Abstract
The immediate-early (IE) proteins of herpes simplex virus (HSV) function on input genomes and affect many aspects of host cell metabolism to ensure the efficient expression and regulation of the remainder of the genome and, subsequently, the production of progeny virions. Due to the many and varied effects of IE proteins on host cell metabolism, their expression is not conducive to normal cell function and viability. This presents a major impediment to the use of HSV as a vector system. In this study, we describe a series of ICP4 mutants that are defective in different subsets of the remaining IE genes. One mutant, d109, does not express any of the IE proteins and carries a green fluorescent protein (GFP) transgene under the control of the human cytomegalovirus IE promoter (HCMVIEp). d109 was nontoxic to Vero and human embryonic lung (HEL) cells at all multiplicities of infection tested and was capable of establishing persistent infections in both of these cell types. Paradoxically, the genetic manipulations that were required to eliminate toxicity and allow the genome to persist in cells for long periods of time also dramatically lowered the level of transgene expression. Efficient expression of the HCMVIEp-GFP transgene in the absence of ICP4 was dependent on the ICP0 protein. In d109-infected cells, the level of transgene expression was very low in most cells but abundant in a small subpopulation of cells. However, expression of the transgene could be induced in cells containing quiescent d109 genomes weeks after the initial infection, demonstrating the functionality of the persisting genomes.
The temporal regulation of herpes simplex virus type 1 (HSV-1) gene expression during permissive infection commences with the induction of the immediate-early (IE) genes by the virion protein VP16 (3, 8). VP16 activates transcription by binding, along with the cellular factors Oct1 and host cell factor (HCF), to the TAATGARAT elements present in all IE promoters (56, 57, 80, 113, 116a). The virus encodes five IE proteins, designated infected cell polypeptides (ICP) 0, 4, 22, 27, and 47 (13, 43, 84). With the exception of ICP47, these IE proteins are known to have regulatory functions demonstrated to affect the coordinated expression of the HSV genome. ICP4 and ICP27 are absolutely essential for virus replication (18, 24, 71, 87, 96, 115), while the growth of ICP0 mutants is significantly impaired (97, 110). These three IE proteins have the most profound effects on subsequent viral gene expression.
ICP4 is the major regulatory protein of the virus, and it is necessary for the transition of viral gene transcription from the IE to the E phase (24, 87, 115). It functions as a repressor (19, 38, 76, 82) or activator (19, 27, 33, 81, 90) of transcription by forming multiple contacts with basal transcription factors (9, 37, 38, 105).
ICP0 is a potent transactivator of viral and cellular promoters in transient assays (27, 33, 81, 90), and it provides for efficient viral gene expression and growth, in vitro and in vivo (5, 7, 62). ICP0 enhances gene expression, in part by increasing the transcription rates of viral genes (50, 99), although the exact mechanism by which ICP0 accomplishes this is not yet clear. These functions most likely underlie the requirement for ICP0 in lytic viral growth and the efficient reactivation of the virus from latent infections as shown in the mouse and rabbit models (5, 14, 28, 35, 62, 95). ICP0 has also been shown to interact with ND10 nuclear structures (29, 67) and components of the transcription (61), translation (51), cell cycle (52), and proteolytic (30) machinery of the cell. Therefore, it has the potential to affect many aspects of host cell metabolism.
ICP27 regulates the processing of viral and cellular mRNAs (39, 73, 74, 102, 106). It may also act by modulating the activities of ICP4 and ICP0 (75, 104), as well as the modification state of ICP4 (75, 92, 112). The combined activities of ICP27 contribute to efficient late-gene expression (71, 96). Recent studies have shown that ICP27 also significantly contributes to elevated levels of early-gene expression, explaining the requirement for ICP27 in viral DNA synthesis (98, 114).
ICP22 and ICP47 can be deleted from the genome without greatly affecting viral growth and viability in most cell types (70, 86, 103). ICP22 promotes efficient late-gene expression in a cell type-dependent manner (103). It has been reported to give rise to a novel phosphorylated form of RNA polymerase II (pol II) (93) as well as to regulate the stability and splicing pattern of the ICP0 mRNA (10). Its regulatory effects may thus be mediated through these activities. The function of ICP47 may be more relevant in vivo, where it may help the virus escape immune surveillance on the basis of its ability to block the presentation of antigenic peptides to CD8+ cells (118).
In ICP4 mutant backgrounds, expression of viral early and late genes is dramatically reduced. This has led to the consideration of such backgrounds as starting points for the construction of gene transfer vehicles. However, despite the limited expression of the HSV genome in ICP4 mutant backgrounds, such mutants are very toxic to cells. This is most likely because of the overexpression of the remaining IE proteins in the absence of ICP4. Several of the IE genes reduce the transformation of cells when cotransfected with a selectable marker, implying that the protein products of the transfected IE genes are not conducive to cell survival (48). Given this observation, and the findings of numerous groups on the effects of IE proteins on different aspects of host cell metabolism, it seems reasonable that the elimination of IE protein activity is essential for the efficient and safe use of HSV as a gene transfer vehicle. Lending support to this hypothesis are the observations that elimination of the VP16 activation function in an ICP4 mutant background (48, 88) and the deletion of subsets of IE genes (99, 117) reduce toxicity. Both ICP0 and ICP22 contribute to toxicity, as demonstrated by the improved survival of infected cells when these activities were individually abrogated in the background of viruses already deleted for ICP4 and ICP27 (99, 117). However, both of these triple mutants remained toxic at high multiplicities of infection (MOI). For one of these studies, we hypothesized that the elimination of ICP22 activity from the ICP4− ICP27− ICP0− background would further reduce toxicity.
Another point of consideration with viral vectors is the degree of transgene expression. The IE proteins of HSV have multiple and varied effects on the expression of viral genes, mostly, but not entirely, acting to increase gene expression. Similar effects are to be expected for transgenes inserted into the viral genome. Therefore, the elimination of viral IE genes may be expected to decrease toxicity but also might significantly reduce transgene expression. In support of this hypothesis, elimination of VP16, ICP4, and ICP0 activity reduced cytotoxicity but also reduced gene expression (88, 89). The same observation was made in studies of an ICP4− ICP27− ICP0− mutant (99). In the latter study it was concluded that ICP0 could significantly increase the transcription rates of genes in the absence of ICP4 and ICP27. The genes of HSV are standard pol II transcription units (15). Therefore, heterologous pol II promoters used to express transgenes may be affected by HSV IE genes in the same way as HSV promoters are affected.
In the current study, we set out to further assess the toxicity of different IE mutant backgrounds and examine the level of gene expression as a consequence of reduced toxicity and elimination of viral trans-acting factors. In all the mutants that are deleted for ICP4 and ICP27, an expression cassette containing the green fluorescent protein (GFP) reporter gene under the control of the human cytomegalovirus (HCMV) IE promoter-enhancer was inserted into the ICP27 deletion locus as a model transgene. The following points were addressed, with the stated general outcomes.
(i) Vector toxicity.
Both ICP0 and ICP22 contribute to the toxicity of a virus deleted for ICP4 and ICP27. Elimination of these functions in addition to ICP47 resulted in a virus that was by several criteria nontoxic to Vero and human embryonic lung (HEL) cells at high MOI.
(ii) Genome persistence.
The genome of the virus (d109) that did not express any IE proteins was capable of long-term persistence in both HEL and Vero cells.
(iii) Gene expression.
The abundant expression of the HCMVIEp-GFP transgene was dependent on the ICP0 protein. In the absence of ICP0, the level of transgene expression was very low in most cells but abundant in a subpopulation of cells. Expression of the transgene could also be induced weeks after infection in cells containing persistent d109 genomes.
MATERIALS AND METHODS
Cells and viruses.
Vero cells and the complementing cell lines E5 (ICP4+) (20), E26 (ICP4+ ICP27+) (98), L7 (ICP0+) (99), and FO6 (ICP4+ ICP27+ ICP0+) (99) were maintained by standard cell culture procedures as previously described (17, 24). Tissue culture medium for maintaining L7 cells was supplemented with 400 μg of G418 (GIBCO-BRL)/ml. For maintaining FO6 cells, the medium was supplemented with 400 μg of G418 and 300 μg of hygromycin B (Boehringer Mannheim)/ml. Viruses carrying single or multiple IE mutations were propagated and subjected to titer determination on the appropriate cell lines. The viruses d92 (ICP4− ICP27−), d96 (ICP4− ICP22−), and d95 (ICP4− ICP27− ICP22−) were described previously (98, 117). The viruses with mutations in capsid genes, including K23Z (22), K5ΔZ (22), KUL26ΔZ (23), and KΔ19C (85), were generously provided by Stanley Person (Johns Hopkins University, Baltimore, Md.). In experiments that compare d107, d106, d104, and d109, the infectivities of the virus stocks were normalized on the basis of the number of genomes in infected nuclei at 6 h postinfection (p.i.) as determined by PCR amplification.
Plasmid constructions.
The plasmid pEBΔAE contains the EcoRI-B genomic fragment with deleted copies of ICP4 (d120) (18) and ICP0 (0Δ) and a wild-type (wt) copy of ICP27. The boundaries of the deletion in the ICP0 gene are defined by the BsmI site at nucleotide (nt) 120142 and the AseI site at nt 124775. pEBΔAE was constructed by inserting the 2.7-kb XbaI fragment isolated from pAE into the unique XbaI site of pEB. Both pEB and pAE derived their cloned HSV sequences from pdlEB, which contains the EcoRI-B genomic fragment isolated from the virus mutant d120 ligated into the unique EcoRI site of pBR325 (21). To construct pEB, pdlEB was cleaved with BsmI and the ends of the resulting fragments were modified by ligation of adapter oligonucleotides (5′-CGTCTAGACGCG-3′) to convert the BsmI sites to XbaI sites. This pool of fragments was digested simultaneously with EcoRI and XbaI and then fractionated in a 0.8% agarose gel to isolate the 10-kb EcoRI-XbaI fragment. After being isolated by electroelution, this 10-kb fragment was cloned into the corresponding sites of puc19 to generate pEB. To construct pAE, pdlEB was cleaved simultaneously with AseI and EcoRI. After the ends of the resulting fragments were filled in with DNA polymerase I Klenow fragment, they were modified with XbaI linkers. From this pool of fragments, a 2.7-kb XbaI fragment was isolated by electroelution and ligated into the XbaI site of puc19 to produce pAE.
