Abstract
The feline homolog of the α-chemokine receptor CXCR4 has recently been shown to support cell-cell fusion mediated by CXCR4-dependent strains of human immunodeficiency virus (HIV) and strains of feline immunodeficiency virus (FIV) that have been selected for growth in the Crandell feline kidney (CrFK) cell line. In this report we demonstrate that expression of CXCR4 alone is sufficient to render cells from diverse species permissive for fusion with FIV-infected cells, suggesting that CXCR4 is the sole receptor for CrFK-tropic strains of FIV, analogous to CD4-independent strains of HIV-2. To identify the regions of CXCR4 involved in fusion mediated by FIV, we screened panels of chimeric CXCR4 molecules for the ability to support fusion with FIV-infected cells. Human CXCR4 supported fusion more efficiently than feline CXCR4 and feline/human CXCR4 chimeras, suggesting that the second and third extracellular loops of human CXCR4 contain a critical determinant for receptor function. Rat/human CXCR4 chimeras suggested that the second extracellular loop contained the principal determinant for receptor function; however, chimeras constructed between human CXCR2 and CXCR4 revealed that the first and third loops of CXCR4 contribute to the FIV Env binding site, as replacement of these domains with the corresponding domains of CXCR2 rendered the molecule nonfunctional in fusion assays. Mutation of the DRY motif and the C-terminal cytoplasmic tail of CXCR4 did not affect the ability of the molecule to support fusion, suggesting that neither signalling via G proteins nor receptor internalization was required for fusion mediated by FIV; similarly, truncation of the N terminus of CXCR4 did not affect the function of the molecule as a receptor for FIV. CXCR4-transfected feline cells were rendered permissive for infection with both the CrFK-tropic PET isolate of FIV and the CXCR4-dependent RF strain of HIV-1, and susceptibility to infection correlated well with ability to support fusion. The data suggest that the second extracellular loop of CXCR4 is the major determinant of CXCR4 usage by FIV.
The initial stage in infection with human immunodeficiency virus (HIV) involves an interaction between the viral envelope glycoprotein (Env) and CD4 on the surface of the target cell. However, fusion of the viral envelope and the plasma membrane of the target cell requires a further interaction between Env and a member of the seven-transmembrane domain (7TM) superfamily of G-protein-coupled receptors. Strains of HIV that form syncytia with the MT-2 cell line (27), formerly classified as syncytium-inducing (SI) strains, have subsequently been shown to interact with the α-chemokine receptor CXCR4 (21), while strains of the virus that fail to form syncytia with MT-2 cells (non-syncytium-inducing [NSI] strains) interact predominantly with the β-chemokine receptor CCR5 (2, 9, 15, 17, 18). Subsequent studies demonstrated that additional members of the 7TM superfamily were able to support infection with HIV, and in some cases simian immunodeficiency virus (SIV), including additional β-chemokine receptors such as CCR2b and CCR3 (9, 17, 43, 47), the receptor encoded by human cytomegalovirus US28 (41), and the orphan receptors STRL33/Bonzo (16, 30), BOB/GPR15 (16, 22), V28 (42, 43), and GPR1 (34). Previous studies have demonstrated that SI variants of HIV appear with disease progression (27, 28, 45) and that the switch to an SI phenotype is accompanied by an increase in the net charge of the V3 loop (14). The discovery that 7TM molecules act as coreceptors for HIV infection has provided a molecular basis for the NSI-to-SI switch. The nature of the coreceptor used by HIV for infection alters with disease progression (5, 11); in early infection CCR5-dependent viruses predominate, while in late infection CXCR4-dependent viruses are more abundant (11). Similarly, the alterations in the V3 loop that generate an SI phenotype select for usage of CXCR4 as a coreceptor (9, 48). The differential usage of chemokine receptors for infection by HIV has prompted the adoption of a new nomenclature in which the nature of the chemokine receptor used by the virus defines the phenotype of the virus: a virus using a CXC receptor would receive the suffix “X,” while a virus using a CC receptor would receive the suffix “C” (4). Thus, an SI virus may be defined as X4, for CXCR4 usage, whereas an NSI virus may be defined as C5, for CCR5 usage.
Variants of feline immunodeficiency virus (FIV) that are able to infect and induce cell fusion in the Crandell feline kidney cell line (CrFK) use the chemokine receptor CXCR4 as cofactor for infection (54, 55). Furthermore, such CrFK-tropic viruses display an increase in charge of the V3 loop (46, 52), analogous to the changes observed in the V3 loop of HIV with the switch from an NSI to an SI phenotype (14). Infection with CrFK-tropic strains of FIV resembles CD4-independent infection with HIV type 2 (HIV-2). CD4-independent infection with HIV-2 is mediated by CXCR4, transfection of cells from diverse species with CXCR4 alone renders them susceptible to infection, and infection is inhibited by the anti-CXCR4 antibody 12G5 (20). Moreover, the envelope glycoprotein from CXCR4-dependent strains of HIV interacts directly with CXCR4 on CD4-negative cells (23). In comparison, human cells are rendered permissive for fusion with FIV-infected cells by transfection with CXCR4 (55), and the envelope glycoprotein from a CrFK-tropic strain of FIV competes with stromal cell-derived factor 1 for binding to feline CXCR4 (25).
