Abstract
The tropical areas in southern and south-western Yunnan are rich in fungal diversity. Additionally, the diversity of seed flora in Yunnan Province is higher than in other regions in China and the abundant endemic species of woody plants provide favourable substrates for fungi. Rubber plantations in Yunnan Province are distributed over a large area, especially in Xishuangbanna. During a survey of rubber-associated fungi in Yunnan Province, China, dead rubber branches with fungal fruiting bodies were collected. Morphological characteristics and multigene phylogenetic analyses (ITS, LSU, SSU, rpb2 and tef1-α) revealed four distinct new species, described herein as Melomastiapuerensis, Nigrogranalincangensis, Pseudochaetosphaeronemalincangensis and Pseudochaetosphaeronemaxishuangbannaensis. Detailed descriptions, illustrations and phylogenetic trees are provided to show the taxonomic placements of these new species.
Key words: Dothideomycetes, four new species, multigene phylogeny, Pará rubber, saprobic fungi, taxonomy
Introduction
Heveabrasiliensis (Pará rubber tree) is native to the Amazon River Basin; however, it shows a pantropical species distribution through introductions (Basik et al. 2021). Pará rubber plantations have increased intensely worldwide in the past few decades, with the global consumption of natural rubber increasing by about 3% in 2019 (Bhattacharjee et al. 2021). Yunnan Province is one of the rubber-producing provinces in China and Xishuangbanna Prefecture (located in the south of Yunnan) contributes up to 77% of the rubber production in the province, representing 37% of the national rubber production (National Bureau of Statistics of China 2011, Statistical Bureau of Yunnan Province 2011).
Besides rubber, Yunnan Province is also rich in fungal diversity (Feng and Yang 2018). Approximately 104,000 fungal species are expected to be discovered in Yunnan; however, around 6,000 fungal species have been reported from the Province, leaving much to be described (Feng and Yang 2018; Bhunjun et al. 2022; Phukhamsakda et al. 2022). Surprisingly, a few fungal species have been described on Pará rubber in China (Senwanna et al. 2021; Xu et al. 2022a, 2022b, 2023; Hyde et al. 2023).
Senwanna et al. (2021) listed 67 orders, 168 families and 513 genera of fungi on Pará rubber and reported eight new taxa, two asexual-sexual linkages, 20 new host records and one reference specimen of saprobic fungi from Thailand. In addition, Senwanna et al. (2021) reported that three species from their collections had previously been reported from Pará rubber in the Amazon Forest (Spaulding 1961) and most of the taxa reported on Pará rubber have been found in Thailand. Moreover, Senwanna et al. (2019) discovered that Muyocoprondipterocarpi may have jumped from its original host. Dipterocarpustuberculatus, to the Pará rubber host and adapted to the new host in Thailand.
Dothideomycetes is the largest class of Ascomycota, currently encompassing more than 25 orders, 110 families and over 19,000 species (Wijayawardene et al. 2022). They can be endophytes, epiphytes, saprobes, lichenised or lichenicolous fungi and are found in terrestrial, freshwater and marine habitats worldwide (Hyde et al. 2013). In Pará rubber, Dothideomycetes are predominant amongst ascomycetes (Senwanna et al. 2021).
Fungi associated with rubber in China were poorly studied compared with other countries in the Greater Mekong Subregion (GMS), especially in Thailand. Moreover, saprobic fungal taxa, described in earlier studies, do not have sequence data. Continuing the fungal diversity studies in the GMS (Chaiwan et al. 2021), this study introduces four new taxa of Dothideomycetes associated with Pará rubber trees in Yunnan Province, China. Morphological characteristics and phylogenetic analyses were conducted to find accurate taxonomic placements of these new taxa.
Materials and methods
Collection, morphological examination and isolation
Dead rubber (Heveabrasiliensis) branches with fungal fruiting bodies were collected from Yunnan Province, China, during the summers of 2021 and 2022. The samples were stored in sealable plastic bags and taken to the mycology laboratory at Qujing Normal University. Morphological observations and single spore isolations were conducted following the methods described by Senanayake et al. (2020). Morphological characteristics were observed using a stereomicroscope Leica S8AP0 and photographed with an OLYMPUS BX53 compound microscope. Measurements were obtained using Tarosoft (R) Image Frame Work software. Adobe Photoshop CC 2017 software was used for preparing photo-plates. Herbarium specimens of the new species were deposited at the Herbarium of Zhongkai University of Agriculture and Engineering (ZHKU), China. The living cultures were deposited at the culture collection of Zhongkai University of Agriculture and Engineering (ZHKUCC), China. Facesoffungi (FoF) numbers and Index Fungorum (IF) numbers were obtained as per Jayasiri et al. (2015) and Index Fungorum (2024).
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted directly from scraped fresh mycelia grown on one-month-old artificial culture media (PDA), using an E.Z.N.A. Forensic DNA Kit (BIO-TEK), in accordance with the manufacturer’s protocol. The different gene regions, primers and protocols used for the amplification are summarised in Table 1. Polymerase chain reaction (PCR) amplifications were conducted using 25 μl PCR mixture containing 8.5 μl ddH2O, 12.5 μl 2 × Master Mix (Bioteke Corporation, Beijing, China), 2 μl DNA template and 1 μl each reverse and forward primer (Tibpromma et al. 2018). Purification and sequencing of PCR products were carried out in Bioteke, P.R. China.
Table 1.
Primers, PCR thermal cycles for SSU, ITS, LSU, rpb2 and tef1-α amplification and reference(s).
| Genes | Primers/Loci | PCR condition | References |
|---|---|---|---|
| ITS | ITS4 | (94 °C: 30 s, 55 °C: 50 s, 72 °C: 90 s) × 35 cycles | White et al. (1990) |
| ITS5 | |||
| LSU | LR0R | (94 °C: 30 s, 55 °C: 50 s, 72 °C: 90 s) × 35 cycles | Vilgalys and Hester (1990) |
| LR5 | |||
| SSU | NS1 | (94 °C: 30 s, 55 °C: 50 s, 72 °C: 90 s) × 35 cycles | White et al. (1990) |
| NS4 | |||
| tef1-α | 983F | (95 °C: 30 s, 55 °C: 50 s, 72 °C: 90 s) × 35 cycles | Carbone and Kohn (1999) |
| 2218R | |||
| rpb2 | fRPB2-5f | (94 °C: 60 s, 58 °C: 60 s, 72 °C: 90 s) × 40 cycles | Liu et al. (1999) |
| fRPB2-7cR |
Phylogenetic analyses
Sequences with high similarities (> 90%) were identified by BLASTn searches to determine the closest match to the taxa. Initial alignments of the sequence data were processed using MAFFT v.7 (http://mafft.cbrc.jp/alignment/server) using default settings (Katoh et al. 2019). The sequences were trimmed using TrimAl V 1.2 with ‘gappyout’ automated trimming option (Capella-Gutiérrez et al. 2009). The alignments were checked visually and improved manually wherever necessary. Multiple genes were concatenated by Sequence Matrix.
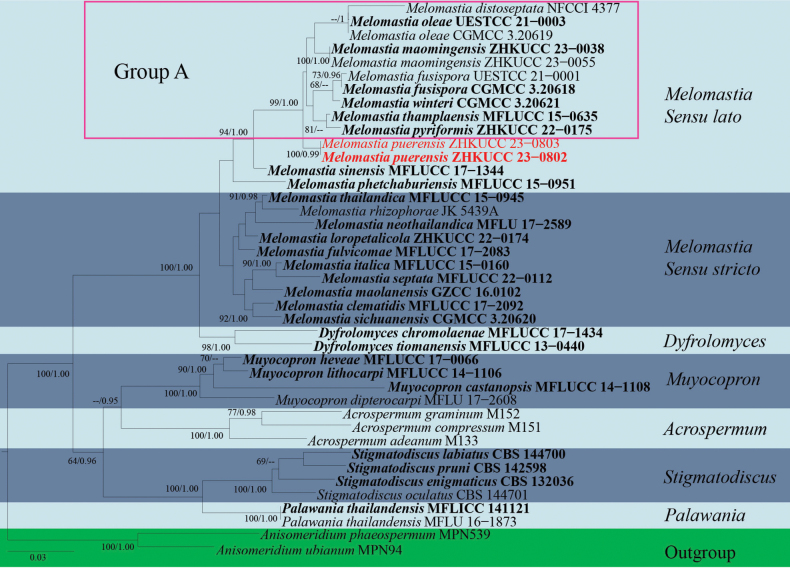
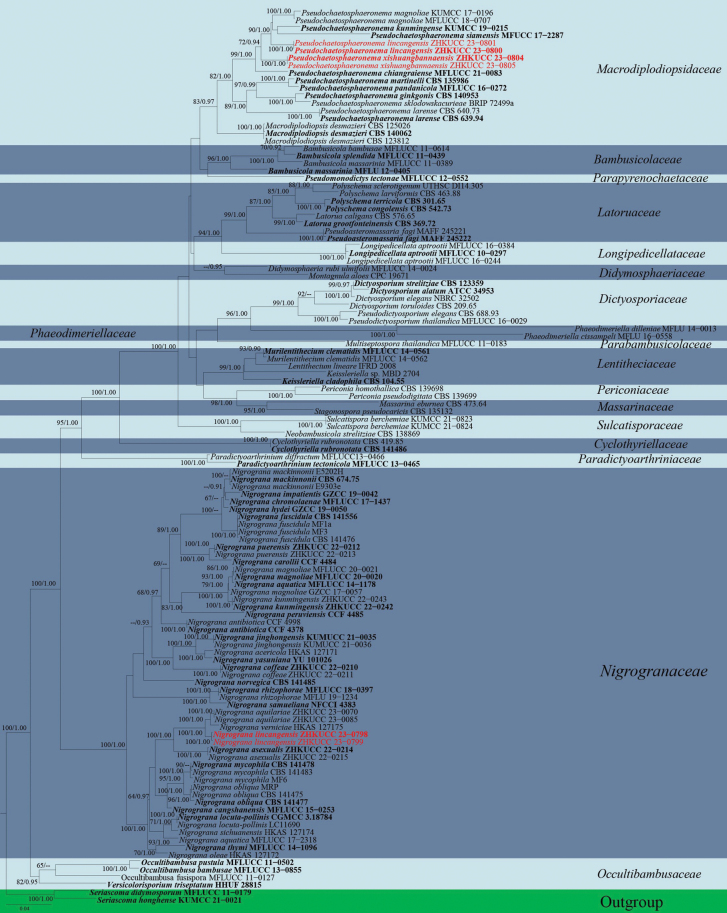
Multigene phylogenetic analyses for the concatenated genes were conducted using Maximum Likelihood (ML) and Bayesian Inference (BI) analyses. The CIPRES Science Gateway portal (Miller et al. 2012) was used to run both RAxML and Bayesian analyses. Maximum Likelihood analysis was made with RAxML-HPC2 on XSEDE v.8.2.10 tool (Stamatakis 2014) employing the GTR+GAMMA model with 1000 bootstrap repetitions. Bayesian analysis was performed by MrBayes v.3.0b4 (Huelsenbeck and Ronquist 2001) with the best-fit model of sequence evolution estimated using MrModelTest 2.2 (Nylander 2004). MrBayes analyses were performed with GTR+I+GAMMA for one million generations, sampling every 100th generation and ending the run automatically when the standard deviation of split frequencies dropped below 0.01 with a 25% burn-in. Phylograms were visualised with the FigTree v.1.4.0 programme (Rambaut 2012) and edited in Microsoft PowerPoint 2021. The final alignments and trees were deposited in TreeBASE, under submission ID 31039 (Fig. 1) and ID 31040 (Fig. 3) (http://www.treebase.org/).
