Abstract
A cytopathogenic variant of hepatitis A virus (HAVcyt/HB1.1) was isolated from persistently infected BS-C-1 cells by serial passages in FRhK-4 cells. This virus shows a rapid replication pattern and high final titers are obtained, which are main characteristics of cytopathogenic HAVs. Sequencing of the nontranslated regions and the coding regions for 2ABC and 3AB revealed that mutations are distributed all over these regions and that certain mutated sites correspond to those in other cytopathogenic HAV variants. Investigating the mechanisms causing the cytopathic effect in FRhK-4 cells infected with this variant, we found that an apoptotic reaction takes place.
Hepatitis A virus (HAV), the only member of the Hepatovirus genus of the picornavirus family, is an important pathogen which causes acute viral hepatitis. In a small number of cases, fatal complications of HAV infections, known as fulminant hepatitis, do occur (13).
HAV is unique among the human picornaviruses with regard to its growth characteristics (9, 11, 16, 31). The virus is hepatotropic in vivo and can infect a variety of primate and nonprimate cell lines in vitro. In contrast to other picornaviruses, HAV exhibits a protracted replication cycle and normally establishes a persistent infection with low virus yields. Although more-rapid replication and higher final virus titers are obtained with cell culture-adapted viruses, even these variants replicate considerably more slowly and less efficiently than other members of the picornavirus family. Replication of cell culture-adapted HAV is not detectable within the first few days after infection. The infection does not induce any visible cytopathic effects (CPE), and there is no evidence that HAV notably interferes with the macromolecular synthesis of its host cell.
However, during the last decade, several cytopathogenic variants of HAV have been described (2, 8, 20, 23, 25, 32). These cytopathogenic variants are highly cell culture adapted and characterized by a rapid replication phenotype. For a variant of an Italian isolate, it is suggested that the CPE is caused by shutoff of host cell protein synthesis, which is mediated by a protease activity of protein 2A via inhibition of cap-dependent translation initiation (22). In contrast, there is no evidence for shutoff of the host cell metabolism by cytopathogenic descendants of HAV strain HM175 (34), despite the virus-induced reduction of cell viability. Morace et al. (23) postulated a primary role for certain mutations in protein 3A in causing the CPE. Data presented by several laboratories indicate that in general the CPE and adaptation of HAV to growth in cell culture are associated with various mutations which are distributed over the 5′ nontranslated region (5′NTR) and the P2 and P3 genomic regions and that therefore the CPE correlates with the overall efficiency of viral replication (23, 34). Apart from the proposed shutdown of host cell protein synthesis by one cytopathogenic variant, the postulated involvement of a mutated 3A protein, and the demonstration that cytopathogenicity of HAV is linked to a rapid replication pattern, the events resulting in the CPE are unknown.
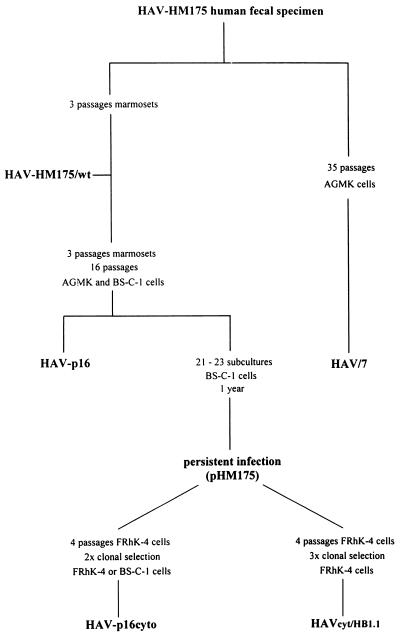
In order to investigate functional properties of cytopathogenic HAV (HAVcyt) and to obtain information on the cause of the virus-induced CPE, we selected a CPE-inducing variant (HAVcyt/HB1.1) of strain HM175, studied its phenotypic and genotypic features, and looked for characteristics of an apoptotic reaction in infected cells as a possible mechanism causing the CPE. As shown in Fig. 1, HAVcyt/HB1.1 was obtained from a virus stock (pHM175) of persistently infected BS-C-1 cells (8) by four serial passages in FRhK-4 cells and by clonal selection three times from agarose overlays of radioimmunofocus assays (19).
FIG. 1.
Passage history of variants of HAV strain HM175. The cytopathogenic variant HAVcyt/HB1.1 was isolated in our laboratory. HAV/7 (7), which was recovered after transfection of FRhK-4 cells with the RNA of the infectious cDNA clone pHAV/7 (7), was used as a reference in our experiments characterizing HAVcyt/HB1.1. Phenotypic and genotypic features of HAVcyt/HB1.1 were compared with those of the p16cyto viruses HM175/18f, -43c, and -24a (20), which were obtained independently from HAVcyt/HB1.1 from the same persistent infection (8).
