Abstract
One of the crucial steps in the assembly of the human immunodeficiency virus type 1 (HIV-1) and other retroviruses is the incorporation and retention of all the key viral enzymes in released virions. The viral enzymes protease, reverse transcriptase, and integrase of HIV-1 are initially synthesized as Gag-Pol fusion polyproteins. It has been shown that the incorporation of Gag-Pol polyproteins during virus assembly requires the Gag domains that are shared by the Gag and Gag-Pol precursors. We now report that truncation of the C-terminal p6 domain of HIV-1 Gag, which is present in the Gag precursor but not in the Gag-Pol precursor, drastically reduced the amount of Pol proteins in the mutant virions. Mutations in the lentivirus conserved motif P(T/S)APP in p6 also drastically reduced the amount of Pol proteins in mutant virions. The steady-state levels of Gag-Pol precursors and cleaved Pol proteins in the transfected cells were not affected by mutations in p6. The incorporation of unprocessed Gag-Pol precursors into p6 mutant virions was detected when the viral protease was mutated, suggesting that the interactions among mutant Gag molecules and Gag-Pol precursors were not significantly affected. These results suggest that the p6 domain of HIV-1 Gag may play an important role in recruiting or retaining cleaved Pol proteins during virus assembly.
The Gag molecule of retroviruses can act alone to direct the assembly and release of immature virus-like particles (15, 35). Various functional domains in Gag have been examined, including several virus assembly domains (15, 35), domains important for packaging of viral genomic RNA (15, 26) and viral Env proteins (15), and domains involved in the early life cycle of the viruses (4–6, 8, 32, 34, 40).
The Gag polyprotein of human immunodeficiency virus type 1 (HIV-1) is first synthesized as a precursor of 55 kDa (Pr55Gag). It is subsequently cleaved by viral protease to yield the following mature proteins (from the N to the C terminus): p17 (MA), p24 (CA), p2 spacer peptide, p7 (NC), p1 spacer peptide, and p6Gag (11). As is true in most retroviruses, the Pol protein of HIV-1 is synthesized as a Gag-Pol (Pr160Gag-Pol) fusion polyprotein (16, 36). There is no independent ribosomal entry site for the pol gene of HIV-1, which is in the −1 reading frame in relation to the upstream gag gene. Instead, 5 to 10% of the ribosomes synthesizing Gag shift to the −1 frame at a special shift site near the 3′ end of the gag gene (see Fig. 1) and continue translation through the pol gene to produce the Gag-Pol fusion polyprotein (16, 36). The advantages associated with generating a polyprotein by nonstandard translation, rather than by splicing, are not clear. One possible advantage is that viral enzymes such as protease (33) and reverse transcriptase (RT) (39) are relatively inactive when they are part of the Gag-Pol polyprotein, although significant RT activity has been detected in the HIV-1 Gag-Pol polyprotein (31). Linkage of Pol to the Gag protein may also facilitate Pol packaging into virions (15, 35).
FIG. 1.
Construction of HIV-1 mutants. (A) The top diagram shows the genome organization of the HIV-1 parental construct HXB2. Mutants were constructed as described in the text. The TAA construct contains a premature stop codon that truncates all but one amino acid of p6. The LTALL construct contains amino acid substitutions of leucine for prolines 7, 10, and 11 in HIV-1 p6, and the amino acid sequences expressed from the overlapping pol open reading frame in LTALL are intact. The constructs PTAP−, PR−, and PR−/PTAP− have been described previously (13). Diagonally striped boxes, Pol domains in Gag-Pol precursor. (B) The nucleotide sequences and corresponding amino acid sequences in Gag (p6) and Pol for the wild type (WT) and the p6-mutant constructs are shown. Nucleotides and amino acids that differ from those in the wild-type sequences are underlined.
