Summary
Nipah virus (NiV) has been recently ranked by the World Health Organization as being among the top eight emerging pathogens likely to cause major epidemics, whereas no therapeutics or vaccines have yet been approved. We report a method to deliver immunogenic epitopes from NiV through the targeting of the CD40 receptor of antigen-presenting cells by fusing a selected humanized anti-CD40 monoclonal antibody to the Nipah glycoprotein with conserved NiV fusion and nucleocapsid peptides. In the African green monkey model, CD40.NiV induces specific immunoglobulin A (IgA) and IgG as well as cross-neutralizing responses against circulating NiV strains and Hendra virus and T cell responses. Challenge experiments using a NiV-B strain demonstrate the high protective efficacy of the vaccine, with all vaccinated animals surviving and showing no significant clinical signs or virus replication, suggesting that the CD40.NiV vaccine conferred sterilizing immunity. Overall, results obtained with the CD40.NiV vaccine are highly promising in terms of the breadth and efficacy against NiV.
Keywords: CD40-targeting vaccine, dendritic cell-based vaccine, Nipah virus, Henipavirus, epitope mapping, neutralizing antibodies, cross-neutralizing responses, germinal center reaction, sterilizing immunity, prophylactic vaccine
Graphical abstract
Highlights
-
•
Vaccine targeting APCs: design for Nipah protection and addressing emerging threats
-
•
CD40-targeting vaccine induces protective responses against Nipah
-
•
Vaccine protection against Nipah by targeting APCs
-
•
Advance in Nipah virus vaccine design by targeting APCs
Pastor et al. describe a Nipah virus vaccine design that leverages an APC-targeting approach, CD40.NiV, demonstrating immunogenicity, neutralization and Henipavirus cross-neutralization, and protection in AGM preclinical models. This strategy holds significance for countering Nipah virus outbreaks and shaping future vaccine designs against emerging infectious threats.
Introduction
Nipah virus (NiV) is an emerging, highly pathogenic, zoonotic paramyxovirus first recognized following a 1998–1999 outbreak of severe febrile encephalitis in Malaysia and Singapore.1 Subsequent outbreaks of NiV occurred in India and have occurred almost annually in Bangladesh. NiV infection is the most severe viral zoonosis to recently emerge from bats, causing serious health and economic problems, and is of particular importance due to its bioweapon potential. NiV is included on the World Health Organization’s Blueprint List of Priority Pathogens, as well as on the Coalition for Epidemic Preparedness Innovations’ Priority Pathogens List.2
Transmission occurs mainly from fruit contaminated by the natural host, the Pteropus fruit bat, to a wide range of mammals such as pigs, horses, or other domestic animals and, finally, humans.3 Phylogenetic analyses indicate that there are at least two strains of NiV.4 Outbreaks from the Malaysian strain (NiV-M) showed high rates of encephalitis, whereas the Bangladesh strain (NiV-B) is also associated with severe respiratory symptoms and human-to-human transmission through close contact with people’s secretions and nosocomial transmission. In addition, NiV-B shows a shorter incubation period and higher mortality rate than NiV-M, reaching 100% in some of outbreaks. The outbreak of 2018 in the South Indian district of Kozhikode, Kerala, showed a fatality rate of 91%,5 and other isolated cases have also been reported since then.6
Several NiV candidate vaccines are at preclinical or early clinical development stages, but no licensed vaccines or therapeutics are available for human use yet.7,8,9,10 Most vaccine candidates target the NiV surface glycoprotein (G) and/or fusion (F) protein as immunogens, as these proteins are required for viral entry and are exposed on virus surface. Here, we took advantage of a method to target sequences from these molecules to dendritic cells (DCs). Targeting vaccine antigens to DCs via surface receptors is an appealing strategy to improve subunit-vaccine efficacy (while reducing the amount of required antigen) that can be used with or without adjuvant.11 Direct delivery of the antigen, which can additionally activate cell receptors, may also evoke a danger signal, stimulating an immune response. Among the various DC receptors tested, including lectins and scavenger receptors, we have reported the superiority of vaccines that target diverse viral antigens to CD40-expressing antigen-presenting cells (APCs) in evoking strong antigen-specific T and B cell responses.12,13,14,15,16,17,18,19,20,21 Drawing from this knowledge, we developed a vaccine that targets selected epitopes from the G, F, and nucleocapsid (N) proteins from NiV-B. Here, we report the immunogenicity and protective efficacy of an anti-CD40 multi-epitope Nipah prophylactic vaccine in African green monkeys (AGMs) exposed to an experimental challenge with the NiV-B virus.
Results
In silico down-selection of NiV predictive immunogenic peptides and CD40.NiV vaccine design
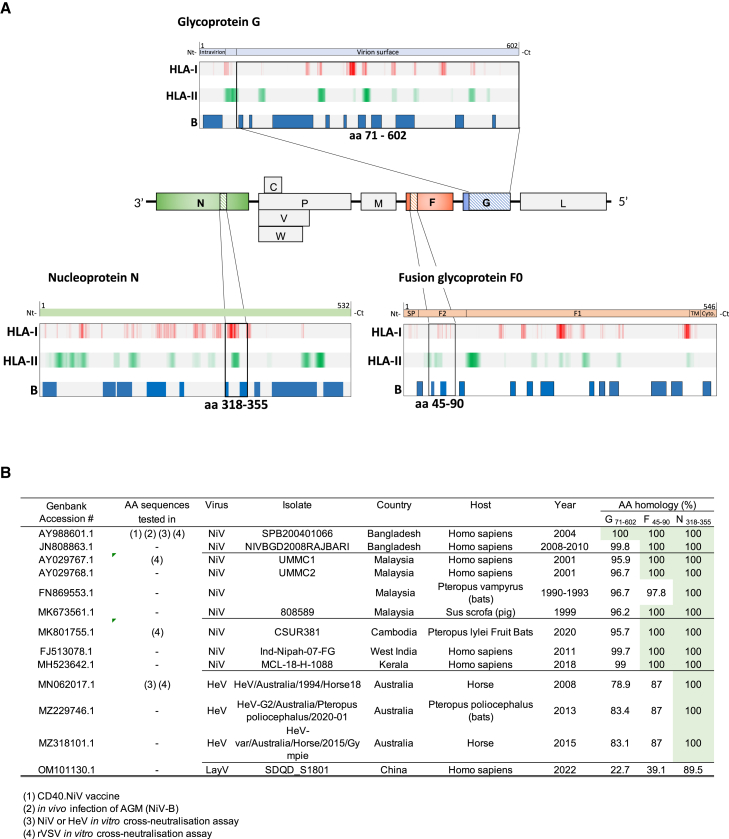
The NiV surface G is a gold-standard antigen for inducing protective humoral responses.22 Other cellular effectors, such as helper and effector T cells, may also participate in host defense.8 We screened the NiV G ectodomain (ECD) to identify vaccine epitopes using the NetMHC 4.0 and NetMHCII 2.3 software, which predict T cell epitopes that bind to a large panel of class I and II HLA molecules, respectively. Linear B cell epitopes were predicted using the BepiPred 2.0 software. The NiV G ECD vaccine region was predicted to contain 3,522 T cell epitopes and 15 B cell epitopes, respectively. Due to their conservation between NiV strains, we further identified vaccine epitopes from the F and N proteins. Regions with strong binder epitopes and the highest HLA coverage were highlighted, as well as the linear B cell epitopes (Figure 1A). The NiV F (aa 45–90) and NiV N (aa 318–355) down-selected peptides contain 356 and 266 predicted T cell epitopes, respectively, as well as three and two linear B cell epitopes, respectively. Globally, these amino acid sequences were screened for homology with other Henipaviruses, obtaining 100% homology between different Nipah strains for the F and N peptides and more than 98% for the NiV G ECD (Figure 1B; see Table S1 for detailed aa sequences).
Figure 1.
Epitope mapping of NiV antigens associated with the CD40 mAb
(A) Open reading frames encoding the G, F, and N proteins in the NiV (Bangladesh strain) genome are highlighted in blue, orange, and red, respectively (center). Full-length amino acid sequences were screened for predicted HLA-I (NetMHC 4.0) HLA-II (NetMHCII 2.3), and linear B cell epitopes (BepiPred 2.0). For HLA-I and -II, the color intensity is representative of the density of predicted epitopes, with coverage of at least 30% of the worldwide HLA. (Top) Ectodomain (ECD) of the G protein and (bottom) F and N down-selected domains associated with the CD40.NiV vaccine.
(B) Amino acid conservation of vaccine antigens.
We next engineered vectors expressing the NiV-B G ECD fused to the C termini of the heavy chains of the anti-human CD40 humanized 12E12 immunoglobulin G4 (IgG4) monoclonal antibody, with additional selected peptides of NiV F and NiV N fused to the C termini of the light chains (named CD40.NiV). Vaccines were produced in CHO cells and controlled for their quality (Figure S1).
We have previously shown that 12E12 anti-CD40 fused to viral antigens enhances CD40-mediated internalization and antigen presentation by mononuclear cells and ex-vivo-generated monocyte-derived DCs.15,19 Similarly, we confirm here the binding of the CD40.NiV vaccine to the human and AGM CD40 receptor in vitro. For this purpose, we used splenocytes from mice transgenic for the human CD40 receptor (CD40Hy/Mu transgenic mice; hCD40Tg) and peripheral blood mononuclear cells (PBMCs) from AGMs, respectively (Figures S2), where binding is demonstrated on B cells, which express CD40, and, to a lesser extent, on monocyte/macrophage populations.
CD40.NiV induces specific T and B cell responses in hCD40Tg mice
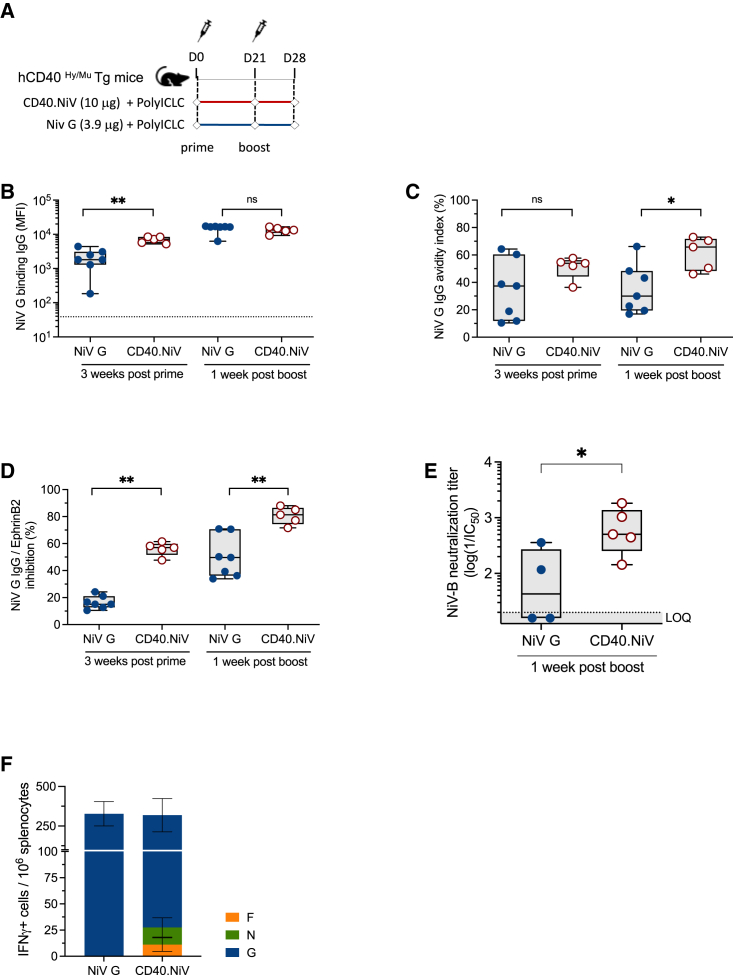
hCD40Tg mice were immunized with two subcutaneous (s.c.) injections of CD40.NiV vaccine (10 μg) or an equivalent amount of NiV G protein (3.9 μg), both with poly-ICLC (50 μg) on days 0 and 21 (Figure 2A). Antibody-mediated immune responses were first evaluated by Luminex assay (Figure 2B). Three weeks post-prime (PP), mice immunized with the CD40-targeted NiV G protein showed significantly higher anti-NiV G IgG levels (p < 0.01), highlighting the benefit conferred by the DC-targeting system, as previously reported for HIV-1 and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) CD40-based vaccines.12,23 We next compared the avidity of NiV G-specific IgG post-boost (PB) using a multiplex immunoassay approach24 (Figure 2C). Strikingly, the avidity index was significantly improved when NiV G was targeted to the CD40 receptor PB (p < 0.05), suggesting an advantage of such targeting for inducing B cell affinity maturation.
Figure 2.
CD40.NiV induces NiV neutralizing IgG and polyepitopic cellular T cell responses in hCD40Tg mice
(A) Schematic representation of the study design, comparing responses to CD40.NiV to the non-targeted NiV G ECD (with poly-ICLC). Blood samplings are represented by diamonds.
(B) IgG titers specific to NiV G ECD were determined by Luminex technology on days 0, 21, and 28 post-immunization with either CD40.NiV (red) or non-targeted NiV G ECD protein (blue), with poly-ICLC. The mean fluorescence intensity (MFI) (±SEM) is represented. Non-parametric Kruskal-Wallis tests with Dunn’s multiple comparison post hoc test, ∗p < 0.05, ∗∗p < 0.01, ns non-significant.
