Abstract
The objective of this study was to identify genes associated with the biodegradation of phenol by Acinetobacter sp. strain DF4 through the use of differential display (DD) methodology. The bacteria were grown in YEPG medium, and total RNA was extracted and analyzed using labeled primers to detect gene expression differences. Three distinctively expressed cDNA bands (ph1, ph2, and ph3) were identified, cloned, and sequenced. DNA analysis involved searching for open reading frames (ORFs), verifying results with the NCBI database, predicting promoter regions, and constructing phylogenetic trees using bioinformatics tools. The ph1 gene displayed a 97% identity with the AraC transcriptional regulator, suggesting its potential role in regulating the ortho-catabolic pathway of phenol. The ph2 gene showed a 98% identity with aspartate semialdehyde dehydrogenase, which is involved in phenol degradation. The ph3 gene had a 93% identity with acetyltransferase. Essential transcription factors, such as TATA, GTGTGT, CACA, and CTTTT, were detected, and the three genes promoter regions were predicted. This study successfully identified functional genes involved in the metabolism of cyclic chemicals, particularly phenol, using the DD technique. These findings provide insights into the biodegradation pathways of phenol by Acinetobacter sp. Strain DF4 and may contribute to the development of more efficient bioremediation strategies for phenol-contaminated environments.
Keywords: Putative genes, Differential Display- Random primers, Phenol catabolic pathway, AraC transcriptional regulator, Dehydrogenase, Acetyltransferase, Transcriptional factors
1. Background
Phenol, a common industrial pollutant, is frequently found in effluents from diverse sectors such as oil refineries, petrochemical plants, coal conversion processes, steel plants, ceramic plants, and phenolic resin industries1. Its presence poses a significant threat to aquatic ecosystems due to its toxicity to aquatic organisms at low concentrations. Furthermore, phenol is a persistent pollutant, resisting biodegradation because of its aromatic structure. Therefore, the development of efficient and sustainable methods for phenol removal is imperative2. In this context, bioremediation emerges as an environmentally friendly and cost-effective solution for phenol removal. To achieve efficient and sustainable treatment methods, a deeper understanding of factors influencing phenol degradation is essential, such as the type of microorganism, phenol concentration, and the presence of other contaminants. The use of mixed cultures and genetically modified microorganisms holds promise for enhancing phenol bioremediation2, 3 Fig. 1.
Fig. 1.
Schematic Diagram of the Differential Display Approach from Culturing to DNA Sequence Analysis.
The aerobic degradation of phenol involves a complex catabolic pathway that transforms phenol into harmless intermediates, allowing their entry into the tricarboxylic acid (TCA) cycle for energy production3. Initiated by phenol hydroxylase, an enzyme catalyzing the hydroxylation of the benzene ring, this process produces catechol—a crucial intermediate acting as a pivotal branching point for two distinct pathways: the ortho pathway and the meta pathway4. In the ortho pathway, catechol undergoes cleavage by catechol-1,2-oxygenase, resulting in the formation of 2-hydroxymuconic semialdehyde. This intermediate undergoes further enzymatic transformations leading to the production of 3-oxoadipate, which readily integrates into the TCA cycle. Conversely, the meta pathway involves the cleavage of catechol by 2,3-dioxygenase, yielding 2-oxo-3-hydroxymuconate. Subsequent enzymatic reactions convert this intermediate into pyruvate and acetaldehyde, both of which can enter the TCA cycle4.
The bacterium Acinetobacter sp. strain DF4 is a proficient phenol-degrading organism, capable of degrading phenol at an impressive rate of 500 mg/L within a mere 15 hours5. Investigations into the catabolic pathway employed by this strain reveal high activities of both catechol 1,2-dioxygenase and phenol hydroxylase, indicating the utilization of the ortho pathway for phenol degradation4. Notably, during the stationary growth phase, the maximum dioxygenase activity observed was 0.68 mg indigo/mg protein/min, highlighting the robust metabolic capacity of this strain for phenol metabolism. To delve deeper into the genetic mechanisms governing phenol degradation in Acinetobacter sp. strain DF4, the present study employed the Differential Display (DD) approach—a technique facilitating the identification of differentially expressed genes under varying environmental conditions. DD identified three novel genes upregulated during phenol degradation in Acinetobacter sp. strain DF4, likely involved in diverse aspects of phenol metabolism. Subsequent exploration of these genes promises valuable insights into the molecular intricacies of phenol degradation in this strain.