Figure 1.
Phylogram generated from Maximum Likelihood analysis, based on combined LSU, SSU and tef1-α sequence data of 41 taxa, which comprised 2836 base pairs (LSU = 902 bp, SSU = 1031 bp, tef1-α = 903 bp). The best scoring RAxML tree with a final likelihood value of -14798.632437 is presented. The matrix had 1013 distinct alignment patterns, with 24.90% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.241740, C = 0.258134, G = 0.292403, T = 0.207722; substitution rates: AC = 0.834723, AG = 2.021967, AT = 1.126143, CG = 1.032150, CT = 7.231944, GT = 1.000000; gamma distribution shape parameter α = 0.320795. Bootstrap support values for ML equal to or greater than 60% and Bayesian Inference analysis values equal to or greater than 0.90 PP are labelled at each node. The tree is rooted with Anisomeridiumphaeospermum (MPN539) and A.ubianum (MPN94). Related sequences were collected following Li et al. (2022), Kularathnage et al. (2023) and Dong et al. (2023). The new isolates are indicated in red and the ex-type strains are in bold. Group A indicates the taxa used to compare the morphology with our new species (Melomastiapuerensis).
Figure 3.
Phylogram generated from Maximum Likelihood analysis based on combined LSU, ITS, SSU, tef1-α and rpb2 sequence data of 119 taxa, which comprised 4399 base pairs (LSU = 908 bp, ITS = 512 bp, SSU = 1000 bp, tef1-α = 925 bp, rpb2 = 1054 bp). The best scoring RAxML tree with a final likelihood value of -38918.764563 is presented. The matrix had 2023 distinct alignment patterns, with 39.00% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.245191, C = 0.247520, G = 0.268228, T = 0.239061; substitution rates: AC = 1.533778, AG = 3.877174, AT = 1.672983, CG = 1.254032, CT = 8.838860, GT = 1.000000; gamma distribution shape parameter α = 0.208600. Bootstrap support values for ML equal to or greater than 60% and Bayesian Inference analysis values equal to or greater than 0.90 PP are labelled at each node. The tree is rooted with Seriascomadidymospora (MFLUCC 11–0179) and S.didymospora (MFLUCC 11–0194). Related sequences were obtained from De Silva et al. (2022), Lu et al. (2022) and Li et al. (2023). The new isolates are indicated in red and the ex-type strains are in bold.
Results
Taxonomy and phylogenetic results
Dothideomycetes O.E. Erikss. & Winka
Dyfrolomycetales Pang, K.D. Hyde & E.B.G. Jones
. Pleurotremataceae
Watson
07931BC7-38F4-5875-960A-F07D4A52175C
Notes.
Pleurotremataceae was introduced by Watson (1929) and it comprises three genera viz. Dyfrolomyces, Melomastia and Pleurotrema (Wijayawardene et al. 2022). Species in this family are saprobes on wood in terrestrial and aquatic habitats (Hongsanan et al. 2020). Pleurotremataceae has been classified in several orders. Pleurotremataceae was excluded from Sordariomycetes and placed in Dothideomycetes, based on morphology and DNA sequences (Maharachchikumbura et al. 2016).
. Melomastia
Nitschke ex Sacc.
8EFACC3C-4048-570A-815E-A64DB3C825D3
Notes.
Melomastia was introduced by Saccardo (1875) with M.mastoidea as the type species (Kang et al. 1999). Melomastia has been recorded with 63 epithets in Index Fungorum (2024). Most Melomastia species have been found in terrestrial, freshwater and marine habitats and they have a wide geographical distribution in Africa, China, Germany, Italy, Japan, Poland and the United States of America (Norphanphoun et al. 2017; Dayarathne et al. 2020; Li et al. 2022; Kularathnage et al. 2023). Melomastia was discovered to be closely related to Dyfrolomyces and their exact relationship is still unknown. Li et al. (2022) reclassified Dyfrolomyces as Melomastia, based on morphology and phylogeny of four newly-introduced species from Olive in Sichuan Province, China. Melomastiatiomanensis and M.chromolaenae exhibit spindle-shape, 6–11-septate ascospores with acute ends. Additionally, the phylogenetic analysis conducted by Kularathnage et al. (2023) showed that M.tiomanensis and M.chromolaenae form a distinct lineage. Thus, M.tiomanensis and M.chromolaenae were moved into Dyfrolomyces and named Dyfrolomycestiomanensis and Dyfrolomyceschromolaenae. Melomastia is characterised by immersed, ostiolate ascomata, multiple layered, dark brown peridium, filamentous pseudoparaphyses, unitunicate, cylindrical, 8-spored asci and ovoid, hyaline, 1–10-septate, fusiform to oblong ascospores with rounded or acute ends, with or without gelatinous sheath (Norphanphoun et al. 2017; Dayarathne et al. 2020; Li et al. 2022; Kularathnage et al. 2023). However, the asexual morph of Melomastia is still unknown (Norphanphoun et al. 2017; Li et al. 2022; Kularathnage et al. 2023).
. Melomastia puerensis
R.F. Xu & Tibpromma sp. nov.
2A8B05E7-481A-522A-B739-D3BF341077D4
Index Fungorum number: IF901419
Facesoffungi number: FoF15195
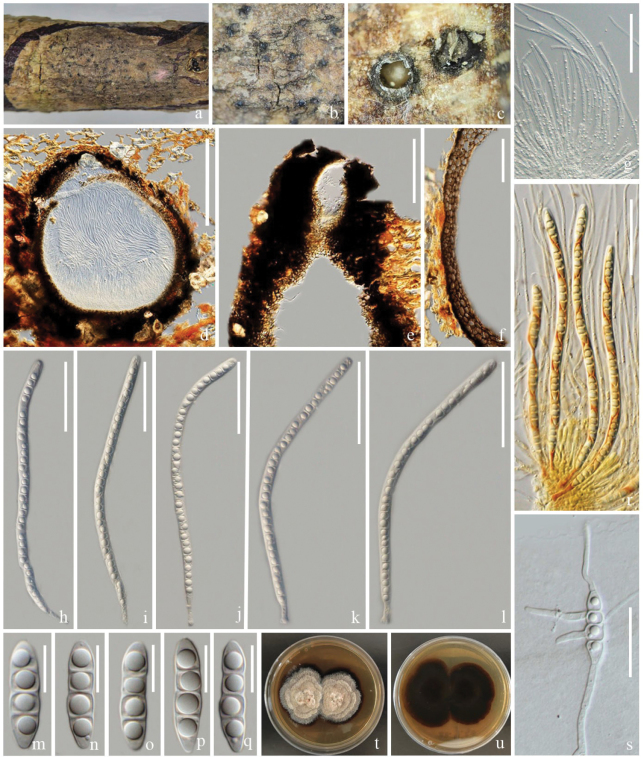
Figure 2.
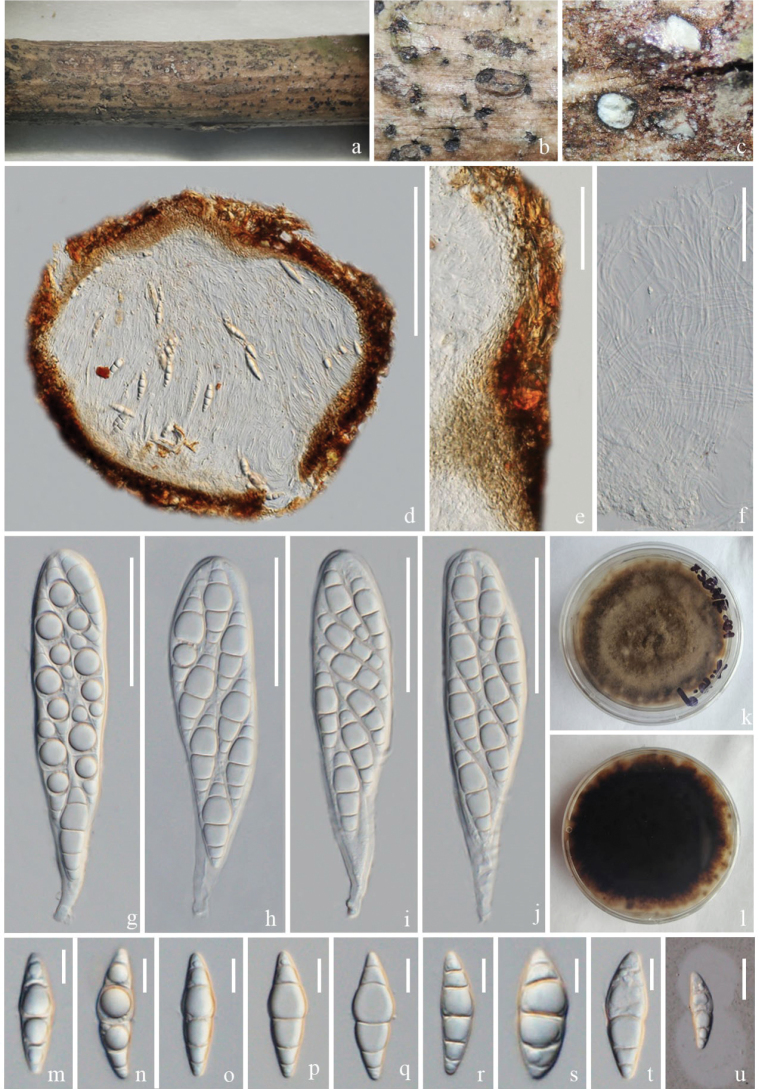
Melomastiapuerensis (ZHKU 23–0106, holotype) a–c appearance of ascomata on host surface d vertical section of an ascoma e vertical section of ostiole f section of peridium g hamathecium h–l asci m–q ascospores r asci stained in Lugol’s iodine s germinated ascospore t, u colonies on PDA (t-front and u-reverse views). Scale bars: 100 µm (d–f); 50 µm (g–l); 10 µm (m–q); 30 µm (s).
Etymology.
The name refers to the location “Pu’er, Yunnan, China”, where the holotype was collected.
Holotype.
ZHKU 23–0106.
Description.
Saprobic on a dead branch of Heveabrasiliensis. Sexual morph: Ascomata 260–720 μm high, 225–850 μm diam. (x‾ = 540 × 520 μm, n = 10), visible as black dots on the host surface, solitary or gregarious, immersed to slightly erumpent, subglobose or pyriform, carbonaceous, dark brown to black, ostiolate, papillate. Ostioles 205–220 × 195–258 µm (x‾ = 233 × 207 μm, n = 5), central, carbonaceous, dark brown to black. Peridium 40–120 µm wide, two-layered, outer layer, thick, carbonaceous, inner layer composed of several layers, brown to pale brown cells of textura angularis. Hamathecium comprises 2–4.5 μm wide, filiform, unbranched, hyaline, aseptate, guttulate, pseudoparaphyses, longer than asci. Asci 175–205 × 6–10 μm (x‾ = 190 × 8, n = 15), 8-spored, hyaline, bitunicate, cylindrical, flexuous, apically obtuse, with an ocular chamber, smooth-walled, short pedicellate. Ascospores 20–30 × 5–8 μm (x‾ = 24 × 7, n = 30), uniseriate, hyaline, fusiform, obtuse or conical ends, narrow towards the apex, 3-septate, constricted at the central septum, with guttulate in each cell. Asexual morph: Undetermined.