The MA, CA, and NC domains of HIV-1 are shared between the Gag and Gag-Pol precursor molecules. However, the p6Gag domain is not present in the Gag-Pol precursor because of the ribosomal frameshifting event that occurs in the upstream p1 region. The role of the p6Gag domain in the HIV-1 life cycle is not fully understood, but it has been suggested to have at least two functions. This domain, and particularly the 30 amino acids at its C terminus, appears to be critical for the incorporation of the accessory viral proteins Vpr (18–20, 24) and Vpx (37) into released HIV-1 and HIV-2 virions, respectively. Previous studies with HIV-1 have also suggested that p6Gag plays a role in efficient virus release from the cell surface (10, 13, 28, 41). When p6Gag is mutated, assembled virus particles are tethered on the surfaces of transfected cells (10, 13, 41), suggesting that the detachment of budding particles (pinching off) is less efficient. It has also been demonstrated that the p6Gag domain can function as a late-budding domain in Rous sarcoma virus to replace the authentic Rous sarcoma virus late-budding domain (22, 38).
By studying point mutations and a truncated form of the HIV-1 Gag molecule, we have found that the p6Gag domain of HIV-1 plays an important role in incorporating or retaining Pol proteins in the released virus particles. Failure to retain Pol proteins was observed in p6Gag mutant virions when HIV-1 protease was active. In contrast, efficient incorporation of the uncleaved Gag-Pol precursor molecules was detected in the p6Gag mutant virions when viral protease was mutated. Collectively, these data suggest that the p6Gag domain of HIV-1 Gag may play a role in retaining Pol in the assembling virus particle when viral protease is activated during virus budding.
Mutant constructs.
The parental HIV-1 construct used for this study (Fig. 1) was derived from the HXB2 clone (25). Several different mutant constructs were studied in order to evaluate the function of p6Gag (Fig. 1). Due to the frameshifting event occurring in the p1 region of the HIV-1 gag sequence, the p6Gag domain is present only in the Gag precursor (Fig. 1A); similarly, the Gag-Pol precursor contains Pol domains that are not present in the Gag precursor. The p6Gag truncation mutant (TAA) contains a premature stop codon immediately after p1 (41). In the LTALL mutant, leucine is substituted for proline at positions 7, 10, and 11 of p6Gag. The constructs PTAP−, PR−, and PR−/PTAP− (13) were obtained from E. Freed in order to compare the effects of mutations in p6Gag in the presence and in the absence of HIV-1 protease activity. The nucleotide sequence changes in TAA, LTALL, and PTAP− constructs are shown in Fig. 1B. Two amino acids expressed from the Pol open reading frame are altered due to mutations introduced into the TAA construct, whereas none have been introduced into the LTALL and PTAP− constructs.
Truncation of p6 resulted in impaired RT activity and aberrant Gag processing of released mutant virions.
COS-7 cells were transiently transfected with the various DNA constructs, and released virions were harvested by ultracentrifugation through a 20% sucrose cushion. Virus yield from the cells transfected with the p6 truncation mutant (TAA) was 70% of that obtained from the cells transfected with wild-type HXB2, as measured by p24 enzyme-linked immunosorbent assay (ELISA) (Fig. 2A). At the same time, the level of virion-associated RT activity in the supernatant of the TAA-transfected cells was 20-fold lower than that from HXB2-transfected cells (Fig. 2B). The results of these two assays suggested that the TAA mutant viruses had a severe defect in particle-associated RT activity. Immunoblotting of extracts from pelletable virions confirmed that there was indeed a defect in the amount of RT molecules in TAA mutant virions (Fig. 2C). Although RTp66, RTp51, and INp32 could be readily detected in the wild-type HXB2 virions by an HIV-1-positive human serum sample (Fig. 2C), none of these proteins were detected in the TAA virions (Fig. 2C). Furthermore, large quantities of unprocessed, p6-truncated Gag precursors (Fig. 2C) as well as partially processed Gag intermediate molecules p41 (p17 plus p24) and p25 (p24 plus p2) were detected in the TAA virions.
FIG. 2.