The interaction between HIV and CXCR4 has been investigated by using a series of chimeric CXCR4 molecules. In studies using chimeric molecules generated between CXCR4 and the related α-chemokine receptor CXCR2, the first and second extracellular loops of CXCR4 were found to be important determinants of the interaction with T-cell-tropic or dualtropic strains of HIV (32). Macrophage-tropic strains could use CXCR4 only when the amino terminus was replaced with that of CCR5 (32). The principal determinant of CXCR4 usage by HIV appeared to be the second extracellular loop in that replacement of the first and third extracellular loops with the corresponding regions of CXCR2 generated molecules that retained the ability to support fusion mediated by some strains of HIV, albeit with a reduced efficiency (32). When chimeras were generated between human CXCR4 and rat CXCR4, similar findings were observed in that the second extracellular loop of CXCR4 was shown to be the key determinant of CXCR4 usage by the NDK strain of HIV-1 (6). Furthermore, chimeras constructed between murine and human CXCR4 have demonstrated that the second loop of human CXCR4 expressed in the context of murine CXCR4 renders canine thymocytes susceptible to infection with HIV-1 strains that are unable to use murine CXCR4 as a receptor (39).
In this study, we investigated the interaction between FIV and CXCR4. We demonstrate that CXCR4 is sufficient to render cells susceptible to both fusion and infection with a CrFK-tropic strain of FIV. Furthermore, using feline CXCR4/human CXCR4, human CXCR2/human CXCR4, and human CXCR4/rat CXCR4 chimeras, we show that the fusogenic determinant consists of a discontinuous epitope formed by the first, second, and third extracellular loops of CXCR4 with the second extracellular loop containing the major determinant of receptor function. Thus, despite their evolutionary divergence, FIV and HIV appear to use similar mechanisms to interact with CXCR4. The results suggest that a conserved structural feature exists in the envelope glycoproteins of both HIV and FIV and that this enables the viruses to bind specifically to CXCR4.
MATERIALS AND METHODS
Antibodies and reagents.
All culture media and supplements were obtained from Life Technologies, Paisley, United Kingdom.
Cell lines and viruses.
U87-T4 cells expressing human CXCR4 or CCR5 (U87-T4/huCXCR4 or U87-T4/huCCR5 cells) were obtained from Dan Littman, Skirball Institute for Biomolecular Medicine, New York University Medical Center. U87 and CrFK cells were maintained in Dulbecco’s modification of minimal essential medium supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, sodium pyruvate (0.11 mg/ml), penicillin (100 IU/ml), and streptomycin (100 μg/ml) (DMEM). The culture media for U87-T4/huCXCR4 and U87-T4/huCCR5 cells were supplemented with puromycin (1 μg/ml; Sigma) and G418 (400 μg/ml; Geneticin, Life Technologies). The CrFK-tropic virus FIVPET was prepared from a culture of CrFK cells persistently infected with FIVPET. CrFK cells persistently infected with the Petaluma (FIVPET) and Glasgow 8 (FIVGL8) isolates of FIV were generated by coculture of CrFK cells with concanavalin A-stimulated feline peripheral blood mononuclear cells infected with the Glasgow 8 and Petaluma isolates of FIV.
Plasmids.
The feline CXCR4/human CXCR4 chimeras were generated by using the HincII site (nucleotide 421 of GenBank entry X71635) of human CXCR4, corresponding to the third transmembrane domain, and the equivalent HincII site at nucleotide 377 of feCXCR4.pcDNA3 (GenBank entry U63558). As feline CXCR4 has a second HincII site at position 523, the appropriate fragments of feline CXCR4 were selected from a HincII partial digest of feCXCR4.pcDNA3. The nucleic acid sequences of the chimeric constructs H5F3 and F5H3 were confirmed by sequencing using Sequenase (Amersham Ltd.). The rat/human CXCR4 and human CXCR2/CXCR4 chimeras have been described previously (6, 32) and were obtained from Stephen Peiper, James Graham Brown Cancer Center, University of Louisville. Bonzo and BOB plasmids were obtained from Dan Littman; GPR1 and GPR15 were from Brian O’Dowd, University of Toronto; CCR1, -2b, -3, -4, and -5, CXCR1 and -2, EBI1 (42), and V28 (42) were obtained from Jacquie Reeves, Chester Beatty Laboratories, London, United Kingdom. Feline CCR5 was a generous gift from John Elder, The Scripps Research Institute, La Jolla, Calif. The molecular cloning and characterization of feline CCR5 by using the rapid amplification of cDNA ends technique will be described elsewhere (29).
Cell fusion assays.
Fusion assays were performed between FIV-infected CrFK cells and cells transfected with feline or human chemokine receptors. Cells to be transfected were seeded in six-well cell culture plates at 1 × 105 to 1.5 × 105 per well in DMEM and allowed to adhere overnight. The cells were then washed twice with serum-free DMEM, and the medium was replaced with Opti-MEM (Life Technologies) containing 2 μg of plasmid DNA and 5 μl of LipofectAMINE (Life Technologies) at 1 ml per well. The cells were incubated with the plasmid-LipofectAMINE mix for 4 h, after which 1 ml of DMEM containing 20% FBS and no antibiotics was added and the cells cultured overnight. The medium was then aspirated and replaced with DMEM containing 10% FBS and no antibiotics. The transfected cells were cultured for a further 24 h, trypsinized, and then seeded in 24-well plates (each well of the original 6-well plate being divided between 4 wells of a 24-well plate) with FIV-infected CrFK cells at 5 × 104 cells per well. Fusion was allowed to proceed for 18 to 24 h at 37°C, and then the cells were fixed and stained with 1% methylene blue–0.2% basic fuchsin in methanol. Syncytia were enumerated by light microscopy using a ×12.5 Leitz periplan eyepiece with a 6.5 × 9 graticule, three separate fields being counted per well, each well in duplicate. Syncytia were scored as cells with five or more nuclei.
