Summary
The immune responses during the initiation and invasion stages of human lung adenocarcinoma (LUAD) development are largely unknown. Here, we generated a single-cell RNA sequencing map to decipher the immune dynamics during human LUAD development. We found that T follicular helper (Tfh)-like cells, germinal center B cells, and dysfunctional CD8+ T cells increase during tumor initiation/invasion and form a tertiary lymphoid structure (TLS) inside the tumor. This TLS starts with an aggregation of CD4+ T cells and the generation of CXCL13-expressing Tfh-like cells, followed by an accumulation of B cells, and then forms a CD4+ T and B cell aggregate. TLS and its associated cells are correlated with better patient survival. Inhibiting TLS formation by Tfh or B cell depletion promotes tumor growth in mouse models. The anti-tumoral effect of the Tfh-dependent TLS is mediated through interleukin-21 (IL-21)-IL-21 receptor signaling. Our study establishes an anti-tumoral role of the Tfh-dependent TLS in the development of LUAD.
Graphical abstract
Highlights
-
•
A time-resolved scRNA-seq map of the immune dynamic during human LUAD development
-
•
During tumor invasion, CD4+ T and B cells increase while NK cells reduce
-
•
Tfh, GC B, and dysfunctional CD8+ T cells form a tertiary lymphoid structure (TLS)
-
•
Tfh cells are required for TLS formation and inhibit tumor growth
A time-resolved immune cell atlas of human lung adenocarcinoma (LUAD) development from Liu et al. shows that Tfh, GC B cells, and dysfunctional CD8+ T cells form a tertiary lymphoid structure (TLS) that is associated with patient outcomes. Further functional studies establish an anti-tumoral role of the Tfh-dependent TLS mediated via IL-21-IL-21R signaling.
Introduction
Tumor development is a multi-step process involving initiation, invasion, progression, and metastasis.1,2 During this process, the malignant cells reciprocally interact with the immune microenvironment, forming a highly complicated and dynamic network of anti- and pro-tumoral immune responses.3,4 Single-cell genomics have extensively explored the human tumor immune microenvironment across cancer types and identified various immune cell types that modulate tumor development at the baseline level and during immunotherapy.5,6,7,8,9 However, the components of the immune microenvironment and their functional roles remain unclear in premalignant, preinvasive, and early-stage cancerous lesions and during their transition.
Lung adenocarcinoma (LUAD) is the most common and lethal type of lung cancer.10,11,12 Early-stage LUAD and its precursors range from atypical adenomatous hyperplasia (AAH), invasive adenocarcinoma in situ (AIS), microinvasive adenocarcinoma (MIA) to, eventually, invasive adenocarcinoma (IAC).13,14 AAH and AIS belong to premalignant lesions, while MIA is the preinvasive stage and IAC is the invasive stage.15 Previous study of early-stage LUAD focused on the immune landscape at the IAC stage, while the immune responses during the initiation and invasion of LUAD in human remain unclear.16,17,18,19
During tumor development, it is believed that transformed cells are first surveilled by innate immune cells such as natural killer (NK) cells and macrophages during tumor initiation, and then CD8+ T cells are activated and act as the major tumor-killing effector cells.20,21,22,23,24 The roles of other immune cells in cancer initiation and progression are relatively much less understood. Emerging studies are revealing the association of CD4+ T follicular helper (Tfh), B cells, and their interactions (tertiary lymphoid structure [TLS]) with clinical outcomes in the tumor microenvironment (TME).25,26,27,28,29,30 In many cancer types, the density and maturation of intra-tumoral TLSs correlate with a favorable prognosis or response to immunotherapy.31,32,33,34,35 However, the functional importance of CXCL13-expressing Tfh cells remains uncertain, as does the importance of their interactions with B cells.36,37 Whether the TLS and its associated cells affect the prognosis of early-stage LUAD and the mechanisms by which it influences clinical outcomes are not well known. Moreover, the cascade of cellular and molecular events of TLS formation during tumor development is still unclear.
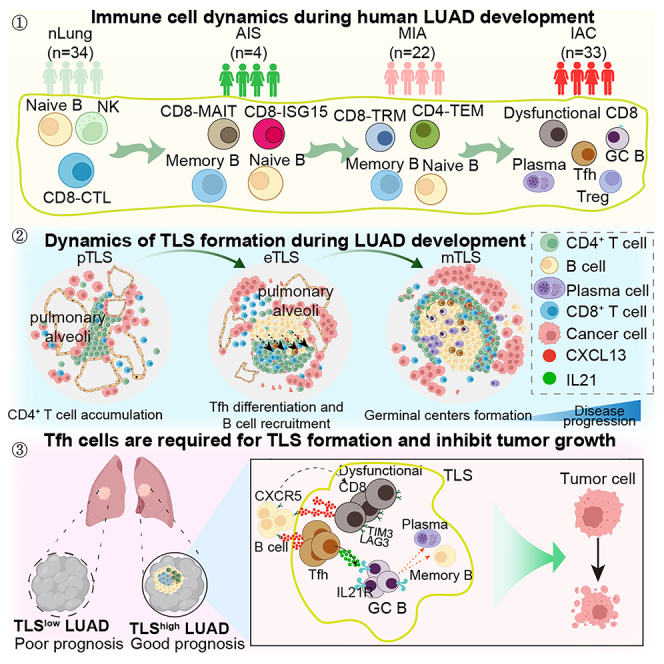
Here, to elucidate the immune dynamics and regulatory mechanisms during LUAD development, we performed time-resolved single-cell RNA sequencing (scRNA-seq) analysis of immune infiltrates from 34 normal lung, 4 AIS, 22 MIA, and 33 IAC samples. We discovered the increase of CD4+ T and B cells during LUAD development due to intra-tumoral proliferation and differentiation. Tfh-like cells, germinal center (GC) B cells, and dysfunctional CD8+ T cells increase during tumor initiation/invasion and form a TLS inside the tumor. The formation of the TLS starts with the initial aggregation of CD4+ T cells around the epithelial cells of the cancerous alveoli, followed by the generation of Tfh-like cells and the accumulation of B cells, and then forms a CD4+ T and B cell aggregate. The TLS and its associated cells are strongly correlated with better survival for patients with IAC. We further demonstrated that inhibiting the formation of the TLS by Tfh or B cell depletion promotes tumor growth in mouse lung cancer models. And the anti-tumoral effect of the Tfh-dependent TLS is likely mediated through interleukin-21 (IL-21)-IL-2 receptor (IL-21R) signaling. Our study establishes an anti-tumoral role of the Tfh-dependent TLS in the development of LUAD.
Results
A time-resolved scRNA-seq map of the immune dynamic during human LUAD development
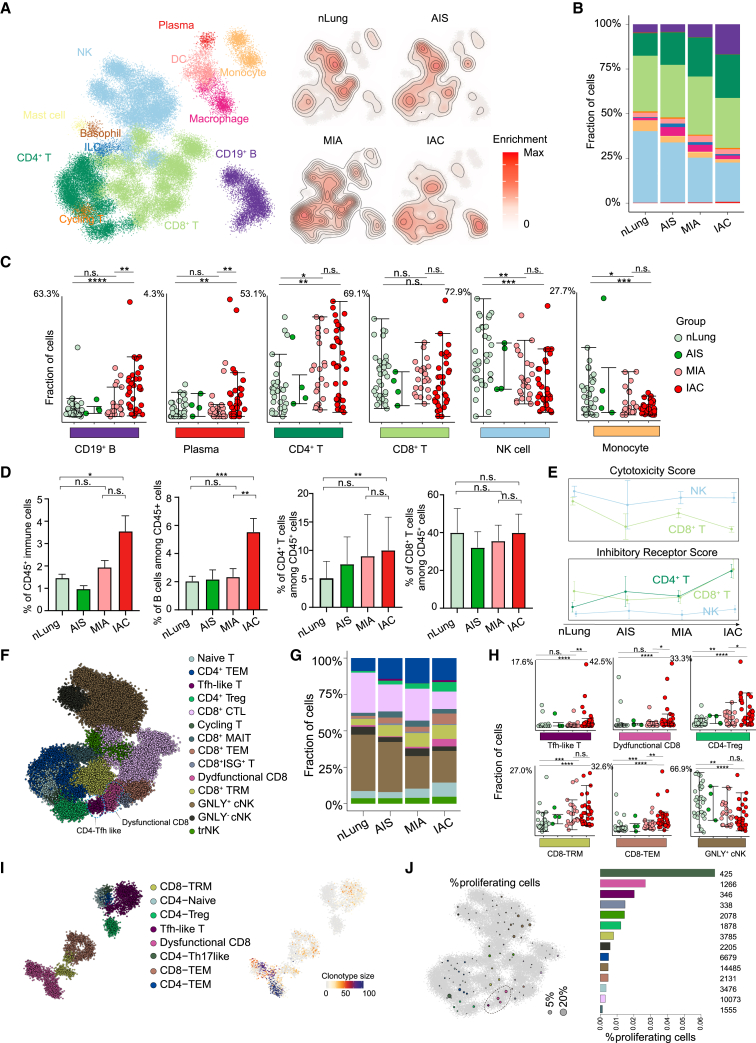
To investigate the immune dynamic during LUAD development, we collected immune cells from 93 clinical samples of 61 patients with LUAD for scRNA-seq. A total of 63,687 quality control (QC)+ immune cells were obtained, covering 34 normal lung, 4 AIS, 22 MIA, and 33 IAC samples, which reflected the initiation, invasion, and progression processes of early-stage LUAD (Figures 1A and S1A–S1E; Tables S1 and S2). We used the metacell algorithm (“metacell”; STAR Methods) to identify homogeneous and robust groups of cells based on the cluster-specific expression patterns of the 1,333 most variable genes, resulting in a detailed map of 324 metacells organized into nine broad lineages, including CD4+ T cells, CD8+ T cells, NK cells, dendritic cells (DCs), macrophages, monocytes, B cells, plasma cells, and innate lymphoid cells (Figures 1B and S1F–S1H; Table S3). We further subdivided each lineage into several subtypes and cellular states, resulting in 32 subtypes/states that represent the transcriptional diversity of the immune landscape during LUAD development (Figures 1C and 1D; Table S5).
Figure 1.
Single-cell atlas of CD45+ immune cells from early-stage LUAD and normal lung tissues
(A) Graphical overview of the experimental setting. Tumor samples of patients with LUAD dissociated into single cells. CD45+ cells were processed by massively parallel single-cell RNA sequencing (MARS-seq) for transcriptional profiling. nLung, normal lung; AIS, invasive adenocarcinoma in situ; MIA, microinvasive adenocarcinoma; IAC, invasive adenocarcinoma.
(B) 2D projection of subclustered nLung and LUAD immune cells. A total of 63,687 immune cells were annotated and are marked by a color code.
(C) Normalized expression of selected genes over the metacell model. Each bar represents one metacell, colored as in (B).
