Abstract
The intricate nature of the brain necessitates the application of advanced probing techniques to comprehensively study and understand its working mechanisms. Neurophotonics offers minimally invasive methods to probe the brain using optics at cellular and even molecular levels. However, multiple challenges persist, especially concerning imaging depth, field of view, speed, and biocompatibility. A major hindrance to solving these challenges in optics is the scattering nature of the brain. This perspective highlights the potential of complex media optics, a specialized area of study focused on light propagation in materials with intricate heterogeneous optical properties, in advancing and improving neuronal readouts for structural imaging and optical recordings of neuronal activity. Key strategies include wavefront shaping techniques and computational imaging and sensing techniques that exploit scattering properties for enhanced performance. We discuss the potential merger of the two fields as well as potential challenges and perspectives toward longer term in vivo applications.
Keywords: complex media, neurophotonics, brain probing, wavefront shaping, computational imaging
Introduction
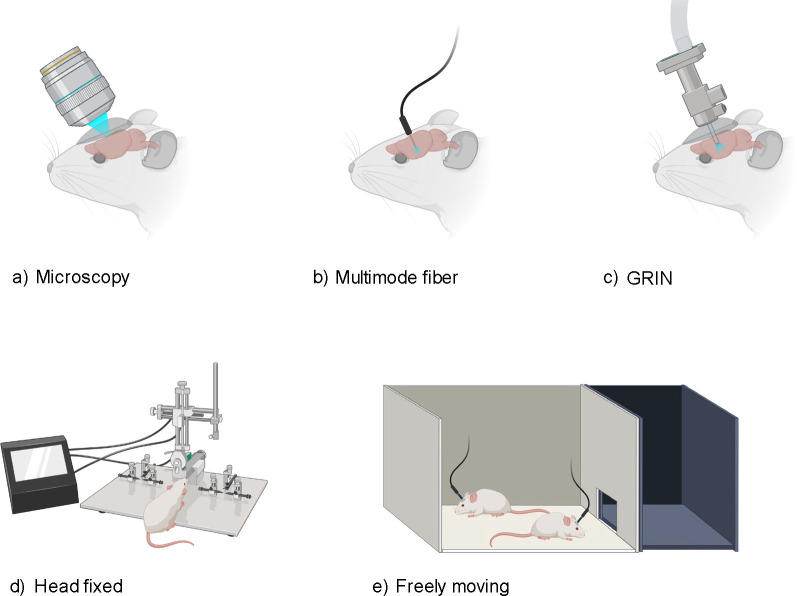
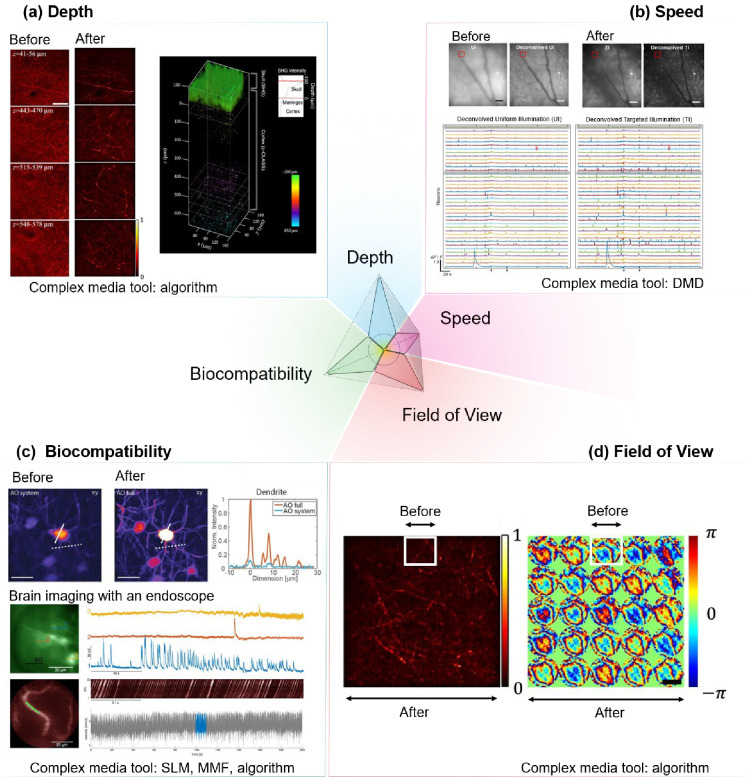
The brain acts as the central regulator in all vertebrate and most invertebrate organisms1. Comprehensive study of its structure and function is not only paramount to our scientific understanding but also crucial for developing interventions for brain-related pathologies2,3. In this context, the field of neurophotonics, a domain that capitalizes on optical tools to study the nervous system (Figure 1), has emerged as a powerful strategy for brain studies. Three defining strengths of optical approaches include: (i) their minimal invasiveness 4–6, (ii) their enhanced specificity when combined with molecular labeling7–11 or label-free optical techniques5,12–16, allowing for targeted imaging at cellular and molecular levels, and (iii) the possibility of chronically recording the same structures of interest, such as neurons, dendrites, and spines17, during development, learning, and sensory deprivation18,19. However, there are persisting challenges that limit the comprehensive use of optical techniques in brain research. In this perspective paper, we specifically focused on optical imaging and sensing tools to probe the brains of animal models. Neuroscientists have proposed a key objective for optical probing of the brain: to develop and integrate advanced optical probing techniques that offer high spatiotemporal resolution, large-scale recording and mapping of neural activity while ensuring safety and minimal invasiveness20–22. Meeting this objective necessitates advancements in: 1) Probing depth, especially important given the size variations of the brain, from larger scales in humans to smaller scales in other species7,23–25 (Figure 2a). 2) Expanding the field of view (FOV), allowing for a more holistic capture and understanding of neuronal networks26–28 (Figure 2d). 3) Improving probing speed to capture and interpret dynamic biological activities in both 2D and 3D contexts9,29–32 (Figure 2b). 4) Ensuring biocompatibility: minimizing phototoxicity and avoiding damage from implanted devices, thereby preserving the brain’s structural and functional integrity during investigations of the brain using optical methods33,34 (Figure 2c).
Figure 1.
Overview of Diverse Brain Probing Techniques: (a) Microscopy: Traditional imaging with direct optical access to the brain. (b) Multimode Fiber: Flexible approach using a fiber optic cable for light delivery and signal collection. (c) GRIN (Gradient Refractive Index) Lens: Minimally invasive imaging through a small-diameter lens. (d) Head Fixed: Apparatus for stable imaging with restrained subject movement. (e) Freely Moving: Setup allowing for natural behavior during imaging with a mobile recording system. Figures (a-e) adapted with BioRender.com.
Figure 2. Representative advances from tools commonly used in the complex media community to address challenges in optical probing of the brain.