(C) Avidity of NiV G-specific IgG measured by Luminex on day 21 (3 weeks post-prime) and 28 (1 week post-boost), comparing mice immunized with non-targeted NiV G versus CD40.NiV, with poly-ICLC. The color legends and statistical analyses are the same as in (B).
(D) Neutralization activity of sera from mice immunized with NiV G ECD versus CD40.NiV (with poly-ICLC) measured first by a Luminex-based inhibition assay. The color legends and statistical analysis are the same as in (B).
(E) In vitro neutralization assay performed on NiV-B-infected cells using serial-diluted sera collected on day 28 post-immunization. The IC50 values were calculated and are reported as titers (log(1/IC50)). The dotted line indicates the limit of quantification (LOQ) for the neutralization titer. Non-parametric Mann-Whitney unpaired t tests; ∗p < 0.05.
(F) IFN-γ T cell responses to the NiV G, F, and N overlapping peptide pools assessed from spleens by ELISpot 1 week post-boost. The total number of spots are reported per million splenocytes (background subtracted). NiV G (blue), N (green), and F (orange). Data are representative of at least two experiments.
We further analyzed the protective capacity of CD40.NiV vaccines by evaluating the neutralizing capacity of the NiV G-specific antibodies, first using a virus-free Luminex-based surrogate neutralization assay (Figure 2D). Of note, this in-house-developed assay, similar to the inhibition assay for testing SARS-CoV-2 neutralizing IgG,25 is an inhibition assay for the binding of NiV G to the Ephrin B2 receptor by the IgG circulating in the sera. We observed significantly higher levels of inhibiting antibodies in mice after CD40.NiV vaccination than in those immunized with the NiV G protein (p < 0.01, PP and PB). The results of NiV G and CD40.NiV vaccination (with poly-ICLC) were further confirmed using a plate-reduction neutralizing test with NiV-B in BSL-4 facilities, confirming the high neutralizing capacity of IgGs induced by two injections of CD40.NiV (Figure 2E). We further confirmed the induced B cell responses in CD40.NiV (+poly-ICLC)-vaccinated mice by detecting germinal center (GC) B cells (Fas+/GL7+, from B220+/IgG+) within the draining lymph nodes (dLNs) (p < 0.001 as compared to the poly-ICLC control group) (Figures S3A and S3B). Furthermore, NiV G-specific GC B cells, gated as biotin+/+ cells, were significantly increased in the dLN of vaccinated animals (p < 0.05) (Figure S3C). To demonstrate the specific T cell responses against the various antigens from CD40.NiV, we performed an interferon γ (IFN-γ) ELISpot assay on splenocytes using pools of overlapping peptides. We detected dose-dependent IFN-γ-producing cells specific for the NiV G ECD, as well as NiV F and N peptides (Figures 2F, S3D, and S3E). Overall, these results demonstrate the immunogenicity of the CD40.NiV vaccine candidate.
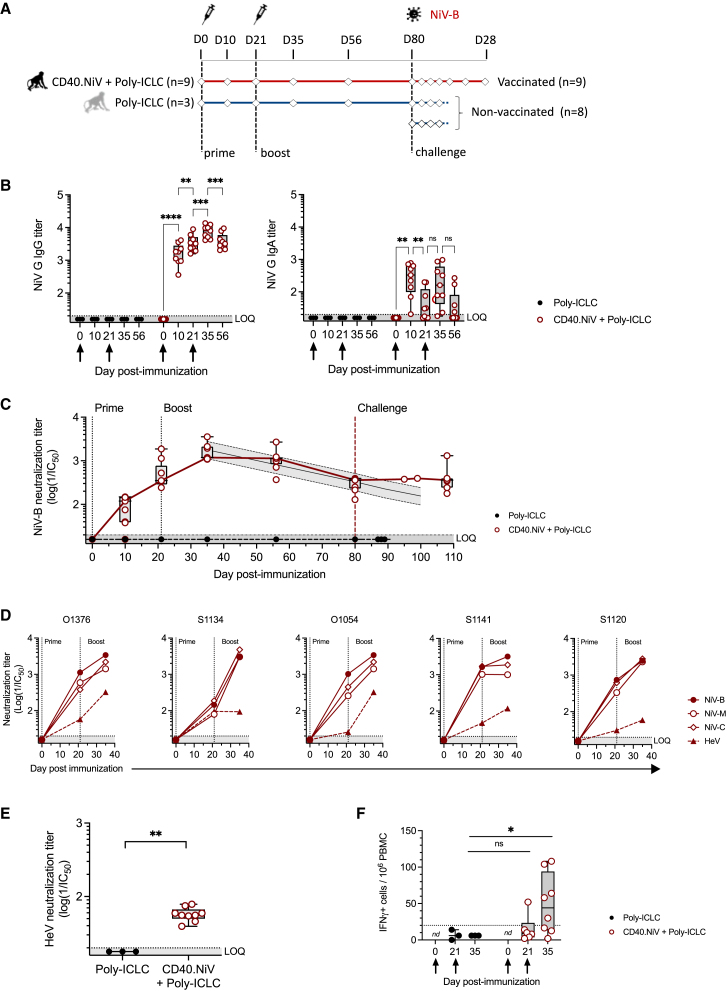
The CD40.NiV vaccine induces early and robust humoral and T cell responses in AGMs
Based on our preliminary results, the CD40.NiV vaccine was selected to be tested in a further AGM challenge study, the most relevant model to test infection by the NiV-B strain.26 Animals received either adjuvant alone (1 mg s.c. poly-ICLC, n = 3) or 200 μg s.c. CD40.NiV with poly-ICLC (n = 9) on days 0 and 21 (Figure 3A). All vaccinated animals showed specific and significant IgG and IgA titers 10 days PP (mean log titer of 3.17 [±0.3] and 2.38 [±0.5] for IgG and IgA, respectively, p < 0.001 compared to controls) (Figure 3B). IgG titers increased over time and then remained the same until day 56, whereas serum IgA levels dropped. Vaccination elicited a neutralization potential of NiV G-specific IgG by day 10 PP, reaching significance 2 weeks PB (mean neutralization titer 3.2 [±0.2], p < 0.001), and remained high until day 56 (Figure 3C). We estimated a significant average decrease of 0.016 log neutralization titer per day, leading to a prediction of the maintenance of a neutralization titer above the detectable threshold (mean log titer 2.2 [±0.1]) up to 100 days following the boost. The neutralizing capacity of specific antibodies was confirmed PP and PB using the Luminex-based surrogate inhibition assay (p < 0.001) (Figure S4). We assessed cross-neutralization to NiV-B, -M (Malaysia), and -C (Cambodia) and to HeV (Hendra virus) using the sera of five vaccinated AGMs by in vitro infection of VeroE6 cells with recombinant (F/G) vesicular stomatitis virus (VSV) particles27,28 (Figure 3D). Two weeks PB, all animals exhibited comparable cross-reactive humoral responses to NiV (F/G) proteins, as well as positive cross-reactive responses to Hendra virus (Figure 3E). IFN-γ ELISpot assays were conducted on PBMCs collected on day 21 (before the boost) and day 35 PP using pools of peptides for each vaccine antigen (Figure 3F). The overall responses were significant 2 weeks PB (p < 0.05) and positive in 5/8 tested animals.
Figure 3.
Cellular and humoral responses of AGMs vaccinated with CD40.NiV
(A) Schematic representation of the study design. Diamonds are representative of sampling.
(B) Blood NiV G-specific IgG (left) and IgA (right) titers, in poly-ICLC- (black) and CD40.NiV (+poly-ICLC)-vaccinated AGMs (red) on days 0, 10, 21, 35, and 56 post-prime. Individual Ig titers (log(1/EC50)) are shown. The whiskers indicate the medians (min-max) for each group and at each time point. The prime and boost are indicated by arrows. The dotted line indicates the LOQ for the Ig titer. Non-parametric Kruskal-Wallis tests with Dunn's multiple comparison post hoc test; ∗ p < 0.05; ∗∗ p < 0.01; ∗∗∗ p < 0.001; ns, non-significant.
(C) In vitro NiV-B neutralization activity of AGM sera measured post-immunization and post-challenge (80 days post-infection [dpi]): black line, poly-ICLC group (n = 3); red line, CD40.NiV with poly-ICLC (n = 9); gray line, modeling of the decrease in neutralizing Abs after the peak of the response (day 35).
(D) Cross-neutralization was assessed with serial dilutions of five AGM sera collected 21 and 35 dpi added to cells infected with VSVΔG-NiVF/G harboring the NiV G and F proteins, either from the Bangladesh (plain circles), Malaysia (open circles), or Cambodia (open diamonds) strain or from the Hendra virus (plain triangles). Results are reported as the log of the IC50. Dotted line: LOQ given by sera from the poly-ICLC group of AGMs. GenBank references of the NiV G and F proteins expressed by rVSV and homology of the amino acid sequence are detailed in Table S1.
(E) In vitro HeV neutralization activity of AGM sera measured 2 weeks post-boost (35 dpi): black dots, poly-ICLC group (n = 3); red dots, CD40.NiV with poly-ICLC (n = 9). Mann-Whitney unpaired t test, ∗∗p < 0.01.
(F) Total specific IFN-γ responses to the NiV G, F, and N antigens assessed in PBMCs on days 21 and 35 post-immunization, as in Figure 2 for the spleen. The number of spots is reported per million PBMCs (background subtracted). The dotted line represents the threshold of positivity, defined by the mean response (±3 SD) of the non-vaccinated AGM group. Non-parametric Kruskal-Wallis tests with Dunn’s multiple comparison post hoc test; ∗p < 0.05; ns non-significant.
Overall, CD40.NiV induced cellular responses associated with strong, rapid, and durable humoral responses in an AGM model, with high titers of neutralizing antibodies.
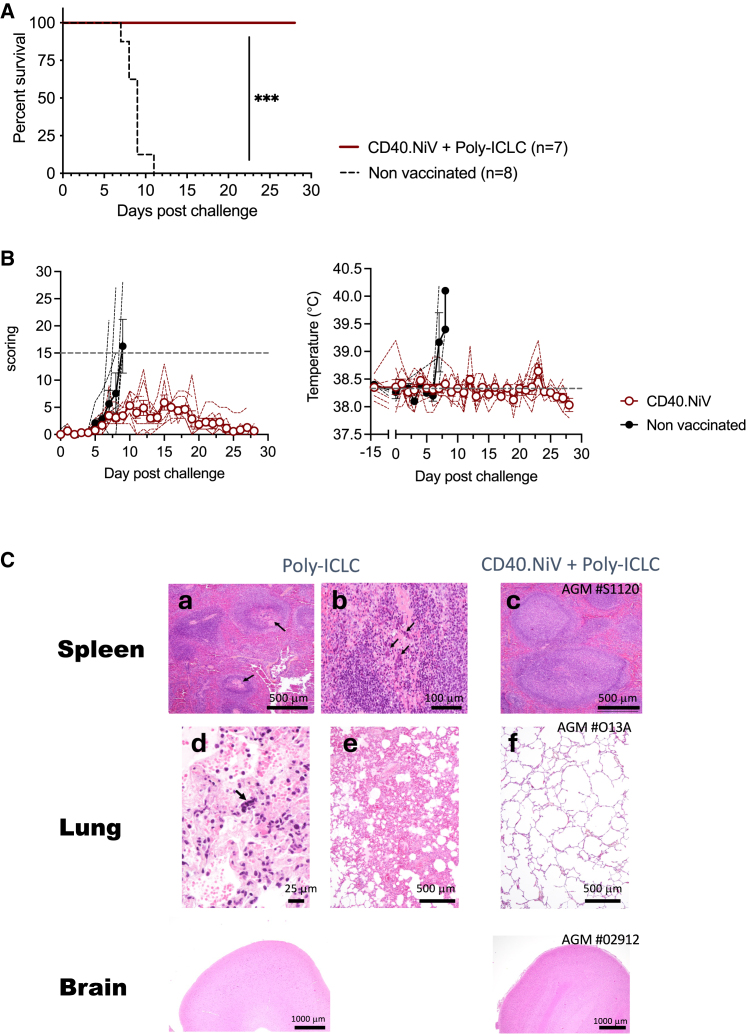
Clinical outcomes and survival for challenged animals
On day 80 post-immunization, the nine vaccinated and three control (poly-ICLC) AGMs were challenged intratracheally with a uniformly lethal dose of 102 plaque-forming units (PFUs) of the NiV-B strain (Figure 3A). The dose for the challenge was selected based on previous experiments that included five non-vaccinated animals infected under the same conditions. These controls were added to the control group for evaluation of the virological and clinical outcomes. Disease symptoms were followed over 28 days post-challenge (dpc), with regular blood draws to perform immunological, viral, and hematobiochemical tests. The CD40.NiV vaccination conferred 100% protection until the end of the follow-up scheduled at 28 dpc. Of note, two vaccinated animals had to be euthanized at 15 and 19 dpc due to injuries and ethical considerations independent of the NiV infection. These animals were excluded from survival study. By contrast, non-vaccinated animals reached the euthanasia criteria related to NiV disease between 7 and 11 dpc (Gehan-Breslow-Wilcoxon test, p < 0.001) (Figure 4A). From a clinical standpoint, only mild and transient clinical signs were evident in the vaccinated animals, with a mean clinical score remaining below 6, mostly attributed to a lack of reactivity (Figure 4B). The control group showed a high-grade clinical score, with apathy, tachypnea, dyspnea, and gastrointestinal symptoms, along with fever (above 38.9°C in all animals). The animals showed various hematological and serum biochemistry changes during the challenge phase (Figure S5). Enzyme activity levels (aspartate aminotransferase; creatine kinase) reflecting hepatic disorders were above normal values during the critical disease period for controls but not the vaccinated animals. We also observed perturbations of the white and red blood counts in the controls, which exhibited lymphopenia and thrombocytopenia, whereas we observed no significant abnormalities in the vaccinees during the post-immunization and challenge phases. Necropsy studies revealed lesions and the pathophysiological process of the NiV-B infection in control animals (Figure 4C). As previously described,8 examination of the lung tissue showed interstitial pneumonia, edema, and vasculitis, with inflammatory cell infiltrates for all controls but rarely for the vaccinated animals (observed in 2 of 9 animals, AGMs #S1134 and #O1376; Figure S6). Non-vaccinees showed follicular depletion in the spleen, whereas eight CD40.NiV-vaccinated AGMs showed follicular hyperplasia, suggesting that a strong adaptive immune response was induced post-challenge. Of note, we observed no relevant lesions in the frontal cortex of any animals. From a clinical point of view, CD40.NiV appears to confer full protection against disease progression and death.