(D) 2D projection of a selected set of marker genes over the metacell model.
(E) Euclidean distances between sample pairs among nLung only (normal-normal), tumor only (AIS-AIS, MIA-MIA, IAC-IAC, IAC-MIA), or between nLung and tumor (MIA-normal, IAC-normal, AIS-normal). Distances between pairs of patient-matched samples were excluded. ∗∗∗p < 0.001, two-sided Mann-Whitney U test.
Notably, a comparison of the distances between AIS-normal, MIA-normal, IAC-normal, and IAC-MIA revealed a gradual increase in distances from normal to IAC stage (Figure 1E; STAR Methods), indicating a potential change in the immune microenvironment before and after tumor invasion through the basement membrane underlying the lung epithelium.
Increased frequency of CD4+ T and B cells is accompanied by NK cell reduction during tumor invasion
We then visualized the alterations of the immune compartment by computing the 2D density of the projected cells from each group (controlling for the number of cells and patients) and observed a global reorganization of immune cell populations during LUAD progression (Figures 2A and S2A). To further understand which cell types were significantly changed, we computed fractional contributions and performed the differential analysis (Figures 2B, 2C, and S2B). We observed the gradual decrease of NK cells and monocytes during disease progression, which is accompanied by an increased frequency of CD19+ B, CD4+ T, and plasma cells, mainly in patients with IAC. Flow cytometry analysis also confirmed that CD45+, CD19+ B, and CD4+ T cells increased significantly during the transition from MIA to IAC stages, indicating the tumor invasion process can induce an abrupt increase of B and CD4+ T cells (Figure 2D). In contrast, no significant changes were observed in CD8+ T cells, macrophages, or DCs (Figures 2C and 2D). These dynamics indicated that B and CD4+ T cells might come into play mainly after tumor invasion. These data suggested that multiple protective immune mechanisms are programmed in a stepwise manner to safeguard the host at different stages of LUAD development.
Figure 2.
Composition of immune cells during LUAD progression
(A) 2D density plots showing immune cell population enrichment over the metacell model in groups of patients with LUAD. (Left) Reference map of nLung and tumor immune populations as in Figure 1B. (Right) Immune cells of nLung and patients with AIS, MIA, and IAC are shown with contour lines indicating the density of projected cells (STAR Methods).
(B) Bar plots showing the immune cell-type composition within the nLung and patients with AIS, MIA, and IAC in each group. Cell types are colored the same as in (A).
(C) Dot plots showing percentages of partial immune cell subpopulation within nLung and patients with AIS, MIA, and IAC. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, two-sided Mann-Whitney U test. Error bars indicate mean ± SEM.
(D) Bar plots showing proportion of CD45+ cells in all live cells and CD19+, CD4+, and CD8+ cells in all CD45+ cells; data from fluorescence-activated cell sorting (FACS) analysis. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, two-sided Mann-Whitney U test. Bars and error bars represent mean +/- standard error.
(E) Line plots showing changes for the activity score of inhibitory receptor genes and cytotoxic genes across four stages (STAR Methods).
(F) 2D projection of subclustered T and NK cells with subpopulations marked by a color code.
(G) Bar plots showing the T and NK cell-type composition within the nLung and patients with AIS, MIA, and IAC in each group. Cell types are colored the same as in (F).
(H) Dot plots showing percentages of partial T and NK cell subpopulation within nLung and patients with AIS, MIA, and IAC. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, two-sided Mann-Whitney U test. Error bars indicate mean ± SEM.
(I) Projection of T cell subclusters and TCR clonotype size from 5′ 10× Genomics data.
(J) Fraction of proliferating cells per metacell calculated by defining a cell as proliferative by its fraction of cell-cycle gene expression out of total expression (STAR Methods). Circle size reflects the percentages of proliferating cells in the metacell using the 2D projection as in (F).
Increase of dysfunctional CD8+ T and Tfh-like cells during tumor invasion
Since the anti-tumoral activity of T and NK cells is tightly regulated by a series of cytotoxicity-related genes and stimulatory/inhibitory receptors, we hence examined their anti-tumoral activity by calculating the cytotoxicity and inhibitory receptor gene signature scores of the CD4+ T, CD8+ T, and NK cells (Figure 2E; Table S4). A decline of the activity scores for cytotoxicity genes and a rise for inhibitory receptor genes was observed, suggesting a transition from an anti-tumoral cellular state toward a dysfunctional cellular state of these cells. We further subdivided the T and NK cells and obtained 14 subclusters based on 29 differentially expressed genes. These included TCF7+ naive T cells, GZMK+ CD8- effector memory T cell (Tem), ZNF683+ CD8- tissue-resident memory T cell (Trm), GZMB+ CD8-cytotoxic T cell (CTL), SLC4A10+ CD8-mucosal-associated invariant T cell (MAIT), interferon-stimulated gene+ CD8+ T cells, FOXP3+ CD4-regulatory T (Treg), CD4-Tem, CXCL13-expressing dysfunctional CD8+ T cells and Tfh-like cells, GNLY+ and GNLY− conventional NK cells, and tissue-resident NK cells (Figures 2F and S2C). Although the frequency of CD8+ T cells stayed relatively constant, we observed a gradual decrease of the cytotoxic, GZMB+ CD8-CTL subtype accompanied by an increase of dysfunctional CD8+ T, Tfh-like, CD8+ Trm, and Treg cells through fraction analysis (Figures 2G, 2H, and S2D; Table S6).
Intra-tumoral proliferation and clonal expansion of dysfunctional CD8+ T and Tfh-like cells
To further understand the proliferation and clonal dynamics of these increased T cell populations, we generated additional paired single-cell RNA and T cell receptor (TCR) sequencing data from two patients with IAC through the 10× Genomics platform. We profiled a total of 5,664 QC+ T cells with TCR sequences and identified 8 T cell subtypes, which were also detected in our MARS-seq dataset. Consistent with their increased frequency in IAC stage, dysfunctional CD8+ T and Tfh-like cells showed larger clonotype sizes in IAC tissues (Figure 2I). To explore whether the observed clonal expansion could be due to T cell proliferation, we computed a proliferation score for each cell by pooling the expression of cell-cycle genes. Except for one metacell we defined as a cycling T cell, a significant fraction of dysfunctional CD8+ T and Tfh-like cells were cycling (in total, 2.02% and 2.69%) (Figure 2J). We also applied Monocle2 to CD8+ T cells. The trajectory analysis showed two differentiation ways containing a path from naive T cells to CD8+ CTL and a path from naive T cells to CD8+ Trm and dysfunctional CD8+ T cells (Figure S3). Further investigation into the distribution across four stages also revealed the increase of dysfunctional CD8+ T cells (Figure S3). Overall, the increase of dysfunctional CD8+ T cells and Tfh-like cells is due to, at least partially, local proliferation in the tumor.
Intra-tumoral proliferation and differentiation of B cells during LUAD progression
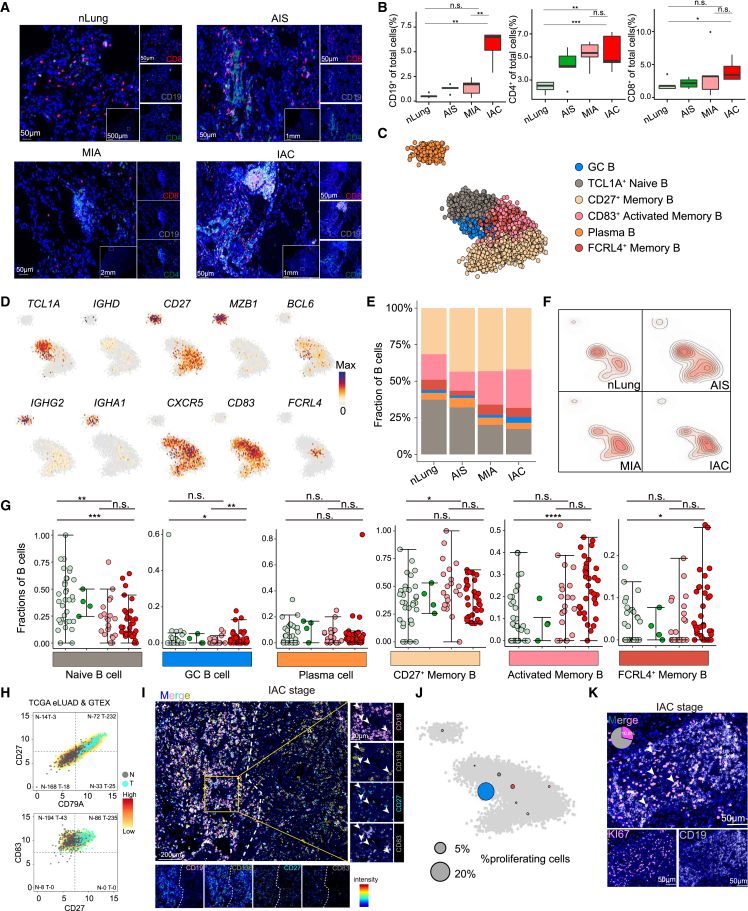
As B and CD4+ T cells showed the most dramatic changes during the invasion process, we thus used multiplex immunostaining to further examine the spatial features of these cells. Results with anti-CD19, anti-CD4, and anti-CD8 antibodies confirmed the increase of B cells from normal lungs to AIS, MIA, and IAC stages, averaging from 0.55% to 1.27%, 1.41%, and 5.59%, and the increase of CD4+ T cells from 2.4% to 4.24%, 5.25%, and 5.37% (Figures 3A and 3B). CD4+ T cells increased from AIS stage prior to B cells, which increased from the more advanced IAC stage, confirming our scRNA-seq and flow cytometry results. Notably, CD4+ T and B cells exhibited an aggregate state rather than being dispersed in the tumor (Figure 3A).
Figure 3.
Characteristics and dynamics of B cells in each group of early-stage LUAD
(A) Tumor sections from patients with LUAD stained with anti-CD4 monoclonal antibody (mAb; green), anti-CD8 mAb (red), anti-CD19 mAb (white), and DAPI (4′,6-diamidino-2-phenylindole; blue). Scale bars, 50 μm, 500 μm, 1 mm, and 2 mm. nLung (n = 11), AIS (n = 6), MIA (n = 10), and IAC (n = 10).
(B) Boxplot showing quantification of CD19+/CD4+/CD8+ cells in total cells, respectively; data from multiplex immunostaining analysis. Data are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, two-sided Mann-Whitney U test.
(C) 2D projection of subclustered of B cells from all nLung and LUAD samples, showing the formation of six main clusters. A total of 6,475 cells were annotated and are marked by color code.
(D) 2D projection of a selected set of B cell marker genes over the metacell model.
(E) Bar plots showing the B cell-type composition within the nLung and patients with AIS, MIA, and IAC in each group. Cell types are colored the same as in (C).
(F) B cells of nLung and patients with AIS, MIA, and IAC are shown with contour lines indicating the density of projected cells.
