Abstract
Our understanding of human immunodeficiency virus type 1 (HIV-1)-induced pathogenesis is hampered by the inability to detect HIV-1 gene expression in infected viable cells. In this report, we describe two HIV-1 reporter constructs that are replication competent and cytopathic in vivo. These constructs contain DNA regions of two different lengths that bear the cDNA for the murine heat-stable antigen in the vpr region of a CXCR4-tropic virus. We used the SCID-hu mouse model and these reporter viruses to perform detailed kinetic studies of HIV-1 infection of human thymocytes in vivo. We document that the CD4+/CD8+ thymocytes are the first to express virus and that this subset demonstrates the most rapid and extensive HIV-1-induced cell depletion. Following depletion of this subset, subsequent virus expression occurs predominantly in phenotypically CD4− cells, suggesting that CD4 down-regulation occurs in HIV-1-infected thymocytes in vivo. These results demonstrate the utility of these HIV-1 reporter constructs to monitor HIV pathogenesis in vitro and in vivo.
The capability to detect human immunodeficiency virus type 1 (HIV-1) gene expression in viable infected cells in vivo would improve our ability to investigate HIV pathogenesis. Current antibodies to HIV-1-encoded proteins on the cell surface either bind with weak affinity or bind to gp120, which is then shed from the cell surface. HIV-1 gene expression can be measured by intracellular staining for the p24 viral Gag protein (7). However, the cells must first be permeabilized; thus, viable cells cannot be assessed. In an attempt to overcome these limitations, cDNAs encoding various reporter proteins have been cloned into HIV-1. Replication of these reporter constructs results in expression of the encoded protein, which can then be detected in viable cells by biochemical means or, in the case of cell surface reporter molecules, by flow cytometry (12, 28). However, in all of these previous constructs, nef, env, or both genes have been functionally deleted, rendering these viruses either replication attenuated in the case of nef-deleted viruses or replication defective in the case of env-deleted viruses.
While nef is required for efficient in vivo replication and cytopathicity of both HIV-1 (15) and simian immunodeficiency virus (9, 19), vpr is not (3, 10, 14). Our laboratory has previously shown that deletion of vpr has little if any effect on the in vivo replication and pathogenicity of HIV-1NL4-3 in human thymic implants in SCID-hu mice (3). To perform detailed studies of HIV infection in the SCID-hu system, we made two HIV-1 reporter constructs by cloning the cell surface molecule, murine heat-stable antigen (HSA) (18), into the vpr gene region of HIV-1NL4-3 (1), a CXCR4-tropic strain. These constructs differ only in the length of the inserted cDNA.
The SCID-hu mouse is a small animal model for HIV-1 pathogenesis (2, 4, 22, 26, 30) and is constructed by surgical implantation of human fetal liver and thymus under the kidney capsule of severe combined immunodeficient (SCID) mice (25, 27). This results in development of a conjoint organ (Thy/Liv) capable of supporting thymopoeisis for up to 1 year (27). We and others have previously demonstrated HIV-1 replication and subsequent depletion of human CD4+ thymocytes after infection of the Thy/Liv implant (2, 4, 15–17, 22, 26, 30).
In this report, we have examined the potential for the reporter constructs to replicate and induce human CD4+ cell loss in vivo in the SCID-hu mouse. We found that these viruses were pathogenic in vivo and induced expression of murine HSA on the surface of human thymocytes. This allowed us to investigate the extent of viral gene expression during various stages of infection and to monitor the spread of virus through various thymocyte subsets as infection progressed.
MATERIALS AND METHODS
Construction of reporter viruses.
To clone murine HSA (CD24) into vpr of the NL4-3 strain of HIV-1, the plasmid pNL4-3 (1) and the plasmid pSL87c4-1 containing the murine HSA cDNA (18) were used. To facilitate cloning, pNL4-3 was digested with NdeI and EcoRI, and the 622-bp fragment containing vpr sequences was then subcloned into Bluescript KS (Stratagene, La Jolla, Calif.). An XbaI site was introduced into the subcloned vpr by changing the nucleotide at position 5625 from T to C, by using PCR-based site-directed mutagenesis kits (Stratagene). This created the plasmid BS KS/r-Xba I.
To potentially optimize expression of the reporter gene, further mutagenesis was performed to silence the start codon of vpr and two potential start codons in the 3′ end of vif. The nucleotides at positions 5559, 5605, and 5611 were mutated from A, A, and G to G, C, and A, respectively, to construct BS KS/r-Xba I/3M.
To allow insertion of the reporter construct, BS KS/r-Xba I/3M was partially digested with XbaI and EcoRI to remove a 119-bp fragment of vpr. The plasmid pSL87c4-1 was fully digested with XbaI and EcoRI, generating a 402-bp fragment containing 231 bp of HSA cDNA and 171 bp of the plasmid pSL87c4-1. This 402-bp fragment was ligated into the digested BS KS/r-Xba I/3M, replacing the original vpr sequences (BS KS/HSAS-Xba I/3M). The subcloned fragment containing HSA in vpr was then liberated from BS KS/HSAS-Xba I/3M by digestion with PflMI and EcoRI and inserted into a pNL4-3 backbone previously digested with the same enzymes. The resulting construct (NL-v-HSAL) is 283 bp longer than pNL4-3 (Fig. 1).
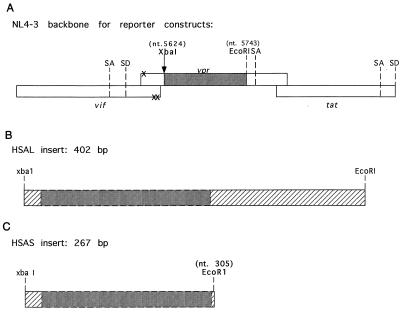
FIG. 1.