(a) Depth: scattering and aberration compensation using computational techniques to enhance reflectance imaging of cortical myelin through the skull in the living mouse brain114. Before: conventional reflectance microscopy through the mouse skull. Left panel: After: computational conjugated adaptive optical corrected reflectance microscopy of cortical myelin in the mouse brain through skull. Right panel: 3D reconstruction of label-free structural information through skull. Scale bar: 40 μm. (b) Speed: Fast 3D volumetric imaging with targeted illumination of neurons in the mouse cortex labelled with a calcium indicator (GCaMP6f) to increase signal-to-noise of recorded neurons. Before: conventional volumetric calcium imaging with electrically tunable lens and extracted traces after deconvolution. After: illumination-targeted volumetric calcium imaging and extracted traces after deconvolution30. Scale bar 50 μm. (c) Biocompatibility: upper panel: enhanced signal given the same laser power enabled by adaptive optics54. Before: low signal-to-background of fluorescence-labelled neurons in the hippocampus around 1 mm depth imaged transcranially by conventional three-photon fluorescence microscopy. After: high signal-to-background neurons in the hippocampus imaged by adaptive optics. Scale bar: 20 μm. Lower panel: brain imaging of deep subcortical neurons labeled with a genetically-encoded calcium indicator GCaMP6s using a multimode fiber-based endoscope combined with wavefront shaping for minimally invasive imaging115. Scale bar: 30 μm. (d) Field of view: enlarged field of view with diffraction-limited high-resolution imaging enabled by computational conjugated adaptive optics (after) compared with computational adaptive optics without conjugation (before, white boxes)114 Left: Image of myelin. Right: Phase pattern for aberration correction. SLM: spatial light modulator, DMD: Digital Micromirror Devices, MMF: multimode fibers. Panel (a) adapted from ref114 under license CC-BY 4.0. Panel (b) adapted from the ref30 under license CC-BY 4.0. Panel (c) the top images adapted from ref54 and the bottom images adapted from ref115 under license CC-BY 4.0. Panel (d) adapted from ref114 under license CC-BY 4.0.
Here, we review recent advances in techniques and devices popularized in the complex media community that have begun to show promise in addressing some of the key challenges (Fig. 2) and discuss our perspectives on moving forward for in vivo applications.
Opportunities: Bridging the Gap
The complex media field studies light propagation in materials with highly inhomogeneous optical properties. Tools developed in this area include advanced computations on light scattering in optically heterogeneous micro- media and algorithm design for shaping light through diffusive materials and image recovery using scattering information35. While rooted in fundamental light scattering, its implications naturally extend to neurophotonics, due to the highly scattering nature of brain tissues.
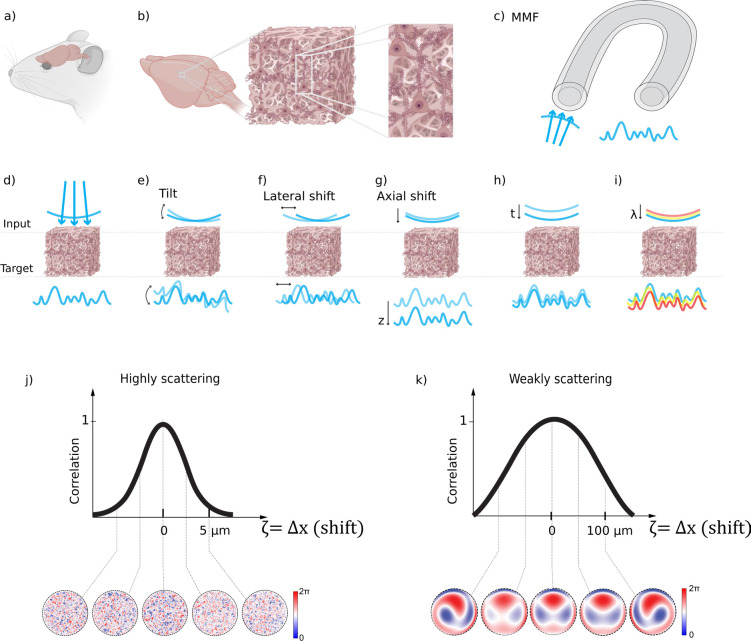
Key techniques in the complex media field can be broadly categorized into two groups: wavefront shaping35–38 through complex media and computational imaging and sensing techniques using complex media39. Wavefront shaping, a technique that modulates the phase and amplitude of incoming light waves using light shaping devices such as spatial light modulator (SLM), is emerging as a promising avenue. Adaptive Optics (AO), a wavefront shaping method focused on spatiotemporal resolution across various optical imaging modalities40–54. Looking ahead, recent wavefront shaping techniques that address scattering (higher-order light distortion)46,55–64 have the potential to further improve signal and resolution, especially at depths where scattering becomes a critical limitation (Fig. 1a). Recent insights into local correlation during scattering events, i.e. the memory effect65, in chromatic66,67, shift68, tilt/angular69,70, and others71 may guide more efficient light manipulation deep within tissues.
The memory effect refers to the phenomenon where the optical fields of scattered light remain correlated when certain properties of the light, such as position, wavevector direction, polarization, or spectrum, change over a specific range. As illustrated in Figure 3: ‘Chromatic’ refers to changes in the light’s wavelength; ‘shift’ pertains to the displacement or angular deviation of light beams; and ‘angular or ‘tilt’’ involves changes in the direction of light propagation. Memory effect enables the prediction of how light’s properties change with scattering, facilitating computational or hardware-based tools for enhanced imaging quality through scattering tissues or interfaces with complex optical properties, such as multimode fibers. For instance, by conjugating the light modulation plane to specific locations within the scattering medium (Fig. 2d), we might find an optimal balance between enhancements of the signal intensity, FOV, and spatial resolution72,73. In the brain, despite the relative dense packing of neurons and vasculature, fluorescence microscopy often reveals a sparser distribution particularly when given at a certain color channel, a result of selective fluorescent labeling targeting specific cellular or vascular components or a sparse expression of fluorescence25,74–76. Leveraging sparse and compressive sampling or scanning techniques, like Acousto-Optic Deflectors77–80 (AOD) and Digital Micromirror Devices30,81–86 (DMD) (Fig. 2b), ensures efficient photon utilization. Such methods not only expedite the imaging process but also preserve the photon budget, setting the stage for up to one order of magnitude increase in the imaging speed (Fig. 2b) and reduction in the laser power (Fig. 2c) for faster and physiologically safer recording given proper guide stars for wavefront shaping87. Guide stars in imaging are akin to its astronomical counterpart; they serves as a reference light source from various contrast mechanisms, such as harmonic, photoacoustic, fluorescence, and scattered light87, within the sample to facilitate the correction of light distortion caused by scattering. By employing the guide stars, we can guide the wavefront shaping process to more precisely manipulate incoming light waves. This improves the efficiency of the photon budget of the incoming field in enhancing the focus intensity and signal at greater penetration depths of imaging systems and in reducing laser intensity, minimizing potential photodamage to biological tissues. They could even help capture faster events such as millisecond action potentials in neurons9,29,30,88–90 (Fig. 2b). Furthermore, compressive random-access sampling with fast light modulators, like AODs, permits to integrate fast temporal sampling and wavefront shaping79,88,89, including adaptive correction of aberrations and scattering over an extended FOV that effectively exceeds the range of the angular memory effect, by taking advantage of the fast AODs’ update rate to correct multiple local aberrations almost synchronously with the progression of a scanning beam, whether in pixel-by-pixel or random-access scan mode91. The primary advantage of employing wavefront shaping in enhancing the capabilities of state-of-the-art optical microscopy lies in the optimization of the photon budget. This technique enables the strategic redistribution of photons to either augment the imaging speed or expand the field of view (FOV), all while maintaining a fixed photon allowance for biological imaging under safe physiological conditions. However, challenges remain in terms of its shaping speed, which needs to be improved to overcome the temporal decorrelation of the scattered light field (Fig. 3h).
