Abstract
The intranuclear assembly of herpesvirus subviral particles remains an incompletely understood process. Previous studies have described the nuclear localization of capsid and tegument proteins as well as intranuclear tegumentation of capsid-like particles. The temporally and spatially regulated replication of viral DNA suggests that assembly may also be regulated by compartmentalization of structural proteins. We have investigated the intranuclear location of several structural and nonstructural proteins of human cytomegalovirus (HCMV). Tegument components including pp65 (ppUL83) and ppUL69 and capsid components including the major capsid protein (pUL86) and the small capsid protein (pUL48/49) were retained within the nuclear matrix (NM), whereas the immediate-early regulatory proteins IE-1 and IE-2 were present in the soluble nuclear fraction. The association of pp65 with the NM resisted washes with 1 M guanidine hydrochloride, and direct binding to the NM could be demonstrated by far-Western blotting. Furthermore, pp65 exhibited accumulation along the nuclear periphery and in far-Western analysis bound to proteins which comigrated with proteins of the size of nuclear lamins. A direct interaction between pp65 and lamins was demonstrated by coprecipitation of lamins in immune complexes containing pp65. Together, our findings provide evidence that major virion structural proteins localized to a nuclear compartment, the NM, during permissive infection of human fibroblasts.
Recent studies have indicated that the human cytomegalovirus (HCMV) virion is composed of a larger number of proteins than previously thought, suggesting that assembly of the infectious particle is extraordinarily complex (3). The description of the architecture of the virion has been simplified to include three distinct structures: the capsid, the envelope, and a poorly characterized region between the capsid and envelope termed the tegument (54, 55). The protein composition of the HCMV tegument has been incompletely defined, but it is thought to be composed of a large number of phosphoproteins (3, 46). Although there is general agreement that the capsid is assembled in the nucleus, considerable controversy continues to surround the identity of the cellular site of envelopment of herpesviruses (4, 19, 30, 53). The assembly pathway of the tegument region remains even less well understood. The distribution of protein components of the HCMV tegument suggests that assembly of this virion structure takes place in both the nucleus and the cytoplasm. Tegument proteins encoded by UL82 (pp71), UL83 (pp65), and UL69 open reading frames appear to localize in the nucleus, while the tegument protein pp28 (ppUL99) is detected in extranuclear compartments of infected cells (24, 26, 38, 58, 61). HCMV pp150 (UL32) has been reported to demonstrate both a nuclear and a cytoplasmic distribution (34), although studies in our laboratory have suggested that pp150 is predominantly a cytoplasmic protein (58). This organization of tegument components suggests that these proteins are incorporated into the virion in an ordered manner and, furthermore, that understanding tegument morphogenesis could provide insight into the pathways of virion assembly and nuclear egress.
To further describe virion maturation, we have begun an investigation of the pathway in which the tegument is assembled around the nucleocapsid. Recent studies of herpes simplex virus (HSV) together with previous reports describing replication centers in the nuclei of infected cells have suggested that herpesviruses not only employ complex regulatory controls of transcription and replication (18, 41, 42, 56) but possibly regulate particle assembly by localizing structural proteins into discrete subnuclear compartments (63, 64). Recent studies by Ward and coworkers have divided the nucleus of HSV-infected cells into different compartments called assemblons based on localization of known proteins of HSV (64). These included compartments for replication and subviral particle formation (64). We have begun a series of experiments to further define the assembly and nuclear egress of HCMV. Specifically, we have examined the distribution of several tegument proteins within the nuclear matrix of infected cells in order to define spatial relationships and potential colocalization of structural proteins late in infection. This compartment of the nucleus was examined initially because it has been defined biochemically and thus represented a nuclear compartment which could be analyzed by both biochemical and imaging techniques.
The nuclear matrix is a proteinaceous network which is tightly associated with the inner nuclear membrane. In many cell systems, the nuclear matrix has been found to be the site of active transcription and replication of cellular DNA (6, 28, 35, 48, 49, 62). Proteins involved in these processes as well as those with regulatory roles in cell division localize to this nuclear scaffold (8, 15, 22, 32, 39, 40, 43, 52). Among DNA viruses, there are numerous examples of viral gene products which associate with the nuclear matrix or nuclear matrix structures (2, 10, 14, 16, 21, 25, 31, 37, 44, 51, 60, 66). This association may serve to compartmentalize products necessary for efficient transcription and replication of the viral genome or to sequester components involved in virion maturation (5, 7, 25, 37, 44, 51, 66). The role of the nuclear matrix in the replicative cycle of herpesviruses, including HCMV, has not been extensively studied. In this report we have described the binding of several HCMV virion structural proteins including pp65 (ppUL83), ppUL69, and the major capsid protein (MCP; pUL86) to the nuclear matrix of HCMV-infected cells. The accumulation of virion components on the nuclear matrix late in infection suggested that this compartment was a potential staging site for virion structural proteins prior to their assembly into subviral particles.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Human foreskin fibroblasts (HF) and monkey BSC-1 cells were maintained in medium 199 with 5% newborn calf serum and antibiotics at 37°C. For in situ nuclear matrix extractions, HF were grown on glass coverslips at 37°C in 5% CO2. Cells were infected with the AD169 strain of HCMV at a multiplicity of infection of 0.1 to 1. The human epidermoid carcinoma cell line HEp-2 and monkey Cos7 cells were maintained at 37°C in 5% CO2 in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum and antibiotics.
The recombinant vaccinia viruses vv-Eco V gB and vv-pp65 were propagated in BSC-1 cells. The construction and characterization of these recombinant vaccinia viruses have been reported elsewhere (9, 13). The vv-Eco V gB construct differs from the vv-gB recombinant previously reported in that it contains the gB gene of AD169 truncated at nucleotide 1950 and the corresponding gB protein terminates at amino acid 650. This protein lacks the carboxy terminus including the transmembrane region and represents a secreted form of HCMV gB. The pp65-green fluorescent protein fusion was constructed by using the EGFP-N2 plasmid (Clontech, Palo Alto, Calif.). A BamHI site was generated at the 5′ end of the pp65 genomic sequence by PCR using the primer 5′ TTTTTTGGATCCATGGAGTCGCGCGGT 3′. The product was fused in frame into the EGFP-N2 vector 3′ to the enhanced green fluorescence protein (EGFP) coding sequence.
HCMV proteins were detected with monoclonal antibodies (MAbs) previously described (1, 12, 50, 65). The MAbs used in this study include those with specific reactivity to IE-1 (p63-27), IE-2 (IE-2-9-5), pp65 (28-19, 65-8, 28-103, 28-77), MCP (28-4), the small capsid protein (SCP) (11-2-23), gB (7-17), UL69 (UL69), UL44 (28-21), and pp28 (41-18). The guinea pig polyclonal serum recognizing pp65 was generated by repeated immunization of guinea pigs with pp65 purified from bacteria. The previously characterized rabbit polyclonal antibody 237 against lamins a, b, and c was a generous gift from Robert Goldman (Department of Cell and Molecular Biology, Northwestern University School of Medicine) (29). The rabbit polyclonal sera against lamins a and c and against lamin b (17) were a generous gift from Nilabh Chaudhary (RPI, Boulder, Colo.). The MAb against lamin B1, NA12, was purchased from Oncogene Sciences (Boston, Mass.). Fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG) and FITC-conjugated goat anti-guinea pig IgG antibodies were obtained from Cappell Laboratories (Raleigh, N.C.). Texas red-conjugated goat anti-rabbit IgG antibody was purchased from Southern Biotechnology Associates (Birmingham, Ala.).
