Abstract
Tumor heterogeneity represents a major challenge in breast cancer, being associated with disease progression and treatment resistance. Precision medicine has been extensively applied to dissect tumor heterogeneity and, through a deeper molecular understanding of the disease, to personalize therapeutic strategies. In the last years, technological advances have widely improved the understanding of breast cancer biology and several trials have been developed to translate these new insights into clinical practice, with the ultimate aim of improving patients’ outcomes. In the era of molecular oncology, genomics analyses and other methodologies are shaping a new treatment algorithm in breast cancer care. In this manuscript, we review the main steps of precision medicine to predict drug sensitivity in breast cancer from a translational point of view. Genomic developments and their clinical implications are discussed, along with technological advancements that could broaden precision medicine applications. Current achievements are put into perspective to provide an overview of the state-of-art of breast cancer precision oncology as well as to identify future research directions.
Key words: precision medicine, breast cancer, biomarkers, genomics, targeted therapy
Highlights
-
•
Precision medicine aims at personalizing cancer treatment through a deeper understanding of tumor biology.
-
•
Genomics and new technological advances are shaping a new treatment algorithm in breast cancer care.
-
•
We review the main steps of precision medicine to predict drug sensitivity in breast cancer from a translational point of view.
-
•
Current achievements are put into perspective to identify future research directions.
Introduction
Breast cancer (BC) is the most common cancer among women, with 2.3 million new cases diagnosed each year. Despite substantial therapeutic improvements, BC remains a major health problem and it is the first cause of female cancer mortality, with 684 996 deaths estimated worldwide each year.1,2
One of the major challenges in BC management is its clinical heterogeneity in terms of treatment response and outcome, which reflects a high molecular and cellular heterogeneity.3 To overcome such biological complexity, technological advances and high-throughput technologies have been widely applied in cancer research, leading to the identification of new prognostic and predictive biomarkers. These efforts have allowed to move forward cancer treatments from an indiscriminate use of cytotoxic agents to a target-guided strategy. Nowadays, recent technologies are rapidly revolutionizing this field with exciting new potentialities, opening more advanced levels of molecular characterization and leading to new encouraging clinical applications.
Since the development of trastuzumab for the HER2-positive disease, and more recently in the era of antibody–drug conjugates (ADCs), BC has represented a paradigm of precision medicine in solid tumors and one of the most studied cancers in this setting.4,5 Several additional biomarkers are now available, such as PIK3CA, ESR1 mutations, and germline BRCA1/2 mutations, while many others are being investigated.6, 7, 8 Aiming to overcome the one-size-fits-all approach and to dissect the biological heterogeneity of BC with a biomarker-based approach, cancer precision medicine has opened the way to a new era of breast oncology. In this rapidly evolving field, new challenges and future needs must be addressed to further move precision oncology in clinical practice.
Modelling Cancer Biology with Genomics
The current approach of precision medicine in metastatic BC (mBC) aims to identify targetable genomic alterations in each patient and to match them with the right therapy.9,10 This strategy presents a double clinical advantage: on the one hand, a biomarker-guided approach could increase treatment efficacy, focusing the therapeutic interventions on tumors harboring specific alterations; and on the other hand, selectively hitting these alterations could improve the safety profile by sparing potential side-effects to patients who are unlikely to derive some benefits.11 In addition, precision medicine could reduce the financial burden on health care systems, allowing a more cost-effective allocation of resources.12
Thus the workflow of precision medicine requires (i) the acquisition of tumor specimens, (ii) the determination of a molecular profile, (iii) data analysis through bioinformatic tools, (iv) the identification of targetable alterations, and (v) the administration of targeted therapy.13 In this context, a genomic approach based on DNA sequencing, namely next-generation sequencing assays, has been extensively studied (Figure 1).
Figure 1.
Milestones of genomics in the path of precision medicine for breast cancer.
Genomic drivers
Characterization of the mechanisms of cancer progression with genomics in mBC
DNA sequencing technologies have allowed the identification of multiple mechanisms involved in cancer development and tumor progression, and genomics has rapidly become a cornerstone of precision oncology.14 Currently, the identification of oncogenic ‘drivers’, defined as the genomic alterations that lead to malignant transformation and cancer progression, is one of the main applications of a genomic-based precision medicine approach and an essential tool for modeling cancer biology and dissecting BC heterogeneity.10
The rationale to look for oncogenic drivers at an individual level derives from large studies on patients with mBC reporting the ability of high-throughput technologies to identify genomic alterations leading to cancer progression. Bertucci et al.15 carried out whole exome sequencing of 617 patients with mBC, identifying recurrent genomic driver alterations guiding cancer progression in a large number of patients. Indeed, genomic mutations widely described in many cancer types, such as TP53 (47%) and PIK3CA (30%), were confirmed, as well as genomic alterations specifically enriched in mBC, such as ESR1 (17%) and KMT2C (10%). Moreover, unexplored targets, such as NF1 (7%), were identified. Subsequent large-scale genomic analyses confirmed a similar shift in the mutational profiles of metastatic disease, with enrichments in potentially targetable drivers.16, 17, 18
Clinical impact of targeting validated genomic drivers
Considering the ability to identify recurrent driver mutations in mBC through DNA sequencing technologies, the clinical impact of a genomic-driven approach was also evaluated, similar to other tumors such as non-small-cell lung cancer and melanoma.19,20 For example, PIK3CA mutations can be identified in ∼40% of hormone receptor (HR)-positive mBC and are associated with chemoresistance and poor outcomes.21,22 SOLAR-1 is a randomized phase III trial that assessed the efficacy of alpelisib, an α-selective phosphoinositide 3-kinase (PI3K) inhibitor, plus fulvestrant in postmenopausal patients with HR-positive/HER2-negative mBC who progressed to aromatase inhibitors. Interestingly, progression-free survival (PFS) was significantly improved by the addition of alpelisib as compared with placebo in the cohort of patients with PIK3CA-mutated mBC [11.0 versus 5.7 months; hazard ratio 0.65, 95% confidence interval (CI) 0.50-0.85; P < 0.001], while no benefit was observed in the subgroup of patients with PIK3CA-wild-type mBC (7.4 versus 5.6 months; hazard ratio 0.85, 95% CI 0.58-1.25; posterior probability of hazard ratio <1.00, 79.4%).6,23 SOLAR1 has been one of the first studies to identify a treatment benefit of an oncogene de-addiction strategy based on the genomic profiling of mBC, demonstrating the possibility of impacting tumor progression by targeting driver alterations.10 Moreover, the lack of efficacy of the targeted agent in the absence of the specific genomic mutation represented a further validation of the molecular driver alteration. These data provided a proof of concept of the clinical utility of genomics and highlighted the need to carry out genomic testing in these patients.
As previously cited, other validated genomic biomarkers are now available based on randomized phase III studies. The EMERALD trial reported a significant PFS improvement with elacestrant compared with standard of care among patients with ESR1-mutated HR-positive/HER2-negative mBC (hazard ratio 0.55, 95% CI 0.39-0.77; P = 0.0005), while no benefit was detected in the subgroup of patients ESR1 wt (hazard ratio 0.86; 95% CI 0.62-1.18; P = 0.3).8 The OlympiAD and EMBRACA trials assessed the efficacy of poly(ADP-ribose) polymerase (PARP) inhibitors in patients with germline BRCA1/2 mutations and HER2-negative mBC, showing an improvement in patient outcome.7,24 Several other potential genomic drivers are currently being investigated in mBC, such as AKT1 mutations, FGFR1/2/3 mutations/fusions, and germline/somatic alterations in homologous recombination repair-related genes. However, available results have yielded a mixed picture, with some drugs matched to the genomic alteration demonstrating notable efficacy, while others showing a limited impact, highlighting limitations of the de-addiction strategy.25, 26, 27 For instance, even tissue-agnostic biomarkers such as high-tumor mutation burden failed to predict immune checkpoint blockade response in mBC.28,29
Challenges of oncogene de-addiction
Despite strong biological rationale and encouraging results, the oncogene de-addiction strategy must still address important challenges in some complex scenarios.
