Abstract
Leptomeningeal carcinomatosis represents a terminal stage and is a devastating complication of cancer. Despite its high incidence, current diagnostic methods fail to accurately detect this condition in a timely manner. This failure to diagnose leads to the refusal of treatment and the absence of clinical trials, hampering the development of new therapy strategies. The use of liquid biopsy is revolutionizing the field of diagnostic oncology. The dynamic and non-invasive detection of tumor markers has enormous potential in cancer diagnostics and treatment. Leptomeningeal carcinomatosis is a condition where invasive tissue biopsy is not part of the routine diagnostic analysis, making liquid biopsy an essential diagnostic tool. Several elements in cerebrospinal fluid (CSF) have been investigated as potential targets of liquid biopsy, including free circulating tumor cells, free circulating nucleic acids, proteins, exosomes, and even non-tumor cells as part of the dynamic tumor microenvironment. This review aims to summarize current breakthroughs in the research on liquid biopsy, including the latest breakthroughs in the identification of tumor cells and nucleic acids, and give an overview of future directions in the diagnosis of leptomeningeal carcinomatosis.
Keywords: brain spine tumors, neuro oncology, brain metastases, leptomeningeal carcinomatosis, liquid biopsy
Introduction and background
Leptomeningeal carcinomatosis (LC) is the metastatic dissemination of cancer cells, mostly originating from breast, lung, and melanoma cancer, to the pia mater, arachnoid, and/or subarachnoid space. It is also known as leptomeningeal metastasis, carcinomatous meningitis, or leptomeningeal disease (LMD) [1,2]. Metastatic cells present in the cerebrospinal fluid (CSF) may invade the central nervous system over a short timeframe, disrupting the brain and spinal cord; on occasion, this can cause sudden neurological deterioration and death. Neoplastic cells may reach the brain through a blood vessel and migrate from the choroid plexus to the CSF. Once they enter the leptomeninges, metastatic cancer cells can circulate with freedom, causing extensive damage to the central nervous system [3]. The current review will highlight the diagnostic frontiers regarding LC and provide a broad panorama of the actual diagnostic methods to detect and monitor disease in the CSF, as well as discuss possible future directions of research on this disease.
Epidemiology and pathophysiology
The true incidence of LC remains unclear, as it ranges from 8% to 35% across studies. The reasons for this broad range are multifactorial, including the tumor histology, location, and size; pial involvement; and the type of surgical resection [4]. Up to 15% of patients with solid tumors are diagnosed with LC, and similar rates are observed in patients with leukemia and lymphoma [5-8].
Resection has become an essential treatment modality for newly diagnosed brain metastases. Nonetheless, studies have suggested an association with a higher risk of developing LC. The latter is due to the disruption of anatomical barriers in the brain tissue and the surgical spillage of tumor cells, resulting in the dissemination of tumor cells.
In a meta-analysis performed by Twarie and collaborators, among 386 patients, 18 risk factors were reported as significantly positively associated with LC occurrence, including, but not limited to, a larger tumor size, infratentorial location of bone metastases (BMs), proximity of BM to cerebrospinal fluid spaces, ventricle violation during surgery, subtotal or piecemeal resection, and postoperative stereotactic radiosurgery. In breast cancer, lymph node metastasis of the primary tumor (HR = 2.73, 95% CI: 2.12-3.52) and multiple BMs (HR = 1.37, 95% CI: 1.18-1.58) were significantly associated with a higher risk of LC occurrence after neurosurgery [9].
The development of LC is thought to be caused by CSF seeding through the leptomeninges by hematogenic, perineural, or direct tumor growth. Thus, an examination of CSF may reveal potentially useful biomarkers for the diagnosis and/or prognosis of LC development.
Current diagnostic workup
Clinical Manifestations
Patients with LC usually present with an existing history of malignancy as well as with a known metastatic disease. A small fraction of the patients has a synchronous or exclusive initial presentation of LC. The great majority of the patients are symptomatic, and these symptoms may be either localizable, due to the involvement of specific neuroanatomic structures, or diffuse, which commonly occurs secondary to the increased intracranial pressure that is frequently associated with communicating hydrocephalus. Localizable symptoms are often multifocal due to the broad involvement of the surface of the brain, spinal cord parenchyma, or other structures that traverse the CSF. This can manifest as multiple cranial neuropathies, especially the involvement of cranial nerves VI, VII, and VIII, or radiculopathies. The chronological presentation is often stable before experiencing an abrupt clinical decline. The natural history of LC is not well delineated, but the evolution from the presentation of the symptoms to death is rather short in most cases [10]. Although LC usually presents as the terminal stage of widely disseminated systemic cancers, up to 10% of patients present with LC as the first cancer manifestation [11].
Diagnostic Evaluation
LC diagnosis is often made through a combination of physical examination, neuroimaging, clinical suspicion, and CSF analysis [10], and the presence of this pathology should be suspected and evaluated more frequently, particularly if there is a known history of malignancy.
The imaging modality of choice for evaluating LC is magnetic resonance imaging (MRI) with contrast [2,9,12,13]. An example of LMD in the brain and spine can be seen in Figure 1. In previous reports, radiographic signs have been reported to be associated with LC that include two different patterns: classic sugar-coating (cLC) and nodular (nLC). The radiographic pattern of LC has been previously demonstrated to be prognostic, with cLC having significantly worse overall survival (OS) compared with nLC [14,15].
Figure 1. Example of LMD with T1 contrast-enhanced lesions in brain (A) and spine (B).
LMD: leptomeningeal disease.
Note: This image is the author's own creation.
Classical leptomeningeal disease (cLMD) includes an enhancement in the cranial nerves, cisterns, cerebellar folia, and sulci [16]; Zuckerguss or diffuse “sugar-coating” enhancement is also observed across the surface of the brain [4,17-19].
The proposed definition of nLC is the presence of a focal nodule or nodules adherent to surfaces with CSF contact, including the dural or pial surface, tentorium, ventricles, and hypervascular dural tail [4].
The downside of the MRI is its limited sensitivity; however, this is secondary to an incomplete understanding of the sensitivity and specificity of other diagnostic modalities, including the best currently available diagnostic modality, i.e., CSF cytology. The MRI protocol should include a contrasted spinal MRI of melanoma and lung cancer who are highly suspected of having CNS metastases.
Other issues concerning the radiological diagnosis also involve discrepancies in the literature in how the radiological findings are defined. Several efforts have been made to unify the different definitions proposed, as well as to be consistent with the Response Assessment in Neuro-Oncology (RANO) criteria for metastases [4,19].
CSF cytologic assessment is currently thought to be the best definitive diagnostic modality. It has a high specificity of up to 100% with a low false-positive rate, although with limited sensitivity with a maximum of 75% [17,20]. The preferred method for collecting CSF for diagnostic purposes is through a lumbar puncture [10,13,20]. However, lumbar punctures should only be performed after proper neuroimaging to avoid performing the procedure in a patient at risk from herniation due to major brain metastases or complications from local bulky disease [21]. It is also possible to encounter unspecific abnormalities during the lumbar puncture; for example, increased opening pressure has been detected in 21-24% of patients [22-24], increased leukocyte count in 39-77.5%, elevated protein in 56-91%, and decreased glucose in 22-38% of patients [24-26] (Table 1).
Table 1. CSF normal reference values in the cytologic assessment.
CSF: cerebrospinal fluid.
| Finding | Values |
| Opening pressure | >200 mm H2O |
| Leukocyte count | >4 per mm3 |
| Protein | >500 mg/L |
| Glucose | <600 mg/L |
Whenever lumbar puncture is not possible, an imaging-guided cisternal puncture should be considered. The current recommendation is to perform the analysis on a large volume (optimally >10 mL, with a minimum of 5 ml) that has been freshly obtained (collected less than 30 minutes before the analysis), accompanied by immunohistochemical staining for epithelial and melanocytic markers to optimize the diagnostic accuracy of the test [10,13]. However, the sensitivity may be affected by the location from which the CSF is obtained (lumbar vs. ventricular) [20].
