Pharmacologic inhibition of KRAS elicits varied responses in pancreatic cancer 2D cell lines, 3D organoid cultures, and xenografts, underscoring the importance of mechanotransduction and the tumor microenvironment in regulating therapeutic responses.
Abstract
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive disease that lacks effective treatment options, highlighting the need for developing new therapeutic interventions. Here, we assessed the response to pharmacologic inhibition of KRAS, the central oncogenic driver of PDAC. In a panel of PDAC cell lines, inhibition of KRASG12D with MRTX1133 yielded variable efficacy in suppressing cell growth and downstream gene expression programs in 2D cultures. On the basis of CRISPR-Cas9 loss-of-function screens, ITGB1 was identified as a target to enhance the therapeutic response to MRTX1133 by regulating mechanotransduction signaling and YAP/TAZ expression, which was confirmed by gene-specific knockdown and combinatorial drug synergy. Interestingly, MRTX1133 was considerably more efficacious in 3D cell cultures. Moreover, MRTX1133 elicited a pronounced cytostatic effect in vivo and controlled tumor growth in PDAC patient-derived xenografts. In syngeneic models, KRASG12D inhibition led to tumor regression that did not occur in immune-deficient hosts. Digital spatial profiling on tumor tissues indicated that MRTX1133-mediated KRAS inhibition enhanced IFNγ signaling and induced antigen presentation that modulated the tumor microenvironment. Further investigation of the immunologic response using single-cell sequencing and multispectral imaging revealed that tumor regression was associated with suppression of neutrophils and influx of effector CD8+ T cells. Together, these findings demonstrate that both tumor cell-intrinsic and -extrinsic events contribute to response to MRTX1133 and credential KRASG12D inhibition as a promising therapeutic strategy for a large percentage of patients with PDAC.
Significance:
Pharmacologic inhibition of KRAS elicits varied responses in pancreatic cancer 2D cell lines, 3D organoid cultures, and xenografts, underscoring the importance of mechanotransduction and the tumor microenvironment in regulating therapeutic responses.
Introduction
In spite of significant effort, pancreatic ductal adenocarcinoma (PDAC) remains largely a therapy recalcitrant disease for which overall 5-year survival remains at ∼11% (1, 2). Genetic analyses indicate that PDAC tumors harbor multiple high-potency oncogenic and tumor suppressive lesions (3, 4). Although a myriad of targeted therapeutic approaches have been tested in clinical trials, in many contexts, rationally targeted therapies exploiting genetic features of PDAC have failed to exhibit superiority to chemotherapy. For example, MEK inhibitors, which would be expected to have broad efficacy in KRAS-driven PDAC, have shown little efficacy (5). Thus, developing new therapeutic strategies that target the genetics of PDAC remain a largely unrealized opportunity.
PDAC is dominated by oncogenic mutation of KRAS occurring in >95% of tumors (6). Mutant KRAS acts through multiple signaling pathways that contribute to deregulated proliferation, invasion, and metastasis (7, 8). The signaling mediated by mutant KRAS initiates at the membrane and is transduced by multiple effectors that represent independent drug targets. In PDAC cell lines, there is a variable response to KRAS depletion, which generally mediates a cytostatic response (8). However, the genetic ablation of KRAS can be tolerated and such genetically modified PDAC cell lines can develop into tumors in xenograft models (9). The targeted deletion of KRAS in genetically engineered mouse models can lead to tumor regression. However, KRAS-independent tumors can evolve, with acquired resistance associated with the activation of bypass pathways (e.g., EGFR, AKT, YAP) that abrogate the dependence (10).
The tumor microenvironment (TME) is particularly relevant in PDAC, where the stromal compartment can contribute to the bulk of the tumor volume (11, 12). This stroma has been proposed to both serve tumor-inhibiting and promoting functions (13, 14). In particular, it is believed that the stroma contributes to exclusion of functional antitumor immune functions and contributes to the lack of efficacy of immune checkpoint inhibitors and other forms of immunotherapy (15, 16). Studies with genetic modulation of KRAS have illustrated that tumor-mediated events contribute to broad ranging effects on the TME (17).
KRAS inhibitors have been developed to the stage of FDA-approval in the context of agents selectively directed against the G12C mutant allele (18). This mutation of KRAS is present at relatively low frequency (1%–2%) in PDAC (7). However, there is evidence that a G12C inhibitor, that is, sotorasib, can have clinical activity in PDAC (19), although there are clearly distinct mechanisms through which resistance can emerge (20, 21). The most prevalent KRAS mutation in PDAC is G12D (7, 22). Recently, the agent MRTX1133 was described as a highly selective and potent inhibitor of G12D and has been shown to have efficacy in PDAC models (23, 24). Here, we explored the functional response to G12D inhibition in a spectrum of models that define features of sensitivity and resistance to this agent and underscore the importance of the TME on therapeutic efficacy.
Materials and Methods
Cell culture and therapeutic agents
Human primary PDAC cell lines 828, 519, 3226, 1222, 1229, and 827, and mouse primary PDAC cell line KC4568 were grown in keratinocyte serum-free media (KSF) supplemented with 2% FBS, EGF (0.2 ng/mL), and bovine pituitary extract (30 μg/mL; Life Technologies) as described previously (25). The tissue culture (TC) dishes were precoated with collagen (Millipore). UM53, ASPC1, and HPAF-II cell lines were kindly provided by Dr. Ethan Abel (Roswell Park Comprehensive Cancer Center, Buffalo, NY) and were grown in RPMI media supplemented with 10% FBS. MIA PaCa-2 cell line was cultured in DMEM media, supplemented with 10% FBS. Mouse KPC PDAC cell line 4662 was obtained from Dr. Vonderheide's laboratory (University of Pennsylvania, Philadelphia, PA) and grown in RPMI media supplemented with 10% FBS. The iKRAS AKB6 cell line was kindly provided by Dr. Prasenjit Dey (Roswell Park Comprehensive Cancer Center) and cultured in RPMI media supplemented with 10% FBS. All cell lines were maintained in 37°C with 5% CO2 and tested to be Mycoplasma free. ASPC1, MIA PaCa-2, and HPAF-II cells were authenticated by STR profiling. MRTX1133 was purchased from Chemietek (catalog no. CT-MRTX1133) and dissolved in DMSO to get a final concentration of 10 mmol/L. For in vivo work, MRTX1133 was kindly provided by Mirati Therapeutics. Gefitinib was purchased from SelleckChem.
Cell proliferation assay
To monitor cell growth, we employed the live-cell imaging systems IncuCyte S3 and CellCyte X. The cell lines were transfected to stably express H2B-GFP and seeded in 96-well TC dishes at a density of 2,500 cell/well in the presence and absence of test agents. GFP counts were measured in real-time that corresponded to cell numbers. The fold change in cell number was calculated by normalizing to the first time point following the exposure to drugs. To determine the efficacy of drug, the fold change at each drug concentration following 5 days of treatment was normalized to the untreated condition. A dose–response curve was generated by plotting the drug efficacy and concentration to calculate the EC50.
Knockdown experiments
Cell lines were reverse transfected with gene-specific RNAi that targeted ITGB1, FOSL1, and EGFR, as described previously (26). On-target plus human RNAi for ITGB1 was purchased from Horizon Discovery (catalog no. L-004506–00–0005). siRNAs for EGFR (catalog no. S563) and FOSL1 (catalog no. S15583) were purchased from Thermo Fisher Scientific. Following transfection, cells were treated with MRTX1133, and cell proliferation was monitored using live-cell imaging.
Western blot analysis
Whole cell extracts were prepared as described in our previous studies (25, 27). The primary antibodies that were purchased from Cell Signaling Technology include, pERK (T202/Y204; catalog no. 9101), ERK (catalog no. 9102), PS6 (S235/236; catalog no. 4858), S6 (catalog no. 2217), pAKT (S473; catalog no. 4060), AKT (catalog no. 4691), YAP/TAZ (catalog no. 8418), cyclin B1 (catalog no. 12231), pRB (S807/811; catalog no. 8516S) and RB (catalog no. 9309). β-Actin (catalog no. MAB8929) and ITGB1 (catalog no. AF1778) antibodies were purchased from R&D Systems. Cyclin A antibody was purchased from Sigma (catalog no. C4710).
Colony formation assay
3226, 827, and UM53 cells were seeded in six-well dishes (1,000 cells/well) and treated with two different KRAS inhibitors, MRTX1133 (500 nmol/L) and MRX849 (500 nmol/L) in combination with ITGB1 RNAi. Colonies were allowed to form for 10 days and stained with Crystal Violet.
Spheroid cell culture
The 96- and 48-well TC dishes were precoated with 40 and 200 μL respectively of 50% Matrigel basement layer and allowed to solidify at room temperature for 30 minutes. H2B-GFP labeled PDAC cell lines were seeded in 96- and 48-well TC dishes at the densities of 4,000 cells/well and 40,000 cells/well, respectively. The seeded cells were allowed to form spheroids for 24 to 48 hours. The spheroids that were cultured in 96-well TC dish was exposed to MRTX1133 and the growth of spheroids were monitored for 5 days using live cell imaging. The change in GFP area was used to determine the growth of spheroids. The spheroids that were cultured in 48-well-TC dish were treated with MRTX1133 for 48 hours for Western blotting. Following 48-hour treatment with MRTX1133, the spheroids were harvested and digested using Trypsin by incubating at 37°C for 15 minutes. The resulting cells following trypsin digestion was subjected to whole protein extraction using RIPA lysis buffer as described in our previous studies (27). To examine the ITGB1 deletion on the growth of spheroids, reverse transfection was performed concurrently when cells were seeded on 50% Matrigel in a 96-well TC dish. The spheroids were allowed to form following the deletion of ITGB1 in the presence and absence of MRTX1133.
Mice and patient-derived xenografts
NSG and C57BL/6 mice were maintained at Roswell Park Comprehensive Cancer Center animal care facilities. All animal care, drug treatments and sacrifice were approved by the Roswell Park Comprehensive Cancer Center Institutional Animal Care and Use Committee in accordance with the NIH guide for the care and use of laboratory animals. Mice were subcutaneously implanted with early passage 828, 1222, and 3226 patient-derived xenograft (PDX) tumor fragments. AKB6 xenografts were developed in both NSG and C57BL/6 strains by subcutaneously injecting 1 × 106 cells/mouse. The entire experimental cohort was comprised of both male and female mice. The mice were always maintained in doxycycline water to induce the oncogenic KRAS. KC4568 xenografts were developed in the C57BL/6 strain by subcutaneously injecting 1 × 106 cells/mouse. Once the tumor reached 200 mm3, mice were randomized in a nonblinded manner into vehicle (n = 5) and MRTX1133 (30 mg/kg; n = 5) cohorts, administered intraperitoneally once per day for 3 weeks. MRTX1133 was dissolved in 50 mmol/L citrate buffer and 10% Captisol. Tumor growth was monitored every other day using digital calipers and the tumor volume was calculated using the formula (length × width × width)/2, as described previously (25). Mice were sacrificed and the tumors were embedded in paraffin for further analysis. Mouse body weight was monitored regularly to indicate any gross adverse effects. Mice that died during the course of treatment were excluded from analysis.
Histologic analysis
Freshly cut tumor tissues were fixed in 10% neutral Formalin solution and subjected to processing and paraffin embedding. The processed embedded tissues were serially sectioned at 4 to 6 μm using the standard procedures. The sectioned tissues were stained with hematoxylin and eosin (H&E) and Masson-Trichrome. The deposition of collagen fibers based on Masson-Trichrome staining was scored on the basis of the proposed four-grading system as defined by focal or extensive depositions in intertrabecular marrow spaces, which result in meshwork of collagen (28). IHC staining was performed using the pERK (T202/Y204) antibody (catalog no. 4370; Cell signaling Technologies). Images were captured using the Vectra Polaris Instrument.
