Graphical abstract
Keywords: Mudskipper, Whole genome sequencing, Chromosome-level genome assembly, Comparative genomics, Water-to-land evolution, Terrestrial adaptation
Highlights
-
•
Provide chromosome-level genome assemblies for BP (Boleophthalmus pectinirostris) and PM (Periophthalmus magnuspinnatus).
-
•
Confirm two specific chromosome fission events in the PM genome by synteny comparison and ancestral chromosome reconstruction.
-
•
Reveal molecular clues from the loss of some important genes (such as SCPP and aanat1a) in certain or all of the three mudskippers for the water-to-land adaptation.
Abstract
Introduction
Mudskippers are a large group of amphibious fishes that have developed many morphological and physiological capacities to live on land. Genomics comparisons of chromosome-level genome assemblies of three representative mudskippers, Boleophthalmus pectinirostris (BP), Periophthalmus magnuspinnatus (PM) and P. modestus (PMO), may be able to provide novel insights into the water-to-land evolution and adaptation.
Methods
Two chromosome-level genome assemblies for BP and PM were respectively sequenced by an integration of PacBio, Nanopore and Hi-C sequencing. A series of standard assembly and annotation pipelines were subsequently performed for both mudskippers. We also re-annotated the PMO genome, downloaded from NCBI, to obtain a redundancy-reduced annotation. Three-way comparative analyses of the three mudskipper genomes in a large scale were carried out to discover detailed genomic differences, such as different gene sizes, and potential chromosomal fission and fusion events. Comparisons of several representative gene families among the three amphibious mudskippers and some other teleosts were also performed to find some molecular clues for terrestrial adaptation.
Results
We obtained two high-quality haplotype genome assemblies with 23 and 25 chromosomes for BP and PM respectively. We also found two specific chromosome fission events in PM. Ancestor chromosome analysis has discovered a common fusion event in mudskipper ancestor. This fusion was then retained in all the three mudskipper species. A loss of some SCPP (secretory calcium-binding phosphoprotein) genes were identified in the three mudskipper genomes, which could lead to reduction of scales for a part-time terrestrial residence. The loss of aanat1a gene, encoding an important enzyme (arylalkylamine N-acetyltransferase 1a, AANAT1a) for dopamine metabolism and melatonin biosynthesis, was confirmed in PM but not in PMO (as previously reported existence in BP), suggesting a better air vision of PM than both PMO and BP. Such a tiny variation within the genus Periophthalmus exemplifies to prove a step-by-step evolution for the mudskippers’ water-to-land adaptation.
Conclusion
These high-quality mudskipper genome assemblies will become valuable genetic resources for in-depth discovery of genomic evolution for the terrestrial adaptation of amphibious fishes.
Introduction
Water-to-land adaptation is one of the most significant events in the evolutionary history of vertebrates. Some teleost fishes had ventured out of water to evolve an amphibious life. Mudskippers, belonging to Oxudercidae of Gobiidae family, are the largest group of such fishes that can adapt to live on mudflats [1]. They have great amphibious capacities, including breathing air [2], aerial vision [3] and walk-like behavior on land [4], [5]. They are a valuable model for comparative studies on the developmental adaptation of vertebrates from water to land.
Mudskippers include ten genera (such as Boleophthalmus, Periophthalmus, Periophthalmodon, Scartelaos, Zappa, and Pseudapocryptes) and 42 species [6]. Boleophthalmus pectinirostris (BP), presenting grey skin with a great number of small and white spots, is widely distributed in coastal mud flats of China and Japan [6]. Periophthalmus magnuspinnatus (PM), owning light brown skin with sky blue spots on its cheeks, opercula, flanks and head, commonly lives in mid-higher intertidal zones of south Korea and the South China Sea [7]. P. modestus (PMO) has grey skin with some dusky stripes and tiny black spots [5]; it is widespread across temperate and tropical intertidal zones, including the northwestern Pacific Ocean from Vietnam to Korea, as well as Japan [6]. Their terrestrial adaptation abilities are largely diverse. For instance, BP takes less time out of water, whereas PM and PMO have more residential time on their terrestrial habitats [8], [9].
In 2014, our team reported the first genome dataset of four mudskipper species by using the Illumina sequencing [8]. These genome assemblies contained long scaffolds but relatively low contigs, without chromosomal details [8]. Yang’s team recently assembled a chromosome-level genome for PMO with 23 chromosomes, which was sequenced by Illumina, PacBio, 10X and Hi-C (high-through-put chromosome conformation capture) methods [9]. However, this PMO annotation has too much redundancy, leading to over 30,000 genes with 10% BUSCO duplicates.
Here, we provided two improved chromosome-level genome assemblies for BP and PM by integrating PacBio, Nanopore, Hi-C sequencing and our previous Illumina [8] data. We also reannotated the PMO genome data [9] to obtain a redundancy-reduced annotation set. This PMO genome resource will be valuable for us to reveal genomic divergence among Periophthalmus species. One of our main goals is to perform three-way comparative analyses of BP, PM and PMO genomes to discover their detailed genomic differences, such as gene changes and chromosomal structure variations. The three chromosome-level assemblies can also help us to construct the mudskipper ancestor genome karyotype. Several representative gene families, such as SCPP (encoding secretory calcium-binding phosphoproteins) and aanat (encoding an important enzyme, arylalkylamine N-acetyltransferase (AANAT), for dopamine metabolism and melatonin biosynthesis [8]; classified into aanat1a, aanat1b and aanat2 in various fishes), among three mudskippers and some other teleosts were compared to reveal some molecular clues for the terrestrial adaptation. Thus, these high-quality mudskipper genome assemblies could be used as good genetic materials for in-depth genomic mining of amphibious fishes during the water-to-land evolution.
Material and methods
Ethics statement, sample collection, and whole genome sequencing
Wild samples of BP (female) and PM (female) were collected from Shenzhen Bay in Shenzhen city, Guangdong province, China. Animal experiments in this study were carried out according to the guidelines of the Animal Ethics Committee of BGI, and they were approved by the Institutional Review Board on Bioethics and Biosafety of BGI (No. 18134). Genomic DNAs of BP and PM were extracted separately from muscle tissues by using a DNeasy Nucleic Acid Kit (Qiagen, Germantown, MD, USA) under the manufacturer’s instructions. The routine strategy of whole genome shotgun sequencing was used to sequence both BP and PM genomes. A SMART Bell library of BP genome with an insert size of 20 kb was constructed based on a PacBio RS II protocol (Pacific Biosciences, Menlo Park, CA, USA), and then it was sequenced by a PacBio SEQUEL II platform. A Nanopore library of PM was processed by using the Ligation Sequencing 1D kit (SQKLSK109, Oxford Nanopore Technologies, Oxford, UK) in accordance with the manufacturer’s instructions, and it was subsequently sequenced by a PromethION instrument (Oxford Nanopore Technologies).
Genome size prediction, genome assembly and evaluation
A 17-mer frequency distribution was employed to predict genome size with the following formula: genome size = Knum / Kdepth, where the Knum represents 17-mer number generated from sequencing reads, and the Kdepth is the peak depth of k-mers at the peak frequency. Reads from Illumina short-insert libraries (500 and 800 bp; from our previous study [8]) were utilized for this prediction.