Figure 4.
Protection assay in AGMs
(A) Survival curve. Nine AGMs were immunized twice with CD40.NiV and challenged with 102 PFUs of NiV-B (intratracheal route) (red line). Eight naive animals were used as controls (black dotted line). Gehan-Breslow-Wilcoxon test, ∗∗∗p < 0.001.
(B) Clinical scores and temperatures of naive (black) and vaccinated AGMs (red). Gray dashed line score threshold for ethical considerations (left); median temperature at day 0 (right).
(C) Hematoxylin and eosin staining of collected tissues from the spleen, lungs, and brain (frontal cortex) at necropsy. Representative staining of naive AGMs is shown in (a) and (b) (follicular degeneration and depletion in spleen) and (d) and (e) (interstitial pneumonia with alveolar edema and syncytial formation in the lungs) and of vaccinated AGMs in (c) (follicular hyperplasia in the spleen) and (f) (normal lung tissue). No obvious changes were observed in the brain (frontal cortex).
Plasma and tissue viral loads in challenged AGMs
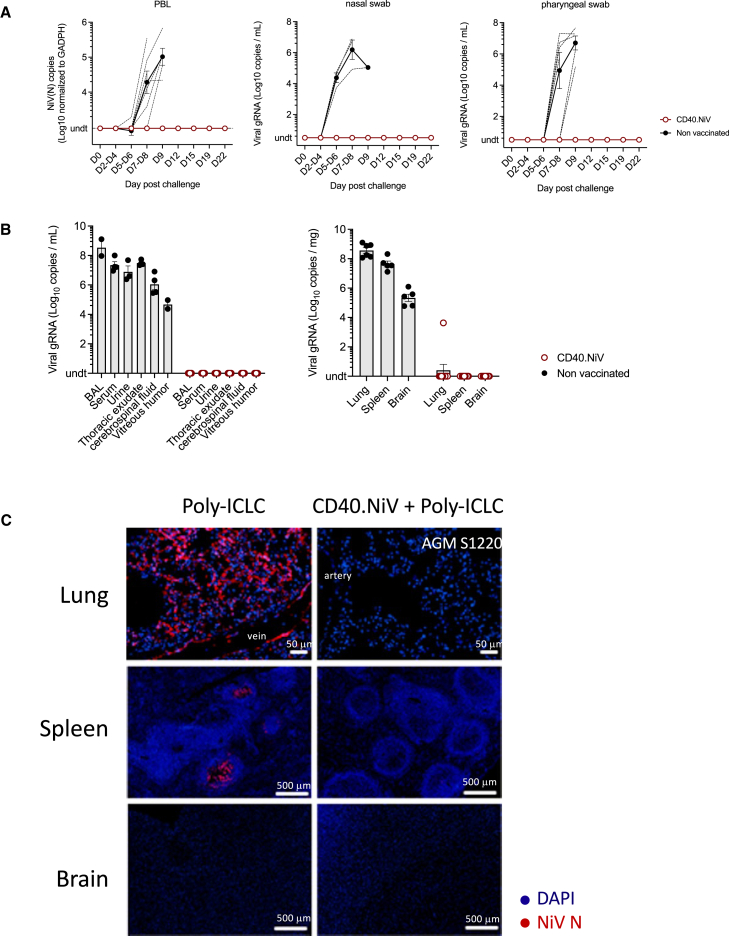
We assessed NiV viremia by RT-qPCR in PBLs from vaccinated and control animals at regular time points and at termination of the study (euthanasia or 28 dpc) for organs, fluids, and swabs. All controls (n = 3 receiving poly-ICLC plus n = 4 naive AGMs) exhibited a high level of NiV-B in PBLs (ranging from 4.5 to 5.5 log10 copies/mL) detectable from 7 to 9 dpc, whereas we detected no virus in vaccinated subjects until 22 dpc (Figure 5A). Except for one animal (AGM #S1134), for which NiV-B transcripts were detected by RT-qPCR in the lung, NiV-B replication in nasal and nasopharyngeal swabs; fluids, including broncho-alveolar lavages, sera, urine, and thoracic exudates; and organs (lung, spleen, and neurological tissue) was undetectable in immunized AGMs, whereas it was detected at high levels in controls (Figure 5B). This strong antiviral effect was confirmed by the lack of expression of the NiV N protein in tissues of the lungs and spleen, as assessed by histo-immunofluorescence (Figure 5C). Viral syncytia were detected in the lungs of non-immunized animals. It is important to note that NiV-B infection of the brain could not be confirmed via histo-immunofluorescence. The infection within blood vessels appears to be heterogeneous, and we cannot rule out the possibility of N protein expression in other areas of the brain. Overall, CD40.NiV vaccination led to sterilizing immunity that limited viral propagation, as well as viral shedding, in AGMs.
Figure 5.
Viral dissemination post-challenge
(A) (Left) Amount of viral NiV RNA quantified by RT-PCR (N gene) in PBLs. Values were normalized against those of the GADPH housekeeping gene. Genomic RNA was measured by RT-qPCR in nasal (middle) and pharyngeal (right) swabs. Mean values (±SEM) are presented for the non-vaccinated (black, n = 7) versus CD40.NiV-vaccinated (red, n = 9) groups.
(B) Same as in (A) for fluids collected at necropsy.
(C) Representative panels of immunofluorescent staining of NiV N protein (red) and DAPI (blue) in the lung, spleen, and brain tissues collected from the poly-ICLC versus CD40.NiV (+poly-ICLC) groups.
Immunological and cytokine features of challenged animals
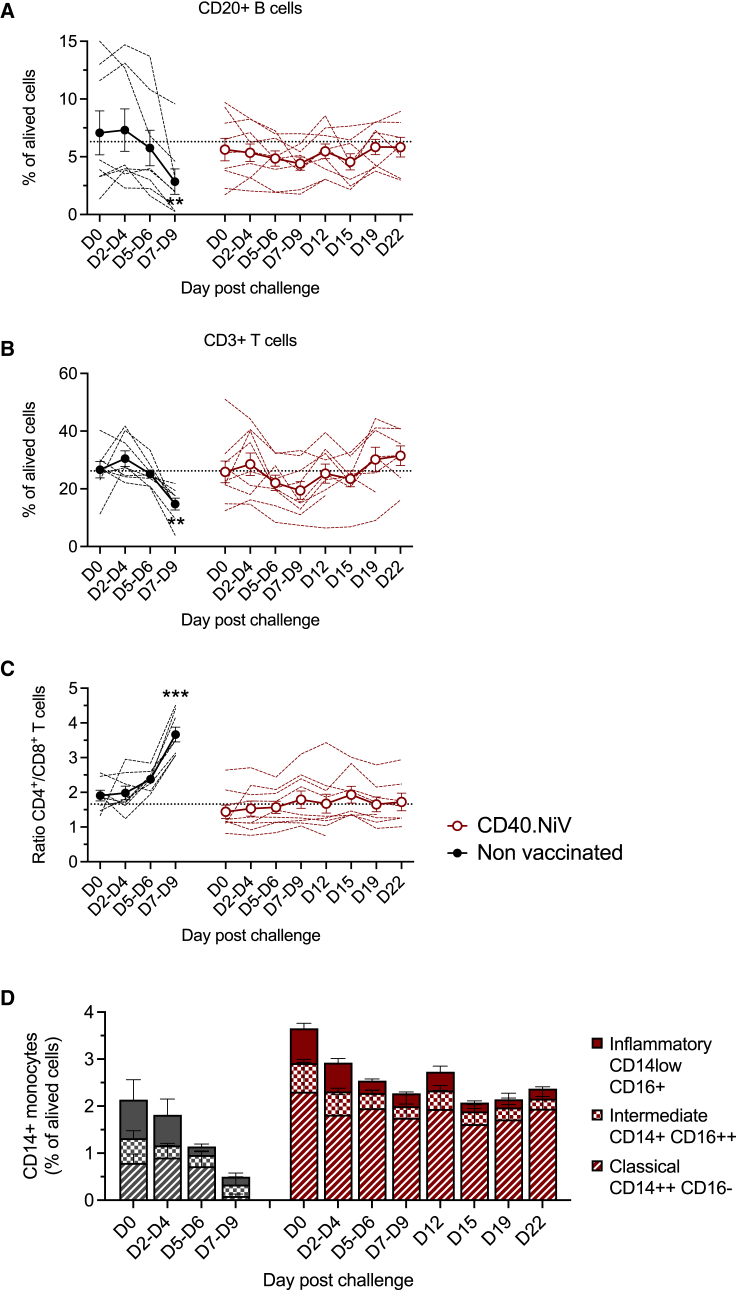
We further characterized the post-challenge immune responses by immunological phenotyping of the cell populations from 0 to 22 dpc. The succumbing animals showed profound and significant defects in the lymphoid populations (CD20+ B and CD3+ T cells) (Figure 6A). Among T cells, the percentage of CD8+ T cells was markedly reduced during NiV infection (Figure 6B). These changes were transitional and not significant in the vaccinated group. Innate immunity was also affected by NiV-B infection, with the almost complete disappearance of circulating monocytes in succumbing animals, whereas the number of inflammatory, intermediate, and classical monocytes in the vaccinees remained stable after the critical period of infection (7–9 dpc) (Figure 6C). Overall, these data indicate that exposure to NiV challenge induces dramatic perturbations of innate and adaptive cellular immunity, which are attenuated and/or non-significant in CD40.NiV-immunized animals.
Figure 6.
Myeloid and lymphoid populations post-challenge
(A) Percentage of CD20+ B cells among live cells determined by flow cytometry for non-immunized (black, n = 5) versus vaccinated AGMs (red, n = 9). Dotted line: median percentage of all animals at day 0. For each time point, percentages are compared to the initial amount in each animal group. Friedman paired t test, ∗∗∗p < 0.001.
(B and C) As in (A), percentage of CD3+ T cells (B) with evolution of CD4/CD8 T cell ratios (C).
(D) CD14+ monocytes exhibiting inflammatory (CD14low CD16+, solid), intermediate (CD14+ CD16hi, square), and classical (CD14hi, CD16−, dashed) phenotypes in non-vaccinated (n = 3, black) versus CD40.NiV AGMs (n = 9, red).
Changes in blood-cell gene expression in vaccinated AGM
To decipher early changes in gene expression associated with the vaccination, we performed RNA sequencing analysis of peripheral whole blood at day 0 (before prime) and 1 day PP (day 1), just before the boost (day 21), and 1 day PB ( day 22) in animals receiving adjuvanted CD40.NiV (see Figure 3A). Given the small number of animals in the adjuvant control group, we analyzed the differentially expressed genes (DEGs) at different time points relative to baseline in the vaccinated animals (VACs). Principal-component analysis showed changes in gene abundance PP and PB relative to baseline (day 0 before injection) (Figure 7A). In total, 773 DEGs were significantly modulated at day 22 (1 day PB). Interestingly, among these DEGs, 437 were noted at days 1 and 22 (1 day PP and PB). An additional 236 DEGs were common between day 1 PP and PB (Figure 7B). The most highly upregulated genes from day 0 to 22 included those that play a major role in antiviral innate and proinflammatory responses (e.g., ISG15, MX1, IFI44, CXCL10, and interleukin-27) and adaptive immunity (e.g., antigen uptake by SIGLEC-1, T cell chemotaxis by CCL8, and CXCL11) (Figure 7C). Most of these transcripts were found to be upregulated PP and PB (Figure S7A). A panel of the most highly down-regulated genes were also associated with a signature of a primary or secondary immune response to the vaccine (e.g., CD1c, CD79A, CCR6, and CXCR4). Globally, the most highly upregulated and down-regulated genes in vaccinated animals were predominantly associated with pathways involved in immune system processes and the defense against viruses and other organisms (Figure 7D). The humoral immune response mediated by circulating Ig was specifically promoted PB (Figure S7B), highlighting a prominent antibody-mediated response after the second injection of CD40.NiV. Overall, the antiviral defense module of genes was the significantly upregulated on day 22 (Figure 7E). The detailed kinetics of the genes involved in this module showed rapid and significant activation of all PP and PB (Figure 7F). Ten transcripts, including IFN-inducible antiviral proteins (RSAD2 and RIG1), were significantly upregulated PB relative to PP. Other gene network modules were also highlighted PP and PB (e.g., response to virus, cytokine signaling, neutrophil degranulation, and regulation of leukocyte activation). Although we could not discriminate between effects of the vaccine and the adjuvant in the vaccinees, our transcriptomic analysis highlights a vaccine-induced gene signature.