Schematic representation of reporter constructs. (A) A diagram of the location of the inserted cDNA (not to scale) and the new XbaI site introduced at nucleotide 5624 is shown. The three nucleotide changes created to silence the start codon of vpr and two potential start codons in the 3′ end of vif are indicated by X. (B and C) The relative sizes of the two HSA cDNAs used to create NL-r-HSAL and NL-r-HSAS are represented. In panel C, the new EcoRI site created at nucleotide 305 is indicated. nt, nucleotide. SA, splice acceptor; SD, splice donor.
To take full advantage of the small size of the HSA coding region, an additional EcoRI site was created in pSL87c4-1 by changing the nucleotides at positions 307, 309, and 310 from C, C, and A to A, T, and C, respectively. Digestion of the mutated pSL87c4-1 with XbaI and EcoRI yielded a 267-bp fragment that contained the HSA cDNA and only 36 extraneous bp of flanking sequences. To insert this fragment into vpr, BS KS/r-Xba I/3M (described above) was digested with PflMI and EcoRI, and the 441-bp fragment containing the mutated HIV sequences was then ligated into pNL4-3, which had been digested with the same enzymes. This created the plasmid pNL4-3-Xba I/3M, which contained a new XbaI site in the vpr gene. pNL4-3-Xba I/3M was then digested with XbaI and EcoRI, and the 267-bp fragment containing the cDNA for HSA was ligated in. The resulting construct, designated NL-r-HSAS (HSAS), is only 148 bp longer than wild-type pNL4-3 (Fig. 1).
The control virus, pNL4-3-Δvpr, was constructed by digestion of BS KS/r-XbaI with PflMI and EcoRI to remove NL4-3 sequences, including vpr and the newly introduced XbaI site (see above). This fragment was ligated into a pNL4-3 backbone that had also been digested with PflMI and EcoRI (pNL4-3-XbaI). pNL4-3-XbaI was then digested with XbaI and EcoRI to remove 119 bp of vpr and was blunt end ligated, resulting in the plasmid pNL4-3-Δvpr.
Preparation of virus stocks.
Stocks of all viruses were made by electroporation of 30 μg of infectious proviral DNA (6) into 107 mycoplasma-free CEM cells. Virus production was quantitated by enzyme-linked immunosorbent assay for p24 Gag, and expression of HSA on the cell surface was determined on day 8 by flow cytometric analysis. Infectious units (IU) were determined by limiting dilution on phytohemagglutinin (PHA)-stimulated human peripheral blood mononuclear cells (PBMC), by using twofold dilutions of virus plated in duplicate. Human PBMC were derived from Red Cross leukopacks, following centrifugation over Ficoll-Hypaque and depletion of macrophages by adherence to plastic for 72 h.
To confirm the DNA sequence of the reporter constructs, virus stocks were used to infect 106 PHA-stimulated PBMC. Three days postinfection, DNA was extracted from the infected cells, and the TD1-TD2 primer pair was used to amplify the proviral DNA in a nonradioactive PCR under the conditions described below (see “Quantitative PCR”). The TD1-TD2 primer pair is specific for nucleotides 5416 to 5448 (GGACGTATAGTTAGTCCTAGGTGTGAATATCAA) and nucleotides 5785 to 5812 (GTCGACATAGCAGAATAGGCGTTACTCG) of HIV-1NL4-3, respectively. These primers flank the inserted HSA cDNA sequences. The PCR products were isolated by use of a Microcon 100 column (Amicon Inc., Beverly, Mass.). Sequencing was performed on an ABI373 automated DNA sequencer (Applied Biosystems Instruments, Foster City, Calif.) by using the VF primer specific for nucleotides 5471 through 5490 of HIV-1NL4-3 (CTCTACAGTACTTGGCACTA).
Construction and infection of SCID-hu mice.
C.B.-17 mice homozygous for the SCID genetic defect (5) were bred at the University of California at Los Angeles, housed in a biosafety level 3 animal facility, and maintained free of antibiotics as approved by the UCLA Animal Research Committee. SCID-hu mice were constructed by coimplantation of pieces of human fetal thymus and liver from a single human fetal donor under the left kidney capsule of SCID mice (2, 15, 27).
Prior to infection, all virus stocks were diluted in RPMI medium with 1% fetal calf serum, and approximately 100 μl was directly injected into the implant (2, 15). Mock infections were performed with supernatants from mock-electroporated CEM cells diluted in the same manner as the virus stocks. Implants were infected with either 52 ng (approximately 210 IU) of NL-r-HSAL or with 20 ng (approximately 570 IU) of NL-r-HSAS. Infections were carried out on a staggered basis, and biopsies were performed on the mice as a group in order to generate both longitudinal and cross-sectional data (17) for the time points indicated.
Sequential biopsy samples of approximately 25% of each implant were obtained at the indicated times, while the animals were sedated. Percent provirus expression shown in Table 1 was calculated (assuming one provirus per cell) by use of the following formula: percent provirus expression = [(% HSA+ cells/100) × 105]/[copies of HIV/105 cells].
TABLE 1.