Culture characteristics.
Colonies on PDA that grow at 28 °C, flat, rough surface, entire edges, culture from above, brownish-grey, forming zonate grey, reverse dark brown, brown at the edge, turning reddish-brown.
Material examined.
China, Yunnan Province, Pu'er on a dead branch of Heveabrasiliensis, 16 September 2021, Rui-Fang Xu, XPER–14 (ZHKU 23–0106, holotype); ex-type ZHKUCC 23–0802, ZHKUCC 23–0803.
GenBank numbers.
ZHKUCC 23–0802 = ITS: OR941077, LSU: OR922309, SSU: OR922340, tef1-α: OR966284; ZHKUCC 23–0803 = ITS: OR941078, LSU: OR922310, SSU: OR922341, tef1-α: OR966285.
Notes.
The phylogenetic analyses showed that Melomastiapuerensis clustered basal to M.distoseptata, M.fusispora, M.maomingensis, M.oleae, M.pyriformis, M.thamplaensis and M.winteri with 99% MP, 1.00 PP support (Fig. 1). We compared the morphology of our collection with closely-related species and the differences are mentioned in Table 2. Our collection has slight differences from other closely-related species by having larger ascomata and wider peridium, but the phylogenetic tree shows that they are different species (Fig. 1, Table 2). Therefore, we introduce M.puerensis as a new species, based on morphology and phylogenetic analyses.
Table 2.
Morphological comparison of M.puerensis and closely-related species viz. M.distoseptata, M.fusispora, M.maomingensis, M.oleae, M.pyriformis, M.thamplaensis and M.winteri.
| Species | Ascomata | Peridium | Pseudoparaphyses | Asci | Ascospores | References |
|---|---|---|---|---|---|---|
| M.distoseptata | 550–630 × 450–600 μm, perithecial, immersed, erumpent neck with pseudoparaphyses, clypeate | 40 μm, with two strata, outer thick, and inner brown and hyaline cells of textura angularis to epidermoidea cells | 1.8–2.1 μm, flamentous, septate, unbranched, dense, longer than asci | 126.7–146.2 × 4.7–6.3 μm, apical ends obtuse, short pedicellate | 19.7–24.9 × 4.3–5 μm, fusoid, obtuse ends, apical ends slightly bent | Hongsanan et al. (2020) |
| M.fusispora | 432–624 × 527–618 μm, cone-shaped structures on the host surface, immersed to erumpent through host tissue, pyriform | 25.5–61.5 µm, two-layered, outer layer of cells of textura intricata, inner layer of textura angularis | 2–2.6 µm, dense, filiform, unbranched, hyaline, aseptate | 200–231 × 7.6–9.2 µm, slightly flexuous, apically round, with well-developed ocular chamber, cylindrical pedicellate | 27.5–32 × 6.5–7.5 µm, fusiform, with rounded to acute ends, narrow towards apex, constricted at the central septum, surrounded by an irregular and thin gelatinous sheath | Li et al. (2022) |
| M.maomingensis | 300–550 µm high × 250–500 µm diam., solitary, semi-immersed to immersed, visible on the host surface as black, obvious, raised spots, black, uni-loculate, globose to subglobose | 35–100 µm wide, comprising dense, thick, brown to dark brown cells of textura angularis, fusion with host tissue | 1.5–3.5 µm wide, comprising numerous, filamentous, hyaline, septate, sometimes branched, longer than asci, attached at the base and between the asci | 175–195 × 7–9 µm, cylindrical pedicel, rounded in apex, J- | (23–)24.5–29 × 6–8 µm, fusiform with acute ends, constricted at the septum, with a large guttule in each cell when mature | Du et al. (2024) |
| M.oleae | 410–440 × 493–520 µm, cone-shaped structures on host surface, semi-immersed, globose to compressed globose | 54–65 µm, two-layered, outer thick and inner composed of 5–6 layers of textura angularis to textura prismatica | 2–2.5 µm, dense, filiform, unbranched, aseptate | 209–237 × 7.5–9 µm, slightly flexuous, apically rounded with ocular chamber, cylindrical pedicellate | 28–34 × 6–7 µm, fusiform with obtuse ends, slightly constricted at the septa | Li et al. (2022) |
| M.puerensis | 260–720 × 225–850 μm, black dot on the host surface, immersed to erumpent to superficial, pyriform | 40–120 µm, two-layered, outer thick, carbonaceous, inner composed of several layers, pale brown to brown cells of textura angularis | 2–4.5 μm, filiform, unbranched, guttulate, pseudoparaphyses, longer than asci | 175–205 × 6–10 μm, flexuous, apical ends obtuse, with ocular chamber, smooth-walled, short pedicellate | 20–30 × 5–8 μm, fusiform, obtuse or conical ends, narrow towards apex, constricted at the central septum, with guttules in each cell | This study |
| M.pyriformis | 330–640 × 275–420 μm, erumpent to superficial when mature, pyriform, papillate, ostiolate | 20–50 μm, thin at the base and become thick towards sides, comprised of brown, thick-walled, cells of textura intricata in sides; and thin-walled, pale brown, cells of textura angularis in base | 1.8–2.5 μm wide, dense, filiform, unbranched, septate, anastomosing between and above the asci | 135–160 × 5.5–7.5 μm, fissitunicate, apically round, with an indistinct ocular chamber, short pedicellate | 20–25 × 4.5–7 μm, , fusiform with acute ends, not constricted at the septa, with guttules in each cell | Kularathnage et al. (2023) |
| M.thamplaensis | 550–630 × 450–600 μm, black spots on the host surface, immersed, clypeate, subglobose to obpyriform, some with a broad, flattened base | 14–49 μm, composed of three strata, an outer stratum, dense, amorphous, thick-walled cells fusing with host tissue, a middle layer of thick-walled, black cells of textura angularis and an inner layer of thin-walled black cells of textura angularis | 1.8–2.1 μm, attached at the base and between the asci, embedded in a gelatinous matrix | 126.7–146.2 × 4.7–6.3 µm, long cylindrical, short-pedicellate, apically rounded with an obvious apical ring | 19.7–24.9 × 4.3–5 μm, fusiform with acute angular ends, constricted at the septum, smooth-walled, containing several guttules when young | Zhang et al. (2017) |
| M.winteri | 340–365 × 364–410 µm, semi-immersed to immersed, globose | 55–62.5 µm, two-layered, outer thick, and inner composed of 3–4 layers of hyaline to lightly brown cells of textura angularis to textura prismatica | 1.5–3.5 µm, dense, filiform, unbranched, septate | 165–189 × 7–8.5 µm, slightly flexuous, apically round, with a distinct ocular chamber, cylindrical pedicellate | 25–30 × 5–6.5 µm, partially overlapping, fusiform with acute ends, deeply constricted at the median septum | Li et al. (2022) |
Pleosporales Luttrell ex M.E. Barr
. Nigrogranaceae
Jaklitsch & Voglmayr
9E017A5B-4E34-573E-8EE4-07D8DE94A085
Notes.
Nigrogranaceae was introduced by Jaklitsch and Voglmayr (2016), with Nigrograna as the type genus. The members of Nigrogranaceae can be found on a wide range of hosts in marine and terrestrial habitats (Dayarathne et al. 2020; Boonmee et al. 2021; Lu et al. 2022; Hyde et al. 2023).
. Nigrograna
Gruyter, Verkley & Crous
7C008AE2-4F41-5071-8E64-AB8BD08F2D46
Notes.
Nigrograna was introduced by De Gruyter et al. (2013) with N.mackinnonii as the type species. Nigrograna has 32 epithets in Index Fungorum (2024). Ahmed et al. (2014) transferred N.mackinnonii to Biatriospora, based on multigene phylogenetic analysis. Kolařík et al. (2017) introduced four new endophytic species viz. B.antibiotica, B.carollii, B.peruviensi, and B.yasuniana in Biatriospora, based on morphology and multigene phylogeny and, later, Kolařík (2018) synonymised these four species under Nigrograna. The sexual morph of Nigrograna is characterised by globose, immersed or less commonly superficial ascomata, bitunicate, fissitunicate 8-spored asci with short stipe and knob-like base, asymmetric, fusoid, 1–3-septate, pale to chocolate brown, smooth or faintly verrucose ascospores (Jaklitsch and Voglmayr 2016). The asexual morph is characterised by globose to subglobose or pyriform pycnidia, solitary terminal phialides conidiophores, ampulliform, lageniform or subcylindrical phialides, oblong, cylindrical or allantoid conidia, sometimes ellipsoid and 1-celled (Jaklitsch and Voglmayr 2016; Lu et al. 2022). In this study, we introduced one new species isolated from rubber tree, based on morphology and phylogeny.
. Nigrograna lincangensis
R.F. Xu & Tibpromma sp. nov.
1AB76FA4-DD2F-5A36-8D5D-3A6BF4295FC0
Index Fungorum number: IF901420
Facesoffungi number: FoF15196
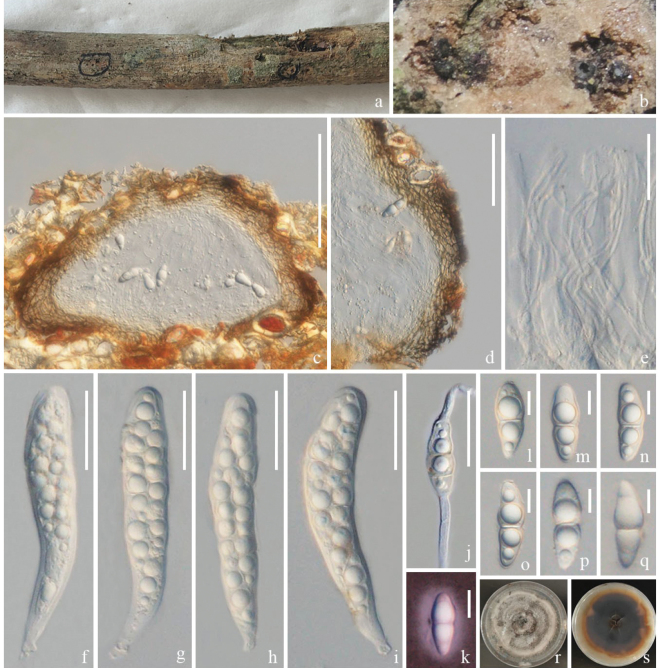
Figure 4.
Nigrogranalincangensis (ZHKU 23–0104, holotype) a–c appearance of ascomata on the host surface d vertical section of an ascoma e vertical section of ostiole f hamathecium and asci g section of peridium h–k asci l ascospores m a germinated ascospore n, o colonies on PDA (n-front and o-reverse views). Scale bars: 100 µm (d); 50 µm (e); 30 µm (f); 200 µm (g); 10 µm (h–k); 5 µm (l); 20 µm (m).
Etymology.
