Abstract
We examined the viral replicative capacity and protease-mediated processing of Gag and Gag-Pol precursors of human immunodeficiency virus (HIV) variants selected for resistance to protease inhibitors. We compared recombinant viruses carrying plasma HIV RNA protease sequences obtained from five patients before protease inhibitor therapy and after virus escape from the treatment. Paired pretherapy-postresistance reconstructed viruses were evaluated for HIV infectivity in a quantitative single-cycle titration assay and in a lymphoid cell propagation assay. We found that all reconstructed resistant viruses had a reproducible decrease in their replicative capacity relative to their parental pretherapy counterparts. The extent of this loss of infectivity was pronounced for some viruses and more limited for others, irrespective of the inhibitor used and of the level of resistance. In resistant viruses, the efficiency of Gag and Gag-Pol precursor cleavage by the protease was impaired to different extents, as shown by the accumulation of several cleavage intermediates in purified particle preparations. We conclude that protease inhibitor-resistant HIV variants selected during therapy have an impaired replicative capacity related to multiple defects in the processing of Gag and Gag-Pol polyprotein precursors by the protease.
Protease inhibitors are the most active antiviral agents used to date for the treatment of human immunodeficiency virus (HIV) infection (8). These drugs inhibit processing of HIV Gag and Gag-Pol polyprotein precursors by the virus-encoded protease into their mature structural or enzymatically active products, resulting in a complete block of particle infectivity (7). Consequently, HIV protease inhibitors exert powerful antiviral activity in infected patients, usually leading to the suppression of detectable viral replication for prolonged periods when used in combination with other antiretroviral agents (12, 20, 22).
In spite of this promising clinical efficacy, the emergence of HIV variants with a reduced sensitivity to protease inhibitors can occur when the treatment fails to achieve a sufficiently profound suppression of viral replication (16). The evolution of HIV toward high-level resistance to protease inhibitors is the result of a gradual accumulation of resistance mutations in the protease (4, 5, 10, 23). Most of the HIV protease mutations that are associated with decreased sensitivity to protease inhibitors are now well identified (4, 5, 14, 15, 21, 26, 27, 29). These mutations are usually not found in isolates that have not been exposed to protease inhibitors (1, 17, 18, 24, 32), implying that they might confer a selective disadvantage to the virus during replication in drug-free conditions. Previous studies conducted on tissue culture-selected or in vitro-mutated viruses indeed found that some protease inhibitor resistance mutations, often located close to the enzyme active site, can decrease protease catalytic activity and viral replicative capacity (2, 6, 13, 15, 19, 30, 31). In some instances, the impairment of HIV replication that results from the selection of these resistance mutations can be partially compensated for by secondary mutations that are usually located outside the enzyme active site (2, 14, 15, 25, 30). Furthermore, for some HIV variants selected for resistance to protease inhibitors in cultures, it has been found that adaptative changes able to partly correct resistance-associated loss of HIV infectivity can emerge outside the protease coding sequence, in the p7-p1 and p1-p6 protease cleavage sites within the Gag polyprotein precursor (6, 9).
Overall, selection for HIV resistance to protease inhibitors follows a complex evolutionary pathway that can differ according to the nature of the inhibitor and to the conditions of the selection. To date, the effects of HIV resistance to protease inhibitors on HIV replicative capacity has not been clearly documented for resistant viruses selected during treatment of infected patients. In this study, we constructed recombinant viruses carrying plasma HIV RNA protease sequences that were obtained from patients and that had escaped therapy by ritonavir or saquinavir. The infectivity of the corresponding particles was compared to that of equivalent molecular clones carrying the parental pretherapy protease from the same patients. We found that all studied recombinant viruses harboring resistant proteases had an impaired viral replicative capacity relative to viruses carrying parental pretherapy proteases. The replicative defect observed in these recombinant viruses was accompanied by abnormalities in the processing of the Gag and Gag-Pol polyprotein precursors and involving several cleavage sites. Analysis of plasma HIV Gag sequences before and after the onset of resistance revealed that significant adaptative changes in Gag cleavage sites had occurred in only one virus, displaying a single mutation in the nucleocapsid (NC)-p1 site.
Taken together, our results reveal that even after in vivo selection for resistance in the course of treatment of infected patients, resistance to protease inhibitors can result in a significant impairment of HIV replicative capacity, due to a decrease in protease cleavage efficiency.
MATERIALS AND METHODS
Patients.
The HIV proteases studied here were amplified from plasma virus obtained from four patients treated with a combination of ritonavir (Abbott) at 1,200 mg/day, dideoxycytosine (zalcitabine; Roche) at 2.25 mg/day, and zidovudine (Glaxo-Wellcome) at 600 mg/day (22) (patients 202, 401, 402, and 506). One patient (patient 246) was treated with saquinavir (Roche; 1,800 mg/day) monotherapy.
Construction of recombinant viral genomes.
