In this study, Fang et al. report that the helicase subunit MCM2, DNA polymerase ε subunits DPB3/4, and histone chaperone FACT complex function collectively to maintain symmetrical parental histone segregation during heterochromatin replication in yeast. They further reveal a basis for leading/lagging strand bias of heterochromatin inheritance and describe the distinct dependence of inheritance efficiency on histone density, providing new insight into the mechanisms of inheritable epigenetic memory.
Keywords: heterochromatin, H3K9 methylation, epigenetic inheritance, histone chaperone, Mcm2, Dpb3, Dpb4, eSPAN, fission yeast, parental histone density
Abstract
Chromatin-based epigenetic memory relies on the accurate distribution of parental histone H3–H4 tetramers to newly replicated DNA strands. Mcm2, a subunit of the replicative helicase, and Dpb3/4, subunits of DNA polymerase ε, govern parental histone H3–H4 deposition to the lagging and leading strands, respectively. However, their contribution to epigenetic inheritance remains controversial. Here, using fission yeast heterochromatin inheritance systems that eliminate interference from initiation pathways, we show that a Mcm2 histone binding mutation severely disrupts heterochromatin inheritance, while mutations in Dpb3/4 cause only moderate defects. Surprisingly, simultaneous mutations of Mcm2 and Dpb3/4 stabilize heterochromatin inheritance. eSPAN (enrichment and sequencing of protein-associated nascent DNA) analyses confirmed the conservation of Mcm2 and Dpb3/4 functions in parental histone H3–H4 segregation, with their combined absence showing a more symmetric distribution of parental histone H3–H4 than either single mutation alone. Furthermore, the FACT histone chaperone regulates parental histone transfer to both strands and collaborates with Mcm2 and Dpb3/4 to maintain parental histone H3–H4 density and faithful heterochromatin inheritance. These results underscore the importance of both symmetric distribution of parental histones and their density at daughter strands for epigenetic inheritance and unveil distinctive properties of parental histone chaperones during DNA replication.
Eukaryotic genomic DNA is folded with histones to form nucleosomes, which serve as the fundamental units of chromatin. Covalent modifications of histones play essential roles in establishing gene expression programs that determine cell identity (Escobar et al. 2021; Du et al. 2022). Histones with certain modifications, especially those associated with repressed states, can be passed down to subsequent generations even after the initial signals for the recruitment of histone-modifying enzymes have disappeared, thereby creating an epigenetic memory (Escobar et al. 2021; Du et al. 2022). The prevailing view is that during DNA replication, the passage of the replication fork disrupts parental nucleosomes, and parental H3–H4 tetramers with pre-existing modifications are equally deposited on both daughter strands at their original locations to guide nucleosome formation (Xu et al. 2010; Gan et al. 2018; Petryk et al. 2018; Yu et al. 2018; Escobar et al. 2019; Schlissel and Rine 2019). In addition, newly synthesized H3–H4, which lack parental histone modifications, also direct nucleosome formation to fill in the gaps created by DNA duplication. The existing modifications on parental histone H3–H4 recruit corresponding modifying enzymes, leading to modifications of nearby newly synthesized histone H3–H4, thereby restoring the original histone modification patterns on both replicated chromatids (Fig. 1A; Escobar et al. 2021; Du et al. 2022).
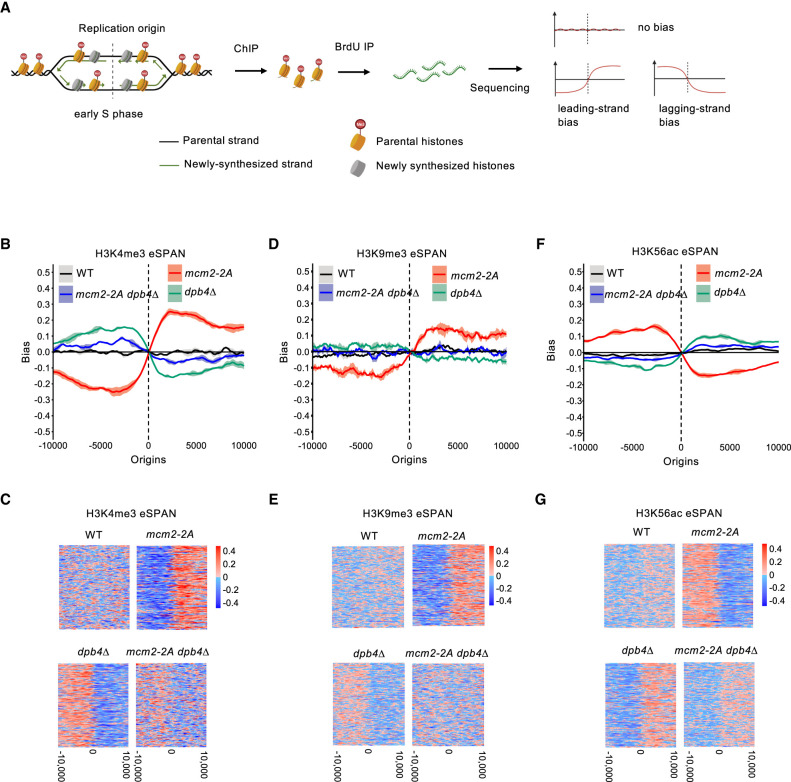
Figure 1.
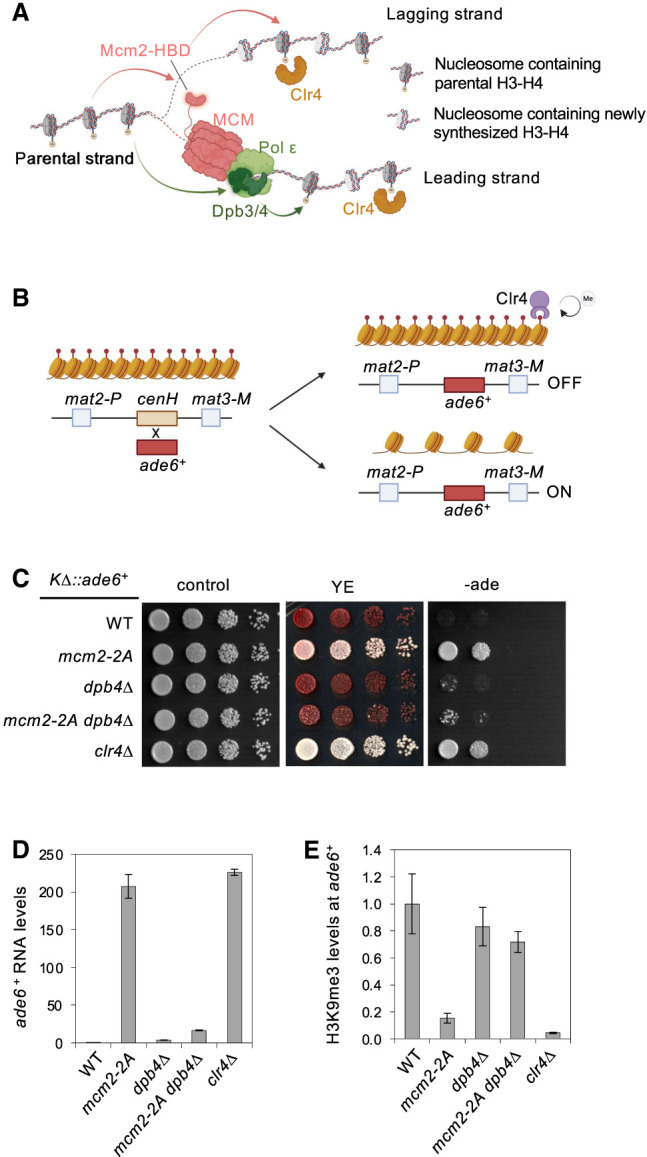
Mcm2 and Dpb3/4 regulate the inheritance of mating-type region heterochromatin. (A) Schematic diagram of parental histone H3–H4 segregation pathways during DNA replication. (B) Schematic diagram of the KΔ::ade6+ reporter. (C) Serial dilution analysis of the indicated strains to measure the expression of KΔ::ade6+. (D) qRT-PCR analyses of ade6+ transcript levels, normalized to act1+. Data are presented as mean ± SD of three technical replicates. (E) ChIP-qPCR analysis of H3K9me3 level at KΔ::ade6+, normalized to act1. Data are presented as mean ± SD of three technical replicates.
Recent advancements in eSPAN (enrichment and sequencing of protein-associated nascent DNA) have provided important insights into the mechanisms controlling parental histone distribution during DNA replication (Yu et al. 2014, 2018; Gan et al. 2018; Petryk et al. 2018; Li et al. 2020). The findings collectively reveal that Mcm2, a component of the replicative helicase, is responsible for transferring parental histone H3–H4 to the lagging strand. Conversely, Dpb3/Dpb4 (POLE4/POLE3 in mammals), part of the leading strand polymerase ε, are responsible for transferring parental histone H3–H4 to the leading strand (Fig 1A).
One critical remaining question is how the misregulation of parental histone segregation affects epigenetic inheritance. According to the aforementioned model, mutations of either Mcm2 or Dpb3/4 histone binding activities are expected to severely impact epigenetic inheritance. However, these mutations have surprisingly mild effects on budding yeast and fission yeast heterochromatin, which has been studied extensively as a heritable chromatin state (He et al. 2017; Yu et al. 2018; Saxton and Rine 2019). In mammalian systems, although defective parental histone segregation affects stem cell maintenance and differentiation, only subsets of the epigenome are affected (Li et al. 2020; Xu et al. 2022; Tian et al. 2023; Wen et al. 2023; Wenger et al. 2023).
One possible reason for the subtle effects of Mcm2 or Dpb3/4 mutations on the epigenome is the existence of additional parental histone segregation pathways. The histone chaperone FACT (facilitates chromatin transcription) is involved in nucleosome disassembly and reassembly during gene transcription, DNA replication, and DNA repair (Formosa and Winston 2020). FACT is required for heterochromatin integrity, which has been attributed to its role in regulating histone turnover and interacting with the heterochromatin machinery (Lejeune et al. 2007; Holla et al. 2020; Murawska et al. 2021; Takahata et al. 2021). Recently, it has been shown that budding yeast FACT has a role in parental histone transfer (Wang et al. 2023). However, how FACT is engaged in parental histone segregation during DNA replication remains largely unknown.
