Abstract
Acarbose is a potent glycosidase inhibitor widely used in the clinical treatment of type 2 diabetes mellitus (T2DM). Various acarbose analogs have been identified while exploring compounds with improved pharmacological properties. In this study, we found that AcbE from Actinoplanes sp. SE50/110 catalyzes the production of acarbose analogs that exhibit significantly improved inhibitory activity towards α-amylase than acarbose. Recombinant AcbE mainly catalyzed the formation of two new compounds, namely acarstatins A and B, using acarbose as substrate. Using high-resolution mass spectrometry, nuclear magnetic resonance, and glycosidase hydrolysis, we elucidated their chemical structures as O-α-d-maltosyl-(1 → 4)-acarbose and O-α-d-maltotriosyl-(1 → 4)-acarbose, respectively. Acarstatins A and B exhibited 1584- and 1478-fold greater inhibitory activity towards human salivary α-amylase than acarbose. Furthermore, both acarstatins A and B exhibited complete resistance to microbiome-derived acarbose kinase 1-mediated phosphorylation and partial resistance to acarbose-preferred glucosidase-mediated hydrolysis. Therefore, acarstatins A and B have great potential as candidate therapeutic agents for T2DM.
Keywords: Acarbose analogs, α-amylase inhibitory activity, Actinoplanes sp., Transglycosylase
1. Introduction
According to the World Diabetes Atlas, the global prevalence of diabetes mellitus is steadily increasing. As of 2021, 537 million individuals worldwide are affected, with most patients suffering from type 2 diabetes mellitus (T2DM) [1]. Glycosidase inhibitors, an important class of drugs used for treating T2DM, delay the breakdown of complex carbohydrates into monosaccharides, thereby suppressing postprandial hyperglycemic symptoms [[2], [3], [4]]. Acarbose, a glycosidase inhibitor widely used in clinical practice, competitively and reversibly inhibits α-amylase, glucoamylase, sucrase, and maltase activities [5,6]. Notably, it significantly inhibits α-amylase, which is responsible for the initial step in complex carbohydrate hydrolysis during digestion [5]. During α-amylase inhibition, acarbose undergoes multiple rounds of hydrolysis and condensation catalyzed by α-amylase, which ultimately leads to the formation of a suicide-type rearrangement product [[7], [8], [9]]. This acarbose rearrangement product serves as an inhibitor at the active site of α-amylase [7,9]. The binding model of the acarbose rearrangement product with α-amylase suggests that nonreducing end-modifications of acarbose-like compounds positively contribute to their biological activities [7,9]; this has been subsequently validated in many studies [10,11]. Acarbose analogs resembling the rearrangement product may therefore be more effective α-amylase inhibitors.
Considering the clinical importance of acarbose in T2DM treatment, developing acarbose analogs with superior pharmacological activity is of great interest. Structural diversity in acarbose can be achieved through microbial bioconversion, enzymatic catalysis, and chemical modification. Various pseudooligosaccharides with structures similar to acarbose have been identified during the bacterial fermentation process, including amylostatin from Streptomyces diastaticus [12,13], acarviostatin from Streptomyces coelicofla [[14], [15], [16]], trestatin from Streptomyces dimorphogenes [17,18], and oligostatin from Streptomyces myxogenes [19]. Acarbose-derived analogs with various modifications have been obtained via catalysis by acarviosyl transferase from Actinoplanes sp. [20], maltogenic amylase from Bacillus stearothermophilus (gseBMSA) [[21], [22], [23], [24]], cyclomaltodextrin glucanyltransferase from Paenibacillus macerans (pmaeCGTase) [10,11], and dextransucrases from Leuconostoc mesenteroides (imeDSR) [25]. In addition, dihydroacarbose [26] and a series of methyl acarviosine analogs [27] have been prepared by chemical modification, most of which exhibited enhanced sucrase and α-glucosidase inhibitory activity [26,27].
Actinoplanes sp. SE50/110 and its derivative strains are important acarbose producers, and previous studies have indicated that SE50/110 has great potential for producing diverse acarbose analogs. The first class of acarbose analogs produced by SE50/110 were the byproducts of fermentation and purification [28]. Among these byproducts, acarviosyl-1,4-glucose-1,1-glucose, denoted as component C, synthesized by maltooligosyltrehalose synthase TreY, was comprehensively investigated [29,30]. The ‘Carbophore Cycle’ has been proposed as a pathway involved in acarbose analog formation, and acbD, acbE and acbZ within acarbose biosynthetic gene cluster (acb BGC) are speculated to participate in the extracellular part of the ‘Carbophore Cycle’. Amylases AcbE and AcbZ are responsible for starch degradation, and acarviosyl transferase AcbD catalyzes acarbose analog production [31,32]. However, the exact role of AcbE in acarbose modification remains unclear.
In addition to direct α-glycosidase inhibition by acarbose, the therapeutic efficacy of acarbose is also associated with its ability to selectively modulate the gut microbiota [[33], [34], [35], [36], [37]]. Most of the orally administered acarbose remains unabsorbed in the gut, and prolonged exposure to acarbose confers a protective advantage for microbes that express acarbose inactivation pathways [38]. Currently, there are two specific pathways for acarbose inactivation encoded by the human microbiome, namely the phosphorylation pathway, mediated by microbiome-derived acarbose kinases (Maks) [38], and the hydrolysis pathway, mediated by acarbose-preferred glucosidase (Apg) [39]. Enrichment of strains harboring these pathways is correlated with a lower response to acarbose treatment in patients with T2DM, thus increasing the prevalence of acarbose resistance in these patients [38,39]. In the clinical application of glycosidase inhibitors such as acarbose, the interaction between the drugs and the gut microbiota should therefore receive more attention to facilitate precision healthcare [39].
In this study, we identified a series of novel acarbose derivatives synthesized by the glycosyltransferase AcbE. We elucidated the chemical structures of two main derivatives, namely acarstatins A and B. Acarstatins A and B exhibited substantially improved inhibitory activity towards α-amylase, as well as resistance to Mak1-and Apg-mediated inactivation pathways.
2. Results
2.1. AcbE-mediated the production of compounds with enhanced inhibitory activity towards porcine pancreatic α-amylase
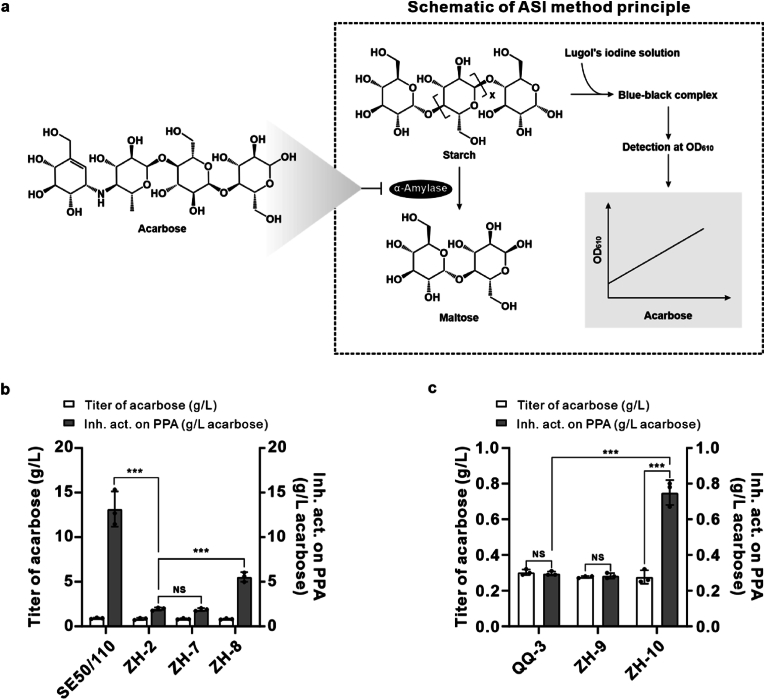
Our previous gene knockout experiment of acbE, located in the acb BGC and encoding an α-amylase, indicated that the ZH-2 mutant strain exhibits a similar acarbose titer to that of the wild-type strain SE50/110. However, ASI (Amylase-Starch-Iodine) assays (Fig. 1a) indicated that ZH-2 culture exhibits an 85% decrease in inhibitory activity towards porcine pancreatic α-amylase compared with the wild-type strain SE50/110 [40]. Therefore, we hypothesized that AcbE participates in the production of compounds with enhanced porcine pancreatic α-amylase inhibitory activity. To elucidate the role of AcbE in the generation of compounds with enhanced pharmacological activity, we performed gene complementation by reintroducing acbE under the control of the kasOp* promoter into strain ZH-2 to generate a recombinant strain ZH-8 (Supplementary Fig. 2). The ratio of ASI-measured inhibitory activity to the actual acarbose titer increased from 2.35 to 6.78 in the ZH-8 strain fermentation broth, which had a similar acarbose titer to strain SE50/110 (Fig. 1b). Subsequently, we conducted a bioconversion experiment using ZH-10, which was constructed by introducing acbE under the control of its native promoter into the acb BGC-deleted strain QQ-3 (Supplementary Fig. 3). During strain ZH-10 fermentation, 0.5 g/L acarbose was incorporated, and the ratio of ASI-measured inhibitory activity of the final cultures to acarbose titer increased to 2.71 from 0.98 for QQ-3 and 1.02 for the control strain ZH-9 (Fig. 1c). These results demonstrated the involvement of AcbE in the formation of compounds with enhanced porcine pancreatic α-amylase inhibitory activity.
