Abstract
Chromosomal instability (CIN) is a hallmark of cancer and is associated with tumor cell malignancy. CIN triggers a chain reaction in cells leading to chromosomal abnormalities, including deviations from the normal chromosome number or structural changes in chromosomes. CIN arises from errors in DNA replication and chromosome segregation during cell division, leading to the formation of cells with abnormal number and/or structure of chromosomes. Errors in DNA replication result from abnormal replication licensing as well as replication stress, such as double-strand breaks and stalled replication forks; meanwhile, errors in chromosome segregation stem from defects in chromosome segregation machinery, including centrosome amplification, erroneous microtubule–kinetochore attachments, spindle assembly checkpoint, or defective sister chromatids cohesion. In normal cells, CIN is deleterious and is associated with DNA damage, proteotoxic stress, metabolic alteration, cell cycle arrest, and senescence. Paradoxically, despite these negative consequences, CIN is one of the hallmarks of cancer found in over 90% of solid tumors and in blood cancers. Furthermore, CIN could endow tumors with enhanced adaptation capabilities due to increased intratumor heterogeneity, thereby facilitating adaptive resistance to therapies; however, excessive CIN could induce tumor cells death, leading to the “just-right” model for CIN in tumors. Elucidating the complex nature of CIN is crucial for understanding the dynamics of tumorigenesis and for developing effective anti-tumor treatments. This review provides an overview of causes and consequences of CIN, as well as the paradox of CIN, a phenomenon that continues to perplex researchers. Finally, this review explores the potential of CIN-based anti-tumor therapy.
Subject terms: Cell biology, Cancer, Oncology
Introduction
Cancer is a widespread and devastating disease, which according to the World Health Organization claimed 10 million lives in 2020.1 Cancer is closely associated with mutations and aberrant expressions of a series of oncogenes and tumor suppressor genes. For many years, studies have focused on identifying genes that influence tumorigenesis, such as oncogenes and tumor suppressor genes.2 However, it has become increasingly clear that the development and progression of tumors do not rely exclusively on the alteration of a single gene.3 Chromosomal instability (CIN), a phenomenon characterized by chromosomal alterations, is observed in over 90% of solid tumors and many blood cancers.4–6 These alterations can result in large-scale changes, rearrangements, or disruptions to cellular genetic information, affecting the expression of numerous genes.2,3,6,7
The maintenance of genomic stability is a fundamental requirement for the normal functioning of cells.8–11 Under normal conditions, cells have developed a series of checkpoints and mechanisms to stringently control the passage of intact and correct genetic information, serving as safeguards that help cells maintain genomic stability and prevent harmful alterations.8–11 Thus, CIN, characterized by chromosomal abnormalities, presents a significant challenge to normal cells, often leading to decreased fitness and cell death.3,12 Interestingly, in simpler organisms such as bacteria and viruses, while excessive genomic instability is also harmful, a certain increase in genomic instability can be beneficial, as it could increase the heterogeneity of the population, thereby promoting the survival and proliferation of cells with specific genetic aberrations that provide a growth advantage in a stressful environment.13 This complex situation reveals that while a moderate level of CIN can be beneficial, extremely high levels result in genetic catastrophe and cell death, highlighting the importance of maintaining a balance.13 This delicate balance is not exclusive to simpler organisms but extends to more complex systems, including mammalian cells.14–20 Interestingly, a similar paradoxical observation emerges when studying tumor cells, where the role of CIN in tumorigenesis exhibits complexity.21,22 Analogous to a double-edged sword, CIN in tumor cells exhibits both tumorigenic and tumor-suppressing effects.21–26 On one hand, CIN can promote tumor progression by increasing heterogeneity, thus playing significant roles in tumor development and influencing treatment outcomes,3,27,28 while on the other hand, excessive CIN can lead to growth arrest and even cell death.29 The precise roles of CIN in tumors remain active areas of research. Elucidating the complex interplay between CIN and tumor progression, as well as treatment response, will not only provide a more comprehensive understanding of a major aspect of tumor development and progression, but also important new perspectives for the development of more effective anti-tumor therapies.
CIN manifests in two distinct forms: numerical CIN and structural CIN (Fig. 1).30 Numerical CIN arises from errors in chromosome segregation due to the defects in the mechanisms that guarantee proper sister chromatid segregation, including mitotic checkpoint, centrosome amplification, and abnormalities in microtubules during cell division, and is characterized by the gain or loss of the entire chromosomes.31 While numerical CIN does not change the nucleotide sequences within the chromosomes, it alters the copy number of chromosomes, thereby changing the genetic landscape.32 In contrast, structural CIN can lead to the gain or loss of chromosomal fragments, which are pieces of chromosomes that have broken off, leading to the alteration in nucleotide sequences of large segments of chromosomes.32 This type of CIN is driven by the amplification or deletion of chromosome segments, the formation of extrachromosomal structures, and complex rearrangements of large nucleotide sequences.31 Its origins are linked to the mechanisms involved in repairing double-strand breaks (DSBs), managing replication stress, and regulating non-allelic homologous recombination.31 Interestingly, structural and numerical CIN often coexist in the majority of tumor cells, thereby creating a complex interplay.33–35
Fig. 1.
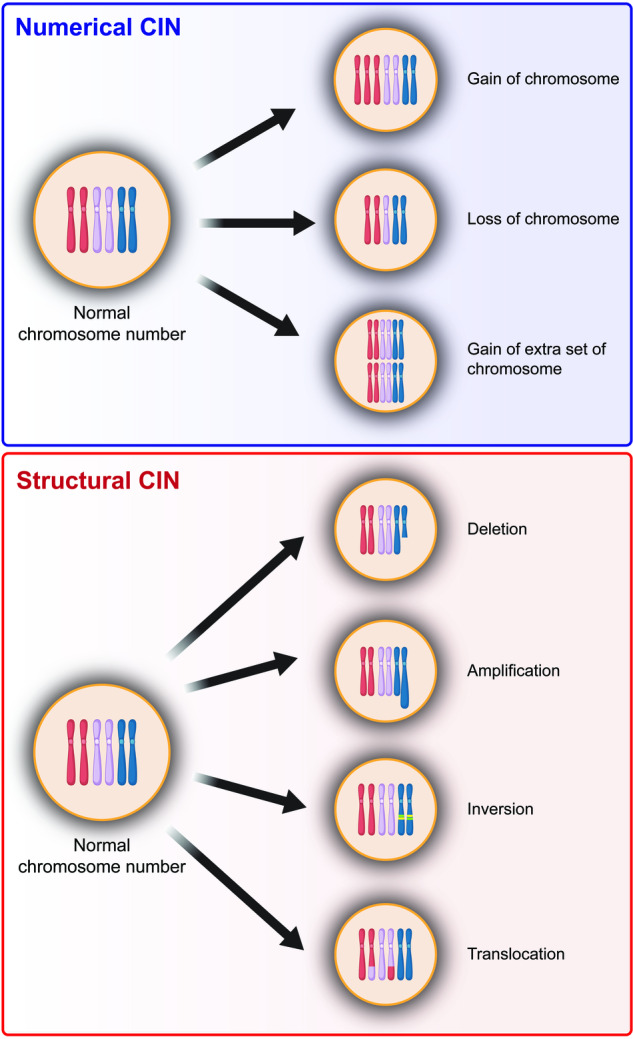
Types of chromosomal instability. CIN is classified into numerical CIN and structural CIN. Numerical CIN corresponds to the gain or loss of whole chromosomes (aneuploidy) or gain of extra set of chromosomes (polyploidy), while structural CIN refers to the gain or loss of chromosome segments due to deletion, amplification, inversion, and translocation
Although the study of CIN has a long history in tumor research, recent advancements in next-generation sequencing technology and a deeper understanding of tumor biology have brought CIN back into the spotlight. From a broader biological perspective, genome diversity is a fundamental aspect of evolution and speciation.36 It provides the raw material upon which natural selection acts, driving the evolution of new species. In the context of tumors, CIN-induced tumor evolution is crucial for creating this genome diversity. The constant reshuffling of the genome creates a vast pool of genetic variants within the tumor population, known as heterogeneity.25 This CIN-induced increased heterogeneity is believed to endow tumors with enhanced evolutionary capabilities due to increased intratumor heterogeneity, facilitating acquisition of malignant phenotypes and adaptive resistance to therapies.23–26,37–39 Moreover, accumulating research has revealed other consequences and associations with CIN, such as its links to metastasis and tumor immune regulation.30 However, despite these advancements, our understanding of CIN remains incomplete. The complex nature of CIN, its causes and consequences, as well as the paradox of CIN, necessitate a systematic review. This is particularly important, given the potential of CIN as a therapeutic target. Revealing the complex nature of CIN is crucial for understanding one of the major causes of tumor progression, as well as for developing more effective anti-tumor treatments. Therefore, in this review, we aim to provide a comprehensive overview of CIN, exploring its research history, causes, paradoxical nature, and multifaceted influence on tumor biology, as well as discussing the potential and progress of CIN-based anti-tumor therapy.
Milestones in CIN research
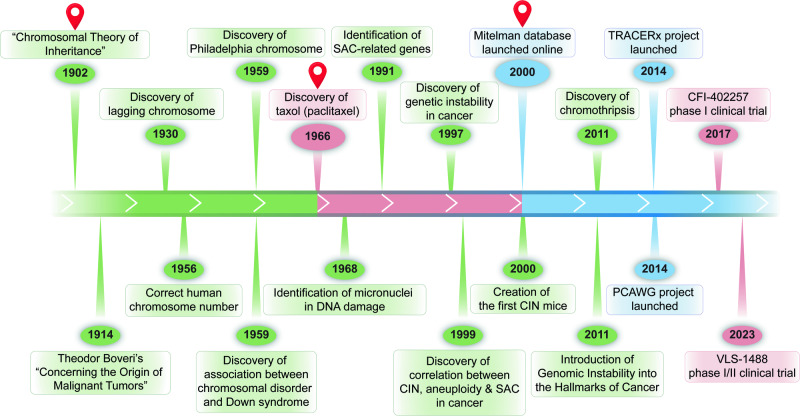
The study of CIN has evolved over a century, marked by pivotal milestones (Fig. 2). The journey began with Theodor Boveri who, in 1902, performed the first systematic analysis of the effects of aneuploidy on cell and organismal physiology in sea urchins. Boveri observed that embryos resulting from eggs fertilized by two sperms exhibited developmental defects and died, concluding that chromosome abnormality leads to defect in development and lethality, marking the first hypothesis that connects between chromosomal abnormality and disease. His subsequent work in 1914, “Concerning the Origin of Malignant Tumors,” linked chromosomal abnormality to cancer, marking the first hypothesis that connected chromosomal abnormality to cancer.40,41 Sixteen years later, Barbara McClintock introduced the terms ‘laggards’ or ‘lagging chromosomes’ to signify chromatin lagging between daughter nuclear masses during anaphase, providing a deeper understanding of chromosomal behavior during cell division.42 The field of clinical cytogenetics was initiated in 1956 when Tjio and Levan discovered that humans have 46 chromosomes. This discovery not only corrected the previously held belief of 48 chromosomes, but also paved the way for the study of chromosomal abnormalities in humans.43 In 1994, Rieder and colleagues performed their classic experiment using laser ablation, which revealed the role of unattached kinetochores in extending the duration of mitosis, providing crucial insights into the mechanisms of mitotic checkpoint control.44
Fig. 2.
Timeline of key milestones in the CIN research. Green represents milestones in basic CIN research, pink represents milestones in CIN clinical translation, and blue represents milestones in CIN meta-analysis and databases
The relation between CIN and diseases was first revealed in 1959, when two significant discoveries were made. Peter Nowell identified the Philadelphia chromosome, a consequence of an abnormal rearrangement between human chromosome 9 and chromosome 22 that could be found in approximately 90% of chronic myeloid leukemia patients, providing the initial evidence of structural chromosome aberration as a malignant factor.45,46 In the same year, Lejeune et al. discovered that an additional copy of chromosome 21, a condition now known as trisomy 21, caused Down syndrome.47 Lejeune et al. continued his research and identified another chromosomal disorder known as cri du chat (cry of the cat) syndrome, a condition that arises when a segment of chromosome 5 is missing.48 These discoveries underscored the detrimental effects associated with chromosome aberrations and emphasized the importance of chromosomal stability.45,47 In 1997, Lengauer et al. quantified CIN in human cancer cell lines, proposing its universality across cancers;49,50 while Angelika Amon’s works from 1999 until 2010 elucidated the molecular aspects of checkpoint crucial in CIN.51,52 These works provided valuable insights regarding the widespread nature of CIN and its molecular mechanisms in cancer, thereby underscoring the significance of CIN in cancer biology. This eventually leads to the recognition of CIN as one of the hallmarks of cancer in 2011.49 Meanwhile, in 2018, Bakhoum et al. provided compelling evidence that CIN could drive tumor metastasis, significantly advancing our understanding of the role of CIN in tumor progression.53
The beginning of the 21st century marked another crucial milestone in CIN research. In 2000, Max Dobles et al. developed the first CIN mice model, enabling in vivo studies of CIN.54 In the same year, Felix Mitelman launched The Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer, which provides a valuable resource for CIN research, marking the opening of the era of meta-analysis in CIN studies.55 This was continued with the initiation of Pan-Cancer Analysis of Whole Genomes (PCAWG) by the International Cancer Genome Consortium (ICGC) in 2014.31,33,56,57 Concurrently, the TRAcking Cancer Evolution through therapy/Rx (TRACERx) clinical study was launched under the guidance of Charles Swanton in 2014. These initiatives marked a significant shift towards large-scale, collaborative efforts in understanding CIN, and paved the way for the development of therapeutic interventions targeting CIN.6,58–62 The discovery of Taxol in 1966 marked a significant milestone in anti-tumor treatment, setting the stage for a novel anti-tumor therapeutic strategy. It was the first drug to successfully demonstrate the potential of targeting mitosis, laying the groundwork for treatments based on CIN.63 Then, half a century later in 2017, this strategy was further realized with the first clinical trial for drugs targeting CIN, starting with the phase I trial of CFI-402257. Although this drug has not yet received full approval, it was granted Fast Track Designation by the FDA in 2023.64–67 To date, two anti-tumor therapies targeting CIN has been approved, while more than 50 are in clinical trial Phase I/II, with the most recent one initiated in October 2023 for VLS-1488.68 These developments have significantly advanced the translation of CIN research into potential therapeutic interventions. To date, research into CIN continues to progress, with each new discovery providing further insight into this complex field and opening up new avenues for potential cancer treatments.
Causes of CIN
CIN is characterized by changes in chromosome structure and number during cell division.30,32,69 There are several key indicators of CIN, including lagging chromosomes, chromosome bridges, micronuclei, aneuploidy, and polyploidy.70–86 As will be discussed below, aberrant spindle assembly checkpoint (SAC) activity, impaired sister chromatid segregation, aberrant centrosome number, and microtubule-kinetochore attachment error could lead to chromosome missegregation.87–112 This could in turn increase the formation of lagging chromosomes, which are chromosome that moves to the poles of the cell during cell division slower than other chromosomes, and chromosome bridges, which are structures formed when part of sister chromatids intertwines and fails to completely segregate.113,114 Lagging chromosomes and chromosome bridges subsequently could lead to the formation of micronucleus, a small, extra-nuclear body that contains chromosomal fragments or whole chromosome that are not incorporated into the main nucleus.71,115–117 Furthermore, chromosome missegregation, along with replication stress, sister chromatid defect, and abnormal centrosome number, could also lead to numerical CIN, as they could promote the occurrence of aneuploidy, a condition where a cell has an abnormal number of individual chromosomes, as well as polyploidy, a condition in where a cell has multiple sets of chromosomes.34,113,114,118,119 These indicators reflect the level of CIN, and are commonly used to assess and study CIN in a cell population (Fig. 3).
Fig. 3.
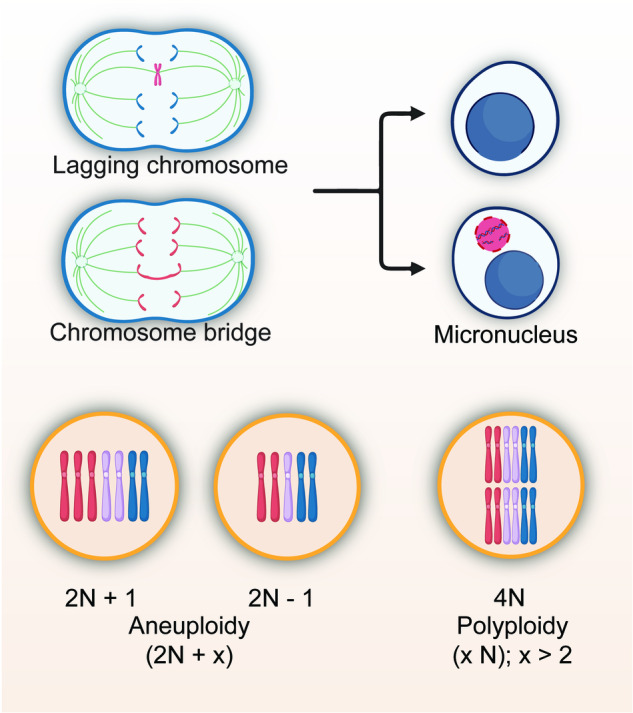
Indicators of CIN. Example of indicators commonly used to assess CIN, including lagging chromosomes, chromosome bridges, micronuclei, aneuploidy, and polyploidy
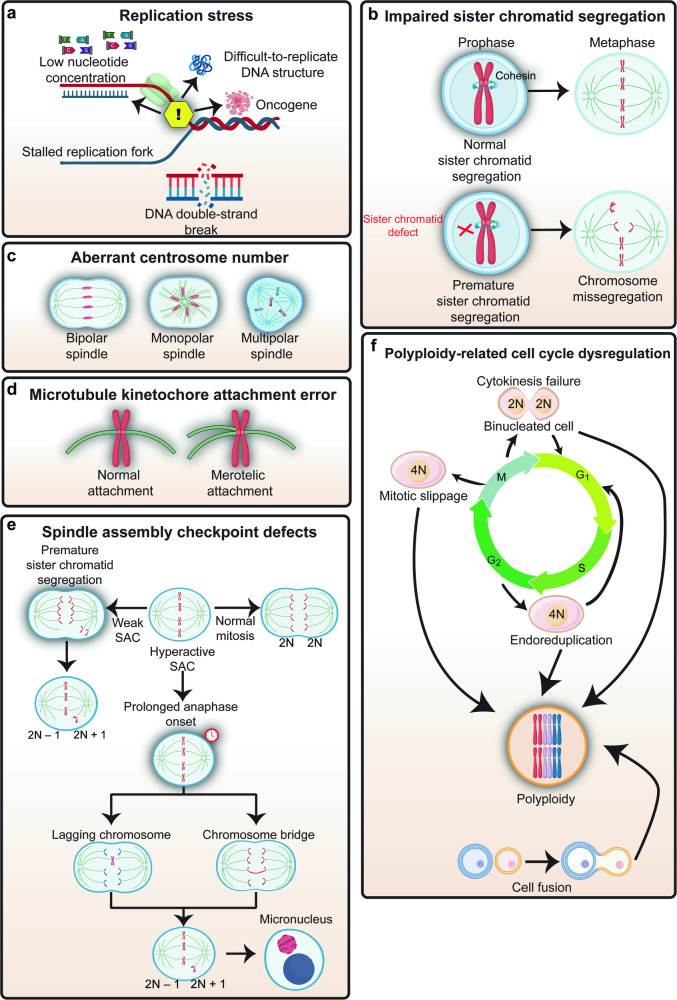
The causes of CIN are multifaceted and can be attributed to a variety of factors (Fig. 4). At its core, CIN is often the result of errors in DNA replication, which can lead to the formation of cells with incomplete or excess genetic material; as well as errors in chromosome segregation during cell division, which can lead to the formation of cells with an abnormal number of chromosomes.120,121 Errors in DNA replication can arise from the abnormal replication licensing as well as replication stress, such as DSBs and stalled replication forks; while errors in chromosome segregation can arise from defects in chromosome segregation machinery, including issues with centrosome amplification, erroneous microtubule-kinetochore attachments, and defects affecting the mitotic checkpoint or impaired sister chromatid segregation. Furthermore, some events induced by CIN could further trigger instability of chromosomes. For instance, chromothripsis is an event of incorporation of chromosome fragments originated from micronuclear chromosome into nuclear chromosome, causing rearrangement of nuclear chromosome.78,115,122–133 Thus, while chromothripsis itself is a consequence of CIN, it could also be a cause of subsequent CIN.
Fig. 4.
Causes of CIN. a Replication stress leads to stalling and collapse of replication forks, which results in DSBs. b Sister chromatid defect allows premature separation of sister chromatids before full alignment, leading to chromosome missegregation. c Aberrant centrosome number such as monopolar spindle and multipolar spindle could lead to aneuploidy. d Microtubule kinetochore attachment error causes failure to form bi-orientation, where each kinetochore is attached to microtubules from only one spindle pole, leading to chromosome missegregation. e Aberrant SAC could lead to aneuploidy, as weakened SAC causes premature chromatid separation, while hyperactivated SAC results in a lagging chromosome. f Extra set of chromosomes as seen in polyploidy could arise from cytokinesis failure, mitotic slippage, or endoreduplication
Replication stress and defective DNA repair
Replication stress is a condition that occurs when the DNA replication machinery is disrupted during the S phase of the cell cycle, leading to a stalled replication fork.134,135 Replication stress can be driven by oncogenes, low nucleotide concentrations, and DNA sequences or structures that are difficult to replicate.136 In response to replication stress, cells activate the DNA damage response (DDR), a cellular response that requires a network of repair proteins. This network includes key proteins such as ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 related (ATR), which are kinases that help the stabilization of the stalled replication fork, preventing it from collapsing.137,138 As has been observed in various precancerous and cancerous lesions, failure in resolving the stalled replication forks can cause DSBs, and subsequently, rearrangements of parts of chromosomes (translocations) or deletions, thus contributing to structural CIN.34,137–144
In addition to the DDR pathway, other DNA repair-related factors, such as Fanconi anemia (FA) proteins are also important for preventing CIN.145–147 The FA pathway is important for repairing interstrand cross-links, which are toxic lesions that prevent DNA strand separation, block replication, and hinder transcription, thereby playing a critical role in responding to replication stress and maintaining chromosome stability.148–150 Defects in the FA pathway, specifically in the Fanconi anemia complementation group D2 (FANCD2), can lead to increased translocations and abnormal chromatin structures, contributing to structural CIN.42,147 Moreover, replication stress can also lead to lagging chromosomes and micronuclei.34,118,151,152 Together, these studies demonstrated that replication stress and defects in DNA repair systems contribute to the generation of structural and numerical CIN.
Impaired sister chromatid segregation
The separation of the chromosomes at anaphase requires the loss of sister cohesion in a timely manner. This is facilitated by the cohesin complex, a multi-protein complex composed of four core subunits: either stromal antigen 1 (STAG1) or stromal antigen 2 (STAG2), structural maintenance of chromosomes 1A (SMC1A), structural maintenance of chromosomes 3 (SMC3), and RAD21 cohesin complex component (RAD21).153–158 During prophase, the bulk of the cohesin complex, which consists of a ring-shaped structure formed by SMC1A, SMC3, RAD21, and STAG1 in the chromosome arm is removed.159,160 This process involves several proteins, including the WAPL cohesin release factor (WAPL), PDS5 cohesin-associated factor (PDS5), and polo-like kinase 1 (PLK1). These proteins assist in opening the cohesin ring, facilitating its removal from the chromosome arms.161 At anaphase, when separase is activated, the cohesin complex consisting of SMC1A, SMC3, RAD21, and STAG2 in the centromere was cleaved at the RAD21 subunit, leading to the opening of the cohesin ring at the centromere and the final separation of sister chromatids.153,159
Genetic alterations of any of the cohesin subunits, including mutations and inactivation, have been associated with CIN in various human tumors, as this dysregulation in turn leads to aberrant chromatid cohesion and allows premature separation of sister chromatids before full alignment, leading to chromosome bridge, lagging chromosome, and micronuclei.153 Defects in cohesin subunits, such as STAG1 or SMC1A, cause premature separation of chromosome arms and increased aneuploidy.87,88 Moreover, WAPL overexpression induces premature separation of chromosome arms, thereby increasing the rate of chromosome bridge and micronuclei;89 while PDS5 defect disrupts the regulation of cohesin ring removal from chromosome arms, leading to increased DSBs through an as-yet-unknown mechanism.90
Meanwhile, mutations in STAG2 as well as RAD21, a core component of cohesin complex at the centromere crucial for holding the centromeres of sister chromatids together from the time of DNA replication in S phase until their segregation in mitosis, also cause premature separation of sister chromatids and subsequently increase aneuploidy.91–93 Interestingly, mutation in STAG2 could also disrupt the interaction between cohesin and the replication machinery, thereby triggering DSBs and subsequently translocation by increasing stalling and collapse of replication forks.94,95 Furthermore, mutation in RAD21, a core component of the cohesin complex that plays a crucial role in holding the centromeres of sister chromatids together from the time of DNA replication in S phase to their segregation in mitosis, induces premature separation of sister chromatids, and increased aneuploid.92 Together, these studies demonstrated that sister chromatid defect contributes to the generation of structural and numerical CIN.
Aberrant centrosome number
CIN can also arise from aberrant centrosome amplification and separation, both of which are critical processes in cell division. Failure in centrosome amplification, for example due to the defects in specific motor proteins such as kinesin family member 2A (KIF2A), kinesin-like protein at 10A (KLP10A), and kinesin-like protein at 67A (KLP67A), or defects in centrosome proteins such as γ-tubulin, gamma complex component 2 (GCP2), and gamma complex component 3 (GCP3), can cause formation of monopolar spindle due to poor centriole separation.162 Monopolar spindle in turn leads to improper sister chromatid separation, as there is only one pole for them to move towards.96 Eventually, all chromosomes end up in a single daughter cell when the cell completely divides, resulting in the formation of polyploid cell.163
Improper timing of centrosome separation prior to cell division, both delayed and accelerated centrosome separation, would also lead to the formation of monopolar spindle.164,165 Loss of ubiquitin-specific peptidase 44 (USP44), a deubiquitinase that localizes at the centrosome, results in incomplete centrosome separation as well as increased monopolar spindle, lagging chromosome, and chromosome bridge. Moreover, USP44 knockout mice are prone to increase in numerical CIN as observed by elevated levels of aneuploidy.166 Defective centrosome separation can also occur in cells with overexpressed kinesin family member 11 (KIF11), a motor protein that drives centrosome separation. KIF11 overexpression can disrupt the normal timing and coordination of centrosome separation, by causing centrosomes to separate too quickly before the completion of centrosome duplication. In such cases, the spindle poles would be formed by imperfectly duplicated centrosomes, leading to the formation of monopolar spindle, and eventually, polyploidy.97
Meanwhile, the mitogen-activated protein kinase (MAPK) pathway, known for its role in cell proliferation, differentiation, and survival, has been implicated in the induction of CIN through centrosome amplification, as constitutive activation of MAPK through rat sarcoma virus (ras)-overexpression resulted in the increase of centrosome amplification, leading to multipolar spindle.167 Furthermore, overexpression of polo-like kinase 4 (PLK4), a master regulator of centrosome amplification, can result in overduplication of centrosome.98 This consequently lead to the formation of cell with multipolar spindles. When anaphase occurs in these cells, the chromosomes are separated abnormally, resulting in aneuploid daughter cells.98 Together, defects in centrosome duplication and separation contribute to the formation of monopolar and multipolar spindles, which are frequently observed in human tumors and are associated with CIN.168–173
Microtubule kinetochore attachment error
The process of spindle microtubules binding to kinetochores is asynchronous and stochastic, occurring at different times and in a random manner. This randomness and lack of synchronization can sometimes lead to erroneous attachments, such as merotelic attachments where a single kinetochore binds to microtubules anchored at both spindle poles, and could lead to chromosome missegregation.113,174–177 Despite the erroneous nature of merotelic attachments, cells often manage to segregate these chromosomes correctly during anaphase. This is due to the cell mechanism that corrects these erroneous kinetochore-microtubule (K-MT) attachments by converting them into bi-oriented attachments, where each kinetochore is attached to microtubules from only one spindle pole.178–180
The efficient correction of merotelic attachments requires the dynamic turnover of K-MT interactions. The optimal stability of K-MT attachments, which is neither too loose nor too hyperstable, is crucial for this process. A decrease in this turnover rate could result in persistent merotelic attachments and increased chromosome segregation errors, such as lagging chromosomes.99,100 For instance, some tumors with CIN are characterized by hyperstable K-MT interactions, a state that is more stable compared to chromosomally euploid cells, leading to failure in correcting merotelic attachments and increased CIN; while reducing the K-MT attachment stability from this hyperstable state can restore normal chromosome segregation in cells with CIN.99,100,181 However, while previous studies have shown that loss of STAG2, a cohesin subunit that has been reported to have roles beyond sister chromatids cohesion, results in hyperstabilized K-MT attachments, the exact molecular mechanisms are not fully understood.182,183 To fully characterize the contribution of microtubule-kinetochore attachment errors in CIN, a more detailed study of the complex and dynamic process of spindle microtubules binding to kinetochores is required.