(G) Dot plots showing percentages of B cell subpopulation within nLung and patients with AIS, MIA, and IAC. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, two-sided Mann-Whitney U test. Error bars indicate mean ± SEM.
(H) Density plots showing the proportions of tumor samples and nLung tissues that express high levels of CD79A and CD27 and CD27 and CD83, respectively, in TCGA stage I LUAD and Genotype-Tissue Expression (GTEx) nLung data. Expression level was measured by log2 (count + 1).
(I) Tumor sections from patients with LUAD stained with anti-CD83 mAb (white), anti-CD138 mAb (yellow), anti-CD27(green), anti-CD19 mAb (red), and DAPI (blue). Scale bars, 20 and 200 μm.
(J) Fraction of proliferating B cells per metacell. Circle size reflects the percentages of proliferating cells in the metacell using the 2D projection as in (C).
(K) Tumor sections from patients with LUAD stained with anti-KI67 mAb (red), anti-CD19 mAb (white), and DAPI (blue). Scale bars, 50 μm.
To further explore the heterogeneity of B cells, we identified six B cell clusters, including TCL1A+ naive B, BCL6+ GC B, FCRL4+ memory B, CD27+ memory B, CD83+CD27+ activated memory B, and immunoglobulin A (IgA)/IgG+ plasma cells (Figures 3C, 3D, and S4A–S4C). Compared to normal lungs, we observed a significant increase of most of the B cell subtypes except for naive B cells in IAC tumors (Figures 3E–3G and S4D). Analysis of the bulk RNA-seq of normal lung and LUAD samples from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression databases further confirmed the increase of CD27+CD83+ activated memory B cells in IAC tumors as compared to the normal lung (Figure 3H). In addition, multiplex immunostaining with anti-CD19, anti-CD27, anti-CD83, and anti-CD138 antibodies from patients with IAC revealed that B cell subtypes, including CD27+ memory B, CD83+CD27+ activated memory B, and plasma cells, were mainly localized inside the B cell aggregate (Figure 3I).
The increase of B cells at the IAC stage can be attributed to local proliferation or recruitment from the periphery or both. We thus performed a proliferation activity analysis on the B cells and observed a significant population of proliferating B cells, mainly the GC B cell subset (Figure 3J). Furthermore, multiplex immunostaining analysis with KI67 expression on the B cell showed that KI67+ B cells account for about 10% of all B cells in the B cell aggregate (Figure 3K). In addition, partition-based graph abstraction analysis suggested a potential differentiation among plasma, memory B, and GC B cells (Figure S3E). These data indicated that the increase of B cells in IAC tumors may be partially due to the local proliferation and differentiation of GC B cells.
To further validate our findings, we integrated scRNA-seq data of LUAD from other studies.7,38,39,40,41,42,43 Our re-analysis revealed that in LUAD, the fraction of Tfh-like (CXCL13+CD4+ T) and dysfunctional CD8+ T (CXCL13+CD8+) cells increased during the early stages but decreased in advanced stages (Figures S5A–S5L). In contrast, the fraction of plasmablasts decreased, while the fraction of IgG+ plasma cells increased, indicating the differentiation of plasma cells (Figures S5M–S5P). In summary, our re-analysis of scRNA-seq data from multiple sources is consistent with our data.
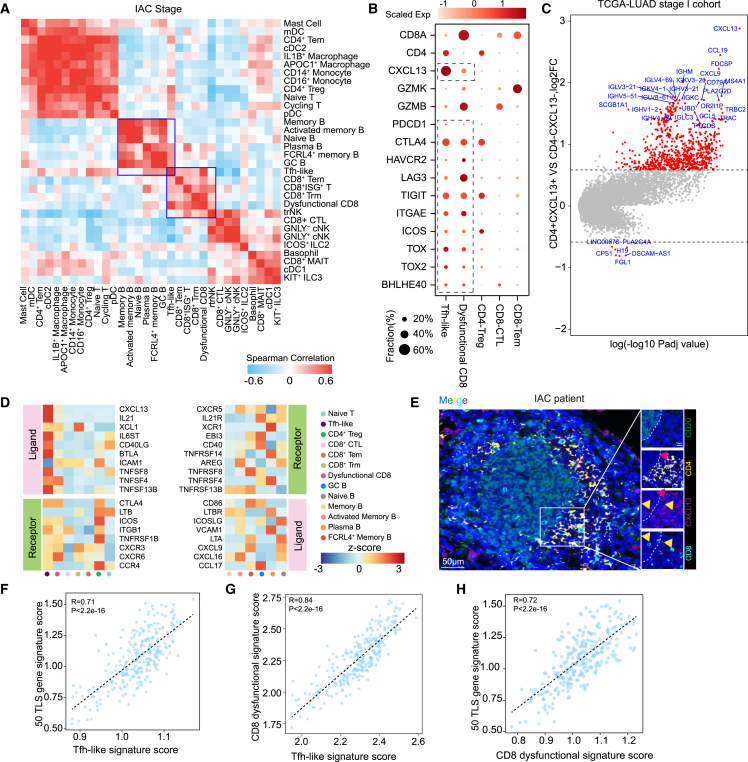
Tfh-like cells, GC B cells, and dysfunctional CD8+ T cells form a TLS
Since cell-cell interaction plays a crucial role in the immune responses toward tumor, we thus correlated lineage-normalized cell-type frequencies across IAC tumors to investigate their interactions in the tumor. Notably, Tfh-like cells showed high correlation with GC B, FCRL4+ memory B, plasma, and dysfunctional CD8+ T cells (Figures 4A and S6A). Tfh-like cells expressed features of both Tfh and T helper type 1 cells, including BHLHE40, TOX, TOX2, and CXCR5, but also exhibited tissue-resident markers such as ITGAE, CD69, and RBPJ (Figures 4B and S6B).
Figure 4.
Tfh-like, dysfunctional CD8+ T, and GC B cells form a TLS
(A) Correlation of lineage-normalized cell-type frequencies in 33 IAC tumors.
(B) Expression of selected genes in IAC-infiltrating T cell clusters.
(C) Volcano plots showing differentially expressed genes (DEGs) of TCGA patients with LUAD in stage I grouped by the expression levels of CD4 and CXCL13 (STAR Methods).
(D) Heatmap showing the expression of ligand-receptor pairs highly expressed in B and T cells.
(E) Tumor sections from patients with IAC stained with anti-CXCL13 mAb (red), anti-CD20 mAb (green), anti-CD4 mAb (yellow), anti-CD8 mAb (cyan), and DAPI (blue). Scale bars, 50 and 10 μm.
(F) Scatterplots showing gene expression of the 50 TLS gene signature scores (y axis) with the difference of Tfh-like signature scores (x axis).
(G) Scatterplots showing gene expression of dysfunctional CD8+ T signature scores (y axis) with the difference of Tfh-like signature scores (x axis).
(H) Scatterplots showing gene expression of the 50 TLS gene signature scores (y axis) with the difference of dysfunctional CD8+ T signature scores (x axis).
We next utilized the bulk-RNA data of LUAD from TCGA to further validate the correlations of these cells observed in our scRNA-seq data. A group of chemokine genes and Ig-associated genes involved in B cell differentiation and maturation were found to be strongly correlated with the expression of CD4 and CXCL13, including MS4A1, CXCR5, CCL19, IGHA, and IGHG (Figures 4C, S6C, and S6D), confirming the observed correlation between Tfh-like and B cell subtypes. Interestingly, we found that multiple ligand-receptor pairs associated with the differentiation and recruitment of B cells were highly expressed between Tfh-like and GC B cells, including CXCL13-CXCR5, IL-21-IL-21R, CD40LG-CD40, and CCL17-CCR4 (Figure 4D). Indeed, multiplex immunostaining analysis showed Tfh-like cells in the T cell zone of the multi-cellular hub, adjacent to the B cell zone, suggesting that Tfh-like cells interact with B cells directly (Figure 4E). To determine whether these multi-cellular hubs were TLSs,31,32 we further stained the LUAD section with anti-CD138, anti-COL1, and anti-CD21 antibodies. We found that CD138+ plasma cells and COL1+ fibroblasts were around the TLS and that CD21+ follicular dendritic cells (FDC) were in the center (Figures S6E–S6G). These results implicated a role of Tfh-like cells in the formation of TLSs that are commonly observed during chronic infection or in cancer. Consistent with this, patients with higher Tfh-like cell gene expression signature had higher TLS-associated gene expression in TCGA data (Figure 4F).
As mentioned above, the abundance of dysfunctional CD8+ T cells is correlated significantly with Tfh-like cells (Figure 4A). Our multiplex immunostaining results also confirmed that the dysfunctional CD8+ T cells co-localized with Tfh-like cells in the T cell zone of the TLS and were more enriched in the TLS as compared to the region outside (Figure 4E). Such association was also confirmed in the bulk RNA-seq data by comparing the signature score of dysfunctional CD8+ T and Tfh-like cells (Figure 4G). Meanwhile, we found that patients with a high dysfunctional CD8+ T gene expression signature also had a higher TLS score in TCGA data (Figure 4H). Together, our analysis demonstrated the cellular crosstalk among key immune cell subsets and suggested the underlying cooperation among Tfh-like, B, and dysfunctional CD8+ T cells in the TLS.
