Abstract
Supplemental material is available for this article.
Keywords: Brain Metastases, Public Datasets, Artificial Intelligence, MRI
Summary
The publicly available University of California San Francisco Brain Metastases Stereotactic Radiosurgery MRI dataset consists of 412 patients with a total of 560 multimodal brain MRI examinations with expert voxelwise annotations of 5136 brain metastases.
Introduction
Public datasets, such as those made available through The Cancer Imaging Archive (1) and multimodal Brain Tumor Segmentation (BraTS [2]) challenges, have been critical in supporting advances in the field of biomedical image segmentation in neuro-oncology, particularly for glioma. Brain metastases are the most common central nervous system tumor (3,4), and new effective treatment methods have led to a dramatic rise in imaging (5). Detection of very small metastases is a major challenge (6–8) that can potentially be overcome by public data sharing initiatives (9).
Here we present the University of California San Francisco Brain Metastases Stereotactic Radiosurgery (UCSF-BMSR) MRI Dataset. This dataset contains 560 multimodal brain MRI examinations with expert annotations of brain metastases in 412 patients undergoing gamma knife radiosurgery. There are voxelwise annotations of 5136 brain metastases, including two sets of annotations in 99 patients.
Materials and Methods
Patient Sample
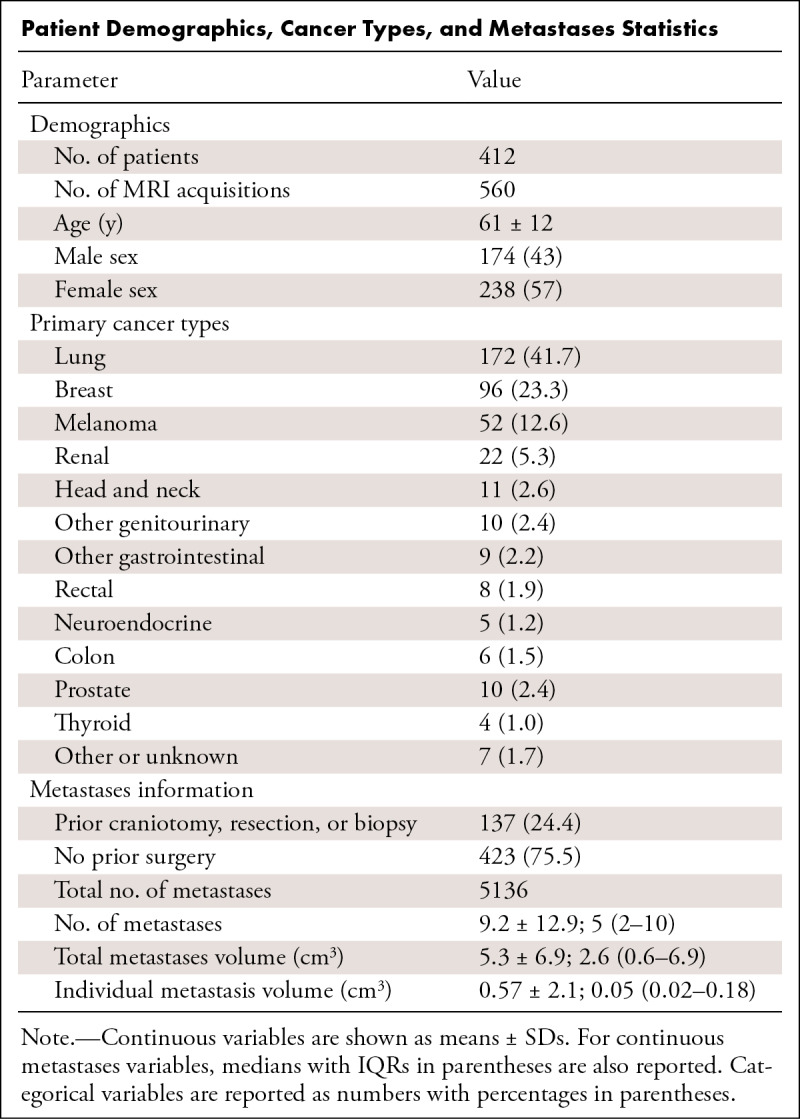
Data collection was performed in accordance with relevant guidelines and regulations and was approved by the UCSF institutional review board with a waiver for consent. The dataset consists of 560 brain MRI examinations from 412 patients (mean age, 61 years ± 12 [SD]; 238 female and 174 male patients [Table 1]) who were undergoing stereotactic radiosurgery planning at the UCSF medical center. MRI examinations were identified through a retrospective search of institutional radiology archives (mPower; Nuance Communications) of stereotactic radiosurgery studies performed between January 1, 2017, and February 29, 2020. Exclusion criteria from initially selected scans included patients without enhancing intracranial metastases (n = 26), only dural-based or leptomeningeal metastases (n = 9), or missing sequences or corrupted data (n = 8). This dataset overlaps with a prior study (8) but also includes the fluid-attenuated inversion recovery (FLAIR) sequence and skull stripping, noting that three examinations from that dataset were not included here given missing FLAIR sequences. Prior craniotomies for resections or biopsies were present in 137 (24.5%) of the scans, noting that none of the patients used in the test set (n = 99) underwent craniotomies. Of the total preoperative training images, 324 were included as an external dataset in the 2023 American Society of Neuroradiology–Medical Image Computing and Computer Assisted Intervention Society (ASNR-MICCAI) BraTS challenge (https://www.synapse.org/#!Synapse:syn51156910/wiki/622553 [9]; with corresponding subject identification in Table S1).