Virus infection assay.
CCC-CD4 cells were seeded at 1.5 × 105 cells per well of six-well tissue culture plates in DMEM and cultured overnight. The cells were then transfected with 2 μg of plasmid per well in duplicate as described above and cultured for 48 h, at which time the cells were trypsinized and plated at 5 × 104 cells per well in 24-well tissue culture plates. After overnight incubation, the cells were infected for 1 h at 37°C with 200 μl of either FIVPET or HIV-1 RF (approximately 200 pg of reverse transcriptase [RT] per well) and washed twice with DMEM; then the medium was replaced with 0.5 ml of DMEM, and the cells were cultured at 37°C. Supernatants were collected at day 7 postinfection and assayed for Mg2+-dependent RT activity by using a nonisotopic assay kit (Lenti-RT kit; Cavidi Tech AB, Uppsala, Sweden). RT levels were calculated relative to the HIV-1 RT standard provided with the kit.
Nucleotide sequence accession number.
The sequence of feline CCR5 has been deposited in GenBank under accession no. U92796.
RESULTS
CXCR4 expression alone is sufficient to confer susceptibility to fusion mediated by FIV.
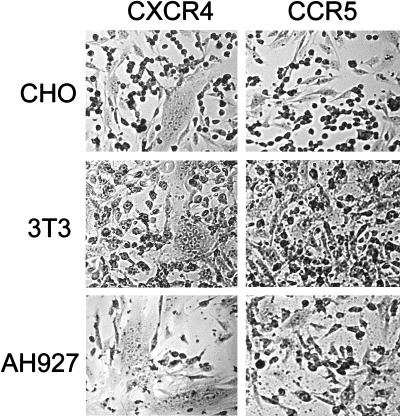
Given that CXCR4-transfected human cells supported fusion mediated by FIV (55), we next asked whether CXCR4 expression alone was sufficient to render cells from diverse species permissive for fusion mediated by FIV. The murine cell line 3T3, hamster cell line CHO, mink cell line Mink S+L-, and CXCR4-negative feline cell lines CCC and AH927 were transfected with either feline or human CXCR4 and examined for the ability to support fusion with FIVPET- and FIVGL8-infected CrFK cells. Irrespective of the species of origin, the CXCR4-transfected cells were rendered permissive for fusion with FIV-infected cells, whereas the CCR5-transfected cells remained refractory to fusion. CXCR4-transfected feline cells CCC and AH927 (Fig. 1) supported fusion with FIV-infected cells, suggesting that the absence of CXCR4 in these cells lines is the principal block to infection with CrFK-tropic strains of FIV. Moreover, the hamster and murine cell lines (CHO and 3T3, respectively) were also rendered permissive for fusion following transfection with CXCR4 (Fig. 1), suggesting either that a highly conserved component of the cell surface acts as the primary receptor for CrFK-adapted FIV or that there is no primary receptor for these viruses and that CXCR4 itself is the viral receptor, analogous to the situation in CD4-independent infection with HIV-2 vcp (20). We compared CXCR4 with feline CCR5 (Fig. 1) or human CCR1, CCR2b, CCR3, CCR4, CCR5, CXCR2, GPR1 (34), GPR15 (22), Bonzo (16), and BOB (16) and the orphan receptors EBI1 (42) and V28 (42) in the same assay system. In each case, fusion was detected only in the CXCR4-transfected cells (data not shown), suggesting that the interaction between FIV and CXCR4 is highly specific. Similar findings were obtained with the CrFK-tropic variant of FIVGL8.
FIG. 1.
Transfection of cells from diverse species with CXCR4 renders the cells permissive for fusion with FIVPET-infected cells. The hamster (CHO), mouse (3T3), and feline (AH927) cell lines were transfected with either CXCR4 or CCR5, incubated for 48 h, and then trypsinized and seeded with FIV-infected CrFK cells. Fusion was allowed to progress for 18 h, at which time the cells were fixed, stained, and photographed.
The high efficiency of human CXCR4 as a cofactor for FIV-dependent fusion is mediated by the second extracellular loop.
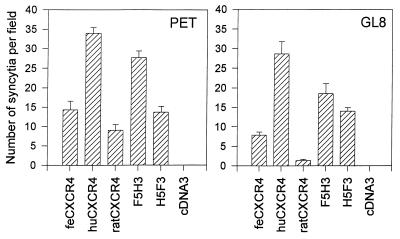
To map the determinants of CXCR4 involved in infection with FIV, we compared feline CXCR4 with human CXCR4 and with chimeric CXCR4 molecules consisting of the N terminus and first loop of feline CXCR4 fused to the second and third loops of human CXCR4 (F5H3) or the converse chimera containing the N terminus and first loop of human CXCR4 fused to the second and third loops of feline CXCR4 (H5F3). Human CXCR4 consistently proved more fusogenic for FIV Env than feline CXCR4 (Fig. 2). Moreover, the F5H3 chimera supported fusion with an efficiency similar to that of human CXCR4, while the H5F3 chimera supported fusion with an efficiency comparable to that of feline CXCR4. The findings were consistent between at least three separate experiments. Furthermore, transfection of 0.5 μg of the green fluorescent protein expression vector GFPN1 in conjunction with each individual plasmid sample confirmed that the ability of each of the constructs to confer permissiveness for FIV-mediated fusion could not be attributed to variability in the relative transfection efficiencies of the plasmids (data not shown). The data suggest that the second and third extracellular loops of human CXCR4 contain a determinant which renders human CXCR4 more fusogenic for FIV Env.
FIG. 2.
