Highlights
-
•
First, we introduce and describe the feasibility of the CC as a screening platform for complex genetic regulation of SARS-Like group 2B coronaviruses pathogenesis.
-
•
Second, we define and describe 2 different CC-derived screening populations (broad F1 screening, and targeted intercrosses) resulting in identification of many significant host susceptibility loci.
-
•
Third, we show how candidate genes can be identified under susceptibility loci and validated for roles in CoV pathogenesis, such as Trim14.
-
•
Lastly, we report that ∼70 % of our identified QTL in mice contain one human GWAS hit, that have been shown to be associated with the development do severe COVID-19, showing potential cross-species mechanisms underlying sarbecovirus pathogenesis.
Keywords: SARS-CoV, SARS-CoV-2, Zoonotic CoV, Pathogenesis, Host susceptibility loci
Abstract
Coronavirus (CoV) cause considerable morbidity and mortality in humans and other mammals, as evidenced by the emergence of Severe Acute Respiratory CoV (SARS-CoV) in 2003, Middle East Respiratory CoV (MERS-CoV) in 2012, and SARS-CoV-2 in 2019. Although poorly characterized, natural genetic variation in human and other mammals modulate virus pathogenesis, as reflected by the spectrum of clinical outcomes ranging from asymptomatic infections to lethal disease. Using multiple human epidemic and zoonotic Sarbecoviruses, coupled with murine Collaborative Cross genetic reference populations, we identify several dozen quantitative trait loci that regulate SARS-like group-2B CoV pathogenesis and replication. Under a Chr4 QTL, we deleted a candidate interferon stimulated gene, Trim14 which resulted in enhanced SARS-CoV titers and clinical disease, suggesting an antiviral role during infection. Importantly, about 60 % of the murine QTL encode susceptibility genes identified as priority candidates from human genome-wide association studies (GWAS) studies after SARS-CoV-2 infection, suggesting that similar selective forces have targeted analogous genes and pathways to regulate Sarbecovirus disease across diverse mammalian hosts. These studies provide an experimental platform in rodents to investigate the molecular-genetic mechanisms by which potential cross mammalian susceptibility loci and genes regulate type-specific and cross-SARS-like group 2B CoV replication, immunity, and pathogenesis in rodent models. Our study also provides a paradigm for identifying susceptibility loci for other highly heterogeneous and virulent viruses that sporadically emerge from zoonotic reservoirs to plague human and animal populations.
1. Material and methods
1.1. Cells and viruses
Recombinant mouse-adapted clade Ia SARS-CoV MA15 (SARS-CoV-Urbani_AY278741), clade II HKU3-SRBD-MA (‘HKU3-MA’, BtCoV HKU3–1_DQ022305), and clade Ib SARS-CoV-2 MA10 (SARS-CoV-2_Wuhan_MN908947) virus were generated as described previously (Becker et al. 2008; Leist, et al. 2020; Roberts, et al. 2007; Schafer, et al. 2022) (Fig. 1). For virus titration, the caudal lobe of the right lung was homogenized in PBS, serial-diluted and inoculated onto confluent monolayers of Vero E6 cells (ATCC, CRL1586), followed by agarose overlay. Plaques were visualized with overlay of Neutral Red dye on day 2 (SARS-CoV MA15, HKU3-MA) or day 3 (SARS-CoV-2 MA10) post infection. All live virologic research was conducted by personnel wearing Tyvek suits, gloves and PAPR who used defined standard operating procedures (SOP) in a BSL3 facility with redundant fans.
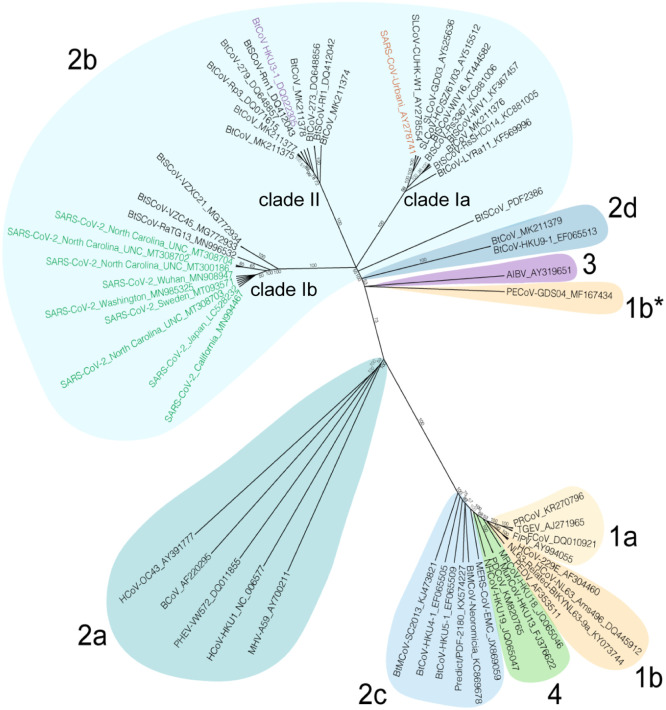
Fig. 1.
Spike phylogeny of representative coronaviruses. The Spike protein sequences of selected coronaviruses were aligned and phylogenetically compared. Coronavirus genera are grouped by classic subgroup designations (1a-b, 2a-d, 3, and 4). PECoV is designated as 1b* because of its distinctive grouping compared with more conserved proteins. Branches in each tree are labeled with consensus support values (in%). The clades for Sarbecoviruses (clades Ia, Ib, and II) are indicated. Sequences were aligned using free end gaps with the Blosum62 cost matrix, and the tree was constructed using the neighbor-joining method based on the multiple sequence alignment in Geneious Prime. Numbers following the underscores in each sequence correspond to the GenBank Accession number. The radial phylogram was exported from Geneious and then rendered for publication using Adobe Illustrator CC 2020.
1.2. Mouse studies and in vivo infections
Mouse studies were performed at the University of North Carolina (Animal Welfare Assurance #A3410–01) using protocols approved by the UNC Institutional Animal Care and Use Committee (IACUC). Animal studies at Washington University were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the IACUC at the Washington University School of Medicine (Assurance number A3381–01).
Mouse studies were divided into three major classes: CC-RIX (Collaborative Cross recombinant intercrosses) experiments, F2 intercross mouse experiments, and inbred wild-type or gene-edited mouse experiments (Fig. 2A). The laboratory of Dr. Pardo Manuel de Villena (FPMV) purchased CC mice from the Systems Genetics Core Facility (SGCF) at UNC between 2012 and 2018. These mice were used to breed F1s between pairs of CC RI strains, hereafter CC-RIXs (Recombinant Inbred Intercross) in the FPMV laboratory, to ensure proper cohorts and batch sizes. CC-RIXs were bred in a ring design such that each CC strain was used both as a dam and a sire in equal portions across all RIXs, and that each CC-RIX had one copy of the MHC H2Bb allele. Mice (115 CC-RIX strains, 3 animals each per timepoint for SARS-CoV MA15 infection; 3 animals per strain for heterologous challenge with HKU3-MA; all mice were females) were transferred at 5–6 weeks of age to the Baric (RSB) laboratory for infection between 9 and 12 weeks of age.