Immunofluorescence staining on tissues
The multispectral immunofluorescent (mIF) staining on formalin-fixed paraffin-embedded (FFPE) tissue sections were performed using the Opal 6-Plex Detection Kit (AKOYA Biosciences, catalog no. NEL821001KT) as described in our previous study (29). The mIF panel consisted of the following antibodies: CD8 (EPR20305, Abcam, 570), Pan Keratin (Wide Spectrum cytokeratin, Abcam, 480), and DAPI. Slides were imaged on the PhenoImager HT Automated Quantitative Pathology Imaging System (AKOYA Biosciences). Further analysis of the slides was performed using inForm Software v2.6.0 (AKOYA Biosciences).
Transcriptome analysis
RNA was extracted using Qiagen RNeasy Plus Kit and analyzed via the RNA6000 Nano assay and with the Agilent 2200 TapeStation (Agilent) for determination of an RNA integrity number (RIN). Only the cases with RIN > 7.0 were included in this study. cDNA synthesis was performed using random hexamers to obtain full-length, strand-specific representation of nonribosomal RNA transcripts. Targeted RNA sequencing libraries were prepared with the DriverMap Human Genome-Wide Gene Expression Profiling Sample Prep Kit hDM18Kv3 (Cellecta Inc.). A defining feature of the DriverMap method is the application of predesigned multiplex PCR primer sets targeted to specific 50 to 75 bp regions of all known protein-coding genes. Notably, each target-specific primer consists of a complementary sequence to specific mRNA targets plus a universal primer binding site (anchor). Ligation of oligonucleotides via PCR amplification introduces adaptors required for sequencing and sample-specific “barcodes” that flank the target sequence and are inserted into standard Illumina adaptors to permit dual-index sequencing and deconvolution of sample-specific reads using standard Illumina software.
Briefly, to mitigate primer dimer formation, anchor PCR was performed with an initial hot start at 95°C for 5 minutes, followed by 15 cycles of (95°C–0.5 minutes, 68°C–1 minute, 72°C–1 minute), and ended with a final 10 minutes extension at 72°C. The reaction products were confirmed on an agarose gel in triplicate to assess replicability. PCR products were then purified by SPRI (Agentcourt, 1:1 sample: reagent ratio) and quantified with the Qubit fluorescence assay (Qubit dsDNA HS Assay Kit; Thermo Fisher Scientific). Target-enriched RNA-seq libraries were analyzed on an Illumina NextSeq 500 sequencer using a NextSeq500/550 High Output v2 Kit (75 cycles) according to the standard manufacturer's protocol (Illumina).
For RNA-seq, alignment was performed using STAR v2.7.10b, and this pipeline outputs gene-level read counts directly (30). Transcript abundance estimates for each sample were performed using Salmon, an expectation–maximization algorithm using the UCSC gene definitions. Raw read counts for all samples were normalized using the “weighted” trimmed mean of M-values (TMM) approach in the Bioconductor package EdgeR (31). After trimming the data [5% for the A values, log ratio 0.3 for the M values to a reference array (the library whose upper quartile is closest to the mean upper quartile)], scaling factors for each sample were generated using the calcNormFactors function. Genes with an average of less than 10 read counts across all samples were excluded from further analysis. Following data integration, systematic bias was corrected using ComBat, as described previously (32). Differentially expressed gene (DEG) analysis was performed with EdgeR and selected based on P value and log2-fold change (log2FC), respectively.
Gene set enrichment analysis
For each cell line, we performed differential gene expression analysis between MRTX1133 treated and control samples using the DESeq2 software package. The log2-fold change was used as input for gene set enrichment analysis (GSEA) preranked analysis to calculate the normalized enrichment scores (NES) and associated P values for each of the 50 gene sets in the hallmark gene sets from Molecular Signature Database (MSigDB, v2022.1.Hs). We used the fgsea Bioconductor package (ref) for this analysis. The NES values as well as the associated adjusted P values for each cell line are merged to make a bubble chart to display the NES and adjusted P values for each gene set and cell line. We ordered the gene sets by the NES values of ASPC1.
CRISPR screening
Human Toronto KnockOut (TKO) CRISPR library version 3, containing 70,948 unique sgRNAs targeting 18,053 genes was packaged in lentivirus according to the previously published protocol (33). ASPC1 and UM53 cells were infected with the TKOv3 lentiviral particles at an MOI ∼0.3 and the positive clones were selected using puromycin to achieve a mutant pool comprising at least 200-fold coverage of the library. The resulting mutant pool was grown in the absence and presence of KRASi at the EC25 concentration for at least five passages. Genomic DNA was extracted and subjected to sequencing library construction by amplifying the gRNA inserts using a 2-step PCR as described in the previously published study (33). Resulting libraries were further sequenced on an Illumina's NextSeq platform. Fastq files were first subjected to adapter removal using Trim Galore (v0.6.7, https://github.com/FelixKrueger/TrimGalore). Next, the MAGeCK (34) pipeline was used to process the adapter trimmed fastq files and the resultant count files were used as input to DrugZ (35) python script for chemogenetic interaction identification. An in-house developed R script was used to create plots for results visualization.
We evaluated the top-scoring sgRNAs that emerged from our screens. To accomplish this, we used a negative binomial distribution model using pseudo counts to adjust for NGS library sizes (36). Specifically, we determined the magnitude (fold change) and significance (P value and FDR) of sgRNA depletion or enrichment. To visualize the differential representation of the individual sgRNAs, a volcano plot was generated where the x-axis indicates the fold change and the y-axis indicates the −log10pVal. The sgRNAs were filtered based on the DrugZ normalized score by setting a threshold with Z-score of <−1.96 and >1.96 and a 95% confidence interval.
10× single-cell RNA sequencing
Tumors excised from AKB6 xenografts that were treated with vehicle (n = 2) and MRTX1133 (n = 2) were digested using Liberase (Sigma, catalog no. 05401020001) to get single cells and were subsequently sent to RPCCC's Genomics Shared Resources for sequencing using 10× Chromium and Illumina's NovaSeq instruments. The raw fastq data were processed using 10X's cellranger pipeline (v6.1.2). The resultant “filtered_feature_bc_matrix.h5” files for treatment and vehicle control, respectively, were used as inputs for a custom R (v4.2.0) script, which utilizes the Seruat (37) R package (v4.2.0) from CRAN. We filtered each dataset with the following criteria: percent.mt < 10, nFeature_RNA between 200 and 4,000, percent.largest.gene < 20. With these parameters, we obtained 9307 and 7204 single cells in the treated and control samples, respectively. Because the treated sample returned more single cells, we down-sampled by randomly selecting the same number of cells (7204) as in the control. The two datasets were then integrated using the FindIntegrationAnchors and IntegrateData function from the Seurat suite. After this step, the standard Seurat workflow was followed to cluster the combined dataset. We identified 20 clusters and annotated the clusters with ScType (38).
For subclustering the cancer clusters (Seurat clusters 3 and 6), we applied the subset function from the Seurat package to extract cells from the original Seurat cluster 3 and 6. For each subset object, we applied the standard workflow (RunPCA, RunUMAP, FindNeighbors, FindClusters). We experimented with the parameter of resolution in the FindClusters function and used the value of 0.3 in the final analysis as this value gave the best partitioning of the cancer cells into subclusters in original Seurat cluster 3. For original Seurat cluster 6, it was not sensitive to this parameter and we used the same value (resolution = 0.3).
Digital spatial profiling
Formalin-fixed paraffin-embedded (FFPE) 5 μm sections were prepared to NanoString specifications on Lecia Apex BOND Superior Adhesive slides (3800040). These slides were then stained following the GeoMX DSP (digital spatial profiling) Automated Slide Preparation user manual (MAN-10151–02). Before the slides were placed on the BOND RXm (Lecia Biosystems), they were baked in a hybridization oven at 60°C for a minimum of 3 hours. All the BOND containers were prepared to manual specifications. (i) Wash Buffer, (ii) 10% NBF, (iii) NBF Stop buffer (containing TRIS base 0.1 M and glycine 0.1 M), (iv) 50% Formamide and 2× SCC solution, (v) Proteinase K (concentration tissue dependent), (vi) Buffer R (NanoString, GeoMx RNA Slide Prep Auto), (vii) GeoMx NGS RNA Auto WTA MM (NanoString, 12140105), (viii) Buffer W (NanoString, GeoMx RNA Slide Prep Auto), and (ix) GeoMx Solid Tumor TME Morphology Kit Mouse RNA FFPE Compatible (NanoString, 121300322), which contains CD45, PanCK, and a nuclear stain. The automated staining process then ran overnight. The following day, slides were removed from the BOND and loaded onto the GeoMX DSP, where 20× fluorescent scans were taken. Regions-of-interest (ROI) were subsequently selected by Dr. Witkiewicz, a clinical pathologist at Roswell Park Comprehensive Cancer Center. On the basis of the distinct fluorescent signals from PanCK and CD45 staining digital masking was automatically performed to demarcate the tumor and immune population. The UV cleavable oligo tags were then collected from PanCK+ and PanCK− regions and separately deposited into a 96-well plate.
We used NanoStrings GeoMx instrument and the Mouse NGS Whole Transcriptome Atlas RNA (version 1.0) Kit for ROI selection and mRNA library preparation. The resulting mRNA samples were subjected to sequencing using Illumina's NovoSeq instrument. Raw fastq files along with the configuration file were used as input to the geomxngspipeline command line tool running on a Linux server. We used NanoString's GeomxTools Bioconductor package (https://www.bioconductor.org/packages/release/bioc/html/GeomxTools.html) for downstream data analyses. These included QC & preprocessing, data normalization, unsupervised clustering, and differential gene expression. For QC, we used these value parameters: minSegmentReads = 1,000, percentTrimmed = 80, percentStitched = 80, percentAligned = 80, percentSaturation = 50, minNegativeCount = 2, maxNTCCount = 1,000, minNuclei = 100, minArea = 5,000, minLOQ = 2. For segments that passed these QC parameters, we utilized Q3 normalization and mixed effect modeling for identifying differentially expressed genes in the PanCK+ segments between treated and control.
Data availability
The datasets for the transcriptome analysis were uploaded in the supplementary files. The raw fastq files for RNA sequencing, CRISPR screening, 10× single-cell sequencing, and DSP analysis are publicly available in the GEO (GSE249541). All additional data generated in this study are available from the corresponding author upon request.
Code availability
No unique code was developed in this study.