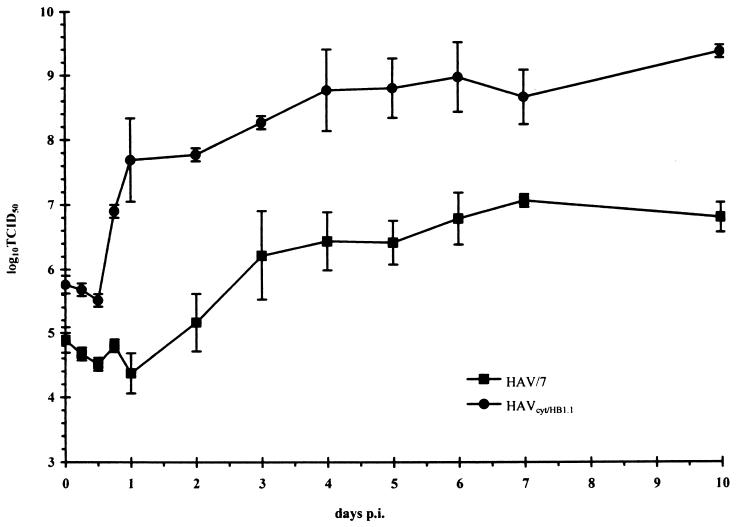
In order to analyze the replication phenotype of HAVcyt/HB1.1, we recorded one-step growth curves (Fig. 2). FRhK-4 cells were infected with a multiplicity of infection (MOI) of 5. The titer was assessed by inoculating cells grown in 96-well microtiter plates, and infection was checked 2 weeks after inoculation by indirect immunofluorescence with the HAV-specific monoclonal antibody 7E7 (Mediagnost, Tübingen, Germany) and a fluorescein-labeled anti-mouse antibody (Kirkegaard and Perry) (9). The maximum total 50% tissue culture infective dose titer of 109 was reached 4 days postinfection (p.i.). The highest titer obtained with the noncytopathogenic variant HAV/7 (6) (Fig. 1), which was used as a reference, was 107 and was detected after an incubation time of 7 days. The data indicate that HAVcyt/HB1.1 is a typical rapidly replicating HAV variant. This is also reflected in the genotype of HAVcyt/HB1.1. We isolated the genomic regions 5′NTR, 2ABC, 3AB, and 3′NTR by PCR (12), because it is suggested that mutations in these regions contribute to the cytopathogenic, rapidly replicating phenotype of HAV in cell culture (34). Three amplified fragments of each region, which were obtained by independent reactions, were analyzed by dideoxy sequencing. The sequence analysis revealed that HAVcyt/HB1.1 contains several mutations from wild-type HAV strain HM175 and the noncytopathogenic variant HAV/7 (Table 1). The mutations are distributed all over the regions mentioned above. Fifteen mutated sites correspond to those found in each of the three other cytopathogenic variants, HM175/18f, -43c, and -24a (Table 1). These three viruses (summarized as p16cyto in Fig. 1), which are sibling clones, originate from the same virus stock (pHM175) as HAVcyt/HB1.1, which was isolated by seven passages independently from them. No other mutations found in HAVcyt/HB1.1 are present in any of these viruses. Using chimeric constructs which contain certain segments of the genome of HM175/18f within the background of the HAV/7 genome, Zhang et al. (34) showed that mutations in the P2 proteins are necessary for the expression of the cytopathogenic phenotype and that P3 region mutations and mutations in the 5′NTR support the CPE. These studies also demonstrated that cytopathogenicity correlates with high replication capacity, and it was supposed that the cytopathogenic phenotype of this HAV variant represents a further adaptation of the virus to cell culture, which results from several interactions between different regions of the viral genome. Because of the similarity of HAVcyt/HB1.1 and HM175/18f, the data presented for this virus concerning the phenotypic features are also applicable to our variant. These data indicate that the identified mutations in the genome of HAVcyt/HB1.1 may act cooperatively to enhance the rate of replication and render the virus cytopathogenic.
FIG. 2.
Replication kinetics under one-step growth curve conditions for HAVcyt/HB1.1 in FRhK-4 cells in comparison with HAV/7. The kinetics show the total titers in the course of 10 days. The cells were infected with an MOI of 5, and at the times indicated the 50% tissue culture infective dose (TCID50) titer was determined in FRhK-4 cells 2 weeks after inoculation by indirect immunofluorescence. Each data point is an average obtained from two separate experiments. Error bars indicate standard deviations of the means.
TABLE 1.