Tumor cells can be detected at diagnosis in up to 80% of LC cases. The CSF cytologic analysis should be reported as positive when the presence of malignant cells in the CSF is observed; equivocal when detecting “suspicious” or “atypical” cells in the CSF; or negative when no malignant or potentially malignant cells are found in the CSF [21].
Thus, a diagnosis of LC can be confirmed in the presence of tumor cells in the CSF or by a positive leptomeningeal biopsy. In the absence of a confirmed diagnosis, the diagnosis of LC can be considered as probable in a patient with a history of histologically proven cancer with a reasonable risk of LC and after discarding other differential diagnoses. The diagnosis is probable when typical clinical findings or typical MRI findings are present without a confirmed CSF test [21].
Genetic Mutations in Leptomeningeal Carcinomatosis
LC is a life-threatening condition that is associated with a poor functional and life prognosis, a consequence often seen with solid tumors, regardless of treatment [27]. Although its current incidence is low, the clinical impact of LC on the patients and its increasing incidence makes the identification of biomarkers and exact molecular mechanisms a priority for the prognosis and the treatment of LMD [3].
The molecular mechanism underlying LC is not yet fully understood. A meta-analysis conducted by Congur et al., which included 16 countries, identified five different genes as the most commonly mutated genes in all three types of cancer included in the study (non-small-cell lung cancer, breast cancer, and melanoma). The genes were TP 53, PTEN, PIK3CA, IL7R, and KMT2D. These genes are associated with the regulation of cell communication and signaling and cell proliferation [3].
Signaling pathways are responsible for cell-to-cell communication, cell turnover, death, and cellular movement. The PI3K-AKT and RAS-ERK pathways regulate processes like cell proliferation and cell-to-cell communication. Alterations in any of the aforementioned genes can therefore affect the signaling pathways controlling cancer progression, causing cells to profusely proliferate [3]. On the other hand, in patients with breast cancer, the HER-2 mutation has been found to decrease the risk of LC development [9].
Unfortunately, as stated before, the prognosis for patients with LC remains significantly poor. Unlike the diagnostic-specific graded prognostic assessment (DS-GPA) [28], there are currently no such strong prognostication systems [28] for LC [10].
Review
Application of liquid biopsies
A better understanding of cancer biology, dynamic system modulation, and cell interaction indicates the importance of approaching cancer as a systemic disease [29]. Moreover, the need for early detection and available screening methods is obvious in preventing and halting the progression of cancer [30]. Nevertheless, the gold-standard method of tumor profiling is still the local-tissue biopsy [31]. However, there is not only the problem of the invasiveness of the procedure but also the inability to dynamically monitor the tumor molecular profile since only one tissue sample can be obtained at any one time [32]. On the other hand, tumor molecular monitoring and the identification of drug resistance markers and escape mutations have drastically improved personalized therapies [33]. These factors have shifted the focus in modern oncology toward less invasive and more dynamic methods of tumor detection [34].
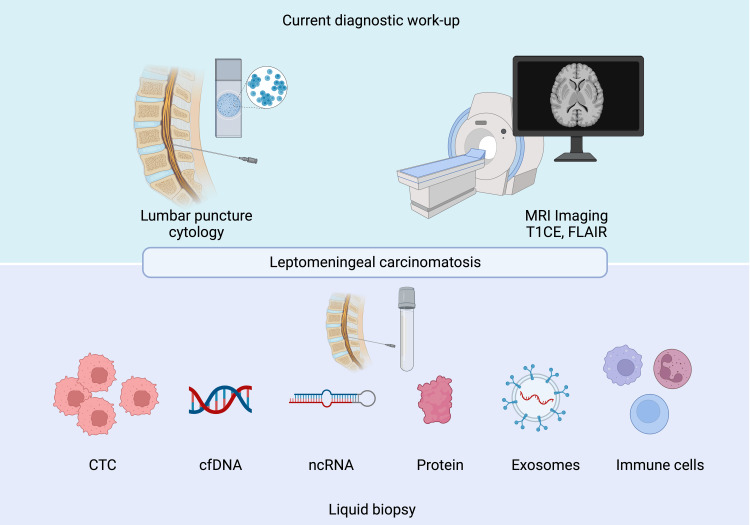
Liquid biopsy (LB) is a concept that was introduced in 1993 in Hamburg [35]. As the name implies, LB is a method for the testing of tumor markers in bodily fluids, such as blood, urine, saliva, and CSF [36]. LB demonstrates several advantages in comparison to traditional biopsy, including non-invasiveness, continuous tumor monitoring, higher sensitivity, and cost-effectiveness [37]. It is well known that tumors release various substances into the circulatory system, which can further migrate throughout the body and reach distant organs [38]. These substances include circulating intact tumor cells, tumor-released nucleic acids, proteins, and tumor-derived extracellular vesicles or exosomes, as well as rearranged immune cells, allowing multiple techniques to be used to analyze these varied groups of structures (Figure 2) [37]. LB appears promising in the diagnosis of LM, demonstrating superiority in terms of accuracy over standard diagnostic methods [39].
Figure 2. Methods to diagnose leptomeningeal carcinomatosis.
Note: This image is created by Biorender.com.
Circulating tumor cells (CTCs)
Discovered in 1869 [40], free circulating tumor cells (CTCs) have become a valuable tool in cancer detection via LB [41]. Not only can CTCs be detected in small fluid volumes [42], but they may also be cultured in vitro for more sophisticated analyses [43]. The number of CTCs in the blood was shown to correlate with increased metastatic burden and overall survival [41,44,45]. CTCs can be detected in the blood in 70% of cases of cancer metastases [46]. The most-used technique, and the only FDA-approved method, is the CellSearch® assay, which examines epithelial cell adhesion molecule (EpCAM) expression, nuclear staining with 4′, 6-diamidino-2-phenylindole (DAPI), and the immunofluorescence detection of cytokeratin and CD45 [47].
The CellSearch® assay (Veridex) consists of a semiautomated system for the preparation of a sample that is enriched for cells expressing EpCAM using antibody-coated magnetic beads. The cells are labeled with the fluorescent nucleic acid dye DAPI, and fluorescently labeled monoclonal antibodies specific for leukocytes (CD45-allophycocyanin) and epithelial cells (cytokeratin 8,18,19-phycoerythrin) are used to distinguish the cells [47]. This test also allows for the additional analysis of specific markers, such as HER2 expression [48], which have proven to be accurate in detecting metastatic cancer.
The number of CTCs strongly correlates with overall survival and tumor progression [41,49-51]. There was an attempt to detect CTCs in the peripheral blood of patients with LM; however, their concentration was significantly lower in comparison to CSF [52], or they could not be detected at all [53]. CSF provides nourishment and waste removal, making it a unique diagnostic material in the investigation of diseases of the central nervous system [54]. Metastatic cells are believed to infiltrate the CSF through Virchow-Robin spaces, the lymphatic system, or via direct infiltration of parenchymal metastases [55-57]. CSF shows low background levels of cell-free DNA, protein, and low lipid content, making it an ideal substance for LC diagnosis [41]. A recent meta-analysis reported a cumulative sensitivity and specificity of the method of 87% and 94%, respectively [58]. A summary of CTC studies in LM research is presented in Table 2.
Table 2. CTCs detection research in CSF in patients with LC.
