Abstract
The nature and stability of the interactions between the gp70 and Pr15E/p15E molecules of murine leukemia virus (MLV) have been disputed extensively. To resolve this controversy, we have performed quantitative biochemical analyses on gp70-Pr15E complexes formed after independent expression of the amphotropic and ecotropic Moloney MLV env genes in BHK-21 cells. We found that all cell-associated gp70 molecules are disulfide linked to Pr15E whereas only a small amount of free gp70 is released by the cells. The complexes were resistant to treatment with reducing agents in vivo, indicating that the presence and stability of the disulfide interaction between gp70 and Pr15E are not dependent on the cellular redox state. However, disulfide-bonded Env complexes were disrupted in lysates of nonalkylated cells in a time-, temperature-, and pH-dependent fashion. Disruption seemed not to be caused by a cellular factor but is probably due to a thiol-disulfide exchange reaction occurring within the Env complex after solubilization. The possibility that alkylating agents induce the formation of the intersubunit disulfide linkage was excluded by showing that disulfide-linked gp70-Pr15E complexes exist in freshly made lysates of nonalkylated cells and that disruption of the complexes can be prevented by lowering the pH. Together, these data establish that gp70 and Pr15E form a stable disulfide-linked complex in vivo.
The envelope (Env) protein of murine leukemia virus (MLV) mediates binding of the virus particle to a specific receptor(s) at the surface of uninfected cells and is responsible for the fusion of the viral and cellular membranes during virus entry (8). In the virion, the Env protein forms a complex that consists of a membrane-anchored (TM) and a surface (SU) molecule. The TM subunit contains an amino-terminal hydrophobic peptide that is thought to mediate membrane fusion (10, 19), whereas the SU subunit bears the receptor binding function (2, 17).
SU and TM are derived from a precursor polypeptide which is inserted into the membrane of the endoplasmic reticulum (ER). The precursor Env protein is proteolytically cleaved into SU and TM by a cellular enzyme at a late stage during its transport to the plasma membrane (PM), where virus assembly takes place (3, 13, 34). In MLV, SU and TM are designated gp70 and Pr15E, respectively, corresponding to their respective molecular weights. During or shortly after virus budding, Pr15E is processed into p15E by the viral protease, which clips off the so-called R peptide, comprising the carboxy-terminal 16 amino acid residues of the molecule (15, 38, 46).
In early studies, it was found that gp70 and Pr15E present in MLV-infected cells and in virions form a disulfide-linked complex (27, 40, 44, 45, 51, 56). Although none of these studies involved a quantitative analysis of the biosynthesis of the Env protein complex, the fraction of gp70 and Pr15E/p15E that was found in the disulfide-bonded form seemed to vary significantly. These differences can be due to the virus strain, experimental variations, inaccurate quantitation, and different labeling methods used. Importantly, the detection of disulfide-linked gp70-Pr15E/p15E complexes in all of the studies was critically dependent on treatment of infected cells and virus particles with thiol-active reagents, such as N-ethylmaleimide (NEM), 2,2′-dithiobis(m-nitropyridine) [DTNP], and iodoacetamide, prior to solubilization.
Findings by Pinter et al. (41) have cast doubt on whether the disulfide linkage between gp70 and Pr15E/p15E actually exists in vivo. It was shown that disruption of virus particles with sodium dodecyl sulfate (SDS) yields only 10% of gp70 in a disulfide-linked complex with p15E. However, when the virions were incubated with Nonidet P-40 (NP-40), before addition of SDS, up to 39% of gp70 was found to be covalently linked to p15E. It has therefore been concluded that only a minor fraction of gp70 and p15E is coupled by disulfide bridges in vivo and that disulfide-linked gp70-p15E complexes are mostly spontaneously formed after solubilization in NP-40 (41). Pretreatment of the virions with NEM or DTNP resulted in a further increase in the yield of disulfide-bonded Env complexes. This was interpreted as indicating that NEM and DTNP activate free sulfhydryls in the Env protein, thereby stimulating the formation of a disulfide linkage between gp70 and p15E (41, 45).
This interpretation has since been changed. Pinter et al. (42) recently showed that treatment of ecotropic, xenotropic, and amphotropic MLV particles with NEM prior to solubilization greatly increases the yield of gp70 engaged in disulfide-linked Env complexes. To explain this effect, these investigators suggested that gp70 and p15E are linked by a labile disulfide bond, which can be stabilized by blocking free thiols, present in intact virions, with NEM. This would prevent thiol-disulfide exchange reactions and thereby inhibit the disruption of the covalent linkage between gp70 and p15E.
A similar theory assumes that the disulfide bond between gp70 and Pr15E/p15E isomerizes readily and reversibly in vivo as well as in cell lysates with a free cysteine thiol group within the Env protein (14). As a consequence, only a fraction of gp70 and Pr15E/p15E is disulfide linked and the proportion of gp70 and Pr15E/p15E involved in covalently linked complexes depends on the redox conditions of the environment. This model is based on the claim of Gliniak et al. (14) that incubation of infected cells with the oxidizing agent diamide increases the amount of gp70 that is disulfide bonded to Pr15E. The possibility that gp70 and Pr15E/p15E are held together by noncovalent interactions has been supported by studies showing that free gp70 is present in the medium of infected cells and that gp70 can be released from virus particles by freeze-thawing (32, 33, 36).
The aim of the work presented here was to elucidate the above-described controversy about the interactions between gp70 and Pr15E/p15E. It is important to establish the nature and stability of these interactions, since structural reorganization or even dissociation of the Env protein complex has been suggested to play a role in the activation of the fusion function of the Env protein during virus entry (42, 43). Therefore, we have expressed the amphotropic and ecotropic Moloney MLV (Mo-MLV) env genes in BHK-21 cells by using recombinant Semliki Forest virus (recSFV) vectors. The recSFV expression system has proven to be particularly useful for the efficient expression of foreign genes in mammalian cells (28, 31, 49). Our laboratory has recently developed a method to produce high-titer stocks of recombinant Mo-MLV in BHK-21 cells by coexpression of the env and gag-pol genes and a recombinant retrovirus genome from separate SFV expression vectors (29). Here we have expressed the env genes in the absence of other Mo-MLV components to investigate the intrinsic properties of the gp70-Pr15E interactions.
MATERIALS AND METHODS
Cells, virus, and antibodies.
BHK-21 cells (American Type Culture Collection, Rockville, Md.) were grown in BHK-21 medium (GIBCO BRL, Life Technologies, Paisley, United Kingdom) containing 5% fetal calf serum, 10% tryptose phosphate broth, 20 mM HEPES, and 2 mM glutamine (BHK medium). The cloning of the ecotropic and amphotropic Mo-MLV env genes into the SFV-1 expression vector (31) has been described previously (29, 49). recSFV genomes were packaged into recSFV particles as described previously (31, 50). The titers of recSFV stocks were determined by indirect immunofluorescence with the gp70-specific rat monoclonal antibody 83A-25 (9), a kind gift of B. W. Chesebro. Polyclonal pig antiserum HC185 against MLV was used for immunoprecipitation of the Env proteins and was purchased from Quality Biotech Inc., Camden, N.J.