HIV protease sequences were amplified from patient plasma virus by nested reverse transcriptase (RT) PCR. The first round of RT PCR was conducted with the ProA+/ProA− primer pair: ProA+, 5′GCTAATTTTTTAGGGAAGATCTG3′, and ProA−, 5′GGCAAATACTGGAGTATTGTATG3′. For sequencing, the second round of amplification was done with nested primers ProB+ and ProB−: ProB+, TTTTTAGGGAAGATCTGGCCTTC, and ProB−, GGAGTATTGTATGGATTTTCAGG. Primer Pro1 (CCCTCTCAGAAGCAGGAG) was used for direct sequencing of the resulting bulk PCR product. For cloning, the second round of PCR was conducted with nested primers PRX+ and PRC−: PRX+, 5′GGAGCCTCTAGACAAGGAACTGTATCCT3′, and PRC−, 5′GTACAGTATCGATAGGACTAATGGGAAA3′.
The resulting nested PCR products were digested with XbaI and ClaI and cloned into pBluescript SKII+, and then the XbaI-ClaI insert was in turn cloned into pNL4-3XC, a previously described modification of the HIV type 1 (HIV-1) proviral molecular clone pNL4-3 in which an XbaI site was inserted immediately upstream of the protease coding sequence together with a ClaI site inserted immediately downstream (27). Clones were sequenced with the Pro1 primer to ensure that the protease sequence was identical to that of the bulk PCR product.
Cell cultures.
HeLa cells and P4 cells (HeLa-CD4 LTR-LacZ) (3) were cultivated in Dulbecco’s modified Eagle’s medium (DMEM). MT4 and HUT78 cells were grown in RPMI 1640. All cultures were supplemented with 10% fetal calf serum and antibiotics. P4 cells were cultured in the presence of 500 μg of G418 per ml.
Protease inhibitor resistance assay.
Subconfluent HeLa cells in 25-cm2 flasks were transfected with 8 μg of HIV proviral plasmid DNA by the calcium phosphate precipitation method. After 24 h, the transfected HeLa cells were trypsinized, split into 1-ml subcultures in 48-well plates, and treated with increasing concentrations of protease inhibitor (0, 1, 5, 25, 125, 625, and 3,125 nM). After 24 h of treatment, 100 μl of viral supernatant from each subculture was used to infect triplicate subconfluent P4 cell cultures in 96-well plates in the presence of 100 μg of DEAE-dextran per ml. In P4 cells, the expression of β-galactosidase is strictly inducible by the HIV transactivator protein Tat, thereby allowing precise quantitation of HIV infectivity based on a single cycle of replication (3). At 24 h after infection of P4 cells, the titer of viruses produced in the presence of inhibitors in a single cycle was determined by quantitation of the β-galactosidase activity in P4 cell lysates with a colorimetric assay based on the cleavage of chlorophenol red-β-d-galactopyranoside (CPRG) by β-galactosidase (adapted from Eustice et al. [11]). Briefly, following elimination of the supernatant, P4 cells were lysed in 100 μl of lysis buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 10 mM MgSO4, 2.5 mM EDTA, 50 mM β-mercaptoethanol, 0.125% Nonidet P-40). After incubation for 5 min at room temperature, 100 μl of reaction buffer (8 mM sodium phosphate buffer [pH 7.4], 10 mM MgCl2, 10 mM β-mercaptoethanol, 6 mM CPRG) was added to the cell lysates and incubated for 1 min to 2 h at 37°C. Colorimetric reactions were irreversibly arrested by freezing the plates at −80°C for at least 2 h. Optical densities in the reaction wells were read at 570 nm with a reference filter set at 690 nm. The susceptibility of the different viruses to protease inhibitors was expressed as the concentration of drug leading to 90% inhibition (IC90).
Infectivity assays.
The single-cycle titer of the recombinant viruses was determined with indicator P4 cells. Briefly, triplicate subconfluent P4 cells in 96-well plates were infected with the equivalent of 5 ng of HIV-1 p24 of the different viruses obtained from transfected HeLa cells in the presence of 100 μg of DEAE-dextran per ml. The infectious titer was measured after 24 h of infection with the CPRG assay.
For analysis of the kinetics of virus replication in T-lymphoid cells, 106 MT4 or HUT78 cells were infected with the equivalent of 10 ng of p24 of each of the recombinant viruses obtained from transfected HeLa cells. Virus production in the infected cultures was monitored with an HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA) (DuPont).
Viral protein analysis.
HeLa cells were grown in DMEM supplemented with 10% fetal calf serum. On the day before transfection, HeLa cells were plated at a seeding concentration of 106 cells per 75-cm2 tissue culture flask. The cultures were transfected with 20 μg of plasmid DNA by the calcium phosphate precipitation technique. Transfected HeLa cells were metabolically labelled with a mixture of [35S]methionine and [35S]cysteine from 48 to 60 h posttransfection. To analyze particle-associated proteins, virions released into the supernatant were pelleted through a 20% sucrose cushion (in phosphate-buffered saline) for 90 min at 4°C and 48,000 rpm in a Beckman TLA-100.3 rotor. Pelleted viral particles were lysed in radioimmunoprecipitation assay buffer (RIPA buffer; 140 mM NaCl, 8 mM NaH2PO4, 1% Nonidet P-40, 0.5% deoxycholate, 0.05% sodium dodecyl sulfate [SDS]), and aliquots were analyzed directly by SDS-polyacrylamide gel electrophoresis (PAGE). After normalization of the particle yield by the HIV-1 p24 ELISA, aliquots of the lysed virions were analyzed by immunoprecipitation with serum from an HIV-1-infected patient prior to electrophoresis.
