Abstract
Wolbachia (phylum Pseudomonadota, class Alfaproteobacteria, order Rickettsiales, family Ehrlichiaceae) is a maternally inherited bacterial symbiont infecting more than half of arthropod species worldwide and constituting an important force in the evolution, biology, and ecology of invertebrate hosts. Our study contributes to the limited knowledge regarding the presence of intracellular symbiotic bacteria in spiders. Specifically, we investigated the occurrence of Wolbachia infection in the spider species Enoplognatha latimana Hippa and Oksala, 1982 (Araneae: Theridiidae) using a sample collected in north-western Poland. To the best of our knowledge, this is the first report of Wolbachia infection in E. latimana. A phylogeny based on the sequence analysis of multiple genes, including 16S rRNA, coxA, fbpA, ftsZ, gatB, gltA, groEL, hcpA, and wsp revealed that Wolbachia from the spider represented supergroup A and was related to bacterial endosymbionts discovered in other spider hosts, as well as insects of the orders Diptera and Hymenoptera. A sequence unique for Wolbachia supergroup A was detected for the ftsZ gene. The sequences of Wolbachia housekeeping genes have been deposited in publicly available databases and are an important source of molecular data for comparative studies. The etiology of Wolbachia infection in E. latimana is discussed.
Subject terms: Bacterial genes, Phylogenetics
Introduction
Wolbachia—a maternally inherited bacterial symbiont—is widespread1–5 and exhibits a spectrum of interactions with its hosts, ranging from mutualistic to parasitic6 and has the ability to manipulate host reproduction to enhance its own transmission7. Studies have shown that Wolbachia can exert both immediate and long-term effects on their hosts. Not only it can act without affecting the host genome8, but, importantly, it can also induce changes in the genome of the invertebrate9, which may be passed on to the next generation with all the implications of these changes10. Therefore, by studying the occurrence of endosymbionts, one can gain a comprehensive understanding of its symbiotic relationships with various hosts. A holistic view of the eukaryotic organism as a holobiont not only has a cognitive aspect, but also allows to trace the path of transmission of microbes and determine their roles within the hosts. This perspective is particularly relevant when studying the interactions between Wolbachia and its host organisms, as these interactions can be highly intricate and dynamic. Firstly, the same bacterial strain may exert various effects depending on the arthropod genotype11, and secondly, the response of an invertebrate host to infection with different Wolbachia strains can also vary12. Additionally, the Wolbachia’s associations with other bacteria within the host microbiota play a significant role in shaping the activity and functions of these microbial communities. By modifying the composition of the microbial community, Wolbachia can indirectly affect important aspects of the host’s physiology, such as nutrition or pathogen resistance13.
Wolbachia constitutes an important force in the evolution14, biology15, and ecology16 of invertebrate hosts. It can cause sex-ratio distortion by inducing several phenotypes in hosts such as feminization17,18, parthenogenesis19, male-killing or cytoplasmic incompatibility20. The microorganism also enhances insect reproduction by providing biotin and vitamin B21,22, leading to increased egg production23 and improved fecundity of invertebrates24. The bacterium may also exert other effects on the host in addition to those related to reproduction. For example, Wolbachia can prevent infections caused by fungal8 or bacterial pathogens25, and reduce pathogenic viral loads in various arthropod species26,27. It can also decrease host susceptibility to different chemical pesticides28,29, which may be associated with changes in metabolism, detoxification gene expression or immune responses in bacterial hosts30.
On the basis of the phylogeny of housekeeping genes31–34 or whole-genome typing methods35,36, the genus Wolbachia has been divided into supergroups and labelled with letters of the alphabet37. An examplary set of genes comprises: coxA coding for cytochrome c oxidase, gatB coding for glutamyl-tRNA(Gln) amidotransferase, hcpA coding for conserved hypothetical protein, ftsZ coding for prokaryotic cell division protein, fbpA coding for fructose-bisphosphate aldolase, and additionally the wsp gene encoding Wolbachia surface protein31, groEL encoding 60 kDa heat-shock protein, and gltA coding for citrate synthase32–34. Sequence-based analysis of bacterial housekeeping genes as a set of genotyping markers can identify and discriminate closely related strains and accurately determine genetic divergence between them38.
Insects are the most comprehensively studied group of invertebrates in terms of the occurrence of endosymbiotic bacteria39. However, unlike insects, there are groups of invertebrates about which knowledge about the frequency and diversity of intracellular microbes is significantly more limited. Spiders are an example. Similarly, the etiology of infection, host specificity and effects of endosymbiotic bacteria in spiders are poorly characterized. Nevertheless, some literature data suggest that spiders may have more diverse microbiome than insects40,41, indicating the potential presence of novel, undiscovered taxa of microorganisms41–44. Spiders are one of the most successful terrestrial colonizers, but the data regarding their endosymbiotic relationships are scarce. Therefore, searching for, describing and understanding the presence of these organisms in spiders is required, especially since it would be interesting to elucidate whether their microbiomes have contributed to the evolutionary success of spiders.
The presence of Wolbachia in spiders has been observed relatively rarely45–58; however, most studies have examined only a few individuals of spiders of certain species40,41,59–63. Bacterial strains occurring in spiders have been classified in supergroup A and B44 together with Wolbachia infected insects, isopods, and mites—carriers of bacteria from supergroup B37. The extent of phenotypic effects induced by microbial endosymbionts in spiders remains largely unknown. Exceptions to this limited knowledge include cases where Wolbachia has been associated with sex ratio imbalances in certain spider species. In Oedothorax gibbosus (Blackwall, 1841), the killing of male embryos is most likely a manipulative effect of Wolbachia50; in Mermessus fradeorum (Berland, 1932), Wolbachia is suspected of causing cytoplasmic incompatibility and feminization64; and lastly, in Pityohyphantes phrygianus (C. L. Koch, 1836), Wolbachia may influence female post-copulatory behavior and sex ratio48. Another effect caused by Wolbachia, not directly related to reproduction, was observed in the spider Hylyphantes graminicola (Sundevall, 1830), where the bacteria beneficially affected host metabolism30, leading to increased enzyme activity and nutrient availability, which contributed to a higher survival rate of the spider under stress65.
The available data on the spread of Wolbachia in spiders are still insufficient, and the diversity of bacterial strains determined by Multilocus Sequence Typing (MLST) in this group of arthropods from Poland is unknown. Therefore, we decided to pursue the issue of intracellular bacteria in spiders. The aim of our study was to determine the distribution and molecular characterization of Wolbachia in these invertebrates, which may contribute to better understanding of host-endosymbiont associations. Here, we report the first detection of Wolbachia in the spider Enoplognatha latimana Hippa and Oksala, 1982 (Araneae: Theridiidae). The Wolbachia strain identified in this spider was examined using MLST and wsp gene analyses. Furthermore, we discuss the etiology of Wolbachia infection in E. latimana.
Materials and methods
Sampling of spiders
Thirty-four E. latimana adult specimens, three juvenile forms, and two egg sacs were collected from nine different locations in the Wielkopolska Voivodeship: (1) coordinates: N 52.46136, E 16.94071; collection date: September 2021; (2) coordinates: N 52.49315, E 16.88068; collection date: July 2021; (3) coordinates: N 52.49315, E 16.88068; collection date: July 2021; (4) coordinates: N 52.49273, E 16.87891; collection date: July 2021; (5) coordinates: N 52.34225, E 18.47713; collection date: August 2021; (6) coordinates: N 52.18284, E 17.746977; collection date: July 2021; (7) coordinates: N 52.64025, E 19.12977; collection date: June 2021; (8) coordinates: N 52.63449, E 19.32619; collection date: June 2021; and (9) coordinates: N 52.47559, E 16.92671; collection date: July 2021.