Figure 3.
Optical access to the mouse brain through a scattering medium: (a) Schematic of a live mouse highlighting the brain area; (b) Inhomogeneous structures within the mouse brain that can cause optical scattering; (c) Multimode fiber (MMF), a frequently studied complex scattering medium in complex media field, is also often utilized for optical access to the brain; (d) Scattering-induced wavefront distortion; (e-i) Various memory effects: (e) Tilt/angular memory effect; (f) Lateral shift memory effect; (g) Axial shift memory effect; (h) Temporal memory effect; (i) Chromatic memory effect; (j) Representative quantitative correlation of wavefront correction pattern for achieving diffraction-limited focusing/imaging in highly scattering brain tissue, demonstrating that the range of the memory effect (defined by the full width at half maximum of the correlation curve) is substantially narrower compared to less scattering scenarios as shown in (k). The patterns for correcting wavefront distortion in highly scattering media (j) are more complex than in weakly scattering media (k). Note in (j,k), ζ could be any of the types of memory effect above in (e-i), but for the illustrative example we chose ζ = Δx (shift). Figures (a-b, d-i) adapted with BioRender.com.
In the realm of computational imaging and sensing techniques through complex media, image reconstruction39,92,93 and signal processing94–96 methods that exploit random or scattering media properties have emerged as potential game-changers. Leveraging the inherently locally correlated nature of scattered light, techniques such as auto-correlation97, cross-correlation98,99, and patch-connecting-based100 image reconstruction methods have been proposed. These aim to directly reconstruct images through highly scattering media, with the potential to achieve a larger FOV at greater depths. Image reconstruction fundamentally is a process of solving optimization problems, which can be categorized into convex and non-convex cases. Convex optimization problems are generally more straightforward to solve because their global minima are easily identifiable. On the other hand, non-convex optimization problems, more common in imaging through scattering tissue, often suffer from multiple local minima, complicating the search for the global minimum. In this challenging landscape, deep learning101,102 emerges as a powerful tool, offering robust methods that learn from data to effectively approximate global optima — opening new exciting avenues for neurophotonics imaging, with significant potential to enhance its capabilities. It provides not only an alternative tool for solving optimization problems, such as optimization as unrolled neural networks98,103,104, but also enhances image reconstruction with deep learning models for better generalization of the scattering problems101. On the other hand, the analysis and understanding of speckle—a highly sensitive interference pattern commonly seen when light propagates through complex media105,106—has proven to be extremely powerful and promising. In the brain, the detected speckle signal can be highly sensitive to various events, such as calcium signaling107, an indirect indicator of voltage fluctuations, and blood flow108.
Computational imaging techniques have shown promising enhanced results in brain imaging. For example, advanced signal processing methods such as non-negative matrix factorization (NMF), have proven instrumental in calcium imaging experiments by effectively removing noise and isolating signal components109,110. Additionally, the utility of computational imaging extends to blood flow estimation and reconstruction from speckle patterns observed in brain tissues111.
Furthermore, computational tools such as constrained NMF112 and DeepCAD113 have greatly enhanced denoising techniques and the ability to retrieve signals from significantly high noiselevels112,113,116–118. These advancements are particularly valuable for imaging through highly scattering tissues, where traditional imaging methods are challenged to provide clear and reliable data. Although still at its early stage, it is anticipated that computational imaging will continue to enhance the clarity and utility of acquired images, enabling more detailed and accurate studies of neural structures and functions in challenging imaging conditions.
Challenges and Limitations Towards Longer-Term In Vivo Applications
Although the progress mentioned earlier has been exciting, the further adoption of these for longer-term in vivo biological studies still faces challenges.
In vivo applications involve imaging and sensing activities within the brain of a living and behaving animal, which raise a first challenge in term of recording artifacts linked to movements. The most common in vivo strategy is to fix the animal’s head under a microscope, allowing for a good control of the sensory stimuli applied to the animal, as well as to accurately measure its behavior. Neurophotonics techniques developed for in vitro samples can be adapted for head-fixed animals provided that motion is taken into account. This encompasses micro- to milli-meter scale motions from heartbeats and respiration, to blood flow, and bulk motions induced by body movements and muscle contractions. These motions cause spatiotemporal noise dynamics in the tissue’s scattering properties. Temporally, these dynamics are observable down to the millisecond range, and spatially, they can be seen down to the micron level. For example, regarding the bulk motion of the brain, in the case of 2-photon imaging experiments in the cortex with a cranial window, motion artifacts were observed to be around 2 to 4 μm in axial direction, which is much shorter than the 150 μm thickness of the optical window79. When implanting an optical fiber, one expects to encounter similar motion artifacts when exploring shallow regions of the brain. Interestingly, however, fewer motion artifacts are observed when a fiber is implanted in deeper brain regions. Indeed, this has been observed for 2-photon imaging with GRIN lenses119,120. From a technical standpoint, this poses concerns regarding the stability and speed of wavefront shaping techniques, as well as noise issues in computational imaging and sensing techniques. These factors underscore the need for adaptive imaging solutions that can recalibrate in real-time, ensuring consistent performance121. However, these hurdles, though significant, are not insurmountable. The way forward may involve a co-design philosophy, harmoniously melding wavefront shaping systems, algorithms, and imaging systems. For head-fixed animals, introducing an ‘animal-in-the-loop’ design could be revolutionary. This innovative approach would use real-time feedback from the animal’s physiological and behavioral changes to continually adapt the imaging process, such as using online motion tracking to adapt in real-time the scanning scheme122 or the heartbeat signal to gate the optical signal and remove heartbeat-related imaging noise54.
A second challenge is improving depth penetration in brain tissues. In brain imaging, the depth achievable with current technologies varies significantly across different microscopy techniques and contrast mechanisms. For example, we have summarized the depth penetration capabilities of some of the most popular fluorescence microscopy techniques, including one-photon, two-photon, and three-photon excited fluorescence, as follows (in the context of in vivo adult mouse brain imaging):
Conventional one-photon (1P) microscopy is limited to depths of approximately 0.3–0.4 mm due to light scattering and absorption in the commonly used visible range of light124,126. Conventional two-photon (2P) microscopy extends this depth to about 0.6–0.8 mm7,128. Conventional three-photon (3P) microscopy further increases imaging depth to 1.2–2.1 mm129,132. The potential depth limits (table 1, column 3) for these imaging methods can be estimated based on effective attenuation lengths125 depending on the excitation and detection method124,128,131.
