Cytidine deaminase reduces replication stress and regulates DNA replication to confer resistance to DNA-damaging drugs in pancreatic cancer, unveiling a molecular vulnerability that could enhance treatment response.
Abstract
Cytidine deaminase (CDA) functions in the pyrimidine salvage pathway for DNA and RNA syntheses and has been shown to protect cancer cells from deoxycytidine-based chemotherapies. In this study, we observed that CDA was overexpressed in pancreatic adenocarcinoma from patients at baseline and was essential for experimental tumor growth. Mechanistic investigations revealed that CDA localized to replication forks where it increased replication speed, improved replication fork restart efficiency, reduced endogenous replication stress, minimized DNA breaks, and regulated genetic stability during DNA replication. In cellular pancreatic cancer models, high CDA expression correlated with resistance to DNA-damaging agents. Silencing CDA in patient-derived primary cultures in vitro and in orthotopic xenografts in vivo increased replication stress and sensitized pancreatic adenocarcinoma cells to oxaliplatin. This study sheds light on the role of CDA in pancreatic adenocarcinoma, offering insights into how this tumor type modulates replication stress. These findings suggest that CDA expression could potentially predict therapeutic efficacy and that targeting CDA induces intolerable levels of replication stress in cancer cells, particularly when combined with DNA-targeted therapies.
Significance:
Cytidine deaminase reduces replication stress and regulates DNA replication to confer resistance to DNA-damaging drugs in pancreatic cancer, unveiling a molecular vulnerability that could enhance treatment response.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) will soon become the second cause of death by cancer worldwide (1). Recent studies demonstrate that 10% to 15% of patients with PDAC show unstable genotypes with chromosomal structural variations characterized by the inactivation of DNA repair genes (BRCA1, BRCA2, PALB2, ATM, and RAD51; refs. 2, 3). Such molecular defects can advocate for promising targeted therapies (4) as transcriptomic signatures of high replication stress predict response to ATR and WEE1 inhibitors in preclinical models (5). More importantly, patients with germline mutations in the BRCA2 gene were recently found to benefit from first-line platinum-based FOLFIRINOX regimen followed by PARP inhibitors treatment (6). Thus, harnessing replication stress is a promising therapeutic avenue in this cancer (4), but the molecular mechanisms that govern replication stress in PDAC remain largely unknown.
Within the pyrimidine salvage pathway, cytidine deaminase (CDA) catalyzes the irreversible hydrolytic deamination of cytidine and deoxycytidine to uridine and deoxyuridine, respectively, for RNA and DNA syntheses (7). Unfortunately, this property can be diverted by tumor cells to deaminate and neutralize deoxycytidine-based therapies. Thus, CDA is now seen as a major contributor of tumor chemoresistance, especially to gemcitabine in PDAC (7). CDA catalytic inhibitors have been developed for cancer-related therapy, but they lack specificity (8, 9) and recently failed to show efficacy in pilot clinical trial in advanced, chemo-refractory PDAC (10). Interestingly, loss of CDA expression in Bloom syndrome cells results in a pyrimidine pool imbalance, which in turn participates to DNA replication stress. This leads to chromosome segregation defects (11) in a genetic disease with one of the strongest known correlation between chromosomal instability and increased risk of malignancy (12). Hence, this study suggests an intriguing connection between CDA expression and DNA replication stress that we aimed to explore in PDAC. Our study demonstrates an association between CDA expression and resistance to DNA-damaging agents. Moreover, when we silenced CDA in patient-derived primary cultures and orthotopic xenografts in vivo, it sensitized tumor cells to oxaliplatin. These findings suggest potential implications for the management of patients with PDAC as CDA expression may serve as an indicator of efficacy for DNA-damaging drugs, whereas targeting CDA presents a novel vulnerability, potentially inducing significant replication stress in cancer cells, particularly in combination with compounds affecting DNA metabolism.
Materials and Methods
Cell lines, culture conditions, and treatments
The MIA PaCa-2 [CRL-1420, DNA damage response (DDR)-proficient], BxPC3 (CRL-1687, DDR-deficient, mutated for POLQ; ref. 4), Capan-1 (HTB-79, DDR-deficient, mutated for BRCA2 and ATM; ref. 4), and Hela S3 (CCL-2.2) human cancer cells were obtained from the ATCC and cultured in DMEM 4.5 g/L glucose (for MIA PaCa-2 and HeLa S3) or RPMI (BxPC3 and Capan-1) containing 10% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin and 250 ng/mL fungizone (AB), and 100 μg/mL normocin (Invivogen) to avoid Mycoplasma contamination. Cells were incubated at 37°C with 5% CO2. We used cell lines under passage 10, with the exception of late passage studies, that were certified Mycoplasma-free using the Mycoplasma detection kit (Invivogen). Pancreatic cancer patient–derived primary cultures PDAC015T and PDAC051T were cultured as described before (13). Cells were incubated at 37°C with 5% CO2. PDAC cells were seeded and 24 hours later were treated with CDA pharmacologic inhibitors (THU, DR) for 72 hours at 100 μmol/L. To induce replicative stress, cells were treated with 0.3 μmol/L aphidicolin or 10 μmol/L oxaliplatin for 24 hours, cells were treated with DMSO as control.
Flow cytometry
Annexin V staining was performed with the Annexin V Apoptosis Detection Kit (BD Biosciences) following the manufacturer's recommendations. For cell-cycle analysis, the protocol was adapted from (14). Data were collected on the MACS Quant Q10 cytometer and analyzed using the MACS Quant software.
Mice models
Experimental procedures performed on mice were approved by the ethical committee of INSERM CREFRE US006 animal facility and authorized by the French Ministry of Research: APAFIS#3600–2015121608386111v3. MIA PaCa-2 cells (2 × 106) stably expressing luciferase and shRNA targeting CDA following lentiviral delivery were engrafted into the pancreas of 6-week-old SCID CB17 mice (Charles River Laboratories) as previously described (15). MIA PaCa-2 cells expressing random siRNAs were used as controls. Tumor progression was monitored once per week by luminescence after intra peritoneal injection of Rediject D-Luciferin (PerkinElmer). Measurement of luminescence was assessed using an IVIS Spectrum apparatus (PerkinElmer). At endpoint, tumors were collected and measured using a caliper, stained for hematoxylin and eosin, and analyzed with an expert pathologist for peripheral necrotic content. For interventional studies, a total of 1×106 PDAC015T cells resuspended in Matrigel were injected subcutaneously in athymic mice (6-week-old SCID CB17 mice from Charles River Laboratories). Tumors were measured with a caliper and tumor volume was calculated using the formula V = (Width2 × Length)/2. Two weeks later, tumors ranging from 150 to 350 mm3 were randomized in two groups (day 0). Tumors were injected intratumorally at days 0 and 5 with 4 μg of siRNA pool targeting CDA (Dharmacon) using in vivo JetPEI (N/P = 6) according to the manufacturer's recommendations (Polyplus). Tumors receiving control siRNA pool (Dharmacon) were used as control. At days 2 and 7, mice received 5 mg/kg of oxaliplatin. Mice were sacrificed on day 10 following cotreatment.
Patient cohorts and clinical sample processing
We used two patient cohorts for CDA expression analysis in normal adjacent and PDAC tissues. The first one includes pancreatic tissue samples obtained from patients receiving pancreatic surgery following the policies and the practices of the facility's ethical committee at the Centre Hospitalo-Universitaire of Toulouse and Bordeaux, and the Cancéropole Grand Sud-Ouest (France), as stated before (16). All patients gave their informed written consent. Histopathology faculty selected cancerous pancreatic tissue with matched adjacent tissue. RNA was extracted and processed as stated before (16). The second cohort was established from pancreatic carcinoma tissues and normal adjacent pancreatic parenchyma collected from 48 patients who had undergone surgical resection for ductal adenocarcinoma of pancreas between 2003 and 2011 at the Bellvitge Hospital (Barcelona, Spain; ref. 17). The study was approved by the Ethical Committee of University Hospital of Bellvitge CEIC 02/04 and written informed consent was obtained from all patients for the use of their tissues. The third and last cohort is composed of 108 patients with histologically proven, locally advanced or metastatic PDAC enrolled in four French referral centers (Clichy, Marseille, Montpellier, and Toulouse) from January, 2005 to April, 2007. Samples were obtained by fine needle aspiration and processed as described elsewhere (18). Informed written consent for analysis was obtained for all patients.
Plasmid cloning
For silencing studies, pLKO.1 puromycin-resistant lentiviral vectors encoding for shRNAs sequences targeting CDA (CCG GCA TGA GAG AGT TTG GCA CCA ACT CGA GTT GGT GCC AAA CTC TCT CAT GTT TTT G) were obtained from Sigma. A lentivector encoding for random shRNA (Sigma) was used as a control. For overexpression studies, we amplified the open reading frame (ORF) of CDA by PCR and subcloned it into Gateway pDONR221 plasmid (Invitrogen). CDAE67Q catalytically inactive mutant (19) was generated by site directed mutagenesis according to the manufacturer's instructions (New England Biolabs). Renilla luciferase 8 (RLuc8) was amplified by PCR from pEF-RLuc8-MCS plasmid and was used as a control (20). Lentiviral expression constructs were obtained by cloning 3xFLAG-CDA, CDAE67Q, and RLuc8 ORFs into pCMV blasticidin DEST vector (Addgene #706–1) using the Gateway strategy (Invitrogen). All constructs were sequence verified. Lentiviral particles production was adapted from (21). Briefly, a total of 4.5 × 106 HEK293T cells were plated in a 100-mm-dish. Twenty-four hours later, cells were transfected with 6 μg of pCMV blasticidin DEST 7vector, 4 μg of psPAX2 and 1.5 μg of pMD2.G and 23 μL of JetPRIME transfection reagent (Polyplus; quantities for one 100-mm-dish). The supernatants were collected 48 hours after transfection, centrifuged 5 minutes at 2000 rpm, filtered (0.45-μm-filter) and concentrated by centrifugation at 2,500 rpm using a 100 kDa cutoff Vivaspin 20 concentrator (Sartorius). For lentiviral transduction of shRNAs and ORF-expressing constructs, 250 ng of p24/5×104 cells were used in Opti-MEM medium (Thermo Fisher Scientific) containing 4 μg/mL protamine sulfate, as previously described (22). Transductions were performed overnight; the medium was changed the next day and transduced cells were selected with 5 μg/mL of blasticidin (Invivogen) 2 days later. Cell cultures were maintained as pools and certified Mycoplasma-free (Lonza).