Infection and metabolic labeling.
Subconfluent monolayers of BHK-21 cells were washed once with phosphate-buffered saline including Ca2+ and Mg2+ (PBS) and inoculated with recSFV in minimal essential medium (MEM; GIBCO BRL, Life Technologies) containing 0.2% bovine serum albumin. After a 1-h incubation at 37°C, the inoculum was replaced with BHK-21 medium. At 5.5 h after inoculation, the cells were starved for 30 min in Dulbecco’s MEM (DMEM) lacking l-cysteine (DMEM − cys). Thereafter, the cells were labeled for the indicated times in DMEM − cys supplemented with 50 to 200 μCi of l-[35S]cysteine (Amersham Corp., Arlington Heights, Ill.). At the end of the labeling, the cells were either directly solubilized (see below) or washed twice with BHK-21 medium supplemented with 2 mM l-cysteine (chase medium) and further incubated in chase medium. When indicated, the chase medium was collected and centrifuged for 5 min at 5,000 rpm and 4°C in an Eppendorf 16F24-11 centrifuge. The cleared media were stored on ice before being used for immunoprecipitation.
Alkylation and solubilization of cells.
Routinely, the cells were put on ice and incubated twice for 1 min with freshly prepared ice-cold PBS containing 20 mM NEM (Sigma-Aldrich Chemie GmbH, Steinheim, Germany) before being solubilized in NP-40 lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1% NP-40) supplemented with 20 mM NEM. The lysates were cleared by centrifugation for 5 min at 6,000 rpm and 4°C in an Eppendorf 16F24-11 centrifuge and the supernatants were stored on ice. When indicated, samples were alkylated after lysis of the cells by adding 1 volume of NP-40 lysis buffer containing 40 mM NEM to the lysate. The low-pH (pH 6.0) NP-40 lysis buffer used in the experiment in Fig. 5C was buffered with 20 mM 2-[N-morpholino]ethanesulfonic acid (Sigma-Aldrich) and 30 mM Tris-HCl.
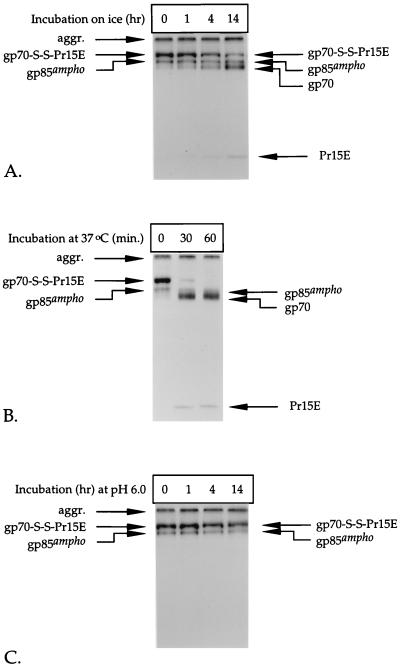
FIG. 5.
Disruption of the disulfide linkage between gp70 and Pr15E after cell lysis depends on temperature and pH. Lysates of metabolically labeled amphotropic Mo-MLV env-expressing cells were prepared in the absence of alkylating agents, either at pH 7.5 (A and B) or at pH 6.0 (C). The lysates were incubated for the indicated times on ice (A and C) or at 37°C (B), after which NEM was added to 20 mM. The Env protein was thereafter immunoprecipitated with the polyclonal anti-MLV serum. Immunoprecipitates were analyzed in nonreducing SDS–12% polyacrylamide gels.
Immunoprecipitation and gel electrophoresis.
For precipitation of the MLV Env proteins, fractions of cell lysates were diluted with NP-40 lysis buffer to 200 μl. The cleared media were mixed (1:1) with NP-40 lysis buffer. Then 4 μl of polyclonal anti-MLV serum or 50 μl of monoclonal antibody against gp70 was added, and the samples were incubated for 30 min on ice. Thereafter, 40 μl of protein A-Sepharose (Pharmacia P-L Biochemicals Inc.) slurry in NP-40 lysis buffer (1:1, vol/vol) was added, and the samples were incubated overnight or for 3 h at 4°C to collect the immune complexes. Protein A-Sepharose-bound immune complexes were then washed twice in 0.2% NP-40–10 mM Tris-HCl (pH 7.5)–150 mM NaCl–2 mM EDTA, twice in 0.2% NP-40–10 mM Tris-HCl (pH 7.5)–0.5 M NaCl–2 mM EDTA, and once in 10 mM Tris-HCl (pH 7.5) and finally suspended in 80 μl of 62.5 mM Tris-HCl (pH 6.8)–2% SDS–10% glycerol (gel sample buffer). For reduction of disulfide bonds prior to gel electrophoresis, dithiothreitol (DTT) was added to the gel sample buffer to a final concentration of 20 or 50 mM. The samples were heated for 5 min at 95°C before being loaded on the gel. In case reduced and nonreduced samples were run in the same gel, the samples were alkylated by adding iodoacetamide (Sigma-Aldrich) to 100 mM. The samples were run in SDS–8 or 12% polyacrylamide gels with the Mighty Small II minigel system (Hoefer Scientific Instruments, San Francisco, Calif.). After electrophoresis, the gels were soaked for 20 to 30 min in sodium salicylate (160 g/liter), dried, and used for autoradiography. Relative amounts of radioactivity in protein bands were measured with a FUJIX BAS 2000 TR phosphorimager (Fuji Photo Film Co.).
Cell surface biotinylation.
Metabolically labeled env-expressing cells were put on ice, incubated twice for 1 min with ice-cold PBS containing 20 mM NEM, and then incubated for 30 min in ice-cold PBS containing 50 μg of NHS-S-S-biotin (Pierce Chemical Co., Rockford, Ill.) per ml. Thereafter, nonbound biotin was inactivated and washed away with PBS containing 50 mM NH4Cl. Cell lysates were prepared as described above, and a one-fifth volume of streptavidin-agarose (Sigma-Aldrich) slurry in NP-40 lysis buffer (1:1 vol/vol) was added to collect the biotinylated proteins. The samples were incubated on a rocker at 4°C for approximately 14 h. Streptavidin-agarose beads were then pelleted, washed as described for the protein A-Sepharose-bound immune complexes, and finally resuspended in gel sample buffer containing 50 mM DTT. The supernatant was used for a second round of precipitation with the polyclonal anti-MLV serum.
RESULTS
Amphotropic Mo-MLV Env protein is processed into a stable, disulfide-linked gp70-Pr15E complex.
Biosynthesis of the amphotropic Mo-MLV Env protein complex has not been studied before. We therefore set out to characterize the synthesis and processing of independently expressed amphotropic Mo-MLV env gene products by pulse-chase analysis. Since several data have been reported on the ecotropic MLV Env protein previously (27, 42), we included expression of the ecotropic Mo-MLV env gene for comparison. recSFV-infected BHK-21 cells expressing these proteins were labeled for 10 min with l-[35S]cysteine at 6 h postinoculation and chased for various periods. The cells were treated with the membrane-permeable alkylating agent NEM before being solubilized, and a polyclonal serum against MLV or a monoclonal antibody specific for gp70 was used to immunoprecipitate the Env protein from the cell lysates and chase media. Immunoprecipitates were analyzed by gel electrophoresis under reducing and nonreducing conditions.