Preparation of nuclear matrix.
Nuclear matrix fractions were prepared by a method similar to that described by Mirkovitch et al. (45). AD169-infected HF, vv-pp65 and vv-Eco V gB-infected BSC-1 cells, or uninfected HEp-2 cells were fractionated by extraction in 0.1 or 0.2% Nonidet P-40 (NP-40) in phosphate-buffered saline (PBS; 137 mM NaCl, 8.1 mM NaH2PO4 · 12H2O, 2.7 mM KCl, 1.8 mM KH2PO4 [pH 7.4]) to yield crude nuclei. Nuclei were resuspended in 1 ml of digestion buffer (20 mM Tris-HCl [pH 7.4], 20 mM KCl, 70 mM NaCl, 10 mM MgCl2, 0.05 mM spermine, 0.125 mM spermidine) with 1 mM phenylmethylsulfonyl fluoride (PMSF) and subjected to digestion with DNase I (0.05 mg/ml) for 15 min at room temperature. Nuclei were then resuspended in digestion buffer with 0.1% digitonin and incubated at room temperature for 10 min. The nuclear material was pelleted and then extracted in high-salt buffer (2 M NaCl, 20 mM HEPES [pH 7.4], 20 mM EDTA) on ice for 5 min. The nuclear material was then pelleted and washed twice in digestion buffer and finally resuspended in sodium dodecyl sulfate (SDS) sample buffer with 5% 2-mercaptoethanol or washed three times in 1 M guanidine hydrochloride in digestion buffer (2 min per wash) before addition of sample buffer (25). For a typical experiment, six 150-cm2 flasks of AD169-infected HF were harvested. For quantitative Western blots, AD169-infected HF cells from 14 150-cm2 flasks were fractionated and protein content was determined by using bicinchoninic acid reagent (Pierce, Rockford, Ill.). The radioactive signal was quantitated on a Molecular Dynamics PhosphorImager. For far-Western blots, nuclear matrix fractions were isolated from four 150-cm2 flasks of HEp-2 cells.
Nuclear matrix extracts for immunoprecipitation were prepared by a method similar to that described by Fey and Penman (23). Briefly, the nuclear matrix material extracted from AD169-infected cells was solubilized in disassembly buffer (8 M urea, 20 mM morpholineethanesulfonic acid [pH 6.6], 1 mM EGTA, 1 mM PMSF, 0.1 mM MgCl2, 1% 2-mercaptoethanol) for 16 h at 4°C. The insoluble material was removed by centrifugation. The urea was removed from the supernatant by step dialysis against Tris-buffered saline (TBS; 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 7.4) at 4°C, and the insoluble material was removed at each step. A mixture of three pp65-specific MAbs (28-19, 28-103, and 65-8) was used to precipitate the dialyzed extract. Immunoprecipitates were collected on protein A-agarose and washed extensively in radioimmunoprecipitation assay buffer (1% NP-40, 1% deoxycholate, and 0.2% SDS in TBS [pH 7.4]). The samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred to nitrocellulose for Western blotting as described previously (12).
For in situ extraction of AD169-infected HF, a different method was used for nuclear matrix preparation. This method was similar to that described by He et al. (33). Infected monolayers grown on glass coverslips were treated with digestion buffer (described above) with 0.1 mM PMSF and 0.5% NP-40 for 3 to 5 min on ice. The monolayers were then treated with digestion buffer containing DNase I (0.05 mg/ml) for 15 min at room temperature. Chromatin was removed by washing monolayers three times with 0.25 M ammonium sulfate in digestion buffer (pH 7.2) at room temperature, 10 min per wash. Cells were then extracted with high-salt buffer (described above) for 5 min on ice. Cell cytoskeletons were carefully rinsed with digestion buffer and then fixed in 2.5% paraformaldehyde in PBS for 20 min at room temperature.
Fluorescence microscopy.
Virus-infected HF grown on glass coverslips were fixed in 2.5% paraformaldehyde in PBS and permeabilized with 0.2% Triton X-100 in PBS for 5 min. After rinsing, cells and extracted cytoskeletons were blocked with 30% goat serum in PBS for 30 min at 37°C. Coverslips were incubated with primary antibody with 1% goat serum for 1 h at 37°C. Coverslips were washed three times in PBS, 5 min per wash, and then incubated with FITC-conjugated and/or Texas red-conjugated secondary antibody for 1 h at 37°C. After washing, coverslips were refixed with 0.5% paraformaldehyde in PBS for 10 min. After rinsing in PBS, coverslips were mounted with SlowFade antifade reagent (Molecular Probes, Eugene, Oreg.), sealed with fingernail polish, and viewed on a Leitz Diavert fluorescence microscope or a Zeiss confocal microscope.
Cos7 cells grown on coverslips were transfected by either the Lipofectin (57) or calcium phosphate (5′ Prime-3′ Prime, Boulder, Colo.) protocol. Cells were transfected with a modified pcDNA3 vector (Invitrogen, San Diego, Calif.) containing the pp65 genomic sequence or a vector encoding a green fluorescent protein-pp65 fusion protein (Clontech). Transfected cells were fixed 36 to 48 h posttransfection, and cells expressing pp65 were reacted with MAb 28-19 followed by FITC-conjugated goat anti-mouse IgG antibody as described above.
Far-Western blots.
pp65 was purified from Escherichia coli transformed with the plasmid trc/hisA (Invitrogen), which contained the complete pp65 open reading frame. After induction with isopropylthio-β-d-galactopyranoside, cultures were collected and the bacterial pellet was resuspended in denaturing lysis buffer (20 mM Tris-HCl, 100 mM NaCl, 8 M urea [pH 8.0]). The suspension was sonicated on ice until translucent. Insoluble material was removed by centrifugation at 10,000 × g for 10 min. The lysate was incubated with Talon metal affinity resin (Clontech) at room temperature with gentle rocking for 30 min. The resin was collected by centrifugation and washed with denaturing lysis buffer three times, 10 min per wash. pp65 was eluted by incubating the resin with denaturing lysis buffer containing 75 mM imidazole four times, 10 min per elution. Fractions were pooled, and the urea was removed by dialysis against TBS. Protein concentration was determined by using bicinchoninic acid reagent (Pierce).
Nuclear matrix samples from HEp-2 cells were subjected to SDS-PAGE as previously described (12). Proteins were blotted to nitrocellulose, and filters were blocked in 5% dry milk in PBS. Strips were incubated with 150 to 200 μg of pp65 per strip in 5% milk overnight at room temperature with gentle rocking (2). The strip was washed in PBS three times, 10 min per wash; following the final wash, it was incubated with the pp65-specific MAb 28-19 for 4 h at 37°C and then washed in PBS as described above. The strip was then incubated with a rabbit anti-mouse IgG secondary antibody for 1 h at 37°C. The filter was washed again and then incubated with 125I-protein A for 30 min at 37°C. After washing, filters were dried and mounted, and bound antibody was detected by autoradiography. For Western blot analysis, nuclear matrix samples were separated electrophoretically and then blotted. Strips were processed as previously described (12).
RESULTS
Tegument and capsid protein components of HCMV are associated with the nuclear matrix.