Three adult male spiders, one adult female spider, and one juvenile form were collected from the same locality characterized in Table 1. The spiders were collected using a sweep net and immediately placed in 96% ethanol. Each specimen was examined for the presence of Wolbachia, as described below. The spiders were also examined for the presence of parasitoid insects by microscopic observation.
Table 1.
Identified Wolbachia gene sequences in the host Enoplognatha latimana and sampling site localities.
| Sample collection locality | Wolbachia gene sequence (GenBank accession number, length) |
|---|---|
| Area with shrubs, shaded with a rich undergrowth of herbaceous vegetation, near municipal waste landfill of the city of Poznań, Wielkopolska Voivodeship |
16S rRNA (OR220066, 1335 bp) coxA (OR227583, 410 bp) fbpA (OR227584, 417 bp) ftsZ (OR227585, 462 bp) gatB (OR227586, 398 bp) gltA (OR227587, 552 bp) groEL (OR227588, 441 bp) hcpA (OR227589, 416 bp) wsp (OR227590, 536 bp) |
| Coordinate | |
| N 52.4931526184082, E 16.88068199157715 | |
| Collection date | |
| 01.07.2021 |
Wolbachia detection
Total DNA was isolated from individual specimens using silica membranes from the commercial Genomic Mini kit for universal genomic DNA isolation (A&A Biotechnology, Gdansk, Poland) according to the manufacturer’s instruction. Wolbachia was identified by PCR using the following Wolbachia-specific primers: 553F_W (5′-CTTCATRYACTCGAGTTGCWGAGT-3′) and 1334R_W (5′-GAKTTAAAYCGYGCAGGBGTT-3′)66, which amplified a 781-bp product of the 16S rRNA gene. The PCR amplification was as follow: 94 °C for 2 min, 35 cycles at 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 45 s, and final elongation at 72 °C for 10 min66.
Analysis of Wolbachia genes
Molecular characterization of Wolbachia was based on sequence analysis of housekeeping genes: 16S rRNA, coxA, fbpA, ftsZ, gatB, gltA, groEL, hcpA, and additionally wsp. Two PCR reactions were conducted for the amplification of the 16S rRNA gene sequence. The first reaction utilized the specific primer EHR16SD67 along with the universal eubacterial primer 1513R68. The second reaction employed the specific primer EHR16SR67 along with the universal eubacterial primer 63F69. Other housekeeping genes included in the analysis were: gatB (glutamyl-tRNA(Gln) amidotransferase), coxA (cytochrome c oxidase), hcpA (conserved hypothetical protein), ftsZ (cell division protein), fbpA (fructose bisphosphate aldolase), wsp (Wolbachia surface protein)31, gltA (citrate synthase)32, and groEL (60-kDa heat-shock protein)70. The primer sequences and PCR amplification conditions are presented in Supplementary Table S1. PCR products were analyzed by electrophoresis on a 1.5% NOVA Mini agarose gel (Novazym) with a Nova 100 bp DNA Ladder (Novazym), sequenced using BigDye Terminator v3.1 with ABI Prism 3130XL (Applied Biosystems) and compared to the GenBank sequence data (International Nucleotide Sequence Database Collaboration) using BLASTn. Wolbachia gene sequences were deposited in GenBank under the accession numbers listed in Table 1.
MLST and phylogenetic analysis using wsp and ftsZ genes
MLST analysis was performed targeting the following eight loci: 16S rRNA, coxA, fbpA, ftsZ, gatB, gltA, groEL, and hcpA. Individual sequences of Wolbachia genes were aligned with sequences of different Wolbachia supergroups deposited in the GenBank database. Phylogenetic trees based on MLST were constructed for single genes, as well as concatenated alignments of the eight bacterial loci, using the maximum-likelihood method in MEGA 11 software71. Additionally, the sequences of Ehrlichia sp. were included as an outgroup. The NCBI accession numbers of the sequences used in the phylogenetic analysis are presented in Supplementary Figs. S1–S8. Sequence alignments were generated using CLUSTAL W software72. The jModelTest 2 software73,74 was applied to select the appropriate sequence evolution model. The HKY + G model was selected for 16S rRNA, coxA, and fbpA sequences, while the TrN + I + G model was chosen for the ftsZ sequence data; the GTR + G model was used for gatB, gltA, and for the concatenated sequence data of eight genes (16S rRNA, coxA, fbpA, ftsZ, gatB, gltA, groEL, and hcpA); the TrN + G model was selected for sequences available for the groEL and hcpA genes. Genetic recombination between strains was detected using the φ test implemented in the SplitsTree4 software75. The maximum likelihood bootstrap support was determined using 1000 bootstrap replicates.
The wsp gene, due to its relatively fast evolutionary rate, experiences significant recombination and diversifying selection, making it unreliable for strain characterization when used alone. However, it can be used as an additional optional strain marker to complement the MLST information31. The wsp gene sequence of Wolbachia from E. latimana was aligned with corresponding sequences of Wolbachia supergroups A and B deposited in GenBank. Additionally, an outgroup of Wolbachia supergroup D sequence was included. The NCBI accession numbers for the sequences used for phylogenetic analysis are shown in Fig. 3. The phylogenetic tree of the wsp gene was reconstructed using the same parameters as described above. The GTR + I + G model was selected for the wsp sequence.
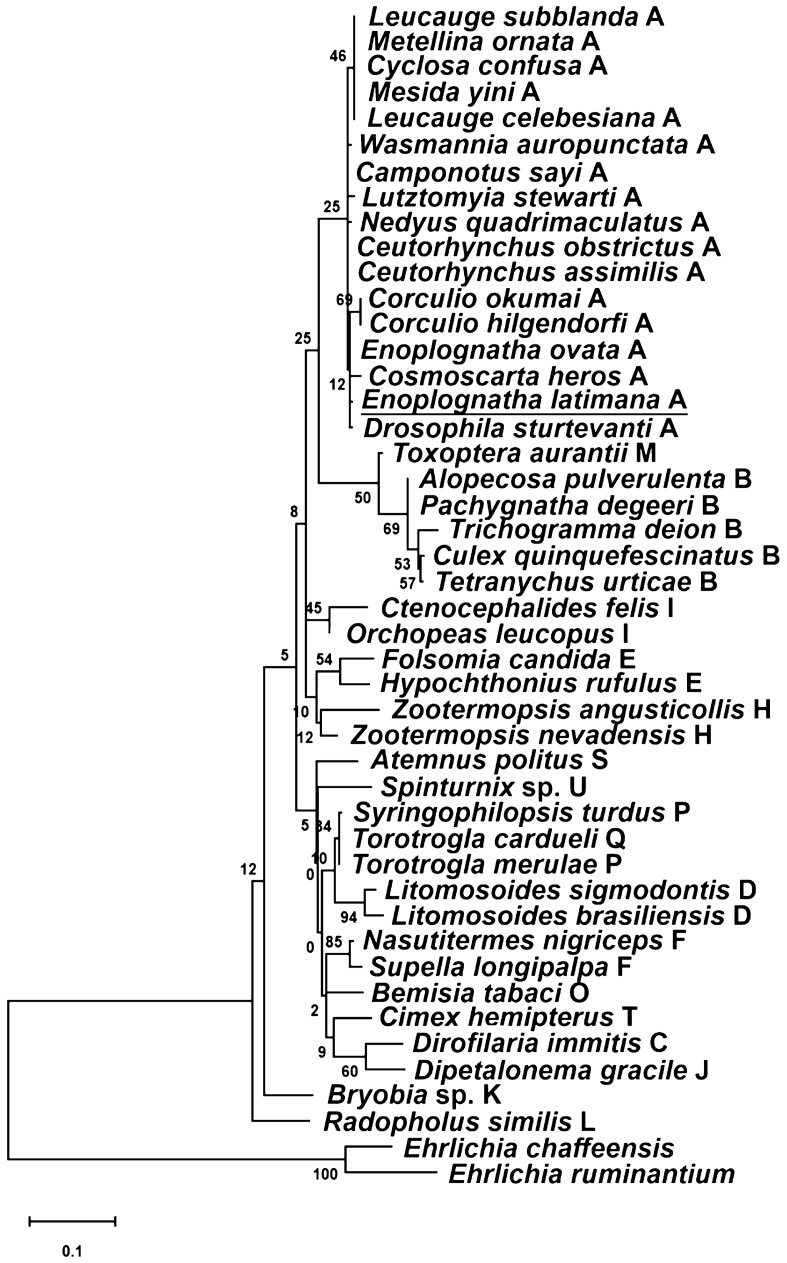
Figure 3.