In plasmid pGFP27, the GFP gene, under the control of the HCMV IE promoter (HCMVIEp), is substituted between the BamHI and SalI sites in the ICP27 locus. pGFP27 was constructed by insertion of the 1.6-kb HCMVIEp-GFP cassette into the unique PacI site of pAT2. The insert corresponded to an AseI-MluI fragment derived from pGFPX whose ends were modified with PacI linkers. pGFPX was derived from pEGFP-C1 (Clontech) by deletion of the multiple cloning sites between the BamHI and BglII restriction enzyme sites. pAT2 resulted from the conversion of the unique XbaI site in pAT1 into a PacI site. Both pAT2 and pAT1 contain the EcoRI-to-SalI (nt 110095 to 120902) HSV sequence with the 1.2-kb deletion between the BamHI and SalI sites in the ICP27 locus. At the site of the ICP27 deletion is a unique XbaI site in pAT1. Construction of pAT1 was done by cloning the 3.2-kb EcoRI-BamHI fragment isolated from pKEB-S1 (4) into the corresponding site in pucSF. The plasmid pucSF contains the 6.4-kb SalI fragment derived from pKSF (4) inserted into the corresponding site in puc19.
The plasmid pTGTΔ contains the joint fragment of the HSV genome (P isotype) extending from the AseI site (nt 124775) 5′ of ICP0 to the proximal BamHI site (nt 136289) 3′ of ICP22, with the d120 mutation and the 270-bp TGTΔ deletion in the ICP22 promoter located in the c′ repeat sequence. The TGTΔ mutation removes the TAATGARAT elements in the ICP22 promoter and is defined by the EcoRI and BssHII restriction enzyme sites. pTGTΔ was generated by insertion of the 2.7-kb XbaI fragment isolated from pAE into the unique XbaI site of pucBNΔXB. To construct pucBNΔXB, pucBN (99) was digested with XbaI and BssHII. The large vector fragment isolated from this digest was then ligated in the presence of the oligonucleotide adapters 5′-CGCGCTTAATTAAT-3′ and 5′-CTAGATTAATTAAG-3′ to allow joining of the XbaI- and BssHII-restricted ends. Both of these restriction enzyme recognition sites remained intact in the resulting plasmid, pucBNΔXB, and the oligonucleotide adapters also introduced a PacI site.
The pucGFP plasmid used as the source for the GFP fragment probe carries the 1.6-kb PacI fragment derived from pGFP27 and inserted into the unique PacI site of pNEB193 (New England Biolabs). The orientation of the insert is such that a HindIII-NheI double digestion results in a 1-kb fragment containing the GFP coding sequence.
Virus constructions.
The DNA-calcium phosphate precipitates used for transfections were prepared essentially as described by Graham and van der Eb (36). A typical mixture used to transfect approximately 106 cells plated in two 60-mm-diameter dishes contained 3 μg of viral DNA, 1 to 3 μg of the linearized plasmid carrying the mutant IE alleles, 1 μg of the plasmid pW3ΔHS8 (20), and 1 μg of pGreen Lantern-1 (GIBCO-BRL). pW3ΔHS8 provides ICP0, which improves the transfection efficiency of recombinant virus mutants deficient in this gene product. The plasmid pGreen Lantern-1 was added to allow convenient monitoring of transfection efficiency. It was not added for the construction of d104. Generation of mutants by crossing two viruses was done by coinfecting cells with approximately 5 PFU of each parent virus per cell.
Southern blot analysis.
DNA, isolated from the IE mutant viruses as previously described (98), was cleaved with HpaI to detect the GFP substitution in ICP27, with PstI and SacI to detect the 0Δ deletion in ICP0, with BamHI and SacI to detect the d120 mutation in ICP4 (18), and with NcoI to detect the TGTΔ mutation in the ICP22 and ICP47 promoters. Digested viral DNA was fractionated by agarose gel electrophoresis, transferred to nitrocellulose, and hybridized to nick-translated probes as previously described (100, 107). Gel-purified DNA fragments that were nick translated and used as probes included the 2.4-kb BamHI-SacI fragment derived from pKHX-BH (4) for detecting ICP27, the 1.3-kb PstI fragment from pEB for detecting ICP0, the 1.8-kb BamHI-Y fragment from pKBY (98) for detecting ICP4, and the 1.6-kb PvuII fragment from pucBNΔXB for detecting the TGTΔ mutation.
Isolation of infected-cell RNA and Northern blot analysis.
Total infected-cell RNA was prepared as previously described (98). For infections performed in the presence of cycloheximide (CHX), the medium was supplemented with the inhibitor (100 μg/ml) 1 h prior to and during infection. Fractionation of the RNA samples in 1.3% agarose–formaldehyde gels and the conditions for blotting, hybridization, and washing have also been previously described (45). Gel-purified DNA fragments that were nick translated and used as probes included the 2-kb HindIII fragment from pucBN (99) for detecting the ICP22 mRNA, the 2.4-kb BamHI-SacI fragment from pKHX-BH (4) for detecting ICP27, and the 1-kb NheI-HindIII fragment from pucGFP for detecting GFP.
Analysis of viral proteins.
Viral polypeptides were radiolabeled by incubating approximately 6 × 105 cells infected at the indicated MOI with 100 μCi of [35S]methionine per ml of medium at 6 to 9 h p.i. The CHX-treated cells were incubated in the presence of the inhibitor (100 μg/ml) for 1 h prior to infection until 6 h p.i. The treated monolayers were then washed twice with Tris-buffered saline and further incubated for 3 h in the presence of actinomycin D (10 μg/ml) and [35S]methionine (100 μCi per plate). The labeled viral proteins were solubilized in sodium dodecyl sulfate (SDS) sample solution and separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (60, 66). For Western blot analysis of GFP, proteins separated by SDS-PAGE were electroblotted to nitrocellulose and analyzed using an anti-GFP (α-GFP) monoclonal antibody (Clontech) diluted 1:500 and a chemiluminiscence Western blotting kit (Boehringer Mannheim).
Colony formation inhibition assays.
Inhibition of colony formation by the different viral mutants was assayed as previously described (117). Briefly, Vero cells infected at different MOI and control uninfected monolayers were trypsinized at 6 h p.i. to prepare single-cell suspensions that were then serially diluted and plated in medium supplemented with 20% fetal bovine serum. Ten to 14 days after plating, the colonies were stained with crystal violet and counted.
PCR analysis.
For normalizing virus stocks, 2 × 106 cells plated in 60-mm-diameter dishes were infected at an MOI of 10 based on titers determined by plaque assay. At 6 h p.i., the cultured cells were washed with cold Tris-buffered saline and scraped from the plates. The cells were pelleted by low-speed centrifugation and resuspended in 0.5 ml of RSB buffer (10 mM NaCl, 10 mM Tris-HCl [pH 7.4], 3 mM MgCl2). Nonidet P-40 (1%, 0.5 ml in RSB) was then added, and the mixture was vigorously vortexed. The infected nuclei were pelleted, resuspended in lysis buffer (1.2% Nonidet P-40 and 0.4 mg of proteinase K per ml, prepared in Tris-EDTA buffer) and incubated overnight at 45°C. The mixtures were then heated at 95°C to inactivate the proteinase. Aliquots (2.5 μl) of these nuclear lysates were used in PCRs. For determining the number of genomes in persistently infected cells, total cell DNA was prepared from the infected cells as previously described (98) and equivalent aliquots from all the samples were used in PCRs.
The PCR conditions, with minor modifications, and the primers used to amplify glycoprotein C (gC) sequences were described previously (55). A typical PCR mixture made up to 100 μl contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 10% glycerol, 3 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 50 pmol of each upstream and downstream primer (gC-1, 5′-GGGTCCGTCCCCCCCAAT-3′; gC-2, 5′-CGTTAGGTTGGGGGCGCT-3′) (55), 2.5 U of Taq DNA polymerase (Boehringer Mannheim), and the sample or standard DNA. Amplifications were done with a Perkin-Elmer GeneAmp PCR System 2400 with an initial denaturation step (94°C, 5 min) followed by 25 cycles of denaturation (94°C, 30 s), annealing (60°C, 30 s), and extension (72°C, 30 s) and a final extension step (72°C, 7 min). For detection of the 109-bp gC PCR product, equal aliquots of each reaction mixture were run on a 1% agarose–1% NuSieve GTG (FMC BioProducts) gel. The gel was treated with 0.4 N NaOH for 30 min and blotted on a GeneScreen Plus membrane (NEN Life Science Products) in 0.4 N NaOH. After being subjected to UV cross-linking for 30 s (UV Stratalinker 2400; Stratagene), the membrane was prehybridized (6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1% SDS, 5× Denhardt’s solution, 8 μg of salmon sperm DNA per ml) at 50°C for 1 h and hybridized (6× SSC, 1% SDS, 105 to 106 cpm/ml probe) at 50°C for 1 h. The end-labeled oligonucleotide probe (gC-3, 5′-TAGAGGAGGTCCTGACGAACA-3′) (55) was used at a concentration of approximately 10 ng/ml of hybridization solution. After hybridization, the blots were washed (6× SSC, 0.001% SDS) with several changes of buffer, rinsed with 2× SSC, allowed to dry, and exposed to film for autoradiography. The radioactive signals were quantitated with an AMBIS 4000 radioanalytic imaging system (AMBIS, Inc., San Diego, Calif.).
Immunofluorescence.
Infected and uninfected cells were prepared on circular coverslips. For detection of promyelocytic leukemia antigen (PML), cells were fixed with methanol as previously described (117). For simultaneous detection of ICP0 or ICP4 with GFP, the infected cells were fixed with paraformaldehyde as previously described (40). Briefly, the cells were washed with phosphate-buffered saline (PBS), incubated in 4% paraformaldehyde solution for 10 min, and then rinsed with PBS. Following paraformaldehyde fixation, the cells were permeabilized with 0.2% Triton X-100 for 2 min and then rinsed twice with PBS. Staining of the cells for PML, ICP0, or ICP4 was done as previously described (117). The monoclonal antibodies against PML (Santa Cruz Biotechnology, Santa Cruz, Calif.), ICP0, and ICP4 (no. 1112 and no. 1011, respectively; Goodwin Institute for Cancer Research, Inc., Plantation, Fla.) were used at dilutions of 1:30, 1:1,000, and 1:1,000, respectively. The stained antigens were visualized at a magnification of ×100 (PML) or ×60 (ICP0 and ICP4) with the appropriate cubes for fluorescent imaging in conjunction with a Nikon FXA photomicroscope.
RESULTS
We have previously described mutants inactivated for ICP22 or ICP0 in an ICP4− ICP27− background (99, 117). Cells infected with d95 (ICP4− ICP27− ICP22−) exhibited prolonged cell survival and high-level expression of ICP0 as well as some early genes (117). Although d95-infected cell monolayers maintained their normal morphology for an extended period of time, the cells stopped dividing and eventually died. Gene expression from the genome was severely restricted upon inactivation of ICP0 in the same ICP4− ICP27− background. In this virus, d97 (ICP4− ICP27− ICP0−), the level of expression of an inserted lacZ transgene under the control of the ICP0 promoter was initially high but declined soon thereafter to low levels that could still be detected up to 2 weeks p.i. The survival of d97-infected cells was improved relative to that of d92- or d95-infected cells, although the virus was still toxic to cells at high MOI. The characteristics of both d95 and d97 demonstrate the contributions of ICP0 and ICP22 to virus toxicity. The studies detailed herein examine genome persistence, vector toxicity, and transgene expression in the absence of IE protein functions.
