Summary
[18F]Fluorodeoxyglucose positron emission tomography (FDG-PET) and computed tomography (CT) are indispensable components in modern medicine. Although PET can provide additional diagnostic value, it is costly and not universally accessible, particularly in low-income countries. To bridge this gap, we have developed a conditional generative adversarial network pipeline that can produce FDG-PET from diagnostic CT scans based on multi-center multi-modal lung cancer datasets (n = 1,478). Synthetic PET images are validated across imaging, biological, and clinical aspects. Radiologists confirm comparable imaging quality and tumor contrast between synthetic and actual PET scans. Radiogenomics analysis further proves that the dysregulated cancer hallmark pathways of synthetic PET are consistent with actual PET. We also demonstrate the clinical values of synthetic PET in improving lung cancer diagnosis, staging, risk prediction, and prognosis. Taken together, this proof-of-concept study testifies to the feasibility of applying deep learning to obtain high-fidelity PET translated from CT.
Keywords: generative AI, image synthesis, synthetic PET, generative adversarial network, radiogenomics, radiomics, screening, lung cancer, prognostic marker, Turing test
Graphical abstract
Highlights
-
•
Deep learning can generate high-fidelity synthetic PET from CT
-
•
Radiologists confirm high imaging fidelity between synthetic and actual PET
-
•
Radiogenomics proves the biological fidelity of synthetic PET
-
•
Synthetic PET improves cancer diagnosis, risk prediction, and prognosis
Salehjahromi et al. develop a GAN-based CT-to-PET translation framework and validate it through thoracic radiologists and radiogenomics analysis. The synthetic PET scans demonstrate potential in enhancing early lung cancer detection, stratification of high-risk populations, and prognostication. Early assessments indicate promising applications in clinical oncology practice.
Introduction
[18F]Fluorodeoxyglucose positron emission tomography (FDG-PET) is widely used to image patients with cancer.1 FDG-PET measures glucose consumption, offering a valuable functional view that is complementary to computed tomography (CT) in multiple clinical settings. For example, PET can improve the accuracy in diagnosing high-risk indeterminate pulmonary nodules detected by low-dose CT screening.2 It has shown superior value over CT in determining the nodal and distant metastasis for cancer staging.3 For treatment response, PET offers a new view to assessment of metabolic tumor response to systemic therapy and beyond as proposed in PERCIST.4 As such, it has become an indispensable component in oncology practice.
Although PET has demonstrated clinical value, it is still not universally applicable compared to CT.5 PET-CT has approximately double the radiation exposure than a standard CT scan, raising concerns about increasing cancer risk,6 especially in those at high risk, including pregnant women, children, and cancer patients with repeated scans. PET is not considered appropriate for lung cancer screening at current dose levels, as the radiation risk outweighs the benefits. This might change with the development of low-dose PET-CT.7 Moreover, PET scanners are expensive and complex to acquire, run, and maintain, which contributes to stark global inequities, with only five PET scanners existing for roughly 50 low-income countries covering a total population of 706 million, based on recent data from the International Atomic Energy Agency.5
One strategy to address these limitations is by using cross-modality imaging synthesis as an alternative way to obtain PET. Deep learning has reshaped the landscape of medical image synthesis.8 Pilot studies have proven the feasibility of machine-learning models to learn inter- and intra-modality mapping functions, with potentially broad applications in radiology. Imaging synthesis may help streamline the clinical workflow and bypass certain imaging procedures, such as synthetic CT, to potentially replace extra CT acquisition for pelvis PET/MR attenuation correction9 and for MRI-based prostate radiation dose planning.10 Currently, cross-modality synthesis is focused on mapping the anatomical findings between CT and MRI,8 where there is a large amount of overlapping information. However, machine-learning-based synthesis remains underexplored for bridging anatomical to functional mapping, given that each modality offers a different view of the underlying physiology. Interestingly, a recent study demonstrated that CT-based lung perfusion images synthesized by a transfer learning framework achieved a strong voxel-wise correlation with single-photon emission CT (SPECT) perfusion images.11 However, most previous studies have assessed synthetic image quality without expert radiologists’ input and have lacked biological and clinical validation.
We hypothesized that deep learning can effectively learn the anatomical-to-metabolic mapping based on paired diagnostic CT and FDG-PET scans, whereby the synthetic PET can preserve biological correlates and add clinical value in lung cancer diagnosis and prognosis. To test this, we performed a comprehensive study on multi-center multi-modal data (imaging, genomics, clinical, and longitudinal data) to develop a computational framework and systematically validate its fidelity based on imaging signal, radiologists’ assessments, and radiogenomics correlates, as well as evaluation of biological and clinical values.
Results
High fidelity of PET images produced by applying conditional GAN on CT with confirmation by thoracic radiologists
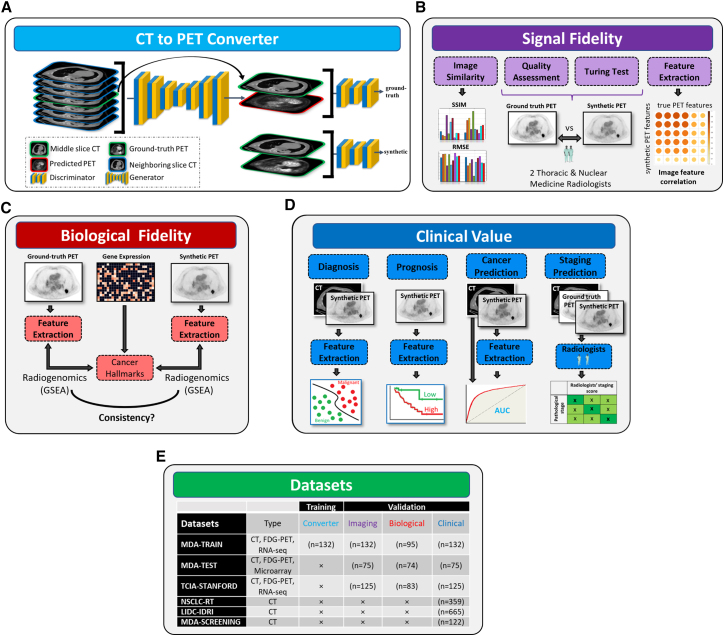
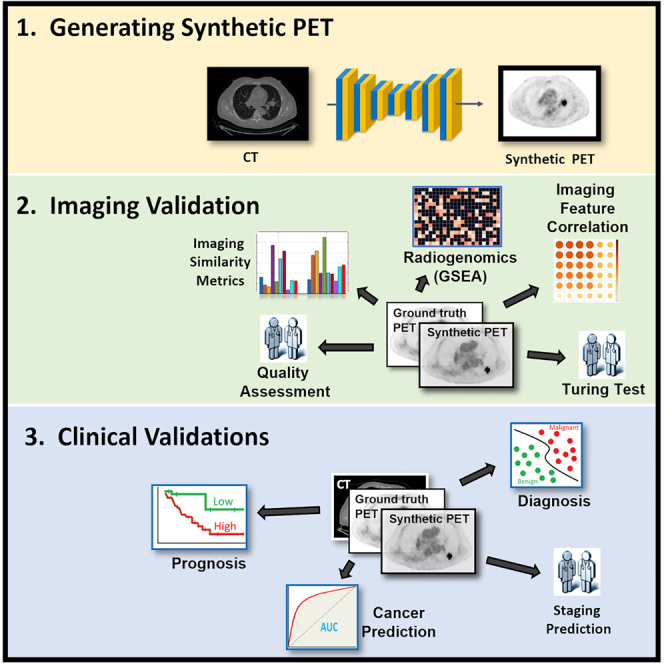
We developed a conditional generative adversarial network (cGAN) model by hybridizing pix2pix and ResUNet++ based on multi-center PET and CT scans (for details see STAR Methods and Figure 1). The model was trained on the MDA-TRAIN cohort and locked for external testing on the MDA-TEST and TCIA-STANFORD cohorts with a head-to-head comparison with acquired PET as ground truth. We displayed the synthetic PET scans from the two testing cohorts to compare them side by side with ground-truth PET and visualize the model output (Figure 2A).
Figure 1.
Overview of the study design
(A) Training the cGAN to predict PET image from CT. The input of the generator is a CT slice along with its six neighboring slices while its output is the synthetic PET image. The discriminator tries to classify between the synthetic and ground-truth PET/CT pairs.
(B) In the imaging validation, similarity metrics including SSIM and RMSE were employed for comparing the synthetic and ground-truth PET images. A group of two thoracic radiologists was enrolled blindly to visually assess the quality of synthetic PET images. Next, they conducted a Turing test on synthetic and ground-truth PET images. Moreover, we analyzed the pairwise similarity between the synthetic and ground-truth PET features.
(C) In the biological validation, we applied radiogenomic analysis using the GSEA method to find the association of cancer hallmarks with extracted features from ground-truth and synthetic PET scans.
(D) In the subsection “synthetic PET complements CT for early lung cancer diagnosis,” we investigated whether the performance of indeterminate pulmonary nodule classification using only main CT-based features can be further improved by adding the extracted features from the synthetic PET. In cancer prediction, we validated the clinical value of synthetic PET in prediction of the development of lung cancer and in staging prediction. In staging prediction, two radiologists assess the synthetic PET capability for accurately staging the lung cancer patients. In the subsection “synthetic PET predicts prognosis after standard of care,” we showed that the extracted nodule features from the synthetic PET are capable of stratifying patients into good and bad survival groups.
(E) The cohorts used in different sections.
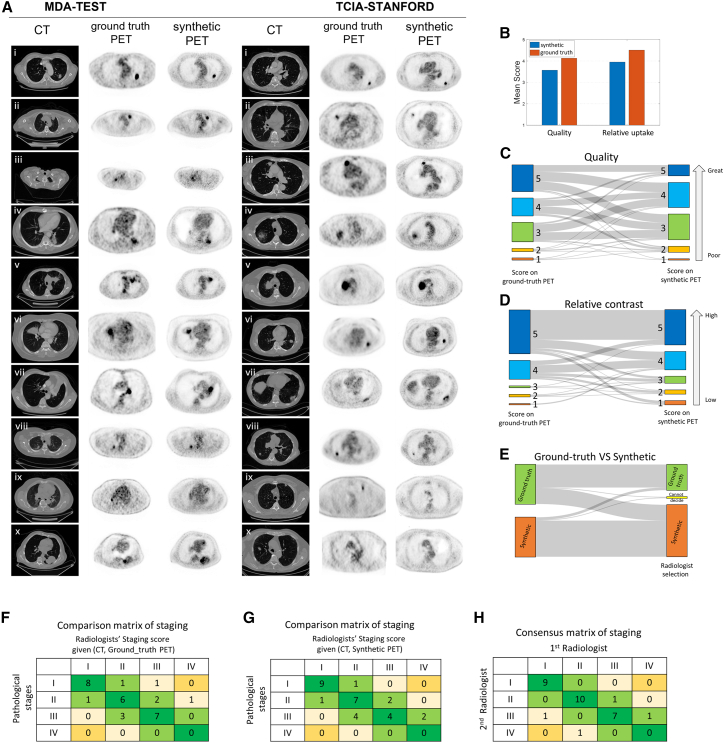
Figure 2.