2. Methods
2.1. Cultivation of Acinetobacter sp. DF4 and total RNA extraction
To cultivate Acinetobacter sp. DF4, we utilized yeast extract peptone glucose (YEPG) medium in 250-ml flasks at 30 °C6. The medium composition included 1.0 g of dextrose per liter, 2 g of Polypeptone per liter, 0.2 g of yeast extract per liter, and 0.2 g of NH4NO3 per liter, with a pH of 7.0. After an overnight incubation, 2 mL of the culture was transferred into 50 mL of fresh YEPG media. The culture was allowed to reach the mid-log phase of growth (OD 600 ca. 0.4). Subsequently, 3 mL of the culture was divided into two flasks, one containing 200 mM phenol and the other without. After 120 min, samples were withdrawn for total RNA isolation using RNeasy columns (Qiagen, Chatsworth, Calif.) following the manufacturer's recommended procedure.
2.2. Differential display (DD) procedure
As outlined in the schematic diagram, the Differential Display (DD) procedure employed an arbitrary set of 10-bp 32P-labeled primers obtained from a commercial source (Genosys Biotechnologies, Woodlands, Tex., USA) to distinguish gene expression differences6. Specific conditions were maintained in the RT-PCR experiment, including the addition of 50 U of Moloney Murine Leukemia Virus (M−MLV) Reverse Transcriptase, 0.2 g of total RNA, 0.4 mM of primer, and 1 mM of M−MLV Buffer in a 20 µL reaction volume. The RT reaction was carried out with a thermocycler (model 480; Perkin-Elmer, Norwalk, Conn.) using the following program: ramp from 50 to 30 °C for 15 min; 37 °C for 1 h; 95 °C for 5 min; and incubation at 4 °C. Following the RT-PCR, denaturing loading dye was added to the products, which were then incubated in a 95 °C water bath for 3 min.
The PCR products were loaded onto a 4.5 % Denaturing Acrylamide Gel and subjected to a two-hour run on an LR Sequencing Apparatus at 2700 V. The gel was exposed to BioMaxMR film for 12 to 24 h at room temperature for visualization and analysis of gene expression patterns influenced by the experimental conditions. The three obtained differential cDNA bands (ph1, ph2, and ph3) were detected by a side-by-side comparison of induced and uninduced PCR products on an autoradiogram. Gel bands were localized and eluted as previously described6. The excised fragments were reamplified and gel-purified, and the identified PCR reamplified segments were cloned into the pCR 2.1 vector (Invitrogen, San Diego, Calif.) following the manufacturer’s protocols. The presence of inserts was checked by electrophoresis into 1 % low-melting-point agarose after restriction with EcoRI.
2.3. DNA sequencing and analysis
Sequences of the expressed bands ph1, ph2, and ph3 were determined with an automated apparatus (model 373a; Applied Biosystems) at the University of Tennessee Molecular Biology Resource Facility. The DNA analysis approach involved searching for open reading frames (ORFs), validating the results using the NCBI database, predicting promoter regions, and constructing phylogenetic trees. Bioinformatics tools, such as ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/) and the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi), were used for precise identification and validation of coding sequences. Promoter regions were predicted using bioinformatics tools available at the website (https://www.fruitfly.org/seq_tools/promoter.html). Conserved motifs and regulatory elements inside each gene were searched according to published literature. DNA sequences were aligned with similar sequences using Clustal W27, and phylogenetic trees were constructed using the neighbor-joining algorithm. The TreeView software8 was used to enhance the visual representation of the phylogenetic trees.