One relevant issue is the co-existence of multiple molecular drivers at the tumor cell level. Indeed, in BC, comutations have been reported in a large number of patients, leading to the need for combination therapies.15 As an illustration, in HER2-positive mBC, the co-existence of ERBB2 amplifications and PIK3CA alterations represents one of the most common scenarios. A combined biomarkers analysis of BOLERO-1 and BOLERO-3 trials showed that the addition of everolimus, a mammalian target of rapamycin (mTOR) inhibitor, to the anti-HER2 antibody trastuzumab plus chemotherapy improved the outcomes of patients with HER2-positive mBC and PI3K alterations. Therefore, despite potential challenges in their development and safety, these data endorse the feasibility and the clinical benefit of combinations of customized agents.30
In addition, drug toxicity remains a major issue in a precision medicine approach, with new side-effects emerging. Indeed, although the primary aim of targeted treatments is to selectively target cancer cells, they have been linked to diverse toxicities, often differing from those associated with cytotoxic chemotherapy. In the SOLAR-1 trial, several patients treated with alpelisib developed G3-4 toxicities (most frequently hyperglycemia in 36.6% of cases, rash in 9.9% of cases, and diarrhea in 6.7% of cases) and 25% permanently discontinued alpelisib due to adverse events.6 Another point to highlight is pharmacogenomics, which assesses polymorphisms of genes that code for drug-metabolizing enzymes, such as tyrosine kinase inhibitors or ADCs, potentially influencing efficacy and tolerability.31, 32, 33, 34 For instance, a safety analysis from the ASCENT trial reported different rates of neutropenia in patients receiving sacituzumab govitecan according to the UGT1A1 genotype.35 A pharmacogenomic approach could therefore guide therapeutic choices and dose modifications, preventing adverse events and improving treatment compliance.
A further issue is the validation of rare genomic alterations, which often remain an unmet medical need. Although the genomic characterization of BC has identified recurrent driver mutations, some oncogenic alterations occur at a very low frequency.15 Given the challenge of conducting randomized trials for rare genomic segments, the development of targeted therapies presents additional difficulties. Nevertheless, rare genomic alterations may behave like driver mutations, showing a major response to targeted therapies. For instance, an outlier sensitivity to capivasertib, an AKT inhibitor, has been reported in patients with mBC and Cowden syndrome (germline PTEN mutation).36 To overcome these limitations, a new clinical research framework should be promoted for rare genomic segments. In the setting of rare genomic alterations, if a large treatment effect can be expected on a hard clinical endpoint, single-arm registration trials should be explored. In this scenario of a single-arm registration trial, there is a need for historical controls. This will be achieved through the development of large clinic-genomics databases such as the American Association for Cancer Research (AACR) project GENIE.37 Finally, efficacy data should be confirmed, after initial regulatory approval, in larger postapproval studies.38
Finally, the necessity of a reorganization of pathways of care is emerging. To identify rare targetable mutations a wide screening is required, and therefore a new model based on a flow from broad access to genomic tests to few reference centers. Many initiatives have promoted an equitable use of comprehensive genomic profiling, as highlighted in Europe’s Beating Cancer Plan and recent guidelines.39 However, the implementation of precision oncology into routine care still faces challenges in many health systems in terms of infrastructure and reimbursements.9
Multigene sequencing and its effect on clinical endpoints
The ability to identify recurrent genomic alterations and the benefit observed by a targeted approach provided the rationale to test multiple genes in individual patients. Indeed, the heterogeneity of oncogenic alterations involved in cancer progression, some of them occurring in a small percentage of patients, requires a molecular selection through wide genomic profiling.40
The feasibility of molecular screening of mBC based on comprehensive multigene sequencing and the clinical utility of targeted agents have been assessed in different clinical trials. The main features of prospective clinical trials (PubMed searching strategy: ‘molecular profiling’ OR ‘molecular screening’ OR ‘personalized medicine’ OR ‘genomic profiling’) are summarized in Table 1.
Table 1.
Clinical trials evaluating biomarker-matched treatments for mBC identified with high-throughput technologies
| Study | NCT registration number | Randomized trial | Patients with BC evaluated, n | Prior lines of treatment | Molecular analyses | Patients with BC with actionable alteration/matched therapy, n (%) | Clinical endpoints | Main results |
|---|---|---|---|---|---|---|---|---|
| Von Hoff et al.41 | NCT00530192 | No | 18a | ≥3 | IHC/FISH and microarray (51 genes) | 18 (100) | PFS ratio compared with the previous line | 44% of patients with BC with a PFS ratio of ≥1.3 |
| Tsimberidou et al.42 | NCT00851032 | No | 143a | >3 for 43% of patients | PCR-based sequencing (10 genes) and IHC/FISH | 87 (60.8) | ORR and PFS and comparisons with patients not treated with matched therapy | ORR of 12% (versus 5%), median TTF of 3.9 months (95% CI 3.4-5.0) versus 2.2 (95% CI 2.0-2.8)b |
| André et al.43 | NCT01414933 | No | 423 | Any | CGH array and Sanger sequencing | 195 (46) | % of patients for whom a targeted therapy could be offered | 55 (13%) patients received targeted treatment based on a genomic alteration |
| Le Tourneau et al.44 | NCT01771458 | Yes | NAa | Between 2 and 5 | NGS (45 genes) | 40 (NA) | PFS in patients treated on genotype-matched and genotype-unmatched trials | A median PFS of 2.3 (95% CI 1.7-3.8) versus 2.0 (95% CI 1.8-2.1) months among matched and unmatched (P = 0.41)b trials |
| Schwaederle et al.45 | NCT02478931 | No | 60a | Median of 3 | NGS (182 or 236 genes) | 45 (75.0) | Disease control rate, PFS, and PFS ratio compared with the previous line | Disease control rate of 33.3%, median PFS of 4.0 (95% CI 3.2-4.8) months, and PFS ratio >1.3 in 54.3% of patients |