Another possible reason is the existence of sequence-dependent pathways that initiate chromatin states regardless of epigenetic inheritance. Therefore, it is critical to examine epigenetic inheritance without the influence of initiation pathways. In the fission yeast Schizosaccharomyces pombe, heterochromatin primarily forms at repetitive DNA elements in the pericentric region, subtelomeres, and silent mating-type region (Grewal and Jia 2007). Nucleosomes within these regions are dimethylated and trimethylated at histone H3 lysine 9 (H3K9me2 and H3K9me3, respectively), which recruits HP1 family proteins to repress transcription (Thon and Verhein-Hansen 2000; Bannister et al. 2001; Nakayama et al. 2001b; Hall et al. 2002; Sadaie et al. 2004). Importantly, initiation and inheritance pathways can be effectively separated (Allshire and Madhani 2018; Grewal 2023; Shan et al. 2023). During the initiation step, the histone H3K9 methyltransferase Clr4 is recruited to specific genomic loci through the RNA interference (RNAi) machinery or DNA binding proteins to initiate H3K9me3. During the subsequent inheritance step, Clr4 restores heterochromatin structure after DNA replication by recognizing pre-existing histone modifications independent of the recruitment signal. The chromodomain of Clr4 plays a crucial role in this process by recognizing H3K9me3, facilitating Clr4 recruitment to parental histones (Zhang et al. 2008). Clr4 then modifies nucleosomes formed by newly synthesized histones during and after DNA replication to restore H3K9me3 levels, known as the “read–write” coupling.
The ability to genetically separate the two steps makes fission yeast heterochromatin assembly a superb model for studying the mechanism of epigenetic inheritance (Shan et al. 2023). For example, at the silent mating-type region (MTR), cenH recruits Clr4 through the RNAi pathway (Hall et al. 2002; Jia et al. 2004). When cenH is replaced by a reporter gene, de novo initiation is mostly disabled, resulting in the reporter exhibiting one of two states: expressed or silenced (Grewal and Klar 1996). The silenced state is efficiently maintained through both mitosis and meiosis, clearly demonstrating epigenetic inheritance. In addition, an ectopic heterochromatin inheritance system is developed by recruiting Clr4 to tetO binding sites through a TetR-Clr4-SET domain (TetR-Clr4-I) fusion protein (Audergon et al. 2015; Ragunathan et al. 2015). The addition of tetracycline removes TetR-Clr4-I from tetO, abolishing any additional initiation events. In the absence of histone H3K9 demethylase Epe1, this ectopic heterochromatin is inherited by subsequent generations through endogenous Clr4. These assays allow precise measurements of heterochromatin inheritance without interference from initiation.
In this study, we used these inheritance-specific assays to demonstrate the crucial role of Mcm2-mediated parental histone H3–H4 deposition to the lagging strand for epigenetic inheritance, whereas Dpb3/4-mediated deposition of parental histone H3–H4 to the leading strand makes a comparatively minor contribution. Importantly, simultaneous mutations of Mcm2 and Dpb3/4 result in stable epigenetic inheritance and reduced parental histone segregation bias, suggesting the importance of a symmetric distribution of parental histones in epigenetic inheritance as well as the involvement of additional histone chaperones in regulating this process. Specifically, we discovered that FACT regulates parental histone H3–H4 segregation independently of DNA strands and collaborates with Mcm2 and Dpb3/4 to maintain parental histone H3–H4 levels. Moreover, we demonstrated that both the symmetric distribution of parental histone H3–H4 and their density at individual daughter strands collectively determine the outcomes of heterochromatin inheritance. Our findings establish a critical link between parental histone segregation and epigenetic inheritance and highlight the cooperative roles of distinct histone chaperone activities in regulating epigenetic inheritance.
Results
Mcm2 and Dpb3/4 have distinct roles in regulating epigenetic inheritance
To examine the role of parental histone segregation in epigenetic inheritance in fission yeast, we first tested whether the functions of the histone chaperones involved in parental histone segregation in budding yeast and mammals—Mcm2 and Dpb3/4—are conserved in fission yeast (Fig. 1A). We generated a GST-tagged version of the histone binding domain of Mcm2 (Mcm2-HBD, amino acids 1–110) and a mutant version containing mutations of two conserved tyrosine residues to alanine (Y81A and Y89A; referred to here as Mcm2-2A) (Supplemental Fig. S1A; Foltman et al. 2013; Huang et al. 2015), which abolish binding of budding yeast and mammalian Mcm2-HBD to the H3–H4 tetramer. We also generated recombinant fission yeast H3 and H4 and assembled H3–H4 tetramers. GST pull-down analyses show that GST-Mcm2-HBD, but not GST or GST-Mcm2-HBD-2A, interacts with H3–H4 tetramers (Supplemental Fig. S1B). Although Dpb3 and Dpb4 are difficult to produce alone as recombinant proteins, we were able to generate a GST-Dpb4/6xHis-Dpb3 heterodimer through the coexpression of GST-Dpb4 and 6xHis-Dpb3, followed by two-step purifications with Talon and GST resins. The purified GST-Dpb3/Dpb4 complex also interacted with H3–H4 tetramers in a GST pull-down assay (Supplemental Fig. S1B). These results suggest that fission yeast Mcm2 and Dpb3/4 interact with H3–H4 tetramers, similar to other systems.
To examine the role of the histone binding activity of Mcm2 in vivo, we introduced the mcm2-2A mutation and a C-terminal 3xFLAG tag at the endogenous mcm2+ locus. Western blot analysis shows that Mcm2-2A is expressed at levels comparable with those of wild-type Mcm2 (Supplemental Fig. S1C). We also generated dpb3Δ and dpb4Δ, which remove the open reading frames of dpb3+ and dpb4+, respectively. We then assessed the effects of mcm2-2A and dpb4Δ on the inheritance-specific reporter KΔ::ade6+, which was created by replacing the cenH sequence at the silent mating-type region (MTR) with an ade6+ gene (Fig. 1B). Loss of cenH disrupts heterochromatin initiation at the MTR, and KΔ::ade6+ can only be maintained as one of two epigenetic states: “ade6-on” or “ade6-off.” Heterochromatin at the MTR is lost in “ade6-on” cells, resulting in white colonies on a low-adenine medium (YE) and robust growth on a medium without adenine (−ade). In contrast, heterochromatin at the MTR is properly inherited in “ade6-off” cells, leading to red colonies on YE medium and poor growth on −ade medium. We used KΔ::ade6+ cells with the “ade6-off” state as a starting point to examine the epigenetic inheritance of heterochromatin. As expected, clr4Δ cells form white colonies on YE medium and grow robustly on −ade medium (Fig. 1C). Strikingly, mcm2-2A and dpb4Δ show distinct phenotypes. mcm2-2A cells produce light-pink/white colonies on YE medium and show robust growth on −ade medium, indicating a severe loss of heterochromatin inheritance (Fig. 1C; Supplemental Fig. S1D). In contrast, dpb4Δ cells form red colonies on YE medium and exhibited minimal growth on −ade medium, indicating efficient heterochromatin inheritance (Fig. 1C). Notably, the absence of KΔ::ade6+ inheritance defects observed in dpb4Δ cells is not due to functional redundancy between Dpb3 and Dpb4, as both dpb3Δ and dpb3Δ dpb4Δ cells show similar phenotypes (Supplemental Fig. S2A). Interestingly, mcm2-2A dpb4Δ and mcm2-2A dpb3Δ cells form red colonies on YE medium and exhibit poor growth on −ade medium (Fig. 1C; Supplemental Fig. S2A), suggesting that dpb3Δ or dpb4Δ suppresses the effects of mcm2-2A on heterochromatin inheritance. Consistent with dilution analysis results, qRT-PCR analyses reveal much higher ade6+ transcript levels in mcm2-2A cells, similar to clr4Δ cells. In contrast, dpb4Δ and mcm2-2A dpb4Δ cells display only slightly higher ade6+ transcript levels than wild-type cells (Fig. 1D). Furthermore, chromatin immunoprecipitation (ChIP) analyses demonstrate that H3K9me3 levels at KΔ::ade6+ are nearly abolished in mcm2-2A and clr4Δ cells, are only mildly affected in dpb4Δ cells, and are at intermediate levels in mcm2-2A dpb4Δ cells (Fig. 1E). Collectively, these findings highlight the involvement of Mcm2 and Dpb3/4 in heterochromatin inheritance.
To further investigate the impact of Mcm2 and Dpb3/4 on epigenetic inheritance, we examined the effects of their mutations on the inheritance of an ectopic heterochromatin. In this system, 10 copies of the tetO binding sites along with a gfp+ reporter gene are inserted into the endogenous ura4+ locus (Fig. 2A; Ragunathan et al. 2015). A Clr4 SET domain is fused with the TetR protein to create TetR-Clr4-I, which is recruited to the tetO binding sites, leading to the formation of an H3K9me3 domain and subsequent silencing of the gfp+ reporter. TetR-Clr4-I can be quickly disassociated from the tetO binding sites by the addition of tetracycline to prevent further initiation. The existing heterochromatin is expected to self-sustain through the action of endogenous Clr4, which binds H3K9me3 through its chromodomain and propagates it through the “read and write” cycle when the H3K9 demethylase Epe1 is absent. Therefore, all strains used for this analysis are in an epe1Δ background.
Figure 2.