Patient Demographics, Cancer Types, and Metastases Statistics
Imaging Data Acquisition
T1-weighted spoiled gradient-echo (T1-pre), T1-weighted spoiled gradient-echo postcontrast (T1-post), and two-dimensional postcontrast T2 FLAIR images are included for each scan. The majority (373; 66%) of studies were acquired with a 1.5-T GE SignaHDxt (GE HealthCare) scanner, 149 (27%) were acquired with a 1.5-T Achieva (Philips Healthcare) scanner, and 38 (7%) were acquired with a 3.0-T GE Discovery MR750 (GE HealthCare). The scanner type, tesla strength, matrix size, in-plane axial dimension, and slice thickness for each scan are available in Table S1.
Although acquisition parameters varied slightly, representative values from a 1.5-T GE SignaHDxt scanner for the T1-pre and T1-post images were as follows: repetition time, 8.8 msec; echo time, 3.3 msec; inversion time, 450 msec; flip angle, 11°; matrix size, 256 × 256 × 106; and voxel size, 0.71 × 0.71 × 1.5 mm. Across all acquisitions, the T1-post image in-plane axial voxel dimension was less than 1 × 1 mm with a slice thickness of less than or equal to 1.5 mm in 498 images (89%). Representative values for the two-dimensional postcontrast FLAIR sequence were as follows: repetition time, 9175 msec; echo time, 109 msec; inversion time, 2600 msec; matrix size, 256 × 256 × 38; and voxel size, 0.98 × 0.98 × 5 mm.
Brain Metastases Annotations
Reference standard brain metastasis voxelwise segmentations were created using ITK-SNAP (http://www.itksnap.org/) (10) by one of two neuroradiology fellows (J.D.R. and B.L.) or one of two attending neuroradiologists (J.E.V.M. and L.P.S., with 5 and 4 years of experience as neuroradiology attending physicians). The associated final radiology report, containing a description of all the metastases, was used as a reference during the annotation process. The T1-pre, T1-post, and subtraction images were available to guide the manual segmentations. The enhancing portions of all metastases were segmented. Areas of T1 intrinsic hyperintensity and central necrosis were not included in the segmentation masks. In studies with resection cavities, the nodular enhancing portion of the resection cavities were included in the segmentation masks. Ninety-nine of the images, representing unique individuals previously used as the test set in a previous study (8), were segmented by both attending neuroradiologists (J.E.V.M. and L.P.S) without reference to the final radiology report. Final reference standard segmentations for these test images were generated by combining and refining the two segmentations with reference to the final radiology report.