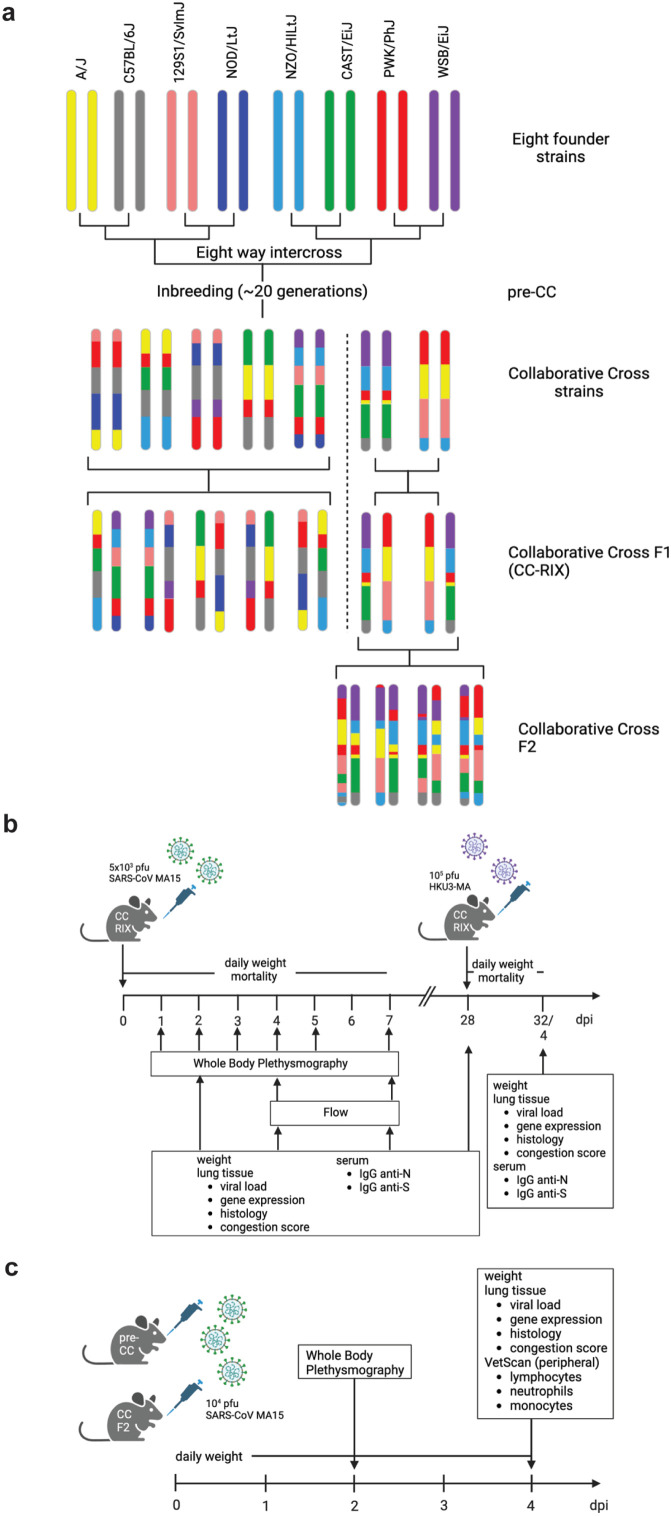
Fig. 2.
Overview of the individual Collaborative Cross derived mapping crosses used in the studies. a. The Collaborative Cross is comprised of eight founder strains: A/J, C57BL/6 J, 129S1/SvlmJ, NOD/ItJ, NZO/HILtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ. An eight-way intercrossing followed by ∼20 sister mating generations resulted in the CC-RI (recombinant inbred) lines. During the sister breeding, so-called pre-CC mice, mice which had not fully reached inbred status yet were used for proof-of-concept experiments. For other mapping studies CC F1 intercrosses (CC-RIX) and CC-F2 crosses were used. The study design for the CC-RIX (b.) and the pre-CC and CC-F2 (c.) mapping studies are shown. Figures were created with Biorender.com.
The details of the F2 intercross between CC003/Unc and CC053/Unc (hereafter, CC003 and CC053, respectively), and CC011/Unc and CC074/Unc hereafter, CC011 and CC074, respectively, have been described previously (Gralinski, et al. 2017; Schafer, et al. 2022).
CC-RIX (females only), CC-F2 mice (both sexes), and Trim14-deficient mice (both sexes) were infected with 5 × 103 plaque-forming units (PFU) (CC-RIX with SARS-CoV MA15), 1 × 104 PFU (CC-F2 with SARS-CoV MA15 and SARS-CoV-2 MA10), and 1 × 105 PFU (CC-RIX with HKU3-MA (reflecting similar ∼5 LD50 doses). C57BL/6 and Trim14Δ47/Δ47 mice were infected intranasally with 1 × 105 PFU with SARS-CoV MA15 and SARS-CoV-2 MA10, respectively, in 50 μl PBS, all between 9 and 12 weeks of age. Higher doses were used because of the more resistant C57BL/6 background. Body weight, mortality, and pulmonary function by whole body plethysmography (Menachery, et al. 2015a) were monitored daily where indicated. At indicated timepoints, mice were euthanized and gross pathology (congestions score) of the lung was assessed and scored on a scale from 0 (no congestion) to 4 (severe congestion affecting all lung lobes). Lung tissue was harvested for titer, histopathology analysis, and analysis of resident and infiltering cells by flow cytometry; and blood samples were harvested to determine antibody composition and for analysis of peripheral immune cells. Samples were stored at −80 °C until homogenized and titered by plaque assay as described above. Serum was prepared and SARS-CoV spike-specific antibody (N- and S-antigen) were quantified by ELISA as previously described (Fig. 2B-C).
1.3. Generation of trim14-deficient mice
The interferon-stimulated gene (ISG) Trim14 is encoded on Chr 4 in the mouse (Kuroda, et al. 2023). Gene-edited Trim14-deficient mice were generated with support from the Genome Engineering and iPSC center and Department of Pathology Micro-Injection Core (Washington University School of Medicine) . A sgRNA targeting exon 4 of Trim14 was selected based on minimal off-target effects in silico and targeting efficiency in vitro. The sgRNA (5′- ACCAATGGACACTCGCCTGANGG-3′) was synthesized, transcribed (HiScribe T7 In vitro Transcription Kit, New England BioLabs), and purified (MEGAclear Transcription Clean-Up Kit, Thermo Fisher). The sgRNA was mixed and co-injected with Cas9 RNA at 5 ng/μl and 10 ng/μl final concentrations into half-day-old C57BL/6 J embryos (E0.5). After next-generation sequencing of founders and two generations of mice backcrossed to C57BL/6 J mice, a mouse line with a 47-nucleotide deletion (5′-GCCTGAAGGAAAGTGAGTTGCCTAAGACCAACTCCAAGTCCTTGCTC-3′) encompassing the 3′ splice site of intron 4 and part of the coding region of exon 4 was generated. These Trim14Δ47/Δ47 mice were bred as homozygotes and used for experiments. Trim14Δ47/Δ47 mice were born in normal Mendelian frequencies and showed no apparent defects in development, growth, or fecundity. Lung tissue from Trim14Δ47/Δ47 were found to lack detectable Trim14 mRNA, likely due to nonsense-mediated decay, as measured by RT-qPCR using a predesigned primer/probe set for Trim14 (IDT, Assay ID Mm.PT.58.286730) and the housekeeping gene GAPDH (IDT, Assay ID Mm.PT.39a.1) (Fig. 5D). Sanger sequencing of a polymerase chain reaction amplicon [5′ GGCACAGCTCAACCCATGG −3′ (forward) and 5′- ACCAGCGAGCTCGTGCTCC −3′ (reverse)] was used for genotyping.
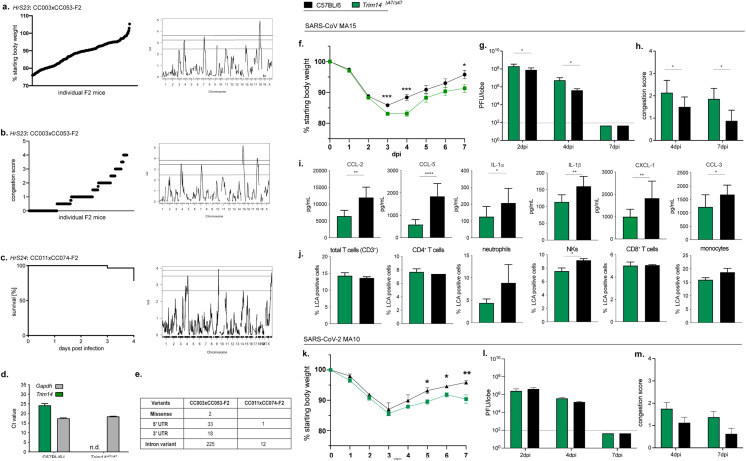
Fig. 5.