Validation of imaging signal fidelity and cancer staging performance by radiologists
(A) Presentation of synthetic PET images with ground-truth PET in MDA-TEST and TCIA-STANFORD testing cohorts. The first three columns from left to right for each cohort correspond to CT, ground-truth, and synthetic PET images. The PET images are shown inversely with the normalized window of SUV in . Therefore, the completely black color in tumors indicates that the tumor had uptake with maximum SUV value of at least 3.
(B) The radiologists’ score on quality and relative uptake of lung region in task 1 of the imaging quality test.
(C) Alluvial plot shows the radiologists’ scoring on imaging quality difference using paired PET scans.
(D) Alluvial plot shows the radiologists’ scoring on tumor contrast difference using paired PET scans.
(E) Alluvial plot shows the radiologists’ reading of ground-truth vs. synthetic using paired PET scans.
(F) Comparison matrix of staging between radiologists reading CT and ground-truth PET and pathological stage.
(G) Comparison matrix of staging between radiologists reading CT and synthetic PET and pathological stage.
(H) Consensus matrix of staging between the two radiologists when one radiologist reads true PET and CT compared to another reading synthetic PET and CT.
Two experienced radiologists evaluated the imaging quality and did a Turing test (see STAR Methods for details). They rated equivalent quality of true and synthetic PET for subjective imaging quality (Figure 2B), with an average score of 4 for true PET and 3.6 for synthetic PET using 5-point system (with 1 as poor, 3 as adequate, and 5 as excellent), as well as for tumor contrast, with an average score of 4.5 for true PET and 4.1 for synthetic PET using a 5-point system (with 1 as low, 3 as equal, and 5 as high). For imaging quality rating and within one score tolerance, radiologists rated 75% cases as equivalent and 19% cases as decreased when comparing synthetic PET to its ground-truth counterpart (Figures 2C and S1A). For lesion contrast evaluation, 83% cases stayed unchanged, and 13% cases decreased within one score tolerance (Figures 2D and S1B). For identification of the synthetic scans in the Turing test, the radiologists achieved overall 75% accuracy and misclassified 7% of synthetic cases and 40% of real cases (Figures 2E and S1C). Also, two radiologists had high agreement regarding their consensus for imaging quality (87%) and lesion contrast evaluation (99%) tolerant within one score difference, while they had 77% agreement in identifying the synthetic PET scans (Figure S1D, with individual radiologists’ performance detailed in Figure S1E).
Quantitative comparison confirmed the high fidelity of the synthetic PET
The generated PET scans were compared with ground-truth PET images quantitatively. As seen in Figure 2A, the model showed good performance (Figures S2A and S2B) for ten presented cases from MDA-TEST (mean structural similarity index measure [SSIM] = 0.88) and TCIA-STANFORD (mean SSIM = 0.81), where the SSIM is between 0 and 1 with a higher value indicating more similarity. We further computed the SSIM and root-mean-square error (RMSE) for a large sample of cases from an internal validation set in MDA-TRAIN as well as in the external testing sets (i.e., MDA-TEST and TCIA-STANFORD). As shown in Figure S2C, the cGAN model had high SSIM values, with a median value in MDA-TRAIN and MDA-TEST around 0.9 and with a narrower deviation for MDA-TRAIN than MDA-TEST. For the TCIA-STANFORD cohort, the median of SSIM was around 0.8. Together, these data indicate high fidelity of synthetic PET with ground-truth PET during validation. The RMSE had a similar trend, with the MDA-TRAIN cohort showing the lowest median and MDA-TEST a lower RMSE compared to TCIA-STANFORD (Figure S2D).
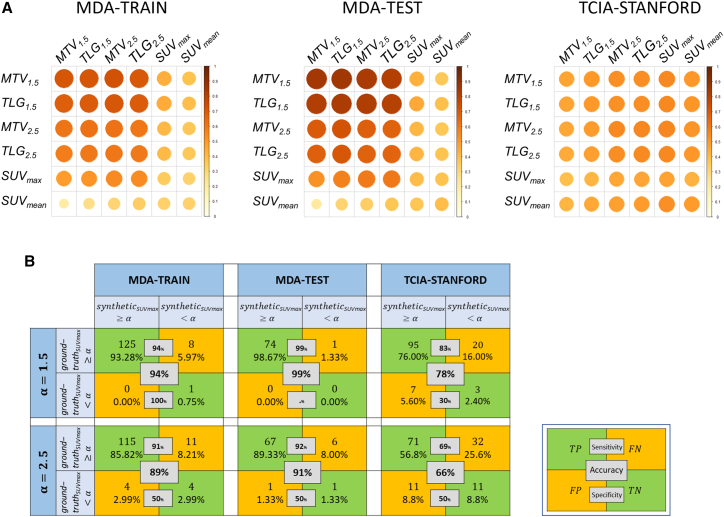
Next, we calculated the pairwise Pearson correlation of imaging metrics (metabolic tumor volume [MTV], total lesion glycolysis [TLG], and mean and maximum standardized uptake values [SUVmean and SUVmax]) between true and generated PET in the MDA-TRAIN, MDA-TEST, and TCIA-STANFORD cohorts (Figure 3A). Relatively high positive correlations were observed for MTV and TLG features consistently in all the cohorts. The highest PET feature correlations were MTV1.5 (ρ = 0.71) for MDA-TRAIN, MTV1.5 (ρ = 0.85) for MDA-TEST, and MTV2.5 (ρ = 0.55) for TCIA-STANFORD. Low correlations were observed for SUVmax and SUVmean features, which may be attributed to the long tail effects of SUV values (refer to STAR Methods). Further, we studied the agreement between predicted and true PET features using Bland-Altman plots (Figure S3). Reasonable agreements were observed across the predicted and true PET consistently in three cohorts for MTV.
Figure 3.
Validation of imaging signal fidelity by PET feature correlation
(A) Pearson correlation for evaluating the pairwise similarity between the synthetic (rows) and ground-truth (columns) PET features in MDA-TRAIN, MDA-TEST, and TCIA-STANFORD cohorts.
(B) Threshold-based confusion matrix for synthetic SUVmax and the ground-truth SUVmax for four different thresholds () in MDA-TRAIN, MDA-TEST, and TCIA-STANFORD cohorts.
Given the observation that synthetic SUVmax had reduced correlation to ground -truth, we computed a threshold-based confusion matrix to evaluate the progressive correlation at different SUV threshold values. As shown in Figure 3B, when the threshold was set at 1.5, the synthetic SUVmax aligned closely with the ground-truth SUVmax. With increasing threshold, SUVmax values of more synthetic PET scans did not pass the threshold, leading to an accuracy of 89%–91% in MDA-TRAIN or MDA-TEST and 66% in TCIA-STANFORD at a high cutoff of 2.5. Taken together, these results systematically validated that a reasonable fidelity of SUVmax exists between the predicted and the ground-truth PET slices.
Furthermore, we assured that the synthetic SUVmax produced by the cGAN model was not solely driven by the tumor/nodule tumor volume. In particular, we computed the correlation between the SUVmax and tumor/nodule volume, with a small correlation for both the ground-truth PET () and synthetic PET () (Figures S4A and S4B). A similar trend was also observed for subgroup analysis of patients with SUVmax < 7 (Figures S4C and S4D). Additionally, we presented a scatterplot illustrating the relationship between ground-truth and synthetic SUVmax in the combined train and test cohorts of MDA-TRAIN, MDA-TEST, and TCIA-STANFORD (Figures S4E and S4F).
Biological inferences are consistent between synthetic and ground-truth PET
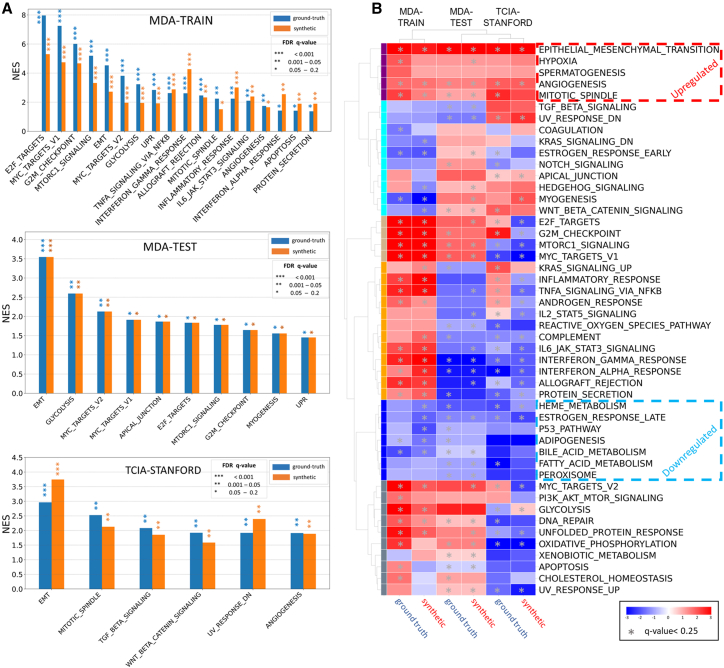
We next performed gene set enrichment analysis (GSEA) to explore radiogenomics correlation12 of an imaging feature (MTV1.5) extracted from true and synthetic PET scans with hallmark pathways. Given the fact that MTV has several advantages over SUVmax, including its robustness to access the spatial extent of metabolically active tumor tissue rather than providing a fragile single value based on the highest activity in a small region,2,13,14 we chose to concentrate our efforts on exploring its radiogenomic correlates. Specifically, we studied MTV1.5, since it shows a higher correlation between synthetic and ground-truth PET scans (Figure 3A). We observed that several cancer-associated hallmark pathways consistently have significant associations with true and predicted MTV1.5, although the enriched pathway lists vary across different cohorts (Figure 4A), which may be attributable to different genes covered in tumors of different cohorts. E2F targets, MYC targets, and G2M checkpoint were the top dysregulated pathways in the MDA-TRAIN cohort; epithelial-mesenchymal transition (EMT), glycolysis, and MYC targets were the top enriched pathways in the MDA-TEST cohort; and EMT, mitotic spindle, and transforming growth factor-β signaling were the top dysregulated pathways in the TCIA-STANFORD cohort. High concordance was observed between synthetic PET and true PET scans regarding the normalized enrichment score of all cancer hallmark pathways calculated, with Pearson correlation of 0.88 (p = 1.3e−16) in MDA-TRAIN for training, 1.0 (p < 1e−16) in MDA-TEST, and 0.74 (p = 1.9e−9) in TCIA-STANFORD, suggesting consistent image-to-genomics alignment.