Analysis of virus production by p24 assay, RT assay, and immunoblot. Virions were purified and analyzed as previously described (40). (A) In the p24 assay, bars represent averages from five replicates and error bars show standard deviations; error bars for TAA are too small to be seen. (B) In the RT assay, bars represent averages from triplicates and error bars show standard deviations; error bars for COS-7 and TAA are too small to be seen. (C and D) For immunoblots, viral lysates were separated by SDS-12% PAGE, transferred onto nitrocellulose filters, and blotted with an HIV-1-positive human serum. (D) Alternatively, lysates were blotted with a MAb against HIV-1 RT (purchased from Biotechnology Transfer, Inc. [Columbia, Md.]) or an antiserum to HIV-1 IN (catalog no. 757; obtained through the AIDS Research Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases). The arrow indicates the position of the p6-truncated Gag precursor molecule. Lane C, viral lysate from COS cells transfected with an HIV-1 protease-mutant virus which contains only unprocessed Pr55Gag and Pr160Gag-Pol precursors.
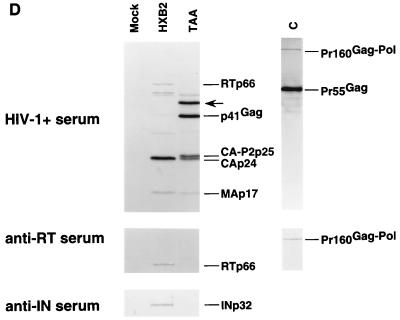
It is less likely that the p24 ELISA overestimated the amount of released TAA mutant viral proteins, as previous studies have suggested that the p24 ELISA tends to underestimate the amount of uncleaved Gag molecules compared to that of cleaved mature p24 molecules (27). To address this question further, we repeated the immunoblotting experiment by using more TAA mutant virion proteins so that the cleaved p17 was comparable to that seen in the wild-type virions (Fig. 2D). Under these conditions, it is likely that more mutant TAA virions than wild-type virions were used, as indicated by significantly more uncleaved or partially cleaved Gag molecules in the TAA mutant. At the same time, significantly reduced levels of RTp66 or INp32 were still observed in the TAA mutant virions compared to those in the wild-type HXB2 virions. No unprocessed Gag-Pol precursors, Pr160Gag-Pol, were detected in the TAA mutant virions by the HIV-1-positive human serum (Fig. 2D, upper panel) or the monoclonal antibody (MAb) against HIV-1 RT (Fig. 2D, middle panel). At the same time, both the HIV-1-positive human serum and the MAb against HIV-1 RT detected the unprocessed Pr160Gag-Pol in virions of an HIV-1 protease mutant (Fig. 2D, lane C), suggesting that the reduced levels of RT and integrase (IN) proteins in TAA mutant virions were not due to reduced processing of Pr160Gag-Pol. All these observations were consistent with the interpretation that lower concentrations of Pol proteins are present in the TAA mutant virions.
It is possible that mutations in p6 reduced the synthesis or stability of the Gag-Pol proteins or reduced the incorporation of Pol proteins into p6 mutant virions. Since p6 is not part of the Gag-Pol precursor, it seemed unlikely that truncation of this domain would affect the stability of the Gag-Pol molecules. Furthermore, the mutations that truncated the p6 domain are downstream from the RNA stem-loop structure that has been shown to be important for ribosomal frameshifting and therefore for synthesis of the Gag-Pol fusion protein (16). However, it was still possible that mutations in TAA had changed the RNA secondary structure and therefore reduced the synthesis of the Gag-Pol proteins.
Truncation of p6 did not affect intracellular expression of Pol proteins.
To determine whether Pol protein expression is reduced by the TAA mutation, we compared the intracellular levels of Pol proteins in TAA- and HXB2-transfected COS-7 cells. Cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, and reacted with the HIV-1-positive human serum or the MAb against HIV-1 RT. Comparable amounts of gp160 and gp120 were detected in the TAA- and HXB2-transfected cells by the HIV-1-positive human serum, suggesting that the transfection efficiencies were comparable (Fig. 3, upper panel). The HIV-1-positive human serum also detected the precursor Gag p55 and intermediate molecules p41 (p17 plus p24) and p24 in the HXB2-transfected COS-7 cells (Fig. 3, upper panel). The same human serum detected a truncated Gag precursor molecule (Fig. 3, upper panel), the intermediate molecule p41 (p17 plus p24), and a significant amount of p25 (p24 plus p2) in the TAA-transfected COS-7 cells. It is noteworthy that a 55-kDa band was detected in the TAA-transfected COS-7 cells. This protein is likely to be a virus-related protein, since it was not detected in the mock-transfected cells. However, this protein did not react with an anti-p6 antibody (data not shown), suggesting that it is not a full-length Gag polyprotein. Also, this protein did not react with the MAb against HIV-1 RT (Fig. 3, lower panel) or with a polyclonal antiserum against HIV-1 IN (data not shown). The precise nature of this protein remains to be characterized.