Cell confluence analysis
For proliferation studies, a total of 6 × 103 cells were seeded in 96-well plates in 100 μL of complete medium. Medium was changed and cells were treated 24 hours after seeding. Confluence was quantified using an Incucyte Zoom apparatus (Sartorius). For silencing studies, siRNA smartpools were purchased from Dharmacon. Cells were transfected with 20 nmol/L of siRNA targeting CDA using JetPrime (Polyplus) following manufacturer's recommendations. Control cells were transfected with control siRNA (Dharmacon). Three days later, cells were treated or not with 10 μmol/L of oxaliplatin and cell confluence was monitored as previously described. Colony formation assays were performed in 6-well plates, with 250 cells initially seeded. Colony presence was revealed at day 10 by cold methanol staining and crystal violet (Sigma) coloration. Density was measured with Image laboratory software (Bio-Rad).
RNA isolation and gene expression analysis
Total RNA was extracted with the RNeasy kit (Qiagen) according to the manufacturer's instructions; quality and quantity were measured on a Nanodrop system (Thermo Fisher Scientific). cDNA synthesis was performed with the Revertaid H minus kit (Thermo Fisher Scientific). cDNA expression analysis was performed using Sybr Green (Bio-Rad) qRT-PCR on a StepOne thermocycler (Thermo Fisher Scientific). CDA forward: GGGG ACA AGT TTG TAC AAA AAA GCA GGC TAT GGC TAT GGC CCA GAA GCG T. CDA reverse: GGGG AC CAC TTT GTA CAA GAA AGC TGG GTT CAC TGA GTC TTC TG.
CDA enzymatic activity assay
The CDA activity assay kit was performed following the manufacturer's instruction with 20 μg of total proteins (BioVision, #K451–100).
RNA-seq data analysis and functional enrichment
RNA-seq data were processed with standard approaches. Briefly, we first converted from Illumina format to fastq using bcl2fastq (https://emea.support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html). Quality control was performed with FastQC and adaptor sequences were trimmed using Trimmomatic. Quality control with FastQC was run again after adaptor removal. Gene expression was quantified at the transcript level by Salmon. Import of Transcript-level abundances and counts from Salmon and name conversion to HUGO gene symbols was performed with the tximport R package. Differential expression analysis was performed with DeSeq2 (23) using control samples as the reference for the sign of the log2-FoldChange. Enrichment analyses were performed using gene set enrichment analysis (GSEA) Hallmarks and Reactome (24) and most other pathways were extracted from MSigDB. No genes were removed during the analysis.
The Cancer Genome Atlas data analysis
HTSeq counts for PDAC were downloaded from the GDC portal. Curated PDAC samples were filtered on the basis of the information from (25). Samples were classified according to low (less than second quartile), normal (between second and third quartile), and high (higher than third quartile) values of CDA expression. Differential expression analysis comparing low CDA versus high CDA samples was performed with DESeq2 using low samples as the reference for the sign of the log2-FoldChange.
Instability signatures and metrics
The CINSARC signature gene list was obtained from (26). Aneuploidy score and Mutational Load score (number of mutations per Mb) data were downloaded from the GDC portal.
Single-cell library preparation and single-cell analysis
The single-cell suspension was loaded onto the Chromium Controller (10x Genomics) and the single-cell libraries preparation was performed using the Chromium Next GEM single Cell 3′ reagent kit v3.1 Dual Index (10x Genomics) according to the manufacturer's instructions. The libraries were qualified with the HS NGS kit for the Fragment Analyzer (Agilent Technologies) and quantified using the KAPA library quantification kit (Roche Diagnostics). The libraries were sequenced on the Illumina NextSeq550 instrument using a High Output 150 cycles kit and the cycling parameters: 28 (read 1), 10 (index 1), 10 (index 2), 90 (read 2) to obtain more than 20,000 read pairs per cell. The sequencing output (bcl2 files) was demultiplexed and aligned on the human reference genome GRCh38 using the software CellRanger v6.1.1 (27). Quality control, normalization, PCA, and t-SNE/UMAP coordinate computation were assessed using the R package Seurat 4.1.0 (28). Quality control was performed using the number of detected genes per cell, and the mitochondrial genes proportion. Enrichment scores were computed using Single-Cell Signature Scorer (29). These encompassed signatures downloaded from MsigDB v7.5.1 (30). For differential pathway analysis, a Student t test or a Mann–Whitney U was applied, if the data followed a normal distribution (validated by the Shapiro–Wilk test for normal distribution). Corrected P values were obtained using a Bonferroni correction. Figures were produced using the ggplot2 (R package). The software used in the present study for single-cell analysis is available here (https://sites.google.com/site/fredsoftwares/home).
Whole-genome sequencing and analysis
MIA PaCa-2 cells were transduced with lentiviral vectors expressing CDA as described before. As controls, cells were transduced with lentiviral vectors encoding for Renilla luciferase 8 or CDAE67Q catalytically inactive mutant. Genomic DNA was harvested using the QIAGEN QIAamp DNA extraction kit at early (passage 3, 5 days of culture) and late (passage 15, 55 days of culture) time points following transduction. Whole-genome sequencing (WGS) was subcontracted to Novogene. Briefly, between 2 and 10 μg of cellular genomic DNA was purified using the DNeasy Tissue kit from Qiagen. The genomic DNA was randomly fragmented by sonication to the size of 350 bp, then DNA fragments were end-polished, A-tailed, and ligated with the full-length adapters of Illumina sequencing, and followed by further PCR amplification. The PCR products were purified with the AMPure XP system. Then libraries were checked for size distribution by Agilent 2100 Bioanalyzer (Agilent Technologies), and quantified by RT-PCR (to meet the criteria of 3 nmol/L). Libraries were sequenced on an Illumina platform (PE150), then quality check and bioinformatic analysis were performed by the provider (Novogene). The percentage of clean reads in all raw reads was more than 99.83% for all samples. The number of reads that mapped to the reference genome and within the expected insert size was more than 98.8% for all samples.
Western blot analysis
Total cell lysate protein extraction for immunoblot analysis was performed using RIPA buffer (Biotech; 50 mmol/L Tris-HCl pH 8, 150 mmol/L NaCl, 0.5% NP40, 0.5% sodium deoxycholate, 0.1% SDS) in the presence of protease inhibitors (Sigma). Extracts were separated by SDS-PAGE under reducing conditions, transferred to a nitrocellulose membrane and analyzed by immunoblotting for cleaved PARP (Bethyl #9541), Cleaved CASPASE-3 (Cell Signaling Technology #9664, 1:1,000), P-CHK1 S345 (Cell Signaling Technology #2348, 1:1,000), CHK1 (Cell Signaling Technology #2360, 1:1,000), CDA (Abclonal A13959, 1:1,000), FLAG (Sigma-Aldrich F1804, 1:5,000), GAPDH (Cell Signaling Technology #5174, 1:5,000), β-actin (Santa Cruz Biotechnology sc-47778, 1:2,000), HSP90 (Cell Signaling Technology #4874, 1:2,000), SP1 (Santa Cruz Biotechnology sc-59 1:5,000), 4EBP1 (Cell Signaling Technology #9644 1:5,000), PCNA (Sigma-Aldrich P8825), and MCM7 (Abcam ab2360). Appropriate horseradish peroxidase (HRP) conjugate secondary antibodies were purchased from Promega. Signal was detected using the ECL system (Bio-Rad) according to the manufacturer's instructions. Densitometry of the bands was done using Chemidoc system (Bio-Rad) and image Laboratory software. Cytoplasmic/nuclear extract isolation was performed as following: Cell pellet was resuspended in 10 mmol/L Tris-HCl, pH 7.4 containing 1.5 mmol/L MgCl2, 5 mmol/L KCl, 0.5 mmol/L dithiothreitol, 0.5% NP-40 and 0.5 mmol/L phenylmethylsulfonyl fluoride complemented with protease inhibitors and incubated on ice for 10 minutes. The mixture was centrifuged for 15 minutes at 4°C at 2,000 rpm. Supernatant was collected (cytoplasmic fraction) and the pellet was washed and centrifuged twice with the same buffer as previously. The pellet was then resuspended in 20 mmol/L Tris-HCl, 0.025% glycerol, 1.5 mmol/L MgCl2, 0.5 mmol/L phenylmethylsulfonyl fluoride, 0.2 mmol/L EDTA, 0.5 mmol/L dithiothreitol and 0.4 mol/L NaCl complemented with protease inhibitors and incubated on ice for 15 minutes. The mixture was centrifuged 20 minutes at 4°C at 12,000 rpm and supernatant was collected (nuclear fraction).
Indirect immunofluorescence protocol
Cells were seeded in 6- or 12-well plates, containing coverslips, in 1–3 mL of complete medium. Medium was changed and cells were treated 24 hours after seeding. For the study of cells in S-phase, EdU 10 μmol/L (5-ethynyl-2′-deoxyuridine, Thermo Fisher Scientific) was added for 10 (MIA PaCa-2), 15 (Capan-1) or 20 minutes (BxPC3). Cells were pre-extracted with 0.2% Triton-X100 for 3 minutes, fixed in 4% PFA for 15 minutes and permeabilized with 0.5% Triton X-100 for 20 minutes. For revealing EdU, the Click-iT EdU Imaging kit (Thermo Fisher Scientific) was used. Samples were blocked with PBS 5% BSA. Primary antibodies were anti–phospho-histone H2AX (Ser139; JBW301, Millipore 05–636, 1:1,000), anti–phospho-53BP1 (S1778; Cell Signaling Technology #2675, 1:1,000), anti–phospho-RPA2 (S4-S8; Bethyl A300–245A, 1:500), anti-RPA70 (Cell Signaling Technology #2267, 1:500), anti-FANCD2 (NOVUSBIO NB100–182, 1:1,000), and anti-53BP1 (Bethyl A300–272A, 1:2,500). DNA was stained with 4,6-diamidino-2-phenylindole (DAPI) and images acquired on Nikon DS-Qi2 fluorescence microscope. ImageJ and CellProfiler software were used to quantify the number of foci and staining intensity per nucleus. For each condition, at least 500 cells were measured. For quantification in early mitotic cells, at least 50 cells in prophase and prometaphase were counted.
