Abstract
The role of calmodulin (CaM) in apoptosis induced by gp160 of human immunodeficiency virus type 1 was investigated with cells undergoing single-cell killing. These cells were found to express, under the control of an inducible promoter, wild-type gp160 or mutant gp160 devoid of various lengths of the carboxyl terminus. Immunoprecipitation accompanied by immunoblotting revealed binding of CaM to wild-type gp160 but not to mutant gp160 bearing a carboxyl terminus with a deletion spanning more than five amino acid residues. A significant coenzyme activity was detected in the CaM bound to gp160 even in the presence of a Ca2+ chelater, EGTA. The cells forming this gp160-CaM complex exhibited an elevated intracellular Ca2+ level followed by DNA fragmentation, which is a hallmark of apoptosis, and finally cell killing, while the cells not forming this complex did not show any significant elevation in Ca2+ level or DNA fragmentation. These results thus indicated that CaM plays a key role in gp160-induced apoptosis.
Apoptosis is an active process of cell death that serves diverse functions in multicellular organisms, and under physiological conditions, it is tightly controlled. Recently, many viruses have been found to induce apoptosis of infected cells, and there is now mounting evidence that such virally induced apoptosis contributes directly to the cytopathogenic effects of these viruses. On the other hand, viruses also have their own antiapoptosis genes, which can block the premature death of infected cells to maximize the production of viral progeny (24). Therefore, an investigation of the mechanism regulating apoptosis in virally infected cells appears to be indispensable to elucidate the pathophysiology of virally induced disease.
Human immunodeficiency virus (HIV) exerts an acute cytopathic effect on single T cells in culture through a mechanism that is independent of syncytium formation. Unlike syncytium formation, single-cell killing is not susceptible to inhibition by soluble CD4 or neutralizing antibodies (2). While the mechanism of single-cell killing by infection with HIV needs further clarification, the event itself may play a crucial role in determining whether cells acutely infected with HIV are killed immediately or are latently infected and thus become reservoirs for the virus. In previous studies (10, 11), we demonstrated that the expression of gp160 of HIV in CD4+ cells causes single-cell killing due to the formation of intracellular gp160 aggregates. In addition, apoptosis accompanied by intracellular Ca2+ elevation has also been shown to be the death mechanism in this system (13, 22). Moreover, a calmodulin (CaM) antagonist blocked the elevation of Ca2+ as well as the following DNA fragmentation, thus suggesting that CaM-dependent intracellular Ca2+ release by gp160 is one of the cardinal events subsequently leading to apoptosis. In the present study, the role of CaM in gp160-mediated apoptosis was investigated with cells undergoing single-cell killing.
MATERIALS AND METHODS
Cell clones.
A gp160-expressing stable cell clone, UE160, was made by the transfection of plasmid pSMTE7-160 into U937 cells, a human CD4+ monocytoid cell line (13). pSMTE7-160 has a human metallothionein IIA gene as the promoter and is thus inducible by the addition of heavy metal ions such as Cd2+. Δ862, Δ859, Δ855, and Δ821 were made by transfection into U937 cells of pSMTE7-160-Δ862 (encoding gp160 with the C-terminal 2 amino acids deleted), -Δ859 (encoding gp160 with the C-terminal 5 amino acids deleted), -Δ855 (encoding gp160 with the C-terminal 9 amino acids deleted), and -Δ821 (encoding gp160 with the C-terminal 43 amino acids deleted), respectively. These mutant gp160-expressing plasmids were constructed by introducing a stop codon into the carboxyl-terminal region of env in pSMTE7-160 by the oligonucleotide-directed mutagenesis method with mismatched 24-mer synthetic DNA primers (21). One base located around the center of these primers is replaced with another base to introduce a stop codon in place of a sense codon, such that the primers for pSMTE7-160-Δ862, -Δ859, -Δ855, and -Δ821 have the changes 8365 T to A, 8355 G to T, 8343 A to T, and 8242 T to A (BH10 numbering [20]) in each 24-mer primer, respectively.
Immunoblotting and immunoprecipitation.
For the immunoblot analysis, the cells were lysed in cold extraction buffer containing 20 mM Tris-HCl (pH 7.5), 0.15 M NaCl, 1% Nonidet P-40, 2 mM phenylmethylsulfonyl fluoride, 10 μM aprotinin, 10 μM leupeptin, and 10−6 M EGTA. A soluble cytoplasmic extract was obtained after sonication and centrifugation at 10,000 × g for 30 min at 4°C. Proteins in the cytoplasmic extract (100 μg for each lane) were separated by sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel electrophoresis under reducing conditions. The separated polypeptides were transferred to a nitrocellulose sheet and treated with anti-envelope glycoprotein monoclonal antibody (MAb) 0.5β (mouse immunoglobulin G1 [IgG1]) (14) and peroxidase-linked second antibody, followed by detection with an enhanced chemiluminescence system (Amersham, Little Chalfont, United Kingdom) according to the manufacturer’s instructions. Densitometric analysis was performed with a chromatoscanner. For immunoprecipitation, cell lysate containing 100 μg of protein was added with a saturating amount (2 μg) of anti-CaM MAb (mouse IgG1) (catalog no. 05-173; Upstate Biotechnology Inc., Lake Placid, N.Y.), incubated at 4°C for 16 h with rotation, and then absorbed with 50 μl of protein G-coupled Sepharose (Pharmacia, Upsala, Sweden) at 4°C for 2 h. These Sepharose beads were washed five times with cold extraction buffer and heated at 95°C with sample buffer containing 0.1 M Tris-HCl (pH 6.8), 2% SDS, 10% 2-mercaptoethanol, 20% glycerol, and 0.001% bromophenol blue to elute absorbed materials. SDS–7.5% polyacrylamide gel electrophoresis was then performed. The separated polypeptides were then analyzed by immunoblotting with 0.5β as described above.
