Abstract
Introduction:
STK11 and KEAP1 mutations (STK11 mutant [STK11 MUT] and KEAP1MUT) are among the most often mutated genes in lung adenocarcinoma (LUAD). Although STK11MUT has been associated with resistance to programmed death-(ligand)1 (PD-[L]1) inhibition in KRASMUT LUAD, its impact on immunotherapy efficacy in KRAS wild-type (KRASWT) LUAD is currently unknown. Whether KEAP1MUT differentially affects outcomes to PD-(L)1 inhibition in KRASMUT and KRASWT LUAD is also unknown.
Methods:
Clinicopathologic and genomic data were collected from September 2013 to September 2020 from patients with advanced LUAD at the Dana-Farber Cancer Institute/Massachusetts General Hospital cohort and the Memorial Sloan Kettering Cancer Center/MD Anderson Cancer Center cohort. Clinical outcomes to PD-(L)1 inhibition were analyzed according to KRAS, STK11, and KEAP1 mutation status in two independent cohorts. The Cancer Genome Atlas transcriptomic data were interrogated to identify differences in tumor gene expression and tumor immune cell subsets, respectively, according to KRAS/STK11 and KRAS/KEAP1 comutation status.
Results:
In the combined cohort (Dana-Farber Cancer Institute/Massachusetts General Hospital + Memorial Sloan Kettering Cancer Center/MD Anderson Cancer Center) of 1261 patients (median age = 61 y [range: 22–92], 708 women [56.1%], 1065 smokers [84.4%]), KRAS mutations were detected in 536 cases (42.5%), and deleterious STK11 and KEAP1 mutations were found in 20.6% (260 of 1261) and 19.2% (231 of 1202) of assessable cases, respectively. In each independent cohort and in the combined cohort, STK11 and KEAP1 mutations were associated with significantly worse progression-free (STK11 hazard ratio [HR] = 2.04, p < 0.0001; KEAP1 HR = 2.05, p < 0.0001) and overall (STK11 HR = 2.09, p < 0.0001; KEAP1 HR = 2.24, p < 0.0001) survival to immunotherapy uniquely among KRASMUT but not KRASWT LUADs. Gene expression ontology and immune cell enrichment analyses revealed that the presence of STK11 or KEAP1 mutations results in distinct immunophenotypes in KRASMUT, but not in KRASWT, lung cancers.
Conclusions:
STK11 and KEAP1 mutations confer worse outcomes to immunotherapy among patients with KRASMUT but not among KRASWT LUAD. Tumors harboring concurrent KRAS/STK11 and KRAS/KEAP1 mutations display distinct immune profiles in terms of gene expression and immune cell infiltration.
Keywords: KRAS, STK11, KEAP1, PD-(L)1 blockade, NSCLC
Introduction
Despite marked improvements in overall survival (OS) with programmed death-(ligand)1 (PD-[L]1) inhibition, most of the patients with metastatic NSCLC do not respond to immune checkpoint inhibition (ICI).1-4 Although tumor cell programmed death-ligand 1 (PD-L1) expression and high tumor mutational burden (TMB) are generally associated with improved benefit from ICI in NSCLC, the ability to discriminate patients who will respond to immunotherapy is limited.2,5-7 Thus, additional biomarkers of response and resistance to immunotherapy are needed to optimize treatment selection for patients with NSCLC.
KRAS mutations identify the largest subset of oncogene-driven lung adenocarcinoma (LUAD),8 and co-occurring genomic alterations in STK11 or KEAP1 genes define a unique subset of KRAS-mutant (KRASMUT) lung cancers with distinct biology and therapeutic vulnerabilities.9-12 The STK11 gene regulates diverse cellular functions including metabolism, growth, and polarity.13 STK11 loss occurs in approximately 15% of LUAD and is associated with a lack of PD-L1 expression, reduced tumor-infiltrating cytotoxic CD8+ T lymphocytes,9,14,15 and resistance to ICI in patients with KRASMUT NSCLC.16
Keap1 is a negative regulator of Nrf2, which is a master regulator of oxidative damage response.17 KEAP1 loss occurs in approximately 20% of NSCLC8 and is associated with an immunosuppressive microenvironment characterized by low infiltration of CD8+ T cells and natural killer cells in mouse models.12,18 Nevertheless, data on the correlation between KEAP1 loss and outcomes to ICI in patients with advanced LUAD are conflicting,19-21 and whether this mutation affects immunotherapy efficacy is in need of further investigation.
Because STK11 and KEAP1 mutations frequently co-occur in NSCLC, we sought to determine whether each gene mutation was independently associated with immunotherapy outcomes in NSCLC and to understand whether this impact was similar in both KRASMUT and KRAS wild-type (KRASWT) NSCLC. To unravel the potential mechanisms by which STK11 and KEAP1 alterations affect outcomes to ICI in LUAD, we also investigated the transcriptomic profiles of tumors harboring these mutations according to KRAS mutation status.
Materials and Methods
Two independent cohorts (Dana-Farber Cancer Institute and Massachusetts General Hospital [DFCI/MGH cohort] and Memorial Sloan Kettering Cancer Center and MD Anderson Cancer [MSKCC/MDACC cohort]) of patients with advanced LUAD who received PD-(L)1 inhibition and whose tumors underwent comprehensive genomic profiling were included. LUADs were characterized as STK11MUT or KEAP1MUT if they harbored loss-of-function alterations, including nonsense, frameshift, insertion/deletion, splice site, or pathogenic missense mutations in these genes (Supplementary Methods).
TMB was determined using the OncoPanel (DFCI) and MSK-IMPACT (MSKCC) next-generation sequencing platforms. TMB distributions were harmonized between the two platforms, as previously described.22 Determination of STK11 and KEAP1 mutation status, gene expression, and cell subset analysis from The Cancer Genome Atlas (TCGA) and detailed statistical analysis are reported in the Supplementary Methods.
Results
Patient Population
We identified a total of 1261 patients with advanced LUAD who received PD-(L)1 inhibition, with 620 (49.2%) in a discovery cohort consisting of cases from the DFCI/MGH cohort and 641 (50.8%) in a validation cohort from the MSKCC/MDACC cohort (Supplementary Table 1). In the combined cohort, co-occurring mutations in KRAS/STK11, KRAS/KEAP1, and STK11/KEAP1 were found in 10.9% (138 of 1261), 8.4% (101 of 1202), and 9.4% (113 of 1202) of KEAP1 assessable cases, respectively (Supplementary Fig. 1).