Image Preprocessing
Images were first converted from Digital Imaging and Communications in Medicine format into Neuroimaging Informatics Technology Initiative (NIfTI) format using dcm2niix. The T1-pre and FLAIR images were registered into the T1-post space by rigid registration (six degrees of freedom) using the FMRIB Software Library tool, FLIRT (11). Subtraction images were generated by subtracting the registered T1-pre images from the T1-post images using fslmaths (11). For the purposes of protecting personal health information, all images were skull stripped using an in-house T1-post skull-stripping nnU-Net (https://github.com/rachitsaluja/UCSF-BMSR-benchmarks). No other image normalization, scaling or resizing, or atlas registration was performed prior to input into the neural network.
Size and Distribution of Brain Metastases
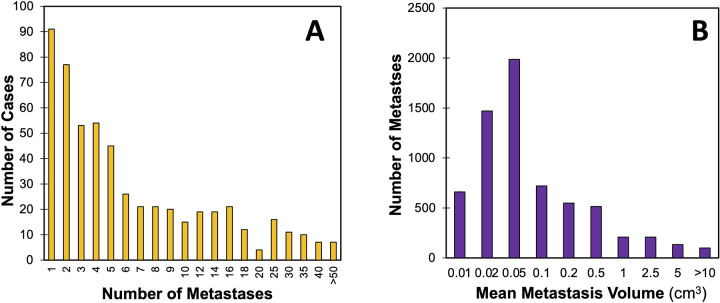
The Table provides an overview of patient characteristics and metastasis number and size distribution, with details for each case provided in Table S1. The most common types of primary cancers were lung cancer, breast cancer, and melanoma. A total of 5136 brain metastases were segmented in the 560 MRI examinations, with a mean of 9.2 metastases ± 12.9 and a median of five metastases (IQR, two to 11). The mean and median total metastasis volumes per MRI were 5.3 cm3 ± 6.9 and 2.6 cm3 (IQR, 0.6–6.9). The mean and median individual metastasis volumes were 0.57 cm3 ± 2.1 and 0.05 cm3 (IQR, 0.02–0.18). The distribution of metastases per scan and metastasis volumes are shown in Figure 1. Example data are shown in Figure 2.
Figure 1:
Distribution of number and size of brain metastases. Chart shows counts of (A) the number of manually segmented brain metastases per case and (B) the average brain metastasis volume.
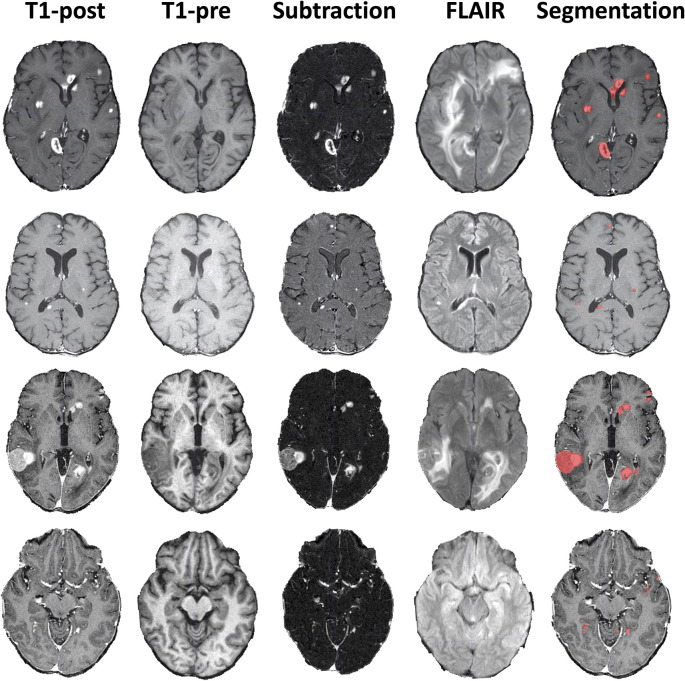
Figure 2:
Example brain metastases data. Four example MRI studies with axial T1 postcontrast images (T1-post), T1 precontrast images (T1-pre), subtraction images, postcontrast fluid-attenuated inversion recovery (FLAIR) images, and ground truth segmentations overlaid on the T1-post images.