IHC
Tumors were harvested and fixed in formalin. Four-micrometer-thick sections were prepared from paraffin-embedded sections and rehydrated. Antigen retrieval was performed using Sodium Citrate Buffer (10 mmol/L Sodium Citrate, 0.05% Tween 20, pH 6.0) and Citric Acid Buffer (10 mmol/L Citric Acid, 0.05% Tween 20, pH 6.0) following routine protocols. Nonspecific signal was blocked using Blocking reagent (DakoCytomation). Slides were incubated overnight at 4°C with anti-Ki67 (SP6; ab16667 Abcam) or Cleaved Caspase-3 (Asp175) antibody #9661 (Cell Signaling Technology) antibodies diluted following the manufacturer's recommendation in Antibody diluent solution (DakoCytomation). Endogenous peroxidase activity was quenched using H2O2, and ffter several washes in PBS, EnVision+ System HRP-labeled polymer anti-rabbit were added as per requested by the manufacturer's (DakoCytomation), followed by HRP streptavidin (dilution 1:500, SA-5004, Vector). Slides were quickly washed twice in PBS and incubated in AEC+ reagent and counterstained with Mayer's hematoxylin (DakoCytomation). After washing in PBS, slides were mounted with Vectashield (Vector). Immunostaining was recorded with an AXIO optical microscope (Zeiss) equipped with a color AXIOCAM camera 105 (Zeiss, Oberkochen, Germany) and quantified using ImageJ. 15 fields per tumor (n = 2 per condition) were analyzed.
DNA fiber assay
Cells were seeded in 6-cm diameter Petri dishes in 3 mL of complete medium. Cells were incubated with 50 μmol/L IdU and 100 μmol/L CldU for 30 minutes at 37°C, with washing between the two pulses. Cells were harvested, resuspended (0.5×106 cells/mL in PBS) and 2 μL were spotted onto microscope slides after and 7 μL of lysis buffer were added (200 mmol/L Tris-HCl pH 7.4, 50 mmol/L EDTA, 0.5% SDS). Glass slides were tilted and dried overnight, DNA spreads were then fixed in ethanol/acetic acid (3:1) 20 minutes at −20°C. Samples were incubated in pepsin buffer (0.5 mg/mL Pepsin, 30 mmol/L HCl) at 37°C for 20 minutes, denatured in HCl 2.5 mol/L at 37°C for 45 minutes and blocked in PBS 1% BSA 0.1% Tween-20. DNA fibers were incubated with mouse-FITC-anti–bromodeoxyuridine (detects IdU, B44, Becton Dickinson, 1:50) and rat-anti–bromodeoxyuridine (detects CldU, Abcam ab6326, 1:100), at 37°C for 1 hour and then with anti-mouse IgG AlexaFluor 488 (Invitrogen A11029, 1:200) and anti-rat IgG AlexaFluor 555 (Invitrogen A21094, 1:200), at 37°C for 1 hour. Images were acquired on a Nikon DS-Qi2 fluorescence microscope and Axio Observer Z1 (Zeiss) and ImageJ was used to measure the length (μm) of at least 100 DNA tracks.
iPOND assay
For EdU-labeled sample preparation, MIA PaCa-2 cells overexpressing CDA-FLAG (∼2 × 108 cells per condition) were incubated with 10 μmol/L of EdU for 15 minutes. For pulse-chase experiments with thymidine, EdU-labeled cells were washed once with warm media to remove the EdU and then incubated with 10 μmol/L thymidine for 2 hours. Next, cells were cross-linked in 2% PFA for 15 minutes, quenched using 0.125 mol/L glycine and washed three times with PBS. Collected cell pellets were frozen in liquid nitrogen and stored at −80°C. Cells were permeabilized with 0.5% Triton-X100 for 30 minutes and Click-it chemistry was used to conjugate biotin–TEG-azide (Eurogentec) to EdU-labeled DNA in PBS containing 10 mmol/L sodium Ascorbate, 10 μmol/L biotin–TEG-azide, 2 mmol/L CuSO4. Cells were resuspended in a lysis buffer (10 mmol/L Hepes-NaOH; 100 mmol/L NaCl; 2 mmol/L EDTA PH8; 1 mmol/L EGTA; 1 mmol/L PMSF; 0.2% SDS; 0.1% Sarkozyl). Sonication was performed using a Qsonica sonicator with the following settings: 30% power, 20 seconds constant pulse and 50 seconds pauses for a total sonication time of 5 minutes on ice with water. Lysates were centrifuged at 13,200 rpm for 10 minutes at room temperature. Supernatants were normalized by DNA quantification using a Nanodrop device. Biotin-conjugated DNA–protein complexes were captured using overnight incubation with magnetic beads coated with streptavidin (Ademtech). Captured complexes were washed with a lysis buffer and 500 mmol/L NaCl. Proteins associated with nascent DNA were eluted under reducing conditions by boiling into SDS sample buffer for 30 minutes at 95°C and analyzed by Western blot.
Statistical analysis
Unpaired Student t or Wilcoxon–Mann–Whitney tests were used to determine the statistical significance of differences between two groups using GraphPad Prism 10 software with the default settings. Methods of statistical analysis are indicated in the figure captions. Values are presented as *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Error bars are s.e.m. unless otherwise stated. The experiments were conducted with a sample size (n) equal to or greater than three replicates. Results from representative experiments were validated through at least two independent repetitions and across multiple cell lines. In animal studies, female mice (n ≥ 5 mice) were used in an age-matched manner. During bioluminescence imaging for tumor growth monitoring, investigators were blinded to group allocation. Experiments were not randomized, and investigators were not blinded to allocation during the experiments or outcome assessment. No data were excluded from the analyses.
Data availability statement
Publicly available The Cancer Genome Atlas (TCGA) data analyzed in this study were obtained from the GDC portal (https://portal.gdc.cancer.gov/projects/TCGA-PAAD). Raw genomic and transcriptomic data for this study were generated at the Cancer Research Center of Toulouse sequencing facility, and deposited in GEO GSE253662 (CDA and CDAE67Q overexpression in cell lines, RNA-seq), GSE252544 (CDA targeting with siRNA in cell lines, RNA-seq), GSE253837 (CDA targeting with siRNA in patient-derived experimental tumors, scRNAseq), and GSE252544 and SRA PRJNA1066503 (CDA and CDAE67Q overexpression in cell lines, WGS). All other raw data are available upon request from the corresponding author.
Results
CDA is overexpressed in PDAC and involved in cell proliferation and tumor growth
CDA expression in tumors has been scarcely investigated to date (7), with only few reports indicative of CDA upregulation in human samples (31) and in PDAC animal models (32). We therefore characterized the expression of CDA in tumor and normal adjacent parenchyma and found that CDA mRNA is significantly overexpressed (5.1 ± 1.5-fold increase, P < 0.0001) in PDAC tissue (Fig. 1A). This finding was confirmed in the curated TCGA_PAAD dataset (Supplementary Fig. S1A; ref. 25). On the contrary, expression of dihydroorotate dehydrogenase (DHODH) from the de novo pyrimidine pathways is downregulated in PDAC tissues, as compared with normal adjacent tissue (Supplementary Fig. S1B). In addition, CDA expression is significantly increased in late-stage tumors as compared with locally advanced tumors from the Bacap cohort (Supplementary Fig. S1C). We next classified PDAC patient samples from TCGA as CDA high-expressing (top 25% quartile) or CDA low-expressing (bottom 25% quartile). We found that CDA expression is positively associated with a shorter survival (P < 0.03; HR, 2.021; Fig. 1B). Transcriptomic analysis recently revealed two main molecular subgroups of PDAC with distinct biology (33); classical tumors show the highest expression of epithelial and adhesion-associated genes, whereas basal-like tumors are associated with a poor differentiation status and a worst outcome (34). Using the Purity Independent Subtyping of Tumors classifier (35), we demonstrate that CDA expression is significantly enriched in basal-like tumors as compared with classical tumors (2.3 ± 0.1-fold increase, P < 0.0001, Supplementary Fig. S1D). We extended this analysis to PDAC primary cultured cells (5) and identified that CDA mRNA is significantly increased (1.2 ± 0.04-fold increase, P < 0.05) in cells with basal-like, squamous molecular subtype (Supplementary Fig. S1E). Thus, our findings indicate that CDA is overexpressed in PDAC. In addition, PDAC tumors displaying elevated CDA expression show association with aggressive molecular characteristics and potentially shorter patient survival.
Figure 1.