Mutations present in selected genomic regions of HAVcyt/HB1.1 and HAV/7 in comparison with wild-type HAV strain HM175
| Genomic region (nucleotides) | Nucleotide position (protein amino acid) | Nucleotide (amino acid)
|
|
|---|---|---|---|
| HAV/7 | HAVcyt/HB1.1 | ||
| 5′NTR (47–734) | 140 | T | |
| 152 | G | Ga | |
| 154/155 | TTGTAAA TATTGATa | ||
| 203–207 | TTTT | TTTTa | |
| 591 | Ga | ||
| 646 | A | ||
| 687 | Ga | ||
| 2A (3027–3243) | 3123 (33) | G (Gly) | |
| 2B (3244–3996) | 3453 (70) | G (Val) | |
| 3464 (74) | Ab | ||
| 3554 (104) | Ab | ||
| 3795 (184) | C (Leu) | ||
| 3812 (190) | Cb | ||
| 3836 (198) | Ab | ||
| 3889 (216) | T (Val) | T (Val)a | |
| 3924 (227) | A (Ile) | ||
| 2C (3997–5000) | 4015 (7) | A (Lys) | |
| 4060 (22) | G (Gly)a | ||
| 4066 (24) | G (Cys)a | ||
| 4085 (30) | Gb | ||
| 4185 (63) | A (Lys) | A (Lys)a | |
| 4194 (66) | G (Val) | ||
| 4222 (76) | C (Ser) | C (Ser)a | |
| 4233 (79) | G (Glu) | ||
| 4251 (85) | G (Val) | ||
| 4419 (141) | C (His)a | ||
| 4511 (172) | Tb | ||
| 4673 (226) | Ab | ||
| 4858 (288) | G (Ser) | ||
| 3A (5001–5222) | 5074 (25) | G (Ser) | |
| 5193 (65) | A (Ser) | ||
| 5204 (68) | A | Aa,b | |
| 3′NTR (7416–7478) | 7429 | Ta | |
| 7430 | Ga | ||
| 7433 | Ca | ||
Mutated sites corresponding to those found in HM175/18f, -43c, and -24a.
Silent mutation.
Besides the demonstration that cytopathogenicity is linked to a rapidly replicating phenotype, the molecular events resulting in the CPE are not known. In some cytopathogenic HAVs, deletions in the N-terminal part of protein 3A which make the protein more hydrophobic have been identified (20, 23). It was shown that the modified protein is able to form pores in membranes (28). Therefore, it was supposed that after integration of this protein into cellular membranes the permeability is increased, finally resulting in cell lysis. However, for HM175/18f it was shown that these mutations were not necessary for the CPE of the virus (34). In accordance with this, HAVcyt/HB1.1 does not have mutations in this region, showing that changes in protein 3A resulting in a more hydrophobic protein are not essential for the cytopathogenicity of HAV. It was also suggested that the CPE is caused by shutoff of host cell protein synthesis mediated by proteolytic activity of protein 2A. For the HM175 variants p16cyto, no mutations in the 2A protein were found. Investigations concerning the function of protein 2A revealed that the 2A protein of HAV lacks proteinase activity, and extensive pattern searches failed to detect identifiable catalytic motifs (27). In cells infected with cytopathogenic variants of HAV strain HM175, a significant shutdown of host cell protein synthesis could not be detected (34). Therefore, 2A does not seem to play a considerable role in the determination of the cytopathogenic phenotype of HM175 descendants. Nüesch et al. (26) presented a functional relationship of the 3′NTR to the HAVcyt phenotype. In a cytopathic infection, down-regulation of viral RNA synthesis, which is seen in persistent infections, did not occur, and it was supposed that this leads to the high viral titers observed. However, Zhang et al. (34) could not demonstrate a role of the mutations in the 3′NTR of HM175/18f for the cytopathogenic feature.
The infection of FRhK-4 cells with HAVcyt/HB1.1, but not of the human hepatoma cell line HepG2, resulted in the development of a CPE, whereas BS-C-1 cells were not infected at all with the variant 3 weeks p.i. A few rounded cells are characteristic for the beginning of the CPE, which occurred within the first week after infection at an MOI of 2. In the course of the proceeding CPE, more and more cells were detached, and finally the whole cell culture was degenerated 14 days after inoculation. These morphological changes could be prevented by preincubation of the inoculum with HAV-specific monoclonal antibody 7E7. As a result of the CPE, the cell culture showed a reduced viability. This has been proved using the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] test (Boehringer Mannheim), which is a colorimetric assay that measures the activity of mitochondrial enzymes (24). FRhK4 cells were infected with HAVcyt/HB1.1 and HAV/7 at an MOI of 4. The MTT test was performed according to the manufacturer’s instructions at days 3, 7, and 10 p.i. In comparison with noninfected cells, HAVcyt/HB1.1-infected cells showed reductions in cell viability, which accompany the developing CPE, of 8% at day 3 p.i., 18% at day 7 p.i., and 43% at day 10 p.i. The corresponding data obtained with HAV/7-infected cells were 1% at day 3 p.i., 3% at day 7 p.i., and 7% at day 10 p.i. The cytopathogenic feature of the virus has also been demonstrated by a conventional plaque assay (not shown) (20). The plaques represented regions with reduced cell growth or dead cells, which could be identified morphologically, but not cell-free areas. Therefore, the morphological characteristics of the CPE did not show that a lytic reaction is responsible for the cell death.