Fig. 1.
The involvement of AcbE in the formation of compounds with enhanced inhibitory activity against porcine pancreatic α-amylase. a, Schematic of ASI detection principle. b, Complementation of the acbE increasing the production of compounds with enhanced inhibitory activity towards porcine pancreatic α-amylase. c, Bioconversion experiment confirming acbE as the core gene to produce compounds with enhanced inhibitory activity towards porcine pancreatic α-amylase. For panels b and c, strains were denoted as follows: SE50/110, wild-type strain; ZH-2, SE50/110ΔacbE; ZH-7, SE50/110ΔacbE::pSET152; ZH-8, SE50/110ΔacbE::acbE; QQ-3, SE50/110Δacb; ZH-9, SE50/110Δacb::pSET152; ZH-10, SE50/110Δacb::acbE. The data are presented as the mean ± standard deviation of three independent experiments. P values are calculated using a two-tailed Student's t-test assuming unequal variance; *, P < 0.01; **, P < 0.005; ***, P < 0.001.
2.2. AcbE converted acarbose into glycosylated analogs with different oligosaccharide substrates
Amino acid sequence alignments revealed that AcbE contains a glycosyl hydrolase family 13 catalytic domain, a glycosyl hydrolase all-β domain, an immunoglobulin-like fold domain, a pullulanase carbohydrate-binding module 41 domain, a pullulanase N2 domain, and a signal peptide region at the N-terminus (Supplementary Fig. 4). To elucidate the biochemical role of AcbE, matured AcbE, designated as AcbE*, was constructed by removing the signal peptide region, heterologously expressed in Escherichia coli Origami B (DE3), and subsequently purified using affinity chromatography (Fig. 2a). Based on biochemical analyses of homologous AcbD and LMZ02_00515 proteins, acarbose (1) and various oligosaccharides, including maltotriose, maltotetraose, maltopentaose, maltooligosaccharides, and α-cyclodextrin, were selected as candidate substrates for AcbE*. The optimized reactions used 10 μM AcbE, 5 mM acarbose, and 20 mM or 20 g/L oligosaccharides at pH 6.5 and 25 °C. ASI assays revealed that reactions with AcbE* and maltotriose, maltotetraose, maltopentaose, maltooligosaccharides, or α-cyclodextrin exhibited 15.15-, 100.42-, 333.14-, 582.06-, and 9.54-fold higher inhibitory activity, respectively, towards porcine pancreatic α-amylase than reaction system with the deactivated AcbE* (Fig. 2b). This suggested that AcbE directly catalyzes the production of compounds with enhanced porcine pancreatic α-amylase inhibitory activity using acarbose and oligosaccharides as substrates, and that AcbE prefers maltopentaose and maltooligosaccharides as substrates.
Fig. 2.
AcbE catalyzed the formation of glycosylated acarbose analogs with different oligosaccharides as the donor substrates. a, SDS-PAGE analysis of His-tagged AcbE*. b, AcbE* catalyzing the formation of products with enhanced inhibitory activity towards porcine pancreatic α-amylase using acarbose and oligosaccharides as substrates. In panel b, the symbols 3,4, 5, O, and C denote maltotriose, maltotetraose, maltopentaose, maltooligosaccharides and α-cyclodextrin, respectively. In the acarbose row, the symbol “+” indicates addition of acarbose. In the AcbE* row, symbols “+” and “-” represent addition of AcbE* or deactivated AcbE*, respectively. The data are presented as the mean ± standard deviation of three independent experiments. P values are calculated using a two-tailed Student's t-test assuming unequal variance; *, P < 0.01; **, P < 0.005; ***, P < 0.001. c, HPLC profiles for the products catalyzed by AcbE* with acarbose and oligosaccharides as substrates. Acar std. Represents standard sample of acarbose. Acar + M3, Acar + M4, Acar + M5, Acar + MO, and Acar + CD represent reactions consisting AcbE*, acarbose and one of oligosaccharides, namely maltotriose, maltotetraose, maltopentaose, maltooligosaccharides and α-cyclodextrin, respectively. d, HRMS confirmation of compounds 2 to 5.
Liquid chromatography coupled with high-resolution mass spectrometry (LC-HRMS) was used to analyze the reactions of AcbE with acarbose as one of the substrates, and maltopentaose or maltooligosaccharides as another substrate. In maltooligosaccharide-containing reaction, four new products were identified with m/z values of 808.3091 [M+H]+ (2), 970.3619 [M+H]+ (3), 1132.4150 [M+H]+ (4), and 1294.4566 [M+H]+ (5). With maltopentaose as substrate, only compounds 3, 4, and 5 were seen (Fig. 2c). Molecular weight analyses identified these products as acarbose derivatives with one to four additional hexose moieties (Fig. 2d). These results indicated that AcbE catalyzes the synthesis of acarbose derivatives with acarbose and oligosaccharides as substrates, some of which exhibit higher inhibitory activity towards porcine pancreatic α-amylase.
2.3. Preparation and structural determination of glycosylated acarbose derivatives
The AcbE-catalyzed acarbose derivatives were prepared and purified to determine their chemical structures. A 300 mL large-scale reaction of AcbE containing 5 mM acarbose, 20 g/L maltooligosaccharide, and 25 mM Na2HPO4 buffer at pH 6.5 was performed at 25 °C for 24 h. Subsequently, the candidate derivatives were purified using a combination of ion-exchange chromatography and gel-filtration chromatography (Supplementary Fig. 5). We successfully isolated two new glycosylated acarbose derivatives, designated as acarstatin A (3; 12 mg) and acarstatin B (4; 2 mg). However, compounds 2 and 5 were failed to obtain for their very low yield in the reactions. HR-MS analyses of 3 and 4 indicated that their molecular weights were 970.3614 [M+H]+ (Fig. 3b) and 1132.4138 [M+H]+ (Fig. 3e), respectively, which were respectively consistent with acarbose derivatives with two and three additional hexose moieties. Subsequently, an integrated approach combining nuclear magnetic resonance (NMR), high-resolution tandem mass spectrometry (HR-MS/MS), and glycosidase hydrolysis was used to elucidate the chemical structures of 3 and 4.
Fig. 3.