Results
Heterogeneous cellular response to MRTX1133 in PDAC models
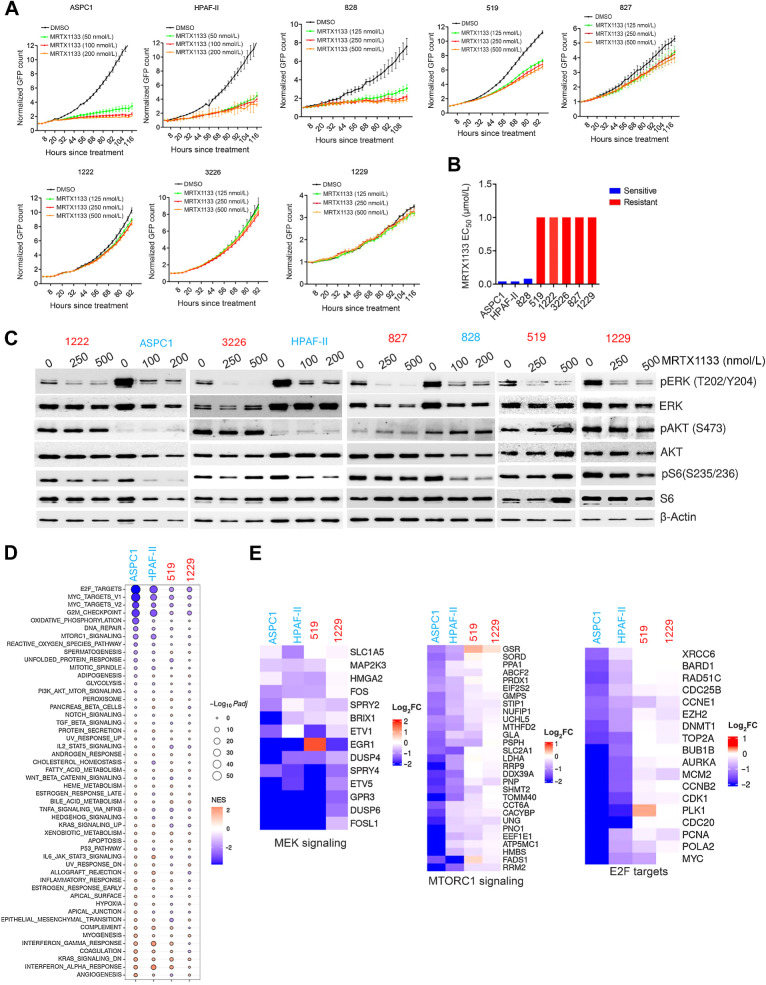
To define the impact of a pharmacologic KRAS inhibitor, MRTX1133, we employed a panel of KRASG12D mutant PDAC cell lines that include both established and patient-derived models (Supplementary Fig. S1A and S1B; ref. 39). The patient-derived PDAC models harbor comutations in other common genes like SMAD4, CDKN2A, and TP53 that drive PDAC tumorigenesis (Supplementary Fig. S1A). MRTX1133 elicited a highly varied response on cell proliferation as monitored by live cell imaging (Fig. 1A). The established PDAC cell lines ASPC1 and HPAF-II, and a patient-derived cell line, 828, were sensitive to MRTX1133 and displayed a robust cytostatic response (Fig. 1A). The 519 cell line elicited a partial response to MRTX1133; however, the impact was relatively moderate as compared with the sensitive lines (Fig. 1A). The other patient derived PDAC cell lines, 827, 1222, 3226 and 1229, were largely resistant, as cellular proliferation continued despite the presence of MRTX1133 (Fig. 1A). On the basis of cell proliferation after 5 days of treatment, the efficacy of MRTX1133 was evaluated and the corresponding EC50 value in sensitive models (ASPC1, HPAF-II, and 828) was determined to be in the range of 25 to 50 nmol/L, whereas in other models (519, 827, 1222, 3226, and 1229), the EC50 was greater than 1 μmol/L (Fig. 1B; Supplementary Fig. S1C). To validate the specificity of MRTX1133 to KRASG12D mutants, we evaluated the anti-proliferative effect of this drug in KRASG12C mutant cell lines MIA PaCa-2 and a patient-derived PDAC cell line, UM53 (Supplementary Fig. S1D). As a control, we included the KRASG12C inhibitor MRTX849. On the basis of growth curves, the proliferation of MIA PaCa-2 cells was robustly inhibited by MRTX849, whereas MRTX1133 had no effect, confirming KRASG12D target specificity (Supplementary Fig. S1D). Although, UM53 cell line possessed modest response to MRTX849 there was a significant difference in response as compared to MRTX1133 (Supplementary Fig. S1D).
Figure 1.
Cellular response to MRTX1133 in a panel of KRASG12D mutant PDAC cell lines. A, Live cell imaging using IncuCyte S3 on the indicated cell lines treated with different concentrations of MRTX1133. Cell proliferation was determined on the basis of the GFP counts. Graph represents mean and SD from triplicates. Experiments were done at two independent times. B, EC50 values of MRTX1133 on different PDAC cell lines were calculated on the basis of the dose–response curve. To determine the drug effect at each dose, the fold change in cell number after 5 days of treatment was normalized to untreated condition. C, Immunoblotting of the indicated cell lines treated to examine the effect of MRTX1133 on the KRAS-mediated signaling pathways following 24 hours treatment at different concentrations. D, Bubble plot depicting the differentially expressed pathways that were ranked on the basis of gene set enrichment scores and their P values on the indicated cell lines in the presence of MRTX1133 (100 nmol/L). E, Heatmap depicting the differential expression of the indicated genes that are involved in MEK signaling, MTOR signaling, and E2F-regulated cell-cycle genes.
To interrogate the impact of MRTX1133 on the KRAS-mediated signaling pathways, biochemical analyses were performed in both sensitive and resistant models. Following 24- and 48-hour exposure with MRTX1133 at the indicated concentrations, ERK phosphorylation was effectively inhibited in all models tested (Fig. 1C; Supplementary Fig. S2A). However, in the HPAF-II cell line, a rebound in phospho-ERK was observed after 48-hour treatment (Supplementary Fig. S2A). In the sensitive lines, ASPC1 and HPAF-II, the intrinsic phosphorylation of AKT was lower compared with the resistant lines, 1222 and 3226 (Fig. 1C). However, this observation was cell line-specific and did not correlate with the response to MRTX1133 as the resistant model 827 displayed lower AKT phosphorylation than a sensitive line, 828 (Fig. 1C). Notably, our data revealed that the impact on S6 phosphorylation was more potent in the sensitive lines (ASPC1, HPAF-II, and 828) whereas a modest-to-no effect was observed in all the resistant models (827, 1222, 3226, and 1229) after 24- and 48-hour treatment with MRTX1133 (Fig. 1C; Supplementary Fig. S2A). Overall, our data indicated that although MRTX1133 inhibits the MEK signaling in all the models that harbor the G12D KRAS mutation, the impact on S6 phosphorylation serves as a key determinant of the anti-proliferative response. To confirm that the inhibition of ERK phosphorylation by MRTX1133 is a consequence of KRASG12D inhibition, we compared its effect with MRTX849 on the MIA PaCa-2 cells. Our data confirmed that inhibition of ERK phosphorylation is not affected by MRTX1133 (Supplementary Fig. S2B).
To investigate the molecular alterations at a global level in an unbiased fashion, transcriptome analysis was carried out in two sensitive lines (ASPC1 and HPAF-II) and two resistant lines (519 and 1229) in the presence and absence of MRTX1133 (100 nmol/L; Supplementary Table S1). On the basis of differential gene expression analysis, the sensitive models ASPC1 and HPAF-II displayed a relatively greater number of genes that were more significantly altered in the presence of MRTX1133 as compared with the resistant models 519 and 1229, indicating resistance to KRAS inhibition (Supplementary Fig. S3A). Gene set enrichment analysis (GSEA) revealed that the top pathways that were differentially expressed following MRTX1133 treatment included E2F targets, MYC targets, G2–M checkpoint and MTOR signaling in all the cell lines (Fig. 1D). Consistent with our biochemical data, some of the critical MEK signature genes, including DUSP6 and SPRY4, were repressed in both the sensitive (ASPC1 and HPAF-II) and resistant (519 and 1229) lines, confirming that, at a minimum, this downstream pathway of KRAS was inhibited (Fig. 1E). However, the impact on genes involved in MTOR signaling was more pronounced in the ASPC1 and HPAF-II cell lines, as compared to 519 and 1229 models (Fig. 1E). Consistent with the inhibitory effect on S6 phosphorylation, the differential impact on MTOR signaling corroborated with the cellular response to MRTX1133 (Fig. 1E). Moreover, MRTX1133 yielded a greater inhibitory effect on E2F target genes in ASPC1 and HPAF-II than in the 519 and 1229 cell lines (Fig. 1E). In addition, bulk RNA sequencing was performed in other resistant models, 1222 and 3226, following treatment with MRTX1133 (100 nmol/L). Our data demonstrated that the genes associated with MEK signaling were repressed more effectively than the genes involved in MTOR pathway and cell cycle (Supplementary Fig. S3B). Because the E2F target genes are known to regulate cell-cycle progression via RB inactivation, biochemical analysis was performed in the PDAC cell lines following treatment with MRTX1133. In the sensitive models ASPC1 and 828, RB phosphorylation was significantly inhibited, which further resulted in the suppression of cyclin A, an E2F target, indicating a durable cell-cycle arrest (Supplementary Fig. S3C). Conversely, in one of the resistant models, 519, RB remained phosphorylated, with modest impact on cyclin A expression after MRTX1133 treatment (Supplementary Fig. S3C). In addition, MRTX1133 significantly inhibited BrdU incorporation in the sensitive models confirming a robust cell-cycle arrest, whereas moderate or no effect was observed in the resistant models (Supplementary Fig. S3D). In conclusion, although different PDAC models harbor oncogenic KRAS G12D mutation, there is heterogeneity in response to the pharmacologic inhibitor, which differentially impacts MTOR signaling and cell cycle.
Genetic modifiers of the response to KRAS inhibitors
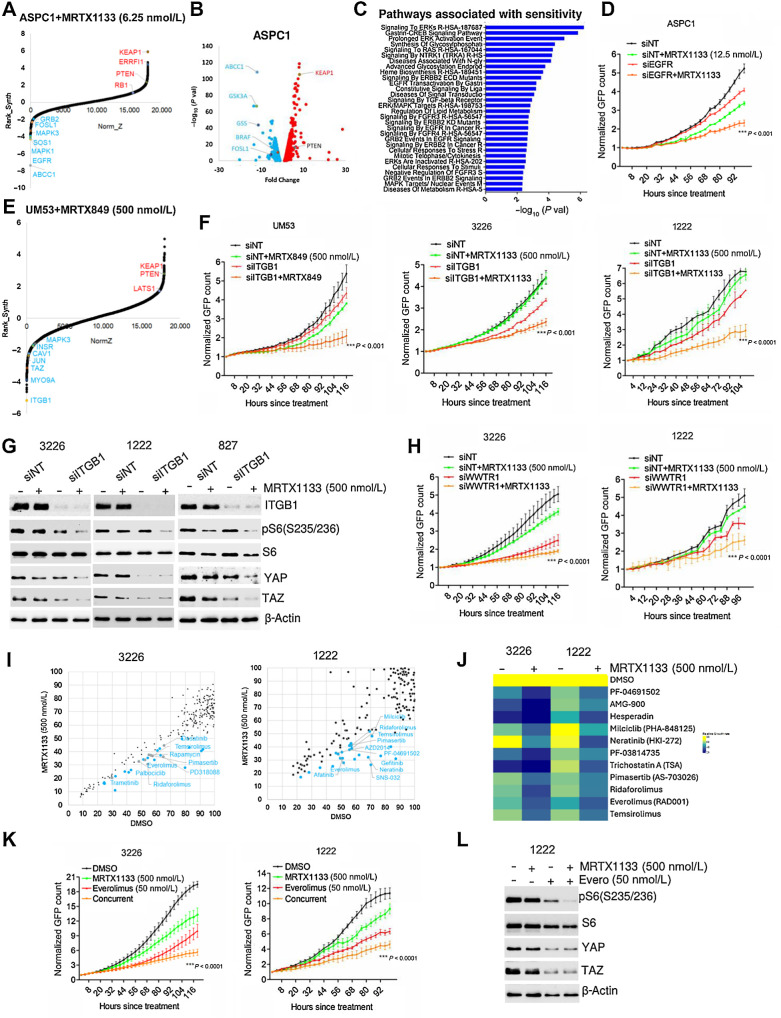
To systematically and comprehensively identify critical genes involved in resistance and sensitivity to KRAS inhibition, we employed a genome-wide loss-of-function CRISPR/Cas9 library screen in ASPC1 cell line. Following infection with the CRISPR/Cas9 pooled library, the cells were grown in the presence and absence of MRTX1133 at a concentration (6.25 nmol/L), where it had modest effect on cell proliferation (Supplementary Fig. S4A). The sgRNAs that were depleted or enriched following the treatment with MRTX1133 imply that the loss of function of their target genes could enhance or attenuate the cytostatic effect of the drug, respectively. As expected, sgRNAs targeting genes that regulate ribosome biogenesis, proteasome degradation, RNA metabolism, and cell cycle were significantly depleted irrespective of drug treatment, indicating the essential function of those genes in cell survival (Supplementary Fig. S4B). Moreover, a pairwise comparison between the replicates indicated a correlation coefficient of approximately 0.9, illustrating the reproducibility of the screen (Supplementary Fig. S4C). On the basis of DrugZ analysis, we identified 235 genes with NormZ values <−1.9 that were significantly depleted in the presence of MRTX1133 (Fig. 2A). The identified genes either directly regulate the function of KRAS, such as SOS1 or GRB2, encompass parallel pathways to drive downstream signaling, such as EGFR, or act as downstream effectors, such as BRAF, MAPK3, and FOSL1, suggesting that these genes contribute resistance to MRTX1133 treatment (Fig. 2A).
