Summary
Helicobacter pylori (H. pylori) infection is a known cause of many digestive diseases, including gastritis, peptic ulcers, and gastric cancer. However, the underlying mechanisms by which H. pylori infection triggers these disorders are still not clearly understood. Gastric cancer is a slow progressing disease, which makes it difficult to study. We have developed an accelerated disease progression mouse model, which leverages mice deficient in the myeloid differentiation primary response 88 gene (Myd88−/−) infected with Helicobacter felis (H. felis). Using this model and gastric biopsy samples from patients, we report that activation of the Toll/interleukin-1 receptor (TIR)-domain-containing adaptor inducing interferon-β (TRIF)-type I interferon (IFN-I) signaling pathway promotes Helicobacter-induced disease progression toward severe gastric pathology and gastric cancer development. Further, results implicated downstream targets of this pathway in disease pathogenesis. These findings may facilitate stratification of Helicobacter-infected patients and thus enable treatment prioritization of patients.
Subject areas: Biological sciences, Microbiology, Clinical microbiology, Cancer
Graphical abstract

Highlights
-
•
Helicobacter-induced gastric cancer (GC) progresses rapidly in absence of MyD88
-
•
Absence of TRIF decreased Helicobacter induced disease pathology
-
•
ISGs upregulated in Helicobacter infected Myd88−/− mice and GC patients
-
•
TRIF-IFN-I pathway plays pivotal role in development of Helicobacter-induced GC
Biological sciences; Microbiology; Clinical microbiology; Cancer
Introduction
Helicobacter pylori (H. pylori) infection is the main risk factor for development of gastric cancer, thus this bacterium is classified as a group I carcinogen.1,2 Gastric cancer, which is often recalcitrant to therapy, remains a significant global health burden and a leading cause of cancer-related deaths worldwide.3,4 Half of the world’s population is infected with H. pylori, contributing significantly to the disease burden.2,5 Infection with H. pylori causes chronic inflammation, leading to the development of preneoplastic lesions (atrophy, intestinal metaplasia, dysplasia), and ultimately to gastric cancer, a progression known as the Correa’s cascade.6 We previously demonstrated that mice deficient in myeloid differentiation primary response gene 88 (MyD88, Myd88−/−) infected with H. felis rapidly developed gastric pathology progressing to high-grade gastric dysplasia,7 a condition considered as gastric cancer in situ.8,9 Mechanisms underlying this phenotype were not fully understood, but our data indicated that the loss of MyD88 led to hyperactivation of the alternate Toll/interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-β (TRIF)-dependent pathway, accelerating gastric cancer progression in response to H. felis infection.7,10 A recent study citing our work revealed that the Toll-like receptor 4 (TLR4)-TRIF-type I interferon (IFN-I) pathway, via the production of IFN-γ, was important for H. suis-induced disease development in a mucosa-associated lymphoid tissue (MALT) lymphoma mouse model.11 MyD88 and TRIF are the main adaptor proteins that mediate TLRs signals.12 TRIF signaling branches into two downstream pathways, one that shares effectors with MyD88 and another that is MyD88-independent, (TRIF/IRF3, IFN regulatory factor 3),13,14,15 leading to the induction of IFN-I genes and IFN-inducible genes.
In this study, we utilized our accelerated animal model for gastric cancer and clinical gastric specimens from patients to establish that the activation of the TRIF-IFN-I signaling pathway promotes Helicobacter-induced disease progression, leading to severe gastric pathology and ultimately resulting in the development of gastric cancer. We made the significant observation that these changes in the gastric epithelium begin early, just one month after Helicobacter infection, and become even more pronounced at 3 months post-infection. This pathway may therefore be an attractive target to intercept the neoplastic process and prevent the development of gastric cancer.
Results
Preneoplastic gastric lesions in response to Helicobacter infection in mice
Our previous in vivo data using Myd88−/− mice demonstrated that infection with H. felis for 25 or 47 weeks resulted in rapid disease progression with severe gastric pathology.7 However, it remained unclear when gastric neoplastic changes were initiated following H. felis infection. To address this, we profiled the gastric epithelium at earlier time points post-infection with H. felis. Wild type (WT) and Myd88−/− mice in the C57BL/6 background were infected with H. felis for 1, 3, or 6 months. Considering the role of the TRIF signaling pathway in H. felis-induced disease progression from our previous study,7 we also infected TrifLps2 and double deficient (Myd88−/− and TrifLps2, DKO) mice with H. felis for 1, 3, and 6 months. All infected mice were colonized, albeit with significant differences in colonization level among strains (Figure S1). Gastric histopathological changes were minimal in TrifLps2 mice in response to infection with H. felis compared to WT, Myd88−/− and DKO mice (Figure 1). In contrast, gastric precancerous lesions developed and progressed more rapidly, with significantly increased severity in H. felis-infected Myd88−/− mice compared to H. felis-infected WT, TrifLps2, and DKO mice. To closely define gastric epithelial changes during H. felis infection, we scored histopathology parameters, including inflammation, epithelial defects, oxyntic gland atrophy, hyperplasia, intestinal metaplasia, and dysplasia, on H&E-stained stomach tissue samples by a pathologist blinded to the study. Differences in histopathologic scores of these parameters were evident in Myd88−/− mice at 1 month following H. felis infection, compared to WT, TrifLps2, and DKO mice, with disease severity increasing with time (Figures S2A‒S2F). There were no significant gastric histopathologic changes in all uninfected mice (Figures S2G‒S2L). Notably, the earliest significant gastric epithelial changes in all infected mice occurred at 3 months. Consequently, we performed comparative analyses in mice at 3 months following H. felis infection. Information gathered at this early phase of infection is ideal for identifying early targets for disease interception as the disease progresses. This is of central importance due to the lack of characteristic symptoms during early disease stages, often leading to diagnosis at a terminal stage with unfavorable outcomes for patients.
Figure 1.
Representative images of hematoxylin & eosin-stained mouse stomach tissue sections
WT, Myd88−/−, TrifLps2, and double deficient (Myd88−/− and TrifLps2, DKO) mice were infected with H. felis for 1 month (WT n = 8; Myd88−/− n = 8; TrifLps2 n = 8; DKO n = 8), 3 months (WT n = 8; Myd88−/− n = 8; TrifLps2 n = 8; DKO n = 7), or 6 months (WT n = 7; Myd88−/− n = 8; TrifLps2 n = 8; DKO n = 8). Scale bar, 500 μm.