SAC defects
The primary goal of a cell undergoing mitosis is to segregate the replicated chromosomes into two new daughter cells. This is achieved through the attachment of chromosomes to microtubules of the mitotic spindle apparatus.184 Chromosomes attach to the ends of microtubules at kinetochores, which are specialized protein structures that bind to the chromatin centromere.184 Normally, each chromosome has two kinetochores, and it is essential for mitotic cells to form bi-orientation.184 This state is achieved when each sister kinetochore binds microtubules oriented toward opposite spindle poles.
A checkpoint mechanism known as the SAC delays the separation of the sister chromatids at anaphase until every kinetochore has correctly attached to spindle microtubules and all sister chromatids have aligned at the metaphase equatorial plate.185–188 Thus, the SAC is a safeguard for guaranteeing chromosome bi-orientation on the mitotic spindle by monitoring the proper kinetochore attachment as well as chromosome alignment. As long as improperly attached or unaligned chromosomes remain, SAC halts cells in mitosis and prevents their progress into the final phases of cell division.185–188 Components of the SAC, including mitotic arrest deficient 2 (MAD2),107,189–192 budding uninhibited by benzimidazoles 1 (BUB1),193–196 budding uninhibited by benzimidazoles 1 beta (BUBR1),110,195,197 and budding uninhibited by benzimidazoles 3 (BUB3),193,194,198–202 migrate to unattached kinetochores and form mitotic checkpoint complex (MCC) along with cell division cycle 20 (CDC20). MCC is a key effector of SAC that inhibits the activation of the CDC20-bound anaphase-promoting complex/cyclosome (APC/CCdc20), an E3 ubiquitin ligase that targets cyclin B and securin for degradation by the proteasome.185–187,203–211 Once sister chromatids have properly attached and aligned, the SAC is inactivated, allowing the MCC to dissociate, freeing CDC20 to activate the APC/C.212 The activation of the APC/CCDC20 triggers securin and cyclin B degradation.203,207,209,213 Securin destruction frees separase, an enzyme that cleaves and inactivates the cohesin complex, allowing sister chromatid separation and the onset of anaphase.203 Meanwhile, cyclin B degradation inactivates cyclin-dependent kinase 1 (Cdk1), allowing the cells to proceed to mitotic exit and complete cell division.185,186,214,215
In eukaryotic cells, SAC plays a crucial role in genomic integrity and its abnormality leads to chromosome segregation errors.10,101 Defects in SAC result in the failure of proper monitoring and controlling the timing of sister chromatid segregation.10,101,102 This in turn leads to increased chromosomal abnormalities, such as chromosome bridge and lagging chromosome, and eventually, errors in equal distribution of genetic material to daughter cells.103–105 Moreover, as described above, lagging chromosome, as well as chromosome bridge, could reassemble and form micronucleus, a nucleus-like structure consisting of a bilayer membrane covering a piece of extrachromosomal DNA.175
Cells lacking MAD2, an SAC component, can proliferate in vitro and in vivo but with increased levels of CIN.103,104 Moreover, weakening the checkpoint in mice by partially reducing the expression of various SAC genes including MAD1, MAD2, BUB1, BUBR1, and BUB3, results in premature separation of sister chromatids, chromosome missegregation, and subsequently, CIN.106–112 In addition to the SAC components, the CDK pathway also plays a significant role in CIN. Gao et al. reported that CUE domain containing 2 (CUEDC2) is phosphorylated by CDK1 during mitosis. This phosphorylated CUEDC2 promotes spindle checkpoint inactivation by promoting MCC dissociation from the APC/C, leading to premature inactivation of SAC and increased CIN.216 Furthermore, chromosome missegregation can be caused by mutations that weaken the SAC, which subsequently results in premature anaphase onset.105 However, mutations in SAC genes are rarely found in human tumors, suggesting that while SAC mutation is one of contributors to CIN in tumor cells, aberrant transcriptional, post-translational modification, and epigenetic regulations might also contribute to SAC defects.121,217–219
Interestingly, while weakened SAC can cause CIN, overactivity of the checkpoint induced by, for example, overexpression of SAC gene such as MAD2, or knockdown of genes involved in SAC silencing pathways such as p31/comet or TRIP3, can also induce CIN.220–224 Similar to SAC defect, SAC hyperactivation could lead to the increase of chromosome bridge, lagging chromosome, and micronucleus.220–225 However, in contrast to weakened SAC which accelerates mitotic progression and tumor cells proliferation, SAC hyperactivation delays the onset of anaphase and prolongs mitotic arrest.220–225 Furthermore, unlike chromosome missegregation induced by SAC defect, which stems from the premature anaphase progression before the erroneous chromosomes-microtubules attachments are corrected, the mechanism of SAC hyperactivation-induced chromosome missegregation is not entirely understood. One possible explanation is that persistent SAC signaling could lead to cohesion fatigue, where the cohesin complexes that hold sister chromatids together become exhausted over time, resulting in aberrant sister chromatid segregation.226,227 Together, while the SAC plays a vital role in ensuring accurate chromosome segregation during mitosis, both its defect and hyperactivation can paradoxically lead to CIN.
Polyploidy-related cell cycle dysregulation
Polyploidy is a condition where a cell has multiple sets of chromosomes and could be both a consequence as well as a cause of CIN.228–232 Polyploidy can occur due to various reasons, including cytokinesis failure, mitotic slippage, endoreduplication, or cell fusion.233 Cytokinesis failure occurs when daughter cells fail to separate after accomplishing telophase.74,234–236 This can happen due to various reasons, such as problems with the contractile ring that separates the two daughter cells or the presence of chromosome bridges that physically prevent the cells from separating.234 When cytokinesis failure occurs, the two daughter cells remain connected and form a binucleated cell with twice of the normal number of chromosomes.234 Furthermore, polyploidy can be caused by mitotic slippage, which is a process of premature mitotic exit. This can occur when the SAC activity is weakened, leading to the misinterpretation that all chromosomes are correctly attached to the spindle and the failure to inhibit the activation of the APC/CCDC20.108,237,238 This failure then promotes the premature degradation of cyclin B1, which in turn leads to a decrease in Cdk1 activity, and, as a consequence, promotes the onset of anaphase and premature exit from mitosis without proper chromosome segregation.108,237,238
Endoreduplication is a process in which cells undergo multiple rounds of DNA replication without mitosis, resulting in cells with multiple copies of their genome.239 Endoreduplication can occur due to various reasons, such as problems with the cell cycle machinery.239 One key cell cycle machinery associated with this endoreduplication is a defect in the pre-replication complex (pre-RC).240 The pre-RC, which includes the origin recognition complex, cell division cycle 6, chromatin licensing and DNA replication factor 1 (CDT1), and minichromosome maintenance complex 2-7, assembles at replication origins during G1 phase to license DNA replication at S phase.240 This complex then dissociates from the replication origins after DNA replication is started to prevent another round of DNA replication before the cell completes cytokinesis, thereby guaranteeing the proper number of chromosomes being passed to daughter cells.240 Hence, dysregulation in pre-RC could lead to whole-genome doubling (WGD), a form of polyploidy.228,241–246
Cell fusion is a process in which two or more cells fuse together to form a single cell with multiple nuclei, also known as a synkaryote.239,247 This can occur due to various reasons, such as exposure to certain viruses or chemicals.239,247 Following fusion, the parental chromosomes mix and redistribute to the fused cells, thereby producing polyploid fused cells.239,247 Therefore, cell fusion can also result in polyploid cells and contribute to CIN.
The increase in polyploidy, therefore, signifies an increase in CIN, underlining the critical role these cellular processes play in causing CIN. However, as mentioned above, besides as a consequence of CIN, polyploidy is also an important cause of CIN. The extra set of chromosomes in a cell can lead to errors in chromosome segregation during subsequent cell division, forming aneuploid cells with abnormal chromosome numbers.74,248–251 For instance, a study using tetraploid cells demonstrated that these cells, which contain twice the number of chromosome sets along with two extra centrosomes, can lead to the formation of multipolar spindles. This, in turn, results in the formation of aneuploid cells.252 On the other hand, cells with CIN can also become polyploid due to errors during cell cycle dysregulation as mentioned above.253 Together, this highlights the complex interplay between polyploidy and CIN.
CIN paradox
Maintaining genomic stability is essential for the normal functioning of the cells, and for ensuring the accurate transmission of genetic information to progeny, thereby preserving the continuity of the species.8–11 Normal cells have intrinsic potentials for maintaining their genome integrity through various mechanisms, such as the ability to repair their damaged DNA, as well as for preventing the passage of damaged, unrepairable DNA to their progenies by triggering apoptosis.254 Given the critical role of genomic stability, any deviation from this state can have deleterious consequences. One such challenge is CIN, which can be detrimental when present in normal cells, as it could decrease cellular fitness.3,12 As observed by Theodor Boveri over a century ago, chromosomal abnormalities are typically intolerable in normal cells, often culminating in cell death.41 However, it is essential to note that CIN is not universally detrimental. There are examples, notably in simpler organisms like bacteria, viruses, and fungi where elevated genomic instability can confer advantages in stressful environments.13
For instance, clinical isolates of the yeast Candida albicans that are resistant to the anti-fungal drug fluconazole carry extra copies of chromosome 5, where genes encoding the drug target, lanosterol 14-α-demethylase (ERG11), and a main regulator of drug efflux pumps, transcription activator of CDR genes 1 (TAC1), are located.255 This indicates that CIN could provide drug resistance through the increased expression of these genes.256 Furthermore, in other yeasts, Saccharomyces cerevisiae and Candida glabrata, increased CIN-induced aneuploidy results in phenotypic advantages by promoting their resistance to fluconazole.257–259 Similarly, in bacteria and viruses, elevated genomic instability benefits the population in stressful environments by promoting the survival and proliferation of cells with specific genetic aberrations that confer a growth advantage.13 This suggests a nuanced perspective on CIN, highlighting its potential benefits under specific circumstances. However, it is important to emphasize that the intensity of CIN cannot exceed certain thresholds. Indeed, in bacteria and viruses, cells with drastic instability never become dominant in a population, as their excessive instability levels exceed the cellular threshold, leading to genetic catastrophe and cell death.13 This observation suggests that while moderate CIN can be beneficial, excessive CIN may lead to genetic catastrophe and is lethal. Therefore, understanding the balance between beneficial and detrimental effects of CIN is crucial to comprehend its role, not only in simpler organisms but also in more complex systems like mammalian cells.14–20
With the advancement of our knowledge regarding tumor biology, a similar observation emerges, where CIN resembles a double-edged sword (Table 1). On one hand, CIN can promote tumorigenesis. As exemplified in yeast and mammalian cells above, CIN could contribute to clonal evolution, providing selective advantages under stressful conditions encountered by tumor cells.257,260,261 This clonal evolution, driven by CIN, can be a key factor in promoting tumorigenesis, as it not only helps tumor cells to survive in harsh environments but can also foster the evolution of the clones with the most tumorigenic phenotypes, that is, clones that have new karyotype that brings them growth advantage and the ability to outcompete others.260 This is also supported by studies using animal models. For example, mice carrying heterozygous deletions of SAC genes, such as MAD1, MAD2, and BUB1B, exhibit increased CIN and develop spontaneous tumors.106–108 Similar evidence comes from human patients with mosaic variegated aneuploidy syndrome (MVA), which is characterized by increased CIN and a predisposition to childhood cancer.109,110,262,263 These results suggest that tumor cells may exploit CIN to harness the potential of clonal evolution for optimal adaptation. However, on the other hand, CIN has also been reported to have anti-proliferative effects,264–266 and can induce cell death,267,268 senescence,269–271 as well as anti-tumor immune response.270,272,273 Clinical observations further complicate the picture, with high CIN signatures in various tumors associated with improved prognosis.274–279 These observations have led to the establishment of the “just-right” hypothesis, proposing a moderate level of CIN that benefits tumorigenesis and tumor progression. However, while the concept of how populations with genetic instability evolve over time has been observed in a study based on mathematical modeling,280,281 experimental evidence to support the “just-right” hypothesis for the relation between CIN level and tumor cell fate determination, as well as the molecular mechanisms underlying it are still lacking. One possible explanation for this, at least in part, is the use of different models to elucidate the role of CIN in cancers, emphasizing the need for further research. In addition, it is noteworthy that the specific context of aneuploidy induced by CIN can also influence tumor cell fate. While tumor cells may be able to tolerate the effects of additional chromosomes, excessive loss of chromosomes that contain essential genes for cell survival is detrimental. Therefore, the balance between gain and loss of chromosomes is crucial for tumor cell viability.282,283
Table 1.
Roles of CIN in cancer
| Phenotype | Model | CIN indicators | Mechanism | Ref |
|---|---|---|---|---|
| Tumor-promoting | AAA-Cdc20 heterozygous MEFs | Increased aneuploidy | Weakened SAC function | 456 |
| Tumor-promoting | ApcMin/+BubR1+/–mice | Increased aneuploidy, polyploidy, sister chromatid premature separation | Weakened SAC function | 457,458 |
| Tumor-promoting | AURKA-overexpressed mice | Increased aneuploidy, polyploidy, chromosome missegregation, sister chromatid premature separation | Centrosome amplification | 459–462 |
| Tumor-promoting | BUB1 haploinsufficient mice | Increased aneuploidy and chromosome missegregation | Weakened SAC function | 463 |
| Tumor-promoting | BUB3 haploinsufficient MEFs | Increased aneuploidy, sister chromatid premature separation | Weakened SAC function | 199 |
| Tumor-promoting | BUBR1 haploinsufficient mice | Increased polyploidy | Weakened SAC function | 464,465 |
| Tumor-promoting | Cyclin B-overexpressed MEFs | Increased aneuploidy, chromosome missegregation | Prolonged mitotic exit | 96 |
| Tumor-promoting | CENP-E heterozygous MEFs | Increased aneuploidy, polyploidy, chromosome missegregation | Microtubule-kinetochore attachment error | 466 |
| Tumor-promoting | HEC1-overexpressed mice | Increased aneuploidy, polyploidy, chromosome breaks | SAC hyperactivation | 467 |
| Tumor-promoting | MAD1 heterozygous deletion mice | Increased aneuploidy | Weakened SAC function | 106 |
| Tumor-promoting | MAD2 haploinsufficient cells | Increased aneuploidy, sister chromatid premature separation, chromosome missegregation | Weakened SAC function | 107 |
| Tumor-promoting | Securin homozygous deletion mice | Increased aneuploidy, polyploidy, sister chromatid premature separation | Premature sister chromatid separation | 468,469 |
| Tumor-promoting | TPX2 heterozygous mice | Increased aneuploidy, chromosome missegregation, | Disrupted normal microtubule polymerization | 470 |
| Tumor-promoting | UbcH10-overexpressed mice | Increased aneuploidy, chromosome missegregation, centrosome amplification | Premature sister chromatid separation | 471,472 |
| Tumor-suppressing | CDH1 heterozygous mice | Increased replication stress, aneuploidy, polyploidy, | Prolonged mitosis | 473 |
| Tumor-suppressing | STAG1 heterozygous deletion mice | Increased aneuploidy, polyploidy, chromosome missegregation | Impaired sister chromatid segregation | 474 |
| Tumor-suppressing | MAD2-overexpressed MEFs | Increased aneuploidy, polyploidy, chromosomal breaks | SAC hyperactivation | 220 |
| Tumor-suppressing | α-GSU PTTG-overexpressed mice | Increased aneuploidy, polyploidy, sister chromatid premature separation | Prolonged mitosis | 475,476 |
| Tumor-suppressing | PLK1 overexpressed mice | Increased polyploidy, lagging chromosome, sister chromatid premature separation, micronucleus, cytokinesis failure | SAC hyperactivation | 477 |
| Tumor-suppressing | MAD2 overexpression | Increased lagging chromosome, micronucleus, polyploidy | SAC hyperactivation | 220 |
| Tumor-suppressing | Ionizing radiation-exposed cells | Increased lagging chromosome, micronucleus | DNA DSBs | 277 |
| Tumor-suppressing | Paclitaxel-treated cells | Increased multipolar spindles, lagging chromosome | Prolonged mitosis | 409 |
Together, the role of CIN in tumorigenesis is complex and paradoxical (Fig. 5), influenced by factors such as the degree of CIN and the specific conditions faced by tumor cells. The intricate balance between the harmful and advantageous effects of CIN underscores the complexity of CIN paradox, emphasizes the need for further research. In the following section, we will discuss further the consequences of CIN and how its seemingly negative effects can, under certain circumstances, confer advantages to tumor cells.
Fig. 5.
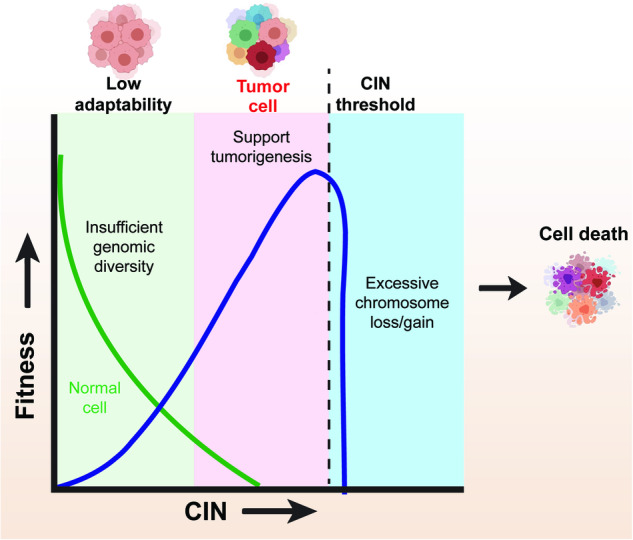
CIN paradox according to the “just-right” model. A moderate, “just-right” level” for CIN could induce a tumor-promoting phenotype by increasing tumor cells adaptability, while excessive CIN is deleterious to tumor cells due to excessive chromosome gain or loss leading to cell death
The multifaceted impacts of CIN on tumor biology
CIN exerts a multifaceted influence on tumor biology, playing significant roles in both tumorigenesis and tumor progression. However, the relationship between CIN and tumorigenesis is complex and resulting in CIN paradox, as mentioned in the previous section. In this section, we will discuss about the diverse consequences of CIN, starting with those closely related to tumorigenesis. These include DNA damage, proteotoxic stress, and metabolic alteration, which potentially have both beneficial and deleterious effects. We will then explore the generally deleterious effects of CIN on cell cycle arrest and senescence. Lastly, we will discuss the beneficial effects of CIN on metastasis, tumor immune regulation, and drug resistance (Fig. 6, Fig. 7, and Table 2).
Fig. 6.
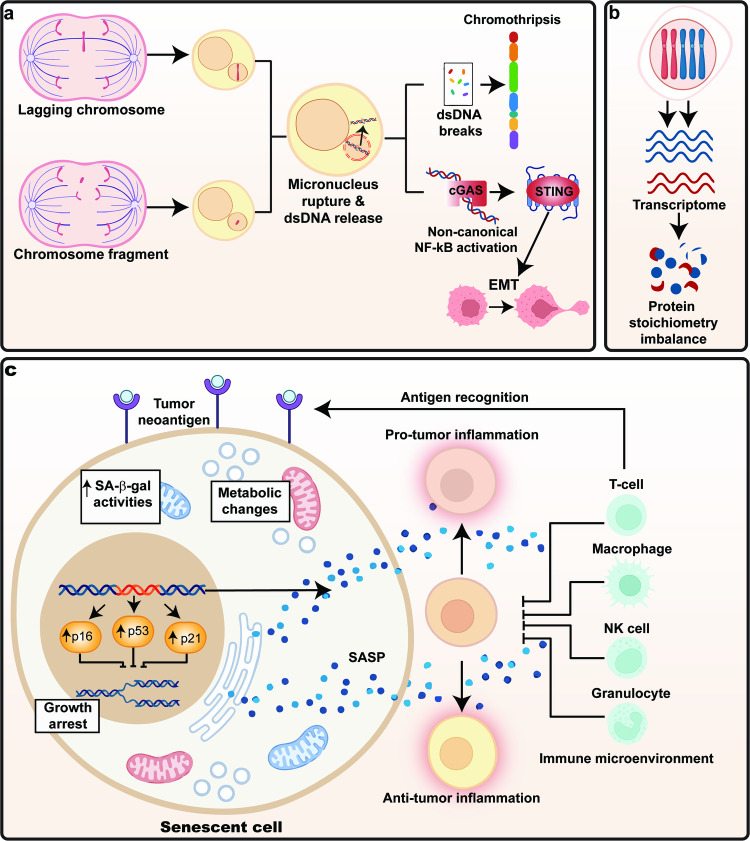
Consequences of CIN. a Chromothripsis, also known as “chromosome shattering” is induced from rupture of micronuclei, followed by fragmentation of micronuclear DNA and its massive rearrangements. b Protein stoichiometry imbalance caused by changes in the copy number of chromosomes, leading to proteotoxic stress. c Other cellular functions altered by CIN, including metabolic alteration, cell cycle arrest, and senescence
Fig. 7.
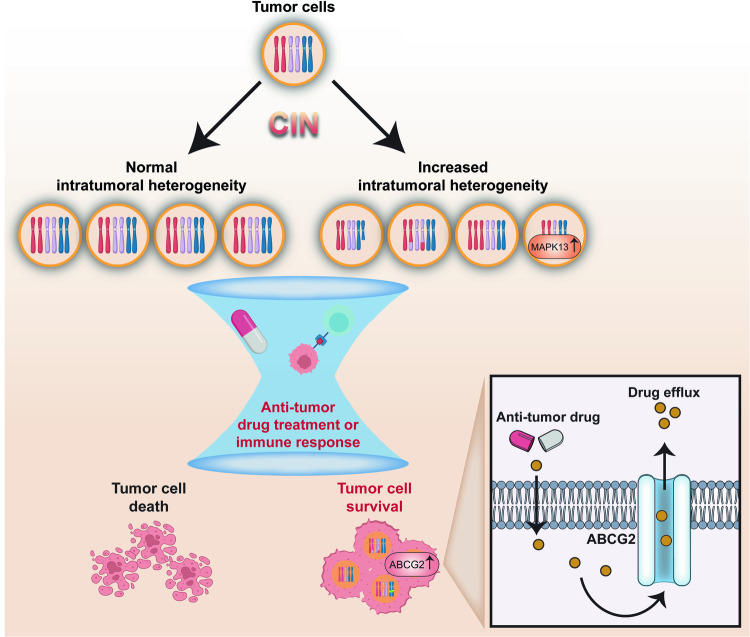
Impact of CIN on drug resistance. CIN confer resistance to anti-tumor drug treatment or immune response through increased intratumor heterogeneity
Table 2.
Consequences of CIN and their impact on tumor biology
| CIN indicators | Consequence of CIN | Impact on tumor biology | Ref |
|---|---|---|---|
| Lagging chromosome | Formation of micronucleus, aneuploid, or polyploid cells | DNA damage, chromothripsis, proteotoxic stress, metabolic alteration | 70,78,115,329 |
| Chromosome bridge | Formation of micronucleus, aneuploid, or polyploid cells | DNA damage, chromothripsis, proteotoxic stress, metabolic alteration | 78,115,329 |
| Micronucleus | DNA damage, chromothripsis | Increased heterogeneity, induction of metastasis through cGAS-STING pathway, tumor immune regulation | 78,115,122–133 |
| Aneuploidy | Changes in chromosome number or structure | Cell cycle arrest, senescence, metastasis, tumor immune regulation, drug resistance | 271,333–336,361 |
| Polyploidy | Changes in chromosome number | Cell cycle arrest, senescence, metastasis, tumor immune regulation, drug resistance | 230,252,271,342 |
DNA damage
CIN can lead to errors in chromosome segregation during cell division, resulting in the formation of lagging chromosomes and chromosome bridges. These lagging chromosome and chromosomal fragments from chromosome bridges often partition into micronuclei, whose nuclear envelope lacks several non-core membrane proteins such as lamin B, lamin B receptor (LBR), and nucleoporins, making them relatively fragile and prone to rupture.116,129,284–286 This rupture exposes the DNA content within the micronuclei to the cytoplasm, leading to chromothripsis, also known as “chromosome shattering”, a process of fragmentation of micronuclear chromosome followed by their massive rearrangements into the main nucleus. Hence, while itself a consequence of CIN as it originates from the micronuclear chromosome, chromothripsis could, at the same time, induce the succeeding CIN, as it could lead to the formation of a karyotype with complex chromosomal rearrangements.78,115,122–133
Chromothripsis has been associated with various tumor types and can lead to both numerical and structural CIN, altering gene expression patterns and driving changes in cellular behavior.122,287–289 Chromothripsis can contribute to tumorigenesis in multiple ways, either beneficial or deleterious. For instance, while DNA damage can be a cause of CIN as discussed in DSBs from replication stress, it is also a significant consequence of CIN, as chromothripsis could induce extensive DNA damage and destabilize tumor cell growth, leading to apoptosis.122 Interestingly, it can also create a tumor-promoting environment under certain circumstances, including rearrangement that results in karyotype with a better survival advantage. For example, reparation of shattered chromosomal fragments can lead to the incorporation of genetic material from the shattered fragments into double minute chromosomes, which are small, circular chromosomes without centrosomes distributed asymmetrically to daughter cells during cell division.290 Double minutes (also known as extrachromosomal DNA) were reported to contain high copy numbers of MYC proto-oncogene (MYC). Previously, Martins et al. reported that high MYC is an early event selected in many tumors with CIN, thereby providing a selective growth advantage to the tumor cells.291–293 Incorrect repair can also lead to a complete loss of gene function, such as the loss of key tumor suppressor gene mothers against decapentaplegic homolog 4 (SMAD4),294 as well as generation of novel oncogenic proteins by chromosome fusion. For example, a novel fusion oncoprotein which could promote AKT signaling activity, ubiquitin specific peptidase 9 X-linked-ES cell expressed Ras (USP9X-ERAS), is formed by chromothripsis involving the US9PX and ERAS genes in colon cancer cells.295
Collectively, chromothripsis is the consequence and at the same time, the cause of further CIN. The cycle of CIN leading to DNA damage, which in turn exacerbates CIN, forms a complex interaction that plays a crucial role in tumor development and progression. Moreover, recent studies suggest that CIN is associated with changes in both chromatin accessibility and transcription resulting from micronuclei formation.296,297 This complex, bidirectional relationship between chromothripsis and CIN, underscores the intricate dynamics of genomic instability in cancer.