Maximum likelihood reconstruction of Wolbachia supergroup phylogeny based on the wsp gene sequences using MEGA 11 software. Strains are designated by their host names. Wolbachia supergroups (A, B, and D) are indicated. Bar, substitutions per nucleotide. Bootstrap values based on 1000 replicates are shown on branches.
The ftsZ gene, which is involved in the regulation of bacterial cell division, contains highly conserved regions76. This characteristic makes it suitable for conducting fine-scale phylogenetic analysis within a bacterial genus77. A phylogenetic network was constructed based on the ftsZ gene sequences of Wolbachia using neighbor-net algorithm distance estimates in SplitsTree4. Unlike traditional phylogenetic trees, a phylogenetic network allows for visualization of multiple connections among examined sequences, which can represent recombination events75,78.
Additionally, the coxA, fbpA, ftsZ, gatB, and hcpA genes were compared with sequences in the PubMLST database (https://pubmlst.org) for generating a MLST allelic profile, determining the sequence type (ST) and the clonal complex.
Results
All collected specimens were screened for the occurrence of Wolbachia and the bacterium was detected in one female only. The infected female was collected together with three adult males and one juvenile from the same population, which were tested negative for Wolbachia infection. In addition, no parasitoid insects were observed upon microscopic examination.
We have successfully detected Wolbachia in E. latimana, marking the first documented occurrence of this bacterium in this spider species. Our analysis involved amplification of the wsp gene and eight housekeeping genes (16S rRNA, coxA, fbpA, ftsZ, gatB, gltA, groEL, and hcpA) of Wolbachia (Table 1).
Comparison of gene sequences
Wolbachia 16S rRNA, coxA, fbpA, ftsZ, gatB, gltA, groEL, and hcpA housekeeping gene sequences housekeeping gene sequences were compared with sequences deposited in GenBank from various invertebrate hosts using BLASTn. The gene sequences of Wolbachia from E. latimana showed the highest identity with Wolbachia from other spiders representing the order Araneae, as well as from insects from the orders Diptera and Hymenoptera.
The 16S rRNA gene sequence of bacteria from E. latimana showed the highest identity with Wolbachia sequence from dipteran insects Aedes albopictus (Skuse, 1894) and Drosophila sturtevanti Duda, 1927 deposited in GenBank under accession numbers CP101657 and CP050531, respectively. Sequence query coverage was 99% and the identity was 99.55%. We also compared the 16S rDNA sequences of Wolbachia infecting E. latimana and Enoplognatha ovata (Clerck, 1757) (accession no. EU333941), since both hosts represented the same genus of spiders. The identity of these sequences was 99.76% with query coverage of 62%.
Sequence analysis of the coxA amplicon using BLASTn showed the highest identity of 98.78% with 100% query coverage with Wolbachia coxA from the spider Mesida yini Zhu, Song and Zhang, 2003 deposited in GenBank under accession no. KX169178. The highest identity (100% with 100% query coverage) was observed between Wolbachia fbpA sequences from E. latimana and the spider Leucauge celebesiana (Walckenaer, 1842) (accession no. KX380749). The ftsZ sequence showed the highest identity of 98.48% with 99% query coverage with Wolbachia gene from D. sturtevanti and the hymenopteran insect Camponotus sayi Emery, 1893 deposited in GenBank under accession numbers CP050531 and DQ266387, respectively. The sequence of the gatB gene of Wolbachia from E. latimana exhibited the highest identity (100%) to the gene of Wolbachia from the spider Metellina ornata (Chikuni, 1955) (accession no. MN202032). The highest identity of 98.36% with 99% query coverage was observed between the sequences of the gltA amplicon detected in Wolbachia from E. latimana and the dipteran insect Sicus ferrugineus (Linnaeus, 1761) (accession no. OX366370). Sequence analysis of the groEL amplicon using BLASTn showed the highest identity of 95.05% with 100% query coverage to Wolbachia groEL from the spider O. gibbosus deposited in GenBank under accession no. OW370537. The highest identity (97.6% with 100% query coverage) was observed between the Wolbachia hcpA sequences from E. latimana and the hymenopteran insect Camponotus pennsylvanicus (De Geer, 1773) (accession no. CP095495). The wsp sequence showed the highest identity of 99.25% with 99% query coverage with the gene of Wolbachia from the spider Trichonephila clavata (L. Koch, 1878) deposited in GenBank under accession no. EF612772.
We have found a unique sequence (5′-GACTTCG-3′) for Wolbachia supergroup A in the ftsZ gene. This sequence has been identified in Wolbachia ftsZ from various species, including D. sturtevanti (accession no. CP050531), C. sayi (accession no. DQ266387), Ceutorhynchus assimilis (Paykull, 1800) (accession no. OU906081), Ceutorhynchus obstrictus (Marsham, 1802) (accession no. HM012590), Cyclosa confusa Bösenberg and Strand, 1906 (accession no. KX380701), L. celebesiana (accession no. KX380698), Leucauge subblanda Bösenberg and Strand, 1906 (accession no. MN202113), Lutzomyia stewarti (Mangabeira Fo and Galindo, 1944) (accession no. KJ174694), M. ornata (accession no. KX380693), M. yini (accession no. KX380706), Nedyus quadrimaculatus (Linnaeus, 1758) (accession no. MG987989), and Wasmannia auropunctata Roger, 1863 (accession no. JX499050). The sequence was not found in the ftsZ gene of other Wolbachia strains representing supergroups B-U used in this study as comparative material. The location of the above nucleotide sequence was determined at positions 673–679 in reference to ftsZ of Wolbachia from D. sturtevanti (accession no. CP050531). An alignment showing the unique ftsZ sequence of Wolbachia supergroup A is presented in Fig. 1.
Figure 1.