FIG. 3.
Immunoblot of intracellular viral proteins in transfected COS-7 cells. Cell lysates from mock-, HXB2-, and TAA-transfected COS-7 cells were electrophoresed and transferred to nitrocellulose filters. Lysates on one filter were blotted with the human HIV-1-positive serum (upper panel), and those on the other were blotted with the MAb against HIV-1 RT (lower panel). Arrow in upper panel points to a truncated Gag precursor molecule.
Similar levels of Pr160Gag-Pol, as well as of mature RTp66 and RTp51, were detected in the TAA- and HXB2-transfected cells by the MAb against HIV-1 RT (Fig. 3, lower panel). In the control samples, Pr160Gag-Pol, RTp66, and RTp51 were not detected in the mock- and ΔPol-transfected cells by the MAb against HIV-1 RT. The ΔPol construct contains a complete deletion of the pol gene, begining downstream of the stop codon in gag. These results indicated that truncation of p6Gag did not decrease the steady-state levels of Gag-Pol polyproteins in TAA-transfected COS-7 cells and did not appreciably inhibit the processing of Pr160Gag-Pol in the transfected COS-7 cells.
Point mutations of the PTAPP motif in p6 also reduced the level of Pol proteins in mutant virions.
Although the lengths of the C-terminal Gag domains (the p6 homologs) vary significantly among the lentiviruses, a conserved motif P(T/S)APP is found in this domain of every lentivirus except equine infectious anemia virus. To examine whether this conserved motif in p6 is required for incorporation or retention of Pol proteins, the three conserved prolines of PTAPP in HIV-1 p6 were changed to leucines (LTALL) (Fig. 1). Wild-type or p6-mutant LTALL virions were obtained from transfected COS-7 cells as described above. The amounts of viral proteins used for immunoblotting were normalized by p24 ELISA. As was observed for the p6 truncation mutant TAA, LTALL mutant virions also displayed reduced cleavage of Gag polyproteins in transfected COS-7 cells as well as in released virions (Fig. 4A). Higher quantities of unprocessed Pr55Gag (Fig. 4A), p41Gag, and p25 were detected in the LTALL mutant virions than in the parental PTAPP virions (Fig. 4A). At the same time, we observed a reduction in RTp66 in the p6 mutant LTALL virions compared to that in the wild-type virions. The virion-associated RT activity of the LTALL mutant was approximately 10-fold lower than that of the parental PTAPP virions when comparable amounts of p24 were used (data not shown).
FIG. 4.
Intracellular and virion-associated viral proteins from transfected COS-7 cells. (A) Cell and viral lysates (left and right panels, respectively) were obtained from mock-, wild-type-, and LTALL-transfected COS-7 cells and were blotted with the human HIV-1-positive serum. (B) Viral lysates were obtained from mock-, wild-type-, and PTAP−-transfected COS-7 cells and blotted with the human HIV-1-positive serum (upper panel) or the MAb against HIV-1 RT (lower panel). (C) Viral lysates were obtained from mock-, PR−-, and PR−/PTAP−-transfected COS-7 cells and were blotted with the human HIV-1-positive serum (upper panel) or the MAb against HIV-1 RT (lower panel). The amounts of viral proteins (p24 equivalent) for PR− and PR−/PTAP− are indicated.
As was observed for the p6Gag truncation mutant TAA and the p6Gag point mutant LTALL, another p6Gag point mutant, PTAP−, which contains four amino acid substitutions (LIRL for PTAP) in p6 (Fig. 1), also showed reduced incorporation of Pol proteins (Fig. 4B). The amounts of viral proteins used for immunoblotting were again normalized by the levels of p24. When both the wild-type and PTAP− constructs were compared, reduced levels of RTp66 and RTp51 in the PTAP− virions were again observed, as indicated by reactivity with the MAb against HIV-1 RT (Fig. 4B, bottom panel).