Human CXCR4 (huCXCR4) is a more efficient receptor for FIV than either feline CXCR4 (feCXCR4) or rat CXCR4. CCC-CD4 cells were transfected with feline, human, or rat CXCR4, the feline/human chimeras F5H3 and H5F3, or the vector pcDNA3 alone; 48 h posttransfection, the cells were trypsinized and mixed with FIVPET- or FIVGL8-infected CrFK cells. Fusion was allowed to proceed for 18 h, at which time the cells were fixed and stained, and the number of syncytia per field was quantified by light microscopy. Results represent the mean number (n = 6) of syncytia per field ± SE.
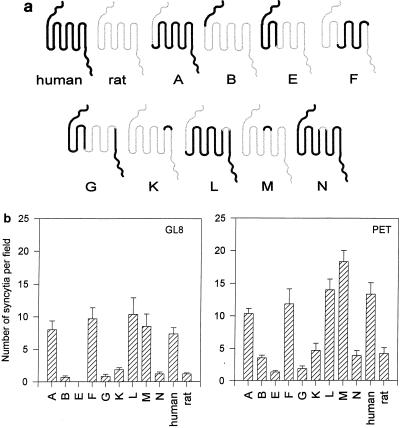
We next asked whether the critical determinant of human CXCR4 for fusion with FIV Env could be localized specifically to the second or third extracellular loop. In a previous study (6), a panel of chimeras between rat and human CXCR4 (Fig. 3a) were screened for the ability to support fusion mediated by HIV-1. While the CXCR4-dependent strain LAI used rat and human CXCR4 with comparable efficiencies for CD4-dependent fusion, replacing the V3 loop of LAI with that of the NDK strain generated a virus with a marked preference for human CXCR4 over rat CXCR4; the critical determinant of the preferential usage of human CXCR4 by HIV-LAI/V3-NDK residing in the second extracellular loop (6). The feline cell line CCC was transfected with each of the rat/human CXCR4 chimeras and plated with FIV-infected CrFK cells. Previous data have demonstrated that the panel of rat/human CXCR4 chimeras are expressed efficiently at the cell surface (6) following transfection. Furthermore, cotransfection of each of the plasmids with 0.5 μg of GFPN1 plasmid confirmed that the ability of each of the constructs to confer permissiveness for FIV-mediated fusion could not be attributed to variability in the relative transfection efficiencies of the plasmids (data not shown). Rat CXCR4 supported fusion less efficiently than either human or feline CXCR4, allowing good discrimination between the individual loops of human CXCR4 in the rat CXCR4 background (Fig. 3b). Chimeras A, F, L, and M supported fusion with an efficiency similar to that of human CXCR4, whereas chimeras B, G, K and N supported fusion with an efficiency similar to that of rat CXCR4. Chimera A contains extracellular loops 1, 2, and 3 of human CXCR4, F contains loops 2 and 3, L contains loops 1 and 2, and M contains the second loop alone in the rat CXCR4 background. Results represent the mean of six estimations ± standard error (SE). Pairwise comparisons between plasmids containing either the second loop of human CXCR4 (A, F, L, M, and human CXCR4) or that of rat CXCR4 (B, E, G, K, N, and rat CXCR4) demonstrated that those containing the second extracellular loop of human CXCR4 supported fusion significantly more efficiently than those containing the second loop of rat CXCR4 (P < 0.001 to 0.005). Comparisons within the two groups of chimeras demonstrated that the human and rat CXCR4 loop 2 constructs supported fusion with similar efficiencies. The data suggest that the second extracellular loop confers the high capacity for FIV-dependent fusion to human CXCR4 and that the N terminus and extracellular loops 1 and 3 can be exchanged with the equivalent regions of rat CXCR4 without adversely affecting the function of the molecule.
FIG. 3.
Receptor function of rat/human CXCR4 chimeras (6) for fusion mediated by FIV. CCC cells were transfected with a panel of rat/human CXCR4 chimeras (a). Thick lines denote regions derived from human CXCR4; thin lines denote regions derived from rat CXCR4. The transfected cells were then assayed for the ability to support fusion with FIVPET- or FIVGL8-infected CrFK cells as described above (b). Results represent the mean (n = 6) number of syncytia per field ± SE.
The second loop of CXCR4 alone is insufficient to support FIV-dependent fusion.