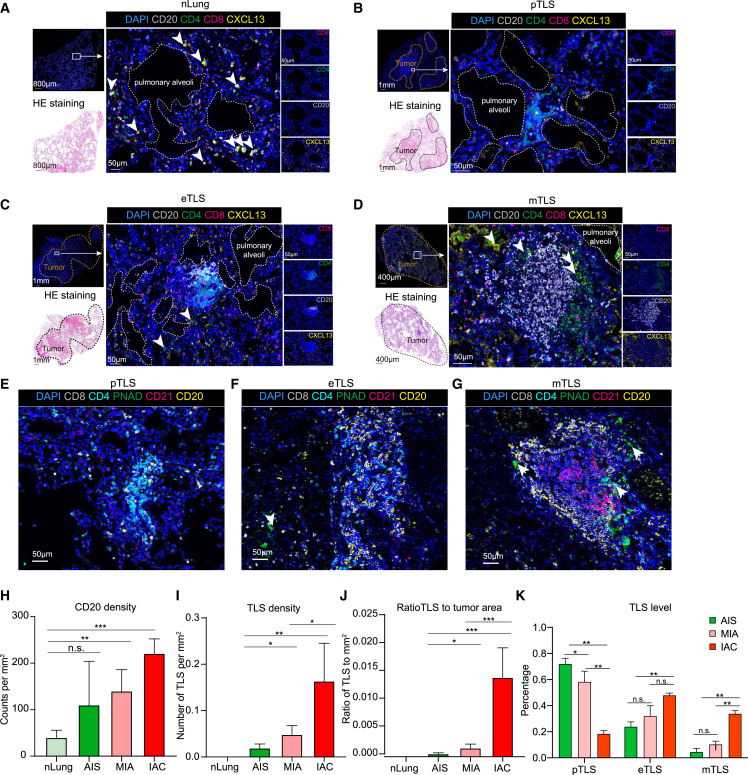
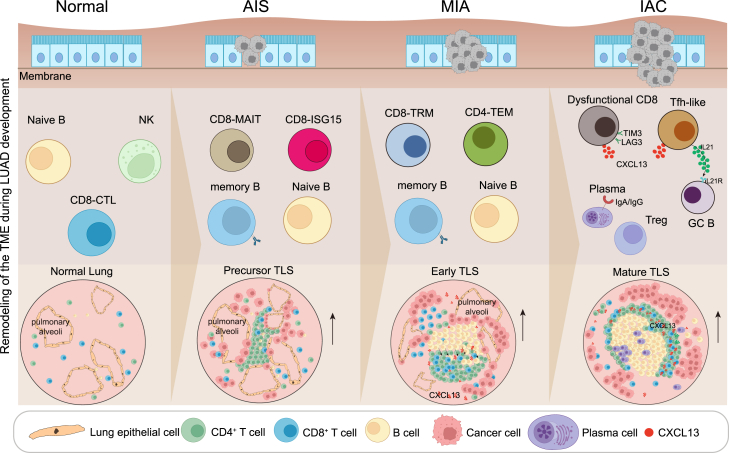
A stepwise formation of the TLS during LUAD initiation and invasion
To find out how the TLS is formed, we then analyzed normal lung, AAH, AIS, MIA, and IAC samples and explored the dynamic of the lymphocyte aggregates by co-staining anti-CD4, anti-CD8, anti-CD20, and anti-CXCL13 antibodies. CD4+ T, CD8+ T, and B cells were scattered in normal lung tissues (Figure 5A), while various phenotypes of lymphocyte aggregates were detected around the alveoli in tumors. We further defined the process of the TLS formation as three stages: precursor, early, and mature. CD4+ T cells were the first to accumulate around the pulmonary alveolar epithelium cells during tumor initiation. In this stage, CD8+ T and B cells are sporadically distributed. We thus termed this stage as precursor of the TLS (pTLS). (Figure 5B). As the tumor invaded, Tfh-like cells appeared, and B cells began to accumulate and aggregate, which was defined as the CD4+ T and B aggregated early TLS (eTLS) (Figure 5C). Subsequently, as the tumor progresses, mature B and T cell regions were formed, which was defined as the mature TLS (mTLS) (Figure 5D). We further confirmed that the mTLS contained GCs, PNAd+ high endothelial venules, and CD21+ follicular DCs, while the pTLS and eTLS contained only sporadic PNAd+ cells and no follicular DCs (Figures 5E–5G). The density of CD20+ B cells and the TLSs, and the ratio of TLSs to tumor area, increased from normal lungs to IAC tumors (Figures 5H–5J). Additionally, we found that pTLSs and eTLSs are more enriched in AIS and MIA tumors, while mTLSs are mostly present in IAC tumors, indicating that the TLS matures as the tumor progresses (Figure 5K). These data are consistent with the above results that the number of CD4+ T cells significantly increased in AIS and MIA tumors compared to the normal lung tissue, whereas B cells did not significantly increase until the IAC stage (Figures 2C, 2D, 3A, and 3B). In summary, during tumor initiation and invasion, CD4+ T cells are the first to accumulate and differentiate into Tfh-like cells, which likely drives the proliferation/differentiation/accumulation of B cells, and eventually form the mTLS.
Figure 5.
The formation of TLS during the progression of early-stage LUAD
(A–D) Multiplex immunostaining analysis of TLSs for the following markers: CD20, CD4, CD8, CXCL13, and DAPI. Scale bars, 1 mm, 800 μm, 400 μm, and 50 μm.
(E–G) Multiplex immunostaining analysis of TLSs for the following markers: CD20, CD4, CD8, CD21, PNAd, and DAPI. The yellow, green, and red arrows refer to B cells, high endothelial venules, and follicular DCs. Scale bars, 50 μm.
(H–J) Quantification of CD20+ B cells, density of TLSs, and ratio of tumor area occupied by TLSs in nLung and patients with AIS, MIA, and IAC. nLung (n = 6), AIS (n = 6), MIA (n = 7), and IAC (n = 9). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, two-sided Mann-Whitney U test. Bars and error bars represent mean +/- standard error.
(K) Quantification of different stages of TLSs in patients with AIS, MIA, and IAC. AIS (n = 6), MIA (n = 7), and IAC (n = 9). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, two-sided Mann-Whitney U test.
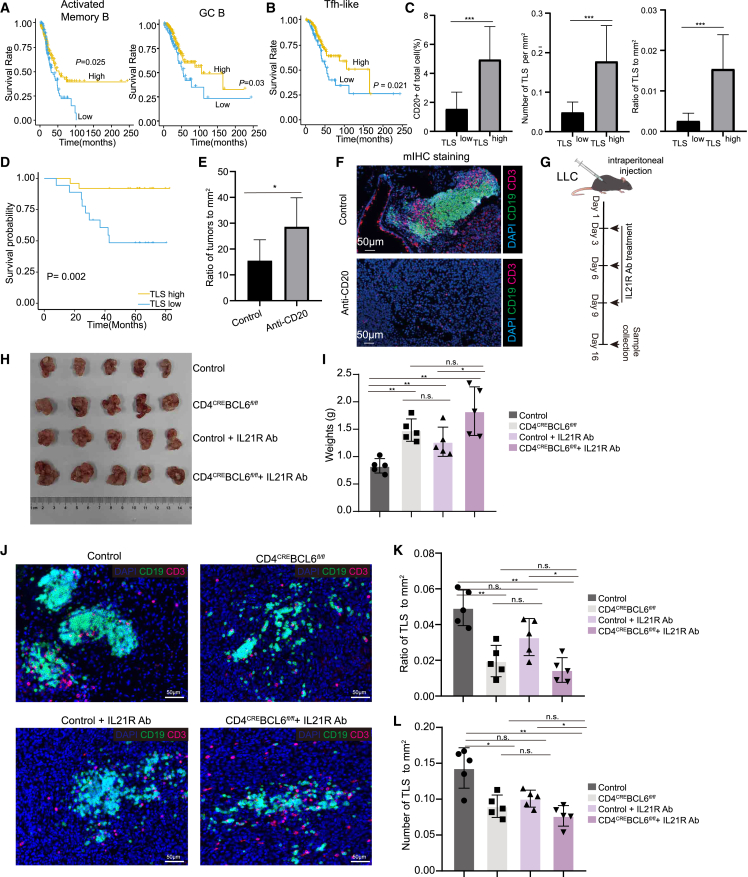
The TLS is associated with better survival in patients with LUAD
To explore the function of the identified TLS in the TME, we first analyzed the role of its associated cells. Analysis of the bulk RNA-seq of LUAD samples from TCGA database revealed that the factions of activated memory B, GC B, and Tfh-like cells were all associated with better survival of patients with early-stage LUAD (Figures 6A and 6B). Consistent with this, by ordering patients with IAC by their percentage of Tfh-like cells, we observed a positive correlation between the infiltration of Tfh-like cells and the expression of effector genes, including GZMB, HAVCR2, LAG3, ENTPD1, and TOX in CD8+ T cells (Figure S7A). Re-analysis of the bulk RNA-seq dataset from Chen et al.44 also confirmed the positive correlation between Tfh-like signature and the expression of effector genes of CD8+ T cells in patients with IAC (Figure S7B). Recent studies indicated Tfh-like cells are tumor reactive, which may explain their positive correlation with better patient survival.45,46,47
Figure 6.
The Tfh-dependent TLS predicts tumor prognosis of patients with early-stage LUAD and inhibits tumor growth
(A) Kaplan-Meier curves showing the overall survival values of activated memory B (left) and GC B (right) cell scores in TCGA database (STAR Methods).
(B) Kaplan-Meier curves showing the overall survival values of Tfh-like cell signature scores in TCGA database.
(C) Quantification of CD20+ cells, density of TLSs, and ratio of tumor area occupied by TLSs in TLShigh and TLSlow patients. TLShigh, n = 27; TLSlow, n = 15. Bars and error bars represent mean +/- standard error.
(D) Kaplan–Meier estimates of overall survival shown by in TLShigh and TLSlow patients. TLShigh, n = 27; TLSlow, n = 15.
(E) Ratio of tumors to mm2 of the lung tissues obtained from anti-CD20-Ab-treated KPC mice. Control (n = 6) and anti-CD20-treated KPC mice (n = 6).
(F) Multiplex immunostaining assay of TLSs for the following markers: CD3, CD19, and DAPI in anti-C20-treated KPC mice. Scale bars, 50 μm. Control (n = 6) and anti-CD20-treated KPC mice (n = 6).
(G) Schematic illustration showing IL-21R Ab neutralization of control and tumor-bearing CD4CRE; Bcl6fl/fl mice.
(H and I) LLC cells were intraperitoneally injected into 8-week-old controls, CD4CRE; Bcl6fl/fl mice, control+IL-21R Ab and CD4CRE; Bcl6fl/fl+IL-21R Ab mice. The image and weight of tumors were obtained at 16 days post-tumor inoculation. Control (n = 5), CD4CRE; Bcl6fl/fl (n = 5), control+IL-21R Ab (n = 5), and CD4CRE; Bcl6fl/fl+ IL-21R Ab mice (n = 5). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, two-sided Mann-Whitney U test. Error bars indicate mean ± SEM.
(J) Multiplex immunostaining assay of TLS for the following markers: CD3, CD19, and DAPI in controls, CD4CRE; Bcl6fl/fl mice, control+IL-21R Ab, and CD4CRE; Bcl6fl/fl+IL-21R Ab mice. Scale bars, 50 μm.
(K and L) Density of TLSs and ratio of tumor area occupied by controls, CD4CRE; Bcl6fl/fl mice, control+IL-21R Ab, and CD4CRE; Bcl6fl/fl+IL-21R Ab mice. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, two-sided Mann-Whitney U test. Error bars indicate mean ± SEM.
To further validate the role of the TLS in predicting patient survival, we stained with anti-CD4, anti-CD8, anti-CD20, and anti-KI67 antibody to define the TLSs in 42 IAC samples and performed prognostic analysis. Patients with IAC were classified as TLShigh or TLSlow based on their percentage of CD20+ B cells, the density of the TLS, and the ratio of TLS to tumor area (Figures 6C and S7C). Overall survival (OS) analysis revealed that TLShigh patients had a better prognosis than TLSlow patients (Figure 6D). Taken together, these data indicated that the TLS and its associated cells play an anti-tumoral role in LUAD.
