Abstract
Background
Choline oxidase, a flavoprotein, is an enzyme that catalyzes the reaction which converts choline into glycine betaine. Choline oxidase started its journey way back in 1933. However, the impact of the high temperature on its structure has not been explored despite the long history and availability of its crystal structure. Both choline oxidase and its product, glycine betaine, have enormous applications spanning across multiple industries. Understanding how the 3D structure of the enzyme will change with the temperature change can open new ways to make it more stable and useful for industry.
Process
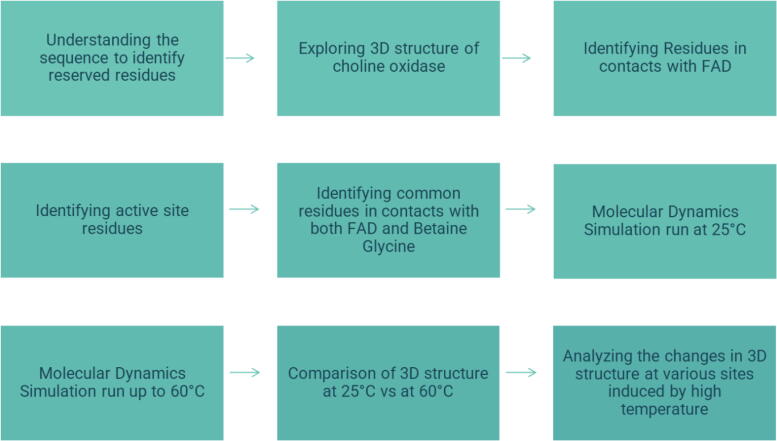
This research paper presents the in-silico study and analysis of the structural changes of A. globiformis choline oxidase at temperatures from 25 °C to 60 °C. A step-wise process is depicted in Fig. 1.
Results
Multiple sequence alignment (MSA) of 11 choline oxidase sequences from different bacteria vs Arthrobacter globiformis choline oxidase showed that active site residues are highly conserved.
The available crystal structure of A. globiformis choline oxidase with cofactor Flavin Adenine Dinucleotide (FAD) in the dimeric state (PDB ID: 4MJW)1 was considered for molecular dynamics simulations. A simulated annealing option was used to gradually increase the temperature of the system from 25 °C to 60 °C. Analysis of the conserved residues, as well as residues involved in Flavin Adenine Dinucleotide (FAD) binding, substrate binding, substate gating, and dimer formationwas done. At high temperatures, the formation of the inter-chain salt bridge between Arg50 and Glu63 was a significant observation near the active site of choline oxidase.
Conclusion
Molecular dynamics studies suggest that an increase in temperature has a significant impact on the extended Flavin Adenine Dinucleotide (FAD) binding region. These changes interfere with the entry of substrate to the active site of the enzyme and make the enzyme inactive.
Keywords: Choline oxidase, Glycine betaine, Structural changes, Molecular dynamics simulation, In-silico
1. Introduction
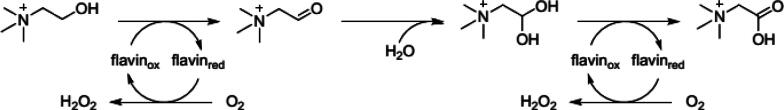
Choline oxidase, an oxidoreductase enzyme, catalyzes the oxidation of choline to glycine betaine, which serves as a ubiquitous osmoprotectant in bacteria, plants, and animals.1 The conversion of choline to glycine betaine oxidase proceeds via two hydride-transfer reactions, in which two flavin reductions are the rate-limiting steps2 as shown in Fig. 2.3
Fig. 1.
Process flow of the in-silico study of the structural changes for A. globiformis choline oxidase at temperatures from 25 °C to 60 °C.
Fig. 2.
The two-step process of conversion of choline to glycine betaine (3).
Choline oxidase plays a key role in the development of transgenic plants against various stresses like salt, drought, salinity, and low temperature.4 Choline oxidase is a very useful enzyme and has wide applicability across fields of biotechnology, chemical warfare, diagnosis, and medical.5
Choline oxidase has a challenge with its thermostability, it loses its catalytic activity rapidly above 37 °C.5 A. globiformis choline oxidase has been reported to work at 37 °C6 and its activity at 50 °C is negligible (SIGMA ALDRICH, Catalog number: SAE0044).
Due to industrial applications of choline oxidase, there is a strong interest in modifying the choline oxidase. In 1992, a USA patent was granted for the production of novel thermostable choline oxidase by actinomycetes.7 Turner and Team8 worked on choline oxidase from Arthrobector cholorphenoculicus. They used structure-guided mutagenesis to improve the enzyme’s substate scope.9 They identified the active sites and the access channels and targeted them for the mutations.