Measurement of intracellular Ca2+ level.
The intracellular Ca2+ level was measured by using a confocal laser microscope equipped with a digital image analyzer and fluorescent Ca2+ indicators, by a method described previously (18). Briefly, cells at 106/ml were loaded with 10 μM 1-[2-amino-5-(2,7-dichloro-6-hydroxy - 3 - oxy - 9 - xanthenyl)phenoxy] - 2 - (2 - amino - 5 - methylphenoxy)ethane - N,N, N′,N′-tetraacetic acid, pentaacetoxymethyl ester (Fluo 3-AM; Dojindo Laboratory, Kumamoto, Japan), and 0.1% p-3000 (Dojindo Laboratory) in a Ca2+-staining buffer (137 mM NaCl, 2.7 mM KCl, 5 mM glucose, 1 mg of bovine serum albumin per dl, 20 mM HEPES [pH 7.4]). Samples were then washed, resuspended in the Ca2+-staining buffer, transferred to a glass-bottom dish, and spun down. Then, the intensity of fluorescence, reflecting the intracellular Ca2+ level, was analyzed with a confocal laser microscope. As a positive control for Ca2+ flux, UE160 cells were stimulated with A23187 (Sigma Chemical Co., St. Louis, Mo.) in Ca2+-staining buffer containing 0.4 mM CaCl2.
Assay of the coenzyme activity of CaM.
The specific activity of CaM as a coenzyme was assayed by determining the ability of CaM to activate cyclic nucleotide phosphodiesterase (PDE) (EC 3. 1. 4. 17). Cell lysates were prepared in the presence of 10−6 M EGTA to prevent Ca2+-mediated activation of CaM from those cell clones after induction. Envelope glycoprotein (Env)-CaM complex for one sample was immunoprecipitated by addition of 2 μg of 0.5β to a cell lysate containing 100 μg of protein and then absorbed to 50 μl of protein G-coupled Sepharose beads as described above. Then, 2 μl of Sepharose beads conjugated with Env-CaM complex was applied to a PDE assay mixture consisting of a 1-ml final volume, according to a previously described method (23). To make a calibration curve between the amount of CaM and the PDE activity in the assay, increasing amounts of highly purified CaM (product no. 1915; Sigma) were added to the assay mixture with nontreated protein G-Sepharose beads.
Determination of apoptosis in cell culture.
Cells of each cell clone adjusted to 2 × 105/ml in RPMI 1640 medium supplemented with 10% fetal calf serum were added with 10 μM CdCl2 on day 0 and cultured thereafter. On each day, the number of viable cells was determined with a hemocytometer and 0.1% trypan blue. Agonistic anti-Fas MAb CH-11 and neutralizing anti-Fas MAb ZB4 were purchased from MBL, Nagoya, Japan, and added to the cell culture on day 0 at concentrations of 25 and 250 ng/ml, respectively. A cell cycle analysis to examine the degree of apoptosis was performed after a 48-h culture by a method reported previously (16). Briefly, the cells were fixed with 2% paraformaldehyde, permeabilized with 0.1% Nonidet P-40, and treated with 0.05 mg of RNase A per ml for 30 min at 37°C. After being washed, the cells were stained with propidium iodide. The cells in a discrete subpopulation of signals under the G0/G1 cell cycle region (subdiploid cells) were thus designated cells undergoing apoptosis.
RESULTS
Complex formation between gp160 and CaM.