| Wheler et al.46 | NCT02437617 | No | NAa | ≥3 for 66% of patients | NGS (236 genes) | 317 (93.5)b | Disease control rate and TTF in patients treated on genotype-matched and genotype-unmatched trials | Disease control rate of 19% versus 8% among matched and unmatched (P = 0.061) trials, TTF of 2.8 (95% CI 2.1–3.5) versus 1.9 (95% CI 1.5-2.3) months (P = 0.001)b |
| Stockley et al.47 | NCT01505400 | No | 341a | Median of 2 | Three costumed panels (23, 48, or 50 genes) | 130 (41.5) | ORR in patients treated on genotype-matched and genotype-unmatched trials | ORR of 19% among genotype-matched versus 9% in genotype-unmatched trials (P = 0.026)b |
| Massard et al.48 | NCT01566019 | No | 135a | Median of 4 | Targeted sequencing, CGH array, RNA sequencing, and whole-exome sequencing | 38 (28) | PFS ratio compared with the previous line | 36% of patients with BC with a PFS ratio of >1.3 |
| Mangat et al.49 | NCT02693535 | No | NAa | ≥3 for most patients | Various NGS platforms | 96 (NA) | ORR | ORR of 0% among 10 BCs with no KRAS, NRAS, or BRAF alterations treated with cetuximab50; ORR of 21% among 39 BCs with high tumor mutation burden treated with pembrolizumab29 |
| Trédan et al.51 | NCT01774409 | No | 275a | >1 | NGS (69 genes) | 138 (50.2) | ORR | ORR of 13% |
| Sicklick et al.52 | NCT02534675 | No | NAa | Median of 2 | NGS (236 or 405 genes), ctDNA, and PD-L1 IHC | 12 (NA) | Disease control rate and PFS | Disease control rate 50%, PFS of 3.5 months (95% CI 0.57-6.43) |
| Rodon et al.53 | NCT01856296 | No | NAa | Median of 3 | NGS (264 genes) and transcriptomics | 4 (NA) | ORR, PFS, and PFS ratio compared with the previous line | ORR 26.2%, PFS of 2.01 months; PFS ratio >1.5 in 22.4%b |
| Réda et al.54 | NCT02840604 | No | 98a | ≥3 for 60% of patients | NGS (317 genes) | 12 (12.2) | PFS and PFS ratio compared with previous treatment | PFS of 2.5 months (95% CI 2.2-3.7) versus 2.4 months with nontargeted therapy (95% CI 2.1-3.3), PFS ratio >1.3 in 26%b |
| Flaherty et al.55 | NCT02465060 | No | 96a | Median of 3 | NGS (143 genes) | 2 (2.1) | NA | NA |
| Turner et al.56 | NCT03182634 | No | 1034 | >1 | ctDNA testing with PCR (for PIK3CA, ESR1, HER2, and AKT1) and NGS panel (73 genes) | 357 (34.5) | ORR | ORR between 85 and 25% according to the study cohort |
| Pierobon et al.57 | NCT01919749 | No | 32 | Median of 4 | NGS (52 genes), RNA-seq, and protein microarray | 29 (90.6) | ORR | ORR of 76% |
| Chen et al.58 | NCT01827384 | Yes | 9a | Median of 4 | Costumed panel (20 genes) | 7 (77.8) | ORR in patients with targeted mutations (versus patients without targeted mutations) | ORR 2% (95% CI 0% to 10.9%) versus 5% in the nontargeted control armb |
| Andre et al.59 | NCT02299999 and NCT03386162 | Yes | 1462 | 1 or 2 | NGS (50 and 65 genes) | 646 (44.2) | PFS compared with maintenance chemotherapy | PFS of 9.1 months (90% CI 7.1-9.8) versus 2.8 (90% CI 2.1-4.8) in patients with ESCAT I/II mutations |
BC, breast cancer; CGH, comparative genomic hybridization; CI, confidence interval; ctDNA, circulating tumor DNA; IHC, immunohistochemistry; mBC, metastatic breast cancer; NA, not available; NCT, National Clinical Trial; NGS, next-generation sequencing; ORR, overall response rate; PD-L1, programmed death-ligand 1; PFS, progression-free survival; TTF, time to treatment failure.
Trials enrolling patients with different cancer types.
Results of the overall population of patients with different cancer types.
These studies provided a proof of concept of the feasibility of using high-throughput technologies for extensively profiling BCs in a timeframe compatible with clinical practice. In addition, most of them showed that a subset of patients derives a clinical benefit from high-throughput genomics. The rate of detection of actionable alterations among patients with mBC was extremely variable, depending on the technologies applied and the number of genes investigated. The benefit showed with target therapy was also highly variable because, for instance, in the SHIVA study matched therapy comprised marketed drugs already approved in other indications. Furthermore, caution should be applied when comparing these data as these studies were conducted at different stages of precision medicine development, during which the availability of target agents has widely changed.
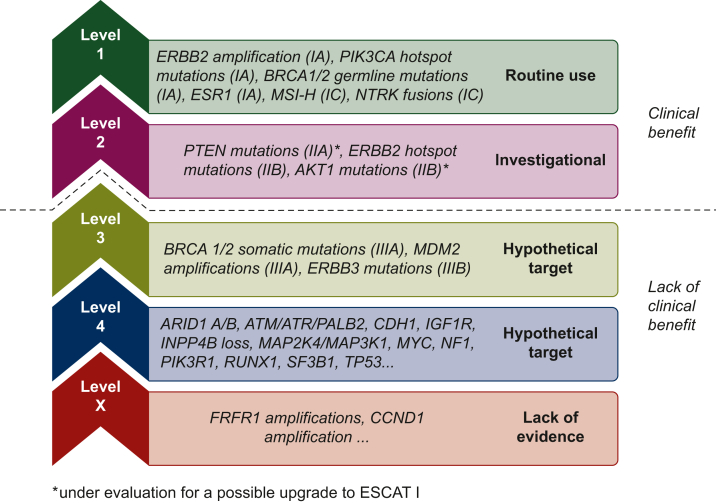
To elucidate the clinical utility of multigene sequencing technologies in daily practice in patients with mBC, a dedicated randomized study was designed. SAFIR02 is a phase II trial that enrolled patients with HER2-negative mBC pretreated with a maximum of one line of chemotherapy (and resistant to endocrine therapy if HR positive). The genomic profile was assessed by next-generation sequencing and comparative genomic hybridization array at baseline. Patients without progressive disease after six to eight cycles of chemotherapy and presenting a targetable genomic alteration were randomized between targeted therapies and maintenance chemotherapy. The primary objective of the study was to evaluate whether targeted therapies improved PFS as compared with maintenance chemotherapy. Out of the 1462 patients included, 646 (44%) had a targetable genomic alteration and 238 (16%) were randomized between maintenance chemotherapy (n = 81) and targeted therapy (n = 157). Among the 115 patients with an ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT) I/II genomic alteration, the median PFS was 9.1 months (90% CI 7.1-9.8) and 2.8 months (90% CI 2.1-4.8) in the targeted therapy and maintenance chemotherapy arms, respectively (P < 0.001), whereas in the overall population there was no significant difference in PFS between the two arms (P = 0.109).59 These data highlight that results of multigene sequencing should be interpreted in the context of a framework of target actionability, such as ESCAT, OncoKB, or the classification of the Association for Molecular Pathology (Figure 2).60, 61, 62
Figure 2.
Genomic alterations according to ESCAT in metastatic breast cancer. ESCAT, ESMO Scale for Clinical Actionability of Molecular Targets.
In this direction, liquid biopsy, through the analysis of circulating tumor DNA (ctDNA), may represent an additional tool to personalize treatment decisions in a dynamic and noninvasive way. In the AURORA trial, Aftimos et al.17 reported the ability of liquid biopsy to identify cancer mutations. In their study, ctDNA successfully detected the majority of actionable alterations in tissues (60% of cases). In addition, there was a noteworthy finding in 11% of cases, where ESCAT I/II mutations were identified in ctDNA but not in tissue samples. Liquid biopsy may therefore represent a promising complementary tool in precision oncology.
