Abstract
Background.
Produce-associated outbreaks of Shiga toxin–producing Escherichia coli (STEC) were first identified in 1991. In April 2018, New Jersey and Pennsylvania officials reported a cluster of STEC O157 infections associated with multiple locations of a restaurant chain. The Centers for Disease Control and Prevention (CDC) queried PulseNet, the national laboratory network for foodborne disease surveillance, for additional cases and began a national investigation.
Methods.
A case was defined as an infection between 13 March and 22 August 2018 with 1 of the 22 identified outbreak-associated E. coli O157:H7 or E. coli O61 pulsed-field gel electrophoresis pattern combinations, or with a strain STEC O157 that was closely related to the main outbreak strain by whole-genome sequencing. We conducted epidemiologic and traceback investigations to identify illness subclusters and common sources. A US Food and Drug Administration–led environmental assessment, which tested water, soil, manure, compost, and scat samples, was conducted to evaluate potential sources of STEC contamination.
Results.
We identified 240 case-patients from 37 states; 104 were hospitalized, 28 developed hemolytic uremic syndrome, and 5 died. Of 179 people who were interviewed, 152 (85%) reported consuming romaine lettuce in the week before illness onset. Twenty subclusters were identified. Product traceback from subcluster restaurants identified numerous romaine lettuce distributors and growers; all lettuce originated from the Yuma growing region. Water samples collected from an irrigation canal in the region yielded the outbreak strain of STEC O157.
Conclusions.
We report on the largest multistate leafy greens–linked STEC O157 outbreak in several decades. The investigation highlights the complexities associated with investigating outbreaks involving widespread environmental contamination.
Keywords: outbreak, Escherichia coli, Romaine Lettuce, foodborne illness, produce safety
Produce-associated outbreaks of Shiga toxin–producing Escherichia coli (STEC) were first identified in 1991, and produce remains a common outbreak vehicle today [1, 2]. STEC is estimated to cause >265 000 infections in the United States annually, with 3600 hospitalizations and 30 deaths [3]. Most outbreaks of STEC infections are caused by STEC O157; 7% of STEC O157 outbreaks during 2003–2012 were associated with leafy greens [4, 5].
In 2006, spinach was implicated as the outbreak vehicle in a large, multistate outbreak of E. coli O157:H7 (STEC O157) [6]. This outbreak resulted in enhanced industry guidance to improve the safety of leafy green vegetables and was one of several factors driving implementation of the Food Safety Modernization Act (FSMA) in 2007 [6, 7].
Symptoms of STEC infection typically begin 2–5 days after infection and include severe diarrhea, often bloody, and abdominal pain. STEC O157 patients might also develop hemolytic uremic syndrome (HUS). Most cases of HUS in the United States result from an STEC O157 infection [8].
In April 2018, local, state, and federal officials began investigating a large multistate outbreak of STEC O157 infections, which was ultimately linked to romaine lettuce from the Yuma growing region. We describe the epidemiologic, laboratory, and traceback investigations that identified romaine lettuce as the source of this outbreak, and the public health implications of the investigation.
METHODS
Outbreak Detection
In April 2018, the New Jersey Department of Health and the Pennsylvania Department of Health notified the Centers for Disease Control and Prevention (CDC) of 2 STEC infection clusters. Before becoming ill, people in both clusters reported eating salads at a national restaurant chain. Pulsed-field gel electrophoresis (PFGE) showed that the STEC O157 isolated from ill people in both states had the same, rare PFGE pattern combination.
Case Identification
Cases were identified in PulseNet, the national molecular subtyping network for foodborne disease surveillance [9]. Cases were included in this outbreak that were related by PFGE or whole-genome sequencing (WGS). Cases were defined as infection with E. coli O157:H7 or E. coli O61 with 1 of the 22 outbreak PFGE pattern combinations, as 1 case-patient was coinfected with both STEC O157 and STEC O61, and 2 additional case-patients shared the same rare STEC O61 PFGE pattern (Table 1), and with illness onset occurring between 13 March 2018 and 22 August 2018. The isolates from these ill people were also found to be highly related (0–40 single-nucleotide polymorphisms [SNPs]) to each other by WGS and were subsequently included in the outbreak. Public heath scientists at federal and state laboratories performed WGS on clinical and nonclinical isolates, using standard methods. The Lyve-SET pipeline was used to perform high-quality SNP analysis using standard setting for E. coli and TW14359 as a reference with phages masked [10].
Table 1.