Figure 4.
Validation of biological fidelity by radiogenomics analysis
(A) Significant hallmark gene sets associated with MTV extracted from ground-truth and synthetic PET for MDA-TRAIN, TCIA-STANFORD, and MDA-TEST cohorts.
(B) The first column shows the unsupervised hierarchical clustering heatmap of hallmark pathways normalized enrichment score for correlation of each pathway with MTV feature from ground-truth and synthetic PET across MDA-TRAIN, TCIA-STANFORD, and MDA-TEST cohorts, where the asterisks represent the significant false discovery rate q value >0.25.
Further unsupervised hierarchical clustering of the pathway enrichment scores from all three cohorts revealed hallmark pathways with consistent positive/negative correlation with MTV1.5 across the cohorts (Figures 4B and S5A). Increased activity reportedly associated with cancer aggressiveness such as EMT, hypoxia, angiogenesis, and mitotic spindle pathways were consistently associated with greater MTV1.5, while biological processes implicated in cancer suppression, such as peroxisome, adipogenesis, heme metabolism, fatty acid metabolism, and bile acid metabolism, were consistently associated with lower MTV1.5 values. Among processes linked to cancer progression, EMT and angiogenesis were the top pathways significantly positively correlated with MTV1.5 values (Figure S5A). Highly similar enrichment plots were observed for the association of each pathway with MTV1.5 from true or synthetic PET in each studied cohort (Figure S6). Interestingly, glycolysis was found to be significantly positively associated with true and synthetic MTV1.5 only in two cohorts, MDA-TRAIN and MDA-TEST (q values from <0.0001 to 0.02). Considering the known association of PET features with glycolysis-related genes in non-small cell lung cancer (NSCLC),15 we further evaluated associations between the true and synthetic MTV1.5 with a glycolytic score representing overall expression of glycolysis-related genes in each cohort. Combining the three cohorts and dichotomizing tumors into glycolysis-high and glycolysis-low groups, we found that the former group has significantly greater MTV1.5 values extracted from either ground-truth or synthetic PET images (Wilcoxon rank-sum test p value 0.03 and 0.05, respectively) (Figure S5B).
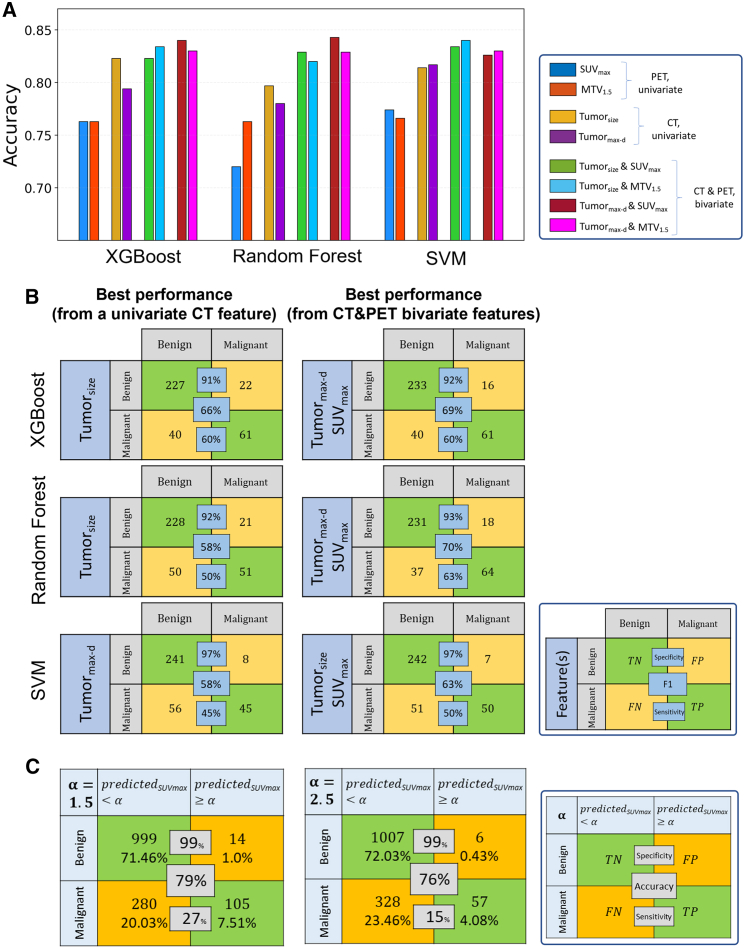
Synthetic PET complements CT for early lung cancer diagnosis
Next, we applied the cGAN model and generated the corresponding PET from the CT scans in the LIDC-IDRI cohort (n = 1,048 for training and n = 350 for testing) to assess whether the synthetic PET could provide additional information to classify the benign vs. malignant nodules. A cutoff value was set at 1.5 for MTV, which is highly correlated with MTV2.5 and TLG features (Figure 3A). We also chose SUVmax, which was less correlated with MTV and TLG-based features (Figure 3A). We first evaluated the accuracy of individual CT features (tumorsize and tumormax-d) or PET features (SUV or MTV), where CT features have demonstrated higher accuracy (training in Figure S7, testing in Figure 5A). Next, we integrated PET and CT features to assess whether PET provides additional value on top of CT and improves the accuracy of lung cancer diagnosis. For the three machine-learning models, we observed consistent augmentation when adding PET features (Figures 5A and S7). For instance, using the XGBoost classifier, the top-ranked models were obtained by adding SUVmax and MTV to tumormax-d and tumorsize, respectively, resulting in training accuracies of 98.1% and 90.8%, and testing accuracies of 84.1% and 83.5%, respectively. These results outperformed the best CT-based prediction, which had a training accuracy of 89.8% and a testing accuracy of 82.3%, when using tumorsize (training accuracy = 89.8%, testing accuracy = 82.3%). Similar patterns were observed for random forest and support vector machine (SVM) models, where PET improved the training accuracy by 3%–7% and testing accuracy by 2%–5%, respectively.
Figure 5.
Validation of clinical value by diagnosing malignant vs. benign from indeterminate pulmonary nodules
(A) Model accuracy in the test cohort (n = 350) obtained from synthetic PET univariate features (MTV, SUVmax), CT univariate features (tumorsize, tumormax-d), and CT and PET bivariate features (tumorsize & MTV, tumorsize & SUVmax, tumormax-d & MTV, tumormax-d & SUVmax) in the LIDC-IDRI cohort.
(B) The first column corresponds to the confusion matrix of the best performance obtained using either of single CT features tumorsize or tumormax-d. The second column corresponds to the confusion matrix of the best performance when adding one synthetic PET feature, SUVmax or MTV.
(C) Threshold-based confusion matrix for synthetic SUVmax and the nodule malignancy at four different thresholds () in the LIDC-IDRI cohort.
Further, we presented the lesion level classification performance by head-to-head comparison of the optimal CT model with the optimal PET-CT model (Figure 5B). In general, during testing the synthetic PET improved the F1 score in lesion detection by 3%, 5%, and 12% in XGBoost, SVM, and random forest, respectively. Also, we observed a 5% and 13% sensitivity increase in SVM and random forest, respectively.
We next explored the relationship of nodule likelihood of malignancy with the synthetic SUVmax (Figure 5C). With the increase of threshold , an increase in the specificity and decrease in the sensitivity were observed as expected. Of note, when we used the cutoff of 1.5, the specificity reached 99% with sensitivity at 27%.
Synthetic PET improves the performance of state-of-the-art CT deep-learning model for predicting lung cancer risks
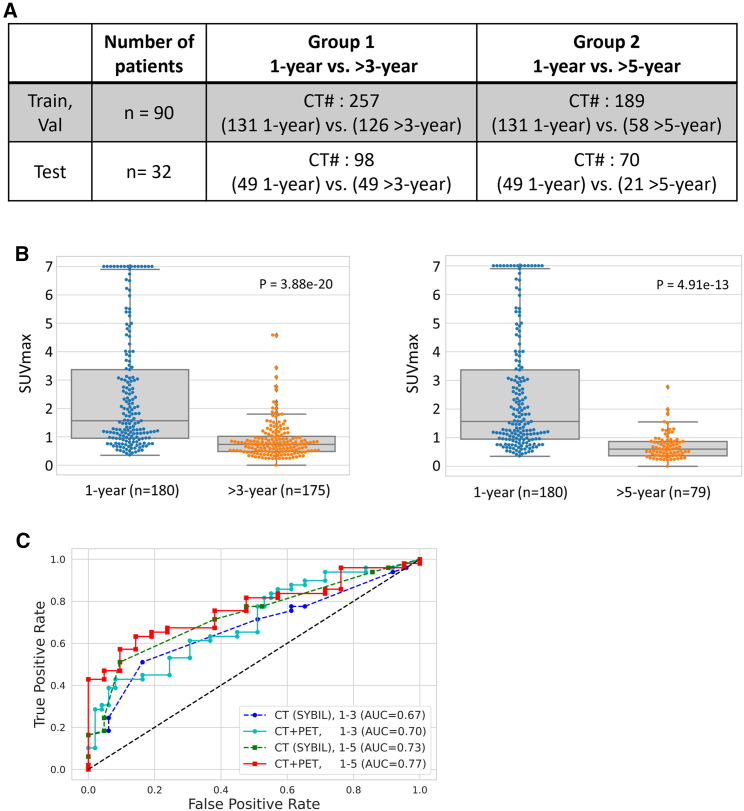
We next sought to test whether synthetic PET scans will add additional value to the CT deep-learning model that has demonstrated high accuracy in predicting lung cancer risks using low-dose CT scan images for lung cancer screening.16 We applied the cGAN model to predict the risk of developing lung cancer in the MDA-SCREENING set (n = 122). We divided this dataset into two subsets. The first subset includes CT scans from patients who developed lung cancer within 1 year vs. patients who were diagnosed with lung cancer after >3 years, consisting of 355 CT scans. The second subset consists of CT scans from patients who were diagnosed with cancer within 1 year vs. patients who were diagnosed with lung cancer after >5 years, totaling 259 CT scans (Figure 6A). We randomly divided the patients into discovery (n = 90) and testing (n = 32). We then obtained the synthetic PETs.
Figure 6.
Validation of clinical value of synthetic PET by predicting the risk of developing lung cancer
(A) Distribution of CT scans in training, validation, and test sets in both group 1 and group 2.
(B) Two box plots were used to compare the synthetic SUVmax values in group 1 (lung cancer diagnosed within 1 year vs. lung cancer diagnosed at >3 years) and group 2 (lung cancer diagnosed within 1 year vs. lung cancer diagnosed at >5 years).
(C) Receiver-operating characteristic curves for CT and CT plus synthetic PET analysis along with their respective AUC values for both group 1 and group 2.