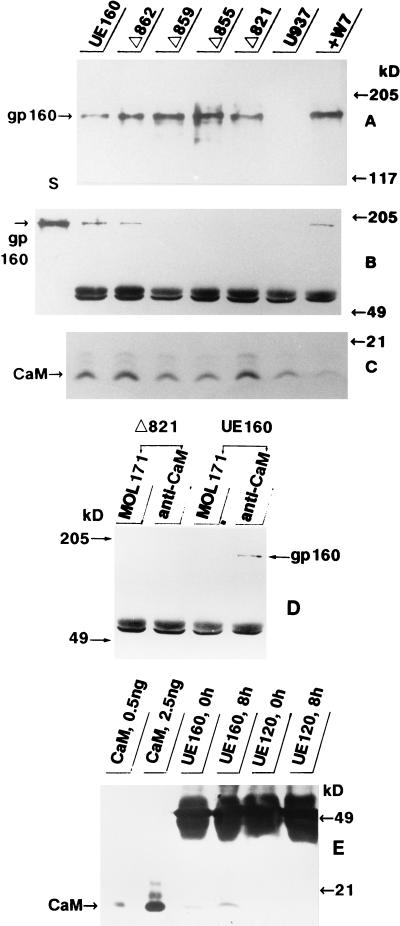
The cell clones were examined for their levels of expression of Env by an immunoblot analysis with an anti-Env MAb, 0.5β, after induction with 10 μM CdCl2. At 4 h after induction, the expression levels for gp160 and mutant gp160s reached their peaks, and the relative expression levels as determined by densitometry were 1.0, 1.8, 1.9, 2.7, and 1.5 in UE160, Δ862, Δ859, Δ855, and Δ821 cells, respectively (Fig. 1A). The UE160 cells already contained a detectable amount of gp160 without induction, whereas at 4 h after induction, the gp160 expression level was more than five times higher, as reported in a previous paper (13). Similar ratios for the Env expression levels among these cell clones were also obtained from another immunoblot analysis with a polyclonal anti-Env antibody (catalog no. 13-204-000; Advanced Biotechnologies, Columbia, Md.) (data not shown). To determine whether Env binds to CaM in these cell clones, lysate from each cell clone at 4 h postinduction was subjected to immunoprecipitation by an anti-CaM MAb. Next, the precipitates were run on a polyacrylamide gel and transferred to a nitrocellulose filter. The resulting blot was developed with the 0.5β MAb and second antibody, followed by detection with an enhanced chemiluminescence system (Fig. 1B). While coprecipitation of gp160 with CaM was detected in UE160 and Δ862 cells, no band corresponding to gp160 was found in Δ859, Δ855, or Δ821 cells by immunoprecipitation with anti-CaM antibody. In a parallel experiment, almost equal amounts of CaM were found in the lysates of UE160 and Δ862 cells, as well as Δ859, Δ855, and Δ821 cells, by an immunoblot analysis with anti-CaM MAb (Fig. 1C). To exclude a nonspecific association of gp160 with contaminating proteins during immunoprecipitation by the anti-CaM MAb, the lysates were subjected to immunoprecipitation with MOL171, a mouse IgG1 MAb recognizing the human Lck protein, which is irrelevant to Env (17). As shown in Fig. 1D, MOL171 did not coprecipitate any band corresponding to gp160, while the anti-CaM MAb specifically coprecipitated this Env from UE160 cell lysate. Essentially the same result was obtained in another series of experiments in which no EGTA was added to the working solution (the data obtained in the presence of EGTA is shown in Fig. 1). To further confirm such complex formation between gp160 and CaM, the lysate from UE160 cells was first immunoprecipitated with 0.5β, and then the precipitate was examined with the anti-CaM MAb after gel electrophoresis (Fig. 1E). At 8 h postinduction, a moderate but significant CaM band was able to be coprecipitated with gp160 in UE160 cells but not at all in UE120 cells expressing gp120. These results thus indicated that a complex between gp160 and CaM is formed in the cells and that the C-terminal three to five amino acid residues of gp160 are crucial for such complex formation.
FIG. 1.
Immunoblot and immunoprecipitation analyses. For the immunoblot analysis, the cell lysate from each cell clone, containing 100 μg of protein, was loaded as indicated at the top and then was examined with an anti-Env MAb (A) or anti-CaM MAb (C). For the detection of the gp160-CaM complex, the cell lysate was either (i) immunoprecipitated with the anti-CaM MAb, electrophoresed on a polyacrylamide gel, and then analyzed by immunoblotting with anti-Env MAb (B) or (ii) subjected to the same procedure with the MAbs reversed (E). The lysate was also subjected to immunoprecipitation with MOL171 or anti-CaM MAb and then analyzed by immunoblotting with 0.5β (D). As the standard for gp160 in panel B, the lysate from UE160 cells containing 100 μg of protein was loaded (lane S). As the standard for CaM in panel E, 0.5 or 2.5 ng of a purified CaM was loaded. The relative densities of bands corresponding to CaM as determined by densitometry were 1.0, 4.4, 0.46, and 1.1 in the first four lanes, respectively. The bands at around 49 kDa in panels B and D represent mouse IgG.
Elevation of intracellular Ca2+ and induction of apoptosis by the gp160-CaM complex.