For the assembly of BP and PM genomes, Platanus (version 1.2.4) [10] with defaulted parameters was employed to generate a de novo assembly by using clean Illumina reads from the short-insert libraries (250, 500 and 800 bp; Tables S1 and S2) from our previous report [8]. Subsequently, PacBio and Nanopore reads of BP and PM respectively (Tables S3 and S4) along with above assembled contigs were used for further assembly by employing the DBG2OLC pipeline (default version) [11] with the following parameters: LD10, MinLen 200, KmerCovTh 6, MinOverlap 80, AdaptiveTh 0.012, and RemoveChimera 1. The PacBio and Nanopore reads of BP and PM were then mapped onto the DBG2OLC assembled contigs by using Minimap2 [12] with default parameters. The DBG2OLC contigs were further corrected with six rounds by Racon (v1.2.1) [13] based on above Minimap2 alignments. After correction, all clean Illumina reads of BP and PM were separately mapped onto the Racon corrected contigs by BWA-0.7.17 [14] and further corrected by Nextpolish [15] with default parameters. SSPACE-LongRead [16] with default parameters was applied to build scaffolds based on above-mentioned Nextpolish contigs and long reads. The BUSCO (Benchmarking Universal Single-Copy Orthologs; University of Geneva Medical School and Swiss Institute of Bioinformatics, Geneva, Switzerland; version 5.22) [17] with actinopterygii_odb10 orthologues was used to evaluate the completeness of BP and PM genome assemblies.
Chromosome construction
Muscle samples from BP and PM individuals were separately fixed by formaldehyde. The restriction enzyme (Mbo I) was used for digestion of extracted genomic DNAs by repairing of the 5′ overhangs with a biotinylated residue. Both Hi-C paired-end libraries of BP and PM were produced, and they were then sequenced by a Hiseq Xten platform (Illumina, San Diego, CA, USA). Hi-C reads were filtered by using the Soapnuke v1.6.5 [18] with parameters: -n 0.01 -l 20 -q 0.1 -i -Q 2 -G -M 2 -A 0.5 -d. These Hi-C reads were aligned onto the assembled scaffolds by using Bowtie2 [19]. And then, scaffold linkage information from the Hi-C pair data were produced by HiC-Pro v2.8.0 [20] with default parameters. Juicer v1.5 [21] and 3d-DNA v170123 [22] were employed to link scaffolds to form chromosomes. Juicebox v1.11.08 [23] was applied to fix error-joins and remove duplicated contigs to draw Hi-C heatmaps.
Repeat annotation and gene structure prediction
Both de novo and homology repetitive identification methods were used to detect repetitive sequences in BP and PM genomes. For the de novo repeat annotation, RepeatModeler v1.0.8 [24] and LTR-FINDER v1.0.6 [25] with default parameters were employed to identify each type of repetitive sequences. Subsequently, RepeatMasker v4.0.6 [26] was used to build a new library based on the Repbase TE v21.01 [27]. Tandem elements were identified by Tandem Repeats Finder [28]. For the homology prediction, RepeatMasker v4.0.6 [26] and RepeatProteinMask v4.0.6 [26] were applied to find repeat sequences in the BP and PM genome assemblies based on the repeat libraries from the de novo repeat annotation.
For analysis of LTR (long terminal repeat) insertion time, pairs of LTRs with complete 5′ and 3′ ends from the LTR-FINDER results were collected, and then they were self-aligned using MUSCLE (V3.8.31). The nucleotide distance K values from each pair of LTRs were calculated by using the Kimura two-parameter model from the EMBOSS package [29]. The LTR insertion time (T) was calculated according to the following formula T = K / 2r, where the mutation rate (r) was set as 3.51 × 10−9 substitution rate per base per year [30].
Homology, de novo and transcriptome annotation pipelines were employed to identify gene structures in BP and PM genomes. For the homology annotation, protein sequences of five representative species, including Danio rerio (zebrafish), Gasterosteus aculeatus (threespine stickleback), Homo sapiens (human), Oryzias latipes (medaka), and Tetraodon nigroviridis (green spotted puffer), were downloaded from the NCBI database (release 95). Each protein set was mapped onto the BP and PM assemblies by tBLASTn [31] with an e-value ≤ 10−5, respectively. Subsequently, GeneWise v2.4.1 [32] was employed to identify gene structures on the tBLASTn alignments. For the transcriptome annotation, pooled RNA-seq reads were mapped onto the assembly by using HISAT2 v2.0.4 [33]. Cufflinks v2.2.1 (https://cole-trapnell-lab.github.io/cufflinks/) was employed to identify gene structures on above RNA-seq alignments. For the de novo annotation, Augustus v3.3.3 [34] was used to predict potential gene structures in BP and PM genomes. Three gene sets from above-mentioned three annotation approaches were merged by MAKER [35] to generate final non-redundant gene sets for BP and PM respectively. Similarly, we used same approaches to annotate the assembled PMO genome (related data were downloaded from the GigaScience database of Yang’s study [9]).
Genome alignments among the three mudskippers
The chromosome assembly of PMO (downloaded from NCBI), the protein sequences of PMO (annotated by us in this study), and chromosome assemblies and protein sets of both BP and PM were used as the input data for genomic comparisons. Whole genome and protein alignments among these three mudskippers were performed by i-ADHoRe v3.0 [36] with default parameters based on protein identity and gene synteny. Highly conserved synteny and strict correspondence of chromosomes were displayed by using SVG in Perl. Detailed chromosome fissions, fusions, and translocations were manually checked.
Reconstruction of ancestral chromosomes
Each protein set of three mudskippers and stickleback was aligned to the protein set of previously predicted ancestor chromosomes [37], [38] respectively by using BLASTp (e-value < 1 × 10−10). We then identified the reciprocal best-hit alignments between each mudskipper and the ancestor chromosomes. Finally, chromosomal fissions and fusions were determined by using SVG in Perl.