We observed that the SUVmax measured on synthetic PET were significantly different between the low-risk vs. high-risk groups (Figure 6B). Next, we employed a DenseNET-based autoencoder to extract its latent features from synthetic PETs. In addition, we obtained the CT deep-learning-based risk prediction based on a pre-trained model (i.e., SYBIL16) risk. By combining the PET features with SYBIL risk, we trained a joint CT and PET model for risk prediction. We observed higher area-under-the-curve (AUC) values when adding the synthetic PET features. Specifically, for predicting cancer development within 1 year vs. >3 years, the AUC increased from 0.67 to 0.70; and for the 1-year vs. >5-year group, the AUC increased from 0.73 to 0.77 (Figure 6C).
Radiologist staging on synthetic PET scans achieves accuracy similar to that of staging on true PET with pathological staging as gold standard
To further assess the potential clinical utility of synthetic PET, two thoracic radiologists directly staged the lung cancer on synthetic PET to compare head-to-head with ground-truth PET based on 30 lung cancer cases randomly selected from the testing group (for details see STAR Methods and Table S1B). The final pathological staging was used as the gold standard. The radiologists reading CT paired with ground-truth PET achieved an overall accuracy of 70% compared to the pathological stage, with 13.3% downstaged and 16.7% upstaged (Figure 2F). When reading CT paired with synthetic PET, their accuracy was 66.7%, with 16.7% downstaged and 16.7% upstaged (Figure 2G). Interestingly, compared to ground-truth PET, radiologists’ reading from synthetic PET is better aligned with pathological stage I and II patients, which dropped for stage III patients. Furthermore, two radiologists exhibited an 87% consensus in staging (Figure 2H) when one read ground-truth PET and another read synthetic PET.
Synthetic PET predicts prognosis after standard of care
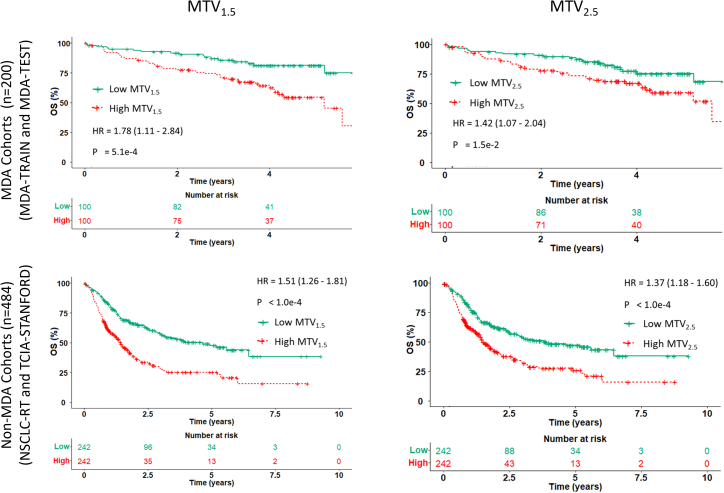
Given the correlation of imaging features obtained from synthetic PET and ground-truth PET (Figure 3) and prior clinical evidence,14,17,18 we next assessed the prognostic performance of MTV1.5 and MTV2.5 to stratify patients’ overall survival (OS) using Kaplan-Meier analysis (Figure 7). We observed that the synthetic MTV1.5 and MTV2.5 can stratify patients’ OS in both MDA and non-MDA cohorts: for MTV1.5, MDA cohort hazard ratio (HR) = 1.78, 95% confidence interval (CI) 1.11–2.84 ) and non-MDA cohort HR = 1.51, 95% CI 1.26–1.81 ); for MTV2.5, MDA cohort HR = 1.42, 95% CI 1.07–2.04 ) and non-MDA cohort HR = 1.37, 95% CI 1.18–1.60 ).
Figure 7.
Validation of clinical value of synthetic PET by predicting overall survival
Kaplan-Meier curves of patients’ overall survival (OS) stratified by MTV1.5 and MTV2.5 features obtained from the synthetic PET images on MDA cohorts (MDA-TRAIN and MDA-TEST) and non-MDA cohorts (TCIA-STANFORD and NSCLC-RT).
Furthermore, we compared the prognostic value of individual features in a head-to-head fashion between synthetic PET and ground-truth PET when it was available (Figure S8A). We observed that the prediction capacity of synthetic MTV declined as the SUV cutoff threshold was raised from to , possibly due to decreasing correlation of the MTV feature extracted from synthetic and true PET for higher SUV cutoff thresholds (Figure 3). While it was expected that the synthetic PET would achieve an inferior performance as measured by concordance index compared to ground-truth PET, it did achieve statistically meaningful prediction in the majority of cases. For SUVmax, we observed a relatively lower prognosis compared to MTV and TLG, especially the cases with low fidelity of ground truth (Figures S8B and S8C). More interestingly, the synthetic PET offered meaningful prediction for stratifying a patient’s prognosis for the NSCLC-RT cohort when true PET data were not available.
Discussion
In this study, we developed and validated a conditional GAN-based pipeline to synthesize PET of high fidelity from diagnostic CT scans from multi-center multi-modal lung cancer datasets (n = 1,478). Our proposed computational framework demonstrates robust performance during external validation assessed by thoracic radiologists, measuring image quality and a Turing test, whereby the synthetic PET scans were found to be of equivalent quality and tumor avidity when compared side by side to the ground-truth FDG-PET scans. The synthetic PET images were also consistent with true PET in radiogenomics validation by demonstrating significant correlation of imaging features with dysregulated biological processes. More importantly, the synthetic PET demonstrated additional diagnostic value in distinguishing malignant from benign lung nodules compared to CT nodule size and maximum diameter measurements, in accurately staging patients compared to pathological staging, in predicting risk of developing lung cancer during follow-up, and in predicting survival of lung cancer patients. Taken together, our proof-of-concept study proves the feasibility of applying deep learning to obtain high-fidelity functional imaging translated from the anatomical imaging. With further tuning and validation, this pipeline may potentially add value in cancer screening, staging, diagnosis, and prognosis.
It is worth clarifying that synthesizing PET is not intended to replace PET scanning but rather to offer complementary value to CT data, which are more popular, especially when PET images are difficult to acquire. A few pilot studies have been carried out to show the feasibility of CT-to-PET conversion in some clinical applications, such as reducing the false-positive rate in detecting malignant liver lesions19 and improving the differentiation between malignant lymph nodes from thyroid tissue in head and neck cancer.20 However, such attempts at clinical translation have been missing in lung cancer, the leading cause of cancer death, where PET-CT is part of the standard of care across different clinical settings. To the best of our knowledge, this is the largest reported CT-to-PET mapping study based on multi-center multi-modality datasets of clinical, radiological, and molecular information. The details of our algorithm development and evaluation according to the newly published guidelines to develop and evaluate artificial intelligence (AI) specifically in nuclear medicine21,22 are detailed in Tables S2 and S3. Our study has made important contributions in the following ways.
At the technical level, we leveraged the 2.5D convolutional neural network that combines neighboring slice information to augment performance and to balance computational cost. Moreover, in contrast to generation of a fake image, which utilizes a fixed-length random vector to generate a photorealistic sample, we used ResUNet++ as backbone for the generator without adding a random vector to learn the definitive CT-to-PET mapping that increases the reproducibility. Collectively, this has also led to our cGAN model outperforming the diffusion model UNSB (unpaired neural Schrödinger bridge),23 which holds a distinct advantage over classical diffusion models by bypassing the limitation imposed by the Gaussian prior assumption (Figure S9).
Beyond the classical imaging signal validation, we have pioneered a systemic validation incorporating different fronts, including image quality and Turing test evaluated by thoracic radiologists and biological validation by radiogenomics design. Contrasting with prior studies, which utilized radiologists to qualitatively assess the image in limited small tumor regions,24 we asked them to quantitatively evaluate the quality and contrast of synthetic PET in the whole field. In addition, we directly fed these synthetic PET scans for cancer staging to radiologists—a common clinical task performed on PET. With pathological staging as ground truth, it is encouraging to see that staging on synthetic PET has achieved an equivalent accuracy with ground-truth PET scans when pairing with CT scans.
Furthermore, we validated synthetic PET at the biological level in three independent cohorts to prove radiogenomics fidelity of cancer-related hallmark pathways between true and synthetic PET. Both true and synthetic PET avid tumors demonstrated upregulated pathways involving EMT, hypoxia, and angiogenesis, corroborating the previous observations of high 18F-labeled FDG activities in cancer cells undergoing EMT, particularly in hypoxic regions.25 In addition, we observed that aggressive tumors with higher MTV, as stratified by true or synthetic PET, were consistently associated with elevated glycolytic activity and downregulated metabolism of fatty acid, bile acid, and heme. This is consistent with a recent study on The Cancer Genome Atlas lung adenocarcinomas showing that high glycolytic and low lipid metabolism exhibited high metastatic potency and poor survival.26
A robust CT-to-PET translation framework would bring several potential impacts on oncology practice. Our data suggested that synthetic PET can confer additional value on top of CT scans to distinguish benign from malignant lung nodules, a clinically unmet need for lung cancer screening and early diagnosis. In addition, it can augment the CT deep-learning model to accurately identify patients at high risk of developing lung cancer.27 We also demonstrated a similar prognostic capacity of synthetic PET scans compared to true PET. With sufficient training data for individual tasks to further improve the model, this technology can be of great potential from a patient safety perspective and financial standpoint. Because of concerns regarding higher radiation doses compared to routine CT scans and radiotracer exposure, PET is used with caution for certain patient populations, including young children and pregnant women. Recently, a strong association between cumulative radiation dose from CT scans and the risk of hematological malignancies was reported for young people.6 Our models can be potentially applied to routine CT scans to extract additional clinically relevant data from these patients and reduce their exposure to radiation. Also, it may reduce the frequency of repetitive PET scans, and the substantially lower cost from routine CT scans may relieve the increasing healthcare cost in western countries. More importantly, the optimized model could be swiftly deployed in low-income countries to fill the gap and improve cancer staging and management. The encouraging results from our study, along with other structure-to-function transfer studies such as hyperpolarized gas MRI ventilation scans derived from CT28 and SPECT perfusion scans based on CT,11 underscore the need for future research investigations. These studies hold the potential to facilitate the development of an all-in-one multi-modality scanner designed to optimize the diagnosis and prognosis prediction of lung cancer. Finally, as PET scans are also widely used in managing other medical conditions such as cardiac and neurological diseases, our proposed tool and pipeline may have a broader impact beyond cancer care.
