Abstract
How bifidobacteria colonize and survive in the intestine is not fully understood. The administration of bifidobacteria to conventional mice can be used to evaluate their ability to colonize the intestine in the presence of endogenous gut microbiota. However, human-derived bifidobacteria do not readily colonize the intestines of conventional mice, and although colonization by Bifidobacterium breve UCC2003 has been achieved, the viability of such populations requires improvement. Therefore, we aimed to establish a colonization system with human-derived bifidobacteria of high viability in conventional mice using Bifidobacterium longum subsp. longum 105-A. Lactose, raffinose, and 1-kestose were identified as the preferred carbohydrate sources for the growth of this strain in culture. The administration of B. longum 105-A to conventional BALB/c mice fed these carbohydrates showed that diets containing 6% (w/w) raffinose or 1-kestose facilitated colonization with >108 colony-forming units/g feces for 2 weeks. The population of this strain was more stable in the raffinose-fed group than in the 1-kestose-fed group. The ingestion of these prebiotics had a greater impact on the composition of the microbiota than the administration of B. longum 105-A. The ingestion of these prebiotics also increased the fecal concentrations of organic acids, which was indicative of greater intestinal fermentation. Collectively, we established a colonization system for B. longum 105-A with high viability in conventional mice by feeding the mice raffinose or 1-kestose. This system should be useful for elucidation of the mechanisms of colonization and survival of bifidobacteria in the intestines in the presence of the endogenous gut microbiota.
Keywords: human-derived bifidobacteria, colonization, conventional mice, raffinose, 1-kestose
INTRODUCTION
Bifidobacteria are one of the major components of the gut microbiota and have various beneficial effects on the health of humans [1]. However, the mechanisms involved in their colonization and survival in the intestine have not been fully established. Several factors are involved in their colonization and survival: type IVb tight adherence pili [2], surface exopolysaccharides [3], and the luxS autoinducer-2-producing gene [4] in Bifidobacterium breve UCC2003; sortase-dependent pili [5], the SiaBb2 sialidase [6], and a housekeeping sortase [7] in Bifidobacterium bifidum; and several housekeeping proteins in the extracellular vesicles of Bifidobacterium longum NCC2705 [8].
The contribution of these factors to intestinal colonization can be demonstrated by generating isogenic mutants of genes that encode these factors in genetically amenable strains [2, 9, 10] and assessing their colonization and survival following their administration to mice. Regarding bifidobacteria, administration of a single strain to germ-free mice [2, 11, 12] and to a gnotobiotic mouse model colonized by representative species of the human gut microbiota has been used to evaluate their function [12,13,14]. The former model is associated with a high level of colonization, and the interaction between the host and the bifidobacteria can be evaluated. However, the competition and interactions with other intestinal bacteria cannot be assessed, owing to the lack of an endogenous gut microbiota. Although the latter model can be used to assess these interactions, both models must be housed in animal facilities suitable for the maintenance of germ-free animals, which is challenging for most microbiology laboratories.
Conventional mice harbor an endogenous gut microbiota and can be managed more easily. Therefore, they may be a useful model for the characterization of the mechanisms involved in the intestinal colonization and survival of bifidobacteria in the presence of gut microbiota. However, it is difficult to induce human-derived bifidobacteria to colonize the intestines of conventional mice [11, 15, 16]. Previously, the feeding of cow’s milk, in place of a solid diet, was shown to enable the maintenance of human-derived bifidobacteria in the mouse intestine [15]. In addition, colonization by human-derived bifidobacteria is facilitated by reducing the endogenous gut microbial population by pretreatment with multiple antibiotics [17]. However, both of these methods have substantial effects on the gut microbiota; therefore, an experimental system that allows human-derived bifidobacteria to colonize mice while maintaining their endogenous gut microbiota as much as possible, should be developed.
The sole successful example of a system to evaluate the intestinal colonization ability of human-derived bifidobacteria using conventional mice established to date is the colonization of the intestines by B. breve UCC2003 in mice fed a polysaccharide-rich diet [2]. This strain can use starch and pullulan as its primary carbohydrate sources [18]. This suggests that the addition of carbohydrates that can be assimilated by human-derived bifidobacteria to the diet permits them to colonize the intestines of conventional mice. However, a limiting issue in this system is the relatively small population of colonized bifidobacteria established, 105–106 colony-forming units (cfu)/g feces [2, 4], which is much smaller than that in the feces of adult humans (>109Bifidobacterium/g feces) [19]. Moreover, when evaluating the ability of mutants of genes encoding possible colonization factors to colonize the intestine, a larger population would permit the identification of more differences in the colonization abilities between the wild type and the mutants.
In the present study, we aimed to establish an experimental system in which human-derived bifidobacteria can colonize the intestines of conventional mice and remain highly viable. For this purpose, we used B. longum subsp. longum 105-A (B. longum 105-A; JCM 31944) [20, 21] as the test strain, identified carbohydrates suitable for the growth of this strain in vitro, and then investigated the effects of the addition of the identified carbohydrates to the diet on the colonization of conventional mouse intestines by this strain.
MATERIALS AND METHODS
Bacterial strains and culture conditions
The bacterial strains and plasmids used in the study are listed in Table 1. Bifidobacterium strains were cultured in an anaerobic chamber (80% N2, 10% CO2, and 10% H2; Coy Laboratory Products, Inc., Grass Lake, MI, USA) at 37°C. Half-strength de Man, Rogosa, and Sharpe medium containing 1% (w/v) glucose, 0.34% (w/v) sodium ascorbate, and 0.02% (w/v) L-cysteine hydrochloride (1/2 MRSCS medium, pH 6.5) was used as the standard medium [22]. The glucose in the 1/2 MRSCS medium was replaced with one of 13 other carbohydrate sources (Supplementary Table 1) at 1% (w/v) to test their assimilation by the Bifidobacterium strain. Bacterial growth was monitored by measuring the optical density of the medium at 660 nm (OD660) using a spectrophotometer (UV-1800, Shimadzu, Kyoto, Japan). The media and 1× PBS (phosphate-buffered saline, pH 7.0) were pre-reduced for at least 24 hr using gas-exchange materials (AnaeroPack Keep, Mitsubishi Gas Chemical Company, Inc., Tokyo, Japan). Chloramphenicol (Cm) was added to the medium at a final concentration of 2.5 or 10 µg/mL when necessary.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Bifidobacterium longum subsp. longum | ||
| 105-A (JCM 31944) | Human fecal isolate, transformation host strain | [20] |
| Blo0145 | 105-A harboring pBFS38 | This study |
| Blo0146 | 105-A harboring pBFS63 | This study |
| Plasmids | ||
| pBFS38 | E. coli-Bifidobacterium shuttle vector, pTB4 ori, p15A ori, Cmr, 5.3 kbp | [23] |
| pBFS63 | E. coli-Bifidobacterium shuttle vector, pTB6 ori, pMB1 ori, Cmr, 3.7 kbp | [22] |
aCmr, chloramphenicol resistance.