Characterization of NL-r-HSAS pathogenesis in vivoa
| Implant type and no. | Days p.i.b | % CD4+/CD8+ cells | No. of HIV DNA copies/105 cells | % HSA+ cells | % Provirus expressionc |
|---|---|---|---|---|---|
| NL-r-HSAS infected | |||||
| 28 | 20 | 79 | 26 | 0.2 | BGd |
| 40 | 20 | 90 | 384 | 0.3 | 68 |
| 34 | 21 | 81 | 7,214 | 0.1 | BG |
| 36 | 21 | 67 | 5,911 | 1.1 | 19 |
| 39 | 21 | 68 | 5,058 | 1.8 | 35 |
| 19 | 22 | 58 | 1,240 | 0.7 | 60 |
| 20 | 22 | 90 | 1,197 | 0.8 | 64 |
| 33 | 22 | 57 | 897 | 0.5 | 58 |
| 16 | 23 | 73 | 34,468 | 3.0 | 9 |
| 15 | 23 | 81 | 6,000 | 1.8 | 30 |
| 2 | 24 | 85 | 3,324 | 1.9 | 58 |
| 5 | 24 | 85 | 3,424 | 1.3 | 39 |
| 8 | 24 | 83 | 5,846 | 2.5 | 43 |
| 30 | 27 | 62 | 39,458 | 2.5 | 6 |
| 28 | 27 | 34 | 45,300 | 7.1 | 16 |
| 34 | 28 | 83 | 1,106 | 0.1 | BG |
| 41 | 28 | 31 | 59,643 | 7.1 | 12 |
| 39 | 28 | 80 | 17,113 | 1.2 | 7 |
| 33 | 29 | 54 | 42,633 | 5.9 | 14 |
| 32 | 29 | 9 | 76,113 | 4.3 | 6 |
| 19 | 29 | 65 | 38,199 | 3.8 | 10 |
| 16 | 30 | 73 | 51,181 | 2.7 | 5 |
| 21 | 30 | 82 | 34,748 | 2.8 | 8 |
| 15 | 30 | 66 | 19,418 | 8.6 | 44 |
| 9 | 31 | 12 | 46,561 | 6.5 | 14 |
| 32 | 32 | 6 | 40,056 | 4.5 | 11 |
| 28 | 34 | 51 | 19,781 | 1.9 | 10 |
| 39 | 35 | 9 | 21,688 | 3.9 | 18 |
| 41 | 35 | 10 | 30,377 | 3.3 | 11 |
| 33 | 36 | 63 | 39,152 | 0.7 | 2 |
| 21 | 37 | 6 | 38,282 | 2.7 | 7 |
| 15 | 37 | 29 | 17,953 | 3.8 | 21 |
| 8 | 38 | 20 | 4,729 | 6.7 | 141 |
| 28 | 41 | 2 | 10,108 | 4.9 | 48 |
| 21 | 44 | 1 | 11,912 | 7.9 | 67 |
| 9 | 45 | 1 | 8,766 | 4.4 | 50 |
| Mock infected | |||||
| 27 | 20 | 85 | NDe | 0.1 | |
| 31 | 29 | 85 | ND | <0.1 | |
| 11 | 31 | 85 | ND | 0.2 | |
| 31 | 36 | 81 | ND | <0.1 | |
| 11 | 38 | 81 | ND | <0.1 | |
| 31 | 43 | 70 | ND | 0.2 | |
| 35 | 43 | 80 | ND | <0.1 | |
Implants from mice infected with NL-r-HSAS or mock infected were assessed by flow cytometry for CD4 and CD8 subset distribution of CD45+ thymocytes and for expression of murine HSA. Proviral burden was determined by quantitative PCR and is shown as copies of HIV DNA per 105 cells. Percent provirus expression was calculated as described in Materials and Methods, assuming one provirus per cell.
p.i., postinfection.
Percent provirus expression was not calculated for mock-infected implants, since the levels of HSA in these implants are considered to be background.
BG, HSA expression was not above the highest level of HSA expression (0.2%) observed in mock-infected implants.
ND, not detectable.
Four-color flow cytometry.
Single-cell suspensions were prepared from biopsy specimens and washed once in phosphate-buffered saline. Cells (106) were then costained with monoclonal antibodies to CD4, CD8, CD45 (Becton Dickinson, Mountain View, Calif.), and murine HSA (Pharmingen, San Diego, Calif.). These antibodies were directly conjugated to allophycocyanin, phycoerythrin, fluorescein isothiocyanate, and biotin, respectively. Streptavidin red 613 (Becton Dickinson) was used as a second-step reagent. Stained cells were fixed in 2% paraformaldehyde. At all time points, mouse immunoglobulin G1 (IgG1; conjugated with allo phycocyanin, phycoerythrin, and fluorescein isothiocyanate) and rat IgG2b(κ) (conjugated with biotin) were used as antibody isotype controls. Ten to 20 thousand events were acquired on a FACStarplus flow cytometer (Becton-Dickinson), and the data were analyzed by using the CELLQuest program (Becton-Dickinson). Forward versus side scatter analysis of mock-infected implants was used to gate on the live thymocyte population. Further gating was performed to include only CD45+ cells, thus excluding any murine cells from the analysis.
Quantitative PCR.
Biopsy samples were washed once in phosphate-buffered saline, and the DNA was extracted by using the QIAamp blood kit (Qiagen, Chatsworth, Calif.). Total nucleic acids obtained from this procedure were then subjected to quantitative PCR, as previously described (2, 32, 33). All quantitative PCR amplifications were performed with one of the primers containing a 32P-end-labeled nucleotide. Briefly, HIV DNA was detected by using the M667-AA55 primer pair specific for the R/U5 region of the viral long terminal repeat. Twenty-five cycles of amplification were used. Standard curves for HIV-1 DNA were generated by using four- or fivefold dilutions of cloned NL-r-HSAS DNA linearized with BamHI in carrier DNA from normal human PBMC. HSA sequences were quantitated by using the TD1-TD2 primer pair specific for nucleotides 5416 to 5448 of HIV-1NL4-3, which flank the inserted HSA sequences (see “Preparation of virus stocks” above). Thirty cycles of amplification were used. To quantitate HIV genomes and copies of HSA cDNA per human cell, replicate samples were analyzed for human DNA with primers specific for nucleotides 14 to 33 and 123 to 104 of the human β-globin gene (23, 32), by using 21 cycles of amplification. Standard curves for human β-globin were generated from 3- and 10-fold dilutions of PBMC DNA. Values were obtained by interpolation from the standard curve, by using an Ambis radioanalytic imager.
RESULTS
Construction of reporter constructs.