Table 1:
Current and potential limits of depth penetration capabilities of one-, two-, three-photon excited fluorescence microscopy. Visible range of light: 380 – 700 nm; near-infrared I light: 700 – 900 nm.
| Fluorescence microscopy | Demonstrated depths so far with high spatial resolution (close to diffraction-limited) | Potential estimated depth limits with high spatial resolution (close to diffraction-limited) |
|---|---|---|
| Excitation: One-photon excited Detection: Widefield | 0.1 ~ 0.2 mm123 (visible range of light) | 0.6 ~ 0.8 mm124,125 (*near-infrared II or short-wave infrared light) |
| Excitation: One-photon excited Detection: Confocal | 0.3 ~ 0.4 mm126 (visible range of light) | 1.5 ~ 2 mm124,125 (*near-infrared II or short-wave infrared light) |
| Excitation: Two-photon excited (temporal focusing) Detection: Wiefield | 0.3 ~ 0.4 mm127 (near-infrared I light) | 0.6 ~ 0.8 mm124,125,128 (*near-infrared II or short-wave infrared light) |
| Excitation: Two-photon excited Detection: single-element detector (e.g. photomultiplier tube, PMT) | 0.6 ~ 0.8 mm7,129 (near-infrared I light) | 1.5 ~ 2 mm125,128 (*near-infrared II or short-wave infrared light) |
| Excitation: Three-photon excited Detection: single-element detector (e.g. photomultiplier tube, PMT) | 1.2 ~ 2.1 mm129,130 (near-infrared II or short-wave infrared light) | 3 ~ 4 mm125,131 (*near-infrared II or short-wave infrared light) |
Indicates optimal imaging windows around 1300 nm and 1700 nm (in the region of near-Infrared II between 1000–1700 nm also called the short-wave infrared range in similar or even broader ranges in some definitions.
Techniques such as wavefront shaping and computational imaging have been developed to mitigate scattering and aberrations, potentially enhancing imaging depth and resolution. These advancements enable more efficient light delivery and collection deep within tissues. Particularly, wavefront shaping can be and has been coupled with 1P-, 2P-, or 3P-excited fluorescence contrasts, thereby having the capability to extend the depth for each modality. For example, in a proof-of-concept multiphoton wavefront shaping experiment, an enhancement of at least one order of magnitude for the 2P signal and a two orders of magnitude gain for the 3P signal were observed52.
One fundamental barrier is the depth beyond which even sophisticated light manipulation or computational imaging strategies become potentially impractical (refer to table 1, column 3) for diffraction-limited focusing and reconstruction. In such cases, minimally invasive fiber optics are the only viable option for high-resolution imaging, especially in scenarios requiring high mobility or minimal interference with the subject (animals). Devices such as miniature multimode fibers can be used to bypass the scattering of tissues. Endoscopes incorporate multimode fibers (MMF)133–136 (Fig. 1b, 2c, 3c), as a relay between the animal and a benchtop wavefront shaping microscope, ensuring minimal invasiveness. Pioneering works, such as MMF-based imaging for mouse brains115,135 and in vivo histology137 as well as deep learning for image reconstruction through MMF138,139, provide glimpses into the potential future of neurophotonics for deep brain imaging.
Advanced wavefront shaping techniques can also help to address a third challenge: imaging in freely behaving configuration, which provides access to a wider range of behaviors, such as social interactions and sleep. Freely-behaving imaging was achieved thanks to the use of wavefront shaping assisted endoscopes based on fiber bundles140,141. Another approach is miniatures microscopes142–146 that allow measuring neuronal activity using conventional widefield142,143, 2-photon144,145 and 3-photon147–149 imaging methods. However, combining these microscopes with wavefront shaping techniques will necessitate miniaturization of beam shaping devices. The ultimate goal would be the development of wireless miniscopes150,151, freeing subjects from physical restraints and promoting natural behaviors.
On the other hand, computational techniques such as machine learning can generally facilitate a more robust search for the global optimum in non-convex optimization problems that exist in probing through scattering tissues. Major application directions involve 1) the reconstruction of high-fidelity images from scattered light patterns, effectively ‘learning’ the tissue’s scattering properties to inversely map the captured signals back to their original, unscattered state; 2) denoising images during high-speed imaging or challenging imaging scenarios; 3) decoding the scattered field/patterns for biomedical insights; 4) predicting correction masks in wavefront shaping.
For instance, neural networks have been utilized to predict the unscattered light path, allowing for real-time correction of distorted images caused by tissue scattering152. This method has the potential to enhance the depth penetration and resolution of imaging modalities such as two-photon and three-photon microscopy, making it possible to visualize neuronal activity deeper within the brain with unprecedented clarity. At the same time, deep learning has enabled enhanced image quality in challenging imaging scenarios by image denoising, such as high-speed voltage imaging153 and high-quality calcium imaging113,117. This advancement has led to improved neural activity traces, facilitating more accurate spike inference. Furthermore, deep learning models have been applied to interpret the speckle patterns resulting from coherent light scattering, extracting meaningful biological signals such as cerebral blood flow from noise and thereby facilitating non-invasive imaging techniques that can monitor brain dynamics154. Deep learning has recently also been applied in predicting scattering or aberration correction patterns in brain imaging155.
Looking ahead, machine learning, with its strengths in generalization and robustness, can be invaluable. Algorithms supported by machine learning can process and interpret vast amounts of data rapidly, ensuring that researchers keep pace with the time-varying optical properties in in vivo environment. As neurophotonics delves deeper into uncharted territories, a symbiotic relationship between industry and academia becomes essential. Industrial stakeholders can develop faster, more stable shapers and sensitive detectors, while academia can push boundaries in algorithmic and system design innovations.
Conclusion
In conclusion, the merging of complex media research with neurophotonics marks the beginning of an era brimming with significant potential and opportunities. Moving forward, there is a need for collaboration and innovation across different disciplines, such as algorithm development, machine learning, optics, and neuroscience. This interdisciplinary approach is essential for overcoming existing technical challenges and unravelling better mechanistic understanding of the brain in the unexplored regime. Given the recent advancements in computational imaging and sensing through complex media, wavefront shaping technology, machine learning tools, and the myriad of chemical and biological tools developed in neuroscience, we believe there lies a tremendous opportunity to synergize these diverse fields.