It is worth clarifying that cross-modality medical imaging synthesis is conceptually different from the fake image generation in computer vision,29 although they share some common technical foundations such as using a GAN algorithm. Fake image generation (e.g., deepfake30) aims to produce photorealistic images to fool people and can be viewed as an interpolation problem to produce non-existing samples. By contrast, cross-modality imaging synthesis, such as the CT-to-PET translation presented here, can be mathematically formulated as a regression problem, which aims to learn a latent transmodality mapping function. This is where deep learning outperforms conventional algorithms, as it efficiently learns any complex non-linear function according to the universal approximation theorem.31 Our study adds important evidence to support the growing utility of deep learning for cross-modal/platform synthesis in radiographic scans,8,11,32 digital pathology for microscopy-based drug discovery,33 and immunohistochemical image quantification.34 When a certain data type is missing or difficult to access, synthetic data will potentially help to fill the coverage gaps in order to build trustworthy AI models.35
Limitations of the study
Our study has several important limitations. First, although we included large multi-center multi-modal datasets, the results need to be interpreted as proof-of-concept and hypothesis-generating research. The primary aim of this pilot study is to establish the feasibility of generating synthetic PET images from CT scans as well as to assess the clinical and biological values of synthetic PET images through radiologist evaluations, radiogenomics correlations, radiomics analyses, and deep-learning experiments. It is important to highlight that while our proof-of-concept study holds promise, it is not intended to supplant conventional PET imaging or alter current clinical practices. Instead, it serves as a critical stepping stone, warranting further investigation using a prospective design with fine-grained lung cancer subtypes to bring this to clinical translation. Second, PET imaging has limited value in lesions with predominant ground-glass opacity36 and nodal immune flare after immunotherapy.37 Unfortunately, a synthetic PET approach would be expected to inherit these intrinsic limitations. Here, we have leveraged limited imaging metrics from synthetic PET scans that have been extensively examined in prior studies, including MTV, SUV, and TLG, to prove the added value. Future efforts are needed to develop next-generation imaging biomarkers beyond conventional metrics to overcome these challenges through radiomics,38,39,40,41 habitat imaging,42,43 and deep learning.27,44 Moreover, with the rapid evolution of generative AI, more sophisticated pipelines45 can be leveraged to improve the quality of synthetic scans. In the end, it remains challenging to integrate complex AI models into clinical workflows, which is beyond the scope of the current study, and future efforts are critical to overcoming these hurdles.46
In conclusion, we have developed and validated a cGAN-based CT-to-PET translation framework based on multi-center PET and CT scans. The synthetic images were extensively validated by clinical thoracic radiologists and biological correlation using a novel radiogenomics analysis. More importantly, the synthetic PET scans demonstrated promising diagnostic values in cancer staging, improving early detection of lung cancer, stratifying lung cancer development in a high-risk population, and cancer prognostication. All things considered, future studies in a prospective setting are warranted to validate and translate these intriguing findings to clinical oncology practice.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| MDA-TRAIN/TEST/SCREENING | This paper | N/A |
| TCIA-STANFORD | Bakr, S. et al.47 | https://wiki.cancerimagingarchive.net/display/Public/NSCLC+Radiogenomics |
| LIDC-IDRI | Armato III, S. G. et al.48 | https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=1966254 |
| NSCLC-RT | Aerts, H. J. et al.49 | https://www.cancerimagingarchive.net/ |
| Software and algorithms | ||
| R 3.6.1. | The R Project for Statistical Computing | https://www.r-project.org |
| Python version 3.8 | Python software | https://www.python.org/ |
| Source code for the deep learning model | This paper | https://github.com/WuLabMDA/Synthetic-PET-from-CT/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Jia Wu (JWu11@mdanderson.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
In our study, the internal datasets from MD Anderson, which contain sensitive patient information, are not publicly available due to privacy and institutional policy. However, the publicly available datasets used in our research can be accessed online, with detailed instructions and links provided in the “patient cohort” section of this paper.
-
•
Source code for the deep learning model is available at: https://github.com/WuLabMDA/Synthetic-PET-from-CT/
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and subject details
Patient cohorts
This study was approved by the Institutional Review Board (IRB) of The University of Texas MD Anderson Cancer Center. We collected 5 multi-center multi-modal datasets from a total of 1478 lung cancer patients. The details of individual datasets and their roles are summarized in Figure 1E and elaborated in the following.
-
•
MDA-TRAIN cohort(n = 132): contains lung cancer patients who underwent surgical resection at MD Anderson (see details elsewhere50). We collected the diagnostic CT, PET/CT, gene expression, and clinicopathological and follow-up data. This cohort was used to train the deep learning model and used for biological and clinical validation. Clinical characteristics of this cohort is presented in Table S1A.
-
•
MDA-TEST cohort(n = 75)& TCIA-STANFORD cohort(n = 125): MDA-TEST contains lung cancer patients who underwent surgical resection at MD Anderson (see details elsewhere51,52), and TCIA-STANFORD cohort contains lung cancer patients treated at Stanford,47 which is publicly shared as NSCLC-Radiogenomics through the TCIA website: https://wiki.cancerimagingarchive.net/display/Public/NSCLC+Radiogenomics. The diagnostic CT, PET/CT, gene expression, and clinicopathological and follow-up data were complied. These cohorts were used to validate the deep learning model, and used for imaging, biological, and clinical validation. Clinical characteristics of these two cohorts are presented in Table S1A.
-
•
LIDC-IDRI cohort(n = 665): LIDC-IDRI contains lung cancer CT screening scans from Lung Image Database Consortium with a total of 1398 nodules with detailed radiologists’ annotation and diagnosis48 (see details at https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=1966254). This dataset contains the lesion annotations and their segmentations for nodule classification, including the nodule information such as CT slice number, malignancy rating, texture information, and the coordinate of the center of the nodules. Four experienced thoracic radiologists independently assigned the malignancy rating from to for all the nodules, where a higher value indicates a higher likelihood of malignancy. To minimize ambiguity, only nodules with malignancy rating of or were selected, while the undifferentiated nodules with malignancy rating of were discarded. This resulted in a total of nodules for further analyses. We have downloaded the screening CT scans, and diagnosis readings. This cohort was used to validate the deep learning model in lung cancer diagnosis.
-
•
NSCLC-RT cohort(n = 359): contains lung cancer patients who received radiation treatment.49 This dataset is publicly shared as NSCLC-Radiomics through the TCIA website (https://www.cancerimagingarchive.net/collection/nsclc-radiomics/). CT scans, and clinical and follow up data were obtained. This cohort was used to validate the prognostic value of the deep learning model.
-
•
MDA-SCREENING (n = 122): comprises longitudinal CT scans obtained from 122 patients who underwent lung cancer screening at MD Anderson, totaling 355 CT scans. This cohort was used to test the predictive value of developing lung cancer based on the deep learning model.
Method details
Study design
Based on the high-quality multimodal data (including FDG-PET and CT, clinicopathological, genomic, and survival information) from different centers, we developed a CT-to-PET deep learning model and evaluated its fidelity as well as value in the clinical context (Figure 1). We first developed a conditional generative adversarial network (cGAN) based model that can generate FDG-PET from CT scans trained from paired CT and PET scans in the MDA-TRAIN cohort. Then, we externally validated the performance of the CT-to-PET model on MDA-TEST and TCIA-STANFORD cohorts by comparing the synthetic PET with acquired PET scans as the ground-truth through the lens of radiologists and quantitative metrics. Next, we biologically validated the model by assessing the degree of consistency of radiogenomics correlates53 with the true PET and synthetic PET scans, leveraging the paired gene expression data in MDA-TRAIN, MDA-TEST and TCIA-STANFORD. Finally, we tested the added clinical value of models for both lung cancer diagnosis in LIDC-IDRI and prognosis in NSCLC-RT.
Imaging data preprocessing
We collected the 18F-FDG PET/CT scans and separate diagnostic CTs. The PET images were reconstructed with an ordered-subset expectation maximization (OSEM) algorithm, using the co-acquired CT data for attenuation correction. We computed standardized uptake value (SUV) maps for the FDG-PET images. The PET SUV map was registered to the diagnostic CT scans using elastix toolbox.54 The registration results were visually checked and manually corrected to mitigate the uncertainties due to respiratory motion and positioning differences when necessary. For MDA-TRAIN and MDA-TEST, radiologist collaborators manually contoured the primary tumor on diagnostic CT scans. For TCIA-STANFORD, LIDC-IDRI, and NSCLC-RT, the tumor contours were provided along with the original imaging data. In addition, we extracted the lung masks based on pre-trained U-net55 to help identify the slices covering lung regions from the original CT images. The CT scans were displayed using lung window/level settings.
Conditional generative adversarial network (cGAN) pixel by pixel maps CT to FDG-PET
A cGAN model was adopted to learn a non-linear mapping function from input CT images in order to output PET images, where it was extended on top of the original pix2pix translation algorithm.56 The objective function in our cGAN is defined as:
| (Equation 1) |
where and are the generator and discriminator, respectively, is the optimized generator, denotes the L1-norm. The first term in Equation 1 is the L1-norm of the pixel-wise difference between the learned PET and ground-truth PET and its goal is to make them closer. is a regularization parameter that balances between the two terms; is the input CT slide(s) and is the ground-truth PET slice. The second term can be extended as follows:
| (Equation 2) |
Essentially, Equation 2 calculates the binary cross entropy (BCE) loss where the discriminator gives higher loss value if it cannot classify the generator’s output as fake and the ground-truth PET output as real. In contrast, the generator learns to generate an output to fool the discriminator with a lower loss value if the discriminator cannot catch its output as fake.
Similar to the pix2pix translation algorithm,56 we did not train our cGAN using a random noise vector as input for the generator . The main reason is that we aim to produce deterministic PET images as the output of generator. Instead, the ResUNet++ architecture was utilized for the generator, which has outperformed U-Net and ResUNet in several image-to-image applications.57 For the discriminator, we employed the original structure used in the image-to-image translation algorithm,56 with a total of five convolution layers with a kernel size of four.
cGAN model configuration
The MDA-TRAIN cohort was utilized for training and internal validation of the deep learning model using a 5-fold cross-validation approach. A total of 120 patients were employed for training, and the remaining 12 patients were used for validation and fine-tuning of the generated PET image quality. Registered diagnostic CT and FDG-PET slices from the lung region were used to train the model. Specifically, the cGAN model was configured with a 2.5-dimentional (2.5D) scheme, which took seven consecutive axial slices of CT () as input to predict the central PET slice () (Figure 1A). Of note, we focused on the lung regions in the current study to mitigate the computational cost. In total, the training dataset was comprised of unique sets of data arrays of seven CT slices and one PET slice.
One challenging aspect of CT to PET translation is related to accurately predicting the dynamic ranges of SUV. As we observed in the training cohort, the SUVmax of primary tumors fluctuated in the range of with a median of . The distribution of SUV scores demonstrated a long tail, with fewer and fewer voxels associated with higher SUV values. Two strategies were used to mitigate the long tail effect. First, we introduced a maximum cutoff (SUV = 7) to clip and normalize the SUV map. Since SUV 2.5 is clinically accepted to distinguish between malignant and benign tumors and also predict patient’s survival,17 the selected cutoff balanced between computational challenges and clinical value. Second, we applied gamma compression with on the SUV intensity maps. Thus, the distribution range for lower SUV values becomes wider to increase its resolution, with a narrower range for the less frequent higher SUV values.
cGAN model training
We applied the common strategy to train the cGAN model, alternating between one step of optimizing and one step of optimizing . The Adam optimizer with and was used in optimizing both and . The initial learning rate was set to for epochs and batch size of 5. For data augmentation, we used on-the-fly augmentation: affine transformation with rotation (−π/4, +π/4), translation , scaling , shearing (−8, +8).