Construction of Cm-resistant B. longum strains
B. longum 105-A was transformed with two types of Escherichia coli-Bifidobacterium shuttle vectors harboring the Cm-resistance gene, pBFS38 [23] and pBFS63 [22], according to the protocol used in our previous study [9]. Transformants were selected and isolated using 1/2 MRSCS agar medium containing 2.5 µg/mL of Cm. Colony PCR was used to verify the introduction of each vector using the following primer pairs: Pr-Blo0192/Pr-Blo0194 for pBFS38 transformants and Pr-Blo0239/Pr-Blo0240 for pBFS63 transformants (see Supplementary Table 2 for the primer sequences). Subsequently, Blo0145 (B. longum 105-A/pBFS38) and Blo0146 (B. longum 105-A/pBFS63) were used to evaluate plasmid stability.
Plasmid stability in B. longum cells was evaluated as follows. Blo0145 and Blo0146 were pre-cultured in 1/2 MRSCS containing 2.5 µg/mL of Cm. After washing the cells with saline, they were inoculated into 1/2 MRSCS medium at an initial OD660 of 0.002 and then sub-cultured in the absence of Cm for 71 generations (seven rounds of 12-hr culture). The estimated generation times were 1.23 hr for Blo0145 and 1.28 hr for Blo0146. A portion of each culture collected at generations 11, 41, and 71 was serially diluted and spread onto 1/2 MRSCS agar medium, and the colonies were replicated on 1/2 MRSCS agar medium and 1/2 MRSCS agar medium containing 2.5 µg/mL of Cm using the replica-plating method. Plasmid stability was compared using a percentage of the number of Cm-resistant colonies compared with the number of replicated colonies.
Mouse administration experiment I
Five-week-old female BALB/c mice (Japan SLC, Shizuoka, Japan) were housed in standard plastic cages in an air-conditioned room at 23 ± 2°C under a 12-hr light/dark cycle. The mice had free access to water and food. An AIN-93G-based control diet (Supplementary Table 3) was fed to the mice during a 2-week acclimation period.
The Cm-resistant strain Blo0146 was administered to the mice. The control diet or AIN-93G-based diet containing one of 1-kestose, raffinose, or lactose at 3% or 6% (w/w), in place of a portion of maltodextrin (Supplementary Table 3), was fed to the mice from the first day until the end of the trial, and the mice were allocated to seven groups according to their diets. This trial was conducted in three rounds, with two mice in each diet group (control group, n=6 in total; each test diet group, n=2). Strain Blo0146 was cultured in 1/2 MRSCS medium containing 10 µg/mL of Cm for 7 hr (to an approximate OD660 of 2.0). The cells were collected and washed with 1× PBS and then resuspended to an estimated dilution of 1.0 × 109 cfu in 200 µL of 1× PBS. This suspension was used as an inoculum. The cells were administered by gavage once daily for 3 consecutive days. Fresh mouse feces were collected every 24 hr following the first Blo0146 administration. They were collected into 500 µL of 1× PBS and weighed, and then a fecal suspension was prepared by homogenization using a sterilized homogenizer. The suspensions were then serially diluted in 1× PBS, and 50 µL of each dilution was inoculated onto 1/2 MRSCS agar medium containing 10 µg/mL of Cm. Agar plates that developed ~300 colonies were used for colony counting, and the number of cfu of Blo0146 in 1 gram of wet feces was calculated.
Mouse administration experiment II
Administration experiment II was conducted like experiment I, with some modifications. The mice were allocated to five groups according to the diet fed and whether they were administered Blo0146 or 1× PBS. The groups were as follows: C group, control diet and Blo0146; KB group, 6% 1-kestose-containing diet and Blo0146; KP group, 6% 1-kestose-containing diet and 1× PBS; RB group, 6% raffinose-containing diet and Blo0146; RP group, 6% raffinose-containing diet and 1× PBS (see Supplementary Table 4 for more details). AIN-93G-based diets containing 1-kestose or raffinose at 6% (w/w) in place of a portion of maltodextrin (Supplementary Table 3) were fed to the mice in the test diet groups every day from the first day of administration. This trial was also conducted in three rounds (Supplementary Table 4); in each round, three mice comprised each of the C groups and test diet groups. However, one of the mice in the KB group died unexpectedly after administration.
The administration of Blo0146, the feeding, and the counting of viable Blo0146 cells in the mouse feces were conducted as described in experiment I. The mice were fed the test diets for 15 days, and on the 15th day following the first administration, they were anesthetized with sevoflurane (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan). They were then killed by exsanguination via the carotid artery, and their ceca were excised. The animal studies were approved by the Institutional Animal Care and Use Committee of National University Corporation Hokkaido University (approval number 17-0050), and the mice were cared for according to the Hokkaido University Manual for Implementing Animal Experimentation.
Analysis of the cecal microbiota
Microbial DNA was extracted from the mouse cecal contents collected in experiment II using a previously described method for rat cecal contents, with some modifications [24]. Briefly, the cecal contents (25 mg) were washed once with 1× PBS and again with washing buffer (200 mM Tris-HCl, 80 mM EDTA, pH 9.0). The pellets were resuspended in 500 µL of TES buffer (30 mM Tris-HCl, 1 mM EDTA, 50 mM NaCl, pH 8.0), 500 µL of 30 mg/mL lysozyme (Fujifilm Wako Pure Chemical Corporation) in 25% (w/v) sucrose solution was added, and the mixture was then incubated at 37°C for 1 hr. Subsequently, 500 µL of 9 mg/mL achromopeptidase (Fujifilm Wako Pure Chemical Corporation) in 12.5% (w/v) sucrose solution was added, and the mixture was incubated at 37°C for 30 min to disrupt the cell walls. Enzyme-treated cells were collected by centrifugation at 20,000 × g for 5 min at 4°C, and the DNA was extracted according to a published protocol [24]. One sample from the C group (C6) was excluded from the analysis of the microbiota because only a small quantity of cecal contents was collected.
The V3–V4 region of the 16S rRNA gene was amplified from the community DNA samples using the primers 347F_MiSeqF and 803R_MiSeqR (Supplementary Table 2) [25], as described previously [24]. The amplicons were purified using a MinElute PCR purification kit (Qiagen, Hilden, Germany). A second PCR was then conducted to add the index sequences, using Nextera XT Index Kit v2 Set D for 96 indexes and 384 samples (FC-131-2004, Illumina, Inc., San Diego, CA, USA). A 50-µL reaction mixture containing 25 µL of 2× PCR buffer, 0.4 mM of each dNTP, 5 µL of each of Nextera XT Index primers 1 and 2, 4.5 µL of the purified first amplicon, and 0.5 U of KOD Fx Neo DNA polymerase (Toyobo Co., Ltd., Osaka, Japan) was prepared. Amplification was performed using the following conditions: initial denaturation at 94°C for 2 min; eight cycles of amplification, comprising denaturation at 98°C for 10 sec, annealing at 55°C for 30 sec, and extension at 68°C for 30 sec; and a final extension at 68°C for 7 min. The addition of the index sequences was confirmed by comparing the sizes of the first and second PCR products by agarose gel electrophoresis. The amplicons were purified as described above, and the DNA concentrations were measured and adjusted to 25 ng/µL. The samples were then mixed in equal molar ratios and purified using a Gene Read Size Selection Kit (Qiagen). The purified samples were processed using an Illumina MiSeq and MiSeq Reagent Kit V3 (600 cycles; Illumina Inc.). The sequences obtained were deposited in the Sequence Read Archive of the DNA Data Bank of Japan (DRA) under the accession number DRA017054.