CDA is overexpressed in PDAC tumors and is essential to cell proliferation and tumor growth. A, Expression of CDA mRNA in pancreatic tumors and matched normal tissues (n = 37) from CHU Toulouse and Idibell cohorts. Each dot represents one sample. B, Kaplan–Meier survival plot for CDA expression in patients with PDAC from TCGA cohort (n = 41). C, Growth analysis of MIA PaCa-2, Capan-1, and BxPC3 cells expressing or not siRNA against CDA. Mean of three independent experiments performed in technical triplicate. D–F, Colony formation assay (D), Annexin V staining (E), or Western blot analysis of cleaved PARP, cleaved caspase-3 (F) of MIA PaCa-2 cells stably expressing or not siRNA against CDA. Results are expressed as mean ± SEM of four independent experiments. GAPDH was used as loading control. G, Individual tumor progression (bioluminescence unit intensity, BLI; fold increase) of Mia PaCa-2 cells expressing control siRNA (n = 7) or CDA siRNA (n = 6). H, Tumor volume at endpoint. I, Immunostaining for Ki67 (top) and cleaved caspase-3 (bottom) in experimental Mia PaCa-2 tumors expressing control or CDA siRNA. Representative of 15 fields from three different tumors. Arrows, cells with positive signal. Scale bar, 150 μm. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
We next addressed the functional importance of CDA in PDAC. We inhibited CDA expression in Mia PACA-2, Capan-1, and BxPC3 cells using stable expression of shRNAs (Supplementary Fig. S1F and S1G) and demonstrated that CDA targeting strongly decreases PDAC cells proliferation (−90% ± 1%, P < 0.0001, −58% ± 9%, P < 0.001, −65% ± 8%, P < 0.001, respectively), as compared with control cells expressing random shRNA (Fig. 1C). In addition, MIA PaCa-2 cells expressing CDA shRNA showed a reduced ability to form colonies when compared with control cells (−65% ± 3%, P < 0.05, Fig. 1D). CDA invalidation also increases cell death by apoptosis as shown by FACS for Annexin V (6.05 ± 3 fold-increase, P < 0.05 Fig. 1E) and western blotting for PARP and caspase-3 cleavage (Fig. 1F). We next engrafted MIA PaCa-2 cells stably expressing CDA shRNA or control shRNA in the pancreas of athymic mice. Figure 1G shows that silencing of CDA in PDAC cells results in significant inhibition of tumor growth, and tumor volume at endpoint (−52% ± 18%, P < 0.005, Fig. 1H). CDA silencing also significantly decreases tumor size (Supplementary Fig. S1H) and weight (−53% ± 16%, P < 0.01, Supplementary Fig. S1I). Last, the Ki67 cell proliferation marker shows fewer positive cells in the CDA shRNA condition compared with the control condition, whereas conversely, the cleavage of caspase-3, synonymous with apoptosis, is more significant in the CDA shRNA group (Fig. 1I). Collectively, our data indicate that PDAC cell proliferation and tumor growth strongly rely on CDA expression.
CDA overexpression increases replication fork speed and promotes restart in response to replication stress
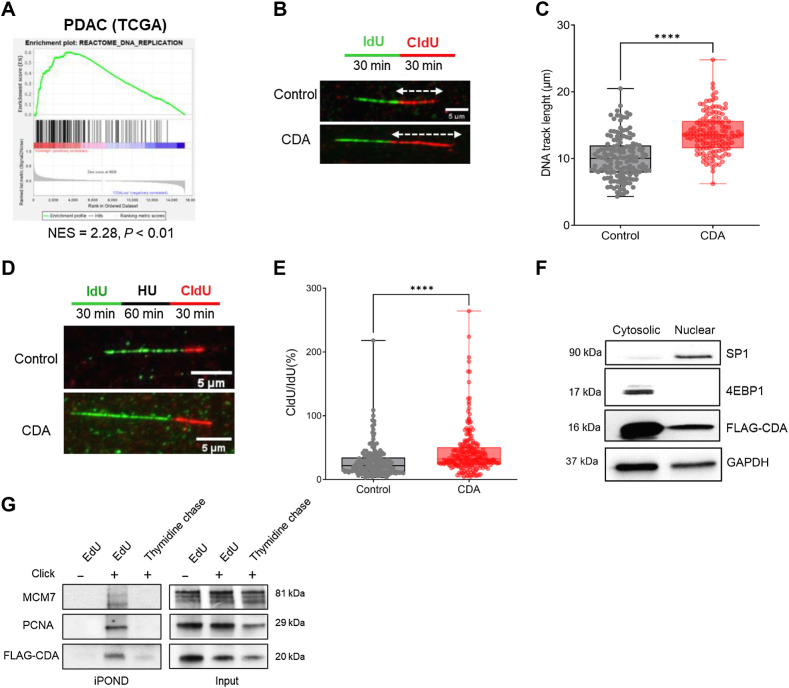
To gain further insights into the role of CDA in PDAC, we analyzed the transcriptome of PDAC samples from TCGA that overexpress this enzyme. As shown in Fig. 2A; Supplementary Table S1, CDA expression positively correlates with the transcriptomic signature of DNA replication (NES = 2.28, P < 0.01). For functional studies, we used lentiviral cDNA delivery to increase CDA mRNA (Supplementary Fig. S2A) and protein (Supplementary Fig. S2B) expression in Mia PACA-2 cells. As control, MIA PaCa-2 cells expressed a catalytically inactive mutant (CDAE67Q) of the enzyme (19), or luciferase (control). We verified that CDA catalytic activity was elevated in CDA-expressing Mia PACA-2 cells, as compared with control cells expressing CDAE67Q or luciferase (Supplementary Fig. S2C).
Figure 2.
CDA increases replication fork speed and restart efficiency in PDAC cells. A, Enrichment plots. DNA replication pathway (GSEA-Reactome) of PDAC samples from TCGA sorted for CDA expression (high, CDA n = 39; low CDA, n = 38). B, Immunofluorescence detection of IdU (green) and CldU (red) DNA tracks after DNA stretching of MIA PaCa-2 cells overexpressing or not CDA. White arrows, measured red tracks. C, Quantification of the length of DNA tracks (μm); at least 100 DNA fibers were measured per condition. D, Immunofluorescence detection of IdU (green) and CldU (red) DNA tracks after DNA stretching of control MIA PaCa-2 cells, or cells overexpressing CDA. HU, hydroxyurea, treatment at 4 mmol/L for 1 hour. E, Ratio of CldU/IdU tracks length after hydroxyurea treatment (μm); at least 70 DNA fibers were measured per condition. Results are representative of three independent transduction pools. F, Western blot analysis of FLAG-CDA, 4EBP1, and SP1 in cytosolic and nuclear fractions of MIA PaCa-2 cells overexpressing CDA (same time exposure). Representative of three independent experiments. G, IPOND experiments in MIA PaCa-2 cells overexpressing FLAG-CDA. PCNA and MCM7 were used as controls. Cells were treated with EdU and revealed or not with Click-it EdU (EdU− and EdU+, respectively). After EdU incorporation, cells were treated with thymidine to chase EdU and to address whether candidate proteins progress with replication fork. Results are representative of four independent experiments. ****, P < 0.0001, Wilcoxon-Mann-Whitney test.
We found that MIA PaCa-2 CDA cells recapitulate the DNA replication signature enrichment (NES = 1.98, P < 0.01) found in tumors (Supplementary Fig. S2D; Supplementary Table S2). Importantly, this enrichment was not found in cells overexpressing CDAE67Q catalytically inactive mutant (NES = 1.19, P = 0.144). We then performed DNA-spreading assays to quantify the progress of individual DNA replication forks (extended Fig. 2E). Figure 2B and C shows that DNA tracks are significantly longer in cells with a high CDA expression as compared with control cells (12.5 ± 0.37 μm vs. 9.4 ± 0.34 μm, P < 0.0001), indicating that the replication fork speed is increased in these cells. This finding is further supported by a slight decrease of the proportion of cells in S-phase (30.2% ± 1.3% vs. 35.8% ± 2.1%, P < 0.05) as compared with control cells (Supplementary Fig. S2F), reminiscent of a shorter S phase, whereas cell doubling time remains unchanged (Supplementary Fig. S2G), indicating that increased DNA replication is not linked to increased proliferation of these cells.
We further investigated whether CDA also rescues stalled replication forks. Cells were treated with 4 mmol/L hydroxyurea to inhibit riboside nucleotide reductase (RNR) and to deplete dNTPs to induce fork stalling. Using DNA-spreading analysis, we found that cells with a high CDA expression show a higher percentage of replication forks restarting DNA synthesis following blockage, as shown by the increased percentage of CldU red fibers (91.7% ± 1.7% vs. 78.7% ± 1.3%, P < 0.0001), when compared with control cells (Supplementary Fig. S2H). In addition, CDA increases the replication fork speed following stalling, as showed by an increased length of DNA tracks and of the CldU/IdU ratio (0.43 ± 0.025 vs. 0.282 ± 0.019, P < 0.0001; Fig. 2D and E). We next investigated whether CDA directly interacts with the DNA replication machinery. Using a FLAG-tagged CDA, we demonstrate that CDA is located both in the cytoplasm and in the nucleus of MIA PaCa-2 cells (Fig. 2F). To determine whether CDA is in closer proximity to DNA, we isolated proteins bound to replication forks using the iPOND (Isolation of Proteins on Nascent DNA) approach described in Supplementary Fig. S2I. Results presented in Fig. 2G show that MCM7 and PCNA, two main components of the replisome, are detected within the EdU-positive fraction, indicating an association with newly synthesized DNA (Fig. 2G, top left). Remarkably, we also detected CDA at the DNA replication fork (Fig. 2G, bottom left).
Using Hela cells, we confirmed that endogenous CDA also locates at the replication fork (Supplementary Fig. S2J). We performed a thymidine-chase to analyze how proteins assemble and disassemble from the nascent DNA segment (Fig. 2G, left, right lane). In this condition, the CDA signal disappears similarly to MCM7 and PCNA, indicating that this protein progresses with the DNA replication machinery, rather than just being a protein constitutively bound on chromatin. To summarize, we present in this study the initial evidence suggesting that CDA potentially enhances DNA replication speed, improves replication fork restart efficiency, and appears to be positioned at the DNA replication fork in PDAC cells.