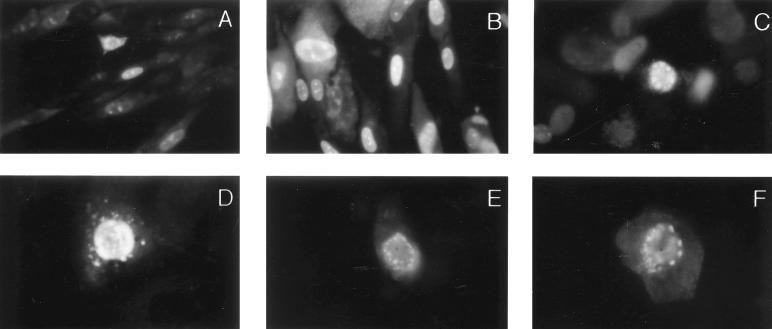
Phosphoprotein 65 (pp65) is one of the most abundant protein components of extracellular virions and dense bodies. Shortly after HCMV infection of human fibroblasts, pp65 can be detected in the nucleus of infected cells, suggesting that it is actively transported to this cellular compartment (26, 61). Immunofluorescence microscopy of AD169-infected HF as well as of Cos7 cells transfected with a plasmid containing the pp65 genomic sequence revealed the accumulation of pp65 into discrete structures in the nucleus (Fig. 1A to D). Similarly, in studies utilizing a pp65-green fluorescent protein fusion, we observed compartmentalization of the protein as well as focal accumulation along the periphery of the nucleus (Fig. 1E and F). Together with previously reported findings which indicated that deletion mutants of pp65 lacking the carboxy-terminal nuclear targeting signals continued to accumulate in the nucleus (26, 61), these results suggested that pp65 might contain additional domains which could mediate nuclear retention by targeting the protein to a specific nuclear structure.
FIG. 1.
HCMV pp65 is a nuclear protein that is detected in subnuclear structures. HF were grown on coverslips and infected with AD169. Three to five days postinfection (100% cytopathic effect), cells were fixed and stained in immunofluorescence assays with the pp65-specific MAb 65-8 (A), MAb 28-19 (B), or polyclonal guinea pig serum against bacterially expressed pp65 (C). Cos7 cells transfected with a vector expressing pp65 (D) or an EGFP-pp65 fusion protein (E and F) were fixed and stained with MAb 28-19 (D only). Magnifications: (A to C) ×400; (D to F) ×1,000.
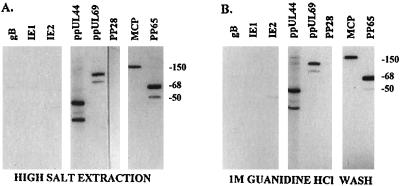
To characterize the interaction of pp65 with subnuclear structures, we investigated the binding of the protein to the nuclear matrix of HCMV-infected HF. Extraction of isolated nuclei with 2 M NaCl resulted in an insoluble pellet containing pp65, as demonstrated by Western blotting using the pp65-specific MAb 28-19 (Fig. 2A). Because pp65 was an abundant structural protein, we examined the possibility that other virion components were also present in the nuclear matrix. Western blot analysis of infected cells revealed that several virion structural proteins were associated with the nuclear matrix. Structural components including the MCP (pUL86) and the tegument protein ppUL69 were also retained in the nuclear matrix fraction. In addition, the nonstructural viral polymerase accessory protein ppUL44 and previously described lower-molecular-weight forms of this protein were associated with the nuclear matrix fraction of infected cells. The binding of these proteins was stable and resisted three washes with 1 M guanidine hydrochloride except for an observable decrease in the 50- to 52-kDa forms of pp65 (Fig. 2B). The interaction of these proteins was also specific, as other virus-encoded proteins previously shown to localize in the nucleus of infected cells, including the 72-kDa IE-1 and 86-kDa IE-2, were not detected in the nuclear matrix (Fig. 2). In addition, two cytoplasmic virion proteins, gB (gpUL55) and pp28 (ppUL99), were not detected in the nuclear matrix of infected cells (Fig. 2).
FIG. 2.
HCMV-encoded structural and nonstructural proteins are retained in the nuclear matrix of HCMV AD169-infected human fibroblasts. Nuclei from AD169-infected HF were isolated by treatment with nonionic detergent and then treated with DNase and high salt to remove soluble components from the nuclear matrix fraction. (A) Nuclear matrix-containing filters were probed with MAbs specific for gB, IE1, IE2, ppUL44, ppUL69, pp28, MCP, and pp65. (B) Filters containing proteins from nuclear matrix fractions washed with 1 M guanidine hydrochloride prior to electrophoretic separation were probed with the antibodies listed above. Sizes are indicated in kilodaltons.
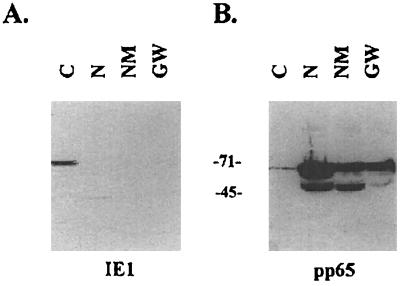
The presence of structural components of the capsid and tegument on the nuclear matrix suggested that this nuclear structure was a potential assembly site for subviral structures. Furthermore, the strength of the association between viral and cellular proteins of the nuclear matrix as reflected by their binding following washes with 1 M guanidine hydrochloride suggested a direct interaction between individual virion proteins and components of the nuclear matrix. Alternatively, individual virion structural proteins could be tethered to the matrix through interactions with other virus-encoded proteins. To gauge the relative strength of these protein-protein interactions, we measured the quantity of pp65 distributed between the different pools collected during the fractionation procedure (Fig. 3). As shown in Fig. 3B and Table 1, only a small amount of the total cellular pp65 can be detected in the soluble fraction isolated by treatment of cells with 0.2% NP-40. This fraction contained the cytoplasm and nuclear proteins released by this treatment, as demonstrated by the presence of IE-1 in Fig. 3A. The nuclear and nuclear matrix fractions contained more pp65 per microgram of protein than the soluble fraction (Fig. 3B; Table 1). In addition, quantitation of signal intensity showed that the relative amount of pp65 was not greatly reduced by the fractionation procedure (Fig. 3B, lanes 2 to 4; Table 1). In fact, there was a relative enrichment of pp65 on the nuclear matrix and in the guanidine hydrochloride-washed pellet (Table 1). Of interest was the apparent loss of the 50- and 52-kDa forms of pp65 during the guanidine hydrochloride wash (Fig. 3; Table 1), suggesting that these products were not as strongly retained as the full-length protein. Consistent with our findings shown in Fig. 2, the IE-1 protein was not enriched in the nuclear matrix fraction and was contained in the soluble fraction (Fig. 3).
FIG. 3.
Quantitative Western blots of nuclear matrix preparations from AD169-infected HF. HCMV-infected cells were fractionated, and protein content in each fraction was determined as described in Materials and Methods. Twenty-five micrograms of each of the fractions was loaded into the lanes (C, cytoplasm, soluble protein; N, detergent-treated nuclei; NM, nuclear matrix pellet; GW, guanidine hydrochloride-washed NM) and transferred to nitrocellulose filters. Filters were reacted with MAb p63-27 against IE-1 (A) or MAb 28-19 against pp65 (B) and processed for autoradiography. Counts for each fraction were determined on a PhosphorImager and were as follows: IE-1, 449,874.4, 13,951.6, 9,332.5, and 9,514.3 for C, N, NM, and GW fractions, respectively; pp65 (68 kDa), 239,720.7, 2,048,547.0, 1,206,468.7, and 1,172,115.9 for C, N, NM, and GW fractions, respectively; pp65 (50-kDa form), 33,155.2, 1,003,560.2, 774,609.0, and 188,991.4 for C, N, NM, and GW fractions, respectively. Sizes are indicated in kilodaltons.