NL4-3 is a CXCR4-tropic molecular clone of HIV-1 (1) that has been well characterized in the SCID-hu mouse model (2, 15–17). It is also one of the fastest replicating viral strains in this model, causing rapid and severe depletion of CD4-bearing thymocytes (16) due to high levels of CXCR4 in this organ (21). For these reasons, NL4-3 was chosen as the HIV-1 backbone for the reporter constructs. Murine HSA was selected as the reporter gene because of the small size of the cDNA (231 nucleotides), the commercial availability of antibodies, and the lack of cross-reactivity of those antibodies with human HSA. HSA has been used successfully in previous HIV reporter constructs (12). vpr was selected as the site for insertion of the reporter gene (Fig. 1) because, unlike nef deletion mutants of NL4-3, which are attenuated for replication and cytopathicity in the SCID-hu mouse model (15), a vpr deletion mutant replicates and depletes CD4-bearing cells with kinetics similar to that of wild-type virus (3). To facilitate cloning into vpr, an XbaI site was introduced at nucleotide 5624, four nucleotides 3′ from the end of vif (Fig. 1) (see Materials and Methods). The existing EcoRI site at nucleotide 5743 was used as the second restriction site.
Two reporter constructs were made. NL-r-HSAL was constructed to contain a 402-bp fragment which includes the 231 bp of HSA cDNA and 171 bp of extraneous plasmid sequences (Fig. 1). NL-r-HSAL is 283 bp longer than wild-type NL4-3. To determine whether the length of the insert affected replication and cytopathicity, a smaller insert was created. To this end, an additional EcoRI site was introduced at nucleotide 305, two nucleotides 3′ of the HSA stop codon, allowing removal of an additional 135 bp of flanking sequences. When ligated into vpr, this reporter construct is 148 bp longer than NL4-3, contains only 36 extraneous bp, and is referred to as NL-r-HSAS (Fig. 1). To eliminate the potential of alternate translation initiation sites and thus maximize the expression of the reporter gene, the vpr start codon and two potential start codons in the 3′ end of vif were mutated in both constructs (Fig. 1). However, constructs without these mutations have not yet been tested to determine whether these alterations enhance reporter gene expression.
In vitro replication of the reporter constructs.
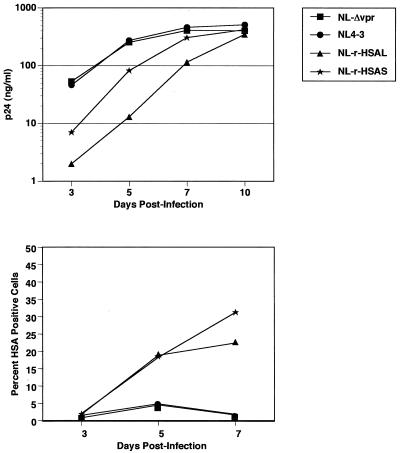
To investigate the kinetics of in vitro replication and HSA expression of NL-r-HSAS and NL-r-HSAL, PHA-stimulated PBMC were infected with 170 ng of p24 Gag of either virus. To determine if deletion of vpr or insertion of HSA cDNA affected replication of the constructs, PBMC were similarly infected with wild-type NL4-3 or NL-Δvpr. As previously reported (3, 11), deletion of vpr did not affect replication kinetics in mitogen-stimulated PBMC (Fig. 2). However, insertion of HSA slowed replication. NL-r-HSAL demonstrated the greatest delay in replication kinetics, whereas NL-r-HSAS replication was somewhat attenuated compared to that of wild-type NL4-3, although not as much as that of NL-r-HSAL. Expression of the reporter gene was observed by flow cytometry in both NL-r-HSAS- and NL-r-HSAL-infected cultures as early as 3 days postinfection (Fig. 2). Similar data were obtained when input virus was adjusted for infectious units (data not shown). These results demonstrate that although the replication of both reporter constructs is somewhat attenuated, both constructs are replication competent in vitro. The results also suggest that the size of the insert may be an important determinant in replication kinetics.
FIG. 2.
In vitro replication kinetics of reporter constructs in PHA-stimulated PBMC. (Upper panel) Viral replication was quantitated by enzyme-linked immunosorbent assay for p24 Gag antigen at the indicated days postinfection. (Lower panel) Cells were stained for expression of HSA on the indicated days postinfection and analyzed by flow cytometry. Gates were set based on isotype control staining of live cells.
Thymocyte subset distribution of the reporter constructs.
To determine whether the reporter gene is expressed in vivo and, if so, which thymocytes express HIV-encoded proteins, implants were infected with either NL-r-HSAS or NL-r-HSAL. Standard infections in our laboratory with wild-type NL4-3 are performed with 100 IU of virus (2, 17). In an attempt to overcome the attenuated replication of the reporter constructs, higher concentrations of virus were used in vivo (approximately two- and sixfold higher IU for NL-r-HSAL and NL-r-HSAS, respectively). We were unable to perform infections with higher than a twofold concentration of NL-r-HSAL, since higher-titer stocks were not available. Although we performed in vivo studies with both reporter viruses, since NL-r-HSAL was more attenuated in vitro, all subsequent data shown are derived from implants infected with NL-r-HSAS.