Impact of KRAS, STK11, and KEAP1 Mutation Status on PD-L1 Expression and TMB
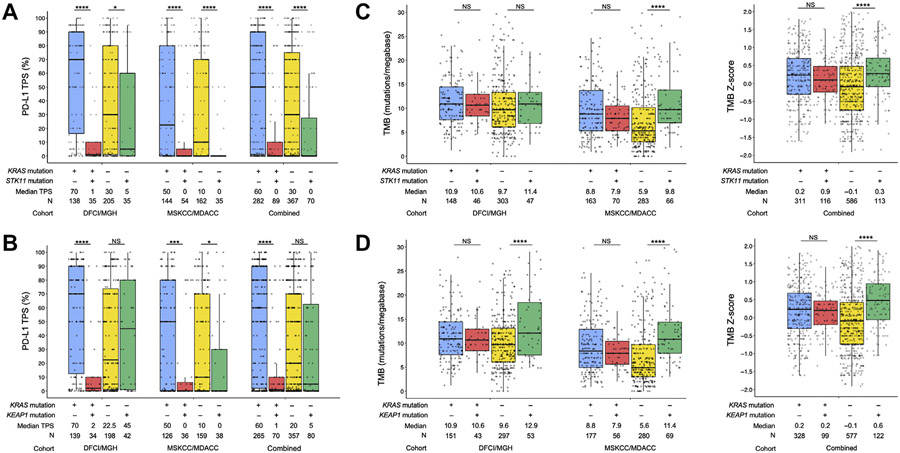
We first analyzed the impact of KRAS, STK11, and KEAP1 mutation status on PD-L1 expression and TMB. STK11MUT and KEAP1MUT LUADs had significantly lower PD-L1 expression in the DFCI/MGH, MSKCC/MDACC, and combined cohorts, whereas KRASMUT LUADs had significantly higher PD-L1 expression only in the DFCI/MGH cohort (Supplementary Fig. 2A-C). When analyzed by KRAS status, STK11 alterations were associated with significantly lower PD-L1 expression among both KRASMUT and KRASWT LUADs, whereas KEAP1 mutations were associated with lower PD-L1 expression predominantly among KRASMUT but not KRASWT cases (Fig. 1A and B). In terms of TMB distributions, we found significantly higher median TMB in KRASMUT and STK11MUT LUADs in the MSKCC/MDACC and in the combined cohort and among KEAP1MUT tumors in all the cohorts evaluated (Supplementary Fig. 2D-F). When TMB distributions were analyzed according to KRAS status, LUADs harboring STK11 mutations had a higher TMB only among KRASMUT but not KRASWT cancers in the MSKCC/MDACC and in the combined cohort, whereas KEAP1 MUT tumors had consistently higher TMB only among KRASMUT but not KRASWT cases, in all the cohorts evaluated (Fig. 1C and D).
Figure 1.
PD-L1 expression according to (A) KRAS/STK11 comutation status and (B) KRAS/KEAP1 comutation status, in the DFCI/MGH, MSKCC/MDACC, and combined cohorts. (C) Tumor mutational burden according to KRAS/STK11 comutation status, in the DFCI/MGH, MSKCC/MDACC, and combined cohorts. (D) Tumor mutational burden according to KRAS/KEAP1 comutation status, in the DFCI/MGH, MSKCC/MDACC, and combined cohorts. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. DFCI, Dana-Farber Cancer Institute; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; MSKCC, Memorial Sloan Kettering Cancer Center; NS, not significant; PD-L1, programmed death-ligand 1; TMB, tumor mutational burden; TPS, tumor proportion score
Impact of STK11 and KEAP1 Mutation Status on Clinical Outcomes to Programmed Cell Death Protein-1 Inhibition in KRASMUT and KRASWT LUAD
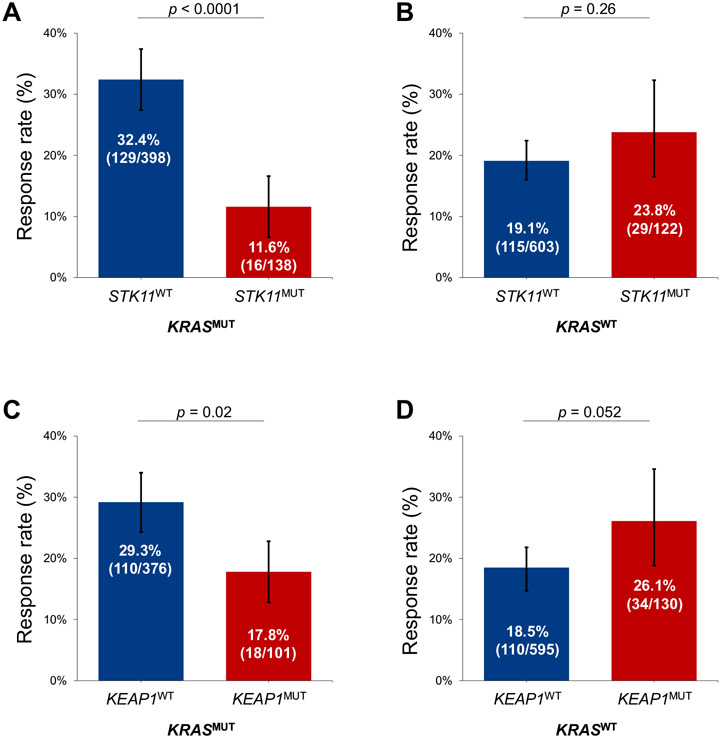
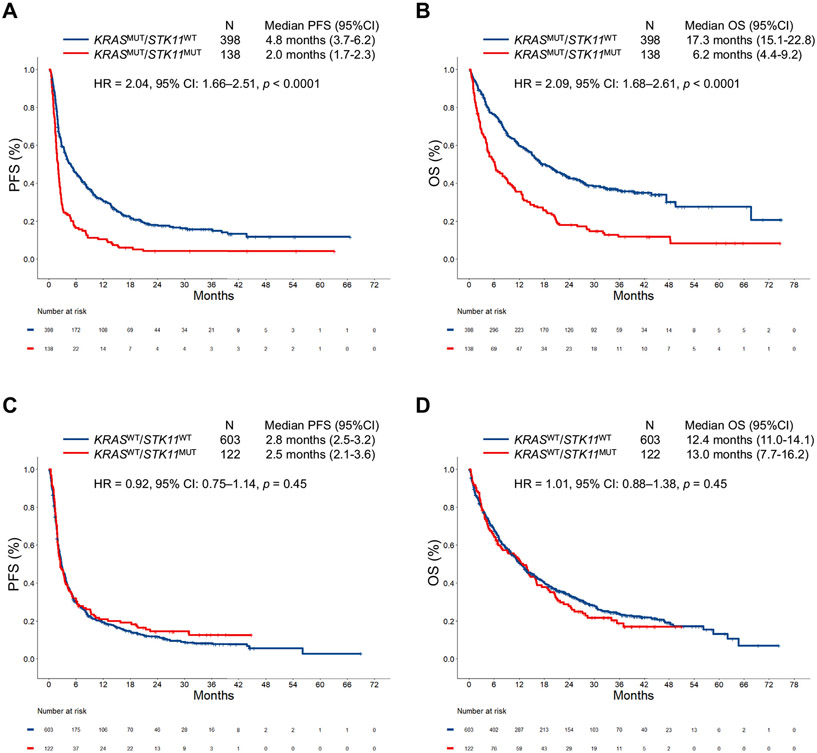
We next analyzed the impact of STK11 mutation on ICI efficacy in all comers with LUAD and in the context of KRAS mutation status. In both the independent cohorts, and in the combined cohort, STK11 mutation was associated with significantly shorter median progression-free survival (mPFS) and median OS (mOS) to ICI (Supplementary Fig. 3A-I). This deleterious effect of STK11 mutations on immunotherapy outcomes was largely driven by the KRASMUT subgroup of LUAD. In the DFCI/MGH and MSKCC/MDACC cohorts (Supplementary Fig. 4A-D), and in the combined cohort, STK11 mutation was associated with significantly worse overall response rate (ORR), mPFS, and mOS among KRASMUT LUADs but not among KRASWT tumors (Figs. 2A and B and 3A-D). STK11 mutation was confirmed to be an independent predictor of shorter PFS (hazard ratio [HR] = 1.46, p = 0.01) and