Mutation of protease resulted in the detection of Gag-Pol precursor in p6Gag mutant virions.
Mutations in p6Gag could inhibit interaction between Gag and the Gag-Pol precursors during virus assembly and therefore reduce the incorporation of Pol proteins into mutant virions. However, if mutations in p6Gag do not affect interaction between Gag and Gag-Pol precursors, one would expect to detect Gag-Pol precursor molecules in the released p6Gag mutant virions. To address this question, we compared the level of Gag-Pol precursor molecules in the released virions of the same p6Gag mutant (PTAP−) in the context of an HIV-1 protease mutation (PR−/PTAP−) to that in an HIV-1 protease mutant that has wild-type p6Gag (PR−). These mutants have been described previously (13).
After transfection of COS-7 cells, both PR− and PR−/PTAP− released virions that contained uncleaved Pr55Gag but not p24 or p17, as detected by the human HIV-1-positive serum (Fig. 4C, top panel), suggesting that HIV-1 protease was inactive. Only Pr160Gag-Pol was detected in the PR− and PR−/PTAP− virions by the human HIV-1-positive serum (Fig. 4C, top panel) or the MAb against HIV-1 RT (Fig. 4C, bottom panel), suggesting that mutation in the protease also prevented Pr160Gag-Pol cleavage. Two different amounts of PR− virions (2 and 1 μg of p24 equivalent) were analyzed to indicate that our immunoblot was not saturated. Comparable levels of Pr160Gag-Pol were detected in the PR− and PR−/PTAP− virions (Fig. 4C, lanes 2 and 4) when Gag molecules were normalized. These data indicated that although a mutation in p6Gag reduced the level of Pol proteins in the mutant virions when viral protease was active, the same p6Gag mutation did not reduce the incorporation of Pr160Gag-Pol precursors when viral protease was inactivated. Since incorporation of Pr160Gag-Pol requires interaction between Pr55Gag and Pr160Gag-Pol precursors, these results suggest that mutations in p6Gag did not significantly affect the interaction between Gag and Gag-Pol precursors when the viral protease was inactivated by mutations.
Conclusion.
In this study we have observed that mutations in a region of HIV-1 Gag that is not shared between the Gag and Gag-Pol precursor molecules can have a drastic effect on the content of Pol proteins in the released virions. Truncation of the entire p6Gag domain or substitutions of conserved amino acids in the p6Gag PTAPP motif drastically reduced Pol proteins in the mutant virions. The defects observed with the p6Gag mutants were not due to reduced expression of Pol proteins, since comparable Pol proteins were detected in wild-type- and p6Gag mutant-transfected COS cells. Although data presented here were derived from COS cells, similar observations have also been made with transfected HeLa cells, suggesting that the phenomenon of reduced Pol proteins in these p6-mutant virions is not unique to COS cells. It is unlikely that mutations in p6 would affect the structure of the Gag-Pol molecule, since p6 is not part of Gag-Pol. Mutations introduced into the TAA construct also changed two amino acids in the Pol region. However, it is less likely that the observed defect in the TAA mutant virions was a result of these changes, since a similar defect was also observed in the LTALL and PTAP− mutant virions, which did not have any amino acid changes in the Pol region.
Mutations in p6 did not inhibit interaction among the mutant Gag molecules themselves, as judged by efficient assembly of mutant Gag particles. It is possible that mutations in p6 changed the Gag structure in a way that affected the interaction between Gag and Gag-Pol molecules. However, this possibility is less likely because when the viral protease was inactivated by mutation, the Gag-Pol precursor was efficiently detected in the p6Gag-mutant virions, suggesting that the interaction between the p6-mutant Gag and the Gag-Pol precursors was not significantly affected. Therefore, viral-protease activity was apparently responsible for the observed reduction in Pol proteins in the p6-mutant virions. A link between the role of p6Gag in particle production and protease activity has also been reported previously (13).