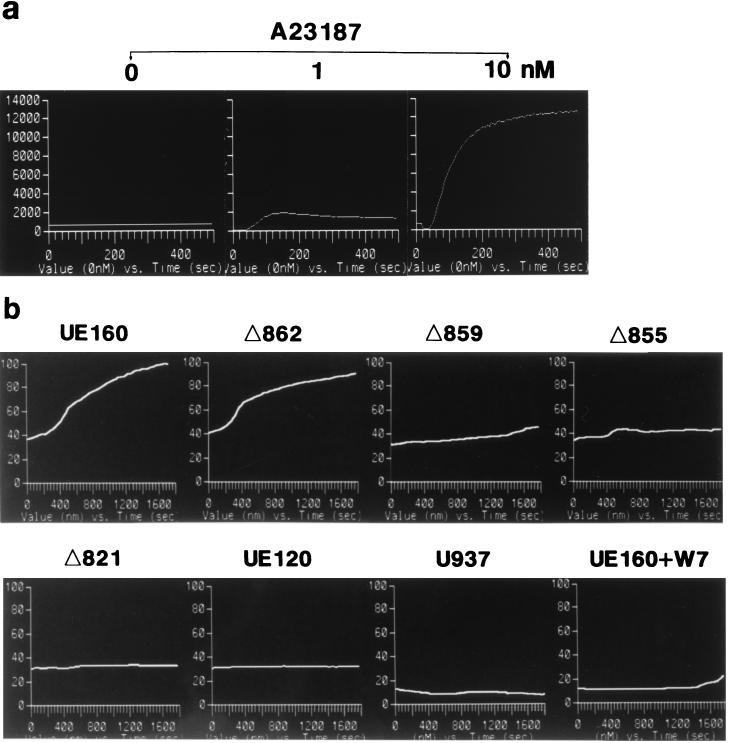
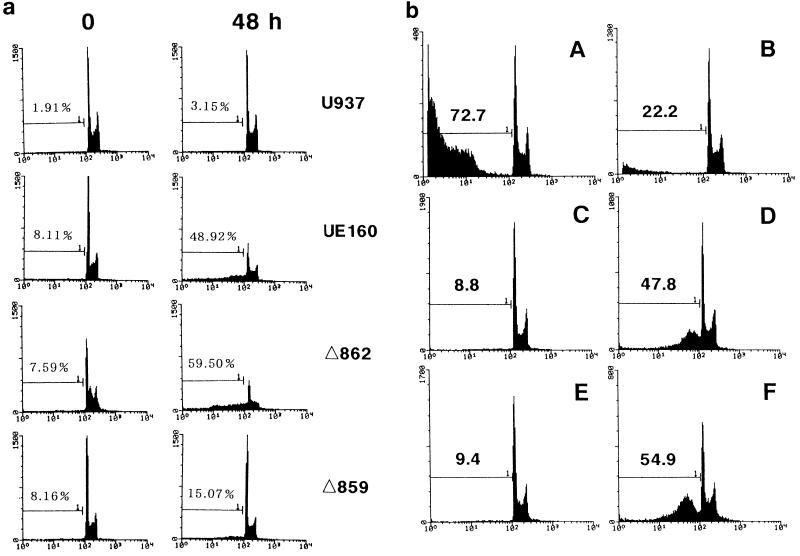
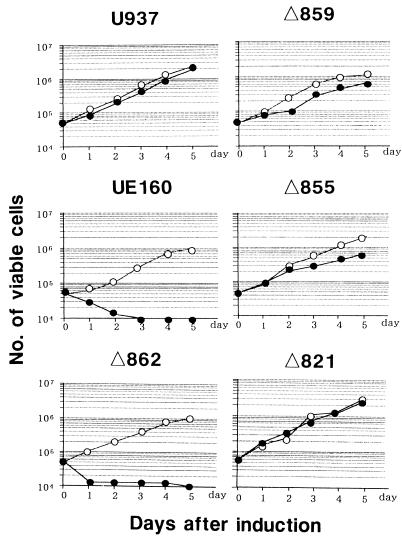
In a previous study (22), we found that an increase in intracellular Ca2+ occurs postinduction before the appearance of DNA fragmentation in UE160 cells. In addition, the Ca2+ increase was considered to play a key role in the occurrence of DNA fragmentation, a hallmark of apoptosis, because chelation of such free intracellular Ca2+ by treatment with O,O′-bis(2-aminophenyl)ethyleneglycol-N,N,N′,N′-tetraacetic acid, tetraacetoxymethyl ester (BAPTA-AM) blocked the appearance of DNA fragmentation after induction. Therefore, in the present study, the intracellular Ca2+ concentration was measured by using a fluorescent Ca2+ indicator (Fig. 2). As a positive control for Ca2+ influx, UE160 cells were stimulated with A23187 at 0 s in the presence of 0.4 mM CaCl2 (Fig. 2a). The Ca2+ influx in UE160 cells was maximal when 10 nM A23187 was added, and its level was more than 100 times higher than that found in the UE160 cells after induction (Fig. 2b). The cell clones were then examined for their Ca2+ level after induction in buffer containing no CaCl2 (Fig. 2b). Δ859, Δ855, Δ821, and UE120 cells, in which the Env expressed is unable to bind to CaM (Fig. 1), did not show any elevation in the intracellular Ca2+ level, whereas UE160 and Δ862 cells, in which the Env expressed is capable of binding to CaM, demonstrated a significant increase of Ca2+ at around 600 s after induction. Moreover, the treatment of UE160 cells with 10 μM W7, a CaM antagonist, completely blocked the elevation of Ca2+ after induction. These results therefore suggested that the binding of gp160 to CaM and subsequent activation of this coenzyme are prerequisites for the elevation of the intracellular Ca2+ level. A severalfold-higher level of Ca2+ in UE160 cells than in U937 cells even at the initial level was thought to be due to leaky expression of gp160 in the UE160 cells without induction, as shown in a previous study (13). To substantiate the gp160-induced activation of CaM, the specific activity of CaM as a coenzyme was assayed by determining the ability of CaM to activate PDE, a representative CaM-dependent enzyme that is transformed to an active form by CaM in the presence of Ca2+. First, cell lysates were prepared in the presence of 10−6 M EGTA to prevent Ca2+-mediated activation of CaM from those cell clones after induction. Second, the CaM bound to Env was immunoprecipitated with 0.5β, and its coenzyme activity in the PDE assay system was examined (Fig. 3). The Envs from UE160 and Δ862 cells, which are able to form a complex with CaM, demonstrated a significant CaM coenzyme activity, while the Env from Δ859 cells, which is unable to form detectable complex with CaM, showed only a minimal coenzyme activity. Moreover, in UE160 cells, gp160 in the absence of W7 exhibited a three- to fivefold-higher coenzyme activity at 10 and 240 min postinduction than did gp160 in the presence of W7, while the amounts of CaM bound to gp160 were almost the same as those for these two samples as shown in Fig. 1B. The coenzyme activity of CaM thus appears to be enhanced by binding to gp160 even in the absence of Ca2+. This event leads to the elevation of the intracellular Ca2+ level through a CaM-mediated system. This CaM-dependent elevation of the intracellular Ca2+ level was followed by DNA