OS (HR = 1.73, p = 0.002) to ICI in multivariable analysis in the combined cohort (Supplementary Table 2). Importantly, STK11 mutation was also associated with significantly worse clinical outcomes among KRASMUT LUADs across different PD-L1 expression level subgroups of less than 1%, 1% to 49%, and greater than or equal to 50%, when analyzed separately (Supplementary Fig. 5). The impact of STK11 mutation on ICI efficacy in the three most common KRASMUT alleles (G12C/V/D) is found in Supplementary Figure 6. Clinicopathologic characteristics of patients with KRASMUT and KRASWT LUADs according to STK11 mutation status are found in Supplementary Tables 3 and 4, whereas multivariate analyses for PFS and OS in the KRASWT group are found in Supplementary Table 5.
Figure 2.
Objective response rate to PD-(L)1 inhibition according to STK11 mutation status among (A) KRASMUT and (B) KRASWT LUADs in the combined cohort. Objective response rate to PD-(L)1 inhibition according to KEAP1 mutation status among (C) KRASMUT and (D) KRASWT LUADs in the combined cohort. LUAD, lung adenocarcinoma; MUT, mutant; PD-(L)1, programmed death-(ligand)1; WT, wild-type.
Figure 3.
(A) PFS and (B) OS to PD-(L)1 inhibition according to STK11 mutation status among KRASMUT LUADs in the combined cohort (DFCI/MGH + MSKCC/MDACC). (C) PFS and (D) OS to PD-(L)1 inhibition according to STK11 mutation status among KRASWT LUADs in the combined cohort (DFCI/MGH + MSKCC/MDACC). CI, confidence interval; DFCI, Dana-Farber Cancer Institute; HR, hazard ratio; LUAD, lung adenocarcinoma; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; MSKCC, Memorial Sloan Kettering Cancer Center; MUT, mutant; OS, overall survival; PD-(L)1, programmed death-(ligand)1; PFS, progression-free survival; WT, wild-type.
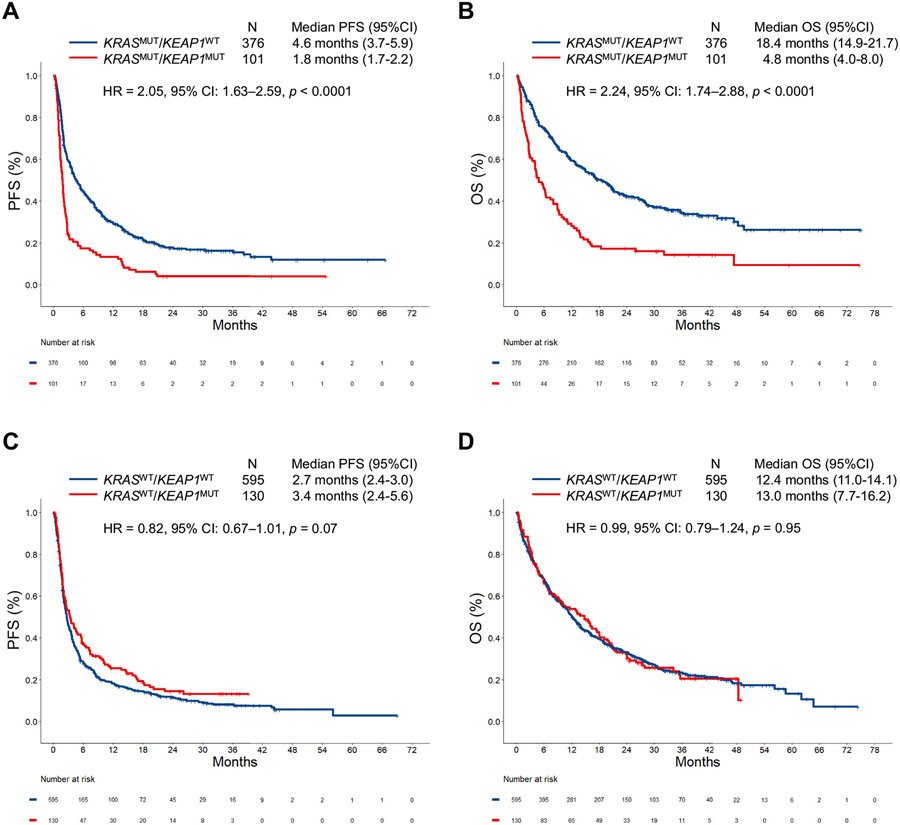
We next analyzed the impact of KEAP1 mutation on immunotherapy efficacy. In all comers with LUAD, KEAP1 mutation was associated with diminished survival from ICI (Supplementary Fig. 7A-I). Here, too, KEAP1 mutation affected immunotherapy efficacy in the KRASMUT subgroup but not among KRASWT cases in the two independent cohorts (Supplementary Fig. 8A-D) and in the combined cohort (Figs. 2C and D and 4A-D). KEAP1 mutation also retained a significant association with shorter PFS (HR = 2.15, p < 0.0001) and OS (HR = 2.44, p < 0.0001) among KRASMUT cases in multivariable models in the combined cohort (Supplementary Table 6). When the impact of KEAP1 mutation was analyzed in the different PD-L1 subgroups, we found that KEAP1 loss was associated with worse outcomes to immunotherapy in LUADs with a PD-L1 tumor proportion score less than 1% and 1% to 49% (Supplementary Fig. 9). The impact of KEAP1 mutation on ICI efficacy according to KRASMUT alleles (G12C/V/D) is found in Supplementary Figure 10. Clinicopathologic characteristics of patients with KRASMUT and KRASWT LUADs according to KEAP1 mutation status are found in Supplementary Tables 7 and 8, whereas multivariate analyses for PFS and OS in the KRASWT group are found in Supplementary Table 9.
