Abstract
Gastrointestinal cancer (GIC) is the most prevalent and highly metastatic malignant tumor and has a significant impact on mortality rates. Nevertheless, the swift advancement of contemporary technology has not seamlessly aligned with the evolution of detection methodologies, resulting in a deficit of innovative and efficient clinical assays for GIC. Given that exosomes are preferentially released by a myriad of cellular entities, predominantly originating from neoplastic cells, this confers exosomes with a composition enriched in cancer-specific constituents. Furthermore, exosomes exhibit ubiquitous presence across diverse biological fluids, endowing them with the inherent advantages of non-invasiveness, real-time monitoring, and tumor specificity. The unparalleled advantages inherent in exosomes render them as an ideal liquid biopsy biomarker for early diagnosis, prognosticating the potential development of GIC metastasis.
In this review, we summarized the latest research progress and possible potential targets on cancer-derived exosomes (CDEs) in GIC with an emphasis on the mechanisms of exosome promoting cancer metastasis, highlighting the potential roles of CDEs as the biomarker and treatment in metastatic GIC.
Keywords: Exosomes, Metastasis, Biomarker, Gastrointestinal cancer
Introduction
According to the latest statistical data on cancer, 1,918,030 new cases and 609,360 cancer-related deaths (CRDs) had been documented. Among them, the GIC ranks second but is responsible for about 28% of cancer-related deaths. Colorectal cancer (CRC) is the most prevalent GIC which accounts for 44% of all cancer cases, followed by pancreatic cancer (PC) which has a 5-year survival rate of less than 11% and is the most lethal cancer [1]. Furthermore, global GIC morbidity and mortality continue to rise [1]. Metastatic cancers are in charge of 90% of CRDs [2, 3], and metastatic GIC is characterized by high aggressiveness and heterogeneity which take primary responsibility for death in patients with malignant GIC [4]. While diverse diagnostic methods, including gastroscope, computed tomography (CT) scan, and pathological examinations [4, 5], are vital for terminal-stage cancer with conspicuous symptoms, the main challenge in GIC diagnosis stems from the restricted sensitivity of these technical approaches to small lesions in pre-metastatic GIC or residual masses post radiotherapy and chemotherapy. In addition, some methods own limitations on incursion and potential of transmission. Current therapeutic targets and diagnostic biomarkers still do not meet the clinical needs of the goal of treatment for GIC. Presently, there is an urgent need for novel diagnostic methods with accurate detection rates and high-quality specificity, especially in cases of pre-metastatic PC.
As the liquid biopsy undergoes continual expansion and refinement, it was not until very recently that exosomes as the biomarkers for cancer metastasis detection have been appreciated. Exosome, initially elucidated in 1983 by Johnstone et al. [6, 7], is a subset of extracellular vesicles (EVs) with sizes ranging from 40 to 160 nm. They are ubiquitously secreted by nearly all cell types, with a particular emphasis on cancer cells [8]. Initially, researchers considered that exosomes are just the lipid bilayer to transport cellular metabolic waste [6]. However, our comprehension of exosomal function has been substantially deepened and broadened over the past decades. As research dug deeper, massive studies revealed that exosomes are essential for the intercellular communication from cancer cells to neighboring stromal and immune cells to promote tumor metastasis via mediating immune evasion, facilitating pre-metastatic niche (PMN) formation, angiogenesis, and epithelial-mesenchymal transition (EMT) [9–12].
In this review, we intend to present a comprehensive overview on how GIC-derived exosomes promote cancer metastasis through participating in PMN formation, immune evasion, angiogenesis, and EMT. Moreover, founded on the culmination of exosomes as discerning indicators within the context of liquid biopsy, we predominantly summarize the exosomes derived from CRC, hepatocellular carcinoma (HCC), gastric cancer (GC), and PC as pivotal prognostic, diagnostic, and predictive biomarkers for metastatic GIC.
The mechanisms of CDEs-related cancer metastasis
In 1889, Stephen Paget proposed a promising and unprecedented hypothesis of metastasis which is “seed and soil” and revealed that the metastasis of cancer is not random but the interactions between 'seeds' (the cancer cells) and the 'soil' (the host microenvironment) [13, 14]. Several studies have intensively illustrated that the pending metastasis site could induce the formation of tumor-friendly microenvironment which are termed PMNs, to suit cancer cell growth [15]. In this process, the medium is exosomes, which means that exosomes carry different cargos to mediate cell-to-cell communication which can assist primary cancer in selectively modifying the target organs of future metastases. Meanwhile, exosomes can reach target organs to increase vascular permeability, remodel stromal cells and extracellular matrix (ECM), and change the immune system before metastasis occurs [14, 16–18].
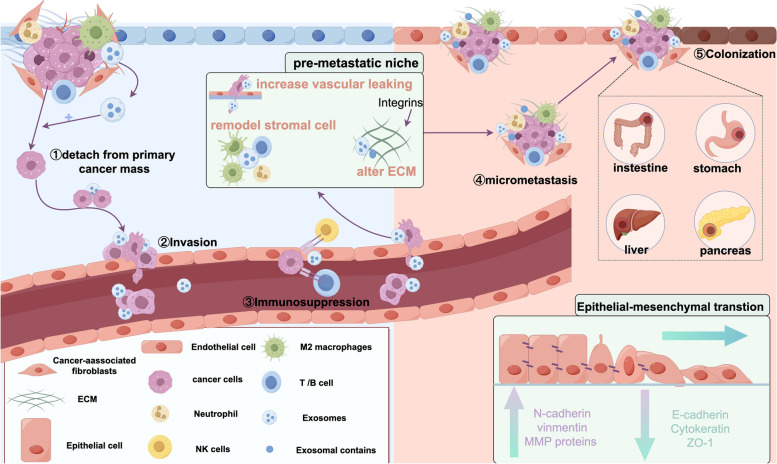
Metastasis, also termed the metastatic cascade, is categorizable into three distinct and overlapping phases—dissemination, dormancy, and colonization [19]. And exosomes assume a pivotal role in the intricate process of tumor metastasis: ①Aided by CDEs, cancer cells detach from the primary cancer mass. ② CDEs can assist cancer cells in passing the bloodstream or lymphatic vessels to infiltrate surrounding or target tissues. ③CDEs induce immunosuppression and escape from immune surveillance by interaction with immune cells [20]. ④Exosomes carry a bunch of proteins, integrins (ITGs), RNA, and DNA to alter the ECM, re-organize the target organ structure and vasculature, recruit stromal cells to facilitate the target organ, and form the tumor-friendly microenvironment or PMNs. ⑤CDEs activate EMT to increase invasion ability and promote migration (Fig. 1).
Fig. 1.
The mechanisms of CDEs in cancer metastasis. CDEs are involved in the processes of assistant tumors to depart from primary site, immune evasion, angiogenesis, increasing vascular permeability, the formation of pre-metastatic niche (PMN), epithelial-mesenchymal transition (EMT), planting on distant organs, and promoting cancer metastasis
These detailed and in-depth studies provide us with a comprehensive reference for understanding the role of exosomes in the metastasis, deterioration, and treatment resistance of tumors. However, the most pivotal questions that remain unresolved are how and to what extent the CDEs work in the process of cancer metastasis between the receptors and distinct sites.
Exosomal biogenesis
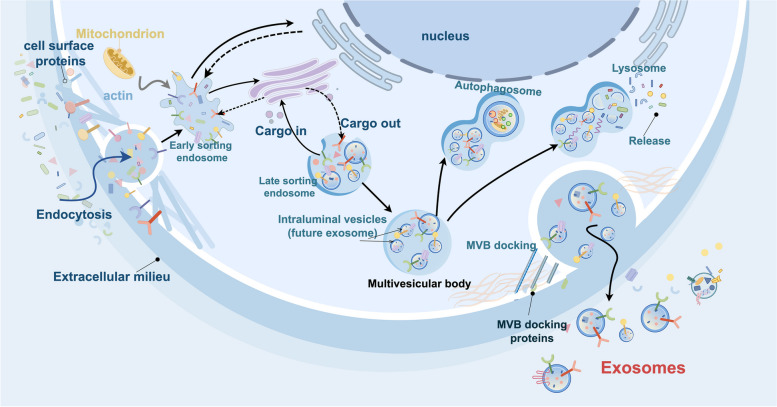
Exosomal biogenesis involves several steps: ①Facilitated by Golgi complex, the plasma membrane undergoes invagination to form the cup-shaped structure called early-sorting endosome (ESE) which contains the receptors, proteins, and others. ②ESEs maturation and conversion to late-sorting endosomes (LSEs). Both ESEs and LSEs can exchange substances with Golgi and Nucleus. ③LSEs then generate multivesicular bodies (MVBs), which are formed by endosomal limiting membrane inward invagination. Because of this process, MVBs contain several intraluminal vesicles (ILVs), the future exosomes. ④MVBs combine with MVB docking proteins to release exosomes; otherwise, the MVBs will degrade if they fuse with lysosomes or autophagosomes [8, 21]. This process whether the exosome will be released or degraded by lysosomes or autophagosomes, is determined by the endosomal sorting complexes required for transport (ESCRT) which contribute to exosome biogenesis [22] (Fig. 2).
Fig. 2.
The biological roles of CDEs in gastrointestinal cancer
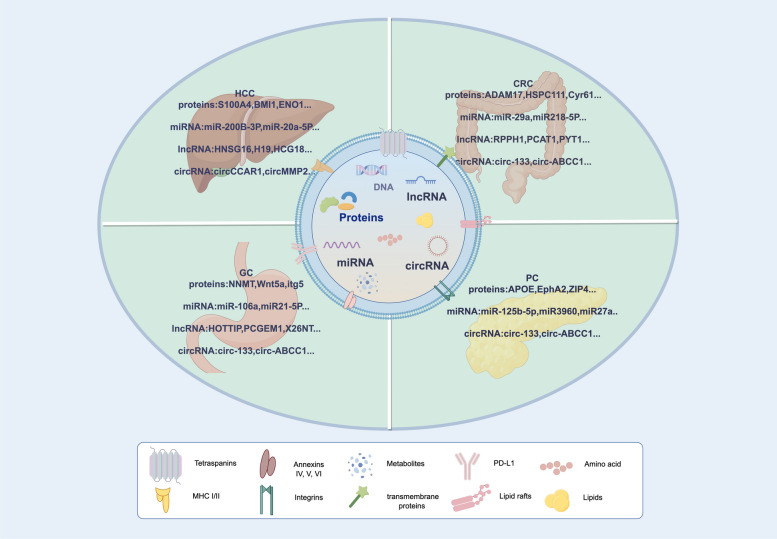
Theoretically, exosomes are produced by the majority of cells [23], but the quantity of CDEs significantly surpasses that of healthy cells [12]. Riches A et al. demonstrated that within 24 h, compared with exosomes released from normal cells ((4.5 ± 2.3) × 108 exosomes per 106 cells), exosomes secreted by cancer cell lines are (53.2 ± 1.6) × 108 per 106 cells, which are obviously higher than normal cells derived exosomes [24]. Moreover, the content of exosomes is highly variable between cancer and normal tissue sources, and depending on cell origin, exosome cargo from cancer cells can be altered [8, 12, 25, 26]. Melo et al. found that exosomes derived from breast cancer cells contain massive microRNAs (miRNAs) which are way more than exosomes released from normal cells [26]. Meanwhile, in different cell types, exosomes might have diverse functions. For instance, some exosomes can interact with host immune cells, including macrophages, B cells, and T cells, to promote cancer metastasis, immune invasion, and transfer antigens to dendritic cells [27, 28]. The bioactive molecules in exosomes mirror the pathological state of the cells and tissues they obtained from, reflecting the composition of the donor cell [23, 29]. Exosomes contain a complex of diverse proteins, including receptors, transcription factors, enzymes, GTPases, annexins, cell-surface proteins, cytosolic proteins, ANNEXIN, RAB proteins, ITGs, tetraspanins (CD9, CD81, CD63, and CD82), heat shock proteins (HSP90, HSP70), and MHC proteins; DNA; different RNAs, including miRNAs, circular RNAs (circRNA), long non-coding RNAs (lncRNAs), as well as ribosomal RNAs; lipids; and metabolites [22, 25, 27, 30, 31] (Fig. 3).
Fig. 3.
The hallmarks of CDEs in gastrointestinal cancer
Pre-metastatic niche
The pre-metastatic niche (PMN) refers to the microenvironment orchestrated by the primary tumor in distant metastases which foster a conducive milieu to the colonization of tumor cells. Research has elucidated that the prerequisite for PMN initiation is that exosome wrap the bioactive substances released by primary tumor cells, breach the tumor invasion barriers, and directly transported through the blood circulation to reach the site of metastasis, thereby instigating local PMN formation [32]. PMN plays a pivotal role in contributing to metastatic colonization, and mainly includes six components: immunosuppression, vascular permeability/angiogenesis, organotropism, lymphangiogenesis, inflammation, and reprogramming [33]. Within this context, the immunosuppression at the metastatic site is regarded as a pivotal initiation step for the establishment of PMN, which primarily formation through ①intricate interplay of CDEs interacting with stromal cells. ②recruit, silence immune cells, downregulate the expression of immune cells surface antibody, and inhibit immune cell activation. CDEs block the T cell signaling cascade to restrain T cell activation [32, 34]. ③CDEs recruit vascular endothelial cell to promote PMN formation to enhanced cancer proliferation and metastasis [35]. ④reprogram ECM. CDEs can carry bioactive factors to promote ECM remodeling [14]. ⑤infiltrate tumor cells (Fig. 1) [19]. Overwhelming evidence shows that exosomes and their contents, especially from highly malignant tumor optimize the pre-metastatic microenvironment for GIC colonization, outgrowth, and migration by helping the metastatic cells to escape immune surveillance, transporting inflammatory factors, and increasing vascular permeability [36–38]. Recently, Shao Y et al. demonstrated that exosomal miR-21 can specifically deliver to the liver and facilitate organ-specific, distant metastasis by establishing an inflammatory PMN [39]. Meanwhile, Bruno et al. reported that the migration inhibitory factor (MIF) is highly expressive in pancreatic ductal adenocarcinoma (PDAC) derived exosomes which will be uptake by liver Kupffer cells (KCs) to release transforming growth factor beta (TGFβ) to activate fibrotic pathways to promote liver PMN formation and metastasis [38]. Zhou et al. reported that miR-105-containing exosomes efficiently destroy the tight junctions and integrity of endothelial monolayers to reach ideal organs to increase vascular permeability, thereby facilitating metastasis [40].
Cancer-associated fibroblasts
It is clear that cancer-associated fibroblasts (CAFs), an integral part of tumor microenvironment (TME), are vital contributors to cancer progression and metastasis [41]. Not only do the CAFs provide physical support for epithelial cells, but they also secrete a variety of cell growth factors, inflammatory ligands, numerous chemokines and cytokines, and ECM proteins, such as HGF, CXCL12, insulin-like growth factor (IGF), epidermal growth factor (EGF), IL-8, IL-6, and IL-11, which are carried by exosome to influence EMT; interact with cancer cells to increase cancer proliferation, migration, invasion; induce immunosuppression and chemotherapy resistance; and remold ECM [42]. Wang et al. found that CAFs tend to uptake miR-146a-5p and miR-155-5p via exosomes to promote CRC metastasis via activating NF-κB/JAK2–STAT3 signaling to upregulate the expression of inflammatory cytokines including tumor necrosis factor-α, interleukin-6, CXCL12, and TFG-β which trigger EMT and facilitate CRC distant metastasis [43]. Recently, a study demonstrates that CAF-derived exosomes containing miR-20a-5p exert functions on regulating the TME of HCC to facilitate HCC metastasis, and EMT procession by targeting the Wnt/β-Catenin signaling pathway [44].
Epithelial-mesenchymal transition
EMT is a biological process that can significantly enhance the ability of cell migration, which means cells lose the characteristics of epithelial cell polarity and intercellular adhesion capacity and transform into mesenchymal phenotype cells with high migration and invasion ability. EMT can implicate both physiological (including embryogenesis, wound healing, tissue regeneration, and fibrosis) and pathological processes (including cancer cell proliferation, invasion, migration, and metastasis) [45]. During the cancer microenvironment, with the alteration of ECM by CAFs, exosomes carry a large cohort of cytokines and chemokines to influence the cell–cell interaction to promote tumor progression and metastasis [46]. Zhang et al. have demonstrated that exosomal circNRIP1 can be propagated by exosomal communication between GC cells and promote cancer proliferation, migration, invasion, and metastasis by activating AKT/mTOR axis which imposes a positive impact on EMT [47]. Of note, a recent study has reported that HCC can enhance cancer proliferation and migration by secreting exosomal circ-0004277 to inhibit ZO-1 and stimulate EMT of peripheral cells through intercellular communication [48].
Integrins
ITGs, which are the large cell adhesion receptor family and contain α and β two transmembrane glycoproteins, relate to the regulation of ECM to interact with ECM proteins and participate in cell adhesion procession. Consolidated evidence has demonstrated that CDEs express particular ITGs, including α1β1, α2, α2β1, αvβ3, αvβ5, αvβ6, α4, α4β1, α5β1, α6β1, α6β4, and α9β1, which can address CDEs into specific organs and target cells in a tissue-specific fashion and interact with ECM proteins to initiate PMNs formation and promote cancer cell proliferation, invasion, migration, and metastasis [49]. Functionally, ITGs educate cancer metastasis with two novel cooperative mechanisms: selection of target tissues to form new tumor niches during metastatic spread by ITGs carried on the exosome’s surface and horizontal transfer of integrin transcripts as vesicle cargo [18, 49, 50].
Plenty of studies have highlighted that exosomal ITGs play a crucial role in cancer progression by directly fusing with receptors in tissue-specific manner, thus proposing PMN formation and mediating organ-specific colonization [18]. Of note, exosomal ITGs α5β1, α2, αvβ3, and αvβ5 were linked to liver metastasis by modifying the local microenvironment. Oliver et al. suggested that inhibiting the expression of integrin α5β1 can significantly decrease tumor cell proliferation and reduce liver metastasis formation by suppressing the activation of endothelial cells [51]. Yoshimura et al. analyzed the primary CRC patients coexisting liver and lung metastasis and revealed that ITGs α2 expression leads to liver metastases by binding to collagen type IV [52].