Figure 7.
Characterization of gene expression profiles induced after prime-boost vaccinations
(A) Principal-component analysis based on the full transcriptomic profile of vaccinated AGMs sampled at day 0 (before vaccination) (yellow); day 1 after prime vaccination (purple); 1 day before the boost, corresponding to day 21 (orange); and 1 day after the boost (day 22) (sky blue).
(B) Venn diagram of differential expression of genes (DEGs) comparing VAC day 1 to 0, 22 to 21, and 22 to 0 (adjusted p value ≤ 0.05, absolute log2 fold change [FC] ≥ 0.58).
(C) Volcano plot of DEGs observed between VAC day 22 and 0. The down- (190) and upregulated (583) genes are shown in blue and red, respectively. The top 35 most upregulated and down-regulated genes based on the log2FC are depicted.
(D) Gene set enrichment analysis was performed on the DEGs with an adjusted p value ≤ 0.05 between VAC day 22 and 0 using the ClusterProfiler v.4.4.4 R package. The density plot shows the top 25 pathways based on the adjusted p value and normalized enrichment score.
(E) The highest ranked network of DEGs (adjusted p value ≤ 0.05, absolute log2FC ≥ 0.58) between VAC days 22 and 0 was obtained from the protein-protein interaction (PPI) network using the MCODE plug-in of Cytoscape. The DEGs in the network are colored based on the log2FC values.
(F) Heatmap of gene expression belonging to the MCODE network at days 0 (green), 1 (salmon), 21 (blue), and 22 (purple). Gene expression levels in the heatmap are shown as Z scores. Significantly differentially expressed genes in post-boost versus post-prime are indicated by a square.
Discussion
In this study, we demonstrate the potency of a DC-targeting vaccine candidate in preventing NiV-B infection in challenge experiments using an AGM model. More than three decades after the discovery of the immunological properties of DCs,29 we provide evidence that targeting viral antigens to professional APCs can be efficiently employed as a prophylactic measure against a lethal viral challenge. The present study reinforces previous findings that DC-based vaccines can efficiently reduce viral loads and/or viral reservoirs when used as a therapeutic approach against HIV-1 infection30,31 or SARS-CoV-2 reinfection.15
The DC-targeting strategy enabled us to design a subunit construct containing immunogenic and cross-reactive epitopes from the F and N NiV proteins, in addition to the NiV G ECD. This is in contrast to most other NiV vaccine platforms.8,9,10,32,33,34,35 These peptides are predicted to contain numerous T cell and B cell epitopes compared to other strategies, such as the sGHeV vaccine,10 and they cover 100% of the worldwide repertoire of HLA molecules. These sequences show 95%–100% conservation between the NiV-M and NiV-B strains, including recent strains that have infected human populations, farm animals, and bat reservoirs in different countries. Notably, the vaccine sequences G71–602, F45–90, and N318–355 exhibit 83%, 87%, and 100% homology with the Hendra virus, respectively (Figure 1B). We have confirmed that neutralizing antibodies induced by CD40.NiV can cross-neutralize various strains of NiV using a VSV-based surrogate neutralization assay (see Figure 3). Whether the CD40.NiV vaccine candidate might also provide cross-protection against various Henipaviruses causing sporadic outbreaks (e.g., the Langya virus36; see Table S1), and thus reduce the risk of the emergence of new pathogenic strains, remains to be tested. However, the epitope-based strategy developed here can be extended to multiple conserved and highly immunogenic peptide vaccines, as proposed for the development of an HIV-1 therapeutic vaccine31 and an SARS-CoV-2 booster vaccine,12 which will advance to a phase 1/2 clinical trial.
The CD40.NiV vaccine induced both NiV G-specific IgG and IgA antibodies as early as 10 days PP in AGMs (see Figure 3). We have also demonstrated that neutralizing responses can be maintained, with a stable estimated mean log titer of approximately 2.2 (±0.1) 100 days after the peak of the antibody response. These responses were shown to cross-neutralize multiple strains of NiV, as well as HeV. Although detectable at a lower level, we found that the CD40.NiV vaccine elicited a T cell response against the NiV G ECD and down-selected F and N peptides in two preclinical models. While the role of specific T cells in this protection was not fully demonstrated, the induction of T cell responses, in addition to the antibody-mediated response, might help to improve cross-protection, reduce shedding, and limit the spread of NiV, as well as enhance the durability of the immune response. Transcriptomic analysis revealed a common set of DEGs 1 day following the prime and boost (days 1 and 22), including genes associated with antiviral innate (ISG15, MX1, IFI44) and adaptive immunity. The DEGs in vaccinated animals were predominantly associated with pathways involved in immune system processes and defense against viruses, involving innate immunity at an earlier time point (day 1, PP), as described in other vaccine strategies.37 Interestingly, the modulation of gene pathways associated with humoral responses was significant PB at day 22. Overall, these results highlight a vaccine signature associated with the protective effect of the vaccine.
AGMs were challenged with a lethal dose of NiV-B 60 days PB, revealing complete protection conferred by the CD40.NiV vaccine candidate. Intratracheal infection with 102 PFUs of NiV-B has been demonstrated to be highly reproducible in previous experiments, resulting in 100% mortality between days 9 and 12 post-challenge. This model better mimics human infection associated with severe respiratory symptoms than NiV-M cases, which are characterized by a high rate of encephalitis. We have demonstrated that CD40.NiV can confer protection against both the disease and the virus, with 100% survival of immunized AGMs up to the end of the study 28 days post-infection. These clinical results were corroborated by the preservation of lymphoid structures and the absence of major abnormalities in the lungs and blood lymphoid populations post-challenge in vaccinated animals relative to controls. The absence of detectable virus in the organs and fluids (RNA or protein) from most of these animals at the time of necropsy demonstrates the effectiveness of CD40.NiV in this model. The limited spread of the virus within the organism, without significant or detectable shedding of the virus through nasal secretions or other fluids, has the potential to limit interindividual viral dissemination and the establishment of a viral reservoir or latent infection. Overall, the absence of illness and detectable viral replication in the vaccinated animals suggests that CD40.NiV induced sterilizing immunity.
From a clinical perspective, the CD40.NiV vaccine was administered to AGMs with the clinical-grade poly-ICLC adjuvant, as previously done for HIV DC-targeting vaccines in preclinical models15,18 and in healthy volunteers enrolled in the first human phase 1 trials to test the CD40.HIVRI.Env vaccine (ClinicalTrials.gov: NCT04842682), demonstrating the safety and immunogenicity of the vaccine.38 However, the capacity to induce protective immunity without requiring an adjuvant14 would accelerate the development of a protein-based vaccine with improved tolerability over adjuvanted vaccines, making it suitable for specifically vulnerable individuals and children. Indeed, licensed subunit vaccines have demonstrated tolerability and safety in diverse population groups, including pregnant women and children.39 Further studies in non-human primates should be conducted to determine the minimal amount of CD40.NiV required to induce protective responses and/or the need for an adjuvant.14
Our study has limitations. First, we did not investigate whether AGM immunized with the non-targeted NiV G ECD at equivalent doses would produce neutralizing antibodies and achieve immune protection from infection. Nevertheless, we have previously reported in several models, including non-human primates, that the targeting of antigen to CD40 receptor induces early integrated (T and B cell responses) and long-lasting responses as compared to the non-targeted antigen. Moreover, the preclinical data in mice presented in Figure 2 demonstrated a significant advantage of CD40 targeting for inducing NiV G-binding IgG, with improved avidity and neutralization capacity. Although we observed GC reaction in spleens of vaccinated mice, the mechanism of action for improved humoral responses by targeting CD40 remains to be elucidated. For ethical considerations, we decided to demonstrate the protection in AGMs with the best-in-class formulation deciphered in mice (CD40.NiV + poly-ICLC). Second, 6 females and 3 males were immunized with CD40.NiV. As depicted in Figures 3, 4, 5, and, all vaccinated animals survived, with no discernible differences noted between genders in terms of clinical, virological, or immunological responses post-challenge. Non-vaccinated animals, comprising 3 from the current study and 5 from previous challenge experiments, included a mix of males and females. All succumbed between days 9 and 12 post-challenge, exhibiting similar virus loads and physiopathological disorders across all animals. Given the balance of males and females and the uniformity of the data, we opted not to delineate responses within each gender group. Third, while we demonstrated that F and N down-selected epitopes contribute to the diversity of T cell responses, we have not asserted either antibody responses induced by these peptides or their eventual adjuvant effect. Finally, our objective was to diversify and enrich the arsenal of vaccines to prevent future Nipah outbreaks. The next stage would be to compare head-to-head candidate vaccines under development; to assess the precocity, durability, magnitude, and cross-reactivity of neutralizing antibodies; and to unravel whether platforms could be associated for improved protection against Nipah.
Our results demonstrating the protective efficacy of the CD40.NiV vaccine represent an important milestone toward clinical development. Various vaccine platforms are under clinical development, including the subunit vaccine composed of the G protein of Hendra virus (ClinicalTrials.gov: NCT04199169), the rVSV-ΔG vector39 (ClinicalTrials.gov: NCT05178901), and mRNA (ClinicalTrials.gov: NCT05398796), but no licensed vaccines are still available for human use. These platforms may represent prime candidates for new “emergency vaccines” to be utilized for the management of outbreaks. However, concerns remain about the safety, development, and inadequate transport/storage in affected areas and the durability of immunity.33,40,41,42 One caveat of subunit vaccines in general is related to constraints of antigen design and the time required to produce large numbers of doses, making these vaccines difficult to develop as first responses for suddenly emerging epidemics. However, the CD40.NiV vaccine, designed to extend the breadth of responses, may enrich the portfolio of countermeasures against Henipaviruses as a prime and/or boost to induce early and durable immune responses to fill the gap of sustainable protection in a clinical situation characterized by unpredictable outbreaks.