Phylogeny, divergence time, and gene expansion/contraction analyses
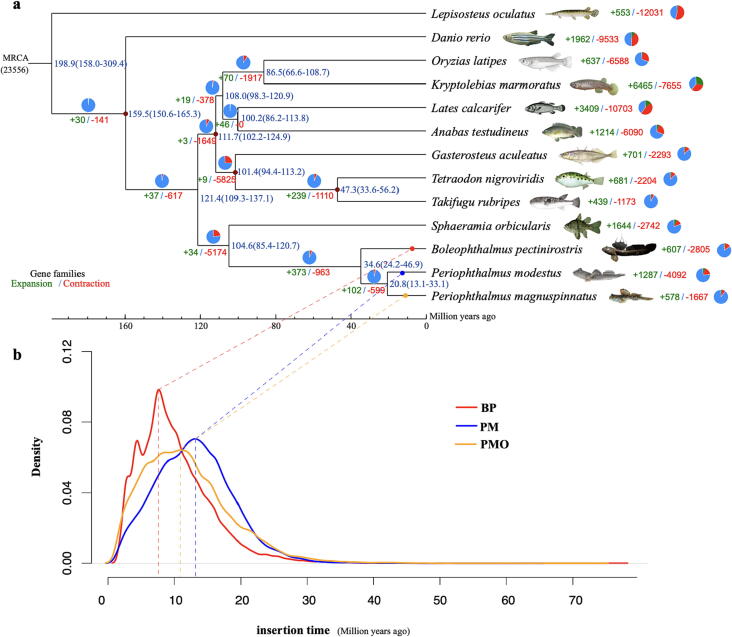
Genome data and protein sets of ten representative teleost species were download from NCBI for a phylogenomic analysis, including Lepisosteus oculatus (spotted gar; GCF_000242695.1), Danio rerio (zebrafish; GCF_000002035.6), Oryzias latipes (medaka; GCF_002234675.1), Kryptolebias marmoratus (Mangrove killifish; GCF_001649575.2), Lates calcarifer (Asian seabass; GCF_001640805.1), Anabas testudineus, (climbing perch; GCF_900324465.2), Gasterosteus aculeatus (threespine stickleback; GCF_016920845.1), Tetraodon nigroviridis (green spotted puffer; GCA_000180735.1), Takifugu rubripes (Japanese pufferfish; GCF_901000725.2) and Sphaeramia orbicularis (orbiculate cardinalfish; GCF_902148855.1). Protein sequences and protein sets of the three mudskippers were aligned with each other by using BLASTp [39] (e-value ≤ 1e-5). OrthoMCL v2.0.92 [40] with default parameters was employed to cluster gene families. The detailed number of multiple-copy, single-copy orthologs, and unique paralogs were summarized in Table S5. We then aligned those single-copy gene families using MUSCLE v3.8.31 [41] and connected the coding sequence (CDS) regions of all single-copy genes. Gblock [42] was applied to determine the conserved regions for constructing a phylogenetic tree by using PhyML v3.0 with the maximum likelihood method [43].
A divergence time analysis was performed by MCMCtree in the PAML package [44]. Fossil records [45] were used to calibrate four branch nodes. In addition, we predicted expansion and contraction gene families by using CAFE v4.2.1 [46], and then annotated these gene families based on Gene Ontology (GO) [47] terms and KEGG [48] pathways (Fig. S1).
Identification of aanat1a and its neighboring genes
Zebrafish genome (GRCz11, GCF_000002035.6) was used as the reference (Table S6) to identify melatonin biosynthesizing genes in three mudskippers. We first employed tBLASTn [31] to search separately against three mudskipper genome assemblies, and then retained the high-identity regions by using Solar v0.9.6 [49], followed by the extraction of exon sequences using Exonerate v2.4 [50].
In order to verify the loss of aanat1a in PM [8], we searched protein sequences of aanat1a and 20 nearby genes to detect the synteny correlations among three mudskipper genomes. BLASTp [31] was performed for the aanat1a along with the adjacent upstream ten genes (including pex6, atl2, acox1, slc22a7, ttbk1a, ttbk1b, ddx5, polg2, srp68, and evpl) and ten downstream genes (ube2o, sphk1, cygb2, myh10, map2k7, pkn2, c19orf67, rh2a, mcoln1, and trappc5) of the aanat1a gene (Table S7). Genomic locations of these genes were collected, and a synteny plot was drawn using the SVG module in Perl.
To obtain a broader view of this region, we aligned the corresponding nucleotide sequences of these target regions among the three mudskipper genomes using LASTZ v1.04.15 [51], and a synteny plot was generated by CIRCOS v.0.69 [52]. Entire chromosomes containing aanat1a genes were also aligned and plotted using the LAST package [53].
Identification of SCPP gene family
The same pipeline was employed to identify the less conserved SCPP genes in three mudskipper genomes, and the following two methods were also used so as to generate a full list of SCPP genes. First, the SCPP proteins of both zebrafish and spotted gar [54], [55] were aligned and indexed by HMMER v3.3.2 [56], and hmmsearch was conducted against the three mudskipper protein sets separately. Secondly, we applied published transcriptome assemblies [8] as the subjects, and those known SCPP proteins as the queries. BLASTp was performed and the EMBOSS tool [29] was then used to predict open reading frames of those aligned mRNA sequences, which were further fed as inputs in Exonerate to extract gene structures from the three mudskipper genome assemblies. All predicted SCPP genes were concatenated and those replicates were discarded manually.
Results
Summary of the three mudskipper genome assemblies
We conducted whole genome sequencing and annotation of both BP and PM genomes, and we also reannotated the PMO genome from Yang’s data [9]. Approximately 39.3 gigabases (Gb) and 55.8 Gb of raw reads were produced from PacBio and Nanopore platforms for BP and PM, respectively. After a series of assembly procedures, we generated genome assemblies for both BP and PM with a total length of 957.8 Mb and 753.0 Mb, respectively. These lengths are close to the estimated values (973.0 Mb for BP and 773.0 Mb for PM; Table 1). Their detailed contig N50 values are 1.1 Mb (BP) and 2.5 Mb (PM), and their BUSCO completeness are about 93.8% and 93.3% respectively (Table 1).
Table 1.
Statistics of the three mudskipper genomes.
| species | B. pectinirostris (BP) | P. magnuspinnatus (PM) | P. modestus (PMO) |
|---|---|---|---|
| Common name | Blue-spotted mudskipper | Giant-fin mudskipper | Shuttles hoppfish |
| Estimated genome size | 973.0 Mb | 773.0 Mb | 729.0 Mb |
| Assembly genome size | 957.8 Mb | 753.0 Mb | 854.4 Mb |
| Scaffold N50 | 40.7 Mb | 32.0 Mb | 32.9 Mb |
| Contig N50 | 1.1 Mb | 2.5 Mb | 0.6 Mb |
| Gene Numbers | 22,685 | 22,272 | 29,442* |
| Repeat elements | 47.4% | 40.4% | 43.8% |
| Genome BUSCO | C:93.8% [S:90.9%, D:2.9%], F:2.0%, M:4.2% | C:93.3% [S:90.5%, D:2.8%], F:2.4%, M:4.3% | C:90.4% [S:89.1%, D:1.3%], F:0.8%, M:8.8% |
| Gene BUSCO | C:96.8% [S:92.9%, D:3.9%], F:1.4%, M:1.8% | C:96.1% [S:92.6%, D:3.5%], F:1.9%, M:2.0% | C:82.4% [S:80.1%, D:2.3%], F:3.4%, M:14.2%* |
These annotation and evaluation data, performed through our pipelines in this study, are improved from those in Yang’s study [9].