Identification of major effect locus on chromosome 4 and of Trim14 as a susceptibility gene during SARS-CoV MA15 and SARS-CoV-2 MA10 infection. a. Phenotypic distribution and genomic scan for HrS23 (4dpi weight loss in CC003xCC053-F2), b.HrS23 (4dpi lung congestion score in CC03xCC053-F2), and c. HrS24 (overall mortality in CC011xCC074-F2). d. Lung tissue from Trim14Δ47/Δ47 were found to lack detectable Trim14 mRNA, likely due to nonsense-mediated decay, as measured by RT-qPCR in comparison to littermates (n = 2 for Trim14Δ47/Δ47 and C57BL/6 J, respectively). e. SNPs in CC003 and CC053 as well as CC011 and CC074 in the Trim14 gene; Contributing haplotypes: CC003, WSB and CC053, CAST; CC011, PWK and CC074, S129; UTR- untranslated region. Infection of Trim14Δ47/Δ47 mice (n = 25; n = 9 for 2 and 4dpi, n = 7 for 7dpi) with SARS-CoV MA15 showed significantly more weight loss (f.) and an increase in viral load (g.) and congestion score in the lung (h.) compared to C57BL/6 J mice (n = 20; n = 10 for 2dpi, n = 6 for 4dpi, n = 4 for 7dpi) over the course of a 7-day infection. i. Cytokine/chemokine distribution in the lung of Trim14Δ47/Δ47 and C57BL/6 J at 4dpi of infection (n = 6 for Trim14Δ47/Δ4 and n = 6 for C57BL/6 J). j. Composition of lung infiltrating immune cells in the lung of Trim14Δ47/Δ4 mice and C57BL/6NJ control mice (n = 3 for each). A similar trend of infection progression was observed in Trim14Δ47/Δ47 mice infected with SARS-CoV-2 MA10; n = 10 Trim14Δ47/Δ47 and n = 10 C57BL/6 J for weight loss (k.), viral load (l.), and congestion score (m.). Data was analyzed via student t-test, *p < 0.05, **p < 0.01, ***p < 0.005).
1.4. Quantitatve trait locus (QTL) mapping and statistical analyses
For the CC-RIX, pre-CC, and the CC-F2 crosses, we used the same genetic mapping pipeline we previously described (Gralinski, et al. 2017; Noll, et al. 2020; Schafer, et al. 2022). Briefly, each CC-RIX had their genome represented as an array of probabilities of each of the 8 CC founder haplotypes. This array was used in the DOQTL R package (Gatti, et al. 2014) to run an 8-allele regression at each of 77,000 markers for our CC-RIX phenotypes. At each marker, a LOD (logarithm of the odds) score is calculated describing the goodness of fit of our trait∼genotype model relative to a null model. Significance was determined by running 1000 permutations scrambling the relationship between phenotypes and haplotypes. In this way, significance is independent of both population allele frequencies, as well as the phenotypic distribution.
2. Introduction
Natural host genetic variation regulates disease severity following most viral infections, yet the specific susceptibility loci and the natural allele variants that regulate differential disease outcomes remain largely unknown (Ge, et al. 2009; McLaren, et al. 2015). Coronaviruses (CoV) are significant human and animal pathogens and six (three human, three swine) novel CoVs have emerged or expanded their geographic range in the 21st century (Chen, et al. 2020; Wang, et al. 2019). Two of the most impactful emergent human CoVs (SARS-CoV and SARS-CoV-2) are group 2B coronaviruses, which likely emerged from bat reservoirs to cause world-wide human epidemics or global pandemics of respiratory illness, leading to substantial morbidity and mortality (Zhang and Holmes 2020; Zhou, et al. 2020). The 2003 SARS-CoV epidemic caused about 8000 infections with a 10 % mortality rate, while the ongoing SARS-CoV-2 pandemic caused >760 million infections and ∼7 million deaths to date (Ahmad, et al. 2020; Enserink and Kupferschmidt 2020). In 2012, Middle East Respiratory Coronaviruses emerged to cause sporadic disease in humans with an ∼35 % mortality rate (Annan, et al. 2013). Importantly, our group and others have shown that many high risk zoonotic group 2B SARS-like and group 2C MERS-like bat CoVs are poised for future human emergence events (Anthony, et al. 2017; Ge, et al. 2013; Hou, et al. 2023; Menachery, et al. 2015b, et al. 2016).
The Sarbecovirus subgenus of group 2B CoVs currently clusters into the clade Ia 2003 SARS-CoV and related high risk SARS-like Bat CoVs (BtCoV), clade II low risk SARS-like BtCoVs, clade Ib SARS-CoV-2, SARS-CoV-2 variants of concern (VoC) and related strains, and less well defined clade III and IV strains (Coronaviridae Study Group of the International Committee on Taxonomy of 2020). Sarbecoviruses vary in their ability to cause human disease although the BtCoV HKU3-related strains have not yet been reported to replicate in humans (Becker et al. 2008; Schafer et al. 2022). Evidence from emerging infections including SARS-CoV, influenza virus, ebolavirus, and flavivirus show that significant inter-host genetic heterogeneity influences disease outcomes in humans and mice (Cameron, et al. 2007; Rasmussen, et al. 2014; Sanchez et al., 2007). Despite a few examples of genes (e.g., Ccr9, Cxcr6, Mx1, and Oas1b) that regulate virus pathogenesis, the conservation of susceptibility loci within a virus family and across species remains understudied (Ferris, et al. 2013; Green, et al. 2017; Schafer, et al. 2022).
Thus, understanding the role of natural host variants in genes that regulate susceptibility and disease severity after Sarbecovirus infection in mice will not only provide insights into pathogenic mechanisms and effective host-based countermeasure strategies, but allow for the identification of any potential conserved genes under these loci between human and mice.
The angiotensin-converting enzyme 2 (ACE2) receptor interacts with the Spike protein (S) receptor binding domains of many but not all Sarbecoviruses (Shang, et al. 2020; Starr, et al. 2022). As many of these strains do not produce disease in mice, we had isolated variant clade Ia SARS-CoV MA15 and clade II HKU3-MA strains that replicate efficiently and produce severe disease in mice (Becker et al. 2008; Roberts, et al. 2007). This strategy has been most recently applied to SARS-CoV-2 (SARS-CoV-2 MA10) (Leist, et al. 2020). Mouse genetic reference populations (GRPs) have been used to identify host susceptibility loci, genes, genetic networks and higher-level genetic interactions that regulate phenotypic variation and disease severity in microbial diseases,(Gralinski, et al. 2015; Noll et al., 2019; Schafer et al., 2014). Among these reference populations, the Collaborative Cross (CC) mouse resource encodes over 44 million common single nucleotide polymorphisms (SNPs), 4 million common insertions and deletions (InDels), as well as several hundred thousand novel variants (both SNPs as well as InDels), present only in single strains (Collaborative Cross 2012; Srivastava, et al. 2017). Recently, we identified a multitrait locus for Sarbecovirus pathogenesis on Chr9 in mice (Schafer, et al. 2022). The defined region, which includes the Cxcr6, Ccr9, Xcr1, Fyco1, Ltztfl1, and Slc6a20 genes is in synteny of a region in humans on Chr3 which has been identified in several GWAS studies as a susceptibility region for hospitalization during SARS-CoV-2 infection and development of severe COVID-19 (Ellinghaus, et al. 2020; Pairo-Castineira, et al. 2020; Schafer, et al. 2022). Here, we more fully probe and synthesize results from a set of studies of CC mapping studies, and integrate them with known human GWAS hits to more fully assess the overlap in infectious disease susceptibility loci between model organism and human populations (Pairo-Castineira, et al. 2023).
3. Results
To identify additional susceptibility loci that regulate Sarbecovirus pathogenesis, we genetically mapped quantitative trait loci in: i) incipient mice from CC strains (partially inbred Pre-CC (Gralinski, et al. 2015), ii) a large panel of 115 F1 crosses between different inbred CC recombinant inbred (RI) strains (CC-RIX; an outbred population, like humans, but reproducible), and iii) two CC-F2 crosses generated from CC RI strains (Fig. 2a, Tablea 1, 2). In the latter example, independent intercross mapping populations (CC-F2) were derived from a cross between two genetically diverse CC strains, with extreme phenotype responses to aspects of SARS-CoV infection (Gralinski, et al. 2017; Schafer, et al. 2022). The overall goal was to use multiple CC mapping platforms to maximize the identification of many broadly relevant loci that control Sarbecovirus pathogenesis and adaptive immune responses.