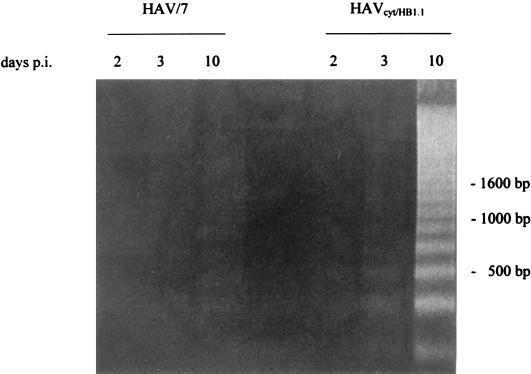
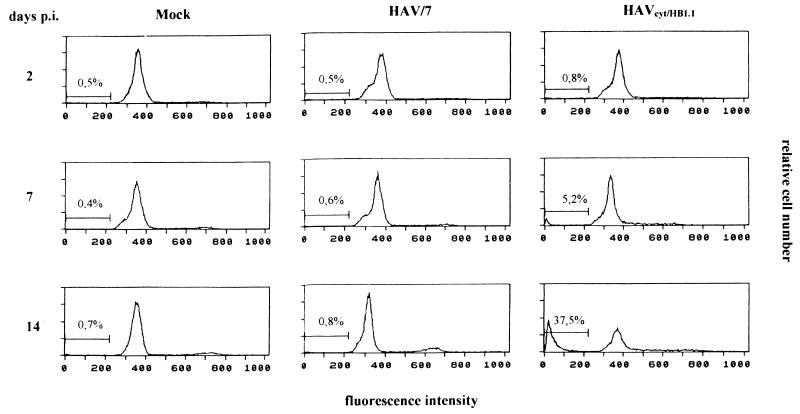
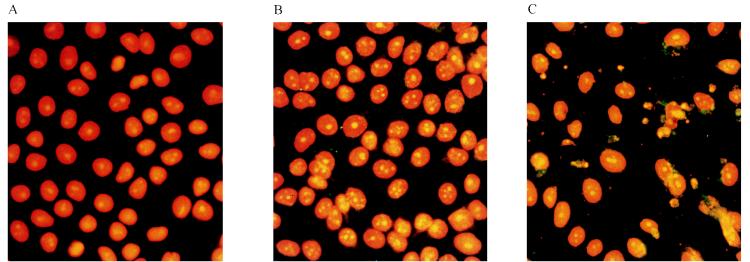
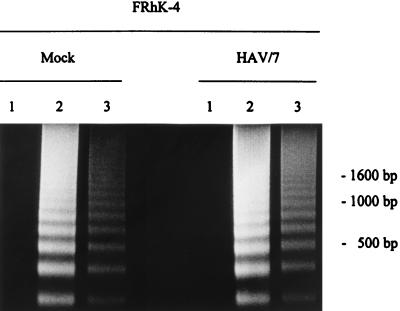
Apoptosis is a fundamental process involved in the development and homeostasis of multicellular organisms (7). It is also a cellular response to viral infections. Among RNA viruses, apoptosis-inducing activities have been reported for alphaviruses (21), myxoviruses (14), arenaviruses (29), retroviruses (1, 3), and picornaviruses (15, 30). Apoptosis is characterized by pronounced morphological changes and internucleosomal DNA degradation resulting in fragments consisting of 180-bp multimers (17, 33). Investigating this, we performed a DNA fragmentation analysis in FRhK-4 cells infected with HAVcyt/HB1.1 and HAV/7 at an MOI of 4. At the times indicated, 2 × 106 trypsinized cells, which contained less than 10% dead cells (ascertained with trypan blue staining), were washed twice with phosphate-buffered saline (PBS) and lysed in 400 μl of lysis buffer (10 mM Tris, 0.5% Triton X-100 [pH 7.5]) for 30 min on ice. Nuclei were removed, and the supernatants were extracted with phenol-chloroform. After precipitation, the samples were treated with RNase A (final concentration, 1 μg/μl) and analyzed electrophoretically (Fig. 3). Samples were prepared at days 2, 3, and 10 p.i. Neither in noninfected cells (not shown) nor in HAV/7-infected cells did DNA fragmentation occur. In the course of the HAVcyt/HB1.1 infection, DNA fragments could be detected in increasing amounts, becoming visible for the first time at 3 days p.i. Infection of the cells was confirmed by indirect immunofluorescence and was evident at 2 days p.i. In cells infected with HAVcyt/HB1.1, the CPE and the DNA fragmentation occurred 3 days p.i. At the same time, 100% of the cells were infected. In order to obtain more quantitative data with regard to apoptotic cells after HAV infection, we analyzed the DNA contents of the cells infected with HAVcyt/HB1.1 and HAV/7 by flow cytometry. For this purpose, FRhK-4 cells were infected with an MOI of 4. At days 2, 3, 5, 7, 10, and 14 p.i., cells were trypsinized, washed twice with PBS, and fixed with 70% ethanol overnight. For nuclear staining, 5 × 105 cells were resuspended in 100 μl of staining solution (50 μg of propidium iodide per ml, 100 U of RNase A per ml, 0.1% glucose in PBS) and incubated for 30 min at room temperature. Apoptotic nuclei were identified as a subdiploid peak (DNA content < 2 N), which is clearly distinguishable from the G0-G1 peak (DNA content = 2 N). The results obtained are shown in Table 2 and Fig. 4. For HAV/7-infected cells, the percentage of apoptotic cells was identical to that for noninfected cells, with values of ≤1.2 in the course of 14 days. However, after infection with HAVcyt/HB1.1, the proportion of apoptotic cells increased with advancing incubation periods, reaching 37.5% at day 14 p.i. These results also show that nuclear changes, which are indicative for apoptosis, are not common for infections with noncytopathogenic HAV. By use of morphological criteria, we were able to strengthen the finding that apoptosis occurs. The nuclei