Chemical structure elucidation of acarstatins A and B. a, Chemical structure of acarstatin A (3). b, HR-MS analysis of acarstatin A (3). c, HR-MS/MS analysis of acarstatin A (3). d, Chemical structure of acarstatin B (4). e, HR-MS analysis of acarstatin B (4). f, HR-MS/MS analysis of acarstatin B (4).
1H NMR analysis of acarstatin A (3) revealed two additional characteristic hydrogens (δ5.34 and δ5.31) on C-1 sites of the six-membered sugar ring compared with that of acarbose, indicating the presence of two additional six-membered sugar rings. Proton spectra indicated a shift in the nonlabile hydrogen at valienol C-4 from δ3.95 of acarbose to δ4.17–δ4.06 of 3, which suggested the additional six-membered sugar ring is connected to C-4 of the valienol. The H–H coupling constant of every junction point at C-1 of the six-membered sugar ring was below 4 Hz, indicating an α-configuration for the glycosidic linkages in 3. Detailed chemical shifts of the critical carbon and hydrogen atoms, as well as with the core H–H coupling constants in 3, were provided in the Supplementary tables (Supplementary Fig. 6, Supplementary Tables 9 and 10). Based on the 1H and 13C NMR data, the chemical structure of 3 could be inferred as O-α-d-maltosyl-(1 → 4)-acarbose (Fig. 3a), with further confirmation provided by two-dimensional NMR techniques including COSY, HSQC, HMBC, and TOCSY NMR (Supplementary Fig. 6). HR–MS/MS analysis of 3 further identified daughter ions with m/z 146.0813 [M+H]+, 304.1392 [M+H]+, 466.1912 [M+H]+, and 628.2453 [M+H]+, which suggested that two additional hexose moieties were substituted at the nonreducing end of acarbose (Fig. 3c). Finally, glycosidase hydrolysis of 3 was used to confirm the glycosylation site and the type of newly formed glycosyl linkages.
Based on the chemical structure analysis of 3, glucoamylase catalyzed the hydrolysis of α-1,4-glycosidic bonds, resulting in the production of glucose-α-1,4-acarbose (6) as the final product (Supplementary Fig. 8a). HR-MS analysis of glucoamylase-treated 3 revealed a hydrolysis product with m/z 808.3083 [M+H]+ (Supplementary Fig. 10a). Further HR–MS/MS analysis of this hydrolysis product revealed m/z values of the daughter ions as 146.0811 [M+H]+, 304.1389 [M+H]+, and 466.1920 [M+H]+, which indicated that the hydrolysis product of the glycosidase reaction was glucose-α-1,4-acarbose (6) (Supplementary Fig. 10b). The β-amylase could catalyze the hydrolysis of α-1,4-glycosyl linkages in 3 from the nonreducing end, resulting in acarbose as the product (Supplementary Fig. 9a). HR-MS analysis following β-amylase treatment of 3 revealed that the m/z value of the hydrolysis product was 646.2560 [M + H]+ (Supplementary Fig. 10c). HR–MS/MS analysis of this hydrolysis product revealed m/z values of the daughter ions as 146.0811 [M+H]+ and 304.1393 [M+H]+, which was consistent with the HR-MS/MS analysis of acarbose (1) (Supplementary Fig. 10d). These results provided further evidence that two additional sugar moieties in 3 are linked at the nonreducing end of acarbose via an α-1,4-glycosidic bond. The chemical structure of 3 was therefore determined as O-α-d-maltosyl-(1 → 4)-acarbose (Fig. 3a).
1H NMR analysis of acarstatin B (4) revealed three additional characteristic hydrogens (δ5.36–δ5.31) on the C-1 sites of the six-membered sugar ring distinct from acarbose, in accordance with the HR-MS results. Compound 4 displayed a shift in the nonlabile hydrogen at valienol C-4 to δ4.17–δ4.06, deviating from the δ3.95 of acarbose, and indicating attachment of a newly introduced sugar ring to C-4 of acarbose. The H–H coupling constant of every junction point at C-1 of the six-membered sugar ring was consistently below 4 Hz, implying an α-configuration for the glycosidic linkages in 4. Combining 1H-, 13C-, COSY, HSQC, HMBC, and TOCSY NMR data, the chemical structure of 4 was therefore suggested to be O-α-d-maltotriosyl-(1 → 4)-acarbose (Supplementary Fig. 7, Supplementary Tables 11 and 12). HR–MS/MS analysis of acarstatin B indicated daughter ions with m/z values of 146.0813 [M+H]+, 304.1392 [M+H]+, 466.1919 [M+H]+, 628.2436 [M+H]+, and 790.2975 [M+H]+, which indicated that additional glucose moieties were located at the nonreducing end of acarbose (Fig. 3f). Following glucoamylase hydrolysis, the hydrolysis product exhibited an m/z value of 830.3081 [M+H]+ (Supplementary Fig. 11a). The treatment with β-amylase yielded a hydrolysis product with an m/z value of 808.3080 [M+H]+ (Supplementary Fig. 11c). HR–MS/MS analysis confirmed that the hydrolysis product of 4 following both glucoamylase and β-amylase treatment was 6 (Supplementary Figs. 11b and d). These results indicated that the three additional sugar moieties of 4 are linked at the nonreducing end of acarbose via α-1,4-glycosidic bonds. The chemical structure of 4 was therefore determined as O-α-d-maltotriosyl-(1 → 4)-acarbose (Fig. 3d).
Compound 2 and 5 with molecular weights of 808.3091 [M+H]+ and 1294.4666 [M+H]+, respectively, were further subjected to HR-MS/MS analysis for elucidating their chemical structures. HR–MS/MS analysis of 2 revealed daughter ions with m/z values of 146.0816 [M+H]+, 304.1379 [M+H]+, and 466.1912 [M+H]+, suggesting that one additional hexose moieties were substituted at the nonreducing end of acarbose. Similarly, HR–MS/MS analysis of 5 indicated daughter ions with m/z values of 146.0812 [M+H]+, 304.1392 [M+H]+, 466.1947 [M+H]+, 628.2483 [M+H]+, 790.2935 [M+H]+, and 952.3534 [M+H]+, which confirmed that four additional hexoses were located at the nonreducing end of acarbose. The chemical structures of 2 and 5 were therefore tentatively determined as O-α-d-glucosyl-(1 → 4)-acarbose and O-α-d-maltotetrosyl-(1 → 4)-acarbose, respectively (Supplementary Fig. 12).
2.4. Acarstatins A and B exhibited enhanced inhibitory activity towards α-amylase
We chose α-amylase from human saliva and porcine pancreas as targets to determine the inhibition constants (Ki) of acarstatins A and B, with acarbose as control. With human salivary α-amylase as the target, acarbose, acarstatin A, and acarstatin B exhibited Ki values of 4.01 μM, 2.53 nM, and 2.72 nM, respectively, indicating that acarstatins A and B have 1584- and 1478-fold greater inhibitory activity towards human salivary α-amylase than acarbose (Table 1, Supplementary Fig. 13). With porcine pancreatic α-amylase as the target, acarbose, acarstatin A, and acarstatin B exhibited Ki values of 6.54 μM, 4.16 nM, and 8.81 nM, respectively, indicating that acarstatins A and B have 1573- and 742-fold greater inhibitory activity towards porcine pancreatic α-amylase than acarbose (Table 1, Supplementary Fig. 14).
Table 1.