At least three possible explanations might account for the observed defects in the p6Gag mutants. First, the Pol proteins may be incorporated into p6Gag-mutant virions and subsequently degraded in the released virions. This occurs only when the viral protease is active. However, we think that this is less likely, since degradation of Pol proteins by viral protease is unprecedented. Also, we did not observe any degraded RT- or IN-related proteins in the p6Gag-mutant virions.
Second, it is possible that the p6Gag domain of HIV-1 Gag interacts directly or indirectly with a region in Pol. Interaction between Gag and Gag-Pol precursors requires shared Gag domains, such as CA, between the two precursors (12, 14, 29, 31). After activation of HIV-1 protease at the plasma membrane (17, 33), the Pol region will be separated from the Gag domain. An interaction between p6Gag and Pol may be required to retain the Pol proteins in the virus assembly complex prior to the completion of virus budding. In the case of the protease mutant, separation of Pol from Gag will not occur during virus assembly. Therefore, an independent interaction between p6Gag and the Pol region will not be necessary. It is worth noting that mutations in RT (2, 21) or IN (1, 7) of HIV-1 have been demonstrated to affect the Pol protein contents in the released virions when the viral protease is active. However, in the absence of viral protease activity, deletion of IN did not affect the incorporation of truncated Gag-Pol into released virions (3). Further study is necessary to determine whether p6Gag interacts directly with a region in Pol.
Third, recent evidence has suggested that the p6Gag region could play a role similar to that of other retroviral late-budding domains, which are thought to interact with various cellular proteins to facilitate efficient virus release (9, 38). Assembly, budding, and maturation of retroviruses are highly dynamic and tightly regulated processes (15, 33, 35). In general, the activation of retroviral protease is dependent on virus assembly and budding at the plasma membrane (33). Interaction between Gag and Gag-Pol molecules in HIV-1 may occur before they are targeted to the plasma membrane, since the nonmyristylated Gag-Pol precursor p160 is incorporated into virus-like particles as efficiently as is the myristylated Gag-Pol precursor (23, 30). However, the viral protease activity must be suppressed until virus assembly and budding are initiated at the plasma membrane. It is possible that the viral protease is activated prematurely in the p6Gag mutants, resulting in the loss of Pol proteins. Alternatively, when virus budding is delayed in the p6Gag mutants, the Pol region is separated from the Gag region of the Gag-Pol precursor prior to the completion of virus budding. If interaction between Gag and Gag-Pol precursors is solely dependent on the Gag regions, such as CA, that are shared between the two precursors (12, 14, 29, 31), the Pol proteins will be excluded from assembling p6Gag-mutant viruses if they have already been cleaved from the Gag-Pol precursor. The loss of viral protease from the assembly complexes due to delayed budding in the p6 mutants may be responsible for the incomplete processing of Gag molecules observed in the p6-mutant virions. The precise mechanism by which mutations in p6Gag reduce incorporation and/or retention of Pol proteins in released virions remains to be determined.
Acknowledgments
We are grateful to Robert Gorelick for supplying us with the HIV-1 p6Gag-mutant constructs from which we generated the LTALL construct and to Mingjun Huang, Malcolm Martin, and Eric Freed for the PTAP−, PR−, and PR−/PTAP− constructs. We thank Richard Markham and David Schwartz for critical reading of the manuscript. The antiserum to HIV-1 integrase (catalog no. 757) was obtained through the AIDS Research Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases.
This work was supported by grant AI-35525 from the National Institutes of Health. M.D. was supported in part by grant ES-07141 from the National Institutes of Health.