of acetone-fixed infected cells were stained with the nuclear dye propidium iodide and studied by fluorescence microscopy (Fig. 5). FRhK-4 cells were infected with HAVcyt/HB1.1 and HAV/7 (MOI, 1). At 10 days p.i., cells were prepared for nuclear staining. In addition, the presence of HAV antigen was proved by indirect immunofluorescence. The medium was removed, and the cells were acetone fixed (90% in PBS), treated with HAV-specific murine monoclonal antibody 7E7, and stained with a fluorescein-labeled anti-mouse antibody. For nuclear staining, propidium iodide (40 μg/ml in PBS) was added and the cells were incubated for an additional 15 min at room temperature. The double staining was visualized by fluorescence microscopy. In comparison with uninfected (Fig. 5A) and HAV/7-infected (Fig. 5B) cells, nuclear changes, which are indicative of apoptosis, are evident in cells infected with HAVcyt/HB1.1 (Fig. 5C). The nuclei are irregularly formed and show protuberances on the surface. Finally, they disintegrate into densely stained nuclear apoptotic bodies. An accumulation of viral antigens in the apoptotic cells is also evident, which points out that there is a correlation between the efficiency of viral replication and cell death. Attempts to study the ultrastructural nuclear changes of the infected cells by electron microscopy were made. However, because of the high degree of convolutions of the nuclear outlines of FRhK-4 cells, which can be particularly clearly observed by this technique, no significant differences in the morphology of the nuclei of the HAVcyt/HB1.1-infected cells and the controls could be detected.
FIG. 3.
DNA fragmentation analysis of FRhK-4 cells infected with HAVcyt/HB1.1. FRhK-4 cells were infected with HAVcyt/HB1.1 or with HAV/7 at an MOI of 4. Cytoplasmic extracts from 2 × 106 cells were prepared at 2, 3, and 10 days p.i., and DNA was extracted with phenol-chloroform. After treatment with RNase A, the samples were analyzed on a 1.6% agarose gel. Two days after infection, no DNA fragments indicative for apoptosis could be seen. From 3 days p.i. onward, increasing amounts of DNA fragments could be detected in HAVcyt/HB1.1-infected cells. In HAV/7-infected cells, no fragmentation occurred.
TABLE 2.
Fraction of HAVcyt/HB1.1-infected FRhK-4 cells with a DNA content of less than 2 N in comparison with mockand HAV/7-infected cells
| Days p.i. | % Apoptotic cells
|
||
|---|---|---|---|
| Mock infected | HAV/7 infected | HAVcyt/HB1.1 infected | |
| 2 | 0.5 | 0.5 | 0.8 |
| 3 | 0.8 | 0.5 | 2.3 |
| 5 | 0.8 | 1.2 | 4.3 |
| 7 | 0.4 | 0.6 | 5.2 |
| 10 | 0.3 | 0.3 | 15.0 |
| 14 | 0.7 | 0.8 | 37.5 |
FIG. 4.
Flow cytometric DNA fluorescence profiles of HAVcyt/HB1.1-infected FRhK-4 cells in comparison with mock- and HAV/7-infected cells. At the days indicated, the cells were trypsinized and fixed with 70% ethanol. For nuclear staining, the cells were incubated with 50 μg of propidium iodide per ml for 30 min at room temperature. Apoptotic nuclei were identified as a subdiploid peak with a DNA content of less than 2 N. The percentages of cells with a DNA content of less than 2 N are indicated (see also Table 2).
FIG. 5.