Immunosuppression
Immune system is the most powerful barrier to cancer metastasis. Massive studies have demonstrated that, within the tumor local inflammatory microenvironment, CDEs can transport proteins similar to parent tumor cells, proinflammatory chemokines, and cytokines, such as IL-35, IL10, TNF-α, IFN-γ, and TGF-β2, genomic DNA, mRNA, and microRNAs to interact with immune cells, including B cells, T cells, natural killer (NK) cells, regulatory T cells (Tregs), and tumor-associated macrophages (TAMs), thus weakening antitumor immune responses and escaping from immune surveillance [53, 54]. CDEs can ①influence immune cell maturity: impair DC maturation by influencing IL-6 expression [55], ②inhibit immune cell proliferation: impact T cell proliferation by targeting TGF-β and NK cells proliferation and cytotoxic functions via downregulating NK associated proteins [56, 57], ③inhibit the functions necessary for antitumor responses, ④induce apoptosis of activated immune cells, ⑤suppress immune cells activity, ⑥interfere with monocyte differentiation, ⑦skew the differentiation of myeloid precursor cells, ⑧polarize immune cells into tumor-promoting phenotypes [53]. Extracellular circRNAs can mediate the interaction between GIC cells and immune cells including neutrophils, NK cells, and TAM [29]. Wang et al. reported that downregulating HCC cell-derived exosomal has_circ_0074854 can inhibit HCC invasion and migration by inhibiting M2 polarization of macrophages [58]. Chen et al. reported that exosomes pack immunosuppressive proteins such as program death ligand 1 (PD-L1) on the surface to suppress CD8 + T cells function and facilitate cancer progress [59]. HCC cell-derived exosomal circTMEM181 contributes to immunosuppressive microenvironment formation, upregulates CD39 expression in macrophages, and is resistant to anti-programmed cell death 1 (PD1) treatment in HCC [60].
Exosomes as prognostic, diagnostic and predictive biomarkers for GIC metastases
Constantly, the gold standard for the diagnosis of GIC and metastasis is image assessment combined with tissue biopsy. To date, for CRC and GC identification the gold standard is colonoscopic tissue biopsy under anesthesia [61]. In the case of HCC and PC, the established gold standard is CT-guided puncture biopsy. Elevated malignancy and the intrinsic tumor heterogeneity constitute the principal determinants underlying GIC recurrence. However, in order to surveillance of disease advancement, iterative biopsies are imperative, so patient compliance may decrease owing to the elevated risk linked to recurrent traumas and medical implantation. For patients who incapable of undergoing anesthesia and confronted with challenges in procuring biopsies from the ileocecum, duodenum, cephalic" and "caudal" regions of the pancreas, and difficult-to-biopsy patients, the feasibility of real-time monitoring of GIC progression is substantially diminished. The traditional diagnostic paradigm is undergoing transformation with the growing recognition of exosomes as indispensable elements in the diagnostic arsenal, furnishing a nuanced and comprehensive comprehension of metastases in GIC. In contrast, the swift progress in cancer examination has underscored limitations in the conventional approach of tissue biopsy. This invasive method, susceptible to advanced metastasis, confines its utility to a singular snapshot of cancer information, thereby inadequately capturing the intricate heterogeneity within the TME. On the opposite, exosomes exhibit notable advantages and stamp significant superiority over other liquid biopsy sources due to ① Consistent with the content mentioned earlier, CDEs transport a substantial protein, non-coding RNA, and DNA, facilitating intercellular communication and fostering cancer progression (Fig. 3) [22]. ② Exosomes exhibit extensive distribution within a variety of bodily fluids, thereby augmenting the practicality of their application in clinical disease assessments [23, 62]. ③ Exosomes possess high stability encapsulated by phospholipid bilayer membrane, which allows the researcher to store for a long time. ④ The approaches to get exosomes are simple, noninvasive, and real-time, so regular examinations do not cause a significant burden for patients, allowing for precise assessment of cancer progression and relapse [63, 64]. ⑤Exosomes show the potential for multicomponent analysis. ⑥Exosomes reflect the proteome and bio-information of the originating cell. ⑦Exosomes can present surface-specific proteins from parental cells or even target cells. ⑧Exosomes show superior performance over traditional serum-based biomarkers [25, 62]. As previously elucidated, established evidence underscores that GIC derived exosomes assume a critical role in mediating CRC, PC, HCC, and GC distant metastasis, especially invasive involvement with the liver, lungs, and brain. CDEs circular in biofluids and carry diverse GIC-specific bioactive constituents, (encompassing proteins, miRNA, circRNA, lncRNA, and active factor) to enhance the migratory capacity of GIC cells [65], assistant PMN and CAF formation which creates a hotbed for GIC colonization [39], cross-talking with immune cells, and destroy vascular endothelial barrier to advance vascular permeability [66]. Table 1 furnishes a comprehensive summary of different exosomal proteins, miRNAs, circRNAs, and lncRNAs between patients with CRC, HCC, GC, and PC serving as the minimally invasive and conspicuous heterogeneity source of potential diagnostic, prognostic, and predictive biomarkers.
Table 1.
Potential exosomal biomarkers in GIC metastasis and their mechanism
| Cancer type | Exosomal Contents | Exosome Source | Exosomal Cargo | Effect | Clinical Significance | Ref |
|---|---|---|---|---|---|---|
| colorectal cancer | miRNA | Cell culture fluid | miR-29a | downregulate the expression of ZO-1, Claudin-5, and Occludin via targeting KLF4 | promote metastasis | [66] |
| Cell culture fluid | miR-218-5p | activate the Ras/ERK/c-Fos pathway | promote metastasis | [110] | ||
| Cell culture fluid | miR-181b-3p | downregulate SNX2 expression | promote metastasis | [111] | ||
| serum | miR-92a-3p | inhibit FBXW7 and MOAP1 | promote metastasis | [104] | ||
| Cell culture fluid | miR-27b-3p | activate STAT3 pathway | promote metastasis | [113] | ||
| Cell culture fluid | miR-203a-3p | inhibit the Src/PKC/GSK-3β | promote metastasis | [108] | ||
| Serum | miRNA132-5p, miRNA6087, and miRNA320d | not mention | promote metastasis | [189] | ||
| Serum | miR-146a-5p and miR-155-5p | activate JAK2–STAT3/NF-κB signaling | promote metastasis | [43] | ||
| Cell culture fluid | miRNA-335-5p | overexpress of RASA1 | promote metastasis | [190] | ||
| Serum | miR-122 | not mention | promote metastasis | [191] | ||
| Serum | miR-141-3p and miR-375 | not mention | promote metastasis | [91] | ||
| Serum | miR-106b-3p | downregulate DLC-1 expression | promote metastasis | [92] | ||
| Serum | miR-1246/92b-3p/27a-3p | target GSK3β and activate the Wnt/β-catenin pathway | promote metastasis | [65] | ||
| Serum | miR-193a and let-7 g | not mention | promote metastasis | [192] | ||
| Serum | miR-128-3p | activate TGF-β/SMAD and JAK/STAT3 signal pathway | promote metastasis | [102] | ||
| Cell culture fluid | miR-106b-5p | downregulate PDCD4 and activate the PI3Kγ/AKT/mTOR signaling pathway | promote metastasis | [193] | ||
| Cell culture fluid | miR-21-5p | suppress VHL/ HIF-1α axis, target TrkA/ERK/ELK1 pathway | promote metastasis | [194] | ||
| Serum | miR-934 | downregulate PTEN expression and activate the PI3K/AKT signaling pathway | promote metastasis | [195] | ||
| Serum | miR-25-3p, miR-130b-3p, miR-425-5p | activate of the CXCL12/CXCR4/PTEN/PI3K/Akt pathway axis | promote metastasis | [196] | ||
| Serum | miR- 221/222 | suppress SPINT1 expression | promote metastasis | [94] | ||
| Serum | miR-1229 | inhibit the expression of HIPK2, activate VEGF pathway | promote metastasis | [197] | ||
| Cell culture fluid | miR-183-5p | suppress FOXO1 expression | promote metastasis | [112] | ||
| circRNA | serum | circPABPC1 | upregulate HMGA2, BMP4 and ADAM19 expression | promote metastasis | [139] | |
| Serum | circ_FMN2 | serve as miR-338-3p Sponge to downregulate MSI1 | promote metastasis | [198] | ||
| Cell culture fluid | circPACRGL | facilitate the TGF-β1 expression, regulate miR-142-3p/miR-506-3p-TGF-β1 axis | promote metastasis | [199] | ||
| Tissue | circLONP2 | not mention | promote metastasis | [140] | ||
| Cell culture fluid | circ-ABCC1 | activate the Wnt/β-catenin pathway | promote metastasis | [200] | ||
| serum | circ-133 | activate the miR-133a/GEF-H1/RhoA axis | promote metastasis | [136] | ||
| serum | circGAPVD1 | not mention | promote metastasis | [142] | ||
| serum | circTUBGCP4 | activate Akt signaling pathway, sponge with miR-146b-3p | promote metastasis | [135] | ||
| serum | hsa-circ-0004771 | not mention | promote metastasis | [201] | ||
| Cell culture fluid | ciRS-122 | ciRS-122-miR-122-PKM2 pathway | promote metastasis | [151] | ||
| serum | circCOG2 | Activate the miR-1305/TGF-β2/SMAD3 pathway | promote metastasis | [202] | ||
| Cell culture fluid | circLPAR1 | suppress the METTL3-eIF3h interaction, decrease the translation of oncogene BRD4 | promote metastasis | [203] | ||
| lncRNA | Cell culture fluid | lnc-HOXB8-1:2 | bind hsa-miR-6825-5p to upregulate CXCR3 expression | promote metastasis | [204] | |
| Cell culture fluid | lncRNA RPPH1 | interact with protein TUBB3 | promote metastasis | [122] | ||
| serum | lncRNA PVT1/VEGFA | downregulate miR-152-3p | promote metastasis | [120] | ||
| Cell culture fluid | lncRNA PCAT1 | activate of the miR-329-3p/Netrin-1-CD146 Complex | promote metastasis | [127] | ||
| Cell culture fluid | lncRNA BANCR | regulate RhoA/ROCK signaling | promote metastasis | [205] | ||
| Cell culture fluid | lncRNA MALAT1 | sponge miR-26a/26b and activate PI3K/Akt/mTOR pathway | promote metastasis | [128] | ||
| Cell culture fluid | lncRNA UCA1 | sponge with miR-143 | promote metastasis | [117] | ||
| proteins | serum | ADAM17 | cleaving the E-cadherin junction | promote metastasis | [69] | |
| serum | HSPC111 | activate CXCL5-CXCR2 axis | promote metastasis | [68] | ||
| serum | KRAS | not mention | promote metastasis | [71] | ||
| serum | Cyr61 | activate αV β5 /FAK/HIF-1α/STAT3/MMP2 signaling | promote metastasis | [83] | ||
| Cell culture fluid | FMNL2 | activate the EGFL6/CKAP4/ERK axis | promote metastasis | [84] | ||
| Cell culture fluid | ADAM17 | not mention | promote metastasis | [70] | ||
| hepatocellular cancer | miRNA | serum | miR-200b-3p | activate JAK/STAT signaling pathway | promote metastasis | [95] |
| liver tissue | miR-20a-5p | downregulate LIMA1-Mediated β-Catenin Pathway | promote metastasis | [44] | ||
| Cell culture fluid | miR-412,36 miR-4286,37 miR-423–5p, and miR-29a-5p, | impair lysosome biogenesis | promote metastasis | [206] | ||
| Cell culture fluid | miR-452-5p | downregulate TIMP3 | promote metastasis | [207] | ||
| serum | miR92a-3 | inhibit PTEN and activating Akt/Snail signaling | promote metastasis | [107] | ||
| Cell culture fluid | miR-21 and miR-10b | not mention | promote metastasis | [208] | ||
| serum | miR-18a, miR-20b, and miR-221 | not mention | promote metastasis | [96] | ||
| serum | miR-1307-5p | downregulate miR-1307-5p/SEC14L2/Akt and miR-1307-5p/ENG signaling pathways | promote metastasis | [97] | ||
| Cell culture fluid | miR-1273f | activate the Wnt/β-catenin signaling, and downregulate LHX6 | promote metastasis | [209] | ||
| Cell culture fluid | miR-100-5p | downregulate CLDN11, activate PI3K/AKT signaling pathway | promote metastasis | [210] | ||
| serum | miR-4661-5p | not mention | promote metastasis | [101] | ||
| circRNA | Cell culture fluid | circ_002136 | downregulate miR-19a-3p/RAB1A pathway | promote metastasis | [211] | |
| serum | circCCAR1 | upregulate WTAP | promote metastasis | [149] | ||
| Cell culture fluid | circ_0003028 | activate miR-498/ODC1 signaling axis, sponge miR-498, | promote metastasis | [212] | ||
| Cell culture fluid | circ_MMP2 | upregulate of its host gene MMP2 via sponging of miR-136-5p | promote metastasis | [213] | ||
| HCC tissues | circTTLL5 | activate miR-136-5p/KIAA1522 axis, sponge miR-136-5p | promote metastasis | [137] | ||
| serum | circRNA-100338 | activate of MMP9, regulate VM formation | promote metastasis | [214] | ||
| serum | circGSE1 | activate the miR‐324‐5p/TGFBR1/Smad3 axis | promote metastasis | [215] | ||
| serum | CircANTXR1 | activate miR-532-5p/XRCC5 axis | promote metastasis | [216] | ||
| HCC tissues | CircPAK1 | bind 14–3–3 ζ with YAP | promote metastasis | [217] | ||
| Cell culture fluid | circPTGR1 | activate miR449a-MET pathway | promote metastasis | [141] | ||
| Cell culture fluid | Circ-0000284 | not mention | promote metastasis | [134] | ||
| plasma | circUHRF1 | inhibit NK cell function | promote metastasis | [150] | ||
| Plasma | hsa_circ_0004277 | inhibit of ZO-1 | promote metastasis | [48] | ||
| Cell culture fluid | hsa_circ_0074854 | mediate macrophage M2 polarization | promote metastasis | [58] | ||
| Cell culture fluid | CircRNA Cdr1as | promote the expression of AFP and sponge with miR-1270 | promote metastasis | [218] | ||
| Cell culture fluid | circTMEM181 | inhibit the ATP-adenosine pathway, increase the expression of CD39 | promote metastasis | [60] | ||
| serum | Circ-ZNF652 | vie miR-29a-3p/guanylyl cyclase domain containing 1 axis | promote metastasis | [219] | ||
| lncRNA | serum | LINC00161 | inhibiting miR-590-3p to activate the ROCK2 signaling pathway | promote metastasis | [119] | |
| serum | lncRNA THEMIS2- 211 | activate THEMIS2-211/miR-940/SPOCK1 axis | promote metastasis | [121] | ||
| serum | lncRNA SNHG16 | activate miR-942-3p/MMP9 axis | promote metastasis | [124] | ||
| Cell culture fluid | lncRNA H19 | activate miR-520a-3p/LIMK1 axis | promote metastasis | [220] | ||
| Cell culture fluid | lncRNA HCG18 | downregulate miR-424-5p/SOX9 axis and PI3K/AKT pathway | promote metastasis | [221] | ||
| serum | lncRNAENSG00000248932.1, ENST00000440688.1 and ENST00000457302.2 | not mention | promote metastasis | [222] | ||
| proteins | Cell culture fluid | CTLA-4 | activate PTEN/CD44 signal pathway | promote metastasis | [85] | |
| Cell culture fluid | S100A4 | activate STAT3 phosphorylation and up-regulating OPN expression | promote metastasis | [74] | ||
| Cell culture fluid | BMI1 | not mention | promote metastasis | [79] | ||
| tumor tissue | LOXL4 | activate the FAK/Src pathway | promote metastasis | [82] | ||
| Cell culture fluid | ENO1 | activate the FAK/Src-p38MAPK pathway | promote metastasis | [81] | ||
| Cell culture fluid | RAB13 | upregulate VEGF and activity of CUX1 | promote metastasis | [223] | ||
| Cell culture fluid | RAB5A | not mention | promote metastasis | [224] | ||
| gastric cancer | miRNA | peritoneal lavage | hsa-let-7 g-3p and hsa-miR-10395-3p | activate the TGFβ signaling pathway | promote metastasis | [225] |
| Cell culture fluid | MiR-374a-5p | activate NF-κB signaling | promote metastasis | [226] | ||
| serum | miR-519a-3p | activate the MAPK/ERK pathway by targeting DUSP2 | promote metastasis | [98] | ||
| Cell culture fluid | miR-106a | inhibit the expression of Smad7 | promote metastasis | [227, 228] | ||
| Cell culture fluid | miR-21-5p | activate TGF-β/Smad pathway | promote metastasis | [229] | ||
| serum | miR-10b-5p, miR-101-3p and miR143-5p | not mention | promote metastasis | [114] | ||
| serum | miR-301a-3p | activate MiR-301a-3p/PHD3/HIF-1α signaling axis | promote metastasis | [230] | ||
| Cell culture fluid | miR-501-5p | downregulate of BLID, subsequent inactivate of caspase-9/-3 and phosphorylate of Akt | promote metastasis | [231] | ||
| serum | miR-379-5p and miR-410-3p | not mention | promote metastasis | [99] | ||
| Cell culture fluid | miR-486-5p | downregulate SMAD2, CDK4, and ACTR3 | promote metastasis | [232] | ||
| serum | hsa‐miR‐148‐3p, hsa‐miR‐335‐5p, hsa‐miR‐3613‐5p, and hsa‐miR‐556‐5p | not mention | promote metastasis | [233] | ||
| Peritoneal lavage fluid | miR-21-5p, miR-92a-3p, miR-223-3p, and miR-342-3p | not mention | promote metastasis | [106] | ||
| serum | miR-196a-1 | inhibit SFRP1 protein expression | promote metastasis | [234] | ||
| serum | miR-1307-3p、piR-019308、piR-004918, piR-018569 | not mention | promote metastasis | [100] | ||
| circRNA | serum | circ_0038138 | downregulate miR-198/EZH2 axis | promote metastasis | [235] | |
| serum | circFCHO2 | activate the JAK1/STAT3 pathway via sponging miR-194-5p | promote metastasis | [236] | ||
| Tumour tissues | circNRIP1 | activate AKT1/mTOR pathway, sponge of miR-149-5p | promote metastasis | [47] | ||
| serum | circNEK9 | activate miR-409-3p/MAP7 axis | promote metastasis | [133] | ||
| serum | Hsa_circ_0000437 | upregulate SRSF3, activate HSPA2-ERK signaling pathway | promote metastasis | [138] | ||