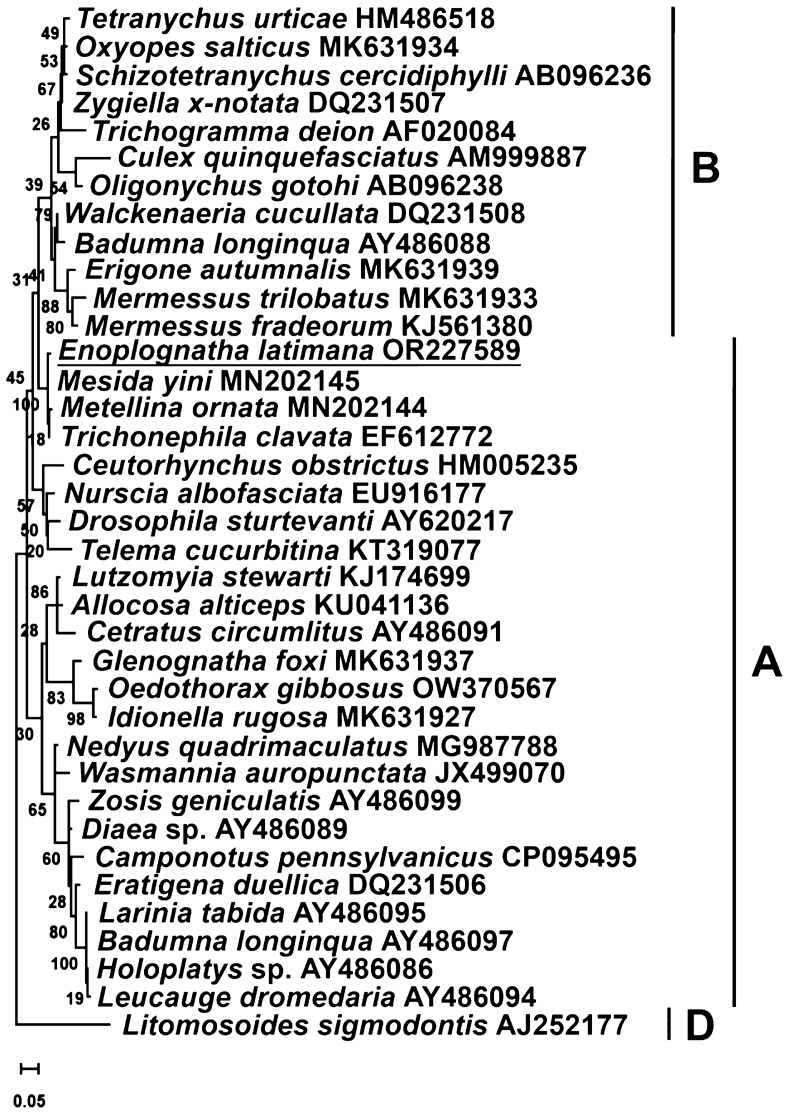
Maximum likelihood reconstruction of Wolbachia supergroup phylogeny based on concatenated sequence alignments of eight bacterial loci (16S rRNA, coxA, fbpA, ftsZ, gatB, gltA, groEL, hcpA) using MEGA 11 software. Strains are designated by their host names, except for outgroup bacteria. Capital letters indicate individual Wolbachia supergroups. Bar, substitutions per nucleotide. Bootstrap values based on 1000 replicates are shown on branches.
MLST and phylogenetic analysis using the wsp and ftsZ genes
Phylogeny based on concatenated MLST sequence data analysis of eight genes (16S rRNA, coxA, fbpA, ftsZ, gatB, gltA, groEL, and hcpA) showed that Wolbachia from the spider E. latimana was related to endosymbionts of other spider hosts from the order Araneae and dipteran and hemipteran insects, representing supergroup A. The analysis of both individual genes (Supplementary Figs. S1–S8 available in the online Supplementary Information), as well as the combined eight-gene analysis (Fig. 2) consistently demonstrated that the bacterium belonged to supergroup A. The absence of statistically significant evidence of recombination (p = 1.0) using the φ test suggested that Wolbachia from E. latimana was not a recombinant between strains of other Wolbachia supergroups.
Figure 2.
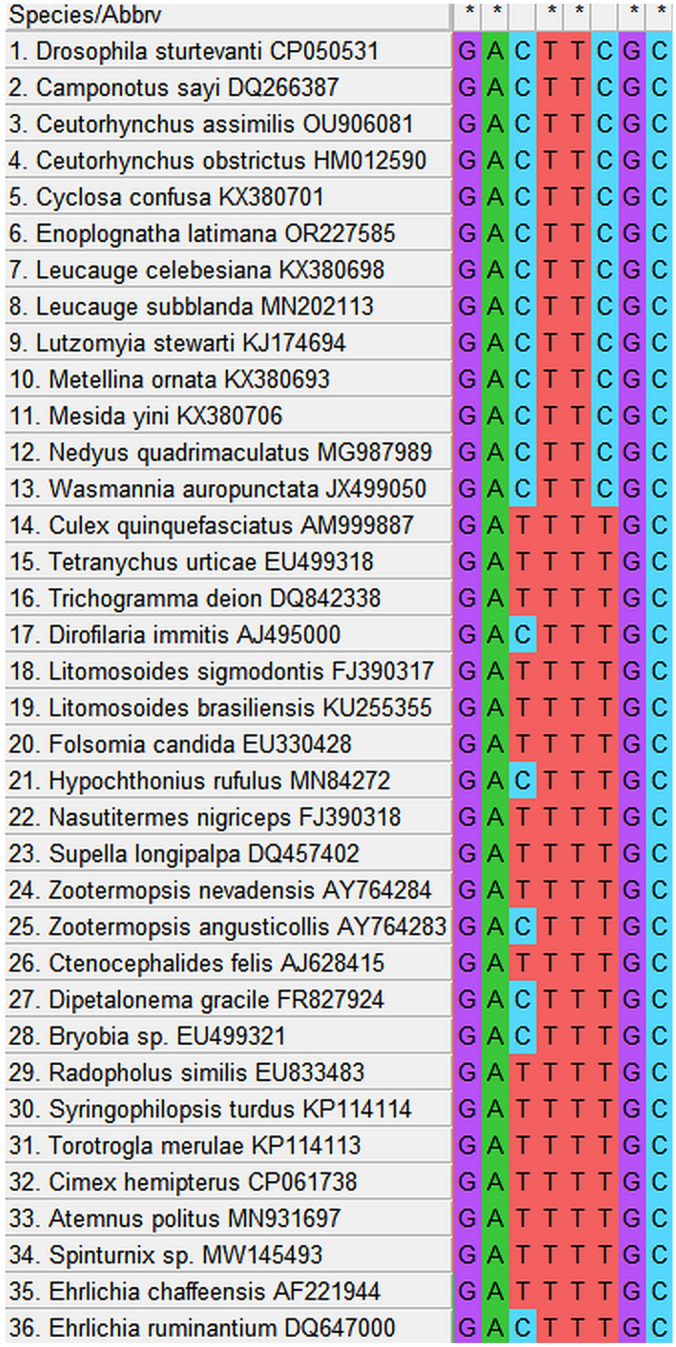
Alignment showing the unique 5′-GACTTCG-3′ sequence from the ftsZ gene of Wolbachia supergroup A from Enoplognatha latimana, Drosophila sturtevanti, Camponotus sayi, Ceutorhynchus assimilis, Ceutorhynchus obstrictus, Cyclosa confusa, Leucauge celebesiana, Leucauge subblanda, Lutzomyia stewarti, Metellina ornata, Mesida yini, Nedyus quadrimaculatus, and Wasmannia auropunctata.
The reconstruction of the phylogenetic tree based on the wsp gene of Wolbachia supergroups A and B has confirmed that the endosymbiont from E. latimana belongs to supergroup A. It formed a cluster with bacteria from three spider species representing the families Tetragnathidae (M. yini and M. ornata) and Araneidae (T. clavata) (Fig. 3).
In addition, a phylogenetic network based on the ftsZ gene sequences of Wolbachia (Fig. 4) revealed the relationship of Wolbachia from E. latimana with representative strains of Wolbachia supergroup A. The network mostly contained very narrow fields, indicating a low level of conflict in the data at the nucleotide level. Wolbachia from E. latimana clearly clustered with supergroup A strains, excluding other supergroups, as confirmed by the φ test results. Moreover, the analysis indicated the diversity within supergroup A, with two noticeable subgroups: (1) Wolbachia from insects L. stewarti, W. auropunctata, C. obstrictus, N. quadrimaculatus, C. assimilis, and (2) Wolbachia from spiders E. latimana, L. celebesiana, M. ornate, C. confuse, M. yini, L. subblanda, and insects C. sayi, D. sturtevanti.
Figure 4.
Median network reconstructed for Wolbachia supergroup A based on sequence polymorphism of the ftsZ genes. Conflicting phylogenetic signals (due to recombination and/or homoplasy) are represented as boxes or parallelograms in the network.
Using the PubMLST database, the MLST allelic profile was generated (Table S2). The allelic profile, ST and clonal complex were new according to the available data in the Wolbachia database.