Figure 4.
(A) PFS and (B) OS to PD-(L)1 inhibition according to KEAP1 mutation status among KRASMUT LUADs in the combined cohort (DFCI/MGH + MSKCC/MDACC). (C) PFS and (D) OS to PD-(L)1 inhibition according to KEAP1 mutation status among KRASWT LUADs in the combined cohort (DFCI/MGH + MSKCC/MDACC). CI, confidence interval; DFCI, Dana-Farber Cancer Institute; HR, hazard ratio; LUAD, lung adenocarcinoma; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; MSKCC, Memorial Sloan Kettering Cancer Center; MUT, mutant; OS, overall survival; PD-(L)1, programmed death-(ligand)1; PFS, progression-free survival; WT, wild-type.
When the impact of STK11 and KEAP1 mutation was explored in the TCGA cohort, there was no significant effect on disease-free survival or OS (Supplementary Figs. 11A and B and 12A and B). To further explore whether STK11 and KEAP1 mutations are negative prognostic markers in the context of KRAS mutation, we also evaluated the effect of these mutations on the ORR and PFS to first-line platinum-doublet chemotherapy in a cohort of 248 patients from DFCI. We found that both STK11 and KEAP1 mutations were associated with a significantly shorter PFS among KRASMUT but not KRASWT LUADs (Supplementary Fig. 13A-D), suggesting STK11 or KEAP1 loss may also influence efficacy of chemotherapy in the setting of KRASMUT but not KRASWT advanced LUAD.
Because LUADs harbor EGFR mutations or ALK rearrangements, and LUADs from never smokers do not typically respond to ICI,2,23,24 to ensure that our findings were not due to an enrichment in EGFR/ALK-positive LUAD or in LUAD from never smokers in the KRASWT cohort (Supplementary Tables 4, 8, and 10), we analyzed the impact of STK11 and KEAP1 mutations on clinical outcomes in KRASWT NSCLC after excluding EGFR/ALK-positive cases and never smokers. We confirmed that both STK11 and KEAP1 mutations had no impact on ORR, mPFS, or mOS among KRASWT/EGFRWT/ALKWT tumors (Supplementary Figs. 14A-C and 15A-C) and among ever smokers (Supplementary Figs. 14D-F and 15D-F).
Because STK11 and KEAP1 mutations tend to co-occur (Supplementary Table 10), we lastly evaluated whether among KRASMUT LUADs, STK11 and KEAP1 mutations affected ICI efficacy in KEAP1WT and STK11WT NSCLCs, respectively. We first analyzed the impact of STK11 mutation among KRASMUT/KEAP1WT LUADs and found that cases harboring and STK11 mutation had a significantly lower ORR (p = 0.004), shorter mPFS (HR = 1.93, p < 0.0001), and mOS (HR = 1.89, p < 0.0001) to ICI compared with KRASMUT/KEAP1WT/STK11WT cases (Supplementary Fig. 16A-C).
When we analyzed the effect of KEAP1 mutation among KRASMUT/STK11WT cases, there was no difference in ORR to immunotherapy between KEAP1MUT and KEAP1WT cases (p = 0.99, Supplementary Fig. 16D). Nevertheless, the mPFS and mOS were significantly shorter among cases with KEAP1 mutation compared with KRASMUT/STK11WT/KEAP1WT cases (PFS HR = 1.80, p = 0.002; mOS HR = 2.06, p = 0.002, Supplementary Fig. 16E and F), suggesting the deleterious impact of KEAP1 mutation on ICI efficacy was independent from the presence of a concurrent STK11 mutation. We confirmed the independent contributions of STK11 and KEAP1 mutations to worse outcome to ICI among KRASMUT cases by lastly testing them in a multivariate model including the interaction between mutations in the two genes (Supplementary Fig. 17A and B). Both STK11 and KEAP1 mutations were associated with lower PFS and OS, whereas the interaction term was not associated with PFS and OS, suggesting an additive effect of STK11 and KEAP1 mutations on immunotherapy outcomes. The impact of concurrent STK11/KEAP1 mutations in these two genes on ORR, mPFS, and mOS to ICI among KRASMUT LUAD is found in Figure 5A-C. The overlay of KRAS status (KRASMUT and KRASWT) with KEAP1, STK11, and concurrent STK11/KEAP1 mutation is found in Supplementary Figure 18A-C.
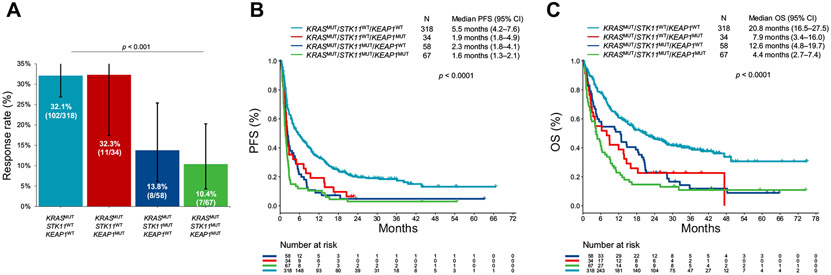
Figure 5.
(A) Objective response rate, (B) PFS, and (C) OS to PD-(L)1 inhibition according to STK11/KEAP1 comutation status, among patients with KRASMUT lung adenocarcinoma in the combined cohort. CI, confidence interval; MUT, mutant; OS, overall survival; PD-(L)1, programmed death-(ligand)1; PFS, progression-free survival; WT, wild-type.
Gene Ontology Analysis Reveals That STK11MUT and KEAP1MUT LUADs Have Different Transcriptomic Profiles According to KRAS Mutation Status
To unravel the potential mechanisms by which the deleterious impact of STK11 and KEAP1 mutations on outcomes to ICI in LUAD is primarily driven by KRAS mutation, we investigated the transcriptomic profiles of tumors harboring these mutations in KRASMUT and KRASWT LUADs. RNA sequencing data of 513 LUADs in the TCGA data set were analyzed according to KRAS/STK11 and KRAS/KEAP1 comutation status (Supplementary Methods).