Sequence data were analyzed using Quantitative Insights Into Microbial Ecology (QIIME, ver. 1.8.0) [26]. Paired reads were merged using paired-end read merger (PEAR) [27], and chimeric sequences were removed using Usearch 61 in QIIME. Operational taxonomic units (OTUs) were identified using the pick_open_reference_otus.py script at a threshold of 97% sequence identity, and 0.001% of the unassigned sequences were randomly picked and used for de novo OTU picking. A representative sequence for each OTU was subsequently analyzed. The taxonomy of each OTU was assigned using the Greengenes database (release gg_13_8). Data were standardized to 21,900 reads/sample, based on the rarefaction plots obtained by α-rarefaction analysis using the alpha_rarefaction.py script. The relative abundance of each taxon and the α- and β-diversity were analyzed according to published protocols [28].
Analysis of fecal organic acid concentrations
Fresh feces were obtained on days 1 and 14 during experiment II. The feces were weighed, and 100 mg of each sample was homogenized in 2-mL screw-cap vials containing 400-µL aliquots of absolute ethanol by three 20-sec cycles of vigorous shaking with metal corn (Yasui Kikai Corp., Osaka, Japan) in a Multi-Beads Shocker (Yasui Kikai Corp.) at 2,500 rpm and 4°C, with a 10-sec break between cycles. The tubes were then centrifuged at 860 × g for 10 min at 4°C. The organic acids in the supernatant were derivatized using labeling reagents containing 2-nitrophenylhydrazine hydrochloride and analyzed using a YMC Fatty Acid Analysis Kit (YMC Co., Ltd., Kyoto, Japan), according to the manufacturer’s instructions. Crotonic acid (500 µM) was added to the samples as an internal standard before derivatization. A standard organic acid mixture, comprising the sodium salts of acetate, propionate, n-butyrate, and isobutyrate; succinic acid; and DL-lactic acid (1 mM each), was also prepared and derivatized similarly. The samples and standard were passed through a 0.22-µm filter (Shimadzu GLC Ltd., Tokyo, Japan) and subjected to high-performance liquid chromatography (HPLC) analysis. The standard solution was serially diluted with filtered methanol up to 1,000 times and used to construct a standard curve.
HPLC analysis was conducted using an L-2000 HPLC System (Hitachi High-Tech Corporation, Tokyo, Japan), according to the instructions accompanying the YMC Fatty acid Analysis Kit. The organic acids were separated using a YMC-Pack FA column (250 × 6.0 mm I.D., YMC Co., Ltd.) at 50°C using isocratic flow (1.2 mL/min) of the eluent (acetonitrile:methanol:water, 30:16:54, v/v, pH 4.5) for 15 min. The derivatized organic acids were detected at 400 nm using an L-2400 UV detector (Hitachi High-Tech Corporation). A standard curve was constructed using the peak area for each standard organic acid, and this was used to calculate the organic acid concentrations in one gram of fecal samples. The data were standardized using the concentration of the internal standard.
Statistical analysis
Differences in the data between samples were basically evaluated using a parametric Tukey–Kramer honestly significant difference (HSD) test. Differences in relative abundances of each phylum and Bifidobacterium OTU were tested using a non-parametric Steel test because those data did not fit a normal distribution. All statistical analyses were conducted using JMP version 14.0 (SAS Institute Inc., Cary, NC, USA). Statistical significance was set at p<0.05.
RESULTS
In vitro selection of carbohydrates that facilitate colonization
The ability of B. longum 105-A to use specific carbohydrates was evaluated to permit the selection of appropriate carbohydrates for efficient colonization of the intestines of conventional mice. Fourteen types of carbohydrates (listed in Supplementary Table 1), comprising five monosaccharides, two disaccharides, and seven oligosaccharides that have been reported to have prebiotic effects on bifidobacteria [29,30,31,32,33,34,35], were used as the sole carbohydrate sources. The growth of B. longum 105-A when cultured in the presence of each carbohydrate was monitored by measuring the OD660 after 12 hr and 24 hr. The effect of each carbohydrate on growth was evaluated as the relative growth compared with that in the presence of glucose (Fig. 1). The bacterial growth in the presence of monosaccharides (xylose, galactose, fructose, and arabinose) and epilactose was less than that in the presence of glucose. In contrast, lactose showed comparable growth to that associated with glucose after 12 hr and superior growth after 24 hr. Among the oligosaccharides, raffinose and 1-kestose showed comparable growth to that associated with glucose both at 12 hr and 24 hr. Consequently, lactose, raffinose, and 1-kestose were selected as candidate carbohydrates for subsequent mouse administration experiments.
Fig. 1.
Relative growth of B. longum 105-A in the presence of various sole carbohydrate sources.
Bacteria were cultured anaerobically at 37°C for 24 hr in 1/2 MRSCS medium containing a sole carbohydrate source (at least n=3), and their growth (OD660) was measured. The relative growth (%) after 12 hr (black bars) and 24 hr (gray bars) was calculated by dividing the mean OD660 value in the presence of each carbohydrate by that in the presence of glucose at the same time point. Glc: glucose; Gal: galactose; Fru: fructose; Xyl: xylose; Ara: arabinose; Lac: lactose; Elac: epilactose; Ltu: lactulose; Raf: raffinose; Kes: 1-kestose; Nys: nystose; Fos: fructooligosaccharides; Gos: galactooligosaccharides; Xos: xylooligosaccharides.
Effects of the selected carbohydrates on bacterial colonization in conventional mice
B. longum 105-A was transformed with plasmids to monitor the viability of the administered strain in the mouse intestine. Two types of plasmids conferring Cm resistance, pBFS38 [23] and pBFS63 [22], were introduced into B. longum 105-A to generate Blo0145 and Blo0146, respectively. An evaluation of the plasmid stability of these strains in the absence of Cm revealed that pBFS63 was stable for 71 generations, whereas pBFS38 was rapidly eliminated from the cells (Supplementary Table 5). Therefore, Blo0146 was used as the test strain for the mouse administration experiments. This strain can be selected from mouse fecal samples using 1/2 MRSCS agar medium containing 10 µg/ml of Cm. No colonies appeared for at least 3 days when a serially diluted slurry of feces collected from mice during the acclimation period was inoculated on this medium.