Discussion
The sequences of 16S rRNA, coxA, fbpA, ftsZ, gatB, gltA, groEL, hcpA, and wsp genes of the bacterial strain found in this study in the spider species E. latimana showed the highest identity with Wolbachia from supergroup A discovered in other spider hosts and in insects of the orders Diptera and Hymenoptera. The phylogeny based on the concatenated dataset of eight housekeeping genes (16S rRNA, coxA, fbpA, ftsZ, gatB, gltA, groEL, hcpA) and the wsp gene confirmed the close relationship between Wolbachia infecting E. latimana and the strains found in other spider and insect hosts, representing the same supergroup carrying the unique 5′-GACTTCG-3′ sequence in the ftsZ gene. Additionally, phylogenetic network analysis of the ftsZ gene of Wolbachia revealed high intragroup diversity of supergroup A, with the supergroup being subdivided into two clades. Wolbachia from E. latimana clustered with bacteria from other spiders, as well as with strains from dipteran and hymenopteran hosts, further supporting their close relationship. The Wolbachia in E. latimana sequences of coxA, fbpA, ftsZ, gatB, and hcpA genes did not show an exact match with previously identified STs in the PubMLST Wolbachia database and the bacterial strain in the spider is new.
A question arises about the etiology of Wolbachia in E. latimana, as only one spider out of 39 tested specimen was infected. Spiders of the genus Enoplognatha feed on insects of different orders79–81, including Diptera and Hymenoptera82. Among them, pollinators and other flower-visiting insects are predominant in the spiders diet. Interestingly, our study revealed that Wolbachia genes in E. latimana exhibited the highest identity and closest relationships to bacteria found in insects, associated with flowering plants, from (1) Diptera: S. ferrugineus83, A. albopictus84, and (2) Hymenoptera: C. sayi85, C. pennsylvanicus86. Other authors have confirmed that the transfer of Wolbachia can occur through the ingestion of remains from infected specimens87, and these insects may be a potential source of Wolbachia infection in E. latimana. It is plausible that Wolbachia identified in E. latimana could be the result of its presence in insect cells found in the spider’s digestive tract, without infecting the spider’s own cells. In this case, the presence of the bacteria in the spider should be considered accidental rather than as a stable and permanent infection of the host. Considering that hymenopteran and dipteran insects are also parasites of spiders88, they could be regarded as potential sources of the bacteria. Some spiders from the genera Trichonephila89,90 and Leucauge91,92 are known to be attacked by Hymenoptera parasitoids. Dipteran insects are also known enemies of Trichonephila sp. and Enoplognatha sp.93. Wolbachia genes discovered in E. latimana showed the highest identity with the corresponding genes of bacterial supergroup A from T. clavata and L. celebesiana. Furthermore, the close relationship between these Wolbachia strains may suggests the potential possibility of bacterial transmission from insect parasitoids to spider hosts, especially that insect parasites can serve as vectors for Wolbachia transmission between hosts94,95. While insect parasitoids typically kill their host upon completion of their larval development and parasitism do not allow hosts to transmit the infection to the progeny88,92,96,97, there have been cases of spiders that were able to get rid of the intruder and survive90. Among the analyzed specimens of E. latimana, no insect parasites were found during microscopic observations. If the role of the parasite in the transfer of Wolbachia to E. latimana may be assumed, one could attempt to speculate that the spider have been temporarily inhabited by the parasite but managed to survive. However, this is not the only potential scenario, as Wolbachia transmission via food cannot be ruled out either. Insects from the orders Diptera and Hymenoptera, infected by Wolbachia with high genetic identity and relatedness to Wolbachia from E. latimana, feed on plant nectars. Examples include the flower and leaf nectar-eating dipteran A. albopictus84,98 or the extrafloral nectar-eating hymenopteran C. sayi85. Although literature data do not indicate plant nectar in the diet of E. latimana and the possibility of acquisition from nectar contaminated by infected insects is unlikely, it may be not excluded, as some species of spiders, especially early instars of web-building spiders, rely on floral and extrafloral nectar as an important component of their food99–103. The latter hypothesis may be supported by the results of other authors, suggesting that food can serve as a medium for Wolbachia transmission among invertebrates with similar feeding habits. Sharing the same plant diet may facilitate horizontal transmission of these bacteria104–106. All of the modes of Wolbachia transfer described above are possible. Although we have not determined the exact etiology of Wolbachia in E. latimana, the endosymbiont is undoubtedly related to those found in other spider species and insects from the orders Diptera and Hymenoptera, and transfer of the microorganism between these hosts cannot be excluded.
Conclusion
In conclusion, we have detected for the first time the bacterium Wolbachia associated with the spider E. latimana. The microorganism was found in only one female and a question arises about the etiology of Wolbachia in E. latimana. Our data are not sufficient to support the stable presence of Wolbachia in the spider species. The high probability of only accidental bacterial presence cannot be excluded. Our study revealed that Wolbachia genes associated with E. latimana exhibited the highest identity and closest relationships to bacteria found in insects from Diptera and Hymenoptera. As the insects are predominant in the spiders diet, the detected Wolbachia could have been present in ingested remains from infected insect specimens.
Our study confirmed the classification of the bacteria associated with E. latimana to Wolbachia supergroup A. These data provide insight into the occurrence of Wolbachia in arthropods. Additionally, we have deposited the sequences of Wolbachia wsp and housekeeping genes in publicly available databases, providing valuable molecular data for future comparative studies in this field.
Supplementary Information
Acknowledgements
We would like to thank our friend Dr. Ziemowit Olszanowski (1961-2019), Associate Professor at the Department of Animal Taxonomy and Ecology of the Adam Mickiewicz University in Poznań, Poland, a specialist in taxonomy, ecology, biology, zoogeography of moss mites (Acari: Oribatida), and co-discoverer of Wolbachia in Oribatida. He inspired our research on endosymbiotic bacteria in spiders and it is thanks to his influence that this study came to fruition. This article is dedicated to the memory of Ziemowit Olszanowski.
Author contributions
E.K. and P.Sz. designed the study. P.Sz. collected samples and identified spiders. E.K. detected bacterial genes and performed phylogenetic analysis. E.K. wrote the manuscript in collaboration with P.Sz. The authors read and approved the final manuscript.
Data availability
Sequencing data generated and analyzed in this study are deposited to NCBI Nucleotide Database (accession nos. OR220066 and OR227583–OR227590).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-57701-y.