Figure 2.
Combinatorial approach to overcome resistance to MRTX1133. A, DrugZ analysis in ASPC1 cells indicating the genes that were negatively selected during the MRTX1133 (6.25 nmol/L) treatment. B, Volcano plot indicating the genes whose sgRNAs are differentially selected in the presence of MRTX1133 (6.25 nmol/L) in ASPC1 cells. The individual guides were filtered with a Z-score <−1.96 and >1.96 (95% CI) based on the DrugZ analysis. C, ENRICHR analysis to determine the pathways that are associated with the negatively selected genes from ASPC1 cells. D, Live cell imaging to monitor the proliferation of ASPC1 cells following EGFR deletion in the absence and presence of MRTX1133. E, DrugZ analysis in UM53 cells indicating the genes that were negatively selected during the MRTX849 (500 nmol/L) treatment. F, Effect of ITGB1 knockdown in UM53, 326, and 1222 cells in the absence and presence of MRTX1133 and MRTX849 at the indicated concentrations on cell proliferation. G, Biochemical analysis in 3226, 1222, and 827 cells on the indicated proteins following ITGB1 knockdown in combination with MRTX1133 (500 nmol/L). H, Effect of WWTR1 deletion on the proliferation of 1222 and 3226 cells treated with MRTX1133 upto 5 days. I, Scatter plot analysis in 1222 and 3226 cell lines following a combinatorial drug screen. Each dot represents a drug from the library. X-axis indicates the effect of individual drugs from the library. Y-axis represents the impact of MRTX1133 combination on the individual drugs from the library. The drug effect was calculated on the basis of the fold change in cell number following 5 days of treatment normalized to the mean vehicle-treated groups in the library. J, Heatmap depicting the effect of the indicated drugs in combination with MRTX1133 on 1222 and 3226 cells. K, Live cell imaging to validate the effect of everolimus in combination with MRTX1133 on the proliferation of 1222 and 3226 until the indicated times. L, Western blotting of the indicated proteins from 1222 cells following the treatment with MRTX1133 in combination with everolimus. Error bars in all the graphs represent mean and SD from triplicates. ***, P < 0.0001 as determined by two-way ANOVA. Experiment was done at two individual times.
As a complementary approach, the differential representation of individual sgRNAs was determined between vehicle- and MRTX1133-treated groups to define the genes that were positively and negatively selected as mentioned in the methods (Fig. 2B). REACTOME pathway analysis indicated that the negatively selected sgRNAs target gene sets were highly enriched in different mitogenic pathways, including EGFR signaling, ERK signaling cascade, and metabolic pathways (Fig. 2C). The individual guides that target EGFR and FOSL1 revealed that at least three sgRNAs were depleted in the presence of MRTX1133 (Supplementary Fig. S4D). To validate the functional roles of EGFR and FOSL1, RNAi-mediated deletion was performed, which significantly cooperated with MRTX1133 treatment and yielded a potent growth suppression in ASPC1 cells (Fig. 2D; Supplementary Fig. S4E). Among the sgRNAs that were positively selected following MRTX1133 treatment, KEAP1 was the most enriched, which is consistent with previous studies (Fig. 2C; refs. 24, 40). Furthermore, sgRNAs targeting, ERRFI1 and PTEN, which inhibits EGFR and mTOR pathways respectively and RB1, a negative regulator of cell cycle was also positively selected (Fig. 2A). To validate these findings, we employed a pharmacologic approach where the ASPC1 cell line was exposed to different EGFR inhibitors in combination with MRTX1133. On the basis of the relative growth rate, it was evident that a subset of EGFR inhibitors cooperated with MRTX1133 treatment to induce a cytostatic response; gefitinib significantly enhanced the efficacy of MRTX1133 in ASPC1 cells (Supplementary Fig. S4F and S4G). Although ASPC1 is intrinsically sensitive to MRTX1133, it is expected that targeting EGFR-mediated signaling pathways could hypersensitize the cells to KRAS inhibition.
To further identify gene sets that could increase sensitivity to KRAS inhibition, CRISPR/CAS9 screening was also performed in the UM53 cell line that is resistant to pharmacologic KRAS inhibition. Because UM53 harbors a G12C KRAS mutation, we used MRTX849, which is a selective G12C inhibitor, for the positive and negative selection of guide sequences. Moreover, the use of this G12C model allowed us to define consistent features of KRAS inhibition with mechanistically distinct agents. Integrin subunit beta 1 (ITGB1) was identified as a negatively selected gene upon MRTX849 treatment (Fig. 2E). In addition, sgRNAs targeting CAV1, which is involved in integrin-mediated mechanotransduction, also dropped out and the individual guides were depleted following MRTX849 treatment in UM53 cells (Fig. 2E; Supplementary Fig. S5A; ref. 41). Similarly, individual guides against ITGB1 in ASPC1 cells were depleted in the presence of MRTX1133 (Supplementary Fig. S5A). Further validation was performed using gene-specific RNAi that targeted ITGB1 in combination with KRAS inhibition in four different PDAC cell lines. In the resistant cell lines UM53, 3226, 1222 and 827, KRAS inhibition delayed cell proliferation following ITGB1 deletion as determined by live-cell imaging and colony formation assay (Fig. 2F; Supplementary Fig. S5B). Prior studies have demonstrated that the integrin family of proteins sense different extracellular mechanical stimuli and transduce them into downstream signaling cascades to elicit specific cellular functions (42, 43). Integrin-mediated mechanotransduction is known to activate the PI3K/MTOR pathway and regulate the levels of YAP and TAZ, which are involved in the transactivation of several genes involved in cell proliferation, differentiation, and focal adhesion, among other cellular processes (44, 45). Consistent with this, our biochemical analysis revealed that deletion of ITGB1 enhanced the impact of MRTX1133 on phospho-S6 levels in the resistant models 1222, 3226, and 827 (Fig. 2G). Moreover, loss of ITGB1 resulted in a cooperative downregulation of YAP and TAZ in 3226 and 827 models and suppressed the endogenous YAP/TAZ levels in 1222 cells (Fig. 2G). In addition, depletion of the WWTR1 gene, which encodes TAZ, resulted in the cooperative inhibition of cell proliferation in 1222, 3226, and 827 cells in combination with MRTX1133 (Fig. 2H; Supplementary Fig. S5C). Mechanistically, WWTR1 deletion enhanced the effect of MRTX1133 on RB activation and the downregulation of cyclin A in 1222 and 3226 cell lines (Supplementary Fig. S5D). Overall our data imply that ITGB1-mediated regulation of TAZ expression potentially plays a role in modulating the cellular response to KRAS inhibition.
To identify pharmacologic agents that could therapeutically target mechanotransduction, we carried out a combinatorial drug screen in the presence and absence of MRTX1133 in the 1222 and 3226 cell lines. Although MEK and EGFR inhibitors enhanced the efficacy of MRTX1133 in a cell line-specific manner, a subset of MTOR inhibitors cooperatively inhibited cell proliferation in both 1222 and 3226 cells, as determined by live-cell imaging (Fig. 2I and J; Supplementary Figs. S6A and S7A). Further validation using gefitinib resulted in durable arrest in combination with MRTX1133 selectively in 519 and 1222 cell lines, with a modest effect in the 3226 and 827 models (Supplementary Fig. S8A). However, inhibition of the MTOR pathway using everolimus inhibited cell proliferation in combination with MRTX1133 in all resistant models tested (1222, 3226, and 827), with intrinsic everolimus sensitivity in 519 (Fig. 2K; Supplementary Fig. S8B). Biochemical analysis in 1222 cells revealed that everolimus enhanced the impact of MRTX1133 on S6 phosphorylation and suppressed YAP/TAZ levels similarly to ITGB1 deletion (Fig. 2L). In summary, these results reveal a molecular crosstalk between the integrin-mediated mechanical signaling and the KRAS pathway that could be therapeutically targeted to enhance the cellular responses to KRAS inhibition.
Potent efficacy of MRTX1133 in 3D cell culture and in vivo settings
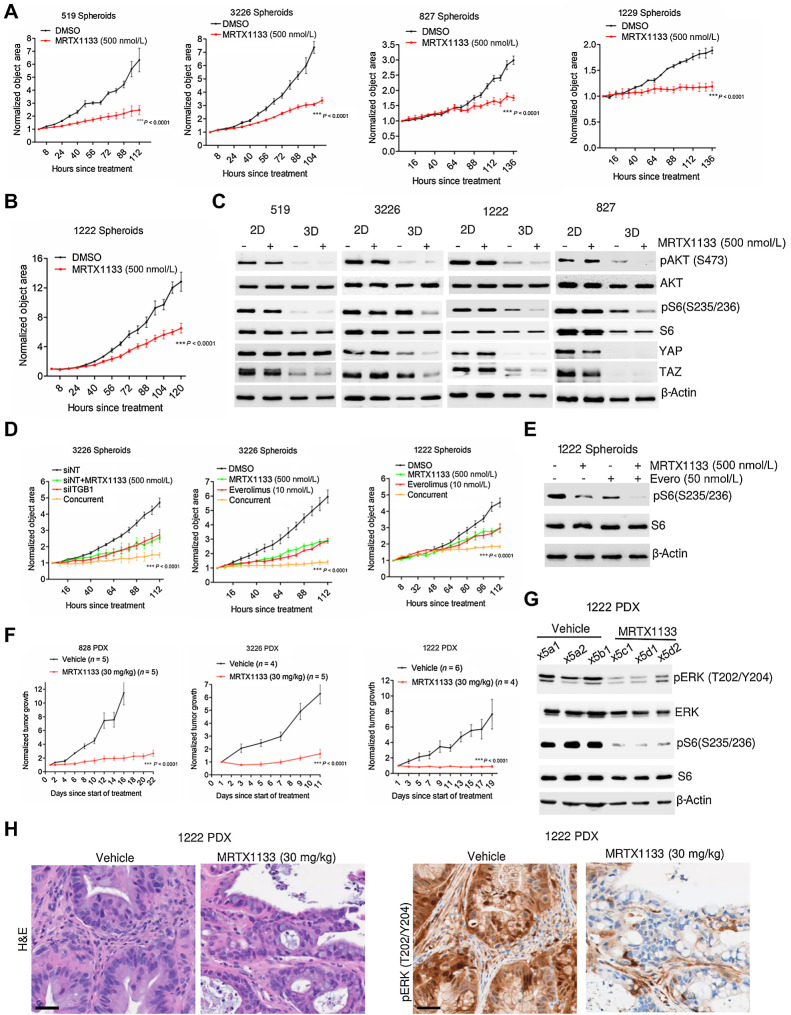
A previous study has established that the anchorage-dependent growth of KRAS mutant models can attenuate the impact of KRAS inhibition (46). Our observations align with this finding as integrins sense mechanical signals arising from cell adhesion to the extracellular matrix (ECM; ref. 42). Therefore, we postulated that the anchorage-dependent growth of cells in two-dimensional (2D) monolayer might lead to increased mechanical stress, which could be alleviated in three-dimensional (3D) culture when grown as spheroids in low ECM stiffness conditions such as Matrigel. To test this hypothesis, we evaluated the impact of MRTX1133 on the growth of spheroids that were derived from the cell lines, 519, 3226, 827, 1229, and 1222, which exhibited intrinsic resistance to KRAS inhibition under 2D culture conditions. Surprisingly, in all the models tested, MRTX1133 significantly inhibited the growth of spheroids even at concentrations that had no effect in 2D culture (Fig. 3A; Supplementary Fig. S9A). Although 1222 spheroids displayed a modest response to MRTX1133, the effect was still more pronounced than the response in 2D culture, suggesting that growing cells as spheroids in Matrigel could enhance the response to MRTX1133 (Fig. 3B).