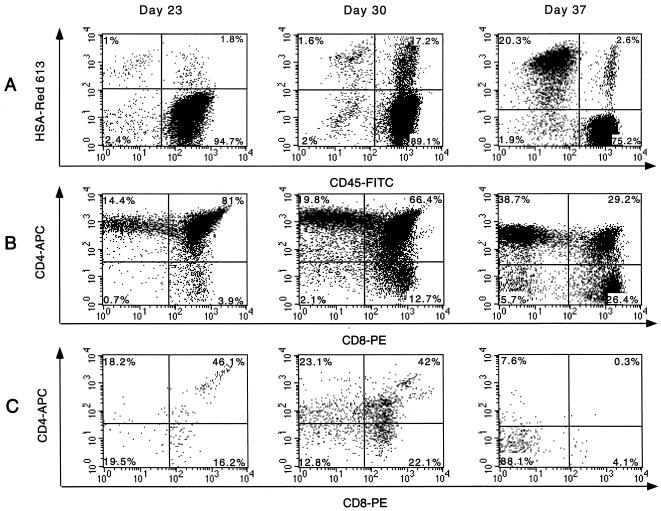
Single-cell suspensions obtained from biopsy samples were costained for the human surface markers CD4, CD8, and CD45, as well as for murine HSA. As shown in Fig. 3A, HSA+ murine cells can be detected in Thy/Liv implants although usually not to the high levels observed in this implant. To ensure that murine cells were not included in the analysis and only human thymocytes were studied, gates were set on CD45+ cells in all tables and figures, with the exception of Fig. 3A. The phenotype of thymocytes expressing HSA was determined by gating on the CD45+/HSA+ population and analyzing these cells for CD4 and CD8 surface markers (Fig. 3C). Reporter gene expression and CD4+ cell loss were observed in implants infected with either construct, although somewhat delayed kinetics were found for NL-r-HSAL-infected implants relative to implants infected with NL-r-HSAS (Table 1, Fig. 3, and data not shown). With both constructs, the earliest thymocytes to express the reporter gene were the immature CD4high/CD8high cells. Minor CD4+ cell loss was observed in several implants concurrent with the earliest detection of HSA expression (Table 1). Reporter gene expression was observed shortly thereafter in the CD4−/CD8low thymocytes (Fig. 3). Together, the CD4high/CD8high and CD4−/CD8low subsets represented the majority of the HSA+ population during both the early and intermediate stages of infection. Later in the course of infection, reporter gene expression shifted into both the CD4low/CD8− and CD4−/CD8− subsets, and when depletion of CD4-bearing cells was severe, the CD4−/CD8− subset often comprised the major subpopulation of HSA+ cells. The CD4−/CD8+ subset, however, also continued to express the reporter construct, often to high percentages (Fig. 3 and data not shown). The emergence of these virally infected subsets over time was slightly more pronounced in implants infected with NL-r-HSAL (data not shown). It is interesting to note that expression of HSA in implant 15 occurred almost exclusively in the CD4−/CD8− subset at 37 days postinfection, despite the obvious presence of CD4+ thymocyte subsets (Fig. 3). It is also interesting to note that with the exception of the CD4high/CD8high population, HSA expression was rarely observed in CD4high cells. This may reflect down-regulation of the CD4 molecule. This phenotype cannot be explained by lower overall surface molecule expression due to cell death, since the majority of these HSA+ cells were CD45high, suggesting that specific mechanisms of down-regulation are operative. While further studies are needed to better understand these observations, it is clear that a large percentage of virus-expressing cells during HIV-1 infection of the Thy/Liv implant are phenotypically CD4− cells.
FIG. 3.
Subset distribution of HSA-expressing thymocytes. Thymocytes obtained from biopsy samples of implant 15 at the indicated times postinfection were costained for human CD4, CD8, and CD45, as well as murine HSA. APC, allophycocyanin; Red 613, streptavidin red 613; PE, phycoerythrin; FITC, fluorescein isothiocyanate. (A) Gating was performed on total live thymocytes. The percentage of each indicated subset is based on relative distribution in total live thymocytes. (B) Gates were set on live CD45+ (human) cells, and the CD4 and CD8 distribution of these CD45+ cells is shown. The percentage of each indicated subset is based on relative distribution in CD45+ thymocytes. (C) Gates were further narrowed to include only the CD45+/HSA+ subset. The profiles in panel C illustrate CD4 and CD8 distribution of these CD45+/HSA+ cells and demonstrate the subsets of human thymocytes expressing HIV genes. Percentages of CD45+/HSA+ cells are shown in each quadrant.
Kinetics of CD4+ cell depletion.
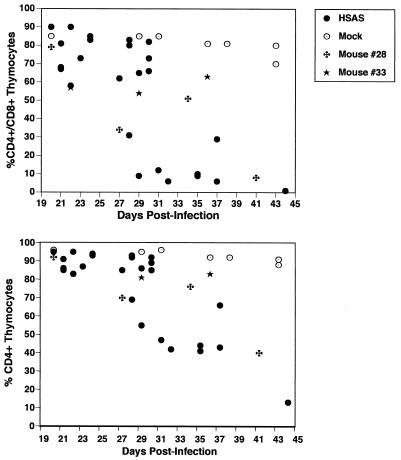
We have previously reported that infection of Thy/Liv implants with 100 IU of HIV-1NL4-3 induces CD4+ cell depletion, beginning approximately 19 days postinfection and resulting in almost complete loss of all CD4+ cells by 30 days postinfection (2, 16, 17). NL-r-HSAL also elicits CD4+ cell loss but with delayed kinetics relative to NL-r-HSAS (data not shown). To examine the extent and kinetics of CD4+ cell loss induced by NL-r-HSAS, CD45+ (human) thymocytes were analyzed by flow cytometry to determine thymocyte subset distribution. Mock-infected implants maintained normal levels of CD4+ thymocytes throughout the course of the experiment (Table 1, Fig. 4, and data not shown). In contrast, depletion of CD4+/CD8+ cells was observed in two of three NL-r-HSAS-infected implants at 21 days postinfection (Table 1). An almost total loss of the immature CD4+/CD8+ cells was first observed in one implant (no. 32) at 29 days postinfection, and by 37 days postinfection, severe depletion of this cell subset was observed in all implants tested. As previously reported for HIV-1NL4-3, this decline in CD4+ cells occurred simultaneously with a drop in proviral burden (Fig. 5). This is most likely due to the loss of potential CD4+ target cells (2, 16, 17).
FIG. 4.
NL-r-HSAS-induced depletion of CD4-bearing cells. Biopsy specimens of mock-infected (open circles) and NL-r-HSAS-infected (solid circles) implants were collected at the indicated times postinfection and assayed by flow cytometry for percent CD4+/CD8+ (upper panel) and total CD4-bearing cells (lower panel). Percent CD4+ thymocytes includes both immature CD4+/CD8+ and mature CD4+/CD8− cells. Each point represents a single implant. The crosses and stars represent multiple biopsy samples from implants 28 and 33, respectively.
FIG. 5.