TABLE 1.
Enrichment of pp65 on the nuclear matrix
| Fractiona | % Total cellular proteinb | % of protein in fractionc
|
||
|---|---|---|---|---|
| IE-1 | pp65 (68 kDa) | pp65 (50 kDa) | ||
| Cytoplasm/soluble protein | 86.3 | 93.2 (1.08d) | 5.1 (0.06) | 1.7 (0.02) |
| Detergent-treated nuclei | 8.9 | 2.9 (0.33) | 43.9 (4.93) | 50.2 (5.64) |
| Nuclear matrix | 3.0 | 1.9 (0.63) | 25.9 (8.63) | 38.7 (12.94) |
| Guanidine-HCl-washed nuclear matrix | 1.8 | 2.0 (1.11) | 25.1 (13.94) | 9.4 (5.22) |
HCMV-infected HF were fractionated into the indicated cellular fractions as described in Materials and Methods.
The amount of total protein represented in each fraction was determined as described in Materials and Methods, and the percentage of total cellular protein was calculated as follows: % = (total protein in fraction/total cellular protein) × 100.
The amount of IE-1, pp65 (68 kDa) or pp65 (50 kDa) in each fraction was determined by phosphorimaging. The results are presented as the percentage of total IE-1, pp65 (68 kDa), or pp65 (50 kDa) in each fraction as calculated by the following formula: % = (counts in fraction/total counts for protein) × 100.
The specific enrichment of each protein in a specific fraction, determined by the following formula: enrichment = % of protein determined in footnote c/% of total cellular protein determined in footnote b.
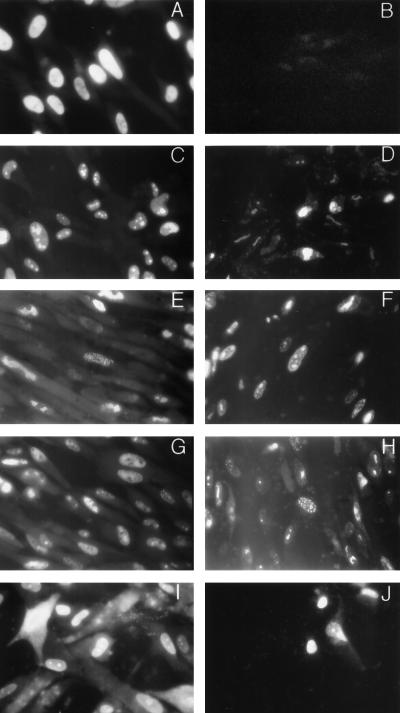
To further examine the compartmentalization of virion structural components on the nuclear matrix, we performed immunofluorescence assays of in situ-extracted, HCMV-infected HF. These assays were also used to estimate the quantity of protein removed during the extraction procedure. As shown in Fig. 4A, the IE-1 protein was readily detected in fixed cells but not in extracted cytoskeletal frameworks following treatment with high salt and DNase (Fig. 4B). The absence of IE-1 in in situ-extracted cells confirmed the results of the Western blot analysis which suggested that a significant amount of IE-1 was not retained on the nuclear matrix. In contrast, ppUL44, MCP, ppUL69, and pp65 were retained in the insoluble nuclear matrix fraction (Fig. 4D, F, H, and J, respectively). Furthermore, the pattern of immunofluorescence suggested that these proteins were localized to subnuclear structures and not evenly distributed throughout the nucleus. In addition, we observed the retention in the nuclear matrix of the minor capsid protein (pUL85) and the small capsid protein, p12 (pUL48/49) (data not shown). Note the lack of reactivity of primary and secondary antibodies with uninfected cells which were present in these preparations demonstrating the specificity of these MAbs.
FIG. 4.
Immunofluorescence assays of in situ-extracted, AD169-infected HF. HF grown on glass coverslips were infected with HCMV AD169 5 days prior to harvesting. Infected cells were untreated (A, C, E, G, and I) or extracted with detergent, DNase, and high salt (B, D, F, H, and J) before fixation with 2.5% paraformaldehyde. Coverslips were then reacted with MAbs specific for IE-1 (A and B), ppUL44 (C and D), MCP (E and F), ppUL69 (G and H), or pp65 (I and J). Antibody binding was detected with FITC-conjugated goat anti-mouse IgG antibody and recorded by conventional fluorescence microscopy. Magnification for all frames is ×348.
The tegument phosphoprotein pp65 binds to the nuclear matrix.
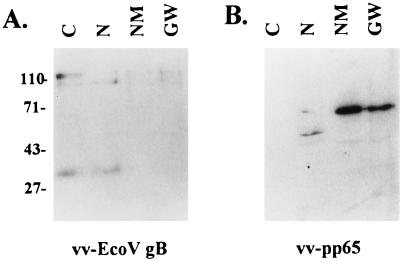
The interaction between pp65 and the nuclear matrix could be explained by either a direct binding of pp65 to a component of the nuclear matrix or an indirect binding through an association with another virus-encoded protein and/or DNase-resistant nucleic acid which was associated with a protein component of the nuclear matrix. We initially addressed this question by examining the association of pp65 with the nuclear matrix of cells infected with a recombinant vaccinia virus expressing pp65. Recombinant pp65 expressed in the absence of other HCMV-encoded proteins was enriched in the nuclear matrix and 1 M guanidine hydrochloride-washed fractions (Fig. 5). As a control for the extraction procedure, similar experiments were performed with cells expressing a truncated form of HCMV gB. We could not detect enrichment of the 130-kDa precursor form of this gB or of its 30-kDa cleavage product in the nuclear matrix or guanidine hydrochloride-washed pellets. These results indicated that pp65 was retained in the nuclear matrix of cells in the absence of other viral proteins.
FIG. 5.
pp65 expressed by recombinant vaccinia virus vv-pp65 is retained in the nuclear matrix of infected BSC-1 monkey cells. Cytoplasmic (C), nuclear (N), nuclear matrix (NM), and guanidine-washed nuclear matrix (GW) fractions were prepared from recombinant vaccinia virus vv-Eco V gb (A)- or vv-pp65 (B)-infected BSC-1 cells as described in Materials and Methods. The samples (10 μg per lane) were analyzed by Western blotting using a gB-specific or pp65-specific MAb and developed with 125I-protein A. Sizes are indicated in kilodaltons.
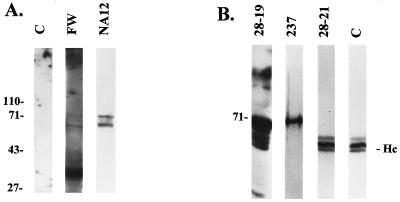
To directly investigate the specificity of the protein-protein interactions between pp65 and nuclear matrix components, we performed far-Western blotting with nuclear matrix material derived from the human cell line HEp-2 as the substrate for binding. A pp65 fusion protein containing a His6 tag at the amino terminus was purified from E. coli and used to probe the nitrocellulose membrane containing electrophoretically separated nuclear matrix proteins. Binding of pp65 to the nuclear matrix proteins was then detected with the pp65-specific MAb 28-19. The results of this experiment indicated a direct interaction between pp65 and at least three cellular proteins which migrated between 50 and 70 kDa and two other proteins which migrated at approximately 30 kDa (Fig. 6A, lane FW). Interestingly, the 50- to 70-kDa bands were of the approximate molecular size of nuclear lamins, which together represent major protein constituents of the nuclear matrix.