3. Results and discussion
The results obtained through RT-PCR and subsequent analysis provides insight into the molecular responses of Acientobacter sp. DF4 to phenol stress. The detection of three differently expressed bands, Ph1, Ph2, and Ph3, suggests that phenol exposure triggers the expression of specific genes in this bacterium. The total nucleotide base pairs of these bands are 890, 527, and 477, respectively. The fact that only primer out of ten (3-CGAAGCGATC- 5) was able to amplify these bands indicates that these genes are specific to phenol stress and may play a crucial role in the bacterium's survival under these conditions. The absence of differentiation bands in the culture without phenol further supports this notion6.
The sequencing and phylogeny analysis (Fig. 2, A, B and C) of the DNA fragments revealed that Ph1 shares a high degree of similarity with the Acientobacter AraC family transcriptional regulator, suggesting that it may be involved in regulating gene expression in response to phenol stress. The AraC family of transcriptional regulators is known to be involved in the regulation of aromatic compound metabolism, and their activation has been linked to the induction of genes involved in detoxification and degradation pathways9, 10. Ph2, on the other hand, exhibited a high degree of similarity with aspartate-semialdehyde dehydrogenase, an enzyme involved in the biosynthesis of lysine, threonine, and methionine. The presence of this gene in response to phenol stress may suggest that the bacterium is utilizing alternative metabolic pathways to cope with the toxic effects of phenol. Finally, Ph3 displayed a high degree of similarity with acetyltransferase, an enzyme involved in the modification of various biomolecules, including proteins and nucleic acids. The role of this gene in phenol stress response is less clear, but it may be involved in the modification of proteins or nucleic acids in response to phenol stress12. Therefore, the results obtained through RT-PCR, sequencing, and phylogeny analysis suggest that phenol stress triggers the expression of specific genes in Acientobacter sp. DF4, including a transcriptional regulator, an enzyme involved in amino acid biosynthesis, and an acetyltransferase. These findings provide insights into the molecular responses of this bacterium to phenol stress and may have implications for understanding the mechanisms by which bacteria cope with environmental stressors13.
Fig. 2.
The phylogenetic tree of differentially expressed DNA segments in Acinetobacter sp. strain DF4 under phenol stress, utilizing the DD approach. The bands, Ph1, Ph2, and Ph3, correspond to A, B, and C, respectively. The N-BLAST tool facilitated sequence comparison against databases, and the resulting phylogenetic tree was crafted through the neighbor-joining method.
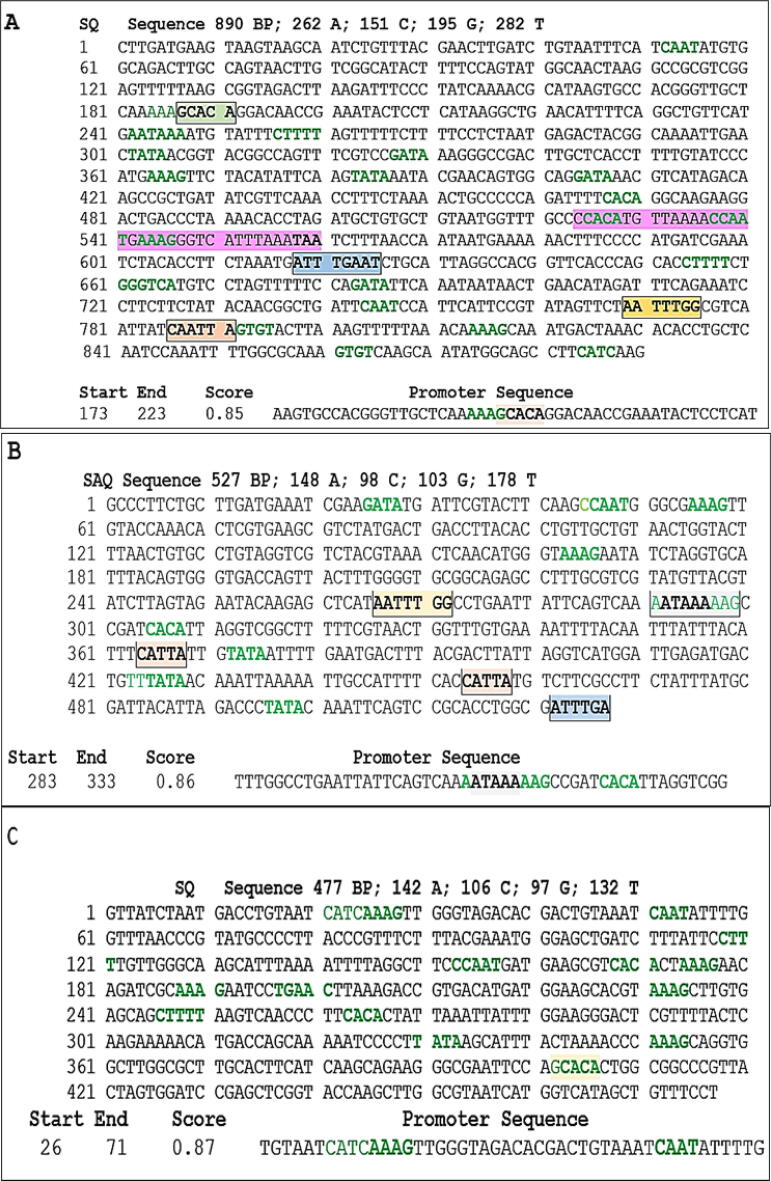