The name refers to the location “Lincang, Yunnan, China”, where the holotype was collected.
Holotype.
ZHKU 23–0104.
Description.
Saprobic on a dead branch of Heveabrasiliensis. Sexual morph: Ascomata 285–360 μm high, 230–307 μm diam. (x‾ = 337 × 272 μm, n = 5), immersed, under the clypeus, sometimes inconspicuous on host surface and small bumps can be seen, solitary, dark brown, globose or ellipsoid, with papilla. Ostioles 117–217 × 68–124 μm (x‾ = 152 × 99 μm, n = 10), central, brown, papillate. Peridium 16–45 μm wide, comprising several layers with dark-brown to dark cells of textura angularis. Hamathecium comprises 1.5–3 μm wide, unbranched, septate, hyaline, pseudoparaphyses. Asci 45–70 × 9–12 μm (x‾ = 57 × 10 μm, n = 10), 8-spored, bitunicate, pedicellate, club shape, cylindrical to clavate, straight or slightly curved, apically rounded, thick-walled. Ascospores 10–15 × 4–6 μm (x‾ = 13 × 4.8 μm, n = 30), 1–2-seriate, initially 1-septate, becoming 3-septate at the maturity, fusoid to narrowly ellipsoid, upper part or second cell slightly wider and tapering towards narrow ends, constricted at the septa, hyaline to yellow-brown to brown with age, guttulate, think-walled. Asexual morph: Undetermined.
Culture characteristics.
Spores germinated within 12 hours, colonies grow on PDA at 28 °C, circular, floppy, entire edge, raised, grey to taupe, reverse dark brown.
Material examined.
China, Yunnan Province, Lincang, on a dead branch of Heveabrasiliensis, 28 July 2022, Rui-Fang Xu, LCR06, (ZHKU 23–0104, holotype); ex-type ZHKUCC 23–0798, ZHKUCC 23–0799.
GenBank numbers.
ZHKUCC 23–0798 = ITS: OR853099, LSU: OR922323, SSU: OR941079, tef1-α: OR966282, rpb2: OR966280; ZHKUCC 23–0799 = ITS: OR853100, LSU: OR922324, SSU: OR941080, tef1-α: OR966283, rpb2: OR966281.
Notes.
In the phylogenetic analyses, Nigrogranalincangensis (ZHKUCC 23–0798) forms a closely-related clade to N.asexualis (ZHKUCC 22–0214), N.aquilariae (ZHKUCC 23–0070) and N.verniciae with 100% ML and 1.00 PP support (Fig. 3). However, we could not compare the morphological characteristics of N.lincangensis and N.asexualis, because N.lincangensis was described only from its sexual morph in nature, while N.asexualis was described by its asexual morph in nature from coffee in China. A comparison of the ITS region of N.lincangensis and N.asexualis revealed 16 base pair differences (3.46%) across 462 nucleotides, 40 base pair differences (4.21%) across 949 nucleotides in tef1-α gene, 124 base pair differences (12%) across 1033 nucleotides in rpb2 gene. Nigrogranaaquilariae and N.verniciae have very similar morphological characteristics, but they can be differentiated by having wider ascomata (285–360 μm vs. 180–270 μm), larger asci (45–70 × 9–12 μm vs. 49–57 × 7–9 μm) and larger ascospores (10–15 × 4–6 μm vs. 10–13 × 3.5–4.5 µm) in N.lincangensis (Du et al. 2024); while N.verniciae has larger ascomata (340–360 × 350–370 μm vs. 85–360 μm × 230–307 μm) and asci with knob-like to furcate pedicels (Li et al. 2023).
Nigrogranalincangensis has similar ascomata, asci and ascospore characteristics similar to other Nigrograna species (Jaklitsch and Voglmayr 2016; Hyde et al. 2017; Tibpromma et al. 2017; Dayarathne et al. 2020; Mapook et al. 2020; Lu et al. 2022). However, N.lincangensis differs from N.cangshanensis by having larger ascomata (285–360 × 230–307 μm vs. 120–135 × 135–155 μm) (Tibpromma et al. 2017). Nigrogranachromolaenae can be distinguished from N.lincangensis in having smaller ascomata (160–280 × 115–130 μm vs. 285–360 × 230–307 μm), smaller asci (40–55 × 7–10 μm vs. 45–70 × 9–12 μm), and greyish-brown to dark brown ascospores (Mapook et al. 2020). Nigrogranacoffeae differs from N.cangshanensis by having smaller ascomata (90–140 × 140–200 μm vs. 285–360 × 230–307 μm), 1-septate ascospores (Lu et al. 2022). Nigrogrananovergica differs from N.lincangensis as it occurs on pseudostromata from the host of Diaporthe sp. (Jaklitsch and Voglmayr 2016). Nigrogranamycophila and N.obliqua are distinct from N.lincangensis by having dark brown ascospores (Jaklitsch and Voglmayr 2016). Nigrogranapuerensis differs from N.lincangensis by having acute apical and basal cells and the apical cells are wider than the basal cells (Lu et al. 2022). Nigrogranasamueliana differs from N.lincangensis by the absence of ostiole (Dayarathne et al. 2020). Nigrogranathymi can be easily distinguished from N.lincangensis in having 4–5 septate (Hyde et al. 2017). Therefore, N.lincangensis is described here as a new species, based on phylogeny and morphology.
. Macrodiplodiopsidaceae
Voglmayr, Jaklitsch & Crous
E7146F67-0544-5AE8-A461-36B87DF91A50
Notes.
Macrodiplodiopsidaceae was introduced by Crous et al. (2015) with Macrodiplodiopsis as the type genus. There are two genera viz. Macrodiplodiopsis and Pseudochaetosphaeronema in this family (Wijayawardene et al. 2022).
. Pseudochaetosphaeronema
Punith.
119D0C27-97DF-57A1-83CF-4CE6FCF4EF22
Notes.
Pseudochaetosphaeronema was introduced by Punithalingam (1979), with P.larense as the type species. The members of this genus have been reported as human pathogens, endophytes and saprobes (Boonmee et al. 2021). Nine epithets are listed in Index Fungorum (2024), i.e. one sexual P.chiangraiense and eight asexual species viz. P.ginkgonis, P.kunmingense, P.larense, P.magnoliae, P.martinelli, P.pandanicola, P.siamense and P.sklodowskacurieae. The asexual morph of Pseudochaetosphaeronema is characterised by globose, conidiomata, monophialidic, cylindrical conidiogenous cells and hyaline, subglobose to oval, aseptate conidia (De Silva et al. 2022). The sexual morph is characterised by immersed, uni-loculate ascomata, peridium with the cells of textura angularis, unbranched, septate pseudoparaphyses, 8-spored, bitunicate, fissitunicate, short distinct pedicel asci with rounded end and fusiform, 1-septate, guttulate ascospores with pointed ends (Boonmee et al. 2021).
. Pseudochaetosphaeronema lincangensis
R.F. Xu & Tibpromma sp. nov.
AA520AD2-DE95-5072-8D54-6E125383EF49
Index Fungorum number: IF901421
Facesoffungi number: FoF15197
Figure 5.
Pseudochaetosphaeronemalincangensis (ZHKU 23–0105, holotype) a, b appearance of ascomata on host substrate c vertical section of an ascoma d section of peridium e pseudoparaphyses f–i asci j a germinated ascospore l–q ascospores k ascospore stained with Indian ink r, s colonies on PDA (r-front and s-reverse views). Scale bars: 100 µm (c); 50 µm (d); 30 µm (e–j); 200 µm (g); 10 µm (k, l–q).
Etymology.
The name refers to the location “Lincang, Yunnan, China”, where the holotype was collected.
Holotype.
ZHKU 23–0105.
Description.
Saprobic on a dead branch of Heveabrasiliensis. Sexual morph: Ascomata 140–245 μm high, 255–290 μm diam., (x‾ = 190 × 267 μm, n = 5), immersed, visible as dark-brown dots on the host surface, solitary, uni-loculate, ampulliform, without ostiole. Peridium 18–50 μm wide, several layers, comprising dark-brown to pale-brown cells of textura angularis. Hamathecium comprises 2–3 μm wide, numerous, hyaline, unbranched, pseudoparaphyses. Asci 90–145 × 15–30 μm (x‾ = 112 × 22 μm, n = 15), 8-spored, bitunicate, cylindrical to clavate, apically rounded, short pedicelate, with a small ocular chamber, thick-walled. Ascospores 25–40 × 8–15 μm (x‾ = 30 × 11 μm, n = 35), overlapping, 2-seriate, fusiform, 1-septum in the middle of cell, widest at the centre and tapering towards narrow ends, constricted at the septum, hyaline, guttulate, with ellipsoid mucilaginous sheath, thick and smooth-walled. Asexual morph: Undetermined.
Culture characteristics.
culture on PDA, colonies slow growing on 28 °C, low convex, entire, smooth, edge is off-white from above, dark brown, edge is orange on reverse side.
Material examined.
China, Yunnan Province, Lincang on a dead branch of Heveabrasiliensis, 28 July 2022, Rui-Fang Xu, LCR07 (ZHKU 23–0105, holotype); ex-type ZHKUCC 23–0800, ZHKUCC 23–0801.
GenBank numbers.
ZHKUCC 23–0800 = ITS: OR853095, LSU: OR922336, SSU: OR922342, tef1-α: OR966290; ZHKUCC 23–0801 = ITS: OR853096, LSU: OR922337, SSU: OR922343, tef1-α: OR966291.
Notes.
In the phylogenetic analyses, Pseudochaetosphaeronemalincangensis clusters distinctly, sister to P.kunmingense, P.magnoliae and P.siamensis with 90% MP, 1.00 PP support (Fig. 3). The base pair differences in ITS, LSU, SSU and tef1-α sequences of our new species are compared with P.kunmingense, P.magnoliae and P.siamensis (Table 3). However, we could not compare the morphological characteristics of the species above, as they were described, based on asexual morphs. Therefore, based on morphology and phylogeny, we introduce Pseudochaetosphaeronemalincangensis as a new species.
Table 3.
Nucleotide differences in the ITS, LSU, SSU and tef1-α of P.lincangensis (ZHKUCC 23–0800) compared with P.kunmingense, P.magnoliae and P.siamensis.
| Strains | ITS | LSU | SSU | tef1-α |
|---|---|---|---|---|
| P.kunmingense (KUMCC 19–0215) | 30/506 (5.93%) | 10/855 (1.16%) | 4/1012 (0.39%) | 38/893 (4.26%) |
| P.magnoliae (KUMCC 17–0196) | 51/539 (9.46%) | 19/854 (2.22%) | 8/939 (0.85%) | 32/899 (3.56%) |
| P.siamensis (MFUCC 17–2287) | 43/480 (8.96%) | 11/848 (1.29%) | 1/1005 (0.09%) | 98/645 (15.19%) |
. Pseudochaetosphaeronema xishuangbannaensis
R.F. Xu & Tibpromma sp. nov.
EAECAD44-38E7-5AAC-9586-CA3E531C6A3B
Index Fungorum number: IF901422
Facesoffungi number: FoF15198
Figure 6.