Proteotoxic stress
Proteins in cells are often composed of more than one subunit. Multi-subunit protein complexes require balanced stoichiometry to function properly.268,298–303 This is achieved by regulating the ratio of protein subunits, and by degrading excessive, unassembled protein subunits.300,304 Unassembled protein subunits must be bound by chaperones to remain in solution until they are degraded by the ubiquitin-proteasome degradation pathway, thus, excessive production of subunits can overwhelm the protein quality control systems, impairing the stoichiometry and homeostasis of proteins in multi-subunit complexes.300,304 Furthermore, the increased demand for degradation puts the cell under proteotoxic stress, a form of cellular stress, and impairs cellular proliferation.268 This is often observed in aneuploid cells, where excessive protein subunits encoded by the altered chromosome could lead to imbalanced protein stoichiometry.305–308
In contrast, polyploid cells, which contain multiple sets of chromosomes, are likely to suffer less from the effects of genetic alteration and stoichiometry imbalance.230 When an essential gene is altered in polyploid cells, they still have more copies of functional genes compared to aneuploid cells. This redundancy provides a buffer against genetic alterations that might otherwise be detrimental. Kuznetsova et al. revealed that tetraploid cells proliferate almost as efficiently as diploid cells, and exhibit only some detrimental phenotypes observed in aneuploid cells.230 One reason for this could be that the multiple sets of chromosomes in polyploid cells help maintain a balance in protein stoichiometry by keeping the ratio of subunits forming a protein complex constant;230 while in aneuploid cells, gain or loss of certain chromosomes disrupts this ratio, eventually causes the dysfunction of protein complexes that require a specific stoichiometry of their subunits.
Imbalance protein stoichiometry could impair specific cellular functions associated with the affected protein complexes. For example, the gain of chromosome 6, which carries the gene encoding β-tubulin, causes lethality in yeast due to excessive β-tubulin production. However, this lethality can be rescued by additional gain of chromosome 13, which carries the α-tubulin gene, thus restoring the stoichiometry of α/β-tubulin dimers.309 Furthermore, imbalanced protein stoichiometry due to aneuploidy can also lead to the misfolding of proteins, which can accumulate and form toxic aggregates.268,298–301,310,311
Paradoxically, aneuploidy could also be beneficial for tumor cells. Aneuploidy could alter protein stoichiometry at the level of the interactome, which is the complete set of molecular interactions within a cell or organism including protein-protein, protein-DNA, and other types of molecular interactions essential for cellular function.312 A study of aneuploid patient tumor samples indicated that MET proto-oncogene, receptor tyrosine kinase (MET) amplification conferred resistance to epidermal growth factor receptor (EGFR) inhibitors, erlotinib and gefitinib.313 Typically, MET does not directly activate kinases downstream of the EGFR due to its low binding affinities with them. However, excessive MET expression can bypass the effect of EGFR inhibitors by directly interacting with these kinases and activating them, thereby counteracting the anti-tumorigenic effect of EGFR inhibitors.313 This finding illustrates that the aberrant protein interactions, which arise from the excessive proteins due to stoichiometry imbalance, could serve as a mechanism through which aneuploidy reshapes the interactome, thereby promoting tumorigenesis in tumor cells.
Metabolic alteration
Metabolism, an essential biological process involving a series of chemical reactions that convert food into energy and building blocks for the cells, is controlled by a finely-tuned coordination of complex metabolic networks, which depends on the precise balance of enzymes and regulators. Introduction of additional gene copies could disrupt this delicate balance, leading to metabolic alterations. Aneuploid cells, whether in yeast, animal models, or human cells, exhibit altered cellular metabolism.265,299,314,315 For instance, amplification of chromosome 4 in yeast results in the increase of amino acid levels, except the levels of aspartate and isoleucine, and various tricarboxylic acid cycle intermediates, leading to defects in cell growth.316 In MEFs, trisomy in either chromosome 1, 3, 16, or 19 leads to alterations in glutamine metabolism, and subsequently proliferation defects.265 Similarly, extra copy of chromosome 3 or 5 can impair human cell proliferation through downregulation in proteins involved in carbohydrate metabolism264 Therefore, CIN can lead to alterations in cellular metabolism, which can have detrimental effects on cells.
However, metabolic alteration, also known as metabolic reprogramming, is a hallmark of cancer that can provide several advantages for tumor cells.317–319 Metabolic alteration in tumor cells was first observed by Otto Warburg, who noted alterations in the glucose metabolism of tumor cells.320 Since then, alterations in other metabolic pathways, such as amino acids and lipid, along with their importance in tumor biology, have also been found.321,322 Normal cells produce energy from glucose effectively in the presence of oxygen by coupling glycolysis with oxidative phosphorylation.320 In contrast, one of the most well-known tumor cells metabolic reprogramming is the Warburg effect, a phenomenon where tumor cells increase their glucose uptake and glycolysis rate, and prefer glycolysis followed by fermentation, or aerobic glycolysis, instead of glycolysis followed by oxidative phosphorylation even when oxygen is sufficient.320,323 This allows tumor cells to cope with the fluctuating oxygen levels often found within tumor tissues.324 Moreover, aerobic glycolysis could also meet the increased demands of rapid cell proliferation for essential building blocks such as nucleotides, amino acids, and lipids.320,325–327 Furthermore, it could fulfill the demand of highly proliferating tumor cells for cellular reductants such as nicotinamide adenine dinucleotide phosphate (NADPH), which are crucial for lipid biosynthesis, drug resistance, and for scavenging reactive oxygen species (ROS) generated by high proliferation.320,323,328
CIN can induce the Warburg effect, triggering metabolic changes that promote tumorigenesis. This is evidenced by a correlation between karyotypic heterogeneity, which serves as an indicator of CIN, and increased consumption of glucose and glutamine, as well as increased production of lactate and glutamate.22,299,329,330 However, the association between other metabolic alterations, such as amino acids metabolic alteration and lipid metabolic alteration, and CIN, except the glutamine and glutamate metabolisms,329 has not been reported and still needs further investigation. In addition to the alterations in nutrient-related metabolism, CIN also impacts the cellular redox state. Cells with CIN exhibit changes in mitochondrial numbers and activity, typically resulting in increased ROS.331 While high levels of ROS can lead to oxidative stress and potential cell death, moderate levels of ROS can promote metastasis.332 Therefore, while CIN initially seems detrimental, it can actually benefit tumor cells by providing them with metabolic advantages. This highlights the complex interplay between CIN, metabolic reprogramming, and tumorigenesis.
Cell cycle arrest
CIN has been reported to cause cell cycle arrest.333–336 Live-cell imaging of human cells with chromosome missegregation demonstrated that missegregation induces cell cycle arrest in a p53-dependent manner.333–335,337 Known as the “guardian of the genome”, p53 plays crucial role in controlling cell cycle progression.338–341 The tumor suppressive function of p53 is closely related with response to CIN and is critical for determining the fate of cells experiencing CIN.333–335,342–346
p53 could suppress the propagation of structural CIN following chromosomal missegregation by inducing cell cycle arrest and apoptosis, thereby limiting the proliferation of aneuploid cells.270,334,347,348 p53 inactivation in tumor cells with CIN results in defects in inducing cell cycle arrest and apoptosis, and eventually, in increased tumor heterogeneity.104,349–351 This tendency is consistent with the findings in clinical non-small cell lung tumors, whereas p53-mutant tumors display more complex karyotypes than their wild-type counterparts.352,353 Furthermore, apoptosis observed in CIN mice model, presumably triggered by increased CIN, was rescued upon depletion of p53.348,354 In line with these, in a CIN model using SAC-deficient mice, reducing p53 level leads to increased aneuploidy and T-cell lymphoma proliferation, and at the same time, decreased survival.104,348,349 The RAS pathway, a critical signaling pathway in cells, is involved in cellular signal transduction, leading to cell growth and division. Overexpression of Harvey rat sarcoma virus (H-RAS) can induce CIN. However, the activation of RAS and the subsequent induction of CIN can be halted by the activation of p53, resulting in reduced transforming potential in mice model.355 This study further highlights the importance of p53 in monitoring and preventing CIN propagation to progenies.
In addition to p53, the stress kinase p38, which is a proline-directed serine/threonine kinases of the MAPK family, also plays a role in controlling the proliferation of aneuploid cells. p38 is activated in response to various stress stimuli, including CIN induction, and can induce cell cycle arrest by several mechanisms, including the upregulation of CDK inhibitors, growth arrest and DNA damage inducible alpha (GADD45α), and cyclin D, as well as the downregulation of CDC25.333,356–358 Moreover, p38 can work side-by-side with p53 to limit the progression of cells with CIN. Upon missegregation events, p38 increases the degradation of MDM2, a negative regulator of p53, through phosphorylation. This leads to the stabilization of p53 protein and apoptosis induction, thereby preventing the proliferation of cells with CIN.333,359 In addition to chromosomal-related events, the p38/p53 axis can also respond to metabolic stress induced by ROS formation as a consequence of CIN.333,334,358,360 These findings highlight the complex interplay between stress pathways, CIN, and cellular responses, underscoring the crucial roles of p53 and p38 in limiting the progression of cells with CIN.
Senescence
Cells with DNA damage and chromosome missegregation often became senescent and acquired the senescence-associated secretory phenotype (SASP). CIN enhanced by the treatment with SAC inhibitor could result in senescence, as indicated by the increase of senescence markers, such as p53, p21, p16, and senescence-associated β-galactosidase activity.271,361–364 The DDR pathway, activated in response to CIN-induced DNA damage plays a crucial role in this process.139 DDR can lead to the upregulation of p53, which in turn activates the expression of p21.137,365,366 This subsequently induces cell cycle arrest as a temporary response to allow DNA repair, or senescence as a permanent state of cell cycle arrest when the DNA damage is too severe to be repaired.367 Furthermore, CIN could also enhance the level of another senescence marker, p16, with a mechanism which is still unclear.
The consequence of CIN-induced senescent is complex. From a cell-autonomous perspective, senescence is a mechanism of tumor suppression in which aneuploid cells that undergo senescence will stop dividing and unlikely to undergo cellular transformation.362,363 In addition, senescent cells could be further cleared through autophagic cell death.368 However, CIN-induced senescence also triggers SASP-like gene expression signature, which might contribute to tumorigenesis.270,369 The unique secretome from SASP contains biologically active factors that are released into the microenvironment.370,371 These factors, including chemokines, cytokines, growth factors, and immune regulators, can induce a positive feedback loop and cause chronic inflammation, which can have dual effects on tumorigenesis.270 On one hand, it can act as a defense mechanism against tumors by promoting an anti-tumor immune response. On the other hand, it can contribute to tumorigenesis, as the secretome could activate key transcription factors involved in tumorigenesis, such as RAS.372 Furthermore, CIN-induced senescent cells can also increase the migration and invasion capacities of the neighboring tumor cells through SASP, thereby contributing to tumor progression.369 Therefore, the effect of CIN-induced senescence and its SASP-like gene expression is complex, resulting in both tumor-suppressive and pro-tumorigenic outcomes.
Metastatic capacity
As supported by pre-clinical and clinical data, changes in chromosome copy number could also influence cell motility, matrix degradation, epithelial-mesenchymal transition (EMT), and other processes necessary for metastatic behavior.373–376 For instance, tumor cells harboring an extra copy of chromosome 5 displayed increased metastatic capacity, as it leads to the silencing of epithelial cell-adhesion genes and thereby activates EMT.377 Meanwhile, the loss of chromosome 16q has been associated with downregulation of E-cadherin (CDH1).378 Interestingly, Gao et al. demonstrated that CIN is also associated with mesenchymal-epithelial-transition (MET), a reverse version of EMT required for extravasation and colonization in different tissues in the process of distant metastasis.378,379 Loss of chromosome 10p results in the loss of the zinc finger E-box-binding homeobox 1 (ZEB1) gene, thus promoting MET, and subsequently metastasis378 Furthermore, CIN could drive metastasis by activating Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway, and through the establishment of local immunosuppressive microenvironment.380,381 Moreover, CIN can also indirectly induce EMT. For instance, the loss of chromosome 8p results in the downregulation of 8p-localized genes, such as N-acylsphingosine amidohydrolase 1 (ASAH1), farnesyl-diphosphate farnesyltransferase 1 (FDFT1), leptin receptor overlapping transcript-like 1 (LEPROTL1), epoxide hydrolase 2 (EPHX2), and BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like (BNIP3L), thereby altering the mevalonate and fatty acid metabolic pathways. Disruption of these lipid metabolic pathways in turn increases the activities of small GTPases, such as Ras homolog family member (RHO), Ras-related C3 botulinum toxin substrate (RAC), and rat sarcoma (RAS), and subsequently promotes invasion and metastasis.378,382,383 Meanwhile, in the clinical setting, longitudinal studies such as TRACERx, which track the progression of cancer from primary disease to metastasis and recurrence, have reported that elevated CIN correlates with an increase in metastasis and worse survival outcomes, and subsequently poor prognosis.384–389 These results demonstrate the positive correlation between CIN and metastasis, as well as poor survival and outcomes. However, it should be noted that a recent study has also reported the anti-metastasis function CIN. For example, changes in chromosome copy number, for example gaining extra copy of chromosome 13 or chromosome 18, could suppress metastasis.377 The underlying mechanisms of how these specific chromosomal changes suppress metastasis remain unclear, highlighting the complex role of CIN in tumor metastatic capacity.
Tumor immune regulation
CIN has dual activity in immune response, as it is capable of inducing either anti-tumor or pro-tumor immune response. In xenograft models, tumor cells with increased aneuploidy and polyploidy tend to form tumors in immunocompromised mice. However, these tumors either fail to grow or grow more slowly in immunocompetent mice, suggesting the anti-tumor role of CIN-induced immune response.390 This could be attributed to genomic alterations that produce neoantigens, which are recognized by the immune cells, and thus activate the adaptive immune response.391 Consequently, CIN can activate anti-tumor immune responses, thereby subjecting tumor cells to the selection pressure imposed by the immune system, which subsequently eliminates them.391
However, CIN could also contribute to pro-tumor immune response. As a contributor to genomic instability, CIN increases intratumor heterogeneity,37 allowing the generation of different tumor cells with variations in antigen presentation, thereby reducing their visibility to the adaptive immune system, which subsequently leads to immune evasion.30 Furthermore, micronucleus, as a product of missegregation of chromosomes, could also trigger CIN-related immune regulation.53,392–396 Exposure of chromosomal double-stranded DNA (dsDNA) in the micronuclei to the cytoplasm activates the cyclic GMP-AMP synthase-stimulator of interferon genes (cGAS-STING)-dependent immune response, a type of innate immune response originally discovered as a sensor of viral dsDNA.53,397–400 Typically, recognition of dsDNA by cGAS activates STING, which could in turn activate anti-tumor immune response through type 1 interferon (IFN) and canonical nuclear factor kappa B (NF-κB) signaling.53,392,401,402 Interestingly, in the context of CIN, STING promotes EMT and the expression of inflammatory genes, enhancing cell migratory capacity, and subsequently promoting metastasis, by activating non-canonical NF-κB signaling.53,392,403 This process indicates that CIN manipulates the innate immune system to promote tumor cells immune evasion.53 Furthermore, this example clearly demonstrates that CIN could alter normal cell function to favor tumor progression. Moreover, the activation of SASP through CIN-induced senescence could also shift the immune response towards pro-tumor.372 Together, these observations suggest that CIN, through increased heterogeneity or by activation of pathways in favor of pro-tumor immune response, can promote tumor cells immune evasion.
Drug resistance
Anti-tumor drugs encompass a wide range, each with its own complex mechanisms. For instance, alkylating agents cause DNA crosslinks that disrupt DNA replication, eventually inhibiting tumor cell division,13,317,401,402 while anti-metabolites can cause improper DNA synthesis by mimicking endogenous molecules.19,127 Irrespective of their mechanism, resistance to anti-tumor drugs has become a significant hurdle in anti-tumor treatment.27,404 While the molecular mechanism of tumor cells drug resistance are complex and have not been totally elucidated, CIN, as a fuel of tumor evolution that causes intratumor heterogeneity, has been assumed as one of the major reason for drug resistance.230,336,405,406 This increase in heterogeneity can be viewed as a survival strategy employed by tumor cells to adapt to unpredictable environments. This strategy, known as biological bet-hedging or “not putting all your eggs in one basket”, allows tumor cells to diversify their phenotypes, spreading the risk and increasing the likelihood of some cells to survive under selective pressures, such as those imposed by anti-tumor drug treatments.23,407,408
Moreover, Ippolito et al. previously reported the link between CIN and drug resistance through the upregulation of ATP binding cassette subfamily G member 2 (ABCG2), a drug efflux pump, due to the amplification of its upstream regulator MAPK13 in topotecan-resistant tumor cells generated from treatment using CIN-inducing drug nocodazole.17 Together, these show that CIN fuels genomic diversity, upon which selection works, leading to the development of drug resistance.
Although the result from the aforementioned study suggests that the generation of intratumor heterogeneity through CIN can shield tumor cells from the selective pressure caused by anti-tumor drugs, the role of CIN in drug response is nevertheless a complex relationship, as CIN could potentially be induced to excessive level, leading to cell death and enhancing the effectiveness of anti-tumor drug treatments. The following section will discuss how the induction of CIN can be leveraged to enhance the efficacy of anti-tumor drugs. A summary of the different types of anti-tumor drugs and their interactions with CIN is provided in Table 3.
Table 3.
Anti-tumor drugs and CIN-related drug resistance mechanism
| Class | Mechanism of Action | Examples | CIN-modulated resistance mechanism | Ref |
|---|---|---|---|---|
| Anti-metabolites | Mimic endogenous molecules, causes improper DNA synthesis | Methotrexate, cladribine, fluorouracil, cytarabine, mercaptopurine, fludarabine | Increased heterogeneity | 19,127 |
| Alkylating agents | Form DNA strand cross-links, induce DNA damage | Myleran, chlorambucil, cisplatin, oxaliplatin | Increased DNA repair mechanisms, delayed cell cycle | 13,317,401,402 |
| Topoisomerase inhibitors | Inhibits DNA unwinding during replication or transcription | Irinotecan, topotecan, etoposide, teniposide | Upregulation of drug efflux pumps | 17,406 |
| Mitotic inhibitors | Inhibits tubulin polymerization | Vincristine, vinblastine, docetaxel, paclitaxel | Alterations in SAC, delayed cell cycle | 336 |
| Anthracyclines | DNA intercalation, induce DNA damage | Doxorubicin, idarubicin, daunorubicin, epirubicin, | Increased DNA repair mechanisms | 18,230,405,406 |
| Protein kinase inhibitors | Blocks the action of protein kinases | Imatinib, dasatinib, nilotinib, gefitinib, vemurafenib, trametinib | Alterations in signaling pathways | 18,272,478,479 |
| Proteasome inhibitors | Induces ER stress due to accumulation of misfolded proteins | Bortezomib, carfilzomib, ixazomib | Upregulation of drug efflux pumps | 480–482 |
| PARP inhibitors | Block PARP-dependent DNA repair mechanism | Olaparib, rucaparib | Alterations in DNA repair mechanisms | 406 |
| Monoclonal antibodies | Blocks specific signaling pathway, trigger anti-tumor immune response | Rituximab, trastuzumab, cetuximab, nivolumab, pembrolizumab | Alterations in antigen expression | 483 |
CIN-based potential anti-tumor therapy
Previous studies indicated that an excessive level of CIN beyond a certain threshold could potentially induce tumor cell death. Thus, enhancing the CIN level has been proposed as a promising approach to target tumor cells, and strategies to exacerbate CIN for anti-tumor therapy have been explored.66,358,406,409–422 For example, taxol could increase the number and severity of chromosome segregation errors in tumor cells, while cells with excessive CIN were more sensitive to low doses of taxol.423 Indeed, combining taxol and monopolar spindle 1 (MPS1) inhibitor could reduce xenograft growth more effectively than either compound alone.411 Furthermore, combining SAC inhibitor and other non-taxol-based compounds that induce CIN can synergistically reduce tumor growth. For example, combining a SAC inhibitor with microtubule-destabilizing drug SKI606 results in the selective killing of tumor cells exhibiting a CIN phenotype.410 Moreover, combining a p38α inhibitor, which interferes with DNA damage response, with taxane-based chemotherapy increased the efficiency of killing breast tumor cells compared to taxanes alone by boosting CIN.358 While they have not yet been used in clinical settings, there are also other compounds that can induce CIN. These include inhibitors of the SAC proteins MAD2 or BUBR1, which can induce tumor cell death. Moreover, a compound that induces CIN by targeting the highly expressed in cancer 1/NIMA-related kinase 2 (Hec1/Nek2)-related mitotic pathway also demonstrates promising results.424–426
While enhancing CIN could potentially be a powerful method to eradicate tumors, the feasibility of such therapies will depend on many factors including CIN status and the capacity of the tumor cells to tolerate CIN.267,427–437 Moreover, it is important to consider that untransformed cells will also be affected by the CIN-inducing agents and thus will suffer from low to moderate CIN rates. This may predispose these cells to become tumorigenic, leading to therapy-induced tumorigenesis subsequently.
To overcome this problem, efforts have been made to develop strategies that more selectively target cells displaying CIN phenotype, either by exploiting specific vulnerabilities of tumor cells with CIN or identifying new weaknesses incurred in tumor cells with new karyotype. For example, a study by Marquis et al., which aims to identify synthetic lethal gene in tumor cells with CIN, has discovered that targeting kinesin family member 18A (KIF18A) is particularly detrimental to aneuploid tumor cells.438 This sensitivity arises from alterations in spindle geometry and microtubule dynamics specific in tumor cell with CIN, which, upon KIF18A knockdown, leads to excessive CIN and reduces tumor cells viability. This suggests that KIF18A could be a promising synthetic lethal candidate for future drug development efforts targeting tumor cells with CIN. Furthermore, Hong et al. found that IL-6/STAT3 signaling axis downstream of cGAS-STING enables the survival of tumor cells with CIN, and blockade of IL-6 signaling by tocilizumab, a clinically used drug that targets the IL-6 receptor (IL-6R), can impair their growth specifically.403 Moreover, aneuploid cells have also been found to contain higher levels of ceramide, and further increasing the levels of ceramide through treatment with N-[2-hydroxy-1-(4-morpholinylmethyl)-2-phenylethyl]-decanamide monohydrochloride (DL-PDMP), an antagonist of UDP-glucose ceramide glucosyltransferase, is significantly more toxic to aneuploid cells compared with diploid cells.439
Another strategy utilizes new weakness that results from gaining specific chromosome. For instance, a gene encoded on chromosome 1, known as uridine-cytidine kinase 2 (UCK2), is required to activate certain pro-drugs, such as RX-3117 and 3-deazauridine. A recent study revealed that cells with an extra copy of chromosome 1 express higher level of UCK2 and are more sensitive to those drugs compared to diploid cells with just two copies,440 suggesting that introducing specific aneuploidies that can exert anti-tumor function, for example using CRISPR-based tools, might also be a potential CIN-based anti-tumor therapeutic strategy.441–447 This strategy is still not yet translated to clinical trial, nevertheless, several compounds targeting different pathways to increase CIN are now in phase I-III clinical trials,448–450 representing promising progress for future research and development in CIN-based anti-tumor therapy (Table 4).
Table 4.