Figure 3.
Efficacy of MRTX1133 on spheroid growth and PDAC PDX models. A, Effect of MRTX1133 on the growth of spheroids derived from the indicated PDAC cell lines in the absence and presence of MRTX1133 (500 nmol/L). Growth of spheroids were determined on the basis of the object area using IncuCyte S3. B, Effect of MRTX1133 on the growth of 1222 spheroids. C, Western blotting to compare the effect of MRTX1133 on the indicated proteins between the cells grown in 2D monolayer and 3D culture as spheroids. D, Effect of ITGB1 knockdown in combination with MRTX1133 on the growth of spheroids derived from 3226 cell line. The impact of everolimus (10 nmol/L) in combination with MRTX1133 (500 nmol/L) on the growth of spheroids derived from 1222 and 3226 cell lines based on live cell imaging. E, Immunoblotting to determine the effect of everolimus in combination with MRTX1133 on the phosphorylation status of S6 in spheroids derived from 1222 cells. F, Normalized tumor growth rate of 828, 3226, and 1222 PDX following vehicle and MRTX1133 (30 mg/kg) treatment, administered intraperitonially (four times a day) for the indicated number of days. G, Biochemical analysis from tumor tissues excised from 1222 PDX treated with MRTX1133 to determine its effect on the KRAS-mediated signaling pathways. H, Representative images for H&E staining and IHC to evaluate the effect of MRTX1133 on the phosphorylation of ERK1/2. Scale bar, 40 μm. Error bars from all growth curves indicate mean and SEM from triplicates and the experiment was done at two independent times. ***, P < 0.0001 as determined by two-way ANOVA.
To investigate the molecular mechanisms, we performed biochemical analyses comparing the impact of MRTX1133 on cells grown in 2D monolayer versus 3D culture. The effect of MRTX1133 in inhibiting spheroid growth corroborated with its ability to block cell cycle as determined by enhanced RB dephosphorylation in 519, 3226, and 827 cells cultured under 3D conditions (Supplementary Fig. S9B). Moreover, in all the models tested, (519, 3226, 1222, and 827), the intrinsic AKT phosphorylation was prominently lower in cells grown in 3D culture, which, in turn, resulted in an enhanced inhibition of S6 phosphorylation in the presence of MRTX1133 (Fig. 3C). The endogenous YAP and TAZ levels were low in spheroids derived from 1222 and 827 models whereas in the 3226, YAP and TAZ expression was significantly suppressed following MRTX1133 treatment selectively in 3D conditions (Fig. 3C). In the 519 model, the basal TAZ levels were lower in 3D culture as compared with 2D monolayer cells (Fig. 3C). Overall, our data indicate that the regulatory mechanism of YAP/TAZ is intrinsically limited in spheroids, which in turn augments the efficacy of MRTX1133. In addition, ITGB1 deletion further inhibited the growth of spheroids derived from 3226 cells and MRTX1133 combination yielded a durable cytostatic response (Fig. 3D). Similarly, pharmacologic inhibition of the MTOR pathway using everolimus significantly enhanced the inhibitory effect of MRTX1133 on the growth of spheroids derived from the 1222 and 3226 models (Fig. 3D). Biochemical analysis confirmed that MRTX1133 in combination with everolimus displayed a cooperative inhibition of S6 phosphorylation, which, in turn, resulted in durable cell-cycle arrest, as determined by RB activation and downregulation of cyclin A and cyclin B1 (Fig. 3E; Supplementary Fig. S9C).
Consistent with its efficacy against spheroid growth, MRTX1133 also significantly inhibited tumor growth in vivo in three different PDX models (828, 3226, and 1222; Fig. 3F; Supplementary Fig. S9D and S9E). The in vivo effect of MRTX1133 treatment on KRAS signaling was examined using 1222 PDX tumor samples following 5 days of treatment. Our findings revealed that MRTX1133 effectively inhibited the phosphorylation of ERK and S6, suggesting that in vivo KRAS inhibition leads to the inactivation of both MEK and MTOR signaling pathways (Fig. 3G). Histologic analysis from the 1222 PDX model revealed a composition of tumor and stromal populations both exhibiting ERK phosphorylation (Fig. 3H). Interestingly, following treatment with MRTX1133, ERK phosphorylation in the tumor was prominently inhibited, whereas the stromal population remained unaffected (Fig. 3H). This observation confirms the specificity of MRTX1133 in inhibiting the intratumoral KRAS, which harbors the G12D mutation.
Heterogeneous cellular response to MRTX1133 in murine models
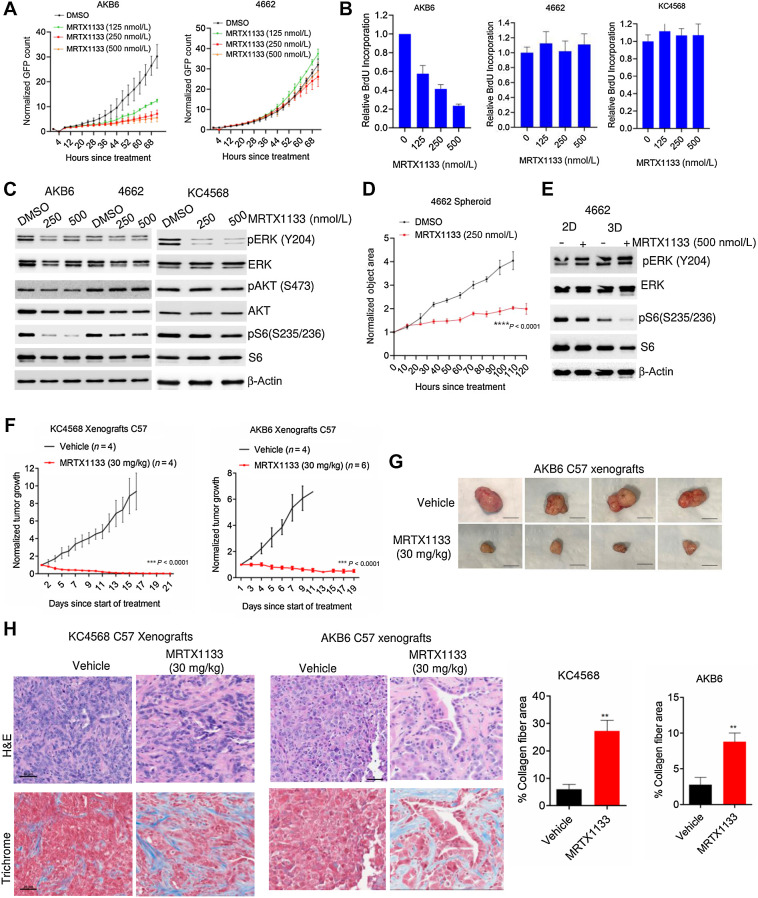
Our data highlight that the environmental niche can modulate the response to KRAS inhibition, which prompted us to further explore the role of the TME on the impact of MRTX1133 treatment. The TME and features of the host immune system play a major role in the pathogenesis of PDAC. Hence, we investigated the effect of MRTX1133 in an immunocompetent setting (17). We employed two syngeneic mouse models that were previously established from KPC mice (4662; ref. 47) and the iKRASG12D model where the oncogenic KRASG12D is induced by doxycycline (AKB6; ref. 48). Similar to the human cell lines, the mouse models displayed differential response to MRTX1133 in cell culture, where the AKB6 underwent growth arrest in a concentration-dependent manner and 4662 continued proliferations (Fig. 4A). The anti-proliferative effect of MRTX1133 in the AKB6 cell line was corroborated by a more potent inhibition on cell cycle, compared with its effects in the 4662 cell line, as demonstrated by BrdU incorporation (Fig. 4B). We developed another syngeneic PDAC model derived from Kras+/LSL-G12D;Pdx1-Cre (KC) mice termed KC4568, which was also resistant to MRTX1133 treatment, as determined by BrdU incorporation (Fig. 4B; Supplementary Fig. S9F). Biochemical analysis revealed that MRTX1133 effectively inhibited ERK phosphorylation in the KC4568 cell line with a modest impact in the AKB6 and 4662 models (Fig. 4C). However, the inhibitory effect on S6 phosphorylation was more pronounced in the sensitive model (AKB6) whereas in the resistant models (4662 and KC4568) it remained unaffected, indicating that the impact on MTOR signaling correlates with the response to KRAS inhibition in mouse cell lines (Fig. 4C). Similar to human cell lines, the 4662 model possessed a more profound response to MRTX1133 in 3D culture as spheroids (Fig. 4D). Despite no impact on ERK phosphorylation in 3D culture, the inhibition of S6 phosphorylation by MRTX1133 was enhanced, suggesting that KRAS inhibition inactivates MTOR signaling, which contributes to delayed spheroid growth (Fig. 4E).
Figure 4.
Differential response to MRTX1133 in PDAC cell lines derived from syngeneic mice models. A, Live cell imaging to examine the effect of MRTX1133 in AKB6 and 4662 mouse cell lines in the presence of different concentrations of MRTX1133. Error bars were determined based on mean and SD. Experiments were done in triplicates. B, Relative BrdU incorporation in three different syngeneic models, AKB6, 4662, and KC4568 following the treatment with different concentrations of MRTX1133 up to 72 hours. Mean and SD were calculated from triplicates and the experiments were done at three independent times. C, Western blotting to determine the effect of MRTX1133 on the indicated proteins following 24 hours treatment in AKB6, 4662, and KC4568 cell lines. D, Effect of MRTX1133 on the growth of spheroids derived from 4662 cell lines. Mean and SEM were used to calculate error bars. The experiment was done in triplicates at two independent times. ****, P < 0.0001 as determined by two-way ANOVA. E, Biochemical analysis to compare the effect of MRTX1133 on 4662 cells cultured in 2D monolayer and 3D conditions. F, Effect of MRTX1133 (30 mg/kg) on the normalized tumor growth rate of xenografts derived from KC4568 and AKB6 in C57BL/6 mice. ***, P < 0.0001 as determined by two-way ANOVA. G, Representative tumor images from AKB6 xenografts. Scale bar, 50 μm. H, Representative images of H&E and Masson's Trichrome staining on tumor tissues excised from KC4568 and AKB6 xenografts that were treated with vehicle and MRTX1133 (30 mg/kg) in C57BL/6 mice. Scale bar, 50 μm. The deposition of collagen fibers was quantified from the vehicle- and MRTX1133-treated tissues. Error bars, mean and SEM. **, P < 0.001 as determined by Student t test.
We further examined the in vivo antitumor effects of MRTX1133 in C57BL/6J mice, bearing tumors from the KC4568 and AKB6 cell lines. KRAS inhibition resulted in durable disease control in both xenograft models. However, unlike the PDX models, MRTX1133 elicited an enhanced antitumor effect, resulting in significant tumor regression (Fig. 4F and G). Histologic analysis on the tumor tissues based on H&E staining indicated a reduced tumor cell population in MRTX1133-treated tissues as compared with vehicle-treated tissues (Fig. 4H). Evaluation of the tissue by Masson's trichrome staining revealed a significant increase in collagen deposition in response to MRTX1133 treatment, shifting from a grade 1 wispy collagen deposition to a grade 3 diffuse connecting meshwork of collagen in both AKB6 and KC4568 models (Fig. 4H; ref. 28). Consistent with this in vivo effect in murine models, MRTX1133 treatment also increased collagen deposition in PDX tissues derived from 1222 and 3226 models (Supplementary Fig. S10A). However, the efficacy of disease control was more profound in the immune-competent setting, suggesting that the host immune constituents contribute to the antitumor efficacy of MRTX1133.