Genome evolution and secondary resistance
Despite our ability to identify recurrent driver mutations and target them, cancer cells eventually develop secondary resistance. Cancer evolution is emerging as a complex and multifactorial process, greatly depending on the acquisition of additional mutations and the subsequent clonal selection under the pressure of endogenous and exogenous factors, such as pharmacological treatments.63 In this context, genomics has also proven to be able to describe the mechanism of genome evolution and, therefore, to characterize the development of resistant clones. For instance, ESR1 mutation has been identified as a mechanism of clinically acquired resistance to previous aromatase inhibitor therapy in patients with HR-positive BC, being present in ∼20% of patients with mBC treated with an aromatase inhibitor.64,65
One of the hot topics in the field of precision medicine is the early identification of resistance mechanisms. This question was addressed in the PADA1 trial, where patients treated with first-line palbociclib plus aromatase inhibitors were monitored for the development of an ESR1 mutation in blood and, in case of detection or increase of the mutation, randomized to switch endocrine therapy to fulvestrant. The trial demonstrated the feasibility and PFS benefit of monitoring resistance-associated mutations by ctDNA analysis in patients with mBC.66
Moreover, the genomic approach may allow identifying the molecular mechanisms behind the mutational processes that lead to treatment resistance. For instance, APOBEC is a family of cytidine deaminase involved in the transformation of cytidine in thymine, therefore acting as a DNA mutator.67 An enrichment in APOBEC mutational signature in HR-positive/HER2-negative mBC compared with early BC was observed, raising the hypothesis that this pathway could be involved in cancer evolution. In the future, the identification of the processes involved in the acquisition of mutations could allow us to block them and delay the occurrence of drug resistance.15
Characterization of the mechanism of immune suppression by genomics
Beyond the identification of targetable driver mutations, genomics can also characterize the mechanism of immune suppression and guide toward the administration of immune checkpoint inhibitors. The SAFIR02-BREAST IMMUNO trial evaluated the efficacy of immune checkpoint inhibitors as maintenance treatment in patients with HER2-negative mBC (n = 199). While a lack of PFS and OS benefit was observed in the overall population, exploratory analyses showed an OS benefit in patients with triple-negative BC (n = 82; hazard ratio 0.54, 95% CI 0.30-0.97; P = 0.04), which became even more important in the subgroup of patients with triple-negative BC (TNBC) plus CD274 gain/amplification, where CD274 is the gene coding for programmed death-ligand 1 (PD-L1; n = 23; hazard ratio 0.18, 95% CI 0.05-0.71; P = 0.01). No significant improvement was shown in patients with TNBC and CD274 normal/loss (n = 32; hazard ratio 1.12, 95% CI 0.42-2.99; P = 0.81).68 CD274 amplification may lead to the overexpression of PD-L1, which may represent a key mechanism of immune suppression in these patients, leading to an outlier sensitivity to anti-PD-L1. Genomics could therefore help identify patients more likely to benefit from immunotherapy and decrease the likelihood of treatment resistance.
Beyond Genomics, Assessing New Dimensions of Cancer Biology
While the sequencing of coding regions of DNA has undoubtedly allowed moving forward in the field of precision medicine, its impact is limited because other components of cancer biology drive cancer progression and/or because new drugs target other biological processes. To further improve the impact of cancer precision medicine, multiple teams have developed new technologies to carry out comprehensive portraits of cancer beyond genomics.
-
•
Cancer cell-related proteins and phosphoproteomics: BC is one of the historical illustrations of the use of protein expression to select patients for targeted therapies. Indeed, ER and HER2 expressions have long been used as criteria for selecting patients for endocrine therapy and HER2-targeted agents, respectively. Phosphoproteomics enables the assessment of protein activation, helping to determine whether a mutation detected through genomics results in pathway activation.69 For instance, the ribosomal protein S6 (pS6) is phosphorylated by the activation of the mTOR pathway, representing a potential predictive biomarker of sensitivity to mTOR inhibitors. In the BOLERO-3 trial, 569 pretreated patients with HER2-positive mBCs were randomized to receive vinorelbine–trastuzumab plus everolimus or a placebo. Despite a modest PFS benefit in the overall population, a translational analysis suggested how high levels of phosphorylated pS6 were associated with a high sensitivity to everolimus.70 Assessing phosphoproteins could be particularly relevant to monitor pharmacodynamics and identify feedback loops in each patient receiving kinase inhibitors, in order to modulate doses, schedule, and develop combinations. Assessment of on-treatment phosphokinome could also be done in circulating tumor cells.71
-
•
Transcriptomics: The study of RNA transcripts and their functions and can be integrated with genomics to correlate DNA alterations with gene expression. Intrinsic subtypes and further subclassifications have allowed to better classify BC beyond immunohistochemistry classification and to predict chemo-endocrine sensitivity.72, 73, 74 For instance, nonluminal subtypes within the HR+/HER2– disease have been associated with a poorer response to endocrine therapy,75,76 while different transcriptomic profiles have been reported among cells resistant to different CdK4/6 inhibitors, which could have relevant clinical implications.77 Therefore transcriptomics represents a powerful tool to understand the phenotypical characteristics of BC and to guide treatment choices. Intrinsic subtypes are now being used as inclusion criteria in several prospective trials (e.g. NCT05207709, NCT04251169, NCT02448420), potentially broadening the use of transcriptomic signatures in clinical practice for mBC. In addition, the analysis of ctDNA through liquid biopsy has been demonstrated to be able to identify a correlation between ctDNA-based signatures and RNA-based intrinsic subtypes, overcoming important limitations in the transcriptomic analysis of mBC.78 The feasibility of using transcriptomic profiling for treatment choice was first explored in the WINTHER trial. Patients were evaluated through a gene expression panel carried out in tumors and matched normal tissues. Among the 107 patients assessable for therapy, 38 (35.5%) were selected based on RNA information, proving how transcriptomics substantially increased the number of patients treated with a target agent.53
-
•
In addition, in the DREAM project gene expression microarrays were associated with the ability to predict drug response in human BC cell lines, and drug sensitivity was improved by the integration of DNA sequencing data.79 In line with these findings, Pradat et al.80 carried out an integrative pan-cancer genomic and transcriptomic analysis of refractory metastatic cancers, including 98 BCs. The combination of genomic and RNA-sequencing data allowed to confirm some validated biomarkers of resistance and sensitivity to treatments, as well as to identify new hypothetical resistance mechanisms. With a similar aim, transcriptomic approaches have been explored to identify predictive immune markers. Paré et al.81 evaluated PD1 messenger RNA expression across multiple cancer types, including BC; the expression of PD1 was associated with anti-programmed cell death protein 1 (PD-1) efficacy both in The Cancer Genome Atlas and in a prospective validation series, confirming the possibility of developing clinically applicable assays associated with immunotherapy response.
-
•
RNA sequencing could be complemented in the near future by epigenomics. It is now feasible to assess chromatin status on a large scale thanks to the ATAC-seq technology.82 ATAC-seq could improve the accuracy of RNA-sequencing to identify dysregulated gene expression. For instance, Corces et al.83 carried out, through ATAC-seq, a systematic chromatin accessibility profiling of 23 cancer types, including 74 samples of BC. BC-specific chromatin patterns were identified, as well as different subgroups of BC according to transposase-accessible DNA elements. Interestingly, it has been shown by several teams that genes coding for methyltransferase (e.g. KMT2C, KMT2D) are altered in patients who are resistant to endocrine therapy15,84 and epigenomic reprogramming has now been established as a hallmark of cancer.85 Drugs targeting epigenetics are being tested in clinical trials, and developing ATAC-seq for clinical use could help drive the development of new drug families.