Clinical trials of CIN-inducing drugs for anti-tumor treatment
| Inhibitors | Drug type | Effect on CIN | Identifier/status/phase | Ref |
|---|---|---|---|---|
| MLN8237 (Alisertib) | AURKA inhibitor | Abnormal spindle poles | NCT00500903 completed I | 484,485 |
| NCT00853307 completed II | 486 | |||
| NCT01653028 completed II | 487 | |||
| NCT01799278 completed II | 488 | |||
| NCT01091428 completed I/II | 489 | |||
| NCT02038647 completed II | 490 | |||
| NCT02109328 completed II | 491 | |||
| NCT01094288 completed I | 492 | |||
| NCT01639911 completed I | 493 | |||
| NCT01601535 completed I | 494 | |||
| NCT02319018 completed II | 495 | |||
| NCT02219789 completed I | 496 | |||
| NCT02293005 ongoing I/II | 497,498 | |||
| NCT01924260 completed I | 499 | |||
| NCT02187991 completed I | 500 | |||
| NCT02719691 completed I | 501 | |||
| NCT04555837 completed I | 502 | |||
| NCT04479306 completed I | 503,504 | |||
| NCT04085315 completedI | 505 | |||
| NCT00697346 completed I | 506 | |||
| NCT01466881 completed II | 507 | |||
| NCT01482962 completed III | 508 | |||
| NCT00807495 completed II | 509 | |||
| NCT01034553 completed I/II | 510,511 | |||
| NCT02560025 completed II | 512 | |||
| NCT01695941 completed I | 510 | |||
| NCT02444884 completed I | 513 | |||
| NCT01154816 completed II | 514 | |||
| NCT02114229 completed II | 515,516 | |||
| ENMD-2076 | AURKA inhibitor | NCT00658671 completed I | 517,518 | |
| NCT01104675 completed I | 519 | |||
| NCT01914510 completed I | 520 | |||
| NCT01719744 completed II | 521 | |||
| NCT01639248 completed II | 522 | |||
| NCT02234986 completed II | 523 | |||
| NCT00904787 completed I | 524 | |||
| LY3295668 | AURKA inhibitor | NCT03092934 completed I | 525 | |
| NCT03955939 completed I/II | 526 | |||
| NCT03898791 completed I | 527 | |||
| NCT04106219 completed I | 528 | |||
| MLN8054 | AURKA inhibitor | NCT00249301 completed I | 529 | |
| NCT00652158 completed I | 530 | |||
| MK-5108 | AURKA inhibitor | NCT00543387 completed I | 531,532 | |
| TAS-119 | AURKA inhibitor | NCT02448589 completed I | 533,534 | |
| NCT02134067 completed I | 535 | |||
| KW-2449 | AURKA inhibitor | NCT00346632 completed I | 536 | |
| NCT00779480 completed I | 497,537 | |||
| AZD1152 (Barasertib) | AURKB inhibitor | Abnormal spindle poles | NCT00338182 completed I | 538,539 |
| NCT00497731 completed I | 540 | |||
| NCT00497991 completed I/II | 541 | |||
| NCT00530699 completed I | 542 | |||
| NCT00952588 completed I | 543 | |||
| NCT01019161 completed I | 544 | |||
| NCT00926731 completed I | 545 | |||
| NCT03217838 completed I/II | 546 | |||
| NCT01354392 completed II | 546 | |||
| BI-831266 | AURKB inhibitor | NCT00756223 completed I | 547 | |
| BI-811283 | AURKB inhibitor | NCT00701324 completed I | 548 | |
| NCT00632749 completed II | 549 | |||
| Chiauranib | AURKB inhibitor | NCT02122809 completed I | 550,551 | |
| NCT03901118 ongoing II | 497 | |||
| NCT03245190 ongoing I/II | 552 | |||
| NCT03974243 ongoing I/II | 553 | |||
| NCT03216343 completed I/II | 554 | |||
| AT9283 | AURKA/B inhibitor | Abnormal spindle poles | NCT00443976 completed I | 555,556 |
| NCT00522990 completed II | 557 | |||
| NCT00985868 completed I | 558 | |||
| Cenisertib | AURKA/B inhibitor | NCT00391521 completed I | 559,560 | |
| NCT01097512 completed I | 561 | |||
| NCT01080664 completed I | 562 | |||
| CYC116 | AURKA/B inhibitor | NCT00560716 completed I | 563 | |
| PF-03814735 | AURKA/B inhibitor | NCT00424632 completed I | 564,565 | |
| TAK-901 | AURKA/B inhibitor | NCT00935844 completed I | 566 | |
| NCT00807677 completed I | 566 | |||
| Danusertib | AURKA/B/C inhibitor | Abnormal spindle poles | NCT00872300 completed II | 567 |
| NCT00766324 completed II | 568 | |||
| GSK1070916 (NIM-900) | AURKB/C inhibitor | NCT01118611 ongoing I | 569 | |
| ABT-348 (Ilorasertib) | AURKA/B/C inhibitor | NCT01110486 completed I | 570,571 | |
| NCT01110473 completed I | 572 | |||
| NCT02478320 completed II | 571 | |||
| AMG-900 | AURKA/B/C inhibitor | NCT00858377 completed I | 573,574 | |
| NCT01380756 completed I | 575 | |||
| BI-847325 | AURKA/B/C inhibitor | NCT01324830 completed I | 576,577 | |
| SNS-314 Mesylate | AURKA/B/C inhibitor | NCT00519662 completed I | 578 | |
| Tozasertib | AURKA/B/C inhibitor | NCT00500006 completed I | 579 | |
| NCT02532868 completed I | 580 | |||
| NCT00111683 completed I | 581 | |||
| GSK923295 | CENP-E inhibitor | Metaphase misalignment | NCT00504790 completed I | 582,583 |
| Ispinesib | KIF11 inhibitor | Monopolar spindle | NCT00169520 completed I/II | 584 |
| NCT00119171 completed I/II | 584 | |||
| NCT00136578 completed I/II | 585 | |||
| NCT00095992 completed I | 586 | |||
| NCT00096499 completed I | 587 | |||
| NCT00095628 completed II | 588 | |||
| NCT00095953 completed II | 589 | |||
| NCT00354250 completed II | 590 | |||
| NCT00097409 completed II | 588 | |||
| NCT00607841 completed I | 591 | |||
| NCT00363272 ongoing I | 592 | |||
| SB-743921 | KIF11 inhibitor | NCT00136513 completed I/II | 593 | |
| NCT00343564 completed I/II | 594 | |||
| Filanesib | KIF11 inhibitor | NCT00462358 completed I | 595 | |
| NCT00637052 completed I/II | 596 | |||
| NCT00821249 completed I/II | 597 | |||
| NCT01248923 completed I | 598 | |||
| NCT01372540 completed I | 599 | |||
| NCT02384083 completed I/II | 600 | |||
| ALN-VSP02 | KIF11 inhibitor | NCT01158079 completed I | 601 | |
| NCT00882180 completed I | 601 | |||
| Litronesib | KIF11 inhibitor | NCT01214629 completed I | 602 | |
| NCT01214642 completed I | 603 | |||
| NCT01358019 completed I | 604 | |||
| 4SC-205 | KIF11 inhibitor | NCT01065025 completed I | 605 | |
| AZD4877 | KIF11 inhibitor | NCT00613652 completed I | 606 | |
| NCT00389389 completed I | 607 | |||
| NCT00486265 completed I | 608 | |||
| NCT00661609 completed II | 609 | |||
| ARQ 621 | KIF11 inhibitor | NCT00825487 completed I | 585 | |
| MK-0731 | KIF11 inhibitor | NCT00104364 completed I | 610 | |
| VLS-1488 | KIF18A inhibitor | NCT05902988 ongoing I/II | 68 | |
| CFI-402257 | Mps1 inhibitor | Chromosome missegregation | NCT02792465 completed I | 64 |
| NCT03568422 completed I | 611 | |||
| BAY1161909 (Empesertib) | Mps1 inhibitor | NCT02138812 completed I | 612 | |
| BAY1217389 | Mps1 inhibitor | NCT02366949 completed I | 613 | |
| S81694 | Mps1 inhibitor | NCT03411161 completed I/II | 614 | |
| BOS172722 | Mps1 inhibitor | NCT03328494 completed I | 615,616 | |
| Olaparib | PARP inhibitor | Disrupted DNA repair system | NCT01623349 completed I | 617 |
| Rucaparib | PARP inhibitor | NCT03654833 completed II | 618 | |
| BI 2536 | Plk1 inhibitor | Premature separation of sister chromatids | NCT00376623 completed I | 619,620 |
| NCT02211833 completed I | 621 | |||
| NCT00710710 completed II | 622 | |||
| NCT00701766 completed I/II | 623 | |||
| NCT00243087 completed I | 624 | |||
| BI6727 (Volastertib) | Plk1 inhibitor | NCT02273388 completed I | 625 | |
| NCT00969553 completed I | 626 | |||
| NCT01348347 completed I | 627 | |||
| NCT01022853 completed I | 628 | |||
| NCT01206816 completed I | 629 | |||
| NCT00969761 completed I | 630 | |||
| NCT01662505 completed I | 631 | |||
| NCT01023958 completed II | 632 | |||
| NCT01121406 completed II | 632 | |||
| NCT00824408 completed II | 633 | |||
| NCT00804856 completed II | 634 | |||
| NCT01721876 completed III | 635,636 | |||
| ON 01910.Na (Rigosertib) | Plk1 inhibitor | NCT01125891 completed I | 637 | |
| NCT01926587 completed I//II | 638 | |||
| NCT03786237 completed I | 612,639 | |||
| NCT04177498 completed I | 612,639 | |||
| NCT04263090 completed I | 640 | |||
| NCT02562443 ongoing III | 641 | |||
| NCT01360853 completed III | 642 | |||
| NCT01241500 completed III | 641 | |||
| NCT01928537 completed III | 643 | |||
| GSK461364 | Plk1 inhibitor | NCT00536835 completed I | 644,645 | |
| MK-1496 | Plk1 inhibitor | NCT00880568 completed I | 612 | |
| TAK-960 | Plk1 inhibitor | NCT01179399 completed I | 646,647 | |
| NMS-1286937 (Onvansertib) | Plk1 inhibitor | NCT01014429 completed I | 648,649 | |
| NCT03303339 completed I/II | 650 | |||
| NCT03829410 completed I/II | 651 | |||
| NCT03414034 completed I/II | 652,653 | |||
| TKM-080301 | Plk1 inhibitor | NCT02191878 completed I/II | 654,655 | |
| NCT01262235 completed I/II | 656 | |||
| NCT01437007 completed I/II | 657,658 | |||
| CYC 140 | Plk1 inhibitor | NCT03884829 ongoing I | 659 | |
| CFI-400945 | Plk4 inhibitor | NCT01954316 completed II | 449,660 | |
| AZD1775 | WEE1 inhibitor | Premature entry into mitosis | NCT03253679 completed II | 661,662 |
| Adavosertib | WEE1 inhibitor | NCT02101775 completed II | 663,664 |
Conclusion
Great progress has been made during this last decade in understanding how chromosome segregation is regulated and what defects might contribute to chromosome missegregation in tumor cells. While the exact source of CIN in tumor cells is complex and diverse, it is clear that a variety of mechanisms can lead to this phenotype. Tumor cells presumably employ different routes to achieve the same CIN phenotype. Therefore, more research to further characterize the mechanisms contributing to CIN is necessary. This will be crucial in laying the foundation for developing strategies that can modulate CIN, with the aim of inhibiting tumor cells ability to adapt to environmental challenges, as well as preventing tumor drug resistance.
From a clinical perspective, although the relative contribution of various mechanisms of CIN has been described in cultured tumor cell lines, whether these models also recapitulate the types of CIN found in primary and metastatic tumors, as well as the epigenetic causes and consequences of CIN, remains to be determined. Enhancing CIN has been proposed as a promising approach to target aneuploid tumor cells. From the perspective of developing new therapeutic strategies, it is crucial to determine whether cells undergoing CIN share common characteristics. It is also important to understand if CIN triggers a common stress response and whether CIN cells need to develop specific adaptations to adapt to their altered genomes. A more comprehensive understanding of these effects and the resulting cellular responses is crucial for the successful exploitation of CIN for therapeutic strategies. Furthermore, while several SAC inhibitors that increase CIN show promising potential for reducing tumor growth and have already entered clinical trial phase, the success of such therapies depends on factors such as CIN status and the capacity of the tumor cells to tolerate CIN.267,427–437 Despite these challenges, recent advances suggest promising possibilities for a new, personalized, CIN-specific approach to anti-tumor therapy.358,406,410,411,423–426,438,451–455
Moreover, considering CIN as the fuel for genomic diversity, it is important to acknowledge the challenges posed by tumor evolution. This suggests that future research could focus on characterizing the common phenotypes that emerge from various CIN routes. These strategies might not directly target the mechanisms of CIN; instead, they could focus on the phenotypes selected during the stages of tumor initiation and progression. Adopting this approach could potentially lead to more effective treatments that are less susceptible to being undermined by tumor adaptation and evolution. Furthermore, it could provide valuable biomarkers for early detection and prognosis, thereby opening up new opportunities for preventive measures.293
Furthermore, while targeting tumor cells with CIN holds promise, it is important to consider the risks. Increasing CIN in untransformed cells could inadvertently lead to therapy-induced tumor due to their potential to become tumorigenic under low to moderate CIN rates. Therefore, future research should aim to selectively target cells with a CIN phenotype, thereby reducing the risk of unintentional transformation of normal cells.
In conclusion, our understanding of CIN and its role in tumorigenesis has greatly improved. Although many questions remain unanswered, and further research is needed to fully understand the mechanisms underlying CIN-related tumorigenesis as well as its potential as a therapeutic target, the study of CIN and its effects on tumor cells have nevertheless laid a promising foundation. These insights could potentially guide the development of new strategies for diagnosing and treating cancer.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32070715, 32270778, 82173029, and 82372655); and the Natural Science Foundation of Chongqing (CSTB2022NSCQ-MSX0612 and CSTB2022NSCQ-MSX0611). Our intention is to summarize the state of art. However, due to space limitations, we would like to apologize to authors whose works are not cited here. Their contributions should not be considered less important than those that are cited.
Author contributions
R.H., S.W., V.K. designed the review; R.H. wrote original draft preparation; R.H. and S.H. drafted original figures preparation; R.H., S.H., and S.N. drafted original tables preparation; R.H., S.W., V.K. revised the manuscript. Corresponding authors S.W. and V.K. provided financial support and supervision. All authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Contributor Information
Shourong Wu, Email: shourongwu@cqu.edu.cn.
Vivi Kasim, Email: vivikasim@cqu.edu.cn.
References
- 1.Wild, C., Weiderpass E., & Stewart B. W. (eds) World Cancer Report Cancer Research for Cancer Prevention (International Agency Research on Cancer, 2020). [PubMed]
- 2.Tijhuis AE, Johnson SC, McClelland SE. The emerging links between chromosomal instability (CIN), metastasis, inflammation and tumour immunity. Mol. Cytogenet. 2019;12:1–21. doi: 10.1186/s13039-019-0429-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasudevan A, et al. Aneuploidy as a promoter and suppressor of malignant growth. Nat. Rev. Cancer. 2021;21:89–103. doi: 10.1038/s41568-020-00321-1. [DOI] [PubMed] [Google Scholar]
- 4.Taylor AM, et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33:676–689.e673. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weaver BAA, Cleveland DW. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Drews RM, et al. A pan-cancer compendium of chromosomal instability. Nature. 2022;606:976–983. doi: 10.1038/s41586-022-04789-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C-Z, Pellman D. Cancer genomic rearrangements and copy number alterations from errors in cell division. Annu. Rev. Cancer Biol. 2022;6:245–268. doi: 10.1146/annurev-cancerbio-070620-094029. [DOI] [Google Scholar]
- 8.Matthews HK, Bertoli C, de Bruin RA. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022;23:74–88. doi: 10.1038/s41580-021-00404-3. [DOI] [PubMed] [Google Scholar]
- 9.Sarkar S, et al. Mitotic checkpoint defects: en route to cancer and drug resistance. Chromosom. Res. 2021;29:131–144. doi: 10.1007/s10577-020-09646-x. [DOI] [PubMed] [Google Scholar]
- 10.Maiato H, Silva S. Double-checking chromosome segregation. J. Cell Biol. 2023;222:e202301106. doi: 10.1083/jcb.202301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khodjakov A, Rieder CL. The nature of cell-cycle checkpoints: facts and fallacies. J. Biol. 2009;8:1–5. doi: 10.1186/jbiol195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chunduri NK, Storchova Z. The diverse consequences of aneuploidy. Nat. Cell Biol. 2019;21:54–62. doi: 10.1038/s41556-018-0243-8. [DOI] [PubMed] [Google Scholar]
- 13.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–M60. doi: 10.1016/S0962-8924(99)01661-X. [DOI] [PubMed] [Google Scholar]
- 14.Lezmi E, Benvenisty N. The tumorigenic potential of human pluripotent stem cells. Stem Cells Transl. Med. 2022;11:791–796. doi: 10.1093/stcltm/szac039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Na J, et al. Aneuploidy in pluripotent stem cells and implications for cancerous transformation. Protein Cell. 2014;5:569–579. doi: 10.1007/s13238-014-0073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbaric I, et al. Time-lapse analysis of human embryonic stem cells reveals multiple bottlenecks restricting colony formation and their relief upon culture adaptation. Stem Cell Rep. 2014;3:142–155. doi: 10.1016/j.stemcr.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ippolito MR, et al. Gene copy-number changes and chromosomal instability induced by aneuploidy confer resistance to chemotherapy. Dev. Cell. 2021;56:2440–2454.e2446. doi: 10.1016/j.devcel.2021.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Lukow DA, et al. Chromosomal instability accelerates the evolution of resistance to anti-cancer therapies. Dev. Cell. 2021;56:2427–2439. doi: 10.1016/j.devcel.2021.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutledge SD, et al. Selective advantage of trisomic human cells cultured in non-standard conditions. Sci. Rep. 2016;6:22828. doi: 10.1038/srep22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-David U, et al. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat. Commun. 2014;5:4825. doi: 10.1038/ncomms5825. [DOI] [PubMed] [Google Scholar]
- 21.Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13:501–514. doi: 10.1038/embor.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukow DA, Sheltzer JM. Chromosomal instability and aneuploidy as causes of cancer drug resistance. Trends Cancer. 2022;8:43–53. doi: 10.1016/j.trecan.2021.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Watkins TB, et al. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature. 2020;587:126–132. doi: 10.1038/s41586-020-2698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrade JR, Gallagher AD, Maharaj J, McClelland SE. Disentangling the roles of aneuploidy, chromosomal instability and tumour heterogeneity in developing resistance to cancer therapies. Chromosom. Res. 2023;31:28. doi: 10.1007/s10577-023-09737-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vitale I, Shema E, Loi S, Galluzzi L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 2021;27:212–224. doi: 10.1038/s41591-021-01233-9. [DOI] [PubMed] [Google Scholar]
- 27.Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature. 2019;575:299–309. doi: 10.1038/s41586-019-1730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dijk E, et al. Chromosomal copy number heterogeneity predicts survival rates across cancers. Nat. Commun. 2021;12:3188. doi: 10.1038/s41467-021-23384-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sansregret L, Vanhaesebroeck B, Swanton C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 2018;15:139–150. doi: 10.1038/nrclinonc.2017.198. [DOI] [PubMed] [Google Scholar]
- 30.Bakhoum SF, Cantley LC. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell. 2018;174:1347–1360. doi: 10.1016/j.cell.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, et al. Patterns of somatic structural variation in human cancer genomes. Nature. 2020;578:112–121. doi: 10.1038/s41586-019-1913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining ‘chromosomal instability'. Trends Genet. 2008;24:64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 33.The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burrell RA, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitelman, F., Johansson, B. & Mertens, F. (eds) Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (National Cancer Institute, 2024); http://mitelmandatabase.isb-cgc.org/.
- 36.Graphodatsky AS, Trifonov VA, Stanyon R. The genome diversity and karyotype evolution of mammals. Mol. Cytogenet. 2011;4:1–16. doi: 10.1186/1755-8166-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakhoum SF, Landau DA. Chromosomal instability as a driver of tumor heterogeneity and evolution. Cold Spring Harb. Perspect. Med. 2017;7:2029611. doi: 10.1101/cshperspect.a029611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bronder D, Bakhoum SF. A CIN ful way to overcome addiction: how chromosomal instability enables cancer to overcome its oncogene addiction. EMBO Mol. Med. 2020;12:e12017. doi: 10.15252/emmm.202012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salgueiro L, et al. Acquisition of chromosome instability is a mechanism to evade oncogene addiction. EMBO Mol. Med. 2020;12:e10941. doi: 10.15252/emmm.201910941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansford S, Huntsman DG. Boveri at 100: Theodor Boveri and genetic predisposition to cancer. J. Pathol. 2014;234:142–145. doi: 10.1002/path.4414. [DOI] [PubMed] [Google Scholar]
- 41.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez-Casanas M, Chan KL. The unresolved problem of DNA bridging. Genes. 2018;9:623. doi: 10.3390/genes9120623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gartler SM. The chromosome number in humans: a brief history. Nat. Rev. Genet. 2006;7:655–660. doi: 10.1038/nrg1917. [DOI] [PubMed] [Google Scholar]
- 44.Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nowell PC. Discovery of the Philadelphia chromosome: a personal perspective. J. Clin. Investig. 2007;117:2033–2035. doi: 10.1172/JCI31771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobrovic A, Peters G, Ford J. Molecular analysis of the Philadelphia chromosome. Chromosoma. 1991;100:479–486. doi: 10.1007/BF00352198. [DOI] [PubMed] [Google Scholar]
- 47.Megarbane A, et al. The 50th anniversary of the discovery of trisomy 21: the past, present, and future of research and treatment of Down syndrome. Genet. Med. 2009;11:611–616. doi: 10.1097/GIM.0b013e3181b2e34c. [DOI] [PubMed] [Google Scholar]
- 48.Lejeune J. Trois cas de délétion partielle du bras court d’un chromosome 5. Comp. Rendus Acad. Sci. 1963;257:3098–3102. [PubMed] [Google Scholar]
- 49.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 50.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 51.Amon A. The spindle checkpoint. Curr. Opin. Genet. Dev. 1999;9:69–75. doi: 10.1016/S0959-437X(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 52.D’Amours D, Amon A. At the interface between signaling and executing anaphase—Cdc14 and the FEAR network. Genes Dev. 2004;18:2581–2595. doi: 10.1101/gad.1247304. [DOI] [PubMed] [Google Scholar]
- 53.Bakhoum SF, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553:467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobles M, et al. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/S0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 55.Denomy C, et al. Banding together: a systematic comparison of the cancer genome atlas and the Mitelman databases. Cancer Res. 2019;79:5181–5190. doi: 10.1158/0008-5472.CAN-19-0585. [DOI] [PubMed] [Google Scholar]
- 56.Weinstein, JN, et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dentro SC, et al. Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell. 2021;184:2239–2254 e2239. doi: 10.1016/j.cell.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jamal-Hanjani M, et al. Tracking genomic cancer evolution for precision medicine: the lung TRACERx study. PLoS Biol. 2014;12:e1001906. doi: 10.1371/journal.pbio.1001906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abbosh C, et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature. 2023;616:553–562. doi: 10.1038/s41586-023-05776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bailey C, et al. Tracking cancer evolution through the disease course. Cancer Discov. 2021;11:916–932. doi: 10.1158/2159-8290.CD-20-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al Bakir M, et al. The evolution of non-small cell lung cancer metastases in TRACERx. Nature. 2023;616:534–542. doi: 10.1038/s41586-023-05729-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGranahan N, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171:1259–1271.e1211. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wani MC, Horwitz SB. Nature as a remarkable chemist: a personal story of the discovery and development of Taxol. Anticancer Drugs. 2014;25:482–487. doi: 10.1097/CAD.0000000000000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mason JM, et al. Functional characterization of CFI-402257, a potent and selective Mps1/TTK kinase inhibitor, for the treatment of cancer. Proc. Natl. Acad. Sci. 2017;114:3127–3132. doi: 10.1073/pnas.1700234114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thu KL, et al. Disruption of the anaphase-promoting complex confers resistance to TTK inhibitors in triple-negative breast cancer. Proc. Natl. Acad. Sci. 2018;115:E1570–E1577. doi: 10.1073/pnas.1719577115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan CY-K, et al. CFI-402257, a TTK inhibitor, effectively suppresses hepatocellular carcinoma. Proc. Natl Acad. Sci. 2022;119:e2119514119. doi: 10.1073/pnas.2119514119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma J, Chan JJ, Toh CH, Yap YS. Emerging systemic therapy options beyond CDK4/6 inhibitors for hormone receptor-positive HER2-negative advanced breast cancer. NPJ Breast Cancer. 2023;9:74. doi: 10.1038/s41523-023-00578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bakhoum SF. Targeting the undruggable. Science. 2023;380:47. doi: 10.1126/science.adg7671. [DOI] [PubMed] [Google Scholar]
- 69.McGranahan N, et al. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–538. doi: 10.1038/embor.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bakhoum SF, et al. The mitotic origin of chromosomal instability. Curr. Biol. 2014;24:R148–R149. doi: 10.1016/j.cub.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He B, et al. Chromosomes missegregated into micronuclei contribute to chromosomal instability by missegregating at the next division. Oncotarget. 2019;10:2660. doi: 10.18632/oncotarget.26853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zasadil LM, Britigan EMC, Weaver BA. 2n or not 2n: aneuploidy, polyploidy and chromosomal instability in primary and tumor cells. Semin. Cell Dev. Biol. 2013;24:370–379. doi: 10.1016/j.semcdb.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nicholson JM, et al. Chromosome mis-segregation and cytokinesis failure in trisomic human cells. elife. 2015;4:e05068. doi: 10.7554/eLife.05068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 75.Passerini V, et al. The presence of extra chromosomes leads to genomic instability. Nat. Commun. 2016;7:10754. doi: 10.1038/ncomms10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ricke RM, van Ree JH, van Deursen JM. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 2008;24:457–466. doi: 10.1016/j.tig.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gantchev, J. et al. Tools used to assay genomic instability in cancers and cancer meiomitosis. J. Cell Commun. Signal. 1–19 (2021). [DOI] [PMC free article] [PubMed]
- 78.Krupina K, Goginashvili A, Cleveland DW. Causes and consequences of micronuclei. Curr. Opin. Cell Biol. 2021;70:91–99. doi: 10.1016/j.ceb.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bolhaqueiro AC, et al. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet. 2019;51:824–834. doi: 10.1038/s41588-019-0399-6. [DOI] [PubMed] [Google Scholar]
- 80.Hoevenaar WHM, et al. Degree and site of chromosomal instability define its oncogenic potential. Nat. Commun. 2020;11:1501. doi: 10.1038/s41467-020-15279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klaasen SJ, et al. Nuclear chromosome locations dictate segregation error frequencies. Nature. 2022;607:604–609. doi: 10.1038/s41586-022-04938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fenech M, et al. Micronuclei as biomarkers of DNA damage, aneuploidy, inducers of chromosomal hypermutation and as sources of pro-inflammatory DNA in humans. Mutat. Res. Rev. Mutat. Res. 2020;786:108342. doi: 10.1016/j.mrrev.2020.108342. [DOI] [PubMed] [Google Scholar]
- 84.Gomes AM, et al. Micronuclei from misaligned chromosomes that satisfy the spindle assembly checkpoint in cancer cells. Curr. Biol. 2022;32:4240–4254. e4245. doi: 10.1016/j.cub.2022.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Toorn M, Gooch A, Boerner S, Kiyomitsu T. NuMA deficiency causes micronuclei via checkpoint-insensitive k-fiber minus-end detachment from mitotic spindle poles. Curr. Biol. 2023;33:572–580.e572. doi: 10.1016/j.cub.2022.12.017. [DOI] [PubMed] [Google Scholar]
- 86.Piemonte KM, et al. Disruption of CDK7 signaling leads to catastrophic chromosomal instability coupled with a loss of condensin-mediated chromatin compaction. J. Biol. Chem. 2023;299:104834. doi: 10.1016/j.jbc.2023.104834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hodges CA, et al. SMC1beta-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat. Genet. 2005;37:1351–1355. doi: 10.1038/ng1672. [DOI] [PubMed] [Google Scholar]
- 88.Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- 89.Ohbayashi T, et al. Unscheduled overexpression of human WAPL promotes chromosomal instability. Biochem. Biophys. Res. Commun. 2007;356:699–704. doi: 10.1016/j.bbrc.2007.03.037. [DOI] [PubMed] [Google Scholar]
- 90.Carvajal-Maldonado D, et al. Perturbing cohesin dynamics drives MRE11 nuclease-dependent replication fork slowing. Nucleic Acids Res. 2019;47:1294–1310. doi: 10.1093/nar/gky519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leylek TR, Jeusset LM, Lichtensztejn Z, McManus KJ. Reduced expression of genes regulating cohesion induces chromosome instability that may promote cancer and impact patient outcomes. Sci. Rep. 2020;10:592. doi: 10.1038/s41598-020-57530-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deardorff MA, et al. RAD21 mutations cause a human cohesinopathy. Am. J. Hum. Genet. 2012;90:1014–1027. doi: 10.1016/j.ajhg.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Solomon DA, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van der Lelij P, et al. STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers. Life Sci. Alliance. 2020;3:e202000725. doi: 10.26508/lsa.202000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.El Beaino M, et al. Loss of Stag2 cooperates with EWS-FLI1 to transform murine mesenchymal stem cells. BMC Cancer. 2020;20:1–14. doi: 10.1186/s12885-019-6465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nam H-J, van Deursen JM. Cyclin B2 and p53 control proper timing of centrosome separation. Nat. Cell Biol. 2014;16:535–546. doi: 10.1038/ncb2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Prigent C, Uzbekov R. Duplication and segregation of centrosomes during cell division. Cells. 2022;11:2445. doi: 10.3390/cells11152445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coelho PA, et al. Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol. 2015;5:150209. doi: 10.1098/rsob.150209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr. Biol. 2009;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell. 2004;7:637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 102.Jallepalli PV, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nat. Rev. Cancer. 2001;1:109–117. doi: 10.1038/35101065. [DOI] [PubMed] [Google Scholar]