Breakdown of Pulsed-field Gel Electrophoresis Patterns and Corresponding Counts
| PFGE Pattern Combination | Total Isolate Count | Nonhuman Samples | Percentage of Total |
|---|---|---|---|
|
| |||
| A | 4 | 4 | 2.2 |
| B | 2 | 0 | 0.6 |
| C | 1 | 0 | 0.3 |
| D | 11 | 11 | 6.1 |
| E | 233 | 16 | 69.6 |
| F | 2 | 0 | 0.6 |
| G | 2 | 0 | 0.6 |
| H | 2 | 0 | 0.6 |
| I | 1 | 1 | 0.6 |
| J | 1 | 0 | 0.3 |
| K | 8 | 3 | 3.1 |
| L | 2 | 0 | 0.6 |
| M | 1 | 0 | 0.3 |
| N | 1 | 0 | 0.3 |
| O | 6 | 5 | 3.1 |
| P | 7 | 7 | 3.9 |
| Q | 4 | 4 | 2.2 |
| R | 3 | 0 | 0.8 |
| S | 1 | 1 | 0.6 |
| T | 4 | 4 | 2.2 |
| U | 1 | 1 | 0.6 |
| V | 2 | 2 | 1.1 |
Abbreviations: PFGE, pulsed-field gel electrophoresis.
Hypothesis Generation
Based on initial reports of food exposures, CDC developed a focused questionnaire, which asked case-patients detailed questions about consumption of leafy greens, where their household purchased groceries, restaurants visited, and specific menu items consumed in the week before illness onset. The proportion of case-patients reporting exposure to specific foods was compared to the proportion of healthy people reporting consumption of that food item in the 2006–2007 Foodborne Disease Active Surveillance Network (FoodNet) population survey [11]. This survey contains data on food consumption frequencies in healthy persons in the days prior to interview. A binomial probability distribution was used to identify food exposures that were reported by a significantly higher proportion of case-patients than healthy people.
Subcluster Identification
We defined outbreak subclusters as 2 or more unrelated case-patients who reported eating at the same restaurant location, different locations of the same restaurant chain, the same institutional setting, or shopping at the same grocery store in the week before becoming ill. These subclusters were used as starting points for traceback activities.
Product Traceback and Environmental Assessment
Locations, menus, meal items, meal dates, and invoices regarding restaurant and institutional subclusters were provided to CDC by state health departments. Individual shopper histories and receipts were also collected. These records were analyzed by the US Food and Drug Administration (FDA) to identify distributors, processors, and farms that supplied produce to the subcluster locations.
An environmental assessment (EA) was initiated in June 2018 to help determine the source of STEC contamination. The EA was conducted to assist FDA in identifying factors that potentially contributed to the introduction and spread of the outbreak strain of STEC O157 that contaminated products associated with this outbreak. The EA was focused on (but not limited to) agricultural water, soil amendments, growing and harvesting practices, animal intrusion, adjacent land use, concentrated animal feeding and manure composting operations, and employee health and hygiene practices [12]. Water, soil, manure, compost, scat, and other environmental samples were collected and analyzed for the outbreak strain of STEC O157 by culture; STEC isolates were genetically characterized by PFGE and WGS. The EA was completed with 2 subsequent trips and additional sampling.
Antimicrobial Susceptibility Testing
CDC’s National Antimicrobial Resistance Monitoring System (NARMS) performed antimicrobial susceptibility testing among selected clinical isolates using standard methods [13]. Drug resistance was defined as resistance to ≥1 antimicrobial, and multidrug resistance was defined as resistance to ≥1 antimicrobial in ≥3 drug classes [14]. We used WGS to identify virulence genes, and predicted resistance based on identification of known resistance genes.
RESULTS
Case Identification
We identified 240 case-patients from 37 states; 238 case-patients were infected with STEC O157, 1 case-patient was coinfected with STEC O157 and STEC O61, and 2 case-patients were infected with only STEC O61. Case-patients were aged 1–93 years (median, 26 years), and 66% (158/239) were female. One hundred four case-patients were hospitalized, 28 developed HUS, and 5 died (Figures 1 and 2). All outbreak isolates were related genetically by WGS (0–40 SNP differences). We identified 2 distinct subclades that included the majority of outbreak isolates, which had isolates with 0–19 SNP differences (Figure 3, clades A and B).
Figure 1.
