Abstract
Glutathione (GSH) is a small-molecule thiol that is abundant in all eukaryotes and has key roles in oxidative metabolism1. Mitochondria, as the major site of oxidative reactions, must maintain sufficient levels of GSH to perform protective and biosynthetic functions2. GSH is synthesized exclusively in the cytosol, yet the molecular machinery involved in mitochondrial GSH import remains unknown. Here, using organellar proteomics and metabolomics approaches, we identify SLC25A39, a mitochondrial membrane carrier of unknown function, as a regulator of GSH transport into mitochondria. Loss of SLC25A39 reduces mitochondrial GSH import and abundance without affecting cellular GSH levels. Cells lacking both SLC25A39 and its paralogue SLC25A40 exhibit defects in the activity and stability of proteins containing iron–sulfur clusters. We find that mitochondrial GSH import is necessary for cell proliferation in vitro and red blood cell development in mice. Heterologous expression of an engineered bifunctional bacterial GSH biosynthetic enzyme (GshF) in mitochondria enables mitochondrial GSH production and ameliorates the metabolic and proliferative defects caused by its depletion. Finally, GSH availability negatively regulates SLC25A39 protein abundance, coupling redox homeostasis to mitochondrial GSH import in mammalian cells. Our work identifies SLC25A39 as an essential and regulated component of the mitochondrial GSH-import machinery.
GSH is commonly recognized as a component of antioxidant defence systems and has a wide range of roles in disulfide bond formation, detoxification of xenobiotics, metabolite transport, formation of iron–sulfur clusters and cellular signalling3. Eukaryotic cells synthesize GSH by the consecutive actions of γ-glutamate–cysteine ligase (GCL) and GSH synthetase—enzymes that are localized in the cytosol. Despite being produced exclusively in the cytosol4, GSH is also abundant in many organelles, including peroxisomes, the nucleus, endoplasmic reticulum and mitochondria5,6. Mitochondria, the most redox-active organelle, contain 10–15% of total cellular GSH7. As mitochondria lack GSH biosynthetic machinery4 and GSH is negatively charged under physiological conditions8, there must be a dedicated transport process for GSH to enter mitochondria2,9, but the molecular machinery involved in mitochondrial GSH transport has remained unknown.
Mitochondrial proteomics under GSH depletion
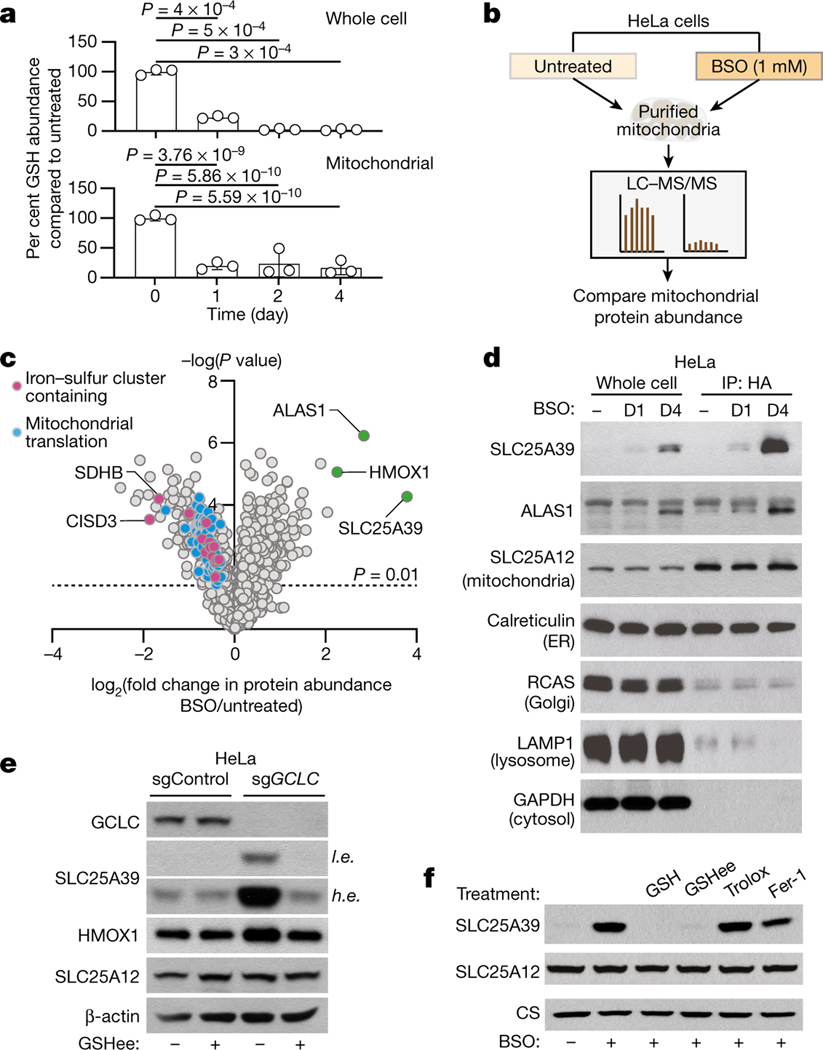
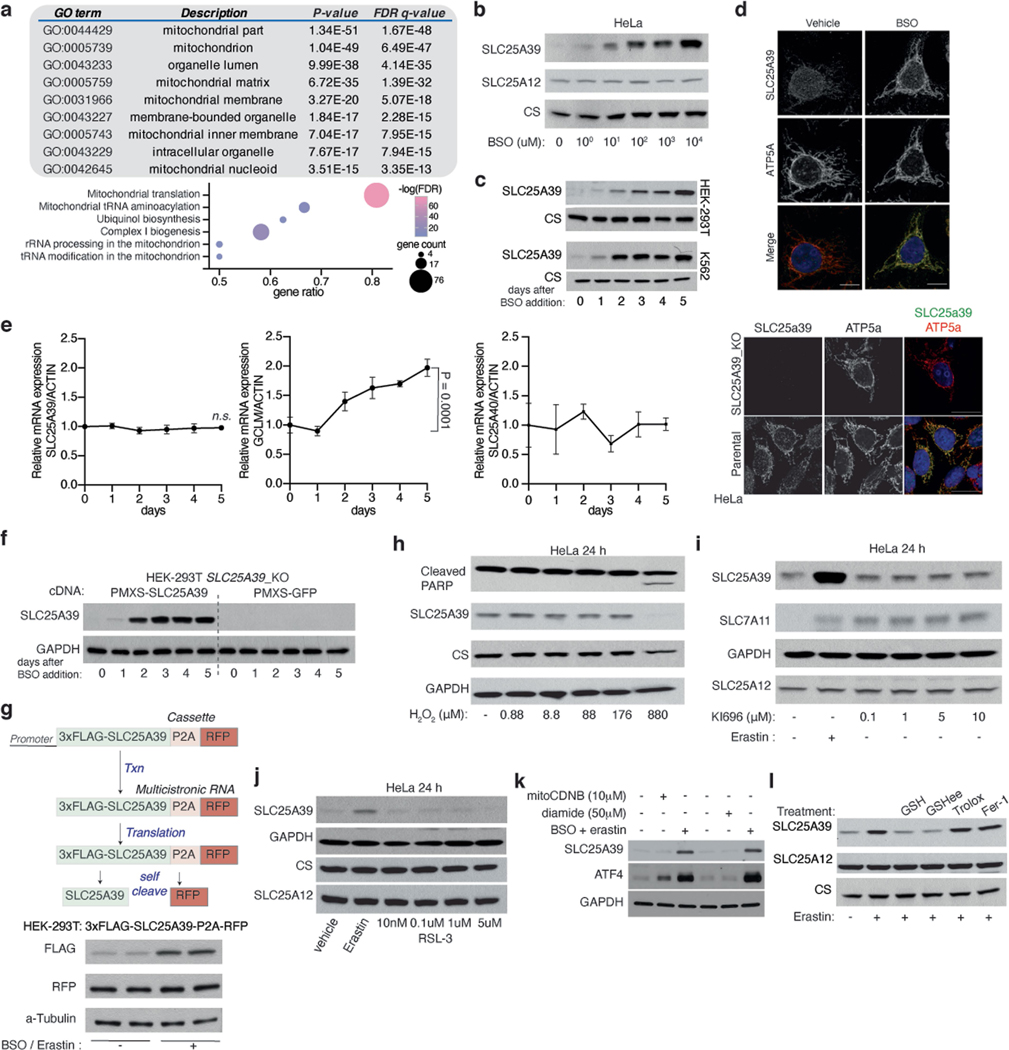
To identify mitochondrial proteins regulated by GSH availability, we applied quantitative proteomics to mitochondria immunopurified from HeLa cells expressing a mitochondrial tag (3×HA–OMP25–mCherry) grown in standard medium or treated with a GSH synthesis inhibitor, buthionine sulfoximine (BSO). BSO treatment decreases GSH levels in whole cells and in purified mitochondria with similar kinetics10 (Fig. 1a). After BSO treatment, compared to untreated cells, 296 proteins exhibited significant alterations in abundance (P < 0.01, change in abundance > 1.5) (Fig. 1b, c, Supplementary Table 1). Gene ontology analysis revealed that many components of the mitochondrial translation machinery were downregulated in response to GSH depletion, a phenotype previously linked to electron transport chain dysfunction11 (Extended Data Fig. 1a). GSH depletion also upregulates downstream metabolic targets (5′-aminolevulinate synthase 1 (ALAS1) and haem oxygenase 1 (HMOX1)) of nuclear factor erythroid 2-related factor 212,13 (NRF2), a transcription factor that induces expression of cellular antioxidant defence genes (Fig. 1c). In individual immunoblotting experiments, we confirmed enrichment of mitochondrial markers and several proteins upregulated in response to BSO as early as 1 day after treatment (Fig. 1d). Among the most upregulated proteins upon GSH depletion was SLC25A39, an uncharacterized mitochondrial membrane transporter (Fig. 1c). Induction of SLC25A39 protein levels was generalizable, as two additional cell lines from distinct tissues of origin (K562 and HEK 293T) exhibited similar responses to BSO (Extended Data Fig. 1b–d). Similar to pharmacologic inhibition of GSH synthesis, loss of GCLC, the rate-limiting enzyme of GSH synthesis, strongly induced SLC25A39 protein expression (Fig. 1e). Given that SLC25A39 was the only mitochondrial membrane transporter of unknown function regulated by GSH depletion, we focused our attention on it.
Fig. 1 |. Global analysis of mitochondrial proteome under GSH depletion.
a, Relative abundance of GSH in whole cells (top) and mitochondria (bottom) immunopurified from HeLa cells treated with BSO (1 mM) for the indicated times compared with untreated controls. Data are mean ± s.d.; n = 3 biologically independent samples. One-way analysis of variance (ANOVA) followed by Bonferroni post hoc analysis. b, Schematic showing proteomics analysis of mitochondria isolated from indicated HeLa cells. c, Volcano plot showing relative fold change (log2) in mitochondrial protein abundance versus –log (P values) from HeLa cells treated with BSO (1 mM) for 4 days compared with untreated. The dotted line represents P = 0.01. Colours denote indicated protein families. Statistical significance was determined by two-tailed unpaired t-test. d, Immunoblot analysis of indicated proteins in whole-cell lysates and mitochondria isolated from HeLa cells (IP: HA) treated with BSO (1 mM) for the indicated days. D1, day 1; D4, day 4. e, Immunoblot of indicated proteins in HeLa cells infected with the indicated sgRNAs. Cells were plated 6 days after transduction and treated for 4 days with GSHee (10 mM). SLC25A12 and β-actin were used as loading controls. GSHee, GSH ethyl ester. f, Immunoblot of indicated proteins in HeLa cells treated with BSO (1 mM) and co-treated with either GSH (10 mM), GSHee (10 mM), trolox (50 μM) or ferrostatin-1 (5 μM) for 48 h. SLC25A12 and citrate synthase (CS) were used as loading controls. For gel source data, see Supplementary Fig. 1.
We first sought to determine the basis for SLC25A39 protein regulation by BSO. Of note, BSO treatment did not affect SLC25A39 mRNA levels, but induced SLC25A39 protein accumulation when an SLC25A39 cDNA lacking untranslated region (UTR) elements was expressed ectopically under a viral promoter (Extended Data Fig. 1e–g), suggesting regulation at the post-translational level. Increasing reactive oxygen species (ROS) or lipid peroxides by treating cells with hydrogen peroxide or a ferroptosis inducer (RSL-3), or activating NRF2 using small molecules (KI696), did not change SLC25A39 protein levels (Extended Data Fig. 1h–j). Similar results were observed with molecules (diamide or mitoCDNB14) that increase thiol oxidation (Extended Data Fig. 1k). These data indicate that SLC25A39 protein levels are regulated independently of NRF2 activation or oxidative stress. Consistent with these findings, supplementation of only GSH or GSH ethyl ester (GSHee), but not other antioxidants (trolox or ferrostatin-1) prevented induction of SLC25A39 protein levels on GSH depletion (Fig. 1e, f, Extended Data Fig. 1l). Collectively, these results suggest that SLC25A39 protein abundance is regulated by cellular GSH availability.