Choline oxidase belongs to the glucose-methanol-choline (GMC) enzyme oxidoreductases superfamily,9 which contains many FAD-dependent enzymes that oxidize various alcohols and have a similar three-dimensional fold.10 Available crystal structure and biochemical studies suggest that choline oxidase exists in a dimeric state.11 Arthrobacter globiformis Choline oxidase is a dimer with 546 amino acids with chains A and B.4 The substrate binding site is occluded from the bulk solvent for choline oxidase. In homotetrameric pyranose-2-oxidase12, 13 and monomeric cholesterol oxidase,14, 15 it has been proposed that entry of the substrate is controlled by a loop and lid mechanism. There is a collection of nonpolar residues acting as a gate in the central portion of the dimer. The gate acts as a way-in for the substrate as well as a way-out for the product from the active site of the enzyme.16
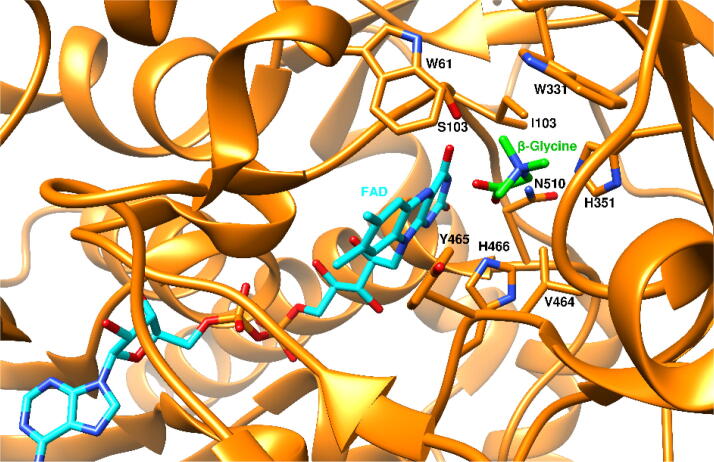
The mechanism of action of Arthrobacter globiformis choline oxidase at the structural level has been studied.17 It showed the significance of active site residues S101,18 E312,17 H35119 V464,20, 21 H466,22 and N510.23 In a study by Xin et al., the corresponding loop region in the choline oxidase remained static over 60 ns molecular dynamics simulation.24 However, residues M62, K65, V355, F357, and M359 showed rapid dynamic motion leading to the opening and closing of the entrance of the substrate entry tunnel.24 The entry of substrate into the active site of choline oxidase is delimited by a hydrophobic cluster of residues.24
Owing to the vast applicability of choline oxidase in the industry, this research work has been carried out to understand the changes in the 3D structure of A. globiformis choline oxidase at various temperatures.
2. Materials and methods
2.1. Multiple Sequence Alignment (MSA)
For understanding the conserved sites, the 12 different protein sequences of choline oxidase from different bacteria were used from the National Center for Biotechnology Information (NCBI) database. While selecting the protein sequences, diversity was ensured and the sequences with 30 to 90 % identity were picked. 12 protein sequence of choline oxidase belonging to Arthrobacter globiformis (AAP68832.1), Lachnellula hyalina (XP_031008190.1), Brevibacterium aurantiacum (SMX63892.1), Streptomyces malaysiensis (PNG98051.1), Amycolatopsis japonica (AIG78012.1), Trichoderma simmonsii (QYT05116.1), Burkholderia vietnamiensis (TCT33723.1), Streptomyces violascens (GHI40600.1), Rhodococcus opacus (ANS30057.1), Nocardia brasiliensis (AFU01643.1), Saccharopolyspora hirsuta (KAA5830183.1) and Streptomyces fradiae (OSY51643.1) were selected for Multiple sequence Alignment (MSA).
Jalview25 was used to align all sequences and to visualize residue conservation at different identity thresholds. The web version of the MUSCLE tool (https://www.ebi.ac.uk/Tools/msa/muscle/)26 was used to generate a percentage identity matrix of aligned sequences.
2.2. Molecular Dynamics Simulation (MSD)
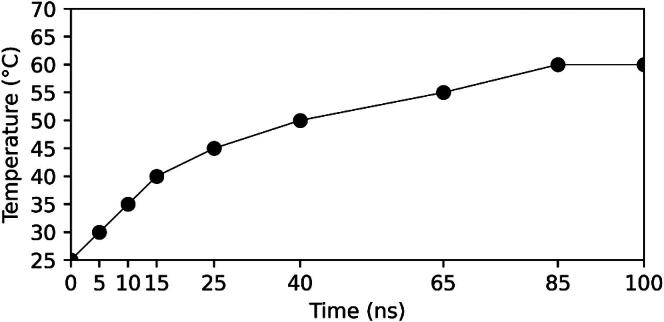
A crystal structure of A. globiformis choline oxidase with cofactor FAD in a dimeric state (PDB ID: 4MJW)1 was considered for molecular dynamics simulations at different temperatures (see Table 1 for more details). All molecular dynamics simulations were performed using GROMACS 2020.9, 10 CHARMM27 forcefield was used to generate the topology of choline oxidase.17, 18 Forcefield parameters of FAD were calculated from the SWISS PARAM server.19 The complex of choline oxidase with FAD was enclosed in a triclinic box. Solvation was carried out using the TIP3P water model.20, 21 The solvated system was neutralized by adding Na+ and Cl- ions at 0.15 mM. The neutralized system was minimized by the steepest descent energy method. After minimization, systems were subjected to constant volume (NVT) and constant pressure of 1 bar (NPT) equilibrations for 2 ns and 3 ns respectively. By setting the random seed values in the NVT.mdp file, two replicas were generated at each temperature. During equilibration, heavy atoms of the complexes were kept fixed by applying position restraints. Pressure and temperature were controlled by the Parrinello-Rahman22 and V-rescale23 methods, respectively. After equilibration, two different approaches were taken for the production runs. In the first approach, production runs were carried out at 25 °C. In the second approach, the simulated annealing option available in the GROMACS was used in the production simulation that allowed the temperature of the system to gradually increase from 25 °C to 60 °C (298 K to 333 K) (Fig. 3). After production runs, trajectories were analysed and visualized using Visual Molecular Dynamics (VMD)2 and UCSF Chimera.27
Table 1.
Various residues of A. globiformis choline oxidase.
| Residues in contact with FAD |
Residues in contact with Betaine Glycine (Active site residues) |
Residues in contact with both FAD and Betaine Glycine |
|---|---|---|
| G20, G22, S23, A24, E44, A45, W61, L65, W71, A88, R89, A90, K91, V92, G95, C96, S97, H99, N100, S101, C102, I103, R231, A232, S268, T269, G270, D273, L277, Y465, H466, D499, A500, N510, P511, N512, V515 | W61, S101, I103, W331, H351, V464, Y465, H466, N510 | W61, S101, I103, Y465, H466, N510 |
Fig. 3.
Temperature setting for a simulated annealing.