-
•
Microenvironment: The tumor microenvironment represents a key component of cancer biology and, since the development of immune checkpoint blockade therapy, different immunological factors, such as immune checkpoint molecules, cytokines, and immune cell composition, have been extensively studied through different approaches.86 PD-L1, interacting with PD-1 on immune cells and leading to tumor immune evasion, is the most studied immune checkpoint and a biomarker for immunotherapy in mBC.87 In addition, a better understanding of the crosstalk between tumoral cells and tumor microenvironment allowed to identify other targetable mechanisms of immune evasion, and other immune checkpoint inhibitors are currently under investigation in clinical trials as potential therapeutic targets to improve antitumoral response, such as lymphocyte activation gene-3 (LAG-3), T-cell immunoglobulin and mucin domain-containing protein 3 (TIM-3), T-cell immunoglobulin and ITIM domain (TIGIT).88 In the future, technological breakthroughs such as single-cell-based technologies and imaging mass cytometry could enable further characterization of immunological features and the identification of new targetable biomarkers, especially in patients deriving less benefit from current immune checkpoint inhibitors.89 Wang et al. showed that multicellular spatial organization assessed by imaging mass cytometry is a determinant of resistance to immune checkpoint inhibitors in patients with localized TNBC.90 Finally, the tumor microenvironment and its crosstalk between cancer cells may play a role in treatment resistance, including targeted agents. For instance, the secretion of interleukin-8 by tumor-associated macrophages and the subsequent activation of epidermal growth factor receptor (EGFR) signaling has been associated with lapatinib resistance,91 while an immunosuppressive microenvironment has been associated with resistance to anti-HER2-based neoadjuvant treatment.92
-
•
Spatial biology: Technological developments have also allowed to overcome some limitations of bulk analyses. Spatial transcriptomic methodologies enable spatial profiling of tumoral features, mapping gene activity up to the single-cell level. This could allow a better understanding of spatial tumoral heterogeneity as well as of tumor microenvironment. In addition, these new platforms could be of major interest in predicting which ADC or combination of ADCs should be given to a specific patient.93
-
•
Patient-derived ex vivo devices: Patient-derived ex vivo devices, such as organoids and spheroids, are one of the most promising tools for functional precision medicine. These assays are able to largely preserve the genomic, epigenetic, and histological phenotype of a patient’s tumor and to faithfully predict responses to systemic therapies.94 Therefore, 3D cell cultures represent an ideal preclinical model not only to study cancer development, but also as personalized platforms for patient-specific drug screening. The potentiality of organoids as a drug screening platform was evaluated with encouraging results,95 and ongoing trials are exploring the possibility of guiding treatment choice according to organoid sensitivity (NCT04931381). In addition, patient-derived xenograft, developed by transplanting tumoral cells into immunodeficient mice, represents a further tool to preclinically analyze tumor features better recapitulating cancer heterogeneity and tumor microenvironment.94,96
New Methods of Data Analysis and Data Integration
Artificial intelligence applied to digital pathology
Artificial intelligence applied to digital pathology could address three challenges in the near future: it could assist the pathologist in the interpretation of markers that are difficult to read, it could detect populations with a high likelihood of presenting a rare molecular alteration, and it could identify new predictors of drug sensitivity. Some proof-of-concept works have already shown how artificial intelligence applied to histological slides may help with relevant oncological questions. For example, Kather et al.97 used hematoxylin and eosin slides of colorectal cancer to detect patients with a high likelihood of microsatellite instability. Similarly, Garberis et al.98, 99, 100 applied a deep learning algorithm to HR-positive/HER2-negative BC treated with upfront surgery to identify features associated with a poor outcome and to predict the risk of distant recurrence within 5 years.
Multimodal data integration
As mentioned in the ‘Introduction’ section, the long-term goal of precision medicine efforts is to reach cancer avatars that recapitulate the tumor biology at the individual scale, through the integration of multiple translational features, the analysis of data from similar large cohorts, and the creation of multidimensional tools that recapitulate cancer and host features (Figure 3). To reach this goal, there is a need to integrate multiple components of biology derived from unidimensional analyses to define a comprehensive molecular and cellular portrait of the cancer.
Figure 3.
Multimodal integration strategy to recreate a comprehensive cancer profile for each patient. In this holistic approach, each component of cancer biology is evaluated and integrated, including biological biomarkers from translational sciences, multidimensional scores, and data from large patient cohorts. Therefore a cancer avatar summarizing cancer biology can be generated and used for a highly personalized approach.
There are two different strategies to generate comprehensive molecular portraits. The first one is a bottom–up approach where all the biomarkers that have a significant impact are selected to reduce the dimensions of the data sets, and are further integrated to define a comprehensive portrait of cancer. For example, estrogen receptor expression and PIK3CA mutations are integrated to define a subset of patients with BC who should receive endocrine therapy plus PI3K inhibitors. This strategy of data integration, more conservative, requires that each biomarker is validated in clinical trials.
The other approach to cancer modeling is a top–down approach. It consists in analyzing simultaneously multiple components of biology thanks to a combination of high-throughput technologies in order to integrate the data types and provide deeper biological insights compared with individual omics.79,101 They can also be used to develop multisource prediction models using new methods of artificial intelligence learning integrative and compact data representation.102 This approach requires large sample sizes and has yet to show its clinical impact (Figure 4).
Figure 4.
Pipeline illustrating the various stages of a comprehensive precision medicine approach for patients with cancer from basic science to clinical practice. At first, high-throughput technologies, new techniques, and patient-derived ex vivo devices are used to identify unidimensional biomarkers for each patient. Data generated in this way are then integrated to obtain a molecular portrait of each specific tumor, which can then be used to offer personalized treatments for each patient. In addition to the improvement of clinical outcomes, cancer profiles and treatment personalization allow to delay the development of resistance, to predict toxicity, and to guarantee optimal access to therapies. Finally, data generated by clinical trials can further enrich the knowledge of cancer biology, alimenting the pipeline of precision oncology.
Perspectives
The advancement in molecular profiling provided by the wider use of biotechnologies has opened new challenges. The ultimate clinical goal of cancer precision medicine is a patient-centered approach with individualized therapies. Although technological improvements will likely allow the identification of targetable driver alterations in most patients, one of the main limitations to face in the next years could be the lack of compounds able to target the identified alterations. In other words, more precise modeling of tumor biology and a deeper fragmentation of cancers in multiple rare subtypes could lead to the conclusion that the needed drug does not exist yet. Therefore the future need to address will be the personalized construction of the specific drugs necessary for each patient with cancer. To overcome the complexity of drug discovery, new tools of clinical pharmacology should be applied to develop new molecules. Ongoing trials are evaluating the feasibility of a patient-personalized treatment through organoids (NCT04931381) or CAR-T cells (NCT03970382), and in the future, the application of artificial intelligence and pharmacogenomics could accelerate this process. To maximize the potentiality of modern biotechnologies in the evolving landscape of precision oncology, a shift of the therapeutic algorithm toward a further level of individualization should be encouraged and should be thoroughly evaluated in clinical trial programs.
Acknowledgements
Figure 3, Figure 4 have been created with BioRender.com.
Funding
None declared.
Disclosure
FM reports receiving consultant fees from Novartis and Pegascy. CL reports research support from imCORE/Genentech. FA received research funding and served as a speaker/advisor (compensated to the hospital) from Roche, AstraZeneca, Daiichi Sankyo, Pfizer, Novartis, and Eli Lilly. The remaining authors declare no competing interests.