Data and Code Availability
Currently, 461 MRI examinations and annotations from the training set are available to download under a noncommercial license at https://imagingdatasets.ucsf.edu/dataset/1. Data from the 99 individuals in the test set will become available after the completion of the 2024 MICCAI challenge.
Discussion
Automated detection and segmentation of brain metastases represents an ideal use case for the translation of artificial intelligence methods into clinical practice. Here, we present the publicly available UCSF-BMSR MRI Dataset which contains 560 multimodal MRI examinations with expert-annotations of over 5000 metastases and associated clinical and image information.
Other publicly available brain metastases datasets include Stanford BrainMetShare (https://aimi.stanford.edu/brainmetshare [6]), NYUmets (https://nyumets.org/ [12]), and the mathematical oncology laboratory [MOLAB] brain metastases dataset (https://molab.es/datasets-brain-metastasis-1) (13). The UCSF-BMSR contains the largest number of annotated brain metastases. The Stanford BrainMetShare contains 105 scans with annotations of 1517 metastases. While the NYUmets is the largest dataset with more than 8000 studies, only a subset of 2367 metastases are segmented as part of radiation treatment planning. The MOLAB dataset contains 637 MRI examinations but only has 593 semiautomatic segmentations available, which may be missing small metastases.
Data sharing efforts and competitions, such as those by BraTS and the Radiological Society of North America, have been critical for the advancement of biomedical image segmentation tasks as they provide large amounts of annotated data and a performance benchmark. The 2023 ASNR-MICCAI BraTS challenge on brain metastases (9) includes data from UCSF-BMSR, Stanford BrainMetShare, NYUmets, and several other sites. Larger and more heterogeneous datasets, including the ASNR-MICCAI BraTS challenge, should allow for improved algorithm performance and generalizability, particularly for difficult to detect small metastases, as well as a clearer comparison of different algorithms. This will be critical in promoting the clinical implementation of automated brain metastasis segmentation algorithms.
In summary, the integration of artificial intelligence tools into clinical workflows should allow for more rapid and precise quantitative assessments of disease burden, ultimately enabling more accurate and efficient diagnosis and treatment. The evaluation and treatment of brain metastases represents an excellent use case for which more public data are needed. We hope that releasing this dataset will lead to the development of algorithms with better segmentation performance and ultimately improved care of patients with intracranial metastatic disease.
Current address: Department of Radiology, Weill Cornell Medicine, Cornell University, New York, NY
Authors declared no funding for this work.
Disclosures of conflicts of interest: J.D.R. Grant from the American Society of Neuroradiology (ASNR Foundation grant in AI) paid to institution; consulting fees from Cortechs.ai; stock in Cortechs.ai and Subtle Medical; editorial board member for Radiology: Artificial Intelligence and the American Journal of Neuroradiology. R.S. No relevant relationships. D.A.W. No relevant relationships. P.N. No relevant relationships. E.C. Research grant from the American Society of Neuroradiology; royalties from Elsevier. J.B.C. No relevant relationships. B.L. No relevant relationships. J.M. Research grant from Siemens; royalties from GE paid through institution; support to Nuance Executive Client Council Meeting; chair of the RSNA Machine Learning Steering Committee. S.B. No relevant relationships. C.P.H. Consulting fees from GE HealthCare and as a member of the Kheiron Medical Technologies medical advisory board; participation of data safety monitoring boards for uniQure Biopharma, Asklepios BioPharmaceutical, and Focused Ultrasound Foundation; chair for the Foundation of the American Society of Neuroradiology. A.M.R. Grant from the Radiological Society of North America paid to institution; consulting fees from Arterys; support from UCSF for attending professional meetings. L.P.S. No relevant relationships. J.E.V.M. No relevant relationships.