Administration experiment I was conducted to determine the effect of the three selected carbohydrates on the intestinal colonization of B. longum 105-A (Supplementary Fig. 1). Blo0146 (mean 3.8 × 108 cfu/administration) was administered to mice consuming diets containing 3% or 6% (w/w) of each selected carbohydrate for 3 consecutive days. In the control diet group, the viability of Blo0146 (i.e., the number of viable Blo0146 cells) in the feces, measured as cfu/g feces, rapidly decreased after day 4 and passed below the detection limit (103 cfu/g feces) on day 7 (Supplementary Fig. 1A). Similar decreases in viability occurred in mice consuming a 3% or 6% lactose-containing diet. In contrast, higher viability was observed during the administration period (from day 1 to day 3) in mice fed raffinose- or 1-kestose-containing diets than in mice in the control diet group (Supplementary Fig. 1B), indicating the proliferation of B. longum 105-A in the mouse intestine. The decrease in viability was also delayed and slower in the 3% raffinose or 1-kestose-containing diet groups than in the control and lactose-containing diet groups. In the 6% raffinose- or 1-kestose-containing diet groups, the number of viable Blo0146 cells remained >105 cfu/g feces for 2 weeks. These results indicate that raffinose and 1-kestose may facilitate the colonization of the intestines of conventional mice by B. longum 105-A.
We performed administration experiment II using a larger number of mice to verify the effects of raffinose and 1-kestose on intestinal colonization (Fig. 2). Blo0146 (mean 1.28 × 109 cfu/administration) was administered to mice consuming diets containing 6% (w/w) raffinose (RB group) or 1-kestose (KB group) for 3 consecutive days. Groups that consumed the same test diet but were administered 1× PBS instead of Blo0146 (RP and KP groups) were used to verify the effects of each carbohydrate on the gut microbiota. There were no significant differences in body mass or total food consumption among the five groups analyzed on day 15 (Supplementary Table 6). However, the test diet groups had significantly higher cecal and cecal content masses. The number of viable Blo0146 cells in feces from the C group reached 107 cfu/g feces on day 2 and then rapidly decreased, passing below the detection limit on day 6 (Fig. 2). As expected based on experiment I, the numbers of viable Blo0146 cells in the RB and KB groups reached 1011 cfu/g feces on day 2 and then stayed at >108 cfu/g feces for at least 2 weeks (Fig. 2). These results demonstrate that the feeding of diets containing 6% raffinose or 1-kestose enables colonization of the intestines of conventional mice by B. longum 105-A for at least 2 weeks.
Fig. 2.
Effects of raffinose and 1-kestose feeding on intestinal colonization by B. longum 105-A in conventional mice.
B. longum 105-A harboring the Cm-resistant plasmid (Blo0146) was administered for 3 consecutive days (day 1 to day 3). Its viability was assessed as cfu per gram of mouse feces every day for 14 days. The mean cfu ± standard deviation values are shown for the C group (n=8, closed circle), KB group (n=5, closed square), and RB group (n=6, closed triangle). The detection limit for colonies was set as 103 cfu/g feces. C: control-diet-fed, B. longum administered; KB: 1-kestose-fed, B. longum administered; RB: raffinose-fed, B. longum administered.
While B. longum 105-A remained in the guts of conventional mice when they were fed diets containing 6% raffinose or 1-kestose (Fig. 2), there were differences in the shedding of viable B. longum cells between mice and between the carbohydrates fed (Supplementary Fig. 2). In the RB group, the number of viable B. longum cells in feces remained >109 cfu/g feces in all the mice until day 8 and then decreased in two mice, though the numbers remained >108 cfu/g feces in the remaining four mice (Supplementary Fig. 2A). In the KB group, four of the five mice analyzed demonstrated a clear downward trend in the shedding of viable B. longum cells in their feces from day 4, and the numbers of viable B. longum cells in fecal samples obtained from these mice on day 14 were 104–7 cfu/g feces (Supplementary Fig. 2B). These results suggest that although there were some variations between the mice analyzed, the feeding of raffinose is associated with superior persistence of B. longum 105-A in the intestine compared with 1-kestose.
Effects of carbohydrate source and B. longum administration on the cecal microbiota
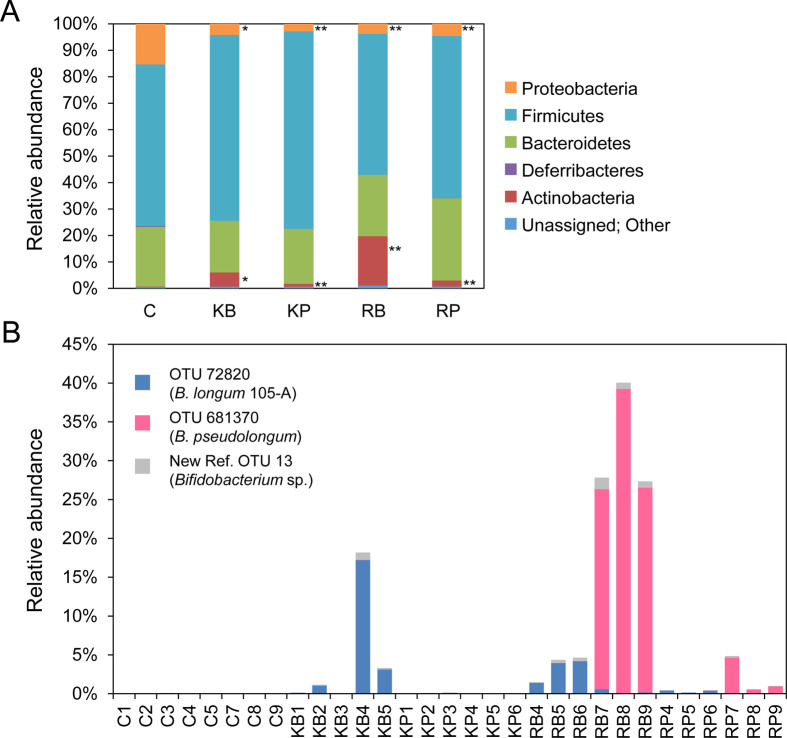
The 16S rRNA gene amplicon sequencing was conducted to clarify the effect of feeding of raffinose or 1-kestose and B. longum 105-A administration on the cecal microbiota of the conventional mice. Cecal contents were collected from the mice on day 15 of experiment II. After extracting the microbial DNA, the V3–V4 region of the 16S rRNA gene was amplified and sequenced by Illumina MiSeq, and then the relative abundances of the identified OTUs and the α- and β-diversities of the cecal microbiota were assessed. The cecal microbial composition of the mice was largely unaltered at the phylum level, except with respect to the Actinobacteria and Proteobacteria, which had significantly higher and lower abundances in the four test groups, respectively, compared with the C group (Fig. 3A and Supplementary Fig. 3).
Fig. 3.
Cecal microbial composition 15 days after the first administration of B. longum 105-A and the selected carbohydrates.