Limitations of the study
Study limitations include not assessing non-targeted NiV G ECD immune responses in AGMs. CD40 targeting showed advantages in inducing early integrated responses. CD40.NiV + poly-ICLC demonstrated protection in AGMs, overcoming ethical concerns. Gender differences were not observed in vaccinated animals post-challenge. The study aimed to enrich Nipah vaccines and proposes comparing candidate vaccines for efficacy, neutralizing antibodies, and potential synergies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CD3-V500, clone SP34-2 | BD Biosciences | Cat# 560770; RRID:AB_1937322 |
| anti-CD3-AF488, clone SP34-2 | BD Biosciences | Cat# 557705; RRID:AB_396814 |
| anti-CD4-FITC, clone L200 | BD Biosciences | Cat# 550628; RRID:AB_393789 |
| anti-CD4-AF700, clone SK3 | BD Biosciences | Cat# 340133; RRID:AB_400007 |
| anti-CD8-BV650, clone RPA-T8 | BD Biosciences | Cat# 563821; RRID:AB_2744462 |
| anti-CD8-BV421, clone SK1 | BD Biosciences | Cat# 568217; RRID:AB_2916846 |
| anti-CD14-AF700, clone M5E2 | BD Biosciences | Cat# 557923; RRID:AB_396944 |
| anti-CD14-APC, clone M5E2 | BD Biosciences | Cat# 555399; RRID:AB_398596 |
| Anti-CD16-PeCy7, clone 3g8 | BD Biosciences | Cat# 557744; RRID:AB_396850 |
| anti-CD20-SB702, clone 2H7 | Thermo Fisher Scientific | Cat# 67-0209-42; RRID:AB_2717137 |
| anti-CD20-APC-H7, clone 2H7 | BD Biosciences | Cat# 560853; RRID:AB_10561681 |
| anti-CD45-PerCP, clone D058-1283 | BD Biosciences | Cat# 558411; RRID:AB_397080 |
| anti-CD163-APC, clone GHI/61 | Biolegend | Cat# 333609; RRID:AB_2291272 |
| anti-HLA-DR-APC-H7, clone L243 | BD Biosciences | Cat# 641393; RRID:AB_1645739 |
| Mouse anti-monkey IgA monoclonal Secondary Antibody, Biotin | Thermo Fisher Scientific | Cat #MA5-16730; RRID:AB_2538222 |
| Goat anti-monkey IgG (H+L) polyclonal Secondary Antibody, HRP | Thermo Fisher Scientific | Cat# PA1-84631; RRID:AB_933605 |
| anti-CD3-AF700, clone 17A2 | Sony Biotechnology | Cat# 1101080; RRID:AB_2935667 |
| anti-CD11c-BV785, clone N418 | BioLegend | Cat# 117335; RRID:AB_11219204 |
| anti-CD45-BV711, clone 30-F11 | BD Biosciences | Cat# 563709; RRID:AB_2687455 |
| anti-CD45-BV605, clone 30-F11 | BD Biosciences | Cat# 564047, RRID:AB_2744403 |
| anti-CD138-PE/Dazzle594, clone 281-2 | BioLegend | Cat# 142528; RRID:AB_2566495 |
| anti-B220-PECy7, clone RA3-6B2 | BD Biosciences | Cat# 561881; RRID:AB_10893024 |
| anti-B220-711, clone RA3-6B2 | BD Biosciences | Cat# 563892; RRID:AB_2738470 |
| anti-FAS-BV421, clone Jo2 | BD Biosciences | Cat# 562633; RRID:AB_2737690 |
| anti-GL7-PE/Cy7, clone GL7 | Biolegend | Cat# 144620; RRID:AB_2800677 |
| anti-IgD-APC-H7, clone 11-26c.2a | BD Biosciences | Cat# 565348; RRID:AB_2739201 |
| anti-IgG-FITC, Poly4060 | Biolegend | Cat# 406001; RRID:AB_315029 |
| anti-mouse IgG-PE, polyclonal secondary | Thermo Fisher Scientific | Cat#12-4010-87; RRID:AB_11044909 |
| anti-MHC-II (I-A/I-E)- PerCP-Cy5.5, clone M5/114.5.2 | BD Biosciences | Cat# 562363; RRID:AB_11153297 |
| anti-biotin APC, clone REA746 | Miltenyi Biotec | Cat# 130-110-952; RRID:AB_2661380 |
| anti-biotin PE, clone 1d4-C5 | BioLegend | Cat# 409004; RRID:AB_10641847 |
| polyclonal rabbit anti-NiV N antibody | This Study | Prof. B Horvat, CIRI |
| goat anti-rabbit IgG, Alexa Fluor 555, polyclonal secondary | Thermo Fisher Scientific, USA | Cat# A21428; RRID:AB_2535849 |
| Bacterial and virus strains | ||
| NiV Bangladesh isolate SPB200401066 | CDC, Atlanta, USA | GenBank: AY988601 |
| Hendra virus/Australia/Horse/1994/Hendra | Porton Down Laboratory, UK | N/A |
| vesicular stomatitis virus (VSV) replicon | This study | N/A |
| Top10 E Coli | Thermo Fisher Scientific, USA | Cat #C404003 |
| Biological samples | ||
| Mouse splenocytes and blood draws | This study | N/A |
| AGM blood draws and PBMCs | This study | N/A |
| AGM nasal and pharyngeal swabs | This study | N/A |
| AGM BAL, Sera, Urine, Thoracic exudate, cerebrospinal fluid, vitreous humor | This study | N/A |
| AGM tissues (Spleen, Lung, Brain) | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| CD40.NiV vaccine | This study | N/A |
| NiV G recombinant protein | This study | N/A |
| Pools of overlapping peptides | JPT Peptide Technologies, Germany | N/A |
| Polyinosinic-polycytidylic acid (Poly-ICLC) | Oncovir, USA | Hiltonol |
| Streptavidin-HRP | Thermo Fisher Scientific | # 21130 |
| DAPI | Thermo Fisher Scientific | #D1306 |
| Critical commercial assays | ||
| TransIT-PRO Transfection Kit | Mirus Bio LLC USA | #MIR 5700 |
| Biotin-Protein Ligase-BIRA kit | Avidity, USA | N/A |
| TMB substrate kit | Thermo Fisher Scientific | # 34021 |
| Bio-Plex Amine Coupling Kit | Bio-Rad, France | # 71406001 |
| IFN-γ ELISpot plates | Mabtech | # 3321-4APT-2 and # 3421M-4APT-2 |
| AF647 labeling kit | Thermo Fisher Scientific, USA | # A20186 |
| Quant-iT RiboGreen RNA Assay Kit | Thermo Fisher Scientific, USA | #R11490 |
| Deposited data | ||
| RNA-Sequencing data | This study | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE245374 |
| NiV-B G, F, and N proteins | NCBI | Genbank: AEZ01396.1 |
| Ephrin-B2 receptor | NCBI | Genbank: NP_004084.1 |
| Experimental models: Cell lines | ||
| mammalian Expi CHO-S cells | Thermo Fisher Scientific, USA | # A29127 |
| VeroE6 cells | ATCC | # CRL-1587 |
| Experimental models: Organisms/strains | ||
| human CD40 transgenic mice | Taconic Biosciences, USA | N/A |
| Chlorocebus aethiops sabaeus of Caribbean origin | Worldwide Primate Resources International, Saint-Kitts, Saint-Kitts&Nevis Isld | N/A |
| Oligonucleotides | ||
| NiV-N Fwd: GTGCTGAGCTATACCCCACC | This study | N/A |
| NiV-N Rev: GAGATAAGCGCCGGACAAGA | This study | N/A |
| GAPDH Fwd: CACCCACTCCTCCACCTTTGAC | This study | N/A |
| GAPDH Rev: GTCCACCACCCTGTTGCTGTAG | This study | N/A |
| Software and algorithms | ||
| NetMHC 4.04 software | DTU Health Tech | https://services.healthtech.dtu.dk/services/NetMHC-4.0/ |
| NetMHCII 2.3 sofware | DTU Health Tech | https://services.healthtech.dtu.dk/services/NetMHCII-2.3/ |
| BepiPred-2.0 | DTU Health Tech | https://services.healthtech.dtu.dk/services/BepiPred-2.0/ |
| Vaxijen 2.0 software | Edward Jenner Institute | http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html |
| Expasy tool | SIS Swiss Institute of Bioinformatics | https://web.expasy.org/protparam/ |
| Bioplex Manager 6.1 software | Bio-Rad, France | N/A |
| FlowJo V.10.8 software | TreeStar, Ashland, OR | N/A |
| Illumina BaseSpace Sequence Hub | Illumina | https://emea.illumina.com/products/by-type/informatics-products/basespace-sequence-hub.html |
| Venn diagrams | VIB/UGent (Belgium) | https://bioinformatics.psb.ugent.be/webtools/Venn |
| METASCAPE web tool | Metascape Foundation | https://metascape.org |
| STRING database | Core Data Resource | http://string-db.org |
| ZEN 2.3 software | Zeiss, Germany | N/A |
| Prism Software, version 9 | Graph-Pad | N/A |
| Salmon v1.9.0 | N/A | https://salmon.readthedocs.io/en/latest/salmon.html |
| MultiQC v1.13 | N/A | https://multiqc.info/ |
| EdgeR v3.38 | N/A | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| dearseq v.1.8.4 | N/A | https://bioconductor.org/packages/release/bioc/html/dearseq.html |
| limma v3.52 | N/A | https://bioconductor.org/packages/release/bioc/html/limma.html |
| FactoMineR v2.7 | N/A | https://www.rdocumentation.org/packages/FactoMineR/versions/2.7 |
| factoextra v1.0.7 | N/A | https://rpkgs.datanovia.com/factoextra/ |
| ClusterProfiler v4.4.4 | N/A | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| pheatmap v1.0.12 | N/A | https://www.rdocumentation.org/packages/pheatmap/versions/1.0.12/topics/pheatmap |
| Other | ||
| FPLC - ÄKTA Pure | Cytiva, USA | N/A |
| Tristar multimode plate reader | Berthold Technologies, France | N/A |
| Auto-2000 cell counter | Nexcelom Bioscience LLC, USA | N/A |
| LSRFortessa5L | BD Biosciences, USA | N/A |
| LSRII flow cytometer | BD Biosciences, USA | N/A |
| Gallios flow cytometer | Beckman Coulter, USA | N/A |
| AID ELISpot reader | Strassberg, Germany | N/A |
| Bioplex-200 plate reader 640 | Bio-Rad, France | N/A |
| PhysioTel digital M10-F2 telemetry devices | Data Science international (DSI), Harvard Bioscience Inc. | N/A |
| Axio Scan.Z1 slide scanner | Zeiss, Germany | N/A |
| X-Cite Fire LED illumination system | Excelitas Technologies, USA) | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sylvain Cardinaud (sylvain.cardinaud@inserm.fr).
Materials availability
All unique reagents generated in this study are listed in the key resources table and available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
-
•
The RNA-Sequencing data generated in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE245374.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and study participant details
Ethics statements
Animal housing and experimental procedures were conducted according to French and European Regulations (Parlement Européen et du Conseil du 22 septembre 2010, Décret n° 2013-118 du 1er février 2013 relatif a la protection des animaux utilisés à des fins scientifiques) and the National Research Council Guide for the Care and Use of Laboratory Animals (National Research Council (U. S.), Institute for Laboratory Animal Research (U.S.), and National Academies Press (U.S.), Eds., Guide for the care and use of laboratory animals, 8th ed. Washington, D.C: National Academies Press, 2011).
Mice
Six-to 24-week-old human CD40 transgenic mice with the human CD40 region BAC inserted into a wildtype C57BL/6 background (Taconic Biosciences, USA), referred to as hCD40Tg mice, were used in this study.11 Mice were housed at the Mondor Institute of Biomedical Research infrastructure facilities (U955 INSERM-Paris Est Créteil University, France) under pathogen-free conditions at 20°C to 24°C and 50% ± 15% humidity with a 12-h light/12-h dark cycle. All procedures were submitted to the Institutional Animal Care and Use Committee approved by the French authorities (APAFIS#25329–2020051119073072 v4).
African green monkeys (AGM)
Twelve Chlorocebus aethiops sabaeus of Caribbean origin (6 males and 6 females, 27 to 35 months old) were obtained from Worldwide Primate Resources International (Saint-Kitts, Saint-Kitts&Nevis Isld). All animals had passed a physical examination and were certified to be healthy by a veterinarian. Animals were quarantined and acclimatized for 40 days, and then included in the immunization and surgery part of the study in AAALAC-certificated BioPRIM facilities (Baziège, France). They were socially housed in pens with perches and platforms, ad libitum access to water and food (SDS OWM 808003 Chunk and daily distribution of fresh fruits), access to toys, and natural light. All procedures were submitted to the local Institutional Animal Care and Use Committee #53 approved by the French authorities (2022051714245912 v2).
AGMs were subsequently transported to the Jean Mérieux BSL4 - facilities (Lyon, France) for Nipah virus challenge. AGMs were housed in groups of three (grouped by gender) in four ventilated cabinets that allowed social interaction. Before challenge, the animals were acclimatized for 14 days in BSL-4 conditions with an enriched sterile environment, access to food (307 extruded pellets, SAFE, St Laurent, France) and water (ad libitum), and light/dark cycles (12 h/12 h). Each cabinet included an elevated perch or platform, and each animal was regularly provided with toys for physical enrichment, as recommended by the Guide for the Care and Use of Laboratory Animals, as well as audio stimulation. Animal housing and experimental procedures were conducted in the BSL-4 laboratory according to the French and European Regulations (Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010). The experimental protocol was approved by the Comité d’Ethique en Expérimentation Animale (CELYNE, n°042) and the French authorities (APAFIS#37780–2022062310304143 v4).
Method details
Immunogens & reagents
CD40.NiV vaccines
The amino acid sequences of the NiV-B G, F, and N proteins (GenBank: AY988601.1) were screened to identify peptides that bind weakly or strongly to HLA-I and -II using NetMHC 4.0443 and NetMHCII 2.344 software, respectively. For global HLA coverage, 9-mer peptides were screened for 81 HLA-A, -B and -C molecules and 15-mer peptides for 54 HLA-DR, -DP, and -DQ molecules. NiV proteins were also examined for linear B-cell epitopes using BepiPred-2.0 software (Sequential B-Cell Epitope Predictor45). The antigenicity and protective capacity of the down-selected peptides were analyzed using Vaxijen 2.0 software (http://www.ddgpharmfac.net/vaxijen/VaxiJen/), with a threshold of 0.4 for viral proteins, considering values >0.4 to be probable protective antigens. Peptides with the highest score were selected for further evaluation and vaccine construction. Physiochemical properties and stability of the multi-epitope vaccine construct were controlled using the Expasy tool (https://web.expasy.org/protparam/). The computed parameters included the molecular weight, theoretical pI, amino acid composition, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index, and grand average of hydrophobicity. The sequences were inserted into expression plasmids in which NiV G ECD was fused to the C-terminus of the heavy chain (HC) of a previously described humanized anti-human CD40 12E12 IgG4 antibody.19 Additional F and N down-selected sequences were fused to the C-terminus of the light chain (LC) to generate the final vaccine candidate (CD40.NiV, 302 kDa).
The vaccines were produced via transient transfection (TransIT-PRO Transfection Kit, Mirus Bio LLC, USA) of mammalian CHO-S cells (Thermo Fisher Scientific, USA). Cells were cultured in Mirus media (Mirus Bio LLC) supplemented with 1% penicillin–streptomycin and the recombinant Abs produced in the supernatants captured on Protein A-affinity columns (Thermo Fisher Scientific) and eluted by FPLC (ÄKTA Pure, Cytiva, USA). Purified mAbs were stored in 125 mM Cavitron cyclodextrin buffer (Ashland, USA) or 1 M arginine buffer (100 mM Tris, pH 7) at −80°C until use. NiV G ECD protein and the Ephrin-B2 receptor (NCBI NP_004084.1) were produced in stably transfected CHO cells and purified on His-tag pre-packed columns (Cytiva). The NiV G ECD protein contains an Avitag and was biotinylated using the Biotin-Protein Ligase-BIRA kit according to the manufacturer’s instructions (Avidity, USA). Quality assurance tests for the proteins and mAbs were performed using the following procedures: i) SDS-PAGE and Coomassie staining, ii) endotoxin level below 0.5 ng/mg of protein, and iii) SEC on a Superdex 200 10/300 column (Cytiva), following the manufacturer’s instructions.