Mitochondrial GSH import requires SLC25A39
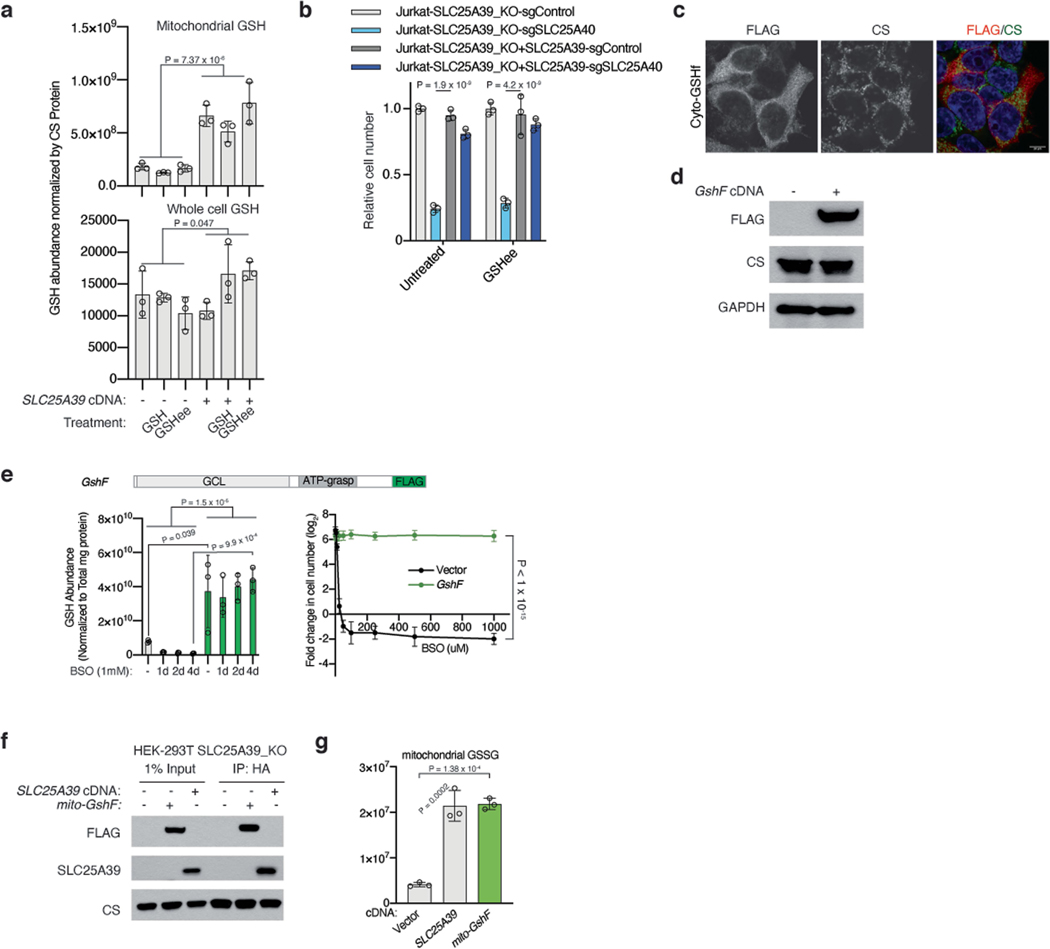
As SLC25A39 is predicted to be a member of the SLC25A family of mitochondrial small molecule transporters15 and its protein levels are regulated by GSH availability, we hypothesized that it may be involved in GSH metabolism through its presumed transport activity. To address this in an unbiased way, we profiled mitochondrial metabolites of three SLC25A39-knockout cell lines and cell lines complemented with SLC25A39 cDNA by liquid chromatography–mass spectrometry (LC–MS). Among all detected metabolites, we observed the largest reductions in the levels of GSH or GSH disulfide in mitochondria of SLC25A39-knockout cells (Fig. 2a, Extended Data Fig. 2a, b). Loss of SLC25A39 did not affect the levels of whole-cell GSH or most other mitochondrial metabolites, but caused a 5- to 10-fold reduction in mitochondrial GSH (Fig. 2b, Extended Data Fig. 2c). These results suggest that SLC25A39 is required specifically for the maintenance of mitochondrial GSH pools in mammalian cells.
Fig. 2 |. SLC25A39 is required for mitochondrial GSH import.
a, Volcano plot showing the log2 fold change in mitochondrial metabolite abundance versus − log P values from SLC25A39-knockout HeLa cells expressing a vector control or SLC25A39 cDNA. GSH and GSSG are shown in red; 3-PGA, 3-phosphoglyceric acid. The dotted line represents P = 0.05. Statistical significance was determined by multiple two-tailed unpaired t-tests with correction for multiple comparisons using the Holm–Šídák method. b, Relative metabolite abundance of indicated whole-cell or mitochondrial metabolites from SLC25A39-knockout K562, HEK 293T and HeLa cell lines compared with those expressing SLC25A39 cDNA. Mitochondrial NAD+ abundance is shown to indicate mitochondrial health. c, Uptake of [13C2,15N]-GSH into mitochondria isolated from SLC25A39-knockout HEK 293T cells expressing a vector control or SLC25A39 cDNA during the indicated times. Metabolite abundance is normalized to citrate synthase protein level. d, Top, structural and sequence comparison of SLC25A39 with the ADP/ATP carrier identifies M133, D226 and K329 as potential substrate contact points. Bottom, sequence alignment of SLC25A39 homologues for indicated residues. Dm, Drosophila melanogaster; Dr, Danio rerio; Hs, Homo sapiens; Mm, Mus musculus; Sc, Saccharomyces cerevisiae. e, Top, uptake of [13C2,15N]-GSH into mitochondria isolated from SLC25A39-knockout HEK 293T cells expressing a vector control or indicated SLC25A39 cDNAs at 5 s and 10 min. Metabolite abundance is normalized to citrate synthase protein level. Bottom, mitochondrial NAD+ abundance is shown to indicate the mitochondrial health. Data are mean ± s.d. (b, c, e); n = 3 biologically independent samples (a–c, e). Two-tailed unpaired t-test (a, b); two-way ANOVA followed by Bonferroni post hoc analysis (c, e). NS, not significant.
Given the strong decrease in the steady-state levels of mitochondrial GSH upon SLC25A39 loss, we reasoned that SLC25A39 may mediate GSH import into mitochondria. To test this possibility, we performed isotope-labelled GSH (GSH-(glycine-13C2,15N))-uptake assays using mitochondria isolated from SLC25A39-knockout cells and those expressing an SLC25A39 cDNA (Extended Data Fig. 2d, e). During a 10-min uptake assay, mitochondria containing SLC25A39 took up sixfold more GSH than those lacking the protein (Fig. 2c). Furthermore, we did not observe any isotope-labelled GSH disulfide (GSSG-(glycines-13C2,15N)) uptake or substantial change in GSH uptake in the presence of a reducing agent, suggesting that GSH may be the major form of GSH transported into mitochondria (Extended Data Fig. 2f, g). To determine whether SLC25A39 transport function is essential for GSH import into mitochondria, we generated a homology model of SLC25A39 based on the structure of the bovine ADP/ATP transporter16 and identified three potential substrate-binding residues: K329, M133 and D226 (Fig. 2d). Owing to their high conservation among SLC25A39 homologues across multiple species, we focused on D226 and K329. Mutating these specific residues to alanine, but not others, completely abolished the uptake of GSH without affecting the levels of NAD+ (Fig. 2e, Extended Data Fig. 2h). These data suggest that SLC25A39 transport function is necessary for mitochondrial GSH import.
Essential role of SLC25A39/40 in proliferation
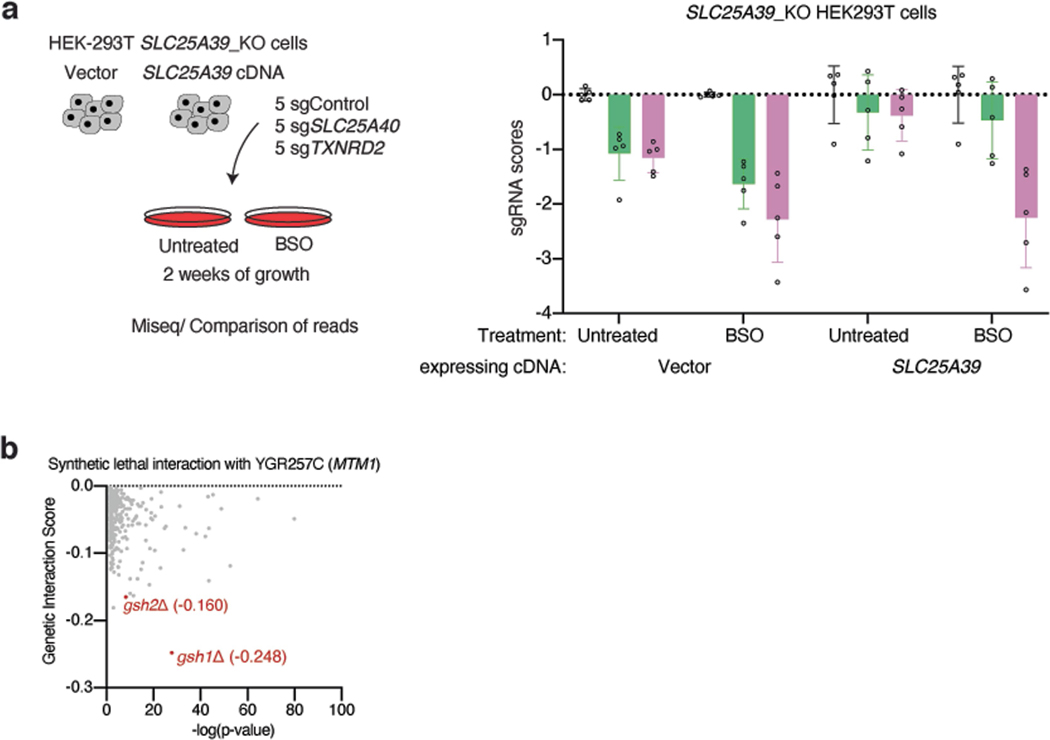
Because SLC25A39 loss did not affect cell proliferation and completely abolish mitochondrial GSH import (Fig. 2c), we hypothesized that other metabolic genes may overcome the decrease in mitochondrial GSH levels. To investigate this, we performed CRISPR–Cas9-based genetic screens in both Jurkat and HEK 293T cells using a metabolism-focused single guide RNA (sgRNA) library17 and screened for genes essential for cellular proliferation in the absence of SLC25A39 (Fig. 3a, Supplementary Table 2). A top-scoring gene from both screens was SLC25A40, the mitochondrial SLC25A family transporter with the highest sequence homology to SLC25A39 (Fig. 3b, Extended Data Fig. 3a–c). A co-evolution analysis for SLC25A39 across a large set of eukaryotes also retrieved SLC25A40 as the top-scoring gene (z score = 9.86), indicating a functional association between these two transporters (Extended Data Fig. 3d, e). Consistent with the screening results, cells lacking both SLC25A39 and SLC25A40 (SLC25A39/40 double-knockout cells) were unable to proliferate under standard culture conditions (Fig. 3c, Extended Data Fig. 3f, g). Furthermore, overexpressing SLC25A40 cDNA in SLC25A39-knockout cells was sufficient to restore mitochondrial GSH levels, suggesting a redundant function in enabling mitochondrial GSH import alongside SLC25A39 (Fig. 3d, Extended Data Fig. 3h, i). Notably, also scoring in HEK 293T cells was the thioredoxin reductase 2 gene (TXNRD2), a component of the thioredoxin (Trx)-dependent antioxidant system specifically localized to mitochondria (Extended Data Fig. 3a, b). This is in line with the presence of the two parallel mitochondrial thiol antioxidant systems based on either GSH or thioredoxin-218.
Fig. 3 |. Mitochondrial GSH import is essential for cell proliferation.
a, Schematic depicting the metabolism-focused CRISPR genetic screens in SLC25A39-knockout Jurkat cells expressing a vector control or SLC25A39 cDNA. b, CRISPR gene scores in SLC25A39-knockout Jurkat cells expressing a vector control or SLC25A39 cDNA. c, Per cent of fold change in cell number of the indicated SLC25A39-knockout Jurkat cells expressing a vector control or SLC25A39 cDNA transduced with the indicated sgRNAs. Cell doublings were normalized to the average for the SLC25A39-knockout cells. d, Steady-state mitochondrial abundance of GSH in HEK 293T SLC25A39-knockout cells expressing the indicated cDNAs. Data are normalized to citrate synthase protein levels. e, Schematic of GSH synthesis in mammals and S. thermophilus. f, Top, schematic of engineered GshF construct targeted to mitochondria (mito-GshF), with regions corresponding to GCL and GSS (ATP-grasp domain). Bottom, immunofluorescence analysis of mito-GshF (Flag, red) and citrate synthase (CS, green) in HeLa cells (bottom). Micrographs are representative of two independent experiments. Scale bar, 20 μm. g, Mitochondrial abundance of GSH in SLC25A39-knockout HEK 293T cells transduced with the indicated cDNAs. Data are normalized to citrate synthase protein level. h, Left, schematic showing the restoration of mitochondrial GSH levels by mito-GshF. Right, log2 fold change in cell number of SLC25A39-knockout Jurkat cells expressing a vector control or mito-GshF transduced with the indicated sgRNA. i, Log2 fold change in cell number of SLC25A39-knockout Jurkat (left) or HEK 293T (right) cells expressing a vector control or SLC25A39 cDNA transduced with the indicated sgRNA. Cells were treated for 4 days with the indicated BSO concentrations. Data are mean ± s.d., n = 3 biologically independent samples (c, d, g–i). One-way ANOVA followed by Bonferroni post hoc analysis (c, d, g, h); two-way ANOVA followed by Bonferroni post hoc analysis (i).
To formally test whether mitochondrial GSH depletion is necessary for cell proliferation, we would need to restore mitochondrial GSH levels upon loss of SLC25A39 and SLC25A40 (hereafter referred to collectively as SLC25A39/40). However, modulating GSH levels specifically in mitochondria through genetic means is challenging. First, GSH synthesis in mammalian cells requires three genes (GCLC, GCLM and GSS) which encode the GCL holoenzyme and GSH synthetase. Second, GSH is a non-allosteric feedback inhibitor of GCL, and impedes efficient GSH production19. Indeed, supplementation of cells with high levels of extracellular GSH did not influence mitochondrial GSH levels or restore proliferation of SLC25A39/40 double-knockout cells (Extended Data Fig. 4a, b).
Several bacterial species including Streptococcus thermophilus express a bifunctional enzyme GshF, which possesses both glutamate–cysteine ligase and GSH synthetase activities20 (Fig. 3e). GshF, unlike the eukaryotic GCLs, does not display feedback inhibition, allowing efficient GSH accumulation. Building on these features, we engineered a GshF construct that could be used to modulate compartmentalized GSH levels (Extended Data Fig. 4c–e). Confirming the activity of the enzyme in mammalian cells, ectopic expression of a codon-optimized GshF cDNA in HEK 293T cells increased cellular GSH levels and conferred resistance to BSO treatment (Extended Data Fig. 4e). Given the presence of GSH synthesis precursors in mitochondria21, we next generated a mitochondrially targeted GshF (mito-GshF) which faithfully localizes to mitochondria and tested whether mito-GshF could functionally complement mitochondrial GSH loss (Fig. 3f, Extended Data Fig. 4f). Expression of mito-GshF completely restored the decrease in mitochondrial GSH levels in SLC25A39-knockout cells (Fig. 3g, Extended Data Fig. 4g) and rescued the anti-proliferative effects of mitochondrial GSH depletion in SLC25A39/40 double-knockout cells (Fig. 3h). Together, these results demonstrate that the essential role of SLC25A39/40 for cell proliferation is to regulate mitochondrial GSH import.
Given the essential function of mitochondrial GSH import by SLC25A39/40 for proliferation, we predict that mitochondrial GSH depletion should further sensitize cells to a decrease in cellular GSH availability. Indeed, cells lacking SLC25A39/40 die on inhibition of GSH synthesis by BSO treatment, an effect rescued by SLC25A39 cDNA (Fig. 3i). Similarly, a small-scale sgRNA competition assay in SLC25A39-knockout and control HEK 293T cells infected with a pool of control and SLC25A40 sgRNAs confirmed these results (Extended Data Fig. 5a). Since SLC25A39/40 is conserved from yeast to higher eukaryotes22 (Extended Data Fig. 3c), we next tested whether this association is present in lower organisms as well. Using a yeast-interaction network (The Cell Map23), we identified gsh1 and gsh2 (yeast glutamylcysteine ligase and GSH synthetase, respectively) as two of the highest scoring synthetic lethal interactions of mtm1, the Saccharomyces cereviase orthologue of SLC25A39 (P = 2.78 × 10−28 (gsh1) and 7.76 × 10−11 (gsh2)) (Extended Data Fig. 5b). These results are consistent with a limiting role of cellular GSH for cell proliferation when mitochondrial GSH import is impaired.