Inhibition constants of acarbose, acarstatin A and acarstatin B towards human salivary α-amylase and porcine pancreatic α-amylase.
| Inhibitors | Human salivary α-amylase |
Porcine pancreatic α-amylase |
||
|---|---|---|---|---|
| Ki | Inhibitory potency | Ki | Inhibitory potency | |
| Acarbose | 4.01 μM | – | 6.54 μM | – |
| Acarstatin A | 2.53 nM | 1584 | 4.16 nM | 1573 |
| Acarstatin B | 2.72 nM | 1478 | 8.81 nM | 742 |
2.5. Acarstatins A and B exhibited resistance to acarbose inactivation pathways
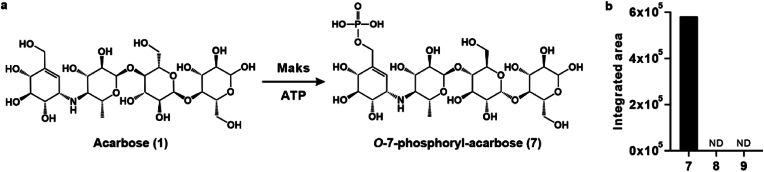
We used two in vitro enzymatic assays to assess the resistance of acarstatins A and B towards acarbose inactivation pathways. Maks, a series of human microbiome-derived acarbose kinases, deactivate acarbose by converting it to O-7-phosphoryl-acarbose. Our results indicated that Mak1 (Supplementary Fig. 15), the most prevalent and abundant Mak, catalyzed the conversion of acarbose to O-7-phosphoryl-acarbose (7), whereas phosphorylated acarstatins A (8) and B (9) were not detected in Mak1-catalyzed reactions (Fig. 4, Supplementary Fig. 16). These data suggested that acarstatins A and B are resistant to the phosphorylation pathway mediated by Mak1.
Fig. 4.
Evaluating resistance of acarstatins A and B of phosphorylation pathway mediated by Mak1. a, Schematic of Maks-catalyzed reaction with acarbose. b, Phosphorylated products detection in Mak1-catalyzed reactions with acarbose, acarstatin A, or acarstatin B.
The Apg-mediated hydrolysis pathway catalyzed the degradation of acarbose (1) into acarviosine-glucose (10) and acarviosine (11), both of which exhibited low inhibitory activity towards α-amylase. Our results showed that 1 is completely converted to 10 and 11 after Apg treatment, whereas incomplete hydrolysis of acarstatin A (3) to O-α-d-maltosyl-(1 → 4)-acarviosine-glucose (12) and O-α-d-maltosyl-(1 → 4)-acarviosine (13), and acarstatin B (4) to O-α-d-maltotriosyl-(1 → 4)-acarviosine-glucose (14) and O-α-d-maltotriosyl-(1 → 4)-acarviosine (15) was seen (Fig. 5, Supplementary Figs. 17, 18, 19). Analysis of the inhibitory activity revealed that the residual reaction retained 31%, 57%, and 71% of the inhibitory activity of acarbose, acarstatin A, and acarstatin B towards porcine pancreatic α-amylase, respectively (Supplementary Fig. 20). These results confirmed that acarstatins A and B exhibited better resistance towards Apg-catalyzed hydrolysis than acarbose.
Fig. 5.
Evaluating resistance of acarstatins A and B of hydrolysis pathway mediated by Apg. a, Schematic of Apg-catalyzed reaction with acarbose. b, Substrate detection in Apg-catalyzed reactions with acarbose (1), acarstatin A (3) or acarstatin B (4). c, First-class hydrolysis products detection of acarviosine-glucose (10), O-α-d-maltosyl-(1 → 4)-acarviosine-glucose (12), and O-α-d-maltotriosyl-(1 → 4)-acarviosine-glucose (14) in Apg-catalyzed reactions with 1, 3, or 4. d, Second-class hydrolysis products detection of acarviosine (11), O-α-d-maltosyl-(1 → 4)-acarviosine (13), and O-α-d-maltotriosyl-(1 → 4)-acarviosine (15) in Apg-catalyzed reactions with 1, 3, or 4.
3. Discussion
The increasing incidence of T2DM [1] and the emergence of acarbose resistance [38,39] necessitate the development of acarbose derivatives with enhanced efficacy. In this study, we isolated two novel acarbose derivatives, acarstatins A and B, with chemical structures of O-α-d-maltosyl-(1 → 4)-acarbose and O-α-d-maltotriosyl-(1 → 4)-acarbose, respectively. Both acarstatins A and B exhibited significantly enhanced inhibitory activity towards human salivary α-amylase compared with acarbose, as well as better resistance to both acarbose inactivation pathways encoded by the human microbiome [38,39].
Additional maltosyl and maltotriosyl groups located at the non-reducing end of acarbose significantly affected inhibitory activity of acarstatins A and B towards α-amylase. Based on previous co-crystal structure studies about α-amylase and acarbose, the following possible mechanism could be likely considered. In the crystal structure of α-amylase, maltosyl-acarviosyl-glucose was revealed as the actual inhibitor binding to the active site of α-amylase [7,9]. Acarstatins A and B share higher structural similarity to maltosyl-acarviosyl-glucose at the non-reducing end due to the presence of maltosyl and maltotriosyl groups. This structural similarity possibly contributes to their increased inhibitory activity towards α-amylase.
AcbE is an α-amylase involved in the previously proposed acarbose ‘Carbophore Cycle’, and may contribute to the viability of the acarbose-producing strain SE50/110 [31,32]. Recent research has confirmed that AcbE acts as an α-amylase to hydrolyze starch to facilitate carbohydrate utilization within its ecological niche [41]. In this study, we demonstrated that AcbE directly catalyzes the conversion of acarbose and oligosaccharides into acarbose derivatives with novel chemical structures, thereby highlighting the dual functionality of AcbE in hydrolysis and sugar moiety delivery. Several carbohydrate-active enzymes, including gseBMSA, pmaeCGTase, and imeDSR, have been shown to catalyze acarbose analog formation in addition to their roles in carbohydrate synthesis and breakdown. To compare AcbE with other enzymes that catalyze the formation of acarbose analogs and evaluate AcbE from an evolutionary perspective, we performed phylogenetic analysis on AcbE, gseBMSA [[21], [22], [23], [24]], pmaeCGTase [10,11], imeDSR [25], as well as AcbD [20], and several α-amylases using the neighbor-joining method. Unexpectedly, we found that these enzymes display considerable evolutionary divergence, and that AcbD or pmaeCGTase, which demonstrate closer evolutionary relationships to AcbE than to the other enzymes, only exhibit 28% and 27% amino acid identity, respectively (Supplementary Fig. 21). The broad array of enzymes expressed in the nature may therefore enrich the structural diversity of acarbose analogs with superior activity.
The chemical structures of acarstatins A and B, whose modifications were catalyzed by AcbE, are similar to compounds whose modifications were catalyzed by CGTase, including O-α-d-maltohexaosyl-(1 → 4)-acarbose, O-α-d-maltododecaosyl-(1 → 4)-acarbose, and O-α-d-maltooctadecaosyl-(1 → 4)-acarbose [10,11]. In the analogs modified by CGTase, maltohexaosyl units were incorporated as the fundamental glycosylation moieties, whereas in analogs modified by AcbE, glucopyranosyl units were incorporated. This enables investigation of the subsite structure of AcbE-catalyzed products more precisely than that of CGTase-catalyzed products [[7], [8], [9]]. While reaction conditions for AcbE have been optimized for the preparation of acarstatins, the yield of acarstatins A and B remained low. To improve the conversion efficiency of acarstatins A and B using enzymological techniques, various approaches to explore AcbE, including crystallography and site-specific mutagenesis, directed evolution, and immobilization, should be considered in future studies.
The interaction between intestinal microbes and ingested drugs is of increasing concern. Two distinct acarbose inactivation pathways, including Maks-mediated phosphorylation and Apg-mediated hydrolysis, are encoded by the human microbiome [38,39]. Donia et al. identified 21 Maks, including 8 acarbose kinases and 13 putative acarbose kinases [38]. Phylogenetic analysis of Apg homologues indicated that Apg-like enzymes are distributed across at least ten genera, including Klebsiella, Salmonella, and Raoultella [39]. In our study, we chose the most representative members of Maks and Apg, namely Mak1 and Klebsiella grimontii TD1 Apg, to evaluate the resistance of the newly identified acarstatins A and B, and found that both compounds exhibit significant resistance to phosphorylation and hydrolysis inactivation. Acarstatins A and B may therefore have potential as candidate therapeutic agents for T2DM.