Abbreviations:
- ASNR-MICCAI
- American Society of Neuroradiology–Medical Image Computing and Computer Assisted Intervention Society
- BraTS
- Brain Tumor Segmentation
- FLAIR
- fluid-attenuated inversion recovery
- T1-post
- T1-weighted spoiled gradient-echo postcontrast examination
- T1-pre
- T1-weighted spoiled gradient-echo examination
- UCSF-BMSR
- University of California San Francisco Brain Metastases Stereotactic Radiosurgery MRI Dataset
Keywords: Brain Metastases, Public Datasets, Artificial Intelligence, MRI
References
- 1. Clark K , Vendt B , Smith K , et al . The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository . J Digit Imaging 2013. ; 26 ( 6 ): 1045 – 1057 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baid U , Ghodasara S , Bilello M , et al . The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification . arXiv 2107.02314 [preprint] https://arxiv.org/abs/2107.02314. Published July 5, 2021. Accessed January 5, 2022. [Google Scholar]
- 3. Ostrom QT , Wright CH , Barnholtz-Sloan JS . Brain metastases: epidemiology . Handb Clin Neurol 2018. ; 149 : 27 – 42 . [DOI] [PubMed] [Google Scholar]
- 4. Arvold ND , Lee EQ , Mehta MP , et al . Updates in the management of brain metastases . Neuro Oncol 2016. ; 18 ( 8 ): 1043 – 1065 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mills SJ , Radon MR , Baird RD , et al . Utilization of volumetric magnetic resonance imaging for baseline and surveillance imaging in Neuro-oncology . Br J Radiol 2019. ; 92 ( 1098 ): 20190059 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grøvik E , Yi D , Iv M , Tong E , Rubin D , Zaharchuk G . Deep learning enables automatic detection and segmentation of brain metastases on multisequence MRI . J Magn Reson Imaging 2020. ; 51 ( 1 ): 175 – 182 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bousabarah K , Ruge M , Brand JS , et al . Deep convolutional neural networks for automated segmentation of brain metastases trained on clinical data . Radiat Oncol 2020. ; 15 ( 1 ): 87 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rudie JD , Weiss DA , Colby JB , et al . Three-dimensional U-Net Convolutional Neural Network for Detection and Segmentation of Intracranial Metastases . Radiol Artif Intell 2021. ; 3 ( 3 ): e200204 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moawad AW , Janas A , Baid U , et al . The Brain Tumor Segmentation (BraTS-METS) Challenge 2023: Brain Metastasis Segmentation on Pre-treatment MRI . arXiv 2306.00838 [preprint] https://arxiv.org/abs/2306.00838. Published June 1, 2023. [Google Scholar]
- 10. Yushkevich PA , Piven J , Hazlett HC , et al . User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability . Neuroimage 2006. ; 31 ( 3 ): 1116 – 1128 . [DOI] [PubMed] [Google Scholar]
- 11. Smith SM , Jenkinson M , Woolrich MW , et al . Advances in functional and structural MR image analysis and implementation as FSL . Neuroimage 2004. ; 23 ( Suppl 1 ): S208 – S219 . [DOI] [PubMed] [Google Scholar]
- 12. Oermann E , Link K , Schnurman Z , et al . Longitudinal deep neural networks for assessing metastatic brain cancer on a massive open benchmark . 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ocaña-Tienda B , Pérez-Beteta J , Villanueva-García JD , et al . A comprehensive dataset of annotated brain metastasis MR images with clinical and radiomic data . Sci Data 2023. ; 10 ( 1 ): 208 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Currently, 461 MRI examinations and annotations from the training set are available to download under a noncommercial license at https://imagingdatasets.ucsf.edu/dataset/1. Data from the 99 individuals in the test set will become available after the completion of the 2024 MICCAI challenge.