Generation of mutants inactivated for multiple IE genes.
Construction of the virus d109 (ICP4− ICP27− ICP0− ICP22− ICP47−) proceeded in a series of steps that generated and utilized a set of mutants defective in sets of IE genes. All mutants deficient in ICP4 carried the d120 allele of the gene. ICP0 was inactivated by a substantial deletion (0Δ) that removed the entire coding region along with the promoter (Fig. 1C). The sequences deleted in ICP27 are the same as those deleted in the 5dl1.2 allele (71) of the gene. In place of the deletion in ICP27, an HCMVIEp-GFP transgene cassette containing the GFP gene under the control of the HCMV IE promoter was inserted (Fig. 1B).
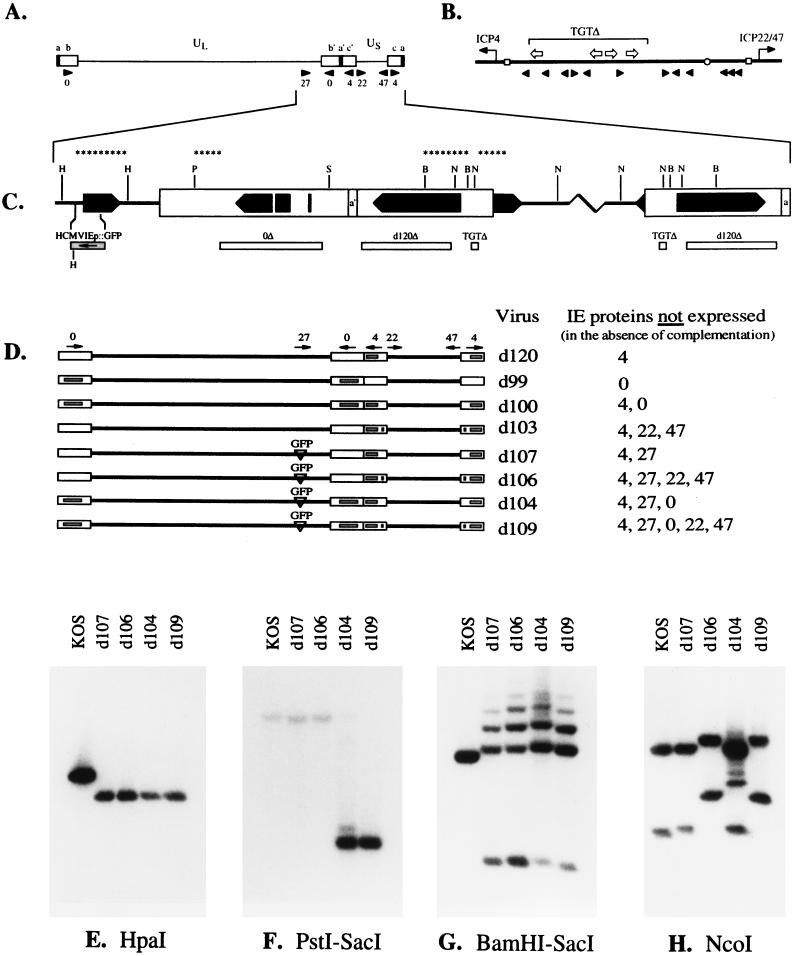
FIG. 1.
Structures of isogenic mutants defective in combinations of IE genes. (A) The viral genome is represented with the unique long (UL) and short (US) regions bounded by terminal and internal repeats (white boxes). The genomic locations and directions of transcription of the IE genes are indicated (arrowheads). (B) The TAATGARAT deletion, TGTΔ (bracketed), introduced into the promoter regions of ICP22/47. The positions of the TAATGARAT elements (open arrows), binding sites for the transcription factor SP1 (arrowheads), TATA boxes (open squares), and oriS (open circle) are shown relative to the transcription start sites of ICP4 and ICP22/47. (C) Expanded map of the right end of the d109 genome, showing the coding sequences of the IE genes (solid arrows) and the deletion mutations (white bars) in ICP4 (d120), ICP0 (0Δ), and the ICP22/47 promoter (TGTΔ). The transgene cassette containing the GFP reporter gene under the control of the HCMV IE promoter substituted into the deletion in ICP27 is represented by a shaded bar, with the arrow inside indicating the direction of transcription. The relevant restriction sites are indicated and are a bbreviated as follows: H, HpaI; P, PstI; S, SacI; B, BamHI; and N, NcoI. The line of asterisks represent the nick-translated DNA fragments used as probes for Southern blot analysis. (D) Structures of the isogenic mutants. The deletion mutations in ICP4, ICP0, and the ICP22/47 promoter present in the different virus mutant strains are indicated by the shaded bars inside the white boxes representing the repeat sequences. The GFP substitution in ICP27 is also shown (inverted triangles). The mutant designations are indicated on the right along with a list of the IE proteins not synthesized by the viruses in the absence of complementation. Viral DNA isolated from cells infected with the indicated viruses were digested with HpaI (E), PstI-SacI (F), BamHI-SacI (G), or NcoI (H) and analyzed by Southern hybridization. The probes used are shown in panel C and described in Materials and Methods.
To inactivate ICP22, a 270-bp sequence containing the TAATGARAT elements was deleted from its promoter (Fig. 1C). Since this promoter is the same as that upstream of the ICP47 gene, the mutation (TGTΔ) affects expression of both ICP22 and ICP47. The mutated promoter, which retains the TATA box and several Sp1 sites, closely resembles some of the early and late promoters activated by ICP4. It was assumed that removal of the TAATGARAT elements would eliminate the responsiveness of the promoter to induction by VP16 but perhaps not to activation by ICP4. The TGTΔ mutation was intended to abrogate ICP22 and ICP47 expression under the noncomplementing conditions in which these experiments are conducted but also to allow expression of functional ICP22 under the complementing conditions used to isolate and grow the mutants. The latter takes into account the apparent growth advantage that ICP22 provides to multiple IE mutants.
The virus d100 (Fig. 1D) was generated by cotransfecting E26 cells with d92 viral DNA and pEBΔAE linearized by HindIII. The d92 virus carries the d120 (ICP4) and 5dl1.2 (ICP27) mutant alleles. The plasmid pEBΔAE contains a genomic fragment that carries the d120 (ICP4) and 0Δ (ICP0) mutations as well as a wt copy of ICP27. Progeny from the cotransfection were plated on FO6 cells, and individual plaques were initially screened by Southern blot analysis for incorporation of the Δ0 allele. Plaque isolates whose ICP0 loci were both converted to the 0Δ allele were further analyzed to determine the state of the ICP27 gene. None of the isolates retained the deletion in ICP27. One isolate carrying the intended deletions in both loci of ICP4 and ICP0 was further purified and designated d100. The virus d99 (Fig. 1D) was derived by rescuing the d120 mutation of d100 to produce a virus that carried only the 0Δ mutation.
To construct d104 (Fig. 1D), FO6 cells were cotransfected with pGFP27 cleaved with HindIII and d100 viral DNA. The plasmid pGFP27 contains the HCMVIEp-GFP transgene inserted in the ICP27 locus. Viruses derived from the cotransfection were plated on FO6 cells, and well-isolated green fluorescent plaques were further purified and analyzed by Southern blotting to confirm incorporation of the GFP insert into the ICP27 locus. The virus resulting from this construction, d104, has the d120 (ICP4) and 0Δ (ICP0) mutations as well as the GFP substitution in ICP27.
To generate virus d103 (Fig. 1D), E5 cells were cotransfected with d96 viral DNA and EcoRI-cleaved pTGTΔ. The d96 virus carries the d120 (ICP4) and n199 (ICP22) mutant alleles. The pTGTΔ plasmid contains a wt ICP22 gene, the d120 mutation, and the TGTΔ deletion that removes the TAATGARAT elements from the ICP22 promoter (Fig. 1C). Progeny from the cotransfection were plated on E5 cells, and individual plaques were screened by Southern hybridization for both the incorporation of the TGTΔ deletion and repair of the n199 nonsense mutation in ICP22. One isolate carrying the d120 mutation in ICP4, the TGTΔ mutation in the ICP22 and ICP47 promoters, and a wt copy of the ICP22 gene was propagated and designated d103.
Viruses d106 and d107 (Fig. 1D) were derived from a cross between d103 and d104. The virus d109 (Fig. 1D) was generated by crossing d106 and d104. Isolated green fluorescent plaques resulting from these coinfections were analyzed by Southern blotting to determine their genotypes. The mutant d107 carried the d120 deletion in ICP4 and the GFP substitution in ICP27. d106 carried both of these mutations in addition to the TGTΔ deletion in the ICP22 and ICP47 promoters. The isolate designated d109 carried the d120 (ICP4), 0Δ (ICP0), and TGTΔ (ICP22 and ICP47 promoters) deletions as well as the GFP substitution in ICP27.
Figure 1E to H shows Southern blot analyses in which restriction digests of KOS, d107, d106, d104, and d109 were probed to demonstrate the structures of the IE genes in d109. Figure 1B shows an expanded map of the right end of the genome, highlighting the sequences used to probe the Southern blots (asterisks), the locations of the coding sequences for the IE genes, relevant restriction sites, and the mutant alleles. To visualize the ICP27 sequences in the viruses, electrophoretically separated HpaI fragments of the viral DNA were probed with the indicated fragment. The substituted HCMVIEp-GFP transgene provides an additional HpaI site (Fig. 1B), and as a consequence of this substitution in the ICP27 locus, all the mutant viruses contained the shortened HpaI fragment (Fig. 1E). To visualize the ICP0 sequences in the viruses, separated PstI-SacI fragments of viral DNA were probed with the indicated fragments. The 4.6-kb deletion (0Δ) in ICP0 present in d104 and d109 resulted in the shorter PstI-SacI fragment relative to the wt allele of the gene (Fig. 1F). To visualize the ICP4 sequences in the viruses, fractionated BamHI-SacI fragments of viral DNA were probed with the indicated fragments. All the mutant viruses carried the d120 allele, as indicated by the fusion of BamHI-Y sequences (corresponding to the probe fragment) to the BamHI-SacI fragment spanning the joint, by virtue of the deletion in ICP4 (Fig. 1B). This 2.2-kb BamHI-SacI fragment spanning the joint is detected running slightly behind the 1.8-kb BamHI fragment characteristic of the wt ICP4 allele (Fig. 1G). Variation in the number of copies of the a′ repeat sequence in a given population produces the multiple bands observed above the 2.2-kb fragment. The smaller, 1.3-kb fragment detected by the probe corresponds to the S-terminal BamHI of d120. To demonstrate the presence of the TGTΔ mutation in the ICP22 and ICP47 promoters of the different viruses, electrophoretically separated NcoI fragments of viral DNA were probed with the indicated fragment. Incorporation of the TGTΔ mutation in d106 and d109 is indicated by the loss of an NcoI site located within the 270-bp deleted sequence (Fig. 1B). As a consequence of this deletion in d106 and d109, two adjacent NcoI fragments are fused, resulting in larger NcoI fragments of 4.6 and 2.8 kb encompassing ICP22 and ICP47, respectively (Fig. 1H). The wt NcoI fragments containing ICP22 and ICP47 are 4.1 and 2.3 kb in size, respectively, and these were not detected in d106 and d109, demonstrating the presence of the TGTΔ mutation in both copies of the c′ repeat. Therefore, d109 contains the d120 (ICP4), 0Δ (ICP0), and TGTΔ (ICP22 and ICP47 promoters) deletions as well as the GFP substitution in ICP27.