Pseudochaetosphaeronemaxishuangbannaensis (ZHKU 23–0107, holotype) a–c appearance of ascomata on host substrate d section of an ascoma e peridium f pesudoparaphyses g–j asci m–t ascospores u ascospore stained with Indian ink k, l colonies on PDA (k-front and l-reverse view). Scale bars: 200 µm (d); 100 µm (e); 50 µm (f–j); 10 µm (m–t); 20 µm (u).
Etymology.
The name refers to the location “Xishuangbanna, Yunnan, China”, where the holotype was collected.
Holotype.
ZHKU 23–0107.
Description.
Saprobic on a dead branch of Heveabrasiliensis. Sexual morph: Ascomata 270–410 μm high, 370–480 μm diam., (x‾ = 350 × 420 μm, n = 5), solitary, scattered, immersed, globose to subglobose, uni-loculate, black. Peridium 40–90 μm wide, thin-walled, composed of several layers of small, brown to pale brown cells of textura intricata. Hamathecium comprises 2–3 μm wide, numerous, dense, filiform, unbranched, hyaline, cellular pseudoparaphyses. Asci 130–180 × 25–35 μm (x‾ = 155 × 32 μm, n = 20), 8-spored, bitunicate, obovoid, short distinct pedicel with conical end, apex rounded with a minute ocular chamber. Ascospores 30–50 × 10–20 μm (x‾ = 42 × 13 μm, n = 30), hyaline, fusiform, with pointed ends, 3–5-septate, larger upper third cell, constricted at the septa, guttulate, thick-walled, with mucilaginous sheath, the sheath constricted at the middle. Asexual morph: Undetermined.
Culture characteristics.
Colony on PDA, colonies slow growing on 28 °C, umbonate, filiform, smooth, edges brown, from above, brown, dark brown on reverse side.
Material examined.
China, Yunnan Province, Xishuangbanna on a dead branch of Heveabrasiliensis, 12 September, 2021, Rui-Fang Xu, XSBNR–41 (ZHKU 23–0107, holotype); ex-type ZHKUCC 23–0804, ZHKUCC 23–0805.
GenBank numbers.
ZHKUCC 23–0804 = ITS: OR853097, LSU: OR922338, SSU: OR922344, tef1-α: OR966286; ZHKUCC 23–0805 = ITS: OR853098, LSU: OR922339, SSU: OR922345, tef1-α: OR966287.
Notes.
In the phylogenetic analyses, Pseudochaetosphaeronemaxishuangbannaensis clusters with P.lincangensis with 99% ML and 1.00 PP support (Fig. 3). Morphologically, P.xishuangbannaensis differs from P.lincangensis in having longer asci (130–180 μm vs. 90–145 μm), 3–5-septate ascospores with sheath constricted at the central septum and brown to dark brown colonies, while P.lincangensis has ascospores with a normal sheath in a circle, 1-septate ascospores with obtuse ends and colonies off-white from the forward edge, orange in reverse. Pseudochaetosphaeronemaxishuangbannaensis shares similar morphologies with P.chiangraiense, but can be differentiated by having the peridium with the cells of textura intricate, larger ascomata (270–410 × 370–480 μm vs. 190–255 × 190–200 μm), longer asci (130–180 μm vs. 50–110 μm), larger (30–50 × 10–20 μm vs. 20–45 × 15–30 μm) and 3–5 septate ascospores with a sheath constricted at the central septum and brown to dark brown colonies. Pseudochaetosphaeronemachiangraiense has textura angularis peridium, ascospores surrounded by a normal sheath in a circle, 1-septum, obtuse ends, from above, greenish-grey in the middle and pale brown at the margin, yellowish-brown on the reverse side (Boonmee et al. 2021). In addition, P.xishuangbannaensis formed a different lineage with P.chiangraiense (Fig. 3). Therefore, P.xishuangbannaensis is described as a new species, based on phylogenetic analyses and morphological comparison.
Discussion
Global fungal diversity is astounding. Although around 155,000 fungal species have been described, up to 19 million have yet to be described (Hyde 2022; Phukhamsakda et al. 2022). Fungi have been classified into five different phyla: Chytridiomycota, Zygomycota, Glomeromycota, Ascomycota and Basidiomycota (Aguilar-Marcelino et al. 2020; Wijayawardene et al. 2022). Fungi play an important role in litter decomposition by breaking down lignin and other refractory components in the litter, thereby affecting the decomposition of terrestrial ecosystems, especially by activities of Basidiomycota and Ascomycota (Osono and Takeda 2002; Bucher et al. 2004; Phukhamsakda et al. 2022). Discovering more saprophytic fungi associated with rubber will enrich our knowledge on saprobic fungi and their functions as litter degraders. Microfungi from warm climates have a more significant decomposition capacity than from cool climates (Osono et al. 2011). Previous studies have reported that Ascomycota, Basidiomycota and Oomycota are abundant on Pará rubber leaf and branch litter (Monkai et al. 2017; Meeboon and Takamatsu 2020; Senwanna et al. 2021). Nizamani et al. (2023) provided a checklist comprising 788 species and 179 taxa identified at the genus level from 57 countries. The taxa listed in the checklist belong to 515 genera, 180 families and 68 orders and more than half of these taxa were isolated from leaf and branch litter.
In Southeast Asia, Pará rubber plantations have been expanding rapidly since the 20th century and, currently, supply over 90% of the world’s natural rubber (Fox and Vogler 2005; Mann 2009; Ziegler et al. 2009). More than one million hectares of lands in Cambodia, Laos, Myanmar, South China, Thailand and Vietnam have been converted into Pará rubber plantations (Li and Fox 2012). In 1904, China planted rubber for the first time in Yingjiang, Dehong, in Yunnan Province (Chapman 1991). Pará rubber is widely cultivated in the Hainan, Guangdong, Guangxi, Fujian and Yunnan Provinces in China as an economically important plant (Wang et al. 2015).
Pará rubber is vulnerable to many pests and diseases, but it is still a mystery why only a few fungal species have been found on rubber (Senwanna et al. 2021). Pará rubber tree was introduced to China from Malaysia, presumably by seed and endemic fungi are unlikely to follow; therefore, new fungi colonise Pará rubber through host-shifting or host-jumping (Roy 2001; Senwanna et al. 2019, 2021). Fungi associated with Pará rubber are found in different life modes such as saprobic, endophytic and pathogenic (Gazis and Chaverri 2010; Monkai et al. 2017; Senwanna et al. 2021). On Pará rubber, Dothideomycetes predominate amongst ascomycetes (Senwanna et al. 2021) and four species described in our study also belong to Dothideomycetes.
Fungal pathogens and endophytes were also isolated from the Pará rubber trees. Additionally, studies have been conducted to analyse the richness and diversity of endophytic fungi in different tissues of Heveabrasiliensis (Martin et al. 2015; Rojas-Jimenez et al. 2016; Araújo et al. 2020). Pathogens cause potential disease threats to Heveabrasiliensis; for example, Corynesporacassiicola causes Corynespora leaf fall disease (Jayasinghe and Fernando 1998), Microcyclusulei causes South American leaf blight (Júnior et al. 2014), the basidiomycete genera Phellinus, Rigidoporus and Ganoderma cause stem- and root-rots (Mohammed et al. 2014). In addition, the estimated richness of endophytic fungi does not significantly differ amongst the leaves, stems and roots; and the fungal diversity is higher in the stems and roots compared to the leaves (Martin et al. 2015; Araújo et al. 2020). Mahendran et al. (2021) revealed that Aspergillusterreus has a good inhibitory potential against Rigidoporusmicroporus and Corynesporacassiicola and has potential for biological control. Therefore, it is important to understand the fungi associated with Pará rubber trees to manage and prevent rubber tree diseases.
Only a few reports are available for the saprobic fungi on Heveabrasiliensis in China and many taxa lack molecular data (Seephueak et al. 2010, 2011; Senwanna et al. 2021). Therefore, a revised taxonomic approach with multi-gene phylogenetic analyses is necessary to understand the fungal diversity associated with Pará rubber. In this study, we introduce four new saprobic fungi from branches and twigs of rubber trees, based on morphology and molecular phylogenetic analyses. This enriches the fungal diversity in Pará rubber and provides information for host jumping.
Supplementary Material
Acknowledgements
Rui-Fang Xu thanks Ying Gao for her support in uploading protein genes to GenBank and Jing-Yi Zhang and Ya-Ru Sun for their help in submitting the alignments to TreeBASE. The authors extend their appreciation to the Researchers Supporting Project Number (RSP2024R56), King Saud University, Riyadh, Saudi Arabia.
Citation
Xu R-F, Karunarathna SC, Phukhamsakda C, Dai D-Q, Elgorban AM, Suwannarach N, Kumla J, Wang X-Y, Tibpromma S (2024) Four new species of Dothideomycetes (Ascomycota) from Pará Rubber (Hevea brasiliensis) in Yunnan Province, China. MycoKeys 103: 71–95. https://doi.org/10.3897/mycokeys.103.117580
Funding Statement
National Natural Science Foundation of China (Numbers NSFC 31760013 and 32260004) High-Level Talent Recruitment Plan of Yunnan Provinces ("Young Talents" and “High-End Foreign Experts” programs) The central government guides local projects of Yunnan Provincial Science and Technology Department (No. 202307AC110003) Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia, for funding this research work through project no. (IFKSUOR3-299-1)
Contributor Information
Xiao-Yan Wang, Email: 527010142@qq.com.
Saowaluck Tibpromma, Email: saowaluckfai@gmail.com.
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
This study was supported by the National Natural Science Foundation of China (Numbers NSFC 32260004 and 31760013) and High-Level Talent Recruitment Plan of Yunnan Province (“Young Talents” and “High-End Foreign Experts” programmes); the central government guides local projects of Yunnan Provincial Science and Technology Department (No. 202307AC110003); Researchers Supporting Project Number (RSP2024R56), King Saud University, Riyadh, Saudi Arabia. This study was partially supported by Chiang Mai University, Thailand.
Author contributions
Conceptualization: SCK, ST. Data curation: RFX. Formal analysis: SCK. Funding acquisition: DQD, SCK, JK. Investigation: ST, SCK, RFX. Methodology: CP, SCK, ST, RFX. Project administration: SCK. Resources: RFX. Software: ST, RFX, CP. Validation: AME, NS. Visualization: RFX. Writing – original draft: RFX. Writing – review and editing: AME, CP, DQD, NS, JK, XYW, RFX, SCK, ST.
Author ORCIDs
Rui-Fang Xu https://orcid.org/0000-0003-1207-8254
Samantha C. Karunarathna https://orcid.org/0000-0001-7080-0781
Chayanard Phukhamsakda https://orcid.org/0000-0002-1033-937X
Dong-Qin Dai https://orcid.org/0000-0001-8935-8807
Abdallah M. Elgorban https://orcid.org/0000-0003-3664-7853
Nakarin Suwannarach https://orcid.org/0000-0002-2653-1913
Jaturong Kumla https://orcid.org/0000-0002-3673-6541
Xiao-Yan Wang https://orcid.org/0009-0009-6430-3637
Saowaluck Tibpromma https://orcid.org/0000-0002-4706-6547
Data availability
All of the data that support the findings of this study are available in the main text.