Imaging signal level validation by thoracic radiologists
We systematically evaluated the fidelity of synthetic PET scans on testing cohorts. First, visual comparison was employed for validation, where we carried out a 2-step imaging Turing test. Two experienced thoracic radiologists qualitatively assessed the scans independently. In task 1, they were blinded to the information that any PET images were synthetic and asked to assess PET scans paired with corresponding CT (randomly sampled from 60 patients’ from the MDA-TEST and TCIA-STANFORD cohorts) for: 1) subjective quality score (5-point system, with 1 as poor, 3 as adequate, 5 as great); 2) uptake of dominant lung lesions relative to mediastinal blood pool (5-point system, with 1 as low, 3 as equal, 5 as high). In task 2, the radiologists were informed of the presence of synthetic PET images and asked to identify whether the PET images (randomly sampled from 30 patients’ from the MDA-TEST and MDA-TRAIN cohorts) were real or synthesized. The questionnaires of tasks 1 and 2 are available online at (https://drive.google.com/drive/folders/13qlGhYc5jl9DrlINPmzAxxRiW8RYBFmW?usp=sharing).
Second, we quantitatively compared the synthetic PET images with the ground-truth PET scans. In particular, we computed the structural similarity index measure (SSIM)58 and the root-mean-square error (RMSE) for both the training cohort (MDA-TRAIN) and two external validation cohorts (MDA-TEST and TCIA-STANFORD), by randomly sampling 195 PET slices from a subset of 13 patients for each cohort. The SSIM index provides a fractional value between 0 and 1 where a higher value indicates more similarity between two images. This index is calculated by weighted multiplications of three global component: luminance, contrast, and structure. While SSIM captures global components, RMSE is calculated based on pixel-to-pixel difference, which makes it very sensitive to misalignment between the two images. Of note, the ground-truth PET scans cannot be perfectly registered to the diagnostic CT images, which were taken on a different date, due to inevitable respiratory motion and positioning differences. Therefore, the RMSE is not expected to be zero even for the ground-truth PET. Nevertheless, RMSE can be used for comparing the model’s performance of training and testing.
Third, we evaluated the consistency of conventional imaging metrics extracted from ground-truth PET and synthetic PET, including SUVmax, SUVmean, metabolic tumor volume (MTV), and total lesion glycolysis (TLG). Beyond directly assessing the clinical value of synthetic PET in clinical workflow by the radiology team, we extend the evaluation of how it would assist cancer diagnosis and prognosis in the context of radiomics analysis. Specifically, we focus on the putative PET metrics, including metabolic tumor volume (MTV), total lesion glycolysis (TLG), in conjunction with SUVmax to provide a comprehensive characterization of tumors, which have been extensively validated in prior studies.2,59 The Pearson’s correlation was used for evaluating the pairwise similarity. Also, Bland–Altman plots used in analyzing the agreement between two different types of PET features. In particular, we assessed the consistency of the SUVmax in the synthetic PET slices. To do so, we introduced a threshold-based confusion matrix (CM) to further examine the change of SUVmax. The elements of the matrix represent the number of cases in which the SUVmax exceeds or falls below a given threshold in the generated and ground-truth PET slices. We calculated this matrix with different SUV thresholds () in MDA-TRAIN, MDA-TEST and TCIA-STANFORD cohorts.
Biological validation using gene expression profiles
We used the gene set enrichment analysis (GSEA, version 4.2.3, default parameters)60 of RNA sequencing data of the MDA-TRAIN and TCIA-STANFORD cohorts and microarray data of the MDA-TEST cohort to identify biologic processes and pathways associated with imaging features extracted from true and synthetic PET scans.
Hallmark gene sets from the Molecular Signatures Database (MSigDB) were tested for associations with ground-truth and synthetic PET features from metabolic tumor volume and total lesion glycolysis in the 3 cohorts. Only known genes, found in all samples in the cohorts, were used for the analysis. The Pearson correlation of log-transformed PET feature values with the gene expression profile across samples was used for ranking genes in each cohort. Gene sets significantly correlated with ground-truth or synthetic PET features were then compared using the normalized enrichment score (NES) and the false discovery rate (FDR) q-value. To discover biological processes that were consistently positively or negatively correlated with PET features across three cohorts, we performed unsupervised hierarchical clustering of all identified NES followed by meta-analysis. Namely, the Fisher’s method implemented in R library “metap” was used to combine the q-value separately for ground-truth PET and synthetic PET. Biological processes with combined q-values less than 0.25 were considered as significantly associated with PET features.
Associations of true and synthetic PET imaging features with the tumor glycolytic activity were evaluated using a glycolytic score15 representing overall expression of glycolysis related genes in the tumor and is calculated as the geometric mean of two genes GPI and GAM4 (log2-transformed). The scores were initially computed separately for each cohort then normalized using the standard score and merged. For tumors having high or low glycolytic activity, the joined set of the glycolytic scores was dichotomized according to the median. Then the Mann-Whitney test was used to determine significance of the difference in values of ground-truth and synthetic PET features, between groups with low versus high glycolytic activity.
Clinical validation: Cancer diagnosis of indeterminant pulmonary nodules
We evaluated the clinical value of the synthetic PET for improving cancer diagnosis in lung cancer screening in LIDC-IDRI cohort. Based on the extracted imaging features from synthetic PET scans, we built different machine learning models (including XGBoost, random forest, and SVM) to classify malignant versus benign nodules, where we randomly split the indeterminate pulmonary nodules (IPNs) into training (n = 1048) and testing (n = 350) sets. To address the added value of synthetic PET on top of CT, we used parsimonious models with limited features to mitigate model overfitting risk. In addition, we also tested the impact of dynamic cutoff values of SUVmax on the prediction accuracy of benign or malignant.
Clinical validation: High-risk patient identification for developing lung cancer
Next, we also applied the deep learning approach to synthetic PET for predicting individual patients’ risk of developing lung cancer. This analysis was carried out on MDA-SCREENING set. DenseNET-based autoencoder was applied to extract PET latent features. Also, we a pre-trained CT deep learning model (i.e., SYBIL16) to obtain the risk of developing lung cancer. For combining PET features and CT-based SYBIL risk, random forest classifier was trained. In order to explore the association between SUVmax from synthetic PET images and the risk of cancer development, we enlisted the assistance of our radiologists, who performed segmentation on the lung lesions of each CT scan for SUVmax extraction.
Clinical validation: Cancer staging by radiologists on synthetic PET with pathology as gold standard
Two radiologists assess the synthetic PET capability for accurately staging the lung cancer patients when replacing the ground truth PET. We randomly selected 30 patients, with 10 patients assigned to each stage of lung cancer (stages I, II, and III) from the testing group (MDA-TEST cohort). For all 30 patients, we collected their pathological staging after surgery, which was used as the ground truth label for evaluating the imaging staging. After obtaining their synthetic PET images, we conducted the experiment in a blind way in which two in-house radiologists were blind of PET image types (synthetic or real). For each lung cancer cases, one radiologist provided the stage reading based on the CT paired with ground-truth PET, and another radiologist independently provided the stage based on the CT paired with the synthetic PET. To mitigate the bias, each radiologist randomly read 50% cases with read PET and 50% cases with synthetic PET (Table S1B).
Clinical validation: Lung cancer survival prediction after standard treatment
We performed survival analysis using the tumor features obtained from the synthetic PET images, and independently tested them on MDA-TRAIN, MDA-TEST, TCIA-STANFORD, and NSCLC-RT cohorts to test whether the synthetic PET can provide clinically relevant information comparable to ground-truth PET scan. In particular, we evaluated the individual feature’s prognostic value to compare between synthetic PET and ground-truth PET.
Quantification and statistical analysis
Statistical analysis
We reported the diagnostic model’s F1 score, sensitivity, specificity, and accuracy when classifying an indeterminate lung nodule into benign versus malignant. Receiver operating characteristic (ROC) curve and area under the curve (AUC) analysis were employed as evaluation metrics to assess the performance and effectiveness in predicting the lung cancer development. For prognostic evaluation, Harrell’s C statistics (C-index) was used to measure the goodness of fit between a PET feature and overall survival (OS) time. The Kaplan-Meier (KM) curves of patients’ OS along with their hazard ratio (HR) and p value were used to show the performance of the tumor features extracted from the individual PET features. The statistical tests were double-sided, with p values less than 0.05 or FDR less than 0.25 assumed statistically significant. All statistical analyses were performed in R 3.6.1.
Acknowledgments
This work was supported by the generous philanthropic contributions to The University of Texas MD Anderson Lung Moon Shot Program and the MD Anderson Cancer Center support grant P30 CA016672. This work was supported by the Tumor Measurement Initiative through the MD Anderson Strategic Initiative Development Program. The research was partially supported by the National Institutes of Health grants R00 CA218667, R01 CA262425, and R01 CA276178. This work was supported by generous philanthropic contributions from Mrs. Andrea Mugnaini and Dr. Edward L.C. Smith, and was further supported by Rexanna’s Foundation for Fighting Lung Cancer.
Author contributions
M.S., J.Z., and J.W. conceived and designed the study. S.J.S., M.B.S., R.B., and M.S. acquired the data. M.S., T.V.K., and M.A. carried out the statistical analyses. M.S. developed, trained, and applied the artificial neural network. T.V.K. implemented the radiogenomics analysis. M.S., T.V.K., G.G., Y.L., M.Q., and C.C.W. carried out the imaging fidelity analysis. M.S., G.G., Y.L., M.Q., C.C.W., J.Z., and J.W. helped with the clinical validation. M.S., T.V.K., S.J.S., and J.W. implemented quality control of data and the algorithms. All authors had access to the data presented in the manuscript. All authors analyzed and interpreted the data. M.S., J.Z., C.C.W., and J.W. prepared the first draft of the manuscript. All authors read and approved the final version of the manuscript. All authors were responsible for the final decision to submit the manuscript for publication.