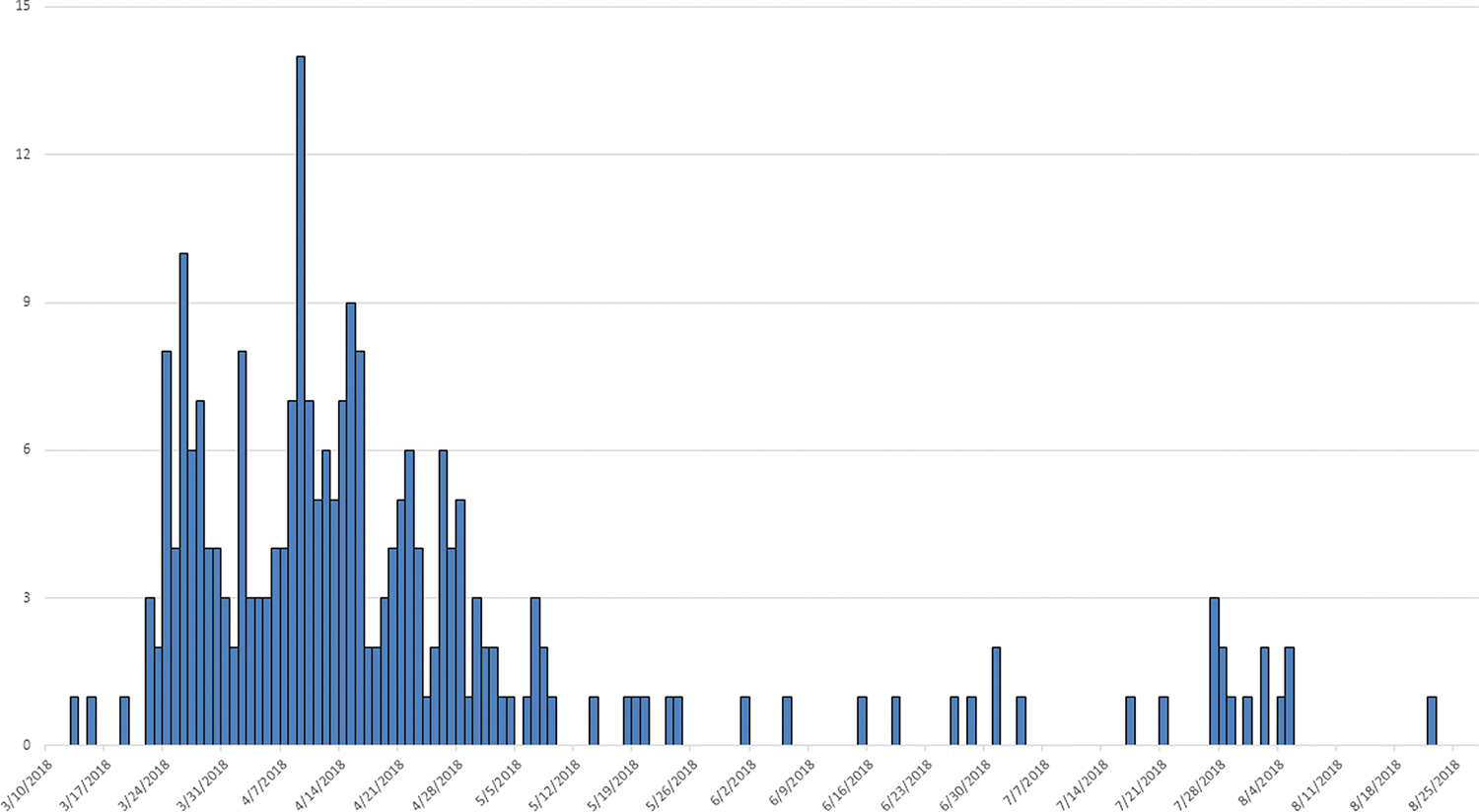
Number of case-patients, according to date of illness onset (month/day/year).
Figure 2.
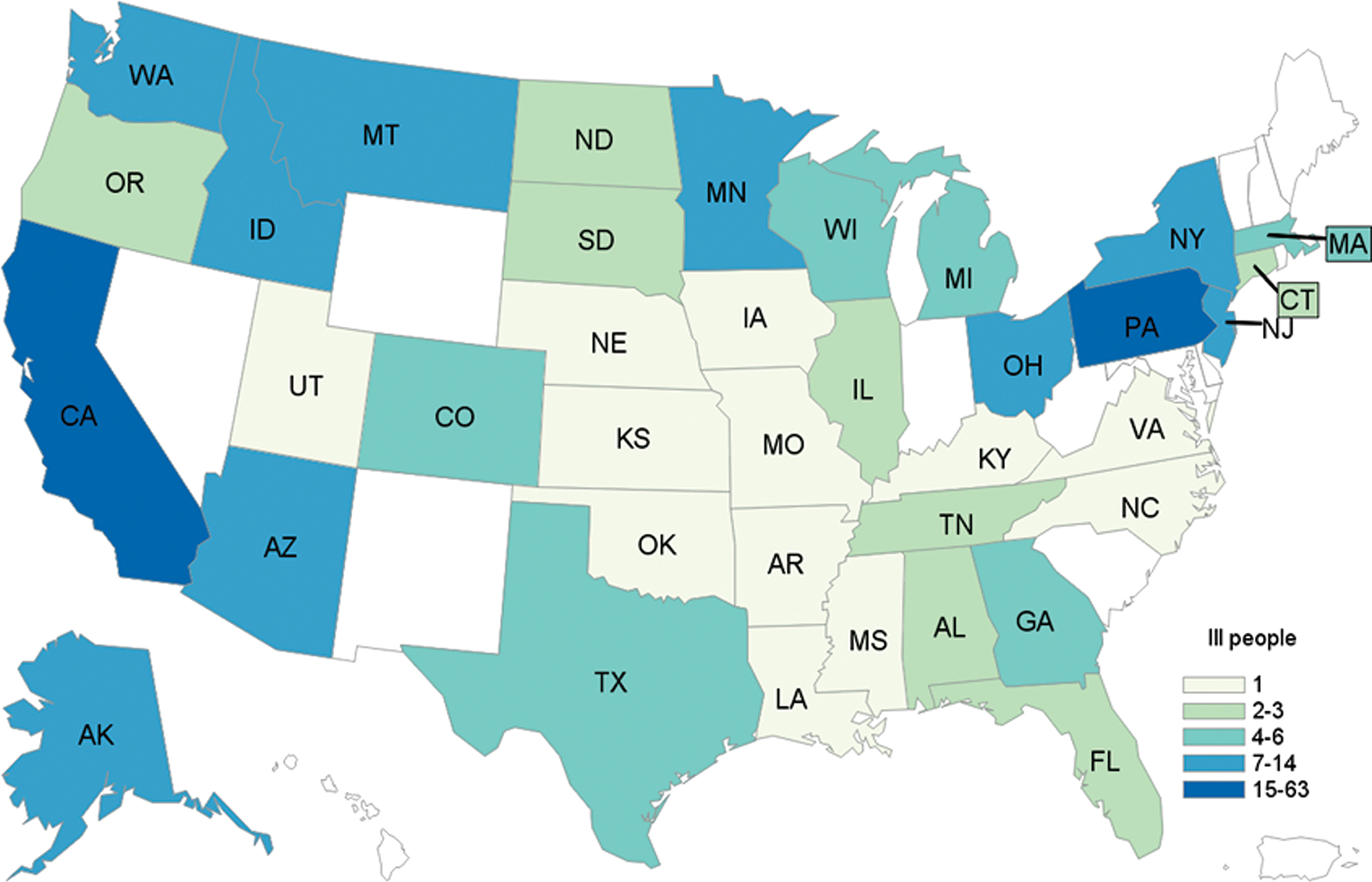
Number of case-patients, according to state of residence (n = 240). Abbreviations: AK, Alaska; AL, Alabama; AR, Arkansas; AZ, Arizona; CA, California; CO, Colorado; CT, Connecticut; FL, Florida; GA, Georgia; IA, Iowa; ID, Idaho; IL, Illinois; KS, Kansas; KY, Kentucky; LA, Louisiana; MA, Massachusetts; MI, Michigan; MN, Minnesota; MO, Missouri; MS, Mississippi; MT, Montana; NC, North Carolina; ND, North Dakota; NE, Nebraska; NJ, New Jersey; NY, New York; OH, Ohio; OK, Oklahoma; OR, Oregon; PA, Pennsylvania; SD, South Dakota; TN, Tennessee; TX, Texas; UT, Utah; VA, Virginia; WA, Washington; WI, Wisconsin.
Figure 3.
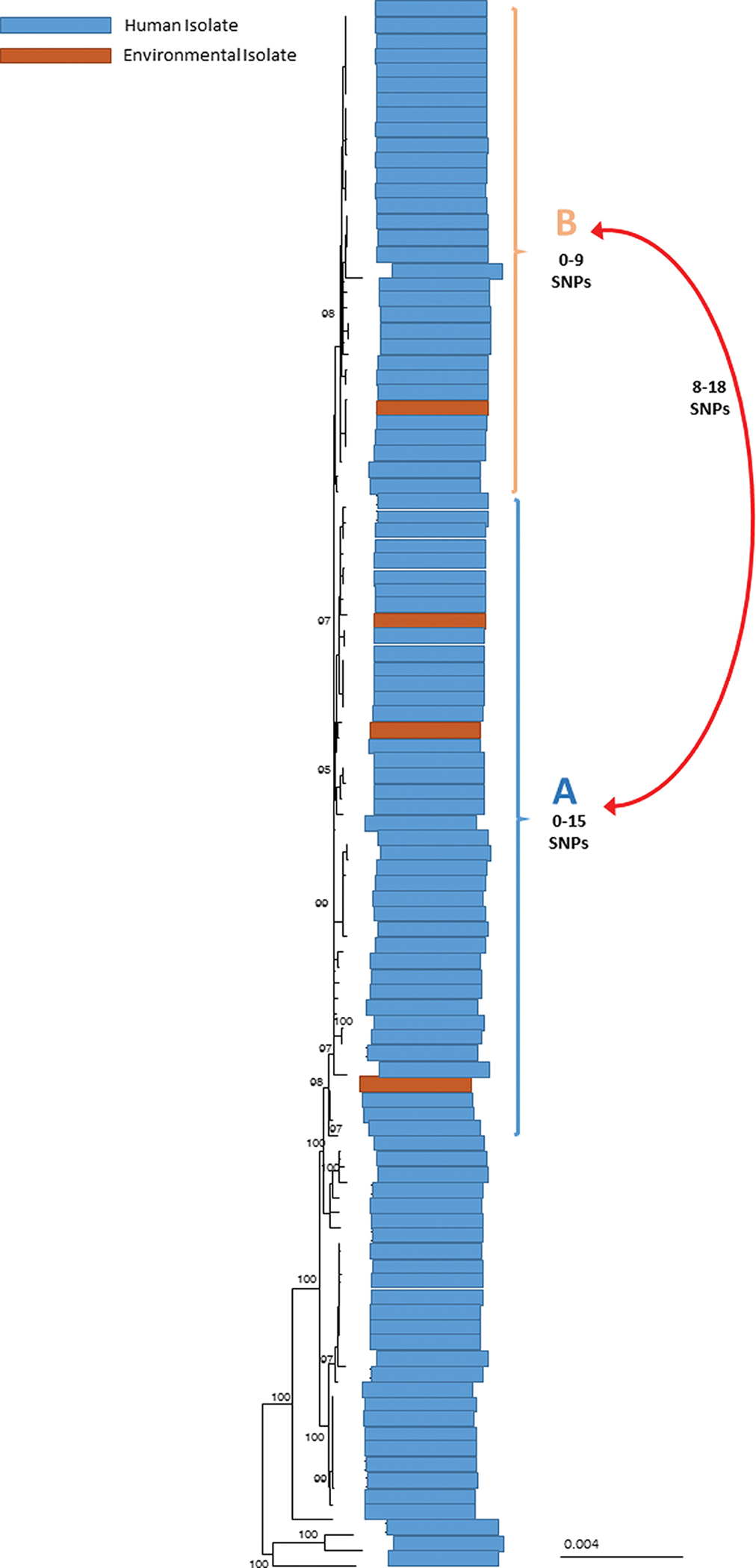
High-quality single-nucleotide polymorphisms (SNP) phylogenetic tree of Shiga toxin–producing Escherichia coli serogroup O157 isolates involved in the outbreak. Human and environmental isolates sequenced for this outbreak are shown above. Human isolates, denoted by blue bars, span the majority of the tree. Environmental isolates are shown in dark orange bars. Clades A and B contain the majority of isolates for this outbreak and differ by 8–18 SNPs. Environmental isolates fall into both clades A and B and are indistinguishable from human isolates.
Hypothesis Generation
Among 179 case-patients who completed the focused questionnaire, 85% reported consuming romaine lettuce in the week prior to illness onset, compared to 47% in the FoodNet Population Survey (P < .001).