Acknowledgements
The authors thank Ruth Sims and Yusaku Hontani for their feedback and proofreading of the manuscript. F. X. and S.G. acknowledge funding from Chan Zuckerberg Initiative (2020-225346); C.R.V and S.G. acknowledge funding from: HFSP project N°RGP0003/2020; W. A., C.V. and L.B acknowledge funding from Institut de Convergence Qlife (ANR-17-CONV-0005); NIH BRAIN Initiative (1U01NS103464); ANR (ALPINS ANR-15-CE19-0011, EXPECT (17-CE37-0022-01) - The program Investissements d’Avenir ANR-10-LABX-54 (Memolife), ANR-11-IDEX-0001-02 (Université PSL) and ANR-10-INSB-04-01 (France-BioImaging infrastructure); C.V. acknowledge ANR MULTIMOD (OP19-121-OTP-01). S.G. acknowledges funding from NIH 1RF1NS113251 and senior fellowship from Institut Universitaire de France. S.G. and H.B.A. acknowledge funding from H2020 Future and Emerging Technologies (grant no. 863203). H.B.A. acknowledges funding from ANR COCOhRICO (ANR-21-CE42-0013).
Footnotes
Disclosures
The authors declare no conflicts of interest.
Code, Data and Materials Availability
Data sharing is not applicable to this article, as no new data was created or analyzed.
References
- 1.Kandel E. R. et al. Principles of neural science. vol. 4 (McGraw-hill; New York, 2000). [Google Scholar]
- 2.Bear M., Connors B. & Paradiso M. A. Neuroscience: exploring the brain, enhanced edition: exploring the brain. (Jones & Bartlett Learning, 2020). [Google Scholar]
- 3.Insel T. R. & Landis S. C. Twenty-five years of progress: the view from NIMH and NINDS. Neuron 80, 561–567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strangman G., Boas D. A. & Sutton J. P. Non-invasive neuroimaging using near-infrared light. Biol. Psychiatry 52, 679–693 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Villringer A. & Chance B. Non-invasive optical spectroscopy and imaging of human brain function. Trends Neurosci. 20, 435–442 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Bandettini P. A. What’s new in neuroimaging methods? Ann. N. Y. Acad. Sci. 1156, 260–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helmchen F. & Denk W. Deep tissue two-photon microscopy. Nat. Methods 2, 932–940 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Pawley J. B. Handbook of biological confocal microscopy: Third edition. Handbook of Biological Confocal Microscopy: Third Edition (2006). doi: 10.1007/978-0-387-45524-2. [DOI] [Google Scholar]
- 9.Marshall J. D. et al. Cell-Type-Specific Optical Recording of Membrane Voltage Dynamics in Freely Moving Mice Resource Cell-Type-Specific Optical Recording of Membrane Voltage Dynamics in Freely Moving Mice. 1650–1662 (2016) doi: 10.1016/j.cell.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watakabe A., Komatsu Y., Ohsawa S. & Yamamori T. Fluorescent in situ hybridization technique for cell type identification and characterization in the central nervous system. Methods 52, 367–374 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Packer A. M., Roska B. & Häusser M. Targeting neurons and photons for optogenetics. Nat. Neurosci. 16, 805–815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson B. C. & Jacques S. L. Optical Reflectance and Transmittance of Tissues: Principles and Applications. IEEE J. Quantum Electron. 26, 2186–2199 (1990). [Google Scholar]
- 13.Matthews T. E. et al. Deep tissue imaging using spectroscopic analysis of multiply scattered light. Optica 1, 105 (2014). [Google Scholar]
- 14.Evans C. L. & Xie X. S. Coherent Anti-Stokes Raman Scattering Microscopy: Chemical Imaging for Biology and Medicine. Annu. Rev. Anal. Chem. 1, 883–909 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Durduran T. & Yodh A. G. Diffuse correlation spectroscopy for non-invasive, micro-vascular cerebral blood flow measurement. Neuroimage 85, 51–63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jermyn M. et al. Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci. Transl. Med. 7, 274ra19–274ra19 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Szalay G. et al. Fast 3D imaging of spine, dendritic, and neuronal assemblies in behaving animals. Neuron 92, 723–738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grewe B. F. & Helmchen F. Optical probing of neuronal ensemble activity. Curr. Opin. Neurobiol. 19, 520–529 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Zepeda A., Arias C. & Sengpiel F. Optical imaging of intrinsic signals: recent developments in the methodology and its applications. J. Neurosci. Methods 136, 1–21 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Jorgenson L. A. et al. The BRAIN Initiative: developing technology to catalyse neuroscience discovery. Philos. Trans. R. Soc. B Biol. Sci. 370, 20140164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alivisatos A. P. et al. The brain activity map project and the challenge of functional connectomics. Neuron 74, 970–974 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang W. et al. Simultaneous Multi-plane Imaging of Neural Circuits. Neuron 89, 284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton N. G. et al. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat. Photonics 7, 205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sholl D. A. The organization of the cerebral cortex. (1956). [PubMed]
- 25.Sunkin S. M. et al. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 41, D996–D1008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sofroniew N. J., Flickinger D., King J. & Svoboda K. A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. Elife 5, e14472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mok A. T. et al. A large field of view two-and three-photon microscope for high-resolution deep tissue imaging. in 2023 Conference on Lasers and Electro-Optics (CLEO) 1–2 (2023). [Google Scholar]
- 28.Yu C.-H., Stirman J. N., Yu Y., Hira R. & Smith S. L. Diesel2p mesoscope with dual independent scan engines for flexible capture of dynamics in distributed neural circuitry. Nat. Commun. 12, 6639 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong Y. et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. 350, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao S., Tseng H., Gritton H., Han X. & Mertz J. Video-rate volumetric neuronal imaging using 3D targeted illumination. Sci. Rep. 8, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber T. D., Moya M. V, Kiliç K., Mertz J. & Economo M. N. High-speed multiplane confocal microscopy for voltage imaging in densely labeled neuronal populations. Nat. Neurosci. 1–9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J. et al. Kilohertz two-photon fluorescence microscopy imaging of neural activity in vivo. Nat. Methods 17, 287–290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Icha J., Weber M., Waters J. C. & Norden C. Phototoxicity in live fluorescence microscopy, and how to avoid it. BioEssays 39, 1700003 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Podgorski K. & Ranganathan G. Brain heating induced by near-infrared lasers during multiphoton microscopy. J. Neurophysiol. 116, 1012–1023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gigan S. et al. Roadmap on wavefront shaping and deep imaging in complex media. J. Phys. Photonics 4, 42501 (2022). [Google Scholar]