2.3. Essential Dynamics (ED)
Biologically relevant low-frequency motions in the proteins can be studied via essential dynamics.28 Principal Component Analysis (PCA) is a method of dimensionality reduction that is widely used to study the low-frequency motions in proteins.29 In PCA, a covariance matrix is constructed from the protein structures aligned to a reference structure, and diagonalization of the covariance matrix yields a set of eigenvectors and eigenvalues. Mostly, the first two principal components explain the majority of structural variation. ProDy package30 available in VMD was used to carry out PCA of the trajectories. Cα atoms of dimeric choline oxidase were used for PCA.
3. Results
3.1. Variation in the conserved residues
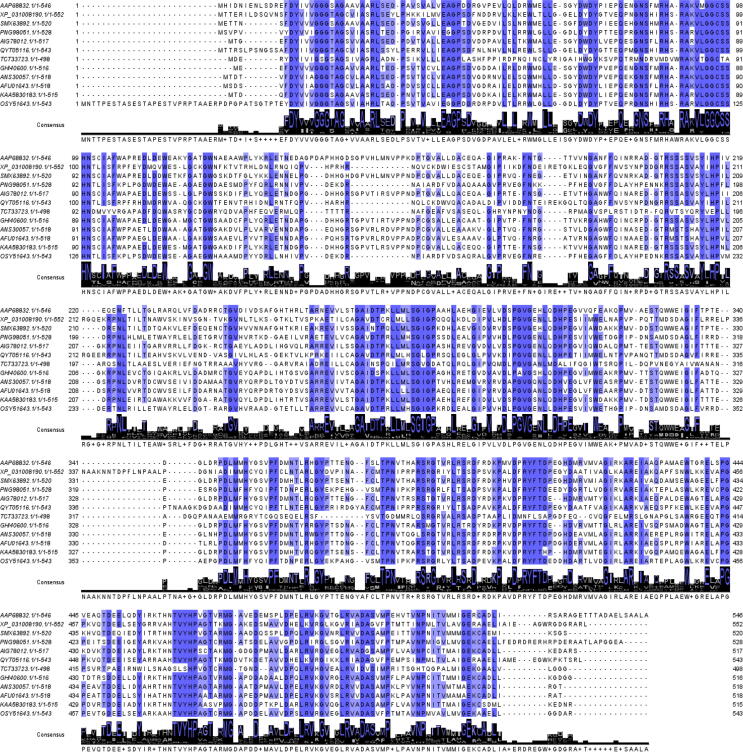
Multiple sequence alignment (MSA) of 12 different choline oxidase sequences (Table S1 and Fig. 4) showed that there are many highly (80 % and 100 %) and moderately conserved residues (60 %) in the sequence of A. globiformis Choline oxidase sequence. (Table S2 and Fig. 4).
Fig. 4.
Multiple sequence alignment (MSA) of 12 choline oxidase sequences from different bacteria. Arthrobacter globiformis choline oxidase was considered as a reference sequence. Residues at 60 % identity and above are highlighted in blue background. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
With the detailed analysis of the available crystal structures of A. globiformis choline oxidase (PDB ID: 4MJW) (1), along with the conserved residues, residues involved in FAD binding, substrate binding, substate gating, and dimer formation were identified. Only 14, 24, and 28 residues are conserved at 100 %, 80 %, and 60 % identity thresholds (Table S2). 36 residues make contact with FAD (Table 1).
Analysis of Multiple Sequence Alignment (MSA) revealed that out of 10 residues that interact with betaine glycine (Table 1 and Fig. 4), only H466 is conserved in all sequences (Table S2). Interestingly S101 which forms an H-bond with FAD was not conserved even at a 60 % identity threshold (Table S2). In the crystal structure of S101A choline oxidase, FAD has identical conformation when compared to the wild-type choline oxidase suggesting that S101 is not a critical residue.31
The 3D structure of A. globiformis, substrate binding pocket is small and consists of polar and non-polar residues (Fig. 5). During catalysis choline converts into glycine betaine, leaving the trimethyl azanium group unaltered. It was assumed that choline and glycine betaine would have almost identical interactions with the residues of the active site. According to Smitherman et al., mutation of H466 results in the inactivation of choline oxidase.32 E312, H351, and N510 appear to be critical (conserved at 80 and 60 % identity threshold). In the crystal structure, E312 did not show close contact with glycine betaine. It has been reported that the positive charge on choline is stabilized by the negatively charged side chain of E312. Mutational study has revealed that H351 has a role in substrate binding as well as in the stabilization of the transition state during hydride transfer to flavin.32 N510 has a role in both reductive and oxidative half-reactions.
Fig. 5.
Residues of A. globiformis choline oxidase show interaction with beta-glycine (product) in green color. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Surprisingly, residues responsible for substrate gating and dimer formation showed overall poor conservation. According to Xin et al., side chains of M62, L65, V355, F357, and M359 form hydrophobic clusters and gate the entry of substrate into the active site.24 Only L65 and F357 showed conservation at 60 % and 80 % identity threshold (Table S2).