CTCs: circulating tumor cells; CSF: cerebrospinal fluid; LC: leptomeningeal carcinomatosis; DAPI: 4′,6-diamidino-2-phenylindole.
| Marker | Technique | Disease | Number of samples | Clinical benefit | Year | Reference |
| EPCAM-PE, DAPI, CD45 | CellSearch® assay | Breast cancer | 5 | Diagnostics, treatment response, tumor burden; number of CTCs correlates with functional status | 2011 | [59] |
| EpCAM, DAPI, Cytokeratin 8/8/19, CD45 | Adapted CellSearch® assay | Breast cancer | 8 | Diagnostics, treatment response | 2012 | [60] |
| EpCAM | Flow cytometry | Epithelial-cell tumors | 78 | Sensitivity 76%, specificity 96% | 2012 | [61] |
| CD146, HMW/MAA, CD34, CD45, DAPI | CellSearch® assay | Melanoma | 2 | Quantitative diagnostic | 2013 | [53] |
| EpCAM, DAPI, Cytokeratin 8/8/19, CD45 | CellSearch® assay | Lung cancer, breast cancer | 51 | Sensitivity 100%, specificity 97% | 2013 | [62] |
| EpCAM, DAPI, Cytokeratin 8/8/19, CD45 | CellSearch® assay | Lung cancer | 18 | Sensitivity 78%, specificity 100% | 2015 | [63] |
| EpCAM, DAPI, Cytokeratin 8/8/19, CD45 | CellSearch® assay | Breast cancer | 38 | Sensitivity 81%, specificity 85% | 2015 | [64] |
| EpCAM, Hoechst, CD45- | Flow-cytometry | Breast cancer, lung cancer, ovarian cancer | 29 | Sensitivity 100%, specificity 100% | 2016 | [65] |
| CD326, CD45, CD33 | Flow-cytometry | - | 6 | Sensitivity 97%, specificity 99% | 2016 | [66] |
| EpCAM, DAPI, Cytokeratin 8/8/19, CD45 | CellSearch® assay | Lung cancer | 21 | Sensitivity 95%, specificity 100% | 2017 | [67] |
| EpCAM, DAPI, Cytokeratin 8/8/19, CD45 | CellSearch® assay | Lung cancer, breast cancer, miscellaneous | 95 | Sensitivity 93%, specificity 95% | 2017 | [68] |
| DAPI/CD45/CK | SEi FISH | Breast cancer | 8 | Monitoring of intrathecal therapy | 2018 | [69] |
| EpCAM, DAPI, Cytokeratin 8/8/19, CD45, CD34, CD146, HMW/MAA | CellSearch® assay | Breast, lung, renal cell cancer, melanoma | 20 | Sensitivity 89%, specificity 100% | 2020 | [70] |
| EpCAM | Flow-cytometry | Breast cancer, lung cancer, ovarian cancer | 72 | Sensitivity 94%, specificity 100% | 2020 | [71] |
| EpCAM, HER2 | CellSearch® assay | Epithelial HER2+ cancers | 34 | Quantification, tumor progression, treatment response | 2020 | [72] |
| EpCAM, DAPI, Cytokeratin 8/8/19, CD45 | CellSearch® assay | Lung cancer, breast cancer, gastrointestinal cancer | 101 | Prediction of survival | 2022 | [73] |
| EpCAM, DAPI, Cytokeratin 8/8/19, CD45, HER2 | CellSearch® assay | Breast cancer | 49 | Sensitivity 100%, specificity 77% | 2022 | [74] |
| Antibody cocktail + ER/HER2 | CNSide® | Breast cancer | 10 | Sensitivity 100%, specificity 83% | 2022 | [75] |
The identification and quantification of CTCs in CSF is a promising diagnostic approach that has the potential to assess therapy response and dynamic monitoring in LC treatment. It shows superiority over classical cytology staining methods and is becoming more accessible in the clinical routine [57]. Some cells, however, are known to lose EpCAM expression, making these methods unsuitable for LC diagnosis [76]. Not only can cells with low or no EpCAM expression not be detected, but they are also either dead or immobilized at the moment of their capture, making further analysis complicated [77]. Additionally, there is a certain level of heterogeneity within this population of cells [78].
cfDNA
Dying tumor cells, or cells undergoing apoptosis, release tumor-cell DNA or cell-free DNA (cfDNA) into the circulatory system [79]. Advances in modern technology allow for the analysis of these fragments via the investigation of mutations, including single nucleotide variants (SNVs), deletions and insertions, gene fusion, copy number variations (CNVs), and large chromosomal rearrangements [80-82]. It is now known that the brain metastasis (BM) genetic profile differs from the one in the primary tumor, recurrent tumor, and even between different metastatic lesions, which makes it essential to analyze the actual metastatic cells themselves [83]. The detection of mutations in real-time allows for the analysis of therapy resistance mechanisms and appropriate clinical decision-making [84,85]. CSF enriched with cfDNA shows a low background of genomic DNA in comparison to blood samples, which, as mentioned above, makes it an optimal resource for sophisticated and precise mutation analysis, and it may be even more sensitive in the case of parenchymal BM [57,86]. Due to the blood-brain barrier, there is a very low enrichment of cfDNA in plasma [87]. It has been shown that CSF is relatively stable at -80°C [88]. Moreover, it more reliably identifies driver mutations compared with plasma [89,90]. Not only are they useful as diagnostic markers, but escape mutations can be detected dynamically before disease progression [91]. The published articles on cfDNA and LM are summarized in Table 3.
Table 3. cfDNA detection research using CSF in patients with LC.
CSF: cerebrospinal fluid; LC: leptomeningeal carcinomatosis; cfDNA: cell-free DNA; EGFR: epidermal growth factor receptor; CTC: circulating tumor cells.
| Marker | Technique | Disease | Number of samples | Clinical benefit | Year | Reference |
| 8q24 gain (from CTC) | Array CGH | Breast cancer | 15 | Early diagnosis, targeted therapy | 2013 | [92] |
| TP53, EGFR, MLL2, NSD1, CREB3L1, TPR, TSC2 | ddPCR | Breast cancer | 1 | Targeted detection of driver mutations | 2015 | [93] |
| BRAFV600E/K | ddPCR | Melanoma | 11 | Quantification, therapy-response monitoring | 2016 | [94] |
| MET amplification, ERBB2 mutation | NGS | Non-small cell lung cancer (NSCLC) | 19 | Diagnostics | 2017 | [67] |
| BRAF600E, NRAS, PIK3CA, ABL1, MET | ddPCR, NGS | Melanoma | 7 | Diagnostics | 2018 | [95] |
| MET/ERBB2/KRAS/ALK/MYC CNV, TP53 LOH, EGFRT790M | NGS | EGFR mutant lung cancer | 26 | Genetic profile of CSF cfDNA, diagnosis, marker of disease progression | 2018 | [89] |
| Mutation rate in 168 lung cancer-related genes, MaxAF, TP53 LOH, EGFR mutations, ALK fusions, and ERBB2 amplification | Capture-based targeted sequencing | Lung cancer | 72 | CSF as a better sample for liquid biopsy than blood | 2018 | [96] |
| EGFR | ddPCR | Lung cancer | 15 | EGFR therapy monitoring | 2019 | [97] |
| EGFR | NGS | Lung cancer | 11 | Targeted detection of driver mutations, not suitable for LM diagnostics | 2020 | [98] |
| EGFR, TP53 | Error-suppressed deep sequencing | Lung cancer | 2 | Targeted detection of driver mutations, not specific for LM diagnostics | 2021 | [99] |
| EGFR C79C7S, MET ampl | Nanowire-based cfDNA | Lung cancer | 11 | EGFR therapy monitoring | 2021 | [100] |
| Cancer fraction using ichorCNA | NGS | - | 22 | Increased sensitivity and specificity in comparison to cytology | 2021 | [101] |
| Genome-wide aneuploidy | NGS, mFAST-SeqS | Breast cancer | 10 | LM prediction, survival prognosis | 2021 | [102] |
| EGFR mutation, T790M | ddPCR | EGFR-mutant lung cancer | 48 | 68.8% positive for EGFR mutation, 14.6% positive for T790M, therapy monitoring | 2021 | [103] |
| TP53, PIK3CA, CCND1 | NGS | Breast cancer | 8 | Targeted detection of driver mutations | 2022 | [75] |
| cfDNA fraction | ulpWGS | Breast cancer | 30 | Quantitative marker for diagnostic and treatment response | 2022 | [104] |
The cfDNA analysis provides a unique opportunity to detect molecular markers that are not present in BM or blood [105]. More and more studies are attempting to avoid targeted marker testing and focus on broad approaches in order to identify new mutations. These would allow one to test for tumor heterogeneity better than by tissue biopsy [106]. The problem, however, is finding a marker specific to LC but not the cfDNA, which arises from coexisting metastatic lesions in the nervous system. A variation in nucleosome positioning in cfDNA was proposed as a method for identifying the tissue origin [107]. Unfortunately, the majority of the current studies are retrospective and small in sample size, limiting their application in a clinical setting. Larger prospective studies investigating broad molecular characteristics are required for the development of a valid, accurate, and reliable diagnostic method.