Table 1.
List of significant QTLs in Pre-CC, CC-RIX, CC011xCC074-F2, and CC003xCC053-F2.
| QTL ID | Population | Chromosome [Mb] | Phenotype(s) (days post infection- dpi) | Haplotype(s)+ | phenotypic variation [%] |
|---|---|---|---|---|---|
| Pre-CC (Gralinski et al., 2015) | |||||
| HrS1 | Pre-CC | chr3:18.3–26.8 | Perivascular cuffing (4dpi) | BH/ACDEFG | 26 % |
| HrS2 | Pre-CC | chr16:31.5–36.7 | Viral titer (4dpi) | G/ABCDEFH | 22 % |
| HrS3 | Pre-CC | chr15:72.2–76.0 | Eosinophilia (4dpi) | A/BCDEFGH | 26 % |
| HrS4 | Pre-CC | chr13:52.7–54.9 | Perivascular cuffing (4dpi) | F/ABCDEGH | 21 % |
| CC-RIX (herein) | |||||
|---|---|---|---|---|---|
| HrS10 | CC-RIX | chr13: 20.3- 23.7 | HKU3-MA mortality | FG/ABCDEH | OR* 6.13 |
| HrS11 | CC-RIX | chr5: 112.9–118.8 | HKU3-MA mortality | C/ABDEFGH | OR* 10.45 |
| HrS12 | CC-RIX | chr15: 51.5- 75.6 | HKU3-MA weight loss (4dpi) | G/ABCDEFH | 3.4 % |
| HrS13 | CC-RIX | chr16:18–51.1 | SARS viral titer (2dpi) | C/G/ABDEFH | 15.94 % |
| HrS14 | CC-RIX | chr11: 9.05- 28.66 | PenH (2dpi) | D/CEH/ABFG | 11.65 % |
| HrS15 | CC-RIX | chr11: 3.26- 28 | Rpef (4dpi) | ABCEFGH/D | 6.48 % |
| HrS17 | CC-RIX | chr17: 27.4- 78.3 chr17: 27.4- 78.3 |
IgG1 (N) (32dpi) Total IgG (N) (32dpi) |
A/BCDEFH/G | 19.97 % 18.53 % |
| HrS18 | CC-RIX | chr16: 18.1- 43.8 | Total IgG (N) (32dpi) | ABCDEFH/G | 22.75 % |
| HrS19 | CC-RIX | chr10: 17.4- 130.5 | Total IgG (N) (32dpi) | ABCDEH/FG | 10.71 % |
| HrS20 | CC-RIX | chr3: 3.1- 35.4 | Total IgG (N) (32dpi) | G/ABCDEFH | 10.83 % |
| HrS22 | CC-RIX | chr6:67.2- 85 | CD8+ DCs [%] (4dpi) | F/ABCDEGH | 8.17 % |
| CC011xCC074-F2 (Schafer et al., 2022) | |||||
|---|---|---|---|---|---|
| HrS24 | CC011xCC074-F2 | chr4:32.95–114.54 | Mortality | CC011: G,H,C CC074:A/H, A/D, D, E, F |
OR* 4.34 |
| HrS25 | CC011xCC074-F2 | chr4: 6.38–17.97 | Weight loss in males (4dpi) | CC011: G, B CC074: A |
12.57 % |
| HrS26 | CC011xCC074-F2 | chr9:74.94–124.06 chr9:117.38–124.07 chr9:116.24–124.07 chr9:111.54–122.63 chr9:111.54–122.63 |
Mortality Hemorrhage (4dpi) PenH (2dpi) Periph. neutrophils (4dpi) Periph. lymphocytes (4dpi) |
CC011: B, G CC074: A |
OR* 3.15 10.24 % 7.76 % 11.8 % 12.39 % |
| HrS27 | CC011xCC074-F2 | chr11:26.44–80.76 chr11:17.89–80.76 |
Periph. neutrophils (4dpi) periph. lymphocytes (4dpi) |
CC011: H, B, H CC074: C |
5.5 % 5.6 % |
| HrS28 | CC011xCC074-F2 | chr15:58.66–74.04 | PenH (2dpi) | CC011: H CC074: B |
8.48 % |
| CC003xCC053-F2 (Gralinski et al., 2017) | |||||
|---|---|---|---|---|---|
| HrS5 | CC003xCC053-F2 | chr18:24.76–51.25 chr18:31.63–51.25 chr18:42.85–73.43 chr18:31.63–65.17 chr18:42.85–65.17 chr18:24.76–51.2 chr18:24.76–51.2 |
Weight loss (3dpi) Weight loss (4dpi) Congestion score (4dpi) Perivascular cuffing (4dpi) Edema (4dpi) Viral titer (4dpi) |
CC003: A, G CC053: C/G, G, B, B/D, B/C, D |
6 %−13.2 % |
| HrS6 | CC003xCC053-F2 | chr9:121.77-end | Weight loss (3dpi) | CC003: B, B/D CC053: H |
7 % |
| HrS7 | CC003xCC053-F2 | chr7:83.45–129.93 chr7:69.79–96.66 |
Weight loss (4dpi) Viral titer (4dpi) |
CC003: B, C, B/C CC053: G/H, H, D |
6.8 % 14.4 % |
| HrS8 | CC003xCC053-F2 | chr12:88.54–103.21 | Viral titer (4dpi) | CC003: D CC053:F/H |
4.27 % |
| HrS9 | CC003xCC053-F2 | chr15:10.7–57.3 | Congestion score (4dpi) | CC003: G, E/G, G CC053: D, C, F |
9.4 % |
| HrS23 | CC003xCC053-F2 | chr4:35.61–104.04 chr4:35.61–104.04 |
Congestion score (4dpi) Weigh loss (4dpi) |
CC003: H, E, G, H CC053: F, G, F |
6.86 % 6.22 % |
QTL influencing mortality calculate an Odds Ratio (OR) for disease rather than a% of variation explained. For both the CC-RIX and the CC-F2, Odds Ratios for specific loci are calculated with a full model of all mortality loci, to better estimate their independent effects.
Haplotype effects are described for each QTL. For the Pre-CC and CC-RIX, the haplotypes are separated based on the allele effect splits between founder haplotypes (A = A/J, B = C57BL/6 J, C = 129S1/SvImJ, D=NOD/ShILtJ, E=NZO/HiLtJ, F=CAST/EiJ, G=PWK/PhJ, H=WSB/EiJ). In CC-F2 crosses, the haplotypes listed are those present in the given parent strains from the proximal to distal ends of each region’
HrS10, HrS11, and HrS12: QTLs for HKU3-MA infection; all other QTLs are for SARS-CoV.
Table 2.
List of suggestive QTLs in CC011xCC074-F2 and CC003xCC053-F2.