Helicobacter induced high expression of interferon-stimulated genes (ISGs) in the absence of MyD88 signaling in mice
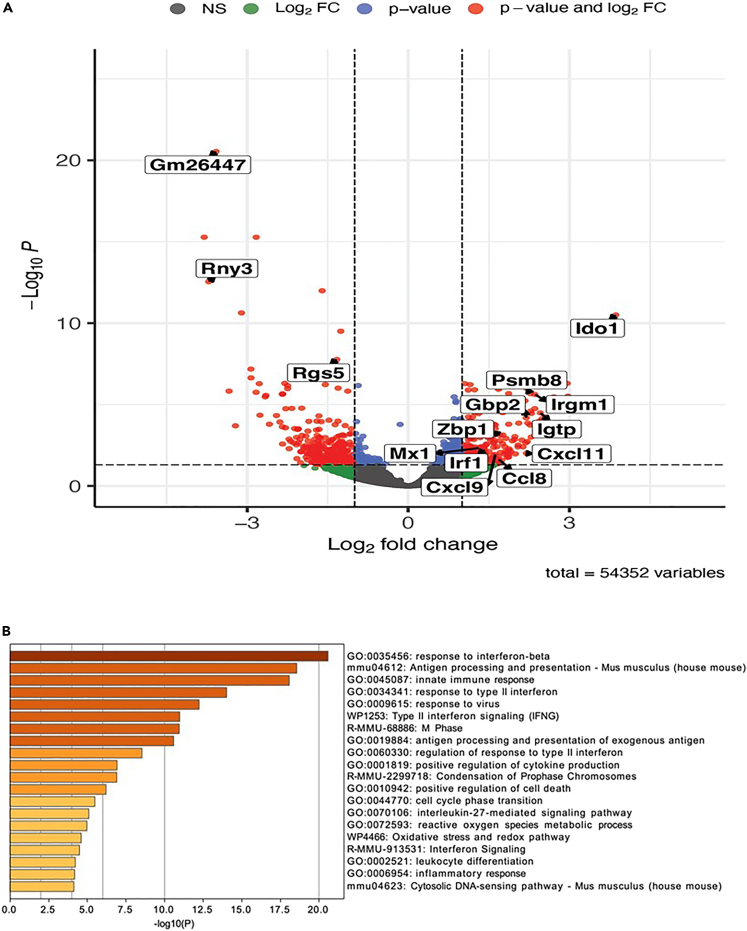
The histopathology data revealed significant changes in the gastric epithelium of Myd88−/−mice, including preneoplastic lesions, following H. felis infection as early as 3 months, compared to WT, TrifLps2, and DKO mice. To identify possible molecular mechanisms driving this disease progression in the absence of MyD88 signaling, we performed RNA-seq to compare gene expression in mouse stomachs between WT and Myd88−/− mice after three months of H. felis infection. We used mouse stomach tissues from 19 mice for RNA-seq, 8 uninfected (WT n = 2; Myd88−/− n = 2; TrifLps2 n = 2; DKO n = 2), and 11 infected (WT n = 3; Myd88−/− n = 2; TrifLps2 n = 3; DKO n = 3) mice. This analysis showed that many of the significant differentially expressed genes between Myd88−/− and WT mice were ISGs, which were highly expressed in gastric tissue samples from Myd88−/− mice (log2 fold change >1, adj. p value <0.05, n = 221, Figure 2A) that exhibited severe gastric pathology. These ISGs were not highly expressed in TrifLps2 (Figure S3A) or DKO (Figure S3B) mice. Pathway enrichment analysis revealed that these ISGs were the dominant upregulated genes in Myd88−/− mice in response H. felis (Figure 2B), and their expression was closely correlated with severe gastric pathology, suggesting their involvement in disease pathogenesis. In contrast, the interferon pathway was not enriched in both stomach tissue samples from TrifLps2 (Figure S4A) or DKO (Figure S4B) mice. Notably, specific ISGs that were upregulated in Myd88−/− mice in response H. felis infection included indoleamine 2,3-dioxygenase 1 (Ido1), guanylate binding protein 2 (Gbp2), transcription factor interferon regulatory factor 1 (Irf1), beta-2 microglobulin (B2m), Z-DNA binding protein 1 (Zbp1), and chemokine (C-X-C motif) ligand 11 (Cxcl11), Cxcl9, Ccl8, MX dynamin-like GTPase 1(Mx1), interferon gamma induced GTPase (Igtp), proteasome subunit beta 8 (Psmb8), and immunity-related GTPase family M member 1 (Irgm1) (Figure 3A). These ISGs were specifically induced in the gastric tissue from H. felis-infected Myd88−/− mice and were not observed in gastric tissue samples from H. felis-infected WT, H. felis-infected TrifLps2, H. felis-infected DKO, or uninfected mice. We further observed that the upregulation of these ISGs in response to H. felis infection was highly specific. ISGs are typically induced through activation of the IFN-I pathway.16 Comparing upregulation of IFN-I targets with that of nuclear factor-kappa B (NF-κB) targets in response to H. felis infection, we found minimal differences in the upregulation of NF-κB targets between mice, as exemplified by the NF-kB target Nfkbia. However, the upregulation of IFN-I targets (i.e., ISGs) were exclusively observed in the absence of MyD88 signaling (Myd88−/− mice) (Figure 3B). This specific association of MyD88-deficiency with a heightened ISG profile strongly suggests the activation of the TRIF-IFN-I signaling pathway.
Figure 2.
Genes expression and pathway analysis in mouse gastric tissue
(A) Volcano plot shows expression of genes following three months of infection with H. felis in WT and Myd88−/− mice. The plot depicts the log2 fold change and adjusted log10 p values for each gene when comparing gene expression values between felis-infected WT and H. felis-infected Myd88−/− mice gastric tissue.
(B) Metascape pathway enrichment analysis of the top 20 pathways and their adjusted log10 p value enrichment values in genes that were upregulated in H. felis-infected Myd88−/− mice gastric tissue samples relative to H. felis-infected WT gastric tissue samples.
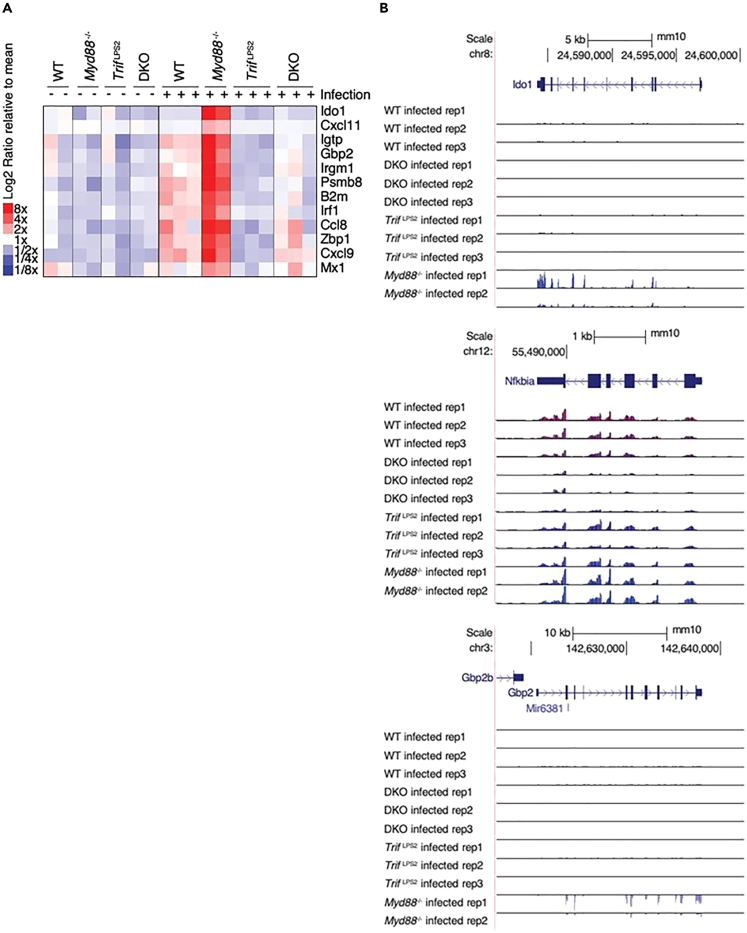
Figure 3.
Helicobacter induced high expression of interferon-stimulated genes (ISGs) in the stomachs of mice in the absence of MyD88 signaling
(A) Relative gene expression values for several ISGs across all replicate experiments for uninfected and H. felis-infected gastric tissue samples from WT, Myd88−/−, TrifLps2, and double deficient (Myd88−/− and TrifLps2, DKO) mice.
(B) The genome tracks show normalized read density for each RNA-seq experiment at the canonical NF-kB target Nfkbia locus, and ISG loci Ido1 and Gbp2 in gastric tissue samples from WT, Myd88−/−, TrifLps2, and double deficient (Myd88−/− and TrifLps2, DKO) mice.