References
- Aguilar-Marcelino L, Mendoza-de-Gives P, Al-Ani LKT, López-Arellano ME, Gómez-Rodríguez O, Villar-Luna E, Reyes-Guerrero DE. (2020) Chapter 26 – Using molecular techniques applied to beneficial microorganisms as biotechnological tools for controlling agricultural plant pathogens and pest. In: Sharma V, Salwan R, Al-Ani LKT. (Eds) Molecular Aspects of Plant Beneficial Microbes in Agriculture.Academic Press, 333–349. 10.1016/B978-0-12-818469-1.00027-4 [DOI]
- Ahmed SA, Van De Sande WWJ, Stevens DA, Fahal A, Van Diepeningen AD, Menken SBJ, De Hoog GS. (2014) Revision of agents of black-grain eumycetoma in the order Pleosporales. Persoonia – Molecular Phylogeny and Evolution of Fungi 33: 141–154. 10.3767/003158514X684744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo KS, Brito VN, Veloso TGR, de Leite TS, Alves JL, da Hora BT Junior, Moreno HLA, Pereira OL, Mizubuti ESG, de Queiroz MV. (2020) Diversity and distribution of endophytic fungi in different tissues of Heveabrasiliensis native to the Brazilian Amazon forest. Mycological Progress 19(10): 1057–1068. 10.1007/s11557-020-01613-4 [DOI] [Google Scholar]
- Basik AA, Sanglier J-J, Yeo CT, Sudesh K. (2021) Microbial degradation of rubber: Actinobacteria. Polymers 13(12): 1989. 10.3390/polym13121989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee A, Bhowmik M, Paul C, Das Chowdhury B, Debnath B. (2021) Rubber tree seed utilization for green energy, revenue generation and sustainable development– A comprehensive review. Industrial Crops and Products 174: 114186. 10.1016/j.indcrop.2021.114186 [DOI]
- Bhunjun CS, Niskanen T, Suwannarach N, Wannathes N, Chen Y-J, McKenzie EHC, Maharachchikumbura SSN, Buyck B, Zhao C-L, Fan Y-G, Zhang J-Y, Dissanayake AJ, Marasinghe DS, Jayawardena RS, Kumla J, Padamsee M, Chen Y-Y, Liimatainen K, Ammirati JF, Phukhamsakda C, Liu J-K, Phonrob W, Randrianjohany É, Hongsanan S, Cheewangkoon R, Bundhun D, Khuna S, Yu W-J, Deng L-S, Lu Y-Z, Hyde KD, Lumyong S. (2022) The numbers of fungi: Are the most speciose genera truly diverse? Fungal Diversity 114(1): 387–462. 10.1007/s13225-022-00501-4 [DOI]
- Boonmee S, Wanasinghe DN, Calabon MS, Huanraluek N, Chandrasiri SKU, Jones GEB, Rossi W, Leonardi M, Singh SK, Rana S, Singh PN, Maurya DK, Lagashetti AC, Choudhary D, Dai Y-C, Zhao C-L, Mu Y-H, Yuan H-S, He S-H, Phookamsak R, Jiang H-B, Martín MP, Dueñas M, Telleria MT, Kałucka IL, Jagodziński AM, Liimatainen K, Pereira DS, Phillips AJL, Suwannarach N, Kumla J, Khuna S, Lumyong S, Potter TB, Shivas RG, Sparks AH, Vaghefi N, Abdel-Wahab MA, Abdel-Aziz FA, Li G-J, Lin W-F, Singh U, Bhatt RP, Lee HB, Nguyen TTT, Kirk PM, Dutta AK, Acharya K, Sarma VV, Niranjan M, Rajeshkumar KC, Ashtekar N, Lad S, Wijayawardene NN, Bhat DJ, Xu R-J, Wijesinghe SN, Shen H-W, Luo Z-L, Zhang J-Y, Sysouphanthong P, Thongklang N, Bao D-F, Aluthmuhandiram JVS, Abdollahzadeh J, Javadi A, Dovana F, Usman M, Khalid AN, Dissanayake AJ, Telagathoti A, Probst M, Peintner U, Garrido-Benavent I, Bóna L, Merényi Z, Boros L, Zoltán B, Stielow JB, Jiang N, Tian C-M, Shams E, Dehghanizadeh F, Pordel A, Javan-Nikkhah M, Denchev TT, Denchev CM, Kemler M, Begerow D, Deng C-Y, Harrower E, Bozorov T, Kholmuradova T, Gafforov Y, Abdurazakov A, Xu J-C, Mortimer PE, Ren G-C, Jeewon R, Maharachchikumbura SSN, Phukhamsakda C, Mapook A, Hyde KD. (2021) Fungal diversity notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 111(1): 1–335. 10.1007/s13225-021-00489-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher VVC, Hyde KD, Pointing SB, Reddy CA. (2004) Production of wood decay enzymes, mass loss and lignin solubilization in wood by marine ascomycetes and their anamorphs. Fungal Diversity 15: 1–14. [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. (2009) trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics (Oxford, England) 25(15): 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn L. (1999) A Method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91(3): 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Chaiwan N, Gomdola D, Wang S, Monkai J, Tibpromma S, Doilom M, Wanasinghe DN, Mortimer PE, Lumyong S, Hyde KD. (2021) An online database providing updated information of microfungi in the Greater Mekong Subregion. Mycosphere 12(1): 1513–1526. 10.5943/mycosphere/12/1/19 [DOI] [Google Scholar]
- Chapman EC. (1991) The Expansion of Rubber in Southern Yunnan, China. The Geographical Journal 157(1): 36–44. 10.2307/635142 [DOI] [Google Scholar]
- Crous PW, Carris LM, Giraldo A, Groenewald JZ, Hawksworth DL, Hemández-Restrepo M, Jaklitsch WM, Lebrun M-H, Schumacher RK, Stielow JB, Van Der Linde EJ, Vilcāne J, Voglmayr H, Wood AR. (2015) The Genera of Fungi – fixing the application of the type species of generic names – G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia. IMA Fungus 6(1): 163–198. 10.5598/imafungus.2015.06.01.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayarathne M, Maharachchikumbura S, Hyde K, Devadatha B, Jones G, Chomnunti P, Khongphinitbunjong K. (2020) Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere: Journal of Fungal Biology 7019(1): 1–188. 10.5943/mycosphere/11/1/1 [DOI] [Google Scholar]
- De Gruyter J, Woudenberg JHC, Aveskamp MM, Verkley GJM, Groenewald JZ, Crous PW. (2013) Redisposition of phoma-like anamorphs in Pleosporales. Studies in Mycology 75: 1–36. 10.3114/sim0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva N, Hyde KD, Lumyong S, Phillips A, Bhat D, Maharachchikumbura S, Thambugala K, Tennakoon D, Suwannarach N, Karunarathna SC. (2022) Morphology, phylogeny, host association and geography of fungi associated with plants of Annonaceae, Apocynaceae and Magnoliaceae. Mycosphere: Journal of Fungal Biology 13(1): 955–1076. 10.5943/mycosphere/13/1/12 [DOI] [Google Scholar]
- Dong W, Hyde KD, Jeewon R, Liao CF, Zhao HJ, Kularathnage ND, Li H, Yang YH, Pem D, Shu YX, Gafforov Y, Manawasinghe IS, Doilom M. (2023) Mycosphere notes 449–468: Saprobic and endophytic fungi in China, Thailand, and Uzbekistan. Mycosphere: Journal of Fungal Biology 14(1): 2208–2262. [Google Scholar]
- Du TY, Tibpromma S, Hyde KD, Dai D-Q, Mapook A, Zhang G-Q, Stephenson SL, Suwannarach N, Elgorban AM, Rajeshkumar KC, Maharachchikumbura SSN, Li Q, Karunarathna SC. (2024) The polyphasic approach reveals twelve novel ascomycota taxa from terrestrial agarwood-producing trees. [Not published]
- Feng B, Yang Z. (2018) Studies on diversity of higher fungi in Yunnan, southwestern China: A review. Plant Diversity 40(4): 165–171. 10.1016/j.pld.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J, Vogler JB. (2005) Land-Use and Land-Cover Change in Montane Mainland Southeast Asia. Environmental Management 36(3): 394–403. 10.1007/s00267-003-0288-7 [DOI] [PubMed] [Google Scholar]
- Gazis R, Chaverri P. (2010) Diversity of fungal endophytes in leaves and stems of wild rubber trees (Heveabrasiliensis) in Peru. Fungal Ecology 3(3): 240–254. 10.1016/j.funeco.2009.12.001 [DOI] [Google Scholar]
- Hongsanan S, Hyde KD, Phookamsak R, Wanasinghe DN, McKenzie EHC, Sarma VV, Lücking R, Boonmee S, Bhat JD, Liu N-G, Tennakoon DS, Pem D, Karunarathna A, Jiang S-H, Jones GEB, Phillips AJL, Manawasinghe IS, Tibpromma S, Jayasiri SC, Sandamali D, Jayawardena RS, Wijayawardene NN, Ekanayaka AH, Jeewon R, Lu Y-Z, Phukhamsakda C, Dissanayake AJ, Zeng X-Y, Luo Z-L, Tian Q, Thambugala KM, Dai D, Samarakoon MC, Chethana KWT, Ertz D, Doilom M, Liu J-K, Pérez-Ortega S, Suija A, Senwanna C, Wijesinghe SN, Niranjan M, Zhang S-N, Ariyawansa HA, Jiang H-B, Zhang J-F, Norphanphoun C, de Silva NI, Thiyagaraja V, Zhang H, Bezerra JDP, Miranda-González R, Aptroot A, Kashiwadani H, Harishchandra D, Sérusiaux E, Abeywickrama PD, Bao D-F, Devadatha B, Wu H-X, Moon KH, Gueidan C, Schumm F, Bundhun D, Mapook A, Monkai J, Bhunjun CS, Chomnunti P, Suetrong S, Chaiwan N, Dayarathne MC, Yang J, Rathnayaka AR, Xu J-C, Zheng J, Liu G, Feng Y, Xie N. (2020) Refined families of Dothideomycetes: Orders and families incertae sedis in Dothideomycetes. Fungal Diversity 105(1): 17–318. 10.1007/s13225-020-00462-6 [DOI] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics (Oxford, England) 17(8): 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hyde KD. (2022) The numbers of fungi. Fungal Diversity 114(1): 1. 10.1007/s13225-022-00507-y [DOI] [Google Scholar]
- Hyde KD, Jones EBG, Liu J-K, Ariyawansa H, Boehm E, Boonmee S, Braun U, Chomnunti P, Crous PW, Dai D-Q, Diederich P, Dissanayake A, Doilom M, Doveri F, Hongsanan S, Jayawardena R, Lawrey JD, Li Y-M, Liu Y-X, Lücking R, Monkai J, Muggia L, Nelsen MP, Pang K-L, Phookamsak R, Senanayake IC, Shearer CA, Suetrong S, Tanaka K, Thambugala KM, Wijayawardene NN, Wikee S, Wu H-X, Zhang Y, Aguirre-Hudson B, Alias SA, Aptroot A, Bahkali AH, Bezerra JL, Bhat DJ, Camporesi E, Chukeatirote E, Gueidan C, Hawksworth DL, Hirayama K, De Hoog S, Kang J-C, Knudsen K, Li W-J, Li X-H, Liu Z-Y, Mapook A, McKenzie EHC, Miller AN, Mortimer PE, Phillips AJL, Raja HA, Scheuer C, Schumm F, Taylor JE, Tian Q, Tibpromma S, Wanasinghe DN, Wang Y, Xu J-C, Yacharoen S, Yan J-Y, Zhang M. (2013) Families of Dothideomycetes. Fungal Diversity 63(1): 1–313. 10.1007/s13225-013-0263-4 [DOI] [Google Scholar]