We first identified genes that were differentially expressed among KRASMUT/STK11WT versus KRASMUT/STK11MUT LUADs and among KRASWT/STK11WT versus KRASWT/STK11MUT cancers. Next, we performed a hierarchical gene ontology analysis only on the subsets of genes which were differentially regulated in KRASMUT/STK11WT tumors versus KRASMUT/STK11MUT but not among KRASWT/STK11WT versus KRASWT/STK11MUT. Among the 22 significant terminal pathways identified (Supplementary Table 11), 13 involved in immune-mediated processes were markedly down-regulated in KRASMUT/STK11MUT compared with KRASMUT/STK11WT LUADs, including the MHC class II protein complex, T-cell activation, immune response-activating signaling, leukocyte migration, leukocyte degranulation, and myeloid leukocyte activation (Fig. 6A). The log2 fold change in mRNA expression of the top 20 individual genes included in the six prioritized pathways which were significantly down-regulated in KRASMUT/STK11MUT tumors compared with KRASMUT/STK11WT LUAD is found in Supplementary Figure 19. We noted that genes of the class II HLA family, including CD74, HLA-DOA, HLA-DRB5, HLA-DRB1, and HLA-DMB, were significantly down-regulated in KRASMUT/STK11MUT LUADs compared with KRASMUT/STK11WT tumors. Also, genes encoding for chemokines and their receptors that are critical for T-cell, natural killer cell, and myeloid-cell recruitment and migration, such as CXCL14, CCL23, CX3CR1, and CCR6, were also significantly down-regulated in KRASMUT/STK11MUT tumors compared with KRASMUT/STK11WT cancers. In addition, SIGLEG-14, an enhancer of inflammasome activation and macrophage interleukin-1β release, exhibited marked down-regulation among KRASMUT/STK11MUT versus KRASMUT/STK11WT LUADs. The full list of genes in the 13 prioritized pathways that are significantly down-regulated in KRASMUT/STK11MUT tumors versus KRASMUT/STK11WT but not among KRASWT/STK11MUT versus KRASWT/STK11WT is found in Supplementary Table 12.
Figure 6.
(A) Bubble plot revealing the 13 prioritized immune-related pathways which are significantly down-regulated in KRASMUT/STK11MUT compared with KRASMUT/STK11WT LUADs, but not in KRASWT/STK11MUT compared with KRASWT/STK11WT LUADs. (B) Bubble plot revealing the 11 prioritized immune-related pathways which are significantly down-regulated in KRASMUT/KEAP1MUT compared with KRASMUT/KEAP1WT LUADs, but not in KRASWT/KEAP1MUT compared with KRASWT/KEAP1WT LUADs. (C) Cell-type enrichment analysis using xCell revealing the cell types that are uniquely enriched in KRASMUT/STK11WT compared with KRASMUT/STK11MUT LUADs, but not in KRASWT/STK11WT compared with KRASWT/STK11MUT LUADs. (D) Cell-type enrichment analysis using xCell revealing the cell types that are uniquely enriched in KRASMUT/KEAP1WT compared with KRASMUT/KEAP1MUT LUADs, but not in KRASWT/KEAP1WT compared with KRASWT/KEAP1MUT LUADs. LUAD, lung adenocarcinoma; MUT, mutant; NK, natural killer; NS, not significant; WT, wild-type. *p < 0.05, **p < 0.01, ***p < 0.001.
# Sum of B cells, CD4+ T cells, CD8+ T cells, Dendritic cells, Eosinophils, Macrophages, Monocytes, Mast cells, Neutrophils, NK cells.
§ Composite score of ImmuneScore + Stroma Score (Adipocytes, Endothelial cells, Fibroblasts)
We next identified genes that were differentially expressed among KRASMUT/KEAP1MUT versus KRASMUT/KEAP1WT LUADs and among KRASWT/KEAP1MUT versus KRASWT/KEAP1WT cancers and performed gene ontology analysis on the subsets of genes which were uniquely up-regulated in KRASMUT/KEAP1WT tumors versus KRASMUT/KEAP1MUT. Among the 13 terminal pathways identified (Supplementary Table 13), 11 were involved in immune-related processes, including the following gene ontology terms: external side of plasma membrane, regulation of T-cell activation, T-cell receptor signaling, defense response to virus, regulation of leukocyte cell-to-cell adhesion, and lymphocyte migration (Fig. 6B). The log2 fold change of the individual top 20 genes included in each of the six prioritized pathways is found in Supplementary Figure 20. Interestingly, we found several genes involved in monocyte, T-cell, and dendritic cell recruitment to be significantly down-regulated in KRASMUT/KEAP1MUT tumors compared with KRASMUT/KEAP1WT cases, including CCL2, CXCL6, CCR1, CCR6, CCR7, and ITGAM. In addition, genes encoding proinflammatory cytokines and their receptors, such as TNF, TNFSF8, TNFRSF9, IL1B, and IL2RA, were also markedly down-regulated in KRASMUT/KEAP1MUT tumors versus KRASMUT/KEAP1WT cancers. Importantly, we also noted positive regulators of type I interferon and other inflammatory cytokine production, such as TMEM173 (STING), DDX58, TLR4, and TLR7, to be markedly down-regulated in KRASMUT/KEAP1MUT versus KRASMUT/KEAP1WT cancers. The full list of genes in the 11 pathways which are significantly down-regulated in KRASMUT/KEAP1MUT tumors versus KRASMUT/KEAP1WT but not among KRASWT/KEAP1MUT versus KRASWT/KEAP1WT is found in Supplementary Table 14.
Cell-Type Enrichment Analysis Reveals That STK11MUT and KEAP1MUT Tumors Have Different Immunophenotypes According to KRAS Mutation Status
We lastly evaluated whether LUADs harboring STK11 or KEAP1 mutations also have distinct immune cell subsets according to the presence or absence of concurrent KRAS mutation; we performed cell-type enrichment analysis by deconvoluting gene expression data into tumor-associated cell population.
First, we evaluated whether STK11 mutation was associated with different cell infiltration according to KRAS mutation status and identified six immune cell types that were significantly enriched in KRASMUT/STK11WT tumors compared with KRASMUT/STK11MUT tumors but not in KRASWT/STK11WT tumors versus KRASWT/STK11MUT cancers, including M1 macrophages (p < 0.01), M2 macrophages (p < 0.01), granulocyte-monocyte progenitors (p = 0.02), CD4+ effector memory cells (p = 0.01), and B cells (p = 0.04) (Fig. 6C). In addition, both the immune score (sum of B cells, T cells, and myeloid-derived cells) and the microenvironment score (composite score of the immune score and stroma cell signatures) were significantly enriched only in KRASMUT/STK11WT tumors compared with KRASMUT/STK11MUT (p < 0.001 and p < 0.01, respectively; Fig. 6C). Conversely, KRASMUT/STK11MUT tumors were significantly enriched in neutrophils (p < 0.01), compared with KRASMUT/STK11WT tumors (Fig. 6C).