To understand the possible pathogenic mechanism of these ISGs in Helicobacter-induced disease, we performed promoter motif enrichment analysis of genes induced in the stomachs of WT, Myd88−/−, TrifLps2, and DKO mice following H. felis infection. In line with the upregulation of ISGs in our H. felis-infected Myd88−/− model, this analysis revealed that the IFN stimulated-response element (ISRE) motif was the single most enriched motif in promoters of genes upregulated in H. felis-infected Myd88−/− mice (Figure 4). In contrast, this motif was not enriched TrifLps2 and DKO mice. This provides strong evidence that IFN-I regulatory pathways and transcription factors play an important role in the severe disease pathology observed in these mice in response to H. felis infection. Since ISGs mainly mediate IFN effects,16 these findings suggest a significant role of the TRIF-IFN-I pathway in driving severe gastric pathology in response to H. felis infection.
Figure 4.
Ranking list of significantly enriched promoter motifs
Top 8 non-redundant transcription factor motifs enriched in the promoters of genes up-regulated in gastric tissue from H. felis-infected Myd88−/− mice relative to H. felis-infected WT, Myd88−/−, TrifLps2, and double deficient (Myd88−/− and TrifLps2, DKO) mice.
Decreased Helicobacter-induced disease pathology in the absence of TRIF signaling in mice
Following infection with H. felis for 3 months, histopathology scoring of gastric tissue from WT, Myd88−/−, TrifLps2, and DKO mice revealed that inflammation was induced in all mouse groups, but it was significantly less severe in TrifLps2 mice compared to WT, Myd88−/−, and DKO mice (Figure 5A). While the disease progressed in WT, Myd88−/−, and DKO mice, leading to oxyntic gland atrophy, TrifLps2 mice did not show evidence of this pathology (Figure 5B). Oxyntic atrophy, representing the loss of parietal cells, is known to be a pivotal step in the progression of lesions, including intestinal metaplasia and dysplasia, eventually leading to gastric cancer.17,18 Importantly, in the absence of TRIF signaling, Helicobacter-induced inflammation did not progress to precancerous lesions, including intestinal metaplasia (Figure 5C) and dysplasia (Figure 5D). The DKO mice with double deficiency in both TRIF and MyD88 signaling showed an intermediate dysplasia score. These findings indicate that deficiency in TRIF signaling mitigates the severe gastric pathology observed in Myd88−/− mice in response to H. felis infection, corroborating our previous studies indicating that the TRIF pathway plays a key role in the development of severe disease pathology. Additionally, a recent study citing our previous work suggested that TLR4-TRIF signaling was important in a MALT lymphoma model involving infection of mice with H. suis.11 Together, our current study and previous work indicate that the TRIF signaling pathway, and consequently downstream activation of IFN-I pathway, may be responsible for early development and accelerated disease progression. This supports the role of downstream target genes of this pathway in promoting disease progression, as we observed a significant association of high-grade dysplasia with the upregulation of ISGs.
Figure 5.
Histopathologic scores in mouse gastric tissue in response to H. felis infection
WT and Myd88−/−TrifLps2, and double deficient (Myd88−/− and TrifLps2, DKO) mice were infected with H. felis (Inf.) for 3 months (WT n = 8; Myd88−/− n = 8; TrifLps2 n = 8; DKO n = 7) or left uninfected (Uninf.) (WT n = 8; Myd88−/− n = 8; TrifLps2 n = 8; DKO n = 6). Mouse stomachs were processed and scored double-blind for histologic disease severity for inflammation (A), gland atrophy (B), intestinal metaplasia (C) and dysplasia (D) on an ascending scale from 0 (no lesions) to 4 (severe lesions). Data are represented as mean ± SEM. Data were analyzed using Mann Whitney U Test. ∗p < 0.01, ∗∗p < 0.001.
Elevated ISG expression in patient gastric tissue samples is closely associated with progression of gastric cancer
The investigation of ISGs in previous studies of H. pylori-induced disease provided valuable insights. Upon reanalyzing microarray data (Gene Expression Omnibus, GEO; Accession number GSE27411),19 which included gastric biopsies of H. pylori-infected and uninfected patients with atrophic gastritis, we observed elevated levels of ISGs ((2′-5′-oligoadenylate synthase 2 (Oas2), B2m, Gbp2, Mx1, Irf1, Ido1, and Cxcl9) in gastric tissue samples from patients with H. pylori-induced chronic atrophic gastritis (Figure 6) similar to our findings observed in Myd88−/− mice following H. felis infection. As chronic atrophic gastritis is considered the initial step in the multi-step pre-neoplasia progression cascade,20 the overexpression of ISGs could signify a transition toward severe disease pathology.
Figure 6.
Heatmap showing expression of ISGs in patient gastric tissue samples
Reanalysis of published microarray dataset of gastric tissue from uninfected and H. pylori-infected patient with atrophic gastritis using publicly available dataset (GSE27411).19 Samples from H. pylori-infected patients are denoted by “+” (n = 6) and uninfected by “-” (n = 4).
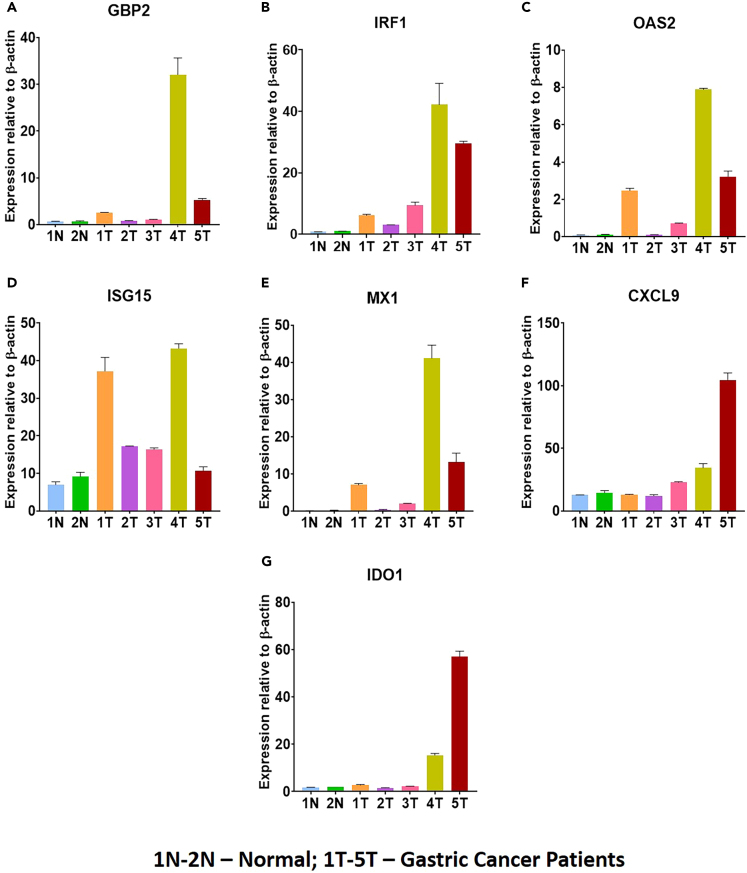
To further validate these findings, we examined available gastric samples obtained from the UCSD Biorepository. Our analysis revealed significant upregulation of several ISGs (Oas2, B2m, ISG product 15 (Isg15), Gbp2, Mx1, Irf1, Ido1, and Cxcl9) in human gastric cancer biopsy samples, mirroring the findings observed in gastric tissue samples from H. felis-infected Myd88−/− mice (Figure 7). This validation of highly expressed ISGs between H. felis-infected Myd88−/− mice and human gastric tissue samples from H. pylori-infected and gastric cancer patients strengthens the evidence for these ISGs being involved in disease progression to gastric cancer.
Figure 7.