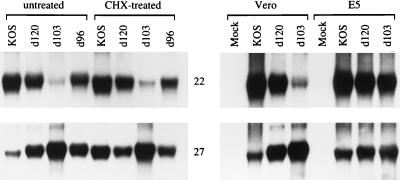
The TGTΔ mutation introduced into the ICP22 and ICP47 promoters was intended to restrict expression of these genes under noncomplementing conditions but also to allow for expression of functional ICP22 under the complementing conditions used to isolate and grow the mutants. Figure 2 shows the effect of the TGTΔ deletion present in d103 on the expression of ICP22. As a consequence of the TGTΔ mutation, accumulation of the ICP22 mRNA in d103-infected Vero cells was significantly reduced compared to that in d120- or d96-infected cells (top left). The levels of the ICP27 mRNA in cells infected in the presence of CHX indicate that the virus input used for the d103 infections was in fact larger (bottom left), thereby suggesting that the actual level of ICP22 mRNA made in d103-infected cells is lower than that observed. Moreover, the d103 mutant still expresses the other IE activator, ICP0. In the absence of ICP0, such as in the mutant d109, expression of ICP22 is expected to be even further reduced. This prediction was borne out at the level of protein synthesis, as shown below. In the presence of ICP4 provided by the complementing cell line E5, ICP22 mRNA expressed by d103 accumulated to levels comparable to those in d120-infected cells (Fig. 2, top right). These results indicate that the TGTΔ mutation can effectively reduce expression of ICP22. In the absence of the TAATGARAT elements, induction by VP16 does not occur and ICP22 is no longer expressed as an IE gene. However, the mutated ICP22 promoter remains responsive to activation by ICP4, so that under complementing conditions in which ICP4 is present, the gene is expressed, perhaps with the same kinetics as an early gene. Consistent with this interpretation, the relative abundance of the ICP22 mRNA in d103-infected Vero and E5 cells is reminiscent of that of thymidine kinase in the two cell types (45). As predicted, expression of functional ICP22 under these conditions gives the virus a growth advantage since d103 can be propagated to higher titers than can d96 (data not shown).
FIG. 2.
Effect of the TGTΔ mutation on expression of ICP22. Total cell RNA, isolated at 6 h p.i. from Vero or E5 cells (ICP4+) infected (MOI = 10) with the indicated viruses, was processed for Northern blot analysis. For infections of Vero cells done in the presence of CHX, the medium was supplemented with the inhibitor 1 h prior to and during infection. The blots were hybridized to probes specific for ICP22 and ICP27 mRNAs as indicated (22 and 27, respectively). KOS is the wt virus control, and d96 (ICP4− ICP22−) (117) carries the d120Δ deletion in ICP4 and the n199 nonsense mutation in ICP22 (93).
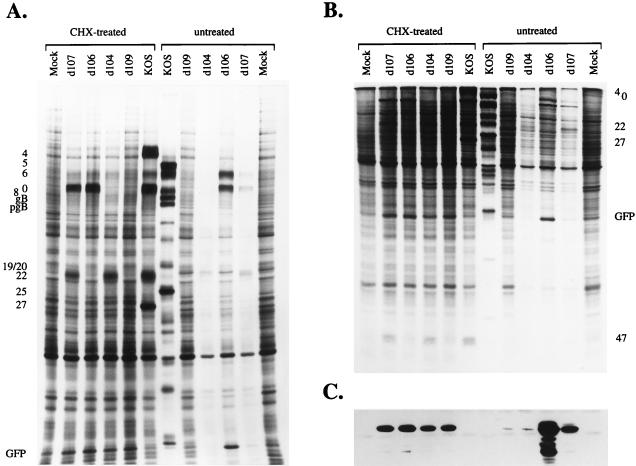
The viruses d106, d107, d104, and d109 carry mutations that should abrogate the expression of specific sets of IE proteins. To demonstrate that this is the case, a CHX reversal experiment (42) was performed on cells infected with the multiple IE mutants. This type of experiment predominantly limits viral gene expression to that of the IE genes. The results of this experiment are shown in Fig. 3. In addition to the SDS–9% polyacrylamide gel routinely used for these experiments (Fig. 3A), the same samples were also run on an 18% gel (Fig. 3B) to allow the detection of the smaller 27-kDa GFP and 11-kDa ICP47 proteins. The mobility of GFP coincides with that of a cellular protein. To verify the identity of the band corresponding to GFP, a Western blot analysis was also performed (Fig. 3C). As expected from the genotypes of these viruses, none of the mutants expressed ICP4 or ICP27. ICP0 was not synthesized in d104- or d109-infected cells. ICP22 and ICP47 were not synthesized in d106- or d109-infected cells. Also, under conditions of CHX reversal, all four mutants synthesized GFP to similar levels. These results show that in infected Vero cells, d109 does not express ICP4, ICP27, ICP0, ICP22, or ICP47, and it synthesizes GFP in the absence of prior viral protein synthesis.
FIG. 3.
Synthesis of viral proteins in cells infected with d109. Vero cells were infected (MOI = 20) with the indicated viruses in the presence or absence of CHX, pulsed with [35S]methionine at 6 to 9 h p.i., and processed for SDS-PAGE. The CHX-treated cultures were incubated in the presence of actinomycin D during labeling. The samples were run on 9% (A) and 18% (B) SDS-polyacrylamide gels (the latter to better visualize the smaller viral proteins). The positions of the IE proteins and GFP, as well as some early and late proteins, are indicated. (C) The same samples were also transferred to a membrane for Western blot analysis with an α-GFP monoclonal antibody. Mock, uninfected cells.
Expression of the viral genome in the absence of IE proteins.
In the absence of regulatory functions encoded by the IE proteins, viral gene expression from d109 was expected to be severely restricted and solely a function of cellular factors. The contribution of the virion component, VP16, in this case is irrelevant since none of the endogenous IE promoters remain intact. Figure 3 shows the results of a parallel experiment conducted in the absence of inhibitors (untreated). Under these conditions, the effects of expressed IE proteins on subsequent gene expression can be observed. A typical permissive pattern of viral protein synthesis from 6 to 9 h p.i. is seen in cells infected with the wt virus KOS. Except for d106, which synthesized abundant amounts of ICP0 and ICP6, none of the mutants expressed the viral gene products to a significant degree. The polypeptide profile of d109 was indistinguishable from that of uninfected cells, thus showing that little or no HSV protein is made in this background.
The GFP transgene was synthesized to abundant levels in d106- or d107-infected cells, in which ICP0 was present (Fig. 3C). In cells infected with a mutant that does not express ICP0 (d104 or d109), the abundance of GFP was greatly reduced. These results clearly indicate that in the absence of ICP4, ICP0 can be a major determinant of gene expression driven from the HCMV IE promoter. Another notable observation is that the amount of GFP made in untreated cells infected with d104 or d109 was very low compared to that expressed in similarly infected CHX-treated cells. Preston and Nicholl previously observed that the heterologous HCMV promoter is active in CHX-treated cells infected with the mutant in1332, which is defective for the viral activators VP16, ICP0, and ICP4 (89). In the absence of the inhibitor, the activity of the HCMV promoter in this background was reduced. These observations suggest that an active mechanism represses the HCMV promoter in the absence of viral activators. This inhibition may involve a cellular factor that is transiently expressed and therefore not made in the presence of CHX.
The greater reduction in the levels of cell and viral proteins seen in d104- or d107-infected cells suggests an increase in shutoff of protein synthesis and correlates with the presence of ICP22 in these viruses. The vhs gene product (UL41) of HSV functions in attenuating host protein synthesis by destabilizing preexisting host mRNAs (31, 91, 111) and ensures the rapid turnover of viral mRNAs after the onset of viral transcription (58, 83). Similar activities have not been described for ICP22, although it is possible that the effect of ICP22 in this case is exerted indirectly. Previous studies with d92 and d97, viruses with IE mutations analogous to those of d107 and d104, respectively, did not show this same effect on cellular protein synthesis (99). However, these previous experiments were conducted at a lower MOI that may be below the threshold level at which the effects of ICP22 are observed. Additional studies are required to determine the contribution of ICP22 to this effect and the status of vhs or its ability to function in these mutant backgrounds.
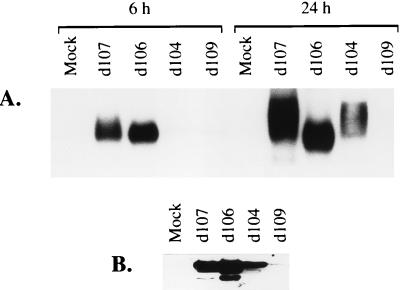
The accumulation of stable GFP mRNA at 6 h p.i. (Fig. 4A) was consistent with the levels of GFP synthesized by the different mutants early in infection (Fig. 3C). GFP mRNA was abundantly expressed in d106- or d107-infected cells, accumulating to a lesser degree in the latter. This minor difference may be due to the higher level of ICP0 synthesized in d106-infected cells (Fig. 3A). In d104- or d109-infected cells, GFP mRNA was barely detectable at 6 h p.i. However, at 24 h p.i., the relative level of GFP mRNA seen in d104-infected cells increased, while in d109-infected cells it remained very low (Fig. 4A). This difference suggests a possible contribution of ICP22 to the transcription of GFP late in infection. The abundance of the GFP mRNA in d106-infected cells relative to that in d107-infected cells at 24 h p.i. is difficult to assess because of the heterogeneity in the size of the mRNA in the latter. This effect was also observed with d104, and it correlates with the presence of ICP22 in these two mutants. The GFP mRNA is not spliced, so this heterogeneity may reflect differential processing of the mRNA at the 3′ end. Rice et al. have previously noted that ICP22 phosphorylates the carboxy-terminal domain (CTD) of pol II, resulting in a novel modified form of the enzyme (93). A possible explanation for the above-described results is suggested by recent findings of McCracken et al. showing a functional linkage between the CTD and 3′ RNA processing (72). They further demonstrated a specific interaction of the CTD with the cleavage-polyadenylation factors CPSF and CstF. An alteration in the polyadenylation process may thus be a consequence of the CTD modification mediated by ICP22. Further studies are necessary to determine the connection between the activity of ICP22 and the coupled processes of transcription and mRNA processing.