RESULTS
Construction of protease inhibitor-resistant recombinant viruses.
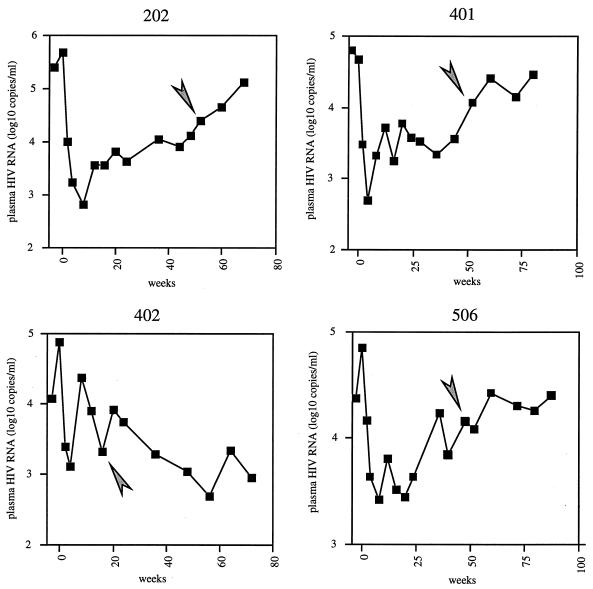
To evaluate the impact of resistance to protease inhibitors on HIV replicative potential, five pairs of viruses carrying pretherapy or postresistance proteases from five patients treated with protease inhibitors were reconstructed with pNL4-3XC, a full-length HIV-1 proviral infectious molecular clone (27). Each pair of molecular clones comprised the PCR-amplified protease obtained from plasma virus shortly before the initiation of the treatment and the protease obtained after evidence that the virus had escaped treatment, as indicated by a rebound in plasma viremia. Four of the patients (patients 202, 401, 402, and 506) were treated with a combination of ritonavir with zidovudine and zalcitabine (22). The profile of viral escape from antiretroviral therapy for these four patients is shown in Fig. 1. In all four cases, the samples were obtained before viremia had reached pretherapy (week-0) levels. One patient (patient 246) was treated with saquinavir as a single agent. Ritonavir escape viruses 202post, 401post, and 506post were harvested 12 months after the initiation of therapy. Ritonavir escape virus 402post was obtained 20 weeks after the onset of therapy, while saquinavir escape virus 246post was harvested after more than 24 months of treatment.
FIG. 1.
Evolution of plasma viremia over time in patients 202, 401, 402, and 506. Week 0 refers to the start of therapy. Arrowheads indicate the time of sampling for the postresistance viruses.
The mutations found in the protease of the escape variant from each patient are shown in Table 1, together with the level of resistance of the corresponding recombinant virus. All of the observed mutations already have been described for patients treated with protease inhibitors (4, 5, 21, 23, 29). Ritonavir escape viruses carried different combinations of typical ritonavir resistance mutations at codons 20, 36, 46, 54, 82, and 90 of the protease. Saquinavir escape virus 246post was mutated at codons 46, 48, and 90, a pattern typical of resistance to saquinavir. The level of resistance of each reconstructed escape virus was measured in a single-cycle system and expressed as the increase in the IC90 relative to that of pretherapy virus. The increase in IC90 ranged from 8.9- to 19-fold, according to the virus studied (Table 1).
TABLE 1.
Genotypic profiles and resistance phenotypes of reconstructed protease inhibitor escape virusesa
| Virus | Protease inhibitor treatment | Mutation in HIV protease
|
Fold increase in IC90 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L10 | K20 | M36 | M46 | G48 | F53 | I54 | L63 | A71 | V82 | I84 | L90 | |||
| 202post | Ritonavir | R | I | Pb | S | 10 | ||||||||
| 246post | Saquinavir | I | V | Pb | M | 10 | ||||||||
| 401post | Ritonavir | R | I | V | A | 8.9 | ||||||||
| 402post | Ritonavir | Ib | R | Ib | I | L | Pb | Vb | A | 14.1 | ||||
| 506post | Ritonavir | R | Pb | A | M | 19 | ||||||||
The amino acid substitutions conferring HIV resistance to saquinavir and ritonavir are indicated in the single-letter code. The IC90 was measured in a single-cycle resistance assay on P4 cells with the protease inhibitor included in the treatment as described in Materials and Methods. The results are expressed as the fold increase in the IC90 compared to the value for a recombinant virus carrying the parental pretherapy protease.
Already present in pretherapy virus.