- 103.Burds AA, Lutum AS, Sorger PK. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc. Natl. Acad. Sci. 2005;102:11296–11301. doi: 10.1073/pnas.0505053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Foijer F, et al. Chromosome instability induced by Mps1 and p53 mutation generates aggressive lymphomas exhibiting aneuploidy-induced stress. Proc. Natl. Acad. Sci. 2014;111:13427–13432. doi: 10.1073/pnas.1400892111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Simon JE, Bakker B, Foijer F. CINcere modelling: what have mouse models for chromosome instability taught us? Recent Results Cancer Res. 2015;200:39–60. doi: 10.1007/978-3-319-20291-4_2. [DOI] [PubMed] [Google Scholar]
- 106.Iwanaga Y, et al. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 107.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 108.Dai W, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–445. doi: 10.1158/0008-5472.CAN-03-3119. [DOI] [PubMed] [Google Scholar]
- 109.Suijkerbuijk SJ, et al. Molecular causes for BUBR1 dysfunction in the human cancer predisposition syndrome mosaic variegated aneuploidy. Cancer Res. 2010;70:4891–4900. doi: 10.1158/0008-5472.CAN-09-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hanks S, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 111.Prinz F, et al. Functional and structural characterization of Bub3·BubR1 interactions required for spindle assembly checkpoint signaling in human cells. J. Biol. Chem. 2016;291:11252–11267. doi: 10.1074/jbc.M115.702142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Carvalhal S, et al. Biallelic BUB1 mutations cause microcephaly, developmental delay, and variable effects on cohesion and chromosome segregation. Sci. Adv. 2022;8:eabk0114. doi: 10.1126/sciadv.abk0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cimini D, Cameron LA, Salmon ED. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr. Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 115.Crasta K, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Orr B, et al. An anaphase surveillance mechanism prevents micronuclei formation from frequent chromosome segregation errors. Cell Rep. 2021;37:109783. doi: 10.1016/j.celrep.2021.109783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wilhelm T, et al. Mild replication stress causes chromosome mis-segregation via premature centriole disengagement. Nat. Commun. 2019;10:3585. doi: 10.1038/s41467-019-11584-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- 120.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 121.Schvartzman J-M, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat. Rev. Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rode A, et al. Chromothripsis in cancer cells: an update. Int. J. Cancer. 2016;138:2322–2333. doi: 10.1002/ijc.29888. [DOI] [PubMed] [Google Scholar]
- 123.Ly P, et al. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 2017;19:68–75. doi: 10.1038/ncb3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Leibowitz ML, Zhang C-Z, Pellman D. Chromothripsis: a new mechanism for rapid karyotype evolution. Annu. Rev. Genet. 2015;49:183–211. doi: 10.1146/annurev-genet-120213-092228. [DOI] [PubMed] [Google Scholar]
- 126.Zhang C-Z, et al. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shoshani O, et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature. 2021;591:137–141. doi: 10.1038/s41586-020-03064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Umbreit NT, et al. Mechanisms generating cancer genome complexity from a single cell division error. Science. 2020;368:eaba0712. doi: 10.1126/science.aba0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Joo YK, et al. ATR promotes clearance of damaged DNA and damaged cells by rupturing micronuclei. Mol. Cell. 2023;83:3642–3658. e3644. doi: 10.1016/j.molcel.2023.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ly P, et al. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat. Genet. 2019;51:705–715. doi: 10.1038/s41588-019-0360-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li Y, et al. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature. 2014;508:98–102. doi: 10.1038/nature13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sanders AD, et al. Single-cell analysis of structural variations and complex rearrangements with tri-channel processing. Nat. Biotechnol. 2020;38:343–354. doi: 10.1038/s41587-019-0366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li, J. S. Z. et al. Chromosomal fragile site breakage by EBV-encoded EBNA1 at clustered repeats. Nature 1–6 (2023). [DOI] [PMC free article] [PubMed]
- 134.Gaillard H, García-Muse T, Aguilera A. Replication stress and cancer. Nat. Rev. Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 135.Sahgal P, et al. Replicative stress in gastroesophageal cancer is associated with chromosomal instability and sensitivity to DNA damage response inhibitors. iScience. 2023;26:108169. doi: 10.1016/j.isci.2023.108169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shaikh N, et al. Replication stress generates distinctive landscapes of DNA copy number alterations and chromosome scale losses. Genome Biol. 2022;23:1–24. doi: 10.1186/s13059-022-02781-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang R-X, Zhou P-K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target. Ther. 2020;5:60. doi: 10.1038/s41392-020-0150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 140.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 141.Garribba L, et al. Short-term molecular consequences of chromosome mis-segregation for genome stability. Nat. Commun. 2023;14:1353. doi: 10.1038/s41467-023-37095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bakhoum SF, et al. Mitotic DNA damage response: at the crossroads of structural and numerical cancer chromosome instabilities. Trends Cancer. 2017;3:225–234. doi: 10.1016/j.trecan.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer. 2016;16:35–42. doi: 10.1038/nrc.2015.4. [DOI] [PubMed] [Google Scholar]
- 144.Li J, et al. The RPA–RNF20–SNF2H cascade promotes proper chromosome segregation and homologous recombination repair. Proc. Natl. Acad. Sci. 2023;120:e2303479120. doi: 10.1073/pnas.2303479120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 147.Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Biol. 2009;11:761–768. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- 148.Howlett NG, et al. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum. Mol. Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 149.Constantinou A. Rescue of replication failure by Fanconi anaemia proteins. Chromosoma. 2012;121:21–36. doi: 10.1007/s00412-011-0349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Madireddy A, et al. FANCD2 facilitates replication through common fragile sites. Mol. Cell. 2016;64:388–404. doi: 10.1016/j.molcel.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Boteva L, et al. Common fragile sites are characterized by faulty condensin loading after replication stress. Cell Rep. 2020;32:108177. doi: 10.1016/j.celrep.2020.108177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Böhly N, et al. Increased replication origin firing links replication stress to whole chromosomal instability in human cancer. Cell Rep. 2022;41:111836. doi: 10.1016/j.celrep.2022.111836. [DOI] [PubMed] [Google Scholar]
- 153.Di Nardo M, Pallotta MM, Musio A. The multifaceted roles of cohesin in cancer. J. Exp. Clin. Cancer Res. 2022;41:1–11. doi: 10.1186/s13046-022-02321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sun Y, et al. RAD21 is the core subunit of the cohesin complex involved in directing genome organization. Genome Biol. 2023;24:1–27. doi: 10.1186/s13059-023-02982-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/S1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 156.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/S0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 157.Cuadrado A, Losada A. Specialized functions of cohesins STAG1 and STAG2 in 3D genome architecture. Curr. Opin. Genet. Dev. 2020;61:9–16. doi: 10.1016/j.gde.2020.02.024. [DOI] [PubMed] [Google Scholar]
- 158.Shi Z, Gao H, Bai X-C, Yu H. Cryo-EM structure of the human cohesin-NIPBL-DNA complex. Science. 2020;368:1454–1459. doi: 10.1126/science.abb0981. [DOI] [PubMed] [Google Scholar]
- 159.Watanabe Y. Sister chromatid cohesion along arms and at centromeres. Trends Genet. 2005;21:405–412. doi: 10.1016/j.tig.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 160.Gerton J. Chromosome cohesion: a cycle of holding together and falling apart. PLoS Biol. 2005;3:e94. doi: 10.1371/journal.pbio.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shintomi K, Hirano T. Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl–Pds5 and Sgo1. Genes Dev. 2009;23:2224–2236. doi: 10.1101/gad.1844309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tillement V, et al. Spindle assembly defects leading to the formation of a monopolar mitotic apparatus. Biol. Cell. 2009;101:1–11. doi: 10.1042/BC20070162. [DOI] [PubMed] [Google Scholar]
- 163.Yoshizawa K, Yaguchi K, Uehara R. Uncoupling of DNA replication and centrosome duplication cycles is a primary cause of haploid instability in mammalian somatic cells. Front. Cell Dev. Biol. 2020;30:721. doi: 10.3389/fcell.2020.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.van Ree JH, et al. Pten regulates spindle pole movement through Dlg1-mediated recruitment of Eg5 to centrosomes. Nat. Cell Biol. 2016;18:814–821. doi: 10.1038/ncb3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Silkworth WT, et al. Timing of centrosome separation is important for accurate chromosome segregation. Mol. Biol. Cell. 2012;23:401–411. doi: 10.1091/mbc.e11-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang Y, et al. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J. Clin. Investig. 2012;122:4362–4374. doi: 10.1172/JCI63084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Saavedra HI, Fukasawa K, Conn CW, Stambrook PJ. MAPK mediates RAS-induced chromosome instability. J. Biol. Chem. 1999;274:38083–38090. doi: 10.1074/jbc.274.53.38083. [DOI] [PubMed] [Google Scholar]
- 168.Chan JY. A clinical overview of centrosome amplification in human cancers. Int. J. Biol. Sci. 2011;7:1122–1144. doi: 10.7150/ijbs.7.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nigg EA, Holland AJ. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 2018;19:297–312. doi: 10.1038/nrm.2017.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PloS One. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cosper PF, et al. HPV16 E6 induces chromosomal instability due to polar chromosomes caused by E6AP-dependent degradation of the mitotic kinesin CENP-E. Proc. Natl. Acad. Sci. 2023;120:e2216700120. doi: 10.1073/pnas.2216700120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bühler M, et al. GPER1 links estrogens to centrosome amplification and chromosomal instability in human colon cells. Life Sci. Alliance. 2023;6:e202201499. doi: 10.26508/lsa.202201499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gregan J, et al. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 2011;21:374–381. doi: 10.1016/j.tcb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Thompson SL, Compton DA. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc. Natl Acad. Sci. 2011;108:17974–17978. doi: 10.1073/pnas.1109720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cimini D. Twenty years of merotelic kinetochore attachments: a historical perspective. Chromosom. Res. 2023;31:18. doi: 10.1007/s10577-023-09727-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kops GJ, Gassmann R. Crowning the kinetochore: the fibrous corona in chromosome segregation. Trends Cell Biol. 2020;30:653–667. doi: 10.1016/j.tcb.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 178.Song X, et al. Dynamic crotonylation of EB1 by TIP60 ensures accurate spindle positioning in mitosis. Nat. Chem. Biol. 2021;17:1314–1323. doi: 10.1038/s41589-021-00875-7. [DOI] [PubMed] [Google Scholar]
- 179.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 180.Navarro AP, Cheeseman IM. Kinetochore assembly throughout the cell cycle. Semin. Cell. Dev. Biol. 2021;117:62–74. doi: 10.1016/j.semcdb.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ertych N, et al. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat. Cell Biol. 2014;16:779–791. doi: 10.1038/ncb2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kabeche L, Compton DA. Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in human cells. Curr. Biol. 2012;22:638–644. doi: 10.1016/j.cub.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kleyman M, Kabeche L, Compton DA. STAG2 promotes error correction in mitosis by regulating kinetochore-microtubule attachments. J. Cell Sci. 2014;127:4225–4233. doi: 10.1242/jcs.151613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chan GK, Liu S-T, Yen TJ. Kinetochore structure and function. Trends Cell Biol. 2005;15:589–598. doi: 10.1016/j.tcb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 185.Musacchio A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 2015;25:R1002–R1018. doi: 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 186.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 187.McAinsh AD, Kops GJ. Principles and dynamics of spindle assembly checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2023;24:543–559. doi: 10.1038/s41580-023-00593-z. [DOI] [PubMed] [Google Scholar]
- 188.London N, Biggins S. Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 2014;15:736–747. doi: 10.1038/nrm3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Liu S, et al. Mad2 promotes Cyclin B2 recruitment to the kinetochore for guiding accurate mitotic checkpoint. EMBO Rep. 2022;23:e54171. doi: 10.15252/embr.202154171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 192.Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 193.Brady DM, Hardwick KG. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 2000;10:675–678. doi: 10.1016/S0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- 194.Basu J, et al. Localization of the Drosophila checkpoint control protein Bub3 to the kinetochore requires Bub1 but not Zw10 or Rod. Chromosoma. 1998;107:376–385. doi: 10.1007/s004120050321. [DOI] [PubMed] [Google Scholar]
- 195.Jablonski SA, et al. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107:386–396. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- 196.Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/S0092-8674(00)80255-X. [DOI] [PubMed] [Google Scholar]
- 197.Chen R-H. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J Cell Biol. 2002;158:487–496. doi: 10.1083/jcb.200204048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lopes CS, et al. The Drosophila Bub3 protein is required for the mitotic checkpoint and for normal accumulation of cyclins during G2 and early stages of mitosis. J. Cell Sci. 2005;118:187–198. doi: 10.1242/jcs.01602. [DOI] [PubMed] [Google Scholar]
- 199.Babu JR, et al. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kalitsis P, Earle E, Fowler KJ, Choo KH. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14:2277–2282. doi: 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhou Z, He M, Shah AA, Wan Y. Insights into APC/C: from cellular function to diseases and therapeutics. Cell Div. 2016;11:9. doi: 10.1186/s13008-016-0021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Izawa D, Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2015;517:631–634. doi: 10.1038/nature13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chen OJ, et al. Germline missense variants in CDC20 result in aberrant mitotic progression and familial cancer. Cancer Res. 2022;82:3499–3515. doi: 10.1158/0008-5472.CAN-21-3956. [DOI] [PubMed] [Google Scholar]
- 206.Tsang MJ, Cheeseman IM. Alternative CDC20 translational isoforms tune mitotic arrest duration. Nature. 2023;617:154–161. doi: 10.1038/s41586-023-05943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Alfieri C, et al. Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature. 2016;536:431–436. doi: 10.1038/nature19083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lara-Gonzalez P, et al. A tripartite mechanism catalyzes Mad2-Cdc20 assembly at unattached kinetochores. Science. 2021;371:64–67. doi: 10.1126/science.abc1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Piano V, et al. CDC20 assists its catalytic incorporation in the mitotic checkpoint complex. Science. 2021;371:67–71. doi: 10.1126/science.abc1152. [DOI] [PubMed] [Google Scholar]
- 210.Fischer ES, et al. Juxtaposition of Bub1 and Cdc20 on phosphorylated Mad1 during catalytic mitotic checkpoint complex assembly. Nat. Commun. 2022;13:6381. doi: 10.1038/s41467-022-34058-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sivakumar S, Gorbsky GJ. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 2015;16:82–94. doi: 10.1038/nrm3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lara-Gonzalez P, Pines J, Desai A. Spindle assembly checkpoint activation and silencing at kinetochores. Semin. Cell. Dev. Biol. 2021;117:86–98. doi: 10.1016/j.semcdb.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kapanidou M, Curtis NL, Bolanos-Garcia VM. Cdc20: at the crossroads between chromosome segregation and mitotic exit. Trends Biochem. Sci. 2017;42:193–205. doi: 10.1016/j.tibs.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 214.Vagnarelli P. Back to the new beginning: mitotic exit in space and time. Semin. Cell. Dev. Biol. 2021;117:140–148. doi: 10.1016/j.semcdb.2021.03.010. [DOI] [PubMed] [Google Scholar]
- 215.Moreno-Andrés D, Holl K, Antonin W. The second half of mitosis and its implications in cancer biology. Semin. Cancer. Biol. 2022;88:1–17. doi: 10.1016/j.semcancer.2022.11.013. [DOI] [PubMed] [Google Scholar]
- 216.Gao Y-F, et al. Cdk1-phosphorylated CUEDC2 promotes spindle checkpoint inactivation and chromosomal instability. Nat. Cell Biol. 2011;13:924–933. doi: 10.1038/ncb2287. [DOI] [PubMed] [Google Scholar]
- 217.Cosper PF, Copeland SE, Tucker JB, Weaver BA. Chromosome missegregation as a modulator of radiation sensitivity. Semin. Radiat. Oncol. 2022;32:54–63. doi: 10.1016/j.semradonc.2021.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.He Z, et al. JMJD5 (Jumonji Domain-containing 5) associates with spindle microtubules and is required for proper mitosis. J. Biol. Chem. 2016;291:4684–4697. doi: 10.1074/jbc.M115.672642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yatskevich S, et al. Molecular mechanisms of APC/C release from spindle assembly checkpoint inhibition by APC/C SUMOylation. Cell Rep. 2021;34:108929. doi: 10.1016/j.celrep.2021.108929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rowald K, et al. Negative selection and chromosome instability induced by Mad2 overexpression delay breast cancer but facilitate oncogene-independent outgrowth. Cell Rep. 2016;15:2679–2691. doi: 10.1016/j.celrep.2016.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sotillo R, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Choi E, Zhang X, Xing C, Yu H. Mitotic checkpoint regulators control insulin signaling and metabolic homeostasis. Cell Rep. 2016;166:567–581. doi: 10.1016/j.cell.2016.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ma HT, Poon RYC. TRIP13 regulates both the activation and inactivation of the spindle-assembly checkpoint. Cell Rep. 2016;14:1086–1099. doi: 10.1016/j.celrep.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 224.Kim DH, et al. TRIP13 and APC15 drive mitotic exit by turnover of interphase-and unattached kinetochore-produced MCC. Nat. Commun. 2018;9:4354. doi: 10.1038/s41467-018-06774-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gong Y, et al. Loss of RanGAP1 drives chromosome instability and rapid tumorigenesis of osteosarcoma. Dev. Cell. 2023;58:192–210. e111. doi: 10.1016/j.devcel.2022.12.012. [DOI] [PubMed] [Google Scholar]
- 226.Sapkota H, Wasiak E, Daum JR, Gorbsky GJ. Multiple determinants and consequences of cohesion fatigue in mammalian cells. Mol. Biol. Cell. 2018;29:1811–1824. doi: 10.1091/mbc.E18-05-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Gorbsky GJ. Cohesion fatigue. Curr. Biol. 2013;23:R986–R988. doi: 10.1016/j.cub.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 228.Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- 229.Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu. Rev. Cell Dev. Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 230.Kuznetsova AY, et al. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle. 2015;14:2810–2820. doi: 10.1080/15384101.2015.1068482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wangsa D, et al. Near-tetraploid cancer cells show chromosome instability triggered by replication stress and exhibit enhanced invasiveness. FASEB J. 2018;32:3502. doi: 10.1096/fj.201700247RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Selmecki AM, et al. Polyploidy can drive rapid adaptation in yeast. Nature. 2015;519:349–352. doi: 10.1038/nature14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhang J, et al. Human cell polyploidization: the good and the evil. Semin. Cancer. Biol. 2022;81:54–63. doi: 10.1016/j.semcancer.2021.04.005. [DOI] [PubMed] [Google Scholar]
- 234.Steigemann P, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 235.Ganem NJ, et al. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 2014;158:833–848. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Darp R, Vittoria MA, Ganem NJ, Ceol CJ. Oncogenic BRAF induces whole-genome doubling through suppression of cytokinesis. Nat. Commun. 2022;13:4109. doi: 10.1038/s41467-022-31899-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ghelli Luserna di Rorà A, Martinelli G, Simonetti G. The balance between mitotic death and mitotic slippage in acute leukemia: a new therapeutic window? J. Hematol. Oncol. 2019;12:1–16. doi: 10.1186/s13045-019-0808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lok TM, et al. Mitotic slippage is determined by p31comet and the weakening of the spindle-assembly checkpoint. Oncogene. 2020;39:2819–2834. doi: 10.1038/s41388-020-1187-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sanz-Gómez N, González-Álvarez M, De Las Rivas J, de Cárcer G. Whole-genome doubling as a source of cancer: how, when, where, and why? Front. Cell Dev. Biol. 2023;11:1209136. doi: 10.3389/fcell.2023.1209136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Coulombe P, et al. The ORC ubiquitin ligase OBI1 promotes DNA replication origin firing. Nat. Commun. 2019;10:2426. doi: 10.1038/s41467-019-10321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Fujita M. Cdt1 revisited: complex and tight regulation during the cell cycle and consequences of deregulation in mammalian cells. Cell Div. 2006;1:1–9. doi: 10.1186/1747-1028-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Vittoria MA, Quinton RJ, Ganem NJ. Whole-genome doubling in tissues and tumors. Trends Genet. 2023;39:954–867. doi: 10.1016/j.tig.2023.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lau TY, Poon RY. Whole-genome duplication and genome instability in cancer cells: double the trouble. Int. J. Mol. Sci. 2023;24:3733. doi: 10.3390/ijms24043733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Prasad K, et al. Whole-genome duplication shapes the aneuploidy landscape of human cancers. Cancer Res. 2022;82:1736–1752. doi: 10.1158/0008-5472.CAN-21-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Bielski CM, et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat. Genet. 2018;50:1189–1195. doi: 10.1038/s41588-018-0165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Gemble S, et al. Genetic instability from a single S phase after whole-genome duplication. Nature. 2022;604:146–151. doi: 10.1038/s41586-022-04578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zhang H, et al. Cell Fusion-related proteins and signaling pathways, and their roles in the development and progression of cancer. Front. Cell Dev. Biol. 2022;9:809668. doi: 10.3389/fcell.2021.809668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–776. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Jamal-Hanjani M, et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 250.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lambuta RA, et al. Whole-genome doubling drives oncogenic loss of chromatin segregation. Nature. 2023;615:925–933. doi: 10.1038/s41586-023-05794-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121:3859–3866. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- 253.Potapova T, Gorbsky GJ. The consequences of chromosome segregation errors in mitosis and meiosis. Biology. 2017;6:12. doi: 10.3390/biology6010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 2015;16:473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- 255.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Selmecki A, et al. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 2008;68:624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 257.Pavelka N, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Rancati G, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482:246–250. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Rancati G, Pavelka N. Karyotypic changes as drivers and catalyzers of cellular evolvability: a perspective from non-pathogenic yeasts. Semin. Cell Dev. Biol. 2013;24:332–338. doi: 10.1016/j.semcdb.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 261.Ben-David U, Amon A. Context is everything: aneuploidy in cancer. Nat. Rev. Genet. 2020;21:44–62. doi: 10.1038/s41576-019-0171-x. [DOI] [PubMed] [Google Scholar]
- 262.Snape K, et al. Mutations in CEP57 cause mosaic variegated aneuploidy syndrome. Nat. Genet. 2011;43:527–529. doi: 10.1038/ng.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Garcia-Castillo H, Vasquez-Velasquez AI, Rivera H, Barros-Nunez P. Clinical and genetic heterogeneity in patients with mosaic variegated aneuploidy: delineation of clinical subtypes. Am. J. Med. Genet. A. 2008;146A:1687–1695. doi: 10.1002/ajmg.a.32315. [DOI] [PubMed] [Google Scholar]
- 264.Stingele S, et al. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 267.López-García C, et al. BCL9L dysfunction impairs Caspase-2 expression permitting aneuploidy tolerance in colorectal cancer. Cancer Cell. 2017;31:79–93. doi: 10.1016/j.ccell.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ohashi A, et al. Aneuploidy generates proteotoxic stress and DNA damage concurrently with p53-mediated post-mitotic apoptosis in SAC-impaired cells. Nat. Commun. 2015;6:7668. doi: 10.1038/ncomms8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Meena JK, et al. Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion. EMBO J. 2017;36:2922–2924. doi: 10.15252/embj.201797470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Santaguida S, et al. Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev. Cell. 2017;41:638–651.e635. doi: 10.1016/j.devcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Andriani GA, et al. Whole chromosome instability induces senescence and promotes SASP. Sci. Rep. 2016;6:35218. doi: 10.1038/srep35218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Vigano C, et al. Quantitative proteomic and phosphoproteomic comparison of human colon cancer DLD-1 cells differing in ploidy and chromosome stability. Mol. Biol. Cell. 2018;29:1031–1047. doi: 10.1091/mbc.E17-10-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Krivega M, et al. Genotoxic stress in constitutive trisomies induces autophagy and the innate immune response via the cGAS-STING pathway. Commun. Biol. 2021;4:831. doi: 10.1038/s42003-021-02278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Birkbak NJ, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71:3447–3452. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zaki BI, et al. Chromosomal instability portends superior response of rectal adenocarcinoma to chemoradiation therapy. Cancer. 2014;120:1733–1742. doi: 10.1002/cncr.28656. [DOI] [PubMed] [Google Scholar]
- 276.Andor N, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 2016;22:105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Bakhoum SF, et al. Numerical chromosomal instability mediates susceptibility to radiation treatment. Nat. Commun. 2015;6:5990. doi: 10.1038/ncomms6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Jamal-Hanjani M, et al. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: a prospective validation cohort study from the TACT trial. Ann. Oncol. 2015;26:1340–1346. doi: 10.1093/annonc/mdv178. [DOI] [PubMed] [Google Scholar]
- 279.Roylance R, et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol. Biomark. Prev. 2011;20:2183–2194. doi: 10.1158/1055-9965.EPI-11-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Laughney AM, Elizalde S, Genovese G, Bakhoum SF. Dynamics of tumor heterogeneity derived from clonal karyotypic evolution. Cell Rep. 2015;12:809–820. doi: 10.1016/j.celrep.2015.06.065. [DOI] [PubMed] [Google Scholar]
