Abstract
PVC-441 murine leukemia virus (MuLV) is a member of the PVC group of Friend MuLV (F-MuLV)-derived neuropathogenic retroviruses. In order to determine the molecular basis for the difference in neuropathogenicity between PVC-441 and the previously characterized PVC-211 MuLVs, the entire nucleotide sequence of PVC-441 MuLV was determined and compared with those of PVC-211 and F-MuLV. The results suggest that PVC-441 and PVC-211 MuLVs were formed as a result of random mutations of F-MuLV and developed differently. The distinct pathogenicities of PVC-441 and PVC-211 MuLVs were maintained in the viruses regenerated from their molecular clones, and the sequences responsible for the pathological differences observed can be localized to the env gene. The amino acid sequence of PVC-441 deduced from its nucleotide sequence revealed a number of differences from PVC-211, the most striking of which was a difference at position 129 of the SU proteins in the two viruses. Host range studies with a brain capillary endothelial cell line (RTEC-6) and Chinese hamster ovary cells (CHO-K1) revealed that PVC-441, like PVC-211, could infect these cells but its efficiency of infection was lower than that of PVC-211. These results may account for the difference in neuropathogenicity between PVC-441 and PVC-211.
PVC murine leukemia viruses (MuLVs) are paralysis-inducing ecotropic virus clones derived from rat-passaged NB-tropic Friend leukemia virus (F-MuLV) (3) that induce spongiform degeneration in the central nervous systems of rodents (2–4). The clones differ in their pathogenicities in rats and mice. PVC-211 is only weakly neuropathogenic in mice but is highly neuropathogenic in rats, causing hind limb paralysis in 3 weeks and death within 1 month after infection (3, 4). In contrast, PVC-441 is more neuropathogenic in mice, causing tremor within 1 month after infection when injected into newborn mice (4), while rats injected with this virus become paralyzed and die around 2 months after infection (3).
To reveal the molecular differences between PVC-441 and PVC-211 MuLVs that are responsible for their biological differences, the extrachromosomal DNA of PVC-441 was molecularly cloned and sequenced so that it could be compared with the previously sequenced PVC-211 and F-MuLV (10, 11).
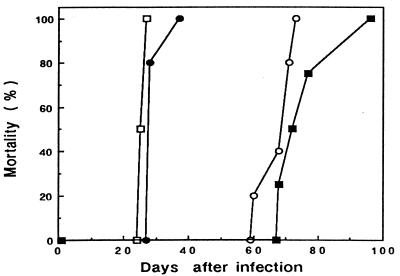
The pathogenicity of the molecularly cloned PVC-441 MuLV clone B5 recovered by transfection was tested in F344 rats in comparison with that of molecularly cloned PVC-211 clone 3d (6). As shown in Fig. 1, the rats infected with PVC-441 clone B5 developed hind leg paralysis and died during the period from 60 to 73 days after infection while those infected with PVC-211 clone 3d developed paralysis and died within 1 month after infection. These results were quite comparable with previous results obtained with biologically cloned viruses (3), and the difference in pathogenicity between PVC-441 and PVC-211 was proved to be maintained in their molecular clones. The pathogenicity of PVC-441 clone B5 was also tested in NFS mice, and the infected mice developed tremor within 1 month after infection, as reported previously (4).
FIG. 1.
Mortality of rats infected with PVC-441 clone B5.c8, PVC-211 clone 3d, or chimeric Lgp2e4 or Lgp4e2 virus. Newborn rats were infected within 24 h of birth with regenerated viruses from the DNAs of PVC-441 clone B5.c8, PVC-211 clone 3d, Lgp2e4, or Lgp4e2 by transfection to normal rat kidney (NRK) cells. ○, PVC-441 (7.6 × 104 PFU/rat); □, PVC-211 (8.1 × 104 PFU/rat); ▪, Lgp2e4 (4.8 × 104 PFU/rat); •, Lgp4e2 (8.8 × 104 PFU/rat).