Tfh cells are required for TLS formation and inhibit tumor growth
Lastly, we sought to demonstrate the anti-tumoral function of the TLS in mouse lung cancers. We first used a subcutaneous model of Lewis lung carcinoma (LLC), and found dispersed B and T cells but no TLSs in this tumor model. Elimination of B cells in this model had no effect on tumor growth (Figures S7D–S7F).
In addition, we further investigated the anti-tumoral function of the TLS using a transgenic KPC (KrasG12D, p53-CKO, Sftpc-creER) mouse lung cancer model. Multiplex immunostaining analysis of anti-CD3 and anti-CD19 antibodies showed that both T and B cells were initially dispersed but became aggregated after 20 weeks of tumor induction. These T/B aggregates resemble the TLS observed in clinical tumor samples but were present at lower abundance in mouse lung cancer (Figure S7G). Elimination of B cells in this orthotopic tumor model significantly promoted tumor growth (Figures 6E, 6F, and S7H).
To further explore the functionality of the TLS, we constructed CD4CRE; Bcl6fl/fl mice, in which Tfh cells can be specifically depleted. In contrast to the subcutaneous LLC model, lymphocyte aggregates/TLSs can be induced when LLC cells are injected intraperitoneally. After intraperitoneal injection of the LLC cells, the tumors were significantly larger in mice without Tfh cells than those in the control group (Figures 6G–6I). Consistent with this, the number of TLSs in the tumor also significantly reduced in Tfh-deficient mice (Figures 6J–6L).
In addition, our ligand-receptor analysis suggested a potential interaction between Tfh and GC B cells via the IL-21-IL-21R signaling pathway (Figure 4D). IL-21 has been implicated in class switching, plasma cell formation, and GC function.48 We found the tumors were significantly larger in mice treated with IL-21R-neutralizing antibodies than those in the control group (Figures 6H and 6I). Meanwhile, the number of lymphocyte aggregates in the tumor also significantly reduced in IL-21R-neutralized mice. However, further neutralizing IL-21R in Tfh-deficient mice showed no significant change in tumor size or the number of lymphocytes aggregates (Figures 6J–6L). Taken together, these results support that the anti-tumoral effect of the Tfh-dependent TLS is likely mediated through IL-21-IL-21R signaling, which is consistent with our analysis with clinical samples.
Discussion
Our single-cell analysis at different stages spanning healthy lungs to premalignant, preinvasive, and then invasive lesions from 61 patients with LUAD provides a comprehensive immune cell atlas of the development of early-stage LUAD. This study filled the gap in our current understanding of the immune response during LUAD initiation and invasion. Our study demonstrated that the immune responses toward tumor occur in a stepwise manner. After tumor invasion, Tfh-like cells recruit and induce the proliferation/differentiation/accumulation of B cells, forming the TLS. The TLS is anti-tumoral, mostly acting after tumor invasion. Our study identified a stage-specific anti-tumoral mechanism of the immune system during LUAD development (Figure 7).
Figure 7.
Summary of immune cell features and dynamics during LUAD initiation and invasion
Schematic illustration showing characteristics and dynamics of key immune cell subsets in different stages. New insights involved in TLS formation during the progression of early-stage LUAD were provided.
B and plasma cells constitute a major population of immune cells in the TME.49 Our results showed that tumor-infiltrating B lymphocytes (TIBs) can be observed in all stages of LUAD development and that their presence differs during tumor initiation, invasion, and progression, suggesting a critical role for B cells during LUAD development. TIBs participate in both humoral and cellular immunity, but their roles in anti-tumor immunity remain controversial.50 Hao et al. found that plasma cells were dominated by IgA1, IgA2, IgG1, and IgG2 isotypes. Relatively high plasma cell signature scores were significantly associated with better OS, better response to anti-PD-1 therapy, a durable clinical benefit, and significantly better progression-free survival.37 Our results revealed that aggregated B cells located in the TLS play an anti-tumor role; however, the underlying mechanism requires further investigation.
Notably, tumors with mTLS and a high density of B and plasma cells, as well as with the presence of antibodies to tumor-associated antigens, are typically associated with favorable clinical outcomes and responses to immunotherapy compared to those lacking these characteristics.51,52,53 We found that the TLS and its associated cells function as tumor suppressors in LUAD. And the anti-tumoral effect of the Tfh-dependent TLS is likely mediated through IL-21-IL-21R signaling. However, the detailed molecular mechanisms underlying the formation and maturation of the TLS and its anti-tumor function require further investigation.
In conclusion, we found that Tfh-like, GC B, and dysfunctional CD8+ T cells increase during tumor initiation/invasion and form a TLS that is dependent on Tfh cells. The anti-tumoral effect of the Tfh-dependent TLS is likely mediated through IL-21-IL-21R signaling. This finding opens avenues for therapeutic strategies that aim at enhancing TLS formation and function, which could result in improved clinical outcomes of patients with LUAD.
Limitations of the study
A potential limitation of our study is primarily the sample size and average cell count per sample of patients with LUAD, especially for AIS-stage patients. Inclusion of a larger sample size could allow for more sufficient statistical analyses. In addition, future studies also need to further explore the formation and maturation of TLSs. Taken together, while our study has some limitations, it establishes a strong foundation for the investigation of an anti-tumoral role of the Tfh-dependent TLS in the development of LUAD.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| DAPI | BioLegend | Cat # 422801 |
| PI | BioLegend | Cat # 421301 |
| anti-human CD45 FITC | BioLegend | Cat # 304054; RRID:AB_2564154 |
| anti-human CD4 BV605 | BioLegend | Cat # 344710; RRID:AB_2044010 |
| anti-human CD19 Pacific blue | BioLegend | Cat # 302224; RRID:AB_493653 |
| anti-human CD11b APC | BioLegend | Cat # 301310; RRID:AB_314162 |
| anti-human CD8 PE-CY7 | BioLegend | Cat # 344712; RRID:AB_2044008 |
| anti-mouse CD45.2 APC | eBioscience | Cat # 17-0454-82; RRID: AB_469400 |
| anti-mouse CD20 PE | Biolegend | Cat # 150410; RRID:AB_2566779 |
| anti-human CD19 | ZSGB-Bio | Cat # ZM-0038 |
| Anti-human CD20 | abcam | Cat #ab9475; RRID: AB_307267 |
| anti-human CD4 | ZSGB-Bio | Cat # ZA-0418; RRID: AB_2890106 |
| anti-human CD8 | ZSGB-Bio | Cat # ZA-0508; RRID:AB_2890107 |
| anti-human CXCL13 | Novus | Cat # NBP2-16041; RRID: AB_2892564 |
| Anti-human KI67 | Gene Tech | Cat # GT209402 |
| Anti-human CD138 | Gene Tech | Cat # GT245102 |
| Anti-human COL1 | Proteintech | Cat # 67288-1; RRID: AB_2882554 |
| Anti-human CD21 | abcam | Cat # ab75985; RRID: AB_1523292 |
| Anti-human PNAD | abcam | Cat # ab111710; RRID: AB_10861722 |
| Anti-mouse IL-21R | bioxcell | Cat # BE0258; RRID: AB_2687737 |
| Anti-mouse CD3 | abcam | Cat # ab16669; RRID: 443425 |
| Anti-mouse CD19 | abcam | Cat # ab245235; RRID: 2895109 |
| Anti-mouse CD20 | bioxcell | Cat # BE0356; RRID: 2894775 |
| Chemicals, peptides, and recombinant proteins | ||
| RiboLock RNase inhibitor | invitrogen | Cat # EO0384 |
| SuperScript™ III Reverse Transcriptase | Invitrogen | Cat # 18080-085 |
| Exonuclease I | NEB | Cat #M0293 |
| Exonuclease I 10x Buffer | NEB | Cat #B0293S |
| NEBNext® mRNA Second Strand Synthesis Module | NEB | Cat #E6111L |
| HiScribe™ T7 High Yield RNA Synthesis Kit | NEB | Cat #E2040S |
| RNA Fragmentation Reagents | Ambion | Cat # AM8740 |
| AffinityScript Multi-Temp RT | Agilent | Cat # 600107 |
| Kapa HiFi HotStart PCR ReadyMix | Kapa Biosystems | Cat # KK2602 |
| ATP solution | thermo | Cat #R0441 |
| T4 RNA Ligase 1 (ssRNA Ligase) | NEB | Cat #M0204L |
| dNTPs | Solarbio | Cat # PC2300 |
| VAHTS ® DNA Clean Beads | Vazyme | Cat #N411-02 |
| Fast SYBR Green master mix | accurate biology (AG) | Cat # AG11719 |
| Absolute ethanol | Sigma | Cat #E7023-1L |
| nuclease-free water | thermo | Cat # AM9932 |
| Equalbit 1 × dsDNA HS Assay Kit | Vazyme | Cat # EQ121-01 |
| TapeStation High Sensitivity D1000 Reagent | Agilent | Cat # 5067-5583 |
| PANO6-plexIHCkit | Panovue | Cat # 102C4100100 |
| Tissue enzymatic reagent | ||
| DNase I | Roche | Cat #10104159001 |
| Collagenase IV | Worthington | Cat # LS004188 |
| Experimental models: Organisms | ||
| KPC (KrasG12D, p53-CKO, Sftpc-creER) mice | Prof. Shun Li | N/A |
| CD4CRE; Bcl6fl/fl mice | Prof. Nengming Xiao | N/A |
| Experimental models: Cell lines | ||
| LLC (Lewis Lung Carcinoma) | ATCC | Cat # CRL-1642; RRID: CVCL_4358 |
| Critical commercial assays | ||
| 10X Chromium Next GEM Single Cell 5′Kit v2 | 10X Genomics | Cat # PN-1000287 |
| 10X Library Construction Kit | 10X Genomics | Cat # PN-1000190 |
| 10X Chromium Next GEM chip K Single Cell Kit | 10X Genomics | Cat # PN-1000287 |
| 10X Dual Index Kit TT Set A | 10X Genomics | Cat # PN-1000215 |
| 10X Chromium Single Cell Human TCR Amplification Kit | 10X Genomics | Cat # PN-1000252 |
| Deposited data | ||
| TCGA LUAD dataset | XENA data portal | http://xena.ucsc.edu/ |
| Invasive and pre-invasive RNA-seq dataset | (Haiquan et al., 2019) | EGAD00001005479 |
| This Paper | NGDC GSA for human | HRA002961 |
| Jermaine_Goveia_2020 (fastq) | ArrayExpress | E-MTAB-6308 |
| He_Fan_2021 (fastq) | NGDC GSA | CRA001963 |
| Kim_Lee_2020 (processed) | GEO | GSE131907 |
| Leader_Merad_2021 (processed) | GEO | GSE154826 |
| Maynard_Bivona_2020 (fastq) | SRA | PRJNA591860 |
| Lambrechts_Thienpont_2018 (fastq) | ArrayExpress | E-MTAB-6653 |
| Stefan_Salcher_2023 (processed) | Zenodo | https://doi.org/10.5281/zenodo.6411867 |
| Software and algorithms | ||
| R (v4.1.3) | (R Core Team, 2013)54 | https://cran.r-project.org/bin/ |
| HISAT2 (v2.1.0) | (Daehwan et al., 2015)55 | http://daehwankimlab.github.io/hisat2 |
| CellRanger (v7.0.0) | 10X Genomics | http://10xgenomics.com |
| Python (v3.8.1) | (Guido et al., 2009)56 | https://www.python.org/downloads/release/python-379/ |
| MetaCell (v0.3.41) | (Baran et al., 2019)57 | https://github.com/tanaylab/metacell/ |
| Scanpy (v1.9.1) | (F. Alexander et al., 2018)58 | https://github.com/scverse/scanpy |
| scvi-tools (v1.1.0) | (Adam Gayoso et al., 2022)59 | https://scvi-tools.org/ |
| Monocle 2 (v2.4.0) | (Xiaojie Qiu et al., 2017)60 | https://github.com/cole-trapnell-lab/monocle-release |
| AUCell (1.4.1) | (Aibar et al., 2017)61 | https://github.com/aertslab/AUCell |
| Immunarch (v0.7.0) | (Vadim et al.,2022)62 | https://immunarch.com/ |
| scRepertoire (v1.0.2) | (Borcherding N et al.,2022)63 | https://github.com/ncborcherding/scRepertoire |
| Toomanycells (v2.2.0.0) | (Gregory et al.,2020)64 | https://github.com/GregorySchwartz/too-many-cells |
| clusterProfiler (v4.4.4) | (Guangchuang et al.,2012)65 | https://github.com/YuLab-SMU/clusterProfiler |
| ssGSEA (GSVA package) (v3.12) | (Hänzelmann et al.,2013)66 | https://www.bioconductor.org/packages/release/bioc/html/GSVA.html |
| QuPath (v0.3.0) | (Humphries et al.,2021)67 | https://qupath.github.io/ |
| Halo (v3.3) | Indica labs | https://www.indicalab.com/halo |
| FACSDiva (v8.0.3) | BD Biosciences | NA |
| FlowJo (v10) | BD Biosciences | NA |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hanjie Li (hj.li@siat.ac.cn).