The analysis of the Ph1 gene sequence reveals a notable abundance of transcription factors dispersed throughout its length. As depicted in Fig. 3(A), the sequence contains several regulatory elements, including the HoxC4 binding site ATTTGAAT, the metal transcription factor binding site GCACA, and the flanking nucleotides CATTA, all of which are crucial for gene regulation14, 15, 16. Additionally, the presence of well-known transcription boxes, such as TATA, GTGT, CTTTT, CATC, CCAAT, AAAG, CACA, CAAT, GGGTCA, TTTGAA, GATA, and AATAAA, further confirms the regulatory potential of the Ph1 gene17.
Fig. 3.
Putative DNA response elements and binding sites for transcription factors present in Ph1, Ph2 and Ph3 DNA segments, respectively. The promoter regions and their transcription factors and/or binding sites are presented below each one of them.
The metal binding site in the Ph1 sequence suggests that it functions as a potent transcription factor that activates gene expression17. Mojica et al.11 have identified the palindromic regulator character GAAC as a short repetition with regular spacing, which is intriguing given its potential role in coherent CRISPR Cas9 cleavage, a crucial process for editing bacterial DNA fragments16. In contrast, the pyrimidine box motif is known to induce hydrolase gene promoters18. The TATA box, on the other hand, is a type of promoter sequence that specifies the starting point of transcription, defines the direction of transcription, and identifies the DNA strand to be read19. Moreover, the CCAAT motif has been shown to be crucial for optimal transcriptional activity in several laboratory studies20. The CATC box, which is believed to be essential for the specific expression of proteins reflecting an increase in the ortho catabolic pathway of phenol, is also present in the Ph1 segment21. Previous research has identified the inverse of the CACA motif to GTGT as the motif with the highest overrepresentation in non-coding areas of T. annulata and T. parva22.
The locations and length of the promoter consensus sequence of Ph1 were also predicted, revealing the presence and overlap of the CACA box (green letters), the metal-responsive binding site GCACA (strong red highlight), and AT-rich sites as part of the regulatory promoter region (Fig. 3A). These findings suggest that the Ph1 gene is subject to complex regulatory mechanisms that contribute to its expression and function. Further research is required to elucidate the precise roles of these transcription factors and regulatory elements in the regulation of the Ph1 gene.
The predicted open reading frames (ORFs) of the Ph1 sequence reveal the presence of a gene with significant similarity to the AraC family of transcriptional regulators found in Acinetobacter. This similarity suggests that the Ph1 gene may also function as a transcriptional regulator. The AraC family of proteins, including MarA, is known to interact with target DNA sequences through helix-turn-helix patterns23, and the Ph1 sequence contains a CCAC-N7-TAA binding domain24 similar to that found in MarA as follow (CCACATGTTAAAACCAATGAAAGGGTCATTTAAATAA). This binding domain is located between positions 523 and 597 in the Ph1 sequence (highlighted in purple in Fig. 3A).