- 281.Komarova NL, Wodarz D. The optimal rate of chromosome loss for the inactivation of tumor suppressor genes in cancer. Proc. Natl Acad. Sci. 2004;101:7017–7021. doi: 10.1073/pnas.0401943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Chunduri NK, Barthel K, Storchova Z. Consequences of chromosome loss: why do cells need each chromosome twice? Cells. 2022;11:1530. doi: 10.3390/cells11091530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Chunduri NK, et al. Systems approaches identify the consequences of monosomy in somatic human cells. Nat. Commun. 2021;12:5576. doi: 10.1038/s41467-021-25288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Karoutas A, Akhtar A. Functional mechanisms and abnormalities of the nuclear lamina. Nat. Cell Biol. 2021;23:116–126. doi: 10.1038/s41556-020-00630-5. [DOI] [PubMed] [Google Scholar]
- 285.Liu S, et al. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature. 2018;561:551–555. doi: 10.1038/s41586-018-0534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Mammel AE, Hatch EM. Genome instability from nuclear catastrophe and DNA damage. Semin. Cell. Dev. Biol. 2022;123:131–139. doi: 10.1016/j.semcdb.2021.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Cortés-Ciriano I, et al. Comprehensive analysis of chromothripsis in 2658 human cancers using whole-genome sequencing. Nat. Genet. 2020;52:331–341. doi: 10.1038/s41588-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Trivedi P, et al. Mitotic tethering enables inheritance of shattered micronuclear chromosomes. Nature. 2023;618:1049–1056. doi: 10.1038/s41586-023-06216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Lin Y-F, et al. Mitotic clustering of pulverized chromosomes from micronuclei. Nature. 2023;618:1041–1048. doi: 10.1038/s41586-023-05974-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Yi E, Chamorro González R, Henssen AG, Verhaak RG. Extrachromosomal DNA amplifications in cancer. Nat. Rev. Genet. 2022;23:760–771. doi: 10.1038/s41576-022-00521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Yamamoto K, et al. Micronuclei‐associated MYC amplification in the form of double minute chromosomes in acute myeloid leukemia. Am. J. Hematol. 2013;88:717–718. doi: 10.1002/ajh.23431. [DOI] [PubMed] [Google Scholar]
- 292.Nones K, et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat. Commun. 2014;5:5224. doi: 10.1038/ncomms6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Martins FC, et al. Clonal somatic copy number altered driver events inform drug sensitivity in high-grade serous ovarian cancer. Nat. Commun. 2022;13:6360. doi: 10.1038/s41467-022-33870-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Notta F, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538:378–382. doi: 10.1038/nature19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Kloosterman WP, et al. A systematic analysis of oncogenic gene fusions in primary colon cancer. Cancer Res. 2017;77:3814–3822. doi: 10.1158/0008-5472.CAN-16-3563. [DOI] [PubMed] [Google Scholar]
- 296.Agustinus AS, et al. Epigenetic dysregulation from chromosomal transit in micronuclei. Nature. 2023;619:176–183. doi: 10.1038/s41586-023-06084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Papathanasiou S, et al. Heritable transcriptional defects from aberrations of nuclear architecture. Nature. 2023;619:184–192. doi: 10.1038/s41586-023-06157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Donnelly N, Storchová Z. Aneuploidy and proteotoxic stress in cancer. Mol. Cell Oncol. 2015;2:e976491. doi: 10.4161/23723556.2014.976491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Zhu J, Tsai H-J, Gordon MR, Li R. Cellular stress associated with aneuploidy. Dev. Cell. 2018;44:420–431. doi: 10.1016/j.devcel.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Oromendia AB, Amon A. Aneuploidy: implications for protein homeostasis and disease. Dis. Model Mech. 2014;7:15–20. doi: 10.1242/dmm.013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 2012;26:2696–2708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Schukken KM, Sheltzer JM. Extensive protein dosage compensation in aneuploid human cancers. Genome Res. 2022;32:1254–1270. doi: 10.1101/gr.276378.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Cheng P, et al. Proteogenomic analysis of cancer aneuploidy and normal tissues reveals divergent modes of gene regulation across cellular pathways. Elife. 2022;11:e75227. doi: 10.7554/eLife.75227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Ishikawa K. Multilayered regulation of proteome stoichiometry. Curr. Genet. 2021;67:883–890. doi: 10.1007/s00294-021-01205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Taggart JC, et al. Keeping the proportions of protein complex components in check. Cell Syst. 2020;10:125–132. doi: 10.1016/j.cels.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.McShane E, et al. Kinetic analysis of protein stability reveals age-dependent degradation. Cell. 2016;167:803–815 e821. doi: 10.1016/j.cell.2016.09.015. [DOI] [PubMed] [Google Scholar]
- 307.Torres EM. Consequences of gaining an extra chromosome. Chromosom. Res. 2023;31:1–19. doi: 10.1007/s10577-023-09732-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Krivega M, Storchova Z. Consequences of trisomy syndromes–21 and beyond. Trends Genet. 2022;39:172–174. doi: 10.1016/j.tig.2022.11.004. [DOI] [PubMed] [Google Scholar]
- 309.Anders KR, et al. A strategy for constructing aneuploid yeast strains by transient nondisjunction of a target chromosome. BMC genet. 2009;10:1–11. doi: 10.1186/1471-2156-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24:390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 311.Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 312.Bowers RR, et al. SWAN pathway-network identification of common aneuploidy-based oncogenic drivers. Nucleic Acids Res. 2022;50:3673–3692. doi: 10.1093/nar/gkac200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Bean J, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc. Natl Acad. Sci. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Lulli M, et al. DNA damage response protein CHK2 regulates metabolism in liver cancer. Cancer Res. 2021;81:2861–2873. doi: 10.1158/0008-5472.CAN-20-3134. [DOI] [PubMed] [Google Scholar]
- 315.Li Y, et al. PDSS2 deficiency induces hepatocarcinogenesis by decreasing mitochondrial respiration and reprogramming glucose metabolism. Cancer Res. 2018;78:4471–4481. doi: 10.1158/0008-5472.CAN-17-2172. [DOI] [PubMed] [Google Scholar]
- 316.Thorburn RR, et al. Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol. Biol. Cell. 2013;24:1274–1289. doi: 10.1091/mbc.e12-07-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 318.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 319.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 320.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat. Rev. Cancer. 2021;21:669–680. doi: 10.1038/s41568-021-00378-6. [DOI] [PubMed] [Google Scholar]
- 322.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 323.Siegel JJ, Amon A. New insights into the troubles of aneuploidy. Annu. Rev. Cell Dev. Biol. 2012;28:189–214. doi: 10.1146/annurev-cellbio-101011-155807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Huang M, et al. Autonomous glucose metabolic reprogramming of tumour cells under hypoxia: opportunities for targeted therapy. J. Exp. Clin. Cancer Res. 2020;39:1–13. doi: 10.1186/s13046-020-01698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Wu S, et al. Transcription factor YY1 promotes cell proliferation by directly activating the pentose phosphate pathway. Cancer Res. 2018;78:4549–4562. doi: 10.1158/0008-5472.CAN-17-4047. [DOI] [PubMed] [Google Scholar]
- 326.Li Z, et al. NeuroD1 promotes tumor cell proliferation and tumorigenesis by directly activating the pentose phosphate pathway in colorectal carcinoma. Oncogene. 2021;40:6736–6747. doi: 10.1038/s41388-021-02063-2. [DOI] [PubMed] [Google Scholar]
- 327.Wang Y, et al. Yin Yang 1 promotes the Warburg effect and tumorigenesis via glucose transporter GLUT3. Cancer Sci. 2018;109:2423–2434. doi: 10.1111/cas.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Ciccarese F, Ciminale V. Escaping death: mitochondrial redox homeostasis in cancer cells. Front. Oncol. 2017;7:117. doi: 10.3389/fonc.2017.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Sheltzer JM. A transcriptional and metabolic signature of primary aneuploidy is present in chromosomally unstable cancer cells and informs clinical prognosis. Cancer Res. 2013;73:6401–6412. doi: 10.1158/0008-5472.CAN-13-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Santaguida S, Vasile E, White E, Amon A. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 2015;29:2010–2021. doi: 10.1101/gad.269118.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Joy J, et al. Proteostasis failure and mitochondrial dysfunction leads to aneuploidy-induced senescence. Dev. Cell. 2021;56:2043–2058. e2047. doi: 10.1016/j.devcel.2021.06.009. [DOI] [PubMed] [Google Scholar]
- 332.Liao Z, Chua D, Tan NS. Reactive oxygen species: a volatile driver of field cancerization and metastasis. Mol. Cancer. 2019;18:1–10. doi: 10.1186/s12943-019-0961-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Simões-Sousa S, et al. The p38α stress kinase suppresses aneuploidy tolerance by inhibiting Hif-1α. Cell Rep. 2018;25:749–760.e746. doi: 10.1016/j.celrep.2018.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Marques JF, Kops GJ. Permission to pass: on the role of p53 as a gatekeeper for aneuploidy. Chromosom. Res. 2023;31:31. doi: 10.1007/s10577-023-09741-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Replogle JM, et al. Aneuploidy increases resistance to chemotherapeutics by antagonizing cell division. Proc. Natl. Acad. Sci. 2020;117:30566–30576. doi: 10.1073/pnas.2009506117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Murai K, et al. p53 mutation in normal esophagus promotes multiple stages of carcinogenesis but is constrained by clonal competition. Nat. Commun. 2022;13:6206. doi: 10.1038/s41467-022-33945-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Huang C, et al. Identification of XBP1-u as a novel regulator of the MDM2/p53 axis using an shRNA library. Sci. Adv. 2017;3:e1701383. doi: 10.1126/sciadv.1701383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Joerger AC, Fersht AR. The p53 pathway: origins, inactivation in cancer, and emerging therapeutic approaches. Annu. Rev. Biochem. 2016;85:375–404. doi: 10.1146/annurev-biochem-060815-014710. [DOI] [PubMed] [Google Scholar]
- 340.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Levine AJ. p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer. 2020;20:471–480. doi: 10.1038/s41568-020-0262-1. [DOI] [PubMed] [Google Scholar]
- 342.Kuffer C, Kuznetsova AY, Storchová Z. Abnormal mitosis triggers p53-dependent cell cycle arrest in human tetraploid cells. Chromosoma. 2013;122:305–318. doi: 10.1007/s00412-013-0414-0. [DOI] [PubMed] [Google Scholar]
- 343.Levine DA, The Cancer Genome Atlas Research Network Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Tang R, et al. Colorectal cancer without high microsatellite instability and chromosomal instability-an alternative genetic pathway to human colorectal cancer. Carcinogenesis. 2004;25:841–846. doi: 10.1093/carcin/bgh074. [DOI] [PubMed] [Google Scholar]
- 345.Adell MAY, et al. Adaptation to spindle assembly checkpoint inhibition through the selection of specific aneuploidies. Genes Dev. 2023;37:171–190. doi: 10.1101/gad.350182.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Hinchcliffe EH, et al. Chromosome missegregation during anaphase triggers p53 cell cycle arrest through histone H3.3 Ser31 phosphorylation. Nat. Cell Biol. 2016;18:668–675. doi: 10.1038/ncb3348. [DOI] [PubMed] [Google Scholar]
- 347.Soto M, et al. p53 prohibits propagation of chromosome segregation errors that produce structural aneuploidies. Cell Rep. 2017;19:2423–2431. doi: 10.1016/j.celrep.2017.05.055. [DOI] [PubMed] [Google Scholar]
- 348.Li M, et al. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc. Natl. Acad. Sci. 2010;107:14188–14193. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Foijer, F. et al. Deletion of the MAD2L1 spindle assembly checkpoint gene is tolerated in mouse models of acute T-cell lymphoma and hepatocellular carcinoma. eLife6 (2017). [DOI] [PMC free article] [PubMed]
- 351.Bao C, et al. Genomic signatures of past and present chromosomal instability in Barrett’s esophagus and early esophageal adenocarcinoma. Nat. Commun. 2023;14:6203. doi: 10.1038/s41467-023-41805-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Marei HE, et al. p53 signaling in cancer progression and therapy. Cancer Cell Int. 2021;21:1–15. doi: 10.1186/s12935-021-02396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Xu D, et al. Acetylation halts missense mutant p53 aggregation and rescues tumor suppression in non-small cell lung cancers. iScience. 2023;26:107003. doi: 10.1016/j.isci.2023.107003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Chi YH, et al. Spindle assembly checkpoint and p53 deficiencies cooperate for tumorigenesis in mice. Int. J. Cancer. 2009;124:1483–1489. doi: 10.1002/ijc.24094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Woo RA, Poon RY. Activated oncogenes promote and cooperate with chromosomal instability for neoplastic transformation. Genes Dev. 2004;18:1317–1330. doi: 10.1101/gad.1165204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Cannell IG, et al. A pleiotropic RNA-binding protein controls distinct cell cycle checkpoints to drive resistance of p53-defective tumors to chemotherapy. Cancer Cell. 2015;28:623–637. doi: 10.1016/j.ccell.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Dolado, I. & Nebreda, A. R. in Stress-Activated Protein Kinases 1st edn, Vol. 20 (eds Posas, F. & Nebreda, R. A.) Ch. 6 (Spinger Berlin, 2008).
- 358.Cánovas B, et al. Targeting p38α Increases DNA damage, chromosome instability, and the anti-tumoral response to taxanes in breast cancer cells. Cancer Cell. 2018;33:1094–1110.e1098. doi: 10.1016/j.ccell.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 359.Zhang T, et al. 125I seed promotes apoptosis in non-small lung cancer cells via the p38 mapk-mdm2-p53 signaling pathway. Front. Oncol. 2021;11:582511. doi: 10.3389/fonc.2021.582511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Clemente-Ruiz M, et al. Gene dosage imbalance contributes to chromosomal instability-induced tumorigenesis. Dev. Cell. 2016;36:290–302. doi: 10.1016/j.devcel.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 361.Maehara K, Takahashi K, Saitoh S. CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol. Cell Biol. 2010;30:2090–2104. doi: 10.1128/MCB.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat. Rev. Cancer. 2010;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Prieur A, Peeper DS. Cellular senescence in vivo: a barrier to tumorigenesis. Curr. Opin. Cell Biol. 2008;20:150–155. doi: 10.1016/j.ceb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 364.Kandala S, et al. Chronic chromosome instability induced by Plk1 results in immune suppression in breast cancer. Cell Rep. 2023;42:113266. doi: 10.1016/j.celrep.2023.113266. [DOI] [PubMed] [Google Scholar]
- 365.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 366.Uetake Y, Sluder G. Activation of the apoptotic pathway during prolonged prometaphase blocks daughter cell proliferation. Mol. Biol. Cell. 2018;29:2632–2643. doi: 10.1091/mbc.E18-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Brady CA, Attardi LD. p53 at a glance. J. Cell Sci. 2010;123:2527–2532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Nassour J, et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature. 2019;565:659–663. doi: 10.1038/s41586-019-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.He Q, et al. Chromosomal instability-induced senescence potentiates cell non-autonomous tumourigenic effects. Oncogenesis. 2018;7:62. doi: 10.1038/s41389-018-0072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Freund A, Orjalo AV, Desprez PY, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol. Med. 2010;16:238–246. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Pawlikowski JS, Adams PD, Nelson DM. Senescence at a glance. J. Cell Sci. 2013;126:4061–4067. doi: 10.1242/jcs.109728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Coppe JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Wörmann SM, et al. APOBEC3A drives deaminase domain-independent chromosomal instability to promote pancreatic cancer metastasis. Nat. Cancer. 2021;2:1338–1356. doi: 10.1038/s43018-021-00268-8. [DOI] [PubMed] [Google Scholar]
- 374.Dongre A, et al. Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res. 2017;77:3982–3989. doi: 10.1158/0008-5472.CAN-16-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Gerstberger S, Jiang Q, Ganesh K. Metastasis. Cell. 2023;186:1564–1579. doi: 10.1016/j.cell.2023.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Shoshani O, et al. Transient genomic instability drives tumorigenesis through accelerated clonal evolution. Genes Dev. 2021;35:1093–1108. doi: 10.1101/gad.348319.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Vasudevan A, et al. Single-chromosomal gains can function as metastasis suppressors and promoters in colon cancer. Dev. Cell. 2020;52:413–428. e416. doi: 10.1016/j.devcel.2020.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Gao C, et al. Chromosome instability drives phenotypic switching to metastasis. Proc. Natl. Acad. Sci. 2016;113:14793–14798. doi: 10.1073/pnas.1618215113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Bakir B, Chiarella AM, Pitarresi JR, Rustgi AK. EMT, MET, plasticity, and tumor metastasis. Trends Cell Biol. 2020;30:764–776. doi: 10.1016/j.tcb.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Li J, et al. Non-cell-autonomous cancer progression from chromosomal instability. Nature. 2023;620:1080–1088. doi: 10.1038/s41586-023-06464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Barrio L, et al. Chromosomal instability-induced cell invasion through caspase-driven DNA damage. Curr. Biol. 2023;33:4446–4457. e4445. doi: 10.1016/j.cub.2023.09.004. [DOI] [PubMed] [Google Scholar]
- 382.Cai Y, et al. Loss of chromosome 8p governs tumor progression and drug response by altering lipid metabolism. Cancer Cell. 2016;29:751–766. doi: 10.1016/j.ccell.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 383.Lu C, et al. Hypoxia-activated neuropeptide Y/Y5 receptor/RhoA pathway triggers chromosomal instability and bone metastasis in Ewing sarcoma. Nat. Commun. 2022;13:2323. doi: 10.1038/s41467-022-29898-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Nguyen B, et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell. 2022;185:563–575. e511. doi: 10.1016/j.cell.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Lengel HB, et al. Genomic mapping of metastatic organotropism in lung adenocarcinoma. Cancer Cell. 2023;41:970–985.e973. doi: 10.1016/j.ccell.2023.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Skakodub A, et al. Genomic analysis and clinical correlations of non-small cell lung cancer brain metastasis. Nat. Commun. 2023;14:4980. doi: 10.1038/s41467-023-40793-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Turajlic S, et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell. 2018;173:581–594.e512. doi: 10.1016/j.cell.2018.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Priestley P, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575:210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Pradat Y, et al. Integrative pan-cancer genomic and transcriptomic analyses of refractory metastatic cancer. Cancer Discov. 2023;13:1116–1143. doi: 10.1158/2159-8290.CD-22-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Senovilla L, et al. An immunosurveillance mechanism controls cancer cell ploidy. Science. 2012;337:1678–1684. doi: 10.1126/science.1224922. [DOI] [PubMed] [Google Scholar]
- 391.Kuang X, Li J. Chromosome instability and aneuploidy as context-dependent activators or inhibitors of antitumor immunity. Front. Immunol. 2022;13:895961. doi: 10.3389/fimmu.2022.895961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Yan H, Lu W, Wang F. The cGAS-STING pathway: a therapeutic target in chromosomally unstable cancers. Signal Transduct. Target. Ther. 2023;8:45. doi: 10.1038/s41392-023-01328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Kwon J, Bakhoum SF. The cytosolic DNA-sensing cGAS–STING pathway in cancer. Cancer Discov. 2020;10:26–39. doi: 10.1158/2159-8290.CD-19-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Samson N, Ablasser A. The cGAS–STING pathway and cancer. Nat. Cancer. 2022;3:1452–1463. doi: 10.1038/s43018-022-00468-w. [DOI] [PubMed] [Google Scholar]
- 395.Ablasser A, Chen ZJ. cGAS in action: expanding roles in immunity and inflammation. Science. 2019;363:eaat8657. doi: 10.1126/science.aat8657. [DOI] [PubMed] [Google Scholar]
- 396.Huang Y, et al. DNAJA2 deficiency activates cGAS-STING pathway via the induction of aberrant mitosis and chromosome instability. Nat. Commun. 2023;14:5246. doi: 10.1038/s41467-023-40952-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Mackenzie KJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548:461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Harding SM, et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548:466–470. doi: 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Vanpouille-Box C, Demaria S, Formenti SC, Galluzzi L. Cytosolic DNA sensing in organismal tumor control. Cancer Cell. 2018;34:361–378. doi: 10.1016/j.ccell.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 400.Hu J, et al. STING inhibits the reactivation of dormant metastasis in lung adenocarcinoma. Nature. 2023;616:806–813. doi: 10.1038/s41586-023-05880-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Boukhaled GM, Harding S, Brooks DG. Opposing roles of type I interferons in cancer immunity. Annu. Rev. Pathol. 2021;16:167–198. doi: 10.1146/annurev-pathol-031920-093932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Wang RW, et al. Aneuploid senescent cells activate NF‐κB to promote their immune clearance by NK cells. EMBO Rep. 2021;22:e52032. doi: 10.15252/embr.202052032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Hong C, et al. cGAS–STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature. 2022;607:366–373. doi: 10.1038/s41586-022-04847-2. [DOI] [PubMed] [Google Scholar]
- 404.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 405.Lee AJX, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71:1858–1870. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Cohen-Sharir Y, et al. Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature. 2021;590:486–491. doi: 10.1038/s41586-020-03114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Brutovský B. Scales of cancer evolution: selfish genome or cooperating cells? Cancers. 2022;14:3253. doi: 10.3390/cancers14133253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Beaumont HJ, et al. Experimental evolution of bet hedging. Nature. 2009;462:90–93. doi: 10.1038/nature08504. [DOI] [PubMed] [Google Scholar]
- 409.Scribano CM, et al. Chromosomal instability sensitizes patient breast tumors to multipolar divisions induced by paclitaxel. Sci. Transl. Med. 2021;13:eabd4811. doi: 10.1126/scitranslmed.abd4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Schukken KM, et al. Altering microtubule dynamics is synergistically toxic with spindle assembly checkpoint inhibition. Life Sci. Alliance. 2020;3:e201900499. doi: 10.26508/lsa.201900499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Jemaà M, et al. Characterization of novel MPS1 inhibitors with preclinical anticancer activity. Cell Death Differ. 2013;20:1532–1545. doi: 10.1038/cdd.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Soria-Bretones I, et al. The spindle assembly checkpoint is a therapeutic vulnerability of CDK4/6 inhibitor–resistant ER+ breast cancer with mitotic aberrations. Sci. Adv. 2022;8:eabq4293. doi: 10.1126/sciadv.abq4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Martino J, et al. Inhibitors of Rho kinases (ROCK) induce multiple mitotic defects and synthetic lethality in BRCA2-deficient cells. Elife. 2023;12:e80254. doi: 10.7554/eLife.80254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Gallo D, et al. CCNE1 amplification is synthetic lethal with PKMYT1 kinase inhibition. Nature. 2022;604:749–756. doi: 10.1038/s41586-022-04638-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Maia ARR, et al. Inhibition of the spindle assembly checkpoint kinase TTK enhances the efficacy of docetaxel in a triple-negative breast cancer model. Ann. Oncol. 2015;26:2180–2192. doi: 10.1093/annonc/mdv293. [DOI] [PubMed] [Google Scholar]
- 416.Maia ARR, et al. Mps1 inhibitors synergise with low doses of taxanes in promoting tumour cell death by enhancement of errors in cell division. Br. J. Cancer. 2018;118:1586–1595. doi: 10.1038/s41416-018-0081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Zasadil LM, et al. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci. Transl. Med. 2014;6:229ra243. doi: 10.1126/scitranslmed.3007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Lucken K, et al. EML4-ALK variant 3 promotes mitotic errors and spindle assembly checkpoint deficiency leading to increased microtubule poison sensitivity. Mol. Cancer Res. 2022;20:854–866. doi: 10.1158/1541-7786.MCR-21-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Bai Z, et al. Perspectives and mechanisms for targeting mitotic catastrophe in cancer treatment. Biochim. Biophys. Acta Rev. Cancer. 2023;1878:188965. doi: 10.1016/j.bbcan.2023.188965. [DOI] [PubMed] [Google Scholar]
- 420.Normandin K, et al. Genetic enhancers of partial PLK1 inhibition reveal hypersensitivity to kinetochore perturbations. PLoS Genet. 2023;19:e1010903. doi: 10.1371/journal.pgen.1010903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Martinez MJ, et al. Inhibition of the serine/threonine kinase BUB1 reverses taxane resistance in prostate cancer. iScience. 2023;26:107681. doi: 10.1016/j.isci.2023.107681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Portelinha A, et al. Synthetic lethality of drug-induced polyploidy and BCL-2 inhibition in lymphoma. Nat. Commun. 2023;14:1522. doi: 10.1038/s41467-023-37216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Janssen A, Kops GJPL, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl Acad. Sci. 2009;106:19108–19113. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Kops GJPL, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl Acad. Sci. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Wu G, et al. Small molecule targeting the Hec1/Nek2 mitotic pathway suppresses tumor cell growth in culture and in animal. Cancer Res. 2008;68:8393–8399. doi: 10.1158/0008-5472.CAN-08-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Hu CM, et al. Novel small molecules disrupting Hec1/Nek2 interaction ablate tumor progression by triggering Nek2 degradation through a death-trap mechanism. Oncogene. 2015;34:1220–1230. doi: 10.1038/onc.2014.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Dhital B, Rodriguez-Bravo V. Mechanisms of chromosomal instability (CIN) tolerance in aggressive tumors: surviving the genomic chaos. Chromosom. Res. 2023;31:15. doi: 10.1007/s10577-023-09724-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Sansregret L, et al. APC/C dysfunction limits excessive cancer chromosomal instability. Cancer Discov. 2017;7:218–233. doi: 10.1158/2159-8290.CD-16-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Dhital B, et al. Harnessing transcriptionally driven chromosomal instability adaptation to target therapy-refractory lethal prostate cancer. Cell Rep. Med. 2023;4:100937. doi: 10.1016/j.xcrm.2023.100937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Gronroos E, Lopez-Garcia C. Tolerance of chromosomal instability in cancer: mechanisms and therapeutic opportunities. Cancer Res. 2018;78:6529–6535. doi: 10.1158/0008-5472.CAN-18-1958. [DOI] [PubMed] [Google Scholar]
- 431.Perelli L, et al. Interferon signaling promotes tolerance to chromosomal instability during metastatic evolution in renal cancer. Nat. Cancer. 2023;4:984–100. doi: 10.1038/s43018-023-00584-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Clarke MN, et al. Adaptation to high rates of chromosomal instability and aneuploidy through multiple pathways in budding yeast. EMBO J. 2023;42:e111500. doi: 10.15252/embj.2022111500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Haase MA, et al. DASH/Dam1 complex mutants stabilize ploidy in histone‐humanized yeast by weakening kinetochore‐microtubule attachments. EMBO J. 2023;42:e112600. doi: 10.15252/embj.2022112600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Du M, et al. Nondiploid cancer cells: Stress, tolerance and therapeutic inspirations. Biochim. Biophys. Acta Rev. Cancer. 2022;1877:188794. doi: 10.1016/j.bbcan.2022.188794. [DOI] [PubMed] [Google Scholar]
- 435.Orr B, et al. Adaptive resistance to an inhibitor of chromosomal instability in human cancer cells. Cell Rep. 2016;17:1755–1763. doi: 10.1016/j.celrep.2016.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Su XA, et al. RAD21 is a driver of chromosome 8 gain in Ewing sarcoma to mitigate replication stress. Genes Dev. 2021;35:556–572. doi: 10.1101/gad.345454.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Schiavoni F, et al. Aneuploidy tolerance caused by BRG1 loss allows chromosome gains and recovery of fitness. Nat. Commun. 2022;13:1731. doi: 10.1038/s41467-022-29420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Marquis C, et al. Chromosomally unstable tumor cells specifically require KIF18A for proliferation. Nat. Commun. 2021;12:1213. doi: 10.1038/s41467-021-21447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Tang Y-C, et al. Aneuploid cell survival relies upon sphingolipid homeostasis. Cancer Res. 2017;77:5272–5286. doi: 10.1158/0008-5472.CAN-17-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Girish V, et al. Oncogene-like addiction to aneuploidy in human cancers. Science. 2023;381:eadg4521. doi: 10.1126/science.adg4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Mays, J. C. et al. KaryoTap enables aneuploidy detection in thousands of single human cells. Preprint at http://www.biorxiv.org/content/10.1101/2023.09.08.555746v1 (2023).