Materials availability
We did not generate new unique reagents in this study.
Data and code availability
Single cell RNA-seq data that support the findings of this study is available for download in the Genome Sequence Archive for Human with accession number: HRA002961 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA002961). This paper does not report any original code. The code that support the findings of this study is available at https://github.com/youwh1996/ealy-LUAD-scRNA. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and subject details
Human samples acquisition
This study involved the scRNA-seq from fresh human LUAD tumor specimens from patients who underwent surgical excision at Shenzhen People’s Hospital. All LUAD patients participated with informed consent, with the research protocol approved by Shenzhen People’s Hospital (Shenzhen, China, IRB approved protocol number KY-LL-2021040-01). The detailed clinical and pathological characteristics of these LUAD patients were summarized in Table S1. The included LUAD patients represented all major subtypes at different pathological stages, such as age, gender, smoking, diagnosis. The use of clinical specimens was completely in compliance with the “Declaration of Helsinki”.
Mice
The KPC (KrasG12D, p53-CKO, Sftpc-creER) mice was a gift from Prof. Shun Li (Shenzhen Majory Biomedical Institute) and CD4CRE; Bcl6fl/fl mice was provived by the lab of Prof. Nengming Xiao. C57BL/6 J (B6) mice were originally from the Jackson Laboratory. Unless otherwise stated, all mice (both sexes) were littermates and were 8–12 weeks old. These mice were raised in the special pathogen-free facilities of Laboratory Animal Center of Xiamen University at a temperature about 22°C and a relative humidity of about 50% for 12 h day/night cycle. They were fed standard mouse feed. All animal protocols were approved by members of the Institutional Animal Care and Use Committee of Shenzhen Institutes of Advanced Technology.
For B cell depletion experiment, mice were injected intraperitoneally with 250ug anti-CD20 murine monoclonal antibody once a month. For IL-21R neutralization experiment, Mice were injected intraperitoneally with 500ug anti-IL21R murine monoclonal antibody each time.
Method details
Sample collection and processing
61 surgically resected sub-solid nodules pathologically as lung adenocarcinoma were collected. Briefly, portions of each resected nodule were immediately used for downstream analysis (freshly resected) and preserved in frozen, and formalin-fixed and paraffin-embedded (FFPE) tissue blocks. Freshly resected tissues used for single cell sequencing were prepared as single cell suspensions and sequenced immediately. Frozen blocks were stored under −80°C and parrffin-embedded blocks were stored under room temperature.
To isolate immune cells from tumors, the tissues were harvested and cut into small piece. The tissue fragments were digested in 2 mL 1640 medium, containing 600U/mL collagenase and 90U/mL DNase I, for 30 min at 37°C. After PBS washing, digested samples were passed through a 70uM cell strainer to obtain a single cell suspension and then washed in FACS buffer.
Flow cytometry (FACS) analysis and single cell sorting of tumor
Immunostaining of single cell suspensions was performed according to standard protocols. Forward and side scatter settings were used to select for immune cells and to exclude doublets. Viable cells were identified by low PI staining, and immune cells were sorted based on CD45 expression. For each tumor, multiple plates were sorted with CD45+ immune cells. Immune cells were single cell sorted using index sorting into 384 wells plates containing 2 mL of lysis solution with barcoded poly(T) and reverse-transcription (RT) primers. Four empty wells were kept in each 384-well plate as ‘no-cell controls’. Plates were briefly centrifuged, snap frozen on dry ice, and stored at −80°. The following fluorescent dye-conjugated anti-human antibodies were used for staining: anti-CD11b, anti-CD19, anti-CD4, anti-CD8a, anti-CD45. Single cell suspensions were stained with antibodies against surface molecules.
Massively parallel single-cell RNA-seq library preparation (MARS-seq 2.0)
Single cell libraries were prepared using a large-scale Parallel Single-Cell RNA-seq method (MARS-seq). In short, mRNA from single cells sorted into cell capture plates was barcoded, converted into cDNA and pooled using an automated pipeline. Subsequently, the collected sample was linearly amplified by T7 in vitro transcription, and resulting RNA was fragmented and converted into a sequencing-ready library by tagging the samples with pool barcodes and illumina sequences during ligation, reverse transcription, and PCR. Each pool of cells was tested for library quality and library concentration was detected.
10X transcriptome sequencing
Cells were loaded onto the 10X Chromium Single Cell Platform (10X Genomics) at a concentration of 1,000 cells per μL (Single Cell 3′ library and Gel Bead Kit v.3) as described in the manufacturer’s protocol. Generation of gel beads in emulsion (GEMs), barcoding, GEM-RT clean-up, complementary DNA amplification and library construction were all performed as per the manufacturer’s protocol. Qubit was used for library quantification before pooling. The final library pool was sequenced on the Illumina NovaSeq 6000 instrument using 150-base-pair paired-end reads.
Multiplex immunostaining
To obtain serial 4-μM-thick sections, 6 primary tumors among the cohort of LUAD patients were immediately fixed into 4% PFA overnight at 4°C, dehydrated and embedded in paraffin. Paraffin slides were first rehydrated to proceed further with antigen retrieval. The sections were blocked with the appropriate Bovine Serum Albumin (BSA) and incubated overnight with the following antibodies: anti-CD8, anti-CD4, anti-CD19, anti-KI67, anti-CD20 anti-CXCL13, anti- CD3 anti-CD19 at a dilution 1:200. Whenever sections were stained in fluorescence, DAPI was used. The morphometric analysis was evaluated by H&E staining, respectively. Images were captured using an Olympus VS200 confocal system (Olympus).
Analysis of single-cell RNA-seq data
scRNA-seq libraries (pooled at equimolar concentration) were sequenced on an Illumina NovaSeq 6000 sequencer. Sequences were mapped to the human genome (hg38), demultiplexed, and filtered as previously described with the following adaptations. Reads were mapped to human reference genome hg38 using HISAT2 (version 2.1.0); reads with multiple mapping positions were excluded. Reads were associated with genes if they were mapped to an exon, using the UCSC genome browser for reference. Exons of different genes that shared genomic position on the same strand were considered a single gene with a concatenated gene symbol.
Metacell modeling for MARS-seq data
To analyze the MARS-seq data from all the samples, we used the MetaCell package57 as previously described.68 Gene features with high variance to mean were selected using the variation-to-mean parameter Tvm >0.08 and minimal total UMI >400. From the gene features, we excluded high abundance lincRNA and genes linked with poorly supported transcriptional models (such as genes annotated with the prefix "AC [0–9]", "AL [0–9]", etc.). Annotation of the metacell model was done using the metacell confusion matrix and analysis of marker genes. We next removed doublets contamination and performed second round clustering on CD45+ immune cells. In the final clustering, the gene feature selection strategy described above retained a total of 1,333 genes from 63717 immune cells for the computation of the Metacell balanced similarity graph. We used 500 bootstrap iterations. Metacell splitting was performed by clustering the cells within each metacell and splitting it if distinct clusters are detected. We examined batch effect by comparing the multiple plates from the same sample processed in the same batch or across different batches and cells from the same population, and confirmed our method displayed no clear batch effect and our analysis is robust. For sub-clustering analysis, the sub-clustering analysis was similar to cluster for all immune cells. From the gene features, we excluded high abundance lincRNA and genes linked with poorly supported transcriptional models (such as genes annotated with the prefix "AC [0–9]", "AL [0–9]", etc.). In the final clustering, the gene feature selection strategy described above retained feature genes from for the computation of the Metacell balanced similarity graph. We used 500 bootstrap iterations. Metacell splitting was performed by clustering the cells within each metacell and splitting it if distinct clusters are detected. Batch effect was removed in this section using KNN methods.
The 2D projection in the metacell modeling for MARS-seq data
The 2D projection is computed using a force-directed layout algorithm on a balanced similarity graph. First, a raw similarity matrix is generated by computing Pearson’s correlations on the transformed UMI counts of features (genes). Next, the raw similarity matrix is used to generate a weighted adjacency matrix for a weighted, directed cell graph G, in which a heavy edge from cell i to cell j indicates strong attraction of the former to the latter. Then, the balanced similarity graph G is partitioned into metacell graph GM using an adaptation of k-means to graphs with bootstrapping. Finally, the coordinates for each metacell in the 2D projection are computed by applying a standard force-directed layout algorithm to the metacell graph GM, and the cells are positioned by averaging the metacell coordinates of their filtered neighbor cells in the original balanced graph G.
Toomanycells clustering
To compare the cellular clustering difference among four pathological stages, we separately performed clustering analysis on samples from four groups by using TooManyCells.64 In detail, we used too-many-cells make-tree function and set draw-collection as “PieRing.”