We next investigated whether KEAP1 mutation was also associated with a distinct pattern of infiltrating cell types in KRASMUT and KRASWT LUADs. We identified four cell types including CD8+ T cells (p < 0.001), CD8+ central memory T cells (p < 0.01), CD8+ naive T cells (p = 0.02), and B cells (p = 0.01) which were uniquely enriched in KRASMUT/KEAP1WT tumors versus KRASMUT/KEAP1MUT but not among KRASWT/KEAP1WT tumors versus KRASWT/KEAP1MUT cancers (Fig. 6D). Instead, mesenchymal stem cells were found to be significantly enriched in only KRASMUT/KEAP1MUT tumors compared with KRASMUT/KEAP1WT tumors (p = 0.02) (Fig. 6D).
Discussion
In this study, we reveal that mutations in STK11 and KEAP1 are frequent and define major subsets of KRASMUT LUADs, characterized by unique immune profiles and poor outcomes to ICI in two independent cohorts. Our results extend previous reports of LUAD with STK11 mutations16 and identify loss-of-function mutations in KEAP1 as a frequent and independent driver of resistance to ICI in patients with advanced KRASMUT LUAD. To gain insights to potential mechanisms by which STK11 and KEAP1 loss exerts deleterious effects on PD-(L)1 inhibition among KRASMUT but not KRASWT LUAD, we found that KRASMUT/STK11MUT tumors had a significant down-regulation of MHC class II compared with KRASMUT/STK11WT, including HLA-DOA, HLA-DRB5, HLA-DRB1, and HLA-DMB. By contrast, STK11 mutation was not associated with MHC class II pathway deregulation among KRASWT cases. The expression of MHC class II-restricted antigens by tumor cells is required for CD4+ T-cell activation to elicit antitumor immune responses,25 and MHC class II expression has been associated with improved PFS and OS in patients treated with ICI in multiple cancer types.26-28
KRASMUT/KEAP1MUT LUAD was also found to have a unique gene expression profile, characterized by significant down-regulation of positive regulators of type I interferon and other inflammatory cytokines, including TMEM173 (STING), DDX58, TLR4, and TLR7. Although STK11 loss has previously been reported to result in marked silencing of STING expression in KRASMUT LUAD, whether a similar mechanism could lead to impaired tumor immunogenicity in KRASMUT/KEAP1MUT LUAD is unknown and deserves additional exploration.
These findings have implications for clinical trial interpretation and design and for treatment selection. Our study suggests that immunotherapy clinical trials should consider using stratification measures to balance randomized groups for STK11 and KEAP1 comutation status and ensure that differences in outcomes are due to therapeutic interventions rather than variations in STK11 or KEAP1 mutation frequency, especially in KRASMUT NSCLC. Our findings could also inform on how to sequence or combine future treatment strategies in KRASMUT LUAD. Preliminary data have revealed that direct KRAS inhibitors can produce responses in approximately 35% to 45% of patients with KRAS G12CMUT NSCLC.29-31 As more effective treatment options become available for KRASMUT LUAD, STK11 and KEAP1 mutation status might be a useful biomarker in determining the optimal treatment sequence, and KRAS G12C inhibitors might be better used before ICI in genomic subsets of NSCLC which are predicted not to respond to PD-1 based regimens. Whether KRAS inhibition could be used in combination with immunotherapy is an area of increasing interest. Preclinical data have revealed that KRAS G12C inhibition reinvigorates the TME with CD8+ T cells, macrophages, and CD103+ cross-presenting dendritic cells, suggesting direct KRAS inhibitors may synergize with ICI,32 particularly among genomically defined LUADs that are not predicted to respond to immunotherapy alone. Phase I/II trials of sotorasib and adagrasib in combination with pembrolizumab in patients with advanced NSCLC with KRAS G12C mutation are currently ongoing (NCT03600883, NCT04613596).
In this study, we also found that patients with KRASMUT LUADs and STK11 or KEAP1 mutation had worse clinical outcomes to platinum-based chemotherapy, which may argue against an only predictive nature of concurrent KRAS/STK11 and KRAS/KEAP1 alterations in LUADs. Consistently with our findings, KEAP1 and STK11 mutations have been previously reported to correlate with inferior clinical outcomes in patients treated with chemotherapy.19,33 Loss of STK11 and constitutive activation of Nrf2 in KEAP1MUT tumors have been found to promote transcription of various cytoprotective genes that are associated with antioxidant and detoxification enzymes and protect cancer cells from ferroptosis, leading to chemoresistance in various cancers.11,34-36 Therefore, STK11 and KEAP1 mutation could potentially be predictive of worse outcomes to both chemotherapy and immunotherapy because of their pleiotropic effects on cancer cell metabolism and immune system engagement. Conversely, STK11 mutation does not seem to affect the efficacy of KRAS G12C inhibition in patients with KRASG12C-mutated NSCLC receiving sotorasib.37
Limitations of this study include the retrospective design and the lack of validation from published randomized clinical trials of ICI versus chemotherapy. In addition, PD-L1 expression was not available in 35.9% of the samples. Nevertheless, to account for the potential selection bias resulting from PD-L1 tumor proportion score missingness, we used an inverse probability weighting in Cox regression analysis. Lastly, it should be acknowledged that KRASWT LUADs are highly heterogeneous in terms of clinicopathologic and genomic features and include subsets of tumors that typically not respond to immunotherapy, such as those harboring EGFR mutations and ALK rearrangements and those from never smokers. To address this bias, we analyzed the impact of STK11 and KEAP1 mutations on clinical outcomes in KRASWT NSCLC after excluding EGFR/ALK-positive cases and excluding never smokers and confirmed that both STK11 and KEAP1 mutation had no impact on immunotherapy efficacy among KRASWT/EGFRWT/ALKWT tumors and among ever smokers.
In conclusion, we reveal that STK11 and KEAP1 mutations confer worse outcomes to immunotherapy among patients with KRASMUT but not among KRASWT LUAD and that tumors harboring concurrent KRAS/STK11 and KRAS/KEAP1 mutations display distinct immune profiles. Preclinical studies are urgently needed to further dissect the molecular mechanism underlying these correlations and identify novel therapeutic vulnerabilities.