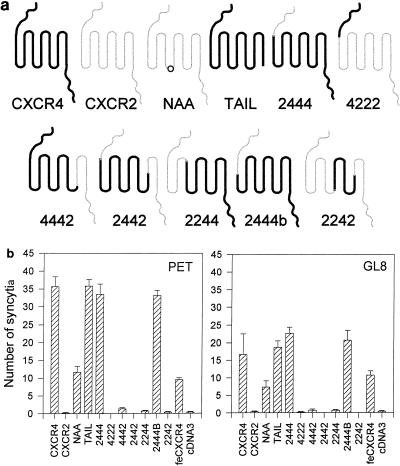
The second extracellular loop of human CXCR4 functioned efficiently in either the feline or the rat CXCR4 background. It is possible that the N terminus and first and third extracellular loops of rat CXCR4 maintain the second loop of human CXCR4 in the correct conformation for it to interact with FIV Env. Therefore, we asked whether it would continue to function as a viral receptor in chimeras between CXCR4 and human CXCR2 (Fig. 4a). While human and rat CXCR4 show 92% similarity at the amino acid level, human CXCR4 and CXCR2 show only 50.7% similarity. In a previous study (32), a series of CXCR2/CXCR4 chimeras was generated and examined for the ability to support CD4-dependent fusion mediated by HIV Env. While the second extracellular loop of CXCR4 appeared to be the principal determinant of CXCR4 usage by HIV, individual strains showed different degrees of dependence on the N terminus and the first or third extracellular loops, exchanging these regions with the corresponding regions of CXCR2 severely compromising the function of the molecule as a fusogenic cofactor. We examined whether CrFK-tropic FIV would tolerate substitutions in the first and third extracellular loops of CXCR4. CCC cells were transfected with those members of the panel of CXCR2/CXCR4 chimeras (32) which had been shown to be expressed at the cell surface following transfection (32) and then plated with FIV-infected CrFK cells (Fig. 4b). CXCR2 did not support fusion mediated by FIV. Replacement of the N terminus of CXCR4 with that of CXCR2 (constructs 2444 and 2444b) did not affect fusion with either FIVPET- or FIVGL8-infected CrFK cells. Similarly, truncation of the C-terminal cytoplasmic domain, preventing internalization of CXCR4 (CXCR4-TAIL), did not affect fusion. Mutation of the DRY motif (CXCR4-NAA) to generate a molecule that is unable to couple to G proteins reduced the efficiency of the molecule in the fusion assay by approximately 60% but did not ablate function completely, suggesting that signalling via G proteins is not required for fusion mediated by FIV. Chimeras in which the third extracellular loop (4442), the N terminus and first extracellular loop (2244), or the N terminus and first and third extracellular loops (2242) of CXCR4 were replaced by the corresponding regions of CXCR2 failed to support fusion mediated by FIV. Given that the first and third loops of human CXCR4 could be replaced with the corresponding regions of rat CXCR4, the data suggest either that the first and third loops of CXCR4 maintain the second loop in a conformation that is essential for CXCR4 to support fusion mediated by FIV Env or that all three extracellular loops contribute to the Env binding site.
FIG. 4.
Receptor function of human CXCR2/human CXCR4 chimeras (32) for fusion mediated by FIV. CCC-CD4 cells were transfected with the panel of CXCR2/CXCR4 chimeras (a). Thick lines denote regions derived from CXCR4; thin lines denote regions derived from CXCR2; a circle denotes the location of the NAA mutation in the DRY motif. The transfected cells were then assayed for the ability to support fusion with FIVPET- or FIVGL8-infected CrFK cells as described above (b). Three fields were counted per well, each well in duplicate. Results represent the mean (n = 6) number of syncytia per field ± SE. feCXCR4, feline CXCR4.
The ability of CXCR4-transfected cells to support fusion correlates with the ability to support infection.
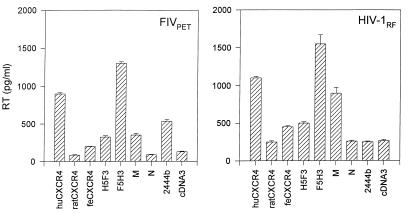
The preceding data demonstrated that the second extracellular loop of CXCR4 contained the principal determinant for fusion with FIV-infected cells. We next asked whether the ability of CXCR4-transfected cells to support fusion correlated with the ability to support infection with cell-free virus. CCC-CD4 cells were transfected with a series of the CXCR4 constructs; expression was allowed to proceed for 2 days, at which time the cells were infected with FIVPET or the CXCR4-dependent RF strain of HIV-1. The cultures were then washed twice and maintained for 7 days, and then the level of Mg2+-dependent RT activity in the culture supernatant was quantified. When RT levels were compared, human CXCR4 and the constructs containing the second loop of human CXCR4 (F5H3 and chimera M) proved the most efficient cofactors for infection with both FIVPET and HIV-1RF (Fig. 5). Feline CXCR4 and chimera H5F3 containing the second loop of feline CXCR4 functioned less efficiently for both viruses, while rat CXCR4 and chimera N (containing the second loop of rat CXCR4) did not differ significantly from the pcDNA3 control. FIVPET used chimera 2444b efficiently for infection, while HIV-1RF was sensitive to replacement of the N terminus with that of CXCR2, in agreement with previous findings (40). Thus, it would appear that the ability of the CXCR4 chimeras to support FIV infection correlates well with fusion assays between transfected cells and FIV-infected cells and that the second extracellular loop of human CXCR4 contains the principal determinant of CXCR4 usage by FIV.
FIG. 5.
Correlation between FIV-mediated cell fusion and virus infection. CCC-CD4 cells were transfected with a selection of plasmids which had been shown previously to support FIV-mediated cell fusion. The cells were then infected with either FIVPET or HIV-1 RF and assayed for Mg2+-dependent RT activity at 7 days postinfection. Results are expressed as mean ± SE (n = 2) and are typical of three separate experiments. hu- and feCXCR4, human and feline CXCR4.