CDA controls the replication stress level of PDAC cells
We next investigated whether CDA may control DNA replication stress in PDAC cells. Using DNA spreading, we found that CDA depletion decreases the length of DNA tracks to the same extent than the inhibitor of DNA replication aphidicolin (Fig. 3A and B, P < 0.0001), suggesting that the replication fork speed is affected. In parallel, we found that CDA inhibition alters the progression in the cell cycle of cancer cells, leading to a decrease in cells in the G1 phase (−31% ± 5%, P < 0.0001, Fig. 3C) and an increase in cells in the S (+195% ± 6%, P < 0.005, Fig. 3C) and G2–M phases (+67% ± 10%, P < 0.01, Fig. 3C). We rescued CDA depletion by supplementing cells with uridine, which not only restored cell-cycle progression (Fig. 3D) but also the proliferation of tumor cells (Supplementary Fig. S3B), as compared with control cells (Supplementary Fig. S3A and S3B). A transcriptomic analysis indicated an enrichment of the ATR response to replicative stress signature in CDA-depleted cells (NES = 1.76, P < 0.01) when compared with control cells (Fig. 3E; Supplementary Table S3). This finding is further supported by the strong activation of the CHK1 effector kinase in MIA PaCa-2 (Fig. 3F), Capan-1 (Supplementary Fig. S3C), and BxPC3 (Supplementary Fig. S3D) cells expressing shRNA against CDA, as compared with control cells. We next measured the number of γH2AX foci in S-phase cells as a canonical marker of DNA breaks and replication stress. As indicated in Fig. 3G and H, CDA overexpression in MIA PaCa-2 cells significantly decreases the number of γH2AX foci in EdU-positive cells (−48% ± 7%, P < 0.0001) compared with control cells. This effect is entirely dependent on CDA deaminase activity, as the expression of a catalytically inactive mutant of CDA has no impact on the number of γH2AX foci (Fig. 3E and F). On the contrary, silencing CDA in BxPC3 cells results in γH2AX foci accumulation in S-phase cells (+100% ± 2%, P < 0.0001, Fig. 3I and J).
Figure 3.
CDA controls replication stress levels in PDAC cells. A, Immunofluorescence detection of IdU (green) and CldU (red) DNA tracks after DNA stretching of Capan-1 cells expressing or not CDA siRNA. White arrows show the measured red tracks. Twenty-four hours treatment of 0.2 μmol/L aphidicolin was used as a positive control of DNA elongation inhibition, IdU, iododeoxyuridine; CldU, chloro-deoxyuridine. B, Quantification of the length of DNA tracks (μm); at least 150 fibers were measured per condition. Results representative of three independent experiments. ****, P < 0.0001 (Mann–Whitney test). C, Cell-cycle analysis of Mia Paca-2 cells depleted or not of CDA (CDA siRNA). Results are mean of three independent experiments performed in triplicates. **, P < 0.01; ***, P < 0.005; ****, P < 0.0001. D, Cell-cycle analysis of Mia Paca-2 cells depleted of CDA (CDA siRNA), supplemented or not with exogenous uridine (25 μg/mL). Results are mean of three independent experiments performed in triplicates. E, Enrichment plot of Reactome activation of ATR in response to replication stress pathway in MIA PaCa-2 cells depleted of CDA (CDA siRNA; NES = 1.76; P < 0.01). Results are representative of three independent transductions. F, Western blotting for P-CHK1 (S345) and CHK1 in MIA PaCa-2 cells depleted for CDA or expressing control siRNA. B-ACTIN protein level was used as loading control. Results represent three independent pools of transduction. G, Immunofluorescence detection of P-H2AX foci (S139; green, γH2AX), DAPI (blue) and EdU (red) in MIA PACA-2 cells overexpressing or not CDA or CDAE67Q. H, Quantification of γH2AX foci in S-phase cells (EdU+) in at least 500 control cells or MIA PACA-2 cells overexpressing or not CDA or CDAE67Q. ***, P < 0.005 (Wilcoxon–Mann–Whitney test). I, Immunofluorescence detection of P-H2AX foci (S139; green, γH2AX), DAPI (blue), and EdU (red) in BxPC3 cells overexpressing or not control siRNA or siRNA-targeting CDA. J, Quantification of γH2AX foci in S-phase cells (EdU+) in at least 500 BxPC3 cells overexpressing or not control siRNA or siRNA-targeting CDA.
We extended this finding to Capan-1 cells expressing siRNA against CDA (Supplementary Fig. S3E) and to MIA PaCa-2 cells treated with pharmacological inhibitors of CDA (Supplementary Fig. S3F) or incubated with cytidine and/or deoxycytidine to recapitulate pyrimidine pool imbalance due to a CDA deficiency (Supplementary Fig. S3G).
We then analyzed several other markers of replication stress in response to CDA expression. Replication protein A (RPA) protects exposed single-stranded DNA during DNA replication and accumulates in response to replicative stress. Remarkably, we found that CDA expression decreases the number of RPA foci (−67% ± 8%, P < 0.0001) in PDAC cells (Supplementary Fig. S3H). The p53-binding protein 1 (53BP1) is a DDR factor recruited to the site of DNA damages following DNA double-strand breaks (DSB). We found that phospho-53BP1 (P-53BP1 and S1778) foci, that mark DSBs, are reduced in MIA PaCa-2 CDA-high cells (−56% ± 5%, P < 0.0001) as compared with control cells (Supplementary Fig. S3I and S3J). Collectively, these results indicate that CDA is involved in the regulation replication stress levels in PDAC cells.
CDA is involved in the control of genome stability of PDAC cells
Residual and unsolved replication stress is a source of genetic instability. For instance, in Bloom syndrome, the loss of CDA expression contributes to alterations of DNA repair, resulting in a high number of chromosome breaks and rearrangements that are responsible for the increased risk for cancer (11). Common fragile sites (CFS) are large chromosomal regions that are preferentially subject to mitotic breakage upon replication stress. They become unstable at the early-stage of cancer development and are hotspots of chromosomal rearrangements in cancer (36). FANCD2 facilitates replication across CFS and can be used as a marker of unstable CSFs in cells with replication stress (37). Hence, under-replicated DNA at CFSs persisting into late mitosis can lead to the formation of ultrafine anaphase bridges (UFB), chromosome nondisjunction, and mitotic catastrophe (38). Here, FANCD2 forms symmetric foci at each end of UFBs and has a role in the resolution of these DNA bridges in mitosis. As shown in Fig. 4A and B, MIA PaCa-2 cells overexpressing CDA show significantly less FANCD2 foci in early mitosis (−56% ± 14%, P < 0.0001). In addition, CDA expression inhibits FANCD2 formation following the treatment of cells with aphidicolin (−21% ± 3%, P < 0.05, Supplementary Fig. S4A). On the contrary, CDA depletion strongly increases the number of damaged CFSs (+76% ± 7%, P < 0.0001, Fig. 4C and D). These results suggest that CDA limits the presence of under-replicated DNA in mitotic PDAC cells. Micronuclei are small sized nuclei that form from one or a few chromosomes or chromatin fragments that are not incorporated into the daughter nuclei during cell division (39).
Figure 4.
CDA participates to the control of genomic stability of PDAC cells. A, Immunofluorescence detection of FANCD2 foci (green), DAPI (blue) in MIA PaCa-2 control cells or MIA PaCa-2 cells overexpressing CDA. B, Quantification of the number of FANCD2 foci in early mitotic cells (prophase, metaphase) in at least 100 MIA PaCa-2 control cells or MIA PaCa-2 cells overexpressing CDA. C, Immunofluorescence detection of FANCD2 foci (green), DAPI (blue) in Capan-1 cells expressing control siRNAs or siRNAs targeting CDA. D, Quantification of the number of FANCD2 foci in early mitotic cells (prophase, metaphase) in at least 100 Capan-1 cells expressing control siRNA or siRNA-targeting CDA. E, Representative captions and percentage of MIA PaCa-2 control cells or MIA PaCa-2 cells overexpressing CDA with at least one micronucleus. Results are expressed as mean ± SEM of three independent experiments. F, Representative captions and percentage of BxPC3 cells expressing control siRNA or siRNA-targeting CDA with at least one micronucleus. Results are expressed as mean ± SEM of three independent experiments. G and H, Immunofluorescence detection (G) and quantification (H) of the number of 53BP1 bodies in G1 cells in at least 1,000 Capan-1 control cells and cells expressing CDA siRNA. Results are expressed as mean ± SEM of four independent experiments. ns, nonsignificant; *, P < 0.05; ****, P < 0.0001, Wilcoxon–Mann–Whitney test.
Micronuclei formation usually serves as an index of genotoxic effects and chromosomal instability. We found that CDA overexpression significantly reduces the number of DAPI-positive micronuclei (−60% ± 13%, P < 0.05) in MIA PaCa-2 cells (Fig. 4E), whereas the overexpression of a catalytically inactive mutant is ineffective. Moreover, targeting CDA increases micronuclei formation in Bx-PC3 (+110% ± 7%, P < 0.05, Fig. 4F–H) and in Capan-1 (Supplementary Fig. S4B) and MIA PaCa-2 (Supplementary Fig. S4C) cells. Recent reports demonstrate that markers of genomic stress, such as remnants of incomplete replication, are inherited by daughter G1 cells and are sequestered in specific G1 nuclear sub-compartments called 53BP1 nuclear bodies (40). We explored this possibility and found that CDA silencing in Capan-1 cells significantly increases the number of G1 cells with 53BP1 bodies (EdU-negative cells, low DAPI intensity; 2.4 ± 0.1-fold, P < 0.0001; Fig. 4G and H). Collectively, these results indicate that CDA controls genomic stability in PDAC cells.
We next measured the consequence of CDA expression on the genome of PDAC cells. Thus, we analyzed genomic DNA from MIA PaCa-2 cells overexpressing or not CDA after 5 (early) and 55 days (late) of culture (Supplementary Fig. S5A). WGS revealed that control cells and cells overexpressing the CDAE67Q mutant show an increase with time in culture in structural variants such as tandem duplication (+16% and +11%, respectively), inversions (+15% and +7%, respectively), translocation (+29% and +16%, respectively), deletions (+8% and +3%, respectively) and insertions (+16% and +5%, respectively, Fig. 5A; Supplementary Fig. S5B and Supplementary Table S4). These cells also show increased number of indels (+3% and +2%, respectively) and of single-nucleotide variants (+1.54% and +1.56%, respectively, Fig. 5B; Supplementary Fig. S5B and Supplementary Table S4) as time in culture increases. Remarkably, we demonstrate that CDA expression significantly decreases the percentage of tandem duplications, inversions, translocations, insertions, and indels, when number of deletions and SNV remained unchanged (Fig. 5A and B; Supplementary Table S5, Supplementary Fig. S5B). This result suggests that CDA expression regulates genetic events in PDAC cells in long-term culture. We next found that tumors with a high level of CDA display a significant enrichment of “activation in the ATR in response to replicative stress” gene set (NES = 1.72, P = 0.01) and in the CINSARC signature of chromosome instability (NES = 2.83, P = 0.01; Fig. 5C and D; ref. 26). We also explored the aneuploidy score and the number of non-silent mutations per megabase of genomic DNA in these tumors. Remarkably, PDAC tumors with high levels of CDA have significantly higher aneuploidy scores (+69% ± 10%, P = 0.01, Fig. 5E) and non-silent mutations per megabase (+43% ± 7%, P < 0.001, Fig. 5F), as compared with tumors with lower CDA expression. Interestingly, the expression of DHODH from the de novo pyrimidine production pathway is not associated with significant changes in tumor genomic instability markers (Fig. 5E and F). In addition, we found that these cells are sensitive to DHODH targeting using leflunomide (Supplementary Fig. S5C). Thus, our results show that CDA influences genetic events in PDAC cells, showing a potential positive association with genetically unstable PDAC tumors.