References
- 1.Malvezzi M., Santucci C., Boffetta P., et al. European cancer mortality predictions for the year 2023 with focus on lung cancer. Ann Oncol. 2023;34:410–419. doi: 10.1016/j.annonc.2023.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Globocan. International Agency for Research on Cancer. 2020. https://gco.iarc.fr/today Available at.
- 3.Lüönd F., Tiede S., Christofori G. Breast cancer as an example of tumour heterogeneity and tumour cell plasticity during malignant progression. Br J Cancer. 2021;125(2):164–175. doi: 10.1038/s41416-021-01328-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slamon D.J., Leyland-Jones B., Shak S., et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 5.Modi S., Saura C., Yamashita T., et al. Trastuzumab deruxtecan in previously treated HER2-positive breast cancer. N Engl J Med. 2020;382(7):610–621. doi: 10.1056/NEJMoa1914510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.André F., Ciruelos E., Rubovszky G., et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380(20):1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 7.Robson M., Im S.A., Senkus E., et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(6):523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 8.Bidard F.C., Kaklamani V.G., Neven P., et al. Elacestrant (oral selective estrogen receptor degrader) versus standard endocrine therapy for estrogen receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: results from the randomized phase III EMERALD trial. J Clin Oncol. 2022;40(28):3246–3256. doi: 10.1200/JCO.22.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mateo J., Steuten L., Aftimos P., et al. Delivering precision oncology to patients with cancer. Nat Med. 2022;28(4):658–665. doi: 10.1038/s41591-022-01717-2. [DOI] [PubMed] [Google Scholar]
- 10.Yates L.R., Seoane J., Le Tourneau C., et al. The European Society for Medical Oncology (ESMO) precision medicine glossary. Ann Oncol. 2018;29(1):30–35. doi: 10.1093/annonc/mdx707. [DOI] [PubMed] [Google Scholar]
- 11.Malone E.R., Oliva M., Sabatini P.J.B., Stockley T.L., Siu L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020;12(1):8. doi: 10.1186/s13073-019-0703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasztura M., Richard A., Bempong N.E., Loncar D., Flahault A. Cost-effectiveness of precision medicine: a scoping review. Int J Public Health. 2019;64(9):1261–1271. doi: 10.1007/s00038-019-01298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnedos M., Vicier C., Loi S., et al. Precision medicine for metastatic breast cancer—limitations and solutions. Nat Rev Clin Oncol. 2015;12(12):693–704. doi: 10.1038/nrclinonc.2015.123. [DOI] [PubMed] [Google Scholar]
- 14.Raphael B.J., Dobson J.R., Oesper L., Vandin F. Identifying driver mutations in sequenced cancer genomes: computational approaches to enable precision medicine. Genome Med. 2014;6(1):5. doi: 10.1186/gm524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertucci F., Ng C.K.Y., Patsouris A., et al. Genomic characterization of metastatic breast cancers. Nature. 2019;569(7757):560–564. doi: 10.1038/s41586-019-1056-z. [DOI] [PubMed] [Google Scholar]
- 16.Angus L., Smid M., Wilting S.M., et al. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat Genet. 2019;51(10):1450–1458. doi: 10.1038/s41588-019-0507-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aftimos P., Oliveira M., Irrthum A., et al. Genomic and transcriptomic analyses of breast cancer primaries and matched metastases in AURORA, the Breast International Group (BIG) molecular screening initiative. Cancer Discov. 2021;11(11):2796–2811. doi: 10.1158/2159-8290.CD-20-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Recio S., Hinoue T., Wheeler G.L., et al. Multiomics in primary and metastatic breast tumors from the AURORA US network finds microenvironment and epigenetic drivers of metastasis. Nat Cancer. 2023;4(1):128–147. doi: 10.1038/s43018-022-00491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan M., Huang L.L., Chen J.H., Wu J., Xu Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct Target Ther. 2019;4:61. doi: 10.1038/s41392-019-0099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo W., Wang H., Li C. Signal pathways of melanoma and targeted therapy. Signal Transduct Target Ther. 2021;6(1):424. doi: 10.1038/s41392-021-00827-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Sáez O., Chic N., Pascual T., et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 2020;22(1):45. doi: 10.1186/s13058-020-01284-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosele F., Stefanovska B., Lusque A., et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol. 2020;31(3):377–386. doi: 10.1016/j.annonc.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 23.André F., Ciruelos E.M., Juric D., et al. Alpelisib plus fulvestrant for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: final overall survival results from SOLAR-1. Ann Oncol. 2021;32(2):208–217. doi: 10.1016/j.annonc.2020.11.011. [DOI] [PubMed] [Google Scholar]
- 24.Litton J.K., Rugo H.S., Ettl J., et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379(8):753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verret B., Bottosso M., Hervais S., Pistilli B. The molecular predictive and prognostic biomarkers in metastatic breast cancer: the contribution of molecular profiling. Cancers (Basel) 2022;14(17):4203. doi: 10.3390/cancers14174203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner N.C., Oliveira M., Howell S.J., et al. Capivasertib in hormone receptor-positive advanced breast cancer. N Engl J Med. 2023;388(22):2058–2070. doi: 10.1056/NEJMoa2214131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tung N.M., Robson M.E., Ventz S., et al. TBCRC 048: phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J Clin Oncol. 2020;38(36):4274–4282. doi: 10.1200/JCO.20.02151. [DOI] [PubMed] [Google Scholar]
- 28.McGrail D.J., Pilié P.G., Rashid N.U., et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol. 2021;32(5):661–672. doi: 10.1016/j.annonc.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alva A.S., Mangat P.K., Garrett-Mayer E., et al. Pembrolizumab in patients with metastatic breast cancer with high tumor mutational burden: results from the Targeted Agent and Profiling Utilization Registry (TAPUR) Study. J Clin Oncol. 2021;39(22):2443–2451. doi: 10.1200/JCO.20.02923. [DOI] [PubMed] [Google Scholar]
- 30.André F., Hurvitz S., Fasolo A., et al. Molecular alterations and everolimus efficacy in human epidermal growth factor receptor 2-overexpressing metastatic breast cancers: combined exploratory biomarker analysis from BOLERO-1 and BOLERO-3. J Clin Oncol. 2016;34(18):2115–2124. doi: 10.1200/JCO.2015.63.9161. [DOI] [PubMed] [Google Scholar]