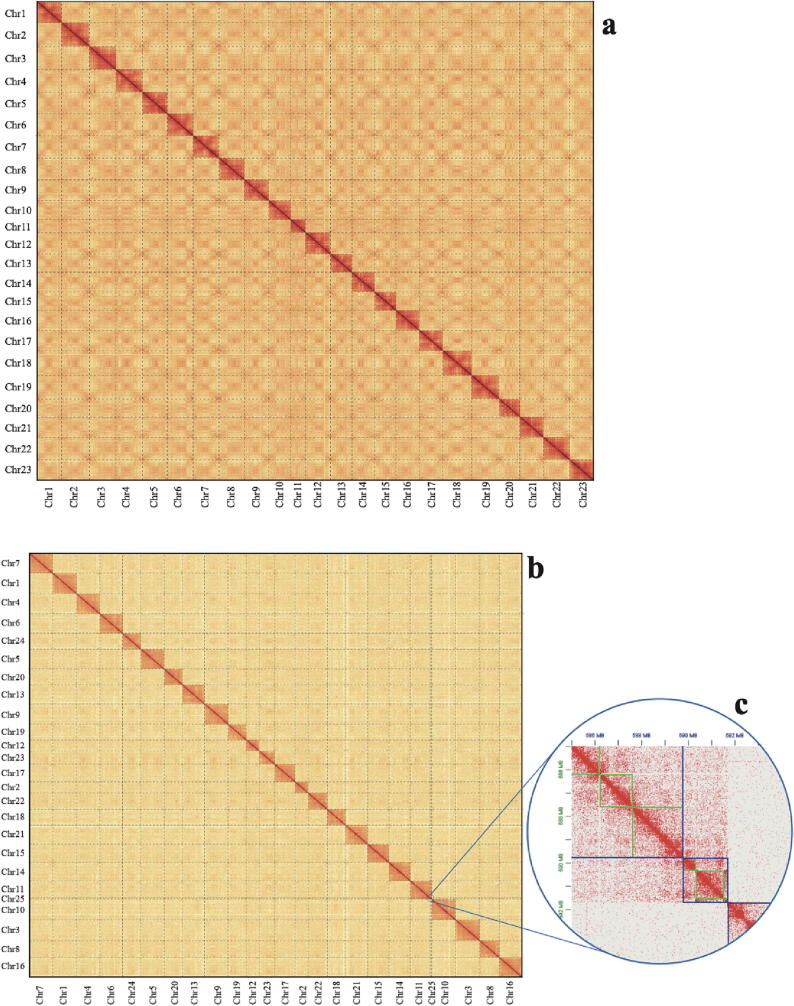
We subsequently anchored their haplotype contigs into 23 and 25 chromosomes (Chr) for BP and PM respectively (Fig. 1a, b). We also performed a series of annotation pipelines to predict gene sets for BP, PM and PMO, and obtained 22,685, 22,272 and 29,442 protein-coding genes with completed BUSCO values of 96.8%, 96.1% and 82.4%, respectively (Table 1). For repeat annotation, we predicted that repeat sequences account for about 47.4%, 40.4% and 43.8% of BP, PM and PMO genomes respectively. Interestingly, BP has the highest ratio of repeats among the three mudskippers, while it is the least terrestrial fish.
Fig. 1.
The Hi-C heatmaps of BP (a) and PM (b) haplotype genomes. The picture in (c) is enlarged of the chromosome 25. Blue boxes represent chromosomes, while green boxes stand for scaffolds. The red plots mark those mapped Hi-C reads. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Genome synteny and ancestor chromosome reconstruction
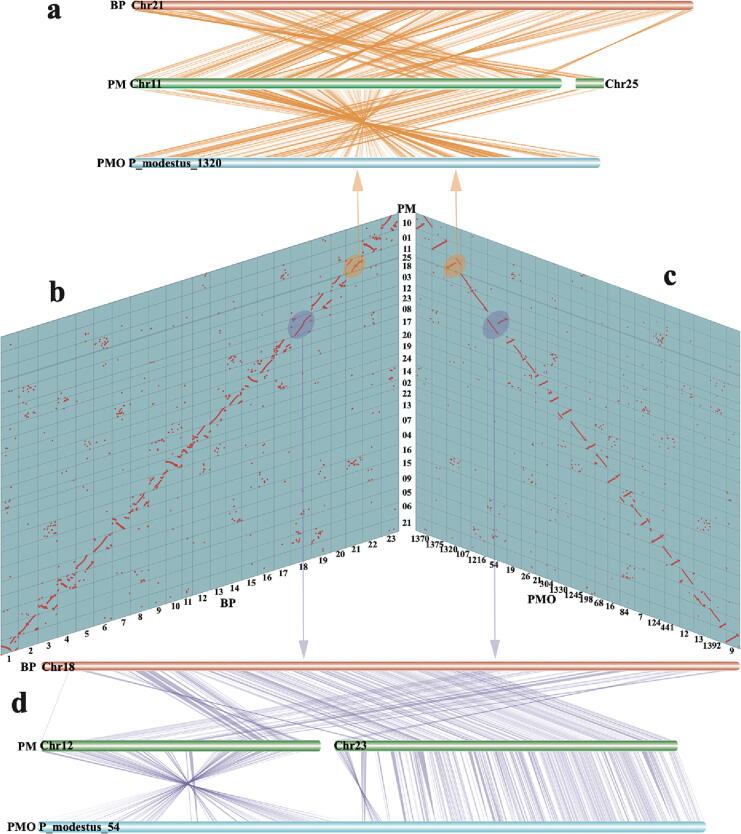
Genome synteny demonstrated that BP chromosomes are one-to-one aligned to PMO chromosomes with identification of 1,912 synteny blocks (Fig. S1). Interestingly, PM has two more chromosomes than BP and PMO, due to chromosome fissions. Detailed synteny shown that Chr11 and Chr25 of PM were well aligned to Chr21 of BP and P_modestus_1320 of PMO (Fig. 2a, b), and Chr12 and Chr25 matched to Chr18 of BP and P_modestus_54 of PMO (Fig. 2b, c). Therefore, we speculate that the two chromosomal breakages were solely presented in the PM lineage after the divergence of PM and PMO.
Fig. 2.
Chromosomal synteny relationships among the three mudskippers. a An amplified synteny block view among BP Chr 21, PM Chr 11 & Chr 25, and PMO P_modestus_1320. b-c Synteny blocks of various chromosomes among BP, PM, and PMO. d An amplified synteny block view among BP Chr 18, PM Chr 12 & Chr 23, and PMO P_modestus_54.
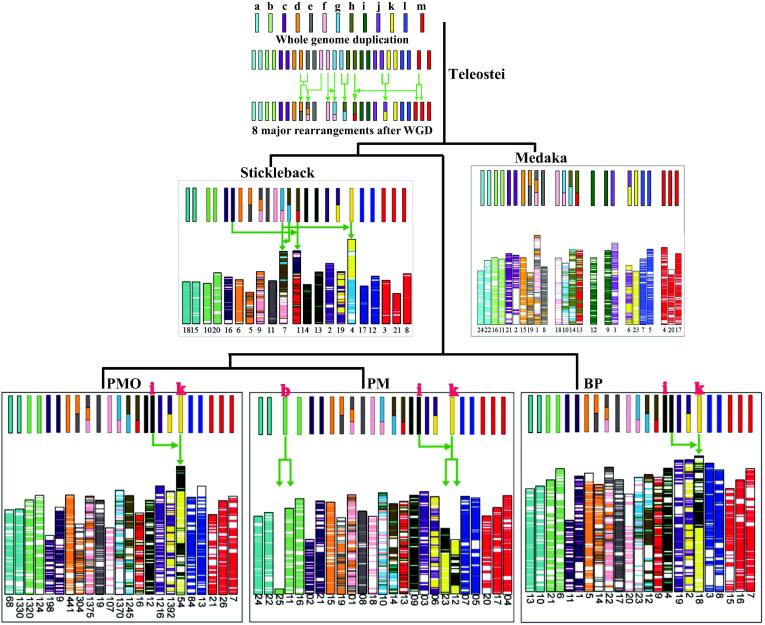
For reconstruction of ancestral chromosomes, we generated 7,639 (BP), 7,956 (PM), 7,207 (PMO) and 7,470 (stickleback) of reciprocal best-hit gene pairs between each of the four species and teleost ancestor protein set. We observed that most mudskipper chromosomes have well retained teleost ancestor chromosome karyotypes (Fig. 3). A common chromosome fusion, appearing in all the three mudskippers, could have occurred from the mudskipper ancestor lineage. However, PM chromosomes had undergone two specific fission events.