Expression of ISGs in patient gastric biopsies
Gastric cancer patients’ gastric tissues obtained from the UCSD Biorepository showed upregulation of ISGs including GBP2 (A), IRF1 (B), OAS2 (C), ISG15 (D), MX1 (E), CXCL9 (F), and IDO1 (G). N, denotes normal and T, tumor. Data are represented as mean ± SEM.
Validation of mouse data in gastric cancer patient biopsy samples using Kaplan-Meier survival analysis
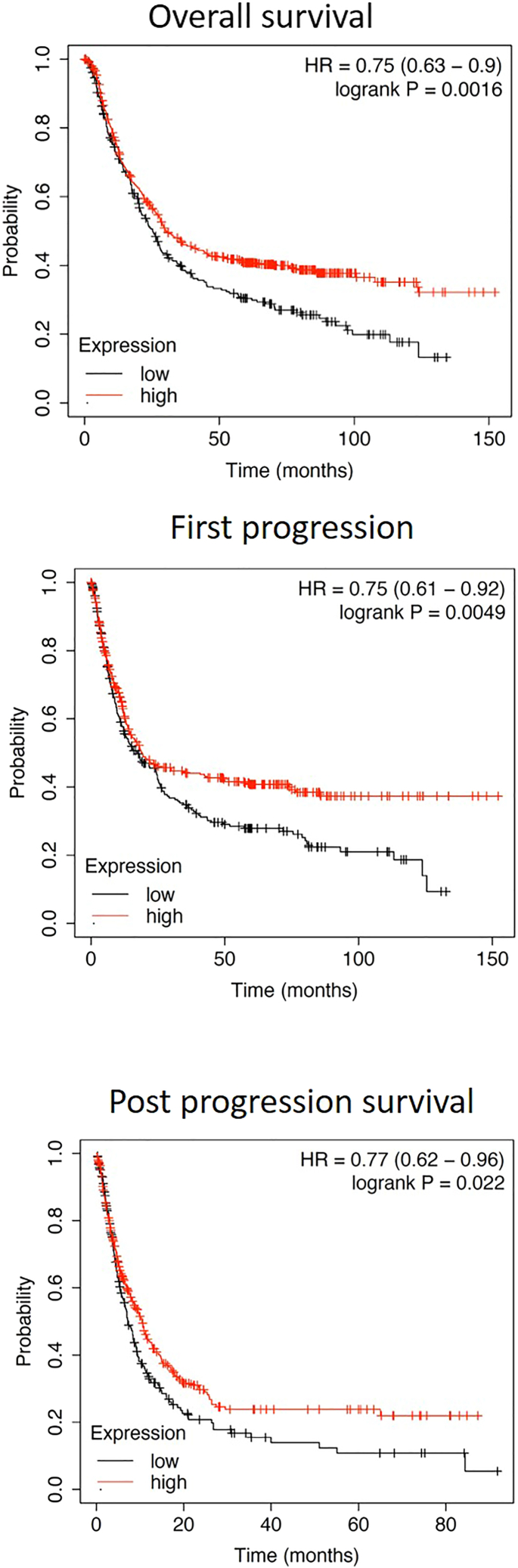
In our previous study, we found that mice deficient in Myd88 showed rapid progression of gastric pathology upon H. felis infection, with more severe gastric pathology including severe gland atrophy, hyperplasia, intestinal metaplasia, and gastric dysplasia than WT mice.7 Two recent clinical datasets of human stomach cancers from The Cancer Genome Atlas (TCGA)21,22 also reported the presence of MYD88 gene deletions and mutations in gastric and esophageal adenocarcinomas. Considering these findings, along with data from our animal model of gastric cancer, we performed Kaplan-Meier analysis of MYD88 expression in gastric cancer patients23 to validate the clinical relevance of our mouse study results. The database provided data from a total of 1,065 gastric cancer patients,23 which we divided into two groups based on their MYD88 expression levels (high and low) based on original criteria described by Busuttil et al. 2014.24 Briefly, the mean expression level of genes was used to divide patient sample into high or low expression groups with the top and bottom 33 percent of the samples defined as high or low, respectively. The resulting survival plots for gastric cancer patients indicated that those with low expression of MYD88 had significantly low survival rates compared to patients with higher expression, as shown in overall survival,OS (p = 0.0016, log rank test), first progression, FP (p = 0.0049, log rank test), and post-progression survival,PPS (p = 0.022, log rank test) analyses (Figure 8). These results establish a clear link between MyD88 deficiency and gastric cancer and suggest that the level of MYD88 expression during Helicobacter infection may serve as an important prognosticator of disease outcome.
Figure 8.
Kaplan-Meier survival curves for MYD88 expression in patients with gastric cancer
This online database includes data from 1065 gastric cancer patients.23 OS, overall survival; FP, first progression; PPS, post-progression survival.
We proceeded to examine the expression of TRIF in human gastric cancer patients using the Kaplan-Meier survival analysis.23 The results revealed that patients with high expression of TRIF had a significantly lower survival rate than that of patients with lower TRIF expression in the OS (log rank p = 0.0077), FP (log rank p = 0.0011), and PPS (log rank p = 0.00039) analyses. This finding strongly suggests that high expression of TRIF is associated with poor prognosis in gastric cancer patients (Figure 9). These clinical observations align with the findings from our mouse studies, indicating that the hyperactivation of the TRIF pathway and subsequent activation of the IFN-I signaling pathway play important roles in promoting the progression of Helicobacter-induced gastric cancer. As a result, targeting the TRIF signaling pathway holds the potential to intercept the development of precancerous lesions, offering a potential approach to mitigate cancer progression.
Figure 9.
Kaplan-Meier survival curves for TRIF expression in patients with gastric cancer
This online database includes data from 1065 gastric cancer patients.23 OS, overall survival; FP, first progression; PPS, post-progression survival.
Discussion
Our previous study using Myd88−/− mice suggested a potential link between TRIF activation and the progression of Helicobacter-induced gastric neoplasia.7 Upon infection with H. felis, these mice exhibited rapid progression of gastric pathology, including severe gland atrophy, hyperplasia, intestinal metaplasia, and dysplasia. However, the underlying molecular mechanism behind this phenotype remained unclear. In a separate study, we had also observed that H. pylori induced higher production of IFNα in bone marrow-derived macrophages (BMDM) from Myd88−/− mice compared to WT mice,25 indicating enhanced IFN-I response in the absence of MyD88 signaling. Based on these findings, we hypothesized that the downstream effects of an activated IFN-I signaling pathway, specifically the induction of ISGs, might play a role in the accelerated disease progression to preneoplastic lesions observed in Myd88−/− mice.7
Recent research using a gastric MALT lymphoma model with H. suis infection also supported the importance of the TRIF-IFN-I signaling pathway in lymphoma development.11 However, the timing of gastric epithelial changes associated with this disease progression and the role of downstream IFN-I target genes, ISGs,16 remained unknown. In the present study, we investigated these aspects and found that gastric epithelial changes occurred as early as one month after H. felis infection, with significant preneoplastic lesions developing at three months. Moreover, upregulation of ISGs was closely associated with severe disease pathology, including high-grade dysplasia, upon H. felis infection, suggesting a potential role for these ISGs in promoting the development and progression of gastric cancer.
Our H. felis-infected Myd88−/− mouse model provided meaningful insights that parallel human disease. Specifically, we observed that ISGs upregulated in the mouse model were also elevated in gastric tissues from H. pylori-infected patients with atrophic gastritis (Figure 6).19 Moreover, our analysis of gastric biopsy samples from our UCSD biorepository showed upregulation of these ISGs in gastric cancer patients (Figure 7). Together, these data suggest that ISGs may be involved in the transition of disease toward malignancy. The upregulation of these ISGs could drive molecular changes associated with Helicobacter-induced disease progression, leading to severe gastric pathology and precancerous lesions.