Mechanistic impact of MRTX1133 in the TME
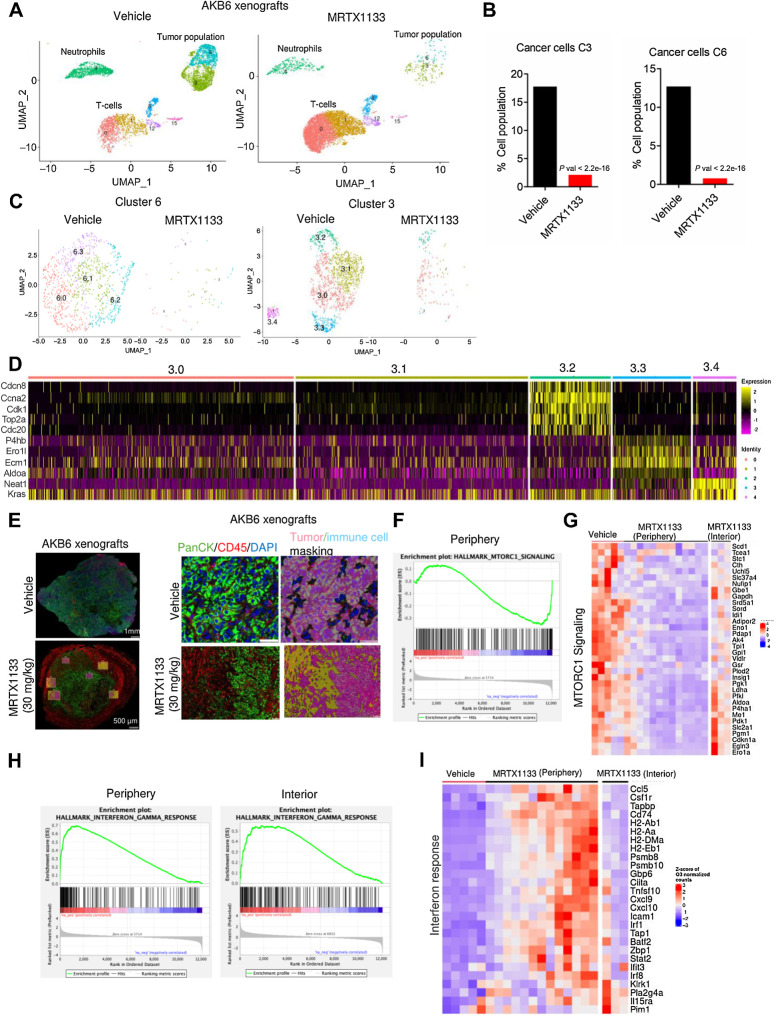
To examine the molecular pathways that are perturbed by MRTX1133 specifically in the tumor component, which could further impact the TME, we performed single-cell RNA sequencing on pooled AKB6 tumor tissues that were treated with vehicle and MRTX1133. Single-cell sequencing resolved multiple cell types including cancer cells, T cells, neutrophils, macrophages, dendritic cells, and cancer-associated fibroblasts (CAF) within the tumors (Supplementary Fig. S11A). Among the different cell clusters, a particularly striking difference was selectively observed in tumor cells, T cells, and neutrophils following MRTX1133 treatment (Fig. 5A; Supplementary Fig. S11B). The tumor cells exhibited distinct clustering into two populations, denoted as cluster 3 and cluster 6. Notably, cluster 6 was predominantly enriched with cells exhibiting an epithelial phenotype, as evident by increased expression of Cdh1, Lamc2, Plec, and Ecm1, which are indicative of epithelial markers (Supplementary Figs. S12A and S13A). However, both tumor populations dramatically decreased following MRTX1333 treatment (Fig. 5B). Because, the tumor population is highly heterogenous, we interrogated whether the efficacy of MRTX1133 varies within the tumor population by further dissecting the clusters 3 and 6 into discrete subclusters (3.0, 3.1, 3.2, 3.3, 3.4, 6.0, 6.1, 6.2, and 6.3; Fig. 5C). Subsequent subclustering revealed a uniform response to MRTX1133 within cluster 6, whereas cluster 3 displayed a degree of heterogeneity (Fig. 5C). Specifically, subcluster 3.4 was completely abrogated following treatment with MRTX1133, whereas a moderate response was observed in subclusters 3.0 and 3.2, as evident by residual tumor cell populations (Fig. 5C). Seurat processing analysis indicated that subcluster 3.2 was enriched in genes regulating cell cycle and mitosis such as Ccna2, Cdk1, and Top2a, which could contribute to the observed attenuated effect of MRTX1133 (Fig. 5D; Supplementary Fig. S13B). Conversely, the subcluster 3.4, which was prominently impacted by MRTX1133 exhibited high expression of Kras and genes associated with MTOR signaling such as Aldoa, P4hb, and Ero1L, indicating that this subpopulation was highly vulnerable to KRAS inhibition (Fig. 5D).
Figure 5.
Mechanistic investigation of in vivo effect of MRTX1133. A, Single-cell clustering of vehicle- and MRTX1133-treated AKB6 tumors to selectively indicate tumor cells, T cells, and neutrophils. B, Bar graph indicating the tumor cell population from the vehicle and MRTX1133 samples. P value was determined by Fisher exact test. C, Subclustering of the tumor population 6 and 3 from the vehicle- and MRTX1133-treated samples. D, Seurat heat map depicting the relative expression of the indicated genes across the subclusters from tumor cluster 3. E, Representative tumor images from AKB6 xenografts that were treated with vehicle (n = 2) and MRTX1133 (n = 4) and stained with PanCK, CD45, and DAPI. Different ROIs that include the periphery and exterior of the tumor tissues were selected for DSP analysis. Representative image of an ROI, demarcating the tumor and stromal population based on PANCK and CD45 staining, respectively, and the masking was performed appropriately. F, GSEA analysis identified a significant enrichment of MTOR signaling, which is downregulated in the MRTX1133-treated tumor in the peripheral region. G, Heat map depicting the expression of the indicated genes from the vehicle-treated tumors across different ROIs and MRTX1133-treated tumors from different ROIs that include both the periphery and the interior regions. H, GSEA analysis comparing the enrichment of IFNγ signaling pathway, which is differentially upregulated between the periphery and interior regions of the tumor following MRTX1133 treatment. I, Heatmap depicting the upregulation of indicated genes from the vehicle- and MRTX1133-treated tumor tissues. The impact of MRTX1133 on those genes were compared between the periphery and interior regions of the tumor.
To further investigate the impact of MRTX1133 in vivo, we performed DSP whole transcriptome analysis on paraffin-embedded tumor tissues. The tissues were stained with fluorescent tagged anti-PanCK and anti-CD45 antibodies that were conjugated with DNA indexing oligonucleotides to selectively recognize the tumor and the immune cell populations, respectively. Based on the fluorescent signal, digital masking was performed that allowed demarcation of the tumor and immune cells (Fig. 5E). Multiple regions of interest (ROI), including the periphery and interior of the tumor, were selected, and illuminated to release the oligos that were digitally counted to determine the differentially expressed genes following MRTX1133 treatment (Fig. 5E; Supplementary Fig. S14A). GSEA uncovered a significant enrichment in MTOR signaling pathway, which was downregulated in the peripheral regions of the tumors treated with MRTX1133 (Fig. 5F). Differential gene expression analysis revealed that MRTX1133 treatment suppressed genes associated with MTOR signaling to a greater extent in the periphery as compared with the interior of the tumor, indicating a heterogenous drug effect depending on the tumor region (Fig. 5G). Among the upregulated pathways, a significant enrichment in the IFNγ signaling was observed and the MRTX1133-mediated upregulation of genes associated with this pathway and antigen presentation was more pronounced in the peripheral region of the tumor as compared with the interior region (Fig. 5H and I). Overall, it is evident that the intratumoral inhibition of KRAS by MRTX1133 results in the inactivation of MTOR pathway and activation of interferon γ signaling. Consistent with the in vivo effect of MRTX1133, transcriptome analysis from in vitro cell culture data indicated that the genes associated with IFNγ signaling and antigen presentation were upregulated in ASPC1 and HPAF-II cells following treatment with MRTX1133 (Supplementary Fig. S14B). These observations demonstrate that KRAS inhibition impacts the immune pathway in tumor cells in both in vitro and in vivo settings.
MRTX1133 broadly modulates the tumor immune component
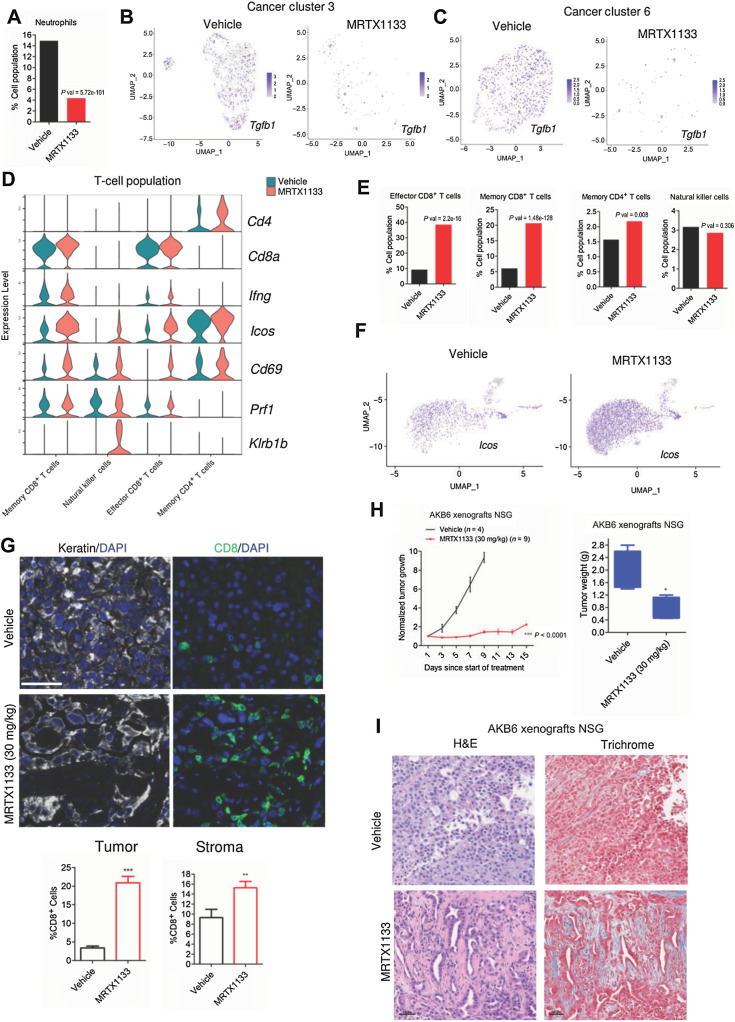
The upregulation of intratumoral antigen presentation and IFN-like signaling pathways are known to enhance immune infiltration to the TME (49–51). Hence, we investigated the effect of MRTX1133 on other immune components in the TME based on single-cell RNA sequencing data. An important immune component that was modulated in the MRTX1133 treated tumor was the neutrophil population, which was prominently decreased in the drug-treated group (Fig. 6A). Previous studies have shown the pivotal role of TGFβ, a cytokine secreted into the TME by cancer cells, which results in the recruitment of neutrophils (52, 53). The immune suppressive function of neutrophils is mediated via the inhibition of CD8+ T cells thereby promoting tumor growth (54, 55). Consistent with these studies, our single cell sequencing data revealed a profound impact on Tgfb1 expression within the tumor population (3 and 6) following the treatment with MRTX1133 (Fig. 6B and C; Supplementary Fig. S15A). The suppression of neutrophil population was associated with the downregulation of metabolic genes such as Hilpda, Ldha, and Eno1, which are required for their survival (Supplementary Fig. S15B). The increase in interferon γ signaling and antigen presentation in tumor cells could also recruit T cells. Consistent with this notion, the T-cell populations that comprise CD4+/ CD8+ T cells were increased in the presence of MRTX1133 (Fig. 6D). Among the different T-cell populations, a prominent increase was observed in CD8+ T cells (Fig. 6E). Increased IFN gamma (Ifng) expression was selectively observed in the CD8+ T-cells following MRTX1133 treatment and the upregulation of Icos and CD69 was observed in CD4+ /CD8+ T cells and NKT cells (Fig. 6D and F; Supplementary Fig. S15C; ref. 17). Moreover, it was evident that Prf1 and Klrb1b were upregulated in the CD8+ T cells and NKT cells (Fig. 6D; Supplementary Fig. S15C). Collectively our observation based on single-cell data underscores that the increased T-cell population following MRTX1133 treatment could facilitate the antitumor response. (Fig. 6D; Supplementary Fig. S15C). To validate this observation, tumor tissues from the vehicle and MRTX1133-treated groups were subjected to multispectral staining that comprised Pan-CK, DAPI, and the T-cell marker, CD8. Consistent with the single-cell sequencing data, MRTX1133-treated tissues exhibited a significantly higher CD8 expression in both the tumor and stroma, confirming that MRTX1133 induces T-cell activation (Fig. 6G).