References
- 1.Ahmed MZ, Ajauro-Jnr EV, Welchm JJ, Kawahara AY. Wolbachia in butterflies and moths: Geographic structure in infection frequency. Front. Zool. 2015;12(16):1–9. doi: 10.1186/s12983-015-0107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinert LA, Araujo-Jnr EV, Ahmed MZ, Welch JJ. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc. R. Soc. B Biol. Sci. 2015;282:20150249. doi: 10.1098/rspb.2015.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sazama EJ, Bosch MJ, Shouldis CS, Ouellette SP, Wesner JS. Incidence of Wolbachia in aquatic insects. Ecol. Evol. 2017;7:1165–1169. doi: 10.1002/ece3.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kajtoch Ł, et al. Using host species traits to understand the Wolbachia infection distribution across terrestrial beetles. Sci. Rep. 2019;9:847. doi: 10.1038/s41598-018-38155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sazama EJ, Ouellette SP, Wesner JS. Bacterial endosymbionts are common among, but not necessarily within, insect species. Environ. Entomol. 2019;48(1):127–133. doi: 10.1093/ee/nvy188. [DOI] [PubMed] [Google Scholar]
- 6.Driscoll TP, et al. Evolution of Wolbachia mutualism and reproductive parasitism: Insight from two novel strains that co-infect cat fleas. PeerJ. 2020;8:e10646. doi: 10.7717/peerj.10646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoang T. Using Wolbachia infected mosquitos to combat arboviral transmission: From molecular mechanisms to global application. UJEMI. 2020;4:1–9. [Google Scholar]
- 8.Zélé F, Altıntaş M, Santos I, Cakmak I, Magalhães S. Population-specific effect of Wolbachia on the cost of fungal infection in spider mites. Ecol. Evol. 2020;10:3868–3880. doi: 10.1002/ece3.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eugénio AT, Marialva MSP, Beldade P. Effects of Wolbachia on transposable element expression vary between Drosophila melanogaster host genotypes. Genome Biol. Evol. 2023;15(3):036. doi: 10.1093/gbe/evad036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Y, Guo J, Li Y. Wolbachia wPip blocks Zika virus transovarial transmission in Aedes albopictus. Microbiol. Spectr. 2022;10(5):e0263321. doi: 10.1128/spectrum.02633-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hague MTJ, Woods FA, Cooper BS. Pervasive effects of Wolbachia on host activity. Biol. Lett. 2021;17(5):20210052. doi: 10.1098/rsbl.2021.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruntenko NE, et al. Various Wolbachia genotypes differently influence host Drosophila dopamine metabolism and survival under heat stress conditions. BMC Evol. Biol. 2017;17(2):252. doi: 10.1186/s12862-017-1104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ourry M, et al. Influential insider: Wolbachia, an intracellular symbiont, manipulates bacterial diversity in its insect host. Microorganisms. 2021;9:1313. doi: 10.3390/microorganisms9061313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruz MA, Magalhães S, Sucena É, Zélé F. Wolbachia and host intrinsic reproductive barriers contribute additively to postmating isolation in spider mites. Evolution. 2021;75(8):2085–2101. doi: 10.1111/evo.14286. [DOI] [PubMed] [Google Scholar]
- 15.Lau MJ, Ross PA, Endersby-Harshman NM, Yang Q, Hoffmann AA. Wolbachia inhibits ovarian formation and increases blood feeding rate in female Aedes aegypti. PLoS Negl. Trop. Dis. 2022;16(11):e0010913. doi: 10.1371/journal.pntd.0010913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quek S, et al. Wolbachia depletion blocks transmission of lymphatic filariasis by preventing chitinase-dependent parasite. PNAS. 2022;119(15):e2120003119. doi: 10.1073/pnas.2120003119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López-Madrigal S, Duarte E. Titer regulation in arthropod-Wolbachia symbioses. FEMS Microbiol. Lett. 2019;366:232. doi: 10.1093/femsle/fnz232. [DOI] [PubMed] [Google Scholar]
- 18.Miyata M, Konagaya T, Yukuhiro K, Nomura M, Kageyama D. Wolbachia-induced meiotic drive and feminization is associated with an independent occurrence of selective mitochondrial sweep in a butterfly. Biol. Lett. 2017;13:20170153. doi: 10.1098/rsbl.2017.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verhulst EC, Pannebakker BA, Geuverink E. Variation in sex determination mechanisms may constrain parthenogenesis-induction by endosymbionts in haplodiploid systems. Curr. Opin. Insect Sci. 2023;56:101023. doi: 10.1016/j.cois.2023.101023. [DOI] [PubMed] [Google Scholar]
- 20.Richardson KM, et al. A male-killing Wolbachia endosymbiont is concealed by another endosymbiont and a nuclear suppressor. PLoS Biol. 2023;21(3):e3001879. doi: 10.1371/journal.pbio.3001879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ju JF, et al. Wolbachia supplement biotin and riboflavin to enhance reproduction in planthoppers. ISME J. 2020;14:676–687. doi: 10.1038/s41396-019-0559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hickin ML, Kakumanu ML, Schal C. Effects of Wolbachia elimination and B-vitamin supplementation on bed bug development and reproduction. Sci. Rep. 2022;12:10270. doi: 10.1038/s41598-022-14505-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Khan J, Zheng XY, Wu Y. Wolbachia increase germ cell mitosis to enhance the fecundity of Laodelphax striatellus. Insect Biochem. Mol. Biol. 2020;127:103471. doi: 10.1016/j.ibmb.2020.103471. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. Wolbachia strain wGri from the tea geometrid moth Ectropis grisescens contributes to its host’s fecundity. Front. Microbiol. 2021;12:694466. doi: 10.3389/fmicb.2021.694466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prigot-Maurice C, et al. Investigating Wolbachia symbiont-mediated host protection against a bacterial pathogen using a natural Wolbachia nuclear insert. J. Invertebr. Pathol. 2023;197:107893. doi: 10.1016/j.jip.2023.107893. [DOI] [PubMed] [Google Scholar]
- 26.Cogni R, Ding SD, Pimentel AC, Day JP, Jiggins FM. Wolbachia reduces virus infection in a natural population of Drosophila. Commun. Biol. 2021;4(1):1327. doi: 10.1038/s42003-021-02838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussain M, Zhang G, Leitner M, Hedges LM, Asgari S. Wolbachia RNase HI contributes to virus blocking in the mosquito Aedes aegypti. Science. 2023;26(1):105836. doi: 10.1016/j.isci.2022.105836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soh LS, Singham GV. Bacterial symbionts influence host susceptibility to fenitrothion and imidacloprid in the obligate hematophagous bed bug, Cimex hemipterus. Sci. Rep. 2022;12:1–16. doi: 10.1038/s41598-022-09015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. Microbiome variation correlates with the insecticide susceptibility in different geographic strains of a significant agricultural pest, Nilaparvata lugens. NPJ Biofilms Microb. 2023;9:2. doi: 10.1038/s41522-023-00369-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, et al. Background-dependent Wolbachia-mediated insecticide resistance in Laodelphax striatellus. Environ. Microbiol. 2020;22(7):2653–2663. doi: 10.1111/1462-2920.14974. [DOI] [PubMed] [Google Scholar]
- 31.Baldo L, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casiraghi M, et al. Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: Clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiology. 2005;151(12):4015–4022. doi: 10.1099/mic.0.28313-0. [DOI] [PubMed] [Google Scholar]
- 33.Glowska E, Dragun-Damian A, Dabert M, Gerth M. New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae) Infect. Gen. Evol. 2015;30:140–146. doi: 10.1016/j.meegid.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 34.Konecka E, Olszanowski Z. Wolbachia supergroup E found in Hypochthonius rufulus (Acari: Oribatida) in Poland. Infect. Genet. Evol. 2021;91:104829. doi: 10.1016/j.meegid.2021.104829. [DOI] [PubMed] [Google Scholar]