- Hyde KD, Norphanphoun C, Abreu VP, Bazzicalupo A, Thilini Chethana KW, Clericuzio M, Dayarathne MC, Dissanayake AJ, Ekanayaka AH, He M-Q, Hongsanan S, Huang S-K, Jayasiri SC, Jayawardena RS, Karunarathna A, Konta S, Kušan I, Lee H, Li J, Lin C-G, Liu N-G, Lu Y-Z, Luo Z-L, Manawasinghe IS, Mapook A, Perera RH, Phookamsak R, Phukhamsakda C, Siedlecki I, Soares AM, Tennakoon DS, Tian Q, Tibpromma S, Wanasinghe DN, Xiao Y-P, Yang J, Zeng X-Y, Abdel-Aziz FA, Li W-J, Senanayake IC, Shang Q-J, Daranagama DA, de Silva NI, Thambugala KM, Abdel-Wahab MA, Bahkali AH, Berbee ML, Boonmee S, Bhat DJ, Bulgakov TS, Buyck B, Camporesi E, Castañeda-Ruiz RF, Chomnunti P, Doilom M, Dovana F, Gibertoni TB, Jadan M, Jeewon R, Jones EBG, Kang J-C, Karunarathna SC, Lim YW, Liu J-K, Liu Z-Y, Plautz Jr HL, Lumyong S, Maharachchikumbura SSN, Matočec N, McKenzie EHC, Mešić A, Miller D, Pawłowska J, Pereira OL, Promputtha I, Romero AI, Ryvarden L, Su H-Y, Suetrong S, Tkalčec Z, Vizzini A, Wen T-C, Wisitrassameewong K, Wrzosek M, Xu J-C, Zhao Q, Zhao R-L, Mortimer PE. (2017) Fungal diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Diversity 87(1): 1–235. 10.1007/s13225-017-0391-3 [DOI] [Google Scholar]
- Hyde K, Norphanphoun C, Hongde Y, Zhang J, Du T, Gao Y, Farias A, Gui H, He S, Yuke H, Cuijinyi L, Lu L, Hongli S, Tang X, Tian X-G. (2023) Mycosphere notes 387–412 – novel species of fungal taxa from around the world. Mycosphere: Journal of Fungal Biology 14(1): 663–744. 10.5943/mycosphere/14/1/8 [DOI] [Google Scholar]
- Index Fungorum (2024) Index Fungorum. https://www.indexfungorum.org [January 15, 2024]
- Jaklitsch WM, Voglmayr H. (2016) Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Studies in Mycology 85(1): 35–64. 10.1016/j.simyco.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasinghe CK, Fernando THPS. (1998) Growth at different temperatures and on fungicide amended media: Two characteristics to distinguish Colletotrichum species pathogenic to rubber. Mycopathologia 143(2): 93–95. 10.1023/A:1006958623733 [DOI] [PubMed] [Google Scholar]
- Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat J, Buyck B, Cai L, Dai Y-C, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R, Jones EBG, Bahkali AH, Karunarathna SC, Liu J-K, Luangsa-ard JJ, Lumbsch HT, Maharachchikumbura SSN, McKenzie EHC, Moncalvo J-M, Ghobad-Nejhad M, Nilsson H, Pang K-L, Pereira OL, Phillips AJL, Raspé O, Rollins AW, Romero AI, Etayo J, Selçuk F, Stephenson SL, Suetrong S, Taylor JE, Tsui CKM, Vizzini A, Abdel-Wahab MA, Wen T-C, Boonmee S, Dai DQ, Daranagama DA, Dissanayake AJ, Ekanayaka AH, Fryar SC, Hongsanan S, Jayawardena RS, Li W-J, Perera RH, Phookamsak R, de Silva NI, Thambugala KM, Tian Q, Wijayawardene NN, Zhao R-L, Zhao Q, Kang J-C, Promputtha I. (2015) The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Diversity 74(1): 3–18. 10.1007/s13225-015-0351-8 [DOI] [Google Scholar]
- Júnior BT da H, Macedo DM de, Barreto RW, Evans HC, Mattos CRR, Maffia LA, Mizubuti ESG. (2014) Erasing the Past: a new identity for the damoclean pathogen causing south American Leaf Blight of Rubber. PLOS ONE 9: e104750. 10.1371/journal.pone.0104750 [DOI] [PMC free article] [PubMed]
- Kang J-C, Hyde K, Kong RYC. (1999) Studies on Amphisphaeriales: The genera excluded from the Amphisphaeriaceae, Cainiaceae and Clypeosphaeriaceae. Fungal Diversity 2: 135–151. 10.1007/BF02464294 [DOI] [Google Scholar]
- Katoh K, Rozewicki J, Yamada K. (2019) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolařík M. (2018) New taxonomic combinations in endophytic representatives of the genus Nigrograna. Czech Mycology 70(2): 123–126. 10.33585/cmy.70202 [DOI] [Google Scholar]
- Kolařík M, Spakowicz DJ, Gazis R, Shaw J, Kubátová A, Nováková A, Chudíčková M, Forcina GC, Kang KW, Kelnarová I, Skaltsas D, Portero CE, Strobel SA, Narváez-Trujillo A. (2017) Biatriospora (Ascomycota: Pleosporales) is an ecologically diverse genus including facultative marine fungi and endophytes with biotechnological potential. Plant Systematics and Evolution 303(1): 35–50. 10.1007/s00606-016-1350-24 [DOI] [Google Scholar]
- Kularathnage N, Tennakoon D, Zhu X, Zhou J, Su B, Xie Y, Chen Q, Calabon M, Kirk P, Senanayake I, Doilom M, Xu B, Dong W, Song J. (2023) Reinstating Dyfrolomyces and introducing Melomastiapyriformis sp. nov. (Pleurotremataceae, Dyfrolomycetales) from Guangdong Province, China. Current Research in Environmental & Applied Mycology 13(1): 13. 10.5943/cream/13/1/16 [DOI] [Google Scholar]
- Li Z, Fox JM. (2012) Mapping rubber tree growth in mainland Southeast Asia using time-series MODIS 250m NDVI and statistical data. Applied Geography (Sevenoaks, England) 32(2): 420–432. 10.1016/j.apgeog.2011.06.018 [DOI] [Google Scholar]
- Li W-L, Maharachchikumbura SSN, Cheewangkoon R, Liu J-K. (2022) Reassessment of Dyfrolomyces and four new species of Melomastia from Olive (Oleaeuropaea) in Sichuan Province, China. Journal of Fungi (Basel, Switzerland) 8(1): 76. 10.3390/jof8010076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-L, Liang R-R, Dissanayake A, Liu J-K. (2023) Mycosphere Notes 413–448: Dothideomycetes associated with woody oil plants in China. Mycosphere: Journal of Fungal Biology 14(1): 1436–1529. 10.5943/mycosphere/14/1/16 [DOI] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16(12): 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Lu L, Karunarathna SC, Dai D, Jayawardena RS, Suwannarach N, Tibpromma S. (2022) Three new species of Nigrograna (Dothideomycetes, Pleosporales) associated with Arabica coffee from Yunnan Province, China. MycoKeys 94: 51–71. 10.3897/mycokeys.94.95751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharachchikumbura SSN, Hyde KD, Jones EBG, McKenzie EHC, Bhat JD, Dayarathne MC, Huang S-K, Norphanphoun C, Senanayake IC, Perera RH, Shang Q-J, Xiao Y, D’souza MJ, Hongsanan S, Jayawardena RS, Daranagama DA, Konta S, Goonasekara ID, Zhuang W-Y, Jeewon R, Phillips AJL, Abdel-Wahab MA, Al-Sadi AM, Bahkali AH, Boonmee S, Boonyuen N, Cheewangkoon R, Dissanayake AJ, Kang J, Li Q-R, Liu JK, Liu XZ, Liu Z-Y, Luangsa-ard JJ, Pang K-L, Phookamsak R, Promputtha I, Suetrong S, Stadler M, Wen T, Wijayawardene NN. (2016) Families of Sordariomycetes. Fungal Diversity 79(1): 1–317. 10.1007/s13225-016-0369-6 [DOI] [Google Scholar]
- Mahendran TR, Thottathil GP, Surendran A, Nagao H, Sudesh K. (2021) Biocontrol potential of Aspergillusterreus, endophytic fungus against Rigidoporusmicroporus and Corynesporacassiicola, pathogens of rubber tree. Archiv für Phytopathologie und Pflanzenschutz 54(15–16): 1014–1032. 10.1080/03235408.2021.1884952 [DOI] [Google Scholar]
- Mann CC. (2009) Addicted to Rubber. Science 325(5940): 564–566. 10.1126/science.325_564 [DOI] [PubMed] [Google Scholar]
- Mapook A, Hyde KD, McKenzie EHC, Jones EBG, Bhat DJ, Jeewon R, Stadler M, Samarakoon MC, Malaithong M, Tanunchai B, Buscot F, Wubet T, Purahong W. (2020) Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaenaodorata (Siam weed). Fungal Diversity 101(1): 1–175. 10.1007/s13225-020-00444-8 [DOI] [Google Scholar]
- Martin R, Gazis R, Skaltsas D, Chaverri P, Hibbett D. (2015) Unexpected diversity of basidiomycetous endophytes in sapwood and leaves of Hevea. Mycologia 107(2): 284–297. 10.3852/14-206 [DOI] [PubMed] [Google Scholar]
- Meeboon J, Takamatsu S. (2020) Hosts of asexual morph of Erysiphequercicola from Thailand. Tropical Plant Pathology 45(2): 122–135. 10.1007/s40858-019-00326-8 [DOI] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the Extreme to the campus and beyond. Association for Computing Machinery, New York, 1–8. 10.1145/2335755.2335836 [DOI]
- Mohammed CL, Rimbawanto A, Page DE. (2014) Management of basidiomycete root- and stem-rot diseases in oil palm, rubber and tropical hardwood plantation crops. Forest Pathology 44(6): 428–446. 10.1111/efp.12140 [DOI] [Google Scholar]
- Monkai J, Hyde KD, Xu J, Mortimer PE. (2017) Diversity and ecology of soil fungal communities in rubber plantations. Fungal Biology Reviews 31(1): 1–11. 10.1016/j.fbr.2016.08.003 [DOI] [Google Scholar]
- National Bureau of Statistics of China (2011) China Statistical Yearbook 2011. http://www.stats.gov.cn/tjsj/ndsj/2011/indexch.htm [Accessed 31 January 2015]
- Nizamani MM, Zhang Q, Zhang H, Wang Y. (2023) Checklist of the fungi associated with the rubber tree (Heveabrasiliensis). Journal of Fungal Biology 13(1): 439–488. [Google Scholar]
- Norphanphoun C, Jeewon R, Mckenzie EHC, Wen T-C, Camporesi E, Hyde KD. (2017) Taxonomic position of Melomastiaitalica sp. nov. and phylogenetic reappraisal of Dyfrolomycetales. Cryptogamie. Mycologie 38(4): 507–525. 10.7872/crym/v38.iss4.2017.507 [DOI] [Google Scholar]
- Nylander J. (2004) MrModeltest V2. Program Distributed by the Author. Bioinformatics (Oxford, England) 24: 581–583. 10.1093/bioinformatics/btm388 [DOI] [Google Scholar]
- Osono T, Takeda H. (2002) Nutrient contents of beech leaf litter decomposed by fungi in Basidiomycota and Ascomycota. Applied Forest Science 11(1): 7–11. 10.20660/applfor.11.1_7 [DOI] [Google Scholar]
- Osono T, Hobara S, Hishinuma T, Azuma J. (2011) Selective lignin decomposition and nitrogen mineralization in forest litter colonized by Clitocybe sp. European Journal of Soil Biology 47(2): 114–121. 10.1016/j.ejsobi.2010.12.002 [DOI] [Google Scholar]
- Phukhamsakda C, Nilsson RH, Bhunjun CS, de Farias ARG, Sun Y-R, Wijesinghe SN, Raza M, Bao D-F, Lu L, Tibpromma S, Dong W, Tennakoon DS, Tian X-G, Xiong Y-R, Karunarathna SC, Cai L, Luo Z-L, Wang Y, Manawasinghe IS, Camporesi E, Kirk PM, Promputtha I, Kuo C-H, Su H-Y, Doilom M, Li Y, Fu Y-P, Hyde KD. (2022) The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Diversity 114(1): 327–386. 10.1007/s13225-022-00502-3 [DOI] [Google Scholar]
- Punithalingam E. (1979) Sphaeropsidales in culture from humans. Nova Hedwigia 31: 119–158. [Google Scholar]
- Rambaut A. (2012) FigTree v1. 4.0. University of Oxford, Oxford, UK.