- 442.Angrisani A, Fachinetti D. The KaryoCreate technology generates specific aneuploid karyotypes on demand. Cell Rep. Meth. 2023;3:100514. doi: 10.1016/j.crmeth.2023.100514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Truong MA, Cané-Gasull P, Lens S. Modeling specific aneuploidies: from karyotype manipulations to biological insights. Chromosom. Res. 2023;31:1–27. doi: 10.1007/s10577-023-09735-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Truong MA, et al. A kinesin‐based approach for inducing chromosome‐specific mis‐segregation in human cells. EMBO J. 2023;15:e111559. doi: 10.15252/embj.2022111559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Lakhani AA, Thompson SL, Sheltzer JM. Aneuploidy in human cancer: new tools and perspectives. Trends Genet. 2023;39:P968–P980. doi: 10.1016/j.tig.2023.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Tovini L, et al. Targeted assembly of ectopic kinetochores to induce chromosome‐specific segmental aneuploidies. EMBO J. 2023;42:e111587. doi: 10.15252/embj.2022111587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Shih J, et al. Cancer aneuploidies are shaped primarily by effects on tumour fitness. Nature. 2023;619:793–800. doi: 10.1038/s41586-023-06266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Dominguez-Brauer C, et al. Targeting mitosis in cancer: emerging strategies. Mol. Cell. 2015;60:524–536. doi: 10.1016/j.molcel.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 449.Veitch ZW, et al. Safety and tolerability of CFI-400945, a first-in-class, selective PLK4 inhibitor in advanced solid tumours: a phase 1 dose-escalation trial. Br. J. Cancer. 2019;121:318–324. doi: 10.1038/s41416-019-0517-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Lorusso P, et al. First-in-human study of the monopolar spindle 1 (Mps1) kinase inhibitor BAY 1161909 in combination with paclitaxel in subjects with advanced malignancies. Ann. Oncol. 2018;29:VIII138. doi: 10.1093/annonc/mdy279.410. [DOI] [Google Scholar]
- 451.Cheng A, et al. A mitotic NADPH upsurge promotes chromosome segregation and tumour progression in aneuploid cancer cells. Nat. Metab. 2023;5:1141–11158. doi: 10.1038/s42255-023-00832-9. [DOI] [PubMed] [Google Scholar]
- 452.Quinton RJ, et al. Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature. 2021;590:492–497. doi: 10.1038/s41586-020-03133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Herman JA, et al. Hyper-active RAS/MAPK introduces cancer-specific mitotic vulnerabilities. Proc. Natl Acad. Sci. 2022;119:e2208255119. doi: 10.1073/pnas.2208255119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Martín A, et al. Mitochondrial RNA methyltransferase TRMT61B is a new, potential biomarker and therapeutic target for highly aneuploid cancers. Cell Death Differ. 2023;30:37–53. doi: 10.1038/s41418-022-01044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Andor N, Altrock PM, Jain N, Gomes AP. Tipping cancer cells over the edge: the context-dependent cost of high ploidy. Cancer Res. 2022;82:741–748. doi: 10.1158/0008-5472.CAN-21-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Li M, et al. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J. Cell Biol. 2009;185:983–994. doi: 10.1083/jcb.200904020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Rao CV, et al. Colonic tumorigenesis in BubR1+/-ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc. Natl Acad. Sci. 2005;102:4365–4370. doi: 10.1073/pnas.0407822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Caldwell CM, Green RA, Kaplan KB. APC mutations lead to cytokinetic failures in vitro and tetraploid genotypes in Min mice. J Cell Biol. 2007;178:1109–1120. doi: 10.1083/jcb.200703186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Anand S, Penrhyn-Lowe S, Venkitaraman AR. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/S1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 461.Zhang D, et al. Cre-loxP-controlled periodic Aurora-A overexpression induces mitotic abnormalities and hyperplasia in mammary glands of mouse models. Oncogene. 2004;23:8720–8730. doi: 10.1038/sj.onc.1208153. [DOI] [PubMed] [Google Scholar]
- 462.Zhang D, et al. Aurora A overexpression induces cellular senescence in mammary gland hyperplastic tumors developed in p53-deficient mice. Oncogene. 2008;27:4305–4314. doi: 10.1038/onc.2008.76. [DOI] [PubMed] [Google Scholar]
- 463.Jeganathan K, et al. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J. Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Wang Q, et al. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103:1278–1285. doi: 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- 465.Baker DJ, et al. Increased expression of BubR1 protects against aneuploidy and cancer and extends healthy lifespan. Nat. Cell Biol. 2013;15:96–102. doi: 10.1038/ncb2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Weaver BA, et al. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 467.Diaz-Rodriguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc. Natl Acad. Sci. 2008;105:16719–16724. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Wang Z, Yu R, Melmed S. Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol. Endocrinol. 2001;15:1870–1879. doi: 10.1210/mend.15.11.0729. [DOI] [PubMed] [Google Scholar]
- 469.Chesnokova V, et al. Pituitary hypoplasia in Pttg-/- mice is protective for Rb+/- pituitary tumorigenesis. Mol. Endocrinol. 2005;19:2371–2379. doi: 10.1210/me.2005-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Aguirre-Portoles C, et al. Tpx2 controls spindle integrity, genome stability, and tumor development. Cancer Res. 2012;72:1518–1528. doi: 10.1158/0008-5472.CAN-11-1971. [DOI] [PubMed] [Google Scholar]
- 471.van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J. Cell Biol. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Fujita T, et al. Overexpression of UbcH10 alternates the cell cycle profile and accelerate the tumor proliferation in colon cancer. BMC Cancer. 2009;9:1–10. doi: 10.1186/1471-2407-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Garcia-Higuera I, et al. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- 474.Remeseiro S, et al. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012;31:2076–2089. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Abbud RA, et al. Early multipotential pituitary focal hyperplasia in the alpha-subunit of glycoprotein hormone-driven pituitary tumor-transforming gene transgenic mice. Mol. Endocrinol. 2005;19:1383–1391. doi: 10.1210/me.2004-0403. [DOI] [PubMed] [Google Scholar]
- 476.Donangelo I, et al. Pituitary tumor transforming gene overexpression facilitates pituitary tumor development. Endocrinology. 2006;147:4781–4791. doi: 10.1210/en.2006-0544. [DOI] [PubMed] [Google Scholar]
- 477.de Cárcer G, et al. Plk1 overexpression induces chromosomal instability and suppresses tumor development. Nat. Commun. 2018;9:3012. doi: 10.1038/s41467-018-05429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Dharanipragada P, et al. Blocking genomic instability prevents acquired resistance to MAPK inhibitor therapy in melanoma. Cancer Discov. 2023;13:880–909. doi: 10.1158/2159-8290.CD-22-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Zerbib, J. et al. Human aneuploid cells depend on the RAF/MEK/ERK pathway for overcoming increased DNA damage. Preprint at http://www.biorxiv.org/content/10.1101/2023.01.27.525822v1 (2023). [DOI] [PMC free article] [PubMed]
- 480.Zhou W, et al. NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell. 2013;23:48–62. doi: 10.1016/j.ccr.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Gu C, et al. CHEK1 and circCHEK1_246aa evoke chromosomal instability and induce bone lesion formation in multiple myeloma. Mol. Cancer. 2021;20:84. doi: 10.1186/s12943-021-01380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Tang X, et al. BUB1B and circBUB1B_544aa aggravate multiple myeloma malignancy through evoking chromosomal instability. Signal Transduct. Target. Ther. 2021;6:361. doi: 10.1038/s41392-021-00746-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355:eaaf8399. doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Manfredi MG, et al. Characterization of Alisertib (MLN8237), an investigational small-molecule inhibitor of aurora A kinase using novel in vivo pharmacodynamic assays. Clin. Cancer Res. 2011;17:7614–7624. doi: 10.1158/1078-0432.CCR-11-1536. [DOI] [PubMed] [Google Scholar]
- 485.Dees EC, et al. Phase I study of aurora A kinase inhibitor MLN8237 in advanced solid tumors: safety, pharmacokinetics, pharmacodynamics, and bioavailability of two oral formulations. Clin. Cancer Res. 2012;18:4775–4784. doi: 10.1158/1078-0432.CCR-12-0589. [DOI] [PubMed] [Google Scholar]
- 486.Matulonis UA, et al. Phase II study of MLN8237 (alisertib), an investigational Aurora A kinase inhibitor, in patients with platinum-resistant or -refractory epithelial ovarian, fallopian tube, or primary peritoneal carcinoma. Gynecol. Oncol. 2012;127:63–69. doi: 10.1016/j.ygyno.2012.06.040. [DOI] [PubMed] [Google Scholar]
- 487.Dickson MA, et al. Phase II study of MLN8237 (Alisertib) in advanced/metastatic sarcoma. Ann. Oncol. 2016;27:1855–1860. doi: 10.1093/annonc/mdw281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Beltran H, et al. A Phase II trial of the aurora kinase A inhibitor Alisertib for patients with castration-resistant and neuroendocrine prostate cancer: efficacy and biomarkers. Clin. Cancer Res. 2019;25:43–51. doi: 10.1158/1078-0432.CCR-18-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Falchook G, et al. Alisertib in combination with weekly paclitaxel in patients with advanced breast cancer or recurrent ovarian cancer: a randomized clinical trial. JAMA Oncol. 2019;5:e183773. doi: 10.1001/jamaoncol.2018.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Necchi A, et al. An open-label, single-arm, phase 2 study of the aurora kinase A inhibitor Alisertib in patients with advanced urothelial cancer. Investig. New Drugs. 2016;34:236–242. doi: 10.1007/s10637-016-0328-9. [DOI] [PubMed] [Google Scholar]
- 491.Owonikoko TK, et al. Randomized phase II study of paclitaxel plus alisertib versus paclitaxel plus placebo as second-line therapy for SCLC: primary and correlative biomarker analyses. J. Thorac. Oncol. 2020;15:274–287. doi: 10.1016/j.jtho.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 492.Graff JN, et al. Open-label, multicenter, phase 1 study of alisertib (MLN8237), an aurora A kinase inhibitor, with docetaxel in patients with solid tumors. Cancer. 2016;122:2524–2533. doi: 10.1002/cncr.30073. [DOI] [PubMed] [Google Scholar]
- 493.Haddad TC, et al. Phase I trial to evaluate the addition of alisertib to fulvestrant in women with endocrine-resistant, ER+ metastatic breast cancer. Breast Cancer Res. Treat. 2018;168:639–647. doi: 10.1007/s10549-017-4616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Goff LW, et al. Phase I study combining the aurora kinase a inhibitor alisertib with mFOLFOX in gastrointestinal cancer. Investig. New Drugs. 2019;37:315–322. doi: 10.1007/s10637-018-0663-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.DuBois SG, et al. Phase II trial of alisertib in combination with irinotecan and temozolomide for patients with relapsed or refractory neuroblastoma. Clin. Cancer Res. 2018;24:6142–6149. doi: 10.1158/1078-0432.CCR-18-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Shah HA, et al. Phase I study of aurora a kinase inhibitor alisertib (MLN8237) in combination with selective VEGFR inhibitor pazopanib for therapy of advanced solid tumors. Am. J. Clin. Oncol. 2019;42:413–420. doi: 10.1097/COC.0000000000000543. [DOI] [PubMed] [Google Scholar]
- 497.Jing XL, Chen SW. Aurora kinase inhibitors: a patent review (2014-2020) Expert Opin. Ther. Pat. 2021;31:625–644. doi: 10.1080/13543776.2021.1890027. [DOI] [PubMed] [Google Scholar]
- 498.Di Noia V, et al. Malignant pleural mesothelioma: is tailoring the second-line therapy really “raising the bar? Curr. Treat. Options. Oncol. 2019;20:23. doi: 10.1007/s11864-019-0616-7. [DOI] [PubMed] [Google Scholar]
- 499.Chen JA, et al. A phase I dose escalation, dose expansion and pharmacokinetic trial of gemcitabine and alisertib in advanced solid tumors and pancreatic cancer. Cancer Chemother. Pharmacol. 2022;90:217–228. doi: 10.1007/s00280-022-04457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.O’Shaughnessy J, et al. Efficacy and safety of weekly paclitaxel with or without oral alisertib in patients with metastatic breast cancer: a randomized clinical trial. JAMA Netw. Open. 2021;4:e214103. doi: 10.1001/jamanetworkopen.2021.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Nguyen TT, Silva FN, Golemis EA. Aurora kinases as therapeutic targets in head and neck cancer. Cancer J. 2022;28:387–400. doi: 10.1097/PPO.0000000000000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Venkadakrishnan VB, et al. Significance of RB loss in unlocking phenotypic plasticity in advanced cancers. Mol. Cancer Res. 2023;21:497–510. doi: 10.1158/1541-7786.MCR-23-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Bain NT, Wang Y, Arulananda S. Minimal residual disease in EGFR-mutant non-small-cell lung cancer. Front. Oncol. 2022;12:1002714. doi: 10.3389/fonc.2022.1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Li S, et al. Emerging targeted therapies in advanced non-small-cell lung cancer. Cancers. 2023;15:2899. doi: 10.3390/cancers15112899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Farnsworth DA, Chen YT, de Rappard Yuswack G, Lockwood WW. Emerging molecular dependencies of mutant EGFR-Driven non-small cell lung cancer. Cells. 2021;10:3553. doi: 10.3390/cells10123553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Kelly KR, et al. Phase I study of MLN8237-investigational Aurora A kinase inhibitor-in relapsed/refractory multiple myeloma, non-Hodgkin lymphoma and chronic lymphocytic leukemia. Investig. New Drugs. 2014;32:489–499. doi: 10.1007/s10637-013-0050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Barr PM, et al. Phase II intergroup trial of alisertib in relapsed and refractory peripheral T-Cell lymphoma and transformed mycosis fungoides: SWOG 1108. J. Clin. Oncol. 2015;33:2399–2404. doi: 10.1200/JCO.2014.60.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Zhang Z, et al. Modulation of oxidative phosphorylation augments antineoplastic activity of mitotic aurora kinase inhibition. Cell Death Dis. 2021;12:893. doi: 10.1038/s41419-021-04190-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Friedberg JW, et al. Phase II study of alisertib, a selective Aurora A kinase inhibitor, in relapsed and refractory aggressive B- and T-cell non-Hodgkin lymphomas. J. Clin. Oncol. 2014;32:44–50. doi: 10.1200/JCO.2012.46.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Kelly KR, et al. Phase I study of the investigational aurora a kinase inhibitor alisertib plus rituximab or rituximab/vincristine in relapsed/refractory aggressive B-cell lymphoma. Clin. Cancer Res. 2018;24:6150–6159. doi: 10.1158/1078-0432.CCR-18-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Rosenthal A, et al. A phase Ib study of the combination of the aurora kinase inhibitor alisertib (MLN8237) and bortezomib in relapsed multiple myeloma. Br. J. Haematol. 2016;174:323–325. doi: 10.1111/bjh.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Brunner AM, et al. Alisertib plus induction chemotherapy in previously untreated patients with high-risk, acute myeloid leukaemia: a single-arm, phase 2 trial. Lancet Haematol. 2020;7:e122–e133. doi: 10.1016/S2352-3026(19)30203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Mosse YP, et al. Pediatric phase I trial and pharmacokinetic study of MLN8237, an investigational oral selective small-molecule inhibitor of Aurora kinase A: a children’s oncology group phase I consortium study. Clin. Cancer Res. 2012;18:6058–6064. doi: 10.1158/1078-0432.CCR-11-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Mosse YP, et al. A phase II study of alisertib in children with recurrent/refractory solid tumors or leukemia: children’s oncology group phase I and pilot consortium (ADVL0921) Clin. Cancer Res. 2019;25:3229–3238. doi: 10.1158/1078-0432.CCR-18-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Zhang C, Li H. Molecular targeted therapies for pediatric atypical teratoid/rhabdoid tumors. Pediatr. Investig. 2022;6:111–122. doi: 10.1002/ped4.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Rechberger JS, Nesvick CL, Daniels DJ. Atypical teratoid rhabdoid tumor (ATRT): disease mechanisms and potential drug targets. Expert. Opin. Ther. Targets. 2022;26:187–192. doi: 10.1080/14728222.2022.2040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Fletcher GC, et al. ENMD-2076 is an orally active kinase inhibitor with antiangiogenic and antiproliferative mechanisms of action. Mol. Cancer Ther. 2011;10:126–137. doi: 10.1158/1535-7163.MCT-10-0574. [DOI] [PubMed] [Google Scholar]
- 518.Diamond JR, et al. Phase I safety, pharmacokinetic, and pharmacodynamic study of ENMD-2076, a novel angiogenic and Aurora kinase inhibitor, in patients with advanced solid tumors. Clin. Cancer Res. 2011;17:849–860. doi: 10.1158/1078-0432.CCR-10-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Matulonis UA, et al. ENMD-2076, an oral inhibitor of angiogenic and proliferation kinases, has activity in recurrent, platinum resistant ovarian cancer. Eur. J. Cancer. 2013;49:121–131. doi: 10.1016/j.ejca.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 520.Lheureux S, et al. A clinical and molecular phase II trial of oral ENMD-2076 in ovarian clear cell carcinoma (OCCC): a study of the princess margaret phase II consortium. Clin. Cancer Res. 2018;24:6168–6174. doi: 10.1158/1078-0432.CCR-18-1244. [DOI] [PubMed] [Google Scholar]
- 521.Veitch Z, et al. A phase II study of ENMD-2076 in advanced soft tissue sarcoma (STS) Sci. Rep. 2019;9:7390. doi: 10.1038/s41598-019-43222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Diamond JR, et al. A phase II clinical trial of the Aurora and angiogenic kinase inhibitor ENMD-2076 for previously treated, advanced, or metastatic triple-negative breast cancer. Breast Cancer Res. 2018;20:82. doi: 10.1186/s13058-018-1014-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Abou-Alfa GK, et al. Phase II multicenter, open-label study of oral ENMD-2076 for the treatment of patients with advanced fibrolamellar carcinoma. Oncol. 2020;25:e1837–e1845. doi: 10.1634/theoncologist.2020-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Yee KW, et al. A phase I trial of the aurora kinase inhibitor, ENMD-2076, in patients with relapsed or refractory acute myeloid leukemia or chronic myelomonocytic leukemia. Investig. New Drugs. 2016;34:614–624. doi: 10.1007/s10637-016-0375-2. [DOI] [PubMed] [Google Scholar]
- 525.Lloyd MR, Spring LM, Bardia A, Wander SA. Mechanisms of resistance to CDK4/6 blockade in advanced hormone receptor-positive, HER2-negative breast cancer and emerging therapeutic opportunities. Clin. Cancer Res. 2022;28:821–830. doi: 10.1158/1078-0432.CCR-21-2947. [DOI] [PubMed] [Google Scholar]
- 526.Chu QS, et al. Aurora kinase A inhibitor, LY3295668 erbumine: a phase 1 monotherapy safety study in patients with locally advanced or metastatic solid tumors. Investig. New Drugs. 2021;39:1001–1010. doi: 10.1007/s10637-020-01049-3. [DOI] [PubMed] [Google Scholar]
- 527.Lum C, Alamgeer M. Technological and therapeutic advances in advanced small cell lung cancer. Cancers. 2019;11:1570. doi: 10.3390/cancers11101570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Moreno L, et al. Accelerating drug development for neuroblastoma: summary of the second neuroblastoma drug development strategy forum from innovative therapies for children with cancer and international society of paediatric oncology europe neuroblastoma. Eur. J. Cancer. 2020;136:52–68. doi: 10.1016/j.ejca.2020.05.010. [DOI] [PubMed] [Google Scholar]
- 529.Dees EC, et al. Phase 1 study of MLN8054, a selective inhibitor of Aurora A kinase in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2011;67:945–954. doi: 10.1007/s00280-010-1377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Macarulla T, et al. Phase I study of the selective Aurora A kinase inhibitor MLN8054 in patients with advanced solid tumors: safety, pharmacokinetics, and pharmacodynamics. Mol. Cancer Ther. 2010;9:2844–2852. doi: 10.1158/1535-7163.MCT-10-0299. [DOI] [PubMed] [Google Scholar]
- 531.Shimomura T, et al. MK-5108, a highly selective Aurora-A kinase inhibitor, shows antitumor activity alone and in combination with docetaxel. Mol. Cancer Ther. 2010;9:157–166. doi: 10.1158/1535-7163.MCT-09-0609. [DOI] [PubMed] [Google Scholar]
- 532.Amin M, et al. A phase I study of MK-5108, an oral aurora a kinase inhibitor, administered both as monotherapy and in combination with docetaxel, in patients with advanced or refractory solid tumors. Investig. New Drugs. 2016;34:84–95. doi: 10.1007/s10637-015-0306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Miura A, et al. TAS-119, a novel selective Aurora A and TRK inhibitor, exhibits antitumor efficacy in preclinical models with deregulated activation of the Myc, beta-Catenin, and TRK pathways. Investig. New Drugs. 2021;39:724–735. doi: 10.1007/s10637-020-01019-9. [DOI] [PubMed] [Google Scholar]
- 534.Robbrecht DGJ, et al. A first-in-human phase 1 and pharmacological study of TAS-119, a novel selective Aurora A kinase inhibitor in patients with advanced solid tumours. Br. J. Cancer. 2021;124:391–398. doi: 10.1038/s41416-020-01100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Sootome H, et al. Aurora A inhibitor TAS-119 enhances antitumor efficacy of taxanes in vitro and in vivo: preclinical studies as guidance for clinical development and trial design. Mol. Cancer Ther. 2020;19:1981–1991. doi: 10.1158/1535-7163.MCT-20-0036. [DOI] [PubMed] [Google Scholar]
- 536.Shiotsu Y, et al. KW-2449, a novel multikinase inhibitor, suppresses the growth of leukemia cells with FLT3 mutations or T315I-mutated BCR/ABL translocation. Blood. 2009;114:1607–1617. doi: 10.1182/blood-2009-01-199307. [DOI] [PubMed] [Google Scholar]
- 537.Cheung CH, et al. Aurora kinase inhibitor patents and agents in clinical testing: an update (2011 - 2013) Expert Opin. Ther. Pat. 2014;24:1021–1038. doi: 10.1517/13543776.2014.931374. [DOI] [PubMed] [Google Scholar]
- 538.Yang J, et al. AZD1152, a novel and selective aurora B kinase inhibitor, induces growth arrest, apoptosis, and sensitization for tubulin depolymerizing agent or topoisomerase II inhibitor in human acute leukemia cells in vitro and in vivo. Blood. 2007;110:2034–2040. doi: 10.1182/blood-2007-02-073700. [DOI] [PubMed] [Google Scholar]
- 539.Schwartz GK, et al. Phase I study of barasertib (AZD1152), a selective inhibitor of Aurora B kinase, in patients with advanced solid tumors. Investig. New Drugs. 2013;31:370–380. doi: 10.1007/s10637-012-9825-7. [DOI] [PubMed] [Google Scholar]
- 540.Boss DS, et al. Clinical evaluation of AZD1152, an i.v. inhibitor of Aurora B kinase, in patients with solid malignant tumors. Ann. Oncol. 2011;22:431–437. doi: 10.1093/annonc/mdq344. [DOI] [PubMed] [Google Scholar]
- 541.Lowenberg B, et al. Phase 1/2 study to assess the safety, efficacy, and pharmacokinetics of barasertib (AZD1152) in patients with advanced acute myeloid leukemia. Blood. 2011;118:6030–6036. doi: 10.1182/blood-2011-07-366930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Tsuboi K, et al. A Phase I study to assess the safety, pharmacokinetics and efficacy of barasertib (AZD1152), an Aurora B kinase inhibitor, in Japanese patients with advanced acute myeloid leukemia. Leuk. Res. 2011;35:1384–1389. doi: 10.1016/j.leukres.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 543.Kantarjian HM, et al. Stage I of a phase 2 study assessing the efficacy, safety, and tolerability of barasertib (AZD1152) versus low-dose cytosine arabinoside in elderly patients with acute myeloid leukemia. Cancer. 2013;119:2611–2619. doi: 10.1002/cncr.28113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Dennis M, et al. Phase I study of the Aurora B kinase inhibitor barasertib (AZD1152) to assess the pharmacokinetics, metabolism and excretion in patients with acute myeloid leukemia. Cancer Chemother. Pharmacol. 2012;70:461–469. doi: 10.1007/s00280-012-1939-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Kantarjian HM, et al. Phase I study assessing the safety and tolerability of barasertib (AZD1152) with low-dose cytosine arabinoside in elderly patients with AML. Clin. Lymphoma Myeloma Leuk. 2013;13:559–567. doi: 10.1016/j.clml.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Collins GP, et al. A phase II trial of AZD1152 in relapsed/refractory diffuse large B-cell lymphoma. Br. J. Haematol. 2015;170:886–890. doi: 10.1111/bjh.13333. [DOI] [PubMed] [Google Scholar]
- 547.Dittrich C, et al. A phase 1 dose escalation study of BI 831266, an inhibitor of Aurora kinase B, in patients with advanced solid tumors. Investig. New Drugs. 2015;33:409–422. doi: 10.1007/s10637-014-0201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Mross K, et al. A phase I study of BI 811283, an Aurora B kinase inhibitor, in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2016;78:405–417. doi: 10.1007/s00280-016-3095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Dohner H, et al. A phase I trial investigating the Aurora B kinase inhibitor BI 811283 in combination with cytarabine in patients with acute myeloid leukaemia. Br. J. Haematol. 2019;185:583–587. doi: 10.1111/bjh.15563. [DOI] [PubMed] [Google Scholar]
- 550.Zhou Y, et al. CS2164, a novel multi-target inhibitor against tumor angiogenesis, mitosis and chronic inflammation with anti-tumor potency. Cancer Sci. 2017;108:469–477. doi: 10.1111/cas.13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Sun Y, et al. Phase I dose-escalation study of chiauranib, a novel angiogenic, mitotic, and chronic inflammation inhibitor, in patients with advanced solid tumors. J. Hematol. Oncol. 2019;12:9. doi: 10.1186/s13045-018-0695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Cao J, Chow L, Dow S. Strategies to overcome myeloid cell induced immune suppression in the tumor microenvironment. Front. Oncol. 2023;13:1116016. doi: 10.3389/fonc.2023.1116016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Gao T, et al. Inhibition of extranodal NK/T-cell lymphoma by chiauranib through an AIF-dependent pathway and its synergy with L-asparaginase. Cell Death Dis. 2023;14:316. doi: 10.1038/s41419-023-05833-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Lv P, et al. Pathogenesis and therapeutic strategy in platinum resistance lung cancer. Biochim. Biophys. Acta Rev. Cancer. 2021;1876:188577. doi: 10.1016/j.bbcan.2021.188577. [DOI] [PubMed] [Google Scholar]
- 555.Howard S, et al. Fragment-based discovery of the pyrazol-4-yl urea (AT9283), a multitargeted kinase inhibitor with potent aurora kinase activity. J. Med. Chem. 2009;52:379–388. doi: 10.1021/jm800984v. [DOI] [PubMed] [Google Scholar]
- 556.Dent SF, et al. NCIC CTG IND.181: phase I study of AT9283 given as a weekly 24 hour infusion in advanced malignancies. Investig. New Drugs. 2013;31:1522–1529. doi: 10.1007/s10637-013-0018-9. [DOI] [PubMed] [Google Scholar]