The sequence of the cleavage sites in Gag that have been described as able to mutate along with or subsequent to the emergence of resistance mutations in the protease (9) was determined for plasma viruses from all five patients before therapy and after escape. For escape viruses from patients 202, 246, 401, and 506, we found only the commonly observed variability described for HIV-1 clade B (1, 24). Therefore, we believe that the replicative behavior of the corresponding reconstructed viruses, which all carried wild-type pNL4-3XC Gag sequences, should not be significantly different from that of the original plasma virus. The only significant change was found for patient 402, in whom a single A→V change at position P2 in the NC-p1 cleavage site was present in the resistant virus at week 20. This same substitution, accompanied by a Q→R substitution immediately upstream, has been described to emerge in HIV variants selected for resistance to protease inhibitors in vitro (9). The potential effects of this sequence change on the resistance and replicative phenotype of resistant virus 402post are currently under investigation.
Effect of resistance to protease inhibitors on HIV replicative capacity.
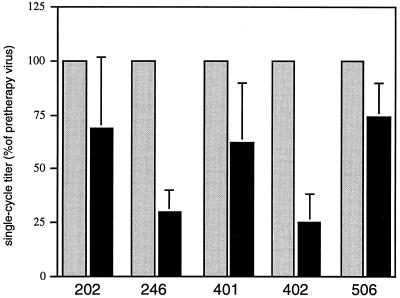
The differences in infectivity between pretherapy and protease inhibitor-resistant viruses were first evaluated by direct titration of HIV in drug-free conditions on the basis of a single cycle of viral infection with the P4 indicator cell line (HeLa-CD4 LTR LacZ) (3). For each tested virus pair, at least three independent titration experiments were conducted, corresponding to three independent transfections. Virus input was normalized by HIV-1 p24 antigen ELISA quantification. For all tested virus pairs, the single-cycle titer of the resistant virus was reproducibly decreased relative to that of its parental pretherapy counterpart (Fig. 2). The relative loss in virus titer was particularly marked for virus from ritonavir-treated patient 402, for which this reduction was consistently more than fourfold, and for virus from saquinavir-treated patient 246, which displayed a highly reproducible three- to fourfold loss of infectivity. For the other viruses (202, 401, and 506), the single-cycle infectious titer of the postresistance virus was 50 to 75% that of the parental pretherapy virus.
FIG. 2.
Single-cycle infectivity of recombinant viruses carrying pretherapy (gray bars) or postresistance (blade bars) HIV protease sequences from patients treated with protease inhibitors. The single-cycle titer of each recombinant virus carrying the postresistance protease was measured by titration on P4 (HeLa-CD4 LTR-LacZ) cells and expressed as a percentage of the titer of the pretherapy virus from the same patient. Postresistance viruses from patients 202, 401, 402, and 506 are resistant to ritonavir; postresistance virus from patient 246 is resistant to saquinavir. Viral input was normalized according to the HIV-1 p24 antigen content of transfected HeLa cell supernatants. The results presented are the mean standard deviation for at least three separate titration experiments for three different transfections.
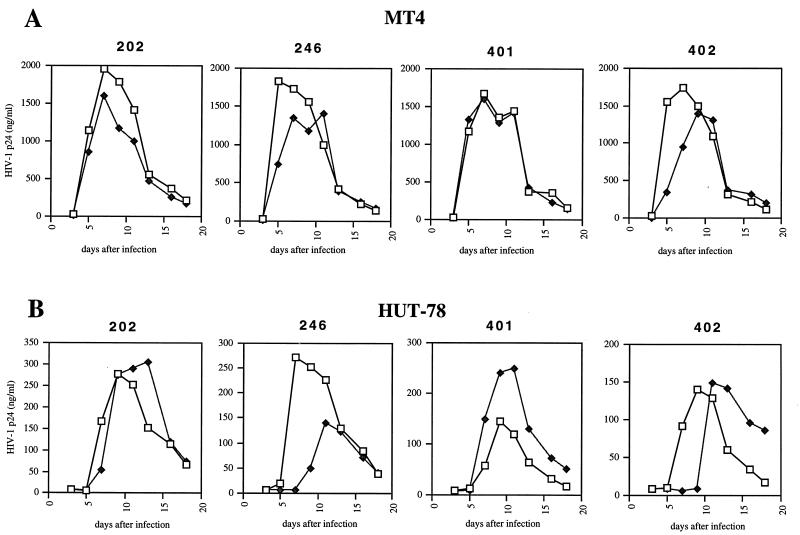
The infectivity of four of the recombinant virus pairs (202, 401, 402, and 246) was also evaluated by analysis of the kinetics of their propagation on T-lymphoid cell lines, in which detectable HIV-1 p24 antigen production results from the accumulation of several cycles of viral replication (Fig. 3). In keeping with the results of the single-cycle titration experiment, there was a significant delay in productive infection of MT4 cells by postresistance viruses from patients 402 and 246 (Fig. 3A). Replication of postresistance virus from patient 202 was only marginally affected in MT4 cells, while the kinetics of propagation of postresistance virus from patient 401 were similar to those of the parental pretherapy virus. Similar results were observed upon infection of HUT78 cells, in which the kinetics of replication of postresistance viruses from patients 402 and 246 displayed a marked delay in peak virus production (Fig. 3B). These results reveal that, in both a quantitative single-cycle titration assay and a virus propagation assay, resistant protease alleles emerging in treated patients can reduce to different degrees the replicative capacity of HIV variants relative to their parental pretherapy counterparts.