Declaration of interests
T.C. reports speaker fees and honoraria from The Society for Immunotherapy of Cancer, Bristol Myers Squibb, Roche, Medscape, and PeerView; having an advisory role or receiving consulting fees from AstraZeneca, Bristol Myers Squibb, EMD Serono, Merck & Co., Genentech, and Arrowhead Pharmaceuticals; and institutional research funding from AstraZeneca, Bristol Myers Squibb, and EMD Serono. L.Y. has grant support from Lantheus Inc. D.L.G. has served on scientific advisory committees for Menarini Ricerche, 4D Pharma, Onconova, and Eli Lilly and has received research support from Takeda, Astellas, NGM Biopharmaceuticals, Boehringer Ingelheim, and AstraZeneca. N.I.V. receives consulting fees from Sanofi, Regeneron, Oncocyte, and Eli Lilly and research funding from Mirati, outside the submitted work. J.D.H. is on the Scientific Advisory Board of Imagion Biosystems. J.Y.C. reports research funding from BMS-MDACC and Siemens Healthcare, and consultation fees from Legion Healthcare Partners. L.Y. has grant support from Lantheus Inc. M.C.B.G. has received research funding from Siemens Healthcare. I.W. has received honoraria from Genentech/Roche, AstraZeneca, Merck, Guardant Health, Flame, Novartis, Sanofi, Daiichi Sankyo, Dava Oncology, Amgen, GlaxoSmithKline, HTG Molecular, Jansen, Merus, Imagene, G1 Therapeutics, Abbvie, Catalyst Therapeutics, Genzyme, Regeneron, Oncocyte, Medscape, Platform Health, Pfizer, Physicians’ Education Resource, HPM Education, and Aptitute Health; additionally, I.W. has received research support from Genentech, Merck, Bristol-Myers Squibb, Medimmune, Adaptive, Adaptimmune, EMD Serono, Pfizer, Takeda, Amgen, Karus, Johnson & Johnson, Bayer, Iovance, 4D, Novartis, and Akoya. D.L.G. has served on scientific advisory committees for Menarini Ricerche, 4D Pharma, Onconova, and Eli Lilly and has received research support from Takeda, Astellas, NGM Biopharmaceuticals, Boehringer Ingelheim, and AstraZeneca. J.V.H. reports being on scientific advisory boards for AstraZeneca, Boehringer Ingelheim, Genentech, GlaxoSmithKline, Eli Lilly, Novartis, Spectrum, EMD Serono, Sanofi, Takeda, Mirati Therapeutics, BMS, and Janssen Global Services; receiving research support from AstraZeneca, Takeda, Boehringer Ingelheim, and Spectrum; and receiving licensing fees from Spectrum. C.C.W. reports research support from Medical Imaging and Data Resource Center from NIBIB/University of Chicago and royalties from Elsevier. J.Z. reports serving on the consulting/advisory board of Bristol-Myers Squibb, AstraZeneca, Novartis, Johnson & Johnson, GenePlus, Innovent, Varian, and Catalyst, and receiving research grants to institution from Merck, Novartis, and Johnson & Johnson. J.W. reports research funding from Siemens Healthcare.
Published: March 11, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101463.
Supplemental information
References
- 1.Fletcher J.W., Djulbegovic B., Soares H.P., Siegel B.A., Lowe V.J., Lyman G.H., Coleman R.E., Wahl R., Paschold J.C., Avril N., et al. Recommendations on the use of 18F-FDG PET in oncology. J. Nucl. Med. 2008;49:480–508. doi: 10.2967/jnumed.107.047787. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Velloso M.J., Bastarrika G., de-Torres J.P., Lozano M.D., Sanchez-Salcedo P., Sancho L., Nuñez-Cordoba J.M., Campo A., Alcaide A.B., Torre W., et al. Assessment of indeterminate pulmonary nodules detected in lung cancer screening: Diagnostic accuracy of FDG PET/CT. Lung Cancer. 2016;97:81–86. doi: 10.1016/j.lungcan.2016.04.025. [DOI] [PubMed] [Google Scholar]
- 3.Shim S.S., Lee K.S., Kim B.-T., Chung M.J., Lee E.J., Han J., Choi J.Y., Kwon O.J., Shim Y.M., Kim S. Non–small cell lung cancer: prospective comparison of integrated FDG PET/CT and CT alone for preoperative staging. Radiology. 2005;236:1011–1019. doi: 10.1148/radiol.2363041310. [DOI] [PubMed] [Google Scholar]
- 4.Wahl R.L., Jacene H., Kasamon Y., Lodge M.A. From RECIST to PERCIST: evolving considerations for PET response criteria in solid tumors. J. Nucl. Med. 2009;50 doi: 10.2967/jnumed.108.057307. 122S-50S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallach M., Mikhail Lette M., Abdel-Wahab M., Giammarile F., Pellet O., Paez D. Addressing global inequities in positron emission tomography-computed tomography (PET-CT) for cancer management: a statistical model to guide strategic planning. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2020;26 doi: 10.12659/MSM.926544. e926544-e926541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch de Basea Gomez M., Thierry-Chef I., Harbron R., Hauptmann M., Byrnes G., Bernier M.-O., Le Cornet L., Dabin J., Ferro G., Istad T.S., et al. Risk of hematological malignancies from CT radiation exposure in children, adolescents and young adults. Nat. Med. 2023;29:3111–3119. doi: 10.1038/s41591-023-02620-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaefferkoetter J.D., Yan J., Sjöholm T., Townsend D.W., Conti M., Tam J.K.C., Soo R.A., Tham I. Quantitative accuracy and lesion detectability of low-dose 18F-FDG PET for lung cancer screening. J. Nucl. Med. 2017;58:399–405. doi: 10.2967/jnumed.116.177592. [DOI] [PubMed] [Google Scholar]
- 8.Wang T., Lei Y., Fu Y., Wynne J.F., Curran W.J., Liu T., Yang X. A review on medical imaging synthesis using deep learning and its clinical applications. J. Appl. Clin. Med. Phys. 2021;22:11–36. doi: 10.1002/acm2.13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torrado-Carvajal A., Vera-Olmos J., Izquierdo-Garcia D., Catalano O.A., Morales M.A., Margolin J., Soricelli A., Salvatore M., Malpica N., Catana C. Dixon-VIBE deep learning (DIVIDE) pseudo-CT synthesis for pelvis PET/MR attenuation correction. J. Nucl. Med. 2019;60:429–435. doi: 10.2967/jnumed.118.209288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Largent A., Barateau A., Nunes J.-C., Mylona E., Castelli J., Lafond C., Greer P.B., Dowling J.A., Baxter J., Saint-Jalmes H., et al. Comparison of deep learning-based and patch-based methods for pseudo-CT generation in MRI-based prostate dose planning. Int. J. Radiat. Oncol. Biol. Phys. 2019;105:1137–1150. doi: 10.1016/j.ijrobp.2019.08.049. [DOI] [PubMed] [Google Scholar]
- 11.Ren G., Li B., Lam S.-k., Xiao H., Huang Y.-H., Cheung A.L.-y., Lu Y., Mao R., Ge H., Kong F.M.S., et al. A Transfer Learning Framework for Deep Learning-Based CT-to-Perfusion Mapping on Lung Cancer Patients. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.883516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J., Mayer A.T., Li R. Integrated imaging and molecular analysis to decipher tumor microenvironment in the era of immunotherapy. Semin. Cancer Biol. 2022;84:310–328. doi: 10.1016/j.semcancer.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massion P.P., Walker R.C. Indeterminate pulmonary nodules: risk for having or for developing lung cancer? Cancer Prev. Res. 2014;7:1173–1178. doi: 10.1158/1940-6207.CAPR-14-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Im H.-J., Bradshaw T., Solaiyappan M., Cho S.Y. Current methods to define metabolic tumor volume in positron emission tomography: which one is better? Nucl. Med. Mol. Imaging. 2018;52:5–15. doi: 10.1007/s13139-017-0493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell K.G., Amini B., Wang Y., Carter B.W., Godoy M.C.B., Parra E.R., Behrens C., Villalobos P., Reuben A., Lee J.J., et al. 18F-fluorodeoxyglucose positron emission tomography correlates with tumor immunometabolic phenotypes in resected lung cancer. Cancer Immunol. Immunother. 2020;69:1519–1534. doi: 10.1007/s00262-020-02560-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikhael P.G., Wohlwend J., Yala A., Karstens L., Xiang J., Takigami A.K., Bourgouin P.P., Chan P., Mrah S., Amayri W. Sybil: a validated deep learning model to predict future lung cancer risk from a single low-dose chest computed tomography. J. Clin. Oncol. 2023;22 doi: 10.1200/JCO.22.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y., Lin W.-Y., Kao C.-H., Yen K.-Y., Chen S.-W., Yeh J.-J. Prognostic value of preoperative metabolic tumor volumes on PET-CT in predicting disease-free survival of patients with stage I non-small cell lung cancer. Anticancer Res. 2012;32:5087–5091. [PubMed] [Google Scholar]
- 18.Liu J., Dong M., Sun X., Li W., Xing L., Yu J. Prognostic value of 18F-FDG PET/CT in surgical non-small cell lung cancer: a meta-analysis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0146195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Cohen A., Klang E., Raskin S.P., Soffer S., Ben-Haim S., Konen E., Amitai M.M., Greenspan H. Cross-modality synthesis from CT to PET using FCN and GAN networks for improved automated lesion detection. Eng. Appl. Artif. Intell. 2019;78:186–194. [Google Scholar]
- 20.Chandrashekar A., Handa A., Ward J., Grau V., Lee R. A deep learning pipeline to simulate fluorodeoxyglucose (FDG) uptake in head and neck cancers using non-contrast CT images without the administration of radioactive tracer. Insights Imaging. 2022;13 doi: 10.1186/s13244-022-01161-3. 45-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradshaw T.J., Boellaard R., Dutta J., Jha A.K., Jacobs P., Li Q., Liu C., Sitek A., Saboury B., Scott P.J.H., et al. Nuclear medicine and artificial intelligence: best practices for algorithm development. J. Nucl. Med. 2022;63:500–510. doi: 10.2967/jnumed.121.262567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jha A.K., Bradshaw T.J., Buvat I., Hatt M., Kc P., Liu C., Obuchowski N.F., Saboury B., Slomka P.J., Sunderland J.J., et al. Nuclear medicine and artificial intelligence: best practices for evaluation (the RELAINCE guidelines) J. Nucl. Med. 2022;63:1288–1299. doi: 10.2967/jnumed.121.263239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim B., Kwon G., Kim K., Ye J.C. Unpaired Image-to-Image Translation via Neural Schrödinger Bridge. arXiv. 2023 doi: 10.48550/arXiv.2305.15086. Preprint at. [DOI] [Google Scholar]
- 24.Chuquicusma M.J., Hussein S., Burt J., Bagci U. 2018 IEEE 15th International Symposium on Biomedical Imaging. IEEE; 2018. How to fool radiologists with generative adversarial networks? A visual Turing test for lung cancer diagnosis; pp. 240–244. [Google Scholar]