The results for the amphotropic Env protein are shown in Fig. 1A through C. The major product that could be detected immediately after the pulse labeling was the precursor Env protein, with a molecular mass of ∼85 kDa (gp85ampho, Fig. 1A). In addition, two other molecular species (seen as one band in this gel) that had migrated faster in the gel were observed. These disappeared rapidly during the chase, suggesting that they were degraded. Because these products virtually lacked N-linked oligosaccharides as well as inter- or intramolecular disulfide bonds (data not shown), they most probably represented nontranslocated, cytoplasmic Env polypeptides. Similar products have been observed previously in MLV infected cells and may result from initiation of translation at an internal AUG site on the mRNA (3).
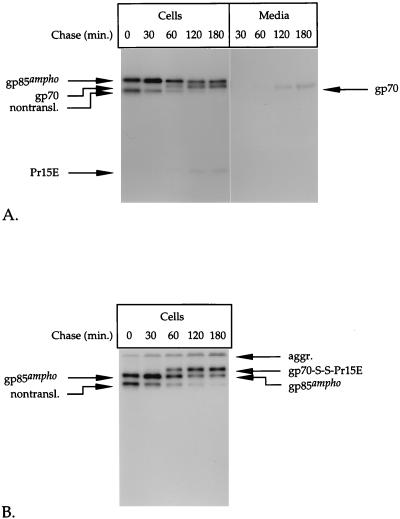
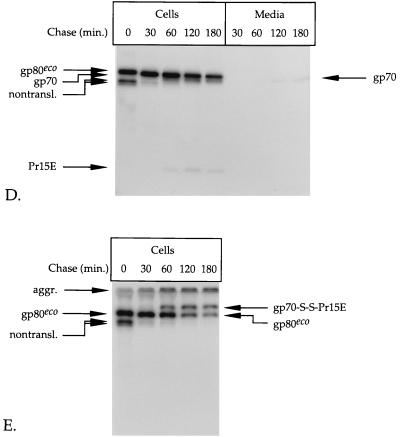
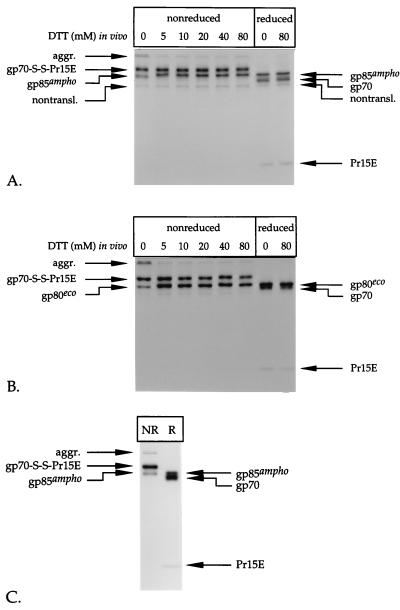
FIG. 1.
Pulse-chase analysis of env-expressing cells. BHK-21 cells infected with recSFV expressing the amphotropic (A through C) or ecotropic (D and E) Mo-MLV env gene were labeled for 15 min with l-[35S]cysteine at 6 h postinoculation and chased for the indicated times. The Env protein was immunoprecipitated from the cell lysates and the media by use of the polyclonal anti-MLV serum (A, B, D, and E). The immunoprecipitates were run in SDS–12% polyacrylamide gels under reducing (A and D) and nonreducing (B and E) conditions. The part of the gel in panel A that contains the samples of media was exposed seven times longer than the part that contains the cell samples. The putative nontranslocated Env molecules (nontransl.) and disulfide-linked aggregates (aggr.) are indicated. A fraction of the lysate of amphotropic env-expressing cells that were chased for 180 min was used for immunoprecipitation with a monoclonal antibody against gp70 (C). The resulting precipitate was analyzed under reducing (R) and nonreducing (NR) conditions in an SDS–12% polyacrylamide gel.
After 30 min of chase, two other proteins with molecular masses of about 70 and 20 kDa started to appear. The increase in their intensity during the chase corresponded to the decrease in the amount of gp85ampho, indicating that they represented the Env protein cleavage products gp70 and Pr15E, respectively. The difference in intensity between gp70 and Pr15E reflects the difference in their cysteine contents; the molecules contain 18 and 4 cysteines, respectively (39). Approximately half of the Env molecules were cleaved within 2 h after synthesis. Cleavage of the Env protein indicates that it had been transported to and through the Golgi complex and the trans-Golgi network (3).
gp70 and Pr15E were not resolved when the immunoprecipitates were analyzed under nonreducing conditions (Fig. 1B). Instead, both products migrated as a disulfide-linked complex with an apparent molecular mass of about 100 kDa, which we refer to as gp70-S-S-Pr15E. That the 100-kDa protein species is composed of both gp70 and Pr15E was verified by immunoprecipitation with a monoclonal antibody against gp70. This antibody was found to precipitate the 100-kDa species in addition to the precursor Env protein, gp85ampho (Fig. 1C, lane NR). The latter was also observed when the immunoprecipitate was reduced with DTT before electrophoresis (second lane). In contrast, the 100-kDa species was not present under these conditions. Instead, gp70 and Pr15E could be seen in the gel, indicating that gp70 was disulfide linked to Pr15E before reduction. Analysis of the immunoprecipitates under nonreducing conditions also revealed the presence of aggregates that stayed at the top of the running gel (Fig. 1B). These aggregates are disulfide bonded because they disappear after reduction with DTT (compare Fig. 1A and B). The observation that the disappearance of this material correlates with an increase in the signal of gp85ampho indicates that the disulfide-linked aggregates contained predominantly precursor Env molecules.
To determine whether the Env protein was released from the cells, we subjected the chase media to immunoprecipitation with the polyclonal anti-MLV serum. Some gp70 was detected in the 1- to 3-h medium samples after prolonged exposure of the autoradiographs (Fig. 1A). Quantitative comparison of the amount of cell-associated and released gp70 revealed that only 2 to 3% of total gp70 was present in the medium after the 2- and 3-h chase periods. Since neither gp85ampho nor Pr15E could be detected in the media, it is most likely that this fraction represents free gp70 that has been shed or secreted from the cells. From all these results, we conclude that the majority of the proteolytically processed amphotropic Env molecules forms stable, disulfide-linked gp70-Pr15E complexes.
The analysis of the ecotropic Env protein is shown in Fig. 1D and E. The precursor form of this protein has a molecular mass of ∼80 kDa (gp80eco) and is somewhat less efficiently processed into gp70 (which migrated very close to gp80eco) and Pr15E than is the amphotropic Env precursor (Fig. 1D). When analyzed under nonreducing conditions (Fig. 1E), ecotropic gp70 and Pr15E also formed a covalently linked complex. Like amphotropic gp70, virtually all cell-associated ecotropic gp70 was engaged in a disulfide-linked complex with Pr15E. Approximately 10 to 15% of total ecotropic gp70 was detected in the medium after the 2- and 3-h chase periods (Fig. 1D).
gp70 and Pr15E are expressed at the cell surface.