Although the role of ISGs in Helicobacter-induced disease pathogenesis is not well understood, some of the upregulated ISGs in mice and patient gastric tissue samples, including Gbp2 and Ido1, have been associated with other human disease conditions. As an example, GBP210 is highly expressed in human esophageal squamous cell carcinoma26 and is associated with increased proliferation,27 which aligns with the enhanced proliferation observed in our Myd88−/− mouse infection model.7 Ido1, another highly expressed ISG in gastric tissues of Myd88−/− mice following H. felis infection, is implicated in immune tolerance and high expression has been correlated with poor clinical outcomes in multiple cancer types.28,29,30,31,32,33,34,35,36,37,38,39 Upregulation of ISG15 expression in colon cancer tissues has been reported to promote proliferation and metastasis and is associated with poor prognosis in colon cancer patients.40 However, additional targeted inhibition studies are needed to directly assess the specific contribution of individual ISGs in disease pathology. Nevertheless, our results strongly indicate that upregulation of ISGs is involved in the severe gastric pathology observed in Myd88−/− mice (Figure 1)7 and in human gastric cancer patients (Figures 6 and 7). Cumulatively, our data show that Helicobacter-induced disease progresses rapidly to severe disease pathology in the presence of an unopposed activated TRIF pathway, leading to the upregulation of ISGs. Activation of the TRIF-IFN-I-dependent signaling pathway by Helicobacter may create a heightened inflammatory microenvironment that disrupts mucosal gastric homeostasis, resulting in the development of a dysplastic epithelium and ultimately gastric cancer. Indeed, gastric biopsy samples from H. pylori-infected patients with atrophic gastritis19 showed higher expression of ISGs compared to uninfected individuals. Furthermore, high expression of TRIF in gastric biopsy samples from gastric cancer patients was associated with poor OS.
In conclusion, our study provides significant evidence, from both in vivo models and patient-derived data, indicating that the TRIF-IFN-I pathway is involved in Helicobacter-induced disease progression to gastric cancer. The findings highlight the central role of this pathway in gastric carcinogenesis, emphasizing the importance of further exploration in this area. Our study demonstrates the importance of examining the mechanisms of Helicobacter-induced disease during the early phase of infection. Early detection of epithelial changes could enhance the identification of drivers, leading to the discovery of effective molecular targets that may halt the progression of disease to neoplastic lesions and ultimately gastric cancer.
Limitations of the study
One limitation of this study is that the H. pylori status of gastric cancer patients from our biorepository and the Kaplan-Meier plotter is often not defined. Infection with H. pylori is the major cause of gastric cancer, as epidemiologic studies estimating that 75% of gastric cancer cases are a result of infection with the bacterium.41,42,43 Thus, there is a possibility that some of the gastric cancer patients in our study may not have been H. pylori positive. Another limitation of the study is the remaining outstanding question of whether MyD88 and TRIF interactively influence Helicobacter-induced gastric carcinogenesis. While we show that disease pathology in DKO mice is moderately severe, we do not yet know whether the MyD88 or TRIF pathway played a more important role. The elucidation of this point will require long-term animal studies to discriminate the key pathways and the impact of interactions between them. Further, while we used male mice to allow for comparison with previous studies that have investigated gastric carcinogenesis using the H. felis infection mouse model that have used only male mice,44,45,46 this limits generalization of the results. Finally, our study used H. felis gene and whole genome reads to estimate colonization, which is not a very quantitative way of assessing colonization. Our prior work had shown no significant differences in H. felis colonization between WT and Myd88−/− mice.7 In the current study, where our primary focus was on disease severity rather than colonization, we found no significant differences in reads either using genes frequently used to assess colonization or whole genome reads between infected mouse strains (Figure S1).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Helicobacter felis, strain CS1 | American Type Culture Collection | ATCC 49179 |
| Biological samples | ||
| Human Gastric Biopsy Tissue Samples | Biorepository, University of California, San Diego | N/A |
| Critical commercial assays | ||
| Direct-zol RNA MiniPrep Kit | Zymo Research | R2050 |
| High Capacity cDNA Reverse Transcription Kit | Thermo Fisher | 4368814 |
| Illumina TruSeq Stranded Total RNA Sample Prep Kit with Ribo-Zero | Illumina | 20020597 |
| Deposited data | ||
| GEO accession number: GSE250438 | This paper | GSE250438 |
| Experimental models: Organisms/strains | ||
| Mouse: WT: C57BL/6 | Jackson Laboratory | C57BL/6J |
| Mouse: Myd88 KO: Myd88 deficient | Dr. Akira’s laboratory | N/A |
| Mouse: Trif KO: Trif deficient | Dr. Akira’s laboratory | N/A |
| Mouse: Myd88/Trif DKO: Myd88/Trif deficient | This paper | N/A |
| Oligonucleotides | ||
| Primers for qPCR | see Table S2 | N/A |
| Software and algorithms | ||
| GraphPad Prism 8.2.0 | GraphPad Software | https://www.graphpad.com/scientificsoftware/prism/ |
| HOMER’s analyzeRepeats.pl | Dr. Benner’s lab | N/A |
| Other | ||
| Nanodrop 2000 | Thermo Fisher | ND-2000 |
| Veriti Thermal Cycler | Applied Biosystems | 4375305 |
| StepOne | Applied Biosystems | 272001471 |
| Illumina/Solexa HiSeq 2500 sequencer | Illumina | SY-401-2501 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Marygorret Obonyo (mobonyo@health.ucsd.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
Single-cell RNA-seq data have been deposited at GEO and is publicly available as of the date of publication. The accession number is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Animals
Six- to ten- week-old wild type (WT) (n=42), MyD88 deficient (Myd88-/-) (n=47), TRIF deficient (TrifLps2) (n=46), double deficient (Myd88-/- and TrifLps2, DKO) (n=37) mice in the C57BL/6 background were used in this study. In this study male mice were used. WT mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Myd88-/- mice were from our breeding colony originally provided by Dr. Akira (Osaka University, Japan) and backcrossed 10 times onto a C57BL/6 background, bred, and maintained at our facility. The DKO mice were bred in our facility by crossing Myd88-/- and TrifLps2. All mice were housed together before and throughout the H. felis infection study for each genotype in a biosafety level II (BSL-2) facility with controlled temperature (23± 2°C) and relative humidity (45-60%) and had full access to food and water. The animal procedures were approved by the Institutional Animal Care and Use Committee at the University of California, San Diego, and conducted following accepted veterinary standards and ARRIVE guidelines.
Human participants
Seven de-identified snap-frozen human gastric biopsy samples were obtained from UCSD’s Biorepository. Prior to sample collection, all patients provided written informed consent and were followed up. Details about the patients and their gastric tumor characteristics including race or ethnicity are provided in Table S1.47 RNA was extracted from each biopsy tissue and subsequently processed for real-time PCR as described below. The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of the University of California, San Diego.
Bacterial growth conditions
Helicobacter felis, strain CS1 (ATCC 49179) was obtained from the American Type Culture collection (Manassas, VA). The bacteria were cultured on solid Columbia agar (Becton Dickinson, MD) supplemented with 5% laked blood under microaerophilic conditions (5% O2, 10% CO2, 85% N2) at 37°C and passaged every 2–3 days, following established protocols.7,48,49,50 Prior to mouse infections, H. felis was cultured in liquid brain heart infusion broth (BHI, Becton Dickinson) supplemented with 10% fetal calf serum and incubated at 37°C under microaerophilic conditions for 48 h. Spiral bacteria were enumerated using a Petroff-Hausser chamber before infections.