- 36.Yu H. et al. Recent advances in wavefront shaping techniques for biomedical applications. Curr. Appl. Phys. 15, 632–641 (2015). [Google Scholar]
- 37.Gigan S. Optical microscopy aims deep. Nat. Photonics 11, 14–16 (2017). [Google Scholar]
- 38.Kubby J., Gigan S. & Cui M. Wavefront shaping for biomedical imaging. Wavefront Shaping for Biomedical Imaging (2019). doi: 10.1017/9781316403938. [DOI] [Google Scholar]
- 39.Tian L. Computational imaging in complex media (Conference Presentation). in Image Sensing Technologies: Materials, Devices, Systems, and Applications V vol. 10656 106560R (2018). [Google Scholar]
- 40.Pérez G. M. & Artal P. Impact of scattering and spherical aberration in contrast sensitivity. 9, 1–10 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Wang K. et al. Direct wavefront sensing for high-resolution in vivo imaging in scattering tissue. Nat. Commun. 6, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodr’iguez C. et al. An adaptive optics module for deep tissue multiphoton imaging in vivo. Nat. Methods 18, 1259–1264 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gould T. J., Burke D., Bewersdorf J. & Booth M. J. Adaptive optics enables 3D STED microscopy in aberrating specimens. Opt. Express 20, 20998–21009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinefeld D. et al. Three-Photon Adaptive Optics for Mouse Brain Imaging. Front. Neurosci. 16, 1–10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Booth M. J. & Patton B. R. Adaptive Optics for Fluorescence Microscopy. Fluorescence Microscopy: Super-Resolution and other Novel Techniques (2014). doi: 10.1016/B978-0-12-409513-7.00002-6. [DOI] [Google Scholar]
- 46.Qin Z. et al. Deep tissue multi-photon imaging using adaptive optics with direct focus sensing and shaping. Nat. Biotechnol. 40, 1663–1671 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Schwertner M., Booth M. J., Neil M. A. A. & Wilson T. Measurement of specimen-induced aberrations of biological samples using phase stepping interferometry. J. Microsc. 213, 11–19 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Hao X., Antonello J., Allgeyer E. S., Bewersdorf J. & Booth M. J. Aberrations in 4Pi microscopy. Opt. Express 25, 14049–14058 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahman S. A., Booth M. J., Rahman S. A. & Booth M. J. microscopy : wave front sensing using. (2018) doi: 10.1117/12.909845. [DOI] [Google Scholar]
- 50.Ji N., Milkie D. E. & Betzig E. Adaptive optics via pupil segmentation for high-resolution imaging in biological tissues. Nat. Methods 7, 141–147 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Ji N., Sato T. R. & Betzig E. Characterization and adaptive optical correction of aberrations during in vivo imaging in the mouse cortex. Proc. Natl. Acad. Sci. U. S. A. 109, 22–27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinefeld D., Paudel H. P., Ouzounov D. G., Bifano T. G. & Xu C. Adaptive optics in multiphoton microscopy: comparison of two, three and four photon fluorescence. Opt. Express 23, 2309–2311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tao X. et al. A three-photon microscope with adaptive optics for deep-tissue in vivo structural and functional brain imaging. Neural Imaging Sens. 10051, 100510R (2017). [Google Scholar]
- 54.Streich L. et al. High-resolution structural and functional deep brain imaging using adaptive optics three-photon microscopy. Nat. Methods 18, 1253–1258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katz O., Ramaz F., Gigan S. & Fink M. Controlling light in complex media beyond the acoustic diffraction-limit using the acousto-optic transmission matrix. Nat. Commun. 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rauer B., de Aguiar H. B., Bourdieu L. & Gigan S. Scattering correcting wavefront shaping for three-photon microscopy. Opt. Lett. 47, 6233–6236 (2022). [DOI] [PubMed] [Google Scholar]
- 57.Vellekoop I. M. & Mosk A. P. Focusing coherent light through opaque strongly scattering media. Opt. Lett. 32, 2309–2311 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Vellekoop I. M., Lagendijk A. & Mosk A. P. Exploiting disorder for perfect focusing. Nat. Photonics 4, 320–322 (2010). [Google Scholar]
- 59.Mosk A. P., Lagendijk A., Lerosey G. & Fink M. Controlling waves in space and time for imaging and focusing in complex media. Nat. Photonics 6, 283–292 (2012). [Google Scholar]
- 60.Dong J., Krzakala F. & Gigan S. Spectral Method for Multiplexed Phase Retrieval and Application in Optical Imaging in Complex Media. in ICASSP, IEEE International Conference on Acoustics, Speech and Signal Processing - Proceedings vols 2019-May 4963–4967 (2019). [Google Scholar]
- 61.Berlage C. et al. Deep tissue scattering compensation with three-photon F-SHARP. Optica 8, 1613–1619 (2021). [Google Scholar]
- 62.May M. A. et al. Fast holographic scattering compensation for deep tissue biological imaging. Nat. Commun. 12, 4340 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pozzi P., Gandolfi D., Porro C. A., Bigiani A. & Mapelli J. Scattering compensation for deep brain microscopy: The long road to get proper images. Front. Phys. 8, 26 (2020). [Google Scholar]
- 64.Papadopoulos I. N., Jouhanneau J.-S., Poulet J. F. A. & Judkewitz B. Scattering compensation by focus scanning holographic aberration probing (F-SHARP). Nat. Photonics 11, 116–123 (2017). [Google Scholar]
- 65.Freund I., Rosenbluh M. & Feng S. Memory effects in propagation of optical waves through disordered media. Phys. Rev. Lett. 61, 2328–2331 (1988). [DOI] [PubMed] [Google Scholar]
- 66.Arjmand P., Katz O., Gigan S. & Guillon M. Three-dimensional broadband light beam manipulation in forward scattering samples. Opt. Express 29, 6563–6581 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Zhu L. et al. Chromato-axial memory effect through a forward-scattering slab. Optica 7, 338–345 (2020). [Google Scholar]
- 68.Judkewitz B., Horstmeyer R., Vellekoop I. M., Papadopoulos I. N. & Yang C. Translation correlations in anisotropically scattering media. Nat. Phys. 11, 684–689 (2015). [Google Scholar]
- 69.Yılmaz H., Kühmayer M., Hsu C. W., Rotter S. & Cao H. Customizing the angular memory effect for scattering media. (2020).