FIG. 4.
Accumulation of GFP mRNA and protein. (A) Total cell RNA, isolated at 6 and 24 h p.i. from Vero cells infected (MOI = 10) with the indicated viruses, was processed for Northern blot analysis with a GFP-specific probe. (B) Proteins extracted at 24 h p.i. from infected cells (MOI = 10) were separated on an SDS–18% polyacrylamide gel and transferred to a membrane for Western blot analysis with an α-GFP monoclonal antibody. Mock, uninfected cells.
The relative levels of GFP synthesized by the different mutants at 24 h p.i. (Fig. 4B) correlated with the amounts of GFP mRNA they express late in infection (Fig. 4A). GFP was abundant in cells infected with d107 or d106 (Fig. 4B). The increased amount of GFP mRNA expressed by d104 late in infection was also reflected proportionally at the protein level. With d109, the amount of GFP expressed was very small. The amount of GFP expressed by the different mutants late in infection was consistent with the level of green fluorescence exhibited by the infected monolayers when viewed under a UV microscope (data not shown). At 24 h p.i., all cells in d106- or d107-infected monolayers fluoresce, with those in the latter exhibiting significant cytopathic effects, consistent with previous observations on analogous mutants d95 and d92, respectively (117). A smaller proportion of fluorescing cells was observed in d104-infected monolayers, and with d109 there were even fewer of these GFP-expressing cells.
Results of the above-described studies show that in the absence of the IE proteins, the level of gene expression from the d109 genome is very low, even from the HCMV promoter, which is generally associated with high levels of transcriptional activity. Possible explanations for this apparent repression of the genome in the absence of viral activators are discussed below.
Toxicity and persistence of d109.
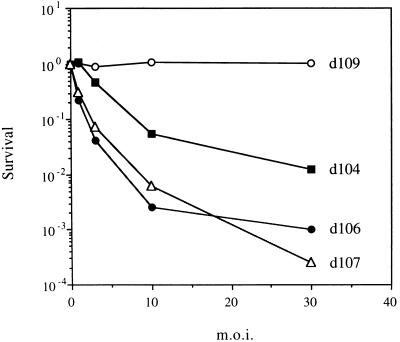
Previous observations suggested that both ICP0 and ICP22 contribute to virus toxicity and that simultaneous elimination of these gene products in an ICP4− ICP27− background would improve the survival of infected cells. To evaluate the toxicity of the different multiple IE mutants, Vero cell monolayers were infected at different MOI. At 6 h p.i., the monolayers were trypsinized to generate single-cell suspensions, and dilutions of these suspensions were plated to enumerate CFU. The abilities of d107 and d106 to inhibit colony formation by infected cells were quite similar (Fig. 5). In comparison, d104-infected cells showed enhanced survival, although virus toxicity was still observed at the higher MOI. These results are consistent with previous observations on the analogous mutants d92, d95, and d97 (99). The survival of d109-infected cells was indistinguishable from that of uninfected cells, even at the highest MOI of 30 PFU/cell. Therefore, from the standpoint of colony-forming ability of the infected cells, d109 is nontoxic.
FIG. 5.
Survival of cells infected with IE mutants. Vero cell monolayers were infected with the IE mutant viruses at the indicated MOI. The monolayers were harvested at 6 h p.i. and plated for determination of CFU. Colonies were counted 10 days after cell plating. Points plotted represent the surviving fractions of the infected cells relative to uninfected cells.
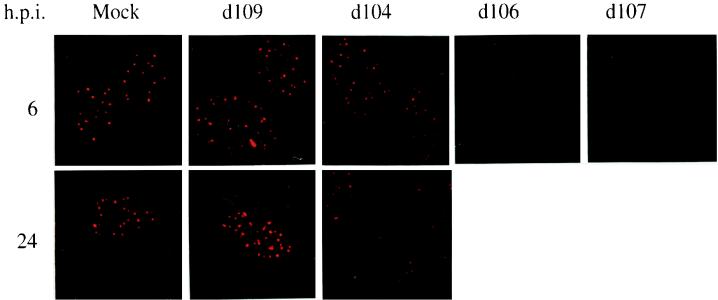
As another potential measure of perturbation of normal host cell function, the different IE mutants were assessed for their ability to disrupt the punctate nuclear bodies known as ND10. Early in infection, ICP0 localizes to these discrete nuclear structures (ND10), which are thought to be involved in the proliferative or differentiation state of the cell (26, 53, 116). As infection progresses, ICP0 perturbs the structure and the composition of these nuclear bodies, which are known to contain a number of cellular proteins, including PML (29, 67, 68). Similar activities have also been described for the IE proteins E4-ORF3 of adenovirus and IE1 of HCMV (25, 54). The association of viral genomes and replication proteins with ND10 early in infection led to the suggestion that these nuclear structures serve as sites where initial stages of HSV replication occur (46, 69). Consistent with previous observations, the discrete PML-containing ND10 structures were not observed in cells infected with mutants that express ICP0 (d107 and d106) (Fig. 6). In contrast, the characteristic punctate staining, similar to that seen in mock-infected cells, was readily observed in d104- or d109-infected cells. There was a small perturbation in the quality and quantity of staining in d104-infected cells late after infection, suggesting that ICP22 may directly or indirectly affect ND10 structure and composition.
FIG. 6.
Nuclear distribution of PML in cells infected with IE mutants. Human fetal lung cells infected (MOI = 10) with the indicated viruses were processed at 6 and 24 h p.i. for immunofluorescence with an α-PML antibody. Shown are stained infected nuclei (magnification, ×100).
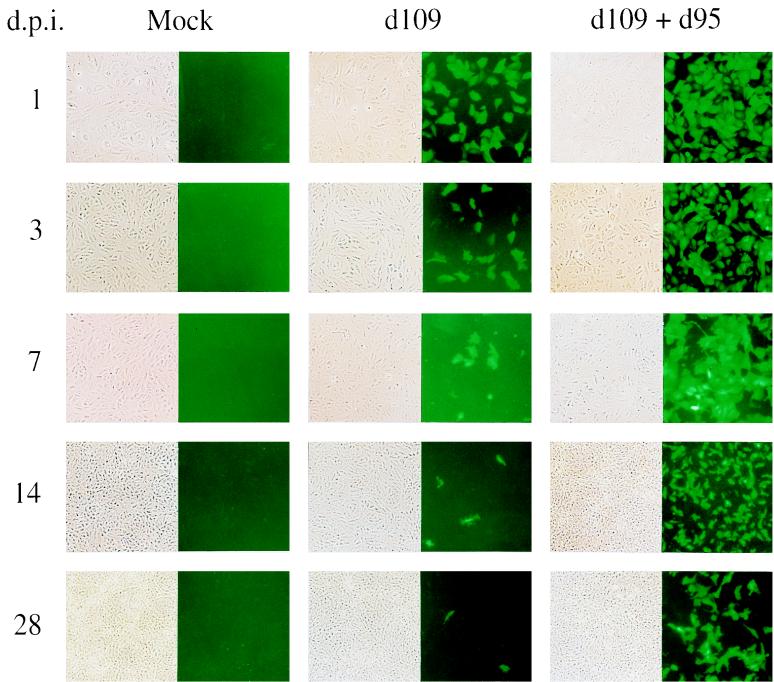
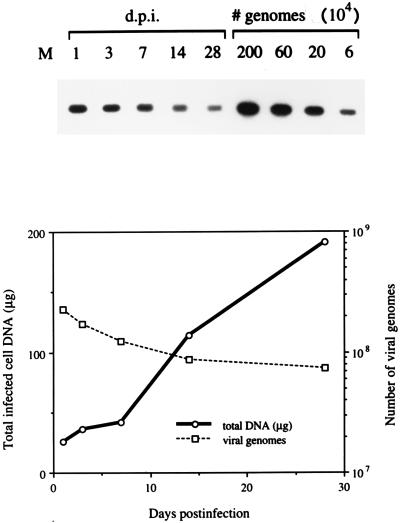
Considering that d109 was nontoxic to cells, we thought it likely that the virus could establish a long-term or persistent infection. To address this possibility, uninfected and d109-infected Vero cells were maintained and monitored for a period of 28 days. Even when infected at an MOI of 20, only a subpopulation of d109-infected monolayers expressed GFP (Fig. 7), and the number of these fluorescing cells decreased with time. Given the abundant expression of GFP in the presence of ICP0, it was reasonable to expect that providing ICP0 to the d109-infected monolayers would induce GFP and thus cause cells harboring d109 to fluoresce. Therefore, a parallel set of d109-infected cultures was superinfected with d95 1 day prior to each time point. The only IE regulatory protein that d95 expresses is ICP0, and this virus does not encode GFP. With the induction of GFP upon superinfection with d95, d109-infected cells were more readily identified, and these could be observed for the duration of the experiment (Fig. 7). Considering that the cells doubled in number about three times during this period, as deduced from the amount of total DNA isolated (Fig. 8), a significant number of d109-infected cells were maintained for up to 28 days p.i. This observation was also seen in HEL cells infected with d109 for up to 6 weeks (data not shown). PCR quantitation of the number of d109 genomes in the nuclei of infected Vero cells showed a gradual decline over time, although the number of viral genomes present at 28 days p.i. still constituted about one-third of the genomes present at 1 day p.i. (Fig. 8).
FIG. 7.
Persistence and induction of d109. Mock-infected (Mock) and d109-infected (MOI = 20) Vero cells were maintained for a period of 28 days in 2% serum at 34°C to limit cell division. At different time points (in days) after infection (indicated on the left), the monolayers were photographed under phase-contrast and fluorescence microscopes. A separate set of d109-infected cultures was superinfected with d95 (MOI = 20) 1 day prior to the time points indicated on the left.
FIG. 8.
Persistence of d109 genomes in infected cells. A separate set of d109-infected monolayers was maintained in parallel to those described in the legend to Fig. 7. At the same time points indicated, total infected cell DNA was isolated. Equivalent aliquots of the DNA samples, along with standards, were used in PCRs to amplify sequences from the gC gene (top). The amounts of total infected cell DNA (in micrograms) isolated at different time points (in days) after infection with d109 are plotted along with the number of viral genomes derived from quantitation of the gC PCR products.