Figure 5.
CDA expression is associated with genetic instability in PDAC tumors. A and B, Whole-genome sequencing was performed for the identification of structural variants (A) or indels and single-nucleotide variants (B). DUP, duplication; INV, inversion; BND, translocation; DEL, deletion; INS, insertion; SNV, single-nucleotide variants. The percentage of change in alteration numbers was quantified in late versus early control cells or cells overexpressing CDA or CDAE67Q. **, P < 0.01; ****, P < 0.0001 (two-way ANOVA). C and D, Enrichment plots for activation of ATR in response to replicative stress (Reactome–GSEA; C) and CINSARC signatures in TCGA PDAC samples expressing high or low CDA mRNA levels (D). E, Quantification of aneuploidy score in TCGA PDAC samples with high and low level of CDA. Aneuploidy score is the total number of chromosome arms containing at least one variation of copy number in an arm, per sample (56). **, P < 0.01 (Wilcoxon–Mann–Whitney test). F, Quantification of the number of non-silent mutations per Mb in TCGA PDAC samples with high and low levels of CDA. ***, P < 0.001 (Wilcoxon–Mann–Whitney test; TCGA PDAC samples: n = 39 high CDA; n = 38 low CDA).
CDA drives the resistance of PDAC cells against DNA-damaging agents
Considering the new role of CDA in the control of DNA replication, we aimed to investigate whether CDA expression could predict DNA-damaging drug efficacy on PDAC cells. We analyzed 26 PDAC cell lines from the cancer cell line encyclopedia database, and we found a significant correlation between CDA expression and resistance to oxaliplatin (R = 0.47, P = 0.014; Fig. 6A) or to the topoisomerase I inhibitor irinotecan (R = 0.41, P = 0.039, Supplementary Fig. S6A). From these 26 cell lines, we selected four cell lines expressing a low (DAN-G, MIA PaCa-2, Capan-1, and KP-4) or a high level of CDA (Capan-2, AsPC-1, BxPC3, and SU8686). As shown in Fig. 6B, PDAC cells with a high level of CDA are 67 ± 0.3 fold (P < 0.01) more resistant to oxaliplatin than cells expressing a low level of the enzyme. We extended this observation to drugs that inhibit the DNA topoisomerase I (SN38), that form DNA adducts (cisplatin), that deposit methyl groups on DNA guanine bases (temozolomide), or cause single- and double-stranded disruptions after intercalating with the DNA molecule (mitoxantrone).
Figure 6.
CDA is associated with resistance to replication stress-inducing drugs in PDAC cells and targeting CDA in vivo sensitizes PDAC tumor to oxaliplatin with molecular evidence of replication stress. A, Correlation plot of CDA expression and log10 IC50 for oxaliplatin in PDAC cell lines from the cancer cell line encyclopedia. B, IC50 value of irinotecan, SN38, cisplatin, temozolomide, and mitoxantrone in PDAC cell lines from CCLE with high and low CDA expressions (n = 4 for each). C, Confluence analysis of MIA PaCa-2 cells expressing or not CDA or CDAE67Q and treated for 72 hours with 10 μmol/L camptothecin. Results are expressed as mean ± SEM of at least three independent experiments. D, Confluence of PDAC051T cells depleted of CDA using siRNA 72 hours after transfection. Results are expressed as mean ± SEM of 20 to 28 individual replicates, respectively. E, Long-term confluence follow-up of PDAC051T cells transfected with control or CDA siRNA, then treated with 10 μmol/L oxaliplatin. F and G, Individual (F) and endpoint (G) tumor growth of PDAC051T experimental tumors treated with control siRNA or siRNA-targeting CDA (red arrow) and receiving oxaliplatin (gray arrow; n = 7). Control tumors received control siRNA and oxaliplatin (n = 6). H, Single-cell signature enrichment of PDAC051T experimental tumors treated with control siRNA or siRNA-targeting CDA and receiving oxaliplatin at endpoint. *, P < 0.05; **, P < 0.01, ***, P < 0.001; ****, P < 0.0001, unpaired t test.
PDAC cells with a high expression of CDA are 5.9 ± 0.3 to 39 ± 0.15-fold (P < 0.05) more resistant to these drugs than cells expressing a low level of the enzyme (Fig. 6B). To obtain direct evidence that CDA protects PDAC cells from DNA-damaging drugs, we treated MIA PaCa-2 cells overexpressing CDA or CDAE67Q catalytically inactive mutant with camptothecin. As shown in Fig. 6C, camptothecin treatment significantly inhibits the proliferation of control cells and of cells overexpressing CDAE67Q (−56% ± 5% and −57% ± 6%, respectively, P < 0.001; Fig. 6C). In contrast, MIA PaCa-2 cells expressing CDA significantly resisted to chemotherapy-induced antiproliferative activity (P < 0.05 and P < 0.01 vs. control cells and cells expressing CDAE67Q, respectively, Fig. 6C). These results demonstrate that CDA expression can predict response to DNA-damaging drug in PDAC cell lines.
We next investigated whether CDA targeting could partner with DNA-damaging drugs to induce irreversible replication stress in preclinical models of PDAC. We selected PDAC051T primary cells (13) that express detectable levels of the enzyme (Supplementary Fig. S6B). We silenced CDA expression using siRNA (Supplementary Fig. S6B) and found that PDAC051T proliferation was significantly inhibited by 1.7-fold (P < 0.0001, Fig. 6D), as compared with control cells. These results were confirmed using PDAC015T primary cultured cells, derived from another PDAC tumor with detectable CDA expression (Supplementary Fig. S6C). These results further support that CDA is important for PDAC cell proliferation. Next, PDAC051T cells were treated with oxaliplatin, a component of FOLFIRINOX, a multi-drug therapy routinely used for PDAC management. As expected, oxaliplatin treatment significantly increases the level of replication stress in primary cultured cells, as measured by the number of γH2AX foci per cell in S-phase (6.3 ± 0.65 vs. 12.7 ± 1.5, P < 0.005, Supplementary Fig. S6D). As shown in Fig. 6E; Supplementary Fig. S6E, CDA silencing followed by oxaliplatin treatment significantly sensitizes PDAC cells to therapy and inhibits cell proliferation, as compared with control cells receiving chemotherapy only (−61% ± 5%, P < 0.0001).
Furthermore, we asked whether targeting CDA may improve response to drugs that induce replication stress in PDAC preclinical models, in vivo. PDAC051T cells were implanted in nude mice and transfected with siRNAs targeting CDA. Control tumors received random siRNAs. Forty-eight hours later, mice were treated with oxaliplatin as previously described (13). The combination treatment was repeated 3 days later (Supplementary Fig. S6G). We found that tumors receiving CDA-targeting siRNA and treated by oxaliplatin showed a robust reduction in tumor growth as compared with control tumors (−29% ± 2%, P < 0.001; Fig. 6F and G; Supplementary Fig. S6G), when tumor weight inhibition at endpoint nearly reached statistical significance (Supplementary Fig. S6H). At the endpoint, mice were sacrificed and tumors were sampled for single cell RNA-seq studies to capture rare events and gene enrichment signatures were analyzed using the Single-Cell Signature Explorer (29). As expected, combining CDA silencing and oxaliplatin treatment reveals signatures indicative of nucleotide metabolism inhibition and replicative stress (Fig. 6H; Supplementary Table S6). This analysis also evidenced cell proliferation inhibition, induction of cell death and autophagy, epithelial differentiation, inflammation and metabolic alterations in treated tumors (Fig. 6H; Supplementary Table S6). Together, our findings suggest for the first time a potential link between CDA expression and the therapeutic effectiveness of DNA-damaging drugs in PDAC. In addition, targeting CDA sensitizes these tumors to drugs that induce replication stress.
Discussion
Exploiting DNA replication stress has recently emerged as a promising strategy for cancer therapy (41) and is starting to be explored as an attractive target for the management of PDAC (6, 42, 43). Notwithstanding the progress, the identification of the molecular mechanisms involved in the dependency of tumor cells on the replication stress response is key to devise future therapies aimed to enhance both endogenous and drug-induced replication stress (4).
In this work, we identify that CDA, a key protein in the pyrimidine salvage pathway, is overexpressed in the most aggressive PDAC tumors, and that CDA silencing provokes tumor cell death by apoptosis and strongly inhibits experimental tumor growth. To our knowledge, this is the first description that CDA is involved in PDAC tumor growth, independently of its role in tumor resistance to pyrimidine-based therapies (7). Others identified that PDAC cells are dependent on a de novo pyrimidine production driven by DHODH (44, 45). However, targeting DHODH would not be selective for PDAC cells, as we found that DHODH is expressed at high levels in normal adjacent tissue.
Exploration of PDAC patient samples from TCGA indicates that CDA expression is positively associated with a transcriptomic DNA replication signature. In PDAC cell lines, we show that a CDA overexpression correlates with efficient DNA replication fork speed and reduces endogenous replication stress, independently of increased cell proliferation. Conversely, the reduction of CDA level in PDAC cell lines slows fork progression and increases DNA damage. These results were recapitulated with CDA inhibitors but not following the overexpression of a catalytically dead CDA mutant, demonstrating that CDA controls replication stress by recycling pyrimidines from nucleotide degradation, through the salvage pathway. This hypothesis is supported by our results showing that targeting CDA disrupts the progression in the cell cycle of cancer cells, which can be rescued by exogenous uridine supplementation. Our results echo with recent findings demonstrating that the inhibition of methylenetetrahydrofolate dehydrogenase/cyclohydrolase (MTHFD2) induces replication stress in cancer cells by inhibiting thymidine production leading to uracil misincorporation into DNA (46).