Overlapping peptide (OVLP) pools
OVLP pools were obtained from JPT Peptide Technologies (Berlin, Germany), with purity >70%. The 15-mer peptides, overlapping by 11 amino acids, were designed to cover the complete selected regions of the NiV G ECD, F, and N peptides separately and were used for IFN-γ ELISpot assays (see Table S2 for detailed aa sequences). The peptide pools were resuspended to 1 mg/mL in DMSO and aliquots conserved at −80°C until use.
Animal immunization and groups
Mice
hCD40Tg mice were immunized SC on days 0 and 21 with 10 μg CD40.NiV vaccine with 50 μg of polyinosinic-polycytidylic acid (poly-ICLC, Hiltonol; Oncovir, USA). Note that the administration of 10 μg of CD40.NiV corresponds to 3.94 μg, 0.34 μg, and 0.27 μg of NiV G ECD, F, and N down-selected peptides, respectively. Groups of mice were also immunized with the same amount of non-targeted NiV G protein. Mice were euthanized seven days after the second immunization. Spleens and blood were collected to assess B- and T cell responses. No animals were excluded from the study.
AGMs
Six male AGMs and three female AGMs were immunized SC on days 0 and 21 with 200 μg CD40.NiV, along with 1 mg poly-ICLC. As a control, a group of three male AGMs were immunized with the vehicle (poly-ICLC) only. Blood was collected on days 0, 10, 21, 35, and 56 for the analysis of B- and T cell responses. No animals were excluded from the study.
Vaccine-induced immune responses
CD40.NiV cellular binding assay
NiV G ECD and CD40.NiV were labeled with AF647 following instructions of the manufacturer (Thermo Fisher Scientific, cat# A20186). Splenocytes from naive hCD40Tg were incubated 30min on ice with 1nM of labeled vaccine and phenotyped using following antibodies: CD45-BV711 (Sony biotechnologies, clone 30-F11), CD11c-BV785 (Sony biotechnologies, clone N418), MHC-II (I-A/I-E)-PerCP-Cy5.5 (Sony biotechnologies M5/114.5.2), CD3-AF700 (Sony Biotechnology, clone 17A2), B220-PECy7 (Sony biotechnologies, clone RA3-6B2). Binding on AGM PBMCs was performed similarly using following phenotypic panel: Anti-CD3-V500 (BD Biosciences, clone SP34-2), CD4-FITC (BD Biosciences, clone L200), CD8-BV650(BD Biosciences, clone RPA-T8), CD20-SB702 (Fisher Scientific, clone 2H7), CD14-AF700 (Biolegend, clone M5E2), CD45-PerCP (Biolegend, clone D058-1283), CD163-APC (Biolegend, clone GHI/61) and HLA-DR-APC-H7 (BD Biosciences, clone L243).
Antibody measurement
ELISA
Anti-NiV G AGM IgG and IgA were measured by ELISA. Briefly, 1 μg/mL of NiV-B G protein (50 μL) was adsorbed into 96-well Maxisorp plates (Nunc cell culture plastic, Thermo Fisher Scientific) in carbonate buffer (pH 9.6) and incubated overnight at 4°C. Plates were washed with phosphate-buffered saline (PBS) - Tween 20 0.05% (PBS-T) and blocked with PBS 3% BSA in PBS for 2 h at room temperature (RT). 3-fold serial dilutions of sera from immunized animals were added to the plates (50 μL), followed by 1 h of incubation at RT. Plates were then washed and goat anti-monkey IgG conjugated with HRP (1/30,000; Thermo Fisher Scientific) was added, followed by 1 h of incubation at RT. For IgA, anti-monkey IgA-biotin secondary antibody (cat#MA5-16730, Thermo Fisher Scientific) was added at 1 μg/mL, followed by 1 h of incubation at RT. Streptavidin-HRP (1/40,000, 1 h RT, Pierce) was used for detection. Plates were washed and revealed using the TMB substrate kit (Thermo Fisher Scientific) and quenched with 2 N H2SO4. Plates were read using a Tristar multimode plate reader (Berthold Technologies, France) at 450 nm (ref. 570 nm).
Multiplex bead-based technology
Mouse IgG was titrated using the Luminex bead-based immunoassay. Luminex beads were prepared by covalent coupling of in-house produced NiV-B G protein with MagPlex beads using the Bio-Plex Amine Coupling Kit (Bio-Rad, France). Briefly, 1 × 106 MagPlex-COOH Microspheres (Bio-Rad) were resuspended in activation buffer containing a freshly prepared solution of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC, 2.5 mg) and N-hydroxysulfosuccinimide (S-NHS, 2.5 mg) (Thermo Fisher Scientific). After 20 min of agitation at RT using a Hula-Mixer (Thermo Fisher Scientific), activated beads were washed in PBS, followed by the addition of 100 μg of protein antigen in a final volume of 1 mL PBS. The coupling reaction was performed at RT for 2 h with bead agitation. Washed beads were resuspended in 500 μL blocking buffer and incubated for 30 min with agitation at RT. Following a final washing step with storage buffer, beads were resuspended in 1 mL storage buffer, counted using an Auto-2000 cell counter (Nexcelom Bioscience LLC, USA), and stored in the dark at 4°C until use. NiV G-coupled beads were diluted to 40,000 beads/mL in PBS and added (50 μL) to Bio-Plex Pro 96-well Flat Bottom Plates (Bio-Rad). Following two washes with PBS-T on a magnetic plate washer (Bio-Rad), 50 μL of the individual serum samples diluted in PBS (1/1,000) were added to the plates. The plates were sealed and agitated (750 rpm) for 1 h in the dark. Beads were then washed three times using a magnetic plate washer and anti-mouse IgG-PE secondary antibody (Thermo Fisher Scientific, cat#12-4010-87) was added to 0.5 μg/mL. After three washes, beads were resuspended in 80 μL Sheath fluid (Bio-Rad), agitated for 5 min, and directly read on a Bioplex-200 plate reader (Bio-Rad) with 50 μL of acquisition volume and DD gate 5000–25000 settings. Median fluorescence intensities (MFI) were exported using Bioplex Manager 6.1 software (Bio-Rad).
Luminex-based antibody avidity assay
To measure the avidity of antigen-specific antibodies, an additional step was added to the Luminex protocol between incubation of the sample with the beads and addition of the detection antibody. After incubating the samples and beads for 1 h with agitation at RT and washing, 1 M sodium thiocyanate (NaSCN) or PBS was added to the plates, followed by incubation for 30 min at RT. After washing, the Luminex protocol was continued. The avidity index was calculated as the ratio of the MFI measured following incubation with NaSCN and the MFI measured following incubation with PBS, expressed as a percentage.
Luminex-based inhibition assay
A surrogate neutralization assay was developed to assess the ability of sera to neutralize the binding of NiV-B G protein to its natural human receptor, EphrinB2 (inhibition assay). Briefly, Luminex beads coupled with NiV G protein were incubated with 50 μL serially diluted sera (2-fold dilutions; 1/30 to 1/1,920) in PBS for 1h at RT with agitation (750 rpm). Following incubation, 1 μg/mL biotinylated EphrinB2 was added (50 μL) and the plates incubated for another 1h at RT with agitation (750 rpm). Plates were then washed three times, streptavidin-PE (Biolegend) added at 1 μg/mL, and the plates incubated for a further 45 min at RT with agitation (750 rpm). Plates were read as described above. The mean MFI of EphrinB2 incubated with the NiV G protein in the absence of serum (PBS) was used as the reference value for 100% binding.
In vitro NiV-B and HeV seroneutralization
Sera was serially diluted (2-fold steps) in DMEM containing 2% FBS and mixed with 50 PFU of NiV-B (isolate SPB200401066, CDC, Atlanta, USA) or HeV (Hendra virus/Australia/Horse/1994/Hendra, kindly provided by Porton Down Laboratory (Porton Down, UK), incubated for 30 min at 37°C, and poured on VeroE6 cells in 96 well-plates for 60min. DMEM containing10% FBS was added after 1 h and the cells further incubated for two days at 37°C. The cytopathic effect (CPE) was evaluated after crystal violet staining and the data analyzed using Prism 8 software to calculate the IC50 values and neutralization titers (non-linear regression, inhibitor vs. response, variable slope fitting).
Cross-neutralization studies
The vesicular stomatitis virus (VSV) replicon, previously described,27,28 was modified to express various fluorescent proteins (CyanFP, Yellow FP, GreenFP, CrimsonE2FP). For pseudotyping, the attachment glycoproteins and fusion proteins of NiV-Bangladesh (GenBank: AY988601), NiV-Cambodia (GenBank: MK801755), NiV-Malaysia (GenBank: AY029767), and Hendra virus (GenBank: MN062017.1) were transfected into HEK293T cells using the JetOptimus Transfection Reagent (Polyplus, France). Cells were infected with rVSVΔG-xFP-VSV-G 8 h post-transfection. After 24 h, the supernatants were discarded, and the virus-producing cells resuspended in 5 mL fresh DMEM and snap-frozen in a −80°C ethanol bath. Lysed cells were centrifuged for 5 min at 2,100 g and the viral particle-containing supernatants collected. Replicons (MOI, multiplicity of infection, 0.15) were mixed with diluted sera (1/60 to 1/3,200) and incubated for 30 min at 37°C. The mix was then added to VeroE6 cells, followed by an 8h incubation. The percentage of infected cells was determined by flow cytometry (BD LSRFortessa5L, BD Biosciences).
NiV G-specific B-cell staining
All cell culture was performed at 37°C in a humid atmosphere containing 5% CO2. The media for all experiments consisted of RPMI containing 10% fetal bovine serum (FBS) (Biowest, France) supplemented with 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. Mouse splenocytes (3 ×106) were resuspended in FACS buffer (PBS, 0.5% BSA, 2.5 nM EDTA) and incubated for 30 min at 4°C with 3 μg biotinylated NiV G protein per 1 × 106 cells. After washing with FACS buffer, cells were incubated for 30 min at 4°C with the Live/Dead Fixable Aqua Stain (Thermo Fisher Scientific, cat# L34975), the antibody phenotyping panel (mCD45-BV605 (BD clone 30-F11), mCD3-AF700 (Sony Biotechnology, clone 17A2), mB220-BV711 (BD, clone RA3-6B2), CD138-PE/Dazzle594 (Sony Biotechnology, clone 281-2), mIgG-FITC (Sony Biotechnology, Poly4060), mIgD-APC-H7 (Sony Biotechnology, clone 11-26c.2a), mFAS-BV421 (BD, clone Jo2) and mGL-7-PC-7 (Biolegend, clone GL7)), and two anti-biotin mAbs (APC, Miltenyi Biotec (France) REA746, and PE, Sony Biotechnology clone 1D4-C5). Cells were fixed (BD fixation kit). Data acquisition was performed using an LSRII flow cytometer (BD). Data were analyzed using FlowJo software (TreeStar, Ashland, OR). All the flow cytometric plots presented in this article were pre-gated on live (using the Live/Dead Stain) and singlet events.
IFN-γ ELISpot assay
IFN-γ-releasing-specific T-cells were detected both among fresh mouse splenocytes and frozen AGM PBMCs using pre-coated IFN-γ ELISpot plates (Mabtech cat#3321-4APT-2 and 3421M-4APT-2, respectively). Cells were seeded at 0.25 ×106 cells/well and stimulated at 37°C for 18 h with 1 μg/mL of each pool of overlapping peptides. Plates were then incubated with detection antibody in PBS-0.5% FBS for 2h at RT following the manufacturer’s instructions. Plates were then washed and streptavidin-ALP (1/1,000 dilution in PBS-0.5% FBS) added, followed by incubation for1 h. Plates were developed using a BCIP/NBT-plus substrate solution. The developed spots were counted using an AID ELISpot reader (Strassberg, Germany). Phorbol myristic acid (PMA)-ionomycin (Sigma-Aldrich, USA) and anti-CD3 mAb provided by the manufacturer were used as positive controls for the mouse and monkey ELISpot assays, respectively.
RNA extraction, library preparation, and sequencing
Whole blood was sampled in Tempus Buffer (Thermo Fisher Scientific). Total RNA was purified from whole blood using the Tempus Spin RNA Isolation Kit (Thermo Fisher Scientific) and then quantified using the Quant-iT RiboGreen RNA Assay Kit (Thermo Fisher Scientific). Quality control performed using a Bioanalyzer (Agilent). RNA samples were depleted of globin mRNA using the Globinclear Kit (Thermo Fisher Scientifc) prior to mRNA library preparation using the NEBNext Kit (New England Biolab, USA). Libraries were sequenced on an Illumina HiSeq 2500 V4 system. Sequencing quality control was performed using Sequence Analysis Viewer (SAV). FastQ files were generated on the Illumina BaseSpace Sequence Hub (https://emea.illumina.com/products/by-type/informatics-products/basespace-sequence-hub.html). Transcript reads were aligned and quantified relative to the “Vero_WHO_p.1.0 Chlorocebus sabaeus (AGM) transcriptome reference from NCBI using Salmon v1.9.046 Quality control of the alignment was performed via MultiQC v1.13.46 Finally, counts were filtered and normalized using the EdgeR v3.38 R package. Weakly expressed genes were excluded using the filterByExpr function with default parameters. The conserved counts were normalized as counts per million, plus 0.0001 to avoid null values, and then log2 transformed.