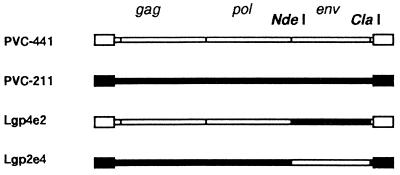
Previous studies with PVC-211 MuLV indicated that the env gene of the virus was the major determinant of its neuropathogenicity (6, 7). In order to determine if pathological differences between PVC-441 MuLV and PVC-211 MuLV were due to differences in their env genes, chimeric viruses were formed between PVC-441 clone B5 and PVC-211 clone 3d. Chimera Lgp2e4, which contains the NdeI-ClaI env gene fragment from PVC-441 on a PVC-211 MuLV background (Fig. 2), induced paralysis in rats and killed them from 68 to 96 days after infection (Fig. 1), while chimera Lgp4e2, which contains the NdeI-ClaI env gene fragment from PVC-211 MuLV on a PVC-441 MuLV background (Fig. 2), induced paralysis in rats and killed them from 28 to 37 days after infection (Fig. 1). These results indicate that the env genes of the parental viruses determine the latency of the disease, although slight differences in latency were observed between viruses containing the same env gene.
FIG. 2.
Chimeras between PVC-441 and PVC-211. Lgp4e2 contains the NdeI-ClaI env fragment of PVC-211, and Lgp2e4 contains the NdeI-ClaI env fragment of PVC-441.
The entire nucleotide sequence of PVC-441 clone B5 was determined by the dideoxynucleotide chain termination method with either a Bca BEST sequencing kit or a TaKaRa Taq cycle sequencing kit and dye-labeled M13 primers (Takara, Kyoto, Japan) on an SQ-3000 DNA sequencer (Hitachi Electronics, Tokyo, Japan).
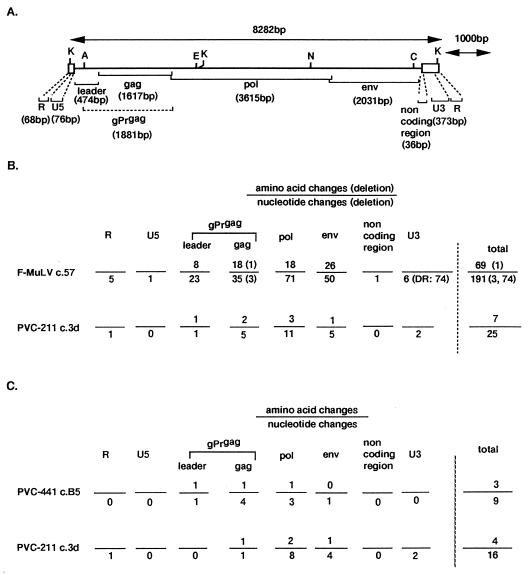
The results of the nucleotide analysis and the deduced amino acid sequence of PVC-441 clone B5 are summarized in Fig. 3. PVC-441 was compared with F-MuLV clone 57 (10) and PVC-211 clone 3d (11). PVC-441 has the same genome size (8,282 bp [Fig. 3A]) as PVC-211. As shown in Fig. 3B, a total of 190 base changes (one base change overlapped in the pol and env regions) were found when PVC-441 was compared with F-MuLV clone 57, including the deletion of 3 bases in the MA protein region of gag and 74 bases in the promoter-enhancer region of the long terminal repeat. Twenty-five base changes were found when PVC-441 was compared with PVC-211 clone 3d. In both cases, the occurrence of base changes was nearly proportional to the size of each viral gene. This fact indicates that both PVC-441 and PVC-211 developed from F-MuLV by random mutation. Although PVC-441 and PVC-211 generally had common nucleotide changes from the sequence of F-MuLV, unique nucleotide changes were also found in their gag and pol gene regions (Fig. 3C). The presence of unique nucleotide changes indicates that these two viruses developed independently from a common ancestor.
FIG. 3.
Summary of results of sequencing of PVC-441 clone B5. (A) Structure of PVC-441 clone B5 genome. The sequence was adjusted to the form of the viral RNA genome arrangement and to an exact size. K, KpnI; A, AatII; E, EcoRI; N, NdeI; C, ClaI. (B) Nucleotide and amino acid changes in every viral genomic region of PVC-441 in comparison with that of either F-MuLV clone 57 or PVC-211 clone 3d. (C) Unique nucleotide and amino acid changes found in PVC-441 or PVC-211.