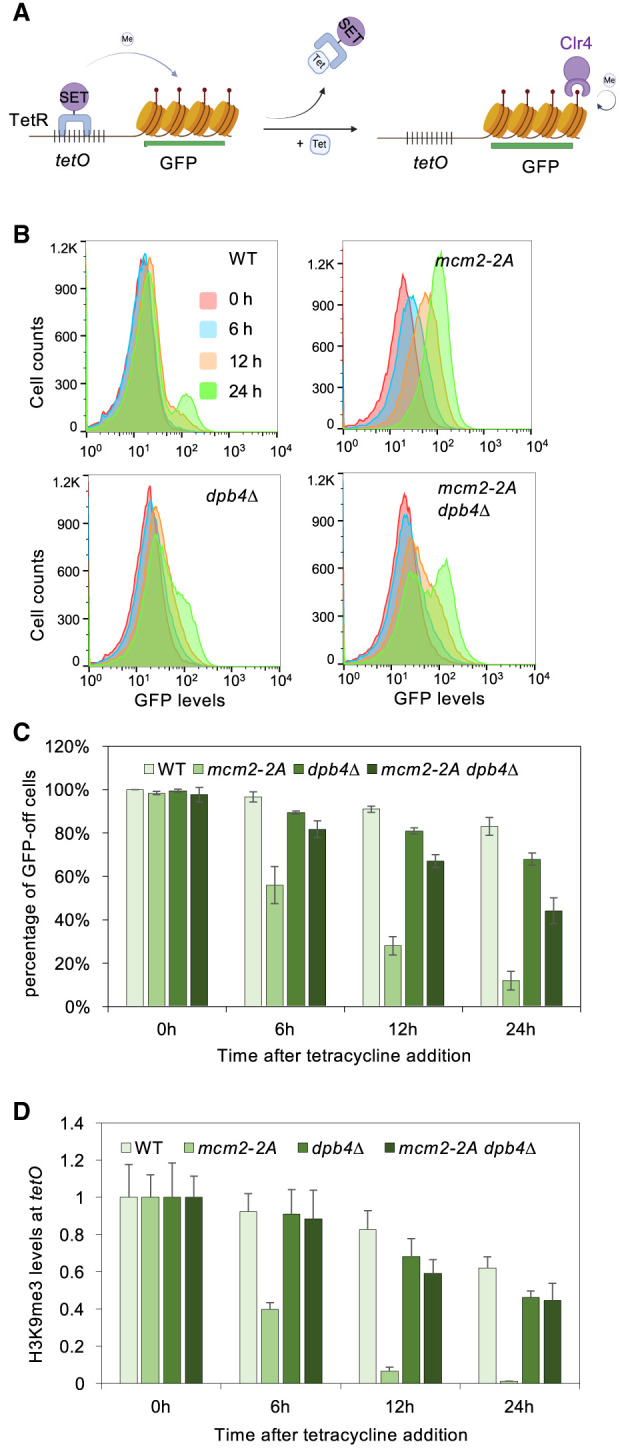
Mcm2 and Dpb4 regulate the inheritance of an ectopic heterochromatin. (A) Schematic diagram of the tetO-gfp+ reporter. (B) Flow cytometry analysis of GFP expression at different time points after tetracycline addition. All strains used are in an epe1Δ background. (C) Quantification of the percentage of cells maintaining low GFP expression at different time points after tetracycline addition. Data are presented as mean ± SD of two biological replicates. All strains are in an epe1Δ background. (D) ChIP-qPCR analysis of H3K9me3 levels at tetO binding sites at different time points after tetracycline addition, normalized to act1. Data are presented as mean ± SD of three technical replicates. All strains are in an epe1Δ background.
To examine the inheritance of this ectopic heterochromatin with high temporal resolution, we used flow cytometry to monitor GFP expression levels (Fig. 2B,C; Supplemental Fig. S2B). The majority of wild-type cells maintain a “GFP-off” state 24 h (∼10 cell generations) following tetracycline addition, with only a minor fraction of cells showing a “GFP-on” state. However, mcm2-2A cells exhibit fast activation of GFP expression compared with dpb4Δ cells, which show only very slow activation of GFP expression. Additionally, mcm2-2A dpb4Δ cells show a mixture of “GFP-off” and “GFP-on” states. Furthermore, ChIP analysis confirms that wild-type cells maintain H3K9me3 levels at the tetO binding sites 24 h after tetracycline addition. However, H3K9me3 levels decline quickly in mcm2-2A cells. Conversely, H3K9me3 exhibits slower decay in both dpb4Δ and mcm2-2A dpb4Δ cells (Fig. 2D). The similarities observed in the phenotypic patterns between the KΔ::ade6+ and tetO-gfp+ systems support the notion that Mcm2 exerts a stronger influence on epigenetic inheritance compared with Dpb3/4. Furthermore, the simultaneous disruption of both Mcm2 and Dpb3/4 leads to reasonably efficient heterochromatin inheritance, suggesting that these two histone-binding proteins function in an antagonistic manner.
Mcm2 and Dpb3/4 have minor roles in pericentric heterochromatin inheritance
We also investigated the roles of Mcm2 and Dpb3/4 in regulating pericentric heterochromatin integrity, where the dh and dg repeats initiate heterochromatin formation through the RNAi pathway (Fig. 3A; Volpe et al. 2002; Verdel et al. 2004). We used the otr::ade6+ reporter, where an ade6+ gene is inserted within the pericentric repeats. In clr4Δ cells, loss of heterochromatin leads to the formation of white colonies on YE medium and strong growth on −ade medium (Fig. 3B). In contrast, mcm2-2A cells give rise to dark-pink colonies on YE medium and show mild growth on −ade medium, while dpb4Δ, dpb3Δ, and mcm2-2A dpb4Δ cells produce red colonies on YE medium and show poor growth on −ade medium (Fig. 3B; Supplemental Fig. S2C). Consistent with the observed phenotypes, qRT-PCR analyses show a slight increase of dh transcript levels in mcm2-2A cells, while dpb4Δ and mcm2-2A dpb4Δ cells exhibit marginally higher dh transcript levels than wild-type cells (Fig. 3C). Moreover, ChIP analyses show that mcm2-2A, dpb4Δ, and mcm2-2A dpb4Δ cells exhibit very minor reduction of H3K9me3 levels compared with wild-type cells (Fig. 3D). It is important to note that their impact on dh transcript levels and H3K9me3 levels at dh are very minor compared with the drastic effects of clr4Δ.
Figure 3.
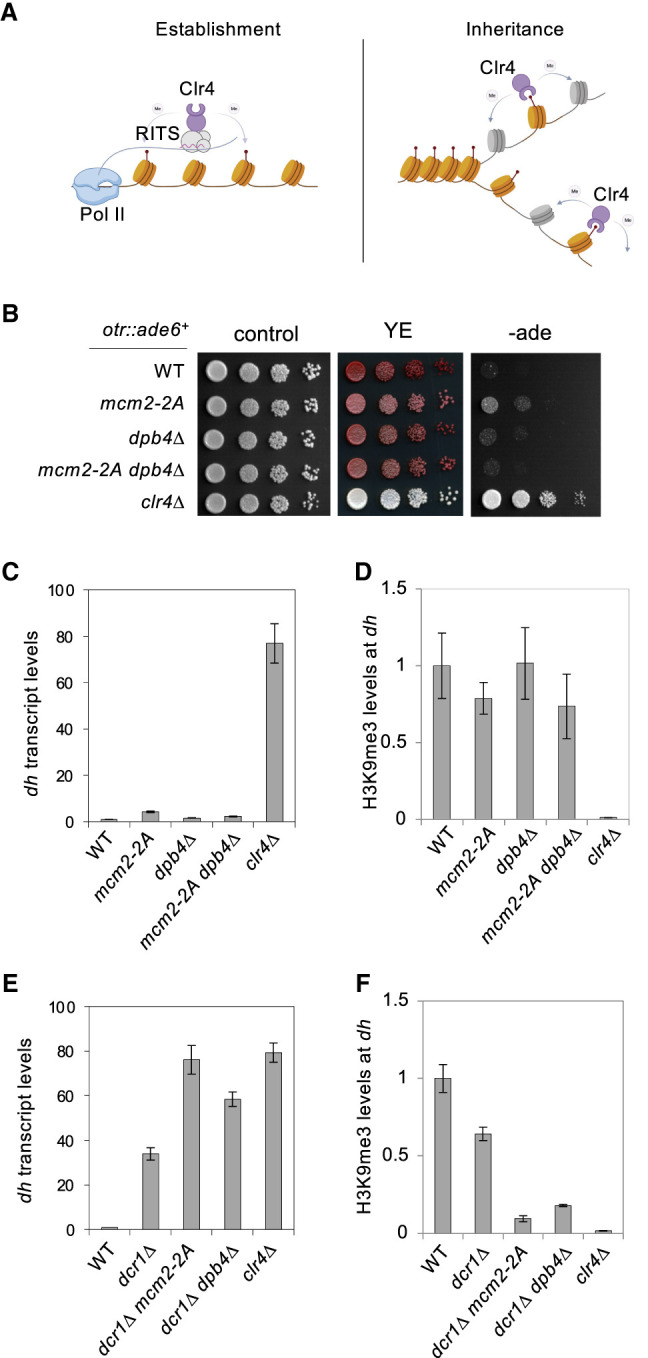
Mcm2 and Dpb4 cooperate with RNAi to regulate pericentric heterochromatin. (A) Schematic diagram of heterochromatin establishment and inheritance pathways at the pericentric region. (B) Serial dilution analysis of the indicated strains to measure the expression of otr::ade6+. (C,E) qRT-PCR analysis of dh transcript levels, normalized to act1+. Data are presented as mean ± SD of three technical replicates. (D,F) ChIP-qPCR analysis of H3K9me3 level at dh, normalized to leu1. Data are presented as mean ± SD of three technical replicates.