Figure 6.
Impact of MRTX1133 on the TME and host immune system. A, Column graph indicating the neutrophils population in the absence and presence of MRTX1133. P value was determined by Fisher exact test. B, Seurat feature plot to illustrate the differential expression of Tgfb1 between the vehicle- and MRTX1133-treated groups from the tumor cluster 3. C, Seurat feature plot to illustrate the differential expression of Tgfb1 between the vehicle- and MRTX1133-treated groups from the tumor cluster 6. D, Violin plots indicating the expression of indicated genes within the neutrophil population in the presence and absence of MRTX1133. E, Column graphs indicating the different T-cell populations that comprise memory CD8+ T cells, Effector CD8+ T cells, NKT cells, and memory CD4+ T cells following MRTX1133 treatment. P values were determined by Fisher exact test. F, Seurat feature plots to illustrate the differential expression of Icos, which is a T-cell marker from the vehicle- and MRTX1133-treated groups. G, Representative images from the multispectral staining on the AKB6 tumors (PanKeratin, CD8, and DAPI). Scale bar, 30 μm. The column graph represents the fraction of CD8-positive cells from the stroma and tumor following the treatment with MRTX1133. Mean and SEM were determined from six ROIs from vehicle (n = 3)- and MRTX1133 (n = 3)-treated tumor tissues. **, P < 0.01; ***, P < 0.0001 as determine by Student t test. H,In vivo effect of MRTX1133 on the tumor growth of AKB6 xenografts in NSG mice. Column graph representing the tumor weights from the AKB6 xenografts derived from NSG strain treated with vehicle and MRTX1133. Mean and SEM are shown. *, P < 0.05 as determined by Student t test. I, Representative images of H&E and Masson's Trichrome staining from AKB6 xenografts derived from NSG strain treated with vehicle and MRTX1133 (30 mg/kg). Scale bar, 50 μm.
The impact of MRTX1133 on the cancer-associated fibroblasts (CAF), which are also the important immune components in the TME, were examined based on our single cell sequencing data. Following MRTX1133 treatment a 10-fold increase in the proportion of CAFs relative to the tumor population was observed (Supplementary Fig. S15D). The expression of CAFs markers, Col1a1 and Col1a2, were prominently increased within the tumor population following MRTX1133 treatment, which is consistent with the collagen deposition (Supplementary Fig. S15E). We also evaluated the effect of MRTX1133 on AKB6 xenografts developed in immune-deficient NSG mice. Similar to the PDX data, MRTX1133 treatment induced a cytostatic response against tumor growth and resulted in increased collagen fibers in the TME (Fig. 6H and I). Overall, these findings suggest that oncogenic KRAS inhibition by MRTX1133 has a major impact on the TME and enhances cytotoxic T-cell infiltration from the host immune system to induce a durable disease control.
Discussion
The mutation of KRAS is considered the initiating oncogenic driver for the pathogenesis of PDAC (6, 56). Although multiple different KRAS mutant isoforms occur in PDAC, the G12D mutation is the most common, occurring in ∼40% of cases (24, 57). Multiple targeted therapies have been interrogated that inhibit KRAS-mediated downstream effector pathways in clinical trials, although the therapeutic benefits are limited (58, 59). Although KRAS was considered undruggable for many years, structure-based drug design approaches have led to the development of multiple selective pharmacological inhibitors (23, 60).
This work demonstrates the cellular response of a potent KRASG12D inhibitor, MRTX1133, in different patient-derived PDAC cell lines and syngeneic models. The efficacy of MRTX1133 in in vitro cell culture assays is heterogeneous among the PDAC models assessed and is characterized as extremely sensitive or mainly refractory. On the basis of transcriptome analysis, the top pathways that were significantly inhibited in the sensitive models include the cell cycle machinery and MTOR signaling, which remains largely unperturbed in the resistant models, suggesting the involvement of parallel pathways that allow the resistant models to bypass the impact of KRAS inhibition. Consistent with this concept, key mediators of KRAS and RTK signaling were identified as cooperating with pharmacologic mutant KRAS inhibitors using CRISPR/Cas9 screening. The genes identified included those directly associated with controlling RAS activity that could represent activity through other RAS genes, as well as downstream effectors (e.g., BRAF and MAPK3). Conversely, EGFR was identified as a key mediator of resistance that could be further targeted pharmacologically to block the ability of cells to bypass mutant KRAS inhibition. Genes whose loss associated with resistance are known to further deregulate signaling pathways (e.g., PTEN) or limit effects on cell cycle (e.g., RB). These findings advance a number of combinatorial strategies that could be deployed with MRTX1133 to enhance cell autonomous activity on tumor cell division.
CRISPR screen analysis further revealed a cadre of genes whose depletion potently cooperated with KRAS inhibition (MRTX1133 and MRTX849) that are associated with adhesion-mediated signaling. This includes integrins (ITGB1 and ITGAV) and genes involved in mechanotransduction (CAV1; refs. 41–43). Mechanistically, depletion of ITGB1 enhanced the inhibitory effect of MRTX1133 on MTOR signaling and limited the expression of YAP and TAZ, thereby enhancing the cellular response to KRAS inhibition. This observation is consistent with recent studies, which highlight a similar approach where pharmacologically interfering the transcriptional functions of YAP/TAZ using novel TEAD inhibitors could overcome intrinsic and adaptive resistance to KRAS inhibitors (61–64). Our study further illustrates that the resistance to MRTX1133 in 2D culture is mediated by anchorage dependent growth that enhances mechanotransduction signaling. This resistance could be alleviated when the cells are grown as spheroids in Matrigel. On the basis of biochemical analysis, it is evident that the phenotypic effect of ITGB1 deletion observed in 2D monolayer is intrinsically exhibited in spheroids that augments the efficacy of MRTX1133. Overall, our data illustrate that the ITGB1-mediated mechanotransduction could overcome the response to KRAS inhibition. It has been posited that cell culture yields enhanced sensitivity to therapeutic agents whereas spheroids or xenografts are considered a more stringent system to assess clinically relevant drug efficacy (65). However, in the context of KRAS inhibition, cells cultured as spheroids are considerably more sensitive to therapeutic targeting (9, 46). Overall, the data here highlights the importance of the 3D cell culture as an in vitro model system to predict the efficacy of KRAS inhibitors more accurately against tumor growth.
Although MRTX1133 inhibited tumor growth in all the G12D mutant models tested, a significant difference in disease control was observed between an immune-deficient NSG strain and an immune-competent C57BL/6 strain. Although loss of KRAS is known to impact the TME, this study illustrates that pharmacologic inhibition of KRAS can have a similar effect (17). Single-cell sequencing revealed highly significant changes in the TME following treatment with MRTX1133. Suppression of the tumor cell population along with increases in cytolytic and effector CD8+/CD4+ T cells and decreases in neutrophils implies that MRTX1133-mediated tumor regression is associated with TME remodeling. In this study we elucidated the mechanism through which MRTX1133 enhances the infiltration of effector T cells by utilizing a more sophisticated, unbiased approach, DSP to determine the differential gene expression selectively on the tumor population. This technique is more advanced than conventional bulk RNA sequencing since it spatially discriminates the tumor population from the stromal population (66). On the basis of our analysis from both single-cell RNA sequencing and DSP, the tumor cells displayed a prominent upregulation of genes involved in interferon γ signaling and MHC class II antigen presentation. These molecular events have been proposed to enhance T-cell recognition and increase activation and infiltration of effector CD8+ T cells to eliminate the cancer cells (49, 67–69). Neutrophils, one of the major constituents of the TME, are known to promote tumorigenesis by suppressing the function of effector T cells (55). The survival of neutrophils is mediated by oxidative mitochondrial metabolism that results in a hypoxic condition and enhances reactive oxygen species production to inhibit effector T-cell activation and suppress the antitumor immunity (54). The effect of MRTX1133 on tumor-related surface molecules (e.g., HLA) and cytokines (e.g., Tgfb1) could impact the TME that lead to a reduced immunosuppressive neutrophil population and an enhanced influx of CD8+ T cells, which results in tumor regression (70).
The in vivo efficacy of MRTX1133 in eliciting disease control is reversible because cessation of treatment leads to tumor recurrence (57). In accordance with this notion, our single-cell sequencing data revealed that the exhaustion markers such as Ctla4 and Lag3 were upregulated that could possibly undermine the antitumor immunity. This underscores the need for exploring new combinatorial treatment modalities involving concurrent targeting of KRAS and immunotherapy to enhance the efficacy of KRAS inhibition (Supplementary Fig. S15F).
In summary, this study highlights the efficacy of a KRASG12D inhibitor, MRTX1133, in PDAC models and demonstrates a differential response in 2D cell culture, spheroids, and the in vivo setting. Although we demonstrate two distinct molecular components, focal adhesion and TME remodeling, that modulate the response to MRTX1133, it is limited to the preclinical setting. Whether the genes involved in those pathways could serve as biomarkers of response or resistance to KRAS inhibitors in patient models needs further investigation.
Supplementary Material
Supplementary information
Transcriptome data
Acknowledgments
The authors thank all members of the laboratory group and colleagues in the discussion and preparation of the manuscript. This work was supported by grants from the NIH, NCI (CA267467, CA211878, and R37CA275961) and S10 Instrumentation Award (S10OD030410). Additionally, this work was supported by National Cancer Institute (NCI) grant P30CA016056 involving the use of Roswell Park Comprehensive Cancer Center's Advanced Tissue imaging (ATISR), Drug Discovery Core (DDCSR), Experimental tumor models (ETM), and Genomics shared resources. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
C. Frangou reports personal fees and nonfinancial support from Cellecta, Inc. outside the submitted work. E.V. Abel reports grants from NCI during the conduct of the study. E.S. Knudsen reports grants from NIH during the conduct of the study; grants from NIH outside the submitted work; Blueprint Medicines sponsored research, Bristol Meyer Squibb sponsored research; and is consultant for Cancer Cell Cycles-LLC. No disclosures were reported by the other authors.
Authors' Contributions
V. Kumarasamy: Conceptualization, data curation, formal analysis, validation, investigation, writing–original draft, writing–review and editing. J. Wang: Data curation, software, formal analysis, investigation. C. Frangou: Data curation, formal analysis, investigation, methodology. Y. Wan: Data curation. A. Dynka: Data curation. H. Rosenheck: Data curation. P. Dey: Conceptualization, resources. E.V. Abel: Conceptualization, resources, funding acquisition. E.S. Knudsen: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, project administration, writing–review and editing. A.K. Witkiewicz: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, methodology, project administration, writing–review and editing.