FIG. 3.
Replication kinetics of recombinant viruses carrying pretherapy (□) or postresistance (⧫) HIV protease sequences in MT4 cells (A) and HUT78 cells (B). MT4 and HUT78 cells were infected with HIV-1 p24-normalized amounts of particles from recombinant viruses carrying pretherapy or postresistance protease sequences from ritonavir-treated patients 202, 401, and 402 and from saquinavir-treated patient 246. The kinetics of virus production in the infected cultures were monitored over time with an HIV-1 p24 ELISA.
Effect of resistance to protease inhibitors on HIV polyprotein processing.
The data reported above demonstrate that the development of resistance to protease inhibitors in treated patients results in a reduction in viral infectivity. To determine whether this impaired viral fitness is associated with reduced enzyme activity, we analyzed the protein composition of viral particles processed by resistant proteases. In particular, we compared the maturation profiles of Gag and Gag-Pol precursors generated by patient-derived resistant proteases with the patterns produced by the corresponding pretherapy proteases.
Infectious proviral clones in which the complete protease domain was substituted by PCR-amplified patient protease coding regions were transfected into HeLa cells. The transfected cells were metabolically labelled with [35S]methionine and [35S]cysteine, and particulate material released into the supernatant was pelleted through 20% sucrose and solubilized in RIPA buffer. The samples were then analyzed both directly by SDS-PAGE and in parallel by immunoprecipitation with a serum sample from a patient infected with HIV-1 after normalization for the p24 antigen content of each pair of samples.
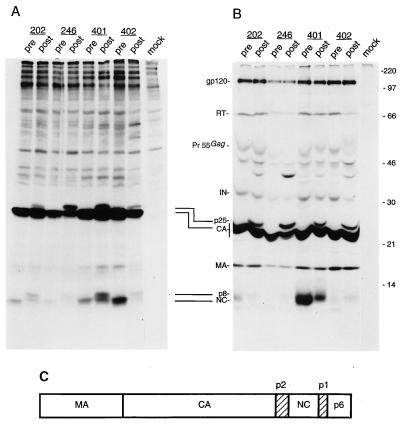
All of the major Gag maturation products were detected in the directly analyzed particle-associated material (Fig. 4A) as well as in the immunoprecipitated aliquots (Fig. 4B). Interestingly, all of the samples obtained from therapy-resistant proteases displayed additional bands which were not detectable in the corresponding pretherapy samples. In particular, a band with the electrophoretic mobility of 25 kDa was reproducibly observed in all of the particle-associated preparations processed by therapy-resistant proteases (Fig. 4A, lanes post). This band was clearly HIV encoded, since it was efficiently immunoprecipitated by a serum sample from an HIV-1-infected patient (Fig. 4B, lanes post); it very likely represents the gag-encoded intermediate cleavage product capsid (CA)-p2 (Fig. 4C). Also, a band with an apparent molecular mass of 8 kDa was reproducibly more prominent in the therapy-resistant protease samples than in the pretherapy counterparts (Fig. 4A, compare lanes pre to lanes post). Such an electrophoretic mobility may have been due to the NC (p7) protein still attached to one of the spacer peptides surrounding the NC protein in the Gag precursor molecule (Fig. 4C).
FIG. 4.
Analysis of the protein content of viral particles produced by recombinant viruses carrying pretherapy (pre) or postresistance (post) protease sequences from patients 202, 246, 401, and 402. (A) Metabolically 35S-labelled viral particles produced by transfected HeLa cells were partially purified by ultracentrifugation through a 20% sucrose cushion, lysed, and directly analyzed by SDS-PAGE. (B) Particle lysates produced and purified under the same conditions as for panel A were immunoprecipitated by serum from an HIV-1-infected patient and subsequently analyzed by SDS-PAGE. The amount of virions analyzed for each pair of pretherapy-postresistance viruses was normalized with an HIV-1 p24 antigen ELISA. Molecular mass markers (in kilodaltons) are shown on the right. On the left, the positions of the prominent HIV-encoded proteins found in virion preparations are indicated: gp120 (outer envelope glycoprotein), RT, IN (integrase), CA, MA, and NC. Pr55Gag indicates the complete, uncleaved Gag polyprotein precursor. Two incomplete Gag cleavage products, found solely in particles produced by recombinant genomes carrying postresistance protease sequences, are also indicated: p25, resulting from incomplete cleavage at the CA-p2 site; and p8, likely to result from incomplete cleavage at the NC-p1 site. (C) Schematic representation of the protein domain organization in the HIV Gag polyprotein precursor. The mature Gag proteins MA, CA, NC, and p6 are shown as open boxes; spacer peptide sequences p2 and p1 are shown as hatched boxes.