The arrival of Env protein at the PM was assayed by cell surface biotinylation. To do this, radiolabeled amphotropic Mo-MLV env-expressing cells were treated with membrane-impermeable biotin on ice. Nonbound biotin was then inactivated and washed away, and the cells were lysed. Biotinylated proteins were collected from the lysates with streptavidin-agarose. The remaining, intracellular pool of proteins was subjected to immunoprecipitation with the polyclonal antibodies against MLV.
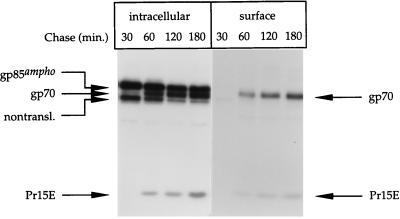
gp70 and Pr15E were the predominant products to be detected by biotinylation, suggesting that they were present at the PM (Fig. 2, lanes surface). Because the precursor protein, gp85ampho, was not biotinylated, we conclude that the biotinylation of gp70 and Pr15E was specific. A significant fraction of gp70 and Pr15E was also precipitated with the MLV-specific antibodies, indicating that not all gp70 and Pr15E molecules were biotinylated. By comparing the amounts of biotinylated and nonbiotinylated gp70 and Pr15E, we estimated that at least 30% of the proteolytically processed Env molecules were present at the cell surface. Similar results were obtained with the ecotropic Env protein (data not shown).
FIG. 2.
Cell surface expression of Env molecules. Amphotropic Mo-MLV env-expressing cells were pulse-labeled and chased as described in the legend to Fig. 1. After the chase, the cells were put on ice and incubated for 30 min with NHS-S-S-biotin. The biotin was then inactivated and washed away. The cells were solubilized in lysis buffer, and the biotinylated proteins (surface) were collected with streptavidin-agarose. The remaining material (intracellular) was subjected to immunoprecipitation with the polyclonal anti-MLV serum. The precipitates were reduced with 50 mM DTT before being subjected to electrophoresis in SDS–12% polyacrylamide gels.
The covalent linkage between gp70 and Pr15E is resistant to reducing agents in vivo.
To test whether the presence of the intermolecular disulfide interaction between gp70 and Pr15E depends on the redox state of the environment, we analyzed the stability of gp70-S-S-Pr15E under reducing conditions in vivo. Parallel cultures of amphotropic or ecotropic Mo-MLV env-expressing cells were therefore labeled and chased for 105 min. Thereafter, the chase media were replaced by fresh media containing the indicated amounts of DTT, and the incubation was continued for 15 min at 37°C before the cells were alkylated and lysed.
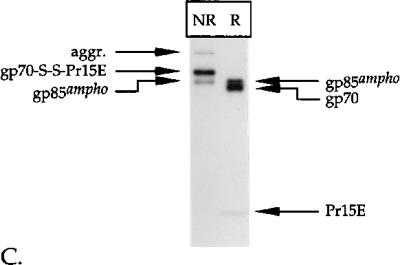
As shown in Fig. 3A and B, treatment with DTT did not cause a decrease in the amount of either amphotropic or ecotropic gp70-S-S-Pr15E, even when very high concentrations of the reducing agent had been applied. The same result was obtained when the cells were treated with up to 160 mM 2-mercaptoethanol (data not shown). This demonstrates that the intermolecular disulfide linkage between gp70 and Pr15E is not sensitive to the redox state in vivo. However, an effect of DTT was seen for the precursor Env molecules. For example, the precursor amphotropic Env protein that was left after the chase in the absence of DTT migrated as a smear (referred to as gp85ox) under nonreducing conditions (Fig. 3C, left lane). In addition, a fraction of Env was present in disulfide-linked aggregates that were found at the top of the gel. The amount of these aggregates was decreased when the cells had been incubated with DTT, while the intensity of monomeric precursor Env molecules (referred to as gp85red) increased (second lane from left). In addition, gp85ox migrated slower when the cells had been treated with DTT, and most precursor Env now comigrated with the in vitro reduced protein (right-hand two lanes), indicating that most of the precursor Env molecules had been reduced. This effect of DTT in living cells was observed previously for other viral membrane proteins (4, 37) and served here as an internal control for the efficacy of the treatment. Similar data were obtained for the ecotropic Env protein (data not shown).
FIG. 3.
Disulfide-bonded gp70-Pr15E complexes are resistant to treatment with reducing agents in vivo. Amphotropic (A) and ecotropic (B) Mo-MLV env-expressing cells were labeled with l-[35S]cysteine for 15 min and chased for 120 min. After 105 min of chase, the media were replaced with fresh media containing the indicated amounts of DTT. The chase was continued for 15 min, and then the cells were alkylated and solubilized. The Env protein was immunoprecipitated from the lysates with the polyclonal anti-MLV serum. The immunoprecipitates were analyzed under reducing and nonreducing conditions in SDS–12% polyacrylamide gels. (C) Samples of the immunoprecipitates of the amphotropic Env protein, derived from cells that had been treated with 0 or 5 mM DTT, were run in an SDS–8% polyacrylamide gel for better separation of the protein bands. The meaning of the designations gp85red and gp85ox is explained in Results. The samples were run under reducing (R) and nonreducing (NR) conditions.
Preservation of the disulfide linkage between gp70 and Pr15E during solubilization requires alkylation.
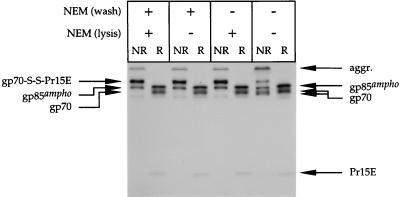
So far, we have analyzed the Env protein that was extracted from cells that had been alkylated before disruption. To test the effects of alkylation, we extracted the amphotropic gp70-Pr15E complexes from cells in the absence of NEM. As a control, we solubilized parallel cultures of env-expressing cells in a procedure in which NEM was included in the wash and/or lysis step.
Figure 4 shows that only a fraction (<50%) of gp70 and Pr15E was recovered in disulfide-linked complexes if the samples had not been alkylated. The results with the controls show that gp70 and Pr15E were quantitatively engaged in the disulfide-linked complexes if NEM was included during sample preparation. Interestingly, the results show that the disulfide interaction between gp70 and Pr15E was preserved even when the cells were only briefly exposed to NEM just before lysis. This was also found when the ecotropic Env protein was used (data not shown). These data indicate that alkylation is required to collect amphotropic and ecotropic gp70 and Pr15E quantitatively in disulfide-linked complexes.
FIG. 4.