| serum | circ-RanGAP1 | activate miR-877-3p/VEGFA axis | promote metastasis | [143] | ||
| serum | circSHKBP1 | activate the miR-582-3p/HUR/VEGF pathway | promote metastasis | [144] | ||
| Cell culture fluid | circular RNA UBE2Q2 | via the circUBE2Q2-miR-370-3p-STAT3 axis | promote metastasis | [237] | ||
| Cell culture fluid | circular RNA circ_0032821 | sponge with miR-515-5p to regulate SOX9 expression | promote metastasis | [238] | ||
| serum | circ-PVT1 | by miR-30a-5p/YAP1 axis | promote metastasis | [239] | ||
| lncRNA | Cell culture fluid | lncRNA HOTTIP | activate microRNA-885-3p/EphB2 axis | promote metastasis | [240] | |
| serum | lnc-SLC2A12-10:1 | not mention | promote metastasis | [241] | ||
| Cell culture fluid | lncRNA PCGEM1 | reduce the degradation of SNAI1 | promote metastasis | [126] | ||
| tumor tissue | lncRNA SPRY4-IT1 | activate SPRY4-IT1/miR-101-3p/AMPK axis | promote metastasis | [118] | ||
| Cell culture fluid | lncRNA TTN-AS1 | activate miR-499a-5p/ZEB1/CDX2 axis | promote metastasis | [125] | ||
| tumor tissue | lncRNA LINC01091 | activate miR-128-3p/ELF4/CDX2 axis | promote metastasis | [109] | ||
| serum | lncRNA X26nt | decrease vascular endothelial cadherin | promote metastasis | [242] | ||
| proteins | peritoneal lavage fluid | NNMT | activate TGF-β/smad2 signal pathway | promote metastasis | [75] | |
| serum | Wnt5a | inhibit of YAP signal pathway | promote metastasis | [72] | ||
| serum | IntegrinB5 | activate PI3K-AKT pathways | promote metastasis | [73] | ||
| pancreatic cancer | miRNA | Cell culture fluid | miR-125b-5p | activate of MEK2/ERK2 signaling | promote metastasis | [243] |
| serum | miR-3960 | suppress TFAP2A/PTEN/AKT signaling pathway | promote metastasis | [244] | ||
| Cell culture fluid | miR-27a | induce angiogenesis by inhibiting BTG2 expression | promote metastasis | [245] | ||
| Cell culture fluid | miR-501-3p | decrease TGFBR3 levels and activate TGF-βpathway | promote metastasis | [246] | ||
| circRNA | serum | circ-IARS | sponge with miR-122 | promote metastasis | [145] | |
| Cell culture fluid | circ-PED8A | upregulate MET, sponge with miR-338, activate MACC/MET/ERK or AKT pathways | promote metastasis | [146] | ||
| serum | circZNF91 | MiRNA sponge for miR-23b-3p | promote metastasis | [152] | ||
| proteins | serum | EphA2 | not mention | promote metastasis | [247] | |
| Cell culture fluid | CD44v6/C1QBP complex | activate of IGF-1 signal | promote metastasis | [76] | ||
| serum | APOE | not mention | promote metastasis | [248] | ||
| Cell culture fluid | Eps8 | not mention | promote metastasis | [78] | ||
| Cell culture fluid | ASPH | activate Notch signaling | promote metastasis | [86] | ||
| serum | DNAJB11 | activate the EGFR/MAPK pathway | promote metastasis | [87] | ||
| serum | CCT8, CTSL, SAA1, IGF2 | not mention | promote metastasis | [88] | ||
| cancer tissue | FGD5-AS1 | activate STAT3/NF-κB signaling pathway | promote metastasis | [89] | ||
| serum | Lin28B | activate the Lin28B/let-7/HMGA2/PDGFB signaling pathway | promote metastasis | [90] | ||
| Cell culture fluid | ZIP4 | not mention | promote metastasis | [249] | ||
| serum | PRKD-1 | increase in F-actin | promote metastasis | [250] | ||
| Cell culture fluid | DYRK1A | stabilize the c-MET receptor through SPRY2, lead to prolonged activate of extracellular signal-regulated kinase signaling | promote metastasis | [77] |
Abbreviations: TGF-β1 transforming growth factor-β1, VM vasculogenic mimicry, VEGF vascular endothelial-derived growth factor, IGF-1 insulin-like growth factor 1, DYRK1A Dual-specificity tyrosine regulated kinase 1A, SPRY2 Sprouty 2, HUR human antigen R
Exosomal proteins as the biomarkers of GIC
Some investigations scrutinizing the biomarkers associated with GIC metastasis have predominantly centered their attention on the impact of exosomal proteins. Conducting proteomic assays and juxtaposing biofluids-derived exosomal proteins from cancer-afflicted individuals with those from cancer-free counterparts reveals substantial disparities in both the quantity and composition of exosomal proteins. Moreover, exosomal proteins sourced from diverse origins and subject to distinct conditions exhibit marked heterogeneity [67]. Upon co-culturing those highly heterogeneous exosomes with tumor cells and nude mice, it was found that exosomal proteins exert a stimulatory influence on the migratory capability of cancer cells, augment the metastatic nodules and activate several pathways usually involved in cancer development. Hence, exosomal proteins serve as dependable prognostic, diagnostic, and predictive biomarkers for the detection of GIC recurrence.
Various investigators have identified that, relative to healthy peoples or non-metastatic patients, five exosomal proteins (exosomal HSPC111, ADAM17, KRAS, Wnt5a, and IntegrinB5) are significantly overexpressed in patients afflicted with highly metastatic CRC and GC [68–73]. And these proteins are downregulating after lumpectomy, escalating the formation of metastatic lesions after injecting into nude mice, and triggering respective downstream signal pathway to facilitate cancer metastasis. The collective observations substantiate that exosomes directly sourced from tumors cells, harboring neoplastic information, thereby implying that those proteins serving as potential diagnostic biomarkers for GIC. In addition to serum, exosomal levels of S100A4, NNMT, CD44v6/C1QBP, DYRK1A, BMI1, and Eps8 derived from tumor tissues are also elevated in highly-metastatic HCC, GC, and PC patients [74–79]. And those exosomal proteins actively participate the aforementioned tumor pre-metastatic process to advance cancer recurrence.
Indeed, the abundance of numerous exosomal proteins manifests as a dynamic and multifaceted profile rather than a static, unidimensional state. This dynamic nature is characterized by elevation across a broad spectrum of metastatic GIC, serving as a diagnostic marker for diverse cancers. For instance, exosomal S100A4 exhibits heightened levels in both metastatic HCC and PC [74, 80], whereas IntegrinB5 is a diagnostic indicator in GC but a prognostic indicator in PC [73, 80]. Li and jiang et al. discerned that the expression of exosomal LOXL4, and ENO1 is also been identified to be overexpressed in HCC patients and intimately associated with the tumor-node-metastasis (TNM) stage, and the protein within serum-derived exosomes is intricately linked to an unfavorable prognosis in HCC patients [81, 82]. Liang et al. found that plasma exosomal Cyr61 was significantly higher in TNM stages III and IV (n = 185) compared with stages I and II (n = 179). Furthermore, in contrast to traditional diagnostic marker carcinoembryonic antigen (CEA) and carbohydrate antigen (CA19-9), serum Cyr61 exhibit superior diagnostic and prognostic efficacy for CRC according to multivariate logistic regression analysis and receiver operating characteristic (ROC) area under the ROC curve (AUC) of 0.933 [83]. Exosomal proteins, with the ability to discern between the early and advanced stages of GIC, emerge as commendable indicators to guide cancer treatment.
Other exosomal proteins have also been identified as important predictive biomarkers of GIC owing to create cancer-friendly condition to promote cancer metastasis, like exosomal Formin-like 2 (FMNL2), cytotoxic T lymphocyte antigen 4 (CTLA-4), RAB5A, aspartate β-hydroxylase, and so on [84–90]. While direct evidence linking them to TNM stage and serous content remains elusive, these exosomal proteins exhibit the capacity to orchestrate conditions and stimulate signaling pathways that facilitate the conversion of the internal environment in cancer patients, making it prone to metastasis, thereby assuming the responsibility of predictive biomarkers for GIC. For instance, CRC FMNL2 overexpression can promote CRC angiogenesis and metastasis by activating the ERK/MMP signal pathway [84], exosomal CTLA-4 could mediate the proliferation, invasion, and metastasis of HCC cells by mediating PTEN/CD44 signal pathway [85], and exosomal DNAJB11 can activate the downstream MAPK signaling pathway, which would enhance PC invasive ability [87, 88]. Another study demonstrated that exosome-derived FGD5-AS1 can activate STAT3/NF-κB pathway to mediate M2 macrophage polarization and advance the malignant behaviors of PC [89].
Exosomal miRNA as the biomarkers of GIC
Proteins represent merely a facet of exosomal carryover and may exhibit limitations as biomarkers for GIC recurrence. Consequently, a multitude of studies have shifted their attention towards on the impact of exosomal miRNA on the induction of cancer progression, which constitutes as the most prevalent constituent within exosomes. Melo et al. found that comparing with normosomes, exosomal miRNAs originating from cancerous sources manifest a surge of up to a 30-fold increment [26]. Other researchers found that some miRNAs, including miR-29a, miR-141-3p, miR-375, miR-106b-3p, miR- 221/222, miR-200b-3p, miR-18a, miR-20b, miR-1307-5p, miR-519a-3p, miR-379-5p, and miR-410-3p are also overexpression in metastatic CRC, HCC, and GC patients. The expression of these miRNAs notably diminishes post-surgical intervention, substantiating their predominant derivation from GIC cells and signifying their role as predictive biomarkers of GIC [66, 91–100]. Tian et al. demonstrated that injection the normal as well as GIC patient serum-derived exosomes into tumor xenograft models, overexpressed miR-200b-3p, miR‐221, and miR‐222 was positively associated with the metastatic foci growth, and increase in the metastatic ratio and diameters of metastatic foci. Sun et al. found that miR-122 is overexpressed in CRC tissue, and miR-122 show discriminatory potential in distinguishing CRC patients with or without liver metastasis (LM) (AUC of 0.89 and 0.81). Moreover, akin to the observation in exosomal proteins, the integration of exosomal biomarkers with existing tumor markers demonstrates the potential to augment precision in monitoring GIC recurrence. Ge et al. revealed that four miRNAs is significantly increased in metastatic GC, and when amalgamated with conventional diagnostic marker CEA and CA-199, the augmentation of the diagnostic accuracy of miR-1307-3p, piR-019308, piR-004918, and piR-018569 is apparent (the AUC separately upregulate from 0.845, 0.820, 0.754 and 0.732 to 0.902, 0.914, 0.859, and 0.868) [100]. Cho et al. elucidated that serval miRNAs are evidently elevated in early‐stage HCC (single tumor < 2 cm), miR‐4661‐5p could diagnose HCC in all stages (AUC = 0.917). The diagnostic accuracy of the serum exosomal miR‐4661‐5p (AUC = 0.923), miR‐1269a (AUC = 0.684), and miR‐25 (AUC = 0.812) in diagnosing early‐stage HCC show markedly superior AUR values than serum AFP (AUC = 0.541) [101]. Clinically, miR-128-3p is overexpressed in patients with later tumor stage (III–IV), and its expressivity is highly associated with perineural invasion, disease stage, and CA 19–9 content in CRC patients (P < 0.05) [102]. While these investigations adopt diverse perspectives on the metastatic process, collectively, they afford contemporaneous insights into the dynamic status of the neoplastic lesion.
Simply classification of RNA into distinct diagnostic and prognostic categories is actually a bit subtle, as certain multiple miRNA species have both diagnostic and prognostic significance. MiR-301a-3p not only increase in the serum of PC patients and its levels positively correlate with invasion depth and advanced TNM stage of PC [103]. Similarly, miR-92a-3p is prominently elevated not solely within the serum of high-metastatic CRC patients, but also HCC and GC. And miR-92a-3p is positively correlate with progressed TNM stage, and can mediate chemotherapy resistance, thereby contributing to an adverse prognosis in patients afflicted with GIC [104–107]. In addition to these two, exosomal miR-1229, miR-1307-5p manifest to this particular phenotype [97]. Apart from the aforementioned miRNAs, other miRNAs have been identified as relevant predictive biomarkers. With the assistant of miR-203a-3p, miR-128-3p, mi-335-5p, miR-106b-5p, miR-218-5p, miR-181b-3p, miR-183-5p, and miR-27b-3p, there is an augmentation in the migratory capability of CRC cells, concomitant with an elevated propensity for metastasis in CRC[92, 93, 102, 108–113]. Analyzing the serum derived exosomal miRNAs, Zhang et al. discovered that the expression profiles of miR-10b-5p, miR 143-5p, and miR 101-3p showed statistically significant up-regulation in individuals diagnosed with GC and concurrent lymph node, liver, and ovarian metastases (p < 0.05, AUC = 0.8919, 0.8247, 0.8905) [114]. While the extensive heterogeneity of biomarkers necessitates additional refinement and standardization, extant evidence highlights the potential of exosomal miRNAs in evaluating metastatic GIC and prognostic outcomes.
Exosomal lncRNA as the biomarkers of GIC
Subsequent to an in-depth investigation of miRNA, researchers contemplated the potential utility of lncRNA, another noncoding RNA subtype, as a prospective biomarker for GIC detection. lncRNA is a large class of transcripts with a length exceeding 200 bp that exert functions in a bunch of biological procedures, including transcriptional, epigenetic, and post-transcriptional levels, and initiate the progression and metastasis of cancers[115]. Substantial evidence suggests that lncRNAs could mediate each step of metastasis such as cell migration, invasion, and distant-site colonization [115]. Empirical investigations disclose the diagnostic aptitude embedded within exosomal lncRNAs, furnishing enlightening perspectives on the detection of metastatic GIC [116]. For instance, luan et al. isolate the exosomal RNA from serum of both CRC patients and normal human. Subsequent analytical assessments found that 569 lncRNAs were up-regulated, concomitant with a downregulation in 475 lncRNAs [117]. LncRNA UCA1 is overexpressed in neoplasms at advanced stages (p = 0.007), distant metastatic patients (p = 0.003), and patients afflicted with tumors > 5 cm (p = 0.005). Analogous observations were reported by cao et al. [118]. LncRNA SPRY4-IT1 is upregulated in GC tissue (p < 0.001), and silencing SPRY4-IT1 results in the attenuation of GC progression. LncRNA UCA1 and lncRNA SPRY4-IT1 have shown considerable potential as prospective diagnostic biomarker for GIC. Likewise, high levels of lncRNA PVT1/VEGFA and LINC00161 were discerned in both serum and tissue of CRC and HCC patients [119, 120]. However, LncRNA THEMIS2-211 and RPPH1 concurrently functions as both a diagnostic and prognostic. within the cohort comprising of 89 HCC patients and 60 normal controls, plasmatic exosomal LncRNA THEMIS2-211 is upregulate in HCC patients (p < 0.001), and the diagnostic efficiency of exosomal THEMIS2-211 experiences a notable improvement when combinate with AFP in diagnosing stage I HCC patients [121]. Additionally, the expression levels of exosomal THEMIS2-211 is way superior in advanced-stage (III, IV stages) than early-stage (I, II stages). Coincidentally, LncRNA RPPH1 is also overexpressed in non-treatment CRC patients but lower after cancer resection, and have a strong connection with advanced TNM stages and poor prognosis [122]. Exosomal RPPH1 bespeak better diagnostic value (AUC = 0.86) when contrasted with CEA and CA199 as well.
Hashemi et al. reported that lncRNA H19 can trigger chemo- and radio-resistance in cancer cells, signifying that lncRNA H19 is a unitary prognostic biomarker of tumor recurrence [123]. The presence of lncRNA SNHG16, TTN-AS1 and PCGEM1 in the tissues of cancer patients accentuates the malignant features of GIC cells, culminating in a diminished survival rate with an unfavorable prognosis. Xu et al. utilized Pearson’s correlation and univariate statistical analysis to examine a cohort of 78 cases with low or high expression of LncSNHG16. Their findings unveiled a positive correlation between heightened LncSNHG16 expression and HCC relapse within a 2-year period (p = 0.000) [124]. Analogous studies have also been proposed by Wang et al. Bioinformatics analysis delineated an augmented expression of lncRNA TTN-AS1 in GC is highly correlation with the poor overall survival (OS) [125]. Other lncRNAs have also been identified as important predictive biomarkers of GIC. Fang et al. identified that lncRNA PCAT1 and PCGEM1 can promote EMT, thereby enhancing the proliferation and migration ability of CRC cells [126]. When knockdown of PCAT1, the metastatic propensity notable attenuated, particularly liver metastasis [127]. Simultaneously, exosomal metastases-associated lung adenocarcinoma transcript 1 (MALAT1) can advance CRC cells with high metastatic and invasive abilities [128]. While these lncRNAs contribute to GIC metastasis in diverse mechanisms, collectively they underscore the heterogeneity inherent of primary tumor cells.