Nuclear morphology in FRhK-4 cells infected with HAVcyt/HB1.1. FRhK-4 cells were infected with HAVcyt/HB1.1 or HAV/7. At 10 days p.i., the cells were prepared for nuclear staining with propidium iodide (orange) and for HAV antigen detection by indirect immunofluorescence (green) with the HAV-neutralizing monoclonal antibody 7E7. Nuclear changes, which are indicative for apoptosis, could not be detected in uninfected (A) and HAV/7-infected (B) cells but were detected in cells infected with HAVcyt/HB1.1 (C). In apoptotic cells, the nuclei are irregularly formed, show protuberances on the surface, and disintegrate into densely stained nuclear apoptotic bodies. The characteristic segregation of chromatin in apoptotic nuclear fragments and nuclear budding are evident. In HAVcyt/HB1.1-infected cells (C), an accumulation of viral antigen was detected.
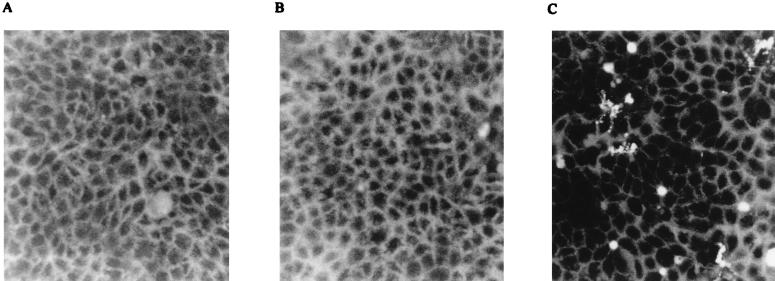
The production of reactive oxygen intermediates (ROI) is a common sign of an apoptotic reaction (5, 18). In order to determine whether FRhK-4 cells infected with HAVcyt/HB1.1 generate increased levels of ROI, we used the dye 2′,7′-dichlorofluorescein diacetate (Molecular Probes), which is permeable to cells and interacts with intracellular ROI to generate fluorescent 2′,7′-dichlorofluorescein (4). Uninfected cells, cells infected with HAVcyt/HB1.1, and the virus control HAV/7 were incubated with 5 μM dichlorofluorescein diacetate for 1 h. The cells were washed twice with PBS and analyzed by fluorescence microscopy. High levels of ROI could be detected as early as 3 days p.i. in cells infected with HAVcyt/HB1.1 but not in the controls (Fig. 6). It should be pointed out that at the same time the first indications of the CPE and of the DNA fragmentation are detectable. Although we could not demonstrate apoptosis by electron microscopy, our data permit the conclusion that HAVcyt/HB1.1 induces an apoptotic reaction in FRhK-4 cells, on the basis of the occurrence of DNA laddering, morphological changes, and accumulation of ROI.
FIG. 6.
Generation of ROI in HAVcyt/HB1.1-infected FRhK-4 cells. Cells were incubated at day 3 p.i. with 2′,7′-dichlorofluorescein diacetate, a fluorescent probe for intracellular ROI, for 1 h and analyzed by fluorescence microscopy. ROI could not be detected in noninfected cells (A) or in HAV/7-infected cells (B) but were detected in cells infected with HAVcyt/HB1.1 (C).
Some cultured cell lines have the intrinsic potential to develop an apoptotic reaction without induction of new RNA or protein species as a response to certain metabolic disturbances (30). In order to investigate this in FRhK-4 cells, we incubated the cells with actinomycin D (1 μg/ml), which inhibits cellular RNA synthesis, and cycloheximide (50 μg/ml), an inhibitor of cellular protein synthesis. Both inhibitors induced a CPE with the same characteristics as in HAVcyt/HB1.1-infected cells and brought about a typical apoptotic reaction, which was revealed by DNA fragmentation after 1 day of incubation (Fig. 7). The development of apoptosis as a response to these inhibitors suggests that the maintenance of the nonapoptotic status in FRhK-4 cells is due to the presence of short-lived mRNA and protein species and that the interplay of apoptosis-promoting and apoptosis-preventing functions is controlled physiologically. It is conceivable that HAVcyt/HB1.1 interferes with this control by competing with the cellular synthesis of macromolecules because of the high replication capacity of the virus. Furthermore, we investigated the ability of noncytopathogenic HAV to interfere with the induction of inhibitor-induced apoptosis in FRhK-4 cells. Incubation of FRhK-4 cell persistently infected with HAV/7 with actinomycin D or cycloheximide, at the concentrations mentioned above, also resulted in apoptosis (Fig. 7). This suggests that, in contrast to poliovirus (30), HAV is not equipped with antiapoptotic functions in this case.
FIG. 7.