Metabolic effects of mitochondrial GSH depletion
We next sought to determine the physiological consequences of mitochondrial GSH depletion in mice. In yeast, mtm1 deficiency results in superoxide dismutase defects24. In flies, loss of shawn, the SLC25A39 homologue, causes neurodegeneration and synaptic defects22. Additionally, SLC25A39 has been shown to co-express with haem synthesis genes25. To determine the organismal function of SLC25A39 in mammals, we generated a full-body Slc25a39-knockout mouse using CRISPR–Cas9 (Extended Data Fig. 6a). Deletion of Slc25a39 was embryonically lethal at embryonic day 13.5 (E13.5), and embryos appeared pale, suggesting a severely anaemic phenotype (Extended Data Fig. 6b). To more precisely examine the extent to which mitochondrial GSH depletion impacts erythropoiesis, we generated an additional mouse model in which mitochondrial GSH import is disrupted specifically in the erythroid lineage (conditional Slc25a39-KO mice, ErGFP-cre;Slc25a39fl/fl) (Extended Data Fig. 6c). Mirroring our observation from full-body knockouts, these mice also exhibited severe anaemia, with a complete lack of Ter119+ cells, iron overload and increased apoptosis in fetal liver cells (Fig. 4a, b, Extended Data Fig. 6d–f). While we observed a robust reduction in the number of erythroblasts at all differentiation stages, we did not detect a defect in other lineages (Extended Data Fig. 6g, h). Notably, SLC25A39 expression was upregulated during red blood cell differentiation in vivo, consistent with it being the major paralogue responsible for GSH import during erythropoiesis (Extended Data Fig. 6i)
Fig. 4 |. Mitochondrial GSH depletion impairs erythropoiesis and iron-sulfur cluster proteins.
a, Representative images of embryos of indicated genotypes at E12.5. Scale bar, 1 mm. b, Immunohistochemical staining of developing hearts (left) and livers (right) of E12.5 embryos for indicated genotypes with a Ter119 antibody (black). Nuclei were counterstained with fast red. Scale bar, 100 μm. c, Volcano plot showing log2 fold change in whole-cell protein abundance versus −log P values for SLC25A39/40 double-knockout Jurkat cells compared to those expressing SLC25A39 cDNA. Colours indicate protein groups whose abundance change significantly upon SLC25A39/40 loss. d, Immunoblot of indicated proteins in SLC25A39-knockout Jurkat cells expressing a vector control or SLC25A39 cDNA transduced with the indicated sgRNA. β-Actin was used as a loading control. e, Aconitase activity (Vmax) in whole-cell lysates of SLC25A39-knockout Jurkat cells expressing a vector control or mito-GshF transduced with the indicated sgRNAs (mean ± s.d., n = 3; mU, milli units). f, Ratios of α-ketoglutarate to succinate in indicated cell lines. Values are normalized to the average for SLC25A39-knockout cells expressing SLC25A39 cDNA. g, Immunoblot of indicated proteins in SLC25A39-knockout Jurkat cells expressing vector control or mito-GshF transduced with the indicated sgRNA. β-Actin was used as a loading control. a, b, n = 3 ErGFP-cre+/− Slc25a39fl/fl, n = 4 Slc25a39fl/fl, n = 4 ErGFP-cre+/− Slc25a39fl/− embryos. e, f, Data are mean ± s.d. c, e, f, n = 3 biologically independent samples. Two-tailed unpaired t-test (c); one-way ANOVA followed by Bonferroni post hoc analysis (e, f). For gel source data, see Supplementary Fig. 1.
Despite the essential role of SLC25A39/40 in cell proliferation and red blood cell development, how mitochondrial GSH import deficiency impairs these cellular functions is unknown. To address this mechanistically, we performed an unbiased proteomics analysis on SLC25A39/40 double-knockout Jurkat cells (Fig. 4c). Gene ontology analysis for downregulated proteins in SLC25A39/40 double-knockout cells revealed a strong enrichment for mitochondrial translation and iron–sulfur cluster-containing proteins (Fig. 4d, Extended Data Fig. 7a–c). Notably, many of these effects could be recapitulated in cysteine desulfurase26 (NFS1)-knockdown cells that have impaired iron–sulfur cluster synthesis, indicating that phenotypes observed in SLC25A39/40-knockout cells are likely to be a consequence of an iron–sulfur cluster deficiency (Extended Data Fig. 7d). Indeed, SLC25A39/40-knockout cells displayed reduced activity of the enzyme aconitase, in which iron–sulfur clusters function in both substrate binding and catalysis (Fig. 4e). Additionally, SLC25A39/40 double-knockout cells exhibit reduction in the protein levels of lipoic acid synthase (LIAS) (Fig. 4d), another iron–sulfur cluster-containing protein involved in the synthesis of lipoate27. As lipoate is essential for the catalytic function of the α-ketoglutarate dehydrogenase (OGDH) and pyruvate dehydrogenase (PDH) complexes, these cells also displayed increased α-ketoglutarate/succinate and pyruvate/citrate ratios (Fig. 4f, Extended Data Fig. 7e). Of note, simply increasing mitochondrial GSH levels by expressing mito-Gshf in SLC25A39/40 double-knockout cells completely restored the reduced levels of mitochondrial translation machinery proteins, iron–sulfur cluster containing proteins and aconitase activity (Fig. 4e, g, Extended Data Fig. 7f). Notably, addition of hemin or pyruvate, a source of NAD+, did not affect these growth phenotypes, suggesting that amelioration of haem defects or inhibition of respiration is not sufficient to overcome the anti-proliferative effects of mitochondrial GSH depletion (Extended Data Fig. 7g). Therefore, we conclude that mitochondrial GSH depletion impairs the activity and stability of proteins containing iron–sulfur clusters.
Discussion
In this study, we identified SLC25A39 and its paralogue SLC25A40 as mitochondrial carriers required for GSH import. Although a previous report suggested that the mitochondrial dicarboxylate and 2-oxoglutarate carriers may transport GSH28, these findings were subsequently not validated29. Similarly, expression of these transporters was unable to restore the decrease in GSH levels in our assays (Extended Data Fig. 8a). Our work strongly argues that the essential role of mitochondrial GSH in mammals is to mediate iron–sulfur cluster biogenesis as opposed to redox buffering. This function is probably conserved, as GSH depletion elicits a similar iron-starvation-like response in yeast30. The critical need for GSH is consistent with its role as a cofactor for mitochondrial glutaredoxins, which are conserved oxidoreductases involved in iron–sulfur cluster biogenesis31.
Given the essential role of SLC25A39 in mitochondrial GSH transport, our results provide a mechanistic explanation for how SLC25A39 loss causes mitochondrial defects, neurodegeneration and anaemia in yeast, flies and mice, respectively. Mutations that impair GSH synthesis are associated with rare forms of anaemia in humans32. Of note, a SLC25A39-associated human phenome analysis using 4,000 traits in the UK Biobank (n = 361,194 subjects) revealed reticulocyte count and haemoglobin concentration as the top significant phenotypes associated with genetically regulated SLC25A39 expression (Extended Data Fig. 8b). SLC25A39 is one of the few mitochondrial carriers that is feedback-regulated at the protein level by a downstream metabolite. Given that this regulation is independent of NRF2, our results suggest the presence of a potential signalling pathway regulated by GSH availability. Biochemical assays with purified SLC25A39 reconstituted in liposomes and structural studies are required to determine the biophysical details of mitochondrial GSH import. Identification of factors regulating SLC25A39 protein levels and mitochondrial GSH import will improve our understanding of how compartmentalized GSH levels may be sensed in mammalian cells.
Methods
Cell lines and reagents
Human cell lines HeLa, Jurkat, K562 and HEK 293T were purchased from the ATCC. Cell lines were verified to be free of mycoplasma contamination and the identities of all were authenticated by STR profiling. Jurkat cells were maintained in RPMI medium (Gibco) containing 2 mM glutamine, 10% fetal bovine serum (FBS), 1% penicillin and streptomycin. HeLa and HEK 293T cells were maintained in either DMEM medium (Gibco) containing 4.5 g l−1 glucose, 110 mg l−1 pyruvate, 4 mM glutamine, 10% FBS, 1% penicillin/streptomycin or in RPMI medium (Gibco) containing 2 mM glutamine, 10% FBS, 1% penicillin and streptomycin.
Antibodies against β-actin (GTX109639, 1:100,000), GAPDH (GTX627408, 1:10,000) and PPAT (GTX102725, 1:1,000) were obtained from GeneTex; lipoic acid antibody (437695, 1:1,000) was obtained from EMD Millipore; SLC25A39 (14963–1-AP, 1:1,000 for western blot, 1:200 for immunofluorescence), ALAS1 (16200–1-AP, 1:1,000), GCLC (12601–1-AP, 1:10,000), HMOX1 (10701–1-AP, 1:1,000), MRM1 (16392–1-AP, 1:10,000), MRPL15 (18339–1-AP, 1:2,000), MRPS7 (26828–1-AP, 1:5,000), FDX1 (12592–1-AP, 1:1,000), FECH (14466–1-AP, 1:1,000) and LIAS (11577–1-AP, 1:1,000) antibodies were from Proteintech; anti-Flag M2 (F1804, 1:1,000) and anti-Flag M2 (F3165, 1:1,000) were from Sigma; NFS1 antibody (sc-365308, 1:1,000) was from Santa Cruz Biotechnology; SLC25A12 (64169S, 1:1,000), calreticulin (12238P, 1:1,000), RCAS1 (12290S, 1:1,000), LAMP1 (9091P, 1:1,000), CS (14309S, 1:1,000), ACO2 (6571S, 1:1,000), IRP2 (37135S, 1:1,000), TFRC (13208S, 1:1,000), FTH1 (3998S, 1:500), cleaved PARP (9546S, 1:1,000), SLC7A11 (12691S, 1:1,000), RPS6 (2217S, 1:2,000) and ATF4 (11815S, 1:1,000) antibodies were from CST; and ATP5A antibody (43–9800, 1:200 for immunofluorescence) was from ThermoFisher Scientific. Anti-mouse IgG–HRP (7076S, 1:5,000) and anti-rabbit IgG–HRP (7074S, 1:5,000) were from Cell Signaling. Antibodies for immunofluorescence staining were donkey anti-rabbit Alexa Fluor 488 (A21206, 1:500) and goat anti-mouse Alexa Fluor 568 (A10037, 1:500) from ThermoFisher. RFP antibody (600–401-379, 1:1,000) was from Rockland. α-Tubulin antibody (AA4.3, 1:2,000) was from DSHB.
Other reagents: isotope-labelled GSH (CNLM-6245–50) and GSSG (CNLM-8782-PK) (Cambridge Isotope Laboratory) (CNLM-6245–50); L(−)-glutathione (reduced form) (0210181401, MP Biomedicals); anti-HA magnetic beads (88837, Thermo Scientific Pierce); DAPI (D1306, ThermoFisher Scientific); sodium pyruvate (P2256), polybrene (H9268), puromycin (P8833), GSH reduced ethyl ester (G1404), hemin (H9039) and BSO (B2515) (Sigma); blasticidin (ant-bl-1, Invivogen); and erastin (5449, Tocris Bioscience).
Generation of knockout, knockdown and cDNA-overexpression cell lines
sgRNAs (listed in ‘Oligonucleotide sequences’) were cloned into lentiCRISPR-v2 (Addgene) (for GCLC) or into lentiCRISPR-v1 (Addgene) (for SLC25A39) linearized with BsmBI by T4 ligase (NEB). sgRNA-expressing vector along with lentiviral packaging vectors Delta-VPR and CMV VSV-G were transfected into HEK 293T cells using the XTremeGene 9 transfection reagent (Roche). Similarly, for over-expression cell lines, gBlocks (IDT) containing the cDNA of interest were cloned into pMXS-IRES-BLAST or pMXS-IRES-GFP linearized with BamHI and NotI by Gibson assembly (NEB). cDNA vectors along with retroviral packaging vectors gag-pol and CMV VSV-G were transfected into HEK 293T cells. The virus-containing supernatant was collected 48 h after transfection and passed through a 0.45-μm filter to eliminate cells. Target cells in 6-well tissue culture plates were infected in medium containing 8 μg ml−1 of polybrene and a spin infection was performed by centrifugation at 2,200 rpm for 1 h. After infection, virus was removed and cells were selected with puromycin (lentiCRISPR-v2), blasticidin (pMXS_IRES_Blast) or by flow sorting of the top 1% GFP+ cells (lentiCRISPR-v1(GFP) or pMXS-IRES-GFP). Clones were validated for loss of the relevant protein via immunoblotting. As no validated antibody is available, sgSLC25A40-transduced cells were validated by ICE analysis (Synthego) of gDNA Sanger sequencing. Unless otherwise stated, double knockout of SLC25A39-KO cells with sgSLC25A40 was lethal and each experiment was done with freshly transduced cells 5 days after transduction for cell proliferation assays and 7 days after transduction for protein assays. For NFS1 knockdown, the following shRNAs were used: shNFS1_1, TRCN0000229753, B4: shNFS1_2, TRCN0000229755, B6 (Addgene).
Oligonucleotide sequences
Human SLC25A39, sg4 F: GATAGGCAGTGAAGTAGATGG, sg4 R: CCAT CTACTTCACTGCCTATC; human GCLC, sg4 F: GTGTAGATGATAGAACTCGGG,sg4R:CCCGAGTTCTATCATCTACAC;humanSLC25A40,sg1F: GGTTGGATTTCCCTTTGGAG, sg1 R: CTCCAAAGGGAAATCCAACC.
Codon-optimized SLC25A39 sequence: ATGGCAGACCAGGACCCCGCGGGCATCTCACCTCTCCAGCAGATGGTCGCATCTGGAACAGGGGCAGTCGTCACAAGTTTGTTCATGACCCCACTTGATGTAGTGAAAGTCCGGCTTCAATCACAACGCCCTAGCATGGCCAGCGAGCTGATGCCGAGCTCCAGGCTCTGGTCACTTTCTTATACGAAGCTTCCCTCTTCTCTCCAGTCTACGGGTAAATGTTTGCTTTATTGTAACGGCGTACTCGAACCTCTGTATTTGTGTCCAAATGGAGCACGCTGCGCCACGTGGTTTCAGGACCCAACTCGATTTACCGGCACAATGGACGCATTTGTCAAGATAGTAAGACACGAGGGTACAAGAACGCTTTGGAGCGGCCTCCCTGCTACGTTGGTGATGACGGTTCCCGCAACGGCCATATACTTTACAGCCTACGACCAGCTGAAGGCCTTTCTGTGTGGTAGGGCACTTACCTCAGACCTTTACGCTCCAATGGTCGCAGGGGCCCTTGCAAGACTTGGTACGGTCACTGTAATAAGTCCGCTCGAACTCATGAGGACAAAACTCCAAGCTCAGCACGTGAGCTACCGGGAACTGGGGGCTTGTGTACGCACAGCGGTCGCGCAAGGCGGCTGGAGGAGTCTGTGGCTGGGTTGGGGGCCCACGGCCCTCCGGGACGTACCGTTTTCTGCGCTTTATTGGTTTAACTACGAGCTTGTGAAATCTTGGCTCAATGGATTCCGGCCGAAAGACCAGACCTCCGTTGGAATGTCTTTCGTCGCCGGGGGCATTTCCGGCACGGTGGCCGCCGTGCTGACCTTGCCATTCGACGTTGTTAAGACCCAGCGACAGGTCGCTTTGGGGGCAATGGAGGCCGTGCGGGTGAACCCACTCCACGTTGACAGTACATGGTTGCTGCTCCGCCGCATCCGGGCCGAAAGCGGAACTAAAGGTCTGTTTGCTGGATTTCTTCCGCGAATCATTAAGGCTGCGCCATCTTGTGCAATCATGATCTCTACATACGAGTTTGGAAAATCCTTCTTTCAGAGGCTTAATCAGGACAGACTGCTCGG AGGGTAA.