RNA
Another element that can be inspected in CSF is cell RNA. DNA mutations do not necessarily lead to downstream changes. Additionally, RNA represents a “snapshot” of genetic changes and disease progression [108]. In addition to coding RNA, long non-coding RNA and microRNA, which are regulatory non-protein-coding transcripts that regulate protein expression, have also been thoroughly investigated in the past few years as potential biomarkers [109]. Due to the low number of cancer cells in CSF and the difficulties in differentiating these from immune cells, the idea of RNA identification was proposed, since even a low concentration of RNA can be detected [110].
Based on the single-cell RNA-sequencing analysis of CSF in five patients with breast cancer and NSCLC, a higher expression of the iron-binding protein lipocalin-2 and its receptor, SCL22A17, was identified in cancer cells as a result of the macrophage cytokine response [90]. A trial of four CSF samples from patients with NSCLC revealed that CEACAM6 and MUC1 were present as cfRNA and that they were associated with disease progression and cell migration [111]. The transcription factor SREBF2 was also found to be activated in the CSF of five patients with breast and lung cancer.
Many physiological processes, including oncogenesis, are regulated by non-coding RNA, such as microRNA [112]. In a recent study, several microRNAs, namely miR-335-5p, miR-34b-3p, and miR-466, were identified in the CSF of 22 LC patients with lung, breast, and ovarian cancer; these differed from those found in primary and metastatic lesions, suggesting them as potential candidates as LC diagnostic markers [113].
Protein markers
Proteins are being widely used in clinical practice as diagnostic markers since they reflect pathophysiological changes under many different conditions [114]. Several unsuccessful attempts have been made to analyze well-known protein biomarkers such as CA15-3 or CEA in CSF, which have proven to be non-specific when found in CSF and in the presence of any intraparenchymal lesions [115-117]. Another commonly used marker, VEGR, when standardized to serum, can be detected in the CSF of LC patients [118]. A higher expression of complement component 3 was observed in the CSF of 37 patients with confirmed LC based on ELISAs. This protein is believed to play a role in disrupting the blood-brain barrier and in cancer growth [119]. Lipocalin 2, an iron-binding protein, and its receptor were exclusively expressed in cancer cells in the CSF of five patients with breast and lung cancer LC, which was confirmed by ELISA and flow cytometry. The authors, moreover, showed inhibition of cancer growth when iron chelation therapy was applied [90]. Increased activities of matrix metalloprotease 9 and A Disintegrin and Metalloproteases (ADAMs) 8 and 17 were detected in the CSF of 12 LM patients as indicators of the breakdown of the blood-brain barrier based on a proteolytic activity matrix assay [120].
Modern research techniques have been expanded from single-protein detection, such as staining for the known protein markers HER2 and BRAF V600E [57], toward more sophisticated proteomic methods that can assess multiple proteins at the same time [121]. Proteomic-based methods are increasingly being used for the identification of CSF markers in different diseases, including LC [122,123]. Mass spectrometry analysis revealed an upregulation of the pathways enrolled in innate immunity and acute-phase response signaling in the CSF of eight LC patients with melanoma, with high-level expression of the TGF-β1 protein confirmed via ELISA [124]. Several proteins specific to lymphoma and leukemia LC, including PTPRC, SERPINC1, sCD44, sCD14, ANPEP, SPP1, FCGR1A, C9, sCD19, and sCD34, were identified in the CSF of 12 patients based on liquid chromatography-tandem mass spectrometry [125].
Tumor microenvironment
The tumor microenvironment (TME) plays an essential role in cell invasion and cancer progression. It consists of immune cells surrounded by the extracellular matrix [126,127]. One of the key components of TME is exosomes, which are lipid bilayer spheroids that contain protein and nucleic acids [128]. Moreover, they have been shown to pass through the blood-brain barrier [129]. The separation of exosomes is possible using ultracentrifugation methods, with subsequent analysis of the marker of interest [130]. The FN1 (fibronectin) exosomal protein, known to be widely expressed in different tumor cells, may serve as a potential diagnostic marker in NSCLC LC diagnosis based on the proteomic analysis of patients with confirmed LC [131]. Another recent study proposed two exosomal microRNA markers, 509-3p and 449a, in the CSF of NSCLC progression as indicators of LC [132]. Exosomal miRNA-483-5p and miRNA-342-5p were found to be significantly upregulated in both the serum and CSF of LM patients with NSCL [133].
It is well known that immune cells are present in the central nervous system and the CSF [134]. In a prospective study, a certain shift pattern in the immune cell presence in CSF was observed in patients with LM. CD8+ T cells were highly present after immune checkpoint inhibitor administration, and the upregulation of IFN-γ signaling was observed [134]. Research on the IFN-γ signaling response may be used in the dynamic assessment of checkpoint inhibitor therapy as an independent marker of tumor cell detection. The dominant presence of M2-polarized macrophages was observed in the CSF of patients with LM based on scRNAseq, displaying immunosuppressive characteristics and showing high levels of ligand-receptor interactions such as MDK‐SORL1 and MDK‐LRP1 [135]. Another interesting phenomenon was observed in seven LM patients with breast cancer. Based on flow cytometry analysis, there was a lower number of CD45+ and CD8+ T-cells and a higher frequency of Treg cells in CSF [136]. Thus, examining the shift in the pattern of immune cells may also be useful in the diagnosis of LC.
Conclusions
Leptomeningeal carcinomatosis in metastatic cancer is a highly morbid and lethal entity in patients with oncologic disease. Currently, there are no effective and reliable methods for early diagnosis, which dampens the opportunity for the optimal management of this group of patients.
Several studies have sought to identify an optimal diagnostic test that is reliable, enables continued monitoring of the disease’s activity, is inexpensive, and, above all, does not significantly increase the morbidity of the already affected oncologic patients. New methods of identifying tumor cells directly or nucleic acid can not only provide an accurate method of diagnostics but also allow monitoring of mutations necessary for targeted therapies. Early detection may lead to new therapeutic approaches, which, consequently, may improve the quality of life of patients. Liquid biopsies remain, however, challenging due to ongoing method establishment and high procedural costs. While the different options seem promising, more combined efforts are still required to obtain more accurate data.
The authors have declared that no competing interests exist.