(A) Relative abundance of bacterial phyla in the cecal contents. The identities of the phyla are shown with the corresponding colors on the right of the figure. The mean relative abundance of each phylum is indicated (C group, n=8; KB group, n=5; others, n=6). *p<0.05, **p<0.01 vs. the C group, by the non-parametric Steel test. See supplementary Fig. 3 for the detailed results of the statistical analysis.
(B) Relative abundances of Bifidobacterium OTUs in the cecal microbiota of individual mice. Bifidobacterium OTUs are indicated in the inset with the corresponding colors. The relative abundances are indicated as follows: blue bars, B. longum 105-A (OTU 72820); magenta bars, B. pseudolongum (OTU 681370); gray bars, Bifidobacterium sp. (New reference OTU 13). C: control-diet-fed, B. longum administered; KB: 1-kestose-fed, B. longum administered; KP: 1-kestose-fed, 1×PBS administered; RB: raffinose-fed, B. longum administered; RP: raffinose-fed, 1×PBS administered; OTU: operational taxonomic unit.
The size of the Bifidobacterium population in the cecal microbiota was assessed to explain the increased size of the Actinobacteria population. Of the 908 OTUs identified in the analysis of the microbiota, three were assigned as Bifidobacterium. A blastn similarity analysis of the representative sequences of the three OTUs revealed that they corresponded to the following Bifidobacterium species, based on sequence identity with the 16S rRNA gene sequences in the non-redundant GenBank database: OTU 72820, B. longum 105-A (accession no. AP014658.1, 100% identity); OTU 681370, Bifidobacterium pseudolongum strain PCK036 (accession no. MN913803.1, 99.8% identity); and New reference OTU 13, uncultured Bifidobacterium sp. (accession no. FJ518688.1, 97.7% identity).
The greater abundance of OTU 72820 (B. longum 105-A) was significant in the KB and RB groups compared with the C group (Supplementary Fig. 4). The relative abundances of this OTU in individual mice were 0.01–4.17% and 0.06–17.2% of the cecal microbiota in the RB and KB groups, respectively (Fig. 3B). These data indicated the presence of inter-mouse differences in the relative abundance of B. longum 105-A, as in the case of the viable cell count of B. longum 105-A in the feces (Supplementary Fig. 2). A strong positive correlation (R2=0.8179) between the relative abundance of B. longum 105-A in the cecal microbiota and its viable cell count in fecal samples on day 14 was obtained, indicating that the size of the population of B. longum 105-A in the feces reflects that in the cecal microbiota. On the other hand, the increase in the relative abundance of OTU 681370 (B. pseudolongum) was not significant in the four test groups, even in the RB group (Supplementary Fig. 4). This OTU accounted for 25–39% of the cecal microbial population of three of the six mice in the RB group (Fig. 3B) and was also more abundant in three mice in the RP group, though to a lesser extent than in the three mice in the RB group. However, a low abundance of this OTU was observed in the other three mice in both the RB and RP groups. These variances in the population are the cause of the non-significant increase in this OTU. Although the relative abundance was small, significant increases in New reference OTU 13 (Bifidobacterium sp.) were observed in the RB and RP groups compared with the C group (Supplementary Fig. 4). These results imply that a larger population of B. longum 105-A mainly accounts for the significantly greater abundance of Actinobacteria in the KB and RB groups.
The microbiota of the four groups of mice that were fed the raffinose or 1-kestose-containing diets commonly showed significantly lower α-diversity indices than those of the C group, with the exception that a significantly lower Shannon index was only present in the RB group (Table 2). There were no significant differences in the α-diversity indices between the test diet-fed groups that were or were not administered B. longum 105-A, indicating that the administration of B. longum 105-A did not affect the α-diversity of the mouse cecal microbiota. The β-diversity of the microbiota of the five groups was also analyzed using the weighted UniFrac distance. In this analysis, shifts in the microbiota from the C group to the test diet-fed groups were identified along the PC2 axis (Fig. 4). However, the microbial compositions of the four test diet-fed groups were not separated clearly according to whether or not they were administered B. longum 105-A. Overall, although an inter-mouse difference in the colonization of B. longum 105-A was observed, the results of the cecal microbiota analysis indicated that the ingestion of raffinose or 1-kestose had a greater impact on the composition of the microbiota than the administration of B. longum 105-A.
Table 2. α-diversity of cecal microbiota in mice among the tested groups.
| α-diversity index | α-diversity in groupsa | ||||
|---|---|---|---|---|---|
| Control diet | 6% 1-kestose diet | 6% raffinose diet | |||
| Blo0146 (C) | Blo0146 (KB) | 1×PBS (KP) | Blo0146 (RB) | 1×PBS (RP) | |
| PD whole tree | 18.58 ± 0.21a | 15.88 ± 0.39b | 15.92 ± 0.23b | 16.01 ± 0.27b | 16.79 ± 0.26b |
| Chao1 | 666.17 ± 14.1a | 525.1 ± 23.13b | 536.02 ± 14.77b | 521.47 ± 16.5b | 572.52 ± 16.2b |
| Observed species | 514.38 ± 11.42a | 413.00 ± 16.23b | 415.32 ± 10.52b | 399.98 ± 9.23b | 429.98 ± 10.86b |
| Shannon | 5.89 ± 0.07a | 5.48 ± 0.14ab | 5.5 ± 0.16ab | 5.17 ± 0.21b | 5.56 ± 0.06ab |
aData are shown as means ± standard error of the mean (C group, n=8; KB group, n=5; others, n=6).
Tukey–Kramer HSD test was conducted as a statistical analysis. Different superscript letters represented significant differences among groups at p<0.05.
C: control-diet-fed, B. longum administered; KB: 1-kestose-fed, B. longum administered; KP: 1-kestose-fed, 1×PBS administered; RB: raffinose-fed, B. longum administered; RP: raffinose-fed, 1×PBS administered; PD: phylogenetic diversity.
Fig. 4.
β-diversity of the cecal microbiota in mice consuming the various diets and being administered B. longum 105-A or 1× PBS.
β-diversity was evaluated using weighted UniFrac distances and is shown as a 2-D plot of the results of principal coordinate analysis. Each sphere corresponds to the microbial composition of one mouse. Groups of mice are distinguished by the colors of the spheres as follows: green, C group; orange, KB group; light orange, KP group; navy, RB group; light blue, RP group. Information regarding the groups is also provided on the right side of the figure. C: control-diet-fed, B. longum administered; KB: 1-kestose-fed, B. longum administered; KP: 1-kestose-fed, 1×PBS administered; RB: raffinose-fed, B. longum administered; RP: raffinose-fed, 1×PBS administered.