FIG. 6.
pp65 binds to a nuclear matrix protein which comigrates with lamin B1 in vitro. (A) Nuclear matrix material was isolated from the human carcinoma cell line HEp-2 as described in Materials and Methods. Proteins were electrophoretically separated and then blotted onto nitrocellulose. Filters were then reacted with pp65-specific MAb 28-19 (lane C), purified pp65 followed by anti-pp65 MAb 28-19 (lane FW), or anti-lamin B1 MAb NA12 (lane NA12). Antibody binding was detected by addition of anti-mouse IgG antibody followed by 125I-protein A and autoradiography. Migration of molecular mass markers is shown in kilodaltons at the left. (B) Coprecipitation of pp65 and lamins from soluble nuclear matrix extracts. Soluble nuclear matrix extracts prepared from AD169-infected HF were immunoprecipitated with MAbs against pp65. Immunoprecipitated proteins were separated by SDS-PAGE and transferred to nitrocellulose for Western blotting with antibodies specific for pp65 (28-19), lamins (237), and a control MAb specific for ppUL44 (28-21). Antibody binding was detected by addition of rabbit anti-mouse IgG antibody (lanes 28-19 and 28-21 only) followed by 125I-protein A and autoradiography. Immunoglobulin heavy chains were detected with a rabbit anti-mouse immunoglobulin antibody followed by addition of 125I-protein A and autoradiography (lane C). Migration of the molecular mass marker (in kilodaltons) and immunoglobulin heavy chains (Hc) is shown in the margins.
In earlier experiments, we observed focal accumulations of pp65 along the periphery of the nucleus (Fig. 1E and F). Together, these findings were consistent with the association of pp65 with proteins of the nuclear lamina, the protein network which provides the structural framework of the nuclear envelope (27, 47). Using a MAb against human lamin B1, we found that the 68-kDa band detected in the far-Western blot comigrated with the 68-kDa band of the lamin B1 Western blot (Fig. 6A, lane NA12). Together, these results suggested a direct interaction between pp65 and proteins of the nuclear matrix, possibly components of the nuclear lamina.
pp65 interacts with lamins in the nuclear matrix fraction.
Direct investigations of protein-protein interactions with nuclear lamins have been complicated by the insolubility of lamins. We approached the study of pp65 interactions with these proteins by preparing soluble nuclear matrix proteins through step dialysis of nuclear matrix extracts from AD169-infected cells. The soluble nuclear matrix proteins were immunoprecipitated with a mixture of pp65-specific MAbs, and the precipitated proteins were separated by SDS-PAGE and then transferred to nitrocellulose membranes. The membranes were probed with the anti-pp65 MAb 28-19, a polyvalent rabbit antiserum specific for lamins (237) (29), and a control MAb, 28-21, which is specific for ppUL44. The pp65-specific MAb detected pp65 and several forms of this protein (Fig. 6B). The lamin-specific antiserum 237 detected lamins in the immunoprecipitated complex (Fig. 6B). The immunoreactive protein(s) migrated at approximately 68 kDa, consistent with the migration of lamins. Antiserum 237 failed to react with recombinant derived pp65 produced in bacteria, indicating that the reactivity for lamins was specific (data not shown). In contrast, MAb 28-21 reacted with three proteins which ranged in size between 50 and 45 kDa (Fig. 6B). Although we cannot rule out the possibility that at least one of these bands represents a ppUL44-related protein, we observed the same three bands in the membrane developed with the pp65-specific MAb, suggesting that these proteins represented reactivity of the anti-mouse IgG second antibody with the different forms of the immunoglobulin heavy chain present in the original immunoprecipitate (Fig. 6B). The identity of these bands was confirmed by probing a filter with rabbit anti-mouse IgG antibody (lane C), which produced the same pattern of reactivity as the control anti-ppUL44 antibody. Together with our findings from the far-Western analysis, these findings indicated that pp65 interacted directly with lamins.
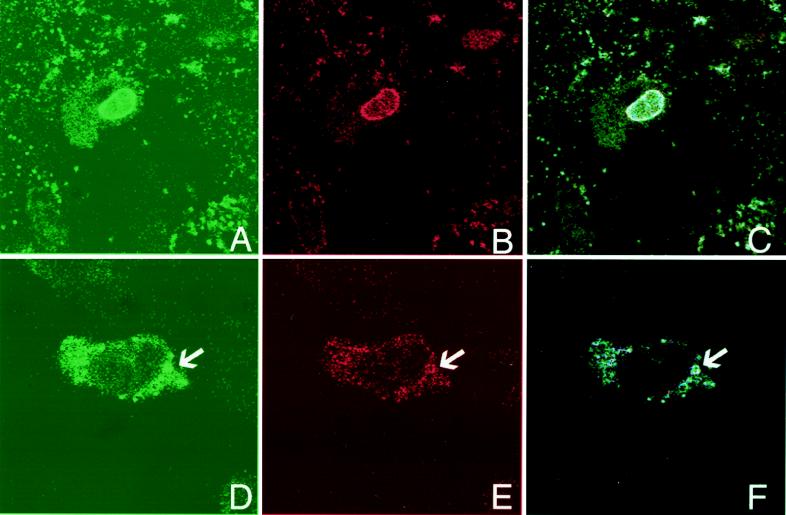
To further examine the interactions between pp65 and lamins, in situ-extracted AD169-infected HF were reacted with an anti-pp65 MAb and a rabbit polyclonal serum against lamins a and c (17). We observed localization of pp65 along the periphery of the nucleus of infected cells (Fig. 7A). Similarly, the distribution of lamins a and c along the nuclear periphery was consistent with previous studies (Fig. 7B) (17, 29). As shown in Fig. 7C, we noted colocalization of pp65 and lamins a and c. We also examined HCMV-infected HF late in infection when extensive cytopathic effects were present. In some cells, we observed extranuclear, vacuole-like structures containing both pp65 and lamin b (Fig. 7D to F). These results were consistent with the biochemical data suggesting a direct interaction between pp65 and proteins of the nuclear lamina.
FIG. 7.
Colocalization of pp65 and lamins. Nuclear matrix-extracted (A to C) or unextracted (D to F) HCMV-infected cells were stained with a murine MAb against pp65 (green; A and D) and a rabbit antiserum against lamins a and c (red; B) or against lamin b (red; E). Colocalization (blue) of pp65 and lamins is shown in panels C and F. Magnification for all panels, ×890.
DISCUSSION
In this report, we have described the subcellular distribution of several structural and nonstructural proteins of HCMV. Specifically, we have investigated the intranuclear localization of protein components of the virion tegument and capsid. Because these proteins were colocalized in a specific nuclear structure, the nuclear matrix, we have proposed that the nuclear matrix is a potential site of particle morphogenesis in the HCMV-infected cell.