The physiological and/or ecological role of acarbose production has been an intriguing topic for many decades. The ‘Carbophore Cycle’ has been proposed as a unique mechanism to improve sugar utilization using acarbose as a vehicle [31,32]. However, in the ‘Carbophore Cycle’, only the functions of acarviosyl transferases AcbD [20] and α-amylase AcbE [41] have been elucidated. Notably, the roles of most of the hypothetical components involved in the ‘Carbophore Cycle’, such as annotated extracellular α-amylase AcbZ, exporters AcbWXY, unknown importers, and intracellular hydrolases, have not been systematically elucidated. In this study, we found for the first time that AcbE can synthesize acarbose analogs with improved inhibitory activity towards α-amylase and that the deletion of acbE substantially decreases the inhibitory activity of the mutant strain. Highly active analogs catalyzed by AcbE in the natural environment may therefore also protect the producers by inhibiting carbohydrate degradation.
4. Methods
Strains, plasmids and media. The detailed information about strains, plasmids, primers, and culture media was listed in Supplementary Tables 1–4. Actinoplanes sp. SE50/110 and its derivative strains were used for the fermentation of acarbose and its analogs. Escherichia coli strains DH10B, ET12567 (pUZ8002) and Origami B (DE3) were used as hosts for gene cloning, E. coli-Actinoplanes bi-parental conjugation and heterologous protein expression, respectively. Integrative plasmid pSET152 and its derivative pLQ648 (containing a constitutive strong promoter kasOp*) were employed to overexpress target genes. Plasmid pET32a was used for heterologous protein expression.
Cultivation and fermentation of Actinoplanes sp. SE50/110 and derivative strains.Actinoplanes sp. SE50/110 and derivative strains were cultured on STY plates at 30 °C for 48 h. The mycelia were then transferred to S1 medium and cultured at 30 °C with shaking at 220 r.p.m. for 36 h. Subsequently, the S1 culture was inoculated into S2 medium to a final concentration of 9% (v/v) and incubated at 30 °C with shaking at 220 r.p.m. for 24 h. Finally, the S2 culture was inoculated into fermentation medium to a final concentration of 10% (v/v) and incubated at 30 °C with shaking at 220 r.p.m. for 72 h before the detection of acarbose and its analogs.
Gene complementation and overexpression in Actinoplanes sp. SE50/110 and derivative strains. The complementation of the acbE mutant was carried out using the integrative plasmid pLQ648. The complete acbE gene was amplified using primers acbE-L and acbE-R. Linearized pLQ648 was obtained by digestion with FastDigest NdeI and EcoRI (Thermo Scientific). The PCR-amplified fragment and the linearized pLQ648 were assembled using the NovoRec® plus One step PCR Cloning Kit (NovoProtein). The resulting construct was transformed into E. coli DH10B by calcium transformation. After verification by sequencing, the constructed plasmid was transferred into E. coli ET12567 (pUZ8002) via calcium transformation and then introduced into ZX-2 by E. coli-Actinoplanes bi-parental conjugation [30].
The overexpression of the acbE was performed using the integrative plasmid pSET152. The DNA fragment, which includes 1188-bp upstream of acbE, the acbE gene, and 275-bp downstream of acbE, was amplified using primers acbE-C-L and acbE-C-R. Linearized pSET152 was obtained by digestion with FastDigest XbaI and EcoRI (Thermo Scientific). The PCR-amplified fragment and the linearized pSET152 were assembled using the NovoRec® plus One step PCR Cloning Kit (NovoProtein). The following steps were similar to those described earlier. The constructed plasmid was ultimately introduced into the QQ-3 by E. coli-Actinoplanes bi-parental conjugation.
The exconjugants were cultured on SFM plate containing 40 μg/mL apramycin and trimethoprim at 30 °C for 5 days. Subsequently, they were transferred to STY plate containing 0.20 μg/mL apramycin and trimethoprim and cultured at 30 °C for 2 days. PCR verification of the exconjugants was performed using primers 152-yz-L and acbE-yz-R.
Quantification of acarbose through high-performance liquid chromatography. In the sample preparation, 2 vol of Milli-Q water were added to the fermentation broth, followed by the addition of 3 vol of chloroform. After vigorous shaking, the mixture was centrifuged at 13,800 g for 2 min. The aqueous phase was filtered through a 0.22-μm filter and subsequently subjected to HPLC analysis. For HPLC determination, samples were analyzed on an Agilent series 1260 system (Agilent Technologies) equipped with an Agilent Original NH2 column (4.6 × 250 mm, 5 μm). It was eluted at a flow rate of 1 mL/min with an elution using 35% A (0.55 g/L K2HPO4, 0.64 g/L NaH2PO4 and 91 mL acetonitrile dissolved in 909 mL Milli-Q water) and 65% B (acetonitrile) for 40 min and detected at 210 nm.
Quantification of acarbose and its analogs through α-amylase-starch-iodine (ASI) colorimetric method. In the ASI assay, the reaction system comprised 10 mM Tris-HCl at pH 7.0, 80 μg/L porcine pancreatic α-amylase, 300 mg/L soluble starch and samples containing acarbose or its analogs. The samples, containing acarbose or its analogs, were mixed with α-amylase in the buffer system and allowed to incubate for 2 min. The reaction was initiated by adding soluble starch. After a 20-min incubation at 25 °C, the reaction was terminated by the addition of 5% Lugol's iodine solution (2% iodide and 4% potassium iodide), followed by absorbance measurement at 610 nm.
Heterologous protein expression and purification. The signal peptide and its cleavage site of AcbE were predicted by SignalP - 6.0 (https://services.healthtech.dtu.dk/services/SignalP-6.0/). It indicates that the signal peptide region of AcbE spans amino acids 1–32, with the cleavage site at A33. The DNA fragment of mature AcbE was amplified using primers AcbE-5-L and AcbE-5-R. Linearized pET32a was obtained by digestion with FastDigest EcoRI and HindIII (Thermo Scientific). The PCR-amplified fragment and the linearized pET32a were assembled using the NovoRec® plus One step PCR Cloning Kit (NovoProtein). The assembled product was transformed into E. coli DH10B by calcium transformation. The plasmid was verification by sequencing and then transferred into E. coli Origami B (DE3) by calcium transformation.
Plasmids for the expression of Mak1 and Apg were ordered from Sangon Biotech. These plasmids were subsequently transferred into E. coli Origami B (DE3) for the expression of Mak1 and Apg proteins.
The heterologous expression strain was cultured in Luria-Bertani medium containing 20 mg/L ampicillin at 37 °C with shaking at 220 r.p.m to an OD of 0.6. IPTG (isopropyl β-d-1-thiogalactopyranoside) was added at a final concentration of 1 mM to induce protein expression, followed by continued cultivation at 16 °C with shaking at 180 r.p.m for 18 h. Cells were harvested by centrifugation at 3000 g and 4 °C for 15 min and then washed once with pre-cold Buffer A containing 25 mM Tris-HCl (pH 7.2) and 300 mM NaCl.
Cells were suspended in pre-cold Buffer A and subjected to sonication. The mixture was centrifuged at 13,800 g for 30 min at 4 °C and the supernatant was collected. Ni-NTA resin (GE HealthCare) was employed for immobilizing the target proteins. Nonspecifically bound proteins were removed by washing with 25 mM and 50 mM imidazole dissolved in Buffer A. The target protein was then eluted using 250 mM imidazole in Buffer A. The eluate was collected, and the imidazole concentration was reduced to below 12.5 mM using ultrafiltration. The purified protein was validated by SDS-PAGE, and the protein concentration was determined using NanoDrop One (Thermo Scientific).