SLC25A39(D226A): ATGGCAGACCAGGACCCCGCGGGCATCTCACCTCTCCAGCAGATGGTCGCATCTGGAACAGGGGCAGTCGTCACAAGTTTGTTCATGACCCCACTTGATGTAGTGAAAGTCCGGCTTCAATCACAACGCCCTAGCATGGCCAGCGAGCTGATGCCGAGCTCCAGGCTCTGGTCACTTTCTTATACGAAGCTTCCCTCTTCTCTCCAGTCTACGGGTAAATGTTTGCTTTATTGTAACGGCGTACTCGAACCTCTGTATTTGTGTCCAAATGGAGCACGCTGCGCCACGTGGTTTCAGGACCCAACTCGATTTACCGGCACAATGGACGCATTTGTCAAGATAGTAAGACACGAGGGTACAAGAACGCTTTGGAGCGGCCTCCCTGCTACGTTGGTGATGACGGTTCCCGCAACGGCCATATACTTTACAGCCTACGACCAGCTGAAGGCCTTTCTGTGTGGTAGGGCACTTACCTCAGACCTTTACGCTCCAATGGTCGCAGGGGCCCTTGCAAGACTTGGTACGGTCACTGTAATAAGTCCGCTCGAACTCATGAGGACAAAACTCCAAGCTCAGCACGTGAGCTACCGGGAACTGGGGGCTTGTGTACGCACAGCGGTCGCGCAAGGCGGCTGGAGGAGTCTGTGGCTGGGTTGGGGGCCCACGGCCCTCCGGGCAGTACCGTTTTCTGCGCTTTATTGGTTTAACTACGAGCTTGTGAAATCTTGGCTCAATGGATTCCGGCCGAAAGACCAGACCTCCGTTGGAATGTCTTTCGTCGCCGGGGGCATTTCCGGCACGGTGGCCGCCGTGCTGACCTTGCCATTCGACGTTGTTAAGACCCAGCGACAGGTCGCTTTGGGGGCAATGGAGGCCGTGCGGGTGAACCCACTCCACGTTGACAGTACATGGTTGCTGCTCCGCCGCATCCGGGCCGAAAGCGGAACTAAAGGTCTGTTTGCTGGATTTCTTCCGCGAATCATTAAGGCTGCGCCATCTTGTGCAATCATGATCTCTACATACGAGTTTGGAAAATCCTTCTTTCAGAGGCTTAATCAGGACAGACTGCTCGGAGGGTAA.
SLC25A39(K329A): gccaccATGGCAGACCAGGACCCCGCGGGCATCTCACCTCTCCAGCAGATGGTCGCATCTGGAACAGGGGCAGTCGTCACAAGTTTGTTCATGACCCCACTTGATGTAGTGAAAGTCCGGCTTCAATCACAACGCCCTAGCATGGCCAGCGAGCTGATGCCGAGCTCCAGGCTCTGGTCACTTTCTTATACGAAGCTTCCCTCTTCTCTCCAGTCTACGGGTAAATGTTTGCTTTATTGTAACGGCGTACTCGAACCTCTGTATTTGTGTCCAAATGGAGCACGCTGCGCCACGTGGTTTCAGGACCCAACTCGATTTACCGGCACAATGGACGCATTTGTCAAGATAGTAAGACACGAGGGTACAAGAACGCTTTGGAGCGGCCTCCCTGCTACGTTGGTGATGACGGTTCCCGCAACGGCCATATACTTTACAGCCTACGACCAGCTGAAGGCCTTTCTGTGTGGTAGGGCACTTACCTCAGACCTTTACGCTCCAATGGTCGCAGGGGCCCTTGCAAGACTTGGTACGGTCACTGTAATAAGTCCGCTCGAACTCATGAGGACAAAACTCCAAGCTCAGCACGTGAGCTACCGGGAACTGGGGGCTTGTGTACGCACAGCGGTCGCGCAAGGCGGCTGGAGGAGTCTGTGGCTGGGTTGGGGGCCCACGGCCCTCCGGGACGTACCGTTTTCTGCGCTTTATTGGTTTAACTACGAGCTTGTGAAATCTTGGCTCAATGGATTCCGGCCGAAAGACCAGACCTCCGTTGGAATGTCTTTCGTCGCCGGGGGCATTTCCGGCACGGTGGCCGCCGTGCTGACCTTGCCATTCGACGTTGTTAAGACCCAGCGACAGGTCGCTTTGGGGGCAATGGAGGCCGTGCGGGTGAACCCACTCCACGTTGACAGTACATGGTTGCTGCTCCGCCGCATCCGGGCCGAAAGCGGAACTAAAGGTCTGTTTGCTGGATTTCTTCCGCGAATCATTGCAGCTGCGCCATCTTGTGCAATCATGATCTCTACATACGAGTTTGGAAAATCCTTCTTTCAGAGGCTTAATCAGGACAGACTGCTCGGAGGGTAA.
SLC25A39(C202S): GCCGGATCTAGCTAGTTAATTAAGCCACCATGGCAGACCAGGACCCCGCGGGCATCTCACCTCTCCAGCAGATGGTCGCATCTGGAACAGGGGCAGTCGTCACAAGTTTGTTCATGACCCCACTTGATGTAGTGAAAGTCCGGCTTCAATCACAACGCCCTAGCATGGCCAGCGAGCTGATGCCGAGCTCCAGGCTCTGGTCACTTTCTTATACGAAGCTTCCCTCTTCTCTCCAGTCTACGGGTAAATGTTTGCTTTATTGTAACGGCGTACTCGAACCTCTGTATTTGTGTCCAAATGGAGCACGCTGCGCCACGTGGTTTCAGGACCCAACTCGATTTACCGGCACAATGGACGCATTTGTCAAGATAGTAAGACACGAGGGTACAAGAACGCTTTGGAGCGGCCTCCCTGCTACGTTGGTGATGACGGTTCCCGCAACGGCCATATACTTTACAGCCTACGACCAGCTGAAGGCCTTTCTGTGTGGTAGGGCACTTACCTCAGACCTTTACGCTCCAATGGTCGCAGGGGCCCTTGCAAGACTTGGTACGGTCACTGTAATAAGTCCGCTCGAACTCATGAGGACAAAACTCCAAGCTCAGCACGTGAGCTACCGGGAACTGGGGGCTTCTGTACGCACAGCGGTCGCGCAAGGCGGCTGGAGGAGTCTGTGGCTGGGTTGGGGGCCCACGGCCCTCCGGGACGTACCGTTTTCTGCGCTTTATTGGTTTAACTACGAGCTTGTGAAATCTTGGCTCAATGGATTCCGGCCGAAAGACCAGACCTCCGTTGGAATGTCTTTCGTCGCCGGGGGCATTTCCGGCACGGTGGCCGCCGTGCTGACCTTGCCATTCGACGTTGTTAAGACCCAGCGACAGGTCGCTTTGGGGGCAATGGAGGCCGTGCGGGTGAACCCACTCCACGTTGACAGTACATGGTTGCTGCTCCGCCGCATCCGGGCCGAAAGCGGAACTAAAGGTCTGTTTGCTGGATTTCTTCCGCGAATCATTAAGGCTGCGCCATCTTGTGCAATCATGATCTCTACATACGAGTTTGGAAAATCCTTCTTTCAGAGGCTTAATCAGG ACAGACTGCTCGGAGGGTAAGCTACGTAAATTCCGCCC.
SLC25A39(C334S): GCCGGATCTAGCTAGTTAATTAAGCCACCATGGCAGACCAGGACCCCGCGGGCATCTCACCTCTCCAGCAGATGGTCGCATCTGGAACAGGGGCAGTCGTCACAAGTTTGTTCATGACCCCACTTGATGTAGTGAAAGTCCGGCTTCAATCACAACGCCCTAGCATGGCCAGCGAGCTGATGCCGAGCTCCAGGCTCTGGTCACTTTCTTATACGAAGCTTCCCTCTTCTCTCCAGTCTACGGGTAAATGTTTGCTTTATTGTAACGGCGTACTCGAACCTCTGTATTTGTGTCCAAATGGAGCACGCTGCGCCACGTGGTTTCAGGACCCAACTCGATTTACCGGCACAATGGACGCATTTGTCAAGATAGTAAGACACGAGGGTACAAGAACGCTTTGGAGCGGCCTCCCTGCTACGTTGGTGATGACGGTTCCCGCAACGGCCATATACTTTACAGCCTACGACCAGCTGAAGGCCTTTCTGTGTGGTAGGGCACTTACCTCAGACCTTTACGCTCCAATGGTCGCAGGGGCCCTTGCAAGACTTGGTACGGTCACTGTAATAAGTCCGCTCGAACTCATGAGGACAAAACTCCAAGCTCAGCACGTGAGCTACCGGGAACTGGGGGCTTGTGTACGCACAGCGGTCGCGCAAGGCGGCTGGAGGAGTCTGTGGCTGGGTTGGGGGCCCACGGCCCTCCGGGACGTACCGTTTTCTGCGCTTTATTGGTTTAACTACGAGCTTGTGAAATCTTGGCTCAATGGATTCCGGCCGAAAGACCAGACCTCCGTTGGAATGTCTTTCGTCGCCGGGGGCATTTCCGGCACGGTGGCCGCCGTGCTGACCTTGCCATTCGACGTTGTTAAGACCCAGCGACAGGTCGCTTTGGGGGCAATGGAGGCCGTGCGGGTGAACCCACTCCACGTTGACAGTACATGGTTGCTGCTCCGCCGCATCCGGGCCGAAAGCGGAACTAAAGGTCTGTTTG CTGGATTTCTTCCGCGAATCATTAAGGCTGCGCCATCTTCTGCAATCAT GATCTCTACATACGAGTTTGGAAAATCCTTCTTTCAGAGGCTTAATCAGGACAGACTGCTCGGAGGGTAAGCTACGTAAATTCCGCCC
These gene fragments were purchased from Twist Biosciences, and cloned directly into linearized pMXS-IRES-BLAST (Cell Biolabs, RTV-016) or pMXS-IRES-GFP (Cell Biolabs,RTV-013).
GshF construct: below are human codon-optimized sequences of S. thermophilus GshF with added mitochondrial targeting sequences (MTS) and Flag tags. To ensure mitochondrial localization of the mito-GshF construct, a tandem MTS consisting of the Lactobacillus brevis COXIV MTS followed by the human ACOI MTS was added to the N-terminus of the GshF sequence, along with an N-terminal Flag-tag.