Intranuclear compartmentalization of proteins from several DNA viruses has been well documented. Studies of adenovirus-infected cells have described the spatial separation of viral transcription and replication sites (51). Virus-encoded proteins involved in replication of the adenovirus genome have been localized to discrete nuclear structures which are distinct from sites of transcription. de Bruyn Kops and Knipe (18) as well as other groups (41, 42, 44) described the spatial organization of viral replication structures in HSV-infected cells and suggested that the arrangement of these structures was defined by preexisting nuclear architecture (18, 44). Similarly, intranuclear HCMV replication compartments were recently characterized by Sarisky and Hayward, who characterized the viral proteins associated with the formation of these sites and for oriLyt-dependent DNA replication (59). A more complete description of intranuclear compartmentalization of HSV proteins was recently reported by Ward et al., who described HSV structures within the nucleus which they termed assemblons (64). These nuclear subcompartments were reported to segregate proteins into groups associated with specific functions including replication of viral DNA and assembly of subviral particles. The mechanisms driving accumulation of proteins into these structures are not clear, but they are likely to involve interaction of viral proteins with the architectural framework of the nucleus, thereby providing spatial organization to an already temporally regulated replicative process.
Previous studies of several DNA viruses have also shown that virion structural proteins localized to the nuclear matrix (5, 7, 37, 66). Recent studies have documented the presence of newly synthesized adenovirus virions on the core filaments of the nuclear matrix, suggesting that this nuclear structure may be a site of adenovirus assembly (66). In this same study, Zhonghe et al. suggested that newly formed adenovirus particles track along 10-nm core filaments of the nuclear matrix, providing some evidence that this filamentous network may also provide a function critical to nuclear egress of progeny virions. HSV proteins have been reported to associate with the nuclear matrix (5, 7). In these studies, proteins comigrating with capsid and DNA-binding proteins of HSV were found in the nuclear matrix fraction (7). In addition, HSV capsids were observed in a filamentous network within the nucleus (5). These findings, together with studies which have shown that the nuclear matrix is an important site of transcription and replication, were consistent with a model in which this nuclear compartment could serve as the site of assembly for viruses which encapsidate nucleic acid in the nucleus (7, 37, 66). Our findings were also in agreement with these previous findings in adenovirus- and HSV-infected cells and suggested that HCMV may also assemble subviral particles in association with the nuclear matrix of infected cells. However, finding virion structural proteins associated with the nuclear matrix does not indicate that in each case there is a direct interaction between individual viral proteins and proteins of the nuclear matrix. In some cases the association could have resulted from virion protein interactions with a limited number of virus-encoded proteins bound to the nuclear matrix; however, the maintenance of this association following washes in 1 M guanidine hydrochloride suggested a very stable interaction.
We focused the majority our studies on the tegument protein pp65 because of its abundance in extracellular particles as well as its nuclear expression shortly after infection of permissive fibroblasts (36, 61). Previous studies have shown that almost immediately after infection, pp65 is transported to the nucleus of fibroblasts, where it accumulates until late in infection (61, data not shown). Targeting of the protein has been attributed to a bipartite nuclear targeting signal at the extreme carboxy terminus of the molecule (61) and more recently to a second signal proximal to this conventional nuclear localization signal (NLS) (26). However, in both of these studies, mutated forms of pp65 which lacked these signals could still be localized to the nucleus when expressed in recombinant systems, suggesting that there were other domains mediating nuclear localization of pp65. Such domains could mediate nuclear retention in addition to nuclear localization associated with previously described NLS (26, 61). As shown in Fig. 1, pp65 was observed in subnuclear structures and also in patches along the periphery of the nucleus. Additional domains within pp65 could therefore mediate retention on a particular nuclear structure such as the nuclear lamina. Such protein interactions could explain the findings of earlier studies which documented the nuclear accumulation of mutant forms of pp65 which lacked NLS but were of such a size as to allow passive diffusion into and out of the nucleus (20, 26, 61).
The nuclear matrix of HCMV-infected fibroblasts was isolated by the method of Mirkovitch et al., which consisted of high-salt extraction of DNase-treated nuclei which were initially isolated by nonionic detergent treatment of viable cells (45). The resulting pellet of nuclear material represented insoluble nuclear proteins and was essentially devoid of DNA. Western blot analysis of nuclear matrix material demonstrated the retention of several HCMV structural proteins which have previously been characterized as nuclear proteins (Fig. 2A). In addition, the polymerase accessory protein ppUL44 and the associated products of this open reading frame were also present in the nuclear matrix pellet. In contrast, the nuclear 72-kDa IE-1 and 86-kDa IE-2 nonstructural proteins were not detected in this assay, suggesting that the association of HCMV proteins with the nuclear matrix was specific. Furthermore, we failed to detect two abundant tegument proteins, pp28 (ppUL99) and pp150 (ppUL32), as well as glycoprotein B (gpUL55) in the nuclear matrix, providing additional evidence for the specificity of the protein-nuclear matrix interactions that we have described (data not shown). These results were confirmed by immunofluorescence of in situ-extracted HCMV-infected cells (Fig. 4). Together, these data suggested that several proteins which were incorporated into nuclear subviral particles were sequestered on the nuclear matrix.
Enrichment of pp65 on the nuclear matrix was demonstrated by quantitative Western blotting (Fig. 3; Table 1). The results from this experiment suggest that pp65 is strongly associated with the nuclear matrix. Moreover, the localization of pp65 to the nuclear matrix in the absence of other HCMV proteins indicated a direct interaction with components of the nuclear matrix. Although initial studies also documented the association of pp65 with the nuclear matrix of monkey cells (Fig. 5) and insect cells (data not shown), we obtained additional evidence of the direct interaction of pp65 with the nuclear matrix by performing far-Western blotting with recombinant-derived pp65 and the nuclear matrix of HEp-2 cells (Fig. 6A). The results of this experiment indicated that pp65 associated with a limited number of protein constituents of the nuclear matrix. The binding of pp65 to a restricted set of nuclear matrix proteins underscored the specificity of the protein-protein interactions and suggested the possibility of a sequence-specific targeting signal for the localization of pp65 to the nuclear matrix. Although the identities of the nuclear matrix proteins detected by far-Western blotting have not been conclusively established, the higher-molecular-weight bands were similar in molecular size to lamins a, b, and c, which together represent major protein components of the nuclear matrix (27, 47). Western blot analysis of the nuclear matrix protein-containing filter showed that one of the proteins detected by far-Western blotting comigrated with lamin B1 (Fig. 6A). Additional biochemical evidence for the interaction between pp65 and lamins was provided by the coprecipitation of pp65 and lamins from a soluble extract of nuclear matrix proteins from HCMV-infected fibroblasts (Fig. 6B). The polymerase accessory protein ppUL44 was not coprecipitated with pp65 and lamins, suggesting that this was a specific interaction (Fig. 6B). Thus, the binding of pp65 to a major constituent of the nuclear matrix and colocalization of pp65 and lamins (Fig. 7) were consistent with the hypothesis that at least one step in nuclear tegumentation of the HCMV capsid might localize to this subnuclear compartment. Finally, we have consistently observed cytoplasmic vacuole-like structures containing pp65 and nuclear lamins in cells transfected with pp65 expression plasmids which are similar to those illustrated in Fig. 7D and E. This observation and the accumulation of pp65 on the nuclear membrane suggest a possible role of pp65 in focal modifications of the nuclear envelope.
In summary, we have provided biochemical and imaging data of the association of nuclear tegument and capsid proteins of HCMV with the nuclear matrix. Together, these findings argued for the nuclear matrix being a potential site for assembly of subviral particles of HCMV and suggested that protein-protein interactions between virus-encoded proteins and this nuclear structure might provide spatial coordination for the highly regulated and temporally coordinated replication of this virus.