The number of d109 genomes present in the nuclei at 1 day p.i. exceeded the expected value by about 10-fold. Jamieson et al. found that the number of viral genomes in the nuclei 1 day after infection with in1833 exceeded the number present at 1 day p.i. because capsid uncoating proceeded relatively slowly (47a). It may be that since our input inoculum was normalized to the number of genomes present in the nuclei at 6 h p.i., the larger number detected at 1 day p.i. reflected additional genomes that had not been uncoated by 6 h p.i. It is also noteworthy that at 28 days p.i. the number of genomes present in the cells exceeded the number of cells in the culture. There were approximately 7 × 107 genomes of d109 (Fig. 8) and 1.5 × 107 cells, assuming that the cell number doubled three times over the 28-day period. However, only about one-third of the cells appeared to express detectable GFP upon infection with d95 at 28 days (Fig. 7). The reason for this is unclear. The cells were infected at an MOI of 20; however, at present we do not know how these genomes partition from the initially infected cell into daughter cells following cell division. It may be that some daughter cells do not contain genomes while others retain multiple genomes. Alternatively, the infection of overly confluent monolayers with d95 may not result in efficient infection, or some of the quiescent genomes may not respond to ICP0. With respect to these last two points, when monolayers of d109-infected (28 days p.i.) HEL cells were infected with d95, nearly all of the cells fluoresced (data not shown). HEL cells are more prone to contact inhibition than are Vero cells and therefore did not divide to the same extent as Vero cells over the 28-day period.
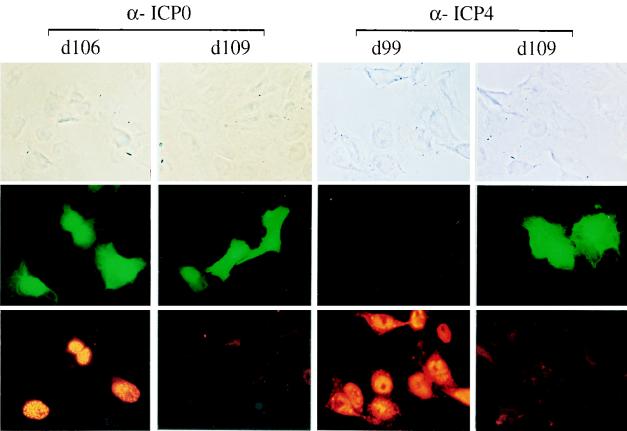
Taken together, the results presented thus far demonstrate that d109 is a noncytotoxic virus that can efficiently establish a persistent infection. Furthermore, the results show that gene expression from the persisting quiescent viral genomes remains inducible after long-term maintenance of the infected cultures. In the absence of induction, the number of green-fluorescing cells in d109-infected monolayers varies, but it is generally low. The reason for the high level of activity of the HCMV promoter regulating GFP expression in this subpopulation of cells is not known at the present time. To rule out the possibility that this small fraction of d109-infected cells expressing GFP was due to a low-level contamination with viruses that synthesize ICP0 or ICP4, Vero cells were infected with d109 and at 24 h p.i. were processed for immunofluorescence staining with α-ICP0 and α-ICP4 monoclonal antibodies. The results show that the d109-infected cells, which expressed GFP, did not express ICP4 or ICP0 (Fig. 9). Therefore, the expression of GFP from the HCMV IE promoter was not due to contamination of the infected cultures by viruses expressing ICP4 or ICP0.
FIG. 9.
Absence of ICP0 and ICP4 in GFP-expressing d109-infected cells. Vero cells were infected with d109 (MOI = 10) and, at 24 h p.i., processed for immunofluorescence with α-ICP0 and α-ICP4 monoclonal antibodies. Also included were d106- and d99-infected cells as positive controls for ICP0 and ICP4 staining, respectively. The secondary antibody used was conjugated to rhodamine. Photomicrographs in each column represent the same field viewed by phase-contrast microscopy (top) or fluorescence microscopy with the appropriate filter blocks for observing green (middle) or red (bottom) fluorescence.
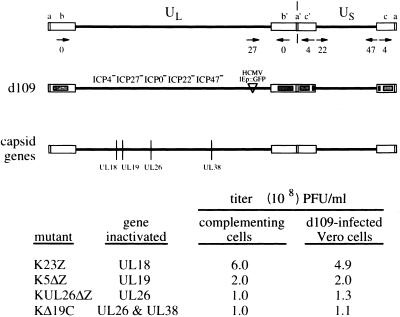
The properties of d109 suggest that it is capable of transducing functional genes into cells with minimal effects on cell survival. As an immediate test of this capability, we explored the possibility that d109-infected monolayers could serve as complementing cells for plaque formation by HSV mutants defective in essential genes. This prediction would hold true assuming that the region of the d109 genome required for complementation is maintained in the infected cells. To address this possibility, Vero monolayers were infected with d109 and, at 24 h p.i., the infected monolayers were used to determine the titers of virus mutants with lesions in capsid genes.
The titers of the capsid mutants obtained on d109-infected Vero cells were not significantly different from their known titers determined on dedicated cell lines that contain and express complementing levels of the genes mutated in the virus (Fig. 10). Induction of GFP from the resident d109 genomes was evident in all of the cells of the plaques (data not shown). It is important to note that the formation of the plaques on the monolayer was possible because d109 is sufficiently nontoxic that the nonsuperinfected areas of the monolayer maintained their integrity. These results further underscore the persistence and subsequent functionality of the d109 genomes resident in noncomplementing cells.
FIG. 10.
Efficiency of plating of capsid gene mutants on d109-infected cells. Vero cell monolayers were infected with d109 (MOI = 10), and at 24 h p.i. the infected monolayers were used to determine the titers of virus mutants with lesions in capsid genes. The structure of the d109 genome is shown, along with the genomic locations of the capsid genes mutated in the viruses whose titers were measured. Virus stock titers of the mutants, determined with the appropriate complementing cell lines, were provided by S. Person. See the legend to Fig. 1 for definitions of symbols and abbreviations.
DISCUSSION
This study was undertaken to determine the effects of the HSV IE proteins expressed from virus mutants defective in ICP4 on cell survival and gene expression. The information gained from these studies is relevant to the activities of the IE genes and the utility of replication-defective viruses as gene transfer vehicles. The results of this study clearly demonstrate that both ICP0 and ICP22 are toxic to cells. We found it necessary to abrogate all IE gene expression to completely eliminate the toxicity associated with HSV infection. Furthermore, both ICP0 and ICP22 affected transgene expression, with ICP0 having the greater effect. One virus, d109, which did not express any IE proteins in noncomplementing cells, was capable of establishing a long-term relationship with the cell, in which the genome persisted in the nucleus in a relatively inactive but potentially functional state.
Toxicity of viral mutants.
The toxicity associated with ICP4 mutants has long been documented. Chromosomal damage occurs (49), and the infected cells die, probably due to both necrotic and apoptotic modes of cell death (49, 63). In the absence of ICP4, the remaining IE proteins are overexpressed. Considering the large number of cellular proteins and metabolic systems that the IE proteins have been found to interact with or modify, it is probable that the observed toxicity is a complex superposition of many deleterious effects.
A UV-irradiated virus is not toxic to cells (78), suggesting, but not proving, that the virus particle itself is not toxic. Elimination of toxicity with the d109 mutant clearly demonstrates that the virion is not toxic to cells. Considering that the virus particle contains enzymatic functions, including a protein involved in the attenuation of host protein synthesis, UL41 or vhs (59, 79, 91), it is surprising that cells infected with d109 at a high multiplicity (30 PFU/cell) are indistinguishable from uninfected cells with respect to morphology, protein synthesis, and the ability to divide. Perhaps d109 virions contain smaller numbers of particular proteins, such as vhs, relative to wt virions. This could occur as a consequence of suboptimal complementation during propagation on FO6 cells. Alternatively, as suggested from the studies with UV-irradiated virus, the effects of the virion proteins in the absence of IE proteins or metabolic inhibitors are not sufficient to adversely affect cell function.
This study complements and extends previous studies from our lab demonstrating that elimination of either ICP22 (117) or ICP0 (99) from an ICP4− ICP27− virus (98) reduced toxicity. While these reports do not elucidate the mechanism by which ICP0 and ICP22 cause cell death, all three studies clearly demonstrate that mutants expressing ICP0 inhibit colony formation, probably by affecting cell cycle progression. The phenotypes of d95 (ICP4− ICP27− ICP22−)- and d106 (ICP4− ICP27− ICP22− ICP47−)-infected cells are similar in that the cells do not divide. They grow in size and continue to be otherwise metabolically active until they die. Further experiments are necessary to determine if and how the ability of ICP0 to affect transcription (50, 99), translation (51), and the activity of DNA-dependent protein kinase (61) and to interact with ubiquitin proteases (30), translation factors (51), cyclins (52), and PML oncogenic domains (PODs) (29, 67, 68) contributes to this phenotype. Considering this, and the observed phenotype of viruses expressing ICP0, such vectors may be of limited utility. Finally, it should be noted that these studies do not directly address the potential contribution of ICP47 to toxicity, since the abrogation of its expression was obligatory to the abrogation of ICP22 expression by construction. However, the results of previous transfection experiments imply that ICP47 does not significantly contribute to toxicity (48).
Gene expression.
ICP0 is a promiscuous transactivator of gene expression (27, 33, 81, 90). Viral gene expression is also generally reduced in ICP0 mutants (5, 6). In accord with these observations, we found that GFP expression from the HCMV IE promoter was highly dependent on ICP0. In the absence of all of the HSV IE proteins, the level of expression was extremely low. Like HSV IE promoters, the HCMV IE promoter can be activated by an IE protein (12, 108) and a virion component (65, 109). Both the HSV and HCMV genomes localize to ND10 structures early in infection, in the absence of viral protein synthesis (47, 69). Both establish latent infections in which the viral genomes, including the IE promoters, are relatively silent (77, 94). HSV and HCMV also express IE proteins, ICP0 by HSV and IE1 by HCMV, which are transactivators that localize to ND10 and dissociate PML from the POD structures. In the case of HSV, ICP0 has been shown to be involved in reactivation events in vivo and in tissue culture model systems (5, 14, 35, 41, 95, 119). Our studies showed that a virus expressing ICP0 will activate a previously silent HCMV promoter on persisting d109 genomes weeks after infection. HCMV infection will also activate GFP expression in d109-infected HEL cells (data not shown). Perhaps the observed low level of HCMV promoter activity in d109-infected cells represents the true “ground state” of genomes localized at ND10 in the absence of viral activator proteins, and it may reflect some of the events that occur in latency. The observations of Preston and Nicholl are consistent with ours, and they have proposed that HSV and HCMV IE promoters are repressed in the absence of VP16, ICP4, and ICP0 (89).