| QTL ID | Chromosome [Mb] | Phenotype(s) (days post infection – dpi) | Haplotype(s)+ |
|---|---|---|---|
| CC011xCC074-F2 suggestive loci (Schafer et al., 2022) | |||
| HrS29 | chr3: 7.2–159.1 | Mortality (4dpi), Congestion score (4dpi) |
CC011: C, C/D, D, A, D, E CC074: D, H, B, B/F, B/D, B |
| HrS30 | chr9: 28.2–124.1 | Weight loss (4dpi) Mortality (4dpi) Weight loss females (4dpi) |
CC011: B, G, B CC074: A |
| HrS31 | chr13: 31.9–90.3 | Congestion score (4dpi) Periph. Monocytes (4dpi) |
CC011: B, C CC074: G, D |
| HrS37 | chr10: 5.9Mb- 86.3Mb | Titer (4dpi) | CC003: C, H CC053: H, B, A |
| HrS32 | chr14: 33.8–114.8 | PenH (2dpi) | CC011: C CC074: E, H, G |
| HrS33 | chrX: 6.2–93.9 | Rpef (2dpi) Periph. Neutrophils (4dpi) |
CC011: E, E/C, E, A, G CC074: G, B |
| CC003xCC053-F2 suggestive loci (herein) | |||
|---|---|---|---|
| HrS34 | chr3: 58- 148.4 | Weight loss (3dpi) | CC003: B, G, H, G CC053: C/D, C, D, A, E |
| HrS35 | chr17: 6- 79 | Weight loss (3dpi) | CC003: C/D, C, C/D, D, B CC053: G, E, G, H, A, G, H |
| HrS36 | chr9: 3.6Mb- 74.9Mb | Weight loss (4dpi), Titer (4dpi), Edema (4dpi) |
CC003: C, B CC053: B, E |
| HrS37 | chr10: 5.9Mb- 86.3Mb | Titer (4dpi) | CC003: C, H CC053: H, B, A |
| HrS38 | chrX: 9.4Mb- 169.5Mb | Titer (4dpi) | CC003: C, G, C/G, C/E, C, E CC053: A, C, B, F |
| HrS39 | chr2: 4.3Mb- 181.8Mb | mortality (4dpi) | CC003: D, B, E CC053: B, E, G, E, A, C, F |
| HrS41 | chr6: 34.2Mb- 114Mb | Interstitial septum (4dpi), Airspace inflammation (dpi4), Eosinophils (4dpi) | CC003: B, E CC053: G, A, A/D, G |
| HrS42 | chr11: 37Mb- 121.6Mb | Edema (4dpi) | CC003: A/C, B/C, A/C, B/H, B, E, H, C, E CC053: B |
Haplotype effects are described for each QTL. For F2 crosses, the haplotypes listed are those present in the given parent strains from the proximal to distal ends of each region (A = A/J, B = C57BL/6 J, C = 129S1/SvImJ, D=NOD/ShILtJ, E=NZO/HiLtJ, F=CAST/EiJ, G=PWK/PhJ, H=WSB/EiJ). In.
We infected the CC-RIX F1 population with two genetically distinct Sarbecoviruses, clade Ia SARS-CoV MA15 and clade II HKU3-MA (Fig. 1) (Becker, et al. 2008; Roberts, et al. 2007; Schafer, et al. 2022). Groups of CC-RIX mice were inoculated with 5 × 103 plaque-forming units (PFU) of SARS-CoV MA15 and virus titers, clinical disease (e.g., weight loss, mortality, and respiratory function), antibody titers, and immune cell infiltrates were measured at multiple timepoints post-infection (ranging from 2 to 32 days) (Fig. 2b, Fig. 3). Using a comparable LD50, on day 28 of SARS-CoV MA15, a cohort of CC-RIX was inoculated with 1 × 105 PFU of HKU3-MA (heterologous challenge) and evaluated for mortality and weight loss through day 4 post infection (Fig. 4). In both cases, virus challenge elicited an array of disease phenotypes, ranging from clinically inapparent infection to severe disease outcomes within the first 4 days of infection. We estimated genetic contributions for many of these traits and determined that heritability for these responses was 44.4 %−80.9 %, estimates that are in line with those for previous CC studies of infectious responses (Aylor, et al. 2011; Ferris, et al. 2013; Gralinski, et al. 2015; Maurizio, et al. 2018; Noll, et al. 2020; Schafer, et al. 2022). Importantly, many of the responses of the CC-RIX mice to various aspects of CoV-induced diseases appeared relatively uncorrelated, with CC-RIXs having a high response in one phenotype, showing divergent outcomes in other phenotypic responses (Fig. 4a-c). This suggests that there are independent genetic factors driving these responses. Similar findings have been reported in human SARS-CoV-2 patients (Ellinghaus, et al. 2020; Pairo-Castineira, et al. 2020), as well as our earlier studies using the CC (Ferris, et al. 2013; Gralinski, et al. 2015; Schafer, et al. 2022). We next conducted genetic mapping, seeking to find significant association between our disease outcomes and the eight CC parental haplotypes across the genome. We identified a set of loci exhibiting clear allele effects (Fig. 3, Fig. 4, Table 1) passing a significance threshold (p < 0.33) in line with community standards (Abiola, et al. 2003; Lander and Kruglyak 1995). These loci were associated with weight loss and mortality (SARS-CoV and HKU3-MA), as well as titer, antibody responses or respiratory function (SARS-CoV only). In total, we identified 11 distinct, high confidence loci impacting susceptibility or immunity to group 2B coronaviruses in our CC-RIX population, including resident CD8+ DC cells which play important roles in differential responsiveness to viral infection, including influenza virus (Beauchamp et al., 2012; Shortman and Heath 2010). Important for modeling disease responses, the effect sizes of these loci varied from 3 to 23 % of the total trait variation (that is, largely moderate effect sizes). These loci were in different genomic regions, had different causal founder alleles, and most loci primarily impacted only one or a few traits in this population. This later point further reinforces our observation (Fig. 4a-c), that disease states are largely uncorrelated during these Sarbecovirus infections. Together, this analysis highlights the genetic complexity driving coronavirus disease outcomes and immunity and the inability of any single animal model of CoV disease to fully address all aspects of the disease response.
Fig. 3.
Phenotypic distribution of disease phenotypes after SARS-CoV MA15 and HKU3-MA infection in the CC-RIX panel. Disease phenotype distribution after infection in the CC-RIX. Groups of 3 mice per CC-RIX strain were infected with either 5 × 103 PFU SARS-CoV MA15 and followed for 28 days to record disease outcomes (a-g). A set of 3 SARS-CoV MA15-infected mice per CC-RIX strain were then used for a heterologous challenged with 1 × 105 PFU HKU3-MA on day 28 post-infection and followed for 4 days for disease (h). a. Percentage survival (0dpi-4dpi, SARS-CoV MA15), b. Lung titer at 2 dpi (SARS-CoV MA15), c. PenH lung function at 2 dpi (respiratory metric for enhanced pause, SARS-CoV MA15), d. Rpef lung function at 2 dpi (respiratory metric for the ratio of time to peak expiratory flow (PEF) relative to total expiratory time, SARS-CoV MA15), total IgG (e) and IgG1 (f) at 32 dpi (SARS-CoV MA15) g. Frequency of lung infiltrating CD8+ Dendritic cells at 4 dpi (SARS-CoV MA15), h. Weight loss at 4 dpi (HKU3-MA), Each dot represents the mean value of an individual CC-RIX strain (n = 3 mice per strain).
Fig. 4.
Phenotypic distributions, genomics scans, and allele effects maps for 3 traits across the CC-RIX. a. (left to right) Log10 SARS-CoV MA15 lung titer at 2 dpi, percentage survival following HKU3-MA infection, and percentage of starting weight at 4 dpi with HKU3-MA. For all 3 panels, the CC-RIX strains are sorted by ascending 2 dpi SARS-CoV MA15 lung titers with each RIX having a row on the x-axis, and the replicate animals from each RIX occupying that row, showing the general lack of correlation between coronavirus disease responses. X-axis legends are over the panels. b. Genome scans showing the LOD traces (see methods), as well as significance thresholds (p = 0.33 (orange) and p = 0.2 (green)) for the same traits listed above. We identified (left to right) HrS13 (Chr16) for SARS-CoV MA15 titer, HrS11 (Chr5) and HrS10 (Chr13) for HKU3-MA mortality, and HrS12 for HKU3-MA weight loss. c. Allele effect plots showing the estimated phenotypic effect of the founder haplotypes at each of the peaks (shown at the bottom as the zoomed in LOD trace for chromosomes where we mapped QTL) at each of these loci showing causal haplotypes for HrS13 (where 129S1/SvImJ (pink) and PWK/PhJ (red) alleles cause a reduced titer, HrS10 where CAST/EiJ (green) and PWK/PhJ (red) alleles cause increased mortality, HrS11 where a 129S1/SvImJ (pink) allele causes increased mortality, and HrS12 where a PWK/PhJ (red) allele causes decreased weight loss. d. We identified relationships between HrS10 and SARS-CoV MA15-related weight loss and clinical disease (shown is weight loss at 4 dpi), where mice which had the PWK or CAST haplotypes were given a score of 1, and all other founder haplotypes given a score of 0), as well as HrS11 and SARS-CoV MA15 titer (shown here is titer at 2 dpi), 0 = low response haplotype, 1 = 1 copy of the high mortality 129s1 haplotype. Each dot represents data from an individual animal.