- 35.Laidoudi Y, et al. An earliest endosymbiont, Wolbachia massiliensis sp. nov., strain PL13 from the bed bug (Cimex hemipterus), type strain of a new supergroup T. Int. J. Mol. Sci. 2020;21(21):8064. doi: 10.3390/ijms21218064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scholz M, et al. Large scale genome reconstructions illuminate Wolbachia evolution. Nat. Commun. 2020;11:5235. doi: 10.1038/s41467-020-19016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konecka E. Fifty shades of bacterial endosymbionts and some of them still remain a mystery: Wolbachia and Cardinium in oribatid mites (Acari: Oribatida) J. Invertebr. Pathol. 2022;189:107733. doi: 10.1016/j.jip.2022.107733. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, et al. Phylogenomic analysis of Wolbachia strains reveals patterns of genome evolution and recombination. Genome Biol. Evol. 2020;12(12):2508–2520. doi: 10.1093/gbe/evaa219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCutcheon JP, Boyd BM, Dale C. The life of an insect endosymbiont from the cradle to the grave. Curr. Biol. 2019;29(11):R485–R495. doi: 10.1016/j.cub.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 40.Duron O, Hurst GDD, Hornett EA, Josling JA, Engelstädter J. High incidence of the maternally inherited bacterium Cardinium in spiders. Mol. Ecol. 2008;17:1427–1437. doi: 10.1111/j.1365-294X.2008.03689.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Yun Y, Hu G, Peng Y. Insights into the bacterial symbiont diversity in spiders. Ecol. Evol. 2018;8:4899–4906. doi: 10.1002/ece3.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Busck MM, et al. Microbiomes and specific symbionts of social spiders: Compositional patterns in host species, populations, and nests. Front. Microbiol. 2020;11:1845. doi: 10.3389/fmicb.2020.01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheffer MM, et al. Tissue- and population-level microbiome analysis of the wasp spider Argiope bruennichi identified a novel dominant bacterial symbiont. Microorganisms. 2020;8(1):8. doi: 10.3390/microorganisms8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halter T, et al. One to host them all: Genomics of the diverse bacterial endosymbionts of the spider Oedothorax gibbosus. Microb. Genom. 2023;9(2):000943. doi: 10.1099/mgen.0.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cordaux R, Michel-Salzat A, Bouchon D. Wolbachia infection in crustaceans: Novel hosts and potential routes for horizontal transmission. J. Evol. Biol. 2001;14:237–243. doi: 10.1046/j.1420-9101.2001.00279.x. [DOI] [Google Scholar]
- 46.Rowley SM, Raven JR, McGraw EA. Wolbachia pipientis in australian spiders. Curr. Microbiol. 2004;49:208–214. doi: 10.1007/s00284-004-4346-z. [DOI] [PubMed] [Google Scholar]
- 47.Baldo L, et al. Insight into the routes of Wolbachia invasion: High levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol. Ecol. 2008;17(2):557–569. doi: 10.1111/j.1365-294X.2007.03608.x. [DOI] [PubMed] [Google Scholar]
- 48.Gunnarsson B, Goodacre SL, Hewitt GM. Sex ratio, mating behaviour and Wolbachia infections in a sheetweb spider. Biol. J. Linn. Soc. 2009;98:181–186. doi: 10.1111/j.1095-8312.2009.01247.x. [DOI] [Google Scholar]
- 49.Wang ZY, Deng Ch, Yun YL, Jian Ch, Peng Y. Molecular detection and the phylogenetics of Wolbachia in Chinese spiders (Araneae) J. Arachnol. 2010;38(2):237–241. doi: 10.1636/JOA_B09-69.1. [DOI] [Google Scholar]
- 50.Vanthournout B, Swaegers J, Hendrickx F. Spiders do not escape reproductive manipulations by Wolbachia. BMC Evol. Biol. 2011;11:15. doi: 10.1186/1471-2148-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yun Y, et al. Wolbachia strains typing in different geographic population spider, Hylyphantes graminicola (Linyphiidae) Curr. Microbiol. 2011;62(1):139–145. doi: 10.1007/s00284-010-9686-2. [DOI] [PubMed] [Google Scholar]
- 52.Yun Y, Peng Y, Liu FX, Lei C. Wolbachia screening in spiders and assessment of horizontal transmission between predator and prey. Neotrop. Entomol. 2011;40(2):164–169. [PubMed] [Google Scholar]
- 53.Vanthournout B, Hendrickx F. Endosymbiont dominated bacterial communities in a dwarf spider. PLoS ONE. 2015;10(2):e0117297. doi: 10.1371/journal.pone.0117297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan Q, et al. Detection and phylogenetic analysis of bacteriophage WO in spiders (Araneae) Folia Microbiol. 2015;60(6):497–503. doi: 10.1007/s12223-015-0393-z. [DOI] [PubMed] [Google Scholar]
- 55.Wang GH, Jia LY, Xiao JH, Huang DW. Discovery of a new Wolbachia supergroup in cave spider species and the lateral transfer of phage WO among distant hosts. Infect. Genet. Evol. 2016;41:1–7. doi: 10.1016/j.meegid.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 56.Bordenstein S, Bordenstein S. Eukaryotic association module in phage WO genomes from Wolbachia. Nat. Commun. 2016;7:13155. doi: 10.1038/ncomms13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivanov V, Lee KM, Mutanen M. Mitonuclear discordance in wolf spiders: Genomic evidence for species integrity and introgression. Mol. Ecol. 2018;27:1681–1695. doi: 10.1111/mec.14564. [DOI] [PubMed] [Google Scholar]
- 58.Kennedy SR, Tsau S, Gillespie R, Krehenwinkel H. Are you what you eat? A highly transient and prey-influenced gut microbiome in the grey house spider Badumna longinqua. Mol. Ecol. 2020;29:1001–1015. doi: 10.1111/mec.15370. [DOI] [PubMed] [Google Scholar]
- 59.Oh HW, et al. Ultrastructural and molecular identification of a Wolbachia endosymbiont in a spider, Nephila clavata. Insect Mol. Biol. 2000;9(5):539–543. doi: 10.1046/j.1365-2583.2000.00218.x. [DOI] [PubMed] [Google Scholar]
- 60.Goodacre SL, Martin OY, Thomas CF, Hewitt GM. Wolbachia and other endosymbiont infections in spiders. Mol. Ecol. 2006;15(2):517–527. doi: 10.1111/j.1365-294X.2005.02802.x. [DOI] [PubMed] [Google Scholar]
- 61.White JA, et al. Endosymbiotic bacteria are prevalent and diverse in agricultural spiders. Microb. Ecol. 2020;79:472–481. doi: 10.1007/s00248-019-01411-w. [DOI] [PubMed] [Google Scholar]
- 62.Dunaj SJ, Bettencourt BR, Garb JE, Brucker RM. Spider phylosymbiosis: Divergence of widow spider species and their tissues` microbiomes. BMC Evol. Biol. 2020;20(104):1664. doi: 10.1186/s12862-020-01664-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tyagi K, Tyagi I, Kumar V. Interspecific variation and functional traits of the gut microbiome in spiders from the wild: The largest effort so far. PLoS ONE. 2021;16(7):e0251790. doi: 10.1371/journal.pone.0251790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Curry MM, Paliulis LV, Welch KD, Harwood JD, White JA. Multiple endosymbiont infections and reproductive manipulations in a linyphiid spider population. Heredity. 2015;115:146–152. doi: 10.1038/hdy.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Su Q, et al. Combined effects of elevated CO2 concentration and Wolbachia on Hylyphantes graminicola (Araneae: Linyphiidae) Ecol. Evol. 2019;9:7112–7121. doi: 10.1002/ece3.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simões PM, Mialdea G, Reiss D, Sagot MF, Charlat S. Wolbachia detection: an assessment of standard PCR protocols. Mol. Ecol. Resour. 2011;11:567–572. doi: 10.1111/j.1755-0998.2010.02955.x. [DOI] [PubMed] [Google Scholar]
- 67.Brown GK, Martin AR, Roberts TK, Aitken RJ. Detection of Ehrlichia platys in dogs in Australia. Aust. Vet. J. 2001;79:554–558. doi: 10.1111/j.1751-0813.2001.tb10747.x. [DOI] [PubMed] [Google Scholar]