fragmentation and cell killing such that UE160 and Δ862 cells, both of which demonstrated an elevation of intracellular Ca2+, showed a high degree of DNA fragmentation 48 h after induction (Fig. 4a) followed by extensive cell killing (Fig. 5), while Δ859 cells, which have only a minimal Ca2+ elevation, exhibited only marginal increases in DNA fragmentation and cell killing. The inhibition of the CaM-mediated elevation of Ca2+ by W7 (Fig. 2b) also blocked the appearance of DNA fragmentation after induction, as shown in a previous study (22). The binding of gp160 to CaM was thus indicated to initiate a series of events, including Ca2+ elevation, which eventually lead the cells to apoptosis. The background levels of apoptosis were somewhat higher in the env-transfected cells than in control U937 cells (Fig. 4a). It is thought that the leaky expression of Env in these transfected cells induced this moderate level of apoptosis without induction. This gp160-CaM-mediated apoptosis also appeared to take place independent of Fas-mediated signal transduction, since a neutralizing anti-Fas MAb could not prevent UE160 cells from undergoing apoptosis after induction (Fig. 4b).
FIG. 2.
Time course of intracellular Ca2+ level. (a) UE160 cells were treated with Fluo 3-AM, and the Ca2+ concentration over time was measured after the addition of various amounts of A23187, as indicated at the top, at 0 s in the presence of 0.4 mM CaCl2. (b) Each cell clone, as indicated at the top of each panel, was treated with Fluo 3-AM, and the Ca2+ concentration over time was measured after the addition of 10 μM CdCl2 at 0 s. In one case, W7 (10 μM) was added 5 min before the induction with CdCl2 (UE160+W7). Each curve represents the mean of the fluorescence curves from 20 to 30 individual cells.
FIG. 3.
CaM activity analyzed with the PDE assay system. Env-CaM complex bound to Sepharose beads was obtained from the cell lysate of each cell clone at 1, 10, and 240 min after induction with CdCl2 as indicated on the left, and its CaM coenzyme activity was examined by using the PDE assay system (23). Where indicated, W7 was added to the cell culture 5 min before induction. As a standard, 0, 1, 2, and 4 ng of purified CaM was added to the PDE assay mixture with nontreated Sepharose beads. ND, not determined. The bars represent the standard errors (n = 3).
FIG. 4.
Detection of apoptosis by cell cycle analysis. (a) The proportion of cells undergoing apoptosis was examined by cell cycle analysis with each cell clone, as indicated to the right, at 0 and 48 h after induction. The numbers indicate the percentages of subdiploid cells and the percentages of apoptotic cells. (b) UE160 cells were treated with CH-11 (A) or CH-11 plus ZB-4 (B) for 24 h in the absence of induction. In addition, UE160 cells were cultured either without (C) or with (D) the addition of CdCl2 as an inducer for 48 h. (E and F) Same experiments as in panels C and D, respectively, except in the presence of ZB-4. The numbers represent the percentages of subdiploid cells.
FIG. 5.
Cell growth after induction. Each cell clone, as indicated at the top of each panel, was adjusted to 5 × 105 cells in 10 ml of RPMI 1640 medium on day 0 and cultured either with (closed circles) or without (open circles) 10 μM CdCl2. On each day, 1 ml of cell suspension was removed from the cultures, and the number of viable cells in the cell suspension was measured.