Mouse infections
This study used a well-characterized cancer mouse model, infecting C57BL/6 mice with H. felis (strain CS1), a close relative of the human gastric pathogen H. pylori. Mice were inoculated with 109 organisms in 300 μL of BHI by oral gavage using a feeding needle.7,48,49,50 Inoculation of mice with H. felis was repeated three times at 2-day intervals. Control mice received BHI only. At 1 month, 3 months, and 6 months post-infection, mice were euthanized, and their stomachs removed under aseptic conditions. The stomach tissue was cut longitudinally, and sections were processed for RNA extraction and histopathology. All H. felis-infected mice were confirmed to be colonized by assessing the expression of the flagella filament B (flab) gene using real-time PCR.7 The amplification conditions consisted of an initial cycle of 95°C for 5 min, annealing at 60°C for 20 sec, and extension at 72°C for 40 sec. Gene expression levels were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). There were no significant differences in the expression of the flab gene among all the infected mice groups.
Method details
Histology
Mouse stomachs were processed for histology according to the following the procedures.7,51 Longitudinal sections of stomach tissue from each mouse were fixed in neutral buffered 10% formalin, embedded in paraffin, and then 5 μm sections were stained with hematoxylin and eosin (H&E). The gastric histopathologies, including inflammation, epithelial defects, gland atrophy, hyperplasia, intestinal metaplasia, and dysplasia, were evaluated and scored for histologic disease severity on a scale from 0 (no lesions) to 4 (severe lesions). The scoring was performed in a double-blind manner by a pathologist (Dr. Estrada) following the criteria outlined by Rogers et al..52 Specifically, the histological characteristics of the parameters were defined as follows: inflammation—infiltration of inflammatory cells into the mucosa and submucosa; epithelial defects—thinned epithelial surface, epithelial and mucosal erosion and multifocal collapse; oxyntic gland atrophy—loss of oxyntic glands (which contain parietal cells and chief cells) in the gastric corpus; hyperplasia—elongation of gastric glands due to increased numbers of surface cells (foveolar cells); intestinal metaplasia—replacement of gastric epithelium by intestinal type gastric epithelium; and dysplasia—abnormal cellular and glandular maturation of the epithelium, with score of 3 indicating carcinoma in situ.52
RNA extraction from mouse and human gastric tissue samples
RNA was extracted from gastric tissue obtained from human biopsy samples (described above) and gastric tissue samples from H. felis-infected and uninfected WT, Myd88-/-, TrifLps, and DKO mice. Gastric tissue samples were homogenized in 1 ml of TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. RNA was extracted using the Direct-zol RNA mini kit (Zymo Research, Irvine CA) as per the manufacturer’s instructions and stored at -70°C until further use. The quality of the extracted RNA was assessed using a Nanodrop system (Thermo Fisher, Waltham MA) by measuring absorbance levels at 260 and 280 nm. Subsequently, the RNA was utilized for RNASeq analysis in the mouse gastric tissue samples and qPCR in the human gastric tissue samples.
cDNA synthesis and quantitative real-time RT-PCR
Real-time RT-PCR (reverse transcription polymerase chain reaction) was conducted the StepOne Real Time PCR system (Applied Biosystems, Carlsbad CA).7,10,50,51 Expression levels of specific downstream genes in the TRIF-IFN-I pathway in human gastric biopsy samples including, indoleamine 2,3-dioxygenase 1 (IDO1), guanylate binding protein 2 (GBP2), transcription factor Interferon regulatory factor 1 (IRF1), 2’-5’-oligoadenylate synthase 2 (OAS2), and chemokine (C-X-C motif) ligand 9 (CXCL9), MX dynamin-like GTPase 1(MX1), and interferon-stimulated gene 15 (ISG15), were determined. RNA (2μg/sample) isolated from gastric tissue samples was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher, Waltham MA). For amplification, 1μl of cDNA was used per well in a total of 10μl reaction mix. The amplification conditions consisted of an initial cycle of 95°C for 5 min, annealing at 60°C for 20 sec, and extension at 72°C for 40 sec. Gene expression levels were normalized to β-actin. The data collected were analyzed using comparative cycle threshold calculations (ΔCT Applied Biosystems) and plotted using GraphPad Prism software. The primers used in this study are listed in Table S2.
RNA sequencing
RNA concentration and integrity were evaluated using an Agilent 2100 Bioanalyzer, and each sample was assigned an RNA integrity number (RIN). Selected samples underwent further processing into RNA-seq libraries using the Illumina TruSeq Stranded Total RNA Sample Prep Kit with Ribo-Zero (Human/Mouse/Rat), which involved ribosomal RNA depletion, fragmentation, cDNA synthesis, and cDNA library preparation. Subsequently, sequencing was performed on an Illumina/Solexa HiSeq 2500 sequencer, generating 75-bp single-end reads for all samples at a depth of approximately 40 million reads per sample.
RNA-seq analysis
For RNA-seq data analysis, the reads were aligned to the combined mouse and H. felis genomes (GRCm38/mm10 and NC_014810.2, respectively) using STAR with default parameters.53 Only reads that aligned uniquely to a single location across genomes with a mapping quality score greater than 10 (MAPQ > 10) were used for downstream analysis. Gene expression levels were determined by counting the number of reads overlapping exons for all genes using HOMER’s analyzeRepeats.pl tool and the transcriptome annotation from GENCODE (version M25).54 Analysis was performed using DESeq2 to identify genes that were differentially expressed from unnormalized read counts.55 Volcano plot visualization was performed using the EnhancedVolcano package from R/Bioconductor. For heatmap visualization, gene expression values were normalized using DESeq2’s rlog function. Normalized genome browser read density plots were generated by creating bedGraph files with HOMER and visualizing them using the UCSC Genome Browser. Gene expression values for genes on the H. felis genome were determined using HOMER’s analyzeRepeats.pl function based on gene annotations associated with NCBI accession number NC_014810.2, and reported as fragments per kilobase per million uniquely mapped fragments (FPKM), normalized to the total number of reads aligning to both the mouse and pathogen genomes.
Pathway and motif enrichment analysis
Pathway enrichment analysis was performed with Metascape,56 a web-based portal that integrates multiple knowledgebases, including the Gene Ontology, to identify overrepresented pathways in sets of regulated genes. Differentially expressed genes with a fold change of at least 2 and an adjusted p-value of 0.05 were used as input for Metascape analysis. The pathway enrichment results reported by Metascape were corrected for multiple hypothesis testing. To analyze transcription factor motifs in mouse stomach tissue sections (RNA-seq data), we used HOMER, a software suite for DNA motif and next-generation sequence (NGS) analysis.54 Known transcription factor motifs from the HOMER database were scanned in mouse promoter sequences defined within a range from -300 to +50 bp relative to the GENCODE-defined transcription start site (TSS) locations. The log p-values reported in the analysis were corrected for multiple hypothesis testing using the Benjamini–Hochberg procedure.
Human gastric interferon-stimulated gene (ISG) expression
Gene expression levels in gastric tissues were reanalyzed using data from the Gene Expression Omnibus (GEO) database, with accession number GSE27411.19 This dataset contains microarray gene expression data from gastric tissue samples obtained from individuals with and without H. pylori infection.19 To ensure consistency, RNA-seq expression values were quantile normalized before visualization. The resulting normalized expression values were then used to generate a heatmap using Java Tree View.57
Survival analyses of MYD88 and TRIF using the Kaplan-Meier plotter
We utilized the Kaplan-Meier plotter bioinformatics platform (https://kmplot.com/analysis/) to analyze the impact of MYD88 and TRIF gene expression on overall survival (OS), first progression (FP), and post-progression survival (PPS) in gastric cancer patients. The data used for analysis was derived from the GEO database23 and included microarray gene expression data from 1065 gastric cancer patients.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism (La Jolla, CA, USA). For normally distributed data, t-test was employed, while non-normally distributed data were analyzed using Mann Whitney U Test. For multiple comparisons, ANOVA with Bonferroni’s correction (for normal distribution) or Kruskal-Wallis with Dunn’s comparison test (for non-normal distribution) were used. Survival analysis was performed using the Kaplan-Meier method, and hazard ratio (HR) with a 95% confidence interval was calculated. Patients were divided into high and low expression groups using the best cut-off, and differences were assessed using the log-rank test. P-values ˂0.05 were considered statistically significant. Differentially expressed genes were identified as those with a fold change of at least 2 and an adjusted p-value of 0.05, determined by DESeq2.