ACKNOWLEDGMENTS
We thank Robert Goldman and Nilabh Chaudhary for the generous gifts of antibody 237 and for the rabbit sera against lamins a/c and b, respectively. We also thank Amy Sears (Department of Microbiology, Emory University, Atlanta, Ga.) for assistance with the confocal microscopic analysis and Scott Swindle and Kenneth Fish for technical advice and helpful discussions. We thank Dana Pinson for assistance with preparation of the manuscript.
P.C.A. was supported by training grant NIH T32 AI07150 and by grant NIH R01 AI20408 to J.A.E. V.S. was supported by a grant supplement to NIH R01 AI30105 and R01 AI35602 to W.J.B.
REFERENCES
- 1.Andreoni M, Faircloth M, Vugler L, Britt W. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J Virol Methods. 1989;23:157–168. doi: 10.1016/0166-0934(89)90129-8. [DOI] [PubMed] [Google Scholar]
- 2.Angeletti P C, Engler J A. Tyrosine kinase dependent release of an adenovirus preterminal protein complex from the nuclear matrix. J Virol. 1996;70:3060–3067. doi: 10.1128/jvi.70.5.3060-3067.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldick C J, Shenk T. Proteins associated with purified human cytomegalovirus particles. J Virol. 1996;70:6097–6105. doi: 10.1128/jvi.70.9.6097-6105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benko D M, Gibson W. Primate cytomegalovirus glycoproteins: lectin-binding properties and sensitivities to glycosidases. J Virol. 1986;59:703–713. doi: 10.1128/jvi.59.3.703-713.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Ze’Ev A, Abulafia R, Bratosin S. Herpes simplex virus and protein transport are associated with the cytoskeletal framework and the nuclear matrix in infected BSC-1 cells. Virology. 1983;129:501–507. doi: 10.1016/0042-6822(83)90190-3. [DOI] [PubMed] [Google Scholar]
- 6.Berezney R, Mortillaro M J, Ma H, Wei X, Samarandu J. The nuclear matrix: a structural milieu of genomic function. Int Rev Cytol. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- 7.Bibor-Hardy V, Pouchelet M, St-Pierre E, Herzberg M, Simard R. The nuclear matrix is involved in herpes simplex virogenesis. Virology. 1982;121:296–306. doi: 10.1016/0042-6822(82)90169-6. [DOI] [PubMed] [Google Scholar]
- 8.Blencowe B J, Issner R, Kim J, McCaw P, Sharp P A. New proteins related to the ser-arg family of splicing factors. RNA. 1995;1:852–865. [PMC free article] [PubMed] [Google Scholar]
- 9.Boppana S B, Britt W J. Recognition of human cytomegalovirus gene products by HCMV-specific cytotoxic T cells. Virology. 1996;222:293–296. doi: 10.1006/viro.1996.0424. [DOI] [PubMed] [Google Scholar]
- 10.Bridge E, Carmo-Fonseca M, Lamond A, Petterson U. Nuclear organization of splicing small nuclear ribonucleoproteins in adenovirus-infected cells. J Virol. 1993;67:5792–5802. doi: 10.1128/jvi.67.10.5792-5802.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Britt W J, Auger D. Synthesis and processing of the envelope gp55-116 complex of human cytomegalovirus. J Virol. 1986;58:185–191. doi: 10.1128/jvi.58.1.185-191.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britt W J, Vugler L. Structural and immunological characterization of the intracellular forms of an abundant 68,000 Mr human cytomegalovirus protein. J Gen Virol. 1987;68:1897–1907. doi: 10.1099/0022-1317-68-7-1897. [DOI] [PubMed] [Google Scholar]
- 13.Britt W J, Vugler L, Butfiloski E J, Stephens E B. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV-recombinant vaccinia virus infected cells in analysis of the human neutralizing antibody response. J Virol. 1990;64:1079–1085. doi: 10.1128/jvi.64.3.1079-1085.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvalho T, Seeler J-S, Ohman K, Jordan P, Petterson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chabot B, Bisotto S, Vincent M. The nuclear matrix phosphoprotein p255 associates with splicing complexes as part of the [U4/U6.U5] tri-snRNP particle. Nucleic Acids Res. 1995;23:3206–3213. doi: 10.1093/nar/23.16.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang Y E, Roizman B. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J Virol. 1993;67:6348–6356. doi: 10.1128/jvi.67.11.6348-6356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhary N, Courvalin J-C. Stepwise reassembly of the nuclear envelope at the end of mitosis. J Cell Biol. 1993;122:295–306. doi: 10.1083/jcb.122.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bruyn Kops A, Knipe D M. Preexisting nuclear architecture defines the intranuclear location of herpesvirus DNA replication structures. J Virol. 1994;68:3512–3526. doi: 10.1128/jvi.68.6.3512-3526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Lazzaro C, Campadelli-Fiume G, Torrisi M R. Intermediate forms of glycoconjugates are present in the envelope of herpes simplex virions during their transport along the exocytic pathway. Virology. 1995;214:619–623. doi: 10.1006/viro.1995.0073. [DOI] [PubMed] [Google Scholar]
- 20.Dingwall C, Laskey R A. Protein import into the cell nucleus. Annu Rev Cell Biol. 1986;2:367–390. doi: 10.1146/annurev.cb.02.110186.002055. [DOI] [PubMed] [Google Scholar]
- 21.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 22.Durfee T, Mancini M A, Jones D, Elledge S J, Lee W-H. The amino-terminal region of the retinoblastoma gene product binds a novel nuclear matrix protein that colocalizes to centers for RNA-processing. J Cell Biol. 1994;127:609–622. doi: 10.1083/jcb.127.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fey E G, Penman S. Nuclear matrix proteins reflect cell type of origin in cultured human cells. Proc Natl Acad Sci USA. 1988;85:121–125. doi: 10.1073/pnas.85.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fish, K. N., and J. A. Nelson. Unpublished results.