3.2. High temperatures induce significant changes in the 3D structure of A. globiformis choline oxidase
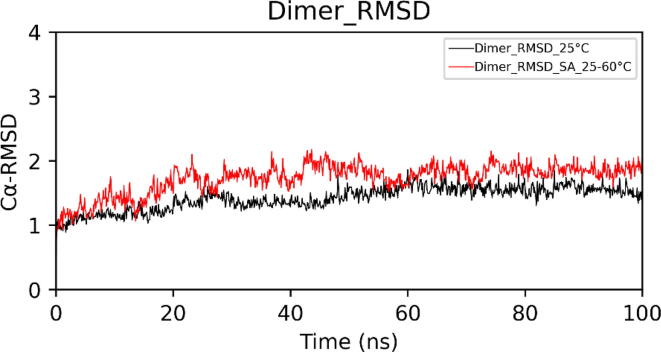
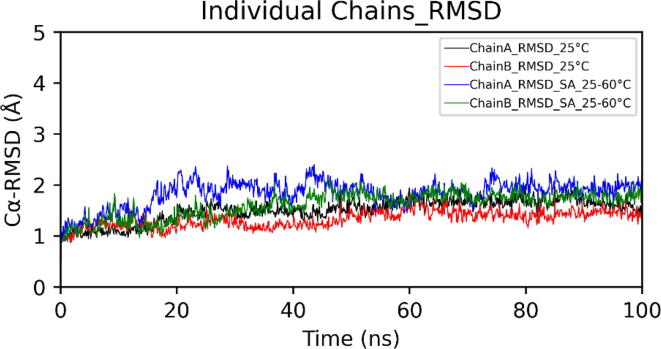
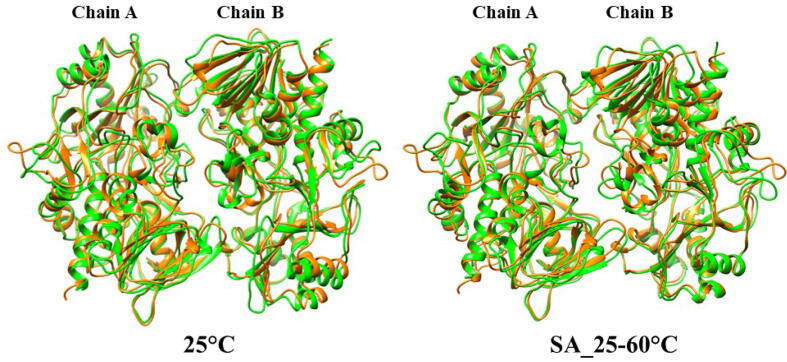
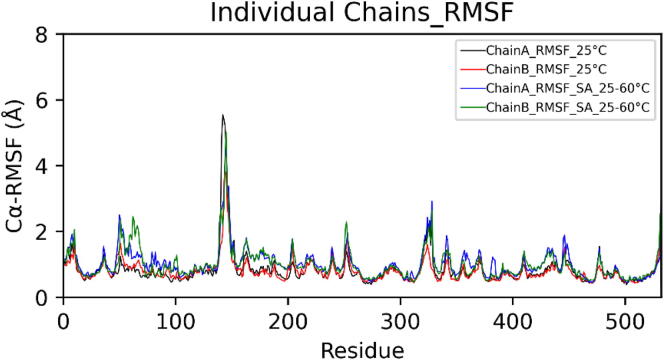
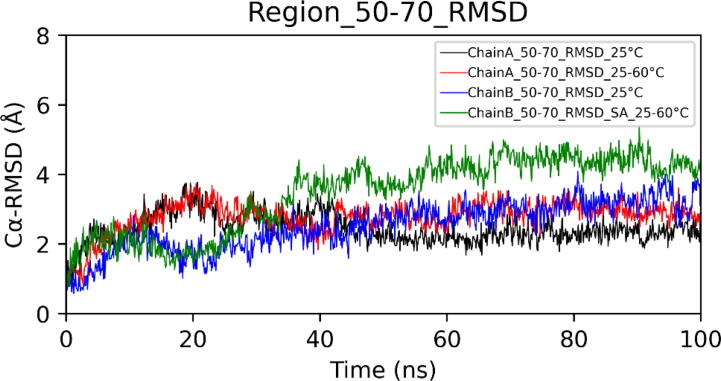
In the case of simulated annealing, the Cα-RMSD (Root Mean Square Deviation) plot of dimeric choline oxidase showed significant structural deviations, suggesting the impact of high temperatures on its 3D structure (Fig. 6). Cα-RMSD of individual chains (Fig. 7) revealed that both chains undergo significant structural changes at high temperatures. The majority of structural deviations were visible in the extended FAD binding region (50–70) and other loop regions (140–157 and 354–371). However, the overall fold remains intact up to 60 °C (Fig. 7). Almost all secondary structure elements were intact at 60 °C. Cα-RMSF plot showed higher fluctuations of many regions of choline oxidase at high temperatures (Fig. 8). Particularly, the extended FAD binding region (50–70) and region 140–157 exhibited higher fluctuations in both chains A and B. We have discussed about residues of extended FAD binding region in the later section (see Fig. 9, Fig. 10, Fig. 11, Fig. 12.
Fig. 6.
Cα-RMSDs of dimeric choline oxidase at 25 °C and in the case of Simulated Annealing (SA_25–60 °C). (A) Run1 and (B) Run2.
Fig. 7.
Cα-RMSDs of chain A and chain B of dimeric choline oxidase at 25 °C and in the case of Simulated Annealing (SA_25–60 °C). (A) Run1 and (B) Run2.
Fig. 8.
Structural comparison of dimeric choline oxidase at 25 °C and in the case of Simulated Annealing (SA_25–60 °C). Orange (structure at 0 ns) and green (structure at 100 ns). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 9.
Cα-RMSDs of chain A and chain B at 25 °C and in the simulated annealing.
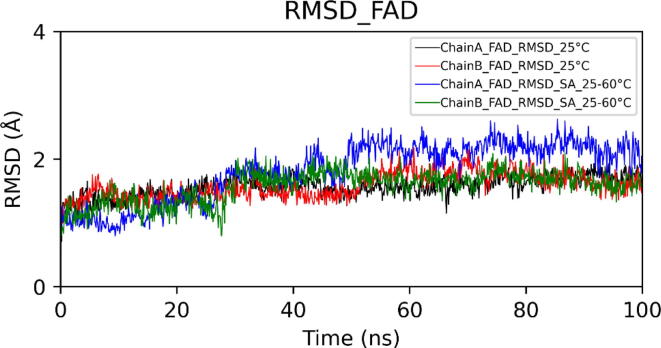
Fig. 10.
Root Mean Square Deviation (RMSD) of FAD in chains A and B.