Twenty illness subclusters, encompassing >50% of case-patients, were identified. The first subclusters were identified at multiple locations of a national restaurant chain by officials in New Jersey. Further investigation identified case-patients who had eaten at locations of the chain in 14 states over the course of the outbreak. Among case-patients who ate at this chain, 90% reported consuming salads containing prechopped romaine lettuce.
Idaho officials identified additional subclusters at 2 Idaho restaurants. Case-patients also reported eating salads containing prechopped romaine prior to illness onset. Alaska identified a subcluster of case-patients at a correctional facility where only whole-head romaine was served prior to illness onset, which was distinct from all other subclusters serving prechopped romaine.
Product Traceback and Environmental Assessment
Product traceback efforts by local health jurisdictions, state health departments, and FDA could not identify a single product lot, processor, or farm as the source of the entire outbreak. In total, 36 fields on 23 farms were identified during the traceback investigation (Figure 4). Only the Alaska correctional facility subcluster could be traced to a single farm. All of the romaine lettuce identified was traced to farms in the Yuma growing region. FDA, CDC, and state partners conducted an EA in the growing region, which consisted of farms in Imperial County, California, and Yuma County, Arizona.
Figure 4.
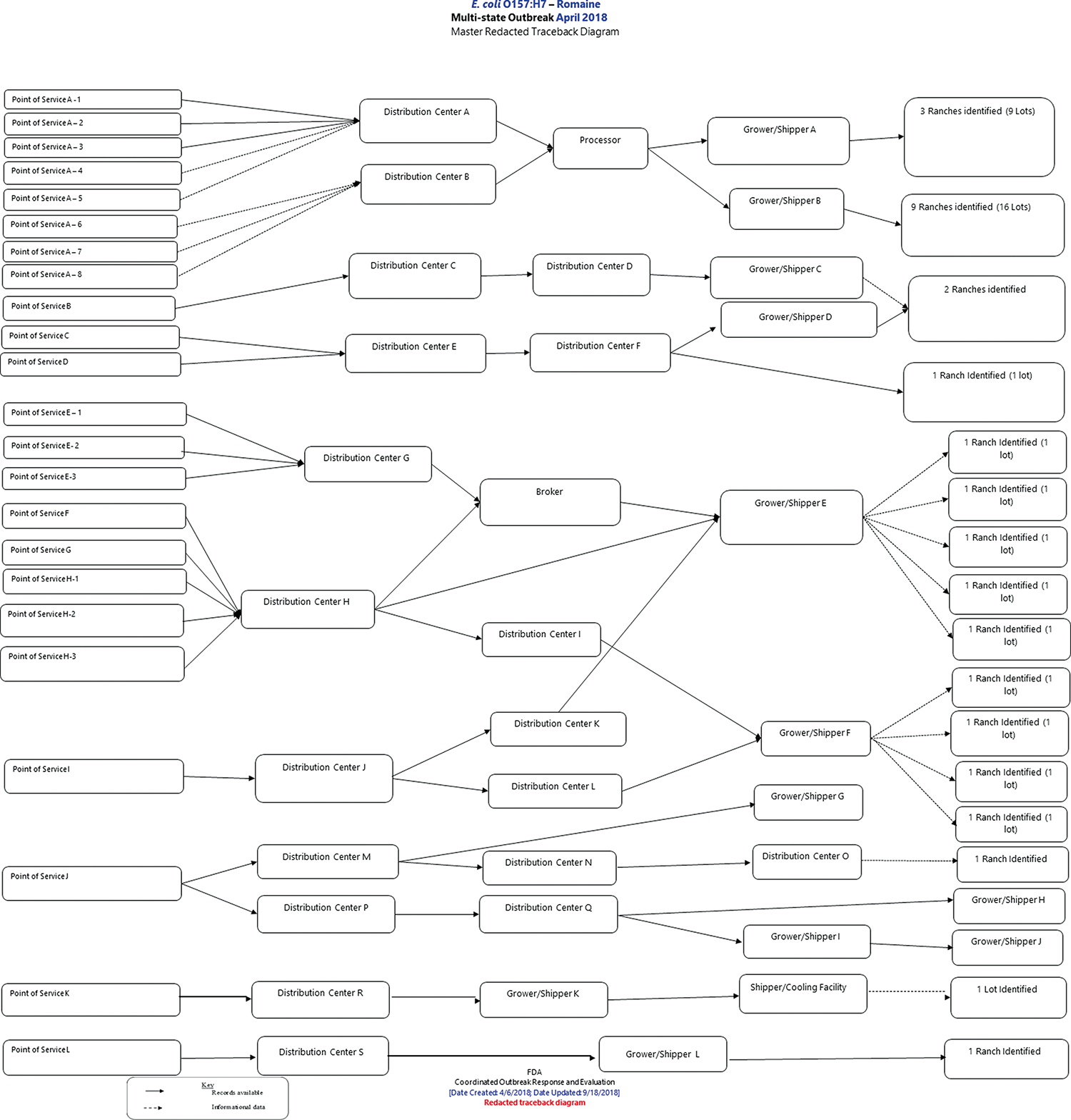
US Food and Drug Administration (FDA) master traceback diagram. Purchases of implicated products are traced from the point of service, through the distribution chain, to shippers. Product originates at the farm, denoted on the right side of the diagram.