REFERENCES
- 1.Ahsari-Lari M A, Dohehower L A, Gibbs R A. Analysis of human immunodeficiency virus type 1 (HIV-1) integrase mutants. Virology. 1995;211:332–335. doi: 10.1006/viro.1995.1412. [DOI] [PubMed] [Google Scholar]
- 2.Ahsari-Lari M A, Gibbs R A. Expression of human immunodeficiency virus type 1 reverse transcriptase in trans during virion release and after infection. J Virol. 1996;70:3870–3875. doi: 10.1128/jvi.70.6.3870-3875.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukovsky A, Gottlinger H. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of active viral protease. J Virol. 1996;70:6820–6825. doi: 10.1128/jvi.70.10.6820-6825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubei A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crawford S, Goff S P. Mutations in gag proteins P12 and P15 of Moloney murine leukemia virus block early stages of infection. J Virol. 1984;49:909–917. doi: 10.1128/jvi.49.3.909-917.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darlix J-L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 7.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franke E K, Yuan H E H, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 9.Garnier L, Wills J W, Verderame M F, Sudol M. WW domain and retrovirus budding. Nature. 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 10.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson L E, Bowers M A, Sowder II R C, Serabyn S A, Johnson D G, Bess J W, Jr, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu H-W, Schwartzberg P, Goff S P. Point mutations in the P30 domain of the gag gene of Moloney murine leukemia virus. Virology. 1984;142:211–214. doi: 10.1016/0042-6822(85)90435-0. [DOI] [PubMed] [Google Scholar]
- 13.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang M, Martin M A. Incorporation of Pr160gag-pol into virus particles requires the presence of both the major homology region and adjacent C-terminal capsid sequences within the Gag-Pol polyprotein. J Virol. 1997;71:4472–4478. doi: 10.1128/jvi.71.6.4472-4478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 16.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-Pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan A H, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondo E, Mammano F, Cohen E A, Göttlinger H G. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J Virol. 1995;69:2759–2764. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y-L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Y-L, Bennett R P, Wills J W, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mak J, Khorchid A, Cao Q, Huang Y, Lowy I, Parniak M A, Prasad V R, Wainberg M A, Kleiman L. Effects of mutations in Pr160gag-pol upon tRNALys3 and Pr160gag-pol incorporation into HIV-1. J Mol Biol. 1997;265:419–431. doi: 10.1006/jmbi.1996.0742. [DOI] [PubMed] [Google Scholar]
- 22.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J, Morrow C D. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J Virol. 1992;66:6304–6313. doi: 10.1128/jvi.66.11.6304-6313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratner L, Haseltine H A, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Jr, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 26.Schlesinger S, Makino S, Linial M L. cis-acting genomic elements and trans-acting proteins involved in the assembly of RNA viruses. Semin Virol. 1994;5:39–49. doi: 10.1006/smvy.1994.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider R, Campbell M, Nasioulas G, Felber B K, Pavlakis G N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz M D, Geraghty R J, Panganiban A T. HIV-1 particle release mediated by Vpu is distinct from that mediated by p6. Virology. 1996;224:302–309. doi: 10.1006/viro.1996.0532. [DOI] [PubMed] [Google Scholar]
- 29.Schwartzberg P, Colicelli J, Gordon M L, Goff S P. Mutations in the gag gene of Moloney murine leukemia virus: effects on production of virions and reverse transcriptase. J Virol. 1984;49:918–924. doi: 10.1128/jvi.49.3.918-924.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith A J, Srinivasakumar N, Hammarskjöld M-L, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srinivasakumar N, Hammarskjöld M-L, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J Virol. 1995;69:6106–6114. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh C T, Sodroski J, Göttlinger H G. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 33.Vogt V M. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–132. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 34.von Schwedler U, Kornbluth R S, Trono D. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc Natl Acad Sci USA. 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wills J W, Craven R C. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Wilson W, Braddock M, Adams S E, Rathjen P D, Kingsman S M, Kingsman A J. HIV express strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 37.Wu X, Conway J A, Kim J, Kappes J C. Localization of the Vpx packaging signal within the C terminus of the human immunodeficiency virus type 2 Gag precursor protein. J Virol. 1994;68:6161–6169. doi: 10.1128/jvi.68.10.6161-6169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang Y, Cameron C E, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu S F, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]
- 40.Yu X-F, Yu Q-C, Lee T-H, Essex M. The C terminus of human immunodeficiency virus type 1 matrix protein is involved in early steps of the virus life cycle. J Virol. 1992;66:5667–5670. doi: 10.1128/jvi.66.9.5667-5670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu X-F, Matsuda Z, Yu Q-C, Lee T-H, Essex M. Role of the C terminus Gag protein in human immunodeficiency virus type 1 virion assembly and maturation. J Gen Virol. 1995;76:3171–3179. doi: 10.1099/0022-1317-76-12-3171. [DOI] [PubMed] [Google Scholar]