References
- 1. Yao W, Maitra A, Ying H. Recent insights into the biology of pancreatic cancer. EBioMedicine 2020;53:102655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, et al. Pancreatic cancer. Nat Rev Dis Primers 2016;2:16022. [DOI] [PubMed] [Google Scholar]
- 3. Witkiewicz AK, McMillan E, Balaji U, Baek G, Cox D, McCue P, et al. Whole exome sequencing of pancreatic cancer:genetic diversity, prognostic features, and potential therapeutic targets. Nat Communication 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. [DOI] [PubMed] [Google Scholar]
- 5. Infante JR, Somer BG, Park JO, Li CP, Scheulen ME, Kasubhai SM, et al. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer 2014;50:2072–81. [DOI] [PubMed] [Google Scholar]
- 6. Huang L, Guo Z, Wang F, Fu L. KRAS mutation: from undruggable to druggable in cancer. Signal Transduct Target Ther 2021;6:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Waters AM, Der CJ. KRAS: The critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med 2018;8:a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nollmann FI, Ruess DA. Targeting mutant KRAS in pancreatic cancer: futile or promising? Biomedicines 2020;8:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Muzumdar MD, Chen PY, Dorans KJ, Chung KM, Bhutkar A, Hong E, et al. Survival of pancreatic cancer cells lacking KRAS function. Nat Commun 2017;8:1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest 2012;122:639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shinkawa T, Ohuchida K, Nakamura M. Heterogeneity of cancer-associated fibroblasts and the tumor immune microenvironment in pancreatic cancer. Cancers (Basel) 2022;14:3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hosein AN, Brekken RA, Maitra A. Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nat Rev Gastroenterol Hepatol 2020;17:487–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell 2014;25:735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 2009;324:1457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li X, Gulati M, Larson AC, Solheim JC, Jain M, Kumar S, et al. Immune checkpoint blockade in pancreatic cancer: Trudging through the immune desert. Semin Cancer Biol 2022;86:14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stott MC, Oldfield L, Hale J, Costello E, Halloran CM. Recent advances in understanding pancreatic cancer. Fac Rev 2022;11:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ischenko I, D'Amico S, Rao M, Li J, Hayman MJ, Powers S, et al. KRAS drives immune evasion in a genetic model of pancreatic cancer. Nat Commun 2021;12:1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakajima EC, Drezner N, Li X, Mishra-Kalyani PS, Liu Y, Zhao H, et al. FDA Approval Summary: Sotorasib for KRAS G12C-mutated metastatic NSCLC. Clin Cancer Res 2022;28:1482–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Strickler JH, Satake H, George TJ, Yaeger R, Hollebecque A, Garrido-Laguna I, et al. Sotorasib in KRAS p.G12C-mutated advanced pancreatic cancer. N Engl J Med 2023;388:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ning W, Marti TM, Dorn P, Peng RW. Non-genetic adaptive resistance to KRAS(G12C) inhibition: EMT is not the only culprit. Front Oncol 2022;12:1004669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xue JY, Zhao Y, Aronowitz J, Mai TT, Vides A, Qeriqi B, et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 2020;577:421–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X, Allen S, Blake JF, Bowcut V, Briere DM, Calinisan A, et al. Identification of MRTX1133, a noncovalent, potent, and selective KRAS(G12D) inhibitor. J Med Chem 2022;65:3123–33. [DOI] [PubMed] [Google Scholar]
- 24. Hallin J, Bowcut V, Calinisan A, Briere DM, Hargis L, Engstrom LD, et al. Anti-tumor efficacy of a potent and selective non-covalent KRAS(G12D) inhibitor. Nat Med 2022;28:2171–82. [DOI] [PubMed] [Google Scholar]
- 25. Knudsen ES, Kumarasamy V, Ruiz A, Sivinski J, Chung S, Grant A, et al. Cell cycle plasticity driven by MTOR signaling: integral resistance to CDK4/6 inhibition in patient-derived models of pancreatic cancer. Oncogene 2019;38:3355–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Knudsen ES, Kumarasamy V, Nambiar R, Pearson JD, Vail P, Rosenheck H, et al. CDK/cyclin dependencies define extreme cancer cell-cycle heterogeneity and collateral vulnerabilities. Cell Rep 2022;38:110448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumarasamy V, Ruiz A, Nambiar R, Witkiewicz AK, Knudsen ES. Chemotherapy impacts on the cellular response to CDK4/6 inhibition: distinct mechanisms of interaction and efficacy in models of pancreatic cancer. Oncogene 2020;39:1831–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kvasnicka HM, Beham-Schmid C, Bob R, Dirnhofer S, Hussein K, Kreipe H, et al. Problems and pitfalls in grading of bone marrow fibrosis, collagen deposition and osteosclerosis - a consensus-based study. Histopathology 2016;68:905–15. [DOI] [PubMed] [Google Scholar]
- 29. Witkiewicz AK, Schultz E, Wang J, Hamilton D, Levine E, O'Connor T, et al. Determinants of response to CDK4/6 inhibitors in the real-world setting. NPJ Precis Oncol 2023;7:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y, Parmigiani G, Johnson WE. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom Bioinform 2020;2:lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hart T, Tong AHY, Chan K, Van Leeuwen J, Seetharaman A, Aregger M, et al. Evaluation and design of genome-wide CRISPR/SpCas9 knockout screens. G3 2017;7:2719–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 2014;15:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Colic M, Wang G, Zimmermann M, Mascall K, McLaughlin M, Bertolet L, et al. Identifying chemogenetic interactions from CRISPR screens with drugZ. Genome Med 2019;11:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alistar A, Morris BB, Desnoyer R, Klepin HD, Hosseinzadeh K, Clark C, et al. Safety and tolerability of the first-in-class agent CPI-613 in combination with modified FOLFIRINOX in patients with metastatic pancreatic cancer: a single-centre, open-label, dose-escalation, phase 1 trial. Lancet Oncol 2017;18:770–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ianevski A, Giri AK, Aittokallio T. Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data. Nat Commun 2022;13:1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Knudsen ES, Balaji U, Mannakee B, Vail P, Eslinger C, Moxom C, et al. Pancreatic cancer cell lines as patient-derived avatars: genetic characterisation and functional utility. Gut 2018;67:508–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zeng H, Castillo-Cabrera J, Manser M, Lu B, Yang Z, Strande V, et al. Genome-wide CRISPR screening reveals genetic modifiers of mutant EGFR dependence in human NSCLC. eLife 2019;8:e50223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pellinen T, Blom S, Sanchez S, Valimaki K, Mpindi JP, Azegrouz H, et al. ITGB1-dependent upregulation of Caveolin-1 switches TGFbeta signalling from tumour-suppressive to oncogenic in prostate cancer. Sci Rep 2018;8:2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cooper J, Giancotti FG. Integrin signaling in cancer: mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell 2019;35:347–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cai X, Wang KC, Meng Z. Mechanoregulation of YAP and TAZ in cellular homeostasis and disease progression. Front Cell Dev Biol 2021;9:673599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Di-Luoffo M, Ben-Meriem Z, Lefebvre P, Delarue M, Guillermet-Guibert J. PI3K functions as a hub in mechanotransduction. Trends Biochem Sci 2021;46:878–88. [DOI] [PubMed] [Google Scholar]
- 45. Rausch V, Hansen CG. The hippo pathway, YAP/TAZ, and the plasma membrane. Trends Cell Biol 2020;30:32–48. [DOI] [PubMed] [Google Scholar]
- 46. Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 2018;172:578–89. [DOI] [PubMed] [Google Scholar]
- 47. Beatty GL, Chiorean EG, Fishman MP, Saboury B, Teitelbaum UR, Sun W, et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science 2011;331:1612–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dey P, Li J, Zhang J, Chaurasiya S, Strom A, Wang H, et al. Oncogenic KRAS-driven metabolic reprogramming in pancreatic cancer cells utilizes cytokines from the tumor microenvironment. Cancer Discov 2020;10:608–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Knudsen ES, Kumarasamy V, Chung S, Ruiz A, Vail P, Tzetzo S, et al. Targeting dual signalling pathways in concert with immune checkpoints for the treatment of pancreatic cancer. Gut 2021;70:127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kalbasi A, Tariveranmoshabad M, Hakimi K, Kremer S, Campbell KM, Funes JM, et al. Uncoupling interferon signaling and antigen presentation to overcome immunotherapy resistance due to JAK1 loss in melanoma. Sci Transl Med 2020;12:eabb0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paschen A, Melero I, Ribas A. Central role of the antigen-presentation and Interferon-γ pathways in resistance to immune checkpoint blockade. Annual Review of Cancer Biology 2022;6:85–102. [Google Scholar]
- 52. Batlle E, Massague J. Transforming growth factor-beta signaling in immunity and cancer. Immunity 2019;50:924–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sanjabi S, Oh SA, MO L. Regulation of the immune response by TGF-beta: from conception to autoimmunity and infection. Cold Spring Harb Perspect Biol 2017;9:a022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rice CM, Davies LC, Subleski JJ, Maio N, Gonzalez-Cotto M, Andrews C, et al. Tumour-elicited neutrophils engage mitochondrial metabolism to circumvent nutrient limitations and maintain immune suppression. Nat Commun 2018;9:5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rogers T, DeBerardinis RJ. Metabolic plasticity of neutrophils: relevance to pathogen responses and cancer. Trends Cancer 2021;7:700–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Uprety D, Adjei AA. KRAS: from undruggable to a druggable cancer target. Cancer Treat Rev 2020;89:102070. [DOI] [PubMed] [Google Scholar]
- 57. Kemp SB, Cheng N, Markosyan N, Sor R, Kim IK, Hallin J, et al. Efficacy of a small-molecule inhibitor of KrasG12D in immunocompetent models of pancreatic cancer. Cancer Discov 2023;13:298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schram AM, Gandhi L, Mita MM, Damstrup L, Campana F, Hidalgo M, et al. A phase Ib dose-escalation and expansion study of the oral MEK inhibitor pimasertib and PI3K/MTOR inhibitor voxtalisib in patients with advanced solid tumours. Br J Cancer 2018;119:1471–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ko AH, Bekaii-Saab T, Van Ziffle J, Mirzoeva OM, Joseph NM, Talasaz A, et al. A multicenter, open-label phase II clinical trial of combined MEK plus EGFR inhibition for chemotherapy-refractory advanced pancreatic adenocarcinoma. Clin Cancer Res 2016;22:61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRAS(G12C) inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov 2020;10:54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hagenbeek TJ, Zbieg JR, Hafner M, Mroue R, Lacap JA, Sodir NM, et al. An allosteric pan-TEAD inhibitor blocks oncogenic YAP/TAZ signaling and overcomes KRAS G12C inhibitor resistance. Nat Cancer 2023;4:812–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mukhopadhyay S, Huang HY, Lin Z, Ranieri M, Li S, Sahu S, et al. Genome-wide CRISPR screens identify multiple synthetic lethal targets that enhance KRASG12C inhibitor efficacy. Cancer Res 2023;83:4095–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Edwards AC, Stalnecker CA, Jean Morales A, Taylor KE, Klomp JE, Klomp JA, et al. TEAD inhibition overcomes YAP1/TAZ-driven primary and acquired resistance to KRASG12C inhibitors. Cancer Res 2023;83:4112–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Johnson CW, Haigis KM. All roads lead to rome: YAP/TAZ activity influences efficacy of KRASG12C inhibitors. Cancer Res 2023;83:4005–7. [DOI] [PubMed] [Google Scholar]
- 65. Tiriac H, Plenker D, Baker LA, Tuveson DA. Organoid models for translational pancreatic cancer research. Curr Opin Genet Dev 2019;54:7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li X, Wang CY. From bulk, single-cell to spatial RNA sequencing. Int J Oral Sci 2021;13:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hu-Lieskovan S, Mok S, Homet Moreno B, Tsoi J, Robert L, Goedert L, et al. Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAF(V600E) melanoma. Sci Transl Med 2015;7:279ra241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jorgovanovic D, Song M, Wang L, Zhang Y. Roles of IFN-gamma in tumor progression and regression: a review. Biomark Res 2020;8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Axelrod ML, Cook RS, Johnson DB, Balko JM. Biological consequences of MHC-II xpression by tumor cells in cancer. Clin Cancer Res 2019;25:2392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yeh C, Park W, Yaeger R. KRAS(G12D) inhibition in pancreatic cancer: Fas expression facilitates immune clearance. Dev Cell 2023;58:1515–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Transcriptome data
Data Availability Statement
The datasets for the transcriptome analysis were uploaded in the supplementary files. The raw fastq files for RNA sequencing, CRISPR screening, 10× single-cell sequencing, and DSP analysis are publicly available in the GEO (GSE249541). All additional data generated in this study are available from the corresponding author upon request.