In June 2018, STEC O157 with PFGE patterns indistinguishable from clinical outbreak isolates and closely related genetically to clinical isolates in both main clades (Figure 3) were detected in irrigation water from the Yuma growing region. The outbreak strains were detected in 3 separate water samples from sites along a 3.5-mile section of the Wellton irrigation canal that runs adjacent to romaine lettuce farms identified during traceback and next to a concentrated animal feeding operation (CAFO) with approximately 105 000 cattle. No STEC O61 was identified from the environmental assessment or water samples collected. No romaine lettuce was available for sampling during this outbreak as harvesting had ended in this region by the time the environmental assessment began.
Antimicrobial Susceptibility Testing
Whole-genome sequencing analysis of case-patient isolates (n = 184) and standard antibiotic resistance testing of case-patient isolates by CDC’s NARMS laboratory (n = 13) identified antibiotic resistance to chloramphenicol, streptomycin, sulfisoxazole, tetracycline, and trimethoprim-sulfamethoxazole. Ampicillin and ceftriaxone resistance genes were also identified in isolates from 4 case-patients. These findings did not affect treatment guidance as antibiotics are generally not recommended for patients with STEC O157 infections.
Public Health Actions
On 10 April 2018, CDC and FDA issued public notices indicating that a multistate STEC O157 outbreak of an unknown source was under investigation. Although the investigation had not yet identified a responsible food vehicle by 13 April, romaine lettuce from the Yuma growing region was the leading hypothesis due to identified subclusters and traceback efforts. Consequently, CDC and FDA advised consumers and retailers on this date not to eat, sell, or serve chopped romaine lettuce from the Yuma growing region. This warning was issued because contaminated lettuce was likely still available in consumers’ homes, stores, and restaurants, as the last harvest of romaine lettuce from Yuma shipped on 16 April 2018, and the product shelf life was 21 days. After the Alaska subcluster was identified, CDC recommended against consuming any romaine lettuce. The results from the environmental assessment, completed almost 2 months after romaine had been identified as the outbreak vehicle, solidified the messaging around romaine lettuce. In total, CDC and FDA issued 11 joint updates, resulting in >1.5 million page views to both agencies’ outbreak webpages.
DISCUSSION
Evidence collected during this investigation demonstrated that this large multistate outbreak was caused by contaminated romaine lettuce from the Yuma growing region. Epidemiologic data showed that a higher than expected proportion of case-patients consumed romaine lettuce prior to illness onset; traceback data showed that the romaine lettuce served at numerous illness subcluster locations was sourced from the Yuma growing region; and water testing identified the outbreak strain of STEC O157 in multiple locations of an irrigation canal that ran adjacent to several romaine fields identified during traceback.