Fig. 3.
Reconstruction of the ancestral chromosomes for the three mudskippers. Different colored bars represent 13 ancestral chromosomes. Gene sequences from the same ancestral chromosomes were presented in the same color. Green arrows mark some translocation, fusion, and fission events. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Gene family clustering and divergence time estimation
We detected 34,292 gene families from 13 examined teleost species, containing 429 common single-copy gene families. For the three mudskippers, there are 14,869, 15,351 and 13,531 gene families clustered from BP, PM and PMO, respectively. Moreover, a total of 11,139 common gene families were detected among the three mudskippers. Gene families shared with each mudskipper species were provided in Fig. S2.
A total of 110,181-bp conserved nucleotide sequences from the single-copy gene families were collected to construct a phylogenetic tree (Fig. 4a). The divergence time between BP and two Periophthalmus mudskippers was predicted at about 34.6 million years ago (Mya), which is consistent with our previous report [8] and Yang’s study [9]. The divergence time of PM and PMO, within the same genus, was about 20.8 Mya.
Fig. 4.
Phylogeny and gene-family analysis of the three mudskippers. a A phylogenetic tree of the three mudskippers (BP, PM and PMO). Estimated divergence times (in the unit of Mya) were marked in blue. Numbers of expanded and contracted gene families were marked in green and red, respectively. MRCA represents the most recent common ancestor. b Density plot of the timing of LTR insertions among the three mudskipper species. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
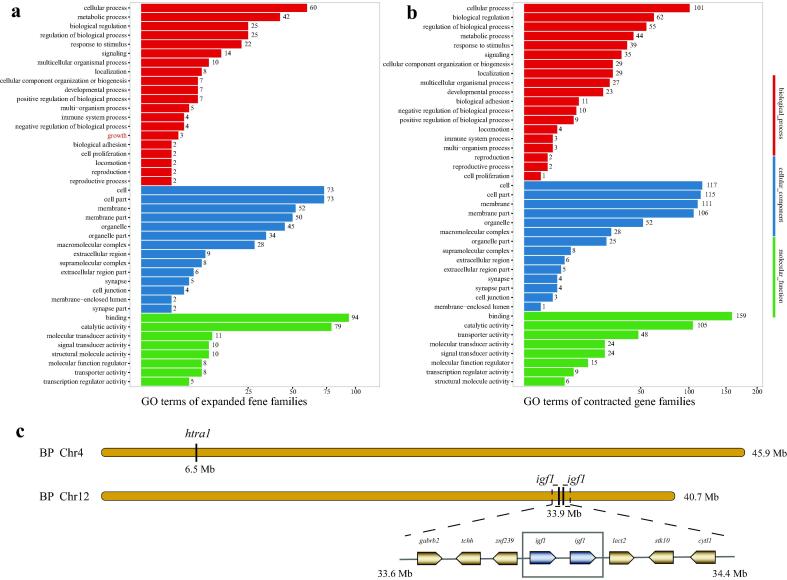
Eventually, we predicted 2,476 expanded gene families and 444 contracted gene families in BP (Fig. 4a), which were enriched into GO terms and KEGG pathways (Fig. 5a, Fig. 5b and Fig. S3). For the enriched expanded gene families, the top five GO terms were “binding”, “catalytic activity”, “cell”, “cell part”, and “cellular process”; the top five KEGG pathways included “infectious disease: viral”, “signal transduction”, “global and overview maps”, “signaling molecules and interaction” and “immune system”. The enriched GO terms and KEGG pathways of PM (Fig. S4) and PMO (Fig. S5) were similar to those of BP. It seems that BP may have developed a good immune system to fight with exogenous pathogens, thereby supporting its daily amphibious life in water and on land. Three genes in GO “growth” were enriched in expanded gene families of BP, including htra1 (encoding serine protease HTRA1) gene, and two copies of igf1 (insulin-like growth factor I). Chromosomal locations of these growth-related genes in the BP genome were provided in Fig. 5c. In brief, the htra1 gene was located at 6.5 Mb on Chr4, while two igf1 genes were tandem duplicated at 33.9 Mb on Chr12.
Fig. 5.
GO enrichments of expanded and contracted gene families and representative tandem duplication genes in BP. a GO enrichment of expanded gene families. b GO enrichment of contracted gene families. c Three expanded genes enriched in the “growth” term. Two tandem duplicated genes (igf1s) and their close neighboring genes were shown in a gray dotted-line box. Abbreviations: cytl1, cytokine-like protein 1; gabrb2, gamma-aminobutyric acid receptor subunit beta-2; lect2, Leukocyte cell-derived chemotaxin-2; stk10, threonine-protein kinase 10; tchh, trichohyalin; znf239, zinc finger protein 239.
SCPP gene variations among the three mudskipper genomes
The scales of air-breathing mudskippers are usually reduced in size and number [57]. We thus investigated SCPP genes, a family encoding a series of P/Q-rich or acidic proteins that are putatively involved in bone mineralization, as well as tooth and scale formation [58], [55], [59]. Our data show that all the three mudskippers had ten SCPP genes (Fig. 6, Table S6), much less than other four representative vertebrates (each with more than 20 SCPP genes). In addition, these SCPP genes distributed in three chromosomes of PM and PMO, namely as scpp1 cluster, spp1 cluster and P/Q-rich-only cluster, respectively (Fig. 6). However, an enam gene, belonging to the SCPP gene family too, located in the fourth chromosome of BP; that is to say, this gene was split from the scpp1 cluster, which is different from the pattern in PM. Interestingly, this enam gene was lost in PMO, but the neighboring gene scpp5 was duplicated (see more details in Fig. 6).
Fig. 6.