Given our more limited assessment of HKU3-MA disease responses, we evaluated whether any of our SARS-CoV MA15 phenotypes were also associated with the haplotypes driving severe HKU3-MA mortality (HrS10 and HrS11, Table 1, Fig. 4d). For these analyses, we binned CC-RIX based on their diplotypes at these loci (e.g. simplifying to susceptible/susceptible, susceptible/resistant, or resistant/resistant diplotypes), then determined whether this binning provided an improved fit to the relationship between SARS-CoV MA15 phenotypes and the CC-RIX IDs themselves. That is, was there any genetic signal at (QTL) HrS10 or HrS11 that was associated with differential SARS-CoV MA15 disease when simplifying the underlying causal model from having eight alleles with unknown effects to asking if traits are significantly associated with 0, 1, or 2 susceptibility alleles - a method we have used to find additional QTLs with small effect sizes in another CC-RIX genetically population (Noll, et al. 2020). HrS10 was associated with enhanced SARS-CoV MA15 wt loss, disease and mortality at day 2 post-infection (2 dpi), whereas HrS11 had moderate associations with SARS-CoV MA15 lung titers at both 2 and 4 dpi (Fig. 4d). These results indicate that common susceptibility loci regulate disease severity across two genetically distinct Sarbecovirus clades and that HrS10 and HrS11 are both associated with HKU3-MA-induced mortality, but likely contribute to virus-induced disease through different mechanisms (an unidentified one for HrS10, and potential inability to control viral replication for HrS11).
We combined our newly discovered QTL in the CC-RIX F1 with data from three prior studies, to provide a comprehensive list of QTLs identified affecting different aspects of Sarbecovirus pathogenesis (Tables 1 and 2). These results comprise i) incipient mice from CC strains (Pre-CC (Gralinski, et al. 2015)), ii) the large panel of 115 CC-RIX, described more fully above, and iii) a pair of CC-F2 mapping crosses we have previously reported on (CC003xCC053-F2 and CC011xCC074-F2) (Gralinski, et al. 2017; Schafer, et al. 2022) (Fig. 2a, Fig. 2c). In total this dataset represents 26 QTL observed across these population. Of note, one QTL for our CC011xCC074-F2 cross (HrS24) had Trim14 as a likely candidate underneath this Chr4 locus (Schafer, et al. 2022). We reinvestigated our CC003xCC053-F2 cross, as these two strains contain a different pair of alleles across this locus (Gralinski, et al. 2017). When controlling for the other loci mapped in the CC003xCC053-F2 cross previously, we found that there was a new genome-wide significant locus (HrS23) overlapping both the location of HrS24 and Trim14 on Chr 4. Both, HrS23 and HrS24, impacted weight loss, mortality, and congestion in the lungs as determined by gross pathology (Fig. 5a-c, Table 1). SNP variation between CC003 and CC053, as well as between CC011 and CC074 in this locus pointed to Trim14 as a likely candidate gene driving these differences in SARS-CoV MA15 disease (Fig. 5e). Previous work identified Trim14 as a key docking platform in the context of MAVS signaling (Tan, et al. 2017; Zhou, et al. 2014). We used CRISPR/Cas9 targeting to edit Trim14 in C57BL/6 J mice, create a functional knockout (Trim14Δ47/Δ47, Fig. 5d), and evaluate its role following infection. Trim14Δ47/Δ47 mice inoculated with 1 × 105 PFU of SARS-CoV MA15 had a modest increase in pathogenesis relative to C57BL/6 J control mice. At 3 and 4 dpi, mice deficient for Trim14 had increased weight loss, which corresponded with increases in viral titer within the lung at 2 and 4 dpi. This result shows that an absence of Trim14 delays viral clearance, suggesting a protective antiviral role for Trim14 during infection. Similarly, Trim14Δ47/Δ47 mice inoculated with SARS-CoV-2 MA10 also sustained modest increases in weight loss and a delayed recovery phenotype when compared to C57BL/6 mice. Further examination of SARS-MA infected Trim14-deficient mice at 4 dpi revealed a global reduction in several inflammatory cytokines and chemokines including CCL2, CCL5, IL1α, IL1β, CXCL1, and CCL3. Analogously, flow cytometry analysis showed a global decrease in immune cell populations in the lung at 4 dpi including NK cells, neutrophils, and monocyte-derived DCs, but not T cell populations. (Fig. 5f-h). However, the difference in viral titer seen at early times post SARS-CoV MA15 infection was not observed with SARS-CoV-2 MA10 (Fig. 5k-m). Together, these data suggest that Trim14 has a shared role in attenuating Sarbecovirus disease potential, but that this effect may vary between clade Ia and clade Ib strains and that common and shared QTLs among group 2B coronaviruses.
Finally, we compared identified QTL regions with recently identified GWAS regions, which have been associated with development of severe COVID-19, hospitalization, and death. A recent study by Pairo-Castineira et al., identified 49 genetic variants underlying critical COVID-19 disease (Table 3) (Pairo-Castineira, et al. 2023). We found several overlaps between human GWAS regions and our QTLs. Besides the multitrait locus on Chr9 region, which we identified and characterized recently as a major susceptibility locus for development of severe COVID-19 disease in mice (Schafer, et al. 2022), we found several other multitrait loci QTLs, which overlap with GWAS studies associated with severe disease. Importantly, we also identified ACE2 and TMPRSS2 as important factors for coronavirus replication in several overlapping QTLs, as well as RAB2a, a priority gene identified under mice and human Chr4 QTL and all three were noted in CRISPR/Cas9 screens as host factors required for SARS-CoV2 infection and CPE (Hoffmann, et al. 2021). Overall, approximately ∼60 % of our identified QTL in mouse contain one of these human candidate GWAS hits, showing potential cross-species mechanisms underlying Sarbecovirus pathogenesis (Table 3).
Table 3.
Common Genes Under Murine and Human QTL after Sarbecovirus Infection.Pairo-Castineira et al., 2023.
| Nearest gene | Chr:pos (GCRm39) | QTL ID |
|---|---|---|
| ELF5 | 2:103,242,443 | HrS39 |
|
HCN3 EFN4 TRIM46 THBS3 |
3:89,054,082 3:89,240,700 3:89,141,476 3:89,122,470 |
HrS29, HrS34 |
| ARHGEF38 | 3:132,818,039 | HrS29, HrS34 |
| AK5 | 3:152,168,461 | HrS29 |
|
RAB2A AQP3 TAC4 |
4:8535,661 4:41,092,724 4:95,152,335 |
HrS24 |
| JAK1 | 4:101,009,171 | HrS23, HrS24 |
| SLC2A5 | 4:150,203,801 | |
| FBRSL1 | 5:110,509,617 | |
| Oas1a | 5:121,034,319 | |
| HIP1 | 5:135,435,350 | |
| ZKSCAN1 | 5:138,083,312 | |
| NRH2 | 7:44,199,040 | |
| FUT2 | 7:45,298,015 | |
| MUC5B | 7:141,392,796 | |
| ATPHA | 8:12,807,016 | |
|
TYK2 PDE4A |
9:21,015,364 9:21,076,998 |
HrS36 |
| PLSCR1 | 9:92,132,265 | HrS30 |
|
SLC6A20 LZTFL1 CCR9 FYCO1 CXCR6 XCR1 |
9:123,465,972 9:123,523,469 9:123,596,276 9:123,618,565 9:123,635,542 9:123,681,380 |
HrS26, HrS30 |
| BCL11A | 11:24,026,498 | HrS14, HrS15 |
| IRF1 | 11:53,660,841 | HrS27, HrS42 |
|
PSMD3 KANSL1 |
11:98,573,380 11:104,224,327 |
HrS42 |
| SFTPD | 14:40,894,169 | HrS32 |
| TMPRSS2 | 16:36,934,306 | HrS2, HrS13, HrS18 |
| NXPE3 | 16:55,660,316 | |
| IFNAR2 | 16:91,166,517 | |
| IL10RB | 16:91,203,166 | |
| ATP5PO | 16:91,722,111 | |
|
LTA CCHCR1 FOXP4 |
17:35,422,141 17:35,829,143 17:48,178,058 |
HrS17, HrS35 |
| DPP9 | 17:56,493,674 | |
| ACE2 | X:162,922,338 | HrS38 |
| ICAM3 | no mouse homolog | |
| ABO | no mouse homolog | |
| HLA-G | no mouse homolog | |
| HLA-DPB1 | no mouse homolog |
4. Discussion
Our study highlights the power of rodent genetic reference populations (GRPs) to understand the role of host genetic variation on microbial disease, as well as to provide new models of disease progression and severity. Detailed studies in humans and mice support the hypothesis that SARS-CoV pathogenesis is a complex trait regulated by the accumulated impact of multiple loci that regulate disease attenuating or pathogenic outcomes. Similar findings have been reported in other human and rodent disease models (Ferris, et al. 2013; Kambhampati, et al. 2016). Together, these data demonstrate that appropriately selected large population screens in either species can be designed to identify highly penetrant genetic variants with varying impacts on specific aspects of disease outcome. One advantage of rodent reference populations is the availability of targeted back-crosses between phenotypically discordant strains to identify more complex networks and loci that are highly penetrant only in the context of specific genetic backgrounds and epistatic interactions. Additionally, private (de novo) mutations in specific mouse strains can also provide insight into relevant disease processes (Smith, et al. 2019; Zhang, et al. 2019).