MarA is a transcriptional activator that controls the Mar regulon in Escherichia coli, and its expression provides resistance to various drugs, oxidative stresses, and chemical solvents25. The regulatory proteins belonging to the AraC family, including MarA, bind target DNA sites in a similar manner, with sequence specificity primarily obtained from interactions between the recognition helices and bases in the major groove, as described by Rhee et al.26. These findings suggest that the sequence similarity between the Ph1 gene and the AraC family raises the possibility that this gene may have evolved through horizontal gene transfer from a related bacterial species. The presence of a CCAC-N7-TAA binding domain in the Ph1 sequence suggests that this gene may have a role in regulating gene expression in response to environmental stresses, similar to the MarA gene in Escherichia coli.
The DNA sequence of the second differentially derived fragment, known as Ph2, showed a similarity of > 98% to the aspartate semialdehyde dehydrogenase gene, which is found in almost all Acientobacter species (Fig. 2 B). Aspartate-semialdehyde dehydrogenase is an enzyme that helps produce various amino acids from aspartate27. Phenol hydroxylase is responsible for converting phenol to catechol. Other enzymes in the cleavage pathway of catechol include aldehyde dehydrogenase, which uses the ring cleavage products from catechol, 4-methyl-2-hydroxymuconic semialdehyde, and 3-methylcatechol as substrates. These products include 2-hydroxymuconic semialdehyde and 2-hydroxy-6-oxo2,4-heptadienoate, which have been shown to be preferred substrates for the dehydrogenase28.
Fig. 3B shows a promoter region in the Ph2 DNA sequence. The gene boxes CTTTT, TATA, CACA, AAAG, AATAAA, and CAAT were detected in the ph2 sequence (highlighted in bold green letters). Additionally, the transcription factors HoxC4 binding site ATTTGAAT (in a blue box), the flanking sequence CATTA (in a light red box), and a cohesive cleavage of the CRISPER Cas9 DNA fragment AATTTGG (in an orange box) were identified11, 12, 13. It is worth noting that three transcription regulatory elements -CCA, AAAG, and AATAAA- are present in the promoter region (Fig. 3B). According to Mehrotra et al.29, the transcription factors that make up the gene component AAAG have a single, highly conserved zinc finger. The entire process of gene expression is aided by polyadenylation, which is used to produce mature mRNA for translation. The fact that it exists demonstrates that the sequence is not a pseudo gene30.
Using the same search parameters as before with Ph1, an analysis of the Ph2 sequence revealed a single open reading frame (ORF) with a size range of 28 to 240 base pairs. This ORF showed high similarity (>97 %) to three proteins: the Asd/ArgC dimerization domain-containing protein from Acinetobacter baumannii (WP_019204636), semialdehyde dehydrogenase from Acinetobacter baumannii 25569_7 (EYS16690), and semialdehyde dehydrogenase from Acinetobacter baumannii 1,465,485 (EXB60716). Protein dimerization plays a crucial role in regulating many physiological processes, including enzyme activation, transcriptional cofactor recruitment, signal transmission, and pathogenic pathways. When organisms are exposed to internal or external stimuli in their natural environment, the regulation of protein dimerization is a critical step for growth and development31. This domain contains enzymes such as N-acetyl-glutamine semialdehyde dehydrogenase (AgrC), which is involved in arginine biosynthesis, and aspartate-semialdehyde dehydrogenase, which is involved in the production of several amino acids from aspartate. The high similarity between the Ph2 ORF and these proteins suggests that the Ph2 gene may also be involved in amino acid biosynthesis or regulation, potentially through protein dimerization32.