Effects of carbohydrate source and B. longum administration on intestinal fermentation
We next evaluated the effects of raffinose and 1-kestose feeding on the intestinal fermentation of the mice by measuring the organic acid concentrations of the fecal samples (Table 3). In the C group, there were no significant differences in the organic acid concentrations between fecal samples collected on day 1, before B. longum 105-A administration commenced, and between day 14 fecal samples, indicating that there was little change in the intestinal fermentation of the mice during the experimental period if they were consuming the control diet. In the raffinose-fed groups (RB and RP), there were significantly higher concentrations of acetate, propionate, n-butyrate, and total organic acids compared with the C group (Table 3). Although not statistically significant, there were similarly high concentrations of these organic acids in the 1-kestose-fed groups (KB and KP) compared with the C group. These results indicate that raffinose or 1-kestose feeding increases intestinal fermentation, which was consistent with the greater cecal content masses of the mice in these feeding groups (Supplementary Table 6). The feeding of raffinose or 1-kestose thus enables B. longum 105-A to colonize the intestines of conventional mice, and this is accompanied by greater carbohydrate fermentation.
Table 3. Organic acid concentrations in the mouse feces.
| Organic acid (μmol/g feces)a | Concentrationon day 1 | Concentration on day 14 | ||||
|---|---|---|---|---|---|---|
| Control diet | 6% 1-kestose diet | 6% raffinose diet | ||||
| Blo0146 (C) | Blo0146 (KB) | 1×PBS (KP) | Blo0146 (RB) | 1×PBS (RP) | ||
| Succinate | 0.04 ± 0.01 | 0.01 ± 0.00 | 0.01 ± 0.01 | 0.02 ± 0.01 | 0.03 ± 0.03 | 0.23 ± 0.21 |
| DL-Lactate | 0.82 ± 0.57 | 0.41 ± 0.07 | 0.53 ± 0.19 | 0.64 ± 0.50 | 1.47 ± 0.69 | 1.28 ± 0.28 |
| Acetate | 2.6 ± 0.4a | 4.07 ± 0.44a | 9.71 ± 1.95ab | 9.65 ± 3.56ab | 18.96 ± 4.99b | 19.63 ± 1.46b |
| Propionate | 3.15 ± 0.26a | 2.83 ± 0.33a | 6.44 ± 1.18ab | 5.67 ± 2.11ab | 9.99 ± 2.39bc | 16.38 ± 3.27c |
| iso-Butyrate | 0.18 ± 0.03 | 0.34 ± 0.07 | 0.22 ± 0.04 | 0.15 ± 0.05 | 0.29 ± 0.06 | 0.23 ± 0.04 |
| n-Butyrate | 1.42 ± 0.21a | 1.76 ± 0.47ab | 6.45 ± 1.64bc | 6.23 ± 1.88abc | 9.12 ± 1.7cd | 12.91 ± 1.5d |
| Total | 8.21 ± 1.1a | 9.42 ± 1.17a | 23.36 ± 4.56ab | 22.35 ± 7.82ab | 39.87 ± 9.48bc | 50.66 ± 3.78c |
aData are shown as means ± standard error of the mean (Day 1, n=10; C group, n=8; others, at least n=4).
Tukey–Kramer HSD test was conducted as a statistical analysis. Different superscript letters represented significant differences among groups at p<0.05.
C: control-diet-fed, B. longum administered; KB: 1-kestose-fed, B. longum administered; KP: 1-kestose-fed, 1×PBS administered; RB: raffinose-fed, B. longum administered; RP: raffinose-fed, 1×PBS administered.
DISCUSSION
The establishment of an experimental system to facilitate the colonization of human-derived bifidobacteria in conventional mice permits further elucidation of the mechanisms of their colonization and survival in the presence of the endogenous gut microbiota. However, human-derived bifidobacteria do not readily colonize the intestines of conventional mice [11, 15, 16]. Although the strains used, viability of the inoculum, and duration of administration varied among previous studies, bifidobacteria typically remained in the intestines during the administration period, but their populations often rapidly decreased in size or their viability fell below the detection limit after the cessation of administration [5, 11, 15, 16, 36]. These results indicate that the use of bifidobacteria is limited by their survival ability in the intestines of conventional mice, mainly because of competition with the endogenous intestinal microbiota for nutrients.
We hypothesized that the feeding of carbohydrates that favor the growth of bifidobacteria would improve the colonization of the intestines of conventional mice. In the present study, three carbohydrates (lactose, raffinose, and 1-kestose) were identified as favoring the growth of B. longum 105-A in culture (Fig. 1). We then showed that the ingestion of diets containing 6% (w/w) raffinose or 1-kestose successfully maintained a highly viable population of B. longum 105-A (108–109 cfu/g feces) in the intestines of conventional mice for at least 2 weeks (Fig. 2), in contrast to mice fed control or lactose-containing diets (Supplementary Fig. 1). Lactose cannot promote the colonization ability of this strain because it has little prebiotic activity due to the wide distribution of the lactose-assimilating enzyme, β-galactosidase, among the intestinal bacteria [37]. These results suggested that both in vitro growth-promoting and prebiotic activities contribute to the successful colonization of B. longum 105-A in the conventional mouse intestine. We cannot conclude the importance of the former activity for colonization because prebiotics that did not promote in vitro growth (e.g., nystose, GOS, FOS) were not tested in the colonization experiments. This point could be clarified by further evaluation; however, it is anticipated that carbohydrates harboring both activities, 1-kestose and raffinose, are advantageous for the colonization of B. longum 105-A compared with those with low-growth promoting activity. Previously, colonization with B. breve UCC2003 has been demonstrated in the intestines of conventional mice fed a polysaccharide-rich diet at 105–106 cfu/g feces [2, 4]. In the present study, the viability of the colonized B. longum 105-A was maintained approximately 103-fold higher than that of the previously documented B. breve UCC2003 [2, 4], although the viability of the administered bifidobacteria and the duration of administration were similar in these studies. Moreover, the viability achieved in the present study was close to that of the bifidobacterial population in adult human feces (>109Bifidobacterium/g feces) [19]. These results indicate that we have developed an effective colonization system involving the simultaneous administration of B. longum and a bifidobacteria-selective prebiotic, raffinose or 1-kestose. During the development and evaluation of the colonization system used in the present study, we analyzed the B. longum 105-A genes that are specifically expressed in the intestines of mice fed a 6% (w/w) 1-kestose-containing diet using recombinase-based in vivo expression technology [22]. In this study, the administered strains were successfully recovered from the feces of the mice 4 days after administration at concentrations of 109 to 1010 cfu/g feces [22]. Recently, repeated administration of a small amount of B. longum BB536 (106 viable bacteria) and 40 mg of 1-kestose was shown to synergistically increase the populations of Bifidobacterium and B. longum in the gut microbiota of C57BL/6 J mice [38]. These data also imply that selective prebiotic ingestion facilitates the colonization of the intestines of conventional mice with human-derived bifidobacteria.