Exosomal circRNA as the biomarkers of GIC
Amidst the surge of circRNA research, some scholarly publications express a profound curiosity regarding the comparability of circRNAs to other non-coding RNAs in their potential to promote tumor metastasis, and the possibility to serve as available biomarkers for cancer recurrence [129, 130]. With deeper investigation, researchers discerned that circRNAs are characterized by rich diversity, a stable structural framework, conserved sequences, and manifest cell- or tissue-specific expression patterns. Furthermore, circRNAs function as miRNA sponges to modulate the microRNA-mRNA regulatory axis [131, 132]. This revelation implies that circRNAs manifest a significant tumor heterogeneity and hold potential as biomarkers for metastatic-GIC. Like the aforementioned non-coding RNA, circ-0000284, circFMN2, circ-ABCC1, circTUBGCP4, circ-133, circTTLL5, circPTGR1, hsa_circ_0004277, circ_0038138, and hsa_circ_0000437 are aberrantly overexpressed in the exosomal circRNAs derived from CRC, HCC, and GC patients when compare with healthy volunteers, especially in patients with distance metastasis (M1) [48, 133–138]. Moreover, the levels of these circRNAs are dramatically descend after removing primary lumps, suggesting that neoplastic cells directly contribute to the pool of circRNAs, thereby potentially serving as diagnostic biomarkers for GIC. Mechanistic analysis has revealed that these circRNAs can sponge with diverse miRNAs to activate downstream to improve the chances of metastatic foci formation in tumor xenograft models. Similar observations are reported by Li et al. The overexpression of circPABPC1 accelerates the advancement of CRC, while the ablation of circPABPC1 attenuates CRC progression [139]. In addition to the above-mentioned non-coding RNA, a number of circRNAs also manifest the diagnostic and prognostic function in detection GIC recurrence. Not only does circLONP2 and circPTGR1 upregulate in tumor patients, but also the OS rate of patients with high expression of these circRNAs is notably unfavorable (p < 0.05) [140, 141]. And elevated expression of circNEK9 (I = 30, p = 0.0099) and circGAPVD1 (n = 78, p = 0.011) are correlate with augmented lump size and advanced TNM stage in GC and CRC patients [133, 142]. Lu et al. conducted an assessment of circ-RanGAP1 expression in a cohort of 97 paired GC samples, and found that circ-RanGAP1 is conspicuously upregulate in stage III than stage I-II [143]. Statistical scrutiny revealed a noteworthy correlation between heightened circ-RanGAP1 expression and large tumor size (p = 0.016) with an advanced clinical stage (p = 0.001). In a comprehensive examination of circSHKBP1 expression across 72-paired GC and normal specimens, Xie et al. found a 2.31-fold upregulation of circSHKBP1 in GC than normal (P < 0.05), and overexpressed circSHKBP1 is highly correlate with poor prognosis and advanced TNM stage, positioning it a potential biomarker for GC [144]. These phenotypic traits are similarly present in circ-IARS, circ-YAP, and circPDE8A [145–147].
Furthermore, few circRNA is just a mono-prognosticator for GIC. For instance, sang et al. found that the expression of circRELL1 is correlated with TNM stage and a poor survival rate, making it an ideal prognostic biomarker of GC [148]. Besides, the expression of circCCAR1 is not only positively associate with CRC grade and TNM stage, but also can induce CRC patients with the resistance to anti-PD1 therapy [149]. It is not a unique instance, but has its counterpart. Overexpression of circUHRF1 and circTMEM181 can attenuate the anti-tumor efficiency of anti-PD1 therapy, consequently diminishing OS rates [60, 150]. Apart from above-mentioned circRNA, ciRS‐122 is higher in the serum of oxaliplatin‐resistant patients (n = 13) than oxaliplatin‐sensitive patients (n = 6). Enhanced ciRS‐122 expression diminishes the oxaliplatin susceptibility both in vivo and vitro, whereas targeted suppression of it can reverse this chemoresistance [151]. likewise, overexpression of circZNF91 can induce Gemcitabine resistance in PC cells [152]. These exosomal circRNAs collectively serve as valuable indicators for guiding the prognosis of GIC. A substantial body of empirical evidence now substantiates the direct origin of exosomes from neoplastic cells, encapsulating a cargo reflective of their cellular origin. These exosomes exhibit flawless heterogeneity, offering the potential to function as evaluative entities and providing insights to assess the severity of disease and monitor the prognosis of GIC in real-time. It is precisely these research results that highlight discernment and therapeutic avenues of metastatic cancer.
The clinic application of CDEs in gastrointestinal cancer
It is well established that CDEs have particularly promising applications as liquid biopsies, because exosomes could be released by all types of cells and widely distributed in all biological fluids, and the components of CDEs reflect the biological state of their origin. Of note, some CDEs show the possibility of mediating the malignant behaviors of cancer. Based on this theory, we can utilize the characteristics of exosomes, not only to monitor the malignant behaviors of cancer but also to target CDEs as a therapeutic approach to cure cancer.
Exosomes can inhibit cancer biogenesis
Besides the aforementioned exosomes that can trigger cancer metastasis, substantial research demonstrates that some exosomes can also inhibit cancer progression. Given this fact, we can harness exosomes or activate relative downstream to forbid cancer metastasis and cure cancer (Table 2).
Table 2.
Exosomal biomarkers for the inhibition of GIC metastasis
| Cancer type | Exosomal Contents | Exosome Source | Exosomal Cargo | Effect | Clinical Significance | Ref |
|---|---|---|---|---|---|---|
| colorectal cancer | miRNA | Cell culture fluid | miR-1827 | downregulate SUCNR1 | inhibit metastasis | [251] |
| Serum | miR-140-3p | upregulate the expression of BCL9 and BCL2 | inhibit metastasis | [155] | ||
| Cell culture fluid | microRNA-3940-5p | upregulate TGF-β1 | inhibit metastasis | [252] | ||
| Cell culture fluid | miR-100 and miR-143 | downregulate miR-100/mTOR/miR-143 axis | inhibit metastasis | [156] | ||
| circRNA | Cell culture fluid | circFNDC3B | inhibit miR‐937‐5p and upregulate TIMP3 | inhibit metastasis | [153] | |
| serum | circRHOBTB3 | secrete outside of cells | inhibit metastasis | [154] | ||
| lncRNA | serum | lncRNA ADAMTS9-AS1 | inhibit the Wnt/β-catenin signaling pathway | inhibit metastasis | [157] | |
| proteins | Cell culture fluid | ANGPTL1 | downregulate MMP9 level in KCs by inhibiting the JAK2-STAT3 signaling pathway | inhibit metastasis | [158] | |
| hepatocellular cancer | miRNA | liver tissue | miR-374c-5p | activate LIMK1-Wnt/β-catenin axis | inhibit metastasis | [160] |
| serum | miR-125b | block TGF-β1/SMAD Pathway, repress protein expression of SMAD2 | Inhibit metastasis | [253] | ||
| serum | microRNA-27a-3p | suppress of Golgi Membrane Protein 1 | inhibit metastasis | [159] | ||
| circRNA | serum | circ-0072089 | sponge with miR-375 and upregulate MMP-16 | inhibit metastasis | [161] | |
| Cell culture fluid | hsa_circ_0004658 | activate hsa_circ_0004658/miR-499b-5p/JAM3 pathway | promote metastasis | [254] | ||
| Plasma | hsa_circ_0051444 | upregulate BAK1 and competitive bound to miR-331-4p | inhibit metastasis | [162] | ||
| gastric cancer | miRNA | Cell culture fluid | miR-29b | not mention | inhibit metastasis | [255] |
| circRNA | serum | CircRNA ITCH | regulate miR-199a-5p/Klotho axis | inhibit metastasis | [165] | |
| Cell culture fluid | CDR1as | activate the CDR1as/miR-876-5p/GNG7 axis | inhibit metastasis | [164] | ||
| Cell culture fluid | circRELL1 | upregulate the expression of EPHB3 | inhibit metastasis | [148] | ||
| pancreatic cancer | miRNA | Cell culture fluid | miR-485-3p | decrease the PAK1 expression | inhibits metastasis | [166] |
| Cell culture fluid | miRNA-29b | downregulate of ROBO1 and SRGAP2 | inhibits metastasis | [256] | ||
| Cell culture fluid | miRNA-339-5p | decrease TGFBR3 levels and activate TGF-β signal | inhibits metastasis | [257] | ||
| circRNA | Cell culture fluid | circ_0030167 | activate miR-338-5p/wnt8/β-catenin axis | inhibit metastasis | [167] | |
| Cell culture fluid | Circ_0006790 | downregulate S100A11, via CBX7-catalyzed DNA hypermethylation, bind to CBX7, MiRNA sponge for miR-144 | inhibit metastasis | [168] |
For CRC, Zeng et al. found that exosomal circFNDC3B can bind to miR-937-5p to upregulate TIMP3, thus inhibiting tumorigenic, metastatic, and angiogenic properties of CRC [153]. Chen et al. observed that circRHOBTB3 acts as a tumor-suppressive circRNA and inhibits CRC growth and metastasis by repressing intracellular ROS production and metabolic pathways in CRC [154]. Another study revealed that exosomal miR-140-3p overexpression suppresses CRC proliferation, β-catenin nuclear translocation, invasion, and migration via targeting BCL9 and BCL2 [155]. Babak et al. found that exosomal miR-143 and miR-100 effectively downregulate mTOR, K-RAS, HK2, and Cyclin D1 while significantly suppressing the expression of MMP9, MMP2, TWIST, and SNAIL; and upregulating p-27 expression, thus hampering CRC proliferation, migration, invasion, and metastasis [156]. Li et al. have shown that upregulation of lncRNA-ADAMTS9-AS1 can suppress β-catenin and mediate Wnt signal pathway to suppress colorectal tumorigenesis [157]. Jiang et al. demonstrated that exosomal ANGPTL1 attenuates LM and impedes vascular leakiness of PMN by downregulating MMP9 expression in KCs and restraining JAK2-STAT3 signaling pathway [158].
For HCC, exosomal microRNA-27a-3p and miR-374c-5p can respectively upregulate GOLM1 and inactivate Wnt/β-catenin pathway to suppress EMT to inhibit HCC progression [159, 160]. Besides, exosomal circ-0072088 can sponge with miR-375 and upregulate MMP-16 to suppress HCC metastasis, and exosomal hsa_circ_0051443 can bound to miR-331-3p to upregulate BAK1 which in turn suppresses HCC progression [161, 162]. Cheng et al. reported that exosomal p120ctn can inhibit expansion of liver cancer stem cells, cell proliferation, and metastasis in HCC by activating STAT3 pathway [163].
For GC, Jiang et al. found that circRNA CDR1as knockdown facilitates GC invasion and migration while the overexpression of circRCDR1as reverses aforementioned phenomena [164]. Another study reported that circRNA ITCH can regulate the miR-199a-5p/Klotho axis to inhibit GC proliferation, migration, invasion, and EMT [165]. Exosomal circRELL1 can sponge miR-637 to indirectly upregulate EPHB3 expression by modulating autophagy to block cancer cell proliferation and migration [148].
For PC, exosomal miR-485-3p suppresses PC cell invasion and migration by targeting p21-activated kinase-1 [166]. Exosomal circ_0030167 significantly reduces PC cell invasion, migration, and proliferation by regulating miR-338-5p, increasing Wif1 expression, and inhibiting the Wnt8/β-catenin pathway [167]. Exosomal hsa_circ_0006790 inhibits immune escape and metastasis in PDAC by inducing translocation of chromobox 7 [168].
Impede production and release of exosomes
In the case of cancer metastasis, blocking exosome release to alleviate cancer migration seems like a practicable and promising approach. Ostrowsk et al. found that Rab27a and Rab27b function as MVE docking at the plasma membrane. So, silencing Rab27a and Rab27b can decrease the release of exosome [169]. For ESCRT independent pathway, silencing Rab31, caveolin-1, and flotillins which contribute to cancer chemoresistance and progression by regulating exosome secretion may exert function on decreasing CDEs production [170–172]. And RBPs (such as MVP, hnRNPA2B1, hnRNPA1, hnRNPH1, and hnRNPK) can regulate exosome generation, so blocking their expression can dwindle exosome production [173]. Vps4A can inhibit β-catenin signal pathway and EMT to suppress exosome release and restrain the proliferation and metastasis of HCC [174]. Parolini et al. demonstrated that CDEs cultured under acidic conditions exhibit more secretion and dangerous delivery activity than those cultured under physiological conditions [175]. In particular, under acidic conditions, the cancer-released exosomes are 3–8 folds compared with those cultured at pH 7.4 conditions [175]. Given this theory, proton pump inhibitors (such as NHE1 inhibitors, CA inhibitors, MCT inhibitors, and V-ATPase inhibitors), altering cellular pH, and alkalizing agents seem to be viable and potential anti-cancer strategies for metastatic patients [176–178].
Datta et al. revealed that Manumycin A seems to be a potential drug candidate to inhibit exosome biogenesis and release by suppressing of Raf/Ras/ERK2/1 signal pathway [179]. Asai et al. demonstrated that the restriction of SMase can decrease exosome secretion [180]. Sun et al. found that hypoxia can affect the composition of CDEs and promote metastasis [181]. Li et al. found that GW4869, the N-SMase inhibitor, can pharmacologically block exosome generation and successfully decrease exosome secretion [82, 149].
Inhibition the interaction of exosome and recipient cell
Based on the process of exosome biogenesis, interrupting CDEs uptake seems another feasible method to avoid cancer metastasis. Sento et al. reported that heparin can remarkably decrease the uptake of CDEs to abrogate tumor progression and metastasis [182]. Hoshino et al. revealed that 4175-LuT exosomes preincubate with HYD-1, and ITGβ5 knockdown in BxPC-3-LiT exosomes and target ITGα6β4 and αvβ5 can respectively diminish exosome uptake in the liver and lung to block cancer metastases [18]. Gong et al. reported that low-pH and hypoxia treatment could significantly evaluate exosome uptake effectiveness by changing the lipid composition in exosome membrane [183]. So, the aforementioned proton pump inhibitors and alkalizing agents can alleviate exosome uptake by reversing the hypoxia and low-pH environment [184].
Exosomes as the drug delivery system in cancer therapy
Given the exosomal hallmarks: ①specificity, safety, and stability which protected by phospholipid bilayer membrane, ②exosome is distributed in all biological fluids which allow exosome to arrive at any site, ③low immune response, ④able to pass the Blood–Brain Barrier, ⑤low systemic toxicity [185]. Harnessing exosomes as the desirable tool to deliver drugs to target cancer sites seems a feasible method. Drugs can directly or indirectly load into exosomes, including through coculture, electroporation, liposomes, sensitive fusogenic peptide, sonication, cationic lipids, and metal–organic nanoparticles coated with exosome [186]. You et al. reported that exosomes can transfer L-PGDS to cancer cells to inhibit gastric cancer progression [187]. Besides, Pascucci et al. found that the exosomes isolated from paclitaxel co-cultured with MSCs exhibit strong anti-tumor activity [188].
Conclusions and future perspectives
Within the past decades, the detection methods for GIC have emerged in an endless stream. In particular, liquid biopsy relies on the advantages of minimal invasiveness, being readily available, and allowing real-time monitoring to hew out a new avenue for GIC pre-metastasis detection. Especially with the proposal of exosome, liquid biopsy reaches the golden period. Exosome gained extensive attention, reshaping our understanding of cancer biology and anti-cancer strategies, and providing new biomarkers for cancer diagnosis. The current studies consistently demonstrate that CDEs play an indispensable role in the biogenesis of GIC invasion and metastasis. Exosomes can not only participate in precancerous processes such as EMT, PMN formation, and immune escape, but also establish a suitable microenvironment for GIC metastasis by rendering exosomes to communicate with pending sites and shuttling proteins and nucleic acids into distant organs.
However, challenges and opportunities coexist in applying exosomes as the biomarker for GIC metastasis detection. There are still several problems to be addressed which may imply some worthwhile directions for future studies. First, there is a lack of standard methodology and standardized criteria for high-purity exosome separation, because different isolation methods may result in high heterogeneity. There is an urgent need for exosome isolation, examination, and production to develop a unified standardization. Second, distinct bioactive molecules contained in exosomes have different functions in promoting cancer metastasis. Although the aforementioned text briefly listed the effects of different components on cancer metastasis, the accurate function of exosome cargo and their association is ethereal. It is essential to identify which contain exosome is responsible for which part and their link when utilizing exosomes in metastatic cancer screening. There are some pivotal questions remain to be answered ①The potentiality of conversing biomarkers of liquid biopsy into clinical practice are still hindered by several limitation. How to transform scientific achievements into clinical applications? ②How to deliver the drug-incubated exosomes into ideal site to inhibit cancer metastasis? ③How to precisely reverse exosome-triggered immune evasion? There is a long way to go before we can convert carcinogens into cancer treatments.
Acknowledgements
The authors apologize to the investigators and research laboratories whose original studies were not cited because of space limitations. Figures were created with Figdraw (www.figdraw.com).
Abbreviations
- GIC
Gastrointestinal cancer
- CDEs
Cancer-derived exosomes
- CRDs
Cancer-related deaths
- CRC
Colorectal cancer
- PC
Pancreatic cancer
- CT
Computed tomography
- EVs
Extracellular vesicles
- EMT
Epithelial-mesenchymal transition
- PMN
Pre-metastatic niche
- ECM
Extracellular matrix
- ITGs
Integrins
- ESE
Early-sorting endosome
- LSEs
Late-sorting endosomes
- ILVs
Intraluminal vesicles
- MVBs
Multivesicular bodies
- ESCRT
Endosomal sorting complexes required for transport
- miRNAs
MicroRNAs
- circRNA
circular RNAs
- lncRNAs
Long non-coding RNAs
- MIF
Migration inhibitory factor
- PDAC
Pancreatic ductal adenocarcinoma
- KCs
Kupffer cells
- TGFβ
Transforming growth factor beta
- CAFs
Cancer-associated fibroblasts
- TME
Tumor microenvironment
- IGF
Insulin-like growth factor
- EGF
Epidermal growth factor
- HIF-1α
Hypoxia-inducible factor 1 alpha
- NK
Natural killer
- Tregs
Regulatory T cells
- TAMs
Tumor-associated macrophages
- PD-L1
Program death ligand 1
- PD1
Programmed cell death 1
- TNM
Tumor-node-metastasis
- CEA
Carcinoembryonic antigen
- CA19-9
Carbohydrate antigen
- ROC
Receiver operating characteristic
- AUC
Area under the ROC curve
- FMNL2
Formin-like 2
- LM
Liver metastasis
- OS
Overall survival
- ZO-1
Zonula occlusion 1
- SNX2
Sorting nexin 2
- PCAT1
Prostate cancer associated transcript 1
- MALAT1
Metastases-associated lung adenocarcinoma transcript 1
- CTLA-4
Cytotoxic T lymphocyte antigen 4
- VM
Vasculogenic mimicry
- HuR
Human antigen R
- SRSF3
Ser/Arg-rich splicing factor 3
- PDCD4
Programmed cell death 4
- ASPH
Aspartate β-hydroxylase
Authors’ contributions
All authors contributed to writing the review. All authors read and approved the final manuscript.