- 25.Sugita S., Yamato M., Hatabu T., Kataoka Y. Involvement of cancer-derived EMT cells in the accumulation of 18F-fluorodeoxyglucose in the hypoxic cancer microenvironment. Sci. Rep. 2021;11:9668–9711. doi: 10.1038/s41598-021-88414-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., Tang L., Deng J., Qi X., Zhang J., Qi H., Li M., Liu Y., Zhao W., Gu Y., et al. Identifying metabolic reprogramming phenotypes with glycolysis-lipid metabolism discoordination and intercellular communication for lung adenocarcinoma metastasis. Commun. Biol. 2022;5:198–213. doi: 10.1038/s42003-022-03135-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saad M.B., Hong L., Aminu M., Vokes N.I., Chen P., Salehjahromi M., Qin K., Sujit S.J., Lu X., Young E. Predicting benefit from immune checkpoint inhibitors in patients with non-small-cell lung cancer by CT-based ensemble deep learning: a retrospective study. The Lancet Digital Health. 2023;5:E404–E420. doi: 10.1016/S2589-7500(23)00082-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Astley J.R., Biancardi A.M., Marshall H., Hughes P.J., Collier G.J., Hatton M.Q., Wild J.M., Tahir B.A. A hybrid model-and deep learning-based framework for functional lung image synthesis from multi-inflation CT and hyperpolarized gas MRI. Med. Phys. 2023;50:5657–5670. doi: 10.1002/mp.16369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu N., Davis L.S., Fritz M. 2019. Attributing fake images to gans: Learning and analyzing gan fingerprints; pp. 7556–7566. [Google Scholar]
- 30.Perov I., Gao D., Chervoniy N., Liu K., Marangonda S., Umé C., Dpfks M., Facenheim C.S., RP L., Jiang J. DeepFaceLab: Integrated, flexible and extensible face-swapping framework. arXiv. 2020 doi: 10.48550/arXiv.2005.05535. Preprint at. [DOI] [Google Scholar]
- 31.Lu L., Jin P., Pang G., Zhang Z., Karniadakis G.E. Learning nonlinear operators via DeepONet based on the universal approximation theorem of operators. Nat. Mach. Intell. 2021;3:218–229. [Google Scholar]
- 32.Conte G.M., Weston A.D., Vogelsang D.C., Philbrick K.A., Cai J.C., Barbera M., Sanvito F., Lachance D.H., Jenkins R.B., Tobin W.O., et al. Generative adversarial networks to synthesize missing T1 and FLAIR MRI sequences for use in a multisequence brain tumor segmentation model. Radiology. 2021;299:313–323. doi: 10.1148/radiol.2021203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong D.R., Conrad J., Johnson N., Ayers J., Laeremans A., Lee J.C., Lee J., Prusiner S.B., Bandyopadhyay S., Butte A.J., et al. Trans-channel fluorescence learning improves high-content screening for Alzheimer’s disease therapeutics. Nat. Mach. Intell. 2022;4:583–595. doi: 10.1038/s42256-022-00490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghahremani P., Li Y., Kaufman A., Vanguri R., Greenwald N., Angelo M., Hollmann T.J., Nadeem S. Deep learning-inferred multiplex immunofluorescence for immunohistochemical image quantification. Nat. Mach. Intell. 2022;4:401–412. doi: 10.1038/s42256-022-00471-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang W., Tadesse G.A., Ho D., Fei-Fei L., Zaharia M., Zhang C., Zou J. Advances, challenges and opportunities in creating data for trustworthy AI. Nat. Mach. Intell. 2022;4:669–677. [Google Scholar]
- 36.Kim T.J., Park C.M., Goo J.M., Lee K.W. Is there a role for FDG PET in the management of lung cancer manifesting predominantly as ground-glass opacity? AJR Am. J. Roentgenol. 2012;198:83–88. doi: 10.2214/AJR.11.6862. [DOI] [PubMed] [Google Scholar]
- 37.Cascone T., Weissferdt A., Godoy M.C.B., William W.N., Leung C.H., Lin H.Y., Basu S., Yadav S.S., Pataer A., Mitchell K.G., et al. Nodal immune flare mimics nodal disease progression following neoadjuvant immune checkpoint inhibitors in non-small cell lung cancer. Nat. Commun. 2021;12:5045–5115. doi: 10.1038/s41467-021-25188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jimenez J.E., Dai D., Xu G., Zhao R., Li T., Pan T., Wang L., Lin Y., Wang Z., Jaffray D., et al. Lesion-based radiomics signature in pretherapy 18F-FDG PET predicts treatment response to ibrutinib in lymphoma. Clin. Nucl. Med. 2022;47:209–218. doi: 10.1097/RLU.0000000000004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J., Li C., Gensheimer M., Padda S., Kato F., Shirato H., Wei Y., Schönlieb C.B., Price S.J., Jaffray D., et al. Radiological tumour classification across imaging modality and histology. Nat. Mach. Intell. 2021;3:787–798. doi: 10.1038/s42256-021-00377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen M.M., Terzic A., Becker A.S., Johnson J.M., Wu C.C., Wintermark M., Wald C., Wu J. Artificial intelligence in oncologic imaging. Eur. J. Radiol. Open. 2022;9 doi: 10.1016/j.ejro.2022.100441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang N., Liang R., Gensheimer M.F., Guo M., Zhu H., Yu J., Diehn M., Loo B.W., Jr., Li R., Wu J. Early response evaluation using primary tumor and nodal imaging features to predict progression-free survival of locally advanced non-small cell lung cancer. Theranostics. 2020;10:11707–11718. doi: 10.7150/thno.50565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J., Gensheimer M.F., Zhang N., Guo M., Liang R., Zhang C., Fischbein N., Pollom E.L., Beadle B., Le Q.-T., Li R. Tumor subregion evolution-based imaging features to assess early response and predict prognosis in oropharyngeal cancer. J. Nucl. Med. 2020;61:327–336. doi: 10.2967/jnumed.119.230037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J., Cao G., Sun X., Lee J., Rubin D.L., Napel S., Kurian A.W., Daniel B.L., Li R. Intratumoral spatial heterogeneity at perfusion MR imaging predicts recurrence-free survival in locally advanced breast cancer treated with neoadjuvant chemotherapy. Radiology. 2018;288:26–35. doi: 10.1148/radiol.2018172462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Tashi Q., Saad M.B., Sheshadri A., Wu C.C., Chang J.Y., Al-Lazikani B., Gibbons C., Vokes N.I., Zhang J., Lee J.J. SwarmDeepSurv: swarm intelligence advances deep survival network for prognostic radiomics signatures in four solid cancers. Patterns. 2023;4:100777. doi: 10.1016/j.patter.2023.100777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H., Fu T., Du Y., Gao W., Huang K., Liu Z., Chandak P., Liu S., Van Katwyk P., Deac A., et al. Scientific discovery in the age of artificial intelligence. Nature. 2023;620:47–60. doi: 10.1038/s41586-023-06221-2. [DOI] [PubMed] [Google Scholar]
- 46.Sahni N.R., Carrus B. Artificial Intelligence in US Health Care Delivery. N. Engl. J. Med. 2023;389:348–358. doi: 10.1056/NEJMra2204673. [DOI] [PubMed] [Google Scholar]
- 47.Bakr S., Gevaert O., Echegaray S., Ayers K., Zhou M., Shafiq M., Zheng H., Benson J.A., Zhang W., Leung A.N.C., et al. A radiogenomic dataset of non-small cell lung cancer. Sci. Data. 2018;5:180202–180209. doi: 10.1038/sdata.2018.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Armato S.G., III, McLennan G., Bidaut L., McNitt-Gray M.F., Meyer C.R., Reeves A.P., Zhao B., Aberle D.R., Henschke C.I., Hoffman E.A., et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med. Phys. 2011;38:915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aerts H.J.W.L., Velazquez E.R., Leijenaar R.T.H., Parmar C., Grossmann P., Carvalho S., Bussink J., Monshouwer R., Haibe-Kains B., Rietveld D., et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014;5:4006–4009. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidt S.T., Akhave N., Knightly R.E., Reuben A., Vokes N., Zhang J., Li J., Fujimoto J., Byers L.A., Sanchez-Espiridion B., et al. Shared Nearest Neighbors Approach and Interactive Browser for Network Analysis of a Comprehensive Non–Small-Cell Lung Cancer Data Set. JCO Clin. Cancer Inform. 2022;6 doi: 10.1200/CCI.22.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cardnell R.J.G., Behrens C., Diao L., Fan Y., Tang X., Tong P., Minna J.D., Mills G.B., Heymach J.V., Wistuba I.I., et al. An integrated molecular analysis of lung adenocarcinomas identifies potential therapeutic targets among TTF1-negative tumors, including DNA repair proteins and Nrf2. Clin. Cancer Res. 2015;21:3480–3491. doi: 10.1158/1078-0432.CCR-14-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cascone T., McKenzie J.A., Mbofung R.M., Punt S., Wang Z., Xu C., Williams L.J., Wang Z., Bristow C.A., Carugo A., et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metabol. 2018;27:977–987.e4. doi: 10.1016/j.cmet.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu J., Tha K.K., Xing L., Li R. Radiomics and radiogenomics for precision radiotherapy. J. Radiat. Res. 2018;59:i25–i31. doi: 10.1093/jrr/rrx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein S., Staring M., Murphy K., Viergever M.A., Pluim J.P.W. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imag. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 55.Isensee F., Jaeger P.F., Kohl S.A.A., Petersen J., Maier-Hein K.H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods. 2021;18:203–211. doi: 10.1038/s41592-020-01008-z. [DOI] [PubMed] [Google Scholar]
- 56.Isola P., Zhu J.-Y., Zhou T., Efros A.A. 2017. Image-to-image translation with conditional adversarial networks; pp. 1125–1134. [Google Scholar]
- 57.Jha D., Smedsrud P.H., Riegler M.A., Johansen D., De Lange T., Halvorsen P., Johansen H.D. 2019 IEEE International Symposium on Multimedia. IEEE; 2019. Resunet++: An advanced architecture for medical image segmentation; pp. 225–2255. [Google Scholar]
- 58.Wang Z., Bovik A.C., Sheikh H.R., Simoncelli E.P. Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 2004;13:600–612. doi: 10.1109/tip.2003.819861. [DOI] [PubMed] [Google Scholar]
- 59.Evangelista L., Panunzio A., Polverosi R., Pomerri F., Rubello D. Indeterminate lung nodules in cancer patients: pretest probability of malignancy and the role of 18F-FDG PET/CT. AJR Am. J. Roentgenol. 2014;202:507–514. doi: 10.2214/AJR.13.11728. [DOI] [PubMed] [Google Scholar]
- 60.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
In our study, the internal datasets from MD Anderson, which contain sensitive patient information, are not publicly available due to privacy and institutional policy. However, the publicly available datasets used in our research can be accessed online, with detailed instructions and links provided in the “patient cohort” section of this paper.
-
•
Source code for the deep learning model is available at: https://github.com/WuLabMDA/Synthetic-PET-from-CT/
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.