Alkylation is required to preserve Env protein complexes upon solubilization. Amphotropic Mo-MLV env-expressing cells were labeled for 15 min with l-[35S]cysteine and chased for 120 min. Before solubilization, the cells were put on ice and washed [NEM (wash)] twice for 1 min with PBS. When indicated, the PBS contained 20 mM NEM. Thereafter, the cells were lysed [NEM (lysis)] in the presence or absence of 20 mM NEM. Immunoprecipitates were analyzed under reducing (R) and nonreducing (NR) conditions in an SDS–12% polyacrylamide gel.
Nonalkylated disulfide-bonded gp70-Pr15E complexes are disrupted in a time-, temperature-, and pH-dependent fashion.
The observation that less gp70-S-S-Pr15E was found in the nonalkylated samples than in the alkylated ones implies either that the formation of disulfide-linked Env complexes is induced by NEM or that NEM prevents disruption of these complexes. To distinguish between these possibilities, we analyzed the effect of adding NEM after solubilization of nonalkylated cells.
Figure 5A shows that the amount of amphotropic gp70-S-S-Pr15E complexes recovered from the lysates decreased when NEM was added after prolonged incubation of the lysate on ice and that the amount of free gp70 and Pr15E increased proportionally with the decrease in gp70-S-S-Pr15E. This clearly indicates that disulfide-bonded Env complexes already existed in the freshly made lysate but that they are disrupted in time unless the process of disruption is blocked by NEM.
The rate at which the intersubunit disulfide interaction was disrupted was largely affected by the temperature. We estimated that the disulfide-linked gp70-Pr15E complexes had a half-life of about 8 h on ice (Fig. 5A). In contrast, the majority of gp70-S-S-Pr15E was converted into free gp70 and Pr15E within 30 min when the lysate was incubated at 37°C (Fig. 5B).
Disruption of the disulfide linkage between gp70 and Pr15E is probably mediated by a thiol-disulfide exchange reaction. Such a reaction can, in principle, be quenched by lowering the pH, since this will lead to protonation of the reactive thiolates (25, 55). Indeed, we found that the disulfide-bonded gp70-Pr15E complexes were less labile at pH values below 7.5. Figure 5C demonstrates that the amount of gp70-S-S-Pr15E did not decrease when the Env protein was extracted and incubated at pH 6.0, indicating that the intersubunit disulfide linkage was stable under these conditions. This shows that low pH can be used as an alternative to alkylating agents to prevent the disruption of disulfide-linked Env complexes after solubilization. Ecotropic gp70-S-S-Pr15E complexes displayed similar behavior under the conditions described in the legend to Fig. 5 (data not shown).
Together, these results strengthen and extend our findings that gp70 and Pr15E are stably linked through disulfide bonding in vivo but that the intersubunit linkage is destabilized upon solubilization unless thiol-disulfide exchange reactions are prevented by alkylation or acidification.
Disruption of the gp70-Pr15E intersubunit disulfide linkage does not involve a soluble cellular factor but is mediated by a thiol-disulfide exchange reaction within the Env complex.
Because the disruption of the disulfide linkage between gp70 and Pr15E seems to be induced by solubilization of the cells, we investigated the possibility that this process is mediated by a cellular factor that is released upon cell lysis. For instance, it is known that the cytoplasm maintains a relatively reductive environment (21); therefore, we reasoned that the intersubunit disulfide linkage might be simply reduced by cytosolic components, such as glutathione and thioredoxin, which are liberated through the disruption of the PM. Alternatively, disruption of the intersubunit disulfide linkage may be mediated by a thiol-disulfide rearrangement occurring within the Env complex itself.
To investigate these possibilities, we analyzed the disruption of disulfide-bonded gp70-Pr15E complexes in a mixture (1:1) of cell lysates that were prepared in the presence and absence of NEM. The rationale of this approach was that the (normally stable) intersubunit disulfide linkage in gp70-Pr15E complexes, derived from the alkylated sample, would be reduced if the nonalkylated sample contained a putative reducing factor. However, if disruption was mediated by a thiol-disulfide exchange reaction within the Env protein itself, the complexes should be stable under these conditions. To monitor the Env protein complexes of the individual lysates, we prepared and mixed extracts of both labeled and nonlabeled amphotropic Mo-MLV env-expressing cells.
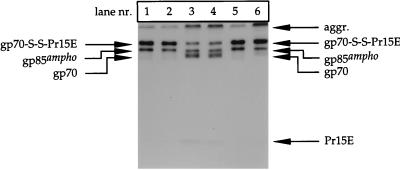
As shown in Fig. 6 (lanes 1 and 2), equal amounts of gp70-S-S-Pr15E were observed when the lysate of alkylated cells was incubated alone or together with a lysate of nonalkylated cells. Neither gp70 nor Pr15E was detected in the gel, indicating that the radiolabeled Env complexes were stable under both conditions. This suggested that the lysate of the nonalkylated cells did not contain a factor that could reduce the disulfide linkage between gp70 and Pr15E. Notably, alkylation of the cells involved only a brief exposure to NEM and extensive washing thereafter, to minimize the chance that free NEM would end up in the lysate. However, to exclude the possibility that residual NEM had prevented the disruption of gp70-S-S-Pr15E in the mixed sample, e.g., by blocking the putative reducing factor from the nonalkylated sample, we monitored the disruption of Env complexes derived from the nonalkylated cells. It was observed that about half of the Env complexes were dissociated into free gp70 and Pr15E when a lysate of nonalkylated cells was incubated alone (lane 3). The same amounts of gp70-S-S-Pr15E as well as free gp70 and Pr15E were found when such lysate was mixed and incubated with the lysate of alkylated cells (lane 4). This proves that the latter lysate did not contain free NEM that could interfere with the disruption of the disulfide-linked complexes. Similar results were obtained when the ecotropic Env protein was used in this assay (data not shown).
FIG. 6.
Disruption of disulfide linkage between gp70 and Pr15E occurs through a thiol-disulfide exchange reaction within the Env complex. Four parallel cultures of BHK-21 cells were infected with recSFV expressing the amphotropic Mo-MLV env gene. Two of these were labeled for 15 min with l-[35S]cysteine and chased for 120 min, whereas the others were not labeled. At the end of the chase, all the cultures were put on ice. One culture of labeled cells and one culture of nonlabeled cells were incubated twice for 1 min with PBS–20 mM NEM and then washed four times in PBS lacking NEM. The remaining cell cultures were washed with PBS lacking NEM. Thereafter, all the cultures were lysed in the absence of NEM. The cell lysates were then incubated for 15 h at 4°C in the following (1:1) mixtures: lysate of labeled, alkylated cells plus lysis buffer lacking NEM (lane 1); lysate of labeled, alkylated cells plus lysate of nonlabeled, nonalkylated cells (lane 2); lysate of labeled, nonalkylated cells plus lysis buffer lacking NEM (lane 3); and lysate of labeled, nonalkylated cells plus lysate of nonlabeled, alkylated cells (lane 4). As a control to check the amount of gp70-S-S-Pr15E at the time of mixing, we diluted lysates of labeled, alkylated cells (lane 5) and labeled, nonalkylated cells (lane 6) with lysis buffer containing NEM to block further disruption of the complexes during incubation. The Env protein was thereafter immunoprecipitated with the polyclonal anti-MLV serum and analyzed under nonreducing conditions in an SDS–12% polyacrylamide gel.