- 31.Reizine N.M., O’Donnell P.H. Modern developments in germline pharmacogenomics for oncology prescribing. CA Cancer J Clin. 2022;72:315–332. doi: 10.3322/caac.21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miteva-Marcheva N.N., Ivanov H.Y., Dimitrov D.K., Stoyanova V.K. Application of pharmacogenetics in oncology. Biomark Res. 2020;8:32. doi: 10.1186/s40364-020-00213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mroz P., Michel S., Allen J.D., et al. Development and implementation of in-house pharmacogenomic testing program at a major academic health system. Front Genet. 2021;12 doi: 10.3389/fgene.2021.712602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaehler M., Cascorbi I. Pharmacogenomics of impaired tyrosine kinase inhibitor response: lessons learned from chronic myelogenous leukemia. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.696960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rugo H.S., Tolaney S.M., Loirat D., et al. Safety analyses from the phase 3 ASCENT trial of sacituzumab govitecan in metastatic triple-negative breast cancer. NPJ Breast Cancer. 2022;8(1):98. doi: 10.1038/s41523-022-00467-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kingston B., Bailleux C., Delaloge S., et al. Exceptional response to AKT inhibition in patients with breast cancer and germline PTEN mutations. JCO Precis Oncol. 2019;3 doi: 10.1200/PO.19.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.AACR Project GENIE Consortium AACR Project GENIE: powering precision medicine through an International Consortium. Cancer Discov. 2017;7(8):818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.André F. Developing anticancer drugs in orphan molecular entities—a paradigm under construction. N Engl J Med. 2018;378(8):763–765. doi: 10.1056/NEJMe1716821. [DOI] [PubMed] [Google Scholar]
- 39.Mosele F., Remon J., Mateo J., et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31:1491–1505. doi: 10.1016/j.annonc.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Andre F., Delaloge S., Soria J.C. Biology-driven phase II trials: what is the optimal model for molecular selection? J Clin Oncol. 2011;29(10):1236–1238. doi: 10.1200/JCO.2010.31.6877. [DOI] [PubMed] [Google Scholar]
- 41.Von Hoff D.D., Stephenson J.J., Rosen P., et al. Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010;28(33):4877–4883. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- 42.Tsimberidou A.M., Wen S., Hong D.S., et al. Personalized medicine for patients with advanced cancer in the phase I program at MD Anderson: validation and landmark analyses. Clin Cancer Res. 2014;20(18):4827–4836. doi: 10.1158/1078-0432.CCR-14-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.André F., Bachelot T., Commo F., et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15(3):267–274. doi: 10.1016/S1470-2045(13)70611-9. [DOI] [PubMed] [Google Scholar]
- 44.Le Tourneau C., Delord J.P., Gonçalves A., et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16(13):1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 45.Schwaederle M., Parker B.A., Schwab R.B., et al. Precision oncology: the UC San Diego Moores Cancer Center PREDICT experience. Mol Cancer Ther. 2016;15(4):743–752. doi: 10.1158/1535-7163.MCT-15-0795. [DOI] [PubMed] [Google Scholar]
- 46.Wheler J.J., Janku F., Naing A., et al. Cancer therapy directed by comprehensive genomic profiling: a single center study. Cancer Res. 2016;76(13):3690–3701. doi: 10.1158/0008-5472.CAN-15-3043. [DOI] [PubMed] [Google Scholar]
- 47.Stockley T.L., Oza A.M., Berman H.K., et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med. 2016;8(1):109. doi: 10.1186/s13073-016-0364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massard C., Michiels S., Ferté C., et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7(6):586–595. doi: 10.1158/2159-8290.CD-16-1396. [DOI] [PubMed] [Google Scholar]
- 49.Mangat P.K., Halabi S., Bruinooge S.S., et al. Rationale and design of the targeted agent and profiling utilization registry (TAPUR) study. JCO Precis Oncol. 2018;2018 doi: 10.1200/PO.18.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fisher J.G., Tait D., Garrett-Mayer E., et al. Cetuximab in patients with breast cancer, non-small cell lung cancer, and ovarian cancer without KRAS, NRAS, or BRAF mutations: results from the targeted agent and profiling utilization registry (TAPUR) study. Target Oncol. 2020;15(6):733–741. doi: 10.1007/s11523-020-00753-7. [DOI] [PubMed] [Google Scholar]
- 51.Trédan O., Wang Q., Pissaloux D., et al. Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: analysis from the ProfiLER trial. Ann Oncol. 2019;30(5):757–765. doi: 10.1093/annonc/mdz080. [DOI] [PubMed] [Google Scholar]
- 52.Sicklick J.K., Kato S., Okamura R., et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med. 2019;25(5):744–750. doi: 10.1038/s41591-019-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodon J., Soria J.C., Berger R., et al. Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat Med. 2019;25(5):751–758. doi: 10.1038/s41591-019-0424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Réda M., Richard C., Bertaut A., et al. Implementation and use of whole exome sequencing for metastatic solid cancer. EBioMedicine. 2020;51 doi: 10.1016/j.ebiom.2019.102624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flaherty K.T., Gray R.J., Chen A.P., et al. Molecular landscape and actionable alterations in a genomically guided cancer clinical trial: national cancer institute molecular analysis for therapy choice (NCI-MATCH) J Clin Oncol. 2020;38(33):3883–3894. doi: 10.1200/JCO.19.03010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner N.C., Kingston B., Kilburn L.S., et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): a multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020;21(10):1296–1308. doi: 10.1016/S1470-2045(20)30444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pierobon M., Robert N.J., Northfelt D.W., et al. Multi-omic molecular profiling guide’s efficacious treatment selection in refractory metastatic breast cancer: a prospective phase II clinical trial. Mol Oncol. 2022;16(1):104–115. doi: 10.1002/1878-0261.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen A.P., Kummar S., Moore N., et al. Molecular profiling-based assignment of cancer therapy (NCI-MPACT): a randomized multicenter phase II trial. JCO Precis Oncol. 2021;5 doi: 10.1200/PO.20.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andre F., Filleron T., Kamal M., et al. Genomics to select treatment for patients with metastatic breast cancer. Nature. 2022;610(7931):343–348. doi: 10.1038/s41586-022-05068-3. [DOI] [PubMed] [Google Scholar]
- 60.Mateo J., Chakravarty D., Dienstmann R., et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) Ann Oncol. 2018;29(9):1895–1902. doi: 10.1093/annonc/mdy263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chakravarty D., Gao J., Phillips S.M., et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li M.M., Datto M., Duncavage E.J., et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vendramin R., Litchfield K., Swanton C. Cancer evolution: Darwin and beyond. EMBO J. 2021;40(18) doi: 10.15252/embj.2021108389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toy W., Shen Y., Won H., et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turner N.C., Swift C., Kilburn L., et al. ESR1 Mutations and overall survival on fulvestrant versus exemestane in advanced hormone receptor-positive breast cancer: a combined analysis of the phase III SoFEA and EFECT trials. Clin Cancer Res. 2020;26(19):5172–5177. doi: 10.1158/1078-0432.CCR-20-0224. [DOI] [PubMed] [Google Scholar]
- 66.Bidard F.C., Hardy-Bessard A.C., Dalenc F., et al. Switch to fulvestrant and palbociclib versus no switch in advanced breast cancer with rising ESR1 mutation during aromatase inhibitor and palbociclib therapy (PADA-1): a randomised, open-label, multicentre, phase 3 trial. Lancet Oncol. 2022;23(11):1367–1377. doi: 10.1016/S1470-2045(22)00555-1. [DOI] [PubMed] [Google Scholar]
- 67.Swanton C., McGranahan N., Starrett G.J., Harris R.S. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015;5(7):704–712. doi: 10.1158/2159-8290.CD-15-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bachelot T., Filleron T., Bieche I., et al. Durvalumab compared to maintenance chemotherapy in metastatic breast cancer: the randomized phase II SAFIR02-BREAST IMMUNO trial. Nat Med. 2021;27(2):250–255. doi: 10.1038/s41591-020-01189-2. [DOI] [PubMed] [Google Scholar]