Codon-optimized GshF: GCCGGATCTAGCTAGTTAATTAAGccaccatgACCCTTAATCAGCTTCTCCAGAAGTTGGAGGCGACTTCCCCCATTCTCCAGGCGAACTTCGGGATAGAAAGGGAGTCATTGAGGGTTGACCGCCAGGGTCAGCTGGTCCACACACCGCACCCCTCATGTCTGGGAGCCCGCAGTTTTCATCCTTACATACAAACCGACTTTTGTGAATTCCAAATGGAACTGATTACACCAGTAGCCAAAAGTACGACGGAGGCCCGACGCTTTCTTGGCGCGATAACTGATGTAGCAGGACGAAGCATTGCAACTGACGAGGTGCTGTGGCCATTGAGTATGCCACCACGACTTAAAGCCGAGGAAATTCAAGTAGCGCAACTCGAGAACGACTTCGAAAGACATTATCGGAACTACTTGGCAGAGAAGTACGGCACCAAATTGCAGGCGATTAGTGGAATTCATTACAATATGGAACTTGGGAAGGACTTGGTTGAGGCGCTTTTTCAAGAGTCAGATCAGACTGACATGATCGCATTTAAAAACGCTCTGTATCTCAAGCTCGCCCAGAACTATTTGAGGTATCGGTGGGTCATTACTTACCTGTTTGGAGCAAGTCCCATTGCAGAACAAGGATTCTTTGACCAAGAAGTGCCGGAGCCTATGCGCTCTTTCCGCAACTCCGACCACGGCTACGTTAACAAGGAAGAGATACAGGTAAGCTTTGTATCCCTTGAAGACTATGTCTCCGCGATCGAGACCTACATCGAGCAGGGTGACCTTATAGCCGAGAAAGAGTTTTACTCAGCCGTGCGCTTTAGAGGACAAAAAGTCAATCGCTCCTTCCTTGATAAGGGTATAACTTATCTGGAGTTCAGAAACTTTGACTTGAACCCATTTGAGAGAATAGGCATCAGTCAAACCACTATGGATACCGTTCACTTGCTCATACTGGCCTTTCTCTGGTTGGATAGTCCGGAAAACGTGGACCAGGCCCTTGCCCAGGGACACGCGCTTAACGAGAAAATAGCCCTCTCCCATCCATTGGAGCCCCTCCCTTCAGAGGCCAAGACACAGGACATCGTGACCGCACTCGACCAGCTCGTACAGCACTTCGGATTGGGAGATTACCACCAGGATCTCGTGAAACAAGTGAAAGCGGCGTTTGCTGATCCGAATCAAACCCTGTCAGCTCAACTTCTTCCTTATATTAAGGACAAGTCACTCGCAGAATTCGCTCTCAATAAGGCACTCGCATATCATGACTATGACTGGACCGCTCACTACGCCCTTAAAGGTTACGAAGAAATGGAGCTCAGTACGCAGATGCTGCTCTTTGATGCTATCCAGAAAGGAATACACTTCGAGATACTCGATGAGCAAGATCAGTTCTTGAAGCTGTGGCATCAAGATCACGTAGAATATGTTAAAAACGGTAATATGACCAGCAAGGATAACTATGTAGTACCTCTCGCAATGGCCAACAAGACTGTTACTAAGAAAATTCTTGCTGACGCTGGGTTCCCTGTTCCGTCCGGGGACGAATTTACTAGCTTGGAGGAGGGACTGGCCTACTACCCGCTTATTAAAGATAAGCAAATTGTAGTAAAGCCAAAGAGCACGAATTTCGGCTTGGGTATCAGCATCTTCCAAGAACCCGCCAGTCTCGACAATTATCAAAAAGCATTGGAAATAGCATTTGCGGAGGACACTAGTGTGCTCGTCGAAGAATTCATTCCAGGCACGGAATACCGATTCTTCATTTTGGACGGACGCTGTGAGGCAGTCCTTTTGAGGGTAGCTGCCAATGTAATAGGGGACGGGAAACACACAATCAGAGAGTTGGTAGCGCAGAAAAACGCAAATCCCCTGCGCGGTAGGGATCATAGATCACCCTTGGAAATCATAGAGCTTGGGGACATAGAGCAACTCATGCTGGCACAGCAGGGTTACACTCCAGATGACATCCTGCCAGAGGGTAAGAAAGTGAATTTGAGGCGGAACAGCAATATTAGTACTGGGGGAGACTCCATAGACGTCACAGAAACAATGGATAGCTCTTATCAAGAACTTGCAGCAGCGATGGCTACCAGTATGGGGGCATGGGCCTGTGGAGTTGATCTGATTATACCCGACGAAACGCAGATTGCCACAAAGGAAAATCCACATTGCACGTGTATTGAACTTAACTTCAACCCCTCCATGTACATGCATACATACTGCGCTGAGGGGCCGGGGCAGGCAATTACAACCAAAATACTCGACAAACTCTTCCCGGAGATCGTGGCCGGACAAACTGGAGGAAGCGGAGGAAGCGGAGGAAGCGATTACAAGGATGACGATGACAAGTAAG CTACGTAAATTCCGCCC
Mito-Gshf: GCCGGATCTAGCTAGTTAATTAAGccaccATGCTCGCTACAAGGGTCTTTAGCCTCGTCGGAAAGAGAGCTATCAGCACCTCCGTCTGCGTGAGAGCTCATatggcgccctacagcctactggtgactcggctgcagaaagctctgggtgtgcggcagtaccatgtggcctcagtcctgtgcGATTACAAGGATGACGATGACAAGGGAGGAAGCGGAGGAAGCGGAGGAAGCACCCTTAATCAGCTTCTCCAGAAGTTGGAGGCGACTTCCCCCATTCTCCAGGCGAACTTCGGGATAGAAAGGGAGTCATTGAGGGTTGACCGCCAGGGTCAGCTGGTCCACACACCGCACCCCTCATGTCTGGGAGCCCGCAGTTTTCATCCTTACATACAAACCGACTTTTGTGAATTCCAAATGGAACTGATTACACCAGTAGCCAAAAGTACGACGGAGGCCCGACGCTTTCTTGGCGCGATAACTGATGTAGCAGGACGAAGCATTGCAACTGACGAGGTGCTGTGGCCATTGAGTATGCCACCACGACTTAAAGCCGAGGAAATTCAAGTAGCGCAACTCGAGAACGACTTCGAAAGACATTATCGGAACTACTTGGCAGAGAAGTACGGCACCAAATTGCAGGCGATTAGTGGAATTCATTACAATATGGAACTTGGGAAGGACTTGGTTGAGGCGCTTTTTCAAGAGTCAGATCAGACTGACATGATCGCATTTAAAAACGCTCTGTATCTCAAGCTCGCCCAGAACTATTTGAGGTATCGGTGGGTCATTACTTACCTGTTTGGAGCAAGTCCCATTGCAGAACAAGGATTCTTTGACCAAGAAGTGCCGGAGCCTATGCGCTCTTTCCGCAACTCCGACCACGGCTACGTTAACAAGGAAGAGATACAGGTAAGCTTTGTATCCCTTGAAGACTATGTCTCCGCGATCGAGACCTACATCGAGCAGGGTGACCTTATAGCCGAGAAAGAGTTTTACTCAGCCGTGCGCTTTAGAGGACAAAAAGTCAATCGCTCCTTCCTTGATAAGGGTATAACTTATCTGGAGTTCAGAAACTTTGACTTGAACCCATTTGAGAGAATAGGCATCAGTCAAACCACTATGGATACCGTTCACTTGCTCATACTGGCCTTTCTCTGGTTGGATAGTCCGGAAAACGTGGACCAGGCCCTTGCCCAGGGACACGCGCTTAACGAGAAAATAGCCCTCTCCCATCCATTGGAGCCCCTCCCTTCAGAGGCCAAGACACAGGACATCGTGACCGCACTCGACCAGCTCGTACAGCACTTCGGATTGGGAGATTACCACCAGGATCTCGTGAAACAAGTGAAAGCGGCGTTTGCTGATCCGAATCAAACCCTGTCAGCTCAACTTCTTCCTTATATTAAGGACAAGTCACTCGCAGAATTCGCTCTCAATAAGGCACTCGCATATCATGACTATGACTGGACCGCTCACTACGCCCTTAAAGGTTACGAAGAAATGGAGCTCAGTACGCAGATGCTGCTCTTTGATGCTATCCAGAAAGGAATACACTTCGAGATACTCGATGAGCAAGATCAGTTCTTGAAGCTGTGGCATCAAGATCACGTAGAATATGTTAAAAACGGTAATATGACCAGCAAGGATAACTATGTAGTACCTCTCGCAATGGCCAACAAGACTGTTACTAAGAAAATTCTTGCTGACGCTGGGTTCCCTGTTCCGTCCGGGGACGAATTTACTAGCTTGGAGGAGGGACTGGCCTACTACCCGCTTATTAAAGATAAGCAAATTGTAGTAAAGCCAAAGAGCACGAATTTCGGCTTGGGTATCAGCATCTTCCAAGAACCCGCCAGTCTCGACAATTATCAAAAAGCATTGGAAATAGCATTTGCGGAGGACACTAGTGTGCTCGTCGAAGAATTCATTCCAGGCACGGAATACCGATTCTTCATTTTGGACGGACGCTGTGAGGCAGTCCTTTTGAGGGTAGCTGCCAATGTAATAGGGGACGGGAAACACACAATCAGAGAGTTGGTAGCGCAGAAAAACGCAAATCCCCTGCGCGGTAGGGATCATAGATCACCCTTGGAAATCATAGAGCTTGGGGACATAGAGCAACTCATGCTGGCACAGCAGGGTTACACTCCAGATGACATCCTGCCAGAGGGTAAGAAAGTGAATTTGAGGCGGAACAGCAATATTAGTACTGGGGGAGACTCCATAGACGTCACAGAAACAATGGATAGCTCTTATCAAGAACTTGCAGCAGCGATGGCTACCAGTATGGGGGCATGGGCCTGTGGAGTTGATCTGATTATACCCGACGAAACGCAGATTGCCACAAAGGAAAATCCACATTGCACGTGTATTGAACTTAACTTCAACCCCTCCATGTACATGCATACATACTGCGCTGAGGGGCCGGGGCAGGCAATTACAACCAAAATACTCGACAAACTCTTCCCGGAGATCGTGGCCGGACAAACTTAAGCTACGTAA ATTCCGCCC
Human codon-optimized GshF was custom synthesized by IDT and directly assembled into pMXS-puro linearized with BamHI and NotI with a Gibson reaction.
To add the MTS, the GshF gene was divided into three fragments: MTS, middle and C-terminal, with 5′ and 3′ regions designed to overlap with the preceding or proceeding fragment or the pMXS vector. Each fragment was custom synthesized by Twist Bioscience. Fragments were amplified by PCR and gel extracted with a Zymo gel extraction kit before being assembled into pMXS-puro linearized with BamHI and NotI with a four-component Gibson reaction.
Primers: MTS_Fwd: 5′-GCCGGATCTAGCTAGTTAATTAAGccacc-3′; MTS_Rev: 5′-CCATTTGGAATTCACAAAAGTCGGTTTGT-3′; Mid_GSHf_Fwd: 5′-ACAAACCGACTTTTGTGAATTCCAAATGG-3′; Mid_GSHf_rev: 5′-GGAAGAAGTTGAGCTGACAGGGTTTGATT-3′; C-GSHf_Fwd: 5′-ATGAAACCCTGTCAGCTGAACTTC-3′; C-GSHf_Rev: 5′-GGGCGGAATTTACGTAGCTTAAGT-3′.
Other constructs in the study: pMXS_IRES_BLAST_SLC25A39; pMXS_IRES_BLAST_SLC25A39_D226A mutant; pMXS_IRES_BLAST_SLC25A39_K329A mutant; pMXS_IRES_GFP_SLC25A39_C202S mutant; pMXS_IRES_GFP_SLC25A39_C334S mutant; pMXS_IRES_BLAST_SLC25A40; pMXS_IRES_PURO_mito-Gshf; pMXS_IRES_PURO_cyto-Gshf; pMXS_IRES_GFP_cyto-Gshf; pMXS_IRES_PURO_control; pMXS_IRES_GFP_mito-Gshf; pMXS_IRES_BLAST_mito-Gshf; pMXS_IRES_BLAST_cyto-Gshf; lentiCRISPR-v1_sg4_SLC25A39 (GFP); lentiCRISPR-v2_sg1_SLC25A40 (PURO); lentiCRISPR-v2_sg4_GCLC (PURO); pMXS_IRES_BLAST_3xHA-OMP25-mCherry; pMXS_IRES_BLAST_3xMyc-OMP25-mCherry; pMXS_IRES_BLAST_3xFlag-SLC25A39-P2A-RFP; pMXS_IRES_BLAST_SLC25A11; pMXS_IRES_BLAST_SLC25A10.
Immunoblotting
Cell pellets were washed twice with ice-cold PBS prior to lysis in RIPA buffer (10 mM Tris-Cl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS) supplemented with protease inhibitors Sigma-Aldrich, 11836170001) and phosphatase inhibitors (Roche, 04906837001). Each lysate was sonicated and, after centrifugation for 5 min at 4 °C and 20,000g, supernatants were collected. Sample protein concentrations were determined by using Pierce BCA Protein Assay Kit (Thermo Scientific) with bovine serum albumin as a protein standard. Samples were resolved on 12% or 10–20% SDS–PAGE gels (Invitrogen) and analysed by standard immunoblotting protocol. In brief, membranes were incubated with primary antibodies at 4 °C overnight with shaking. Secondary antibodies, including anti-mouse IgG–HRP linked (Cell Signaling, 7076) and anti-rabbit IgG–HRP linked (Cell Signaling, 7074), were used at 1:5,000 dilution. Membranes were incubated with secondary antibodies at room temperature for 30 min. SRX-101A Film Processor (Konica Minolta) and Premium autoradiography Films (Thomas Scientific (1141J52) were used for developing.
Cell proliferation assays
For cell proliferation assays with cell counting, cells were plated in triplicates in 24-well plates at 10,000 cells per well. Cells were collected after 4 days and counted by Z2 Coulter Counter (Beckman). For cell proliferation assays with fold change in cell number, cells were cultured in triplicate in 96-well plates at 1,000 (suspension) or 150 (adherent) cells per well in 200 μl RPMI or DMEM base medium under the conditions described in each experiment, and a separate group of 3 wells was also plated for each cell line with no treatment for an initial time point. Adherent cells were allowed 1 day to adhere. Initially (untreated cells for initial time point) or after 4 days (with varying treatment conditions), 40 μl of Cell Titer Glo reagent (Promega) was added to each well, mixed briefly, and the luminescence read on a luminometer (Molecular Devices). For each well, the fold change in luminescence relative to the initial luminescence was measured and reported on a log2 scale as the number of cell doublings. Cell culture images were taken using a Zeiss Primovert equipped with Axio 503 mono camera and Zen 2 blue edition 2011 software.
Whole-cell and mitochondrial proteomics
After the indicated culture conditions, cell pellets were processed as per immunoblot protocol and lysed with lysis buffer (50 mM Tris-Cl pH 7.5, 150 mM NaCl, 1 mM EDTA, 2% Triton X-100) supplemented with protease inhibitors and phosphatase inhibitors (Roche).
Digestion.
Samples were dried and dissolved in 8 M urea, 50 mM triethylammonium bicarbonate (TEAB) and 10 mM dithiothreitol (DTT), and disulfide bonds were reduced for 1 h at room temperature. Alkylation was performed using iodoacetamide (IAA) for 1 h at room temperature in the dark. Proteins were precipitated by Wessel/Flügge extraction33 and pellets were dissolved in 100 mM TEAB with endopeptidase LysC (2% w/w, enzyme/substrate) and incubated at 37 °C for 2–3 h. Sequencing-grade modified trypsin (2 % w/w, enzyme/substrate) was added, and digestion proceeded overnight.
Labelling and fractionation.
Peptide solutions were labelled with 270 μg aliquots of TMTpro (Thermo Scientific) for 1 h at room temperature and subsequently quenched with hydroxylamine for 15 min. An aliquot from each sample was combined for a ratio check, according to which the samples were mixed. The pooled sample was purified using a high-capacity reverse phase cartridge (Oasis HLB, Waters) and the eluate was fractionated using high pH reverse phase spin columns (Pierce) according to manufacturer specifications, yielding eight fractions.
LC-MS/MS.
Fractionated peptides were analysed using an Easy-nLC 1200 HPLC equipped with a 250 mm × 75 μm Easyspray column connected to a Fusion Lumos mass spectrometer (all Thermo Scientific) operating in synchronous precursor selection (SPS)-MS3 mode34 (10 SPS events). Solvent A was 0.1% formic acid in water and solvent B was 80% acetonitrile, 0.1% formic acid in water. Peptides from the mitochondrial immunoprecipitation were separated across a 90-min linear gradient and peptides from the whole-cell lysate were separated across a 120-min linear gradient going from 7 to 33% solvent B at 300 nl min−1. Precursors were fragmented by CID (35% CE) and MS2 ions were measured in the ion trap. MS2 ions were fragmented by HCD (65% CE) and MS3 reporter ions were measured in the orbitrap at 50K resolution. Data were analysed using Skyline v.20.1.1.158.
Data analysis.
Raw files were searched through Proteome Discoverer v.2.3 (Thermo Scientific) and spectra were queried against the human proteome (database downloaded from uniprot.org on 2 December 2019, containing 73662 sequences) using Sequest HT with a 1 % false discovery rate (FDR) applied. Oxidation of M was applied as a variable modification and carbamylation of C was applied as a static modification. A maximum isolation interference of 50% was allowed and 80% matching SPS ions were required. Protein abundance values were used for further statistical analysis. A complete list of identified peptides is provided in Supplementary Table 3.
Subsequent statistical analysis was performed within the Perseus framework35. All values were log2-transformed and normalized to the median intensity within each sample. An FDR-corrected t-test (q = 0.05) was used to test for significant differences between sample groups.
For mitochondrial proteomics, mitochondria were rapidly immunopurified from HeLa cells expressing a mitotag (3×HA–OMP25–mCherry) grown in indicated cell culture conditions, according to the rapid mitochondrial purification for metabolite profiling protocol. At the final step after the third KPBS wash, the entire bead volume was lysed in 70 μl of lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 with protease inhibitors (Roche)). Approximately 50 μg of protein extracted from mitochondria was submitted for proteomic analysis. Peptides were analysed similarly to whole-cell proteomics and a complete list of identified peptides is provided in Supplementary Table 1. Owing to high isolation interference, the SLC25A39 peptide TMTpro-GLFAGFLPR (m/z 641.3848) underwent a targeted MS3 strategy. MS2 fragmentation was performed as described in the method section, but only the singly charged b7-ion (m/z 1010.5) was targeted for MS3 fragmentation. Owing to the low reporter ion intensities resulting from fragmentation of a low-abundant peptide and selecting only one fragment, the reporter ion intensities were summed across the chromatographic elution profile of the peptide. Missing values were imputed with random low-abundance numbers from a normal distribution and the result was incorporated into the volcano plot in Fig. 1c.