Author Contributions
Concept and design: Maria Goldberg, Amir Kaywan Aftahy, Bernhard Meyer
Acquisition, analysis, or interpretation of data: Maria Goldberg, Michel G. Mondragon-Soto, Ghaith Altawalbeh
Drafting of the manuscript: Maria Goldberg
Critical review of the manuscript for important intellectual content: Maria Goldberg, Amir Kaywan Aftahy, Michel G. Mondragon-Soto, Ghaith Altawalbeh, Bernhard Meyer
Supervision: Amir Kaywan Aftahy
References
- 1.Leptomeningeal metastasis from systemic cancer: review and update on management. Wang N, Bertalan MS, Brastianos PK. Cancer. 2018;124:21–35. doi: 10.1002/cncr.30911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leptomeningeal carcinomatosis: molecular landscape, current management, and emerging therapies. Bhambhvani HP, Rodrigues AJ, Umeh-Garcia MC, Hayden Gephart M. Neurosurg Clin. 2020;31:613–625. doi: 10.1016/j.nec.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meta-analysis of commonly mutated genes in leptomeningeal carcinomatosis. Congur I, Koni E, Onat OE, Tokcaer Keskin Z. PeerJ. 2023;11:0. doi: 10.7717/peerj.15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nodular leptomeningeal disease-a distinct pattern of recurrence after postresection stereotactic radiosurgery for brain metastases: a multi-institutional study of interobserver reliability. Turner BE, Prabhu RS, Burri SH, et al. Int J Radiat Oncol Biol Phys. 2020;106:579–586. doi: 10.1016/j.ijrobp.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CSF analysis for protein biomarker identification in patients with leptomeningeal metastases from CNS lymphoma. Galicia N, Dégano R, Díez P, González-González M, Góngora R, Ibarrola N, Fuentes M. Expert Rev Proteomics. 2017;14:363–372. doi: 10.1080/14789450.2017.1307106. [DOI] [PubMed] [Google Scholar]
- 6.Intrathecal chemotherapy for treatment of leptomeningeal dissemination of metastatic tumours. Beauchesne P. Lancet Oncol. 2010;11:871–879. doi: 10.1016/S1470-2045(10)70034-6. [DOI] [PubMed] [Google Scholar]
- 7.Experimental treatments for leptomeningeal metastases from solid malignancies. Sahebjam S, Forsyth PA, Smalley KS, Tran ND. Cancer Control. 2017;24:42–46. doi: 10.1177/107327481702400106. [DOI] [PubMed] [Google Scholar]
- 8.Therapy of leptomeningeal metastasis in solid tumors. Mack F, Baumert BG, Schäfer N, Hattingen E, Scheffler B, Herrlinger U, Glas M. Cancer Treat Rev. 2016;43:83–91. doi: 10.1016/j.ctrv.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Predicting leptomeningeal disease spread after resection of brain metastases using machine learning. Tewarie IA, Senko AW, Jessurun CA, et al. J Neurosurg. 2022;1:1–9. doi: 10.3171/2022.8.JNS22744. [DOI] [PubMed] [Google Scholar]
- 10.Leptomeningeal metastases: the future is now. Lukas RV, Thakkar JP, Cristofanilli M, et al. J Neurooncol. 2022;156:443–452. doi: 10.1007/s11060-021-03924-2. [DOI] [PubMed] [Google Scholar]
- 11.Leptomeningeal metastasis in solid tumors with a special focus on lung cancer. Joshi A, Ghosh J, Noronha V, Parikh PM, Prabhash K. Indian J Cancer. 2014;51:410–413. doi: 10.4103/0019-509X.175351. [DOI] [PubMed] [Google Scholar]
- 12.Identification of risk factors associated with leptomeningeal disease after resection of brain metastases. Morshed RA, Saggi S, Cummins DD, et al. J Neurosurg. 2023;139:402–413. doi: 10.3171/2022.12.JNS221490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leptomeningeal metastasis from solid tumours: EANO-ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Le Rhun E, Weller M, van den Bent M, et al. ESMO Open. 2023;8:101624. doi: 10.1016/j.esmoop.2023.101624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leptomeningeal disease and neurologic death after surgical resection and radiosurgery for brain metastases: a multi-institutional analysis. Prabhu RS, Turner BE, Asher AL, et al. Adv Radiat Oncol. 2021;6:100644. doi: 10.1016/j.adro.2021.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.A multi-institutional analysis of presentation and outcomes for leptomeningeal disease recurrence after surgical resection and radiosurgery for brain metastases. Prabhu RS, Turner BE, Asher AL, et al. Neuro-Oncology. 2019;21:1049–1059. doi: 10.1093/neuonc/noz049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MR imaging of leptomeningeal metastases: comparison of three sequences. Singh SK, Leeds NE, Ginsberg LE. http://ajnr.org/content/23/5/817. AJNR Am J Neuroradiol. 2002;23:817–821. [PMC free article] [PubMed] [Google Scholar]
- 17.Imaging of spinal metastatic disease. Shah LM, Salzman KL. Int J Surg Oncol. 2011;2011:769753. doi: 10.1155/2011/769753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neuroimaging and cerebrospinal fluid cytology in the diagnosis of leptomeningeal metastasis. Freilich RJ, Krol G, DeAngelis LM. Ann Neurol. 1995;38:51–57. doi: 10.1002/ana.410380111. [DOI] [PubMed] [Google Scholar]
- 19.Leptomeningeal metastases: a RANO proposal for response criteria. Chamberlain M, Junck L, Brandsma D, et al. Neuro Oncol. 2017;19:484–492. doi: 10.1093/neuonc/now183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A comparison between ventricular and lumbar cerebrospinal fluid cytology in adult patients with leptomeningeal metastases. Chamberlain MC, Kormanik PA, Glantz MJ. Neuro-Oncology. 2001;3:42–45. doi: 10.1093/neuonc/3.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.EANO-ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up of patients with brain metastasis from solid tumours. Le Rhun E, Guckenberger M, Smits M, et al. Ann Oncol. 2021;32:1332–1347. doi: 10.1016/j.annonc.2021.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Clinicopathological and treatment‐associated prognostic factors in patients with breast cancer leptomeningeal metastases in relation to tumor biology. Griguolo G, Pouderoux S, Dieci MV, et al. Oncologist. 2018;23:1289–1299. doi: 10.1634/theoncologist.2018-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leptomeningeal carcinomatosis in non-small-cell lung cancer patients: impact on survival and correlated prognostic factors. Lee SJ, Lee JI, Nam DH, et al. J Thorac Oncol. 2013;8:185–191. doi: 10.1097/JTO.0b013e3182773f21. [DOI] [PubMed] [Google Scholar]
- 24.Impact of multimodality approach for patients with leptomeningeal metastases from solid tumors. Kwon J, Chie EK, Kim K, et al. J Korean Med Sci. 2014;29:1094–1101. doi: 10.3346/jkms.2014.29.8.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osimertinib improves overall survival in patients with EGFR-mutated NSCLC with leptomeningeal metastases regardless of T790M mutational status. Lee J, Choi Y, Han J, et al. J Thorac Oncol. 2020;15:1758–1766. doi: 10.1016/j.jtho.2020.06.018. [DOI] [PubMed] [Google Scholar]
- 26.Breast cancer and leptomeningeal disease (LMD): hormone receptor status influences time to development of LMD and survival from LMD diagnosis. Yust-Katz S, Garciarena P, Liu D, et al. J Neurooncol. 2013;114:229–235. doi: 10.1007/s11060-013-1175-6. [DOI] [PubMed] [Google Scholar]
- 27.Leptomeningeal metastases from non-small cell lung cancer: state of the art and recent advances. Pellerino A, Internò V, Muscolino E, et al. J Cancer Metastasis Treat. 2020;6:41. [Google Scholar]
- 28.Survival in patients with brain metastases: summary report on the updated diagnosis-specific graded prognostic assessment and definition of the eligibility quotient. Sperduto PW, Mesko S, Li J, et al. J Clin Oncol. 2020;38:3773–3784. doi: 10.1200/JCO.20.01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.How Can We Treat Cancer Disease Not Cancer Cells? Kim KW, Lee SJ, Kim WY, Seo JH, Lee HY. Cancer Res Treat. 2017;49:1–9. doi: 10.4143/crt.2016.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer screening in the United States, 2019: a review of current American Cancer Society guidelines and current issues in cancer screening. Smith RA, Andrews KS, Brooks D, Fedewa SA, Manassaram-Baptiste D, Saslow D, Wender RC. CA Cancer J Clin. 2019;69:184–210. doi: 10.3322/caac.21557. [DOI] [PubMed] [Google Scholar]
- 31.Application of cell-free DNA analysis to cancer treatment. Corcoran RB, Chabner BA. N Engl J Med. 2018;379:1754–1765. doi: 10.1056/NEJMra1706174. [DOI] [PubMed] [Google Scholar]