Ph3 is the third gene fragment obtained using the DD technique, sharing 90–93 % similarity with the Acinetobacter acetyltransferase gene family. Its sequence contains transcriptional binding sites, such as CTTTT, TATA, CCAAT, CACA, CATC, and AAAG, as well as a metal transcription activation site, GCACA (Fig. 3C). The promoter region (Fig. 3C) contains essential elements for optimal transcriptional activity, including CAAT and the highly conserved zinc finger AAAG, as well as potential regulatory components CATC. With an identity of less than 95 %, the Ph3-positive stranded ORF (9–477 bp) belongs to the putative acetyltransferase YihG of Acinetobacter baumannii (AXU43955) and Acinetobacter pitti (AMX18444). Gu et al.33 discovered that acetyl-CoA acetyltransferase, which converts b-ketoadipate into succinyl, is a significant phenol catabolic enzyme in the catechol branch of the ortho-cleavage pathway in Acinetobacter DW-1. Therefore, Ph3 is a potential gene involved in Acinetobacter phenol degradation pathway.
The gene Ph3, encoding a stranded ORF (9–477 bp), belongs to the putative acetyltransferase YihG in Acinetobacter baumannii (AXU43955) and Acinetobacter pitti (AMX18444), with a low identity of less than 95 %. Acetyl-CoA acetyltransferase, which converts β-ketoadipate into succinyl, is a crucial enzyme in the catechol branch of the ortho-cleavage pathway for phenol degradation in Acinetobacter DW-133. Therefore, Ph3 is a strong candidate for involvement in the phenol degradation pathway in Acinetobacter, as highlighted through a partial characterization of the pathway using a differential display strategy based on gene-associated activities in Acinetobacter sp. Strain DF4.
Based on the results of differential display (DD) analysis, three fragments, Ph1, Ph2, and Ph3, were identified as potentially involved in the ortho-catabolic pathway for phenol degradation. To further investigate these fragments, two approaches can be taken: (1) making assumptions about the functions of gene regions, known as gene function boxes, or (2) grouping sequences based on similarity and evaluating their similarity to one another34. By using the latter approach, transcriptional factors were identified in the putative genes Ph1, Ph2, and Ph3, which are likely shared signals for most transcriptional and translational regulators. This suggests that screening for transcription factors and identifying stress-responsive binding sites may be useful techniques for estimating a species' capacity for environmental plasticity and niche-specific adaptation. The presence of upregulation elements, such as the CACA consensus sequence and CATC boxes, suggests that they may play a role in overall gene regulation rather than being exclusive to the expression of specific genes. The presence of upregulated components supports the idea that phenol biodegradation occurs through the ortho-cleavage of the ring.35.
The study identified three genes, Ph1, Ph2, and Ph3, which play a role in the expression of pathways related to phenol catabolism in Acinetobacter. Ph1 encodes the AraC transcriptional regulator, which is involved in regulating the expression of genes in the phenol ortho-catabolic pathway. Ph2 encodes Aspartate Semialdehyde Dehydrogenase, an enzyme that catalyzes the conversion of aspartate semialdehyde to aspartate in the phenol ortho-catabolic pathway. Ph3 encodes an Acetyltransferase, which may also be involved in the phenol ortho-catabolic pathway. These genes were successfully functionalized using the Directed Discovery (DD) approach, which allows for the identification and characterization of expressed genes involved in the metabolism of cyclic compounds, particularly phenol.
4. Conclusion
In this study, we employed RT-PCR, sequencing, and phylogeny analysis to delve into the molecular response of Acinetobacter sp. DF4 to phenol stress. Utilizing the differential display technique (DD), we uncovered three key genes—Ph1, Ph2, and Ph3—that exhibited varied expression levels in the presence of phenol. Through sequence analysis, we identified Ph1 as a member of the AraC family transcriptional regulator, indicating its potential role in regulating gene expression in response to phenol stress. Ph2, resembling aspartate-semialdehyde dehydrogenase, suggests the activation of alternative metabolic pathways. The examination of promoter regions for Ph1 and Ph2 revealed a complex gene regulation network involving various transcription factors. Furthermore, Ph3, akin to acetyltransferase genes, hints at its involvement in the phenol degradation pathway. In summary, this study offering valuable insights into the intricate molecular mechanisms at play. These findings significantly contribute to our understanding of bacterial adaptation to environmental stress and hold promise for potential applications in phenol biodegradation and bioremediation strategies.
Declarations.
Abbreviations: Not applicable.