In the present study, the feeding of raffinose or 1-kestose resulted in some modifications to the gut microbiota of the mice. Lower α-diversity (Table 3) and differences between the prebiotic-fed and C groups concerning β-diversity (Fig. 3) were identified. Colonization by human-derived bifidobacteria without any alteration in the composition of the gut microbiota is an ideal scenario. However, successful colonization requires compensatory alterations to the endogenous mouse gut microbiota. Although comparable microbial composition data were unavailable, it is anticipated that the effects of prebiotic feeding on the gut microbiota would be smaller than those associated with the ingestion of cow’s milk [15] or prior treatment of mice with a combination of antibiotics [17]. Therefore, the system established in the present study can be considered to reflect the competition between B. longum and other intestinal bacteria during colonization.
One issue with the established system is the variability in the numbers of viable B. longum 105-A cells in individual mice within the same groups (Supplementary Fig. 2). This phenomenon could be attributed to competition with the endogenous intestinal bacteria, the composition of which can be different between mice. Although raffinose and 1-kestose are effective prebiotics for bifidobacteria, other intestinal bacteria can also use them. For example, Lactobacillus-related species use raffinose [39]. Superior growth rates of Prevotella were recently shown in the presence of raffinose or fructooligosaccharides in human fecal batch culture [40]. Some butyrate-producing bacterial species, such as Faecalibacterium prausnitzii and Anaerostipes, have also been reported to use 1-kestose [41]. Ensuring stable intestinal colonization by exogenous bifidobacteria is difficult because multiple factors, including the population sizes of competing bacteria, their metabolic activities, and changes in these parameters over time, affect the level of competition. Conversely, reducing the competition with endogenous intestinal bacteria can lead to stable colonization by exogenous bifidobacteria. This might be facilitated by feeding Bifidobacterium-specific, next-generation prebiotics, such as galactosyl-β1,4-L-rhamnose [42], as an improved colonization system.
The colonization system developed in this study achieves a high degree of colonization by administering raffinose and 1-kestose as prebiotics, and thus there is a limitation in that the contribution of carbohydrate utilization genes to colonization cannot be evaluated. On the other hand, this system can be used to assess the contribution of genes involved in metabolic pathways other than carbohydrates, regulatory genes, and genes involved in adhesion to the intestinal epithelium or mucus layer. There is a concern that the contribution of the target genes to colonization might be masked due to the high degree of colonization if their contribution is weak. In such a situation, controlling the degree of colonization may be necessary by adjusting the proportions of the prebiotics in the diets. The application of the colonization system we have established to other Bifidobacterium species should be attempted in the future because raffinose and 1-kestose are well-known prebiotics for other Bifidobacterium species [33, 34]. Therefore, the established system may be versatile and could be used for other Bifidobacterium species, especially for genetically amenable strains, to clarify the functions of key genes in intestinal colonization.
In conclusion, we established an experimental system that enables intestinal colonization by B. longum subsp. longum with high viability in conventional mice by feeding them raffinose or 1-kestose. This system should help elucidate the mechanisms by which bifidobacteria survive and colonize the intestine in the presence of endogenous gut microbiota.
CONFLICTS OF INTEREST
The authors declare they have no conflicts of interest associated with this manuscript.
Supplementary Material
Acknowledgments
We would like to thank the following companies and individuals for the donation of carbohydrates: epilactose, Dr. Wataru Saburi (Research Faculty of Agriculture, Hokkaido University, Sapporo, Japan); lactulose, Morinaga Milk Industry, Co., Ltd., Tokyo, Japan; raffinose pentahydrate, Nippon Beet Sugar Manufacturing Co., Ltd., Tokyo, Japan; and 1-kestose and xylooligosaccharides, B Food Science Co., Ltd., Aichi, Japan. This research was supported in part by a Grant-in-Aid for Scientific Research (B) from the Japan Society for the Promotion of Science (grant number 25450090, to S.F.). We also thank Mark Cleasby, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
REFERENCES
- 1.Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sánchez B, Margolles A. 2017. Bifidobacteria and their health-promoting effects. Microbiol Spectr 5: BAD-0010-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O’Brien F, Flynn K, Casey PG, Munoz JAM, et al. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci USA 108: 11217–11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, Motherway MO, Shanahan F, Nally K, Dougan G, et al. 2012. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci USA 109: 2108–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christiaen SEA, O’Connell Motherway M, Bottacini F, Lanigan N, Casey PG, Huys G, Nelis HJ, van Sinderen D, Coenye T. 2014. Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PLoS One 9: e98111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turroni F, Serafini F, Foroni E, Duranti S, O’Connell Motherway M, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, et al. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci USA 110: 11151–11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishiyama K, Yamamoto Y, Sugiyama M, Takaki T, Urashima T, Fukiya S, Yokota A, Okada N, Mukai T. 2017. Bifidobacterium bifidum extracellular sialidase enhances adhesion to the mucosal surface and supports carbohydrate assimilation. mBio 8: e00928-e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikawa E, Yamada T, Yamaji K, Serata M, Fujii D, Umesaki Y, Tsuji H, Nomoto K, Ito M, Okada N, et al. 2021. Critical roles of a housekeeping sortase of probiotic Bifidobacterium bifidum in bacterium-host cell crosstalk. iScience 24: 103363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishiyama K, Takaki T, Sugiyama M, Fukuda I, Aiso M, Mukai T, Odamaki T, Xiao JZ, Osawa R, Okada N. 2020. Extracellular vesicles produced by Bifidobacterium longum export mucin-binding proteins. Appl Environ Microbiol 86: e01464-e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirayama Y, Sakanaka M, Fukuma H, Murayama H, Kano Y, Fukiya S, Yokota A. 2012. Development of a double-crossover markerless gene deletion system in Bifidobacterium longum: functional analysis of the α-galactosidase gene for raffinose assimilation. Appl Environ Microbiol 78: 4984–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469: 543–547. [DOI] [PubMed] [Google Scholar]