Acknowledgments
This work is supported by funding from the Department of Defense (DOD), award W81XWH-20-1-0675 to M.O. The project was also partially funded by NIH grants, UL1TR001442 and P30 DK120515.
Author contributions
M.O. supervised and designed the study concept and research; C.B. supervised data analysis; P.B., I.L.-P., and J.H. collected and analyzed data; M.V.E. performed mouse histopathologic scoring; M.C. supervised mouse breeding; M.A.T. and M.B. acquired UCSD biorepository patient information; P.B. and I.L.-P. wrote the draft of the manuscript; C.B. and M.O. revised the manuscript. All authors read and approved the manuscript.
Declaration of interests
All authors declare that no conflict of interest exists.
Published: March 8, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109457.
Supplemental information
References
- 1.IARC Schistomes, liver flukes and Helicobacter pylori. IARC Monographs on the evaluation of carcinogenesis risks to humans. IARC Sci. Publ. 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 2.Peek R.M., Jr., Blaser M.J. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 3.SEER Cancer statistics review (CSR) 1975-2014. 2017. https://seer.cancer.gov/csr/1975 2014/
- 4.Torre L.A., Siegel R.L., Ward E.M., Jemal A. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol. Biomarkers Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 5.Fox J.G., Wang T.C. Inflammation, atrophy, and gastric cancer. J. Clin. Invest. 2007;117:60–69. doi: 10.1172/JCI30111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correa P., Piazuelo M.B. The gastric precancerous cascade. J. Dig. Dis. 2012;13:2–9. doi: 10.1111/j.1751-2980.2011.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerjee A., Thamphiwatana S., Carmona E.M., Rickman B., Doran K.S., Obonyo M. Deficiency of the myeloid differentiation primary response molecule MyD88 leads to an early and rapid development of Helicobacter-induced gastric malignancy. Infect. Immun. 2014;82:356–363. doi: 10.1128/IAI.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee C.W., Rickman B., Rogers A.B., Ge Z., Wang T.C., Fox J.G. Helicobacter pylori eradication prevents progression of gastric cancer in hypergastrinemic INS-GAS mice. Cancer Res. 2008;68:3540–3548. doi: 10.1158/0008-5472.CAN-07-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stenström B., Zhao C.M., Rogers A.B., Nilsson H.O., Sturegård E., Lundgren S., Fox J.G., Wang T.C., Wadström T.M., Chen D. Swedish moist snuff accelerates gastric cancer development in Helicobacter pylori-infected wild-type and gastrin transgenic mice. Carcinogenesis. 2007;28:2041–2046. doi: 10.1093/carcin/bgm071. [DOI] [PubMed] [Google Scholar]
- 10.Lozano-Pope I., Sharma A., Matthias M., Doran K.S., Obonyo M. Effect of myeloid differentiation primary response gene 88 on expression profiles of genes during the development and progression of Helicobacter-induced gastric cancer. BMC Cancer. 2017;17:133. doi: 10.1186/s12885-017-3114-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto K., Kondo Y., Ohnishi S., Yoshida M., Sugiyama T., Sakamoto N. The TLR4-TRIF-type 1 IFN-IFN-gamma pathway is crucial for gastric MALT lymphoma formation after Helicobacter suis infection. iScience. 2021;24 doi: 10.1016/j.isci.2021.103064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai T., Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 13.Hoebe K., Du X., Georgel P., Janssen E., Tabeta K., Kim S.O., Goode J., Lin P., Mann N., Mudd S., et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 14.Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 16.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Zimaity H.M.T., Ota H., Graham D.Y., Akamatsu T., Katsuyama T. Patterns of gastric atrophy in intestinal type gastric carcinoma. Cancer. 2002;94:1428–1436. doi: 10.1002/cncr.10375. [DOI] [PubMed] [Google Scholar]
- 18.Goldenring J.R., Nam K.T. Oxyntic atrophy, metaplasia, and gastric cancer. Prog. Mol. Biol. Transl. Sci. 2010;96:117–131. doi: 10.1016/B978-0-12-381280-3.00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nookaew I., Thorell K., Worah K., Wang S., Hibberd M.L., Sjövall H., Pettersson S., Nielsen J., Lundin S.B. Transcriptome signatures in Helicobacter pylori-infected mucosa identifies acidic mammalian chitinase loss as a corpus atrophy marker. BMC Med. Genom. 2013;6:41. doi: 10.1186/1755-8794-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah S.C., Piazuelo M.B., Kuipers E.J., Li D. AGA Clinical Practice Update on the Diagnosis and Management of Atrophic Gastritis: Expert Review. Gastroenterology. 2021;161:1325–1332.e7. doi: 10.1053/j.gastro.2021.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network, Analysis Working Group: Asan University, BC Cancer Agency, Brigham and Women’s Hospital, Broad Institute, Brown University, Case Western Reserve University, Dana-Farber Cancer Institute, Duke University, Greater Poland Cancer Centre, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szász A.M., Lánczky A., Nagy Á., Förster S., Hark K., Green J.E., Boussioutas A., Busuttil R., Szabó A., Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Busuttil R.A., George J., Tothill R.W., Ioculano K., Kowalczyk A., Mitchell C., Lade S., Tan P., Haviv I., Boussioutas A. A signature predicting poor prognosis in gastric and ovarian cancer represents a coordinated macrophage and stromal response. Clin. Cancer Res. 2014;20:2761–2772. doi: 10.1158/1078-0432.CCR-13-3049. [DOI] [PubMed] [Google Scholar]
- 25.Obonyo M., Sabet M., Cole S.P., Ebmeyer J., Uematsu S., Akira S., Guiney D.G. Deficiencies of myeloid differentiation factor 88, Toll-like receptor 2 (TLR2), or TLR4 produce specific defects in macrophage cytokine secretion induced by Helicobacter pylori. Infect. Immun. 2007;75:2408–2414. doi: 10.1128/IAI.01794-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guimarães D.P., Oliveira I.M., de Moraes E., Paiva G.R., Souza D.M., Barnas C., Olmedo D.B., Pinto C.E., Faria P.A., De Moura Gallo C.V., et al. Interferon-inducible guanylate binding protein (GBP)-2: a novel p53-regulated tumor marker in esophageal squamous cell carcinomas. Int. J. Cancer. 2009;124:272–279. doi: 10.1002/ijc.23944. [DOI] [PubMed] [Google Scholar]
- 27.Gorbacheva V.Y., Lindner D., Sen G.C., Vestal D.J. The interferon (IFN)-induced GTPase, mGBP-2. Role in IFN-gamma-induced murine fibroblast proliferation. J. Biol. Chem. 2002;277:6080–6087. doi: 10.1074/jbc.M110542200. [DOI] [PubMed] [Google Scholar]