DISCUSSION
In this study, we demonstrated that ectopic expression of CXCR4 on feline, murine, human, hamster, or mink cells was sufficient to render the cells permissive for fusion mediated by CrFK-tropic strains of FIV, suggesting that infection with these strains of virus is analogous to infection with CD4-independent strains of HIV-2 (20). Furthermore, human CXCR4 supports both infection and cell fusion with FIV more efficiently than either feline or rat CXCR4. Using this property, we demonstrated that the second extracellular loop of CXCR4 contains a critical determinant for the function of CXCR4 as a cofactor for infection with FIV. These findings are similar to those observed with HIV-1 LAI/V3-NDK, in which the V3 loop of LAI was replaced with that of the NDK strain, but differ significantly from those observed with the parent LAI strain itself (6). The LAI strain of HIV-1 fused human and rat CXCR4-expressing cells with comparable efficiencies, whereas the HIV-1 LAI/V3-NDK chimeric virus showed preferential fusion of cells expressing either human CXCR4 or chimeric molecules carrying the second extracellular loop of human CXCR4 in a rat CXCR4 background (6). In a similar study, a chimeric murine CXCR4 in which the second loop was exchanged with that of human CXCR4 was found to support infection with the 89.6 strain of HIV-1 (39) whereas murine CXCR4 itself did not support infection. In contrast to the LAI and NDK strains of HIV-1, CrFK-tropic FIV was not sensitive to deletions of the amino-terminal region of CXCR4. In a previous study, the BH8, 89.6, RF, and BK132 strains of HIV-1 and the SBL6669 strain of HIV-2 were shown to be sensitive to replacement of the N terminus of CXCR4 with the corresponding domain of CXCR2 (32). Given that these strains of HIV are CD4 dependent, it is possible that these viruses require an additional interaction with the N terminus of CXCR4 prior to the interaction with the extracellular loops for fusion to occur.
Chimeras in which either the first or third extracellular loop of CXCR4 was replaced with the corresponding domain of CXCR2 (chimeras 2442, 2242, 4442, and 2244) failed to support cell fusion mediated by FIV. These data suggest that although the second extracellular loop of CXCR4 is the principal determinant of function as a receptor for FIV, the adjacent loops either maintain the second loop in the appropriate conformation for a direct interaction between CXCR4 and the viral envelope (25) or interact directly themselves with the envelope glycoprotein. These findings are reminiscent of studies of the interaction between gibbon ape leukemia virus and feline leukemia virus subgroup B (FeLV-B) and their receptor Pit1 (8, 49), amphotropic murine leukemia virus (A-MLV) and Pit2, and ecotropic murine leukemia virus (E-MLV) and MCAT-1 (1). Chimeric MCAT-1 receptors constructed between the murine and human MCAT-1 molecules indicated that the third extracellular loop contained the principal determinant of receptor usage by E-MLV (1). However, the interaction between FeLV-B and A-MLV and Pit1 and Pit2 appears more complex, with recent data suggesting that loops 4 and 5 form the initial binding site for the viral envelope and that a subsequent interaction with loop 2 is essential for infection (49). By comparing the interaction between FIV and CXCR4 with the interaction between the mammalian type C retroviruses and multitransmembrane domain molecules such as Pit1, Pit2, and MCAT-1, it may be possible to determine whether a common mechanism of interaction is employed by all retroviruses as has been proposed (49).
While CD4-dependent infection would appear to be the principal mechanism of infection with HIV and SIV, several examples of CD4-independent infection have been described (10, 26, 36, 50). At present, the role of CD4-independent infection in the pathogenesis of AIDS remains unclear; however, productive infection of CD4-negative cells has been observed in tissues from HIV-infected individuals (3, 31, 37) and SIV-infected nonhuman primates in advanced disease states (13, 33). The consequences of high-affinity interactions between the viral envelope glycoprotein and the chemokine receptor may extend beyond viral tropism. Recent studies have demonstrated that binding of HIV envelope glycoprotein to either CXCR4 or CCR5 results in the transduction of a signal through receptor-coupled G proteins (12). Where binding is CD4 dependent, the viral envelope may thus mimic the effect of chemokines on CD4+ cells. However, envelope glycoproteins of HIV and SIV that interact directly with either CXCR4 (20, 23) or CCR5 (35) in the absence of CD4 have been identified. It is possible that the envelope glycoproteins from these viruses have more diverse effects, mimicking the effects of chemokines on CD4-negative cells such as cells of neuronal origin (23) and perhaps contributing to the neuropathology associated with AIDS. The majority of CD4-independent infection appears to occur relatively inefficiently and is only weakly cytopathic. In contrast, infection with the HIV-2 strains ROD-B and vcp occurs efficiently on CD4-negative cell lines (10, 20), while strains such as HIV-2 CBL-22 will infect efficiently following pretreatment with soluble CD4 (10). The resemblance between CrFK-tropic strains of FIV and CD4-independent infection with HIV-2 is striking. Adaptation of FIV to a CrFK-tropic phenotype is accompanied by an increase in charge of the V3 loop (typically by an E-to-K mutation [46, 52]), while the Q310K mutation in the V3 loop of HIV-2 ROD-B enhances the CD4-independent phenotype (44) of the virus. An M751T mutation in the TM protein of FIV is associated with the CrFK-tropic phenotype (51), and a similar A526T mutation in the TM protein of HIV-2 ROD is observed upon adaptation to a CD4-independent phenotype (44); in each case the mutations are located adjacent to the region of TM thought to be involved in the formation of a coiled-coil domain involved in oligomerization, viral entry, and neutralization (7, 19, 24, 38, 53). While it is evident that mutations in other regions of the env gene are required to confer a CrFK-tropic phenotype on FIV (46, 51, 52) and a CD4-independent phenotype on HIV-2 (44), the two viruses adapt similarly to the use of pathways of infection that ultimately result in the use of CXCR4 alone as a viral receptor. Future studies of infection with CXCR4-independent strains of FIV may provide an insight into the role of CXCR4-dependent infection in the pathogenesis of AIDS.