DNA fragmentation analysis of FRhK-4 cells and cells persistently infected with HAV/7 after treatment with actinomycin D and cycloheximide. FRhK-4 cells as well as cells persistently infected with HAV/7 were incubated with actinomycin D (1 μg/ml) (lanes 2) and cycloheximide (50 μg/ml) (lanes 3). Both inhibitors of macromolecular synthesis induced an apoptotic reaction in FRhK-4 cells and HAV/7-infected cells, which was revealed by DNA fragmentation after 1 day of incubation. No fragmentation occurred in cells not treated with the inhibitors (lanes 1).
HAVcyt/HB1.1 should be useful for future studies concerning the molecular mechanisms leading to the CPE in cells infected with cytopathogenic HAV. In addition, it is intriguing to speculate on the participation of HAVcyt variants and the involvement of apoptosis in the clinical course of fulminant hepatitis, because there is histological evidence that no inflammatory response occurs (10, 13).
Acknowledgments
This work was supported by the Tönjes-Vagt-Stiftung.
We thank Mediagnost, Tübingen, Germany, for providing monoclonal anti-HAV antibody 7E7.
Footnotes
This work is dedicated to H.-J. Gerth on the occasion of his 70th birthday.
REFERENCES
- 1.Ameisen J C, Capron A. Cell dysfunction and depletion in AIDS: the programmed cell death hypothesis. Immunol Today. 1991;12:102–105. doi: 10.1016/0167-5699(91)90092-8. [DOI] [PubMed] [Google Scholar]
- 2.Anderson D A. Cytopathology, plaque assay, and heat inactivation of hepatitis A virus strain HM 175. J Med Virol. 1987;22:35–44. doi: 10.1002/jmv.1890220106. [DOI] [PubMed] [Google Scholar]
- 3.Antoni B A, Sabbatini P, Rabson A B, White E. Inhibition of apoptosis in human immunodeficiency virus-induced cells enhances virus production and facilitates persistent infection. J Virol. 1995;69:2384–2392. doi: 10.1128/jvi.69.4.2384-2392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busciglio J, Yankner B A. Apoptosis and increased generation of oxygen species in Down’s syndrome neurons in vitro. Nature. 1995;378:776–779. doi: 10.1038/378776a0. [DOI] [PubMed] [Google Scholar]
- 5.Buttke T M, Sandstrom P A. Oxidative stress as a mediator of apoptosis. Immunol Today. 1994;15:7–10. doi: 10.1016/0167-5699(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 6.Cohen J I, Rosenblum B, Ticehurst J R, Daemer R, Feinstone S M, Purcell R H. Complete nucleotide sequence of an attenuated hepatitis A virus: comparison with wild-type virus. Proc Natl Acad Sci USA. 1987;84:2497–2501. doi: 10.1073/pnas.84.8.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins M K L, Rivas A L. The control of apoptosis in mammalian cells. Trends Biochem Sci. 1993;18:307–309. doi: 10.1016/0968-0004(93)90042-l. [DOI] [PubMed] [Google Scholar]
- 8.Cromeans T, Sobsey M D, Fields H A. Development of a plaque assay for a cytopathic, rapidly replicating isolate of hepatitis A virus. J Med Virol. 1987;22:45–56. doi: 10.1002/jmv.1890220107. [DOI] [PubMed] [Google Scholar]
- 9.Dotzauer A, Feinstone S M, Kaplan G. Susceptibility of nonprimate cell lines to hepatitis A virus infection. J Virol. 1994;68:6064–6068. doi: 10.1128/jvi.68.9.6064-6068.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fagan E, Yousef G, Brahm J, Garelick H, Mann G, Wolstenhome A, Portmann B, Harrison T, Mowbray J F, Mowat A, Zuckerman A, Williams R. Persistence of hepatitis A virus in fulminant hepatitis and after liver transplantation. J Med Virol. 1990;30:131–136. doi: 10.1002/jmv.1890300210. [DOI] [PubMed] [Google Scholar]
- 11.Gauss-Müller V, Deinhardt F. Effect of hepatitis A virus infection on cell metabolism in vitro. Proc Soc Exp Biol Med. 1984;175:10–15. doi: 10.3181/00379727-175-41757. [DOI] [PubMed] [Google Scholar]
- 12.Graff J, Normann A, Feinstone S M, Flehmig B. Nucleotide sequence of wild-type hepatitis A virus GBM in comparison with two cell culture-adapted variants. J Virol. 1994;68:548–554. doi: 10.1128/jvi.68.1.548-554.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gust I D, Feinstone S M. Hepatitis A. Boca Raton, Fla: CRC Press, Inc.; 1988. [Google Scholar]