- 11.Grimm V, Radulovic K, Riedel CU. 2015. Colonization of C57BL/6 mice by a potential probiotic Bifidobacterium bifidum strain under germ-free and specific pathogen-free conditions and during experimental colitis. PLoS One 10: e0139935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugahara H, Odamaki T, Fukuda S, Kato T, Xiao JZ, Abe F, Kikuchi J, Ohno H. 2015. Probiotic Bifidobacterium longum alters gut luminal metabolism through modification of the gut microbial community. Sci Rep 5: 13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, et al. 2011. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med 3: 106ra106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woting A, Pfeiffer N, Hanske L, Loh G, Klaus S, Blaut M. 2015. Alleviation of high fat diet-induced obesity by oligofructose in gnotobiotic mice is independent of presence of Bifidobacterium longum. Mol Nutr Food Res 59: 2267–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitsuoka T, Kaneuchi C. 1977. Ecology of the bifidobacteria. Am J Clin Nutr 30: 1799–1810. [DOI] [PubMed] [Google Scholar]
- 16.Singh N, Arioli S, Wang A, Villa CR, Jahani R, Song YS, Mora D, Guglielmetti S, Comelli EM. 2013. Impact of Bifidobacterium bifidum MIMBb75 on mouse intestinal microorganisms. FEMS Microbiol Ecol 85: 369–375. [DOI] [PubMed] [Google Scholar]
- 17.Rossini V, Nally K. 2021. Model for murine gut colonization by bifidobacteria, In Bifidobacteria: Methods and Protocols, van Sinderen, D, Ventura, M (eds), Springer US, New York, pp. 131–139. [DOI] [PubMed] [Google Scholar]
- 18.Ryan SM, Fitzgerald GF, van Sinderen D. 2006. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl Environ Microbiol 72: 5289–5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K, Tanaka R. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol 70: 167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumura H, Takeuchi A, Kano Y. 1997. Construction of Escherichia coli-Bifidobacterium longum shuttle vector transforming B. longum 105-A and 108-A. Biosci Biotechnol Biochem 61: 1211–1212. [DOI] [PubMed] [Google Scholar]
- 21.Kanesaki Y, Masutani H, Sakanaka M, Shiwa Y, Fujisawa T, Nakamura Y, Yokota A, Fukiya S, Suzuki T, Yoshikawa H. 2014. Complete genome sequence of Bifidobacterium longum 105-A, a strain with high transformation efficiency. Genome Announc 2: e01311–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koguchi H, Ishigami N, Sakanaka M, Yoshida K, Hiratou S, Shimada M, Fukiya S, Sonoyama K, Yokota A. 2020. Application of recombinase-based in vivo expression technology to Bifidobacterium longum subsp. longum for identification of genes induced in the gastrointestinal tract of mice. Microorganisms 8: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakanaka M, Tamai S, Hirayama Y, Onodera A, Koguchi H, Kano Y, Yokota A, Fukiya S. 2014. Functional analysis of bifidobacterial promoters in Bifidobacterium longum and Escherichia coli using the α-galactosidase gene as a reporter. J Biosci Bioeng 118: 489–495. [DOI] [PubMed] [Google Scholar]
- 24.Lee JY, Shimizu H, Hagio M, Fukiya S, Watanabe M, Tanaka Y, Joe GH, Iwaya H, Yoshitsugu R, Kikuchi K.2020. 12α-Hydroxylated bile acid induces hepatic steatosis with dysbiosis in rats. Biochim Biophys Acta Mol Cell Biol Lipids 1865: 158811. [DOI] [PubMed] [Google Scholar]
- 25.Nossa CW, Oberdorf WE, Yang L, Aas JA, Paster BJ, DeSantis TZ, Brodie EL, Malamud D, Poles MA, Pei Z. 2010. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J Gastroenterol 16: 4135–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30: 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navas-Molina JA, Peralta-Sánchez JM, González A, McMurdie PJ, Vázquez-Baeza Y, Xu Z, Ursell LK, Lauber C, Zhou H, Song SJ, et al. 2013. Advancing our understanding of the human microbiome using QIIME. In Methods in Enzymology, Volume 531, DeLong, EF (eds), Academic Press, Cambridge, pp. 371–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe J, Nishimukai M, Taguchi H, Senoura T, Hamada S, Matsui H, Yamamoto T, Wasaki J, Hara H, Ito S. 2008. Prebiotic properties of epilactose. J Dairy Sci 91: 4518–4526. [DOI] [PubMed] [Google Scholar]
- 30.Sakai Y, Seki N, Hamano K, Ochi H, Abe F, Masuda K, Iino H. 2019. Prebiotic effect of two grams of lactulose in healthy Japanese women: a randomised, double-blind, placebo-controlled crossover trial. Benef Microbes 10: 629–639. [DOI] [PubMed] [Google Scholar]
- 31.Bali V, Panesar PS, Bera MB, Panesar R. 2015. Fructo-oligosaccharides: production, purification and potential applications. Crit Rev Food Sci Nutr 55: 1475–1490. [DOI] [PubMed] [Google Scholar]
- 32.Walton GE, van den Heuvel EGHM, Kosters MHW, Rastall RA, Tuohy KM, Gibson GR. 2012. A randomised crossover study investigating the effects of galacto-oligosaccharides on the faecal microbiota in men and women over 50 years of age. Br J Nutr 107: 1466–1475. [DOI] [PubMed] [Google Scholar]
- 33.Dinoto A, Marques TM, Sakamoto K, Fukiya S, Watanabe J, Ito S, Yokota A. 2006. Population dynamics of Bifidobacterium species in human feces during raffinose administration monitored by fluorescence in situ hybridization-flow cytometry. Appl Environ Microbiol 72: 7739–7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ose R, Hirano K, Maeno S, Nakagawa J, Salminen S, Tochio T, Endo A. 2018. The ability of human intestinal anaerobes to metabolize different oligosaccharides: Novel means for microbiota modulation? Anaerobe 51: 110–119. [DOI] [PubMed] [Google Scholar]
- 35.Finegold SM, Li Z, Summanen PH, Downes J, Thames G, Corbett K, Dowd S, Krak M, Heber D. 2014. Xylooligosaccharide increases bifidobacteria but not lactobacilli in human gut microbiota. Food Funct 5: 436–445. [DOI] [PubMed] [Google Scholar]
- 36.Hidalgo-Cantabrana C, Algieri F, Rodriguez-Nogales A, Vezza T, Martínez-Camblor P, Margolles A, Ruas-Madiedo P, Gálvez J. 2016. Effect of a ropy exopolysaccharide-producing Bifidobacterium animalis subsp. lactis strain orally administered on DSS-induced colitis mice model. Front Microbiol 7: 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He T, Priebe MG, Vonk RJ, Welling GW. 2005. Identification of bacteria with β-galactosidase activity in faeces from lactase non-persistent subjects. FEMS Microbiol Ecol 54: 463–469. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe A, Teragaki Y, Kitaura Y, Tochio T. 2023. The synergistic synbiotic potential of 1-kestose and Bifidobacterium longum in the mouse gut. J Funct Foods 101: 105403. [Google Scholar]
- 39.Gänzle MG, Follador R. 2012. Metabolism of oligosaccharides and starch in lactobacilli: a review. Front Microbiol 3: 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Li Z, Wang X, Fan B, Deng F, D Yu H, Ze X, Zhu L, Yin Y, Chen Y, et al. 2022. Isomaltooligosaccharides sustain the growth of Prevotella both in vitro and in animal models. Microbiol Spectr 10: e0262121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanno H, Fujii T, Hirano K, Maeno S, Tonozuka T, Sakamoto M, Ohkuma M, Tochio T, Endo A. 2021. Characterization of fructooligosaccharide metabolism and fructooligosaccharide-degrading enzymes in human commensal butyrate producers. Gut Microbes 13: e1869503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirano R, Sakanaka M, Yoshimi K, Sugimoto N, Eguchi S, Yamauchi Y, Nara M, Maeda S, Ami Y, Gotoh A, et al. 2021. Next-generation prebiotic promotes selective growth of bifidobacteria, suppressing Clostridioides difficile. Gut Microbes 13: e1973835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.