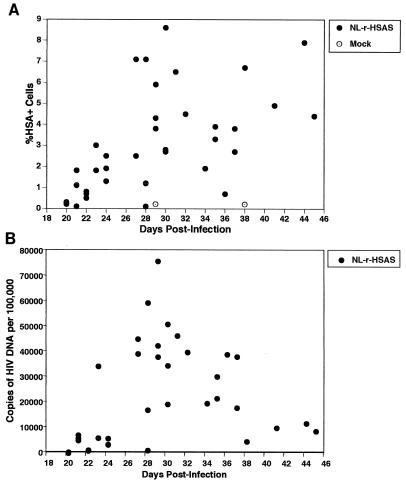
In vivo replication and expression of NL-r-HSAS. The percentages of HSA+ cells (A) and the copies of NL-r-HSAS proviral DNA (B) at the indicated times postinfection are shown. Each point represents an individual implant. Percentages of HSA+ cells and CD4+/CD8+ thymocytes were determined by flow cytometry. Copy numbers of NL-r-HSAS DNA per 105 cells were determined by quantitative PCR.
Although depletion of the CD4+/CD8+ subset began at 21 days postinfection, several implants either did not deplete further by the time of the second biopsy (implants 16, 34, and 33) or showed a slight increase in this subset (implants 19 and 39). One implant (no. 28) showed a dramatic drop in CD4+/CD8+ cells by 27 days postinfection, but this subset had increased again at the time of biopsy 7 days later. Subsequently, this implant lost an average of approximately 7% CD4+/CD8+ cells per day between days 34 and 41 postinfection (Table 1 and Fig. 4). Implant 39 showed a similar pattern, where a transient increase in CD4+/CD8+ cells was followed by a loss of approximately 10% CD4+/CD8+ cells per day between 28 and 35 days postinfection (Table 1). This transient increase in CD4+/CD8+ cells is consistent with our earlier reports that the SCID-hu mouse is a dynamic system, and depletion of CD4+ cells therefore represents a balance between HIV-induced depletion and the regeneration of new thymocytes. To this end, we have previously reported a resurgence of thymopoiesis in an HIV-1NL4-3-infected implant in which CD4+ cells had been severely depleted and proviral burden had decreased (15) and in HIV-1NL4-3-infected implants treated with antiretroviral drugs to halt virus replication (31). The observed transient increase of immature thymocytes may therefore reflect the attenuated phenotype of the NL-r-HSAS construct, which allows for ongoing thymopoiesis to temporarily increase thymocyte numbers. However, once thymocyte depletion resumed, depletion of CD4+/CD8+ thymocytes occurred rapidly, with a kinetics similar to that observed for the loss of CD4+ cells in wild-type HIV-1NL4-3-infected implants (17). Unlike HIV-1NL4-3 (17), infection of Thy/Liv implants with NL-r-HSAS did not result in rapid depletion of CD4+/CD8− cells. However, as demonstrated by implant 28 (Table 1 and Fig. 4), depletion of this subset is likely to occur with sufficient time.
Kinetics of in vivo replication and reporter gene expression.
Infections and biopsies were performed to provide both cross-sectional and longitudinal data, allowing a detailed kinetic study of the relationship between HIV-1 replication and gene expression. Thymocytes obtained by biopsy were subjected to quantitative PCR to assess proviral burden and by flow cytometry to assess reporter gene expression. As shown in Table 1 and Fig. 5B at 21 days postinfection, all NL-r-HSAS-infected implants tested (three of three) contained significant levels of proviral DNA. Expression of the reporter gene was clearly above background levels in two of three implants (Table 1 and Fig. 5A). Both viral DNA burden and HSA gene expression continued to increase until approximately day 27, demonstrating in vivo replication of this reporter virus (Table 1 and Fig. 5). Between 27 and 37 days postinfection, proviral burden remained relatively constant, with a median proviral load of 38,300 copies of HIV-1 DNA per 105 cells. This proviral burden is similar to that previously observed in Thy/Liv implants infected with wild-type HIV-1NL4-3, although the peak of viral replication occurs somewhat later with NL-r-HSAS (16, 17). Between days 27 and 37, a median of 4% of all CD45+ cells expressed the reporter gene, although up to 8.6% expression was observed in an individual implant (implant 15, day 30). As previously reported for wild-type HIV-1NL4-3, proviral load then began to decrease (2, 16, 17), and a median of 9,000 copies of HIV-1 DNA per 105 cells was observed between 38 and 45 days postinfection (Table 1 and Fig. 5). However, the levels of HSA expression did not drop after day 37, with a median of 6% of cells expressing HSA.
Relative expression of provirus.
To determine relative amounts of the proviral DNA expressed at each time point, the percent provirus expression was calculated by assuming one copy of proviral DNA per cell (Table 1). Provirus expression was high early in infection (days 20 through 24), with as many as 60% (a median of 39%) of infected cells expressing the reporter gene. By day 27, when proviral DNA was at the highest levels, relative proviral expression dropped and stayed low through day 37, with a median of 10% proviral expression. Late in infection (days 38 through 45), provirus expression rose again to a median of 58%, reflecting the drop in proviral DNA and the maintenance of high HSA expression. It is not clear whether the increase in the levels of proviral expression represents waves of virus replication or accumulation of cells more resistant to virally induced cell death.
Taken together, these data clearly demonstrate that NL-r-HSAS replicates in vivo, is cytopathic for CD4+ cells, and efficiently expresses the reporter gene, making this a useful virus for investigating the pathogenic process in an in vivo model.
Retention of HSA DNA.
The long time frame and complicated selection pressures operating in vivo could easily select for viruses that delete reporter sequences. To ensure that deletion of HSA sequences was not occurring in vivo, copies of HSA DNA were quantitated by DNA PCR in 19 biopsy samples from six Thy/Liv implants, at time points ranging from 23 to 41 days postinfection. Following PCR amplification using the TD1-TD2 primer pair, no evidence of deletion was observed, in that PCR products smaller than the predicted size of 543 bp were not detected. In addition, the copy number of HSA-containing sequences was not less than the number of viral long terminal repeat sequences quantitatively amplified from the same sample (data not shown). These data are consistent with the high levels of proviral expression observed up to 37 days postinfection and demonstrate that no detectable selection of HSA-deleted virions occurred in vivo.