Differential gene expression analysis
Differential gene expression analysis was performed using the dearseq v.1.8.4 package and a mixed model with variance component test47 to calculate the adjusted p values and the limma v3.52 package to obtain the fold-change in expression. The principal component analysis (PCA) based on gene expression of the samples was performed using with FactoMineR v2.7 and factoextra v1.0.7 R packages. The Venn diagram comparing the DEGs between the various gene expression analyses was created using the web application (https://bioinformatics.psb.ugent.be/webtools/Venn/). The volcano plots of the DEGs were created using the ggplot2 v3.4.1 package (https://ggplot2.tidyverse.org/).48 The genes with an adjusted p value (false discovery rate) ≤ 0.05 were subjected to gene set enrichment analysis using the ClusterProfiler v4.4.4 R package.49 DEGs were also analyzed for pathway enrichment using the METASCAPE web tool (https://metascape.org).50 Gene expression heatmaps were created using the pheatmap R package 1.0.12.
Construction of the protein–protein interaction (PPI) network
The PPI network between proteins encoded by the DEGs, with adjusted p values (false discovery rate) ≤ 0.05 and absolute fold-change in expression ≥ Log2 0.58, was established by importing genes into the STRING database (http://string-db.org)51 application using Cystoscope version 3.9.1.52 The Molecular Complex Detection (MCODE) plug-in Cytoscape, with the default parameters, was used to detect the highest computed score network of DEGs (adjusted p value ≤0.05, absolute log2FC ≥ 0.58) from the PPI network, based on highly interconnected genes.
Nipah virus challenge
Nipah virus infection
Wildtype NiV Bangladesh isolate SPB200401066 (CDC, Atlanta, USA; GenBank AY988601) (wt NiV) was prepared in Vero-E6 cells negative for Mycoplasma spp. as previously described. NiV infections were carried out at the INSERM Jean Mérieux BSL4 laboratory in Lyon, France.
The viral inoculum was prepared at 102 PFU in DMEM media and tittered before and after infection. AGMs were infected intratracheally (IT) in the BSL4 laboratory with 102 PFU of Bangladesh NiV in 2 mL DMEM under anesthesia with Zolétil 100 (Virbac, France). Animals were followed daily and kept under video-camera observation. Blood sampling was performed every two to three days using EDTA-containing tubes for hematological analysis and heparin lithium-containing tubes for biochemical analysis, RNA extraction, and B- and T cell response analysis. Samples were centrifuged at 1500 × g for 10 min and plasma samples were stored at −80 °C.
Euthanasia was performed when a humane endpoint was reached or at the end of the study, 27 or 28 days after infection, by intracardiac injection of 5 mL Euthasol (Dechra, UK). Necropsies were performed and the organs fixed using 4% formaldehyde (Merck, USA) during two series of seven days in BSL4 conditions and then processed for histopathological analysis. Of note, two vaccinated animals had to be euthanized at day 15 and 19 post-challenge due to injuries and ethical considerations independent of the NiV infection. These animals were excluded from survival study.
Clinical and hematobiochemical follow-up
Clinical exams were performed everyday post-challenge and the following parameters were scored: temperature, weight, dehydration, breath, reactivity, feces exams and neurological symptoms. Animals were euthanized when the total score reached >15 for ethical reasons.
PhysioTel digital M10-F2 telemetry devices (DSI) were surgically implanted for surveillance of the animals after the challenge in BSL-4 facilities. Surgical implantation of the devices was performed by the NHP (non-human primates) supplier (Bioprim, Baziège, France). The units transmitted blood pressure, temperature, and activity beginning from the day of arrival of the animals until the day of euthanasia. During acquisition, data was transmitted from the implant to a TRX-1 receiver mounted in the room connected via a communications link controller (CLC, DSI) to allow digital multiplexing and simultaneous collection of the signals from all animals. Digital data were then captured, reduced, and stored using Ponemah Acquisition Software.
Blood collection was performed under sedation via the femoral route using a Vacutainer system (Vacutest, Becton-Dickinson, France). Full hematological analysis was performed directly on whole blood stored in EDTA tubes using an MS9-5 device (Melet Schloesing Laboratories) with an MS9 pack and a control sample tube (Melet Schloesing Laboratories). Biochemistry analysis was performed directly on whole blood stored in heparin lithium tubes using an MScan-e unit (Melet Schloesing Laboratories) and the 20 parameter rotors (Melet Schloesing Laboratories). Several additional parameters of the plasma samples were analyzed using a Pentra C200 device (HORIBA Medical PENTRA C200).
Virology
RNA extraction from infected AGM PBL
Blood samples from infected AGMs collected in EDTA tubes were diluted in Pharmalyse buffer, vortexed, and incubated in the dark for 15 min. They were then centrifuged at 200 x g for 5 min and the pellets washed in 2 mL PBS 1% FBS, 2 mM EDTA. Finally, pellets were resuspended in 600 μL RLT lysis buffer and purified following the manufacturer’s instructions (Macherey Nagel). Samples were stored at −80°C till use.
RT-qPCR
Viral RNA was extracted using a Qiamp Viral RNA Kit (Qiagen) for serum, swabs, urine, BAL, vitreous humor, and thoracic exudate samples and a Nucleospin Kit (Macherey Nagel) for PBMCs and organs. Viral load was evaluated by one-step RT-qPCR (NEB Luna Universal One-Step RT-qPCR kit) using NiV-N-specific primers (NiV-N Fwd: GTGCTGAGCTATACCCCACC and NiV-N Rev: GAGATAAGCGCCGGACAAGA) and GAPDH primers (GAPDH Fwd: CACCCACTCCTCCACCTTTGAC, GAPDH Rev: GTCCACCACCCTGTTGCTGTAG), if necessary. PCR amplification was recorded on a Step One Plus apparatus (Thermo Fisher Scientific). All samples were run in duplicates and the results analyzed using ABI StepOne software v2.1 (Applied Biosystems, Thermo Fisher Scientific).
Post-challenge immune responses
Peripheral blood leukocyte isolation and fluid collection
One milliliter of blood was collected in EDTA tubes from each AGM and added to 10 mL 1X PharmLyse (BD Biosciences) to eliminate red blood cells. The tube was mixed by successive inversion and incubated for 15 min at RT in the dark. The mix was then centrifuged at 200 x g for 5 min at RT, the supernatants removed, and cell pellets resuspended in 1 mL 1X PBS +1% FBS before starting the staining procedure. Oral and nasopharyngeal swabs were sampled every two or three days and conditioned in preservation medium (UTM medium, Labelians).
Blood FACS phenotyping
Following isolation, PBLs were washed once in 1X PBS +1% FBS, resuspended in 150 μL 1X PBS +1% FBS containing the antibody cocktail (anti- CD3-AF488 (BD Biosciences, clone SP34-2), CD4-AF700 (BD Biosciences, clone SK3), CD8-BV421 (BD Biosciences, clone SK1), CD20-APC-H7 (BD Biosciences, clone 2H7), CD14-APC (BD Biosciences, clone m5e2), and CD16-PeCy7 (BD Biosciences, clone 3g8), and incubated for 20 min at 4°C in the dark. Cells were then washed twice with 1 mL 1X PBS +1% FBS, centrifuged for 5 min at 530 g, and fixed with 150 μL 4% methanol-free formaldehyde for 15 min at 4°C in the dark. Finally, 1 mL 1X PBS was added and the fixed cells centrifuged at 1,470 g for 5 min. The final pellets were resuspended in 500 μL 1X PBS and the data acquired using a Gallios flow cytometer (Beckman Coulter) and analyzed using Flowjo V.10.8 (Treestar).
Histology and immunohistochemistry
Formalin-fixed samples were embedded in paraffin. Then, 4 μm-thick tissue sections were cut, prepared according to standard procedures, and stained with hematoxylin – eosin – saffron (HES) for histopathological evaluation.
For immunohistochemistry, 4 μm-thick tissue sections were deparaffinized and rehydrated. The slides were incubated for 30 min at RT with 5% Fetal calf serum (Eurobio Scientific, France) diluted in PBS, followed by overnight incubation at 4°C with a polyclonal rabbit anti-NiV N antibody (1/500, courtesy of Prof Branka Horvat, CIRI) diluted in a 5% FBS, 0.1% Tween 20 (Sigma-Aldrich), PBS solution. Then the slides were rinsed and incubated for 30 min at RT with secondary antibody (goat anti-rabbit IgG, Alexa Fluor 555, 1/300; Thermo Fisher Scientifc, cat # A21428) and DAPI (1/1000; Thermo Fisher Scientifc, cat #D1306), diluted in 5% FBS, 0.1% Tween 20, PBS solution. Finally, the slides were rinsed prior to mounting with an in-house preparation of Mowiol (Merck). Whole slide images were acquired using an Axio Scan.Z1 slide scanner (Zeiss, Germany) with an ×10 objective (Plan Apochromat, Zeiss) and an X-Cite Fire LED illumination system (Excelitas Technologies, USA) for the following emission passbands: 445/50 (DAPI), 525/50 (autofluorescence), and 605/70 (Alexa Fluor 555). The whole slide scans were visualized using ZEN 2.3 software (Zeiss). This protocol was also performed on tissue sections from an animal that was not exposed to Nipah virus as a negative control and quality controls were performed by omission of the primary antibody.
Quantification and statistical analysis
We studied the dynamics of the neutralization response after the peak observed at 35 days and before the challenge. We used linear mixed models to consider the correlation of repeated measures within individuals (including one random intercept to capture inter-individual variability). For statistical analyses of the immune responses of the mice and AGMs, Kruskal-Wallis tests with Dunn’s multiple comparison post-hoc test were used for multiple comparisons. For phenotypic analysis of B-cells by FACS, groups were compared using unpaired Mann Whitney t-tests (ns: non-significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). The Gehan-Breslow-Wilcoxon test was used to assess survival curves. Error bars depict the mean ± the standard error of the mean (SEM) in all bar graphs shown. Whiskers define the minimum and maximum values of the data presented, with the medians shown as bars. GraphPad Prism statistical analysis software (Graph-Pad Prism Software, version 9) was used throughout.
Acknowledgments
We thank Dr. Andres M. Salazar (Oncovir, Inc.) for providing the poly-ICLC (Hiltonol), Valérie Favede and Edoardo Pizzioli for preparing BSL-4 material, and members of Bioprim and the BSL-4 animal facility for handling AGMs. We thank the Center for Disease Control and Prevention (Atlanta, GA, USA) for the NiV-B isolate. This work received funding from (1) INSERM and the Investissements d’Avenir program, Vaccine Research Institute (VRI), managed by the ANR under reference ANR-10-LABX- 77, and (2) the French Ministry of Higher Education, Research and Innovation for the Nipah Virus Project and the SARS-CoV-2 Vaccine Project.
Author contributions
Conceptualization, E.M., M.M., S.B., E.C., H.R., B.H., H.H., V.G., S.C., and Y.L.; mouse experimentation, Y.P., O.R., M.I., M.S., F.P., L.D., and A.H.; AGM and BSL-4 experimentation, Y.P., O.R., M.I., M.S., N.E.-J., C.L., E.B., S.R., S.D., C.G., L.A., A.-S.G., R.M., and R.T.; funding acquisition, B.H., V.G., S.C., and Y.L.; project administration, M.C.; writing – original draft, Y.P., S.C., and Y.L. All authors participated in discussions of experimental results and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 11, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101467.
Contributor Information
Yves Lévy, Email: yves.levy@inserm.fr.
Sylvain Cardinaud, Email: sylvain.cardinaud@inserm.fr.