The above results suggest a minor role of Mcm2 and Dpb3/4 on pericentric heterochromatin integrity. One possible reason is that RNAi-mediated heterochromatin initiation preserves pericentric heterochromatin integrity even when heterochromatin inheritance is compromised. To test this idea, we examined the requirement of Mcm2 and Dpb3/4 on pericentric heterochromatin integrity when RNAi is compromised. The ribonuclease Dicer (Dcr1) is a critical component of the RNAi pathway and is required for heterochromatin establishment at pericentric repeats (Volpe et al. 2002; Verdel et al. 2004). qRT-PCR analyses show that dh transcripts levels increase in dcr1Δ cells and further increase in dcr1Δ mcm2-2A and dcr1Δ dpb4Δ cells (Fig. 3E). Moreover, ChIP analyses show that H3K9me3 levels at dh repeats decrease in dcr1Δ cells but further decrease in mcm2-2A dcr1Δ and dpb4Δ dcr1Δ cells (Fig. 3F). These findings suggest that RNAi and Mcm2-Dpb3/4 cooperate to regulate the integrity of pericentric heterochromatin.
The dpb4Δ mutant displays more pronounced effects on pericentric heterochromatin in the dcr1Δ background compared with the KΔ::ade6+ and tetO-gfp+ systems. Despite the expectation that dcr1Δ primarily affects heterochromatin establishment, it causes robust silencing defects at pericentric repeats as a result of a higher histone turnover rate, mediated by factors such as Epe1, the Mst2 histone acetyltransferase, the INO80 chromatin remodeling complex, and the Paf1 complex (Reddy et al. 2011; Aygün et al. 2013; Sadeghi et al. 2015; Wang et al. 2015b; Shan et al. 2020). Therefore, dcr1Δ might create a more sensitized background for the effects of dpb4Δ to be observed.
We also directly tested the effects of Mcm2 and Dpb3/4 on heterochromatin establishment by using genetic crosses to introduce the otr::ade6+ reporter from a clr4Δ background (Supplemental Fig. S3). We found that neither mcm2-2A nor dpb4Δ exhibits any difference in the silencing status of otr::ade6+ compared with strains in which otr::ade6+ is introduced from a clr4+ background. These results demonstrate that Mcm2 and Dpb3/4 do not affect heterochromatin establishment.
Mcm2 and Dpb3/4 regulate parental histone H3–H4 segregation during DNA replication
We then investigated the molecular mechanisms through which Mcm2 and Dpb3/4 influence heterochromatin inheritance. Considering their roles as regulators of parental histone segregation in budding yeast and mammals, we examined their impact on parental histone segregation during DNA replication in fission yeast using eSPAN (Fig. 4A; Yu et al. 2014, 2018; Gan et al. 2018). We first synchronized yeast cultures at the G2/M phase by growing a cdc25-22 temperature-sensitive mutant at restrict temperature. After release into permissive temperature, BrdU was added before S phase to label newly synthesized DNA (Hodson et al. 2003; Sivakumar et al. 2004), and cells were fixed at early S phase (Supplemental Fig. S4A). All mutants used had a similar septation index profile (Supplemental Fig. S4A) and were not sensitive to hydroxyurea (HU) (Supplemental Fig. S4B), a ribonucleotide reductase inhibitor, indicating that they did not have major DNA replication defects. Chromatin immunoprecipitation (ChIP) was then performed using antibodies against H3K4me3 or H3K9me3, representing parental histones in euchromatin and heterochromatin, respectively. A subsequent immunoprecipitation with an antibody against BrdU allowed us to the isolate newly replicated single-strand DNA (ssDNA) for sequencing. We calculated the partition ratio of Watson (W) and Crick (C) strands with the formula
at selected replication origins to determine the eSPAN bias (Fig. 4A; Yu et al. 2014, 2018; Gan et al. 2018). To ensure methodological consistency, we selected 162 replication origins that exhibited consistent usage in the wild-type samples and had been annotated as origins in the Pombase for our analysis.
Figure 4.
Mcm2 and Dpb4 regulate parental histone H3–H4 deposition at the replication fork. (A) Schematic diagram of eSPAN workflow and expected results. (B,D,F) eSPAN analysis of H3K4me3, H3K9me3, or H3K56ac bias levels at replication origins. The shading of the bias line plot is the 95% confidence interval of mean value of at least two biological replicates, which is mean ± twofold of the standard error. (C,E,G) Heat maps of H3K4me3, H3K9me3, and H3K56 eSPAN bias at each of the 162 replication origins analyzed.
We initially focused on examining the segregation of H3K4me3-containing parental histones during DNA replication due to its high enrichment at replication origins. While wild-type cells displayed no H3K4me3 eSPAN bias, mcm2-2A cells exhibited a strong leading strand bias, and dpb4Δ cells showed a strong lagging strand bias (Fig. 4B,C). These results align with previous findings in budding yeast and mammals (Gan et al. 2018; Petryk et al. 2018; Yu et al. 2018; Li et al. 2020), highlighting the conservation of Mcm2 and Dpb3/4 in regulating parental histone deposition. Interestingly, mcm2-2A dpb4Δ cells showed reduced bias levels compared with each of the single mutants (Fig. 4B,C).
Next, we examined the segregation of H3K9me3-containing parental histones during DNA replication. As H3K9me3 is not typically enriched at replication origins, the data were less robust. Nevertheless, H3K9me3 eSPAN in different genetic backgrounds displayed a pattern similar to that of H3K4me3 eSPAN, indicating that the segregation of parental histone H3–H4 was likely independent of the modifications they carried (Fig. 4D,E). Considering the more robust H3K4me3 eSPAN results, we focused on H3K4me3 eSPAN for our subsequent analyses of parental histone H3–H4.
To validate our findings, we also investigated the segregation of newly synthesized histones, which are enriched with acetylated histone H3 lysine 56 (H3K56ac) (Masumoto et al. 2005). The H3K56ac eSPAN results show that wild-type cells did not exhibit bias, mcm2-2A cells displayed a lagging strand bias, and dpb4Δ cells showed a leading strand bias. Additionally, mcm2-2A dpb4Δ cells showed a reduction in bias, approaching levels similar to wild-type cells (Fig. 4F,G). The H3K56ac eSPAN results were the opposite of those of H3K4me3 eSPAN, suggesting that defects in parental histone transfer are partially compensated for by the deposition of newly synthesized histones.
Collectively, these results demonstrate that Mcm2 and Dpb3/4 are critical for parental histone H3–H4 segregation to the lagging and leading strands, respectively, highlighting the conservation of parental histone segregation pathways.
Parental histone H3–H4 density is critical for epigenetic inheritance.
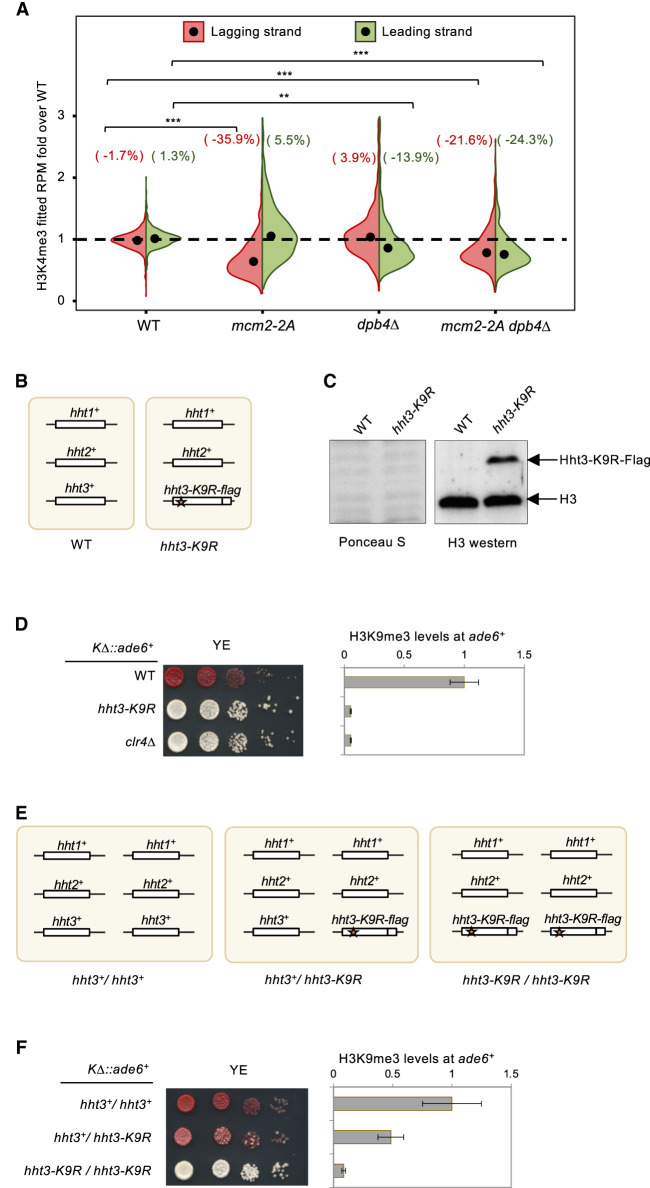
To further investigate the connections between parental histone H3–H4 segregation and epigenetic inheritance of heterochromatin, we analyzed parental histone density on daughter strands for each genotype. To make a quantitative comparison, we first normalized raw H3K4me3 eSPAN coverage to the total number of reads of each sample and then normalized against BrdU input to derive a density score. The normalized density value for each genotype was then divided by the corresponding wild-type (WT) density to determine the fold change. In wild-type cells, the daughter strands exhibited similar H3K4me3 density, consistent with balanced parental histone H3–H4 deposition (Fig. 5A). In mcm2-2A cells, the lagging strand displayed a strong decrease in H3K4me3 density, while the leading strand showed a slight increase. Conversely, in dpb4Δ cells, the leading strand showed a decrease in H3K4me3 density, while the lagging strand remained almost unchanged. Interestingly, the mcm2-2A dpb4Δ double mutant demonstrated a reduction in H3K4me3 density on both strands, consistent with the disruption of both deposition pathways (Fig. 5A).
Figure 5.