Our study could aid in revisiting the proposed mechanism of action described in this study, as the control of the dUTP to dTTP balance, in which CDA is involved, is essential in the therapeutic efficacy of the MTHFD2 inhibitor. Importantly, CDA activity occurs upstream of the reduction of ribonucleoside diphosphates to deoxyribonucleoside diphosphates by RNR. Thus, our results might facilitate re-examination of the central position of RNR in controlling DNA replication in PDAC cells (47). Notably, we found that CDA, which is generally considered as a cytoplasmic enzyme (7), can also be nuclear and associated with the replication fork through a yet unclear mechanism. In protein interaction databases, CDA is shown to interact with MCM2 in a high-throughput affinity Capture MS experiment (7), but we failed to demonstrate such association by coimmunoprecipitation assays in our models. Nevertheless, our findings may revive the “dNTP-channeling hypothesis” according to which a functional complex, known as “replitase” (48), could concentrate dNTP in close proximity with replication forks for maximal efficacy. Replitase is thought to contain enzymes that are involved in nucleotide precursor synthesis and DNA replication, such as RNR (49), NDP-kinase and DNA polymerase α (50). Interestingly, as deoxyribonucleotides are detected in newly synthesized DNA much faster than in the dNTP pool (51), this strongly suggests that the salvage pathway may also actively participate to channel dNTPs to the fork environment.
Replicative stress and genomic instability can provide multiple variants upon which selection could act, constitutes a driving force of cancer development and heterogeneity, and is generally associated with poor prognosis. In contrast, excessive replication stress and genomic instability are obviously deleterious for cell fitness and have been demonstrated to correlate with bad outcome for cancer cells (52). This suggests that an appropriate threshold of replicative stress and genetic instability is required for tumor viability and that any mechanism that prevents such excessive genome instability is a potential therapeutic target. Here, we found that CDA is involved in reducing the endogenous replication stress that ultimately limits genetic alterations in PDAC cells in long-term culture conditions. Furthermore, we observed that CDA is increased in genetically unstable tumors, particularly in primary cultured cells from the basal-like molecular subtype, which is closely linked to replication stress (5). This suggests a potential role for CDA in potentially participating in managing replicative stress in PDAC.
Another crucial question raised by our study is why CDA from the pyrimidine salvage pathway is essential to DNA replication in PDAC cells, when the de novo pyrimidine synthesis is fully functional. Many tumors, including PDAC, have very poor blood supply, so that cancer cells must get the nutrients they need to grow from other sources. One option for tumor cells is to find new ways to use what they already have. One example is the ability of PDAC cells to face glutamine shortage by inducing the expression of NAGK (N-acetylglucosamine kinase), to salvage N-acetylglucosamine to generate UDP-N-acetylglucosamine (53).
Thus, it is tempting to speculate that PDAC cells favor the CDA salvage pathway to recover nucleosides or bases formed during DNA or RNA degradation when resources are scarce. This could be more efficient than soliciting the de novo pathway that comes with a higher energetic cost. As the pyrimidine salvage pathway is not essential in normal cells, targeting CDA may create a synthetic lethality situation for PDAC tumors when cotreated with chemotherapies such as oxaliplatin, which may benefit to a large number of patients diagnosed with this cancer.
Finally, our work may have important implications for the treatment of patients, as it could influence future clinical studies. With only a handful of mechanism-based therapies, the future of PDAC treatment lies in carefully chosen therapy combinations for durable tumor control in patients. Our study presents a compelling scientific basis to consider using CDA expression as a potential marker for resistance to DNA-damaging drugs like SN38 and oxaliplatin, commonly used in routine clinical practice as part of FOLFIRINOX chemotherapy. In addition, our findings indicate that targeting CDA may potentially increase replicative stress in tumors and enhance sensitivity to oxaliplatin treatment, observed both in vitro and in vivo. This is evident through signs of senescence, checkpoint activation, DNA repair, and a potential inhibition of replication initiation in experimental tumors post-treatment. Importantly, this strategy may reveal to be safe as CDA is not essential to mice development or fertility (54), and because CDA is poorly expressed in normal pancreas (55). Interestingly, Dreyer and colleagues (5) proposed an elegant matrix for therapeutic decision using inhibitors of the DDR pathway and/or cell-cycle checkpoint, based on DDR deficiency and on the level of replication stress in PDAC cells. Unfortunately, half of the samples that are DDR-proficient, but with low levels of replication stress, do not respond to any of these molecules. During this study, we found that CDA effectively induced replication stress in DDR-proficient (Mia PACA-2) and DDR-deficient (BxPC3, Capan-1) PDAC models, respectively (4). According to our findings, it could be valuable to consider assessing CDA expression in the majority of PDAC tumors that presently evade DDR-based targeted therapies, as a potential step toward further interventional studies aimed at enhancing replication stress in cancer cells. Following eligibility criteria, these tumors might be subsequently treated with ATR inhibitors, especially considering our observation that CDA invalidation is linked to an increased ATR response to replicative stress both in vitro and in vivo. In the case of DDR-deficient tumors, the concurrent use of PARP inhibitors might present a promising molecular-based therapeutic approach for patients with PDAC.
Supplementary Material
Supplemetary table
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Acknowledgments
The authors thank the financial support of the “Fondation Toulouse Cancer Santé“ (to A. Frances), “Inserm“ and “Région Occitanie“ (to A. Lumeau), “Université Paul Sabatier UT3“ (to M. Gayral), “INSERM, Pierre Fabre and Fondation Toulouse Cancer Santé chair of Bioinformatics in Oncology of the CRCT” (to V. Pancaldi and M. Madrid-Mencia), “Fondation de France (n°00097692)” (to N. Bery), and “Ligue Nationale Contre le Cancer“ (to A. Lumeau). We would like to thank the animal facility team (UMS006/CREFRE, Plateforme Anexplo, Toulouse) for their support and their technical assistance. We thank Emilie Martin and Catherine Zanibellato for technical assistance and Jacobo Solorzano for data analysis. We are grateful to the GenoToul bioinformatics platform Toulouse Midi-Pyrenees (GenoToul Bioinfo) for providing computing resources. This work was granted access to the HPC resources of CALMIP supercomputing center under the allocation P19043. We warmly thank Jérôme Cros, Drs. F. Chibon, and M. Amor-Guéret for helpful discussions.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
V. Pancaldi reports grants from SANOFI, Pierre Fabre, Janssen, and Evotec outside the submitted work. P. Cordelier reports grants from Fondation Toulouse Cancer Santé, Région Occitanie, Inserm, Ligue Nationale Contre le Cancer, Université, Paul Sabatier, Fondation de France, and INSERM, Pierre Fabre and Fondation Toulouse Cancer Santé Chair of Bioinformatics in Oncology of the CRCT during the conduct of the study; as well as reports a patent for 16/084830 issued. No disclosures were reported by the other authors.
Authors' Contributions
A. Lumeau: Conceptualization, investigation, methodology, writing–original draft, writing–review and editing. N. Bery: Conceptualization, writing–review and editing. A. Francès: Investigation. M. Gayral: Investigation. G. Labrousse: Investigation. C. Ribeyre: Supervision, investigation, writing–original draft. C. Lopez: Investigation. A. Nevot: Investigation. A. El Kaoutari: Investigation. N. Hanoun: Investigation. E. Sarot: Investigation, writing–original draft. M. Perrier: Investigation, writing–original draft. F. Pont: Investigation, writing–original draft. J.-P. Cerapio: Investigation, writing–original draft. J.-J. Fournié: Investigation. F. Lopez: Supervision, methodology. M. Madrid-Mencia: Investigation. V. Pancaldi: Supervision, investigation, writing–original draft, writing–review and editing. M.-J. Pillaire: supervision. V. Bergoglio: Supervision. J. Torrisani: Supervision, writing–original draft, writing–review and editing. N. Dusetti: Conceptualization, writing–original draft, writing–review and editing. J.-S. Hoffmann: Conceptualization, supervision, funding acquisition, writing–original draft, writing–review and editing. L. Buscail: Conceptualization, supervision, writing–review and editing. M. Lutzmann: Conceptualization, supervision, writing–review and editing. P. Cordelier: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, methodology, writing–original draft, writing–review and editing.