DISCUSSION
This is the first report of HIV-1 reporter constructs that are pathogenic in vivo. Both NL-r-HSAL and NL-r-HSAS contain full-length nef, giving these constructs an in vivo replication advantage over other reporter viruses which are nef deleted (12, 28). However, the in vitro replication kinetics of NL-r-HSAL, when compared to NL-r-HSAS, suggest that in addition to the site of gene insertion, the length of the insert may also contribute to an attenuated phenotype.
The greatest CD4+ cell depletion due to these viruses was observed in the immature CD4+/CD8+ subset. These thymocytes express the highest levels of the coreceptor, CXCR4, in the thymus (21) and are the first subset to express virus both in vivo (Fig. 3) and in thymocytes cultured in vitro (21). Thus, it is likely that high levels of both primary and coreceptor molecules likely contribute to the efficient infection of this subset. The mature CD4+/CD8− thymocyte subset expresses lower levels of CXCR4 (21), does not express high levels of virus during any stage of the infection in vivo, and is not depleted as rapidly in vivo as the immature thymocytes. While lower CXCR4 expression in CD4+/CD8− cells may contribute to these differences, it is not likely to be the sole factor. vpr is important for the infection of nondividing cells (8, 13). The absence of vpr in NL-r-HSAS may inhibit productive infection of the mature CD4+ cells, reflecting their quiescent status. Alternatively, differences in transcription factors or other cell functions between these two thymocyte subsets may contribute to this phenomenon. However, it is clear that some CD4+/CD8− thymocytes can be infected, and the data strongly suggest that the number of CD4+/CD8− cells that express virus is underrepresented due to CD4 down-regulation. Aside from the CD4high/CD8high subset, HSA was rarely detected in CD4high cells. CD4low and CD4− thymocyte subsets emerge as infection progresses, giving a “falling rain” appearance in the CD4 versus CD8 profiles of infected implants (Fig. 3B). These subsets are not observed in mock-infected implants and are the same subsets which express virus (Fig. 3C), strongly supporting the theory of CD4 down-regulation. The pronounced emergence of these subsets with NL-r-HSAS infection may reflect the slightly more attenuated killing of infected mature cells by this virus than by HIV-1NL4-3. Taken together, these data suggest that infection of CD4+ mature cells does occur but that CD4 down-regulation occurs in vivo and masks the true phenotype of infected cells.
Virus expression in CD4− thymocytes is also consistent with infection of an immature CD4+ population, followed by subsequent differentiation into a CD4− mature thymocyte. To this end, we have recently shown that CD4+/CD8+ thymocytes infected in vitro with a nef-deleted reporter construct can differentiate into CD4−/CD8+ cells and express HIV (20). As we have previously suggested (20), export of virally infected CD8+ cells from the thymus could account for the presence of virally infected CD8+ cells in the periphery of HIV-infected individuals (24, 29).
In this study, we also examined the relationship between proviral burden and viral gene expression. The observed median of 6% HSA+ cells is consistent with our previous determination that approximately 10% of thymocytes in SCID-hu thymic implants harbor productive wild-type provirus (17). While our previous data were obtained by coculturing infected thymocytes with PHA-stimulated PBMC for 7 days, the 6% level of HSA+ expression in the SCID-hu mouse represents only a snapshot of viral replication and is therefore likely to underrepresent the number of productively infected cells.
Understanding the pathogenic mechanisms of HIV-1 in a lymphoid organ is critical to the development of vaccines and therapeutic strategies. With these pathogenic HIV reporter constructs, we will be better able to dissect the mechanism(s) of HIV-1-induced cell death, to examine the viral and cellular factors involved in cellular tropism, and to explore the relationship between viral replication and gene expression. These viruses may also be valuable tools to help evaluate the efficacy of antiviral therapeutic strategies.
ACKNOWLEDGMENTS
We thank R. Keith Humphries for the pSL87c4-1 plasmid containing murine HSA and Amelia Kacena, Ruth Cortado, Junli Zha, and Greg Bristol for excellent technical assistance.