Maintaining parental histone H3–H4 density on daughter strands during DNA replication is critical for epigenetic inheritance. (A) Violin plot of H3K4me3 density on the leading and lagging strands at replication origins. Data are the average of at least two biological repeats for each genotype. The numbers represent change over WT for each strand. (B,E) Diagram of histone H3 genes and the hht3-K9R mutation. (C, left) Ponceau S stain of the membrane used for Western blot analysis. (Right) Western blot analysis of histone H3 levels in cell extracts. Hht3-K9R has a FLAG tag at its C terminus, resulting in a higher molecular weight. (D,F, left) Serial dilution analysis of the indicated strains to measure the expression of KΔ::ade6+. (Right) ChIP analysis of H3K9me3 levels at KΔ::ade6+, normalized to leu1. Data are presented as mean ± SD of three technical replicates.
We then investigated whether directly modulating the density of parental histones containing H3K9me3 affects heterochromatin inheritance. In fission yeast, H3 is encoded by three genes: hht1+, hht2+, and hht3+. We introduced a K9R mutation together with a FLAG tag into hht3+ (hh3-K9R), which is expected to result in about one-third of histones lacking H3K9me3 (Fig. 5B). Western blot analysis with an H3 antibody shows that H3K9R is ∼36% ± 6% (n = 3) of total histone H3 (Fig. 5C). As expected, this mutation impaired the silencing of KΔ::ade6+ and the inheritance of tetO-gfp+ after TetR-Clr4-I release (Fig. 5D; Supplemental Fig. S5), consistent with the idea that high parental histone density is required for proper epigenetic inheritance. Importantly, hht3-K9R only marginally affects pericentric heterochromatin (Shan et al. 2016), suggesting that it does not significantly affect heterochromatin initiation.
To further determine the histone density threshold required for proper heterochromatin inheritance, we generated diploids with either one or two copies of hht3-K9R (Fig. 5E). When two copies of hht3-K9R were present, there were severe silencing defects of KΔ::ade6+, as indicated by white colonies on YE medium and a strong reduction of H3K9me3 levels at the reporter. In contrast, the presence of one copy of hht3-K9R exhibited only mild silencing defects of KΔ::ade6+, marked by mostly red colonies on YE medium and an intermediate decrease in H3K9me3 levels (Fig. 5F). Therefore, we conclude that the parental histone threshold required for heterochromatin inheritance falls within the range of 64%–82%. This threshold also aligns with the observed relationship between silencing defects and H3K4me3 density in our eSPAN analysis.
FACT regulates parental histone H3–H4 segregation to both daughter strands
The observation that mcm2-2A dpb4Δ cells exhibit considerable leading strand H3K4me3 eSPAN bias suggests the potential contribution of other histone chaperones to parental histone deposition when both Mcm2 and Dpb3/4 are absent. We turned our focus to the histone chaperone complex FACT, which is involved in nucleosome dynamics during DNA replication (Formosa and Winston 2020).
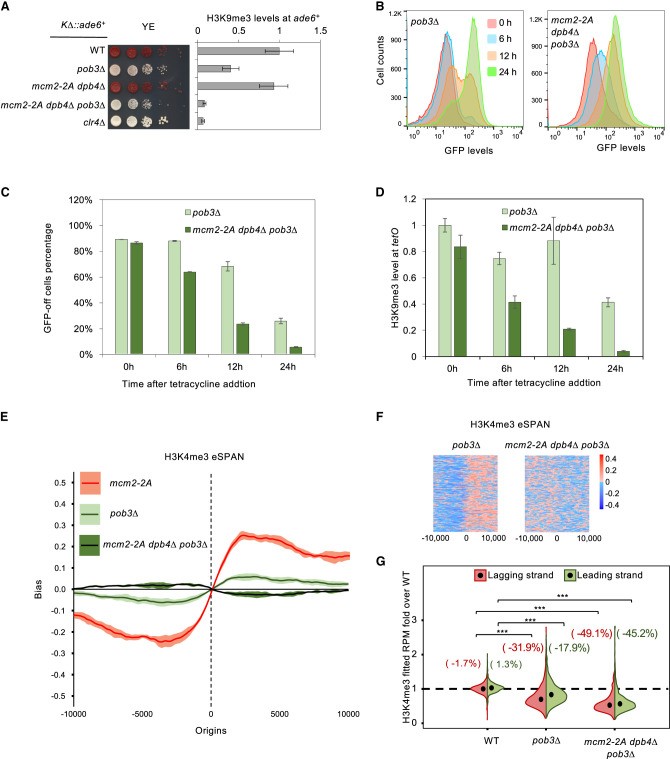
The yeast FACT complex consists of Spt16 and Pob3 (Formosa and Winston 2020). While Spt16 is essential for cell survival, pob3Δ is viable for further genetic analysis. The inheritance of KΔ::ade6+ is compromised in pob3Δ cells, as evidenced by light-pink colonies on YE medium, and is further compromised in mcm2-2A dpb4Δ pob3Δ cells, resulting in white colonies on YE medium (Fig. 6A). ChIP analyses confirmed that H3K9me3 levels at KΔ::ade6+ are reduced in pob3Δ cells and further decrease in mcm2-2A dpb4Δ pob3Δ cells (Fig. 6A). Similarly, pob3Δ cells showed a quick activation of GFP expression and loss of H3K9me3 at tetO after tetracycline addition to remove TetR-Clr4-I. In addition, mcm2-2A dpb4Δ pob3Δ cells showed an even faster rate of GFP activation and H3K9me3 loss at tetO compared with pob3Δ cells (Fig. 6B–D).
Figure 6.
FACT regulates parental histone deposition and heterochromatin inheritance. (A, left) Serial dilution analysis of the indicated genotypes to measure the expression of KΔ::ade6+. (Right) ChIP-qPCR analysis of H3K9me3 levels KΔ::ade6+, normalized to leu1. Data are presented as mean ± SD of three technical replicates. (B) Flow cytometry analysis of GFP expression at different time points after tetracycline addition. All strains are in an epe1Δ background. (C) Quantification of the percentage of cells maintaining low GFP expression at different time points after tetracycline addition. Data are presented as mean ± SD of two biological replicates. All strains are in an epe1Δ background. (D) ChIP-qPCR analysis of H3K9me3 levels at tetO at different time points after tetracycline addition, normalized to leu1. Data are presented as mean ± SD of three technical replicates. All strains are in an epe1Δ background. (E) eSPAN analysis of H3K4me3 bias levels at replication origins of the indicated strains. The shading of the bias line plot is the 95% confidence interval of mean value of at least two biological replicates, which is approximately mean ± twofold of the standard error. (F) Heat maps of H3K4me3 bias at each of the 162 replication origins analyzed. (G) Violin plot of H3K4me3 density on leading and lagging strands at replication origins. Data are the average of at least two biological repeats for each genotype. The numbers represent changes over WT for each strand.
We then investigated the role of FACT in parental histone segregation. H3K4me3 eSPAN revealed a mild leading strand bias in pob3Δ cells, and mcm2-2A dpb4Δ pob3Δ cells displayed even milder H3K4me3 eSPAN bias, although directed toward the lagging stand (Fig. 6E,F). However, pob3Δ cells showed strong reductions in histone density on both strands, and pob3Δ mcm2-2A dpb4Δ cells displayed an even more pronounced reduction in density on both strands (Fig. 6G). Similar to what has been observed in other mutant backgrounds, H3K56 eSPAN mirrors that of H3K4me3 eSPAN (Supplemental Fig. S6). These findings suggest that the FACT complex acts in concert with Mcm2 and Dpb3/4 for parental histone segregation and heterochromatin inheritance.
Discussion
One of the central questions of chromatin-based epigenetic inheritance is how parental histone H3–H4 tetramers are distributed to daughter strands, which serve as seeds to restore parental histone modification patterns. The histone binding activities of two replisome components, Mcm2 and Dpb3/4, are responsible for distributing parental histone H3–H4 to the lagging and leading strands, respectively. However, despite their strong effects on parental histone H3–H4 segregation, mutations of Mcm2 and Dpb3/4 have only mild effects on heterochromatin stability in yeast (Foltman et al. 2013; He et al. 2017; Yu et al. 2018; Saxton and Rine 2019). In this study, we investigated the role of effects of mutating the histone binding activity of Mcm2 (mcm2-2A) and Dpb3/4 (dpb3Δ or dpb4Δ) in heterochromatin inheritance in fission yeast using inheritance-specific reporter assays. Surprisingly, mcm2-2A abolishes heterochromatin inheritance, dpb3Δ and dpb4Δ have only minor defects, and mcm2-2A dpb3Δ and mcm2-2A dpb4Δ have intermediate defects. We also found that these mutants have mild effects on native pericentric heterochromatin because initiation and inheritance pathways function together to regulate pericentric heterochromatin integrity, and Mcm2 and Dpb3/4 are only required for heterochromatin inheritance. These results align with recent studies in mammalian systems indicating that the disruption of parental histone H3–H4 segregation pathways can impact cellular processes in specific contexts, potentially due to defective epigenetic inheritance (Li et al. 2020, 2023; Xu et al. 2022; Tian et al. 2023; Wen et al. 2023; Wenger et al. 2023).
We also developed eSPAN in fission yeast to measure parental histone H3–H4 segregation. We found that H3K4me3 eSPAN and H3K9me3 eSPAN generally agree with each other, suggesting that the segregation of parental histone H3–H4 is independent of their modifications. This is consistent with the fact that histone chaperones, such as Mcm2-HBD, interact with the core histone fold rather than histone tails, where these modifications occur (Huang et al. 2015; Richet et al. 2015; Wang et al. 2015a). We observed that H3K9me3 eSPAN is less robust compared with H3K4me3 eSPAN, which we attribute to much higher levels of H3K4me3 than H3K9me3 around replication origins. However, given the interaction between heterochromatin proteins and parental histone chaperones within the DNA replication machinery (Nakayama et al. 2001a; Motamedi et al. 2008; Fischer et al. 2009; Takahata et al. 2021; Li et al. 2023), it is conceivable that differences may arise in the deposition of H3K9me3-containing parental histones, particularly in specialized contexts. For example, in mouse embryonic stem cells, H3K9me3 is preferentially transferred to the leading strand at long interspersed nuclear element (LINE) retrotransposons, mediated by the interaction of the human silencing hub (HUSH) complex and DNA polymerase ε (Li et al. 2023).