References
- 1. Rahib L, Wehner MR, Matrisian LM, Nead KT. Estimated projection of US cancer incidence and death to 2040. JAMA Netw Open 2021;4:e214708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. [DOI] [PubMed] [Google Scholar]
- 3. Waddell N, Pajic M, Patch A-M, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stoof J, Harrold E, Mariottino S, Lowery MA, Walsh N. DNA damage repair deficiency in pancreatic ductal adenocarcinoma: preclinical models and clinical perspectives. Front Cell Dev Biol 2021;9:749490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dreyer SB, Upstill-Goddard R, Paulus-Hock V, Paris C, Lampraki E-M, Dray E, et al. Targeting DNA damage response and replication stress in pancreatic cancer. Gastroenterology 2021;160:362–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hammel P, Vitellius C, Boisteau É, Wisniewski M, Colle E, Hilmi M, et al. Maintenance therapies in metastatic pancreatic cancer: present and future with a focus on PARP inhibitors. Ther Adv Med Oncol 2020;12:1758835920937949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frances A, Cordelier P. The emerging role of cytidine deaminase in human diseases: a new opportunity for therapy? Mol Ther 2020;28:357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou L, Cheng X, Connolly BA, Dickman MJ, Hurd PJ, Hornby DP. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J Mol Biol 2002;321:591–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Funamizu N, Lacy CR, Fujita K, Furukawa K, Misawa T, Yanaga K, et al. Tetrahydrouridine inhibits cell proliferation through cell-cycle regulation regardless of cytidine deaminase expression levels. PLoS ONE 2012;7:e37424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sohal D, Krishnamurthi S, Tohme R, Gu X, Lindner D, Landowski TH, et al. A pilot clinical trial of the cytidine deaminase inhibitor tetrahydrouridine combined with decitabine to target DNMT1 in advanced, chemorefractory pancreatic cancer. Am J Cancer Res 2020;10:3047–60. [PMC free article] [PubMed] [Google Scholar]
- 11. Gemble S, Ahuja A, Buhagiar-Labarchède G, Onclercq-Delic R, Dairou J, Biard DSF, et al. Pyrimidine pool disequilibrium induced by a cytidine deaminase deficiency inhibits PARP-1 activity, leading to the under replication of DNA. PLoS Genet 2015;11:e1005384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flanagan M, Cunniff CM. Bloom Syndrome. In:Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJ, Mirzaa G, et al. editors. GeneReviews. Seattle (WA): University of Washington, Seattle; 1993. Available from: https://www.ncbi.nlm.nih.gov/books/NBK1398/. [Google Scholar]
- 13. Fraunhoffer NA, Abuelafia AM, Bigonnet M, Gayet O, Roques J, Telle E, et al. Evidencing a pancreatic ductal adenocarcinoma subpopulation sensitive to the proteasome inhibitor carfilzomib. Clin Cancer Res 2020;26:5506–19. [DOI] [PubMed] [Google Scholar]
- 14. Larrieu D, Brunet M, Vargas C, Hanoun N, Ligat L, Dagnon L, et al. The E3 ubiquitin ligase TRIP12 participates in cell-cycle progression and chromosome stability. Sci Rep 2020;10:789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sicard F, Gayral M, Lulka H, Buscail L, Cordelier P. Targeting miR-21 for the therapy of pancreatic cancer. Mol Ther 2013;21:986–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. du Rieu MC, Torrisani J, Selves J, Al Saati T, Souque A, Dufresne M, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem 2010;56:603–12. [DOI] [PubMed] [Google Scholar]
- 17. Diaz-Riascos ZV, Ginesta MM, Fabregat J, Serrano T, Busquets J, Buscail L, et al. Expression and role of MicroRNAs from the miR-200 family in the tumor formation and metastatic propensity of pancreatic cancer. Mol Ther Nucleic Acids 2019;17:491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bournet B, Pointreau A, Souque A, Oumouhou N, Muscari F, Lepage B, et al. Gene expression signature of advanced pancreatic ductal adenocarcinoma using low density array on endoscopic ultrasound-guided fine needle aspiration samples. Pancreatology 2012;12:27–34. [DOI] [PubMed] [Google Scholar]
- 19. Cambi A, Vincenzetti S, Neuhard J, Costanzi S, Natalini P, Vita A. Identification of four amino acid residues essential for catalysis in human cytidine deaminase by site-directed mutagenesis and chemical modifications. Protein Eng 1998;11:59–63. [DOI] [PubMed] [Google Scholar]
- 20. Bery N, Cruz-Migoni A, Bataille CJ, Quevedo CE, Tulmin H, Miller A, et al. BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein-protein interactions. eLife 2018;7:e37122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bery N, Miller A, Rabbitts T. A potent KRAS macromolecule degrader specifically targeting tumours with mutant KRAS. Nat Commun 2020;11:3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ravet E, Lulka H, Gross F, Casteilla L, Buscail L, Cordelier P. Using lentiviral vectors for efficient pancreatic cancer gene therapy. Cancer Gene Ther 2010;17:315–24. [DOI] [PubMed] [Google Scholar]
- 23. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gillespie M, Jassal B, Stephan R, Milacic M, Rothfels K, Senff-Ribeiro A, et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res 2022;50:D687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nicolle R, Raffenne J, Paradis V, Couvelard A, de Reynies A, Blum Y, et al. Prognostic biomarkers in pancreatic cancer: avoiding errata when using the TCGA dataset. Cancers 2019;11:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lagarde P, Pérot G, Kauffmann A, Brulard C, Dapremont V, Hostein I, et al. Mitotic checkpoints and chromosome instability are strong predictors of clinical outcome in gastrointestinal stromal tumors. Clin Cancer Res 2012;18:826–38. [DOI] [PubMed] [Google Scholar]
- 27. Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pont F, Tosolini M, Fournié JJ. Single-cell signature explorer for comprehensive visualization of single cell signatures across scRNA-seq datasets. Nucleic Acids Res 2019;47:e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zauri M, Berridge G, Thézénas M-L, Pugh KM, Goldin R, Kessler BM, et al. CDA directs metabolism of epigenetic nucleosides revealing a therapeutic window in cancer. Nature 2015;524:114–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hessmann E, Patzak MS, Klein L, Chen N, Kari V, Ramu I, et al. Fibroblast drug scavenging increases intratumoural gemcitabine accumulation in murine pancreas cancer. Gut 2018;67:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Collisson EA, Bailey P, Chang DK, Biankin AV. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol 2019;16:207. [DOI] [PubMed] [Google Scholar]
- 34. Puleo F, Nicolle R, Blum Y, Cros J, Marisa L, Demetter P, et al. Stratification of Pancreatic ductal adenocarcinomas based on tumor and microenvironment features. Gastroenterology 2018;155:1999–2013. [DOI] [PubMed] [Google Scholar]
- 35. Rashid NU, Peng XL, Jin C, Moffitt RA, Volmar KE, Belt BA, et al. Purity independent subtyping of tumors (PurIST), a clinically robust, single-sample classifier for tumor subtyping in pancreatic cancer. Clin Cancer Res 2020;26:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simpson BS, Pye H, Whitaker HC. The oncological relevance of fragile sites in cancer. Commun Biol 2021;4:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Madireddy A, Kosiyatrakul ST, Boisvert RA, Herrera-Moyano E, García-Rubio ML, Gerhardt J, et al. FANCD2 facilitates replication through common fragile sites. Mol Cell 2016;64:388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 2009;11:753–60. [DOI] [PubMed] [Google Scholar]
- 39. Ye CJ, Sharpe Z, Alemara S, Mackenzie S, Liu G, Abdallah B, et al. Micronuclei and genome chaos: changing the system inheritance. Genes 2019;10:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bertolin AP, Hoffmann J-S, Gottifredi V. Under-replicated DNA: the byproduct of large genomes? Cancers 2020;12:2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ubhi T, Brown GW. Exploiting DNA replication stress for cancer treatment. Cancer Res 2019;79:1730–9. [DOI] [PubMed] [Google Scholar]
- 42. Hammel P, Zhang C, Matile J, Colle E, Hadj-Naceur I, Gagaille M-P, et al. PARP inhibition in treatment of pancreatic cancer. Expert Rev Anticancer Ther 2020;20:939–45. [DOI] [PubMed] [Google Scholar]
- 43. Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med 2019;381:317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Buettner R, Morales C, Wu X, Sanchez JF, Li H, Melstrom LG, et al. Leflunomide synergizes with gemcitabine in growth inhibition of PC cells and impairs c-Myc signaling through PIM kinase targeting. Mol Ther Oncolytics 2019;14:149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koundinya M, Sudhalter J, Courjaud A, Lionne B, Touyer G, Bonnet L, et al. Dependence on the pyrimidine biosynthetic enzyme DHODH is a synthetic lethal vulnerability in mutant KRAS-driven cancers. Cell Chem Biol 2018;25:705–17. [DOI] [PubMed] [Google Scholar]
- 46. Bonagas N, Gustafsson NMS, Henriksson M, Marttila P, Gustafsson R, Wiita E, et al. Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress. Nat Cancer 2022;3:156–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hu C-M, Tien S-C, Hsieh P-K, Jeng Y-M, Chang M-C, Chang Y-T, et al. High glucose triggers nucleotide imbalance through O-GlcNAcylation of key enzymes and induces KRAS mutation in pancreatic cells. Cell Metab 2019;29:1334–49. [DOI] [PubMed] [Google Scholar]
- 48. Técher H, Koundrioukoff S, Nicolas A, Debatisse M. The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat Rev Genet 2017;18:535–50. [DOI] [PubMed] [Google Scholar]
- 49. Alabert C, Bukowski-Wills J-C, Lee S-B, Kustatscher G, Nakamura K, de Lima Alves F, et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 2014;16:281–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Prem veer Reddy G, Pardee AB. Multienzyme complex for metabolic channeling in mammalian DNA replication. Proc Natl Acad Sci USA 1980;77:3312–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. veer Reddy GP, Pardee AB. Coupled ribonucleoside diphosphate reduction, channeling, and incorporation into DNA of mammalian cells. J Biol Chem 1982;257:12526–31. [PubMed] [Google Scholar]
- 52. Sansregret L, Patterson JO, Dewhurst S, López-García C, Koch A, McGranahan N, et al. APC/C Dysfunction limits excessive cancer chromosomal instability. Cancer Discov 2017;7:218–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Campbell S, Mesaros C, Izzo L, Affronti H, Noji M, Schaffer BE, et al. Glutamine deprivation triggers NAGK-dependent hexosamine salvage. eLife 2021;10:e62644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Onclercq-Delic R, Buhagiar-Labarchède G, Leboucher S, Larcher T, Ledevin M, Machon C, et al. Cytidine deaminase deficiency in mice enhances genetic instability but limits the number of chemically induced colon tumors. Cancer Lett 2023;555:216030. [DOI] [PubMed] [Google Scholar]
- 55. Segerstolpe Å, Palasantza A, Eliasson P, Andersson E-M, Andréasson A-C, Sun X, et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab 2016;24:593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018;33:676–89.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemetary table
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Figure S6
Data Availability Statement
Publicly available The Cancer Genome Atlas (TCGA) data analyzed in this study were obtained from the GDC portal (https://portal.gdc.cancer.gov/projects/TCGA-PAAD). Raw genomic and transcriptomic data for this study were generated at the Cancer Research Center of Toulouse sequencing facility, and deposited in GEO GSE253662 (CDA and CDAE67Q overexpression in cell lines, RNA-seq), GSE252544 (CDA targeting with siRNA in cell lines, RNA-seq), GSE253837 (CDA targeting with siRNA in patient-derived experimental tumors, scRNAseq), and GSE252544 and SRA PRJNA1066503 (CDA and CDAE67Q overexpression in cell lines, WGS). All other raw data are available upon request from the corresponding author.