We have leveraged large-scale population, as well as specific mouse strain mapping populations, to better characterize the genetic susceptibility landscape to Sarbecovirus infection (Gralinski, et al. 2015, et al. 2017; Schafer, et al. 2022). We have shown that a large number of polymorphic loci regulate the host disease responses to this subgroup of viruses. Further, we have identified specific genes like Trim55 (Gralinski, et al. 2015), Ticam2 (Gralinski, et al. 2017), Ccr9 and Cxcr6 (Schafer, et al. 2022), and herein Trim14, which encode naturally occurring polymorphisms that alter aberrant disease responses or virus replication. Another fundamental question in virology is whether genetically related viruses encode intrinsic properties that are regulated by similar natural variation in host genes in a species-dependent or independent manner. Importantly, we demonstrate that HrS10 and HrS11 influence disease severity following both clade I SARS-CoV MA15 and clade II HKU3-MA infection in the CC-RIX, supporting the hypothesis that intrinsic properties encoded within the Sarbecoviruses are subject to similar susceptibility loci in mammals. Likewise, CCR9 and CXCR6 alter the pathogenesis of clade Ia, clade Ib and clade II Sarbecoviruses (Schafer, et al. 2022). These findings are consistent with the discovery that group I and II human noroviruses infection and pathogenesis are heavily regulated by polymorphisms in fucosyltransferase 2 (Fut2) (Lindesmith, et al. 2003) while flavivirus and influenza virus infections are modulated by Oas1 and Mx1, respectively (Ferris, et al. 2013; Green, et al. 2017).
While empirical studies designed to validate the role of genes/alleles in infectious disease severity are restricted in human populations, no such restrictions exist in studying the role of host genes and allele variants in regulating disease severity across rodent models. Importantly, a wealth of cross species susceptibility loci, like the ones described herein, enable the identification of common QTL/genes across human and rodent genetic mapping populations (Ellinghaus, et al. 2020; Kuroda, et al. 2023; Pairo-Castineira, et al. 2023). Although speculative, the identification of ∼60 % concordance between common human and rodent genes under disease regulating QTL provides for focused studies in the mouse that are germane to human disease outcomes. For example, a variety of genes on human Chr3 and rodent Chr9 suggest the presence of conserved signaling pathways that regulate protective or pathogenic inflammatory/innate immune signaling networks across species. While our own data argue that CCR9 and CXCR6 are important drivers of disease severity, other groups have identified other cytokine signaling pathways, such as CCR2 which also reside under the Chr9 QTL in rodents (Schafer, et al. 2022; Vanderheiden, et al. 2021). Most Sarbecoviruses use ACE2 and TMPRSS2 to mediate docking and entry and a variety of human allele variants have been identified which regulate virus entry efficiency (Hoffmann, et al. 2020; Iwata-Yoshikawa, et al. 2022; Starr, et al. 2022). In our studies a variety of silent and coding mutations exist in these genes which could also influence virus entry and replication outcomes. A third example is Trim14, which is an important driver of disease across both species. Trim14 is an interferon-stimulated gene, which plays a role in innate immunity like interferon signaling and inflammatory cytokine production. During acute coronavirus infection, Trim14 had a protective function, as mice deficient for Trim14 lost significantly more weight and had significantly higher viral loads in their lungs. A study by Aquino Y et al. showed that Trim14, as a major player in IFN-mediated antiviral immunity, was targeted by signals of rapid adaption during SARS-CoV-2 infection (Aquino, et al. 2023). Also, a similar protective role for Trim14 has been demonstrated during EBOV infection (Kuroda, et al. 2023). Future studies are in process to identify the mechanism by which Trim14 mediates alterations in Sarbecovirus disease.
In addition to identifying a set of conserved genes across mammalian hosts with which to prioritize future studies, the unexplained heritability, and suggestive loci in rodent resource populations, suggests that CoV disease and immunity are even more complex and polygenic than had been previously suspected, but provides a platform for evaluating the role of such genes/alleles in rodent models. Our research findings suggest that common and unique susceptibility loci regulate Sarbecovirus pathogenesis, although the mechanistic details may differ across clades. As two or more contemporary group 2a human betacoronavirus and two group 1b alphacoronaviruses co-circulate in human populations, our data would predict that newly emergent Sarbecoviruses may cause similar disease phenotypes and be regulated by a subset of similar host loci across humans and mice, informing animal model development and host-based treatment regimens. Although predictive, additional research is clearly needed to validate the role of many of these commonly identified genes in humans and rodent populations, especially since these studies use the most highly correlative allele variant in human genetic databases to identify common genetic targets across species. As several common loci encode more than one common gene, detailed validation studies will be required to demonstrate genetic concordance across species. Moreover, new experimental crosses in rodent and human populations could provide greater mapping resolution and insight into the genetics of disease pathogenesis that informs the development of host-targeting therapeutics against CoVs.
CRediT authorship contribution statement
Alexandra Schäfer: Conceptualization, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft. Lisa E. Gralinski: Conceptualization, Formal analysis, Investigation, Validation, Visualization, Writing – review & editing. Sarah R. Leist: Investigation, Methodology, Validation, Visualization, Writing – review & editing. Brea K. Hampton: Investigation, Methodology, Visualization, Writing – review & editing. Michael A. Mooney: Funding acquisition, Investigation, Project administration, Resources, Writing – review & editing. Kara L. Jensen: Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. Rachel L. Graham: Formal analysis, Investigation, Methodology, Visualization, Writing – review & editing. Sudhakar Agnihothram: Investigation, Methodology, Writing – review & editing. Sophia Jeng: Investigation, Methodology, Writing – review & editing. Steven Chamberlin: Investigation, Methodology, Writing – review & editing. Timothy A. Bell: Investigation, Methodology, Writing – review & editing. D. Trevor Scobey: Investigation, Methodology, Writing – review & editing. Colton L. Linnertz: Investigation, Methodology, Writing – review & editing. Laura A. VanBlargan: Investigation, Methodology, Resources, Writing – review & editing. Larissa B. Thackray: Investigation, Methodology, Resources, Writing – review & editing. Pablo Hock: Investigation, Methodology, Writing – review & editing. Darla R. Miller: Investigation, Methodology, Resources, Writing – review & editing. Ginger D. Shaw: Investigation, Methodology, Resources, Writing – review & editing. Michael S. Diamond: Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. Fernando Pardo Manuel de Villena: Conceptualization, Funding acquisition, Investigation, Resources, Writing – review & editing. Shannon K. McWeeney: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Writing – review & editing. Mark T. Heise: Conceptualization, Formal analysis, Funding acquisition, Investigation, Writing – review & editing. Vineet D. Menachery: Conceptualization, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. Martin T. Ferris: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Writing – original draft. Ralph S. Baric: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by grants in aid from the National Institutes of Health, Allergy and Infectious Diseases: AI100625 and AI149644 (R.S.B., M.T.H., M.T.F., and F.P.M.V.), AI157253 (M.H. and R.S.B), AI167966 (R.S.B.), R00AG049092 (V.D.M.), R21AI145372 (L.E.G.), a contract from the NIH (U19 AI109761 to M.S.D and R.S.B), and support from NCATS (UL1TR002369 to M.A.M. and S.K.M.). We would like to thank the Systems Genetics Core Facility (UNC) for maintaining and distributing Collaborative Cross mice.