GFP expression in d109-infected cells was not uniform, being abundant in a subpopulation of cells and undetectable in most cells. It may be that the HCMV promoter is active in the absence of ICP0 only in cells which are in a particular stage of the cell cycle or state of activation by signaling pathways. Cai and Schaffer have shown that at a particular point in the cell cycle, the intracellular environment reduces the requirement for ICP0 (6). It is possible that the occurrence of GFP-expressing d109-infected cells is a reflection of cells in this state. Alternatively, the fluorescent cells may represent those in which the appropriate signaling pathways for specific targets in the HCMV promoter are activated. The HCMV promoter contains sites for NF-κB, CREB, retinoic acid receptor, and AP-1 (1, 34, 44, 101), all of which are components of regulated pathways. It remains to be tested whether the HCMV promoter on the d109 genome can be activated by one of these pathways.
d109 as a gene transfer vector.
From the standpoints of toxicity, genome persistence, and efficiency of gene delivery, d109 is clearly an optimal replication-defective herpesvirus vector. However, the very manipulations that conferred these attributes resulted in low-level and perhaps repressed gene expression. It may be that this level of gene expression is sufficient for some applications. Most cellular genes are not expressed at the level of an ICP0-induced viral gene. However, it is likely that systems for cell- or tissue-specific expression of transgenes will have to be incorporated into d109.
It is interesting that the development of adenovirus vectors has proceeded along parallel lines, with analogous results. E1-deleted adenovirus exhibits high-level expression of transgenes from the HCMV promoter; however, other viral proteins are also expressed, and the virus is somewhat toxic to cells (16). The additional deletion of the E4 region resulted in a virus that was less toxic and persisted in cells for long periods of time (2, 32). However, expression from the HCMV promoter was greatly reduced. E4 contains seven open reading frames (64). One of them, Orf3, like ICP0, perturbs ND10 structures (11, 25). The adenovirus genome also localizes to ND10 structures in the absence of viral protein synthesis (96). Considering these parallels as well as the similar phenotypes of E1-E4-deleted adenovirus and d109, it is likely that approaches for promoting or regulating gene expression from the vectors will have similar outcomes in both viruses.
The development of HSV strains has systematically progressed (18, 98, 99, 117) to the point where we now have a virus that has no detectable harmful effects on cells, does not express its own genes, and efficiently delivers the genome to the nucleus, where it persists for a prolonged period of time in a functional form. The lack of toxicity does not appear to be constrained by the input MOI. The results of this study allow future efforts to be aimed at understanding the association between persisting HSV genomes and the cell nucleus and determining how this association affects transcription. These considerations have obvious relevance to expression of transgenes from HSV vectors and may reflect events occurring during latency, when the genome is relatively silent but persists in a potentially functional state.
ACKNOWLEDGMENTS
This work was supported by NIH grants DK44935 and AI30612.
We are grateful to Stanley Person for providing the capsid gene mutants and to Colton A. Smith and Michael J. Carrozza for reviewing the manuscript. We also thank Patricia Bates for supplying the specific PCR conditions and Arthur Webb for the construction of pAT1 and pAT2.
REFERENCES
- 1.Angulo A, Suto C, Heyman R A, Ghazal P. Characterization of the sequences of the human cytomegalovirus enhancer that mediate differential regulation by natural and synthetic retinoids. Mol Endocrinol. 1996;10:781–793. doi: 10.1210/mend.10.7.8813719. [DOI] [PubMed] [Google Scholar]
- 2.Armentano D, Zabner J, Sacks C, Sookdeo C C, Smith M P, St. George J A, Wadsworth S C, Smith A E, Gregory R J. Effect of the E4 region on the persistence of transgene expression from adenovirus vectors. J Virol. 1997;71:2408–2416. doi: 10.1128/jvi.71.3.2408-2416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batterson W, Roizman B. Characterization of the herpes simplex virion-associated factor responsible for the induction of α genes. J Virol. 1983;46:371–377. doi: 10.1128/jvi.46.2.371-377.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond V C, Person S. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology. 1984;132:368–376. doi: 10.1016/0042-6822(84)90042-4. [DOI] [PubMed] [Google Scholar]
- 5.Cai W, Astor T L, Liptak L M, Cho C, Coen D M, Schaffer P A. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol. 1993;67:7501–7512. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai W, Schaffer P A. A cellular function can enhance gene expression and plating efficiency of a mutant defective in the gene for ICP0, a transactivating protein of herpes simplex virus type 1. J Virol. 1991;65:4078–4090. doi: 10.1128/jvi.65.8.4078-4090.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai W, Schaffer P A. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol. 1992;66:2904–2915. doi: 10.1128/jvi.66.5.2904-2915.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell M E, Palfreyman J W, Preston C M. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate-early transcription. J Mol Biol. 1984;180:1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- 9.Carrozza M J, DeLuca N A. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Mol Cell Biol. 1996;16:3085–3093. doi: 10.1128/mcb.16.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter K L, Roizman B. Alternatively spliced mRNAs predicted to yield frame-shift proteins and stable intron 1 RNAs of the herpes simplex virus 1 regulatory gene α0 accumulate in the cytoplasm of infected cells. Proc Natl Acad Sci USA. 1996;93:12535–12540. doi: 10.1073/pnas.93.22.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho T, Seeler J-S, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherrington J M, Mocarski E S. Human cytomegalovirus ie1 transactivates the α promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clements J B, Watson R J, Wilkie N M. Temporal regulation of herpes simplex virus type 1 transcription: location of transcripts on the viral genome. Cell. 1977;12:275–285. doi: 10.1016/0092-8674(77)90205-7. [DOI] [PubMed] [Google Scholar]
- 14.Clements J B, Stow N D. A herpes simplex virus type 1 mutant containing a deletion within immediate-early gene 1 is latency competent in mice. J Gen Virol. 1989;70:2501–2506. doi: 10.1099/0022-1317-70-9-2501. [DOI] [PubMed] [Google Scholar]
- 15.Costanzo F, Campadelli-Fiume G, Foà-Tomasi L, Cassai E. Evidence that herpes simplex virus DNA is transcribed by cellular RNA polymerase B. J Virol. 1977;21:996–1001. doi: 10.1128/jvi.21.3.996-1001.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dedieu J-F, Vigne E, Torrent C, Jullien C, Mahfouz I, Caillaud J-M, Aubailly N, Orsini C, Guillaume J-M, Opolon P, Delaère P, Perricaudet M, Yeh P. Long-term delivery into the livers of immunocompetent mice with E1/E4-defective adenoviruses. J Virol. 1997;71:4626–4637. doi: 10.1128/jvi.71.6.4626-4637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLuca N A, Courtney M A, Schaffer P A. Temperature-sensitive mutants in herpes simplex virus type 1 ICP4 permissive for early gene expression. J Virol. 1984;52:767–776. doi: 10.1128/jvi.52.3.767-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLuca N A, McCarthy A M, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLuca N A, Schaffer P A. Activation of immediate-early, early, and late promoters by temperature-sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Mol Cell Biol. 1985;5:1997–2008. doi: 10.1128/mcb.5.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeLuca N A, Schaffer P A. Activities of herpes simplex virus type 1 (HSV-1) ICP4 genes specifying nonsense peptides. Nucleic Acids Res. 1987;15:4491–4511. doi: 10.1093/nar/15.11.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLuca, N. A., and P. A. Schaffer. Unpublished data.
- 22.Desai P, DeLuca N A, Glorioso J C, Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J Virol. 1993;67:1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai P, Watkins S C, Person S. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J Virol. 1994;68:5365–5374. doi: 10.1128/jvi.68.9.5365-5374.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon R A F, Schaffer P A. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J Virol. 1980;36:189–203. doi: 10.1128/jvi.36.1.189-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 26.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocytic-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 27.Everett R D. trans-activation of transcription by herpesvirus products: requirements of two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Everett R D. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 29.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenwick M L, McMenamin M M. Early virion-associated suppression of cellular protein synthesis by herpes simplex virus is accompanied by inactivation of mRNA. J Gen Virol. 1984;65:1225–1228. doi: 10.1099/0022-1317-65-7-1225. [DOI] [PubMed] [Google Scholar]
- 32.Gao G-P, Yang Y, Wilson J M. Biology of adenovirus vectors with E1 and E4 deletions for liver-directed gene therapy. J Virol. 1996;70:8934–8943. doi: 10.1128/jvi.70.12.8934-8943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gelman I H, Silverstein S. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc Natl Acad Sci USA. 1985;82:5265–5269. doi: 10.1073/pnas.82.16.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghazal P, DeMattei C, Giulietti E, Kliewar S A, Umesuno K, Evans R M. Retinoic acid receptors initiate induction of the cytomegalovirus enhancer in embryonal cells. Proc Natl Acad Sci USA. 1992;89:7630–7634. doi: 10.1073/pnas.89.16.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon Y J, McKnight J L C, Ostrove J M, Romanowski E, Araullo-Cruz T. Host species and strain differences affect ability of an HSV-1 ICP0 deletion mutant to establish latency and spontaneously reactivate in vivo. Virology. 1990;178:469–477. doi: 10.1016/0042-6822(90)90344-q. [DOI] [PubMed] [Google Scholar]
- 36.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus type 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 37.Gu B, DeLuca N. Requirements for activation of the herpes simplex virus glycoprotein C promoter in vitro by the viral regulatory protein ICP4. J Virol. 1994;68:7953–7965. doi: 10.1128/jvi.68.12.7953-7965.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu B, Rivera-Gonzalez R, Smith C A, DeLuca N A. Herpes simplex virus infected cell polypeptide 4 preferentially represses Sp1-activated over basal transcription from its own promoter. Proc Natl Acad Sci USA. 1993;90:9528–9532. doi: 10.1073/pnas.90.20.9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harlow E, Lane D. Antibodies, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 41.Harris R A, Everett R D, Zhu X, Silverstein S, Preston C M. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex type 2 in an in vitro system. J Virol. 1989;63:3513–3515. doi: 10.1128/jvi.63.8.3513-3515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honess R W, Roizman B. Regulation of herpes virus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci USA. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunninghake G W, Monick M M, Liu B, Stinski M F. The promoter-regulatory region of the major immediate-early gene of human cytomegalovirus responds to T-lymphocyte stimulation and contains functional cyclic AMP-response elements. J Virol. 1989;63:3026–3033. doi: 10.1128/jvi.63.7.3026-3033.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imbalzano A N, Coen D M, DeLuca N A. Herpes simplex virus transactivator ICP4 operationally substitutes for the cellular transcription factor Sp1 for efficient expression of the viral thymidine kinase gene. J Virol. 1991;65:565–574. doi: 10.1128/jvi.65.2.565-574.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Jamieson D S R, Robinson L H, Daksis J I, Nichol M J, Preston C M. Quiescent viral genomes in human fibroblasts after infection with herpes simplex virus type 1 Vmw65 mutants. J Gen Virol. 1995;76:1417–1431. doi: 10.1099/0022-1317-76-6-1417. [DOI] [PubMed] [Google Scholar]