From the above experiments, we conclude that the disulfide linkage between gp70 and Pr15E is not reduced after solubilization by a soluble cellular factor. Rather, the Env protein itself contains a free thiol(s) which can initiate a thiol-disulfide rearrangement that eliminates the disulfide linkage between gp70 and Pr15E upon solubilization.
DISCUSSION
The major conclusion of this paper is that the gp70 and Pr15E subunits of the amphotropic and ecotropic Mo-MLV Env protein complex are held together by a stable disulfide linkage. Essentially all gp70 molecules were found to be engaged in disulfide-linked complexes with Pr15E within and at the surface of the cells. In addition, the covalent interaction between gp70 and Pr15E has proven to be highly stable under reductive conditions in vivo. Only under the conditions used for analysis, i.e., after solubilization of the Env protein, was the intersubunit gp70-Pr15E disulfide linkage found to be disrupted if disulfide exchange reactions were not blocked by alkylation or acidification.
In contrast to previous reports (41), we found that the disulfide linkage between gp70 and Pr15E is not spontaneously formed in nonionic detergents. Instead, the results in Fig. 5A show unambiguously that (nonalkylated) gp70-S-S-Pr15E complexes lose their intersubunit disulfide linkage in an NP-40 lysate. Furthermore, the findings that a substantial fraction of gp70 and Pr15E remains disulfide linked in the absence of alkylating agents (Fig. 4) and that disruption of gp70-S-S-Pr15E can be blocked just by lowering the pH exclude the possibility that NEM induces the formation of the intersubunit disulfide linkage. Together, these data resolve the suspicion that the disulfide linkage between gp70 and Pr15E is formed artificially. Previous data showing that solubilization of virions in NP-40 before the addition of SDS increases the yield of disulfide-linked Env protein complexes (41) should therefore be put in a new perspective. Although these findings are difficult to interpret, we can only guess that Env complexes solubilized in NP-40 assume a conformation that is partially resistant to SDS.
The disulfide linkage between gp70 and Pr15E appeared not to be affected by the redox potential of the cellular milieu, because the Env complexes were stable under both normal and reducing conditions. This finding contradicts the results of Gliniak et al. (14), who claimed that incubation of cells with the oxidizing agent diamide increased the proportion of gp70 that is disulfide bonded to Pr15E. The effect of reducing agents was not investigated in their study. We could not confirm the effect of diamide since, in our case, all cell-associated gp70 was disulfide linked to Pr15E even under normal conditions. However, we have observed that Env complexes are stable after solubilization of cells that have been briefly treated with 20 mM diamide instead of NEM (data not shown). In other words, diamide can replace alkylating agents to prevent the disruption of the intersubunit disulfide linkage upon cell lysis. This can be explained by the fact that diamide, when added to cells in high concentrations, oxidizes not only glutathione but also thiols present in proteins (26). Thus, diamide probably inactivates the thiols that attack the disulfide bond(s) between gp70 and Pr15E after solubilization.
The finding that the presence and stability of the disulfide linkage between gp70 and Pr15E do not depend on the redox state invalidates the argument for the model of Gliniak et al. (14), which assumes that the intersubunit disulfide bond(s) isomerizes readily and reversibly in vivo. According to this model, only a fraction of gp70 and Pr15E would be disulfide linked. In contrast, our quantitation of the radiolabeled Env products showed that almost all gp70 and Pr15E molecules form a stable disulfide-linked complex. This was supported by the fact that only a minor amount of free gp70 could be detected in the medium. We therefore conclude that the disulfide linkage between gp70 and Pr15E should be regarded as stable.
In this work, we have analyzed Env complexes that contain unprocessed Pr15E. In the virion, however, a large fraction of Pr15E is cleaved into p15E, a process which is believed to be required for fusion (22, 53). Pinter et al. (42) have demonstrated that disulfide-linked Env complexes also exist in ecotropic and amphotropic MLV particles. Nevertheless, their data also show that a significant fraction of gp70 is not disulfide linked to Pr15E/p15E even when the virions had been treated with alkylating agents before solubilization (Fig. 1 in reference 42). This result clearly differs from our observations that virtually all cell-associated gp70 is involved in stable disulfide-bonded complexes with Pr15E. Therefore, it might be that cleavage of Pr15E destabilizes the intersubunit disulfide linkage. We are currently investigating this possibility by quantitative biochemical analysis of Env protein complexes in virus particles.
Disruption of the disulfide interaction between gp70 and Pr15E after cell lysis seems to be an intrinsic property of the Env complex, because this process appears not to involve a cellular factor. We therefore think that the disulfide linkage between gp70 and Pr15E rearranges with a free thiol within the Env protein itself after solubilization. Rearrangement of the intersubunit disulfide linkage has been proposed previously, although in a different context (14, 42). The reaction would in principle be possible, because the Env polypeptide contains an odd number of cysteines in its ectodomain, which implies that at least one of the sulfhydryls in the gp70-Pr15E complex is unoxidized. Why such a disulfide rearrangement would be induced upon solubilization of the Env protein is not known. Perhaps the Env protein complex undergoes conformational changes in the presence of detergents which make this reaction favorable.
Based on the analysis of tryptic fragments of the Friend MLV Env protein, Pinter et al. (42) recently proposed that the cysteine(s) of a CWLC sequence at the carboxy terminus of gp70 is involved in disulfide bonding with p15E. Interestingly, this peptide, which is highly conserved in the Env protein of MLV and other type C and D retroviruses (23, 47), resembles the active site of cellular enzymes engaged in thiol-disulfide exchange reactions (7). Our results are consistent with a model in which the CWLC peptide is involved in the rearrangement of the disulfide linkage between gp70 and Pr15E after solubilization. It has also been hypothesized that the CWLC sequence facilitates functional disulfide isomerization reactions necessary for the folding and activity of the Env protein in vivo (42).
The formation of disulfide bonds in the Env protein plays an important role in the establishment of its three-dimensional, functional structure (11, 12, 16, 32, 33, 52). Most, if not all, of the disulfide bonds in the Env molecule are settled co- and posttranslationally in the ER (14). It is likely that the disulfide linkage between gp70 and Pr15E has already been made in the precursor polypeptide before this polypeptide is transported to the cell surface and cleaved. This strategy ensures that the structural and functional integrity of the Env protein is maintained upon its cleavage despite the possibility that the cleavage changes the protein structure. Accordingly, the majority of the proteolytically processed Env molecules forms a stable disulfide-bonded complex. Nevertheless, a small amount of gp70 is not linked to Pr15E but is released from the cells. A fraction of the disulfide-linked Env complexes may be disrupted in vivo, e.g., through disulfide rearrangement, which would result in shedding of gp70. However, it is also possible that free gp70 is formed by cleavage of incorrectly oxidized Env precursor molecules that have not established the disulfide linkage between the gp70 and Pr15E domains. In this case, gp70 and Pr15E may not form a complex, and as a result, gp70 is released from cells as an ordinary secretory protein. Incorrectly oxidized and therefore possibly incompletely folded Env molecules are probably recognized and retained by the ER quality control system (20). Nevertheless, there are examples of misfolded or partially assembled membrane proteins that can escape from the ER and can be found at the cell surface (1). Indeed, it has been shown that abnormally folded glycosylation mutants of the MLV Env protein are transported to the plasma membrane (23, 30). Interestingly, several of these mutant Env proteins produced elevated levels of gp70, which was freely released from the cells (30).