It is intriguing that despite the evolutionary divergence between the amino acid sequences of FIV and HIV, the structure of the envelope glycoprotein is conserved sufficiently that both viruses can interact with CXCR4. Moreover, no other human 7TM molecules that we have tested to date will support fusion mediated by FIV, suggesting that the interaction is extremely specific. Feline CCR5 failed to support fusion or infection of CCC-CD4 cells with the CrFK-tropic virus FIVPET, and preliminary data (not shown) suggest that CCR5 expression alone is insufficient to confer susceptibility to infection with a primary isolate of FIVGL8 and that CCR5 expression does not correlate with susceptibility to infection with primary isolates of FIV (29). The primary binding receptor for primary strains of FIV remains to be discovered; until the nature of this elusive molecule is determined, the role of 7TM molecules in infection with primary isolates of virus will prove difficult to resolve. We have found no evidence for inhibition of infection with primary isolates of FIV by a range of human β-chemokines, while CrFK infection by FIVPET is inhibited efficiently by human stromal cell-derived factor 1 (25). However, until the feline homologs of the β-chemokines are available, we cannot discount a role for β-chemokine receptors such as CCR5 in infection with FIV.
ACKNOWLEDGMENTS
This work was supported by The Wellcome Trust (B.J.W., K.A., and M.J.H.).
We thank J. H. Elder, D. R. Littman, P. R. Clapham, T. N. Wells, and J. Hesselgesser for the provision of reagents and helpful discussions.
REFERENCES
- 1.Albritton L M, Kim J W, Tseng L, Cunningham J M. Envelope-binding domain in the cationic amino-acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Bagasra O, Lavi E, Bobroski L, Khalili K, Pestaner J P, Pomerantz R. Cellular reservoirs of HIV-1 in the central nervous system of HIV-infected individuals: identification by combination of in situ PCR and immunochemistry. AIDS. 1996;10:573–585. doi: 10.1097/00002030-199606000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Berger E A, Doms R W, Fenyo E M, Korber B T M, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 5.Bjorndal A, Deng H K, Jansson M, Fiore J R, Colognesis C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyo E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J Virol. 1997;71:4744–4751. doi: 10.1128/jvi.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao J, Bergeron L, Helseth E, Thali M, Pepke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudry G J, Eiden M V. Mutational analysis of the proposed gibbon ape leukemia virus binding site in Pit1 suggests that other regions are important for infection. J Virol. 1997;71:8078–8081. doi: 10.1128/jvi.71.10.8078-8081.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 10.Clapham P R, McKnight A, Weiss R A. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J Virol. 1992;66:3531–3537. doi: 10.1128/jvi.66.6.3531-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis C B, Dikic I, Unutmaz D, Hill C M, Arthos J, Siani M A, Thompson D A, Schlessinger J, Littman D R. Signal transduction due to HIV-1 envelope interactions with chemokine receptors CXCR4 or CCR5. J Exp Med. 1997;186:1793–1798. doi: 10.1084/jem.186.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean G A, Reubel G H, Moore P F, Pedersen N C. Proviral burden and infection kinetics of feline immunodeficiency virus in lymphocyte subsets of blood and lymph node. J Virol. 1996;70:5165–5169. doi: 10.1128/jvi.70.8.5165-5169.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Jong J J, de Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng H, Liu R, Ellmeir W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 16.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 17.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion co-factors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 18.Dragic T, Litwin T, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 19.Dubay J W, Roberts S J, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 21.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry co-factor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 22.Heiber M, Marchese A, Nguyen N Y, Heng H, George S, O’Dowd B. A novel human gene encoding a G-protein-coupled receptor (gpr15) is located on chromosome 3. Genomics. 1996;32:462–465. doi: 10.1006/geno.1996.0143. [DOI] [PubMed] [Google Scholar]
- 23.Hesselgesser J, HalksMiller M, DelVecchio V, Peiper S C, Hoxie J, Kolson D L, Taub D, Horuk R. CD4-independent association between HIV-1 gp120 and CXCR4: functional chemokine receptors are expressed in human neurons. Curr Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- 24.Ho D D, Sarngadharan M G, Hirsch M S, Schooley R T, Rota T R, Kennedy R C, Chanh T C, Sato V L. Human immunodeficiency virus neutralizing antibodies recognize several conserved domains on the envelope glycoprotein. J Virol. 1987;61:2024–2028. doi: 10.1128/jvi.61.6.2024-2028.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosie M J, Broere N, Hesselgesser J, Turner J D, Hoxie J A, Neil J C, Willett B J. Modulation of feline immunodeficiency virus infection by stromal cell-derived factor. J Virol. 1998;72:2097–2104. doi: 10.1128/jvi.72.3.2097-2104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikeuchi K, Kim S, Byrn R A, Goldring S R, Groopman J E. Infection of nonlymphoid cell by human immunodeficiency virus type 1 or type 2. J Virol. 1990;64:4226–4231. doi: 10.1128/jvi.64.9.4226-4231.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsson A, Parsmyr K, Sandstrom E, Fenyo E M, Albert J. MT-2 cell tropism as a prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J Clin Microbiol. 1994;32:364–370. doi: 10.1128/jcm.32.2.364-370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koot M, Keet I P M, Vos A H V, De Goede R E Y, Roos M T L, Coutinho R A, Miedema F, Schellekens P T A, Tersmette M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 29.Lerner, D. L., and J. H. Elder. 1997. Unpublished data.