Polar metabolite profiling
After the indicated culture conditions, cells were washed with 1 ml cold 0.9% NaCl. Polar metabolites were extracted in 0.5 ml cold 80% methanol containing internal standards (Cambridge Isotope Laboratories). After extraction, samples were nitrogen-dried and stored at −80 °C until analysis by LC–MS. Analysis was conducted on a QExactive benchtop orbitrap mass spectrometer equipped with an Ion Max source and a HESI II probe, which was coupled to a Dionex UltiMate 3000 UPLC system (Thermo Fisher Scientific). External mass calibration was performed using the standard calibration mixture every 7 days.
Dried polar samples were resuspended in 100 μl water and 2 μl were injected into a ZIC-pHILIC 150 × 2.1 mm (5 μm particle size) column (EMD Millipore). Chromatographic separation was achieved using the following conditions: buffer A was 20 mM ammonium carbonate, 0.1% ammonium hydroxide; buffer B was acetonitrile. The column oven and autosampler tray were held at 25 °C and 4 °C, respectively. The chromatographic gradient was run at a flow rate of 0.150 ml min−1 as follows: 0–20 min linear gradient from 80% to 20% B; 20–20.5 min: linear gradient from 20% to 80% B; 20.5–28 min hold at 80% B. The mass spectrometer was operated in full-scan, polarity switching mode with the spray voltage set to 3.0 kV, the heated capillary held at 275 °C, and the HESI probe held at 350 °C. The sheath gas flow was set to 40 units, the auxiliary gas flow was set to 15 units, and the sweep gas flow was set to 1 unit. The MS data acquisition was performed in a range of 70–1000 m/z, with the resolution set at 70,000, the AGC target at 106, and the maximum injection time at 20 ms. Relative quantitation of polar metabolites was performed with XCalibur QuanBrowser 2.2 (Thermo Fisher Scientific) using a 5 ppm mass tolerance and referencing an in-house library of chemical standards. Metabolite levels were normalized to the total protein amount for each condition.
Rapid mitochondrial purification for metabolite profiling
Mitochondria were purified from HEK 293T, HeLa or K562 cells expressing 3×HA–OMP25–mCherry (mitochondrial isolation) or 3×Myc–OMP25–mCherry (background control) according to a previously described protocol. In brief, 25–30 million cells were collected and washed twice with cold saline (0.9% NaCl), scraped into 1 ml of cold KPBS, and pelleted via centrifugation at 1,000g for 1.5 min at 4 °C. Cells were resuspended in 1 ml of KPBS, 10 μl of cells were transferred into 40 ml of 1% Triton lysis buffer for a whole-cell protein sample and 10 μl of cells were transferred into 50 ml of 80% methanol, containing heavy labelled amino acid standards, for direct extraction of whole-cell metabolites. With one set of 20 strokes and another set of 10 strokes, the remaining sample was homogenized using a 2-ml homogenizer. After centrifugation, the homogenate was incubated with 200 ml of KPBS pre-washed anti-HA magnetic beads (Thermo Scientific Pierce 88837) on a rotator shaker for 5 min at 4 °C. Beads were washed three times in cold KPBS, then 10% of bead volume was lysed with 1% Triton buffer for protein extracts and the remaining 90% was extracted in 80% methanol containing heavy labelled amino acid standards (Cambridge Isotope Labs) on a rotator shaker for 10 min at 4 °C. Samples were spun down at 20,000g to remove potential cellular debris or bead contamination. Samples were subjected to LC-MS polar metabolite profiling without drying. Data were normalized to citrate synthase protein level (western blot) or NAD+ abundance (see details in figure legends).
GSH uptake assay in immunopurified mitochondria
Mitochondria were purified from HEK 293T cells expressing 3×HA–OMP25–mCherry (mitochondrial isolation) or 3×Myc–OMP25–mCherry (background control) according to a previously described protocol21. In brief, 30 million cells were collected and washed twice with cold saline (0.9% NaCl), scraped into 1 ml cold KPBS, and pelleted via centrifugation at 1,000g for 1.5 min at 4 °C. Cells were resuspended in 1 ml of KPBS and 10 μl of cells were transferred into 40 ml of 1% Triton lysis buffer for a whole-cell protein sample. With one set of 20 strokes and another set of 10 strokes, the remaining sample was homogenized using a 2-ml homogenizer. After centrifugation, the homogenate was incubated with 200 ml of KPBS pre-washed anti-HA magnetic beads on a rotator shaker for 5 min at 4 °C. Following incubation, beads were washed once with cold KPBS (pH 7.25) before being incubated in 200 μl transport buffer (KPBS, 10 mM HEPES, 0.5 mM EGTA) at room temperature containing indicated doses of [13C2,15N]-GSH (Cambridge Isotope Laboratory, CNLM-6245–50) or GSSG (Cambridge Isotope Laboratory, CNLM-8782-PK) for the indicated times. It is important to maintain the pH between 7.3 and 7.4 by adding 0–2 μl 2 M KOH per ml of transport buffer. Uptake was stopped with the addition of cold KPBS and beads were subsequently washed three more times in cold KPBS. Following the third wash, 10% of the bead volume was lysed with 1% Triton buffer for protein extracts and the remaining 90% was extracted in 80% methanol containing heavy labelled amino acid standards on a rotator shaker for 10 min at 4 °C. Samples were spun down at 20,000g and the uptake of [13C2,15N]-GSH into mitochondria is measured by LC–MS.
Aconitase assay
Assay was carried out with the Aconitase Activity Assay Kit (Abcam ab109712). Cell lines were generated as indicated and each condition was collected in triplicates of 1,000,000 cells each. Cell pellets were washed with ice-cold PBS prior to lysis in 1 ml aconitase preservation solution with detergent. Lysates were kept on ice for 30 min and centrifuged for 10 min at 4 °C and 20,000g. 50 μl of the supernatant was transferred to a well on the assay plate and 200 μl of assay buffer was added to each well. Plate was read on a plate reader (Molecular Devices) according to manufacturer’s protocol with a kinetic program of 45 s intervals.
CRISPR-based genetic screens
The metabolism-focused human sgRNA library was designed and screens performed as previously described36. Oligonucleotides for sgRNAs were synthesized by CustomArray and amplified by PCR. A complete list of differential gene scores for each screen is provided in Supplementary Table 2. In brief, plasmid pool was used to generate a lentiviral library, which was transfected into HEK 293T cells and used to generate viral supernatant as described above. SLC25A39-knockout cells expressing a vector control or SLC25A39 cDNA, in both Jurkat and HEK 293T, were infected at a multiplicity of infection of 0.7 and selected with puromycin. An initial sample of 30 million cells was harvested and infected cells were cultured for approximately 14 population doublings. Final samples of 30 million cells were collected. DNA was extracted (DNeasy Blood and Tissue kit, Qiagen). sgRNA inserts were PCR-amplified and barcoded by PCR, using primers unique to each condition. PCR amplicons were purified and sequenced (NextSeq 500, Illumina). Screens were analysed using Python (v.2.7.13), R (v.3.3.2) and Unix (v.4.10.0–37-generic x86_64). The gene score for each gene was defined as the median log2 fold change in the abundance of each sgRNA targeting that gene. All gene scores for each screen are provided in Supplementary Table 2.
Immunofluorescence
For SLC25A39 localization immunofluorescence assays, 100,000 HeLa cells were seeded onto coverslips. Twenty-four hours after plating, cells were treated with vehicle or BSO (1mM). After 24 h of treatment, cells were fixed for 10 min at −20 °C with ice-cold acetone. After three PBS washes, cells were blocked with 0.5% BSA in PBS, and incubated with the anti-SLC25A39 (Proteintech, 14693–1-AP, 1:200) and anti-ATP5A (Thermo Fisher Scientific, 439800, 1:200) primary antibodies in blocking solution for 1 h. Cells were washed 3 times with PBS and incubated with a donkey anti-mouse Alexa Fluor 568 (ThermoFisher Scientific, A10037) and donkey anti-rabbit Alexa Fluor 488 (ThermoFisher Scientific, A21206) secondary antibodies (1:400) for 1 h in the dark. Cells were washed 3 times with PBS, and incubated with a 200 nM solution of DAPI in the dark. Cells were washed three final times, and the coverslip was mounted onto slides with Prolong Gold antifade mounting medium (Invitrogen). Confocal images were acquired with a Zeiss inverted LSM 789 laser scanning confocal microscope (Zeiss) using a 100×/1.4 DIC Plan-Apochromat oil immersion objective. Four representative fields were captured for each condition under identical exposure times. Images were obtained with excitation and emission wavelengths as follows: DAPI 420–498 nm, Alexa Fluor 568 578–650 nm, Alexa Fluor 488 498–552 nm. The images are 512 × 512 pixels with a pixel depth of 12-bit, a pixel size of 0.08 μm per pixel, a dwell time of 6.30 μs, a pinhole size of 60 μm (1 Airy unit), and a line averaging of 1.
For mito- and cyto-GshF localization immunofluorescence assays, 100,000 HEK 293T cells were seeded onto coverslips coated with poly-D-lysine. After 24 h, the cells were fixed at room temperature for 10 min with 4% paraformaldehyde in PBS. After three PBS washes, cells were permeabilized with 0.1% Triton-X for 10 min, shaking. After three additional PBS washes, cells were blocked in 5% normal donkey serum (NDS) for 1 h at room temperature, shaking. The coverslips were subsequently incubated with anti-Flag-M2 (Sigma, F3165, 1:400) and anti-citrate synthase (Cell Signaling, 14309S, 1:200) for 16h at 4 °C before washing 3 times with PBS. Coverslips were then incubated with secondary (Alexa Flour 568 anti-mouse and Alexa Flour 488 anti-rabbit) 1:500 in 5% NDS for 1 h at room temperature in the dark and washed 3 times with PBS. Cells were incubated with a solution of 200 nM DAPI in the dark before and washed an additional 3 times with PBS. Finally, coverslips were mounted onto slides with Prolong Gold Antifade mounting medium (Invitrogen). Images were obtained with a Nikon A1R MP+ multiphoton confocal microscope (Nikon) using a 60×/1.4 DIC Plan-Apochromat oil immersion objective. Images were obtained with excitation wavelengths as follows: DAPI 425–475 nm, Alexa Fluor 488 500–550 nm, Alexa Fluor 568 570–620 nm. The images are 1,024 × 1,024 pixels with a pixel depth of 12-bit, a pixel size of 0.07 μm, a dwell time of 0.5 s, a pinhole size of 39.59 μm (1.2 Airy units), and a line averaging of 4.
Generation of Slc25a39-KO and conditional KO mice
All animal studies and procedures were conducted according to a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at the Rockefeller University. All mice were maintained on a standard light:dark cycle with food and water ad libitum. All treatment studies were randomized and injections were performed by blinded investigators.
CRISPR guide RNAs were designed using CHIRPOR.org37 and were used as two-part synthetic CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA) (Alt-RTM CRISPR guide RNA, Integrated DNA Technologies). Cas9 protein, crRNA and tracrRNA were assembled to ctRNP using protocols described previously38. All guide RNAs were validated in mouse embryos for their efficiency of generating indels on target genomic sequence, and two sgRNAs targeting intron 1 and intron 2 were selected for preparing the microinjection mix. sg1, AAT-GTGCTCACTTTGCCGAGGGG; sg2, TCTATGATCATGGGTAACCCAGG.
The Slc25a39-flox-E2-pUC57 plasmid was constructed to include a floxed Slc25a39 exon2 and 400 bp of homologous sequence on each side. To prepare the long single-stranded editing donor DNA (lssDNA), the 1.5 kb insert was amplified from Slc25a39-flox-E2-Puc57 and processed to long single-stranded DNA (lssDNA) using exonucleolytic hydrolysis of its 5′-phosphorylated DNA strand as described previously39.
The final injection mix was made of 0.6 μM of each guide RNA (crRNA + tracrRNA), 0.3 μM of Cas9 protein, and 10 ng μl−1 of ssDNA according to protocols described. The injection mix was then delivered to 0.5 days of fertilized C57BL/6J mouse embryos using well-established pronuclear injection and surgical protocols38. All animal experiments followed the approved IACUC protocols.
Histology and immunohistochemistry
Paraformaldehyde-fixed and paraffin-embedded tissues were deparaffinized and rehydrated, then incubated with anti-Ter119 antibody (Biolegend, 116201, 1:200) at 4 °C overnight and with secondary Goat anti-Rat IgG (VECTOR, PI-9401) for 1 h at room temperature. The reaction was developed with diaminobenzidine (DAB) staining (VECTOR, SK-4100). Prussian Blue Stain was performed using the Iron Stain Kit (ab150674) on paraffin-embedded whole embryo sessions and following the user manual. Slides were then counterstained in nuclear fast red. Stained slides were analysed in a blinded fashion. Images were taken using Revolve microscope (Echo), equipped with 10× and 100×Olympus UplanFL N objectives.
Cell profiling of fetal liver cells
E12.5 fetal liver cells were resuspended in 2% FBS and 10 mM glucose in PBS. For analysis of erythroid progenitor cells, fetal liver cells were stained with PE–anti mouse Ter119 (Biolegend, 116207, 1:200), PE–Cy7–anti CD44 (Biolegend, 103029, 1:200) and DAPI. For analysis of stem cells, fetal liver cells were stained with the following antibodies at 1:100 dilution: PE–Cy5–B220 (eBioscience 15–0452-83), PE–Cy5–CD3 (eBioscience 15–0031-83), PE–Cy5–Mac1 (eBioscience 15–00112-83), PE–Cy5–Gr1 (eBioscience 15–5931-82), PE–Cy5–Ter119 (eBioscience 15–5921-83), PE–Cy5–CD4 (eBioscience 15–0041-83), PE–Cy5–CD8 (eBioscience 15–0081-83), Pacific Blue-Sca1 (Biolegend 122520), APC–Cy7–Ckit (Biolegend 105826), FITC–CD34 (eBioscience 11–0341-85), PE–Cy7–cd16/cd32 (BD Pharmingen 560829); myeloid cells, PE–Cy5–Ter119 (eBioscience 15–5921-83), FITC–CD71 (eBioscience 11–0711-82), PE–CD41 (BD Pharmingen 558040), Pacific Blue–Mac1 (Biolegend 101224), APC–Cy7–GR1 (Biolegend 108424); lymphoid cells, Pacific Blue–CD3 (eBioscience 48–0031-82), APC–IgM (Biolegend 406509), PECy7–B220 (BD Pharmingen 552772). Flow cytometry was performed using the LSRII (Beckton Dickinson) and Attune NxT Flow Cytometer. Data were analysed using FlowJo 10.6.1 software (Beckton Dickinson) and FCS Express 7.