Differential gene expression between two cell populations
We used the rich single cell gene expression information and performed differential gene expression analysis by comparing cells of two subclusters. To account for the variable cell numbers between individual patients, the same number of cells were sampled from each patient for a given cell population. In order to obtain comparable results across clusters, we also controlled the total number of cells we sampled for a given population. A z-statistics was computed for each gene using Wilcoxon rank-sum test on the down-sampled UMI counts from the cells of two cluster. Z-statistics Zmean for each gene was calculated by randomly repeating 100 times the sampling procedure. Zmean were converted into p values, and then we performed multiple testing correction on the p values with a standard Benjamini-Hochberg procedure and controlled for false discovery rate (FDR) q-value <0.05.
Analysis of 10x genomics 5′ single cell immune profiling data
Raw reads were demultiplexed to FASTQ files using Cell Ranger mkfastq (v7.0.0, 10X Genomics), and the Cumulus cellranger-workflow implementation of Cell Ranger multi (v7.0.0) was used to align reads to the human reference genome (GRCh38-2020-A). The filtered contig annotations, which contained high-level annotations of each high-confidenT cellular contig, were further filtered by removing records with ‘raw_consensus_id’ as ‘none’. Differences in clonality and Morisita indices between different stages and cohorts were evaluated by two-sided t-test. The Immunarch (v0.7.0, R package) was used to calculate the size distribution of tumor TCR clonotypes by using ‘repClonality’ function.
Analysis of public scRNA-seq data of LUAD
The data for the public LUAD immune atlas was from previously published NSCLC studies.7,38,39,40,41,42,43 Individual datasets were merged into a h5ad-format data. Since genome annotations partly differed between the datasets, we transferred the gene symbol to the same hg38 version using alias2SymbolUsingNCBI function in the limma R package. Gene symbols that were missing in more than 25% cells were excluded. We integrated the datasets using the scVI algorithm,59 as These methods have been shown to perform well for integration of scRNA-seq data, especially when dealing with complex batch effects and integrating atlas-level data. Raw counts were used as input for scVI model. We computed the UMAP embeddings for visualization using the embedding learned from scVI. Unsupervised clustering was also carried out using this learnt embedding using the Leiden algorithm as implemented in Scanpy.
Spatially resolved cell quantification framework
H&E and multiplex immunostaining images were visualized using QuPath and Halo software. For multi-color images, cell detection algorithm used on QuPath was “positive cell detection”. In order to accommodate for potential variability in signal intensities among cell types and slides, the parameters of these cell detection algorithms were tuned with visual inspection in each slide and for each cell type.
Quantification and statistical analysis
Statistical analysis
Statistical analysis Data was presented as mean (±SEM) of three independent experiments. Comparisons between two groups of samples were evaluated using the Mann–Whitney U test. All P-values reported were two tailed and statistical significance was defined as P less than 0.05. All statistical analyses were conducted using R software (R Foundation for Statistical Computing, Vienna, Austria) and GraphPad prism 8.0.2. The Univariate and multivariate analyses were performed with the Cox proportional hazards model to identify independent prognostic factors affecting OS of the LUAD patients.
Acknowledgments
We would like to express our gratitude to the donors and their families for their kind support and all the participating clinical study research teams in this study. We are also very grateful for the mouse resources provided by Prof. Nengming Xiao and Prof. Shun Li. This research is supported by the National Key R&D Program of China (2019YFA0906100), the National Natural Science Foundation of China (32170919), the Shenzhen Science and Technology Program (KQTD20210811090115019), the Shenzhen Institute of Synthetic Biology Scientific Research Program (JCHZ20210003), the National Natural Science Foundation of China (82071767 and 82230059), and the Jiangsu Provincial Key Research Development Program of China (BE2022770).
Author contributions
W.L., W.Y., Z.L., Y.R., Y.C., C.C., G.W., and H.L. conceived the project and designed the experiments. W.L., Z.L., S.G., and H.L. performed the experiments. W.Y., S.L., and H.L. analyzed the data. W.L., Z.L., S.G., G.W., and H.L. were involved in patient inclusion and sample acquisition. W.L., W.Y., and H.L. wrote the manuscript. H.L. supervised the project.
Declaration of interests
The authors declare no competing interests.
Published: March 7, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101448.
Contributor Information
Yun Chen, Email: chenyun@njmu.edu.cn.
Guangsuo Wang, Email: wang.guangsuo@szhospital.com.
Chang Chen, Email: changchenc@tongji.edu.cn.
Hanjie Li, Email: hj.li@siat.ac.cn.
Supplemental information
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber R.D., Old L.J., Smyth M.J. Cancer Immunoediting: Integrating Immunity's Roles in Cancer Suppression and Promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 3.Altorki N.K., Markowitz G.J., Gao D., Port J.L., Saxena A., Stiles B., McGraw T., Mittal V. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat. Rev. Cancer. 2019;19:9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian Y., Li Q., Yang Z., Zhang S., Xu J., Wang Z., Bai H., Duan J., Zheng B., Li W., et al. Single-cell transcriptomic profiling reveals the tumor heterogeneity of small-cell lung cancer. Signal Transduct. Target. Ther. 2022;7:346. doi: 10.1038/s41392-022-01150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelka K., Hofree M., Chen J.H., Sarkizova S., Pirl J.D., Jorgji V., Bejnood A., Dionne D., Ge W.H., Xu K.H., et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell. 2021;184:4734–4752.e20. doi: 10.1016/j.cell.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo X., Zhang Y., Zheng L., Zheng C., Song J., Zhang Q., Kang B., Liu Z., Jin L., Xing R., et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 7.Leader A.M., Grout J.A., Maier B.B., Nabet B.Y., Park M.D., Tabachnikova A., Chang C., Walker L., Lansky A., Le Berichel J., et al. Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification. Cancer Cell. 2021;39:1594–1609.e12. doi: 10.1016/j.ccell.2021.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azizi E., Carr A.J., Plitas G., Cornish A.E., Konopacki C., Prabhakaran S., Nainys J., Wu K., Kiseliovas V., Setty M., et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell. 2018;174:1293–1308.e36. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaitin D.A., Kenigsberg E., Keren-Shaul H., Elefant N., Paul F., Zaretsky I., Mildner A., Cohen N., Jung S., Tanay A., Amit I. Massively Parallel Single-Cell RNA-Seq for Marker-Free Decomposition of Tissues into Cell Types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 11.Wei W., Zeng H., Zheng R., Zhang S., An L., Chen R., Wang S., Sun K., Matsuda T., Bray F., He J. Cancer registration in China and its role in cancer prevention and control. Lancet Oncol. 2020;21:E342–E349. doi: 10.1016/S1470-2045(20)30073-5. [DOI] [PubMed] [Google Scholar]
- 12.Cascone T., William W.N., Weissferdt A., Leung C.H., Lin H.Y., Pataer A., Godoy M.C.B., Carter B.W., Federico L., Reuben A., et al. Neoadjuvant nivolumab or nivolumab plus ipilimumab in operable non-small cell lung cancer: the phase 2 randomized NEOSTAR trial. Nat. Med. 2021;27:504–514. doi: 10.1038/s41591-020-01224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J., Yang H., Teo A.S.M., Amer L.B., Sherbaf F.G., Tan C.Q., Alvarez J.J.S., Lu B., Lim J.Q., Takano A., et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 2020;52:177–186. doi: 10.1038/s41588-019-0569-6. [DOI] [PubMed] [Google Scholar]
- 14.Detterbeck F.C., Boffa D.J., Kim A.W., Tanoue L.T. The Eighth Edition Lung Cancer Stage Classification. Chest. 2017;151:193–203. doi: 10.1016/j.chest.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Nicholson A.G., Tsao M.S., Beasley M.B., Borczuk A.C., Brambilla E., Cooper W.A., Dacic S., Jain D., Kerr K.M., Lantuejoul S., et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 2022;17:362–387. doi: 10.1016/j.jtho.2021.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Chen W.W., Liu W., Li Y., Wang J., Ren Y., Wang G., Chen C., Li H. Deciphering the Immune-Tumor Interplay During Early-Stage Lung Cancer Development via Single-Cell Technology. Front. Oncol. 2021;11:716042. doi: 10.3389/fonc.2021.716042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavin Y., Kobayashi S., Leader A., Amir E.A.D., Elefant N., Bigenwald C., Remark R., Sweeney R., Becker C.D., Levine J.H., et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell. 2017;169:750–765.e17. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carbone D.P., Reck M., Paz-Ares L., Creelan B., Horn L., Steins M., Felip E., van den Heuvel M.M., Ciuleanu T.E., Badin F., et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017;376:2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reck M., Rodríguez-Abreu D., Robinson A.G., Hui R., Csőszi T., Fülöp A., Gottfried M., Peled N., Tafreshi A., Cuffe S., et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 20.Greten F.R., Grivennikov S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cong J., Wang X., Zheng X., Wang D., Fu B., Sun R., Tian Z., Wei H. Dysfunction of Natural Killer Cells by FBP1-Induced Inhibition of Glycolysis during Lung Cancer Progression. Cell Metab. 2018;28:243–255.e5. doi: 10.1016/j.cmet.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 22.Cassetta L., Pollard J.W. Targeting macrophages: therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018;17:887–904. doi: 10.1038/nrd.2018.169. [DOI] [PubMed] [Google Scholar]