Ethics approval and consent to participate: Not applicable.
Consent for publication: This research paper is original of the author and no co-authors associated.
Availability of data and material: The availability of the data is open and free for all public.
Competing interests: There are no competing of interest.
Funding: Not Applicable.
Authors' contributions: Prof. Dr. Desouky Abd-El-Haleem conduct and wrote this paper by himself.
CRediT authorship contribution statement
Desouky Abd-El-Haleem: Conceptualization, Data curation, Formal analysis, Investigation, Methodology.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was done during my visit to the University of Tennessee's Center for Environmental Biotechnology in the United States. I want to express my gratitude to Prof. Dr. Gary Sayler, Aaron Nagel and James Fleming for their unwavering support during the practical part. The vist was a grant from the US-Egypt joint fund program.
References
- 1.Deng Z.H., Li N., Jiang H.L., et al. Pretreatment techniques and analytical methods for phenolic endocrine disrupting chemicals in food and environmental samples. Trends Anal Chem. 2019 doi: 10.1016/j.trac.2019.07.003. [DOI] [Google Scholar]
- 2.Abdel-El-Haleem D. Acinetobacter: environmental and biotechnological applications. Afr J Biotechnol. 2003;2(4):71–74. [Google Scholar]
- 3.Wu L., Ali D.C., Liu P., et al. Degradation of phenol via ortho-pathway by Kocuria sp. strain TIBETAN4 isolated from the soils around Qinghai Lake in China. PLoS One. 2018 doi: 10.1371/journal.pone.0199572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaki S. “Detection of meta- and ortho-cleavage dioxygenases in bacterial phenol-degraders.” J Appl Sci. Environ Manage. 2006 doi: 10.4314/jasem.v10i3.17323. [DOI] [Google Scholar]
- 5.Abd-El-Haleem D., Moawad H., Zaki E., et al. Molecular characterization of phenol-degrading bacteria isolated from different Egyptian ecosystems. Microb Ecol. 2003 doi: 10.1007/s00248-002-2003-2. [DOI] [PubMed] [Google Scholar]
- 6.Fleming J.T., Yao W.H., Sayler G.S. Optimization of differential display of prokaryotic mRNA: application to pure culture and soil microcosms. Appl Environ Microbiol. 1998 doi: 10.1128/AEM.64.10.3698-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Page R.D. TreeView: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12(4):357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 9.Steffen D., Schleif R. Overproducing araC protein with lambda-arabinose transducing phage. Mol Gen Genet. 1985 doi: 10.1007/BF00268671. [DOI] [PubMed] [Google Scholar]
- 10.Shen X., Liu S. Key enzymes of the protocatechuate branch of the beta-ketoadipate pathway for aromatic degradation in Corynebacterium glutamicum. Curr Microbiol. 2001 doi: 10.1007/BF03183617. [DOI] [PubMed] [Google Scholar]
- 11.Mojica F.J., Juez G., Rodríguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol Microbiol. 1993 doi: 10.1111/j.1365-2958.1993.tb01721.x. [DOI] [PubMed] [Google Scholar]
- 12.García-Fontana C, Narváez-Reinaldo JJ, Castillo F, González-López J, Luque I, Manzanera M. “A New Physiological Role for the DNA Molecule as a Protector against Drying Stress in Desiccation-Tolerant Microorganisms.” Front Microbiol. 2016;7:2066. doi: 10.3389/fmicb.2016.02066. [DOI] [PMC free article] [PubMed]
- 13.Johnson K., Charles I., Dougan G., et al. The role of a stress-response protein in Salmonella typhimurium virulence. Mol Microbiol. 1991;5(2):401–407. doi: 10.1111/j.1365-2958.1991.tb02122.x. [DOI] [PubMed] [Google Scholar]
- 14.Park S.Y., Fung P., Nishimura N., et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin S., Lee J. Determinants of DNA Bending in the DNA−Cyclic AMP Receptor Protein Complexes in Escherichia coli. Biochemistry. 2003 doi: 10.1021/bi027259. [DOI] [PubMed] [Google Scholar]