In contrast to base changes, amino acid changes were not proportional to gene size. When compared to F-MuLV, more changes, based on gene sizes, were seen in the gag leader region of PVC-441, followed in declining order by the env, gag, and pol regions. Changes in the gag leader and gag regions were most prominent when PVC-441 was compared to PVC-211 (Fig. 3B). Unique amino acid changes were also found in the leader, gag, and pol regions of PVC-441 and in the gag, pol and env regions of PVC-211. Interestingly, we found only one amino acid difference, located in the receptor-binding region of the SU protein, between the env gene products of PVC-441 and PVC-211 (Glu129 in PVC-441 and Lys129 in PVC-211), as shown in Table 1.
TABLE 1.
Comparison of amino acids in the env gene products of PVC-441 MuLV, PVC-211 MuLV, and F-MuLVa
| Positionb
|
Amino acid (charge) in:
|
Structural elementc | |||
|---|---|---|---|---|---|
| env | SU | PVC-441 | PVC-211 | F-MuLV | |
| 7 | Ser | Ser | Pro | ||
| 74 | 40 | Asp (−1) | Asp (−1) | Val | |
| 95 | 61 | Arg (+1) | Arg (+1) | Gln | I |
| 113 | 79 | Asn | Asn | Ser | I |
| 114 | 80 | Arg (+1) | Arg (+1) | Ser | I |
| 118 | 84 | Ala | Ala | Ser | I |
| 150 | 116 | Gly | Gly | Glu (−1) | Hinge |
| 163 | 129 | Glu (−1) | Lys (+1) | Glu (−1) | II |
| 186 | 152 | Ala | Ala | Val | II |
| 206 | 172 | Asn | Asn | Ser | |
| 209 | 175 | Ala | Ala | Val | |
| 237 | 203 | Ile | Ile | Thr | |
| 250 | 216 | Gln | Gln | Arg (+1) | |
| 261 | 227 | Lys (+1) | Lys (+1) | Arg (+1) | |
| 283 | 249 | Phe | Phe | Leu | |
| 285 | 251 | Leu | Leu | Arg (+1) | |
| 297 | 263 | Ser | Ser | Pro | |
| 311 | 277 | Ala | Ala | Thr | |
| 353 | 319 | Ala | Ala | Gly | |
| 378 | 344 | Ala | Ala | Val | |
| 379 | 345 | Gly | Gly | Ala | |
| 392 | 358 | Gln | Gln | Arg (+1) | |
| 413 | 379 | Thr | Thr | Ile | |
| 414 | 380 | Gly | Gly | Asp (−1) | |
| 424 | 390 | Ala | Ala | Thr | |
| 427 | 393 | Met | Met | Thr | |
| 676 | Gln | Gln | TERd | ||
Amino acid sequences of the env product of PVC-441, PVC-211, and F-MuLV were deduced from the nucleotide sequences and compared.
The amino-terminal methionine of the envelope precursor protein is numbered 1 (env), and the amino-terminal alanine of the SU protein is numbered 1 (SU).
Structural elements are described by Linder et al. (5).
TER, termination codon.
When the amino acid sequences of the env gene products of PVC-441, PVC-211, and F-MuLV are compared (Table 1), we can see some characteristic features in the SU proteins of the PVC virus group in comparison with that of F-MuLV. Since the change of charged amino acids may affect the three-dimensional structure of the SU protein, the charge of each amino acid is indicated in parentheses in Table 1. Two changes, from a neutral amino acid to a basic amino acid, were found in the region upstream of position 129 of the PVC group, whereas three of the four basic amino acids in the region downstream of position 129 of F-MuLV changed to neutral amino acids in the PVC group. One change, from Val40 to Asp40, in PVC viruses was found in the upstream region. On the other hand, Asp380 in F-MuLV changed to Gly380 in the PVC viruses. Also, since the amino acid at position 116 is located in the hinge region between elements I and II (5), the change from Glu116 in F-MuLV to Gly116 in the PVC viruses may affect the three-dimensional structure significantly. Finally, since elements I and II are fixed by intramolecular disulfide bonds (5), the effects of individual changes in charge at positions 61, 80, and 129 may be limited, although the total charge of an element may affect the three-dimensional structure. Thus, the main structural difference between the SU proteins of the PVC virus group and that of F-MuLV seems to be in the three-dimensional folding of the protein, and the only difference between the SU proteins of PVC-441 MuLV and PVC-211 MuLV is that that of PVC-211 has a change from Glu129 to Lys129 in the receptor binding region.