Funding
This work is supported by Key Research and Development Project of Zhejiang Province (2020C03122).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Danyang Zhong, Ziyuan Wang and Zhichao Ye contributed equally to this work.
Contributor Information
Yifan Wang, Email: anwyf@zju.edu.cn.
Xiujun Cai, Email: srrsh_cxj@zju.edu.cn.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Obenauf AC, Massague J. Surviving at a Distance: Organ-Specific Metastasis. Trends Cancer. 2015;1:76–91. doi: 10.1016/j.trecan.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng S, Gu J, Jiang Z, Cao Y, Mao F, Xue Y, Wang J, Dai K, Qin L, Liu K, et al. Application of nanotechnology in the early diagnosis and comprehensive treatment of gastrointestinal cancer. J Nanobiotechnology. 2022;20:415. doi: 10.1186/s12951-022-01613-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang FH, Shen L, Li J, Zhou ZW, Liang H, Zhang XT, Tang L, Xin Y, Jin J, Zhang YJ, et al. The Chinese Society of Clinical Oncology (CSCO): clinical guidelines for the diagnosis and treatment of gastric cancer. Cancer Commun (Lond) 2019;39:10. doi: 10.1186/s40880-019-0349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.JOHNSTONE RM, ADAM M, PAN BT. The fate of the transferrin receptor during maturation of sheep reticulocytes in vitro. Can J Biochem Cell biol. 1984;62(62):1246–1254. doi: 10.1139/o84-159. [DOI] [PubMed] [Google Scholar]
- 7.Trams EG, Lauter CJ, Jr, NS, Heine U, Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochem Biophys Acta. 1981;645:63–70. doi: 10.1016/0005-2736(81)90512-5. [DOI] [PubMed] [Google Scholar]
- 8.Kalluri R, LeBleu VS. he biology, function, and biomedical applications of exosomes. Science. 2020; 367(6478):eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vermeer PD. Exosomal Induction of Tumor Innervation. Cancer Res. 2019;79:3529–3535. doi: 10.1158/0008-5472.CAN-18-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-Mediated Metastasis: Communication from a Distance. Dev Cell. 2019;49:347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Wang M, Su Z, Amoah Barnie P. Crosstalk among colon cancer-derived exosomes, fibroblast-derived exosomes, and macrophage phenotypes in colon cancer metastasis. Int Immunopharmacol. 2020;81:106298. doi: 10.1016/j.intimp.2020.106298. [DOI] [PubMed] [Google Scholar]
- 12.Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015;219:278–294. doi: 10.1016/j.jconrel.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 13.Paget S. The distribution of secondary growths in cancer of the breast. Cancer Metastasis Rev. 1889;1989(8):98–101. [PubMed] [Google Scholar]
- 14.Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, Psaila B, Kaplan RN, Bromberg JF, Kang Y, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer. 2017;17:302–317. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- 15.Mazumdar A, Urdinez J, Boro A, Arlt MJE, Niederost B, Jaeger PK, Moschini G, Muff R, Fuchs B, et al. Exploring the Role of Osteosarcoma-Derived Extracellular Vesicles in Pre-Metastatic Niche Formation and Metastasis in the 143-B Xenograft Mouse Osteosarcoma Model. Cancers (Basel) 2020;12(11):3457. doi: 10.3390/cancers12113457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014;16:717–727. doi: 10.1038/ncb3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weidle UH, Birzele F, Kollmorgen G, Ruger R. The Multiple Roles of Exosomes in Metastasis. Cancer Genomics Proteomics. 2017;14:1–15. doi: 10.21873/cgp.20015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerstberger S, Jiang Q, Ganesh K. Metastasis Cell. 2023;186:1564–1579. doi: 10.1016/j.cell.2023.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krakhmal NV, Zavyalova MV, Denisov EV, Vtorushin SV, Perelmuter VM. Cancer Invasion: Patterns and Mechanisms. Acta Naturae. 2015;7:17–28. doi: 10.32607/20758251-2015-7-2-17-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim Biophys Acta Rev Cancer. 2019;1871:455–468. doi: 10.1016/j.bbcan.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 23.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 24.Riches A, Campbell E, Borger E, Powis S. Regulation of exosome release from mammary epithelial and breast cancer cells - a new regulatory pathway. Eur J Cancer. 2014;50:1025–1034. doi: 10.1016/j.ejca.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 25.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 28.Chen H, Chengalvala V, Hu H, Sun D. Tumor-derived exosomes: Nanovesicles made by cancer cells to promote cancer metastasis. Acta Pharm Sin B. 2021;11:2136–2149. doi: 10.1016/j.apsb.2021.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, Li R, Jiang J, Qian H, Xu W. Exosomal circRNAs: Novel biomarkers and therapeutic targets for gastrointestinal tumors. Biomed Pharmacother. 2023;157:114053. doi: 10.1016/j.biopha.2022.114053. [DOI] [PubMed] [Google Scholar]
- 30.Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. 2019;18:75. doi: 10.1186/s12943-019-0991-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons M, Raposo G. Exosomes–vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Feng W, Dean DC, Hornicek FJ, Shi H, Duan Z. Exosomes promote pre-metastatic niche formation in ovarian cancer. Mol Cancer. 2019;18:124. doi: 10.1186/s12943-019-1049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Cao X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell. 2016;30:668–681. doi: 10.1016/j.ccell.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 34.Kelleher RJ, Jr, Balu-Iyer S, Loyall J, Sacca AJ, Shenoy GN, Peng P, Iyer V, Fathallah AM, Berenson CS, Wallace PK, et al. Extracellular Vesicles Present in Human Ovarian Tumor Microenvironments Induce a Phosphatidylserine-Dependent Arrest in the T-cell Signaling Cascade. Cancer Immunol Res. 2015;3:1269–1278. doi: 10.1158/2326-6066.CIR-15-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma Z, Wei K, Yang F, Guo Z, Pan C, He Y, Wang J, Li Z, Chen L, Chen Y, Xia Y. Tumor-derived exosomal miR-3157-3p promotes angiogenesis, vascular permeability and metastasis by targeting TIMP/KLF2 in non-small cell lung cancer. Cell Death Dis. 2021;12:840. doi: 10.1038/s41419-021-04037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lobb RJ, Lima LG, Moller A. Exosomes: Key mediators of metastasis and pre-metastatic niche formation. Semin Cell Dev Biol. 2017;67:3–10. doi: 10.1016/j.semcdb.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Robado de Lope L, Alcíbar OL, Amor López A, Hergueta-Redondo M, Peinado H: Tumour–adipose tissue crosstalk: fuelling tumour metastasis by extracellular vesicles. Philos Trans R Soci B Biol Sci. 2017;373. [DOI] [PMC free article] [PubMed]
- 38.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao Y, Chen T, Zheng X, Yang S, Xu K, Chen X, Xu F, Wang L, Shen Y, Wang T, et al. Colorectal cancer-derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis. 2018;39:1368–1379. doi: 10.1093/carcin/bgy115. [DOI] [PubMed] [Google Scholar]
- 40.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biffi G, Tuveson DA. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol Rev. 2021;101:147–176. doi: 10.1152/physrev.00048.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi H, Enomoto A, Woods SL, Burt AD, Takahashi M, Worthley DL. Cancer-associated fibroblasts in gastrointestinal cancer. Nat Rev Gastroenterol Hepatol. 2019;16:282–295. doi: 10.1038/s41575-019-0115-0. [DOI] [PubMed] [Google Scholar]
- 43.Wang D, Wang X, Song Y, Si M, Sun Y, Liu X, Cui S, Qu X, Yu X. Exosomal miR-146a-5p and miR-155-5p promote CXCL12/CXCR7-induced metastasis of colorectal cancer by crosstalk with cancer-associated fibroblasts. Cell Death Dis. 2022;13:380. doi: 10.1038/s41419-022-04825-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi Y, Wang H, Zhang Q, Liu Z, Wang T, Wu Z, Wu W. CAF-Released Exosomal miR-20a-5p Facilitates HCC Progression via the LIMA1-Mediated beta-Catenin Pathway. Cells. 2022;11(23):3857. doi: 10.3390/cells11233857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manfioletti G, Fedele M. Epithelial-Mesenchymal Transition (EMT) 2021. Int J Mol Sci. 2022;23(10):5848. doi: 10.3390/ijms23105848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J, Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu C, Su Y, Liu L, Wang S, Liu Y, Wu J. Circular RNA hsa_circ_0004277 Stimulates Malignant Phenotype of Hepatocellular Carcinoma and Epithelial-Mesenchymal Transition of Peripheral Cells. Front Cell Dev Biol. 2020;8:585565. doi: 10.3389/fcell.2020.585565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paolillo M, Schinelli S. Integrins and Exosomes, a Dangerous Liaison in Cancer Progression. Cancers (Basel) 2017;9(8):95. doi: 10.3390/cancers9080095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai S, Wei Y, Liu R, Xu R, Xiang L, Du J. Role of tumour-derived exosomes in metastasis. Biomed Pharmacother. 2022;147:112657. doi: 10.1016/j.biopha.2022.112657. [DOI] [PubMed] [Google Scholar]
- 51.Stoeltzing O, Liu W, Reinmuth N, Fan F, Parry GC, Parikh AA, McCarty MF, Bucana CD, Mazar AP, Ellis LM. Inhibition of integrin alpha5beta1 function with a small peptide (ATN-161) plus continuous 5-FU infusion reduces colorectal liver metastases and improves survival in mice. Int J Cancer. 2003;104:496–503. doi: 10.1002/ijc.10958. [DOI] [PubMed] [Google Scholar]
- 52.Yoshimura K, Meckel KF, Laird LS, Chia CY, Park JJ, Olino KL, Tsunedomi R, Harada T, Iizuka N, Hazama S, et al. Integrin alpha2 mediates selective metastasis to the liver. Cancer Res. 2009;69:7320–7328. doi: 10.1158/0008-5472.CAN-09-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whiteside TL. Exosomes and tumor-mediated immune suppression. J Clin Invest. 2016;126:1216–1223. doi: 10.1172/JCI81136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang J, He J, Feng C, Tu C. Exosomal MiRNAs in Osteosarcoma: Biogenesis and Biological Functions. Front Pharmacol. 2022;13:902049. doi: 10.3389/fphar.2022.902049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skokos D, Botros HG, Demeure C, Morin J, Peronet R, Birkenmeier G, Boudaly S, Mecheri S. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J Immunol. 2003;170:3037–3045. doi: 10.4049/jimmunol.170.6.3037. [DOI] [PubMed] [Google Scholar]
- 56.Whiteside TL. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes) Biochem Soc Trans. 2013;41:245–251. doi: 10.1042/BST20120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clayton A, Mitchell JP, Court J, Linnane S, Mason MD, Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression. J Immunol. 2008;180:7249–7258. doi: 10.4049/jimmunol.180.11.7249. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, Li S. Downregulation of hsa_circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting with HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization. Int J Nanomedicine. 2021;16:2803–2818. doi: 10.2147/IJN.S284560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu JC, Zhang PF, Huang XY, Guo XJ, Gao C, Zeng HY, Zheng YM, Wang SW, Cai JB, Sun QM, et al. Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J Hematol Oncol. 2021;14:200. doi: 10.1186/s13045-021-01207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raza A, Khan AQ, Inchakalody VP, Mestiri S, Yoosuf Z, Bedhiafi T, El-Ella DMA, Taib N, Hydrose S, Akbar S, et al. Dynamic liquid biopsy components as predictive and prognostic biomarkers in colorectal cancer. J Exp Clin Cancer Res. 2022;41:99. doi: 10.1186/s13046-022-02318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y, Shao Y, Zheng S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther. 2020;5:144. doi: 10.1038/s41392-020-00258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thakur A, Qiu G, Ng SP, Guan J, Yue J, Lee Y, Wu CL. Direct detection of two different tumor-derived extracellular vesicles by SAM-AuNIs LSPR biosensor. Biosens Bioelectron. 2017;94:400–407. doi: 10.1016/j.bios.2017.03.036. [DOI] [PubMed] [Google Scholar]
- 64.Pi C, Zhang MF, Peng XX, Zhang YC, Xu CR, Zhou Q. Liquid biopsy in non-small cell lung cancer: a key role in the future of personalized medicine? Expert Rev Mol Diagn. 2017;17:1089–1096. doi: 10.1080/14737159.2017.1395701. [DOI] [PubMed] [Google Scholar]
- 65.Guo S, Chen J, Chen F, Zeng Q, Liu WL, Zhang G. Exosomes derived from Fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut. 2022;71(2):e1–e3. doi: 10.1136/gutjnl-2020-321187corr1. [DOI] [PubMed] [Google Scholar]
- 66.Liu K, Dou R, Yang C, Di Z, Shi D, Zhang C, Song J, Fang Y, Huang S, Xiang Z, et al. Exosome-transmitted miR-29a induces colorectal cancer metastasis by destroying the vascular endothelial barrier. Carcinogenesis. 2023;44(4):356–367. doi: 10.1093/carcin/bgad013. [DOI] [PubMed] [Google Scholar]
- 67.Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, Zambirinis CP, Rodrigues G, Molina H, Heissel S, et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell. 2020;182(1044–1061):e1018. doi: 10.1016/j.cell.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang C, Wang XY, Zhang P, He TC, Han JH, Zhang R, Lin J, Fan J, Lu L, Zhu WW, et al. Cancer-derived exosomal HSPC111 promotes colorectal cancer liver metastasis by reprogramming lipid metabolism in cancer-associated fibroblasts. Cell Death Dis. 2022;13:57. doi: 10.1038/s41419-022-04506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun J, Lu Z, Fu W, Lu K, Gu X, Xu F, Dai J, Yang Y, Jiang J. Exosome-Derived ADAM17 Promotes Liver Metastasis in Colorectal Cancer. Front Pharmacol. 2021;12:734351. doi: 10.3389/fphar.2021.734351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cardenes B, Clares I, Toribio V, Pascual L, Lopez-Martin S, Torres-Gomez A, de la Cuesta Sainz R, Lafuente EM, Lopez-Cabrera M, Yanez-Mo M, Cabanas C. Cellular Integrin alpha5beta1 and Exosomal ADAM17 Mediate the Binding and Uptake of Exosomes Produced by Colorectal Carcinoma Cells. Int J Mol Sci. 2021;22(18):9938. doi: 10.3390/ijms22189938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lucchetti D, Zurlo IV, Colella F, Ricciardi-Tenore C, Di Salvatore M, Tortora G, De Maria R, Giuliante F, Cassano A, Basso M, et al. Mutational status of plasma exosomal KRAS predicts outcome in patients with metastatic colorectal cancer. Sci Rep. 2021;11:22686. doi: 10.1038/s41598-021-01668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang M, Zhao X, Qiu R, Gong Z, Huang F, Yu W, Shen B, Sha X, Dong H, Huang J, et al. Lymph node metastasis-derived gastric cancer cells educate bone marrow-derived mesenchymal stem cells via YAP signaling activation by exosomal Wnt5a. Oncogene. 2021;40:2296–2308. doi: 10.1038/s41388-021-01722-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Q, Peng K, Chen E, Jiang H, Wang Y, Yu S, Li W, Yu Y, Liu T. IntegrinB5 upregulated by HER2 in gastric cancer: a promising biomarker for liver metastasis. Ann Transl Med. 2020;8:451. doi: 10.21037/atm.2020.03.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun H, Wang C, Hu B, Gao X, Zou T, Luo Q, Chen M, Fu Y, Sheng Y, Zhang K, et al. Exosomal S100A4 derived from highly metastatic hepatocellular carcinoma cells promotes metastasis by activating STAT3. Signal Transduct Target Ther. 2021;6:187. doi: 10.1038/s41392-021-00579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu AK, Shan YQ, Zhang J, Liu XC, Ying RC, Kong WC. Exosomal NNMT from peritoneum lavage fluid promotes peritoneal metastasis in gastric cancer. Kaohsiung J Med Sci. 2021;37:305–313. doi: 10.1002/kjm2.12334. [DOI] [PubMed] [Google Scholar]
- 76.Xie Z, Gao Y, Ho C, Li L, Jin C, Wang X, Zou C, Mao Y, Wang X, Li Q, et al. Exosome-delivered CD44v6/C1QBP complex drives pancreatic cancer liver metastasis by promoting fibrotic liver microenvironment. Gut. 2022;71:568–579. doi: 10.1136/gutjnl-2020-323014. [DOI] [PubMed] [Google Scholar]
- 77.Luna J, Boni J, Cuatrecasas M, Bofill-De Ros X, Nunez-Manchon E, Gironella M, Vaquero EC, Arbones ML, de la Luna S, Fillat C. DYRK1A modulates c-MET in pancreatic ductal adenocarcinoma to drive tumour growth. Gut. 2019;68:1465–1476. doi: 10.1136/gutjnl-2018-316128. [DOI] [PubMed] [Google Scholar]
