expression domain reveals the Meristematic nature of Arabidopsis gynoecial medial domain. Plant Physiol. 2016:171(1):42–61. 10.1104/pp.15.01845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang F, Rode S, Chin KK, Ko EE, Kim J, Iyer VR, Qiao H. Ethylene induces combinatorial effects of histone H3 acetylation in gene expression in Arabidopsis. BMC Genomics. 2017:18(1):538. 10.1186/s12864-017-3929-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn AN, Rueschhoff EE, Franks RG. Transcriptomic characterization of a synergistic genetic interaction during carpel margin meristem development in arabidopsis thaliana. PLoS One. 2011:6(10):e26231. 10.1371/journal.pone.0026231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Pollack D, Nieves E, Winchell A, Callaway M, Vigodner M. Can your protein be sumoylated? A quick summary and important tips to study SUMO-modified proteins. Anal Biochem. 2015:477:95–97. 10.1016/j.ab.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SX, Zhou LW, Hu LQ, Jiang YT, Zhang YJ, Feng SL, Jiao Y, Xu L, Lin W-H. Asynchrony of ovule primordia initiation in Arabidopsis. Development. 2020:147(24):dev196618. 10.1242/dev.196618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014:42(W1):W325–W330. 10.1093/nar/gku383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Yang S, Chen C-Y, Li C, Shan W, Lu W, Cui Y, Liu X, Wu K. Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture. PLoS Genet. 2015:11(3):e1005125. 10.1371/journal.pgen.1005125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Zhang L, Duan J, Miki B, Wu K. Histone Deacetylase19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell. 2005:17(4):1196–1204. 10.1105/tpc.104.028514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JJ, Cho JS, Han H, Blitz IL, Wang W, Cho Ken WY. Histone deacetylase 1 maintains lineage integrity through histone acetylome refinement during early embryogenesis. eLife. 2023:12. 10.7554/eLife.79380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Tan B, Luo M, Li Y, Liu C, Chen C, Yu C-W, Yang S, Dong S, Ruan J. HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell. 2013:25(1):134–148. 10.1105/tpc.112.096313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X, Zhang Y. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci U S A. 2010:107(31):13960–13965. 10.1073/pnas.1002828107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga-Mayo VM, Gómez-Felipe A, Herrera-Ubaldo H, de Folter S. Gynoecium development: networks in Arabidopsis and beyond. J Exp Bot. 2019:70(5):1447–1460. 10.1093/jxb/erz026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data are available under the accession number PRJNA1031145 (www.ncbi.nlm.nih.gov/bioproject/PRJNA1031145).