- 25.Fredman J N, Engler J A. Adenovirus precursor to terminal protein interacts with the nuclear matrix in vivo and in vitro. J Virol. 1993;67:3384–3395. doi: 10.1128/jvi.67.6.3384-3395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallina A, Percivalle E, Simoncini L, Revello M G, Gerna G, Milanesi G. Human cytomegalovirus pp65 lower matrix phosphoprotein harbours two transplantable nuclear localization signals. J Gen Virol. 1996;77:1151–1157. doi: 10.1099/0022-1317-77-6-1151. [DOI] [PubMed] [Google Scholar]
- 27.Gerace L, Burke B. Functional organization of the nuclear envelope. Annu Rev Cell Biol. 1988;4:335–374. doi: 10.1146/annurev.cb.04.110188.002003. [DOI] [PubMed] [Google Scholar]
- 28.Getzenberg R H. Nuclear matrix and the regulation of gene expression: tissue specificity. J Cell Biochem. 1994;55:22–31. doi: 10.1002/jcb.240550105. [DOI] [PubMed] [Google Scholar]
- 29.Goldman A E, Moir R D, Montag-Lowy M, Stewart M, Goldman R D. Pathway of incorporation of microinjected lamin A into the nuclear envelope. J Cell Biol. 1992;119:725–735. doi: 10.1083/jcb.119.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Granzow H, Weiland F, Jons A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenfield I, Nickerson J, Penman S, Stanley M. Human papillomavirus 16 E7 protein is associated with the nuclear matrix. Proc Natl Acad Sci USA. 1991;88:11217–11221. doi: 10.1073/pnas.88.24.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo B, Odgren P R, Van Wijnen A J, Last T J, Nickerson J, Penman S, Lian J B, Stein J L, Stein G S. The nuclear matrix protein NMP-1 is the transcription factor YY1. Proc Natl Acad Sci USA. 1995;92:10526–10530. doi: 10.1073/pnas.92.23.10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He D, Nickerson J A, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hensel G, Meyer H, Gärtner S, Brand G, Kern H F. Nuclear localization of the human cytomegalovirus tegument protein pp150 (UL32) J Gen Virol. 1995;76:1591–1601. doi: 10.1099/0022-1317-76-7-1591. [DOI] [PubMed] [Google Scholar]
- 35.Hozac P, Hassan A B, Jackson D A, Cook P R. Visualization of replication factories attached to a nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- 36.Irmiere A, Gibson W. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology. 1983;30:118–133. doi: 10.1016/0042-6822(83)90122-8. [DOI] [PubMed] [Google Scholar]
- 37.Khittoo G, Delorme L, Dery C V, Tremblay M L, Weber J M, Bibor-Hardy V, Simard R. Role of the nuclear matrix in adenovirus maturation. Virus Res. 1986;5:391–403. doi: 10.1016/0168-1702(86)90085-7. [DOI] [PubMed] [Google Scholar]
- 38.Landini M P, Severi B, Furlini G, Badiali De Giorgi L. Human cytomegalovirus structural components: intracellular and intraviral localization of p28 and p65-69 by immunoelectron microscopy. Virus Res. 1987;8:15–23. doi: 10.1016/0168-1702(87)90036-0. [DOI] [PubMed] [Google Scholar]
- 39.Lauber A H, Sandhu N P, Schuchard M, Subramanian M, Spelsberg T C. Nuclear matrix acceptor binding sites for steroid hormone receptors: a candidate nuclear matrix acceptor protein. Int Rev Cytol. 1995;162B:337–375. doi: 10.1016/s0074-7696(08)62621-2. [DOI] [PubMed] [Google Scholar]
- 40.Liao H, Winkfein R J, Mack G, Rattner J B, Yen T J. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liptak L, Uprichard S L, Knipe D M. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J Virol. 1996;70:1759–1767. doi: 10.1128/jvi.70.3.1759-1767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukonis C J, Weller S K. Characterization of nuclear structures in cells infected with herpes simplex virus type 1 in the absence of viral DNA replication. J Virol. 1996;70:1751–1758. doi: 10.1128/jvi.70.3.1751-1758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mancini M, Shan B, Nickerson J A, Penman S, Lee W-H. The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc Natl Acad Sci USA. 1994;91:418–422. doi: 10.1073/pnas.91.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start site of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 45.Mirkovitch J, Mirault M E, Laemmli U K. Organization of the higher-order chromatin loop: specific DNA attachment sites on the nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 46.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Strauss S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 47.Newport J W, Forbes D J. The nucleus: structure, function, and dynamics. Annu Rev Biochem. 1987;56:535–565. doi: 10.1146/annurev.bi.56.070187.002535. [DOI] [PubMed] [Google Scholar]
- 48.Nickerson J A, Blencowe B, Penman S. The architectural organization of nuclear metabolism. Int Rev Cytol. 1995;162A:67–123. doi: 10.1016/s0074-7696(08)61229-2. [DOI] [PubMed] [Google Scholar]
- 49.Pienta K J, Hoover C N. Coupling of cell structure to cell metabolism and function. J Cell Biochem. 1994;55:16–21. doi: 10.1002/jcb.240550104. [DOI] [PubMed] [Google Scholar]
- 50.Plachter B, Britt W, Vornhagen R, Stamminger T, Jahn G. Analysis of proteins encoded by IE regions 1 and 2 of human cytomegalovirus using monoclonal antibodies generated against recombinant antigens. Virology. 1993;193:642–652. doi: 10.1006/viro.1993.1172. [DOI] [PubMed] [Google Scholar]
- 51.Pombo A, Ferreira J, Bridge E, Carmo-Fonseca M. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 1994;13:5075–5085. doi: 10.1002/j.1460-2075.1994.tb06837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radha V, Nambirajan S, Swarup G. Association of Lyn tyrosine kinase with the nuclear matrix and cell cycle-dependent changes in matrix-associated tyrosine kinase activity. Eur J Biochem. 1996;23:352–359. doi: 10.1111/j.1432-1033.1996.00352.x. [DOI] [PubMed] [Google Scholar]
- 53.Roffman E, Albert J P, Goff J P, Frenkel N. Putative site for the acquisition of human herpesvirus 6 virion tegument. J Virol. 1990;64:6308–6313. doi: 10.1128/jvi.64.12.6308-6313.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Strauss S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2221–2230. [Google Scholar]
- 55.Roizman B, Furlong D. The replication of herpesviruses. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. Vol. 3. New York, N.Y: Plenum Press; 1974. pp. 229–403. [Google Scholar]
- 56.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Strauss S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 57.Rose J K, Buonocore L, Whitt M A. A new liposome reagent mediating nearly quantitative transfection of animal cells. Bio/Technology. 1991;10:520–525. [PubMed] [Google Scholar]
- 58.Sanchez V, Greis K, Akimoto Y, Hart G, Britt W J. Abstracts from the 21st Herpesvirus Workshop. 1996. Assembly of HCMV tegument occurs in different cellular compartments. [Google Scholar]
- 59.Sarisky R T, Hayward G S. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J Virol. 1996;70:7398–7413. doi: 10.1128/jvi.70.11.7398-7413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schirmbeck R, Deppert W. Nuclear subcompartmentalization of simian virus 40 large T antigen: evidence for in vivo regulation of biochemical activities. J Virol. 1989;63:2308–2316. doi: 10.1128/jvi.63.5.2308-2316.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmolke S, Drescher P, Jahn G, Placter B. Nuclear targeting of the tegument protein pp65 (UL83) of human cytomegalovirus: an unusual bipartite nuclear localization signal functions with other portions of the protein to mediate its efficient nuclear transport. J Virol. 1995;69:1071–1078. doi: 10.1128/jvi.69.2.1071-1078.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stein G S, van Wijnen A J, Stein J L, Lian J B, Bidwell J P, Montecino M. Nuclear architecture supports integration of physiological regulatory signals for transcription of cell growth and tissue-specific genes during osteoblast differentiation. J Cell Biochem. 1994;55:4–15. doi: 10.1002/jcb.240550103. [DOI] [PubMed] [Google Scholar]
- 63.Ward P L, Barker D E, Roizman B. A novel herpes simplex virus 1 gene, UL43.5, maps antisense to the UL43 gene and encodes a protein which colocalizes in nuclear structures with capsid proteins. J Virol. 1996;70:2684–2690. doi: 10.1128/jvi.70.5.2684-2690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ward P L, Ogle W O, Roizman B. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J Virol. 1996;70:4623–4631. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winkler M, Rice S A, Stamminger T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhonghe Z, Nickerson J A, Krochmalnic G, Penman S. Alterations in nuclear matrix structure after adenovirus infection. J Virol. 1987;61:1007–1018. doi: 10.1128/jvi.61.4.1007-1018.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]