This outbreak was the largest multistate STEC O157 outbreak in several decades, eclipsing in magnitude a 2006 outbreak linked to fresh spinach [6]. As there are an estimated 26 unreported illnesses for every STEC O157 case reported to PulseNet, the true size of this outbreak was likely much larger than the 240 illnesses reported through PulseNet, suggesting that thousands of people were actually sickened in this outbreak [3]. This outbreak was also much larger than any of the 28 multistate leafy green outbreaks reported from 1998 to 2016, which saw a mean size of 40 case-patients [15]. It is unclear why the size of the current outbreak exceeded that of previous similar outbreaks, but the volume of contaminated product may have played a role. This is supported by the fact that the outbreak strain was identified along multiple points of a 3.5-mile stretch of an irrigation canal and that traceback led to 36 romaine fields.
In addition to this outbreak being unusually large, the case-patient clinical course was unusually severe. The proportion of case-patients developing HUS (12.7%) was twice as high as previous outbreaks of STEC O157 (6.3%) [16]. There were 5 deaths in this outbreak (2.2%), which is almost 4 times higher than expected (0.6%) [16]. This could be explained by the strain’s stx2a toxin subtype, which produces more virulent toxins than other types. Outbreaks with stx2 toxin are more likely to result in increased rates of HUS [16]. Communicating unusually severe outcomes associated with STEC infections during an outbreak to healthcare providers is important. Clinicians should be made aware of ongoing outbreaks and test patients exhibiting diarrhea, especially bloody, with low or absent fever. Clinicians should not administer antibiotics to these patients as it increases the risk of severe outcomes, even if testing indicates the bacteria may be susceptible to antibiotics. Increased vigilance among children <5 years of age, adults 65 years and older, and immunocompromised patients can reduce the likelihood of progression to severe illness.
Creating clear and actionable messaging for consumers and retailers during this outbreak was challenging because a single source of contaminated romaine lettuce was never identified. Advice to consumers and retailers focused on avoiding romaine lettuce from a particular growing region, but leafy greens packaging rarely included growing region information, making it difficult to identify a specific product of concern. Many consumers and retailers may have simply avoided eating, selling, and serving any romaine lettuce during the outbreak because of this confusion.
Several factors complicated the traceback investigation during this outbreak. Many case-patients reported frequent consumption of romaine lettuce prior to their illness onset. Since the majority of traceable information was provided through restaurant or institutional subclusters, the case-patients had limited knowledge of the provenance of their romaine, including brand or harvest location, and traceback relied heavily on information provided from suppliers. Many of the restaurants identified during the traceback investigation were part of regional or national chains, which handled produce in regional distribution centers before shipment to specific retail locations. Due to this practice, these restaurants could not identify their specific romaine sources. Traceback was further complicated by the fact that a single production lot of chopped romaine might have contained lettuce from multiple farms. Although the FSMA Produce Safety Rule established specific record-keeping requirements for farms, records specific to traceability of produce along the supply chain are not required [17]. During this outbreak, leafy greens were not classified as an FDA high-risk food, thus not subject to enhanced record-keeping. Romaine served at the Alaskan correctional facility was the only product that was traced to a single farm due to the unique circumstance of being the only cluster that was associated with whole-head romaine lettuce from a single distributor to Alaska. In response to this outbreak, shipment origin labeling was voluntarily added to romaine packages by produce packers in December 2018. Increased detail about the growing region and limiting product lots to a single farm could alleviate the burden of traceback and identify a source more quickly in the future.
The ultimate source of STEC O157 for this outbreak is unknown, but reasonable hypotheses exist that explain how romaine lettuce could have become contaminated. The STEC O157 outbreak strain was detected at multiple points in a Yuma growing region irrigation canal, and the contaminated canal water may have subsequently contacted romaine lettuce in several ways, including direct application to the crop through aerial or land-based spray chemicals diluted with canal water. However, how and when STEC was introduced into the irrigation canal is unknown, as well as why the majority of illnesses were associated with romaine when other types of lettuce were grown in the same area during the time of interest. A CAFO was located adjacent to the irrigation canal where the outbreak strain was found, but no obvious route for contamination from the facility was identified. Environmental contamination could have been caused by the adjacent CAFO, as cattle are a well-documented reservoir for pathogenic STEC [18–20]. More research into the ecology of pathogenic E. coli in concentrated animal settings may help determine if the proximity of CAFOs to lettuce fields and produce irrigation water is a concern for the produce industry. After the outbreak, several task forces were formed to address problems identified, including the Leafy Greens Food Safety Task Force and the Romaine Task Force. The Leafy Greens Food Safety Task Force recommended increasing the buffer zones between CAFOs and romaine farms. The Romaine Task Force recommended traceability improvements in addition to changes in agricultural water practices. Ongoing work from both of these task forces will be used in the future to help prevent additional outbreaks.