Contributor Information
Alexandra Schäfer, Email: aschaefe@email.unc.edu.
Lisa E. Gralinski, Email: lgralins@email.unc.edu.
Martin T. Ferris, Email: mtferris@email.unc.edu.
Ralph S. Baric, Email: rsbaric@email.unc.edu.
Data availability
Data will be made available on request.
References
- Abiola O., et al. The nature and identification of quantitative trait loci: a community's view. Nat. Rev. Genet. 2003;4(11):911–916. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad T., et al. Coronavirus Disease 2019 (COVID-19) pandemic and economic impact. Pak. J. Med. Sci. 2020;36(COVID19–S4):S73–S78. doi: 10.12669/pjms.36.COVID19-S4.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annan A., et al. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19(3):456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S.J., et al. Further evidence for bats as the evolutionary source of middle east respiratory syndrome Coronavirus. mBio. 2017;8(2) doi: 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino Y., et al. Dissecting human population variation in single-cell responses to SARS-CoV-2. Nature. 2023;621(7977):120–128. doi: 10.1038/s41586-023-06422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylor D.L., et al. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 2011;21(8):1213–1222. doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp N.M., Yammani R.D., Alexander-Miller M.A. CD8 marks a subpopulation of lung-derived dendritic cells with differential responsiveness to viral infection and toll-like receptor stimulation. J. Virol. 2012;86(19):10640–10650. doi: 10.1128/JVI.01413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.M., et al. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. u S. a. 2008;105(50):19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., et al. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81(16):8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., et al. Overview of lethal human coronaviruses. Signal. Transduct. Target. Ther. 2020;5(1):89. doi: 10.1038/s41392-020-0190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., et al. Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink M., Kupferschmidt K. With COVID-19, modeling takes on life and death importance. Science (1979) 2020;367(6485):1414–1415. doi: 10.1126/science.367.6485.1414-b. [DOI] [PubMed] [Google Scholar]
- Ferris M.T., et al. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS. Pathog. 2013;9(2) doi: 10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D.M., et al. Quantitative trait locus mapping methods for diversity outbred mice. G3. (Bethesda) 2014;4(9):1623–1633. doi: 10.1534/g3.114.013748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D., et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- Ge X.Y., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., et al. Genome wide identification of SARS-CoV Susceptibility loci using the collaborative cross. PLoS. Genet. 2015;11(10) doi: 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., et al. Allelic Variation in the toll-like receptor adaptor protein ticam2 contributes to SARS-Coronavirus pathogenesis in mice. G3. (Bethesda) 2017;7(6):1653–1663. doi: 10.1534/g3.117.041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R., et al. Oas1b-dependent immune transcriptional profiles of west nile virus infection in the collaborative cross. G3. (Bethesda) 2017;7(6):1665–1682. doi: 10.1534/g3.117.041624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H.H., et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host. Microbe. 2021;29(2):267–280. doi: 10.1016/j.chom.2020.12.009. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., et al. Host range, transmissibility and antigenicity of a pangolin coronavirus. Nat. Microbiol. 2023;8(10):1820–1833. doi: 10.1038/s41564-023-01476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., et al. Essential role of TMPRSS2 in SARS-CoV-2 infection in murine airways. Nat. Commun. 2022;13(1):6100. doi: 10.1038/s41467-022-33911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambhampati A., et al. Host genetic susceptibility to enteric viruses: a systematic review and metaanalysis. Clin. Infect. Dis. 2016;62(1):11–18. doi: 10.1093/cid/civ873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., et al. An antiviral role for TRIM14 in Ebola virus infection. J. Infect. Dis. 2023 doi: 10.1093/infdis/jiad325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E., Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 1995;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Leist Sarah R., et al. A mouse-adapted SARS-CoV-2 induces acute lung injury (ALI) and mortality in standard laboratory Mice. Cell. 2020 doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L., et al. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003;9(5):548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- Maurizio P.L., et al. Bayesian Diallel analysis reveals Mx1-dependent and Mx1-independent effects on response to influenza a virus in Mice. G3. (Bethesda) 2018;8(2):427–445. doi: 10.1534/g3.117.300438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren P.J., et al. Polymorphisms of large effect explain the majority of the host genetic contribution to variation of HIV-1 virus load. Proc. Natl. Acad. Sci. u S. a. 2015;112(47):14658–14663. doi: 10.1073/pnas.1514867112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., et al. New metrics for evaluating viral respiratory pathogenesis. PLoS. One. 2015;10(6) doi: 10.1371/journal.pone.0131451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21(12):1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. u S. a. 2016;113(11):3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll K.E., Ferris M.T., Heise M.T. The collaborative cross: a systems genetics resource for studying host-pathogen interactions. Cell Host. Microbe. 2019;25(4):484–498. doi: 10.1016/j.chom.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll K.E., et al. Complex genetic architecture underlies regulation of influenza-a-virus-specific antibody responses in the collaborative cross. Cell Rep. 2020;31(4) doi: 10.1016/j.celrep.2020.107587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., et al. Genetic mechanisms of critical illness in Covid-19. Nature. 2020 doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Pairo-Castineira E., et al. GWAS and meta-analysis identifies 49 genetic variants underlying critical COVID-19. Nature. 2023;617(7962):764–768. doi: 10.1038/s41586-023-06034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A.L., et al. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science (1979) 2014;346(6212):987–991. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS. Pathog. 2007;3(1):e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A., Wagoner K.E., Rollin P.E. Sequence-based human leukocyte antigen-B typing of patients infected with Ebola virus in Uganda in 2000: identification of alleles associated with fatal and nonfatal disease outcomes. J. Infect. Dis. 2007;196(Suppl 2):S329–S336. doi: 10.1086/520588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A., Baric R.S., Ferris M.T. Systems approaches to Coronavirus pathogenesis. Curr. Opin. Virol. 2014;6:61–69. doi: 10.1016/j.coviro.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A., et al. A Multitrait Locus Regulates Sarbecovirus Pathogenesis. mBio. 2022;13(4) doi: 10.1128/mbio.01454-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., et al. Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS. Pathog. 2020;16(3) doi: 10.1371/journal.ppat.1008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortman K., Heath W.R. The CD8+ dendritic cell subset. Immunol. Rev. 2010;234(1):18–31. doi: 10.1111/j.0105-2896.2009.00870.x. [DOI] [PubMed] [Google Scholar]
- Smith C.M., et al. Functionally Overlapping Variants Control Tuberculosis Susceptibility in Collaborative Cross Mice. mBio. 2019;10(6) doi: 10.1128/mBio.02791-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., et al. Genomes of the mouse collaborative cross. Genetics. 2017;206(2):537–556. doi: 10.1534/genetics.116.198838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., et al. ACE2 binding is an ancestral and evolvable trait of sarbecoviruses. Nature. 2022;603(7903):913–918. doi: 10.1038/s41586-022-04464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P., et al. Assembly of the WHIP-TRIM14-PPP6C mitochondrial complex promotes rig-i-mediated antiviral signaling. Mol. Cell. 2017;68(2):293–307. doi: 10.1016/j.molcel.2017.09.035. e5. [DOI] [PubMed] [Google Scholar]
- Vanderheiden A., et al. CCR2 signaling restricts SARS-CoV-2 infection. mBio. 2021;12(6) doi: 10.1128/mBio.02749-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., et al. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019;34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. A loss-of-function mutation in the integrin alpha L (Itgal) gene contributes to susceptibility to salmonella enterica serovar typhimurium infection in collaborative cross strain CC042. Infect. Immun. 2019;88(1) doi: 10.1128/IAI.00656-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181(2):223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., et al. TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I-like receptor-mediated innate immune response. Proc. Natl. Acad. Sci. u S. a. 2014;111(2):E245–E254. doi: 10.1073/pnas.1316941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.