It should be noted that free gp70 may accumulate in the medium to high concentrations during long-term incubation of virus-producing cells. This could be the reason why it was previously thought that gp70 and Pr15E are linked noncovalently (36). Comparing the amounts of free gp70 and gp70 present in virus-associated Env complexes could easily raise the suggestion that the disulfide-bonded gp70-Pr15E/p15E complexes are unstable, whereas this is not necessarily true. Soluble gp70 molecules may compete with virus particles in binding to the receptor (24, 35) and therefore may cause a potential problem for large-scale production of high-titer stocks of, for instance, recombinant retrovirus vectors used in gene therapy.
Recently, it has been shown that fragments corresponding to the core of the ectodomain of retroviral TM molecules (6, 11, 54) form a structure that resembles the low-pH-induced conformation of the influenza virus hemagglutinin protein (5). It is not yet known whether the retroviral TM molecules exhibit such a conformation throughout their lifetime or whether they assume an alternative conformation before membrane fusion takes place during virus entry. It is possible that the membrane fusion peptide is buried within the SU-TM complex, thus being protected from premature interactions, and exposed only after conformational changes which may be induced upon receptor binding (18, 43, 48). Further characterization of the fusion mechanism of retroviral Env proteins thus requires determination of the structure of Env in its prefusion state. Many retroviral Env proteins consist of a noncovalently linked SU-TM complex, whose integrity is difficult to maintain during purification. However, as we have shown here, the cell-associated MLV Env complex is stable in vivo and can be extracted without disruption; therefore, it represents an interesting candidate for X-ray crystallographic analysis.
ACKNOWLEDGMENTS
We thank B. W. Chesebro for the 83A-25 monoclonal antibody against gp70, and we are grateful to Kristina Wallengren and Ke-Jun Li for providing the DNA copies of the SFV expression vector containing the ecotropic and amphotropic Mo-MLV env genes, respectively. In addition, we thank José Casasnovas and Mathilda Sjöberg for critical reading of the manuscript.
REFERENCES
- 1.Barth B U, Garoff H. The nucleocapsid-binding spike subunit E2 of Semliki Forest virus requires complex formation with the E1 subunit for activity. J Virol. 1997;71:7857–7865. doi: 10.1128/jvi.71.10.7857-7865.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battini J-L, Rodrigues P, Müller R, Danos O, Heard J M. Receptor-binding properties of a purified fragment of the 4070A amphotropic murine leukemia virus envelope glycoprotein. J Virol. 1996;70:4387–4393. doi: 10.1128/jvi.70.7.4387-4393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedgood R M, Stallcup M R. A novel intermediate in processing of murine leukemia virus envelope glycoprotein. J Biol Chem. 1992;267:7060–7065. [PubMed] [Google Scholar]
- 4.Braakman I, Helenius J, Helenius A. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 1992;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 6.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope protein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 7.Chivers P T, Prehoda K E, Raines R T. The CXXC motif: a rheostat in the active site. Biochemistry. 1997;36:4061–4066. doi: 10.1021/bi9628580. [DOI] [PubMed] [Google Scholar]
- 8.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1767–1847. [Google Scholar]
- 9.Evans L H, Morrison R P, Malik F G, Portis J, Britt W J. A neutralizable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J Virol. 1990;64:6176–6183. doi: 10.1128/jvi.64.12.6176-6183.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fass D, Kim P S. Dissection of a retrovirus envelope protein reveals structural similarity to influenza hemagglutinin. Curr Biol. 1995;5:1377–1383. doi: 10.1016/s0960-9822(95)00275-2. [DOI] [PubMed] [Google Scholar]
- 11.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7Å resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 12.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 13.Freed E O, Risser R. The role of envelope glycoprotein processing in murine leukemia virus infection. J Virol. 1987;61:2852–2856. doi: 10.1128/jvi.61.9.2852-2856.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gliniak B C, Kozak S L, Jones R T, Kabat D. Disulfide bonding controls the processing of retroviral glycoproteins. J Biol Chem. 1991;266:22991–22997. [PubMed] [Google Scholar]
- 15.Green N, Shinnick T M, Witte O, Ponticelli A, Sutcliffe J G, Lerner R A. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu J, Parthasarathi S, Varela-Echavarría A, Ron Y, Dougherty J P. Mutations of conserved cysteine residues in the CWLC motif of the oncoretrovirus SU protein affect maturation and translocation. Virology. 1995;206:885–893. doi: 10.1006/viro.1995.1011. [DOI] [PubMed] [Google Scholar]
- 17.Heard J M, Danos O. An amino-terminal fragment of Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J Virol. 1991;65:4026–4032. doi: 10.1128/jvi.65.8.4026-4032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughson F M. Enveloped viruses: a common mode of membrane fusion? Curr Biol. 1997;7:565–569. doi: 10.1016/s0960-9822(06)00283-1. [DOI] [PubMed] [Google Scholar]
- 19.Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 20.Hurtley S M, Helenius A. Protein oligomerization in the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- 21.Hwang C, Sinskey A J, Lodish H F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 22.Jones J S, Risser R. Cell fusion induced by the murine leukemia virus envelope glycoprotein. J Virol. 1993;67:67–74. doi: 10.1128/jvi.67.1.67-74.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kayman S, Kopelman R, Kiney D, Projan S, Pinter A. Mutational analysis of N-linked glycosylation sites of the Friend murine leukemia virus envelope proteins. J Virol. 1991;65:5323–5332. doi: 10.1128/jvi.65.10.5323-5332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitagawa M, Aizawa S, Kamisaku H, Ikeda H, Hirokawa K, Sado T. Cell-free transmission of Fv-4 resistance gene product controlling Friend leukemia virus-induced leukemogenesis: a unique mechanism for interference with viral infection. Blood. 1995;86:1557–1563. [PubMed] [Google Scholar]
- 25.Konishi Y, Ooi T, Scheraga H A. Regeneration of ribonuclease A from the reduced protein. Rate-limiting steps. Biochemistry. 1982;21:4734–4740. doi: 10.1021/bi00262a033. [DOI] [PubMed] [Google Scholar]
- 26.Kosower N S, Kosower E M. Diamide: an oxidant probe for thiols. Methods Enzymol. 1995;251:123–133. doi: 10.1016/0076-6879(95)51116-4. [DOI] [PubMed] [Google Scholar]