In vitro reaction of AcbE. The AcbE reaction contained 25 mM Na2HPO4 at pH 6.5, 10 μM AcbE*, 5 mM acarbose, and 10 mM linear oligosaccharides (maltotriose, maltotetraose, or maltopentaose), or 10 mg/mL of other sugars (maltooligosaccharides or α-cyclodextrin). The reaction was carried out at 25 °C for 12 h and then terminated with methanol, which was added in a volume twice that of the reaction mixture. In control experiments, AcbE was deactivated by heating at 95 °C for 30 min, while other conditions remained the same.
For the quantitative assessment of inhibitory activity using the ASI method, the reaction mixture was evaporated at 40 °C to remove methanol and re-dissolved in distilled water for analysis. For the detection of reaction products by MALDI-TOF and LC-MS, the reaction mixture containing methanol was centrifuged at 13,800 g for 10 min, and the resulting supernatant was filtered through a 0.22-μm filter and subsequently subjected to MALDI-TOF and LC-MS analysis.
Qualitative analysis of acarbose and its analogs by MALDI-TOF. In the preparation of the matrix solution, 2,5-dhydroxybenzoic acid was dissolved at a concentration of 100 mg/mL in TA30 (30% acetonitrile, 70% Milli-Q water, and 0.1% trifluoroacetic acid, v/v). A mixture of 0.5 μL of the sample and 0.5 μL of the matrix solution was spotted onto MTP 384 target plate ground steel BC (8,280,784) ID 210 (Bruker) and allowed to dry. For MALDI-TOF determination, samples were analyzed on an ultrafleXtreme (Bruker) using reflectron mode and positive polarity.
Qualitative detection of acarbose derivatives via thin-layer chromatography. The chromatography plate used was TLC Silica gel 60 F254 Alu (Merck). The mobile phase consisted of n-butanol: ethanol: water = 9: 7: 4 (v/v/v), with the addition of 0.1% formic acid. Samples were spotted on the plates, 0.5 cm from the lower edge, and allowed to develop in the mobile phase for 45 min. A hair dryer was used for rapid drying of the chromatography plate. Subsequently, a staining solution, comprising 2.5 g of ammonium molybdate tetrahydrate, 1 g of ammonium cerium sulfate, 10 mL of sulfuric acid, and 90 mL of distilled water, was evenly sprayed onto the chromatography plate, followed by heating to 105 °C for visualization.
Preparation and purification of acarbose derivatives. Acarbose derivatives were prepared using an in vitro enzymatic reaction catalyzed by AcbE. The reaction system consisted of 25 mM Na2HPO4 at pH 6.5, 10 μM AcbE*, 5 mM acarbose and 20 g/L maltooligosaccharides, with a total volume of 300 mL. The reaction was performed at 25 °C for 12 h and terminated by adding 600 mL methanol.
Following solvent removal by rotary evaporation at 40 °C, the sample was reconstituted in 500 mL of distilled water and adjusted to pH 5.0. The sample was then slowly injected into a Dowex 50WX8 (Sigma-Aldrich) column at a flow rate of 1 mL/min. Subsequently, 1000 mL of distilled water was introduced through the column at a rate of 1 mL/min to collect the effluent. After solvent removal by rotary evaporation at 40 °C, the sample was redissolved in 10 mL of a solution containing 30% methanol and 70% Milli-Q water. The 10 mL sample was loaded onto a Sephadex LH-20 (GE Healthcare) column and eluted at a rate of 0.1 mL/min using a mobile phase consisting of 30% methanol and 70% Milli-Q water. Fractions containing acarbose derivatives were collected, combined, and then subjected to solvent removal by rotary evaporation at 40 °C. The resulting residue was redissolved in 10 mL of Milli-Q water. Then 5 mL sample was injected into an UniRPC30SCX column (NanoMicro Tech) at a flow rate of 1 mL/min. Subsequently, 50 mL of Milli-Q water was introduced through the column at a flow rate of 1 mL/min, followed by elution with 50 mL of 0.5 M hydrochloric acid at the same flow rate. Fractions containing acarbose derivatives from the hydrochloric acid eluate were collected, combined, and then subjected to solvent removal by rotary evaporation at 40 °C. The sample was redissolved in 2 mL of a solution consisting of 30% methanol and 70% Milli-Q water. Subsequently, the 2 mL sample was loaded onto a Sephadex LH-20 (GE Healthcare) column and eluted at a flow rate of 0.05 mL/min using a mobile phase consisting of 30% methanol and 70% Milli-Q water. Fractions containing different acarbose derivatives were collected separately. During the purification process, fractions containing acarbose derivatives were determined using thin-layer chromatography or MALDI-TOF.
Qualitative analysis of acarbose and its analogs by LC-MS. For LC-MS determination, samples were analyzed on an Agilent series 1290-MS 6546 system (Agilent Technologies) equipped with an Agilent Original NH2 column (4.6 × 250 mm, 5 μm). It was eluted at a flow rate of 0.4 mL/min with a mobile phase of 35% A (Milli-Q water) and 65% B (acetonitrile) for 60 min and detected at 210 nm. The ion source was an AJS ESI model with positive polarity. The gas temperature, flow and nebulizer pressure were 325 °C, 8 L/min and 35 psi, respectively. The fragmentor, skimmer and OCT 1 RF Vpp were set at 175, 65 and 750 V, respectively.
Elucidation of chemical structures of acarbose derivatives through glycosidase hydrolysis. The glucoamylase reaction contained 100 mM sodium acetate-acetic acid buffer at pH 5.0, 1 U/mL glucoamylase and 1 mM acarbose derivatives. The reaction was carried out at 50 °C for 3 h and then terminated with methanol, which was added in a volume twice that of the reaction mixture. In control experiments, glucoamylase was deactivated by heating at 95 °C for 30 min, while other conditions remained the same.
The β-amylase reaction contained 100 mM sodium acetate-acetic acid buffer at pH 4.5, 1 U/mL β-amylase and 1 mM acarbose derivatives. The reaction was conducted at 55 °C for 3 h and then terminated with methanol, which was added in a volume twice that of the reaction mixture. In control experiments, β-amylase was deactivated by heating at 95 °C for 30 min, while other conditions remained the same.
The sample preparation processes for ASI, MALDI-TOF and LC-MS analysis were consistent with the previously described methods.
Determination of Ki values for acarbose derivatives. In the determination of Ki values for acarbose and its derivatives on human salivary α-amylase, reactions contained 10 mM Tris-HCl at pH 7.0, 10 mg/L α-amylase, starch as the substrate, and acarbose or its derivatives as the inhibitor. Starch concentrations included 0, 0.58, 1.16, 1.69, 2.23, 3.24, 4.42, and 5.55 g/L. Acarbose concentrations were 0, 3, 6, and 9 μM. Acarstatin A concentrations were 0, 3, 6, and 9 nM. Acarstatin B concentrations were 0, 4, 8, and 12 nM. The initial reaction rate (V0) was determined by quantifying of maltose production with 3,5-dinitrosalicylic acid (DNS) method, which was used for the colorimetric quantification of reducing sugars.
In porcine pancreatic α-amylase assay, reactions contained 10 mM Tris-HCl at pH 7.0, 80 μg/L α-amylase, starch and acarbose or its derivatives. Starch concentrations included 0, 0.58, 1.16, 1.69, 2.23, 3.24, and 4.42 g/L. Acarbose concentrations were 0, 1, 2, and 3 μM. Acarstatin A concentrations were 0, 0.3, 0.6, and 0.9 nM. Acarstatin B concentrations were 0, 0.3, 0.6, and 0.9 nM. The initial reaction rate (V0) was determined by quantifying of maltose production.
The substrate concentrations [S], inhibitor concentrations [I], and their corresponding initial reaction rates [V0] for different groups were recorded, and inhibition constants Ki were fitted using Graphpad Prism.
In vitro reaction of Mak1. The Mak1-catalyzed reactions contained 100 mM Tris-HCl at pH 7.0, 10 μM Mak1, 0.5 mM acarbose or its derivatives, 2 mM ATP, and 10 mM MgCl2. The reaction was carried out at 30 °C for 2 h and then terminated with methanol, which was added in a volume twice that of the reaction mixture. In control experiments, Mak1 was deactivated by heating at 95 °C for 30 min, while other conditions remained the same. The sample preparation processes for ASI and LC-MS analysis were consistent with the previously described methods.