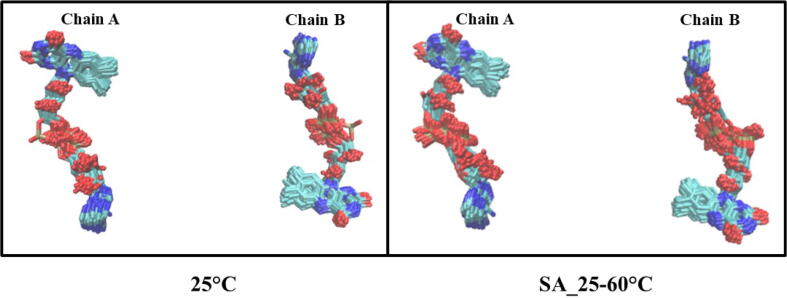
Fig. 11.
Ensemble of FAD conformations at 25 °C and in the simulated annealing.
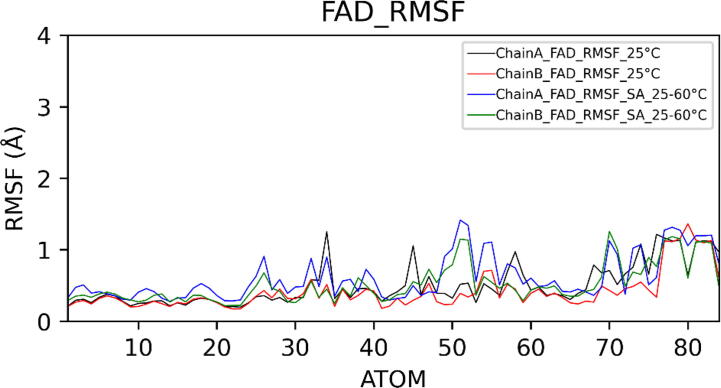
Fig. 12.
Root Mean Square Fluctuations (RMSF) of FAD.
3.3. FAD and catalytic residues remain organized at high temperature
In choline oxidase, FAD is covalently linked to His99. RMSD plot of FAD showed that it exhibited more conformational deviations at high temperatures showing average RMSD (for both chains) of 1.28 ± 0.15 Å at 25 °C and 1.62 ± 0.30 Å in the case of simulated annealing. FAD deviated more in chain A than in chain B. Compared to the isoalloxazine ring, the adenine ring of FAD exhibited very low fluctuation. We also investigated the relative positioning of catalytic residues concerning the isoalloxazine ring of FAD. Orientations of side chains of all catalytic residues except His496 remained unaltered. His496 showed flipping of the side chain at 25 °C as well as at high temperatures.
3.4. High temperatures induce significant conformational change in extended FAD binding region
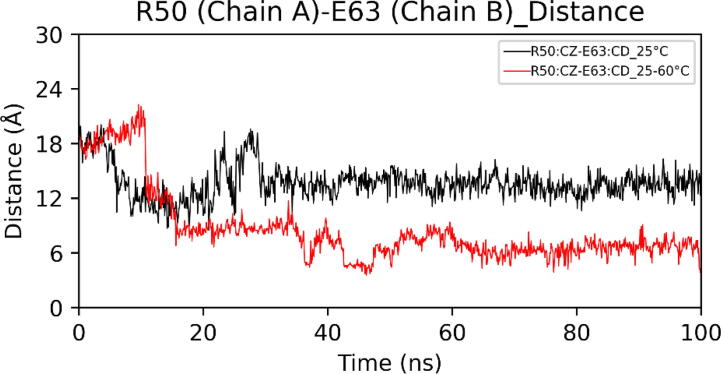
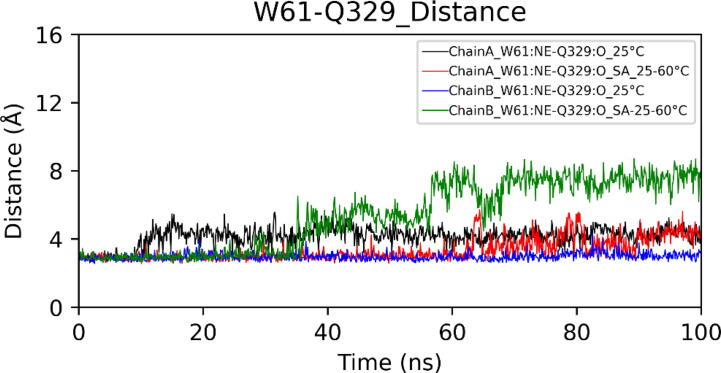
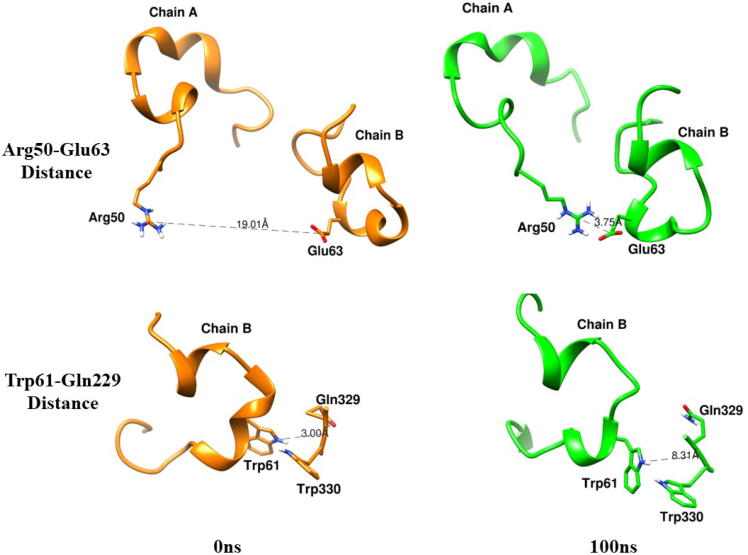
Extended FAD binding region is important as it harbors residues that have been reported to control the entry of substrate in the active site.24 At high temperatures, the extended FAD binding region (50–70) in chain B showed a significant high deviation from the initial conformation (Fig. 13). Principal Component Analysis (PCA) also showed increased motion of the above region at high temperatures (Figure S1). In the crystal structure, the distance between Arg50 (Chain A) and Glu63 (Chain B) is 19.36 Å. Molecular dynamics (MD) simulation at 25 °C showed that the average distance is 13.68 Å. However, in the case of simulated annealing, the average distance decreases to 8.57 Å (Fig. 14). We also observed an increase in distance between Trp61 and Gln329 (Fig. 15). Conformational change in the extended FAD binding region was due to the formation of a salt bridge between side chains of Arg50 (Chain A) and Glu63 (Chain B) (Fig. 16). In the distance plot, before 40 °C, side chains of Arg50 and Glu63 remained far apart but after 40 °C, a sudden decrease in the distance between them led to the increased chances of salt bridge formation. In the last frame of the simulated annealing trajectory, we observed that the Arg50-Glu63 distance decreased to 3.75 Å. Trp61 is situated in the extended FAD binding region and its hydrophobic sidechain is in the vicinity of the isoalloxazine ring of FAD. We studied the change in the orientation of Trp61 by measuring the distance between the side chain atom of Trp61 and the main chain atom of Gln329. We observed that a decrease in Arg50-Glu63 distance leads to an increase in Trp61-Glu329 distance. These changes might interfere with the entry of the substrate inside the active site.