Immunoprecipitation of the particulate material allowed the detection of additional precursor molecules, which were clearly more prominent in the protease inhibitor-resistant samples than in their pretherapy counterparts (Fig. 4B). A band of approximately 39 kDa accumulated in all of the protease inhibitor-resistant virus particles (Fig. 4B, lanes post) and could have corresponded either to the matrix (MA)-CA-p2 intermediate cleavage product (28) or to a portion of the Gag precursor encompassing CA, NC, and p6. Of particular interest, in virus 402post the intensity of the RT p66 band was drastically reduced with respect to that of its pretherapy counterpart (Fig. 4B), indicating that a functionally impaired protease can result in a significant alteration in the RT content of virions. Remarkably, some bands seemed to accumulate specifically in association with certain proteases (Fig. 4B, lanes 246post and 402post). In addition, the relative accumulation of the full-length Gag precursor with respect to the mature product was variable in unrelated samples (Fig. 4B).
DISCUSSION
In this study, we showed that recombinant HIV proviral clones carrying protease sequences from viruses selected for resistance to protease inhibitors during the treatment of infected patients displayed a significant reduction in replicative capacity relative to that of their parental pretherapy counterparts. Our data are in agreement with the conclusions of other studies conducted on viruses that were selected for resistance under tissue culture conditions, but to our knowledge, this is the first report of a resistance-associated loss of viral fitness as a result of protease inhibitor resistance in treated patients. We found that the extent of this loss of fitness could be dramatically different from one resistant virus to another. In some reconstructed resistant viruses (carrying plasma protease sequences from patients 401, 202, and 506), the extent of the loss of fitness was limited. In reconstructed viruses from two other patients, a more dramatic loss of viral fitness was observed. In virus selected by saquinavir treatment of patient 246 and carrying mutations M46I, G48V, and L90M, the marked reduction in viral infectivity was visible in both the single-cycle and the viral propagation experiments.
Among the studied viruses, resistant virus from patient 402 was the most affected in its replicative capacity. Like virus 246post, virus 402post was characterized by a marked loss of fitness in both the P4 single-cycle assay and the MT4 and HUT78 propagation assays. The similarity of the impact of protease inhibitor resistance on virus replication kinetics in these two cell lines reinforces the results of the single-cycle titration experiments. The protease from this virus carries eight previously described resistance mutations: L10I, K20R, M36I, M46I, F53L, L63P, A71V, and V82A; four of these (L10I, M36I, L63P, and A71V) existed before ritonavir therapy. Remarkably, this heavy resistance mutation pattern was acquired in less than 20 weeks. The individual roles of particular resistance mutations in the marked loss of fitness that characterizes this resistant virus, as well as the evolution of its phenotype over time, are currently being evaluated.
Analysis of the pattern of particle-associated polyprotein precursor cleavage in the reconstructed resistant viruses revealed that even in the case of a modest loss of fitness, remarkable abnormalities in Gag precursor cleavage were consistently observed, a sign of a decrease in the cleavage efficiency of the resistant protease. The most evident abnormality involved the accumulation of p25 protein, with a PAGE migration profile slightly slower than that of the CA p24 protein and likely to correspond to incomplete cleavage of Gag at the CA-p2 site, immediately downstream of CA. Since this site has been described as being subject to the slowest rate of cleavage by the HIV protease, a small reduction in the global efficiency of cleavage by the protease could be more perceptible at this site than at others. However, other Gag cleavage abnormalities were observed in resistant viruses. In particular, all resistant variants displayed an abnormal maturation of the NC protein, characterized by the accumulation of a protein of 8 kDa which was likely to result from incomplete cleavage at the NC-p1 junction. More Gag maturation abnormalities were also observed, leading to the accumulation of small but significant amounts of the full-length p55 Gag precursor and the accumulation of a 39-kDa intermediate precursor, which could correspond either to incomplete cleavage between MA and CA or to an uncleaved segment of the Gag precursor comprising CA, NC, and p6. Interestingly, virus 246post displayed a pattern of Gag precursor accumulation that was strikingly different from that of the other viruses. Indeed, this virus was characterized by a marked inhibition of cleavage at the CA-p2 site, as revealed by the accumulation of large amounts of p25 protein. Whether this pattern reflects a globally higher level of impairment in protease cleavage efficiency or whether it is specific to the particular mutation profile of the virus 246post protease remains to be determined.
A more detailed investigation of the consequences of protease inhibitor resistance on Gag precursor cleavage is in progress. We have found that the consequences of resistance-associated impairment of protease function are not limited to abnormal cleavage of Gag precursors. In virus 402post, which appeared to be the most fitness impaired, the amounts of virion-associated RT p66 were significantly reduced relative to those in its pretherapy counterpart. It is likely that the reduction in the amounts of particle-associated RT plays an important role in the marked reduction of fitness that characterizes this virus.