- 16.Wu Q., Shou J. Toward precise CRISPR DNA fragment editing and predictable 3D genome engineering. J Mol Cell Biol. 2021 doi: 10.1093/jmcb/mjaa060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egg M., Hckner M., Brandsttter A., et al. Structural and bioinformatic analysis of the Roman snail Cd-metallothionein gene uncovers molecular adaptation towards plasticity in coping with multifarious environmental stress. Mol Ecol. 2009 doi: 10.1111/j.1365-294X.2009.04191. [DOI] [PubMed] [Google Scholar]
- 18.Shigemi K., Kuniya A., Misao S., et al. A 900 bp genomic region from the mouse dystrophin promoter directs lacZ reporter expression only to the right heart of transgenic mice. Transgenic Res. 2001 doi: 10.1046/j.1440-169X. [DOI] [PubMed] [Google Scholar]
- 19.Mena M., Francisco C., Ines I., et al. A Role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol. 2002 doi: 10.1104/pp.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weimin B., Ling W., Françoise C., et al. DNA Binding Specificity of the CCAAT-binding Factor CBF/NF-Y. J Biol Chem. 1997 doi: 10.1074/jbc.272.42.26562. [DOI] [Google Scholar]
- 21.Viggor S., Jõesaar M., Soares-Castro P., et al. Microbial metabolic potential of phenol degradation in wastewater treatment plant of crude oil refinery: Analysis of metagenomes and characterization of isolates. Microorganisms. 2020;8(5):652. doi: 10.3390/microorganisms8050652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo X., Silva J.C. Properties of non-coding DNA and identification of putative is-regulatory elements in Theileria parva. BMC Genomics. 2008;9:582. doi: 10.1186/1471-2164-9-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Busby S., Ebright R.H. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79(5):743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 24.Egan S.M. Growing repertoire of AraC/XylS activators. J Bacteriol. 2002;184(20):5529–5532. doi: 10.1128/JB.184.20.5529-5532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alekshun M.N., Levy S.B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41(10):2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee S., Martin R.G., Rosner J.L., et al. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci U S a. 1998;95(18):10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teakel S.L., Fairman J.W., Muruthi M.M., et al. Structural characterization of aspartate-beta-semialdehyde dehydrogenase from Escherichia coli, a key enzyme in the aspartate family of amino acid biosynthesis. Sci Rep. 2022;12:14010. doi: 10.1038/s41598-022-17384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powlowski J., Shingler V. Genetics and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biotechnol Genet Eng Rev. 1994;11(1):237–256. doi: 10.1007/BF00696461. [DOI] [PubMed] [Google Scholar]
- 29.Mehrotra R., Jain V., Shekhar C., et al. “Genome-wide analysis of Arabidopsis thaliana reveals high frequency of AAAGN7CTTT motif”. Meta Gene. 2014 doi: 10.1016/j.mgene.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLauchlan J., Gaffney D., Whitton J.L., et al. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang D.T. Molecular Approaches to Protein Dimerization: Opportunities for Supramolecular Chemistry. Front Chem. 2022;10 doi: 10.3389/fchem.2022.829312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadfield A., Kryger G., Ouyang J., et al. Structure of aspartate-beta-semialdehyde dehydrogenase from Escherichia coli, a key enzyme in the aspartate family of amino acid biosynthesis. J Mol Biol. 1999;289(4):991–1002. doi: 10.1006/jmbi.1999.2828. [DOI] [PubMed] [Google Scholar]
- 33.Gu Q, Chen M, Zhang J, et al. “Genomic Analysis and Stability Evaluation of the Phenol-Degrading Bacterium Acinetobacter sp. DW-1 During Water Treatment,” Front Microbiol. 2021;12:687511. doi: 10.3389/fmicb.2021.687511. [DOI] [PMC free article] [PubMed]
- 34.Joshi C.P. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987;15(16):6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karigar C.S., Rao S.S. Role of microbial enzymes in the bioremediation of pollutants: a review. Enzyme Res. 2011;2011 doi: 10.4061/2011/805187. [DOI] [PMC free article] [PubMed] [Google Scholar]