Arrangement of SCPP genes in mudskippers and other vertebrates. Each polygon denotes a gene. Blue, yellow, and red pentagons represent P/Q-rich, acidic, and sparc SCPP genes, respectively. Orthologs are linked with shadows. A cluster of P/Q-rich SCPPs in zebrafish is translocated to the downstream of scpp1 on the Dre1 chromosome. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
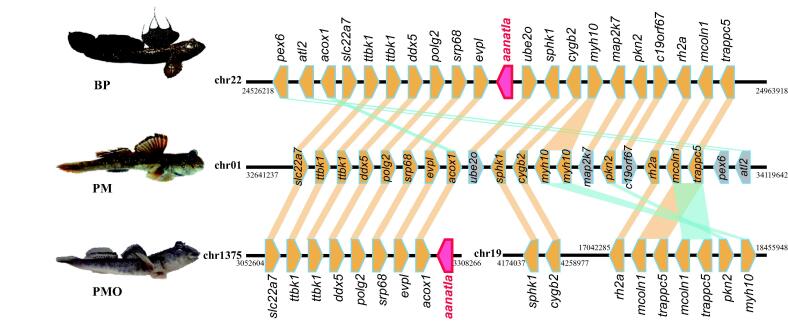
Validation of the loss of aanat1a in PM but not in PMO
Our previous study [8] revealed that the scaffold-level genome assembly of PM has lost aanat1a, an important gene encoding the most crucial enzyme AANAT for melatonin biosynthesis [60], [61] and dopamine metabolism, while BP retains all the three aanat genes (aanat1a, aanat 1b and aanat2) as most teleosts [8], [60]. To further verify this interesting loss, we screened ten core genes for melatonin biosynthesis in our improved chromosome-level genome assemblies of BP, PM (this study) and PMO [9]. Obviously, we confirmed that PM lost aanat1a, while PMO in the same genus and BP both kept the three aanat genes (Fig. 7, Table S7). This consistence suggests that the loss of aanat1a is species-specific rather than lineage shared.
Fig. 7.
Identification of aanat1a gene and genomic comparison of its neighboring genes among the three mudskippers. Each polygon denotes a gene, and its right or left direction represents the forward or backward transcription. Red polygons mark the aanat1a genes. Yellow polygons refer to those aligned genes, whereas blue and gray ones represent translocated genes and no hits, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
To figure out how aanat1a was lost in PM, we performed a synteny comparison of the target genomic regions with this gene among the three mudskipper genomes (Table S8). It seems that both BP and PM shared a well-conserved suite of genes around aanat1a disregarding the presence or absence of this gene, while this region was split into two clusters right after aanat1a in PMO, and the downstream cluster was translocated to another chromosome with relative variability (Fig. 7). Although the whole chromosome alignments show extensive synteny and collinearity between the two Periophthalmus species due to their closer relationship, the aanat1a region was less conserved within this genus (Fig. S6).
Discussion
Genomic differences among the three mudskippers
In the present study, we obtained two high-quality chromosome-level genome assemblies for BP and PM with 23 and 25 chromosomes respectively. Both assemblies and previously reported PMO assembly enable us to perform genomic comparisons among the three mudskippers. Their genome sizes are largely diverse. BP’s genome is about 100 ∼ 200 Mb larger than other twos. A total length of repeat sequences of BP (454.0 Mb) is also longer than the other two mudskippers (304.2 Mb for PM and 374.2 Mb for PMO). After identifying the detailed types of repeat sequences, we found that the total length of LTRs in BP (105.1 Mb) were about two-fold as long as those in PM (47.5 Mb) and PMO (59.0 Mb). After calculating their LTR insertion time, we observed that their LTRs appeared to be accumulated gradually over time (Fig. 4b). A remarkable LTR accumulation and insertion period in BP genome was between 5 and 10 Mya (Fig. 4b), which is more recent than PM’s and PMO’s. A big scale of LTR explosion in the BP lineage could be one major reason to interpret its larger genome than the other two mudskippers.
Moreover, when comparing various gene families among the three mudskippers, we obtained a GO term of “growth” that contained three genes in expanded gene family of BP (Fig. 5a). Chromosomal locations of these genes were summarized in Fig. 5c for detailed comparisons. Interestingly, one gene on Chr4 was annotated as htra1, and other two genes were both annotated as igf1 with tandem duplication on Chr10. The igf1 is known to be crucial for bone growth [62] and tissue physiology [63]. It seems that expansions of these growth-related genes may be related, at least in part, to the largest body size of BP among the three mudskippers.
Chromosomal evolution of three mudskippers and their common ancestor
Another surprising difference among the three mudskippers is that PM has 25 chromosomes that are two more than both BP and PMO. We clearly observed four separate squares with densely mapped Hi-C reads for four important chromosomes (Chr12 and Chr23, Chr11 and Chr 25) in the Hi-C heatmap (Fig. 1b and Fig. 1c). Synteny blocks also clearly demonstrated that Chr11 and Chr25 of PM were split from one ancestor chromosome that was subsequently well retained to be the Chr21 of BP and P_modestus_1320 of PMO (Fig. 2a and Fig. 2b). Similarly, Chr12 and Chr25 of PM were derived from one ancestor chromosome corresponding to Chr18 of BP and P_modestus_54 of PMO (Fig. 2b and Fig. 2c).
To our best knowledge, our present work is the first report of the mudskipper ancestor chromosome karyotype. Previous studies have predicted that the ancestral teleost karyotype possibly contained 13 pairs of chromosomes (represented as a ∼ m), subsequently experienced teleost specific genome duplication, underwent chromosome translocation or variation and returned to diploid by losing some functionally redundant genes and segments, and then gradually shaped the recent teleost karyotype [37], [38]. Our ancestor chromosome data revealed that an ancestor chromosome (i) and another ancestor chromosome (k) had undergone a fusion event in the mudskipper ancestor chromosome karyotype (Fig. 3). Therefore, BP and PMO present a similar fused chromosome (i.e., Chr18 and P_modestus_54 respectively). However, in PM we clearly observed that its ancestor chromosomes (i) and (k) had merged into one original chromosome (mudskipper ancestor chromosome), and then they were split to form two independent chromosomes (Chr12 and Chr23). In addition, a specific fission event was also identified in PM’s Chr11 and Chr25, which were directly split from the ancestor chromosome (b) in the PM lineage (Fig. 3).
Gene families potentially related to water-to-land adaptation
The air-breathing mudskippers deal with the dried condition on land through some relatively developed or modified structures, such as thick skin with abundant blood vessels [2] and a reduction or even absence of scales [57], to improve gas exchange. Histology of the skin of BP [64], [65], PM [57] and PMO [66] reveals that they all present a reduction of scales, and their minute scales are located under the epidermis. It is therefore reasonable to propose difference of certain genes involved in scale formation between these mudskippers and other teleosts with normal scales. For example, the SCPP gene family encodes a series of P/Q-rich or acidic proteins that are involved in bone mineralization, as well as tooth and scale formation [58], [55].
We thus investigated these SCPPs, a gene family with many reports for involvement in scale formation [58]. Our result shows that the mudskippers’ SCPP gene repertoires are considerably smaller (Fig. 6) than those of normal-scaled zebrafish [58], the modified elasmoid-scaled bowfin with enamel-covered teeth [67], and the ganoid-scaled spotted gar that has the largest known SCPP family (38 genes) [54], the denticle-scaled cartilage sturgeons and paddlefish [68], [69], and the diverse tetrapods [59]. In addition, the spp1 cluster is supposed to play major roles in scale formation [67]. Interestingly, this cluster was split apart and reduced to only three P/Q-rich genes in the mudskippers (Fig. 6), and it was reduced to only one gene (scpp8) in scaleless channel catfish [67], [70], [71]. We hence hypothesize that the loss of some specific SCPP genes may lead to the reduction of mudskipper scales, which is definitely beneficial to the terrestrial life of these mudskippers.