- 557.Foran J, et al. A phase I and pharmacodynamic study of AT9283, a small-molecule inhibitor of aurora kinases in patients with relapsed/refractory leukemia or myelofibrosis. Clin. Lymphoma Myeloma Leuk. 2014;14:223–230. doi: 10.1016/j.clml.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Moreno L, et al. A phase I trial of AT9283 (a selective inhibitor of aurora kinases) in children and adolescents with solid tumors: a Cancer Research UK study. Clin. Cancer Res. 2015;21:267–273. doi: 10.1158/1078-0432.CCR-14-1592. [DOI] [PubMed] [Google Scholar]
- 559.Peter B, et al. Drug-induced inhibition of phosphorylation of STAT5 overrides drug resistance in neoplastic mast cells. Leukemia. 2018;32:1016–1022. doi: 10.1038/leu.2017.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Mita M, et al. A phase l study of three different dosing schedules of the oral aurora kinase inhibitor MSC1992371A in patients with solid tumors. Target Oncol. 2014;9:215–224. doi: 10.1007/s11523-013-0288-3. [DOI] [PubMed] [Google Scholar]
- 561.Raymond E, et al. A phase I schedule dependency study of the aurora kinase inhibitor MSC1992371A in combination with gemcitabine in patients with solid tumors. Investig. New Drugs. 2014;32:94–103. doi: 10.1007/s10637-013-9950-y. [DOI] [PubMed] [Google Scholar]
- 562.Graux C, et al. A phase I dose-escalation study of MSC1992371A, an oral inhibitor of aurora and other kinases, in advanced hematologic malignancies. Leuk. Res. 2013;37:1100–1106. doi: 10.1016/j.leukres.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 563.Kim JT, et al. The discovery of aurora kinase inhibitor by multi-docking-based virtual screening. Int. J. Mol. Sci. 2014;15:20403–20412. doi: 10.3390/ijms151120403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Jani JP, et al. PF-03814735, an orally bioavailable small molecule aurora kinase inhibitor for cancer therapy. Mol. Cancer Ther. 2010;9:883–894. doi: 10.1158/1535-7163.MCT-09-0915. [DOI] [PubMed] [Google Scholar]
- 565.Schoffski P, et al. Phase I, open-label, multicentre, dose-escalation, pharmacokinetic and pharmacodynamic trial of the oral aurora kinase inhibitor PF-03814735 in advanced solid tumours. Eur. J. Cancer. 2011;47:2256–2264. doi: 10.1016/j.ejca.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 566.Farrell P, et al. Biological characterization of TAK-901, an investigational, novel, multitargeted Aurora B kinase inhibitor. Mol. Cancer Ther. 2013;12:460–470. doi: 10.1158/1535-7163.MCT-12-0657. [DOI] [PubMed] [Google Scholar]
- 567.Carpinelli P, et al. PHA-739358, a potent inhibitor of Aurora kinases with a selective target inhibition profile relevant to cancer. Mol Cancer Ther. 2007;6:3158–3168. doi: 10.1158/1535-7163.MCT-07-0444. [DOI] [PubMed] [Google Scholar]
- 568.Meulenbeld HJ, et al. Randomized phase II study of danusertib in patients with metastatic castration-resistant prostate cancer after docetaxel failure. BJU Int. 2013;111:44–52. doi: 10.1111/j.1464-410X.2012.11404.x. [DOI] [PubMed] [Google Scholar]
- 569.Adams ND, et al. Discovery of GSK1070916, a potent and selective inhibitor of Aurora B/C kinase. J. Med. Chem. 2010;53:3973–4001. doi: 10.1021/jm901870q. [DOI] [PubMed] [Google Scholar]
- 570.Glaser KB, et al. Preclinical characterization of ABT-348, a kinase inhibitor targeting the aurora, vascular endothelial growth factor receptor/platelet-derived growth factor receptor, and Src kinase families. J. Pharmacol. Exp. Ther. 2012;343:617–627. doi: 10.1124/jpet.112.197087. [DOI] [PubMed] [Google Scholar]
- 571.Maitland ML, et al. Clinical pharmacodynamic/exposure characterisation of the multikinase inhibitor ilorasertib (ABT-348) in a phase 1 dose-escalation trial. Br. J. Cancer. 2018;118:1042–1050. doi: 10.1038/s41416-018-0020-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Garcia-Manero G, et al. Phase 1 dose escalation trial of ilorasertib, a dual Aurora/VEGF receptor kinase inhibitor, in patients with hematologic malignancies. Investig. New Drugs. 2015;33:870–880. doi: 10.1007/s10637-015-0242-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Payton M, et al. Preclinical evaluation of AMG 900, a novel potent and highly selective pan-aurora kinase inhibitor with activity in taxane-resistant tumor cell lines. Cancer Res. 2010;70:9846–9854. doi: 10.1158/0008-5472.CAN-10-3001. [DOI] [PubMed] [Google Scholar]
- 574.Carducci M, et al. A phase 1, first-in-human study of AMG 900, an orally administered pan-Aurora kinase inhibitor, in adult patients with advanced solid tumors. Investig. New Drugs. 2018;36:1060–1071. doi: 10.1007/s10637-018-0625-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Kantarjian HM, et al. A phase 1 study of AMG 900, an orally administered pan-aurora kinase inhibitor, in adult patients with acute myeloid leukemia. Am. J. Hematol. 2017;92:660–667. doi: 10.1002/ajh.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Sini P, et al. Pharmacological Profile of BI 847325, an Orally Bioavailable, ATP-Competitive Inhibitor of MEK and Aurora Kinases. Mol. Cancer Ther. 2016;15:2388–2398. doi: 10.1158/1535-7163.MCT-16-0066. [DOI] [PubMed] [Google Scholar]
- 577.Schoffski P, et al. A phase I study of two dosing schedules of oral BI 847325 in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2016;77:99–108. doi: 10.1007/s00280-015-2914-5. [DOI] [PubMed] [Google Scholar]
- 578.Oslob JD, et al. Discovery of a potent and selective aurora kinase inhibitor. Bioorg. Med. Chem. Lett. 2008;18:4880–4884. doi: 10.1016/j.bmcl.2008.07.073. [DOI] [PubMed] [Google Scholar]
- 579.Harrington EA, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat. Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 580.Traynor AM, et al. Phase I dose escalation study of MK-0457, a novel Aurora kinase inhibitor, in adult patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2011;67:305–314. doi: 10.1007/s00280-010-1318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.Giles FJ, et al. MK-0457, an Aurora kinase and BCR-ABL inhibitor, is active in patients with BCR-ABL T315I leukemia. Leukemia. 2013;27:113–117. doi: 10.1038/leu.2012.186. [DOI] [PubMed] [Google Scholar]
- 582.Wood KW, et al. Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc. Natl Acad. Sci. 2010;107:5839–5844. doi: 10.1073/pnas.0915068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Chung V, et al. First-time-in-human study of GSK923295, a novel antimitotic inhibitor of centromere-associated protein E (CENP-E), in patients with refractory cancer. Cancer Chemother. Pharmacol. 2012;69:733–741. doi: 10.1007/s00280-011-1756-z. [DOI] [PubMed] [Google Scholar]
- 584.Blagden SP, et al. A phase I trial of ispinesib, a kinesin spindle protein inhibitor, with docetaxel in patients with advanced solid tumours. Br. J. Cancer. 2008;98:894–899. doi: 10.1038/sj.bjc.6604264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Garcia-Saez I, Skoufias DA. Eg5 targeting agents: From new anti-mitotic based inhibitor discovery to cancer therapy and resistance. Biochem. Pharmacol. 2021;184:114364. doi: 10.1016/j.bcp.2020.114364. [DOI] [PubMed] [Google Scholar]
- 586.Beer TM, et al. Southwest Oncology Group phase II study of ispinesib in androgen-independent prostate cancer previously treated with taxanes. Clin. Genitourin. Cancer. 2008;6:103–109. doi: 10.3816/CGC.2008.n.016. [DOI] [PubMed] [Google Scholar]
- 587.Tang PA, et al. Phase II study of ispinesib in recurrent or metastatic squamous cell carcinoma of the head and neck. Investig. New Drugs. 2008;26:257–264. doi: 10.1007/s10637-007-9098-8. [DOI] [PubMed] [Google Scholar]
- 588.Lee CW, et al. A phase II study of ispinesib (SB-715992) in patients with metastatic or recurrent malignant melanoma: a National Cancer Institute of Canada Clinical Trials Group trial. Investig. New Drugs. 2008;26:249–255. doi: 10.1007/s10637-007-9097-9. [DOI] [PubMed] [Google Scholar]
- 589.Knox JJ, et al. A phase II and pharmacokinetic study of SB-715992, in patients with metastatic hepatocellular carcinoma: a study of the National Cancer Institute of Canada Clinical Trials Group (NCIC CTG IND.168) Investig. New Drugs. 2008;26:265–272. doi: 10.1007/s10637-007-9103-2. [DOI] [PubMed] [Google Scholar]
- 590.Lee RT, et al. A University of Chicago consortium phase II trial of SB-715992 in advanced renal cell cancer. Clin. Genitourin. Cancer. 2008;6:21–24. doi: 10.3816/CGC.2008.n.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Gomez HL, et al. Phase I dose-escalation and pharmacokinetic study of ispinesib, a kinesin spindle protein inhibitor, administered on days 1 and 15 of a 28-day schedule in patients with no prior treatment for advanced breast cancer. Anticancer Drugs. 2012;23:335–341. doi: 10.1097/CAD.0b013e32834e74d6. [DOI] [PubMed] [Google Scholar]
- 592.Souid AK, et al. A pediatric phase I trial and pharmacokinetic study of ispinesib: a Children’s Oncology Group phase I consortium study. Pediatr. Blood Cancer. 2010;55:1323–1328. doi: 10.1002/pbc.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.Holen KD, et al. A first in human study of SB-743921, a kinesin spindle protein inhibitor, to determine pharmacokinetics, biologic effects and establish a recommended phase II dose. Cancer Chemother. Pharmacol. 2011;67:447–454. doi: 10.1007/s00280-010-1346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.O’Connor OA, et al. The addition of granulocyte-colony stimulating factor shifts the dose limiting toxicity and markedly increases the maximum tolerated dose and activity of the kinesin spindle protein inhibitor SB-743921 in patients with relapsed or refractory lymphoma: results of an international, multicenter phase I/II study. Leuk. Lymphoma. 2015;56:2585–2591. doi: 10.3109/10428194.2015.1004167. [DOI] [PubMed] [Google Scholar]
- 595.LoRusso PM, et al. First-in-human phase 1 study of filanesib (ARRY-520), a kinesin spindle protein inhibitor, in patients with advanced solid tumors. Investig. New Drugs. 2015;33:440–449. doi: 10.1007/s10637-015-0211-0. [DOI] [PubMed] [Google Scholar]
- 596.Khoury HJ, et al. A phase 1 dose-escalation study of ARRY-520, a kinesin spindle protein inhibitor, in patients with advanced myeloid leukemias. Cancer. 2012;118:3556–3564. doi: 10.1002/cncr.26664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Shah JJ, et al. A Phase 1 and 2 study of Filanesib alone and in combination with low-dose dexamethasone in relapsed/refractory multiple myeloma. Cancer. 2017;123:4617–4630. doi: 10.1002/cncr.30892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Chari A, et al. A phase 1 dose-escalation study of filanesib plus bortezomib and dexamethasone in patients with recurrent/refractory multiple myeloma. Cancer. 2016;122:3327–3335. doi: 10.1002/cncr.30174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Lee HC, et al. A phase 1 study of filanesib, carfilzomib, and dexamethasone in patients with relapsed and/or refractory multiple myeloma. Blood Cancer J. 2019;9:80. doi: 10.1038/s41408-019-0240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Ocio EM, et al. Filanesib in combination with pomalidomide and dexamethasone in refractory MM patients: safety and efficacy, and association with alpha 1-acid glycoprotein (AAG) levels. Phase Ib/II Pomdefil clinical trial conducted by the Spanish MM group. Br. J. Haematol. 2021;192:522–530. doi: 10.1111/bjh.16788. [DOI] [PubMed] [Google Scholar]
- 601.Tabernero J, et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- 602.Ye XS, et al. A novel Eg5 Inhibitor (LY2523355) causes mitotic arrest and apoptosis in cancer cells and shows potent antitumor activity in xenograft tumor models. Mol. Cancer Ther. 2015;14:2463–2472. doi: 10.1158/1535-7163.MCT-15-0241. [DOI] [PubMed] [Google Scholar]
- 603.Infante JR, et al. Two phase 1 dose-escalation studies exploring multiple regimens of litronesib (LY2523355), an Eg5 inhibitor, in patients with advanced cancer. Cancer Chemother. Pharmacol. 2017;79:315–326. doi: 10.1007/s00280-016-3205-5. [DOI] [PubMed] [Google Scholar]
- 604.Wakui H, et al. A phase 1 and dose-finding study of LY2523355 (litronesib), an Eg5 inhibitor, in Japanese patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2014;74:15–23. doi: 10.1007/s00280-014-2467-z. [DOI] [PubMed] [Google Scholar]
- 605.Masanas M, et al. The oral KIF11 inhibitor 4SC-205 exhibits antitumor activity and potentiates standard and targeted therapies in primary and metastatic neuroblastoma models. Clin. Transl. Med. 2021;11:e533. doi: 10.1002/ctm2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Esaki T, et al. Phase I study to assess the safety, tolerability and pharmacokinetics of AZD4877 in Japanese patients with solid tumors. Arch. Drug Inf. 2011;4:23–31. doi: 10.1111/j.1753-5174.2011.00034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Infante JR, et al. A phase I study to assess the safety, tolerability, and pharmacokinetics of AZD4877, an intravenous Eg5 inhibitor in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2012;69:165–172. doi: 10.1007/s00280-011-1667-z. [DOI] [PubMed] [Google Scholar]
- 608.Jones R, et al. Phase II study to assess the efficacy, safety and tolerability of the mitotic spindle kinesin inhibitor AZD4877 in patients with recurrent advanced urothelial cancer. Investig. New Drugs. 2013;31:1001–1007. doi: 10.1007/s10637-013-9926-y. [DOI] [PubMed] [Google Scholar]
- 609.Kantarjian HM, et al. Phase I/II multicenter study to assess the safety, tolerability, pharmacokinetics and pharmacodynamics of AZD4877 in patients with refractory acute myeloid leukemia. Investig. New Drugs. 2012;30:1107–1115. doi: 10.1007/s10637-011-9660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Holen K, et al. A phase I trial of MK-0731, a kinesin spindle protein (KSP) inhibitor, in patients with solid tumors. Investig. New Drugs. 2012;30:1088–1095. doi: 10.1007/s10637-011-9653-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.Zheng L, et al. Tyrosine threonine kinase inhibition eliminates lung cancers by augmenting apoptosis and polyploidy. Mol. Cancer Ther. 2019;18:1775–1786. doi: 10.1158/1535-7163.MCT-18-0864. [DOI] [PubMed] [Google Scholar]
- 612.Novais P, Silva PMA, Amorim I, Bousbaa H. Second-Generation Antimitotics in Cancer Clinical Trials. Pharmaceutics. 2021;13:1011. doi: 10.3390/pharmaceutics13071011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Wengner AM, et al. Novel Mps1 Kinase Inhibitors with Potent Antitumor Activity. Mol. Cancer Ther. 2016;15:583–592. doi: 10.1158/1535-7163.MCT-15-0500. [DOI] [PubMed] [Google Scholar]
- 614.Schoffski P, et al. First-in-man, first-in-class phase I study with the monopolar spindle 1 kinase inhibitor S81694 administered intravenously in adult patients with advanced, metastatic solid tumours. Eur. J. Cancer. 2022;169:135–145. doi: 10.1016/j.ejca.2022.04.001. [DOI] [PubMed] [Google Scholar]
- 615.Woodward, H. L. et al. Introduction of a methyl group curbs metabolism of Pyrido[3,4- d]pyrimidine Monopolar Spindle 1 (MPS1) inhibitors and enables the discovery of the phase 1 clinical candidate N(2)-(2-Ethoxy-4-(4-methyl-4 H-1,2,4-triazol-3-yl)phenyl)-6-methyl- N(8)-neopentylpyrido[3,4- d]pyrimidine-2,8-diamine (BOS172722). J. Med. Chem. 61, 8226–8240 (2018). [DOI] [PMC free article] [PubMed]
- 616.Anderhub SJ, et al. High proliferation rate and a compromised spindle assembly checkpoint confers sensitivity to the MPS1 inhibitor BOS172722 in triple-negative breast cancers. Mol. Cancer Ther. 2019;18:1696–1707. doi: 10.1158/1535-7163.MCT-18-1203. [DOI] [PubMed] [Google Scholar]
- 617.Batalini F, et al. Phase 1b clinical trial with alpelisib plus olaparib for patients with advanced triple-negative breast cancer. Clin. Cancer Res. 2022;28:1493–1499. doi: 10.1158/1078-0432.CCR-21-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.Fennell DA, et al. Rucaparib in patients with BAP1-deficient or BRCA1-deficient mesothelioma (MiST1): an open-label, single-arm, phase 2a clinical trial. Lancet Respir. Med. 2021;9:593–600. doi: 10.1016/S2213-2600(20)30390-8. [DOI] [PubMed] [Google Scholar]
- 619.Lenart P, et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 620.Sebastian M, et al. The efficacy and safety of BI 2536, a novel Plk-1 inhibitor, in patients with stage IIIB/IV non-small cell lung cancer who had relapsed after, or failed, chemotherapy: results from an open-label, randomized phase II clinical trial. J. Thorac. Oncol. 2010;5:1060–1067. doi: 10.1097/JTO.0b013e3181d95dd4. [DOI] [PubMed] [Google Scholar]
- 621.Ellis PM, et al. A phase I open-label dose-escalation study of intravenous BI 2536 together with pemetrexed in previously treated patients with non-small-cell lung cancer. Clin. Lung Cancer. 2013;14:19–27. doi: 10.1016/j.cllc.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 622.Mross K, et al. A randomised phase II trial of the Polo-like kinase inhibitor BI 2536 in chemo-naive patients with unresectable exocrine adenocarcinoma of the pancreas - a study within the central european society anticancer drug research (CESAR) collaborative network. Br. J. Cancer. 2012;107:280–286. doi: 10.1038/bjc.2012.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Muller-Tidow C, et al. A randomized, open-label, phase I/II trial to investigate the maximum tolerated dose of the Polo-like kinase inhibitor BI 2536 in elderly patients with refractory/relapsed acute myeloid leukaemia. Br. J. Haematol. 2013;163:214–222. doi: 10.1111/bjh.12518. [DOI] [PubMed] [Google Scholar]
- 624.Vose JM, et al. The Plk1 inhibitor BI 2536 in patients with refractory or relapsed non-Hodgkin lymphoma: a phase I, open-label, single dose-escalation study. Leuk. Lymphoma. 2013;54:708–713. doi: 10.3109/10428194.2012.729833. [DOI] [PubMed] [Google Scholar]
- 625.Schoffski P, et al. A phase I, dose-escalation study of the novel Polo-like kinase inhibitor volasertib (BI 6727) in patients with advanced solid tumours. Eur. J. Cancer. 2012;48:179–186. doi: 10.1016/j.ejca.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 626.Nokihara H, et al. Phase I trial of volasertib, a Polo-like kinase inhibitor, in Japanese patients with advanced solid tumors. Investig. New Drugs. 2016;34:66–74. doi: 10.1007/s10637-015-0300-0. [DOI] [PubMed] [Google Scholar]
- 627.Lin CC, et al. A phase I study of two dosing schedules of volasertib (BI 6727), an intravenous polo-like kinase inhibitor, in patients with advanced solid malignancies. Br. J. Cancer. 2014;110:2434–2440. doi: 10.1038/bjc.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628.de Braud F, et al. A phase I, dose-escalation study of volasertib combined with nintedanib in advanced solid tumors. Ann. Oncol. 2015;26:2341–2346. doi: 10.1093/annonc/mdv354. [DOI] [PubMed] [Google Scholar]
- 629.Machiels JP, et al. A phase I study of volasertib combined with afatinib, in advanced solid tumors. Cancer Chemother. Pharmacol. 2015;76:843–851. doi: 10.1007/s00280-015-2860-2. [DOI] [PubMed] [Google Scholar]
- 630.Awada A, et al. Phase I trial of volasertib, a Polo-like kinase inhibitor, plus platinum agents in solid tumors: safety, pharmacokinetics and activity. Investig. New Drugs. 2015;33:611–620. doi: 10.1007/s10637-015-0223-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 631.Kobayashi Y, et al. Phase I trial of volasertib, a Polo-like kinase inhibitor, in Japanese patients with acute myeloid leukemia. Cancer Sci. 2015;106:1590–1595. doi: 10.1111/cas.12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Stadler WM, et al. An open-label, single-arm, phase 2 trial of the Polo-like kinase inhibitor volasertib (BI 6727) in patients with locally advanced or metastatic urothelial cancer. Cancer. 2014;120:976–982. doi: 10.1002/cncr.28519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Ellis PM, et al. A randomized, open-label phase II trial of volasertib as monotherapy and in combination with standard-dose pemetrexed compared with pemetrexed monotherapy in second-line treatment for non-small-cell lung cancer. Clin. Lung Cancer. 2015;16:457–465. doi: 10.1016/j.cllc.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 634.Dohner H, et al. Randomized, phase 2 trial of low-dose cytarabine with or without volasertib in AML patients not suitable for induction therapy. Blood. 2014;124:1426–1433. doi: 10.1182/blood-2014-03-560557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Tontsch-Grunt U, et al. Synergistic activity of BET inhibitor BI 894999 with PLK inhibitor volasertib in AML in vitro and in vivo. Cancer Lett. 2018;421:112–120. doi: 10.1016/j.canlet.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 636.Kadia TM, Ravandi F, Cortes J, Kantarjian H. New drugs in acute myeloid leukemia. Ann. Oncol. 2016;27:770–778. doi: 10.1093/annonc/mdw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.Ma WW, et al. Phase I study of Rigosertib, an inhibitor of the phosphatidylinositol 3-kinase and Polo-like kinase 1 pathways, combined with gemcitabine in patients with solid tumors and pancreatic cancer. Clin. Cancer Res. 2012;18:2048–2055. doi: 10.1158/1078-0432.CCR-11-2813. [DOI] [PubMed] [Google Scholar]
- 638.Navada SC, et al. Rigosertib in combination with azacitidine in patients with myelodysplastic syndromes or acute myeloid leukemia: Results of a phase 1 study. Leuk. Res. 2020;94:106369. doi: 10.1016/j.leukres.2020.106369. [DOI] [PubMed] [Google Scholar]
- 639.Tartaglia G, Cao Q, Padron ZM, South AP. Impaired wound healing, fibrosis, and cancer: the paradigm of recessive dystrophic epidermolysis bullosa. Int. J. Mol. Sci. 2021;22:5104. doi: 10.3390/ijms22105104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Veluswamy R, et al. KRAS G12C-mutant non-small cell lung cancer: biology, developmental therapeutics, and molecular testing. J. Mol. Diagn. 2021;23:507–520. doi: 10.1016/j.jmoldx.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 641.Garcia-Manero G, et al. Rigosertib versus best supportive care for patients with high-risk myelodysplastic syndromes after failure of hypomethylating drugs (ONTIME): a randomised, controlled, phase 3 trial. Lancet Oncol. 2016;17:496–508. doi: 10.1016/S1470-2045(16)00009-7. [DOI] [PubMed] [Google Scholar]
- 642.O’Neil BH, et al. A phase II/III randomized study to compare the efficacy and safety of rigosertib plus gemcitabine versus gemcitabine alone in patients with previously untreated metastatic pancreatic cancer. Ann. Oncol. 2015;26:1923–1929. doi: 10.1093/annonc/mdv264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643.Navada SC, et al. A phase 1/2 study of rigosertib in patients with myelodysplastic syndromes (MDS) and MDS progressed to acute myeloid leukemia. Leuk. Res. 2018;64:10–16. doi: 10.1016/j.leukres.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 644.Gilmartin AG, et al. Distinct concentration-dependent effects of the polo-like kinase 1-specific inhibitor GSK461364A, including differential effect on apoptosis. Cancer Res. 2009;69:6969–6977. doi: 10.1158/0008-5472.CAN-09-0945. [DOI] [PubMed] [Google Scholar]
- 645.Olmos D, et al. Phase I study of GSK461364, a specific and competitive Polo-like kinase 1 inhibitor, in patients with advanced solid malignancies. Clin. Cancer Res. 2011;17:3420–3430. doi: 10.1158/1078-0432.CCR-10-2946. [DOI] [PubMed] [Google Scholar]
- 646.Nie Z, et al. Discovery of TAK-960: an orally available small molecule inhibitor of polo-like kinase 1 (PLK1) Bioorg. Med. Chem. Lett. 2013;23:3662–3666. doi: 10.1016/j.bmcl.2013.02.083. [DOI] [PubMed] [Google Scholar]
- 647.Hikichi Y, et al. TAK-960, a novel, orally available, selective inhibitor of polo-like kinase 1, shows broad-spectrum preclinical antitumor activity in multiple dosing regimens. Mol. Cancer Ther. 2012;11:700–709. doi: 10.1158/1535-7163.MCT-11-0762. [DOI] [PubMed] [Google Scholar]
- 648.Valsasina B, et al. NMS-P937, an orally available, specific small-molecule polo-like kinase 1 inhibitor with antitumor activity in solid and hematologic malignancies. Mol. Cancer Ther. 2012;11:1006–1016. doi: 10.1158/1535-7163.MCT-11-0765. [DOI] [PubMed] [Google Scholar]
- 649.Weiss GJ, et al. Phase I dose escalation study of NMS-1286937, an orally available Polo-Like Kinase 1 inhibitor, in patients with advanced or metastatic solid tumors. Investig. New Drugs. 2018;36:85–95. doi: 10.1007/s10637-017-0491-7. [DOI] [PubMed] [Google Scholar]
- 650.Zeidan AM, et al. A phase Ib study of onvansertib, a novel oral PLK1 inhibitor, in combination therapy for patients with relapsed or refractory acute myeloid leukemia. Clin. Cancer Res. 2020;26:6132–6140. doi: 10.1158/1078-0432.CCR-20-2586. [DOI] [PubMed] [Google Scholar]
- 651.Garcia IA, Garro C, Fernandez E, Soria G. Therapeutic opportunities for PLK1 inhibitors: Spotlight on BRCA1-deficiency and triple negative breast cancers. Mutat. Res. 2020;821:111693. doi: 10.1016/j.mrfmmm.2020.111693. [DOI] [PubMed] [Google Scholar]
- 652.Patterson JC, et al. Plk1 inhibitors and abiraterone synergistically disrupt mitosis and kill cancer cells of disparate origin independently of androgen receptor signaling. Cancer Res. 2023;83:219–238. doi: 10.1158/0008-5472.CAN-22-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653.Wang D, et al. A novel PLK1 inhibitor onvansertib effectively sensitizes MYC-driven medulloblastoma to radiotherapy. Neuro. Oncol. 2022;24:414–426. doi: 10.1093/neuonc/noab207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.El Dika I, et al. An open-label, multicenter, phase I, dose escalation study with phase ii expansion cohort to determine the safety, pharmacokinetics, and preliminary antitumor activity of intravenous TKM-080301 in subjects with advanced hepatocellular carcinoma. Oncologist. 2019;24:747–e218. doi: 10.1634/theoncologist.2018-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Zhou LY, et al. Current RNA-based therapeutics in clinical trials. Curr. Gene Ther. 2019;19:172–196. doi: 10.2174/1566523219666190719100526. [DOI] [PubMed] [Google Scholar]
- 656.Uchino K, Ochiya T, Takeshita F. RNAi therapeutics and applications of microRNAs in cancer treatment. Jpn. J. Clin. Oncol. 2013;43:596–607. doi: 10.1093/jjco/hyt052. [DOI] [PubMed] [Google Scholar]
- 657.Yim H. Current clinical trials with polo-like kinase 1 inhibitors in solid tumors. Anticancer Drugs. 2013;24:999–1006. doi: 10.1097/CAD.0000000000000007. [DOI] [PubMed] [Google Scholar]
- 658.Kim HJ, Kim A, Miyata K, Kataoka K. Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv. Drug Deliv Rev. 2016;104:61–77. doi: 10.1016/j.addr.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 659.Ramos Perez J, Montalban-Bravo G. Emerging drugs for the treatment of chronic myelomonocytic leukemia. Expert Opin. Emerg. Drugs. 2020;25:515–529. doi: 10.1080/14728214.2020.1854224. [DOI] [PubMed] [Google Scholar]
- 660.Kawakami M, et al. Polo-like kinase 4 inhibition produces polyploidy and apoptotic death of lung cancers. Proc. Natl Acad. Sci. 2018;115:1913–1918. doi: 10.1073/pnas.1719760115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661.Fu S, et al. Multicenter Phase II Trial of the WEE1 inhibitor adavosertib in refractory solid tumors harboring CCNE1 amplification. J. Clin. Oncol. 2023;41:1725–1734. doi: 10.1200/JCO.22.00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 662.Vakili-Samiani S, et al. Targeting Wee1 kinase as a therapeutic approach in hematological malignancies. DNA Repair. 2021;107:103203. doi: 10.1016/j.dnarep.2021.103203. [DOI] [PubMed] [Google Scholar]
- 663.Elbaek CR, Petrosius V, Sorensen CS. WEE1 kinase limits CDK activities to safeguard DNA replication and mitotic entry. Mutat. Res. 2020;819-820:111694. doi: 10.1016/j.mrfmmm.2020.111694. [DOI] [PubMed] [Google Scholar]
- 664.Chandrasekaran A, Elias KM. Synthetic lethality in ovarian vancer. Mol. Cancer Ther. 2021;20:2117–2128. doi: 10.1158/1535-7163.MCT-21-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]