- 48.Johnson P A, Wang M J, Friedmann T. Improved cell survival by the reduction of immediate-early gene expression in replication-defective mutants of herpes simplex virus type 1 but not by mutation of the virion host shutoff function. J Virol. 1994;68:6347–6362. doi: 10.1128/jvi.68.10.6347-6362.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson P A, Miyanohara A, Levine F, Cahill T, Friedmann T. Cytotoxicity of a replication-defective mutant of herpes simplex virus type 1. J Virol. 1992;66:2952–2965. doi: 10.1128/jvi.66.5.2952-2965.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jordan R, Schaffer P A. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J Virol. 1997;71:6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koken M H M, Puvion-Dutilleul F, Guillemin M, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, Degos L, Puvion E, de The H. The t(15;17) translocation alters a nuclear body in a retinoic acid reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korioth F, Maul G G, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 55.Kramer M F. Quantification of herpes simplex virus DNA and transcripts during latent infection in mouse trigeminal ganglia by reverse transcriptase PCR. Ph.D. thesis. Cambridge, Mass: Harvard University; 1995. [Google Scholar]
- 56.Kristie T M, LeBowitz J H, Sharp P A. The octamer-binding proteins form multi-protein-DNA complexes with the HSV alpha TIF regulatory protein. EMBO J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kristie T M, Roizman B. Host cell proteins bind to the cis-acting site required for virion mediated induction of herpes simplex virus 1 α genes. Proc Natl Acad Sci USA. 1987;84:71–75. doi: 10.1073/pnas.84.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwong A D, Frenkel N. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc Natl Acad Sci USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwong A D, Kruper J A, Frenkel N. Herpes simplex virus virion host shutoff function. J Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laemmli U K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1979;227:680–684. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 61.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leopardi R, Roizman B. The herpes simplex virus major regulatory protein ICP4 blocks apoptosis induced by the virus or by hyperthermia. Proc Natl Acad Sci USA. 1996;93:9583–9587. doi: 10.1073/pnas.93.18.9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leppard K N. E4 gene functions in adenovirus, adenovirus vector and adeno-associated virus infections. J Gen Virol. 1997;78:2131–2138. doi: 10.1099/0022-1317-78-9-2131. [DOI] [PubMed] [Google Scholar]
- 65.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manservigi R, Spear P, Buchan A. Cell fusion induced by herpes simplex virus is promoted and suppressed by different viral glycoproteins. Proc Natl Acad Sci USA. 1977;74:3913–3917. doi: 10.1073/pnas.74.9.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 68.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate-early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 69.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type 1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 70.Mavromara-Nazos P, Ackermann M, Roizman B. Construction and properties of a viable herpes simplex virus 1 recombinant lacking coding sequences of the α47 gene. J Virol. 1986;60:807–812. doi: 10.1128/jvi.60.2.807-812.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCarthy A M, McMahan L, Schaffer P A. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J Virol. 1989;63:18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 73.McGregor F, Phelan A, Dunlop J, Clements J B. Regulation of herpes simplex virus poly(A) site usage and the action of immediate-early protein IE63 in the early-late switch. J Virol. 1996;70:1931–1940. doi: 10.1128/jvi.70.3.1931-1940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McLauchlan J, Phelan A, Loney C, Sandri-Goldin R M, Clements J B. Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate viral mRNA 3′ processing. J Virol. 1992;66:6939–6945. doi: 10.1128/jvi.66.12.6939-6945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McMahan L, Schaffer P A. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions and are required to modulate viral gene expression very early in infection. J Virol. 1990;64:3471–3485. doi: 10.1128/jvi.64.7.3471-3485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Michael N, Roizman B. Repression of the herpes simplex virus 1 alpha 4 gene by its gene product occurs within the context of the viral genome and is associated with all three identified cognate sites. Proc Natl Acad Sci USA. 1993;90:2286–2290. doi: 10.1073/pnas.90.6.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mocarski E S., Jr . Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 78.Munyon W, Kraiselburd E, Davis D, Mann J. Transfer of thymidine kinase to thymidine kinaseless L cells by infection with ultraviolet-irradiated herpes simplex virus. J Virol. 1971;7:813–820. doi: 10.1128/jvi.7.6.813-820.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nishioka Y, Silverstein S. Alterations in the protein synthetic apparatus of Friend erythroleukemia cells infected with vesicular stomatitis virus or herpes simplex virus. J Virol. 1978;25:422–426. doi: 10.1128/jvi.25.1.422-426.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Hare P, Goding C R, Haigh A. Direct combinatorial interaction between a herpes simplex virus regulatory protein and a cellular octamer-binding factor mediates specific induction of virus immediate-early gene expression. EMBO J. 1988;7:4231–4238. doi: 10.1002/j.1460-2075.1988.tb03320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Hare P, Hayward G S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O’Hare P, Hayward G S. Three trans-acting regulatory proteins of herpes simplex virus modulate immediate-early gene expression in a pathway involving positive and negative feedback regulation. J Virol. 1985;56:723–733. doi: 10.1128/jvi.56.3.723-733.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oroskar A A, Read G S. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J Virol. 1989;63:1897–1906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pereira L, Wolff M H, Fenwick M, Roizman B. Regulation of herpes virus macromolecular synthesis. V. Properties of polypeptides made in HSV-1 and HSV-2 infected cells. Virology. 1977;77:733–749. doi: 10.1016/0042-6822(77)90495-0. [DOI] [PubMed] [Google Scholar]
- 85.Person, S., and P. Desai. Unpublished data.
- 86.Post L E, Roizman B. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus type 1 is not essential for growth. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 87.Preston C M. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsk. J Virol. 1979;29:275–284. doi: 10.1128/jvi.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Preston C M, Mabbs R, Nicholl M J. Construction and characterization of herpes simplex type 1 mutants with conditional defects in immediate early gene expression. Virology. 1997;229:228–239. doi: 10.1006/viro.1996.8424. [DOI] [PubMed] [Google Scholar]
- 89.Preston C M, Nicholl M J. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J Virol. 1997;71:7807–7813. doi: 10.1128/jvi.71.10.7807-7813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Quinlan M P, Knipe D M. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol Cell Biol. 1985;5:957–963. doi: 10.1128/mcb.5.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Read G S, Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of α (immediate early) viral polypeptides. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rice S A, Knipe D M. Gene-specific transactivation by herpes simplex virus type 1 alpha protein ICP27. J Virol. 1988;62:3814–3823. doi: 10.1128/jvi.62.10.3814-3823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rice S A, Long M C, Lam V, Schaffer P A, Spencer C A. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J Virol. 1995;69:5550–5559. doi: 10.1128/jvi.69.9.5550-5559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 95.Russell J, Stow N D, Stow E C, Preston C M. Herpes simplex virus genes involved in latency in vitro. J Gen Virol. 1987;68:3009–3018. doi: 10.1099/0022-1317-68-12-3009. [DOI] [PubMed] [Google Scholar]
- 96.Sacks W R, Greene C C, Aschman D P, Schaffer P A. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J Virol. 1985;55:796–805. doi: 10.1128/jvi.55.3.796-805.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Samaniego L A, Webb A L, DeLuca N A. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J Virol. 1995;69:5705–5715. doi: 10.1128/jvi.69.9.5705-5715.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Samaniego L A, Wu N, DeLuca N A. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J Virol. 1997;71:4614–4625. doi: 10.1128/jvi.71.6.4614-4625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 101.Sambucetti L C, Cherrington J M, Wilkinson G W G, Mocarski E S. NF-κB activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 1989;8:4251–4258. doi: 10.1002/j.1460-2075.1989.tb08610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sandri-Goldin R M, Mendoza G E. A herpes virus regulatory protein appears to act posttranscriptionally by affecting mRNA processing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 103.Sears A E, Halliburton I W, Meignier B, Silver S, Roizman B. Herpes simplex virus 1 mutant deleted in the α22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J Virol. 1985;55:338–346. doi: 10.1128/jvi.55.2.338-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sekulovich R E, Leary K, Sandri-Goldin R M. The herpes simplex virus type 1 α protein ICP27 can act as a trans-repressor or a trans-activator in combination with ICP4 and ICP0. J Virol. 1988;62:4510–4522. doi: 10.1128/jvi.62.12.4510-4522.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smith C A, Bates P, Rivera-Gonzales R, Gu B, DeLuca N A. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J Virol. 1993;67:4676–4687. doi: 10.1128/jvi.67.8.4676-4687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smith I L, Hardwicke M A, Sandri-Goldin R M. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- 107.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 108.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol. 1990;64:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stinski M F, Roehr T J. Activation of the major immediate early gene of human cytomegalovirus by cis-acting elements in the promoter-regulatory sequence and by virus-specific trans-acting components. J Virol. 1985;55:431–441. doi: 10.1128/jvi.55.2.431-441.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stow N D, Stow E C. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate-early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 111.Strom T, Frenkel N. Effects of herpes simplex virus on mRNA stability. J Virol. 1987;61:2198–2207. doi: 10.1128/jvi.61.7.2198-2207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Su L, Knipe D M. Herpes simplex virus alpha protein ICP27 can inhibit or augment viral gene transactivation. Virology. 1989;170:496–504. doi: 10.1016/0042-6822(89)90441-8. [DOI] [PubMed] [Google Scholar]
- 113.Triezenberg S J, LaMarco K L, McKnight S L. Evidence of DNA:protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes Dev. 1988;2:730–742. doi: 10.1101/gad.2.6.730. [DOI] [PubMed] [Google Scholar]
- 114.Uprichard S L, Knipe D M. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J Virol. 1996;70:1969–1980. doi: 10.1128/jvi.70.3.1969-1980.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Watson R J, Clements J B. Characterization of transcription-deficient temperature-sensitive mutants of herpes simplex virus type 1. Virology. 1978;91:364–379. doi: 10.1016/0042-6822(78)90384-7. [DOI] [PubMed] [Google Scholar]
- 116.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, DeJean A. Retinoic acid regulates aberrant nuclear localization of PML-RARα in acute promyelocytic leukaemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 116a.Wilson A C, LaMarco K, Peterson M G, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 117.Wu N, Watkins S C, Schaffer P A, DeLuca N A. Prolonged gene expression and cell survival after infection by a herpes simplex virus mutant defective in the immediate-early genes encoding ICP4, ICP27, and ICP22. J Virol. 1996;70:6358–6369. doi: 10.1128/jvi.70.9.6358-6369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.York A I, Roop C, Andrews D W, Ridell S R, Graham F L, Johnson D C. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 119.Zhu X, Chen J, Young C S H, Silverstein S. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant IE-0 gene products. J Virol. 1990;64:4489–4498. doi: 10.1128/jvi.64.9.4489-4498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]