Supplementary Material
Acknowledgments
The work of Dr. Ricciuti was supported by the International Association for the Study of Lung Cancer Fellowship Award 2020 and by the 2020 Conquer Cancer Foundation of ASCO Young Investigator Award. The Mark Foundation for Cancer Research (Grant # 19-029 MIA) supported in part the collection, management, analysis, and interpretation of the data at the MD Anderson Cancer Center (MDACC). The MD Anderson Cancer Center Support Grant P30 CA01667 supported in part the collection, management, analysis, and interpretation of the data at the MDACC. The generous philanthropic contributions to The University of Texas MD Anderson Lung Moon Shot Program supported in part the collection, management, analysis, and interpretation of the data at the MDACC.
Disclosure:
Dr. Hellmann reports receiving research support from Bristol-Myers Squibb; has been a compensated consultant for Merck, Bristol-Myers Squibb, AstraZeneca, Genentech/Roche, Nektar, Syndax, Mirati, Shattuck Labs, Immunai, Blueprint Medicines, Achilles, and Arcus; receiving travel support/honoraria from AstraZeneca, Eli Lilly, and Bristol-Myers Squibb; has options from Shattuck Labs, Immunai, and Arcus; and has a patent filed by his institution related to the use of tumor mutation burden to predict response to immunotherapy (PCT/US2015/062208), which has received licensing fees from Personal Genome Diagnostics. Dr. Lin has served as a compensated consultant or received honorarium from Chugai Pharma, Boehringer Ingelheim, Pfizer, C4 Therapeutics, Nuvalent, Turning Point Therapeutics, and Genentech; received institutional research funds from Hengrui Therapeutics, Turning Point Therapeutics, Neon Therapeutics, Relay Therapeutics, and Novartis; received CME funding from OncLive, MedStar Health, and Northwell Health; and received travel support from Pfizer. Dr. Nishino reports serving as consultant to Daiichi Sankyo and AstraZeneca; receiving research grant from Merck, Canon Medical Systems, AstraZeneca, and Daiichi Sankyo; receiving honorarium from Roche; and being supported by R01CA203636 and U01CA209414 (NCI). Dr. Arbour reports serving as a consultant for AstraZeneca and Iovance Biotherapeutics and receiving research support through her institution on her behalf from Novartis, Takeda, and Nektar. Dr. Gainor reports serving as a compensated consultant or received honoraria from Bristol-Myers Squibb, Genentech, Ariad/Takeda, Loxo Oncology/Eli Lilly, Blueprint, Oncorus, Regeneron, EMD Serono, Gilead, AstraZeneca, Pfizer, Incyte, Novartis, Merck, Agios, Amgen, and Array; receiving research support from Novartis, Genentech/Roche, and Ariad/Takeda; receiving institutional research support from Bristol-Myers Squibb, Tesaro, Moderna, Blueprint, Jounce, Array Biopharma, Merck, Adaptimmune, Novartis, and Alexo; and having an immediate family member who is an employee of Ironwood Pharmaceuticals. Dr. Awad reports serving on the consultant/advisory board for Bristol-Myers Squibb, AstraZeneca, Achilles, AbbVie, Neon, Maverick, Nektar, Hegrui, Syndax, and Gritstone and receiving research funding from Bristol-Myers Squibb, AstraZeneca, Eli Lilly, and Genentech. Dr. Sholl reports serving as consultant for Foghorn Therapeutics. Dr. Barbie reports serving as consultant for N of One/Qiagen and Tango Therapeutics; receiving research grants from Bristol-Myers Squibb, Novartis, Eli Lilly, and Gilead Sciences; and being a cofounder and serving on the scientific advisory board of Xsphera Biosciences Inc. Dr. Zhang reports receiving grants from Merck and Johnson and Johnson and personal fees from Bristol-Myers Squibb, AZ, GenePlus, and Innovent. Dr. Heymach reports serving as a consultant to AstraZeneca, Bristol-Myers Squibb, GlaxoSmithKline, Guardant Health, Kairos Venture Investments, BrightPath Biotherapeutics, Hengrui Therapeutics, Eli Lilly, Spectrum, EMD Serono, Roche, and Foundation One Medicine; receiving research grant from the National Institutes of Health/National Cancer Institute, American Cancer Society, Cancer Prevention & Research Institute of Texas, American Association for Cancer Research Johnson & Johnson Lung Cancer, AstraZeneca, Spectrum, and Checkmate Pharmaceuticals; and having royalties from Bio-Tree Systems, Inc. Dr. Skoulidis reports receiving personal fees from Tango Therapeutics and grants from Amgen. Dr. Altan reports receiving research funding from Genentech, Nektar Therapeutics, Merck, GlaxoSmithKline, Novartis, Jounce Therapeutics, Bristol-Myers Squibb, Eli Lilly, and Adaptimmune and serving on the advisory board of GlaxoSmithKline and Shattuck Lab. The remaining authors declare no conflict of interest.
Footnotes
CRediT Authorship Contribution Statement
Biagio Ricciuti, Kathryn C. Arbour, Matthew D. Hellmann, Mark M. Awad: Conceptualization.
Biagio Ricciuti, Kathryn C. Arbour, Jessica J. Lin, Gonzalo Recondo, Giuseppe Lamberti, Lingzhi Hong, Jianjun Zhang, Deepti Venkatraman, Joao V. Alessi, Victor R. Vaz, Hira Rizvi, Mizuki Nishino, Jacklynn Egger, Andrew J. Plodkowski, Sara Khosrowjerdi, Subba Digumarthy, Hyesun Park, Nuno Vaz: Data curation.
Biagio Ricciuti, Amir Vajdi, Michael Y. Tolstorukov, Yvonne Y. Li, Liam F. Spurr, Andrew D. Cherniack, Xinan Wang: Formal analysis.
Biagio Ricciuti, Kathryn C. Arbour, Jessica J. Lin, Amir Vajdi, Natalie Vokes, Lingzhi Hong, Jianjun Zhang, Michael Y. Tolstorukov, Yvonne Y. Li, Liam F. Spurr, Andrew D. Cherniack, Gonzalo Recondo, Giuseppe Lamberti, Xinan Wang, Deepti Venkatraman, Joao V. Alessi, Victor R. Vaz, Hira Rizvi, Jacklynn Egger, Andrew J. Plodkowski, Sara Khosrowjerdi, Subba Digumarthy, Hyesun Park, Nuno Vaz, Mizuki Nishino, Lynette M. Sholl, David Barbie, Mehmet Altan, John V. Heymach, Ferdinandos Skoulidis, Justin F. Gainor, Matthew D. Hellmann, Mark M. Awad: Writing - original draft.