- 70.Schott S., Bertolotti J., Léger J.-F., Bourdieu L. & Gigan S. Characterization of the angular memory effect of scattered light in biological tissues. Opt. Express 23, 13505–13516 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Osnabrugge G., Horstmeyer R., Papadopoulos I. N., Judkewitz B. & Vellekoop I. M. Generalized optical memory effect. Optica 4, 886 (2017). [Google Scholar]
- 72.Hampson K. M. et al. Closed-loop multiconjugate adaptive optics for microscopy. in Progress in Biomedical Optics and Imaging - Proceedings of SPIE vol. 11248 (2020). [Google Scholar]
- 73.Papadopoulos I. N. et al. Dynamic conjugate F-SHARP microscopy. Light Sci. & Appl. 9, 110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang X. et al. High-resolution mapping of brain vasculature and its impairment in the hippocampus of Alzheimer’s disease mice. Natl. Sci. Rev. 6, 1223–1238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deng X. & Gu M. Penetration depth of single-, two-, and three-photon fluorescence microscopic imaging through human cortex structures: Monte Carlo simulation. Appl. Opt. 42, 3321–9 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Nicholson C. Diffusion and related transport mechanisms in brain tissue. Reports Prog. Phys. 64, 815 (2001). [Google Scholar]
- 77.Chong E. Z., Barreiros I., Li B., Kohl M. M. & Booth M. J. Fast multiplane functional imaging combining acousto-optic switching and remote focusing. in Progress in Biomedical Optics and Imaging - Proceedings of SPIE vol. 10070 (2017). [Google Scholar]
- 78.Blochet B., Akemann W., Gigan S. & Bourdieu L. Fast wavefront shaping for two-photon brain imaging with large field of view correction. bioRxiv 2009–2021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akemann W. et al. Fast optical recording of neuronal activity by three-dimensional custom-access serial holography. Nat. Methods 19, 100–110 (2022). [DOI] [PubMed] [Google Scholar]
- 80.Salomé R. et al. Ultrafast random-access scanning in two-photon microscopy using acousto-optic deflectors. J. Neurosci. Methods 154, 161–174 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Otsu Y. et al. Optical monitoring of neuronal activity at high frame rate with a digital random-access multiphoton (RAMP) microscope. J. Neurosci. Methods 173, 259–270 (2008). [DOI] [PubMed] [Google Scholar]
- 82.De Aguiar H. B. H. B., Gigan S. & Brasselet S. Enhanced nonlinear imaging through scattering media using transmission-matrix-based wave-front shaping. Phys. Rev. A 94, 1–9 (2016). [Google Scholar]
- 83.Gentner C., Burri S., Charbon E., Bruschini C. & de Aguiar H. B. Compressive Raman microspectroscopy parallelized by single-photon avalanche diode arrays. arXiv Prepr. arXiv2301.07709 (2023). [Google Scholar]
- 84.Soldevila F., Dong J., Tajahuerce E., Gigan S. & DE AGUIAR H. B. Fast compressive Raman bio-imaging via matrix completion. Optica 6, 341–346 (2019). [Google Scholar]
- 85.Sturm B. et al. High-Sensitivity High-Speed Compressive Spectrometer for Raman Imaging. ACS Photonics 6, 1409–1415 (2019). [Google Scholar]
- 86.Zheng C. et al. De-scattering with excitation patterning enables rapid wide-field imaging through scattering media. Sci. Adv. 7, eaay5496 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Horstmeyer R., Ruan H. & Yang C. Guidestar-assisted wavefront-shaping methods for focusing light into biological tissue. Nat. Photonics 9, 563–571 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu Z. et al. Sustained deep-tissue voltage recording using a fast indicator evolved for two-photon microscopy. Cell 185, 3408–3425 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Villette V. et al. Ultrafast two-photon imaging of a high-gain voltage indicator in awake behaving mice. Cell 179, 1590–1608 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sims R. R. et al. Scanless two-photon voltage imaging. Res. Sq. (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blochet B., Akemann W., Gigan S. & Bourdieu L. Fast wavefront shaping for two-photon brain imaging with multipatch correction. Proc. Natl. Acad. Sci. 120, e2305593120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhuang H., He H., Xie X. & Zhou J. High speed color imaging through scattering media with a large field of view. Sci. Rep. 6, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Popoff S., Lerosey G., Fink M., Boccara A. C. & Gigan S. Image transmission through an opaque material. Nat. Commun. 1, (2010). [DOI] [PubMed] [Google Scholar]
- 94.Moretti C. & Gigan S. Readout of fluorescence functional signals through highly scattering tissue. Nat. Photonics 14, 361–364 (2020). [Google Scholar]
- 95.Soldevila F. et al. Functional imaging through scattering medium via fluorescence speckle demixing and localization. Opt. Express 31, 21107–21117 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rimoli C. V. et al. Demixing fluorescence time traces transmitted by multimode fibers. arXiv Prepr. arXiv2306.00695 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Katz O., Small E. & Silberberg Y. Looking around corners and through thin turbid layers in real time with scattered incoherent light. Nat. Photonics 6, 549–553 (2012). [Google Scholar]
- 98.d’Arco A., Xia F., Boniface A., Dong J. & Gigan S. Physics-based neural network for non-invasive control of coherent light in scattering media. Opt. Express 30, 30845–30856 (2022). [DOI] [PubMed] [Google Scholar]
- 99.Zhu L. et al. Large field-of-view non-invasive imaging through scattering layers using fluctuating random illumination. Nat. Commun. 13, 1447 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y. et al. Deep imaging inside scattering media through virtual spatiotemporal wavefront shaping. arXiv Prepr. arXiv2306.08793 (2023). [Google Scholar]
- 101.Li Y., Xue Y. & Tian L. Deep speckle correlation: a deep learning approach toward scalable imaging through scattering media. Optica 5, 1181–1190 (2018). [Google Scholar]
- 102.Antipa N. et al. DiffuserCam: lensless single-exposure 3D imaging. Optica 5, 1–9 (2018). [Google Scholar]
- 103.Monakhova K. et al. Unrolled, model-based networks for lensless imaging. in NeurIPS 2019 Workshop on Solving Inverse Problems with Deep Networks (2019). [Google Scholar]
- 104.Monakhova K. et al. Learned reconstructions for practical mask-based lensless imaging. Opt. Express 27, 28075–28090 (2019). [DOI] [PubMed] [Google Scholar]
- 105.Goodman J. W. Speckle phenomena in optics: theory and applications. (Roberts and Company Publishers, 2007). [Google Scholar]
- 106.Goodman J. W. Some fundamental properties of speckle. JOSA 66, 1145–1150 (1976). [Google Scholar]
- 107.Moretti C. & Gigan S. Readout of fluorescence functional signals through highly scattering tissue. arXiv (2019). [Google Scholar]
- 108.Briers J. D. Laser speckle contrast imaging for measuring blood flow. XXXVII, 14 (2007). [Google Scholar]
- 109.Carbonero D., Noueihed J., Kramer M. A. & White J. A. Non-Negative Matrix Factorization for Analyzing State Dependent Neuronal Network Dynamics in Calcium Recordings. bioRxiv (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Friedrich J. et al. Fast constrained non-negative matrix factorization for whole-brain calcium imaging data. in NIPS workshop on statistical methods for understanding neural systems (2015). [Google Scholar]
- 111.Qureshi M. M. et al. Quantitative blood flow estimation in vivo by optical speckle image velocimetry. Optica 8, 1092–1101 (2021). [Google Scholar]
- 112.Pnevmatikakis E. A. et al. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89, 299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li X. et al. Reinforcing neuron extraction and spike inference in calcium imaging using deep self-supervised denoising. Nat. Methods 18, 1395–1400 (2021). [DOI] [PubMed] [Google Scholar]