- 14.Hinshaw V S, Olsen C W, Dybdahl-Sissoko N, Evans D. Apoptosis: a mechanism of cell killing by influenza A and B viruses. J Virol. 1994;68:3667–3673. doi: 10.1128/jvi.68.6.3667-3673.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jelachich M L, Lipton H L. Theiler’s murine encephalomyelitis virus kills restrictive but not permissive cells by apoptosis. J Virol. 1996;70:6856–6861. doi: 10.1128/jvi.70.10.6856-6861.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia X-Y, Tesar M, Summers D F, Ehrenfeld E. Replication of hepatitis A viruses with chimeric 5′ nontranslated regions. J Virol. 1996;70:2861–2868. doi: 10.1128/jvi.70.5.2861-2868.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerr J F R, Searle J, Harmon B V, Bishop C J. Apoptosis. In: Potten C S, editor. Perspectives on mammalian cell death. Oxford, England: Oxford University Press; 1987. pp. 93–128. [Google Scholar]
- 18.Kroemer G, Zamzami N, Susin S A. Mitochondrial control of apoptosis. Immunol Today. 1997;18:44–51. doi: 10.1016/s0167-5699(97)80014-x. [DOI] [PubMed] [Google Scholar]
- 19.Lemon S M, Jansen R W. A simple method for clonal selection of hepatitis A virus based on recovery of virus from radioimmunofocus overlays. J Virol Methods. 1985;11:171–176. doi: 10.1016/0166-0934(85)90040-0. [DOI] [PubMed] [Google Scholar]
- 20.Lemon S M, Murphy P C, Shields P A, Ping L-H, Feinstone S M, Cromeans T, Jansen R W. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol. 1991;65:2056–2065. doi: 10.1128/jvi.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine B, Huang Q, Isaacs J T, Reed J C, Griffin D E, Hardwick J M. Conversion of lytic to persistent alphavirus infection by the bcl-2 cellular oncogene. Nature. 1993;361:739–742. doi: 10.1038/361739a0. [DOI] [PubMed] [Google Scholar]
- 22.Macchia S, Latorre P, Pagnotti P, Perez-Bercoff R. Abstracts of the 10th International Congress of Virology. 1996. Inhibition of cap-dependent protein synthesis induced by protein 2A of hepatitis A virus, abstr. PW02-2; p. 95. [Google Scholar]
- 23.Morace G, Pisani G, Benoduce F, Divizia M, Panà A. Mutations in the 3A genomic region of two cytopathic strains of hepatitis A virus isolated in Italy. Virus Res. 1993;28:187–194. doi: 10.1016/0168-1702(93)90135-a. [DOI] [PubMed] [Google Scholar]
- 24.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 25.Nasser A M, Metcalf T G. Production of cytopathology in FRhK-4 cells by BS-C-1-passaged hepatitis A virus. Appl Environ Microbiol. 1987;53:2967–2971. doi: 10.1128/aem.53.12.2967-2971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nüesch J P F, Weitz M, Siegl G. Proteins specifically binding to the 3′ untranslated region of hepatitis A virus RNA in persistently infected cells. Arch Virol. 1993;128:65–79. doi: 10.1007/BF01309789. [DOI] [PubMed] [Google Scholar]
- 27.Palmenberg A C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 28.Pisani G, Beneduce F, Gauss-Müller V, Morace G. Recombinant expression of hepatitis A virus protein 3A: interaction with membranes. Biochem Biophys Res Commun. 1995;211:627–638. doi: 10.1006/bbrc.1995.1859. [DOI] [PubMed] [Google Scholar]
- 29.Razvi E S, Welsh R M. Programmed cell death of T lymphocytes during acute viral infection: a mechanism for virus-induced immune deficiency. J Virol. 1993;67:5754–5765. doi: 10.1128/jvi.67.10.5754-5765.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tolskaya E A, Romanova L I, Kolesnikova M S, Ivannikova T A, Smirnova E A, Raikhlin N T, Agol V I. Apoptosis-inducing and apoptosis-preventing functions of poliovirus. J Virol. 1995;69:1181–1189. doi: 10.1128/jvi.69.2.1181-1189.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vallbracht A, Hofmann L, Wurster K G, Flehmig B. Persistent infection of human fibroblasts by hepatitis A virus. J Gen Virol. 1984;65:609–615. doi: 10.1099/0022-1317-65-3-609. [DOI] [PubMed] [Google Scholar]
- 32.Venuti A, Di Russo C, Del Grosso M, Patti A-M, Ruggeri F, De Stasio P R, Martiniello M G, Pagnotti P, Degener A M, Midulla M, Panà A, Perez-Bercoff R. Isolation and molecular cloning of a fast-growing strain of human hepatitis A virus from its double-stranded replicative form. J Virol. 1985;56:579–588. doi: 10.1128/jvi.56.2.579-588.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyllie A H. Glucocorticoid-induced thymocyte apoptosis with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Chao S-F, Ping L-H, Grace K, Clarke B, Lemon S M. An infectious cDNA clone of a cytopathic hepatitis A virus: genomic regions associated with rapid replication and cytopathic effect. Virology. 1995;212:686–697. doi: 10.1006/viro.1995.1526. [DOI] [PubMed] [Google Scholar]