In viruses selected for resistance to protease inhibitors in tissue cultures, mutations in the Gag NC-p1-p6 cleavage sites have been found to partially compensate for a resistance-associated loss of fitness (9). The sequence of these cleavage sites was examined in plasma viruses from all the samples that were used here for the reconstruction of recombinant resistant viruses. Only native virus 402post was found to harbor a change in the NC-p1 site, which emerged along with protease inhibitor resistance mutations 20 weeks after the onset of therapy. However, in this significantly replication-impaired virus, the maturation of Gag was not markedly more affected than in the other viruses. Instead, the main cleavage defect in virus 402post involved the maturation of RT. Therefore, in this native virus, the NC-p1 site substitution might have had only a limited fitness-corrective effect. An extensive examination over time of the phenotypic and genotypic changes in Gag, RT, and protease in viruses from patient 402 is currently under way. The adaptative changes in Gag described to date for protease inhibitor-resistant HIV variants involve only two cleavage sites that border the p1 spacer peptide, located between NC and p6. We report here that the impairment in viral fitness that results from resistance is associated with a loss of protease cleavage efficiency at several other sites in Gag and at some sites in Pol. Therefore, the possibility of coevolution of all of these cleavage sites together with the protease, as well as the capacity of the corresponding changes to fully or only partly compensate for a resistance-associated loss of viral fitness, remains to be evaluated.
The potential clinical benefit of a resistance-associated loss of viral fitness needs to be fully investigated. It is possible that resistant viruses displaying a marked impairment in replicative capacity exhibit a reduction in pathogenic potential. In this case, the evolution of HIV disease beyond the emergence of resistance to protease inhibitors could adopt a slower pace in spite of a rebound in plasma viremia. It should be emphasized that none of the patients for whom viremia was regularly monitored over time had regained plasma viremia levels above pretherapy levels. In patient 402, in whom the most significant loss of viral fitness was observed, viremia remained remarkably low in spite of high levels of resistance to ritonavir. Resistance-associated loss of viral fitness could also explain the results of preliminary surveys of viruses escaping protease inhibitor therapy, which have revealed in some cases a strong dissociation between a rebound in plasma viremia and CD4 counts that remain surprisingly high. However, further studies on similar cases are required before any correlation can be attempted. Furthermore, it should be emphasized again that the loss of fitness observed in the resistant viruses studied here could be gradually corrected over time during the course of therapy, following the emergence of adaptative changes inside or outside the protease, as discussed above. Extensive virological and clinical follow-up of patients treated with protease inhibitors is required to ascertain the overall impact of a resistance-associated loss of fitness on the long-term effects of antiretroviral treatments for HIV.
ACKNOWLEDGMENTS
We thank Françoise Brun-Vézinet and Diane Descamps (Laboratoire de Virologie, Hôpital Bichat-Claude Bernard, Paris, France) for viruses from patient 246, Frédéric Subra for the modified CPRG protocol, Jacques Leibowitch for interest in this work, and Luc Montagnier for support.
This work was supported in part by the Agence Française de Recherches sur le Sida (ANRS). F.M. is the recipient of a fellowship from ANRS.
REFERENCES
- 1.Barrie K A, Perez E E, Lamers S L, Farmerie W G, Dunn B M, Sleasman J W, Goodenow M M. Natural variation in HIV-1 protease, Gag p7 and p6, and protease cleavage sites within Gag/Pol polyproteins: amino acid substitutions in the absence of protease inhibitors in mothers and children infected by human immunodeficiency virus type 1. Virology. 1996;219:407–416. doi: 10.1006/viro.1996.0266. [DOI] [PubMed] [Google Scholar]
- 2.Borman A, Paulous S, Clavel F. Resistance of HIV-1 to protease inhibitors: selection of resistance mutations in the presence and in the absence of the drug. J Gen Virol. 1996;77:419–426. doi: 10.1099/0022-1317-77-3-419. [DOI] [PubMed] [Google Scholar]
- 3.Charneau P, Mirambeau G, Roux P, Paulous S, Buc H, Clavel F. HIV-1 reverse transcription: a termination step at the center of the genome. J Mol Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 4.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to Indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Hobbins H L, Roth E, Shivaprakash M, Titus D L, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 6.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debouck C. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res Hum Retroviruses. 1992;8:153–164. doi: 10.1089/aid.1992.8.153. [DOI] [PubMed] [Google Scholar]
- 8.Deeks S G, Smith M, Holodniy M, Kahn J O. HIV-1 protease inhibitors: a review for clinicians. JAMA. 1997;277:145–153. [PubMed] [Google Scholar]
- 9.Doyon L, Poulin F, Pilote L, Clouette C, Thibeault D, Croteau G, Lamarre D. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson J W. The not-so-great escape. Nat Struct Biol. 1995;2:523–529. doi: 10.1038/nsb0795-523. [DOI] [PubMed] [Google Scholar]