Gene ontology, co-evolution analysis and homology modelling
Genes with phylogenetic profiles similar to SLC25A40 was determined using Phylogene40. Phylogenetic tree for SLC25A39 is constructed using Treefam41. Gene ontology analysis for proteomics results has been performed with PANTHER and GORILLA42.
A homology model of human SLC25A39 was generated using the SWISS-MODEL server homology modelling pipeline with the bovine ATP–ADP carrier (PDB: 1OKC) as a scaffold43,44. Substrate-binding residues in human SLC25A39 were predicted based on biochemical and genetic analyses of the related ATP–ADP carrier using the ClustalW algorithm in MegAlign Pro 16 (DNASTAR)16,45. Structural depictions of the SLC25A39 homology model were prepared in Chimera 1.1446.
RNA extraction and real-time PCR
RNA was extracted from 293T using the Qiagen RNeasy mini kit according to the manufacturer’s instructions. One microgram of RNA was reverse transcribed using the Superscript III Reverse Transcriptase kit (Invitrogen) according to the manufacturer’s instructions. Quantitative real-time PCR (qPCR) was performed using SYBR green master mix and ACTB was used as a control. Data were collected by QuantStudio 6 Flex Applied Biosystems (Life Technologies) and QuantStudio Real-Time Software v1.1. The primer sequences are as below: ACTB F: CATGTACGTTGCTATCCAGGC, R: CTCCTTAATGTCACGCACGAT; SLC25A39 F: TGCC CTTCTCAGCCCTGTA, R: GGTTCACTCTCACAGCCTCC; GCLM F: TGTCTTG-GAATGCACTGTATCTC; R: CCCAGTAAGGCTGTAAATGCTC; SLC25A40 F: ATGGAGTGGCCTTCCTCCTA; R: CCCCTCCAAAGGGAAATCCA.
Cell-mixing competition assays
A small competition sgRNA library targeting SLC25A40 and TXNRD2 (5 sgRNAs each) in addition to 5 sgRNAs controls was designed from a larger focused sgRNA metabolism library. Oligos (IDT) were cloned into lentiCRISPR-v1 (SLC25A40 and TXNRD2) or lentiCRISPR-v2 (controls) using T4 ligase (NEB). Ligation products were cloned into Escherichia coli (NEB) and plasmids were isolated by Miniprep (Qiagen). This plasmid pool was used to generate a lentiviral library, which was transfected into HEK 293T cells and used to generate viral supernatant as described above. HEK 293T SLC25A39-knockout cells expressing a vector control or SLC25A39 cDNA were infected at a multiplicity of infection of 0.7 and selected with puromycin. An initial sample of 30 million cells washarvested and infected cells were cultured for approximately 14 population doublings in either standard cell culture conditions or treated with 20 μM BSO. Final samples of 30 million cells were collected. DNA was extracted (DNeasy Blood and Tissue kit, Qiagen). sgRNA inserts were PCR-amplified and barcoded by PCR, using primers unique to each condition. PCR amplicons were purified and sequenced (NextSeq 500, Illumina). Screens were analysed using Python (v.2.7.13), R (v.3.3.2) and Unix (v.4.10.0–37-generic x86_64). The gene score for each gene was defined as the median log2 fold change in the abundance of each sgRNA targeting that gene.
Oligo sequences for competition library: human SLC25A40 sg1 F: caccGGTTGGATTTCCCTTTGGAG, sg1 R: aaacCTCCAAAGGGAAATCCAACC, sg5 F: caccgAAGAGGGAGGCAACAAACTA, sg5 R: aaacTAGTTTGTTGCCTCCCTCTTc, sg6 F: caccgAAGAAGCCAGGAAATTTCCA, sg6 R: aaacTGGAAATTTCCTGGCTTCTTc, sg7 F: caccGGTACATCTCTAAGAACAGT, sg7 R: aaacACTGTTCTTAGAGATGTACC, sg8 F: caccgCTATGTGTCTGTGAAGAGGG, sg8 R: aaacCCCTCTTCACAGACACATAGc, human TXNRD2 sg2 F: caccgTAAACCACTGGAGTTCACGG, sg2 R: aaacCCGTGAACTCCAGTGGTTTAc, sg3 F: caccGATTAGGAGGGCGCTTCCGG, sg3 R: aaacCCGGAAGCGCCCTCCTAATC, sg6 F: caccgCAAGAAGCTGATGCAC-CAGG, sg6 R: aaacCCTGGTGCATCAGCTTCTTGc, sg7 F: caccGCACTACCTGGTCGAAGCCG, sg7 R: aaacCGGCTTCGACCAGGTAGTGC, sg8 F: caccGGAGGACAGCACCACCGGCA, sg8 R: aaacTGCCGGTGGTGCTGTCCTCC, control sg5 F: GCGGATTAGAGGTAATGCGG, sg5 R: CCGCATTACCTCTAATCCGC, sg6 F: GGAGCCATGGTAGAGCGTAT, sg6 R: ATACGCTCTACCATGGCTCC, sg11 F: GGCGCTCGATTAAGACTCAG, sg11 R: CTGAGTCTTAATCGAGCGCC, sg12F:GTTACGCGTGCTGTCGCTTC, sg12 R: GAAGCGACAGCACGCGTAAC, sg13 F: GTTCCTTCTGCAGGCACGCT, sg13 R: AGCGTGCCTGCAGAAGGAAC.
Analysis of TWAS data
We analysed transcriptome-wide association studies (TWAS) results on 4,000 traits for SLC25A39 in the UK Biobank47 (n = 361,194) using the summary-statistics-based, joint-tissue PrediXcan48,49. Application of PrediXcan to a large-scale biobank has been shown to enable discovery of gene-level effects on the medical phenome50. Here, for the gene expression models, we leveraged the GTEx V851 transcriptome panel. This approach regresses each phenotype on the principal components of the estimated genetically determined SLC25A39 expression data across the 49 tissues (Supplementary Table 4). We applied Bonferroni adjustment for multiple testing correction (adjusted P < 0.05). We report the most significant results (that is, for haematological traits) from the unbiased analysis across the entire phenomic dataset.
Statistics and reproducibility
GraphPad PRISM 7 and Microsoft Excel 15.21.1 software were used for statistical analysis. XCalibur QuanBrowser 2.2 (Thermo Fisher Scientific) and Skyline (v.20.1.1.158) were used for metabolomic analyses and ImageJ (NIH, Version 1.52 and 2.1.0) were used for image analysis. Error bars, P values and statistical tests are reported in the figure captions. All experiments (except full metabolite-profiling experiments, CRISPR screens and proteomics, which were done once) were performed at least twice with similar results. Both technical and biological replicates were reliably reproduced. Comparison of two mean values was evaluated by two-tailed unpaired t-test. Comparison of multiple mean values was evaluated by one-way ANOVA followed by post hoc Bonferroni test. Comparison of multiple mean values under different conditions was evaluated by two-way ANOVA.
Extended Data
Extended Data Fig. 1 |. SLC25A39 protein levels are directly regulated by cellular GSH availability.
a, Gene enrichment analysis (Gorilla) for proteomics data from the immunopurified mitochondria of HeLa cells (top). Gene ontology analysis of proteins altered by BSO treatment (bottom). Log (FDR), log false discovery rate (bottom). Statistical significance was determined by Fisher’s exact or binomial distribution test. b, Immunoblot of SLC25A39, SLC25A12 and CS in HeLa cells treated with the indicated doses of BSO for 24 h. CS was used as the loading control. c, Immunoblot of indicated proteins in HEK-293T (top) and K562 (bottom) cells treated with BSO (1 mM;10 μM, respectively) for the indicated days. CS and GAPDH were used as loading controls. d, Immunofluorescence analysis of indicated proteins in HeLa cells treated with vehicle or BSO (1 mM) for 24 h. Micrographs are representative images. Scale bar, 10 μm (top). Immunofluorescence analysis of SLC25A39 in parental and SLC25A39 knockout HeLa cells treated with BSO (1 mM) and erastin (5 uM) for 24 h (bottom). Micrographs are representative images of three independent experiments. Scale bar, 20 μm. e, Relative abundance of SLC25A39, GCLM and SLC25A40 transcripts in HEK-293T cells treated with BSO (1 mM) for the indicated days using quantitative reverse transcription PCR (RT-qPCR), normalized to transcripts of β-ACTIN. Error bars represent mean ± s.d.; n = 3 biologically independent samples. Statistical significance was determined by one-way ANOVA followed by Bonferroni post-hoc analysis. f, Immunoblot of indicated proteins in HEK-293T SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA treated with BSO (1 mM) for the indicated times. GAPDH was used as the loading control. g, The design of 3xFLAG- SLC25A39-P2A-RFP construct (top). Immunoblot of indicated proteins in HEK-293T cells expressing 3xFLAG- SLC25A39-P2A-RFP treated with BSO (1 mM) and erastin (5 μM) for 48 h (bottom). α-Tubulin was used as the loading control. h, Immunoblot of indicated proteins in HeLa cells treated with indicated doses of H2O2 for 24 h. CS and GAPDH were used as loading controls. i, Immunoblot of indicated proteins in HeLa cells treated with erastin (5 μM) or indicated doses of KI696 (NRF2 activator) for 24 h. GAPDH were used as loading controls. j, Immunoblot of indicated proteins in HeLa cells treated with erastin (5 μM) or indicated doses of RSL-3, an inhibitor of glutathione peroxidase 4 (GPX4), for 24 h. CS was used as a loading control. k, Immunoblot of SLC25A39 and ATF4 proteins in HeLa cells treated with indicated doses of mitoCDNB, diamide and BSO (1 mM)/erastin (5 μM) for 24 h. GAPDH was used as loading control. l, Immunoblot of indicated proteins in HeLa cells treated with erastin (5 μM) and co-treated with either GSH (10 mM), GSHee (10 mM), Trolox (50 μM) or Ferrostatin-1 (5 μM) for 48 h. CS was used as a loading control. GSHee, glutathione ethyl ester.
Extended Data Fig. 2 |. Mitochondrial GSH import is mediated by SLC25A39.
a, Volcano plots showing the fold change in mitochondrial metabolite abundance (log2) versus P values (-log) from K562 and HEK-293T SLC25A39 knockout cell lines expressing a vector control or SLC25A39 cDNA. Red data points highlight GSH and GSSG. The dotted line represents P = 0.05. Statistical significance was determined by multiple two-tailed unpaired t-tests with correction for multiple comparisons using the Holm-Šídák method. b, Immunoblot of indicated proteins in whole cell lysates and mitochondria isolated from HEK-293T (top) and HeLa (bottom) SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA. c, Mitochondrial GSH and GSSG abundance (top and middle panels) and total intracellular GSH levels in HEK-293T parental or SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA. Data are normalized by citrate synthase (CS) protein levels. d, Uptake of indicated concentrations of [13C2,15N]-GSH into mitochondria isolated from parental HEK-293T mitotag cells for 10 min. Data are normalized to the NAD+ abundance. e, Uptake of indicated concentrations of [13C2,15N]-GSH or GSH into mitochondria isolated from parental HEK-293T mitotag cells for 10 min. Data are normalized to the NAD+ abundance. f, Uptake of [13C2,15N]-GSH (5 mM, top) or [13C4,15N2]-GSSG (0.5 mM, bottom) into mitochondria isolated from HEK-293T SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA for 10 min. Data are normalized by citrate synthase (CS) protein levels. g, Uptake of [13C2,15N]-GSH (5 mM) into mitochondria isolated from HEK-293T SLC25A39 knockout cells expressing SLC25A39 cDNA for 10 min in the presence or absence of DTT (100 mM). Data are normalized by citrate synthase (CS) protein levels. h, Mitochondrial GSH abundance in HeLa SLC25A39 knockout cells expressing indicated cDNAs (top). Data are normalized by citrate synthase (CS) protein levels. Immunoblot of indicated proteins in HeLa SLC25A39 knockout cells expressing indicated cDNAs (bottom). α-Tubulin was used as a loading control. c, d, e, f, g, h, Bars represent mean ± s.d.; a, c, d, e, f, g, h, n = 3 biologically independent samples. Statistical significance in c, d, f, g and h was determined by one-way ANOVA followed by Bonferroni post-hoc analysis; a, e by two-tailed unpaired t-test.
Extended Data Fig. 3 |. SLC25A40, the mammalian paralog for SLC25A39 can compensate SLC25A39 loss.
a, Individual sgRNA scores of SLC25A40 and TXNRD2 from the CRISPR screens in indicated cell lines from Fig. 3b. b, CRISPR gene scores in indicated SLC25A39 knockout HEK-293T cells expressing a vector control or SLC25A39 cDNA. SLC25A40 data point is highlighted in blue (top). Top 15 scoring genes differentially required for the proliferation of HEK-293T SLC25A39 knockout cells (bottom). c, Phylogenetic tree of SLC25A39 homologs across model organisms. d, Top ranked (z-scores) co-evolved genes with SLC25A40 across species. e, Relative conservation of SLC25A40 and SLC25A39 in different species compared to their human homologs. f, Representative bright-field micrographs of Jurkat SLC25A39 knockout cells transduced with the indicated sgRNA at the end of the cell proliferation assay. Scale bar, 50 μm (right). g, Relative fold change in cell number of the indicated HEK-293T SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA transduced with the indicated sgRNAs. Cells were cultured for 4 days (mean ± SD, n = 3). Cell doublings were normalized to the average of the SLC25A39 knockout cells expressing SLC25A39 cDNA. Statistical significance was determined by one-way ANOVA followed by Bonferroni post-hoc analysis. h, Volcano plots of the fold change in mitochondrial metabolite abundance (log2) versus P values (-log) from HEK-293T SLC25A39 knockout cell line expressing a vector control or SLC25A40 cDNA. Red data points highlight GSH and GSSG. The dotted line represents P = 0.05. i, Immunoblot of indicated proteins in HEK-293T cells expressing an SLC25A40 cDNA in the presence or absence of BSO. CS and β-actin were used as loading controls.
Extended Data Fig. 4 |. Expression of S.thermophilus GshF can modulate cellular GSH levels in mammalian cells.
a, Mitochondrial abundance of GSH in HEK-293T SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA treated with GSH (5 mM) or GSHee (5 mM) for 24 h. Data are normalized by CS protein levels. b, Relative cell number of indicated SLC25A39 knockout Jurkat cells transduced with indicated sgRNAs. Cells were cultured for 4 days with or without GSHee (10 mM). Cell numbers were normalized to the average of the SLC25A39 knockout cells in each treatment condition. c, Immunofluorescence analysis of GshF (FLAG, red) and CS (green) in HeLa cells. Micrographs are representative images. Scale bar, 20 μm. d, Immunoblots of indicated proteins in HEK-293T cells expressing a vector control or GshF cDNA. GAPDH was used as the loading control. e, Schematic of engineered GshF construct for mammalian expression and the domains of the protein with GCL and GS function (top). Whole cell GSH abundance (bottom left) and fold change in cell number (log2) of HEK-293T cells expressing a vector control or engineered GshF cDNA treated with indicated BSO concentrations for 5 days (bottom right). f, Immunoblot analysis of indicated proteins in whole-cell lysates and mitochondria isolated from SLC25A39 knockout HEK-293T cells expressing a vector control, mito-GshF or SLC25A39 cDNA. g, Mitochondrial abundance of GSSG in HEK-293T SLC25A39 knockout cells transduced with the indicated cDNAs. Data are normalized by CS protein levels. a, b, e, g Bars represent mean ± s.d.; a, b, e, g n = 3 biologically independent samples. Statistical significance in a, g were determined by one-way ANOVA followed by Bonferroni post-hoc analysis; b, e were determined by two-way ANOVA followed by Bonferroni post-hoc analysis.