- 27.Leamnson R N, Shander M H M, Halpern M S. A structural protein complex in Moloney leukemia virus. Virology. 1977;76:437–439. doi: 10.1016/0042-6822(77)90318-x. [DOI] [PubMed] [Google Scholar]
- 28.Lebedeva I, Fujita K, Nihrane A, Silver J. Infectious particles derived from Semliki Forest virus vectors encoding murine leukemia virus envelopes. J Virol. 1997;71:7061–7067. doi: 10.1128/jvi.71.9.7061-7067.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li K-J, Garoff H. Production of infectious recombinant Moloney murine leukemia virus particles in BHK cells using Semliki Forest virus-derived RNA expression vectors. Proc Natl Acad Sci USA. 1996;93:11658–11663. doi: 10.1073/pnas.93.21.11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Pinter A, Kayman S C. The critical N-linked glycan of murine leukemia virus envelope protein promotes both folding of the C-terminal domains of the precursor polyprotein and stability of the postcleavage envelope complex. J Virol. 1997;71:7012–7019. doi: 10.1128/jvi.71.9.7012-7019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liljeström P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 32.Linder M, Linder D, Hahnen J, Schott H-H, Stirm S. Localization of the intrachain disulfide bonds of the envelope glycoprotein 71 from Friend murine leukemia virus. Eur J Biochem. 1992;203:65–73. doi: 10.1111/j.1432-1033.1992.tb19828.x. [DOI] [PubMed] [Google Scholar]
- 33.Linder M, Wenzel V, Linder D, Stirm S. Structural elements in glycoprotein 70 from polytropic Friend mink cell focus-inducing virus and glycoprotein 71 from ecotropic Friend murine leukemia virus, as defined by disulfide-bonding pattern and limited proteolysis. J Virol. 1994;68:5133–5141. doi: 10.1128/jvi.68.8.5133-5141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Machida C A, Kabat D. Role of partial proteolysis in processing murine leukemia virus membrane envelope glycoproteins to the cell surface. J Biol Chem. 1982;257:14018–14022. [PubMed] [Google Scholar]
- 35.Nihrane A, Fujita K, Willey R, Lyu M S, Silver J. Murine leukemia virus envelope protein in transgenic-mouse serum blocks infection in vitro. J Virol. 1996;70:1882–1889. doi: 10.1128/jvi.70.3.1882-1889.1996. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Nihrane A, Lebedeva I, Lyu M S, Fujita K, Silver J. Secretion of a murine retroviral Env associated with resistance to infection. J Gen Virol. 1997;78:785–793. doi: 10.1099/0022-1317-78-4-785. [DOI] [PubMed] [Google Scholar]
- 37.Opstelten D-J E, de Groote P, Horzinek M C, Vennema H, Rottier P J M. Disulfide bonds in folding and transport of mouse hepatitis coronavirus glycoproteins. J Virol. 1993;67:7394–7401. doi: 10.1128/jvi.67.12.7394-7401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oroszlan S, Luftig R B. Retroviral proteinases. Curr Top Microbiol Immunol. 1990;157:153–185. doi: 10.1007/978-3-642-75218-6_6. [DOI] [PubMed] [Google Scholar]
- 39.Ott D, Friedrich R, Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990;64:757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinter A, Fleissner E. The presence of disulfide-linked gp70-p15E complexes in AKR murine leukemia virus. Virology. 1977;83:417–422. doi: 10.1016/0042-6822(77)90187-8. [DOI] [PubMed] [Google Scholar]
- 41.Pinter A, Lieman-Hurwitz J, Fleissner E. The nature of the association between the murine leukemia virus envelope proteins. Virology. 1978;91:345–351. doi: 10.1016/0042-6822(78)90382-3. [DOI] [PubMed] [Google Scholar]
- 42.Pinter A, Kopelman R, Li Z, Kayman S C, Sanders D A. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J Virol. 1997;71:8073–8077. doi: 10.1128/jvi.71.10.8073-8077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider J, Hunsmann G. Surface expression of murine leukemia virus structural polypeptides on host cells and the virion. Int J Cancer. 1978;22:204–213. doi: 10.1002/ijc.2910220215. [DOI] [PubMed] [Google Scholar]
- 45.Schneider J, Falk H, Hunsmann G. Envelope polypeptides of Friend leukemia virus: purification and structural analysis. J Virol. 1980;33:597–605. doi: 10.1128/jvi.33.2.597-605.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultz A M, Rein A. Maturation of murine leukemia virus env proteins in the absence of other viral proteins. Virology. 1985;145:335–339. doi: 10.1016/0042-6822(85)90168-0. [DOI] [PubMed] [Google Scholar]
- 47.Sitbon M, d’Auriol L, Ellerbrok H, André C, Nishio J, Perryman S, Pozo F, Hayes S F, Wehrly K, Tambourin P, Galibert F, Chesebro B. Substitution of leucine for isoleucine in a sequence highly conserved among retroviral envelope surface glycoproteins attenuates the lytic effect of the Friend murine leukemia virus. Proc Natl Acad Sci USA. 1991;88:5932–5936. doi: 10.1073/pnas.88.13.5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stamatatos L, Cheng-Mayer C. Structural modulations of the envelope gp120 glycoprotein of human immunodeficiency virus type 1 upon oligomerization and differential V3 loop epitope exposure of isolates displaying distinct tropism upon virion-soluble receptor binding. J Virol. 1995;69:6191–6198. doi: 10.1128/jvi.69.10.6191-6198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suomalainen M, Garoff H. Incorporation of homologous and heterologous proteins into the envelope of Moloney murine leukemia virus. J Virol. 1994;68:4879–4889. doi: 10.1128/jvi.68.8.4879-4889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suomalainen M, Hultenby K, Garoff H. Targeting of Moloney murine leukemia virus Gag precursor to the site of virus budding. J Cell Biol. 1996;135:1841–1852. doi: 10.1083/jcb.135.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takemoto L J, Fox C F, Jensen F C, Elder J H, Lerner R A. Nearest-neighbor interactions of the major RNA tumor virus glycoprotein on murine cell surfaces. Proc Natl Acad Sci USA. 1978;75:3644–3648. doi: 10.1073/pnas.75.8.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas A, Roth M J. Analysis of cysteine mutations on the transmembrane protein of Moloney murine leukemia virus. Virology. 1995;211:285–289. doi: 10.1006/viro.1995.1402. [DOI] [PubMed] [Google Scholar]
- 53.Thomas A, Gray K D, Roth M J. Analysis of mutations within the cytoplasmic domain of the Moloney murine leukemia virus transmembrane protein. Virology. 1997;227:305–313. doi: 10.1006/viro.1996.8333. [DOI] [PubMed] [Google Scholar]
- 54.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 55.Weissman J S, Kim P S. The pro region of BPTI facilitates folding. Cell. 1992;71:841–851. doi: 10.1016/0092-8674(92)90559-u. [DOI] [PubMed] [Google Scholar]
- 56.Witte O N, Tsukamoto-Adey A, Weissman I L. Cellular maturation of oncornavirus glycoproteins: topological arrangement of precursor and product forms in cellular membranes. Virology. 1977;76:539–553. doi: 10.1016/0042-6822(77)90236-7. [DOI] [PubMed] [Google Scholar]