- 114.Kwon Y. et al. Computational conjugate adaptive optics microscopy for longitudinal through-skull imaging of cortical myelin. Nat. Commun. 14, 105 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stibuurek M. et al. 110 $μ$m thin endo-microscope for deep-brain in vivo observations of neuronal connectivity, activity and blood flow dynamics. Nat. Commun. 14, 1897 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li X. et al. Real-time denoising enables high-sensitivity fluorescence time-lapse imaging beyond the shot-noise limit. Nat. Biotechnol. 41, 282–292 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang Y. et al. Rapid detection of neurons in widefield calcium imaging datasets after training with synthetic data. Nat. Methods 20, 747–754 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou P. et al. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. Elife 7, e28728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bocarsly M. E. et al. Minimally invasive microendoscopy system for in vivo functional imaging of deep nuclei in the mouse brain. Biomed. Opt. Express 6, 4546–4556 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Meng G. et al. High-throughput synapse-resolving two-photon fluorescence microendoscopy for deep-brain volumetric imaging in vivo. Elife 8, e40805 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Valzania L. & Gigan S. Online learning of the transmission matrix of dynamic scattering media. Optica 10, 708–716 (2023). [Google Scholar]
- 122.Griffiths V. A. et al. Real-time 3D movement correction for two-photon imaging in behaving animals. Nat. Methods 17, 741–748 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li Z. et al. Fast widefield imaging of neuronal structure and function with optical sectioning in vivo. Sci. Adv. 6, eaaz3870 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xia F. et al. Short-wave infrared confocal fluorescence imaging of deep mouse brain with a superconducting nanowire single-photon detector. ACS Photonics 8, 2800–2810 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang M. et al. Comparing the effective attenuation lengths for long wavelength in vivo imaging of the mouse brain. Biomed. Opt. Express 9, 3534–3543 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schain A. J., Hill R. A. & Grutzendler J. Label-free in vivo imaging of myelinated axons in health and disease with spectral confocal reflectance microscopy. Nat. Med. 20, 443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Papagiakoumou E., Ronzitti E. & Emiliani V. Scanless two-photon excitation with temporal focusing. Nat. Methods 17, 571–581 (2020). [DOI] [PubMed] [Google Scholar]
- 128.Theer P. & Denk W. On the fundamental imaging-depth limit in two-photon microscopy. J. Opt. Soc. Am. A 23, 3139 (2006). [DOI] [PubMed] [Google Scholar]
- 129.Wang T. et al. Quantitative analysis of 1300-nm three-photon calcium imaging in the mouse brain. Elife 9, e53205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu H. et al. In vivo deep-brain structural and hemodynamic multiphoton microscopy enabled by quantum dots. Nano Lett. 19, 5260–5265 (2019). [DOI] [PubMed] [Google Scholar]
- 131.Akbari N., Rebec M. R., Xia F. & Xu C. Imaging deeper than the transport mean free path with multiphoton microscopy. Biomed. Opt. Express 13, 452–463 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Takasaki K., Abbasi-Asl R. & Waters J. Superficial bound of the depth limit of two-photon imaging in mouse brain. Eneuro 7, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Turcotte R., Schmidt C. C., Booth M. J. & Emptage N. J. Volumetric two-photon fluorescence imaging of live neurons using a multimode optical fiber. Opt. Lett. 45, (2020). [DOI] [PubMed] [Google Scholar]
- 134.Sych Y., Chernysheva M., Sumanovski L. T. & Helmchen F. High-density multi-fiber photometry for studying large-scale brain circuit dynamics. Nat. Methods 16, 553–560 (2019). [DOI] [PubMed] [Google Scholar]
- 135.Turtaev S. et al. High-fidelity multimode fibre-based endoscopy for deep brain in vivo imaging. Light Sci. & Appl. 7, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sivankutty S. et al. Ultra-thin rigid endoscope: two-photon imaging through a graded-index multi-mode fiber. Opt. Express 24, 825–841 (2016). [DOI] [PubMed] [Google Scholar]
- 137.Wen Z. et al. Single multimode fibre for in vivo light-field-encoded endoscopic imaging. Nat. Photonics 17, 679–687 (2023). [Google Scholar]
- 138.Resisi S., Popoff S. M. & Bromberg Y. Image transmission through a dynamically perturbed multimode fiber by deep learning. Laser & Photonics Rev. 15, 2000553 (2021). [Google Scholar]
- 139.Borhani N., Kakkava E., Moser C. & Psaltis D. Learning to see through multimode fibers. Optica 5, 960–966 (2018). [Google Scholar]
- 140.Accanto N. et al. A flexible two-photon fiberscope for fast activity imaging and precise optogenetic photostimulation of neurons in freely moving mice. Neuron 111, 176–189 (2023). [DOI] [PubMed] [Google Scholar]
- 141.Szabo V., Ventalon C., De Sars V., Bradley J. & Emiliani V. Spatially selective holographic photoactivation and functional fluorescence imaging in freely behaving mice with a fiberscope. Neuron 84, 1157–1169 (2014). [DOI] [PubMed] [Google Scholar]
- 142.Ghosh K. K. et al. Miniaturized integration of a fluorescence microscope. Nat. Methods 8, 871–878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Aharoni D. & Hoogland T. M. Circuit investigations with open-source miniaturized microscopes: past, present and future. Front. Cell. Neurosci. 141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zong W. et al. Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nat. Methods 14, 713–719 (2017). [DOI] [PubMed] [Google Scholar]
- 145.Zong W. et al. Large-scale two-photon calcium imaging in freely moving mice. Cell 185, 1240–1256 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Andresen E. R., Sivankutty S., Tsvirkun V., Bouwmans G. & Rigneault H. Ultrathin endoscopes based on multicore fibers and adaptive optics: a status review and perspectives. J. Biomed. Opt. 21, 121506 (2016). [DOI] [PubMed] [Google Scholar]
- 147.Zhao C. et al. Miniature three-photon microscopy maximized for scattered fluorescence collection. Nat. Methods 20, 617–622 (2023). [DOI] [PubMed] [Google Scholar]
- 148.Klioutchnikov A. et al. A three-photon head-mounted microscope for imaging all layers of visual cortex in freely moving mice. Nat. Methods 20, 610–616 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Klioutchnikov A. et al. Three-photon head-mounted microscope for imaging deep cortical layers in freely moving rats. Nat. Methods 17, 509–513 (2020). [DOI] [PubMed] [Google Scholar]
- 150.Barbera G., Liang B., Zhang L., Li Y. & Lin D.-T. A wireless miniScope for deep brain imaging in freely moving mice. J. Neurosci. Methods 323, 56–60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang Y. et al. Cable-free brain imaging with miniature wireless microscopes. bioRxiv 2006–2022 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tahir W., Wang H. & Tian L. Adaptive 3D descattering with a dynamic synthesis network. Light Sci. & Appl. 11, 42 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Platisa J. et al. High-speed low-light in vivo two-photon voltage imaging of large neuronal populations. Nat. Methods 1–9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pan Y. et al. Dynamic 3D imaging of cerebral blood flow in awake mice using self-supervised-learning-enhanced optical coherence Doppler tomography. Commun. Biol. 6, 298 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hu Q. et al. Universal adaptive optics for microscopy through embedded neural network control. Light Sci. Appl. 12, 270 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article, as no new data was created or analyzed.