- 32.Emerging concepts in liquid biopsies. Perakis S, Speicher MR. BMC Med. 2017;15:75. doi: 10.1186/s12916-017-0840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Personalized cancer therapy-leveraging a knowledge base for clinical decision-making. Dumbrava EI, Meric-Bernstam F. Cold Spring Harb Mol Case Stud. 2018;4:0. doi: 10.1101/mcs.a001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liquid biopsy: monitoring cancer-genetics in the blood. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Nat Rev Clin Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 35.Differential expression of proliferation-associated molecules in individual micrometastatic carcinoma cells. Pantel K, Schlimok G, Braun S, et al. J Natl Cancer Inst. 1993;85:1419–1424. doi: 10.1093/jnci/85.17.1419. [DOI] [PubMed] [Google Scholar]
- 36.The future of liquid biopsy. Alix-Panabières C. Nature. 2020;579:0. doi: 10.1038/d41586-020-00844-5. [DOI] [PubMed] [Google Scholar]
- 37.Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Lone SN, Nisar S, Masoodi T, et al. Mol Cancer. 2022;21:79. doi: 10.1186/s12943-022-01543-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Detection and localization of surgically resectable cancers with a multi-analyte blood test. Cohen JD, Li L, Wang Y, et al. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Comparison of the diagnostic value of liquid biopsy in leptomeningeal metastases: a systematic review and meta-analysis. Wang H, Wang L, Fang C, Li C, Zhang L. Front Oncol. 2022;12:1079796. doi: 10.3389/fonc.2022.1079796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Ashworth TR. https://www.scirp.org/reference/ReferencesPapers?ReferenceID=2005935 Aust Med J. 1869;14:146. [Google Scholar]
- 41.Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Allard WJ, Matera J, Miller MC, et al. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 42.Circulating tumor cell isolation for cancer diagnosis and prognosis. Deng Z, Wu S, Wang Y, Shi D. EBioMedicine. 2022;83:104237. doi: 10.1016/j.ebiom.2022.104237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Circulating tumor cells (CTCs): a unique model of cancer metastases and non-invasive biomarkers of therapeutic response. Liu J, Lian J, Chen Y, et al. Front Genet. 2021;12:734595. doi: 10.3389/fgene.2021.734595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. De Bono JS, Scher HI, Montgomery RB, et al. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 45.Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. Cohen SJ, Punt CJ, Iannotti N, et al. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 46.Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Bidard FC, Peeters DJ, Fehm T, et al. Lancet Oncol. 2014;15:406–414. doi: 10.1016/S1470-2045(14)70069-5. [DOI] [PubMed] [Google Scholar]
- 47.Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Riethdorf S, Fritsche H, Müller V, et al. Clin Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 48.Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Riethdorf S, Müller V, Zhang L, et al. Clin Cancer Res. 2010;16:2634–2645. doi: 10.1158/1078-0432.CCR-09-2042. [DOI] [PubMed] [Google Scholar]
- 49.Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D, Heller G. Lancet Oncol. 2009;10:233–239. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Circulating tumor cells: a useful predictor of treatment efficacy in metastatic breast cancer. Liu MC, Shields PG, Warren RD, et al. J Clin Oncol. 2009;27:5153–5159. doi: 10.1200/JCO.2008.20.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Circulating tumor cells, disease progression, and survival in metastatic breast cancer. Cristofanilli M, Budd GT, Ellis MJ, et al. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 52.Diagnostic value of cerebrospinal fluid human epididymis protein 4 for leptomeningeal metastasis in lung adenocarcinoma. Li X, Ruan H, Guan M. Front Immunol. 2024;15 :1339914. doi: 10.3389/fimmu.2024.1339914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Detection and quantification of CSF malignant cells by the CellSearch technology in patients with melanoma leptomeningeal metastasis. Le Rhun E, Tu Q, De Carvalho Bittencourt M, et al. Med Oncol. 2013;30:538. doi: 10.1007/s12032-013-0538-3. [DOI] [PubMed] [Google Scholar]
- 54.Central nervous system disease in hematologic malignancies: historical perspective and practical applications. Pui CH, Thiel E. Semin Oncol. 2009;36:0. doi: 10.1053/j.seminoncol.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Biology and therapy of neoplastic meningitis. Mammoser AG, Groves MD. Curr Oncol Rep. 2010;12:41–49. doi: 10.1007/s11912-009-0079-2. [DOI] [PubMed] [Google Scholar]
- 56.Treatment of central nervous system metastases: parenchymal, epidural, and leptomeningeal. Taillibert S, Hildebrand J. Curr Opin Oncol. 2006;18:637–643. doi: 10.1097/01.cco.0000245323.19411.d7. [DOI] [PubMed] [Google Scholar]
- 57.Liquid biopsy in central nervous system metastases: a RANO review and proposals for clinical applications. Boire A, Brandsma D, Brastianos PK, et al. Neuro-oncology. 2019;21:571–584. doi: 10.1093/neuonc/noz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diagnostic accuracy of cerebrospinal fluid liquid biopsy and MRI for leptomeningeal metastases in solid cancers: a systematic review and meta-analysis. Nakasu Y, Deguchi S, Nakasu S, Yamazaki M, Notsu A, Mitsuya K, Hayashi N. Neuro-oncol Adv. 2023;5:0. doi: 10.1093/noajnl/vdad002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Identification and enumeration of circulating tumor cells in the cerebrospinal fluid of breast cancer patients with central nervous system metastases. Patel AS, Allen JE, Dicker DT, Peters KL, Sheehan JM, Glantz MJ, El-Deiry WS. Oncotarget. 2011;2:752–760. doi: 10.18632/oncotarget.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Development of a new method for identification and quantification in cerebrospinal fluid of malignant cells from breast carcinoma leptomeningeal metastasis. Le Rhun E, Massin F, Tu Q, Bonneterre J, Bittencourt MDC, Faure GC. BMC Clin Pathol. 2012;12:21. doi: 10.1186/1472-6890-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Role of flow cytometry immunophenotyping in the diagnosis of leptomeningeal carcinomatosis. Subirá D, Serrano C, Castañón S, et al. Neuro-Oncology. 2012;14:43–52. doi: 10.1093/neuonc/nor172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rare cell capture technology for the diagnosis of leptomeningeal metastasis in solid tumors. Nayak L, Fleisher M, Gonzalez-Espinoza R, et al. Neurology. 2013;80:1598–1605. doi: 10.1212/WNL.0b013e31828f183f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.CellSearch technology applied to the detection and quantification of tumor cells in CSF of patients with lung cancer leptomeningeal metastasis. Tu Q, Wu X, Le Rhun E, et al. Lung Cancer. 2015;90:352–357. doi: 10.1016/j.lungcan.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 64.Detection of cerebrospinal fluid tumor cells and its clinical relevance in leptomeningeal metastasis of breast cancer. Lee JS, Melisko ME, Magbanua MJ, Kablanian AT, Scott JH, Rugo HS, Park JW. Breast Cancer Res Treat. 2015;154:339–349. doi: 10.1007/s10549-015-3610-1. [DOI] [PubMed] [Google Scholar]
- 65.EpCAM-based flow cytometry in cerebrospinal fluid greatly improves diagnostic accuracy of leptomeningeal metastases from epithelial tumors. Milojkovic Kerklaan B, Pluim D, Bol M, et al. Neuro-oncology. 2016;18:855–862. doi: 10.1093/neuonc/nov273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Screening of carcinoma metastasis by flow cytometry: a study of 238 cases. Acosta M, Pereira J, Arroz M. Cytometry Part B. 2016;90:289–294. doi: 10.1002/cyto.b.21258. [DOI] [PubMed] [Google Scholar]
- 67.Detection of driver and resistance mutations in leptomeningeal metastases of NSCLC by next-generation sequencing of cerebrospinal fluid circulating tumor cells. Jiang BY, Li YS, Guo WB, et al. Clin Cancer Res. 2017;23:5480–5488. doi: 10.1158/1078-0432.CCR-17-0047. [DOI] [PubMed] [Google Scholar]