Supplemental information
References
- 1.Chua K.B., Bellini W.J., Rota P.A., Harcourt B.H., Tamin A., Lam S.K., Ksiazek T.G., Rollin P.E., Zaki S.R., Shieh W., et al. Nipah Virus: A Recently Emergent Deadly Paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 2.Sweileh W.M. Global research trends of World Health Organization’s top eight emerging pathogens. Global. Health. 2017;13:9. doi: 10.1186/s12992-017-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luby S.P., Gurley E.S., Hossain M.J. Transmission of Human Infection with Nipah Virus. Clin. Infect. Dis. 2009;49:1743–1748. doi: 10.1086/647951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelissier R., Iampietro M., Horvat B. Recent Advances in the Understanding of Nipah Virus Immunopathogenesis and Anti-viral Approaches. F1000Research. 2019;8 doi: 10.12688/f1000research.19975.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thayyil J., Padmanabhan A., Gangadharan A., Salim S., Jayakrishnan T. Nipah outbreak in Kerala, South India: Ethical challenges in the deployment of healthcare workers. Indian J. Med. Ethics. 2020;V:1–9. doi: 10.20529/IJME.2020.078. [DOI] [PubMed] [Google Scholar]
- 6.Yadav P.D., Sahay R.R., Balakrishnan A., Mohandas S., Radhakrishnan C., Gokhale M.D., Balasubramanian R., Abraham P., Gupta N., Sugunan A.P., et al. Nipah Virus Outbreak in Kerala State, India Amidst of COVID-19 Pandemic. Front. Public Health. 2022;10 doi: 10.3389/fpubh.2022.818545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ithinji D.G., Buchholz D.W., Ezzatpour S., Monreal I.A., Cong Y., Sahler J., Bangar A.S., Imbiakha B., Upadhye V., Liang J., et al. Multivalent viral particles elicit safe and efficient immunoprotection against Nipah Hendra and Ebola viruses. npj Vaccines. 2022;7 doi: 10.1038/s41541-022-00588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woolsey C., Borisevich V., Fears A.C., Agans K.N., Deer D.J., Prasad A.N., O’Toole R., Foster S.L., Dobias N.S., Geisbert J.B., et al. Recombinant vesicular stomatitis virus–vectored vaccine induces long-lasting immunity against Nipah virus disease. J. Clin. Invest. 2023;133 doi: 10.1172/JCI164946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoneda M., Georges-Courbot M.C., Ikeda F., Ishii M., Nagata N., Jacquot F., Raoul H., Sato H., Kai C. Recombinant Measles Virus Vaccine Expressing the Nipah Virus Glycoprotein Protects against Lethal Nipah Virus Challenge. PLoS One. 2013;8 doi: 10.1371/journal.pone.0058414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bossart K.N., Rockx B., Feldmann F., Brining D., Scott D., LaCasse R., Geisbert J.B., Feng Y.R., Chan Y.P., Hickey A.C., et al. A Hendra virus G glycoprotein subunit vaccine protects African green monkeys from Nipah virus challenge. Sci. Transl. Med. 2012;4:146ra107. doi: 10.1126/scitranslmed.3004241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ceglia V., Kelley E.J., Boyle A.S., Zurawski S., Mead H.L., Harms C.E., Blanck J.-P., Flamar A.-L., Kirschman J.H., Ogongo P., et al. A Framework to Identify Antigen-Expanded T Cell Receptor Clusters Within Complex Repertoires. Front. Immunol. 2021;12:735584–735614. doi: 10.3389/fimmu.2021.735584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coléon S., Wiedemann A., Surénaud M., Lacabaratz C., Hue S., Prague M., Cervantes-Gonzalez M., Wang Z., Ellis J., Sansoni A., et al. Design, immunogenicity, and efficacy of a pan-sarbecovirus dendritic-cell targeting vaccine. EBioMedicine. 2022;80 doi: 10.1016/j.ebiom.2022.104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J., Zurawski G., Zurawski S., Wang Z., Akagawa K., Oh S., Hideki U., Fay J., Banchereau J., Song W., Palucka A.K. A novel vaccine for mantle cell lymphoma based on targeting cyclin D1 to dendritic cells via CD40. J. Hematol. Oncol. 2015;8:35. doi: 10.1186/s13045-015-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastor Y., Ghazzaui N., Hammoudi A., Centlivre M., Cardinaud S., Lévy Y. Refining the DC-targeting vaccination for preventing emerging infectious diseases. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.949779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marlin R., Godot V., Cardinaud S., Galhaut M., Coleon S., Zurawski S., Dereuddre-Bosquet N., Cavarelli M., Gallouët A.S., Maisonnasse P., et al. Targeting SARS-CoV-2 receptor-binding domain to cells expressing CD40 improves protection to infection in convalescent macaques. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-25382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandre M., Marlin R., Prague M., Coleon S., Kahlaoui N., Cardinaud S., Naninck T., Delache B., Surenaud M., Galhaut M., et al. Modelling the response to vaccine in non-human primates to define SARS-CoV-2 mechanistic correlates of protection. Elife. 2022;11 doi: 10.7554/eLife.75427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flamar A.L., Bonnabau H., Zurawski S., Lacabaratz C., Montes M., Richert L., Wiedemann A., Galmin L., Weiss D., Cristillo A., et al. HIV-1 T cell epitopes targeted to Rhesus macaque CD40 and DCIR: A comparative study of prototype dendritic cell targeting therapeutic vaccine candidates. PLoS One. 2018;13 doi: 10.1371/journal.pone.0207794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zurawski G., Shen X., Zurawski S., Tomaras G.D., Montefiori D.C., Roederer M., Ferrari G., Lacabaratz C., Klucar P., Wang Z., et al. Superiority in Rhesus Macaques of Targeting HIV-1 Env gp140 to CD40 versus LOX-1 in Combination with Replication-Competent NYVAC-KC for Induction of Env-Specific Antibody and T Cell Responses. Journal of Virology. 2017;91:10–1128. doi: 10.1128/JVI.01596-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin W., Gorvel L., Zurawski S., Li D., Ni L., Duluc D., Upchurch K., Kim J., Gu C., Ouedraogo R., et al. Functional Specialty of CD40 and Dendritic Cell Surface Lectins for Exogenous Antigen Presentation to CD8 + and CD4 + T Cells. EBioMedicine. 2016;5:46–58. doi: 10.1016/j.ebiom.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin W., Duluc D., Joo H., Xue Y., Gu C., Wang Z., Wang L., Ouedraogo R., Oxford L., Clark A., et al. Therapeutic HPV cancer vaccine targeted to CD40 elicits effective CD8+ T-cell immunity. Cancer Immunol. Res. 2016;4:823–834. doi: 10.1158/2326-6066.CIR-16-0128. [DOI] [PubMed] [Google Scholar]
- 21.Kervevan J., Bouteau A., Lanza J.S., Hammoudi A., Zurawski S., Surenaud M., Dieudonné L., Bonnet M., Lefebvre C., Hocini H., et al. Targeting human langerin promotes HIV-1 specific humoral immune responses. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., Amaya M., Addetia A., Dang H.V., Reggiano G., Yan L., Hickey A.C., DiMaio F., Broder C.C., Veesler D. Architecture and antigenicity of the Nipah virus attachment glycoprotein. Science. 2022;375:1373–1378. doi: 10.1126/science.abm5561. [DOI] [PubMed] [Google Scholar]
- 23.Flamar A.-L., Contreras V., Zurawski S., Montes M., Dereuddre-Bosquet N., Martinon F., Banchereau J., Le Grand R., Zurawski G., Lévy Y. Delivering HIV Gagp24 to DCIR Induces Strong Antibody Responses In Vivo. PLoS One. 2015;10 doi: 10.1371/journal.pone.0135513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor D.W., Bobbili N., Kayatani A., Tassi Yunga S., Kidima W., Leke R.F.G. Measuring antibody avidity to Plasmodium falciparum merozoite antigens using a multiplex immunoassay approach. Malar. J. 2020;19:171. doi: 10.1186/s12936-020-03243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fenwick C., Turelli P., Ni D., Perez L., Lau K., Herate C., Marlin R., Lana E., Pellaton C., Raclot C., et al. Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys. Nature Microbiology Jul. 2022;25 doi: 10.1038/s41564-022-01198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geisbert T.W., Daddario-DiCaprio K.M., Hickey A.C., Smith M.A., Chan Y.-P., Wang L.-F., Mattapallil J.J., Geisbert J.B., Bossart K.N., Broder C.C. Development of an Acute and Highly Pathogenic Nonhuman Primate Model of Nipah Virus Infection. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynard O., Volchkov V.E. Characterization of a Novel Neutralizing Monoclonal Antibody Against Ebola Virus GP. J. Infect. Dis. 2015;212:S372–S378. doi: 10.1093/infdis/jiv303. [DOI] [PubMed] [Google Scholar]
- 28.Gaudino M., Aurine N., Dumont C., Fouret J., Ferren M., Mathieu C., Reynard O., Volchkov V.E., Legras-Lachuer C., Georges-Courbot M.-C., Horvat B. High Pathogenicity of Nipah Virus from Pteropus lylei Fruit Bats, Cambodia. Emerg. Infect. Dis. 2020;26:104–113. doi: 10.3201/eid2601.191284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 30.Lévy Y., Thiébaut R., Montes M., Lacabaratz C., Sloan L., King B., Pérusat S., Harrod C., Cobb A., Roberts L.K., et al. Dendritic cell-based therapeutic vaccine elicits polyfunctional HIV-specific T-cell immunity associated with control of viral load. Eur. J. Immunol. 2014;44:2802–2810. doi: 10.1002/eji.201344433. [DOI] [PubMed] [Google Scholar]
- 31.Cheng L., Wang Q., Li G., Banga R., Ma J., Yu H., Yasui F., Zhang Z., Pantaleo G., Perreau M., et al. TLR3 agonist and CD40-targeting vaccination induces immune responses and reduces HIV-1 reservoirs. J. Clin. Invest. 2018;128:4387–4396. doi: 10.1172/JCI99005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geisbert T.W., Bobb K., Borisevich V., Geisbert J.B., Agans K.N., Cross R.W., Prasad A.N., Fenton K.A., Yu H., Fouts T.R., et al. A single dose investigational subunit vaccine for human use against Nipah virus and Hendra virus. npj Vaccines. 2021;6 doi: 10.1038/s41541-021-00284-w. 23–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foster S.L., Woolsey C., Borisevich V., Agans K.N., Prasad A.N., Deer D.J., Geisbert J.B., Dobias N.S., Fenton K.A., Cross R.W., Geisbert T.W. A recombinant VSV-vectored vaccine rapidly protects nonhuman primates against lethal Nipah virus disease. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2200065119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loomis R.J., Stewart-Jones G.B.E., Tsybovsky Y., Caringal R.T., Morabito K.M., McLellan J.S., Chamberlain A.L., Nugent S.T., Hutchinson G.B., Kueltzo L.A., et al. Structure-Based Design of Nipah Virus Vaccines: A Generalizable Approach to Paramyxovirus Immunogen Development. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohammed A.A., Shantier S.W., Mustafa M.I., Osman H.K., Elmansi H.E., Osman I.A.A., Mohammed R.A., Abdelrhman F.A., Elnnewery M.E., Yousif E.M., et al. Epitope-Based Peptide Vaccine against Glycoprotein G of Nipah Henipavirus Using Immunoinformatics Approaches. J. Immunol. Res. 2020;2020 doi: 10.1155/2020/2567957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X.-A., Li H., Jiang F.-C., Zhu F., Zhang Y.-F., Chen J.-J., Tan C.-W., Anderson D.E., Fan H., Dong L.-Y., et al. A Zoonotic Henipavirus in Febrile Patients in China. N. Engl. J. Med. 2022;387:470–472. doi: 10.1056/NEJMc2202705. [DOI] [PubMed] [Google Scholar]
- 37.Hagan T., Gerritsen B., Tomalin L.E., Fourati S., Mulè M.P., Chawla D.G., Rychkov D., Henrich E., Miller H.E.R., Diray-Arce J., et al. Transcriptional atlas of the human immune response to 13 vaccines reveals a common predictor of vaccine-induced antibody responses. Nat. Immunol. 2022;23:1788–1798. doi: 10.1038/s41590-022-01328-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lelièvre J.-D., Moog C., Wiedemann A., Lacabaratz C., Candotti F., Durand M., Rieux V., Hardel L., Diallo A., Ding S., et al. CROI; 2023. CD40.HIVRI.ENV Vaccine Induces Strong and Durable Immune Responses: ANRS/VRI06 Trial. [Google Scholar]
- 39.Schiller J.T., Lowy D.R. Raising Expectations For Subunit Vaccine. J. Infect. Dis. 2015;211:1373–1375. doi: 10.1093/infdis/jiu648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collier A.R.Y., Yu J., McMahan K., Liu J., Chandrashekar A., Maron J.S., Atyeo C., Martinez D.R., Ansel J.L., Aguayo R., et al. Differential Kinetics of Immune Responses Elicited by Covid-19 Vaccines. N. Engl. J. Med. 2021;385:2010–2012. doi: 10.1056/NEJMc2115596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.PREVAC Study Team. Kieh M., Richert L., Beavogui A.H., Grund B., Leigh B., D'Ortenzio E., Doumbia S., Lhomme E., Sow S., et al. Randomized Trial of Vaccines for Zaire Ebola Virus Disease. N. Engl. J. Med. 2022;387:2411–2424. doi: 10.1056/NEJMoa2200072. [DOI] [PubMed] [Google Scholar]
- 42.Agnandji S.T., Huttner A., Zinser M.E., Njuguna P., Dahlke C., Fernandes J.F., Yerly S., Dayer J.-A., Kraehling V., Kasonta R., et al. Phase 1 Trials of rVSV Ebola Vaccine in Africa and Europe. N. Engl. J. Med. 2016;374:1647–1660. doi: 10.1056/NEJMoa1502924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreatta M., Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32:511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jensen K.K., Andreatta M., Marcatili P., Buus S., Greenbaum J.A., Yan Z., Sette A., Peters B., Nielsen M. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018;154:394–406. doi: 10.1111/imm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gauthier M., Agniel D., Thiébaut R., Hejblum B.P. dearseq: a variance component score test for RNA-seq differential analysis that effectively controls the false discovery rate. NAR Genom. Bioinform. 2020;2 doi: 10.1093/nargab/lqaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickham H. 2nd ed. Springer International Publishing : Imprint: Springer; 2016. ggplot2: Elegant Graphics for Data Analysis. [DOI] [Google Scholar]
- 49.Yu G., Wang L.-G., He Q.-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 50.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The RNA-Sequencing data generated in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE245374.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.