Dissociation of surface CD4 downregulation from apoptosis induction.
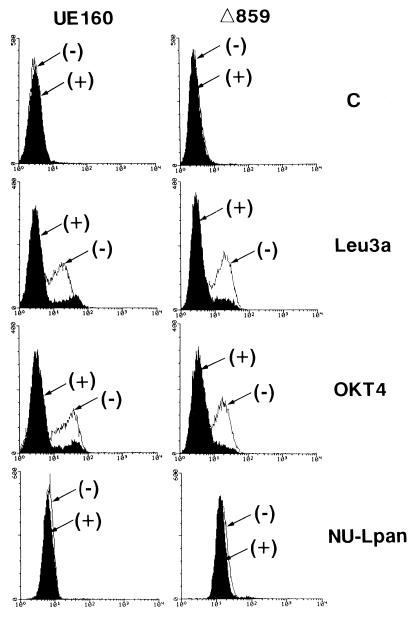
In our previous study (11), we showed that the intracellular expression of gp160 in CD4+ cells preceded the downregulation of surface CD4 and single-cell killing. An analysis of these cells demonstrated that the intracellular sequestration of CD4 by gp160 causes such downregulation of surface CD4. This finding also suggested the possible participation of such a gp160-CD4 complex in single-cell killing accompanied by the CD4 downregulation. In the present study, however, in view of the downregulation of surface CD4 after induction, it appears unlikely that such complex formation between gp160 and CD4 is directly involved in cell killing due to apoptosis, because not only UE160 cells but also Δ859 cells, which do not succumb to apoptosis after induction, exhibited a complete downregulation of surface CD4 after induction (Fig. 6). The possibility that such downregulation is an artifact of CdCl2 treatment was eliminated in a previous study (10).
FIG. 6.
Fluorescence-activated cell sorter analysis of surface CD4. UE160 or Δ859 cells were cultured for 4 h either without (−) or with (+) the addition of 10 μM CdCl2. The cells were then examined for their levels of expression of surface CD4 with a FACScan with MAb Leu3a (anti-CD4; Becton Dickinson, San Jose, Calif.), OKT4 (anti-CD4; Ortho Diagnostic Inc., Raritan, N.J.), or NU-Lpan (anti-CD45; Nichirei Corp., Tokyo, Japan) as indicated on the right. The amounts of these MAbs which bound to the cell surface were then revealed by secondary staining with fluorescein isothiocyanate–anti-mouse IgG antibody (Tago Inc., Burlingame, Calif.). Cells treated only with fluorescein isothiocyanate–anti-mouse IgG antibody were used as an autofluorescence control (C).
DISCUSSION
Fisher et al. (7) reported a variant of HIV type 1 (HIV-1) that replicates well but does not kill normal human T cells in vitro. This variant, designated X10-1, was derived from the genome of a cytopathic human T-cell leukemia virus type 3 clone (pXHB2D) by excising the portion of the envelope gene encoding the C-terminal five amino acid residues. As a result, the possibility was raised that the carboxyl terminus of the envelope protein of HIV-1 plays a direct role in T-cell killing by this virus. The env gene of pXH2BD is essentially identical to that used in our present study. This means that UE160 cells express the Env protein of cytopathic wild-type HIV-1, while Δ859 cells express that of noncytopathic X10-1 HIV-1. Taken together, the results of the present study supported the hypothesis of Fisher et al. (7) that the carboxyl terminus of Env plays a key role in Env-mediated cell killing of HIV infection, and they also indicate that the binding of CaM to the carboxyl terminus and subsequent apoptosis induction therefore constitute the causal mechanism of such cell killing in those cells infected with HIV-1.
CaM is a multifunctional Ca2+-binding protein that is ubiquitous in eukaryotes. It has no intrinsic enzyme activity, but it mediates the control of a wide spectrum of enzymes by Ca2+ (9). Amphipathic helical segments have been identified as an important structural motif in the recognition of CaM by different CaM-activated enzymes (5). The carboxyl-terminal domains of the Envs of HIV and simian immunodeficiency viruses contain regions that can fold into amphipathic helical segments, which closely resemble the amphipathic segments found in CaM-activated enzymes. Srinivas et al. (25) showed by a gel overlay assay that purified HIV-1 gp160 binds to CaM in either the presence or absence of Ca2+, although binding was greatly diminished in the absence of Ca2+. In contrast, gp120, which lacks putative amphipathic helical segments, did not bind to CaM. In line with their findings, the present in vivo study also demonstrated the binding of gp160 to CaM in the cells by its carboxyl terminus. Miller et al. (15) reported that a synthetic peptide homolog encompassing 27 amino acid residues of the carboxyl terminus is critical for gp160-CaM interaction, since this peptide binds efficiently to purified CaM and inhibits the CaM-mediated stimulation of phosphodiesterase activity in vitro. The present study, on the other hand, revealed that only a segment spanning three to five amino acid residues of the carboxyl terminus is crucial for gp160 to bind to CaM and activate CaM in the cells. Such CaM-bound gp160 exerted a significant level of coenzyme activity even in the presence of EGTA, thus suggesting that the gp160-mediated activation of CaM can occur either in the absence Ca2+ of at a very low Ca2+ concentration.