- 11.Eustice D, Feldman P, Colberg-Poley A, Buckery R, Neubauer R. A sensitive method for the detection of β-galactosidase in transfected mammalian cells. BioTechniques. 1991;11:739–743. [PubMed] [Google Scholar]
- 12.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 13.Gulnick S V, Suvorov L I, Liu B, Yu B, Anderson B, Mitsuya H, Erickson J W. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 14.Ho D, Toyoshima T, Mo H, Kempf D, Norbeck D, Chen C, Wideburg N, Burt S, Erickson J, Singh M. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan A, Michael S, Wehbie R, Knigge M, Paul D, Everitt L, Kempf D, Norbeck D, Erickson J, Swanstrom R. Selection of multiple human immunodeficiency virus type 1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc Natl Acad Sci USA. 1994;91:5597–5601. doi: 10.1073/pnas.91.12.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kempf D, Rode R, Xu Y, Sun E, Japour A, Danner S, Boucher C, Leonard J, Molla A. International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication. 1997. The durability of response to protease inhibitor therapy is predicted by viral load, abstr. 62. [Google Scholar]
- 17.Kozal M, Shah N, Shen N, Yang R, Fucini R, Merigan T, Richman D, Morris D, Hubbell E, Chee M, Gingeras T. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 18.Lech W, Wang G, Yang Y, Chee Y, Dorman K, McCrae D, Lazzeroni L, Erickson J, Sinsheimer J, Kaplan A. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J Virol. 1996;70:2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y, Lin X, Hong L, Foundling S, Heinrickson R L, Thaisrivongs S, Leelamanit W, Raterman D, Shah M, Dunn B M, Tang J. Effect of point mutations on the kinetics and the inhibition of human immunodeficiency virus type 1 protease: relationship to drug resistance. Biochemistry. 1995;34:1143–1152. doi: 10.1021/bi00004a007. [DOI] [PubMed] [Google Scholar]
- 20.Markowitz M, Cao Y, Hurley A, et al. Abstracts of the XI International Conference on AIDS. 1996. Triple therapy with AZT, 3TC and ritonavir in 12 subjects newly infected with HIV-1, abstr. Th. B. 933. [Google Scholar]
- 21.Markowitz M, Mo H, Kempf D, Norbeck D, Bhat T, Erickson J, Ho D. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J Virol. 1995;69:701–706. doi: 10.1128/jvi.69.2.701-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathez D, de Truchis P, Gorin I, Katlama C, Pialoux G, Saimot A G, Tubiana R, Chauvin J P, Bagnarelli P, Clementi M, Leibowitch J. Third Conference on Retroviruses and Opportunistic Infections. 1996. Ritonavir, AZT, ddC, as a triple combination in AIDS patients, abstr. 285. [Google Scholar]
- 23.Molla A, Kempf D, Korneyeva M, Gao Q, Shipper P, Mo H, Markowitz M, Vasavanonda S, Chernyavskyi T, Niu P, Lyons N, Hsu A, Granneman G, Ho D, Boucher C, Leonard J, Norbeck D. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 24.Myers G, Wain-Hobson S, Henderson L, Korber B, Jeang K, Pavlakis G. Human retroviruses and AIDS. Los Alamos, N.Mex: Los Alamos National Laboratory; 1994. [Google Scholar]
- 25.Nijhuis M, Schuurman R, Schipper P, de Jong D, van Bommel T, de Groot T, Molla A, Borleffs J, Danner S, Boucher C. Abstracts of the International Discussion Meeting on HIV Population Dynamics, Variation and Drug Resistance. 1997. Reduced replication potential of HIV-1 variants initially selected under Ritonavir therapy is restored upon selection of additional substitutions. [Google Scholar]
- 26.Partaledis J, Yamagushi K, Tisdale M, Blair E, Falcione C, Maschera B, Myers R, Pazhanisamy S, Futer O, Cullinan A, Stuver C, Byrn R, Livingston D. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J Virol. 1995;69:5228–5235. doi: 10.1128/jvi.69.9.5228-5235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patick A, Rose R, Greytok J, Bechtold C, Hermsmeier M, Chen P, Barrish J, Zahler R, Colonno R, Lin P. Characterization of a human immunodeficiency virus type 1 variant with reduced sensitivity to an aminodiol protease inhibitor. J Virol. 1995;69:2148–2152. doi: 10.1128/jvi.69.4.2148-2152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettit S, Moody M, Wehbie R, Kaplan A, Nantermet P, Klein C, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts N A. Drug-resistance patterns of saquinavir and other HIV proteinase inhibitors. AIDS. 1995;9:S27–S32. [PubMed] [Google Scholar]
- 30.Rose R, Gong Y, Greytok J, Bechtold C, Terry B, Robinson B, Alam M, Colonno R, Lin P. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc Natl Acad Sci USA. 1996;93:1648–1653. doi: 10.1073/pnas.93.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sardana V V, Schlabach A J, Graham P, Bush B L, Condra J H, Culberson J C, Gotlib L, Graham D J, Kohl N E, LaFemina R L, Schneider C L, Wolanski B S, Wolfgang J A, Emini E A. Human immunodeficiency virus type 1 protease inhibitors: evaluation of resistance engendered by amino-acid substitutions in the enzyme substrate binding site. Biochemistry. 1994;33:2004–2010. doi: 10.1021/bi00174a005. [DOI] [PubMed] [Google Scholar]
- 32.Winslow D L, Stack S, King R, Scarnati H, Bincsik A, Otto M J. Limited sequence diversity in the HIV type 1 protease gene from clinical isolates and in vivo susceptibility to HIV protease inhibitors. AIDS Res Hum Retroviruses. 1995;11:107–113. doi: 10.1089/aid.1995.11.107. [DOI] [PubMed] [Google Scholar]