Evaluation of acarbose and its analogs phosphorylation by LC-MS. For LC-MS determination, samples were analyzed on an Agilent series 1290-MS 6546 system (Agilent Technologies) equipped with an Agilent ZORBAX SB-Aq column (4.6 × 250 mm, 5 μm). It was eluted at a flow rate of 0.4 mL/min with 99.5% A (Milli-Q water) and 0.5% B (methanol) for 30 min. The ion source was an AJS ESI model with negative polarity. The gas temperature, gas flow, nebulizer pressure, fragmentor, skimmer and OCT 1 RF Vpp parameters aligned with the previous described method.
In vitro reaction of Apg. The Apg-catalyzed reactions contained 50 mM Tris-HCl at pH 7.9, 10 μM Apg, 1 mM acarbose or its derivatives, 100 mM NaCl. The reaction was carried out at 37 °C for 2 h and then terminated with methanol, which was added in a volume twice that of the reaction mixture. In control experiments, Apg was deactivated by heating at 95 °C for 30 min, while other conditions remained the same. The sample preparation processes for ASI and LC-MS analysis were consistent with the previously described methods.
CRediT authorship contribution statement
Xin Zhang: Conceptualization, Investigation, Methodology, Data curation, Formal analysis, Writing – original draft, Writing – review & editing. Qungang Huang: Data curation, Formal analysis, Writing – original draft. Ziyue Guo: Methodology, Data curation, Writing – review & editing. Feifei Cai: Methodology, Formal analysis, Writing – review & editing. Qianjin Kang: Methodology, Data curation, Writing – original draft, Writing – review & editing. Linquan Bai: Conceptualization, Methodology, Data curation, Funding acquisition, Supervision, Writing – review & editing.
Declaration of competing interest
The author is an editorial board member for Synthetic and Systems Biotechnology and was not involved in the editorial review or the decision to publish this article. All authors declare that there are no competing interests.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (grant No. 2021YFC2100600), National Natural Science Foundation of China (grant No. 31830104), and Science and Technology Commission of Shanghai Municipality (grant Nos. 19JC1413000 and 19430750600) to L.B. We thank the Core Facility and Technical Service Center for SLSB and the Instrumental Analysis Center in SJTU for data collection. We thank Xiaobing Chang for helping with NMR data analysis. We thank Dr. Qinqin Zhao and Dan Zhang for helping with compound purification. We thank Dr. Wei Zhang and Dr. Wei Sun for suggestions on analysis of LC-MS and MALDI-TOF data.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2024.03.006.
Contributor Information
Qianjin Kang, Email: qjkang@sjtu.edu.cn.
Linquan Bai, Email: bailq@sjtu.edu.cn.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Magliano D.J., Boyko E.J. tenth ed. 2021. IDF diabetes Atlas. Brussels. [Google Scholar]
- 2.Walton R.J., Sherif I.T., Noy G.A., Alberti K.G. Improved metabolic profiles in insulin-treated diabetic patients given an α-glucosidehydrolase inhibitor. Br Med J. 1979;1(6158):220–221. doi: 10.1136/bmj.1.6158.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hillebrand I., Boehme K., Frank G., Fink H., Berchtold P. The effects of the α-glucosidase inhibitor BAY g 5421 (Acarbose) on meal-stimulated elevations of circulating glucose, insulin, and triglyceride levels in man. Res Exp Med. 1979;175(1):81–86. doi: 10.1007/BF01851236. [DOI] [PubMed] [Google Scholar]
- 4.Hillebrand I., Boehme K., Frank G., Fink H., Berchtold P. The effects of the α-glucosidase inhibitor BAY g 5421 (Acarbose) on postprandial blood glucose, serum insulin, and triglyceride levels: dose-time-response relationships in man. Res Exp Med. 1979;175(1):87–94. doi: 10.1007/BF01851237. [DOI] [PubMed] [Google Scholar]
- 5.Caspary W.F., Graf S. Inhibition of human intestinal α-glucosidehydrolases by a new complex oligosaccharide. Res Exp Med. 1979;175(1):1–6. doi: 10.1007/BF01851228. [DOI] [PubMed] [Google Scholar]
- 6.Hiele M., Ghoos Y., Rutgeerts P., Vantrappen G. Effects of acarbose on starch hydrolysis. Study in healthy subjects, ileostomy patients, and in vitro. Dig Dis Sci. 1992;37(7):1057–1064. doi: 10.1007/BF01300287. [DOI] [PubMed] [Google Scholar]
- 7.Qian M., Haser R., Buisson G., Duee E., Payan F. The active center of a mammalian α-amylase. Structure of the complex of a pancreatic α-amylase with a carbohydrate inhibitor refined to 2.2-Å resolution. Biochemistry. 1994;33(20):6284–6294. doi: 10.1021/bi00186a031. [DOI] [PubMed] [Google Scholar]
- 8.Brayer G.D., Sidhu G., Maurus R., Rydberg E.H., Braun C., Wang Y., et al. Subsite mapping of the human pancreatic α-amylase active site through structural, kinetic, and mutagenesis techniques. Biochemistry. 2000;39(16):4778–4791. doi: 10.1021/bi9921182. [DOI] [PubMed] [Google Scholar]
- 9.Li C., Begum A., Numao S., Park K.H., Withers S.G., Brayer G.D. Acarbose rearrangement mechanism implied by the kinetic and structural analysis of human pancreatic α-amylase in complex with analogues and their elongated counterparts. Biochemistry. 2005;44(9):3347–3357. doi: 10.1021/bi048334e. [DOI] [PubMed] [Google Scholar]
- 10.Yoon S.H., Robyt J.F. Addition of maltodextrins to the nonreducing-end of acarbose by reaction of acarbose with cyclomaltohexaose and cyclomaltodextrin glucanyltransferase. Carbohydr Res. 2002;337(6):509–516. doi: 10.1016/s0008-6215(02)00018-6. [DOI] [PubMed] [Google Scholar]
- 11.Yoon S.H., Robyt J.F. Study of the inhibition of four α amylases by acarbose and its 4IV-α-maltohexaosyl and 4IV-α-maltododecaosyl analogues. Carbohydr Res. 2003;338(19):1969–1980. doi: 10.1016/s0008-6215(03)00293-3. [DOI] [PubMed] [Google Scholar]
- 12.Murao S., Ohyama K. New amylase inhibitor (S-AI) from Streptomyces diastaticus var. amylostaticus No. 2476. Agric Biol Chem. 1975;39(11):2271–2273. doi: 10.1080/00021369.1975.10861952. [DOI] [Google Scholar]
- 13.Fukuhara K-i, Murai H., Murao S. Isolation and structure-activity relationship of some amylostatins (F-lb fraction) produced by Streptomyces diastaticus subsp. amylostaticus No. 9410. Agric Biol Chem. 1982;46:1941–1945. doi: 10.1080/00021369.1982.10865360. [DOI] [Google Scholar]
- 14.Geng P., Bai G. Two novel aminooligosaccharides isolated from the culture of Streptomyces coelicoflavus ZG0656 as potent inhibitors of α-amylase. Carbohydr Res. 2008;343(3):470–476. doi: 10.1016/j.carres.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Geng P., Qiu F., Zhu Y., Bai G. Four acarviosin-containing oligosaccharides identified from Streptomyces coelicoflavus ZG0656 are potent inhibitors of α-amylase. Carbohydr Res. 2008;343(5):882–892. doi: 10.1016/j.carres.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 16.Geng P., Sun T., Zhong Q., Li X., Shi L., Bai F., et al. Two novel potent α-amylase inhibitors from the family of acarviostatins isolated from the culture of Streptomyces coelicoflavus ZG0656. Chem Biodivers. 2013;10(3):452–459. doi: 10.1002/cbdv.201100451. [DOI] [PubMed] [Google Scholar]