Recently, Masuda et al. revealed that PVC-211 was able to extend its infectivity to the brain capillary endothelial cell line RTEC-6 of the F344 rat and to Chinese hamster ovary (CHO-K1) cells due to amino acid changes in its SU protein from that of F-MuLV (Glu116 to Gly and Glu129 to Lys) (8, 9). PVC-441 has the change from Glu116 to Gly but retains the Glu129 found in F-MuLV (Table 1). Thus, we were interested in testing the infectivity of PVC-441 MuLV on RTEC-6 or CHO-K1 cells in comparison with those of PVC-211 and F-MuLV. The infectivity test was carried out with the N2 virus transduction assay (1, 8, 9), with the N2 virus pseudotyped with PVC-441, PVC-211, or F-MuLV (Table 2), and virus titer was evaluated by counting G418-resistant colonies. Since individual virus preparations may differ from each other in titer, relative infectivity was determined on NIH 3T3, Rat-1, RTEC-6, and CHO-K1 cells. As shown in Table 2, PVC-441 had essentially the same infectivity as PVC-211 and F-MuLV on NIH 3T3 and Rat-1 cells. However, PVC-441 was slightly less infectious (0.66 and 0.067, respectively) than PVC-211 on RTEC-6 and CHO-K1 cells, although PVC-441 was far more infectious (30- and >30,000-fold, respectively) than F-MuLV on RTEC-6 cells and CHO-K1 cells. These results are in good agreement with those of previous comparisons of the infectivities of PVC-211 MuLV and F-MuLV on these cells (8, 9). The results suggest that the amino acid at position 129 of the SU protein (Glu129 in PVC-441 and Lys129 in PVC-211) affects the efficiency of infection but not the host range on RTEC-6 and CHO-K1 cells and that the amino acid at position 116 of the SU protein (Gly116 in PVC-441 and PVC-211 and Glu116 in F-MuLV) may determine the host range. This conclusion may explain the observation that PVC-441 MuLV-induced neurological disease has a longer latency in rats than PVC-211 MuLV-induced disease (Fig. 1) (3).
TABLE 2.
Transduction efficiencies of PVC-441, PVC-211, and F-MuLV on NIH 3T3, Rat-1, RTEC-6, and CHO-K1 cellsa
| Virus | Transduction efficiency (G418r CFU/ml) onb:
|
|||
|---|---|---|---|---|
| NIH 3T3 | Rat-1 | RTEC-6 | CHO-K1 | |
| PVC-441 | 2.47 × 105 (1.0) | 5.25 × 105 (2.12) | 6.25 × 102 (0.0025) | 7.25 × 104 (0.29) |
| PVC-211 | 2.02 × 106 (1.0) | 4.38 × 106 (2.17) | 7.75 × 103 (0.0038) | 8.75 × 106 (4.33) |
| F-MuLV | 2.64 × 105 (1.0) | 5.50 × 105 (2.08) | 2.25 × 101 (0.000085) | <2.5 (<0.0000095) |
Cells were seeded at a density of 105 per 60-mm-diameter (NIH 3T3 and RTEC-6) or 5 × 104 per 35-mm-diameter (Rat-1 and CHO-K1) culture dish. Twenty-four hours after infection with N2 vector pseudotyped with each virus in the presence of Polybrene (5 μg/ml), the cultures were treated with G418 (400 μg/ml for NIH 3T3, Rat-1, and RTEC-6; 1 mg/ml for CHO-K1), and incubated for 12 days, including two replacements with fresh medium containing G418. CFU of G418r virus per milliliter were counted after two independent titrations.
Relative infectivity of each virus when the titer on NIH 3T3 was set as 1.0 is shown in parentheses.
Recently, Takase-Yoden and Watanabe reported an F-MuLV-derived neuropathogenic MuLV (A8) that, like PVC-441, was more weakly pathogenic than PVC-211 MuLV in rats (12). Like PVC-441, A8 did not undergo the change from Glu to Lys at position 129 that occurred in PVC-211, lending further support to the idea that this change is necessary for inducing rapid neurological changes in rats.