- 68.Weisburg WGS, Barns M, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fredriksson NJ, Hermansson M, Wilen BM. The choice of PCR primers has great impact on assessments of bacterial community diversity and dynamics in a wastewater treatment plant. PLoS ONE. 2013;8:e76431. doi: 10.1371/journal.pone.0076431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ros VID, Fleming VM, Feil EJ, Breeuwer JAJ. How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae) Appl. Environ. Microbiol. 2009;75(4):1036–1043. doi: 10.1128/AEM.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tamura K, Stecher G, Kumar S. MEGA 11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guindon S, Gascuel OA. Simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 74.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 76.Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 2005;6(11):862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Werren JH, Zhang W, Guo LR. Evolution and phylogeny of Wolbachia: Reproductive parasites of arthropods. Proc. R. Soc. B Biol. Sci. 1995;261(1360):55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- 78.Bryant D, Moulton V. Neighbor-net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 79.Aghdam HR. Natural enemies of Colorado potato beetle, Leptinotarsa decemlineata (Say) and population fluctuation of green lacewing, Chrysoperla carnea Stephen in potato fields of Ardabil plain, Iran. Agroecol. J. 2013;9(1):41. [Google Scholar]
- 80.Gavish-Regev E, Rotkopf R, Lubin Y, Coll M. Consumption of aphids by spiders and the effect of additional prey: Evidence from microcosm experiments. BioControl. 2009;54(3):341–350. doi: 10.1007/s10526-008-9170-0. [DOI] [Google Scholar]
- 81.Luo S, Naranjo SE, Wu K. Biological control of cotton pests in China. Biol. Control. 2014;68:6–14. doi: 10.1016/j.biocontrol.2013.06.004. [DOI] [Google Scholar]
- 82.Scott CE, McCann S. They mostly come at night: Predation on sleeping insects by introduced candy-striped spiders in North America. Ecol. 2023;104(5):e4025. doi: 10.1002/ecy.4025. [DOI] [PubMed] [Google Scholar]
- 83.Wiatrowska B, et al. Linear scaling: Negative effects of invasive Spiraea tomentosa (Rosaceae) on wetland plants and pollinator communities. NeoBiota. 2023;81:63–90. doi: 10.3897/neobiota.81.95849. [DOI] [Google Scholar]
- 84.Cui G, et al. Aedes albopictus life table: Environment, food, and age dependence survivorship and reproduction in a tropical area. Parasit. Vectors. 2021;14:568. doi: 10.1186/s13071-021-05081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chamberlain SA, Holland JN. Body size predicts degree in ant–plant mutualistic networks. Funct. Ecol. 2009;23:196–202. doi: 10.1111/j.1365-2435.2008.01472.x. [DOI] [Google Scholar]
- 86.Hussain M, et al. Distribution patterns of insect pollinator assemblages at Deva Vatala National Park, Bhimber, Azad Jammu and Kashmir. Pak. J. Zool. 2023;2023:1–7. [Google Scholar]
- 87.Brown AN, Lloyd VK. Evidence for horizontal transfer of Wolbachia by a Drosophila mite. Exp. Appl. Acarol. 2015;66(3):301–311. doi: 10.1007/s10493-015-9918-z. [DOI] [PubMed] [Google Scholar]
- 88.Durkin ES, et al. Parasites of spiders: Their impacts on host behavior and ecology. J. Arachnol. 2021;49(3):281–298. doi: 10.1636/JoA-S-20-087. [DOI] [Google Scholar]
- 89.Gauld I. The re-definition of pimpline genus Hymenoepimecis (Hymenoptera: Ichneumonidae) with a description of a plesiomorphic new Costa Rican species. J. Hymenopt. Res. 2000;9:213–219. [Google Scholar]
- 90.Gonzaga MO, Sobczak JF, Penteado-Dias AM, Eberhard WG. Modification of Nephila clavipes (Araneae: Nephilidae) webs induced by the parasitoids Hymenoepimecis bicolor and H. robertsae (Hymenoptera: Ichneumonidae) Ethol. Ecol. Evol. 2010;22:151–165. doi: 10.1080/03949371003707836. [DOI] [Google Scholar]
- 91.Gaione-Costa A, Galvão De Pádua D, Delazari ÍM, Santos ARS, Kloss TG. Redescription and oviposition behavior of an orb-weaver spider parasitoid Hymenoepimecis cameroni Townes, 1966 (Hymenoptera: Ichneumonidae) Zootaxa. 2022;5134(3):415–425. doi: 10.11646/zootaxa.5134.3.5. [DOI] [PubMed] [Google Scholar]
- 92.Kloss TG, et al. A new darwin wasp (Hymenoptera: Ichneumonidae) and new records of behavioral manipulation of the host spider Leucauge volupis (Araneae: Tetragnathidae) Neotrop. Entomol. 2022;51:821–829. doi: 10.1007/s13744-022-00991-6. [DOI] [PubMed] [Google Scholar]
- 93.Gillung JP, Borkent ChJ. Death comes on two wings: A review of dipteran natural enemies of arachnids. J. Arachnol. 2017;45(1):1–19. doi: 10.1636/JoA-S-16-085.1. [DOI] [Google Scholar]
- 94.Dedeine F, Ahrens M, Calcaterra L, Shoemaker DD. Social parasitism in fire ants (Solenopsis spp.): A potential mechanism for interspecies transfer of Wolbachia. Mol. Ecol. 2005;14(5):1543–1548. doi: 10.1111/j.1365-294X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- 95.Ahmed MZ, et al. The intracellular bacterium Wolbachia uses parasitoid wasps as phoretic vectors for efficient horizontal transmission. PLoS Pathog. 2015;11(2):e1004672. doi: 10.1371/journal.ppat.1004672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Messas YF, Sobczak JF, Vasconcellos-Neto J. An alternative host of Hymenoepimecis japi (Hymenoptera, Ichneumonidae) on a novel family (Araneae, Araneidae), with notes on behavioral manipulations. J. Hymenopt. Res. 2017;60:111–118. doi: 10.3897/jhr.60.14817. [DOI] [Google Scholar]
- 97.Souza-Santiago BK, Messas YF, de Pádua DG, Santos AJ, Vasconcellos-Neto J. Taking care of the enemy: Egg predation by the Darwin wasp Tromatobia sp. (Ichneumonidae) on the cobweb spider Chrysso compressa (Araneae, Theridiidae) J. Hymenopt. Res. 2023;95:103–112. doi: 10.3897/jhr.95.97029. [DOI] [Google Scholar]
- 98.Guégan M, et al. Who is eating fructose within the Aedes albopictus gut microbiota? Environ. Microbiol. 2020;22:1193–1206. doi: 10.1111/1462-2920.14915. [DOI] [PubMed] [Google Scholar]
- 99.Nawab M, Lashari MH. Life cycle, survivorship and life expectancy of foliage spider (Marpissa bengalensis) in citrus orchard. Ciênc. Rural. 2018;48(03):e20170531. doi: 10.1590/0103-8478cr20170531. [DOI] [Google Scholar]
- 100.Benhadi-Marín J, Pereira JA, Sousa JP, Santos SAP. Spiders actively choose and feed on nutritious non-prey food resources. Biol. Control. 2019;129:187–194. doi: 10.1016/j.biocontrol.2018.10.017. [DOI] [Google Scholar]
- 101.Saini S, Raina M. Spider nectivory from extranuptial nectaries of Urena labata L.: A case from Indian subcontinent. Nat. Acad. Sci. Lett. 2022;45:531–535. doi: 10.1007/s40009-022-01176-w. [DOI] [Google Scholar]
- 102.Wiggins WD, Wilder SM. Carbohydrates complement high-protein diets to maximize the growth of an actively hunting predator. Ecol. Evol. 2022;12:e9150. doi: 10.1002/ece3.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hesselberg T, Boyd KM, Styrsky JD, Gálvez D. Host plant specificity in web-building spiders. Insects. 2023;14:229. doi: 10.3390/insects14030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chrostek E, Pelz-Stelinski K, Hurst GDD, Hughes GL. Horizontal transmission of intracellular insect symbionts via plants. Front. Microbiol. 2017;8:2237. doi: 10.3389/fmicb.2017.02237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li SJ, et al. Plant-mediated horizontal transmission of Wolbachia between whiteflies. ISME J. 2017;11(4):1019–1028. doi: 10.1038/ismej.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cardoso A, Gómez-Zurita J. Food resource sharing of alder leaf beetle specialists (Coleoptera: Chrysomelidae) as potential insect–plant interface for horizontal transmission of endosymbionts. Environ. Entomol. 2020;49(6):1402–1414. doi: 10.1093/ee/nvaa111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data generated and analyzed in this study are deposited to NCBI Nucleotide Database (accession nos. OR220066 and OR227583–OR227590).