Fig. 13.
(A) Cα RMSD of region 50–70.
Fig. 14.
Distances between R50 (chain A) and E63 (chain A) during MD simulations.
Fig. 15.
Distances between W61 (chain A) and Q329 (chain B) during Molecular dynamics (MD) simulations.
Fig. 16.
Distances between Arg50 and Glu63 (upper) and Trp61-Gln329 (lower) in initial (orange) and final (green) frames. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Choline oxidase is an enzyme of high industrial importance. The optimum pH and temperature of A. globiformis is 7 and 37 °C respectively. A. globiformis Choline oxidase loses its activity rapidly above 37 °C. Choline oxidase from Alcaligenes species becomes inactive in the pH ranges 3–6 and 9–11 due to irreversible changes in structure. Although it has crystal structures of A. globiformis choline oxidase in protein data banks are available in a homodimeric state (PDB ID: 2JBV and 4MJW), there is very limited information available for the effect of high temperatures on its structure. The present study explores the conformational changes in A. globiformis choline oxidase at high temperatures with in-silico approaches.
In the crystal structures of choline oxidase, FAD is covalently bound to H99. In the present study, choline oxidase contains non-covalently bound FAD. In choline oxidase with H99N mutation, FAD is no longer covalently linked but experimental data suggested that mutant choline oxidase is mechanistically similar to the wild-type choline oxidase.33 Covalent binding of FAD ensures its optimal positioning within the active site for catalysis. Hence, consideration of non-covalently linked FAD may not influence our understanding. FAD and important catalytic residues maintained their relative orientations at 60 °C indicating intact active site. However, significant conformational change in the extended FAD binding region may interfere with substrate entry in the active site. In the dimeric structures of A. globiformis choline oxidase, loop 250–255 covers the substrate entry gate and a narrow tunnel is visible in the betaine glycine-bound choline oxidase. Loop 250–255 is involved in dimerization and the orientation of F253 of the loop determines the opening and closing of the tunnel leading to the active site.34 The side chain of F253 assumed different orientations during MD simulations at 25 °C as well as at high temperatures. However, we did not observe the closing of the tunnel by F253. According to Xin et al., a cluster of hydrophobic residues guards the entrance to the active site of choline oxidase and determines the opening and closing of the gate.24 They proposed that opening and closing of the gate could be a rate-limiting step in the catalytic process. Met62 is a part of the hydrophobic cluster and it acts as a gate of the tunnel leading to the active site. Due to conformational change in the extended FAD binding region, side chains of Trp61 and Met62 showed significant deviation from the initial orientations. In the crystal structure, the hydrophobic side chain of Trp61 faces towards the trimethyl group of glycine betaine, suggesting its role in providing a hydrophobic environment to the substrate choline. Another residue in the extended FAD binding region is Glu63, which has been reported to attract and guide the positively charged substrate to the active site. In the case of simulated annealing, we observed that the side chain of Glu63 deviates significantly from the original orientation and no longer lines the substrate entry channel. According to Daniel and Danson,35 at a given temperature, intermediate inactive conformation (not denatured) exists in equilibrium with the active conformation of the enzyme. Local changes in the active site of the enzyme may populate the inactive conformation and further increase in temperature may lead to the irreversible denatured state. The significant changes near the active site of choline oxidase were observed at high temperatures.
5. Conclusion
In the present study, 12 bacterial choline oxidase sequences were compared to find out the conserved residues, and molecular dynamics studies were carried out at different temperatures to understand the conformational changes in the structure of A. globiformis Choline oxidase. At high temperatures, the formation of the inter-chain salt bridge between Arg50 and Glu63 led to significant changes near the active site of choline oxidase. The above changes may interfere with the entry of substrate inside the active site and may lead to the inactivation of choline oxidase.
6. Future prospect
There has been a lot of work done on improving the thermostability of many industrial useful enzymes.36 Glucose Oxidase is one such enzyme, used widely in food and wine industries to contain microbial contamination during the fermentation process.37 Holland and the team induced various site-directed mutagenesis.38 The variant showed improved catalytic function and stability.38 Further work on the identification of the residues suitable for the site-directed mutagenesis and introducing the mutations at the targeted sites can be helpful in the design of thermostable choline oxidase.