- 69.Harsha H.C., Pandey A. Phosphoproteomics in cancer. Mol Oncol. 2010;4(6):482–495. doi: 10.1016/j.molonc.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.André F., O’Regan R., Ozguroglu M., et al. Everolimus for women with trastuzumab-resistant, HER2-positive, advanced breast cancer (BOLERO-3): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Oncol. 2014;15(6):580–591. doi: 10.1016/S1470-2045(14)70138-X. [DOI] [PubMed] [Google Scholar]
- 71.Sinkala E., Sollier-Christen E., Renier C., et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat Commun. 2017;8 doi: 10.1038/ncomms14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perou C.M., Sørlie T., Eisen M.B., et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 73.Bareche Y., Venet D., Ignatiadis M., et al. Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann Oncol. 2018;29(4):895–902. doi: 10.1093/annonc/mdy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lehmann B.D., Bauer J.A., Chen X., et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Prat A., Lluch A., Turnbull A.K., et al. A PAM50-based chemoendocrine score for hormone receptor-positive breast cancer with an intermediate risk of relapse. Clin Cancer Res. 2017;23(12):3035–3044. doi: 10.1158/1078-0432.CCR-16-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prat A., Chaudhury A., Solovieff N., et al. Correlative biomarker analysis of intrinsic subtypes and efficacy across the MONALEESA phase III studies. J Clin Oncol. 2021;39(13):1458–1467. doi: 10.1200/JCO.20.02977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Navarro-Yepes J., Kettner N.M., Rao X., et al. Abemaciclib is effective in palbociclib-resistant hormone receptor–positive metastatic breast cancers. Cancer Res. 2023;83(19):3264–3283. doi: 10.1158/0008-5472.CAN-23-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prat A., Brasó-Maristany F., Martínez-Sáez O., et al. Circulating tumor DNA reveals complex biological features with clinical relevance in metastatic breast cancer. Nat Commun. 2023;14(1):1157. doi: 10.1038/s41467-023-36801-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Costello J.C., Heiser L.M., Georgii E., et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol. 2014;32(12):1202–1212. doi: 10.1038/nbt.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pradat Y., Viot J., Yurchenko A.A., et al. Integrative pan-cancer genomic and transcriptomic analyses of refractory metastatic cancer. Cancer Discov. 2023;13(5):1116–1143. doi: 10.1158/2159-8290.CD-22-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paré L., Pascual T., Seguí E., et al. Association between PD1 mRNA and response to anti-PD1 monotherapy across multiple cancer types. Ann Oncol. 2018;29(10):2121–2128. doi: 10.1093/annonc/mdy335. [DOI] [PubMed] [Google Scholar]
- 82.Grandi F.C., Modi H., Kampman L., Corces M.R. Chromatin accessibility profiling by ATAC-seq. Nat Protoc. 2022;17(6):1518–1552. doi: 10.1038/s41596-022-00692-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Corces M.R., Granja J.M., Shams S., et al. The chromatin accessibility landscape of primary human cancers. Science. 2018;362(6413) doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Razavi P., Chang M.T., Xu G., et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34(3):427–438.e6. doi: 10.1016/j.ccell.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 86.Debien V., De Caluwé A., Wang X., et al. Immunotherapy in breast cancer: an overview of current strategies and perspectives. NPJ Breast Cancer. 2023;9(1):7. doi: 10.1038/s41523-023-00508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cortes J., Rugo H.S., Cescon D.W., et al. Pembrolizumab plus chemotherapy in advanced triple-negative breast cancer. N Engl J Med. 2022;387(3):217–226. doi: 10.1056/NEJMoa2202809. [DOI] [PubMed] [Google Scholar]
- 88.Gaynor N., Crown J., Collins D.M. Immune checkpoint inhibitors: key trials and an emerging role in breast cancer. Semin Cancer Biol. 2022;79:44–57. doi: 10.1016/j.semcancer.2020.06.016. [DOI] [PubMed] [Google Scholar]
- 89.Xu Y., Su G.H., Ma D., Xiao Y., Shao Z.M., Jiang Y.Z. Technological advances in cancer immunity: from immunogenomics to single-cell analysis and artificial intelligence. Signal Transduct Target Ther. 2021;6(1):312. doi: 10.1038/s41392-021-00729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang X.Q., Danenberg E., Huang C.S., et al. Spatial predictors of immunotherapy response in triple-negative breast cancer. Nature. 2023;621:868–876. doi: 10.1038/s41586-023-06498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ahmed S., Mohamed H.T., El-Husseiny N., et al. IL-8 secreted by tumor associated macrophages contribute to lapatinib resistance in HER2-positive locally advanced breast cancer via activation of Src/STAT3/ERK1/2-mediated EGFR signaling. Biochim Biophys Acta Mol Cell Res. 2021;1868(6) doi: 10.1016/j.bbamcr.2021.118995. [DOI] [PubMed] [Google Scholar]
- 92.Hu X., Liu Y., Zhang X., et al. The anti-B7-H4 checkpoint synergizes trastuzumab treatment to promote phagocytosis and eradicate breast cancer. Neoplasia. 2020;22(11):539–553. doi: 10.1016/j.neo.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duan H., Cheng T., Cheng H. Spatially resolved transcriptomics: advances and applications. Blood science (Baltimore, Md) 2023;5(1):1–14. doi: 10.1097/BS9.0000000000000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van Renterghem A.W.J., van de Haar J., Voest E.E. Functional precision oncology using patient-derived assays: bridging genotype and phenotype. Nat Rev Clin Oncol. 2023;20(5):305–317. doi: 10.1038/s41571-023-00745-2. [DOI] [PubMed] [Google Scholar]
- 95.Kastner C., Hendricks A., Deinlein H., et al. Organoid models for cancer research—from bed to bench side and back. Cancers (Basel) 2021;13(19):4812. doi: 10.3390/cancers13194812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abdolahi S., Ghazvinian Z., Muhammadnejad S., Saleh M., Asadzadeh Aghdaei H., Baghaei K. Patient-derived xenograft (PDX) models, applications and challenges in cancer research. J Transl Med. 2022;20(1):206. doi: 10.1186/s12967-022-03405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Echle A., Ghaffari Laleh N., Quirke P., et al. Artificial intelligence for detection of microsatellite instability in colorectal cancer-a multicentric analysis of a pre-screening tool for clinical application. ESMO Open. 2022;7(2) doi: 10.1016/j.esmoop.2022.100400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garberis I.J., Gaury V., Aubert V., et al. 147P Blind validation of an AI-based tool for predicting distant relapse from breast cancer HES stained slides. Ann Oncol. 2022;33:S607. [Google Scholar]
- 99.Garberis I., Gaury V., Saillard C., et al. Deep learning allows assessment of risk of metastatic relapse from invasive breast cancer histological slides. bioRxiv. 2022;2022 http://biorxiv.org/content/early/2022/12/05/2022.11.28.518158.abstract Available at. [Google Scholar]
- 100.Garberis I.J., Saillard C., Drubay D., et al. 1124O Prediction of distant relapse in patients with invasive breast cancer from deep learning models applied to digital pathology slides. Ann Oncol. 2021;32:S921. [Google Scholar]
- 101.Rahnenführer J., De Bin R., Benner A., et al. Statistical analysis of high-dimensional biomedical data: a gentle introduction to analytical goals, common approaches and challenges. BMC Med. 2023;21(1):182. doi: 10.1186/s12916-023-02858-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Benkirane H., Pradat Y., Michiels S., Cournède P.H. CustOmics: a versatile deep-learning based strategy for multi-omics integration. PLoS Comput Biol. 2023;19(3) doi: 10.1371/journal.pcbi.1010921. [DOI] [PMC free article] [PubMed] [Google Scholar]