Extended Data Fig. 5 |. Mitochondrial GSH depletion sensitizes cells to BSO treatment.
a, Scheme of the in vitro sgRNA competition assay performed in HEK-293T SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA transduced with a pool of 5 control sgRNAs (sgControl, gray) and sgRNAs targeting TXNRD2 (pink) and SLC25A40 (green) (left). Differential guide scores in the indicated cell lines upon treatment with 20 μM BSO (right). Bars represent mean ± s.d. b, Synthetic lethal genetic interactions of mtm1 with other genes in S. cerevisiae.
Extended Data Fig. 6 |. Slc25a39 is essential for embryonic development and red cell differentiation in vivo.
a, Targeting scheme for Slc25a39 knockout mice. b, Gross appearance of E12.5 embryos of the indicated genotypes. Genotyping of Slc25a39 E12.5 embryos from heterozygous mating. PCR of wild type allele and targeted allele result in ~1,500bp and ~800bp bands, respectively (left). The number of viable pups with indicated genotypes is shown (right). c, Targeting scheme for Slc25a39 conditional knockout (Slc25a39fl/fl) mice in which two loxp sites were inserted in the indicated intronic regions. The resultant Slc25a39fl/fl mice were mated with erythroid-lineage specific Cre-recombinase (ErGFP-cre) mice to generate an erythroid-specific conditional knockout mice. d, The number of viable pups with indicated genotypes is shown resulting from the indicated mating. e, Representative images of H&E staining of fetal liver cells from E12.5 Slc25a39fl/fl and ErGFP-cre+/− Slc25a39fl/fl embryos. The bottom images are the boxed area of the top images. Arrows show that many haematopoietic cells in the fetal liver display nuclear fragmentation and cellular shrinkage. Bars, 50 μm (top panel), 12.5 μm (bottom panel). f, Percent of Prussian blue positive peripheral blood cells from indicated E12.5 embryos (top). Representative images of Prussian blue staining of indicated E12.5 whole embryo sections (bottom). Blue and orange arrows indicate blood cells in developing heart with negative or positive staining, respectively. Bar, 360 μm. g, Gating strategy and quantification of erythroblast populations at different differentiation stages using surface markers: Ter119 and CD44, and FSC (size). Percentage of cells of indicated populations among total fetal liver cells (left). Gating strategy (right). n = 3, ErGFP-cre+/− Slc25a39fl/fl; n = 4 Slc25a39fl/fl embryos. h, Profiling of cells of myeloid lineage (top), myeloid progenitor cells (middle) and lymphoid lineage (bottom). Data are presented as the population among total fetal liver cells. E12 embryos: n = 12 Slc25a39fl/fl or Slc25a39fl/-, n = 7 ErGFP-cre+/− Slc25a39fl/-, n = 4 ErGFP-cre+/− Slc25a39fl/fl. i, Relative RPKM values of SLC25A39, FTL and HBB genes in human erythroblasts at differing stages of terminal differentiation. f, g and h, Bars represent mean ± s.d.; statistical significance in f and h was determined by one-way ANOVA followed by Bonferroni post-hoc analysis; g was determined by two-tailed unpaired t-test.
Extended Data Fig. 7 |. Loss of SLC25A39/40 decreases the steady state levels of iron-sulfur containing proteins and phenocopies iron-sulfur cluster deficiency.
a, Comparison of proteomics data from BSO treated HeLa cells and SLC25A39/40 double knockout Jurkat cells. The criteria are indicated on the Venn diagram. b, Gene ontology analysis of significantly downregulated genes in SLC25A39/40 double knockout Jurkat cells. FDR, false discovery rate. c, Immunoblot of indicated proteins in the indicated HEK-293T SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA transduced with the indicated sgRNA. β-ACTIN was used as a loading control. d, Immunoblot of indicated proteins in indicated HEK-293T cells transduced with shRNA targeting either GFP as a control or NFS1 (two different shRNAs). Proteins were extracted from cells 6 days after transduction. GAPDH was used as a loading control. e, Schematic showing how the iron-sulfur cluster-containing protein LIAS enables OGDH and PDH activity by transferring lipoic acid as a cofactor to their enzyme complexes (left). Heatmap showing fold change in metabolite levels (log2) of the indicated cell lines relative to the average of those in SLC25A39 knockout cells complemented with SLC25A39 cDNA (middle). Metabolites were extracted 5 days after transduction with indicated sgRNAs. Values were normalized to the protein concentration of each cell line (left). Relative ratios of pyruvate to citrate metabolite levels of each indicated cell line from the metabolomics analysis in Fig. 4e (middle). Ratios were normalized to the average of the SLC25A39 knockout cells expressing SLC25A39 cDNA. Immunoblot with antibody against lipoic acid in the indicated Jurkat SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA transduced with the indicated sgRNA. Arrows indicate E2 complexes of PDH and OGDH containing lipoic acid (right). f, Representative bright-field micrographs of Jurkat SLC25A39 knockout cells transduced with the indicated sgRNA at the end of the cell proliferation assay. Scale bar, 50 μm. g, Relative cell number of the indicated Jurkat SLC25A39 knockout cells expressing a vector control or SLC25A39 cDNA transduced with the indicated sgRNA. Cells were cultured for 4 days with or without hemin (1 μM) and pyruvate (1 mM). Cell numbers were normalized to the average of the SLC25A39 knockout cells in each treatment condition. e, g, Bars represent mean ± s.d.; e, g, n = 3 biologically independent samples. Statistical significance in e was determined by one-way ANOVA followed by Bonferroni post-hoc analysis; g was determined by two-way ANOVA followed by Bonferroni post-hoc analysis.
Extended Data Fig. 8 |. Expression of DIC/OGC does not complement the decrease of mitochondrial GSH availability in SLC25A39 knockout cells.
a, Uptake of [13C2,15N]-GSH into mitochondria isolated from HEK-293T SLC25A39 knockout cells expressing a vector control, SLC25A39 cDNA, SLC25A10 (DIC, mitochondrial dicarboxylate carrier) or SLC25A11 (OGC, mitochondrial oxoglutarate carrier) cDNA at 5 s and 10 min. Bars represent mean ± s.d.; n = 3 biologically independent samples. Statistical significance was determined by two-way ANOVA followed by Bonferroni post-hoc analysis. b, PrediXcan-based TWAS design in UK biobank and summary graph of SLC25A39-associated phenotypes. Bonferroni-adjusted p-values are reported.
Supplementary Material
Acknowledgements
We thank all members of the Birsoy laboratory for helpful suggestions. Y.W. is supported by UL1 TR001866 from the National Center for Advancing Translational Sciences (NCATS, National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) programme. F.S.Y. and R.W. are supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the NIH under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program. G.U. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2431-21). E.R.G. was supported by NIH/NHGRI R01HG011138 and R35HG010718. H.L. is funded by NIH K99 DK128602-01. M.G.K. is a Scholar of the Leukemia and Lymphoma Society and is supported by the US NIH NIDDK R01-DK101989-01A1; NCI 1R01CA193842-01, 1R01CA193842-06A1, 5R01CA186702-07, 1R01DK1010989-06A1, R01HL135564 and R01CA225231-01; NYSTEM 0266-A121-4609, the Alex’s Lemonade Stand A Award. R.H. is supported by NIH/NCI Cancer Center Support Grant (P30 CA008748) and the Searle Scholars Program. We thank R. Vaughan, Director of Biostatistics at the Rockefeller University for his assistance with statistics used in this manuscript. K.B. is supported by the NIH/NCI (DP2 OD024174-01), NIH/NIDDK (R01 DK123323-01), Pershing Square Sohn Foundation and Mark Foundation Emerging Leader Award; and is a Searle and Pew-Stewart Scholar.
Footnotes
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-021-04025-w.
Competing interests K.B. is scientific advisor to Nanocare Pharmaceuticals and Barer Institute.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-021-04025-w.
Data availability
Proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD027673. Other data generated are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
- 1.Meister A. & Anderson ME Glutathione. Annu. Rev. Biochem 52, 711–760 (1983). [DOI] [PubMed] [Google Scholar]
- 2.Mårtensson J, Lai JC & Meister A. High-affinity transport of glutathione is part of a multicomponent system essential for mitochondrial function. Proc. Natl Acad. Sci. USA 87, 7185–7189 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deponte M. The incomplete glutathione puzzle: just guessing at numbers and figures? Antioxid. Redox Signal 27, 1130–1161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffith OW & Meister A. Origin and turnover of mitochondrial glutathione. Proc. Natl Acad. Sci. USA 82, 4668–4672 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang C, Sinskey AJ & Lodish HF Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257, 1496–1502 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Meredith MJ & Reed DJ Status of the mitochondrial pool of glutathione in the isolated hepatocyte. J. Biol. Chem 257, 3747–3753 (1982). [PubMed] [Google Scholar]
- 7.Deneke SM & Fanburg BL Regulation of cellular glutathione. Am. J. Physiol. Lung Cell. Mol. Physiol 257, L163–L173 (1989). [DOI] [PubMed] [Google Scholar]
- 8.Jones DP Redox potential of GSH/GSSG couple: assay and biological significance. Methods Enzymol. 348, 93–112 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Kurosawa K, Hayashi N, Sato N, Kamada T. & Tagawa K. Transport of glutathione across the mitochondrial membranes. Biochem. Biophys. Res. Commun 167, 367–372 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Griffith OW & Meister A. Potent and specific inhibition of glutathione synthesis by buthionine sulfoximine (S-n-butyl homocysteine sulfoximine). J. Biol. Chem 254, 7558–7560 (1979). [PubMed] [Google Scholar]
- 11.Münch C. & Harper JW Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature 534, 710–713 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee H-R et al. Adaptive response to GSH depletion and resistance to l-buthionine-(S,R)-sulfoximine: involvement of Nrf2 activation. Mol. Cell. Biochem 318, 23–31 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Sun X. et al. Activation of the p62–Keap1–NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 63, 173–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Booty LM et al. Selective disruption of mitochondrial thiol redox state in cells and in vivo. Cell Chem. Biol 26, 449–461.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruprecht JJ & Kunji ERS The SLC25 mitochondrial carrier family: structure and mechanism. Trends Biochem. Sci 45, 244–258 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pebay-Peyroula E. et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Zhu XG et al. CHP1 regulates compartmentalized glycerolipid synthesis by activating GPAT4. Mol. Cell 74, 45–58.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Go Y-M & Jones DP Mitochondrial thioredoxin-2/peroxiredoxin-3 system functions in parallel with mitochondrial GSH system in protection against oxidative stress. Arch. Biochem. Biophys 465, 119–126 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Seelig GF, Simondsen RP & Meister A. Reversible dissociation of gamma-glutamylcysteine synthetase into two subunits. J. Biol. Chem 259, 9345–9347 (1984). [PubMed] [Google Scholar]
- 20.Li W, Li Z, Yang J. & Ye Q. Production of glutathione using a bifunctional enzyme encoded by gshF from Streptococcus thermophilus expressed in Escherichia coli.J. Biotechnol 154, 261–268 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Chen WW, Freinkman E, Wang T, Birsoy K. & Sabatini DM Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell 166, 1324–1337.e11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slabbaert JR et al. Shawn, the Drosophila homolog of SLC25A39/40, is a mitochondrial carrier that promotes neuronal survival. J. Neurosci 36, 1914–1929 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Usaj M. et al. TheCellMap.org: a web-accessible database for visualizing and mining the global yeast genetic interaction network. Genes Genomes Genet. 7, 1539–1549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luk E, Carroll M, Baker M. & Culotta VC Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proc. Natl Acad. Sci. USA 100, 10353–10357 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson R. et al. Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metab. 10, 119–130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biederbick A. et al. Role of human mitochondrial Nfs1 in cytosolic iron–sulfur protein biogenesis and iron regulation. Mol. Cell. Biol 26, 5675–5687 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayr JA, Feichtinger RG, Tort F, Ribes A. & Sperl W. Lipoic acid biosynthesis defects. J. Inherit. Metab. Dis 37, 553–563 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Chen Z. & Lash LH Evidence for mitochondrial uptake of glutathione by dicarboxylate and 2-oxoglutarate carriers. J. Pharmacol. Exp. Ther 285, 608–618 (1998). [PubMed] [Google Scholar]
- 29.Booty LM et al. The mitochondrial dicarboxylate and 2-oxoglutarate carriers do not transport glutathione. FEBS Lett. 589, 621–628 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar C. et al. Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control. EMBO J. 30, 2044–2056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez-Manzaneque MT, Tamarit J, Bellí G, Ros J. & Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell 13, 1109–1121 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almusafri F. et al. Clinical and molecular characterization of 6 children with glutamate– cysteine ligase deficiency causing hemolytic anemia. Blood Cells. Mol. Dis 65, 73–77 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Wessel D. & Flügge UI A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem 138, 141–143 (1984). [DOI] [PubMed] [Google Scholar]
- 34.McAlister GC et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyanova S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Birsoy K. et al. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Concordet J-P & Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shola DTN, Yang C, Han C, Norinsky R. & Peraza RD in Mouse Genetics: Methods and Protocols (eds. Singh SR, Hoffman RM & Singh A) 1–27 (Springer US, 2021). [Google Scholar]
- 39.Murgha YE, Rouillard J-M & Gulari E. Methods for the preparation of large quantities of complex single-stranded oligonucleotide libraries. PLoS ONE 9, e94752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadreyev IR, Ji F, Cohen E, Ruvkun G. & Tabach Y. PhyloGene server for identification and visualization of co-evolving proteins using normalized phylogenetic profiles. Nucleic Acids Res. 43, W154–W159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruan J. et al. TreeFam: 2008 update. Nucleic Acids Res. 36, D735–D740 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas PD et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Studer G. et al. ProMod3—a versatile homology modelling toolbox. PLOS Comput. Biol 17, e1008667 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pebay-Peyroula E. et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Robinson AJ & Kunji ERS Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc. Natl. Acad. Sci 103, 2617–2622 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettersen EF et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Bycroft C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barbeira AN et al. Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet. 15, e1007889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou D. et al. A unified framework for joint-tissue transcriptome-wide association and Mendelian randomization analysis. Nat. Genet 52, 1239–1246 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unlu G. et al. Phenome-based approach identifies RIC1-linked Mendelian syndrome through zebrafish models, biobank associations and clinical studies. Nat. Med 26, 98–109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aguet F. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD027673. Other data generated are available from the corresponding author upon reasonable request. Source data are provided with this paper.