The elevation of Ca2+ by the expression of Envs was abrogated by deleting the CaM-binding domain from them or in the presence of a CaM antagonist. Nicotera et al. (19) reported that the incubation of isolated rat liver nuclei with ATP and Ca2+ led to the uptake of Ca2+ into the nuclei. Such an accumulation of Ca2+ in the nuclei was attributed to the activity of the Ca2+ pump located in the nuclear envelope, and activation of CaM was required for the start of this Ca2+ pump. This finding raises the possibility that a CaM-dependent Ca2+ pump is also responsible for the elevation of Ca2+ in gp160-expressing cells, while Ca2+ elevation in these cells seems to be confined to the nuclear region, as reported previously (22); however, this cannot yet be definitely concluded.
The biochemical hallmark of apoptosis is the cleavage of chromatin into nucleosomal fragments. It appears likely that Ca2+ signaling is a critical event leading to such DNA fragmentation by gp160, because the blocking of Ca2+ elevation by BAPTA-AM or W7 also inhibited the appearance of fragmented DNA in UE160 cells, as reported in a previous study (22). However, it remains unclear how Ca2+ signaling leads to apoptosis events, including DNA fragmentation. Multiple lines of evidence indicate that apoptosis can be triggered by the activation of a family of cysteine proteases designated caspases. Recently, Liu et al. (12) identified a 45-kDa heterodimer protein, designated DNA fragmentation factor (DFF), which functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. DFF itself showed no DNase activity, and it is therefore likely that DFF activates a nuclease(s) that resides in the nuclei. One candidate is a Ca2+/Mg2+-dependent nuclease which has been described previously (8), and Ca2+ signaling evoked by gp160-CaM interaction may participate in this stage of the apoptosis pathway.
Cao et al. (3) reported that the expression of Env from HIV-1 in T-cell lines resulted in single-cell lysis, as shown in our present study with a monocytoid cell line, U937. In their system, however, the single-cell lysis involved primarily necrosis, possibly mediated by Env. Moreover, the cytoplasmic tail of the gp41 transmembrane Env, which is the crucial part for induction of CaM-mediated cell killing, was neither necessary nor sufficient for single-cell lysis. In this study, on the other hand, we aimed to elucidate the role of gp160-CaM interaction in the single-cell killing by using U937 cells, which have CD4 but not CD4 bound to Lck, a tyrosine kinase predominant in T cells (17). Lck is reported to play an important role in HIV-induced cell killing by binding to CD4 and acting as an adapter to anchor other proteins to transduce the death signal (4). Such an Lck-mediated death pathway is thus suggested to prevail over the CaM-mediated death signal in the system used by Cao et al. (3), with T-cell lines.
Monocytes and macrophages can be infected with HIV; however, HIV does not induce a significant cytopathic effect in these cells (6). We used U937 cells as the target cells for transfection of env plasmids in the present study because this vector expresses Env very efficiently in this cell line. The U937 cell line used in this study is a subclone derived from the parental U937 cell clone, a human monocytoid tumor cell line, and is named U937 clone 2, as previously reported (11). This subclone has cytological properties different from those of the U937 parental clone, such as higher levels of surface CD4 expression and activities of several intracellular enzymes (1). This change in cytological properties may thus make U937 clone 2 susceptible to the cytopathic effects of Env, whereas this cell line is originally derived from human monocytes that are resistant to such cytopathic effects.