Biagio Ricciuti, Kathryn C. Arbour, Jessica J. Lin, Amir Vajdi, Natalie Vokes, Lingzhi Hong, Jianjun Zhang, Michael Y. Tolstorukov, Yvonne Y. Li, Liam F. Spurr, Andrew D. Cherniack, Gonzalo Recondo, Giuseppe Lamberti, Xinan Wang, Deepti Venkatraman, Joao V. Alessi, Victor R. Vaz, Hira Rizvi, Jacklynn Egger, Andrew J. Plodkowski, Sara Khosrowjerdi, Subba Digumarthy, Hyesun Park, Nuno Vaz, Mizuki Nishino, Lynette M. Sholl, David Barbie, Mehmet Altan, John V. Heymach, Ferdinandos Skoulidis, Justin F. Gainor, Matthew D. Hellmann, Mark M. Awad: Writing - review & editing.
Supplementary Data
Note: To access the supplementary material accompanying this article, visit the online version of the Journal of Thoracic Oncology at www.jto.org and at https://doi.org/10.1016/j.jtho.2021.10.013.
References
- 1.Mok TSK, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393:1819–1830. [DOI] [PubMed] [Google Scholar]
- 2.Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. [DOI] [PubMed] [Google Scholar]
- 3.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389:255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguilar EJ, Ricciuti B, Gainor JF, et al. Outcomes to first-line pembrolizumab in patients with non-small-cell lung cancer and very high PD-L1 expression. Ann Oncol. 2019;30:1653–1659. [DOI] [PubMed] [Google Scholar]
- 6.Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378:2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biton J, Mansuet-Lupo A, Pécuchet N, et al. TP53, STK11, and EGFR mutations predict tumor immune profile and the response to anti-PD-1 in lung adenocarcinoma. Clin Cancer Res. 2018;24:5710–5723. [DOI] [PubMed] [Google Scholar]
- 11.Galan-Cobo A, Sitthideatphaiboon P, Qu X, et al. LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in KRAS-mutant lung adenocarcinoma. Cancer Res. 2019;79:3251–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero R, Sayin VI, Davidson SM, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med. 2017;23:1362–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koyama S, Akbay EA, Li YY, et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res. 2016;76:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamberti G, Spurr LF, Li Y, et al. Clinicopathological and genomic correlates of programmed cell death ligand 1 (PD-L1) expression in nonsquamous non-small-cell lung cancer. Ann Oncol. 2020;31:807–814. [DOI] [PubMed] [Google Scholar]
- 16.Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kansanen E, Kuosmanen SM, Leinonen H, Levonenn AL. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Best SA, De Souza DP, Kersbergen A, et al. Synergy between the KEAP1/NRF2 and PI3K pathways drives non-small-cell lung cancer with an altered immune microenvironment. Cell Metab. 2018;27:935–943.e4. [DOI] [PubMed] [Google Scholar]
- 19.Arbour KC, Jordan E, Kim HR, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin Cancer Res. 2018;24:334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Yang Y, Liu X, et al. NFE2L2/KEAP1 mutations correlate with higher tumor mutational burden value/PD-L1 expression and potentiate improved clinical outcome with immunotherapy. Oncologist. 2020;25:e955–e963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Su C, Ren S, Zhou C, Jiang T. Pan-cancer analysis of KEAP1 mutations as biomarkers for immunotherapy outcomes. Ann Transl Med. 2020;8:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vokes NI, Liu D, Ricciuti B, et al. Harmonization of tumor mutational burden quantification and association with response to immune checkpoint blockade in non-small-cell lung cancer. JCO Precis Oncol. 2019;3. PO.19.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gainor JF, Shaw AT, Sequist LV, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: A retrospective analysis. Clin Cancer Res. 2016;22:4585–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gainor JF, Rizvi H, Jimenez Aguilar E, et al. Clinical activity of programmed cell death 1 (PD-1) blockade in never, light, and heavy smokers with non-small-cell lung cancer and PD-L1 expression ≥50. Ann Oncol. 2020;31:404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alspach E, Lussier DM, Miceli AP, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature. 2019;574:696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson DB, Estrada MV, Salgado R, et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat Commun. 2016;7:10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roemer MGM, Redd RA, Cader FZ, et al. Major histocompatibility complex class II and programmed death ligand 1 expression predict outcome after programmed death 1 blockade in classic Hodgkin lymphoma. J Clin Oncol. 2018;36:942–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson DB, Bordeaux J, Kim JY, et al. Quantitative spatial profiling of PD-1/PD-L1 interaction and HLA-DR/IDO-1 predicts improved outcomes of anti-PD-1 therapies in metastatic melanoma. Clin Cancer Res. 2018;24:5250–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Skoulidis F, Falchook G, et al. PS01.07 registrational phase 2 trial of sotorasib in KRAS p.G12C mutant NSCLC: first disclosure of the Codebreak 100 primary analysis. J Thorac Oncol. 2021;16(suppl):S61. [Google Scholar]
- 30.Hong DS, Bang YJ, Barlesi F, et al. MO01.31 durability of clinical benefit and biomarkers in patients with advanced non-small cell lung cancer (NSCLC) treated with sotorasib, a KRAS(G12C) inhibitor. J Thorac Oncol. 2021;16(suppl):S28–S29. [Google Scholar]
- 31.Riely G, Ou SI, Rybkin I, et al. 99O_PR -KRYSTAL-1: activity and preliminary pharmacodynamic (PD) analysis of adagrasib (MRTX849) in patients (Pts) with advanced non-small cell lung cancer (NSCLC) harboring KRASG12C mutation. J Thorac Oncol. 2021;16(suppl 4):S751–S752. [Google Scholar]
- 32.Canon J, Rex K, Saiki AY, et al. The clinical KRAS (G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. [DOI] [PubMed] [Google Scholar]
- 33.Papillon-Cavanagh S, Doshi P, Dobrin R, Szustakowski J, Walsh AM. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open. 2020;5:e000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wohlhieter CA, Richards AL, Uddin F, et al. Concurrent mutations in STK11 and KEAP1 promote ferroptosis protection and SCD1 dependence in lung cancer. Cell Rep. 2020;33:108444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.No JH, Kim YB, Song YS. Targeting nrf2 signaling to combat chemoresistance. J Cancer Prev. 2014;19:111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeddi F, Soozangar N, Sadeghi MR, Somi MH, Samadi N. Contradictory roles of Nrf2/Keap1 signaling pathway in cancer prevention/promotion and chemoresistance. DNA Repair (Amst). 2017;54:13–21. [DOI] [PubMed] [Google Scholar]
- 37.Skoulidis F, Li BT, Dy GK, et al. Sotorasib for lung cancers with KRAS p. G12C Mutation. N Engl J Med. 2021;384:2371–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.