- 68.Cerebrospinal fluid circulating tumor cells: a novel tool to diagnose leptomeningeal metastases from epithelial tumors. Lin X, Fleisher M, Rosenblum M, et al. Neuro-Oncology. 2017;19:1248–1254. doi: 10.1093/neuonc/nox066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clinical significance of detecting CSF-derived tumor cells in breast cancer patients with leptomeningeal metastasis. Li X, Zhang Y, Ding J, et al. Oncotarget. 2018;9:2705–2714. doi: 10.18632/oncotarget.23597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Integration of rare cell capture technology into cytologic evaluation of cerebrospinal fluid specimens from patients with solid tumors and suspected leptomeningeal metastasis. Torre M, Lee EQ, Chukwueke UN, Nayak L, Cibas ES, Lowe AC. J Am Soc Cytopathol. 2020;9:45–54. doi: 10.1016/j.jasc.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 71.Circulating epithelial tumor cell analysis in CSF in patients with leptomeningeal metastases. van Bussel MT, Pluim D, Milojkovic Kerklaan B, et al. Neurology. 2020;94:521–528. doi: 10.1212/WNL.0000000000008751. [DOI] [PubMed] [Google Scholar]
- 72.Cerebrospinal fluid circulating tumor cells as a quantifiable measurement of leptomeningeal metastases in patients with HER2 positive cancer. Malani R, Fleisher M, Kumthekar P, et al. J Neurooncol. 2020;148:599–606. doi: 10.1007/s11060-020-03555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quantitative assessment of circulating tumor cells in cerebrospinal fluid as a clinical tool to predict survival in leptomeningeal metastases. Diaz M, Singh P, Kotchetkov IS, et al. J Neurooncol. 2022;157:81–90. doi: 10.1007/s11060-022-03949-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Detection of circulating tumor cells in cerebrospinal fluid of patients with suspected breast cancer leptomeningeal metastases: a prospective study. Darlix A, Cayrefourcq L, Pouderoux S, et al. Clin Chem. 2022;68:1311–1322. doi: 10.1093/clinchem/hvac127. [DOI] [PubMed] [Google Scholar]
- 75.Diagnosis of leptomeningeal metastasis in women with breast cancer through identification of tumor cells in cerebrospinal fluid using the CNSide™ assay. Wooster M, McGuinness JE, Fenn KM, et al. Clin Breast Cancer. 2022;22:457–462. doi: 10.1016/j.clbc.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Hyun KA, Koo GB, Han H, et al. Oncotarget. 2016;7:24677–24687. doi: 10.18632/oncotarget.8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Label-free ferrohydrodynamic cell separation of circulating tumor cells. Zhao W, Cheng R, Jenkins BD, et al. Lab Chip. 2017;17:3097–3111. doi: 10.1039/c7lc00680b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Analysis of intrapatient heterogeneity of circulating tumor cells at the single-cell level in the cerebrospinal fluid of a patient with metastatic gastric cancer. Cho JH, Sim MH, Kim SY, et al. J Cancer Res Ther. 2021;17:1047–1051. doi: 10.4103/jcrt.JCRT_108_19. [DOI] [PubMed] [Google Scholar]
- 79.Role of circulating cell-free DNA in cancers. Aarthy R, Mani S, Velusami S, Sundarsingh S, Rajkumar T. Mol Diagn Ther. 2015;19:339–350. doi: 10.1007/s40291-015-0167-y. [DOI] [PubMed] [Google Scholar]
- 80.Enhanced detection of circulating tumor DNA by fragment size analysis. Mouliere F, Chandrananda D, Piskorz AM, et al. Sci Transl Med. 2018;10:0. doi: 10.1126/scitranslmed.aat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Sun K, Jiang P, Cheng SH, et al. Genome Res. 2019;29:418–427. doi: 10.1101/gr.242719.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cell-free DNA technologies for the analysis of brain cancer. Mair R, Mouliere F. Br J Cancer. 2022;126:371–378. doi: 10.1038/s41416-021-01594-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Brastianos PK, Carter SL, Santagata S, et al. Cancer Discov. 2015;5:1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Advances on liquid biopsy analysis for glioma diagnosis. Skouras P, Markouli M, Kalamatianos T, Stranjalis G, Korkolopoulou P, Piperi C. Biomedicines. 2023;11:2371. doi: 10.3390/biomedicines11092371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Euskirchen P, Bielle F, Labreche K, et al. Acta Neuropathol. 2017;134:691–703. doi: 10.1007/s00401-017-1743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. De Mattos-Arruda L, Mayor R, Ng CKY, et al. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Detection of circulating tumor DNA in early- and late-stage human malignancies. Bettegowda C, Sausen M, Leary RJ, et al. Sci Transl Med. 2014;6:224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stability of cell-free DNA from maternal plasma isolated following a single centrifugation step. Barrett AN, Thadani HA, Laureano-Asibal C, Ponnusamy S, Choolani M. Prenat Diagn. 2014;34:1283–1288. doi: 10.1002/pd.4468. [DOI] [PubMed] [Google Scholar]
- 89.Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of EGFR-mutant non-small-cell lung cancer: a new medium of liquid biopsy. Li YS, Jiang BY, Yang JJ, et al. Ann Oncol. 2018;29:945–952. doi: 10.1093/annonc/mdy009. [DOI] [PubMed] [Google Scholar]
- 90.Cancer cells deploy lipocalin-2 to collect limiting iron in leptomeningeal metastasis. Chi Y, Remsik J, Kiseliovas V, et al. Science. 2020;369:276–282. doi: 10.1126/science.aaz2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Advances in the diagnosis, evaluation, and management of leptomeningeal disease. Sener U, Kumthekar P, Boire A. Neurooncol Adv. 2021;3:86–95. doi: 10.1093/noajnl/vdab108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Molecular profiling of tumor cells in cerebrospinal fluid and matched primary tumors from metastatic breast cancer patients with leptomeningeal carcinomatosis. Magbanua MJM, Melisko M, Roy R, et al. Cancer Res. 2013;73:7134–7143. doi: 10.1158/0008-5472.CAN-13-2051. [DOI] [PubMed] [Google Scholar]
- 93.Brain tumor mutations detected in cerebral spinal fluid. Pan W, Gu W, Nagpal S, Gephart MH, Quake SR. Clin Chem. 2015;61:514–522. doi: 10.1373/clinchem.2014.235457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Quantification of tumor-derived cell free DNA(cfDNA) by digital PCR (DigPCR) in cerebrospinal fluid of patients with BRAFV600 mutated malignancies. Momtaz P, Pentsova E, Abdel-Wahab O, et al. Oncotarget. 2016;7:85430–85436. doi: 10.18632/oncotarget.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Evaluating circulating tumor DNA from the cerebrospinal fluid of patients with melanoma and leptomeningeal disease. Ballester LY, Glitza Oliva IC, Douse DY, et al. J Neuropathol Exp Neurol. 2018;77:628–635. doi: 10.1093/jnen/nly046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Unique genomic profiles obtained from cerebrospinal fluid cell-free DNA of non-small cell lung cancer patients with leptomeningeal metastases. Ying S, Ke H, Ding Y, et al. Cancer Biol Ther. 2019;20:562–570. doi: 10.1080/15384047.2018.1538614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Digital PCR-based detection of EGFR mutations in paired plasma and CSF samples of lung adenocarcinoma patients with central nervous system metastases. Huang R, Xu X, Li D, et al. Target Oncol. 2019;14:343–350. doi: 10.1007/s11523-019-00645-5. [DOI] [PubMed] [Google Scholar]
- 98.Detection of circulating tumor DNA from non-small cell lung cancer brain metastasis in cerebrospinal fluid samples. Ma C, Yang X, Xing W, Yu H, Si T, Guo Z. Thorac Cancer. 2020;11:588–593. doi: 10.1111/1759-7714.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tumor DNA mutations from intraparenchymal brain metastases are detectable in CSF. Cheok SK, Narayan A, Arnal-Estape A, et al. JCO Precis Oncol. 2021;5:163–172. doi: 10.1200/PO.20.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.A nanowire-based liquid biopsy method using cerebrospinal fluid cell-free DNA for targeted management of leptomeningeal carcinomatosis. Choi W, Cho Y, Park SY, Hwang KH, Han JY, Lee Y. J Cancer Res Clin Oncol. 2021;147:213–222. doi: 10.1007/s00432-020-03324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Detection of leptomeningeal disease using cell-free DNA from cerebrospinal fluid. White MD, Klein RH, Shaw B, et al. JAMA Netw Open. 2021;4:0. doi: 10.1001/jamanetworkopen.2021.20040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Detection of aneuploidy in cerebrospinal fluid from patients with breast cancer can improve diagnosis of leptomeningeal metastases. Angus L, Deger T, Jager A, et al. Clin Cancer Res. 2021;27:2798–2806. doi: 10.1158/1078-0432.CCR-20-3954. [DOI] [PubMed] [Google Scholar]