We also performed H3K56ac eSPAN to measure the deposition of newly synthesized histone H3–H4. Interestingly, it anticorrelated with H3K4me3 eSPAN. These results confirm that our procedure effectively distinguishes histone segregation into the two daughter strands. Therefore, we focused on H3K4me3 eSPAN to study parental histone H3–H4 segregation.
Our H3K4me3 eSPAN analysis revealed that mcm2-2A cells display a leading strand bias, while dpb4Δ cells exhibit a lagging strand bias. These results are consistent with previous studies in budding yeast and mammals (Gan et al. 2018; Petryk et al. 2018; Yu et al. 2018; Li et al. 2020; Wen et al. 2023; Wenger et al. 2023), suggesting a highly conserved mechanism of parental histone H3–H4 segregation (Fig. 7A–C). Interestingly, mcm2-2A dpb4Δ cells show an intermediate level of both parental histone H3–H4 segregation bias and heterochromatin inheritance. These results are consistent with the idea that a symmetric distribution of parental histone H3–H4 is important for heterochromatin inheritance.
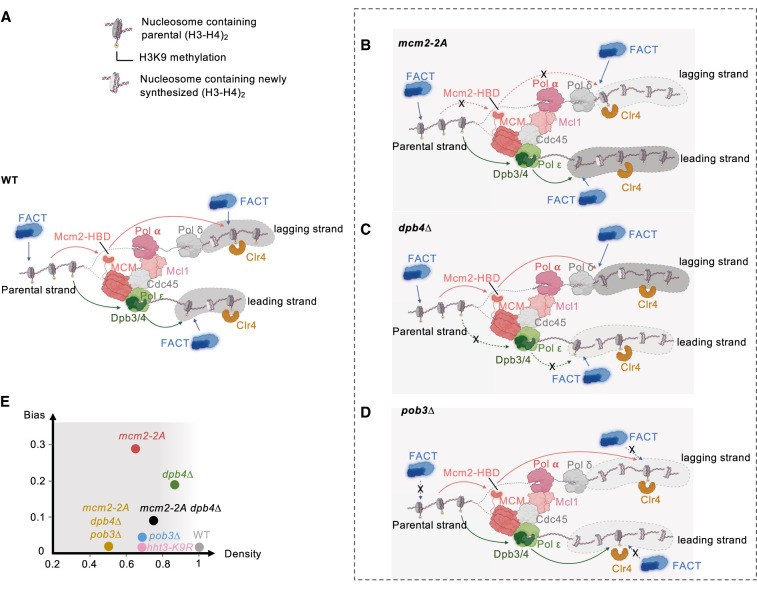
Figure 7.
Model of the cooperation of histone chaperones in regulating parental histone segregation to daughter DNA strands and epigenetic inheritance of heterochromatin. (A) Schematic diagram illustrating parental histone H3–H4 deposition pathways during DNA replication in wild-type cells. (B) Impaired parental histone H3–H4 deposition to the lagging strand in mcm2-2A cells leads to a high H3K4me3 eSPAN bias and low H3K4me3 density on the lagging strand. (C) Impaired parental histone H3–H4 deposition to the leading strand in dpb4Δ cells results in an intermediate H3K4me3 eSPAN bias and a moderate decrease in H3K4me3 density on the leading strand. (D) Impaired parental histone H3–H4 deposition to both strands in pob3Δ cells causes a low H3K4me3 eSPAN bias and reduced H3K4me3 density on both daughter strands. (E) Plotting H3K4me3 eSPAN bias and H3K4me3 density on the lower-density strand. The gradients of gray indicate the severity of the silencing defects.
In addition to the symmetric distribution of parental histone H3–H4, our subsequent analyses indicate that heterochromatin inheritance outcomes also depend on parental histone H3–H4 density on daughter strands (Fig. 7D). This is because while a high bias consistently indicates low density on one strand, a low bias fails to differentiate between high density on both strands or low density on both strands. We propose that the relationship between parental histone H3–H4 density and heterochromatin inheritance is akin to the “weakest link theory,” where each daughter strand represents an individual “link” of epigenetic inheritance, and the strand with the lower parental histone levels dictates the inheritance efficiency (Fig. 7E). The profound heterochromatin inheritance defects in mcm2-2A cells correspond with the notably low density of parental histone H3–H4 on its lagging strand, whereas the mild defects in dpb4Δ cells align with a slight density decrease on its leading strand. Our results are consistent with earlier studies that suggest that low histone turnover rates are critical for epigenetic inheritance (Reddy et al. 2011; Holla et al. 2020; Shan et al. 2020; Cutter DiPiazza et al. 2021), and our model extends this understanding by proposing a discernible threshold of inherited parental histone H3–H4 density that determines the efficiency of epigenetic inheritance.
Heterochromatin formation involves positive feedback loops where, for instance, the H3K9me3-Clr4 “read–write cycles” not only duplicate heterochromatin during DNA replication but also promote heterochromatin spreading (Zhang et al. 2008). These positive feedback cycles have the potential to transform subtle gradient changes into binary switches. When the parental histone H3–H4 density falls below a certain threshold, cells become more susceptible to losing epigenetic information. This prediction is substantiated by the introduction of the H3K9R mutant histone, which effectively reduces the proportion of parental histones containing H3K9me3. Notably, when H3K9R constitutes only 18% of total histone H3, cells can maintain heterochromatin, albeit less effectively than in wild-type cells (Fig. 5F). However, with an increase of H3K9R levels to 36% of total histone H3, a more pronounced loss of heterochromatin occurs, consistent with the existence of a threshold of parental histone H3–H4 density required for epigenetic inheritance (Fig. 5D,F).
It is intriguing to observe that the parental histone H3–H4 density on the lagging strand in mcm2-2A dpb4Δ cells is higher than that in mcm2-2A cells. This result suggests that the deposition of histones by Mcm2 and Dpb3/4 is competitive. In mcm2-2A cells, deposition to the lagging strand fails to compete with Dpb3/4-mediated deposition on the leading strand, resulting in a substantial reduction in lagging strand parental histone H3–H4 density (Fig. 5A). Dpb3/4 captures a portion of released parental histone H3–H4 originally destined for Mcm2 and deposits them to the leading strands. The modest increase in parental histone H3–H4 on the leading strand may be attributed to the fact that Dpb3/4 is either nearing its capacity or facing competition with the deposition of newly synthesized histone H3–H4. However, the simultaneous impairment of Mcm2 and Dpb3/4 recalibrates this balance. First, vacancies emerge for histone H3–H4 deposition in both strands due to the loss of two major parental histone H3–H4 segregation pathways. Second, a larger pool of parental histone H3–H4 becomes available, allowing their incorporation into both strands with the assistance of alternative parental histone segregation pathways. Consequently, parental histone H3–H4 can be recycled on both strands, although with less efficiency than in wild-type cells. This scenario could result in a higher density of parental histone H3–H4 on the lagging strand in mcm2-2A dpb4Δ cells compared with mcm2-2A cells. However, we could not exclude the possibility that the apparent increase in histone density in double mutant cells is due to normalization artifacts.
Given the established roles of FACT in regulating histone dynamics during DNA replication and heterochromatin inheritance, we investigated its potential contribution to parental histone H3–H4 segregation (Lejeune et al. 2007; Formosa and Winston 2020; Holla et al. 2020; Murawska et al. 2021; Takahata et al. 2021). Indeed, we found that a mutation in FACT (pob3Δ) results in only a minor leading strand H3K4me3 eSPAN bias yet elicits a profound impact on H3K4me3 density on both strands. Moreover, mcm2-2A dpb4Δ pob3Δ cells show a greater loss of silencing compared with mcm2-2A dpb4Δ or pob3Δ cells alone, suggesting redundancy in their functions.
It is worth noting that pob3Δ is unlikely to entirely abolish FACT function, given the nonessential nature of Pob3 compared with the essential status of Spt16. Therefore, FACT might play a crucial role in all mechanisms of parental histone H3–H4 transfer, with Mcm2 or Dpb3/4 guiding strand-specific parental histone H3–H4 transfer. Moreover, mcm2-2A harbors point mutations within the HBD, leaving the possibility of residual histone binding activity, and it is plausible that dpb4Δ or dpb3Δ indirectly enhances the activity of Mcm2-HBD. Furthermore, the H3K4me3 eSPAN bias levels of dpb4Δ are smaller than those of mcm2-2A. Despite dpb3Δ and dpb4Δ being null mutations, there may be other histone chaperone activities that collaborate with Dpb3/4 to facilitate parental histone transfer to the leading strand.
Mcm2, Dpb3/4, and FACT are the major histone H3–H4 chaperones with established roles in regulating parental histone segregation. Here we demonstrate that they have distinct properties: While Mcm2 and Dpb3/4 guide parental histone H3–H4 to the lagging and leading strands, respectively, FACT primarily maintains parental histone H3–H4 to both strands. These cooperative actions collectively ensure the disposition of critical levels of parental histone H3–H4 onto both daughter strands, thus safeguarding the fidelity of epigenetic inheritance.
Materials and methods
Fission yeast strains and genetic analyses
Yeast strains containing mcm2+-FLAG and mcm2-2A-FLAG were generated by a PCR-based module method. The FLAG epitope tag was integrated at the endogenous mcm2+ locus so that the transgene was under the control of its native promoters. Deletion strains dpb3Δ and dpb4Δ were derived from the Bioneer deletion library and confirmed by PCR analysis, and pob3Δ was constructed by a PCR-based module method. All other strains were constructed by genetic crosses. A list of fission yeast strains used in this study is in Supplemental Table S1. For serial dilution analysis, 10-fold dilutions of mid-log stage fission yeast cultures were plated on the indicated media and incubated for 3 d at 30°C.