- 30.Liao F, Alkhatib G, Peden K W C, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livingstone W J, Moore M, Innes D, Bell J E, Simmonds P, Whitelaw J, Wyld R, Robertson J R, Brettle R P. Frequent infection of peripheral-blood CD8-positive T-lymphocytes with HIV-1. Lancet. 1996;348:649–654. doi: 10.1016/s0140-6736(96)02091-0. [DOI] [PubMed] [Google Scholar]
- 32.Lu Z H, Berson J F, Chen Y H, Turner J D, Zhang T Y, Sharron M, Jenks M H, Wang Z X, Kim J, Rucker J, Hoxie J A, Peiper S C, Doms R W. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mankowski J L, Spelman J P, Ressetar H G, Strandberg J D, Laterra J, Carter D L, Clements J E, Zink M C. Neurovirulent simian immunodeficiency virus replicates productively in endothelial cells of the central nervous system in vivo and in vitro. J Virol. 1994;68:8202–8208. doi: 10.1128/jvi.68.12.8202-8208.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchese A, Docherty J, Nguyen N Y, Heiber M, Cheng R, Heng H, Tsui L, Shi X, George S, O’Dowd B. Cloning of human genes encoding novel G protein-coupled receptors. Genomics. 1994;23:609–618. doi: 10.1006/geno.1994.1549. [DOI] [PubMed] [Google Scholar]
- 35.Martin K A, Wyatt R, Farzan M, Choe H, Marcon L, Desjardins E, Robinson J, Sodroski J, Gerard C, Gerard N P. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 36.McKnight A, Clapham P R, Weiss R A. HIV-2 and SIV infection of non-primate cell lines expressing human CD4: restrictions to replication at distinct stages. Virology. 1994;201:8–18. doi: 10.1006/viro.1994.1260. [DOI] [PubMed] [Google Scholar]
- 37.Moses A V, Bloom F E, Pauza C D, Nelson J A. Human immunodeficiency virus infection of human brain capillary endothelial cells occurs via a CD4/galactosylceramide-independent mechanism. Proc Natl Acad Sci USA. 1993;90:10474–10478. doi: 10.1073/pnas.90.22.10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parolin C, Borsetti A, Choe H, Farzan M, Kolchinsky P, Heesen M, Ma Q, Gerard C, Palu G, Dorf M E, Springer T, Sodroski J. Use of murine CXCR-4 as a second receptor by some T-cell-tropic human immunodeficiency virus. J Virol. 1998;72:1652–1656. doi: 10.1128/jvi.72.2.1652-1656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Picard L, Wilkinson D A, McKnight A, Gray P W, Hoxie J A, Clapham P R, Weiss R A. Role of the amino-terminal extracellular domain of CXCR-4 in human immunodeficiency virus type 1 entry. Virology. 1997;231:105–111. doi: 10.1006/viro.1997.8506. [DOI] [PubMed] [Google Scholar]
- 41.Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 42.Raport C J, Schweickart V L, Eddy R L, Shows T B, Gray P W. The orphan G-protein-coupled receptor-encoding gene V28 is closely related to genes for chemokine receptors and is expressed in lymphoid and neural tissues. Gene. 1995;163:295–299. doi: 10.1016/0378-1119(95)00336-5. [DOI] [PubMed] [Google Scholar]
- 43.Reeves J D, McKnight A, Potempa S, Simmons G, Gray P W, Power C A, Wells T, Weiss R A, Talbot S J. CD4-independent infection by HIV-2 (ROD/B): use of the 7-transmembrane receptors CXCR-4, CCR-3, and V28 for entry. Virology. 1997;231:130–134. doi: 10.1006/viro.1997.8508. [DOI] [PubMed] [Google Scholar]
- 44.Reeves J D, Schultz T F. The CD4-independent tropism of human immunodeficiency virus type 2 involves several regions of the envelope protein and correlates with a reduced activation threshold for envelope-mediated fusion. J Virol. 1997;71:1453–1465. doi: 10.1128/jvi.71.2.1453-1465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuitemaker H, Koot M, Koostra N A, Dercksen M W, De Goede R E Y, Van Steenwijk R P, Lange J M, Eeftink Schattenkerk J K, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siebelink K H J, Karlas J A, Rimmelzwaan G F, Osterhaus A D M E, Bosch M L. A determinant of feline immunodeficiency virus involved in CrFK tropism. Vet Immunol Immunopathol. 1995;46:61–69. doi: 10.1016/0165-2427(94)07006-s. [DOI] [PubMed] [Google Scholar]
- 47.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tailor C S, Kabat D. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J Virol. 1997;71:9383–9391. doi: 10.1128/jvi.71.12.9383-9391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tateno M, Gonzalez-Scarano F, Levy J A. The human immunodeficiency virus can infect CD4− human fibroblastoid cells. Proc Natl Acad Sci USA. 1989;86:4287–4290. doi: 10.1073/pnas.86.11.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vahlenkamp T W, Verschoor E J, Schuurman N N M P, van Vliet A L W, Horzinek M C, Egberink H F, de Ronde A. A single amino acid substitution in the transmembrane envelope glycoprotein of feline immunodeficiency virus alters cellular tropism. J Virol. 1997;71:7132–7135. doi: 10.1128/jvi.71.9.7132-7135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verschoor E J, Boven L A, Blaak H, van Vliet A R W, Horzinek M C, de Ronde A. A single mutation within the V3 envelope neutralization domain of feline immunodeficiency virus determines its tropism for CRFK cells. J Virol. 1995;69:4752–4757. doi: 10.1128/jvi.69.8.4752-4757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wild C, Greenwell T, Matthew T. A synthetic peptide from HIV-1 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res Hum Retroviruses. 1993;9:1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 54.Willett B J, Hosie M J, Neil J C, Turner J D, Hoxie J A. Common mechanism of infection by lentiviruses. Nature. 1997;385:587. doi: 10.1038/385587a0. [DOI] [PubMed] [Google Scholar]
- 55.Willett B J, Picard L, Hosie M J, Turner J D, Adema K, Clapham P R. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J Virol. 1997;71:6407–6415. doi: 10.1128/jvi.71.9.6407-6415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]