Meanwhile, we validated the loss of aanat1a gene in PM again (Fig. 7), but it was retained in both PMO and BP. The AANAT1a protein has been proven to acetylate dopamine in retina, which brings down the retina dopamine concentration thus causes myopia for most fishes [8], [71]. Therefore, the loss of aanat1a is speculated to keep high dopamine levels in retina, and therefore help the mudskippers retain a good aerial vision [8] so as to be able to see clearly on land for escape or predation [70]. Although both Periophthalmus species spend more time on land than BP [8], [72], the loss of aanat1a gene in PM but not in PMO (as previously reported existence in BP) suggests that PM may have a better air vision than both PMO and BP [8]. Such a tiny variation within the genus Periophthalmus exemplifies to prove a step-by-step evolution for the mudskippers’ water-to-land adaptation.
Conclusion
In this study, we generated high-quality chromosome-level genome assemblies for BP and PM respectively. High completeness of assembled genome and annotation sets of both BP and PM can be widely used as the reference genomes for Gobiidae family. We also discovered two specific chromosome fission events in PM leading to shape its 25 chromosomes, which is two more than both BP and PMO (with 23 chromosomes). Combined our assemblies with the previously reported PMO genome assembly, we constructed ancestor chromosomes of mudskippers and observed a common fusion event in the predicted mudskipper ancestor. Interestingly, some SCPP genes were lost in the three mudskipper genomes, which could potentially lead to the reduction of scales and further improve air-breathing of mudskippers; the loss of aanat1a gene was confirmed in PM but not in PMO, suggesting a better air vision of the more terrestrial PM and a step-by-step evolution for the mudskippers’ water-to-land adaptation. These complicated mudskipper genome data will become valuable genetic resources for in-depth investigations on water-to-land transition and comparative genomics in various vertebrates.
CRediT authorship contribution statement
Chao Bian: Conceptualization, Methodology, Investigation, Writing – original draft. Yu Huang: Methodology, Investigation, Writing – original draft. Ruihan Li: Methodology, Writing – original draft. Pengwei Xu: Methodology, Formal analysis. Xinxin You: Formal analysis. Yunyun Lv: Methodology. Zhiqiang Ruan: Formal analysis. Jieming Chen: Methodology. Junmin Xu: Methodology, Formal analysis. Qiong Shi: Conceptualization, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (no. 2022YFE0139700), and Shenzhen Science and Technology Innovation Program for International Cooperation (no. GJHZ20190819152407214).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.05.005.
Contributor Information
Chao Bian, Email: bianchao@genomics.cn.
Qiong Shi, Email: shiqiong@genomics.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Chakrabarty P. The biology of gobies. Copeia. 2012;2012(2):359–360. doi: 10.1643/ot-11-184.1. [DOI] [Google Scholar]
- 2.Wright P.A. Cutaneous respiration and osmoregulation in amphibious fishes. Comp Biochem Physiol A Mol Integr Physiol. 2021;253 doi: 10.1016/j.cbpa.2020.110866. [DOI] [PubMed] [Google Scholar]
- 3.Tytler P., Vaughan T. Thermal ecology of the mudskippers, Periophthalmus koelreuteri (Pallas) and Boleophthalmus boddarti (Pallas) of Kuwait Bay. J Fish Biol. 2010;23(3):327–337. doi: 10.1111/j.1095-8649.1983.tb02912.x. [DOI] [Google Scholar]
- 4.Pace C.M., Gibb A.C. Mudskipper pectoral fin kinematics in aquatic and terrestrial environments. J Exp Biol. 2009;212(Pt 14):2279–2286. doi: 10.1242/jeb.029041. [DOI] [PubMed] [Google Scholar]
- 5.Harris V.A. On the locomotion of the mudskipper Periophthalmus koelreuteri (Pallas): (Gobiidae) P Zool Soc London. 2009;134:107–135. doi: 10.1111/j.1469-7998.1960.tb05921.x. [DOI] [Google Scholar]
- 6.Murdy E.O. A taxonomic revision and cladistic analysis of the oxudercine gobies (Gobiidae, Oxudercinae) Rec Aus Mus. 1989;Suppl 11 doi: 10.3853/j.0812-7387.11.1989.93. [DOI] [Google Scholar]
- 7.Lee Y.J., Choi Y., Ryu B.S. A taxonomic revision of the genus Periophthalmus (Pisces: Gobiidae) from Korea with description of a new species. Korean J Ichthyol. 1995;7(2):120–127. https://koreascience.kr/article/JAKO199527236817896.pdf [Google Scholar]
- 8.You X., Bian C., Zan Q., Xu X., Liu X., Chen J., et al. Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat Commun. 2014;5:5594. doi: 10.1038/ncomms6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Yoo J.Y., Baek S.H., Song H.Y., Jo S., Jung S.H., et al. Chromosome-level genome assembly of the shuttles hoppfish, Periophthalmus modestus. GigaScience. 2022;11(1):giab089. doi: 10.1093/gigascience/giab089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kajitani R., Toshimoto K., Noguchi H., Toyoda A., Ogura Y., Okuno M., et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24(8):1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye C., Hill C.M., Wu S., Ruan J., Ma Z.S. DBG2OLC: Efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci Rep. 2016;6:31900. doi: 10.1038/srep31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H. Minimap and miniasm: Fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 2015;32(14):103–110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaser R., Sovic I., Nagarajan N., Sikic M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27(5):737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J., Fan J., Sun Z., Liu S. NextPolish: A fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 2019;36(7):2253–2255. doi: 10.1093/bioinformatics/btz891. [DOI] [PubMed] [Google Scholar]
- 16.Boetzer M., Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinf. 2014;15:211. doi: 10.1186/1471-2105-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simao F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y., Chen Y., Shi C., Huang Z., Zhang Y., Li S., et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience. 2018;7(1):1–6. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.-J., Vert J.-P., et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16(1):259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durand N.C., Shamim M.S., Machol I., Rao S.S., Huntley M.H., Lander E.S., et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016;3(1):95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudchenko O., Batra S.S., Omer A.D., Nyquist S.K., Hoeger M., Durand N.C., et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356(6333):92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durand N.C., Robinson J.T., Shamim M.S., Machol I., Mesirov J.P., Lander E.S., et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016;3(1):99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abrusán G., Grundmann N., DeMester L., Makalowski W. TEclass—a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics. 2009;25(10):1329–1330. doi: 10.1093/bioinformatics/btp084. [DOI] [PubMed] [Google Scholar]
- 25.Xu Z., Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35(suppl 2):W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarailo-Graovac M., Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 2009;25(1):4.10. 1–4:4. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 27.Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110(1–4):462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 28.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice P., Longden I., Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 30.Li WH and Graur D. Fundamentals of Molecular Evolution. 2000:284-447. Sinauer Associates Inc.,Sunderland, MA, USA.