- 78.Ohshima K, Hatakeyama K, Kanto K, Ide T, Watanabe Y, Moromizato S, Wakabayashi-Nakao K, Sakura N, Yamaguchi K, Mochizuki T. Comparative proteomic analysis identifies exosomal Eps8 protein as a potential metastatic biomarker for pancreatic cancer. Oncol Rep. 2019;41:1019–1034. doi: 10.3892/or.2018.6869. [DOI] [PubMed] [Google Scholar]
- 79.Liu Z, Hu C, Zheng L, Liu J, Li K, Li X, Wang Y, Mu W, Chen T, Shi A, et al. BMI1 promotes cholangiocarcinoma progression and correlates with antitumor immunity in an exosome-dependent manner. Cell Mol Life Sci. 2022;79:469. doi: 10.1007/s00018-022-04500-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Emmanouilidi A, Paladin D, Greening DW, Falasca M. Oncogenic and Non-Malignant Pancreatic Exosome Cargo Reveal Distinct Expression of Oncogenic and Prognostic Factors Involved in Tumor Invasion and Metastasis. Proteomics. 2019;19:e1800158. doi: 10.1002/pmic.201800158. [DOI] [PubMed] [Google Scholar]
- 81.Jiang K, Dong C, Yin Z, Li R, Mao J, Wang C, Zhang J, Gao Z, Liang R, Wang Q, Wang L. Exosome-derived ENO1 regulates integrin alpha6beta4 expression and promotes hepatocellular carcinoma growth and metastasis. Cell Death Dis. 2020;11:972. doi: 10.1038/s41419-020-03179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li R, Wang Y, Zhang X, Feng M, Ma J, Li J, Yang X, Fang F, Xia Q, Zhang Z, et al. Exosome-mediated secretion of LOXL4 promotes hepatocellular carcinoma cell invasion and metastasis. Mol Cancer. 2019;18:18. doi: 10.1186/s12943-019-0948-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liang Z, Liu H, Zhang Y, Xiong L, Zeng Z, He X, Wang F, Wu X, Lan P. Cyr61 from adipose-derived stem cells promotes colorectal cancer metastasis and vasculogenic mimicry formation via integrin alpha(V) beta(5) Mol Oncol. 2021;15:3447–3467. doi: 10.1002/1878-0261.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He G, Li W, Zhao W, Men H, Chen Q, Hu J, Zhang J, Zhu H, Wang W, Deng M, et al. Formin-like 2 promotes angiogenesis and metastasis of colorectal cancer by regulating the EGFL6/CKAP4/ERK axis. Cancer Sci. 2023;114:2014–2028. doi: 10.1111/cas.15739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y, Li P, Mao S, Mo Z, Cao Z, Luo J, Zhou M, Liu X, Zhang S, Yu L. Exosome CTLA-4 Regulates PTEN/CD44 Signal Pathway in Spleen Deficiency Internal Environment to Promote Invasion and Metastasis of Hepatocellular Carcinoma. Front Pharmacol. 2021;12:757194. doi: 10.3389/fphar.2021.757194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ogawa K, Lin Q, Li L, Bai X, Chen X, Chen H, Kong R, Wang Y, Zhu H, He F, et al. Prometastatic secretome trafficking via exosomes initiates pancreatic cancer pulmonary metastasis. Cancer Lett. 2020;481:63–75. doi: 10.1016/j.canlet.2020.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu P, Zu F, Chen H, Yin X, Tan X. Exosomal DNAJB11 promotes the development of pancreatic cancer by modulating the EGFR/MAPK pathway. Cell Mol Biol Lett. 2022;27:87. doi: 10.1186/s11658-022-00390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu P, Kong L, Jin H, Wu Y, Tan X, Song B. Differential secretome of pancreatic cancer cells in serum-containing conditioned medium reveals CCT8 as a new biomarker of pancreatic cancer invasion and metastasis. Cancer Cell Int. 2019;19:262. doi: 10.1186/s12935-019-0980-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He Z, Wang J, Zhu C, Xu J, Chen P, Jiang X, Chen Y, Jiang J, Sun C. Exosome-derived FGD5-AS1 promotes tumor-associated macrophage M2 polarization-mediated pancreatic cancer cell proliferation and metastasis. Cancer Lett. 2022;548:215751. doi: 10.1016/j.canlet.2022.215751. [DOI] [PubMed] [Google Scholar]
- 90.Zhang YF, Zhou YZ, Zhang B, Huang SF, Li PP, He XM, Cao GD, Kang MX, Dong X, Wu YL. Pancreatic cancer-derived exosomes promoted pancreatic stellate cells recruitment by pancreatic cancer. J Cancer. 2019;10:4397–4407. doi: 10.7150/jca.27590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meltzer S, Bjornetro T, Lyckander LG, Flatmark K, Dueland S, Samiappan R, Johansen C, Kalanxhi E, Ree AH, Redalen KR. Circulating Exosomal miR-141-3p and miR-375 in Metastatic Progression of Rectal Cancer. Transl Oncol. 2019;12:1038–1044. doi: 10.1016/j.tranon.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu H, Liu Y, Sun P, Leng K, Xu Y, Mei L, Han P, Zhang B, Yao K, Li C, et al. Colorectal cancer-derived exosomal miR-106b-3p promotes metastasis by down-regulating DLC-1 expression. Clin Sci (Lond) 2020;134:419–434. doi: 10.1042/CS20191087. [DOI] [PubMed] [Google Scholar]
- 93.Mannavola F, Pezzicoli G, Tucci M. DLC-1 down-regulation via exosomal miR-106b-3p exchange promotes CRC metastasis by the epithelial-to-mesenchymal transition. Clin Sci (Lond) 2020;134:955–959. doi: 10.1042/CS20200181. [DOI] [PubMed] [Google Scholar]
- 94.Tian F, Wang P, Lin D, Dai J, Liu Q, Guan Y, Zhan Y, Yang Y, Wang W, Wang J, et al. Exosome-delivered miR-221/222 exacerbates tumor liver metastasis by targeting SPINT1 in colorectal cancer. Cancer Sci. 2021;112:3744–3755. doi: 10.1111/cas.15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu Y, Luan G, Liu F, Zhang Y, Li Z, Liu Z, Yang T. Exosomal miR-200b-3p induce macrophage polarization by regulating transcriptional repressor ZEB1 in hepatocellular carcinoma. Hepatol Int. 2023;17(4):889–903. doi: 10.1007/s12072-023-10507-y. [DOI] [PubMed] [Google Scholar]
- 96.Huang C, Tang S, Shen D, Li X, Liang L, Ding Y, Xu B. Circulating plasma exosomal miRNA profiles serve as potential metastasis-related biomarkers for hepatocellular carcinoma. Oncol Lett. 2021;21:168. doi: 10.3892/ol.2021.12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eun JW, Seo CW, Baek GO, Yoon MG, Ahn HR, Son JA, Sung S, Kim DW, Kim SS, Cho HJ, Cheong JY. Circulating Exosomal MicroRNA-1307–5p as a Predictor for Metastasis in Patients with Hepatocellular Carcinoma. Cancers (Basel) 2020;12(12):3819. doi: 10.3390/cancers12123819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qiu S, Xie L, Lu C, Gu C, Xia Y, Lv J, Xuan Z, Fang L, Yang J, Zhang L, et al. Gastric cancer-derived exosomal miR-519a-3p promotes liver metastasis by inducing intrahepatic M2-like macrophage-mediated angiogenesis. J Exp Clin Cancer Res. 2022;41:296. doi: 10.1186/s13046-022-02499-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu X, Chu KM. Exosomal miRNAs as circulating biomarkers for prediction of development of haematogenous metastasis after surgery for stage II/III gastric cancer. J Cell Mol Med. 2020;24:6220–6232. doi: 10.1111/jcmm.15253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ge L, Zhang N, Li D, Wu Y, Wang H, Wang J. Circulating exosomal small RNAs are promising non-invasive diagnostic biomarkers for gastric cancer. J Cell Mol Med. 2020;24:14502–14513. doi: 10.1111/jcmm.16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cho HJ, Baek GO, Seo CW, Ahn HR, Sung S, Son JA, Kim SS, Cho SW, Jang JW, Nam SW, et al. Exosomal microRNA-4661-5p-based serum panel as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. Cancer Med. 2020;9:5459–5472. doi: 10.1002/cam4.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bai J, Zhang X, Shi D, Xiang Z, Wang S, Yang C, Liu Q, Huang S, Fang Y, Zhang W, et al. Exosomal miR-128-3p Promotes Epithelial-to-Mesenchymal Transition in Colorectal Cancer Cells by Targeting FOXO4 via TGF-beta/SMAD and JAK/STAT3 Signaling. Front Cell Dev Biol. 2021;9:568738. doi: 10.3389/fcell.2021.568738. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 103.Wang X, Luo G, Zhang K, Cao J, Huang C, Jiang T, Liu B, Su L, Qiu Z. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kgamma to Promote Pancreatic Cancer Metastasis. Cancer Res. 2018;78:4586–4598. doi: 10.1158/0008-5472.CAN-17-3841. [DOI] [PubMed] [Google Scholar]
- 104.Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, Song FY, Wang FF, Zhu XH, Liao WJ, et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18:91. doi: 10.1186/s12943-019-1019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gu J, Chu X, Huo Y, Liu C, Chen Q, Hu S, Pei Y, Ding P, Pang S, Wang M. Gastric cancer-derived exosomes facilitate pulmonary metastasis by activating ERK-mediated immunosuppressive macrophage polarization. J Cell Biochem. 2023;124:557–572. doi: 10.1002/jcb.30390. [DOI] [PubMed] [Google Scholar]
- 106.Ohzawa H, Kumagai Y, Yamaguchi H, Miyato H, Sakuma Y, Horie H, Hosoya Y, Kawarai Lefor A, Sata N, Kitayama J. Exosomal microRNA in peritoneal fluid as a biomarker of peritoneal metastases from gastric cancer. Ann Gastroenterol Surg. 2020;4:84–93. doi: 10.1002/ags3.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang B, Feng X, Liu H, Tong R, Wu J, Li C, Yu H, Chen Y, Cheng Q, Chen J, et al. High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene. 2020;39:6529–6543. doi: 10.1038/s41388-020-01450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu H, Lan Q, Huang Y, Zhang Y, Zeng Y, Su P, Chu Z, Lai W, Chu Z. The mechanisms of colorectal cancer cell mesenchymal-epithelial transition induced by hepatocyte exosome-derived miR-203a-3p. BMC Cancer. 2021;21:718. doi: 10.1186/s12885-021-08419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Q, Zhang C, Cao S, Zhao H, Jiang R, Li Y. Tumor-derived exosomes orchestrate the microRNA-128–3p/ELF4/CDX2 axis to facilitate the growth and metastasis of gastric cancer via delivery of LINC01091. Cell Biol Toxicol. 2022;39(2):519–536. doi: 10.1007/s10565-022-09728-y. [DOI] [PubMed] [Google Scholar]
- 110.Chang Y, Huang Z, Hou F, Liu Y, Wang L, Wang Z, Sun Y, Pan Z, Tan Y, Ding L, et al. Parvimonas micra activates the Ras/ERK/c-Fos pathway by upregulating miR-218-5p to promote colorectal cancer progression. J Exp Clin Cancer Res. 2023;42:13. doi: 10.1186/s13046-022-02572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jiang Y, Qiu Q, Jing X, Song Z, Zhang Y, Wang C, Liu K, Ye F, Ji X, Luo F, Zhao R. Cancer-associated fibroblast-derived exosome miR-181b-3p promotes the occurrence and development of colorectal cancer by regulating SNX2 expression. Biochem Biophys Res Commun. 2023;641:177–185. doi: 10.1016/j.bbrc.2022.12.026. [DOI] [PubMed] [Google Scholar]
- 112.Shang A, Wang X, Gu C, Liu W, Sun J, Zeng B, Chen C, Ji P, Wu J, Quan W, et al. Exosomal miR-183-5p promotes angiogenesis in colorectal cancer by regulation of FOXO1. Aging (Albany NY) 2020;12:8352–8371. doi: 10.18632/aging.103145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dou R, Liu K, Yang C, Zheng J, Shi D, Lin X, Wei C, Zhang C, Fang Y, Huang S, et al. EMT-cancer cells-derived exosomal miR-27b-3p promotes circulating tumour cells-mediated metastasis by modulating vascular permeability in colorectal cancer. Clin Transl Med. 2021;11:e595. doi: 10.1002/ctm2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Y, Han T, Feng D, Li J, Wu M, Peng X, Wang B, Zhan X, Fu P. Screening of non-invasive miRNA biomarker candidates for metastasis of gastric cancer by small RNA sequencing of plasma exosomes. Carcinogenesis. 2020;41:582–590. doi: 10.1093/carcin/bgz186. [DOI] [PubMed] [Google Scholar]
- 115.Liu SJ, Dang HX, Lim DA, Feng FY, Maher CA. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. 2021;21:446–460. doi: 10.1038/s41568-021-00353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Scalia M, Magnano A, Caltabiano R, Barbagallo D, Biondi A, Cappellani A, et al. LncRNA UCA1, Upregulated in CRC Biopsies and Downregulated in Serum Exosomes, Controls mRNA Expression by RNA-RNA Interactions. Mol Ther Nucleic Acids. 2018;12:229–241. doi: 10.1016/j.omtn.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Luan Y, Li X, Luan Y, Zhao R, Li Y, Liu L, Hao Y, Oleg Vladimir B, Jia L. Circulating lncRNA UCA1 Promotes Malignancy of Colorectal Cancer via the miR-143/MYO6 Axis. Mol Ther Nucleic Acids. 2020;19:790–803. doi: 10.1016/j.omtn.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cao S, Lin L, Xia X, Wu H. lncRNA SPRY4-IT1 Regulates Cell Proliferation and Migration by Sponging miR-101-3p and Regulating AMPK Expression in Gastric Cancer. Mol Ther Nucleic Acids. 2019;17:455–464. doi: 10.1016/j.omtn.2019.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 119.You LN, Tai QW, Xu L, Hao Y, Guo WJ, Zhang Q, Tong Q, Zhang H, Huang WK. Exosomal LINC00161 promotes angiogenesis and metastasis via regulating miR-590-3p/ROCK axis in hepatocellular carcinoma. Cancer Gene Ther. 2021;28:719–736. doi: 10.1038/s41417-020-00269-2. [DOI] [PubMed] [Google Scholar]
- 120.Lai SW, Chen MY, Bamodu OA, Hsieh MS, Huang TY, Yeh CT, Lee WH, Cherng YG. Exosomal lncRNA PVT1/VEGFA Axis Promotes Colon Cancer Metastasis and Stemness by Downregulation of Tumor Suppressor miR-152-3p. Oxid Med Cell Longev. 2021;2021:9959807. doi: 10.1155/2021/9959807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yao J, Hua X, Shi J, Hu X, Lui K, He K, Mai J, Lan T, Lu M. LncRNA THEMIS2-211, a tumor-originated circulating exosomal biomarker, promotes the growth and metastasis of hepatocellular carcinoma by functioning as a competing endogenous RNA. FASEB J. 2022;36:e22238. doi: 10.1096/fj.202101564R. [DOI] [PubMed] [Google Scholar]
- 122.Liang ZX, Liu HS, Wang FW, Xiong L, Zhou C, Hu T, He XW, Wu XJ, Xie D, Wu XR, Lan P. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019;10:829. doi: 10.1038/s41419-019-2077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hashemi M, Moosavi MS, Abed HM, Dehghani M, Aalipour M, Heydari EA, Behroozaghdam M, Entezari M, Salimimoghadam S, Gunduz ES, et al. Long non-coding RNA (lncRNA) H19 in human cancer: From proliferation and metastasis to therapy. Pharmacol Res. 2022;184:106418. doi: 10.1016/j.phrs.2022.106418. [DOI] [PubMed] [Google Scholar]
- 124.Xu Y, Luan G, Li Z, Liu Z, Qin G, Chu Y. Tumour-derived exosomal lncRNA SNHG16 induces telocytes to promote metastasis of hepatocellular carcinoma via the miR-942-3p/MMP9 axis. Cell Oncol (Dordr) 2023;46:251–264. doi: 10.1007/s13402-022-00746-w. [DOI] [PubMed] [Google Scholar]
- 125.Wang Y, Jiang R, Zhao H, Li F, Li Y, Zhu M. TTN-AS1 delivered by gastric cancer cell-derived exosome induces gastric cancer progression through in vivo and in vitro studies. Cell Biol Toxicol. 2023;39(2):557–571. doi: 10.1007/s10565-022-09762-w. [DOI] [PubMed] [Google Scholar]
- 126.Piao HY, Guo S, Wang Y, Zhang J. Exosome-transmitted lncRNA PCGEM1 promotes invasive and metastasis in gastric cancer by maintaining the stability of SNAI1. Clin Transl Oncol. 2021;23:246–256. doi: 10.1007/s12094-020-02412-9. [DOI] [PubMed] [Google Scholar]
- 127.Fang X, Xu Y, Li K, Liu P, Zhang H, Jiang Y, Tang J, Li Y. Exosomal lncRNA PCAT1 Promotes Tumor Circulating Cell-Mediated Colorectal Cancer Liver Metastasis by Regulating the Activity of the miR-329-3p/Netrin-1-CD146 Complex. J Immunol Res. 2022;2022:9916228. doi: 10.1155/2022/9916228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xu J, Xiao Y, Liu B, Pan S, Liu Q, Shan Y, Li S, Qi Y, Huang Y, Jia L. Exosomal MALAT1 sponges miR-26a/26b to promote the invasion and metastasis of colorectal cancer via FUT4 enhanced fucosylation and PI3K/Akt pathway. J Exp Clin Cancer Res. 2020;39:54. doi: 10.1186/s13046-020-01562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16:292–301. doi: 10.7150/ijms.28047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shen B. Circular RNAs an emerging landscape in tumor metastasis. Am J Cancer Res. 2019;9(4):630–643. [PMC free article] [PubMed] [Google Scholar]
- 132.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 133.Yu L, Xie J, Liu X, Yu Y, Wang S. Plasma Exosomal CircNEK9 Accelerates the Progression of Gastric Cancer via miR-409-3p/MAP7 Axis. Dig Dis Sci. 2021;66:4274–4289. doi: 10.1007/s10620-020-06816-z. [DOI] [PubMed] [Google Scholar]
- 134.Wang S, Hu Y, Lv X, Li B, Gu D, Li Y, Sun Y, Su Y. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin Sci (Lond) 2019;133:1935–1953. doi: 10.1042/CS20190589. [DOI] [PubMed] [Google Scholar]
- 135.Chen C, Liu Y, Liu L, Si C, Xu Y, Wu X, Wang C, Sun Z, Kang Q. Exosomal circTUBGCP4 promotes vascular endothelial cell tipping and colorectal cancer metastasis by activating Akt signaling pathway. J Exp Clin Cancer Res. 2023;42:46. doi: 10.1186/s13046-023-02619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang H, Zhang H, Yang Y, Wang X, Deng T, Liu R, Ning T, Bai M, Li H, Zhu K, et al. Hypoxia induced exosomal circRNA promotes metastasis of Colorectal Cancer via targeting GEF-H1/RhoA axis. Theranostics. 2020;10:8211–8226. doi: 10.7150/thno.44419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu C, Ren C, Guo L, Yang C, Yu Q. Exosome-mediated circTTLL5 transfer promotes hepatocellular carcinoma malignant progression through miR-136-5p/KIAA1522 axis. Pathol Res Pract. 2023;241:154276. doi: 10.1016/j.prp.2022.154276. [DOI] [PubMed] [Google Scholar]