- 23.van der Leun A.M., Thommen D.S., Schumacher T.N. CD8(+) T cell states in human cancer: insights from single-cell analysis. Nat. Rev. Cancer. 2020;20:218–232. doi: 10.1038/s41568-019-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schenkel J.M., Herbst R.H., Canner D., Li A., Hillman M., Shanahan S.L., Gibbons G., Smith O.C., Kim J.Y., Westcott P., et al. Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1(+) CD8(+) T cells in tumor-draining lymph nodes. Immunity. 2021;54:2338–2353.e6. doi: 10.1016/j.immuni.2021.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mintz M.A., Cyster J.G. T follicular helper cells in germinal center B cell selection and lymphomagenesis. Immunol. Rev. 2020;296:48–61. doi: 10.1111/imr.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaud D., Steward C.R., Mirlekar B., Pylayeva-Gupta Y. Regulatory B cells in cancer. Immunol. Rev. 2021;299:74–92. doi: 10.1111/imr.12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui C., Wang J., Fagerberg E., Chen P.M., Connolly K.A., Damo M., Cheung J.F., Mao T., Askari A.S., Chen S., et al. Neoantigen-driven B cell and CD4 T follicular helper cell collaboration promotes anti-tumor CD8 T cell responses. Cell. 2021;184:6101–6118.e13. doi: 10.1016/j.cell.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fridman W.H., Meylan M., Pupier G., Calvez A., Hernandez I., Sautès-Fridman C. Tertiary lymphoid structures and B cells: An intratumoral immunity cycle. Immunity. 2023;56:2254–2269. doi: 10.1016/j.immuni.2023.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Goubet A.G., Lordello L., Alves Costa Silva C., Peguillet I., Gazzano M., Mbogning-Fonkou M.D., Thelemaque C., Lebacle C., Thibault C., Audenet F., et al. Escherichia coli-Specific CXCL13-Producing TFH Are Associated with Clinical Efficacy of Neoadjuvant PD-1 Blockade against Muscle-Invasive Bladder Cancer. Cancer Discov. 2022;12:2280–2307. doi: 10.1158/2159-8290.CD-22-0201. [DOI] [PubMed] [Google Scholar]
- 30.Zander R., Kasmani M.Y., Chen Y., Topchyan P., Shen J., Zheng S., Burns R., Ingram J., Cui C., Joshi N., et al. Tfh-cell-derived interleukin 21 sustains effector CD8(+) T cell responses during chronic viral infection. Immunity. 2022;55:475–493.e5. doi: 10.1016/j.immuni.2022.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meylan M., Petitprez F., Becht E., Bougoüin A., Pupier G., Calvez A., Giglioli I., Verkarre V., Lacroix G., Verneau J., et al. Tertiary lymphoid structures generate and propagate anti-tumor antibody-producing plasma cells in renal cell cancer. Immunity. 2022;55:527–541.e5. doi: 10.1016/j.immuni.2022.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Helmink B.A., Reddy S.M., Gao J., Zhang S., Basar R., Thakur R., Yizhak K., Sade-Feldman M., Blando J., Han G., et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petitprez F., de Reyniès A., Keung E.Z., Chen T.W.W., Sun C.M., Calderaro J., Jeng Y.M., Hsiao L.P., Lacroix L., Bougoüin A., et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577:556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 34.Patil N.S., Nabet B.Y., Müller S., Koeppen H., Zou W., Giltnane J., Au-Yeung A., Srivats S., Cheng J.H., Takahashi C., et al. Intratumoral plasma cells predict outcomes to PD-L1 blockade in non-small cell lung cancer. Cancer Cell. 2022;40:289–300.e4. doi: 10.1016/j.ccell.2022.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida H., Nomizo T., Ozasa H., Tsuji T., Funazo T., Yasuda Y., Ajimizu H., Yamazoe M., Kuninaga K., Ogimoto T., et al. PD-L1 polymorphisms predict survival outcomes in advanced non-small-cell lung cancer patients treated with PD-1 blockade. Eur. J. Cancer. 2021;144:317–325. doi: 10.1016/j.ejca.2020.11.035. [DOI] [PubMed] [Google Scholar]
- 36.Yang M., Lu J., Zhang G., Wang Y., He M., Xu Q., Xu C., Liu H. CXCL13 shapes immunoactive tumor microenvironment and enhances the efficacy of PD-1 checkpoint blockade in high-grade serous ovarian cancer. J. Immunother. Cancer. 2021;9:e001136. doi: 10.1136/jitc-2020-001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao D., Han G., Sinjab A., Gomez-Bolanos L.I., Lazcano R., Serrano A., Hernandez S.D., Dai E., Cao X., Hu J., et al. The Single-Cell Immunogenomic Landscape of B and Plasma Cells in Early-Stage Lung Adenocarcinoma. Cancer Discov. 2022;12:2626–2645. doi: 10.1158/2159-8290.CD-21-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goveia J., Rohlenova K., Taverna F., Treps L., Conradi L.C., Pircher A., Geldhof V., de Rooij L.P.M.H., Kalucka J., Sokol L., et al. An Integrated Gene Expression Landscape Profiling Approach to Identify Lung Tumor Endothelial Cell Heterogeneity and Angiogenic Candidates. Cancer Cell. 2020;37:421. doi: 10.1016/j.ccell.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Kim N., Kim H.K., Lee K., Hong Y., Cho J.H., Choi J.W., Lee J.I., Suh Y.L., Ku B.M., Eum H.H., et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 2020;11:2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., Bassez A., Decaluwé H., Pircher A., Van den Eynde K., et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 41.Maynard A., McCoach C.E., Rotow J.K., Harris L., Haderk F., Kerr D.L., Yu E.A., Schenk E.L., Tan W., Zee A., et al. Therapy-Induced Evolution of Human Lung Cancer Revealed by Single-Cell RNA Sequencing. Cell. 2020;182:1232–1251.e22. doi: 10.1016/j.cell.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salcher S., Sturm G., Horvath L., Untergasser G., Kuempers C., Fotakis G., Panizzolo E., Martowicz A., Trebo M., Pall G., et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell. 2022;40:1503–1520.e8. doi: 10.1016/j.ccell.2022.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu F., Fan J., He Y., Xiong A., Yu J., Li Y., Zhang Y., Zhao W., Zhou F., Li W., et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat. Commun. 2021;12:2540. doi: 10.1038/s41467-021-22801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H., Carrot-Zhang J., Zhao Y., Hu H., Freeman S.S., Yu S., Ha G., Taylor A.M., Berger A.C., Westlake L., et al. Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat. Commun. 2019;10:5472. doi: 10.1038/s41467-019-13460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veatch J.R., Lee S.M., Shasha C., Singhi N., Szeto J.L., Moshiri A.S., Kim T.S., Smythe K., Kong P., Fitzgibbon M., et al. Neoantigen-specific CD4(+) T cells in human melanoma have diverse differentiation states and correlate with CD8(+) T cell, macrophage, and B cell function. Cancer Cell. 2022;40:393–409.e9. doi: 10.1016/j.ccell.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen M., Giladi A., Barboy O., Hamon P., Li B., Zada M., Gurevich-Shapiro A., Beccaria C.G., David E., Maier B.B., et al. The interaction of CD4(+) helper T cells with dendritic cells shapes the tumor microenvironment and immune checkpoint blockade response. Nat. Cancer. 2022;3:303–317. doi: 10.1038/s43018-022-00338-5. [DOI] [PubMed] [Google Scholar]
- 47.Cho J.W., Park S., Kim G., Han H., Shim H.S., Shin S., Bae Y.S., Park S.Y., Ha S.J., Lee I., Kim H.R. Dysregulation of T(FH)-B-T(RM) lymphocyte cooperation is associated with unfavorable anti-PD-1 responses in EGFR-mutant lung cancer. Nat. Commun. 2021;12:6068. doi: 10.1038/s41467-021-26362-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis M.R., Zhu Z., Hansen D.M., Bai Q., Fang Y. The role of IL-21 in immunity and cancer. Cancer Lett. 2015;358:107–114. doi: 10.1016/j.canlet.2014.12.047. [DOI] [PubMed] [Google Scholar]
- 49.Sautès-Fridman C., Roumenina L.T. B cells and complement at the forefront of chemotherapy. Nat. Rev. Clin. Oncol. 2020;17:393–394. doi: 10.1038/s41571-020-0376-0. [DOI] [PubMed] [Google Scholar]
- 50.Lu Y., Zhao Q., Liao J.Y., Song E., Xia Q., Pan J., Li Y., Li J., Zhou B., Ye Y., et al. Complement Signals Determine Opposite Effects of B Cells in Chemotherapy-Induced Immunity. Cell. 2020;180:1081–1097.e24. doi: 10.1016/j.cell.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 51.Fridman W.H., Meylan M., Petitprez F., Sun C.M., Italiano A., Sautès-Fridman C. B cells and tertiary lymphoid structures as determinants of tumour immune contexture and clinical outcome. Nat. Rev. Clin. Oncol. 2022;19:441–457. doi: 10.1038/s41571-022-00619-z. [DOI] [PubMed] [Google Scholar]
- 52.Sautès-Fridman C., Petitprez F., Calderaro J., Fridman W.H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer. 2019;19:307–325. doi: 10.1038/s41568-019-0144-6. [DOI] [PubMed] [Google Scholar]
- 53.Zhang C., Li J., Cheng Y., Meng F., Song J.W., Fan X., Fan H., Li J., Fu Y.L., Zhou M.J., et al. Single-cell RNA sequencing reveals intrahepatic and peripheral immune characteristics related to disease phases in HBV-infected patients. Gut. 2023;72:153–167. doi: 10.1136/gutjnl-2021-325915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.R Core Team . 2013. “R: A language and environment for statistical computing.”; pp. 275–286. [Google Scholar]
- 55.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perez, Fernando, Brian E., Granger, Hunter John D. “Python: an ecosystem for scientific computing.”. Computing in Science & Engineering. 2010;13.2:13–21. [Google Scholar]
- 57.Baran Y., Bercovich A., Sebe-Pedros A., Lubling Y., Giladi A., Chomsky E., Meir Z., Hoichman M., Lifshitz A., Tanay A. MetaCell: analysis of single-cell RNA-seq data using K-nn graph partitions. Genome Biol. 2019;20:206. doi: 10.1186/s13059-019-1812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolf F., Alexander, Philipp A., Fabian J. “SCANPY: large-scale single-cell gene expression data analysis.”. Genome biology. 2018;19:1–5. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gayoso A., Lopez R., Xing G., Boyeau P., Valiollah Pour Amiri V., Hong J., Wu K., Jayasuriya M., Mehlman E., Langevin M., et al. A Python library for probabilistic analysis of single-cell omics data. Nat. Biotechnol. 2022;40:163–166. doi: 10.1038/s41587-021-01206-w. [DOI] [PubMed] [Google Scholar]
- 60.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Aerts S. SCENIC: single-cell regulatory network inference and clustering. Nature methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nazarov V., Tsvetkov V., Fiadziushchanka S., Rumynskiy E., Popov A., Balashov I., Samokhina M. immunarch. Bioinformatics Analysis of T-Cell and B-Cell Immune Repertoires. 2023 [Google Scholar]
- 63.Borcherding, Nicholas, Nicholas L., Bormann, Kraus G. “scRepertoire: An R-based toolkit for single-cell immune receptor analysis.”. F1000Research. 2020:9. doi: 10.12688/f1000research.22139.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwartz G.W., Zhou Y., Petrovic J., Fasolino M., Xu L., Shaffer S.M., Pear W.S., Vahedi G., Faryabi R.B. TooManyCells identifies and visualizes relationships of single-cell clades. Nat. Methods. 2020;17:405–413. doi: 10.1038/s41592-020-0748-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Yu G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The innovation. 2021;2 doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC bioinformatics. 2013;14:1–15. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Humphries M.P., Maxwell P., Salto-Tellez M. QuPath: The global impact of an open source digital pathology system. Computational and Structural Biotechnology Journal. 2021;19:852–859. doi: 10.1016/j.csbj.2021.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H., van der Leun A.M., Yofe I., Lubling Y., Gelbard-Solodkin D., van Akkooi A.C.J., van den Braber M., Rozeman E.A., Haanen J.B.A.G., Blank C.U., et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell. 2020;181:747. doi: 10.1016/j.cell.2020.04.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single cell RNA-seq data that support the findings of this study is available for download in the Genome Sequence Archive for Human with accession number: HRA002961 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA002961). This paper does not report any original code. The code that support the findings of this study is available at https://github.com/youwh1996/ealy-LUAD-scRNA. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.