List of Tools used.
| Tools | Description of the Tool | Use of the tool |
|---|---|---|
| CHARMM27 forcefield | CHARMM (Chemistry at HARvard Macromolecular Mechanics) is a both a set of force fields and a software package for molecular dynamics simulations and analysis. | To generate topology of choline oxidase |
| GROMACS 2020 | GROningen MAchine for Chemical Simulations (GROMACS) is mainly used to simulate and study the properties of biomolecules such as proteins, lipids, and nucleic acids. | For molecular dynamics simulation |
| Jalview | Jalview is a piece of bioinformatics software that is used to look at and edit multiple sequence alignments. | To visualize residues conservation at different identity threshold |
| MUSCLE | MUltiple Sequence Comparison by Log-Expectation (MUSCLE) is computer software for multiple sequence alignment of protein and nucleotide sequences. | For multiple sequence alignment |
| PCA | Principal component analysis (PCA) is a method of dimensionality reduction which is widely used to study the low frequency motions in the proteins. | To study the low frequency motions in the proteins |
| ProDy | Protein Dynamics and Sequence Analysis(ProDy) is a open-source Python package for protein structure, dynamics, and sequence analysis. | For PCA of the trajectories |
| SWISS PARAM | SwissParam is a fully automatic server that provides topology and parameters for small organic molecules, compatible with the CHARMM all atoms force field, for use with CHARMM and GROMACS | Forcefield parameters of FAD calculation |
| TIP3P water model | The TIP3P water model as implemented in CHARMM (MacKerell) specifies a 3-site rigid water molecule with charges and Lennard-Jones parameters assigned to each of the 3 atoms. | For solvation |
| UCSF Chimera | UCSF Chimera is an extensible program for interactive visualization and analysis of molecular structures and related data | To visualize and analyse trajectories |
| VMD | Visual Molecular Dynamics (VMD) is a molecular modelling and visualization computer program. | To analyse trajectories |
| Web version of MUSCLE | MUltiple Sequence Comparison by Log-Expectation (MUSCLE)is a tool that performs multiple sequence alignments of nucleotide or amino acid sequences. https://www.ebi.ac.uk/Tools/msa/muscle/ | To generate percentage identity matrix of aligned sequences |
CRediT authorship contribution statement
Sonia Kaushik: Writing – original draft. Rashmi Rameshwari: Conceptualization, Formal analysis, Supervision, Writing – review & editing. Shilpa S. Chapadgaonkar: Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgeb.2023.100348.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Bremer E., Krämer R. Responses of Microorganisms to Osmotic Stress. Annu Rev Microbiol. 2019;73:313–334. doi: 10.1146/annurev-micro-020518-115504. [DOI] [PubMed] [Google Scholar]
- 2.Fan F., Gadda G. On the catalytic mechanism of choline oxidase. J Am Chem Soc. 2005;127(7):2067–2074. doi: 10.1021/ja044541q. [DOI] [PubMed] [Google Scholar]
- 3.Wahart A.J., Staniland J., Miller G.J., Cosgrove S.C. Oxidase enzymes as sustainable oxidation catalysts. R Soc Open Sci. 2022 Jan 12;9(1) doi: 10.1098/rsos.211572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaushik S, Rameshwari R, Chapadgaonkar SS. Choline oxidase: An enzyme of immense industrial potential. AsPac J. Mol. Biol. Biotechnol. 2022 Vol. 30 (1) : 37-50. 10.35118/apjmbb.2022.030.1.04. [DOI]
- 5.Kaushik S., Rameshwari R., Chapadgaonkar S.S. Potential rational methods for improving thermostability of choline oxidase. J Basic Sci. 2023;23(1):928–950. doi: 10.37896/JBSV23.1/1723. [DOI] [Google Scholar]
- 6.Ikuta S., Imamura S., Misaki H., Horiuti Y. Purification and characterization of choline oxidase from Arthrobacter globiformis. J Biochem. 1977;82(6):1741–1749. doi: 10.1093/oxfordjournals.jbchem.a131872. [DOI] [PubMed] [Google Scholar]
- 7.Furuoya I., Suzuki T., Takahashi T. Novel choline oxidase and method for producing the same. Google Patents. 1992 [Google Scholar]
- 8.Heath R.S., Birmingham W.R., Thompson M.P., Taglieber A., Daviet L., Turner N.J. An engineered alcohol oxidase for the oxidation of primary alcohols. Chembiochem. 2019;20(2):276–281. doi: 10.1002/cbic.201800556. [DOI] [PubMed] [Google Scholar]
- 9.Cavener D.R. GMC oxidoreductases: A newly defined family of homologous proteins with diverse catalytic activities. J Mol Biol. 1992;223(3):811–814. doi: 10.1016/0022-2836(92)90992-S. [DOI] [PubMed] [Google Scholar]
- 10.Salvi F., Gadda G. Human choline dehydrogenase: Medical promises and biochemical challenges. Arch Biochem Biophys. 2013;537(2):243–252. doi: 10.1016/j.abb.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan F., Ghanem M., Gadda G. Cloning, sequence analysis, and purification of choline oxidase from Arthrobacter globiformis: A bacterial enzyme involved in osmotic stress tolerance. Arch Biochem Biophys. 2004;421:149–158. doi: 10.1016/j.abb.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Bannwarth M., Bastian S., Heckmann-Pohl D., Giffhorn F., Schulz G.E. Crystal structure of pyranose 2-oxidase from the white-rot fungus Peniophora sp. Biochemistry. 2004;43(37):11683–11690. doi: 10.1021/bi048609q. [DOI] [PubMed] [Google Scholar]
- 13.Kujawa M., Ebner H., Leitner C., et al. Structural basis for substrate binding and regioselective oxidation of monosaccharides at C3 by pyranose 2-oxidase. J Biol Chem. 2006;281(46):35104–35115. doi: 10.1074/jbc.M604718200. [DOI] [PubMed] [Google Scholar]