- 138.Shen X, Kong S, Ma S, Shen L, Zheng M, Qin S, Qi J, Wang Q, Cui X, Ju S. Hsa_circ_0000437 promotes pathogenesis of gastric cancer and lymph node metastasis. Oncogene. 2022;41:4724–4735. doi: 10.1038/s41388-022-02449-w. [DOI] [PubMed] [Google Scholar]
- 139.Li Y, Hu J, Wang M, Yuan Y, Zhou F, Zhao H, Qiu T, Liang L. Exosomal circPABPC1 promotes colorectal cancer liver metastases by regulating HMGA2 in the nucleus and BMP4/ADAM19 in the cytoplasm. Cell Death Discov. 2022;8:335. doi: 10.1038/s41420-022-01124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Han K, Wang FW, Cao CH, Ling H, Chen JW, Chen RX, Feng ZH, Luo J, Jin XH, Duan JL, et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer. 2020;19:60. doi: 10.1186/s12943-020-01184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, Zhang Y, Li H, Zhang Q, Yang Y, Chen G. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432–445. doi: 10.1016/j.ebiom.2018.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li T, Zhou T, Wu J, Lv H, Zhou H, Du M, Zhang X, Wu N, Gong S, Ren Z, et al. Plasma exosome-derived circGAPVD1 as a potential diagnostic marker for colorectal cancer. Transl Oncol. 2023;31:101652. doi: 10.1016/j.tranon.2023.101652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038. [DOI] [PubMed] [Google Scholar]
- 144.Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, Ma P, Jiang H, Wu X, Shu Y, Xu T. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19:112. doi: 10.1186/s12943-020-01208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X, Li X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, Zhao X, Zhang Y, Zhen P, Zhu J, Li X. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250. doi: 10.1016/j.canlet.2018.04.035. [DOI] [PubMed] [Google Scholar]
- 147.Zeng K, Peng J, Xing Y, Zhang L, Zeng P, Li W, Zhang W, Pan Z, Zhou C, Lin J. A positive feedback circuit driven by m(6)A-modified circular RNA facilitates colorectal cancer liver metastasis. Mol Cancer. 2023;22:202. doi: 10.1186/s12943-023-01848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sang H, Zhang W, Peng L, Wei S, Zhu X, Huang K, Yang J, Chen M, Dang Y, Zhang G. Exosomal circRELL1 serves as a miR-637 sponge to modulate gastric cancer progression via regulating autophagy activation. Cell Death Dis. 2022;13:56. doi: 10.1038/s41419-021-04364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y, Jiang J, Wang L, Mang Y, Gao Y, et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer. 2023;22:55. doi: 10.1186/s12943-023-01759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, Cai JB, Ke AW. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19:110. doi: 10.1186/s12943-020-01222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, Liu R, Fan Q, Zhu K, Li J, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539–555. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zeng Z, Zhao Y, Chen Q, Zhu S, Niu Y, Ye Z, Hu P, Chen D, Xu P, Chen J, et al. Hypoxic exosomal HIF-1alpha-stabilizing circZNF91 promotes chemoresistance of normoxic pancreatic cancer cells via enhancing glycolysis. Oncogene. 2021;40:5505–5517. doi: 10.1038/s41388-021-01960-w. [DOI] [PubMed] [Google Scholar]
- 153.Zeng W, Liu Y, Li WT, Li Y, Zhu JF. CircFNDC3B sequestrates miR-937-5p to derepress TIMP3 and inhibit colorectal cancer progression. Mol Oncol. 2020;14:2960–2984. doi: 10.1002/1878-0261.12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen C, Yu H, Han F, Lai X, Ye K, Lei S, Mai M, Lai M, Zhang H. Tumor-suppressive circRHOBTB3 is excreted out of cells via exosome to sustain colorectal cancer cell fitness. Mol Cancer. 2022;21:46. doi: 10.1186/s12943-022-01511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu D, Chen C, Cui M, Zhang H. miR-140-3p inhibits colorectal cancer progression and its liver metastasis by targeting BCL9 and BCL2. Cancer Med. 2021;10:3358–3372. doi: 10.1002/cam4.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jahangiri B, Khalaj-Kondori M, Asadollahi E, Purrafee Dizaj L, Sadeghizadeh M. MSC-Derived exosomes suppress colorectal cancer cell proliferation and metastasis via miR-100/mTOR/miR-143 pathway. Int J Pharm. 2022;627:122214. doi: 10.1016/j.ijpharm.2022.122214. [DOI] [PubMed] [Google Scholar]
- 157.Li N, Li J, Mi Q, Xie Y, Li P, Wang L, Binang H, Wang Q, Wang Y, Chen Y, et al. Long non-coding RNA ADAMTS9-AS1 suppresses colorectal cancer by inhibiting the Wnt/beta-catenin signalling pathway and is a potential diagnostic biomarker. J Cell Mol Med. 2020;24:11318–11329. doi: 10.1111/jcmm.15713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jiang K, Chen H, Fang Y, Chen L, Zhong C, Bu T, Dai S, Pan X, Fu D, Qian Y, et al. Exosomal ANGPTL1 attenuates colorectal cancer liver metastasis by regulating Kupffer cell secretion pattern and impeding MMP9 induced vascular leakiness. J Exp Clin Cancer Res. 2021;40:21. doi: 10.1186/s13046-020-01816-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bongolo CC, Thokerunga E, Yan Q, Yacouba MBM, Wang C. Exosomes Derived from microRNA-27a-3p Overexpressing Mesenchymal Stem Cells Inhibit the Progression of Liver Cancer through Suppression of Golgi Membrane Protein 1. Stem Cells Int. 2022;2022:9748714. doi: 10.1155/2022/9748714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ding B, Lou W, Fan W, Pan J. Exosomal miR-374c-5p derived from mesenchymal stem cells suppresses epithelial-mesenchymal transition of hepatocellular carcinoma via the LIMK1-Wnt/beta-catenin axis. Environ Toxicol. 2023;38:1038–1052. doi: 10.1002/tox.23746. [DOI] [PubMed] [Google Scholar]
- 161.Lin Y, Zheng ZH, Wang JX, Zhao Z, Peng TY. Tumor Cell-Derived Exosomal Circ-0072088 Suppresses Migration and Invasion of Hepatic Carcinoma Cells Through Regulating MMP-16. Front Cell Dev Biol. 2021;9:726323. doi: 10.3389/fcell.2021.726323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, Chen L, Liu Q, He P, Ye Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. doi: 10.1016/j.canlet.2020.01.022. [DOI] [PubMed] [Google Scholar]
- 163.Cheng Z, Lei Z, Yang P, Si A, Xiang D, Tang X, Guo G, Zhou J, Huser N. Exosome-transmitted p120-catenin suppresses hepatocellular carcinoma progression via STAT3 pathways. Mol Carcinog. 2019;58:1389–1399. doi: 10.1002/mc.23022. [DOI] [PubMed] [Google Scholar]
- 164.Jiang J, Li R, Wang J, Hou J, Qian H, Xu W. Circular RNA CDR1as Inhibits the Metastasis of Gastric Cancer through Targeting miR-876-5p/GNG7 Axis. Gastroenterol Res Pract. 2021;2021:5583029. doi: 10.1155/2021/5583029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang Y, Wang H, Zheng R, Wu P, Sun Z, Chen J, Zhang L, Zhang C, Qian H, Jiang J, Xu W. Circular RNA ITCH suppresses metastasis of gastric cancer via regulating miR-199a-5p/Klotho axis. Cell Cycle. 2021;20:522–536. doi: 10.1080/15384101.2021.1878327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li M, Zhou J, Zhang Z, Li J, Wang F, Ma L, Tian X, Mao Z, Yang Y. Exosomal miR-485-3p derived from pancreatic ductal epithelial cells inhibits pancreatic cancer metastasis through targeting PAK1. Chin Med J (Engl) 2022;135:2326–2337. doi: 10.1097/CM9.0000000000002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yao X, Mao Y, Wu D, Zhu Y, Lu J, Huang Y, Guo Y, Wang Z, Zhu S, Li X, Lu Y. Exosomal circ_0030167 derived from BM-MSCs inhibits the invasion, migration, proliferation and stemness of pancreatic cancer cells by sponging miR-338-5p and targeting the Wif1/Wnt8/beta-catenin axis. Cancer Lett. 2021;512:38–50. doi: 10.1016/j.canlet.2021.04.030. [DOI] [PubMed] [Google Scholar]
- 168.Gao G, Wang L, Li C. Circ_0006790 carried by bone marrow mesenchymal stem cell-derived exosomes regulates S100A11 DNA methylation through binding to CBX7 in pancreatic ductal adenocarcinoma. Am J Cancer Res. 2022;12:1934–1959. [PMC free article] [PubMed] [Google Scholar]
- 169.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 170.Robinson H, Ruelcke JE, Lewis A, Bond CS, Fox AH, Bharti V, Wani S, Cloonan N, Lai A, Margolin D, et al. Caveolin-1-driven membrane remodelling regulates hnRNPK-mediated exosomal microRNA sorting in cancer. Clin Transl Med. 2021;11:e381. doi: 10.1002/ctm2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W, Zhang R, Wu Y, Gao S, Kang T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021;31:157–177. doi: 10.1038/s41422-020-00409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gauthier-Rouviere C, Bodin S, Comunale F, Planchon D. Flotillin membrane domains in cancer. Cancer Metastasis Rev. 2020;39:361–374. doi: 10.1007/s10555-020-09873-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Han QF, Li WJ, Hu KS, Gao J, Zhai WL, Yang JH, Zhang SJ. Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer. Mol Cancer. 2022;21:207. doi: 10.1186/s12943-022-01671-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Han Q, Lv L, Wei J, Lei X, Lin H, Li G, Cao J, Xie J, Yang W, Wu S, et al. Vps4A mediates the localization and exosome release of beta-catenin to inhibit epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 2019;457:47–59. doi: 10.1016/j.canlet.2019.04.035. [DOI] [PubMed] [Google Scholar]
- 175.Logozzi M, Mizzoni D, Angelini DF, Di Raimo R, Falchi M, Battistini L, Fais S. Microenvironmental pH and Exosome Levels Interplay in Human Cancer Cell Lines of Different Histotypes. Cancers (Basel) 2018;10(10):370. doi: 10.3390/cancers10100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Federici C, Petrucci F, Caimi S, Cesolini A, Logozzi M, Borghi M, D'Ilio S, Lugini L, Violante N, Azzarito T, et al. Exosome release and low pH belong to a framework of resistance of human melanoma cells to cisplatin. PLoS ONE. 2014;9:e88193. doi: 10.1371/journal.pone.0088193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhao H, Achreja A, Iessi E, Logozzi M, Mizzoni D, Di Raimo R, Nagrath D, Fais S. The key role of extracellular vesicles in the metastatic process. Biochim Biophys Acta Rev Cancer. 2018;1869:64–77. doi: 10.1016/j.bbcan.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hamaguchi R, Isowa M, Narui R, Morikawa H, Wada H. Clinical review of alkalization therapy in cancer treatment. Front Oncol. 2022;12:1003588. doi: 10.3389/fonc.2022.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Datta A, Kim H, Lal M, McGee L, Johnson A, Moustafa AA, Jones JC, Mondal D, Ferrer M, Abdel-Mageed AB. Manumycin A suppresses exosome biogenesis and secretion via targeted inhibition of Ras/Raf/ERK1/2 signaling and hnRNP H1 in castration-resistant prostate cancer cells. Cancer Lett. 2017;408:73–81. doi: 10.1016/j.canlet.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Asai H, Ikezu S, Tsunoda S, Medalla M, Luebke J, Haydar T, Wolozin B, Butovsky O, Kugler S, Ikezu T. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat Neurosci. 2015;18:1584–1593. doi: 10.1038/nn.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sun H, Meng Q, Shi C, Yang H, Li X, Wu S, Familiari G, Relucenti M, Aschner M, Wang X, Chen R. Hypoxia-Inducible Exosomes Facilitate Liver-Tropic Premetastatic Niche in Colorectal Cancer. Hepatology. 2021;74:2633–2651. doi: 10.1002/hep.32009. [DOI] [PubMed] [Google Scholar]
- 182.Sento S, Sasabe E, Yamamoto T. Application of a Persistent Heparin Treatment Inhibits the Malignant Potential of Oral Squamous Carcinoma Cells Induced by Tumor Cell-Derived Exosomes. PLoS ONE. 2016;11:e0148454. doi: 10.1371/journal.pone.0148454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gong C, Zhang X, Shi M, Li F, Wang S, Wang Y, Wang Y, Wei W, Ma G. Tumor Exosomes Reprogrammed by Low pH Are Efficient Targeting Vehicles for Smart Drug Delivery and Personalized Therapy against their Homologous Tumor. Adv Sci (Weinh) 2021;8:2002787. doi: 10.1002/advs.202002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Goh WJ, Zou S, Ong WY, Torta F, Alexandra AF, Schiffelers RM, Storm G, Wang JW, Czarny B, Pastorin G. Bioinspired Cell-Derived Nanovesicles versus Exosomes as Drug Delivery Systems: a Cost-Effective Alternative. Sci Rep. 2017;7:14322. doi: 10.1038/s41598-017-14725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bunggulawa EJ, Wang W, Yin T, Wang N, Durkan C, Wang Y, Wang G. Recent advancements in the use of exosomes as drug delivery systems. J Nanobiotechnology. 2018;16:81. doi: 10.1186/s12951-018-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.You B, Jin C, Zhang J, Xu M, Xu W, Sun Z, Qian H. MSC-Derived Extracellular Vesicle-Delivered L-PGDS Inhibit Gastric Cancer Progression by Suppressing Cancer Cell Stemness and STAT3 Phosphorylation. Stem Cells Int. 2022;2022:9668239. doi: 10.1155/2022/9668239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pascucci L, Cocce V, Bonomi A, Ami D, Ceccarelli P, Ciusani E, Vigano L, Locatelli A, Sisto F, Doglia SM, et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: a new approach for drug delivery. J Control Release. 2014;192:262–270. doi: 10.1016/j.jconrel.2014.07.042. [DOI] [PubMed] [Google Scholar]
- 189.Wang Y, Chen X, Lin H, Sun X, Fong WP, Wu X, Pan Z, Yuan Y, Liang J, Wang D, et al. A Prehepatectomy Circulating Exosomal microRNA Signature Predicts the Prognosis and Adjuvant Chemotherapeutic Benefits in Colorectal Liver Metastasis. Cancers (Basel) 2021;13(17):4258. doi: 10.3390/cancers13174258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sun X, Lin F, Sun W, Zhu W, Fang D, Luo L, Li S, Zhang W, Jiang L. Exosome-transmitted miRNA-335-5p promotes colorectal cancer invasion and metastasis by facilitating EMT via targeting RASA1. Mol Ther Nucleic Acids. 2021;24:164–174. doi: 10.1016/j.omtn.2021.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sun L, Liu X, Pan B, Hu X, Zhu Y, Su Y, Guo Z, Zhang G, Xu M, Xu X, et al. Serum exosomal miR-122 as a potential diagnostic and prognostic biomarker of colorectal cancer with liver metastasis. J Cancer. 2020;11:630–637. doi: 10.7150/jca.33022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Cho WC, Kim M, Park JW, Jeong SY, Ku JL. Exosomal miR-193a and let-7g accelerate cancer progression on primary colorectal cancer and paired peritoneal metastatic cancer. Transl Oncol. 2021;14:101000. doi: 10.1016/j.tranon.2020.101000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yang C, Dou R, Wei C, Liu K, Shi D, Zhang C, Liu Q, Wang S, Xiong B. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol Ther. 2021;29:2088–2107. doi: 10.1016/j.ymthe.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Han S, Wang D, Huang Y, Zeng Z, Xu P, Xiong H, Ke Z, Zhang Y, Hu Y, Wang F, et al. A reciprocal feedback between colon cancer cells and Schwann cells promotes the proliferation and metastasis of colon cancer. J Exp Clin Cancer Res. 2022;41:348. doi: 10.1186/s13046-022-02556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, Zhang Z, Cai S, Xu Y, Li X, et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol. 2020;13:156. doi: 10.1186/s13045-020-00991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang D, Wang X, Si M, Yang J, Sun S, Wu H, Cui S, Qu X, Yu X. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020;474:36–52. doi: 10.1016/j.canlet.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 197.Hu HY, Yu CH, Zhang HH, Zhang SZ, Yu WY, Yang Y, Chen Q. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int J Biol Macromol. 2019;132:470–477. doi: 10.1016/j.ijbiomac.2019.03.221. [DOI] [PubMed] [Google Scholar]
- 198.Yu Q, Zhang Y, Tian Y, Peng A, Cui X, Ding B, Yang L, Liu Y, Ju Y, Gao C. Exosomal Circ_FMN2 Derived from the Serum of Colorectal Cancer Patients Promotes Cancer Progression by miR-338–3p/MSI1 Axis. Appl Biochem Biotechnol. 2023;195(12):7322–7337. doi: 10.1007/s12010-023-04456-3. [DOI] [PubMed] [Google Scholar]