CONCLUSIONS
Although this outbreak investigation conclusively identified romaine lettuce as the responsible food vehicle, questions remain about how to further enhance produce safety in the United States. This outbreak highlighted challenges in quickly determining the source and route of outbreak pathogens contaminating leafy greens, and difficulties in rapidly providing actionable guidance to consumers and retailers during leafy greens–associated outbreaks. This STEC O157 outbreak strain was first identified in humans in 2015, and in environmental samples as part of a recreational swimming–associated outbreak in a northern California lake in 2017. It is unclear how the contaminated water found in this previous outbreak might be linked to the contaminated Yuma growing region irrigation water identified during the current outbreak. This raises concern that longstanding environmental or zoonotic reservoirs exist for this STEC strain.
More research is needed to better understand STEC’s ecology in regions that grow ready-to-eat foods, including the role of CAFOs and other domesticated and wild animal populations. Efforts to assure agricultural water is monitored appropriately will also require ongoing evaluation and research. Improved labeling and record-keeping may also improve the speed and accuracy of future traceback investigations, especially for produce-associated outbreaks. Enhanced collaboration between health and agriculture officials, food industries, and academia is needed to fully understand the etiology of this outbreak and, more importantly, how to prevent other, similar outbreaks.
Acknowledgments.
The authors thank all of the outbreak investigation team members in jurisdictions affected by the outbreak who contributed to this investigation, including all local and state partners, partners who assisted with the environmental assessment, and partners at the US Food and Drug Administration and Centers for Disease Control and Prevention (CDC).
Footnotes
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC.
References
- 1.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg Infect Dis 2005; 11:603–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dewey-Mattia D, Manikonda K, Hall AJ, Wise ME, Crowe SJ. Surveillance for foodborne disease outbreaks—United States, 2009–2015. Morb Mortal Wkly Rep Surveill Summ 2018; 67:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 2011; 17:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heiman KE, Mody RK, Johnson SD, Griffin PM, Gould LH. Escherichia coli O157 outbreaks in the United States, 2003–2012. Emerg Infect Dis 2015; 21:1293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. National Shiga toxin–producing Escherichia coli (STEC) surveillance overview. Atlanta, GA: CDC, 2012. [Google Scholar]
- 6.Sharapov UM, Wendel AM, Davis JP, et al. Multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of fresh spinach: United States, 2006. J Food Prot 2016; 79:2024–30. [DOI] [PubMed] [Google Scholar]
- 7.US Food and Drug Administration. Food bill aims to improve safety. FDA consumer health information. 2010. Available at: http://nebula.wsimg.com/8ee97f7c3350a526ef70b45b3335f7a9?AccessKeyId=38B64D84C06ACB080889&disposition=0&alloworigin=1. Accessed 13 February 2019. [Google Scholar]
- 8.Centers for Disease Control and Prevention. General information: Escherichia coli. Available at: https://www.cdc.gov/ecoli/general/index.html. Accessed 13 February 2019.
- 9.Centers for Disease Control and Prevention. PulseNet home page. Available at: https://www.cdc.gov/pulsenet/. Accessed 13 February 2019.
- 10.Katz LS, Griswold T, Williams-Newkirk AJ, et al. A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Front Microbiol 2017; 8:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. Foodborne Diseases Active Surveillance Network (FoodNet): population survey. Available at: https://www.cdc.gov/foodnet/surveys/population.html. Accessed 13 February 2019.
- 12.US Food and Drug Administration. Environmental assessment; Yuma 2018 E. coli O157:H7 outbreak associated with romaine lettuce. Silver Spring, MD: FDA, 2018. [Google Scholar]
- 13.Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS). Available at: https://www.cdc.gov/narms/antibiotics-tested.html. Accessed 13 February 2019.
- 14.Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates surveillance report for 2015 (final report). Atlanta, GA: CDC, 2018. [Google Scholar]
- 15.Centers for Disease Control and Prevention. National Outbreak Reporting System (NORS) dashboard. Available at: https://wwwn.cdc.gov/norsdashboard/. Accessed 13 February 2019.
- 16.Gould LH, Demma L, Jones TF, et al. Hemolytic uremic syndrome and death in persons with Escherichia coli O157:H7 infection, Foodborne Diseases Active Surveillance Network sites, 2000–2006. Clin Infect Dis 2009; 49:1480–5. [DOI] [PubMed] [Google Scholar]
- 17.US Food and Drug Administration. Food Safety Modernization Act (FSMA) final rule on produce safety. Available at: https://www.fda.gov/Food/GuidanceRegulation/FSMA/ucm334114.htm. Accessed 13 February 2019.
- 18.Beauvais W, Gart EV, Bean M, et al. The prevalence of Escherichia coli O157:H7 fecal shedding in feedlot pens is affected by the water-to-cattle ratio: a randomized controlled trial. PLoS One 2018; 13:e0192149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karmali MA. Factors in the emergence of serious human infections associated with highly pathogenic strains of Shiga toxin–producing Escherichia coli. Int J Med Microbiol 2018; 308:1067–72. [DOI] [PubMed] [Google Scholar]
- 20.Wilson D, Dolan G, Aird H, et al. Farm-to-fork investigation of an outbreak of Shiga toxin–producing Escherichia coli O157. Microb Genom 2018; 4:e000160. [DOI] [PMC free article] [PubMed] [Google Scholar]