- Rojas-Jimenez K, Hernandez M, Blanco J, Vargas LD, Acosta-Vargas LG, Tamayo G. (2016) Richness of cultivable endophytic fungi along an altitudinal gradient in wet forests of Costa Rica. Fungal Ecology 20: 124–131. 10.1016/j.funeco.2015.12.006 [DOI] [Google Scholar]
- Roy BA. (2001) Patterns of association between crucifers and their flower-mimic pathogens: Host jumps are more common than coevolution or cospeciation. Evolution. International Journal of Organic Evolution 55(1): 41–53. 10.1111/j.0014-3820.2001.tb01271.x [DOI] [PubMed] [Google Scholar]
- Saccardo P. (1875) Conspectus generum pyrenomycetum italicorum additis speciebus fungorum Venetorum novis vel criticis, systemate carpologico dispositorum. Atti della Società Veneto-Trentina di Scienze Naturali 4: 77–100. [Google Scholar]
- Seephueak P, Petcharat V, Phongpaichit S. (2010) Fungi associated with leaf litter of para rubber (Heveabrasiliensis). Mycology 1(4): 213–227. 10.1080/21501203.2010.536594 [DOI] [Google Scholar]
- Seephueak P, Phongpaichit S, Hyde K, Petcharat V. (2011) Diversity of saprobic fungi on decaying branch litter of the rubber tree (Heveabrasiliensis). Mycosphere: Journal of Fungal Biology 2: 307–330. [Google Scholar]
- Senanayake IC, Rathnayaka AR, Marasinghe DS, Calabon MS, Gentekaki E, Lee HB, Hurdeal VG, Pem D, Dissanayake LS, Wijesinghe SN, Bundhun D, Nguyen TT, Goonasekara ID, Abeywickrama PD, Bhunjun CS, Jayawardena RS, Wanasinghe DN, Jeewon R, Bhat DJ, Xiang MM. (2020) Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere: Journal of Fungal Biology 11(1): 2678–2754. 10.5943/mycosphere/11/1/20 [DOI] [Google Scholar]
- Senwanna C, Hongsanan S, Phookamsak R, Tibpromma S, Cheewangkoon R, Hyde KD. (2019) Muyocopronheveae sp. nov. and M.dipterocarpi appears to have host-jumped to rubber. Mycological Progress 18(5): 741–752. 10.1007/s11557-019-01484-4 [DOI] [Google Scholar]
- Senwanna C, Mapook A, Samarakoon MC, Karunarathna A, Wang Y, Tang A, Haituk S, Suwannarach N, Hyde K, Cheewangkoon R. (2021) Ascomycetes on Para rubber (Heveabrasiliensis). Mycosphere: Journal of Fungal Biology 12(1): 1334–1512. 10.5943/mycosphere/12/1/18 [DOI] [Google Scholar]
- Spaulding P. (1961) Foreign Diseases of Forest Trees of the World: An Annotated List. U.S. Department of Agriculture, 372 pp.
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statistical Bureau of Yunnan Province. (2011) Yunnan Statistical Yearbook 2011. http://tongji.cnki.net/kns55/brief/result.aspx?stab=shuzhi&t=1&f=0&tt=%E6%A9%A1%E8%83%B6&areaname=%E8%A5%BF%E5%8F%8C%E7%89%88%E7%BA%B3%E5%82%A3%E6%97%8F%E8%87%AA%E6%B2%BB%E5%B7%9E [Accessed 21 September 2014]
- Tibpromma S, Hyde KD, Jeewon R, Maharachchikumbura SSN, Liu J-K, Bhat DJ, Jones EBG, McKenzie EHC, Camporesi E, Bulgakov TS, Doilom M, de Azevedo Santiago ALCM, Das K, Manimohan P, Gibertoni TB, Lim YW, Ekanayaka AH, Thongbai B, Lee HB, Yang J-B, Kirk PM, Sysouphanthong P, Singh SK, Boonmee S, Dong W, Raj KNA, Latha KPD, Phookamsak R, Phukhamsakda C, Konta S, Jayasiri SC, Norphanphoun C, Tennakoon DS, Li J, Dayarathne MC, Perera RH, Xiao Y, Wanasinghe DN, Senanayake IC, Goonasekara ID, de Silva NI, Mapook A, Jayawardena RS, Dissanayake AJ, Manawasinghe IS, Chethana KWT, Luo Z-L, Hapuarachchi KK, Baghela A, Soares AM, Vizzini A, Meiras-Ottoni A, Mešić A, Dutta AK, de Souza CAF, Richter C, Lin C-G, Chakrabarty D, Daranagama DA, Lima DX, Chakraborty D, Ercole E, Wu F, Simonini G, Vasquez G, da Silva GA, Plautz Jr HL, Ariyawansa HA, Lee H, Kušan I, Song J, Sun J, Karmakar J, Hu K, Semwal KC, Thambugala KM, Voigt K, Acharya K, Rajeshkumar KC, Ryvarden L, Jadan M, Hosen MI, Mikšík M, Samarakoon MC, Wijayawardene NN, Kim NK, Matočec N, Singh PN, Tian Q, Bhatt RP, de Oliveira RJV, Tulloss RE, Aamir S, Kaewchai S, Marathe SD, Khan S, Hongsanan S, Adhikari S, Mehmood T, Bandyopadhyay TK, Svetasheva TY, Nguyen TTT, Antonín V, Li W-J, Wang Y, Indoliya Y, Tkalčec Z, Elgorban AM, Bahkali AH, Tang AMC, Su H-Y, Zhang H, Promputtha I, Luangsa-ard J, Xu J, Yan J, Ji-Chuan K, Stadler M, Mortimer PE, Chomnunti P, Zhao Q, Phillips AJL, Nontachaiyapoom S, Wen T-C, Karunarathna SC. (2017) Fungal diversity notes 491–602: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 83(1): 1–261. 10.1007/s13225-017-0378-0 [DOI] [Google Scholar]
- Tibpromma S, Hyde KD, McKenzie EHC, Bhat DJ, Phillips AJL, Wanasinghe DN, Samarakoon MC, Jayawardena RS, Dissanayake AJ, Tennakoon DS, Doilom M, Phookamsak R, Tang AMC, Xu J, Mortimer PE, Promputtha I, Maharachchikumbura SSN, Khan S, Karunarathna SC. (2018) Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Diversity 93(1): 1–160. 10.1007/s13225-018-0408-6 [DOI] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang X, Tao L, Bi B, Niu G, Han X, Deng J. (2015) The potential development value of rubber seed. Journal of Yunnan Agricultural University 30: 642–647. [Google Scholar]
- Watson W. (1929) The Classification of Lichens. The New Phytologist 28(1): 1–36. 10.1111/j.1469-8137.1929.tb06745.x [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR Protocols.Elsevier, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wijayawardene NN, Hyde KD, Dai D-Q, Sánchez-García M, Goto B, Saxena R, Erdoğdu M, Selçuk F, Rajeshkumar K, Aptroot A, Błaszkowski J, Boonyuen N, Da Silva G, De Souza F, Dong W, Ertz D, Haelewaters D, Jones E, Karunarathna S, Kirk P, Kukwa M, Kumla J, Leontyev D, Lumbsch H, Maharachchikumbura S, Marguno F, Martínez-Rodríguez P, Mešić A, Monteiro J, Oehl F, Pawłowska J, Pem D, Pfliegler W, Phillips A, Pošta A, He M, Li J, Raza M, Sruthi O, Suetrong S, Suwannarach N, Tedersoo L, Thiyagaraja V, Tibpromma S, Tkalčec Z, Tokarev Y, Wanasinghe D, Wijesundara D, Wimalaseana S, Madrid H, Zhang G, Gao Y, Sánchez-Castro I, Tang L, Stadler M, Yurkov A, Thines M. (2022) Outline of Fungi and fungus-like taxa – 2021. Mycosphere: Journal of Fungal Biology 13(1): 53–453. 10.5943/mycosphere/13/1/2 [DOI] [Google Scholar]
- Xu R-F, Thiyagaraja V, Dai D-Q, Karunarathna SC, Tibpromma S. (2022a) Additions to Fitzroyomyces (Stictidaceae, Ascomycota) from Yunnan Province, China. Phytotaxa 548(2): 253–266. 10.11646/phytotaxa.548.2.8 [DOI] [Google Scholar]
- Xu R-F, Hyde K, Karunarathna S, Xu J-C, Mortimer P, Tibpromma S. (2022b) Morphology and multi-gene phylogeny reveal a new fungal genus and species from Heveabrasiliensis latex in Yunnan, China. Phytotaxa 530(1): 65–76. 10.11646/phytotaxa.530.1.5 [DOI] [Google Scholar]
- Xu R-F, Phukhamsakda C, Dai D-Q, Karunarathna SC, Tibpromma S. (2023) Kirschsteiniotheliaxishuangbannaensis sp. nov. from pará rubber (Heveabrasiliensis) in Yunnan, China. Current Research in Environmental & Applied Mycology 13(1): 34–56. 10.5943/cream/13/1/3 [DOI] [Google Scholar]
- Zhang J, Liu J-K, Hyde KD, Chen Y-Y, Liu Y-X, Liu Z-Y. (2017) Two new species of Dyfrolomyces (Dyfrolomycetaceae, Dothideomycetes) from karst landforms. Phytotaxa 313: 267. 10.11646/phytotaxa.313.3.4 [DOI]
- Ziegler AD, Fox JM, Xu J. (2009) The Rubber Juggernaut. Science 324(5930): 1024–1025. 10.1126/science.1173833 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available in the main text.