- 103.Utility of cerebrospinal fluid cell-free DNA in patients with EGFR-mutant non-small-cell lung cancer with leptomeningeal metastasis. Chiang CL, Lee CC, Huang HC, et al. Target Oncol. 2021;16:207–214. doi: 10.1007/s11523-021-00791-9. [DOI] [PubMed] [Google Scholar]
- 104.Assessing CSF ctDNA to improve diagnostic accuracy and therapeutic monitoring in breast cancer leptomeningeal metastasis. Fitzpatrick A, Iravani M, Mills A, et al. Clin Cancer Res. 2022;28:1180–1191. doi: 10.1158/1078-0432.CCR-21-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. De Mattos-Arruda L, Mayor R, Ng CKY, et al. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Beyond the blood: CSF-derived cfDNA for diagnosis and characterization of CNS tumors. McEwen AE, Leary SE, Lockwood CM. Front Cell Dev Biol. 2020;8:45. doi: 10.3389/fcell.2020.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell. 2016;164:57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Nat Rev Genet. 2016;17:257–271. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: first steps from bench to bedside. Ratti M, Lampis A, Ghidini M, Salati M, Mirchev MB, Valeri N, Hahne JC. Target Oncol. 2020;15:261–278. doi: 10.1007/s11523-020-00717-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.The microRNA spectrum in 12 body fluids. Weber JA, Baxter DH, Zhang S, et al. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Comprehensive RNA analysis of CSF reveals a role for CEACAM6 in lung cancer leptomeningeal metastases. Li Y, Polyak D, Lamsam L, et al. NPJ Precis Oncol. 2021;5:90. doi: 10.1038/s41698-021-00228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Ling H, Fabbri M, Calin GA. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Exploratory profiling of extracellular micrornas in cerebrospinal fluid comparing leptomeningeal metastasis with other central nervous system tumor statuses. Im JH, Kim TH, Lee KY, et al. J Clin Med. 2021;10:4860. doi: 10.3390/jcm10214860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Quantitative protein biomarker panels: a path to improved clinical practice through proteomics. Hartl J, Kurth F, Kappert K, Horst D, Mülleder M, Hartmann G, Ralser M. EMBO Mol Med. 2023;15:0. doi: 10.15252/emmm.202216061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.The clinical relevance of locally produced carcinoembryonic antigen in cerebrospinal fluid. Jacobi C, Reiber H, Felgenhauer K. J Neurol. 1986;233:358–361. doi: 10.1007/BF00313922. [DOI] [PubMed] [Google Scholar]
- 116.Diagnostic value of cerebrospinal fluid level of carcinoembryonic antigen in patients with leptomeningeal carcinomatous metastasis. Kang SJ, Kim KS, Ha YS, Huh SY, Lee JH, Kim JK, Kim MJ. J Clin Neurol. 2010;6:33–37. doi: 10.3988/jcn.2010.6.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Measurements of CSF biochemical tumor markers in patients with meningeal carcinomatosis and brain tumors. Nakagawa H, Kubo S, Murasawa A, Nakajima S, Nakajima Y, Izumoto S, Hayakawa T. J Neurooncol. 1992;12:111–120. doi: 10.1007/BF00172659. [DOI] [PubMed] [Google Scholar]
- 118.Detection of cancer cells in the cerebrospinal fluid: current methods and future directions. Weston CL, Glantz MJ, Connor JR. Fluids Barriers CNS. 2011;8:1–9. doi: 10.1186/2045-8118-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Complement component 3 adapts the cerebrospinal fluid for leptomeningeal metastasis. Boire A, Zou Y, Shieh J, Macalinao DG, Pentsova E, Massagué J. Cell. 2017;168:1101–1113. doi: 10.1016/j.cell.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Profiling of metalloprotease activities in cerebrospinal fluids of patients with neoplastic meningitis. Conrad C, Dorzweiler K, Miller MA, Lauffenburger DA, Strik H, Bartsch JW. Fluids Barriers CNS. 2017;14:22. doi: 10.1186/s12987-017-0070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Revisiting biomarker discovery by plasma proteomics. Geyer PE, Holdt LM, Teupser D, Mann M. Mol Syst Biol. 2017;13:942. doi: 10.15252/msb.20156297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Proteomic biomarker identification in cerebrospinal fluid for leptomeningeal metastases with neurological complications. Galicia N, Díez P, Dégano RM, Guest PC, Ibarrola N, Fuentes M. Adv Exp Med Biol. 2017;974:85–96. doi: 10.1007/978-3-319-52479-5_5. [DOI] [PubMed] [Google Scholar]
- 123.Array-based profiling of proteins and autoantibody repertoires in CSF. Pin E, Sjöberg R, Andersson E, et al. Methods Mol Biol. 2019;2044:303–318. doi: 10.1007/978-1-4939-9706-0_19. [DOI] [PubMed] [Google Scholar]
- 124.Proteomic analysis of CSF from patients with leptomeningeal melanoma metastases identifies signatures associated with disease progression and therapeutic resistance. Smalley I, Law V, Wyatt C, et al. Clin Cancer Res. 2020;26:2163–2175. doi: 10.1158/1078-0432.CCR-19-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Deciphering biomarkers for leptomeningeal metastasis in malignant hemopathies (lymphoma/leukemia) patients by comprehensive multipronged proteomics characterization of cerebrospinal fluid. Juanes-Velasco P, Galicia N, Pin E, et al. Cancers (Basel) 2022;14:449. doi: 10.3390/cancers14020449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Targeting the tumor microenvironment in brain metastasis. Giridharan N, Glitza Oliva IC, O'Brien BJ, Parker Kerrigan BC, Heimberger AB, Ferguson SD. Neurosurg Clin N Am. 2020;31:641–649. doi: 10.1016/j.nec.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 127.Proteoglycans in the pathogenesis of hormone-dependent cancers: mediators and effectors. Tzanakakis G, Giatagana EM, Kuskov A, Berdiaki A, Tsatsakis AM, Neagu M, Nikitovic D. Cancers (Basel) 2020;12:2401. doi: 10.3390/cancers12092401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Exosomes: proteomic insights and diagnostic potential. Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 129.New windows into the brain: central nervous system-derived extracellular vesicles in blood. Shi M, Sheng L, Stewart T, Zabetian CP, Zhang J. Prog Neurobiol. 2019;175:96–106. doi: 10.1016/j.pneurobio.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Théry C, Amigorena S, Raposo G, Clayton A. Curr Protoc Cell Biol. 2006;30:3–22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 131.Cerebrospinal fluid exosomal protein alterations via proteomic analysis of NSCLC with leptomeningeal carcinomatosis. Hou L, Chen X, Qiu G, Qi X, Zou Y, He J, Bu H. J Neurooncol. 2023;164:367–376. doi: 10.1007/s11060-023-04428-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cerebrospinal fluid exosomal microRNAs as biomarkers for diagnosing or monitoring the progression of non-small cell lung cancer with leptomeningeal metastases. Li H, Xia M, Zheng S, et al. Biotechnol Genet Eng Rev. 2023;2023:1–22. doi: 10.1080/02648725.2023.2183613. [DOI] [PubMed] [Google Scholar]
- 133.Serum exosomal miRNA might be a novel liquid biopsy to identify leptomeningeal metastasis in non-small cell lung cancer. Xu Q, Ye L, Huang L, et al. OncoTargets Ther. 2021;14:2327–2335. doi: 10.2147/OTT.S291611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Genomic and transcriptomic correlates of immunotherapy response within the tumor microenvironment of leptomeningeal metastases. Prakadan SM, Alvarez-Breckenridge CA, Markson SC, et al. Nat Commun. 2021;12:5955. doi: 10.1038/s41467-021-25860-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Single-cell RNA sequencing reveals the characteristics of cerebrospinal fluid tumour environment in breast cancer and lung cancer leptomeningeal metastases. Ruan H, Wang Z, Sun Z, et al. Clin Transl Med. 2022;12:0. doi: 10.1002/ctm2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.High-dimensional immune cell profiling of cerebrospinal fluid from patients with metastatic breast cancer and leptomeningeal disease. Im KW, Huppert LA, Malevanchik L, et al. NPJ Breast Cancer. 2023;9:22. doi: 10.1038/s41523-023-00526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]