This work was supported by the Universitywide AIDS Research Program (R96-LA-139) (B.D.J.) and by NIH grants AI36059 and AI DK36554 (J.A.Z.). J.A.Z. is an Elizabeth Glaser Scientist supported by the Pediatric AIDS Foundation.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldrovandi G M, Feuer G, Gao L, Jamieson B, Kristeva M, Chen I S Y, Zack J A. The SCID-hu mouse as a model for HIV-1 infection. Nature. 1993;363:732–736. doi: 10.1038/363732a0. [DOI] [PubMed] [Google Scholar]
- 3.Aldrovandi G M, Zack J A. Replication and pathogenicity of human immunodeficiency virus type 1 accessory gene mutants in SCID-hu mice. J Virol. 1996;70:1505–1511. doi: 10.1128/jvi.70.3.1505-1511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonyhadi M L, Rabin L, Salimi S, Brown D A, Kosek J, McCune J M, Kaneshima H. HIV induces thymus depletion in vivo. Nature. 1993;363:728–736. doi: 10.1038/363728a0. [DOI] [PubMed] [Google Scholar]
- 5.Bosma G C, Custer R P, Bosma M J. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 6.Cann A J, Koyanagi Y, Chen I S Y. High efficiency transfection of primary human lymphocytes and studies of gene expression. Oncogene. 1988;3:123–128. [Google Scholar]
- 7.Chassagne J, Verrelle P, Dionet C, Clavel F, Barre-Sinoussi F, Chermann J C, Montagnier L, Gluckman J C, Klatzman D. A monoclonal antibody against LAV gag precursor: use for viral protein analysis and antigenic expression in infected cells. J Immunol. 1986;136:1442–1443. [PubMed] [Google Scholar]
- 8.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 9.Daniel M D, Kirchoff F, Czajak S C, Sehgal P K, Desrosiers R C. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 10.Gibbs J S, Lackner A A, Lang S M, Simon M A, Sehgal P K, Daniel M D, Desrosiers R C. Progression to AIDS in the absence of a gene for vpr or vpx. J Virol. 1995;69:2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs J S, Regier D A, Desrosiers R C. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res Human Retroviruses. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- 12.He J, Choe S, Walker R, di Marzio P, Morgan D O, Landau N R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M-A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoch J, Lang S M, Weeger M, Stahl-Henning C, Coulibaly C, Dittmer U, Hunsmann G, Fuchs D, Muller J, Sopper S, Fleckenstein B, Uberia K T. vpr deletion mutant of simian immunodeficiency virus induces AIDS in rhesus monkeys. J Virol. 1995;69:4807–4813. doi: 10.1128/jvi.69.8.4807-4813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamieson B D, Aldrovandi G M, Planelles V, Jowett J B M, Gao L, Bloch L M, Chen I S Y, Zack J A. Requirement of human immunodeficiency virus type 1 nef for in vivo replication and pathogenicity. J Virol. 1994;68:3478–3485. doi: 10.1128/jvi.68.6.3478-3485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamieson B D, Pang S, Aldrovandi G M, Zha J, Zack J A. In vivo pathogenic properties of two clonal human immunodeficiency virus type 1 isolates. J Virol. 1995;69:6259–6264. doi: 10.1128/jvi.69.10.6259-6264.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamieson B D, Uittenbogaart C H, Schmid I, Zack J A. High viral burden and rapid CD4+ cell depletion in human immunodeficiency virus type 1-infected SCID-hu mice suggest direct viral killing. J Virol. 1997;71:8245–8253. doi: 10.1128/jvi.71.11.8245-8253.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kay R, Takei F, Humphries R K. Expression cloning of a cDNA encoding M1/J11D heat-stable antigens. J Immunol. 1990;145:1952–1959. [PubMed] [Google Scholar]
- 19.Kestler H W, III, Ringler D J, Kazuyasu M, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 20.Kitchen S G, Uittenbogaart C H, Zack J A. Mechanism of human immunodeficiency virus type 1 localization in CD4-negative thymocytes: differentiation from a CD4-positive precursor allows productive infection. J Virol. 1997;71:5713–5722. doi: 10.1128/jvi.71.8.5713-5722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitchen S G, Zack J A. CXCR4 expression during lymphopoiesis: implications for human immunodeficiency virus type 1 infection of the thymus. J Virol. 1997;71:6928–6934. doi: 10.1128/jvi.71.9.6928-6934.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kollmann T R, Pettoello-Mantovani M, Zhuang X, Kim A, Hachamovitch M, Smarnworawong P, Rubinstein A, Goldstein H. Disseminated human immunodeficiency virus type 1 (HIV-1) infection in SCID-hu mice after peripheral inoculation with HIV-1. J Exp Med. 1994;179:513–522. doi: 10.1084/jem.179.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawn R M, Efstratindus A, O’Connell C, Maniatis T. The nucleotide sequence of the human β-globin gene. Cell. 1980;21:647–651. doi: 10.1016/0092-8674(80)90428-6. [DOI] [PubMed] [Google Scholar]
- 24.Livingstone W J, Moore M, Innes D, Bell J E, Simmonds P. Frequent infection of peripheral blood CD8-positive T-lymphocytes with HIV-1. Edinburgh Heterosexual Transmission Study Group. Lancet. 1996;348:649–654. doi: 10.1016/s0140-6736(96)02091-0. [DOI] [PubMed] [Google Scholar]
- 25.McCune J M, Namikawa R, Kaneshima H, Shultz L D, Lieberman M, Weissman I L. The SCID-hu mouse: murine model for the analysis of human hematolymphoid differentiation and function. Science. 1988;241:1632–1639. doi: 10.1126/science.241.4873.1632. [DOI] [PubMed] [Google Scholar]
- 26.Namikawa R, Kaneshima H, Lieberman M, Weissman I L, McCune J M. Infection of the SCID-hu mouse by HIV-1. Science. 1988;242:1684–1686. doi: 10.1126/science.3201256. [DOI] [PubMed] [Google Scholar]
- 27.Namikawa R, Weilbaecher K N, Kaneshima H, Yee E J, McCune J M. Long-term human hematopoiesis in the SCID-hu mouse. J Exp Med. 1990;172:1055–1063. doi: 10.1084/jem.172.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Planelles V, Haislip A, Withers-Ward E S, Stewart S A, Xie Y, Shah N P, Chen I S Y. A new reporter system for detection of viral infection. Gene Ther. 1995;2:369–376. [PubMed] [Google Scholar]
- 29.Semenzato G, Agostini C, Ometto L, Zambello R, Trentin L, Chieco-Bianchi L, De Rossi A. CD8+ T lymphocytes in the lung of acquired immunodeficiency syndrome patients harbor human immunodeficiency virus type 1. Blood. 1995;85:2308–2314. [PubMed] [Google Scholar]
- 30.Stanley S K, McCune J M, Kaneshima H, Justement J S, Sullivan M, Boone E, Baseler M, Adelsberger J, Bonyhadi M, Orenstein J, Fox C H, Fauci A S. Human immunodeficiency virus infection of the human thymus and disruption of the thymic microenvironment in the SCID-hu mouse. J Exp Med. 1993;178:1151–1163. doi: 10.1084/jem.178.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Withers-Ward E S, Amado R G, Koka P S, Jamieson B D, Kaplan A H, Chen I S Y, Zack J A. Transient renewal of thymopoiesis in HIV-infected human thymic implants following antiviral therapy. Nat Med. 1997;3:1102–1109. doi: 10.1038/nm1097-1102. [DOI] [PubMed] [Google Scholar]
- 32.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 33.Zack J A, Haislip A, Krogstad P, Chen I S Y. Incompletely reverse transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retrovirus life cycle. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]