- 14.Yue Q.K., Kass I.J., Sampson N.S., Vrielink A. Crystal structure determination of cholesterol oxidase from Streptomyces and structural characterization of key active site mutants. Biochemistry. 1999;38(14):4277–4286. doi: 10.1021/bi982497j. [DOI] [PubMed] [Google Scholar]
- 15.Chen X., Wolfgang D.E., Sampson N.S. Use of the parallax-quench method to determine the position of the active-site loop of cholesterol oxidase in lipid bilayers. Biochemistry. 2000;39(44):13383–13389. doi: 10.1021/bi001407j. [DOI] [PubMed] [Google Scholar]
- 16.Gadda G. Choline oxidases. The Enzymes. 2020 Jan;1(47):137–166. doi: 10.1016/bs.enz.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Quaye O., Lountos G.T., Fan F., Orville A.M., Gadda G. Role of Glu312 in binding and positioning of the substrate for the hydride transfer reaction in choline oxidase. Biochemistry. 2008;47(1):243–256. doi: 10.1021/bi7017943. [DOI] [PubMed] [Google Scholar]
- 18.Gadda G., Yuan H. Substitutions of S101 decrease proton and hydride transfers in the oxidation of betaine aldehyde by choline oxidase. Arch Biochem Biophys. 2017;634:76–82. doi: 10.1016/j.abb.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Rungsrisuriyachai K., Gadda G. On the role of histidine 351 in the reaction of alcohol oxidation catalyzed by choline oxidase. Biochemistry. 2008;47(26):6762–6769. doi: 10.1021/bi800650w. [DOI] [PubMed] [Google Scholar]
- 20.Finnegan S., Agniswamy J., Weber I.T., Gadda G. Role of Valine 464 in the Flavin Oxidation Reaction Catalyzed by Choline Oxidase. Biochemistry. 2010;49(13):2952–2961. doi: 10.1021/bi902048c. [DOI] [PubMed] [Google Scholar]
- 21.Gadda G. Oxygen Activation in Flavoprotein Oxidases: The Importance of Being Positive. Biochemistry. 2012;51:2662–2669. doi: 10.1021/bi300227d. [DOI] [PubMed] [Google Scholar]
- 22.Ghanem M., Gadda G. On the Catalytic Role of the Conserved Active Site Residue His 466 of Choline Oxidase †. Biochemistry. 2005;44:893–904. doi: 10.1021/bi048056j. [DOI] [PubMed] [Google Scholar]
- 23.Rungsrisuriyachai K., Gadda G. Role of asparagine 510 in the relative timing of substrate bond cleavages in the reaction catalyzed by choline oxidase. Biochemistry. 2010;49(11):2483–2490. doi: 10.1021/bi901796a. [DOI] [PubMed] [Google Scholar]
- 24.Xin Y., Gadda G., Hamelberg D. The Cluster of Hydrophobic Residues Controls the Entrance to the Active Site of Choline Oxidase. Biochemistry. 2009;48(40):9599–9605. doi: 10.1021/bi901295a. [DOI] [PubMed] [Google Scholar]
- 25.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan F., Gadda G. Oxygen- and temperature-dependent kinetic isotope effects in choline oxidase: correlating reversible hydride transfer with environmentally enhanced tunneling. J Am Chem Soc. 2005;127(50):17954–17961. doi: 10.1021/ja0560377. [DOI] [PubMed] [Google Scholar]
- 28.Amadei A., Linssen A.B., Berendsen H.J. Essential dynamics of proteins. Proteins. 1993;17(4):412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 29.David C., Jacobs D. Principal Component Analysis: A Method for Determining the Essential Dynamics of Proteins. Methods in Molecular Biology (clifton, NJ). 2014;1084:193–226. doi: 10.1007/978-1-62703-658-0_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakan A., Meireles L.M., Bahar I. ProDy: Protein Dynamics Inferred from Theory and Experiments. Bioinformatics. 2011;27(11):1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finnegan S., Yuan H., Wang Y.F., Orville A.M., Weber I.T., Gadda G. Structural and kinetic studies on the Ser101Ala variant of choline oxidase: catalysis by compromise. Arch Biochem Biophys. 2010;501(2):207–213. doi: 10.1016/j.abb.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 32.Smitherman C., Rungsrisuriyachai K., Germann M.W., Gadda G. Identification of the catalytic base for alcohol activation in choline oxidase. Biochemistry. 2015;54(2):413–421. doi: 10.1021/bi500982y. [DOI] [PubMed] [Google Scholar]
- 33.Quaye O., Cowins S., Gadda G. Contribution of flavin covalent linkage with histidine 99 to the reaction catalyzed by choline oxidase. J Biol Chem. 2009;284(25):16990–16997. doi: 10.1074/jbc.M109.003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salvi F., Wang Y.F., Weber I.T., Gadda G. Structure of choline oxidase in complex with the reaction product glycine betaine. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 2):405–413. doi: 10.1107/S1399004713029283. [DOI] [PubMed] [Google Scholar]
- 35.Daniel R.M., Danson M.J. Temperature and the catalytic activity of enzymes: a fresh understanding. FEBS Lett. 2013;587(17):2738–2743. doi: 10.1016/j.febslet.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 36.Rigoldi F., Donini S., Redaelli A., Parisini E., Gautieri A. Review: Engineering of thermostable enzymes for industrial applications. APL Bioeng. 2018;2(1) doi: 10.1063/1.4997367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Röcker J., Schmitt M., Pasch L., Ebert K., Grossmann M. The use of glucose oxidase and catalase for the enzymatic reduction of the potential ethanol content in wine. Food Chem. 2016;210:660–670. doi: 10.1016/j.foodchem.2016.04.093. [DOI] [PubMed] [Google Scholar]
- 38.Holland J.T., Harper J.C., Dolan P.L., et al. Rational redesign of glucose oxidase for improved catalytic function and stability. PLoS One. 2012;7(6):e37924. doi: 10.1371/journal.pone.0037924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.