REFERENCES
- 1.Asjo B, Ivhed I, Gidlund M, Fuerstenberg S, Fenyo E M, Nilsson K, Wigzell H. Susceptibility to infection by the human immunodeficiency virus correlates with T4 expression in a parental monocytoid cell line and its subclones. Virology. 1987;157:359–365. doi: 10.1016/0042-6822(87)90278-9. [DOI] [PubMed] [Google Scholar]
- 2.Bergeron L, Sodroski J. Dissociation of unintegrated viral DNA accumulation from single-cell lysis induced by human immunodeficiency virus type 1. J Virol. 1992;66:5777–5787. doi: 10.1128/jvi.66.10.5777-5787.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao J, Park I-W, Cooper A, Sodroski J. Molecular determinants of acute single-cell lysis by human immunodeficiency virus type 1. J Virol. 1996;70:1340–1354. doi: 10.1128/jvi.70.3.1340-1354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbeil J, Tremblay M, Richman D D. HIV-induced apoptosis requires the CD4 receptor cytoplasmic tail and is accelerated by interaction of CD4 with p56lck. J Exp Med. 1996;183:39–48. doi: 10.1084/jem.183.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox J A, Comte M, Fitton J E, Degrado W F. The interaction of calmodulin with amphiphilic peptides. J Biol Chem. 1985;260:2527–2534. [PubMed] [Google Scholar]
- 6.Fauci A S. The human immunodeficiency virus: infectivity and mechanisms of pathogenesis. Science. 1988;239:617–622. doi: 10.1126/science.3277274. [DOI] [PubMed] [Google Scholar]
- 7.Fisher A G, Ratner L, Mitsuya H, Marselle L M, Harper M E, Broder S, Gallo R C, Wong-Staal F. Infectious mutants of HTLV-III with changes in the 3′ region and markedly reduced cytopathic effects. Science. 1986;233:655–659. doi: 10.1126/science.3014663. [DOI] [PubMed] [Google Scholar]
- 8.Gaido M L, Cidlowski J A. Identification, purification, and characterization of a calcium-dependent endonuclease (NUC18) from apoptotic rat thymocytes. J Biol Chem. 1991;266:18580–18585. [PubMed] [Google Scholar]
- 9.Klee C B, Crouch T H, Richman P G. Calmodulin. Annu Rev Biochem. 1980;49:489–515. doi: 10.1146/annurev.bi.49.070180.002421. [DOI] [PubMed] [Google Scholar]
- 10.Koga Y, Nakamura K, Sasaki M, Kimura G, Nomoto K. The difference in gp160 and gp120 of HIV type 1 in the induction of CD4 downregulation preceding single-cell killing. Virology. 1994;201:137–141. doi: 10.1006/viro.1994.1274. [DOI] [PubMed] [Google Scholar]
- 11.Koga Y, Sasaki M, Yoshida H, Wigzell H, Kimura G, Nomoto K. Cytopathic effect determined by the amount of CD4 molecules in human cell lines expressing envelope glycoprotein of HIV-1. J Immunol. 1990;144:94–102. [PubMed] [Google Scholar]
- 12.Liu X, Zou H, Slaughter C, Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 13.Lu Y-Y, Koga Y, Tanaka K, Sasaki M, Kimura G, Nomoto K. Apoptosis induced in CD4+ cells expressing gp160 of human immunodeficiency virus type 1. J Virol. 1994;68:390–399. doi: 10.1128/jvi.68.1.390-399.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsushita S, Robert-Guroff M, Rusche J, Koito A, Hattori T, Hoshino H, Javaherian K, Takatsuki K, Putney S. Characterization of a human immunodeficiency virus neutralizing monoclonal antibody and mapping of the neutralizing epitope. J Virol. 1988;62:2107–2114. doi: 10.1128/jvi.62.6.2107-2114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller M A, Mietzner T A, Cloyd M W, Robey W G, Montelaro R C. Identification of a calmodulin-binding and inhibitory peptide domain in the HIV-1 transmembrane glycoprotein. AIDS Res Hum Retroviruses. 1993;9:1057–1066. doi: 10.1089/aid.1993.9.1057. [DOI] [PubMed] [Google Scholar]
- 16.Mori T, Ando K, Tanaka K, Ikeda Y, Koga Y. Fas-mediated apoptosis of the hematopoietic progenitor cells in mice infected with murine cytomegalovirus. Blood. 1997;89:3565–3573. [PubMed] [Google Scholar]
- 17.Moroi Y, Koga Y, Nakamura K, Ohtsu M, Kimura G, Nomoto K. Accumulation of p60lck in HTLV-1-transformed T cell lines detected by an anti-Lck monoclonal antibody, MOL171. Jpn J Cancer Res. 1991;82:909–915. doi: 10.1111/j.1349-7006.1991.tb01920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neylon C B, Hoyland J, Mason W T, Irvine R F. Spatial dynamics of intracellular calcium in agonist-stimulated vascular smooth muscle cells. Am J Physiol. 1990;259:C675–C686. doi: 10.1152/ajpcell.1990.259.4.C675. [DOI] [PubMed] [Google Scholar]
- 19.Nicotera P, McConkey D J, Jones D P, Orrenius S. ATP stimulates Ca2+ uptake and increases the free Ca2+ concentration in isolated rat liver nuclei. Proc Natl Acad Sci USA. 1989;86:453–457. doi: 10.1073/pnas.86.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Jr, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature (London) 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 15.74–15.79. [Google Scholar]
- 22.Sasaki M, Uchiyama J, Ishikawa H, Matsushita S, Kimura G, Nomoto K, Koga Y. Induction of apoptosis by calmodulin-dependent intracellular Ca2+ elevation in CD4+ cells expressing gp160 of HIV. Virology. 1996;224:18–24. doi: 10.1006/viro.1996.0502. [DOI] [PubMed] [Google Scholar]
- 23.Schiefer S. Calmodulin. In: Bergmeyer H U, Bergmeyer J, Grable M, editors. Methods of enzymatic analysis. Vol. 9. Deerfield Beach, Fla: VCH Publishers; 1986. pp. 317–331. [Google Scholar]
- 24.Shen Y, Shenk T E. Viruses and apoptosis. Curr Opin Genet Dev. 1995;5:105–111. doi: 10.1016/s0959-437x(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 25.Srinivas S K, Srinivas R V, Anantharamaiah, R. G M, Compans W, Segrest J P. Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J Biol Chem. 1993;268:22895–22899. [PubMed] [Google Scholar]