Flow cytometry
Fission yeast strains with the tetO-ura4-gfp+ reporter were grown in liquid culture and maintained at mid-log phase. Cells were harvested at the indicated time points after the addition of tetracycline to a final concentration of 2.5 μg/mL. Cells were resuspended in cold PBS (10 mM Na2HPO4, 1.8 mM KH2PO4 at pH 7.4, 137 mM NaCl, 2.7 mM KCl). Cold ethanol was added to a final concentration of 70%, and cells were fixed for 15 min on ice. Cells were then washed twice with PBS and resuspended in a flow cytometry tube (Corning 352008). GFP expression levels were measured by FACSCelesta (Becton Dickinson), and excitation was achieved by using an argon laser emission of 488 nm. Data collection was performed using Cellquest (Becton Dickinson), and a primary gate based on physical parameters (forward and side light scatter) was set to exclude dead cells and debris. Typically, 50,000 cells were analyzed for each strain at each time point. Raw data were processed and analyzed with FlowJo (10.6.2; Becton Dickinson).
Chromatin immunoprecipitation (ChIP)
Log-phase yeast cultures (OD595 ∼0.5) were cross-linked with 1% formaldehyde for 20 min at room temperature with shaking. Glycine (125 mM) was added for an additional 5 min to neutralize formaldehyde. Cells were harvested and washed with cold PBS (10 mM Na2HPO4, 1.8 mM KH2PO4 at pH 7.4, 137 mM NaCl, 2.7 mM KCl). Cell pellets then were resuspended in ChIP lysis buffer (50 mM HEPES-KOH at pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% deoxycholate, 1 mM PMSF). Cold glass beads were added, and the mixture was shaken vigorously in a MiniBeadBeater (Biospec Products) four times for 30 sec each. The lysates were collected and fragmented with a Bioruptor Pico sonication system (Diagenode) for 12 cycles (30 sec on/30 sec off). After the lysates were clarified by centrifuge at 13,000 rpm for 15 min, released chromatin was immunoprecipitated with the corresponding antibodies—H3K4me3 (EMD Millipore 07-473) and H3K9me3 (Active Motif 39161)—overnight at 4°C. The DNA–antibody mixtures were incubated with Protein G agarose beads (Sigma 11243233001) for 2 h at 4°C. Beads were washed twice with ChIP lysis buffer and once each with ChIP lysis buffer containing 0.5 M NaCl, wash buffer (10 mM Tris at pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% deoxycholate, 1 mM EDTA), and TE (50 mM Tris at pH 8.0, 1 mM EDTA). Bead-bound chromatin was eluted with TES (50 mM Tris at pH 8.0, 1 mM EDTA, 1% SDS) at 65°C. The cross-linking was reversed by overnight incubation at 65°C. The DNA–protein mixtures were treated with Proteinase K (Invitrogen 10005393) to release the DNA fragments, and DNA was purified by two phenol:chloroform extractions and precipitated by ethanol.
Quantitative PCR (qPCR) was conducted to calculate ChIP enrichment values with primers specific to the indicated heterochromatin regions. qPCR was performed with Luna Universal qPCR master mix (NEB M3003S) in a StepOne Plus real-time PCR system (Applied Biosystems). A list of DNA oligos used is in Supplemental Table S2.
RNA analysis
Total cellular RNA was isolated from log-phase cells using the MasterPure yeast RNA purification kit (Epicenter) according to the manufacturer's protocol. Quantification with real-time RT-PCR was performed with Power SYBR Green RNA-to-CT one-step kit (Invitrogen 4389986). RNA serial dilutions were used as templates to generate a standard curve of amplification for each pair of primers, and the relative concentration of the target sequence was calculated accordingly. An act1 fragment served as a reference to normalize the concentration of samples. The concentration of each target gene in the wild type was arbitrarily set to 1 and served as a reference for other samples.
eSPAN
Yeast strains containing the cdc25-22 temperature-sensitive mutant were first grown at the permissive temperature (25°C) until the early–mid log phase (OD 0.2∼0.4). Cultures were then incubated for 4 h at 36°C to arrest cells at the G2 phase of the cell cycle. Cells were then shifted to 25°C to allow them to enter the S phase synchronously. BrdU (Sigma-Aldrich B5002) was added to a final concentration of 650 μM at 25 min after the temperature shift. Cells were cross-linked at 60 min after temperature shift with the addition of 1% formaldehyde and incubated for 20 min at 25°C with shaking. Immunoprecipitation was performed as described above in “Chromatin Immunoprecipitation (ChIP).” Cross-linking was reversed by Chelex-100 (Bio-Rad 142-1253).
BrdU IP was conducted as previously described (Hodson et al. 2003; Yu et al. 2018). Briefly, total and ChIP DNA were incubated for 5 min at 100°C and then immediately cooled for 5 min on ice. DNA was diluted 10 times with BrdU IP buffer (PBS, 0.0625% [v/v] Triton X-100) and incubated with BrdU antibody (BD Bioscience 555627) for 2 h at 4°C. Next, Sepharose Protein G beads (Cytiva 17-0618-01) were added and incubated for an additional 1 h at 4°C. The beads were then washed three times with BrdU IP buffer and once with TE. Finally, DNA was eluted with TES and purified with the Qiagen MinElute PCR purification kit (Qiagen 28004). qPCR was used to test the eSPAN with primers specific to ARS2004 (an early replication origin) and distal locus ARS2004 + 25 kb. The ssDNA libraries were prepared with xGen ssDNA and low-input DNA library preparation kit (IDT 10009817).
Protein extraction and Western blot analysis
Log-phase yeast cultures were collected and washed with cold PBS (10 mM Na2HPO4, 1.8 mM KH2PO4 at pH 7.4, 137 mM NaCl, 2.7 mM KCl). Cell pellets were then resuspended in ChIP lysis buffer (50 mM HEPES-KOH at pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% deoxycholate, 1 mM PMSF). Cold glass beads were added, and the mixture was shaken vigorously in a MiniBeadBeater (Biospec Products). The lysates were mixed with 2× SDS loading buffer and resolved by SDS-PAGE. The proteins were transferred to a PVDF membrane, and Western blot analysis was performed with FLAG (Sigma F7425) or H3 antibodies.
Sequencing data analysis
The ChIP, BrdU IP, and eSPAN ssDNA libraries were sequenced by paired-end sequencing under Illumina NextSeq platforms at Columbia University Irving Medical Center, supported by the Herbert Irving Comprehensive Cancer Center. The raw reads were first trimmed by Trim Galore! (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore) to remove adapters and low-quality reads and then mapped to the S. pombe genome using Bowtie2 (Langmead and Salzberg 2012). PCR duplicate reads were filtered by Sambamba (Tarasov et al. 2015). The genome-wide read coverage on Watson and Crick strands was calculated at the bin of 1 bp using deepTools bamCoverage (Ramírez et al. 2016).
The bias of read coverage between Watson and Crick strands was calculated at each bin surrounding the origins using the formula
where W and C are the read coverages on the Watson and Crick strands, respectively. For the calculation of bias, we used the bin of a 100-bp sliding window in the [−10 kb, 10 kb] region surrounding the origins using the code from https://github.com/clouds-drift/eSPAN-bias (Li et al. 2021). For each strain, both the eSPAN bias and the BrdU bias were calculated. To avoid the background influence from BrdU, the eSPAN bias was further normalized with the corresponding BrdU bias by subtraction.
To quantitively compare the eSPAN signals of parental histones for each DNA strand among samples, the raw eSPAN coverage was first normalized to the total number of reads of each sample. To mitigate potential variation stemming from replication synchronization, BrdU samples from the same genotype were used to compute the coefficients at each origin. Subsequently, the signals surrounding each origin [−2500 bp, 2500 bp] in the corresponding eSPAN sample were fitted by the BrdU coefficients. This approach minimized the impact of variation in BrdU incorporation on eSPAN samples, enabling accurate quantification of density. Statistical significance of single-strand density differences between mutants and the wild type was assessed using a two-sample t-test.
Statistical analysis
The statistical test of sequencing data was performed using R software.
Data and material availability
Sequencing data have been deposited to NCBI GEO under accession number GSE237770.
Supplementary Material
Acknowledgments
We thank Ke Zhang, Susan Forsburg, Nick Rhind, Danesh Moazed, and Kaushik Ragunathan for yeast strains; Lauren Clarissa Tang, Kehan Bao, Albert Serra-Cardona, and Hunter King for assistance; and Rodney Rothstein and Lorraine Symington for discussions. This work was supported by National Institutes of Health grants R35GM126910 (to S.J.), R35GM118015 (to Z.Z.), and R35GM149572 (to F.Q.).