- 17.Yokose K., Ogawa K., Sano T., Watanabe K., Maruyama H.B., Suhara Y. New α-amylase inhibitor, trestatins. I. Isolation, characterization and biological activities of trestatins A, B and C. J Antibiot (Tokyo) 1983;36(9):1157–1165. doi: 10.7164/antibiotics.36.1157. [DOI] [PubMed] [Google Scholar]
- 18.Yokose K., Ogawa K., Suzuki Y., Umeda I., Suhara Y. New α-amylase inhibitor, trestatins. II. Structure determination of trestatins A, B and C. J Antibiot (Tokyo) 1983;36(9):1166–1175. doi: 10.7164/antibiotics.36.1166. [DOI] [PubMed] [Google Scholar]
- 19.Itoh J., Omoto S., Shomura T., Ogino H., Iwamatsu K., Inouye S., et al. Oligostatins, new antibiotics with amylase inhibitory activity. I. Production, isolation and characterization. J Antibiot (Tokyo) 1981;34(11):1424–1428. doi: 10.7164/antibiotics.34.1424. [DOI] [PubMed] [Google Scholar]
- 20.Hemker M., Stratmann A., Goeke K., Schroder W., Lenz J., Piepersberg W., et al. Identification, cloning, expression, and characterization of the extracellular acarbose-modifying glycosyltransferase, AcbD, from Actinoplanes sp. strain SE50. J Bacteriol. 2001;183(15):4484–4492. doi: 10.1128/JB.183.15.4484-4492.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cha H.J., Yoon H.G., Kim Y.W., Lee H.S., Kim J.W., Kweon K.S., et al. Molecular and enzymatic characterization of a maltogenic amylase that hydrolyzes and transglycosylates acarbose. Eur J Biochem. 1998;253(1):251–262. doi: 10.1046/j.1432-1327.1998.2530251.x. [DOI] [PubMed] [Google Scholar]
- 22.Park K.H., Kim M.J., Lee H.S., Han N.S., Kim D., Robyt J.F. Transglycosylation reactions of Bacillus stearothermophilus maltogenic amylase with acarbose and various acceptors. Carbohydr Res. 1998;313(3–4):235–246. doi: 10.1016/s0008-6215(98)00276-6. [DOI] [PubMed] [Google Scholar]
- 23.Kim M.J., Lee S.B., Lee H.S., Lee S.Y., Baek J.S., Kim D., et al. Comparative study of the inhibition of alpha-glucosidase, α-amylase, and cyclomaltodextrin glucanosyltransferase by acarbose, isoacarbose, and acarviosine-glucose. Arch Biochem Biophys. 1999;371(2):277–283. doi: 10.1006/abbi.1999.1423. [DOI] [PubMed] [Google Scholar]
- 24.Lee S.B., Park K.H., Robyt J.F. Inhibition of β-glycosidases by acarbose analogues containing cellobiose and lactose structures. Carbohydr Res. 2001;331(1):13–18. doi: 10.1016/s0008-6215(01)00016-7. [DOI] [PubMed] [Google Scholar]
- 25.Yoon S.H., Robyt J.F. Synthesis of acarbose analogues by transglycosylation reactions of Leuconostoc mesenteroides B-512FMC and B-742CB dextransucrases. Carbohydr Res. 2002;337(24):2427–2435. doi: 10.1016/s0008-6215(02)00350-6. [DOI] [PubMed] [Google Scholar]
- 26.Hayashida M., Sakairi N., Kuzuhara H., Yajima M. Synthesis of "dihydroacarbose", an α-D-glucosidase inhibitor having a pseudo-tetrasaccharide structure. Carbohydr Res. 1989;194:233–246. doi: 10.1016/0008-6215(89)85022-0. [DOI] [PubMed] [Google Scholar]
- 27.Shibata Y., Kosuge Y., Mizukoshi T., Ogawa S. Chemical modification of the sugar part of methyl acarviosin: synthesis and inhibitory activities of nine analogues. Carbohydr Res. 1992;228(2):377–398. doi: 10.1016/0008-6215(92)84132-c. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Dai K., Xie K., Weng C. Biosynthesis and regulatory mechanism of acarbose and its structural analogs: a review. Chin J Biotechnol. 2022;38(2):605–619. doi: 10.13345/j.cjb.210248. [DOI] [PubMed] [Google Scholar]
- 29.Lee J.S., Hai T., Pape H., Kim T.J., Suh J.W. Three trehalose synthetic pathways in the acarbose-producing Actinoplanes sp. SN223/29 and evidence for the TreY role in biosynthesis of component C. Appl Microbiol Biotechnol. 2008;80(5):767–778. doi: 10.1007/s00253-008-1582-3. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Q., Xie H., Peng Y., Wang X., Bai L. Improving acarbose production and eliminating the by-product component C with an efficient genetic manipulation system of Actinoplanes sp. SE50/110. Synth Syst Biotechnol. 2017;2(4):302–309. doi: 10.1016/j.synbio.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wehmeier U.F., Piepersberg W. Biotechnology and molecular biology of the α-glucosidase inhibitor acarbose. Appl Microbiol Biotechnol. 2004;63(6):613–625. doi: 10.1007/s00253-003-1477-2. [DOI] [PubMed] [Google Scholar]
- 32.Alanzi A.R., Demessie A.A., Mahmud T. Biosynthesis and metabolic engineering of pseudo-oligosaccharides. Emerg Top Life Sci. 2018;2(3):405–417. doi: 10.1042/ETLS20180010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su B., Liu H., Li J., Sunli Y., Liu B., Liu D., et al. Acarbose treatment affects the serum levels of inflammatory cytokines and the gut content of bifidobacteria in Chinese patients with type 2 diabetes mellitus. J Diabetes. 2015;7(5):729–739. doi: 10.1111/1753-0407.12232. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X., Fang Z., Zhang C., Xia H., Jie Z., Han X., et al. Effects of acarbose on the gut microbiota of prediabetic patients: a randomized, double-blind, controlled crossover trial. Diabetes Ther. 2017;8(2):293–307. doi: 10.1007/s13300-017-0226-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu Y., Wang X., Li J., Zhang Y., Zhong H., Liu R., et al. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat Commun. 2017;8(1):1785. doi: 10.1038/s41467-017-01682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao L., Zhang F., Ding X., Wu G., Lam Y.Y., Wang X., et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359(6380):1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- 37.Santilli A.D., Dawson E.M., Whitehead K.J., Whitehead D.C. Nonmicrobicidal small molecule inhibition of polysaccharide metabolism in human gut microbes: a potential therapeutic avenue. ACS Chem Biol. 2018;13(5):1165–1172. doi: 10.1021/acschembio.8b00309. [DOI] [PubMed] [Google Scholar]
- 38.Balaich J., Estrella M., Wu G., Jeffrey P.D., Biswas A., Zhao L., et al. The human microbiome encodes resistance to the antidiabetic drug acarbose. Nature. 2021;600(7887):110–115. doi: 10.1038/s41586-021-04091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian J., Li C., Dong Z., Yang Y., Xing J., Yu P., et al. Inactivation of the antidiabetic drug acarbose by human intestinal microbial-mediated degradation. Nat Metab. 2023;5(5):896–909. doi: 10.1038/s42255-023-00796-w. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X., Zhao Q., Bai L. Functions of genes involved in extracellular acarbose metabolic pathway from Actinoplanes sp. SE50/110. Genom Appl Biol. 2022;41(5):1006–1016. doi: 10.13417/j.gab.041.001006. [DOI] [Google Scholar]
- 41.Tanoeyadi S., Tsunoda T., Ito T., Philmus B., Mahmud T. Acarbose may function as a competitive exclusion agent for the producing bacteria. ACS Chem Biol. 2023;18(2):367–376. doi: 10.1021/acschembio.2c00795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.