- 199.Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-beta1 axis. Mol Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhao H, Chen S, Fu Q. Exosomes from CD133(+) cells carrying circ-ABCC1 mediate cell stemness and metastasis in colorectal cancer. J Cell Biochem. 2020;121:3286–3297. doi: 10.1002/jcb.29600. [DOI] [PubMed] [Google Scholar]
- 201.Pan B, Qin J, Liu X, He B, Wang X, Pan Y, Sun H, Xu T, Xu M, Chen X, et al. Identification of Serum Exosomal hsa-circ-0004771 as a Novel Diagnostic Biomarker of Colorectal Cancer. Front Genet. 2019;10:1096. doi: 10.3389/fgene.2019.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gao L, Tang X, He Q, Sun G, Wang C, Qu H. Exosome-transmitted circCOG2 promotes colorectal cancer progression via miR-1305/TGF-beta2/SMAD3 pathway. Cell Death Discov. 2021;7:281. doi: 10.1038/s41420-021-00680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zheng R, Zhang K, Tan S, Gao F, Zhang Y, Xu W, Wang H, Gu D, Zhu L, Li S, et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol Cancer. 2022;21:49. doi: 10.1186/s12943-021-01471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Li X, Lan Q, Lai W, Wu H, Xu H, Fang K, Chu Z, Zeng Y. Exosome-derived lnc-HOXB8-1:2 induces tumor-associated macrophage infiltration to promote neuroendocrine differentiated colorectal cancer progression by sponging hsa-miR-6825-5p. BMC Cancer. 2022;22:928. doi: 10.1186/s12885-022-09926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ding AX, Wang H, Zhang JM, Yang W, Kuang YT. lncRNA BANCR promotes the colorectal cancer metastasis through accelerating exosomes-mediated M2 macrophage polarization via regulating RhoA/ROCK signaling. Mol Cell Biochem. 2024;479(1):13–27. [DOI] [PubMed]
- 206.Jiang TY, Shi YY, Cui XW, Pan YF, Lin YK, Feng XF, Ding ZW, Yang C, Tan YX, Dong LW, Wang HY. PTEN Deficiency Facilitates Exosome Secretion and Metastasis in Cholangiocarcinoma by Impairing TFEB-mediated Lysosome Biogenesis. Gastroenterology. 2023;164:424–438. doi: 10.1053/j.gastro.2022.11.025. [DOI] [PubMed] [Google Scholar]
- 207.Zongqiang H, Jiapeng C, Yingpeng Z, Chuntao Y, Yiting W, Jiashun Z, Li L. Exosomal miR-452-5p Induce M2 Macrophage Polarization to Accelerate Hepatocellular Carcinoma Progression by Targeting TIMP3. J Immunol Res. 2022;2022:1032106. doi: 10.1155/2022/1032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tian XP, Wang CY, Jin XH, Li M, Wang FW, Huang WJ, Yun JP, Xu RH, Cai QQ, Xie D. Acidic Microenvironment Up-Regulates Exosomal miR-21 and miR-10b in Early-Stage Hepatocellular Carcinoma to Promote Cancer Cell Proliferation and Metastasis. Theranostics. 2019;9:1965–1979. doi: 10.7150/thno.30958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yu Y, Min Z, Zhou Z, Linhong M, Tao R, Yan L, Song H. Hypoxia-induced exosomes promote hepatocellular carcinoma proliferation and metastasis via miR-1273f transfer. Exp Cell Res. 2019;385:111649. doi: 10.1016/j.yexcr.2019.111649. [DOI] [PubMed] [Google Scholar]
- 210.Wang L, Chen X, Meng F, Huang T, Wang S, Zheng Z, Zheng G, Li W, Zhang J, Liu Y. alpha2,6-Sialylation promotes hepatocellular carcinoma cells migration and invasion via enhancement of nSmase2-mediated exosomal miRNA sorting. J Physiol Biochem. 2023;79:19–34. doi: 10.1007/s13105-022-00917-1. [DOI] [PubMed] [Google Scholar]
- 211.Yuan P, Song J, Wang F, Chen B. Exosome-transmitted circ_002136 promotes hepatocellular carcinoma progression by miR-19a-3p/RAB1A pathway. BMC Cancer. 2022;22:1284. doi: 10.1186/s12885-022-10367-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zhang T, Sun Q, Shen C, Qian Y. Circular RNA circ_0003028 regulates cell development through modulating miR-498/ornithine decarboxylase 1 axis in hepatocellular carcinoma. Anticancer Drugs. 2023;34:507–518. doi: 10.1097/CAD.0000000000001457. [DOI] [PubMed] [Google Scholar]
- 213.Liu D, Kang H, Gao M, Jin L, Zhang F, Chen D, Li M, Xiao L. Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol Oncol. 2020;14:1365–1380. doi: 10.1002/1878-0261.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, Zhou J, Tang ZY. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39:20. doi: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Huang M, Huang X, Huang N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 2022;113:1968–1983. doi: 10.1111/cas.15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Huang C, Yu W, Wang Q, Huang T, Ding Y. CircANTXR1 Contributes to the Malignant Progression of Hepatocellular Carcinoma by Promoting Proliferation and Metastasis. J Hepatocell Carcinoma. 2021;8:1339–1353. doi: 10.2147/JHC.S317256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hao X, Zhang Y, Shi X, Liu H, Zheng Z, Han G, Rong D, Zhang C, Tang W, Wang X. CircPAK1 promotes the progression of hepatocellular carcinoma via modulation of YAP nucleus localization by interacting with 14-3-3zeta. J Exp Clin Cancer Res. 2022;41:281. doi: 10.1186/s13046-022-02494-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, Qi F. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY) 2019;11:8183–8203. doi: 10.18632/aging.102312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Li Y, Zang H, Zhang X, Huang G. Exosomal Circ-ZNF652 Promotes Cell Proliferation, Migration, Invasion and Glycolysis in Hepatocellular Carcinoma via miR-29a-3p/GUCD1 Axis. Cancer Manag Res. 2020;12:7739–7751. doi: 10.2147/CMAR.S259424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wang D, Xing N, Yang T, Liu J, Zhao H, He J, Ai Y, Yang J. Exosomal lncRNA H19 promotes the progression of hepatocellular carcinoma treated with Propofol via miR-520a-3p/LIMK1 axis. Cancer Med. 2020;9:7218–7230. doi: 10.1002/cam4.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ni Q, Zhang H, Shi X, Li X. Exosomal lncRNA HCG18 contributes to cholangiocarcinoma growth and metastasis through mediating miR-424-5p/SOX9 axis through PI3K/AKT pathway. Cancer Gene Ther. 2023;30:582–595. doi: 10.1038/s41417-022-00500-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lu Y, Duan Y, Xu Q, Zhang L, Chen W, Qu Z, Wu B, Liu W, Shi L, Wu D, et al. Circulating exosome-derived bona fide long non-coding RNAs predicting the occurrence and metastasis of hepatocellular carcinoma. J Cell Mol Med. 2020;24:1311–1318. doi: 10.1111/jcmm.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Huang XY, Zhang JT, Li F, Li TT, Shi XJ, Huang J, Huang XY, Zhou J, Tang ZY, Huang ZL. Exosomal proteomics identifies RAB13 as a potential regulator of metastasis for HCC. Hepatol Commun. 2023;7:e0006. doi: 10.1097/HC9.0000000000000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Gorji-Bahri G, Moghimi HR, Hashemi A. RAB5A effect on metastasis of hepatocellular carcinoma cell line via altering the pro-invasive content of exosomes. Exp Mol Pathol. 2021;120:104632. doi: 10.1016/j.yexmp.2021.104632. [DOI] [PubMed] [Google Scholar]
- 225.Luo J, Jiang L, He C, Shi M, Yang ZY, Shi M, Lu S, Li C, Zhang J, Yan M, et al. Exosomal hsa-let-7g-3p and hsa-miR-10395-3p derived from peritoneal lavage predict peritoneal metastasis and the efficacy of neoadjuvant intraperitoneal and systemic chemotherapy in patients with gastric cancer. Gastric Cancer. 2023;26:364–378. doi: 10.1007/s10120-023-01368-3. [DOI] [PubMed] [Google Scholar]
- 226.Ji R, Lin J, Gu H, Ma J, Fu M, Zhang X. Gastric cancer derived mesenchymal stem cells promote the migration of gastric cancer cells through miR-374a-5p. Curr Stem Cell Res Ther. 2023;18(6):853–863. doi: 10.2174/1574888X18666221124145847. [DOI] [PubMed] [Google Scholar]
- 227.Zhu M, Zhang N, He S, Lu X. Exosomal miR-106a derived from gastric cancer promotes peritoneal metastasis via direct regulation of Smad7. Cell Cycle. 2020;19:1200–1221. doi: 10.1080/15384101.2020.1749467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhu M, Zhang N, Ma J, He S. Integration of exosomal miR-106a and mesothelial cells facilitates gastric cancer peritoneal dissemination. Cell Signal. 2022;91:110230. doi: 10.1016/j.cellsig.2021.110230. [DOI] [PubMed] [Google Scholar]
- 229.Li Q, Li B, Li Q, Wei S, He Z, Huang X, Wang L, Xia Y, Xu Z, Li Z, et al. Exosomal miR-21-5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition. Cell Death Dis. 2018;9:854. doi: 10.1038/s41419-018-0928-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Xia X, Wang S, Ni B, Xing S, Cao H, Zhang Z, Yu F, Zhao E, Zhao G. Hypoxic gastric cancer-derived exosomes promote progression and metastasis via MiR-301a-3p/PHD3/HIF-1alpha positive feedback loop. Oncogene. 2020;39:6231–6244. doi: 10.1038/s41388-020-01425-6. [DOI] [PubMed] [Google Scholar]
- 231.Liu X, Lu Y, Xu Y, Hou S, Huang J, Wang B, Zhao J, Xia S, Fan S, Yu X, et al. Exosomal transfer of miR-501 confers doxorubicin resistance and tumorigenesis via targeting of BLID in gastric cancer. Cancer Lett. 2019;459:122–134. doi: 10.1016/j.canlet.2019.05.035. [DOI] [PubMed] [Google Scholar]
- 232.Lin XM, Wang ZJ, Lin YX, Chen H. Decreased exosome-delivered miR-486-5p is responsible for the peritoneal metastasis of gastric cancer cells by promoting EMT progress. World J Surg Oncol. 2021;19:312. doi: 10.1186/s12957-021-02381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Chen Y, Feng H, Wu Y, Wang R, Li Z, Chen J, Yan W, Chen W. Evaluation of plasma exosomal microRNAs as circulating biomarkers for progression and metastasis of gastric cancer. Clin Transl Med. 2020;10:e171. doi: 10.1002/ctm2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Exosomal miR-196a-1 promotes gastric cancer cell invasion and metastasis by targeting SFRP1. [DOI] [PubMed]
- 235.Zheng Y, Li P, Ma J, Yang C, Dai S, Zhao C. Cancer-derived exosomal circ_0038138 enhances glycolysis, growth, and metastasis of gastric adenocarcinoma via the miR-198/EZH2 axis. Transl Oncol. 2022;25:101479. doi: 10.1016/j.tranon.2022.101479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zhang Z, Sun C, Zheng Y, Gong Y. circFCHO2 promotes gastric cancer progression by activating the JAK1/STAT3 pathway via sponging miR-194-5p. Cell Cycle. 2022;21:2145–2164. doi: 10.1080/15384101.2022.2087280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yang J, Zhang X, Cao J, Xu P, Chen Z, Wang S, Li B, Zhang L, Xie L, Fang L, Xu Z. Circular RNA UBE2Q2 promotes malignant progression of gastric cancer by regulating signal transducer and activator of transcription 3-mediated autophagy and glycolysis. Cell Death Dis. 2021;12:910. doi: 10.1038/s41419-021-04216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhong Y, Wang D, Ding Y, Tian G, Jiang B. Circular RNA circ_0032821 contributes to oxaliplatin (OXA) resistance of gastric cancer cells by regulating SOX9 via miR-515-5p. Biotechnol Lett. 2021;43:339–351. doi: 10.1007/s10529-020-03036-3. [DOI] [PubMed] [Google Scholar]
- 239.Yao W, Guo P, Mu Q, Wang Y. Exosome-Derived Circ-PVT1 Contributes to Cisplatin Resistance by Regulating Autophagy, Invasion, and Apoptosis Via miR-30a-5p/YAP1 Axis in Gastric Cancer Cells. Cancer Biother Radiopharm. 2021;36:347–359. doi: 10.1089/cbr.2020.3578. [DOI] [PubMed] [Google Scholar]
- 240.Xin Y, Li X, Zhang M, Shang Z, Luo Z, Wang Y, Gui X, Liu Q, Li T, Zeng S, et al. Fusobacterium nucleatum-induced exosomal HOTTIP promotes gastric cancer progression through the microRNA-885–3p/EphB2 axis. Cancer Sci. 2023;114(6):2360–2374. doi: 10.1111/cas.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zheng P, Zhang H, Gao H, Sun J, Li J, Zhang X, Gao L, Ma P, Li S. Plasma Exosomal Long Noncoding RNA lnc-SLC2A12-10:1 as a Novel Diagnostic Biomarker for Gastric Cancer. Onco Targets Ther. 2020;13:4009–4018. doi: 10.2147/OTT.S253600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Chen X, Zhang S, Du K, Zheng N, Liu Y, Chen H, Xie G, Ma Y, Zhou Y, Zheng Y, et al. Gastric cancer-secreted exosomal X26nt increases angiogenesis and vascular permeability by targeting VE-cadherin. Cancer Sci. 2021;112:1839–1852. doi: 10.1111/cas.14740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wu M, Tan X, Liu P, Yang Y, Huang Y, Liu X, Meng X, Yu B, Wu Y, Jin H. Role of exosomal microRNA-125b-5p in conferring the metastatic phenotype among pancreatic cancer cells with different potential of metastasis. Life Sci. 2020;255:117857. doi: 10.1016/j.lfs.2020.117857. [DOI] [PubMed] [Google Scholar]
- 244.Wu J. Pancreatic Cancer-Derived Exosomes Promote the Proliferation, Invasion, and Metastasis of Pancreatic Cancer by the miR-3960/TFAP2A Axis. J Oncol. 2022;2022:3590326. doi: 10.1155/2022/3590326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Shang D, Xie C, Hu J, Tan J, Yuan Y, Liu Z, Yang Z. Pancreatic cancer cell-derived exosomal microRNA-27a promotes angiogenesis of human microvascular endothelial cells in pancreatic cancer via BTG2. J Cell Mol Med. 2020;24:588–604. doi: 10.1111/jcmm.14766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yin Z, Ma T, Huang B, Lin L, Zhou Y, Yan J, Zou Y, Chen S. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-beta signaling pathway. J Exp Clin Cancer Res. 2019;38:310. doi: 10.1186/s13046-019-1313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wei Q, Wei L, Zhang J, Li Z, Feng H, Ren L. EphA2-enriched exosomes promote cell migration and are a potential diagnostic serum marker in pancreatic cancer. Mol Med Rep. 2020;22:2941–2947. doi: 10.3892/mmr.2020.11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Tang P, Tao L, Yuan C, Zhang L, Xiu D. Serum Derived Exosomes From Pancreatic Cancer Patients Promoted Metastasis: An iTRAQ-Based Proteomic Analysis. Onco Targets Ther. 2019;12:9329–9339. doi: 10.2147/OTT.S229494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Stefanius K, Servage K, de Souza Santos M, Gray HF, Toombs JE, Chimalapati S, Kim MS, Malladi VS, Brekken R, Orth K. Human pancreatic cancer cell exosomes, but not human normal cell exosomes, act as an initiator in cell transformation. Elife. 2019;8:e40226. [DOI] [PMC free article] [PubMed]
- 250.Armacki M, Polaschek S, Waldenmaier M, Morawe M, Ruhland C, Schmid R, Lechel A, Tharehalli U, Steup C, Bektas Y, et al. Protein Kinase D1, Reduced in Human Pancreatic Tumors, Increases Secretion of Small Extracellular Vesicles From Cancer Cells That Promote Metastasis to Lung in Mice. Gastroenterology. 2020;159(1019–1035):e1022. doi: 10.1053/j.gastro.2020.05.052. [DOI] [PubMed] [Google Scholar]
- 251.Chen J, Li Z, Yue C, Ma J, Cao L, Lin J, Zhu D, An R, Lai J, Guo Y, Gu B. Human umbilical cord mesenchymal stem cell-derived exosomes carrying miR-1827 downregulate SUCNR1 to inhibit macrophage M2 polarization and prevent colorectal liver metastasis. Apoptosis. 2023;28:549–565. doi: 10.1007/s10495-022-01798-x. [DOI] [PubMed] [Google Scholar]
- 252.Li T, Wan Y, Su Z, Li J, Han M, Zhou C. Mesenchymal Stem Cell-Derived Exosomal microRNA-3940-5p Inhibits Colorectal Cancer Metastasis by Targeting Integrin alpha6. Dig Dis Sci. 2021;66:1916–1927. doi: 10.1007/s10620-020-06458-1. [DOI] [PubMed] [Google Scholar]
- 253.Kim HS, Kim JS, Park NR, Nam H, Sung PS, Bae SH, Choi JY, Yoon SK, Hur W, Jang JW. Exosomal miR-125b Exerts Anti-Metastatic Properties and Predicts Early Metastasis of Hepatocellular Carcinoma. Front Oncol. 2021;11:637247. doi: 10.3389/fonc.2021.637247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zhang L, Zhang J, Li P, Li T, Zhou Z, Wu H. Exosomal hsa_circ_0004658 derived from RBPJ overexpressed-macrophages inhibits hepatocellular carcinoma progression via miR-499b-5p/JAM3. Cell Death Dis. 2022;13:32. doi: 10.1038/s41419-021-04345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Kimura Y, Ohzawa H, Miyato H, Kaneko Y, Kuchimaru T, Takahashi R, Yamaguchi H, Kurashina K, Saito S, Hosoya Y, et al. Intraperitoneal transfer of microRNA-29b-containing small extracellular vesicles can suppress peritoneal metastases of gastric cancer. Cancer Sci. 2023;114(7):2939–2950. doi: 10.1111/cas.15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wang L, Yang L, Zhuang T, Shi X. Tumor-Derived Exosomal miR-29b Reduces Angiogenesis in Pancreatic Cancer by Silencing ROBO1 and SRGAP2. J Immunol Res. 2022;2022:4769385. doi: 10.1155/2022/4769385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Yu Z, Zhao S, Wang L, Wang J, Zhou J. miRNA-339-5p Plays an Important Role in Invasion and Migration of Pancreatic Cancer Cells. Med Sci Monit. 2019;25:7509–7517. doi: 10.12659/MSM.917038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.