Author contributions: S.J., Z.Z., and F.Q. conceived the project. S.J. designed the experiments. Y.F., S.J., C.-M.S., and T.T. performed the experiments. X.H. performed the eSPAN analysis. S.J., Z.Z., and F.Q. supervised the research. Y.F. and S.J. wrote the manuscript with input from all authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.351278.123.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Allshire RC, Madhani HD. 2018. Ten principles of heterochromatin formation and function. Nat Rev Mol Cell Biol 19: 229–244. 10.1038/nrm.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audergon PN, Catania S, Kagansky A, Tong P, Shukla M, Pidoux AL, Allshire RC. 2015. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science 348: 132–135. 10.1126/science.1260638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygün O, Mehta S, Grewal SI. 2013. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat Struct Mol Biol 20: 547–554. 10.1038/nsmb.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. 10.1038/35065138 [DOI] [PubMed] [Google Scholar]
- Cutter DiPiazza AR, Taneja N, Dhakshnamoorthy J, Wheeler D, Holla S, Grewal SIS. 2021. Spreading and epigenetic inheritance of heterochromatin require a critical density of histone H3 lysine 9 tri-methylation. Proc Natl Acad Sci 118: e2100699118. 10.1073/pnas.2100699118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Shi G, Shan CM, Li Z, Zhu B, Jia S, Li Q, Zhang Z. 2022. Mechanisms of chromatin-based epigenetic inheritance. Sci China Life Sci 65: 2162–2190. 10.1007/s11427-022-2120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar TM, Oksuz O, Saldaña-Meyer R, Descostes N, Bonasio R, Reinberg D. 2019. Active and repressed chromatin domains exhibit distinct nucleosome segregation during DNA replication. Cell 179: 953–963.e11. 10.1016/j.cell.2019.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar TM, Loyola A, Reinberg D. 2021. Parental nucleosome segregation and the inheritance of cellular identity. Nat Rev Genet 22: 379–392. 10.1038/s41576-020-00312-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SI. 2009. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci 106: 8998–9003. 10.1073/pnas.0813063106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltman M, Evrin C, De Piccoli G, Jones RC, Edmondson RD, Katou Y, Nakato R, Shirahige K, Labib K. 2013. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep 3: 892–904. 10.1016/j.celrep.2013.02.028 [DOI] [PubMed] [Google Scholar]
- Formosa T, Winston F. 2020. The role of FACT in managing chromatin: disruption, assembly, or repair? Nucleic Acids Res 48: 11929–11941. 10.1093/nar/gkaa912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H, Serra-Cardona A, Hua X, Zhou H, Labib K, Yu C, Zhang Z. 2018. The Mcm2–Ctf4–polα axis facilitates parental histone H3–H4 transfer to lagging strands. Mol Cell 72: 140–151.e3. 10.1016/j.molcel.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SIS. 2023. The molecular basis of heterochromatin assembly and epigenetic inheritance. Mol Cell 83: 1767–1785. 10.1016/j.molcel.2023.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. 2007. Heterochromatin revisited. Nat Rev Genet 8: 35–46. 10.1038/nrg2008 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Klar AJ. 1996. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell 86: 95–101. 10.1016/S0092-8674(00)80080-X [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237. 10.1126/science.1076466 [DOI] [PubMed] [Google Scholar]
- He H, Li Y, Dong Q, Chang AY, Gao F, Chi Z, Su M, Zhang F, Ban H, Martienssen R, et al. 2017. Coordinated regulation of heterochromatin inheritance by Dpb3–Dpb4 complex. Proc Natl Acad Sci 114: 12524–12529. 10.1073/pnas.1712961114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson JA, Bailis JM, Forsburg SL. 2003. Efficient labeling of fission yeast Schizosaccharomyces pombe with thymidine and BUdR. Nucleic Acids Res 31: 134e. 10.1093/nar/gng134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holla S, Dhakshnamoorthy J, Folco HD, Balachandran V, Xiao H, Sun LL, Wheeler D, Zofall M, Grewal SIS. 2020. Positioning heterochromatin at the nuclear periphery suppresses histone turnover to promote epigenetic inheritance. Cell 180: 150–164.e15. 10.1016/j.cell.2019.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Strømme CB, Saredi G, Hödl M, Strandsby A, González-Aguilera C, Chen S, Groth A, Patel DJ. 2015. A unique binding mode enables MCM2 to chaperone histones H3–H4 at replication forks. Nat Struct Mol Biol 22: 618–626. 10.1038/nsmb.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI. 2004. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976. 10.1126/science.1099035 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune E, Bortfeld M, White SA, Pidoux AL, Ekwall K, Allshire RC, Ladurner AG. 2007. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr Biol 17: 1219–1224. 10.1016/j.cub.2007.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hua X, Serra-Cardona A, Xu X, Gan S, Zhou H, Yang WS, Chen CL, Xu RM, Zhang Z. 2020. DNA polymerase α interacts with H3–H4 and facilitates the transfer of parental histones to lagging strands. Sci Adv 6: eabb5820. 10.1126/sciadv.abb5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hua X, Serra-Cardona A, Xu X, Zhang Z. 2021. Efficient and strand-specific profiling of replicating chromatin with enrichment and sequencing of protein-associated nascent DNA in mammalian cells. Nat Protoc 16: 2698–2721. 10.1038/s41596-021-00520-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Duan S, Hua X, Xu X, Li Y, Menolfi D, Zhou H, Lu C, Zha S, Goff SP, et al. 2023. Asymmetric distribution of parental H3K9me3 in S phase silences L1 elements. Nature 623: 643–651. 10.1038/s41586-023-06711-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436: 294–298. 10.1038/nature03714 [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D. 2008. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell 32: 778–790. 10.1016/j.molcel.2008.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawska M, Greenstein RA, Schauer T, Olsen KCF, Ng H, Ladurner AG, Al-Sady B, Braun S. 2021. The histone chaperone FACT facilitates heterochromatin spreading by regulating histone turnover and H3K9 methylation states. Cell Rep 37: 109944. 10.1016/j.celrep.2021.109944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Allshire RC, Klar AJ, Grewal SI. 2001a. A role for DNA polymerase α in epigenetic control of transcriptional silencing in fission yeast. EMBO J 20: 2857–2866. 10.1093/emboj/20.11.2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. 2001b. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113. 10.1126/science.1060118 [DOI] [PubMed] [Google Scholar]
- Petryk N, Dalby M, Wenger A, Stromme CB, Strandsby A, Andersson R, Groth A. 2018. MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361: 1389–1392. 10.1126/science.aau0294 [DOI] [PubMed] [Google Scholar]
- Ragunathan K, Jih G, Moazed D. 2015. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 348: 1258699. 10.1126/science.1258699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BD, Wang Y, Niu L, Higuchi EC, Marguerat SB, Bähler J, Smith GR, Jia S. 2011. Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev 25: 214–219. 10.1101/gad.1993611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richet N, Liu D, Legrand P, Velours C, Corpet A, Gaubert A, Bakail M, Moal-Raisin G, Guerois R, Compper C, et al. 2015. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res 43: 1905–1917. 10.1093/nar/gkv021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J. 2004. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J 23: 3825–3835. 10.1038/sj.emboj.7600401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi L, Prasad P, Ekwall K, Cohen A, Svensson JP. 2015. The Paf1 complex factors Leo1 and Paf1 promote local histone turnover to modulate chromatin states in fission yeast. EMBO Rep 16: 1673–1687. 10.15252/embr.201541214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton DS, Rine J. 2019. Epigenetic memory independent of symmetric histone inheritance. Elife 8: e51421. 10.7554/eLife.51421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel G, Rine J. 2019. The nucleosome core particle remembers its position through DNA replication and RNA transcription. Proc Natl Acad Sci 116: 20605–20611. 10.1073/pnas.1911943116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan CM, Wang J, Xu K, Chen H, Yue JX, Andrews S, Moresco JJ, Yates JR, Nagy PL, Tong L, et al. 2016. A histone H3K9M mutation traps histone methyltransferase Clr4 to prevent heterochromatin spreading. Elife 5: e17903. 10.7554/eLife.17903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan CM, Bao K, Diedrich J, Chen X, Lu C, Yates JR III, Jia S. 2020. The INO80 complex regulates epigenetic inheritance of heterochromatin. Cell Rep 33: 108561. 10.1016/j.celrep.2020.108561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan CM, Fang Y, Jia S. 2023. Leaving histone unturned for epigenetic inheritance. FEBS J 290: 310–320. 10.1111/febs.16260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S, Porter-Goff M, Patel PK, Benoit K, Rhind N. 2004. In vivo labeling of fission yeast DNA with thymidine and thymidine analogs. Methods 33: 213–219. 10.1016/j.ymeth.2003.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S, Chida S, Ohnuma A, Ando M, Asanuma T, Murakami Y. 2021. Two secured FACT recruitment mechanisms are essential for heterochromatin maintenance. Cell Rep 36: 109540. 10.1016/j.celrep.2021.109540 [DOI] [PubMed] [Google Scholar]
- Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. 2015. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31: 2032–2034. 10.1093/bioinformatics/btv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Verhein-Hansen J. 2000. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155: 551–568. 10.1093/genetics/155.2.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Zhou J, Li X, Gao Y, Wen Q, Kang X, Wang N, Yao Y, Jiang J, Song G, et al. 2023. Impaired histone inheritance promotes tumor progression. Nat Commun 14: 3429. 10.1038/s41467-023-39185-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. 10.1126/science.1093686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- Wang H, Wang M, Yang N, Xu RM. 2015a. Structure of the quaternary complex of histone H3–H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2. Protein Cell 6: 693–697. 10.1007/s13238-015-0190-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Reddy BD, Jia S. 2015b. Rapid epigenetic adaptation to uncontrolled heterochromatin spreading. Elife 4: e06179. 10.7554/eLife.06179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tang Y, Xu J, Leng H, Shi G, Hu Z, Wu J, Xiu Y, Feng J, Li Q. 2023. The N-terminus of Spt16 anchors FACT to MCM2-7 for parental histone recycling. Nucleic Acids Res 51: 11549–11567. 10.1093/nar/gkad846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Q, Zhou J, Tian C, Li X, Song G, Gao Y, Sun Y, Ma C, Yao S, Liang X, et al. 2023. Symmetric inheritance of parental histones contributes to safeguarding the fate of mouse embryonic stem cells during differentiation. Nat Genet 55: 1555–1566. 10.1038/s41588-023-01477-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger A, Biran A, Alcaraz N, Redó-Riveiro A, Sell AC, Krautz R, Flury V, Reverón-Gómez N, Solis-Mezarino V, Völker-Albert M, et al. 2023. Symmetric inheritance of parental histones governs epigenome maintenance and embryonic stem cell identity. Nat Genet 55: 1567–1578. 10.1038/s41588-023-01476-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. 2010. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science 328: 94–98. 10.1126/science.1178994 [DOI] [PubMed] [Google Scholar]
- Xu X, Hua X, Brown K, Ren X, Zhang Z. 2022. Mcm2 promotes stem cell differentiation via its ability to bind H3–H4. Elife 11: e80917. 10.7554/eLife.80917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Gan H, Han J, Zhou ZX, Jia S, Chabes A, Farrugia G, Ordog T, Zhang Z. 2014. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell 56: 551–563. 10.1016/j.molcel.2014.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Gan H, Serra-Cardona A, Zhang L, Gan S, Sharma S, Johansson E, Chabes A, Xu RM, Zhang Z. 2018. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361: 1386–1389. 10.1126/science.aat8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI. 2008. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–388. 10.1038/nsmb.1406 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.