Although the difference between the envelope proteins of PVC-441 and PVC-211 may account for the difference in neuropathogenicity between the two viruses, we cannot rule out the possibility that differences found between the gag regions of the two viruses are also involved. We are currently carrying out studies utilizing additional chimeras between PVC-441 and PVC-211 to resolve this question.
Nucleotide sequence accession number.
The sequence described herein has been assigned the EMBL, GenBank, and DDBJ accession no. Y13893.
Acknowledgments
We thank Allen Oliff (Merck Research Laboratories, West Point, Pa.) for providing a molecular clone of F-MuLV clone 57 DNA.
REFERENCES
- 1.Eglitis M A, Kantoff P, Gilboa E, Anderson W F. Gene expression in mice after high efficiency retroviral-mediated gene transfer. Science. 1985;230:1395–1398. doi: 10.1126/science.2999985. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann P M, Cimino E F, Robbins D S, Broadwell R D, Powers J M, Ruscetti S K. Cellular tropism and localization in the rodent nervous system of a neuropathogenic variant of Friend murine leukemia virus. Lab Investig. 1992;67:314–321. [PubMed] [Google Scholar]
- 3.Kai K, Furuta T. Isolation of paralysis-inducing murine leukemia viruses from Friend virus passaged in rats. J Virol. 1984;50:970–973. doi: 10.1128/jvi.50.3.970-973.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kai K, Mitsuno K, Goto N, Ami Y, Ando S, Kanoe M. Factors affecting induction of neurological disorders in mice by paralysis-inducing Friend-related PVC viruses. J Vet Med Sci. 1996;58:285–290. doi: 10.1292/jvms.58.285. [DOI] [PubMed] [Google Scholar]
- 5.Linder M, Wenzel V, Linder D, Stirm S. Structural elements in glycoprotein 70 from polytropic Friend mink cell focus-inducing virus and glycoprotein 71 from ecotropic Friend murine leukemia virus, as defined by disulfide-bonding pattern and limited proteolysis. J Virol. 1994;68:5133–5141. doi: 10.1128/jvi.68.8.5133-5141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masuda M, Remington M P, Hoffman P M, Ruscetti S K. Molecular characterization of a neuropathogenic and nonerythroleukemogenic variant of Friend murine leukemia virus PVC-211. J Virol. 1992;66:2798–2806. doi: 10.1128/jvi.66.5.2798-2806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuda M, Hoffman P M, Ruscetti S K. Viral determinants that control the neuropathogenicity of PVC-211 murine leukemia virus in vivo determine brain capillary endothelial cell tropism of the virus in vitro. J Virol. 1993;67:4580–4587. doi: 10.1128/jvi.67.8.4580-4587.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masuda M, Hanson C A, Alvord W G, Hoffman P M, Ruscetti S K, Masuda M. Effects of subtle changes in the SU protein of ecotropic murine leukemia virus on its brain capillary endothelial cell tropism and interference properties. Virology. 1996;215:142–151. doi: 10.1006/viro.1996.0017. [DOI] [PubMed] [Google Scholar]
- 9.Masuda M, Masuda M, Hanson C A, Hoffman P M, Ruscetti S K. Analysis of the unique hamster cell tropism of ecotropic murine leukemia virus PVC-211. J Virol. 1996;70:8534–8539. doi: 10.1128/jvi.70.12.8534-8539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliff A I, Hager G L, Chang E H, Scolnick E M, Chang H W, Lowy D R. Transfection of molecularly cloned Friend murine leukemia virus DNA yields a high leukemogenic helper-independent type C virus. J Virol. 1980;33:475–486. doi: 10.1128/jvi.33.1.475-486.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Remington M P, Hoffman P M, Ruscetti S K, Masuda M. Complete nucleotide sequence of a neuropathogenic variant of Friend murine leukemia virus PVC211. Nucleic Acids Res. 1992;20:3249–3249. doi: 10.1093/nar/20.12.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takase-Yoden S, Watanabe R. Unique sequence and lesional tropism of a new variant of neuropathogenic Friend murine leukemia virus. Virology. 1997;233:411–422. doi: 10.1006/viro.1997.8619. [DOI] [PubMed] [Google Scholar]