- 31.Mount D.W. Using the basic local alignment search tool (BLAST) CSH Protoc. 2007;2007(7):pdb. top17 doi: 10.1101/pdb.top17. [DOI] [PubMed] [Google Scholar]
- 32.Birney E., Clamp M., Durbin R. GeneWise and genomewise. Genome Res. 2004;14(5):988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanke M., Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):ii215-225 doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 35.Cantarel B.L., Korf I., Robb S.M., Parra G., Ross E., Moore B., et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18(1):188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proost S., Fostier J., De Witte D., Dhoedt B., Demeester P., Van de Peer Y., et al. i-ADHoRe 3.0–fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 2012;40(2):e11. doi: 10.1093/nar/gkr955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kasahara M., Naruse K., Sasaki S., Nakatani Y., Qu W., Ahsan B., et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447(7145):714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- 38.Bian C., Hu Y., Ravi V., Kuznetsova I.S., Shen X., Mu X., et al. The Asian arowana (Scleropages formosus) genome provides new insights into the evolution of an early lineage of teleosts. Sci Rep. 2016;6:24501. doi: 10.1038/srep24501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGinnis S., Madden T.L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32(Web Server issue):W20–25 doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, et al. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics. 2011; Chapter 6:Unit 6.12.1-9. doi:10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed]
- 41.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 43.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate Maximum-Likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z., Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol Biol Evol. 2006;23(1):212–226. doi: 10.1093/molbev/msj024. [DOI] [PubMed] [Google Scholar]
- 45.Benton M.J., Donoghue P.C. Paleontological evidence to date the tree of life. Mol Biol Evol. 2007;24(1):26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- 46.De Bie T., Cristianini N., Demuth J.P., Hahn M.W. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 47.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(Database issue):D258–261 doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li R., Fan W., Tian G., Zhu H., He L., Cai J., et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463(7279):311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slater G.S., Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinf. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harris R. The Pennsylvania State University; 2007. Improved Pairwise Alignment of Genomic DNA. PhD Thesis, [Google Scholar]
- 52.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kielbasa S.M., Wan R., Sato K., Horton P., Frith M.C. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21(3):487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Braasch I., Gehrke A.R., Smith J.J., Kawasaki K., Manousaki T., Pasquier J., et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 2016;48(4):427–437. doi: 10.1038/ng.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawasaki K., Mikami M., Nakatomi M., Braasch I., Batzel P., Postlethwait J.H., et al. SCPP genes and their relatives in gar: Rapid expansion of mineralization genes in Osteichthyans. J Exp Zool B Mol Devl Evol. 2017;328(7):645–665. doi: 10.1002/jez.b.22755. [DOI] [PubMed] [Google Scholar]
- 56.Wheeler J., Eddy, Sean and R. nhmmer T. DNA homology search with profile HMMs. Bioinformatics. 2013;29(19):2487–2489. doi: 10.1093/bioinformatics/btt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park J.Y. Structure of the skin of an air-breathing mudskipper. Periophthalmus magnuspinnatus J Fish Biol. 2002;60:1543–1550. doi: 10.1111/j.1095-8649.2002.tb02446.x. [DOI] [Google Scholar]
- 58.Lv Y., Kawasaki K., Li J., Li Y., Bian C., Huang Y., et al. A genomic survey of SCPP family genes in fishes provides novel insights into the evolution of fish scales. Int J Mol Sci. 2017;18(11):2432. doi: 10.3390/ijms18112432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawasaki K., Keating J., Nakatomi M., Welten M., Mikami M., Sasagawa I., et al. Coevolution of enamel, ganoin, enameloid, and their matrix SCPP genes in osteichthyans. iScience. 2021;24 doi: 10.1016/j.isci.2020.102023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang Y., Li J., Bian C., Li R., You X., Shi Q. Evolutionary genomics reveals multiple functions of arylalkylamine N-acetyltransferase in fish. Front Genet. 2022;13 doi: 10.3389/fgene.2022.820442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.You X., Sun M., Bian C., Chen J., Yi Y., Yu H., et al. Mudskippers and Their Genetic Adaptations to an Amphibious Lifestyle. Animals. 2018;8(2):24. doi: 10.3390/ani8020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yakar S., Werner H., Rosen C.J. Insulin-like growth factors: actions on the skeleton. J Mol Endocrinol. 2018;61(1):T115–T137. doi: 10.1530/jme-17-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al-Samerria S., Radovick S. The role of insulin-like growth factor-1 (IGF-1) in the control of neuroendocrine regulation of growth. Cells. 2021;10(10):2664. doi: 10.3390/cells10102664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu H.H., Sun Q., Jiang Y.T., Fan M.H., Wang J.X., Liao Z. In-depth proteomic analysis of Boleophthalmus pectinirostris skin mucus. J Proteomics. 2019;200:74–89. doi: 10.1016/j.jprot.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 65.Kurita M., Noma S., Ishimatsu A. Morphology of the respiratory vasculature of the mudskipper Boleophthalmus pectinirostris (Gobiidae: Oxudercinae) J Morphol. 2021;282(10):1557–1568. doi: 10.1002/jmor.21404. [DOI] [PubMed] [Google Scholar]
- 66.Park J.Y., Kim I.S., Lee Y.J., Kim S.Y. Histology and morphometrics of the epidermis of the fins and sucking disc of the mudskipper, Periophtholmus modestus (Pisces, Gobiidae) Korean J Biol Sci. 2004;8:111–1115. doi: 10.1080/12265071.2004.9647742. [DOI] [Google Scholar]
- 67.Thompson A.W., Hawkins M.B., Parey E., Wcisel D.J., Ota T., Kawasaki K., et al. The bowfin genome illuminates the developmental evolution of ray-finned fishes. Nat Genet. 2021;53(9):1373–1384. doi: 10.1038/s41588-021-00914-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng P., Huang Y., Du H., Li C., Lv Y., Ruan R., et al. Draft genome and complete Hox-cluster characterization of the sterlet (Acipenser ruthenus) Front Genet. 2019;10:776. doi: 10.3389/fgene.2019.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng P., Huang Y., Lv Y., Du H., Ruan Z., Li C., et al. The American paddlefish genome provides novel insights into chromosomal evolution and bone mineralization in early vertebrates. Mol Biol Evol. 2021;38(4):1595–1607. doi: 10.1093/molbev/msaa326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X., Zhong L., Bian C., Xu P., Qiu Y., You X., et al. High-quality genome assembly of channel catfish, Ictalurus punctatus. GigaScience. 2016;5(1):39. doi: 10.1186/s13742-016-0142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Z., Liu S., Yao J., Bao L., Zhang J., Li Y., et al. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat Commun. 2016;7:11757. doi: 10.1038/ncomms11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hidayat S., Wicaksono A., Raharjeng A., Jin D., Alam P., Retnoaji B. The Morphologies of mudskipper pelvic fins in relation to terrestrial and climbing behaviour. P Zool Soc. 2022;75:83–93. doi: 10.1007/s12595-021-00422-1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.