- 28.Corm S., Berthon C., Imbenotte M., Biggio V., Lhermitte M., Dupont C., Briche I., Quesnel B. Indoleamine 2,3-dioxygenase activity of acute myeloid leukemia cells can be measured from patients' sera by HPLC and is inducible by IFN-gamma. Leuk. Res. 2009;33:490–494. doi: 10.1016/j.leukres.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 29.Creelan B.C., Antonia S., Bepler G., Garrett T.J., Simon G.R., Soliman H.H. Indoleamine 2,3-dioxygenase activity and clinical outcome following induction chemotherapy and concurrent chemoradiation in Stage III non-small cell lung cancer. OncoImmunology. 2013;2 doi: 10.4161/onci.23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferns D.M., Kema I.P., Buist M.R., Nijman H.W., Kenter G.G., Jordanova E.S. Indoleamine-2,3-dioxygenase (IDO) metabolic activity is detrimental for cervical cancer patient survival. OncoImmunology. 2015;4 doi: 10.4161/2162402X.2014.981457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inaba T., Ino K., Kajiyama H., Yamamoto E., Shibata K., Nawa A., Nagasaka T., Akimoto H., Takikawa O., Kikkawa F. Role of the immunosuppressive enzyme indoleamine 2,3-dioxygenase in the progression of ovarian carcinoma. Gynecol. Oncol. 2009;115:185–192. doi: 10.1016/j.ygyno.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Ino K., Yamamoto E., Shibata K., Kajiyama H., Yoshida N., Terauchi M., Nawa A., Nagasaka T., Takikawa O., Kikkawa F. Inverse correlation between tumoral indoleamine 2,3-dioxygenase expression and tumor-infiltrating lymphocytes in endometrial cancer: its association with disease progression and survival. Clin. Cancer Res. 2008;14:2310–2317. doi: 10.1158/1078-0432.CCR-07-4144. [DOI] [PubMed] [Google Scholar]
- 33.Speeckaert R., Vermaelen K., van Geel N., Autier P., Lambert J., Haspeslagh M., van Gele M., Thielemans K., Neyns B., Roche N., et al. Indoleamine 2,3-dioxygenase, a new prognostic marker in sentinel lymph nodes of melanoma patients. Eur. J. Cancer. 2012;48:2004–2011. doi: 10.1016/j.ejca.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Wainwright D.A., Balyasnikova I.V., Chang A.L., Ahmed A.U., Moon K.S., Auffinger B., Tobias A.L., Han Y., Lesniak M.S. IDO expression in brain tumors increases the recruitment of regulatory T cells and negatively impacts survival. Clin. Cancer Res. 2012;18:6110–6121. doi: 10.1158/1078-0432.CCR-12-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ino K., Yoshida N., Kajiyama H., Shibata K., Yamamoto E., Kidokoro K., Takahashi N., Terauchi M., Nawa A., Nomura S., et al. Indoleamine 2,3-dioxygenase is a novel prognostic indicator for endometrial cancer. Br. J. Cancer. 2006;95:1555–1561. doi: 10.1038/sj.bjc.6603477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okamoto A., Nikaido T., Ochiai K., Takakura S., Saito M., Aoki Y., Ishii N., Yanaihara N., Yamada K., Takikawa O., et al. Indoleamine 2,3-dioxygenase serves as a marker of poor prognosis in gene expression profiles of serous ovarian cancer cells. Clin. Cancer Res. 2005;11:6030–6039. doi: 10.1158/1078-0432.CCR-04-2671. [DOI] [PubMed] [Google Scholar]
- 37.Ricciuti B., Leonardi G.C., Puccetti P., Fallarino F., Bianconi V., Sahebkar A., Baglivo S., Chiari R., Pirro M. Targeting indoleamine-2,3-dioxygenase in cancer: Scientific rationale and clinical evidence. Pharmacol. Ther. 2019;196:105–116. doi: 10.1016/j.pharmthera.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Brochez L., Chevolet I., Kruse V. The rationale of indoleamine 2,3-dioxygenase inhibition for cancer therapy. Eur. J. Cancer. 2017;76:167–182. doi: 10.1016/j.ejca.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 39.Ferdinande L., Decaestecker C., Verset L., Mathieu A., Moles Lopez X., Negulescu A.M., Van Maerken T., Salmon I., Cuvelier C.A., Demetter P. Clinicopathological significance of indoleamine 2,3-dioxygenase 1 expression in colorectal cancer. Br. J. Cancer. 2012;106:141–147. doi: 10.1038/bjc.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou S., Ren M., Xu H., Xia H., Tang Q., Liu M. Inhibition of ISG15 Enhances the Anti-Cancer Effect of Trametinib in Colon Cancer Cells. OncoTargets Ther. 2019;12:10239–10250. doi: 10.2147/OTT.S226395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrera V., Parsonnet J. Helicobacter pylori and gastric adenocarcinoma. Clin. Microbiol. Infect. 2009;15:971–976. doi: 10.1111/j.1469-0691.2009.03031.x. [DOI] [PubMed] [Google Scholar]
- 42.Parkin D.M. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 43.Polk D.B., Peek R.M., Jr. Helicobacter pylori: gastric cancer and beyond. Nat. Rev. Cancer. 2010;10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ericksen R.E., Rose S., Westphalen C.B., Shibata W., Muthupalani S., Tailor Y., Friedman R.A., Han W., Fox J.G., Ferrante A.W., Jr., Wang T.C. Obesity accelerates Helicobacter felis-induced gastric carcinogenesis by enhancing immature myeloid cell trafficking and TH17 response. Gut. 2014;63:385–394. doi: 10.1136/gutjnl-2013-305092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai X., Carlson J., Stoicov C., Li H., Wang T.C., Houghton J. Helicobacter felis eradication restores normal architecture and inhibits gastric cancer progression in C57BL/6 mice. Gastroenterology. 2005;128:1937–1952. doi: 10.1053/j.gastro.2005.02.066. [DOI] [PubMed] [Google Scholar]
- 46.Höök-Nikanne J., Aho P., Kärkkäinen P., Kosunen T.U., Salaspuro M. The Helicobacter felis mouse model in assessing anti-Helicobacter therapies and gastric mucosal prostaglandin E2 levels. Scand. J. Gastroenterol. 1996;31:334–338. doi: 10.3109/00365529609006406. [DOI] [PubMed] [Google Scholar]
- 47.Hernandez J., Turner M.A., Bali P., Hosseini M., Bouvet M., Kelly K., Obonyo M. Genomically Silent Refractory Gastric Cancer in a Young Patient Exhibits Overexpression of CXCL5. Curr. Oncol. 2022;29:4725–4733. doi: 10.3390/curroncol29070375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bali P., Coker J., Lozano-Pope I., Zengler K., Obonyo M. Microbiome Signatures in a Fast- and Slow-Progressing Gastric Cancer Murine Model and Their Contribution to Gastric Carcinogenesis. Microorganisms. 2021;9:189. doi: 10.3390/microorganisms9010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bali P., Lozano-Pope I., Pachow C., Obonyo M. Early detection of tumor cells in bone marrow and peripheral blood in a fastprogressing gastric cancer model. Int. J. Oncol. 2021;58:388–396. doi: 10.3892/ijo.2021.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mejías-Luque R., Lozano-Pope I., Wanisch A., Heikenwälder M., Gerhard M., Obonyo M. Increased LIGHT expression and activation of non-canonical NF-kappaB are observed in gastric lesions of MyD88-deficient mice upon Helicobacter felis infection. Sci. Rep. 2019;9:7030. doi: 10.1038/s41598-019-43417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Obonyo M., Rickman B., Guiney D.G. Effects of myeloid differentiation primary response gene 88 (MyD88) activation on Helicobacter infection in vivo and induction of a Th17 response. Helicobacter. 2011;16:398–404. doi: 10.1111/j.1523-5378.2011.00861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogers A.B., Taylor N.S., Whary M.T., Stefanich E.D., Wang T.C., Fox J.G. Helicobacter pylori but not high salt induces gastric intraepithelial neoplasia in B6129 mice. Cancer Res. 2005;65:10709–10715. doi: 10.1158/0008-5472.CAN-05-1846. [DOI] [PubMed] [Google Scholar]
- 53.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saldanha A.J. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
Single-cell RNA-seq data have been deposited at GEO and is publicly available as of the date of publication. The accession number is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.