Abstract
We investigated the role of human CRM1 (hCRM1) (exportin 1) in the function of Rex protein encoded by human T-cell leukemia virus type 1. hCRM1 promoted the export of Rex protein from the nucleus to the cytoplasm. A Rex protein with a mutation in the activation domain, RexM90, lost both the ability to bind to hCRM1 and the ability to multimerize. The overexpression of hCRM1 complemented the functional defects of RexM64, which contains a mutation in the multimerization domain of Rex. A dominant-negative mutant of Rex which sequesters cofactors of Rex abrogated multimerization as well as the activity of the wild-type Rex protein. These two functions were simultaneously restored by the overexpression of hCRM1. Taken together, these results suggest that hCRM1 plays important roles in the multimerization and export of Rex protein.
The Rex protein of human T-cell leukemia virus type 1 acts posttranscriptionally to induce the cytoplasmic expression of incompletely spliced or unspliced mRNAs encoding viral structural proteins (16, 18, 34). Although the exact mechanism of action of Rex has not been elucidated, it is generally accepted that Rex shuttles between the nucleus and the cytoplasm (4, 20) in a process that involves the binding of Rex to specific regions of viral RNAs and their subsequent escort into the cytoplasm (1, 3, 14, 15, 32). Rex acts through cis-acting elements, referred to as Rex response elements (RXRE), within the 3′ long terminal repeat of human T-cell leukemia virus type 1 (32). Direct interaction between Rex and RXRE has been shown by various in vitro binding assays, and this interaction has turned out to be essential for their in vivo activity (1, 3, 14, 15). A discrete region in the amino terminus of the Rex protein which is rich in basic amino acids has been shown to mediate binding to RXRE (1, 3, 14). This region also works as a nuclear and nucleolar targeting signal (NLS) (33). A second essential region, in the carboxy-terminal portion of the Rex protein, contains a leucine-rich activation domain which was shown to function as a nuclear export signal (NES) by binding to cellular cofactors (4, 20). Human immunodeficiency virus (HIV) type 1 contains the regulatory protein Rev, which is functionally equivalent to Rex (7, 8). Rev possesses two functional domains comparable to those of Rex (6, 24–27, 41, 43).
Mutational analysis of Rev and Rex revealed a third domain responsible for the multimerization of these transactivator proteins (2, 26). Multimerization is generally agreed to be critical for Rev and Rex function (26). Although there has been some inconsistency in the mapping of the domain(s) involved in multimerization, all studies have revealed the importance of the N-terminal region of the Rev protein, comprising tyrosine23, serine25, and asparagine26 (2, 26, 37). The domain containing amino acids near positions 60 to 70 of the Rex protein has been shown to functionally replace the amino-terminal region of the Rev protein (42). Interestingly, cellular cofactors have been proposed to be involved in multimerization on the basis of the failure of Rev activation domain mutants to oligomerize in two kinds of two-hybrid assays in mammalian cells (2, 23). However, the involvement of the activation domain of Rev in multimerization was contraindicated by observations showing the mislocalization of wild-type Rev when coexpressed with the activation domain mutants, a result that suggested that heterooligomers had been formed (17, 36, 37). However, the involvement of the Rex activation domain in multimerization has not been extensively studied. To further pursue this subject, it seems necessary to examine the nature of the cellular cofactor(s) directly.
Recently, human CRM1 (hCRM1) (exportin 1) was found to be a receptor for various NES sequences, including the activation domain of Rev. hCRM1 belongs to the importin β family, the members of which act as carriers to transport proteins between the cytoplasm and the nucleus (11, 13, 22, 28, 29, 35, 39). Recently, hCRM1 was shown to form a complex with NES and GTP-loaded Ran, the predominant form found in the nucleus, but not with NES in the presence of GDP-loaded Ran, the predominant form found in the cytoplasm (11). Although these studies clearly indicated that hCRM1 is a major cofactor functioning in the export of Rev, they were done with experimental model systems, such as yeast cells (13, 22, 28, 35), amphibian oocytes (11, 13), and artificially constructed reporter proteins containing both NLS and NES (29). Thus, only a few of the functions carried out by hCRM1 have been adequately addressed.
Dominant-negative (DN) mutants, which inhibit the function of coexpressed wild-type Rev and Rex, have been very useful for investigating the molecular mechanisms underlying Rev and Rex function (2, 19, 24, 27, 40). For example, the RevM10 mutant protein, a prototype of a DN mutant, provided an important clue in the identification of the activation domain (24, 40). More recently, TAgRex, which has the NLS of simian virus 40 T antigen in place of the normal RNA binding domain of the Rex protein, was constructed (19). Since TAgRex inhibits Rev and Rex function by sequestering a cellular cofactor(s), it can be used to classify the routes by which various mRNAs and proteins are exported from the nucleus to the cytoplasm (10, 19, 21, 30, 31). TAgRex may also supply a means to approve cellular cofactors of Rex, as such factors would be expected to abrogate the DN effect of TagRex.
The present study was conducted to investigate the roles of hCRM1 in Rex function by use of TAgRex in human cells. We show that hCRM1 is involved in both the export and the multimerization of the Rex protein.
MATERIALS AND METHODS
Plasmid construction.
To construct pSRαhCRM1, the hCRM1 gene containing the entire coding region was amplified from a human placental cDNA library (Clontech Co. Ltd.) by the PCR technique with primers 5′-GTTCAATCTCTGGTAATCTATGCCAGC-3′ (CRMF1) and 5′-CCCAGCCACAAAAATGGGCATGAAG-3′ (CRMR1). The PCR product was gel purified and used as a template for a second PCR with primers 5′-AACGGTACCCGCACTAGTCACACATTTCTTCTGGAATCTCATGTGGAT-3′ and CRMF1. The second PCR product was blunt ended by Pfu polymerase treatment, digested with KpnI, and cloned into pSRα296 (38) which had been digested with PstI, blunt ended with Pfu polymerase, and then digested with KpnI. The resultant plasmid was named pSRαhCRM1. The integrity of the plasmid sequence was ascertained by sequencing according to the manufacturer’s instructions (Applied Biosystem Co. Ltd.).
To generate pGAL-CRM1, the coding sequence of the CRM1 gene was amplified by PCR with the primer pair 5′-GGAAGATCTTTCAATCTCTGGTTATCTATGCCAGC-3′ and CRM2 and with the first PCR product described above as a template. The PCR product was treated with Pfu polymerase, digested with BglII, and cloned in frame downstream of the GAL4 DNA binding domain sequence in pSGGALVP (12), which had been digested with BamHI, blunt ended, and then digested with BglII. To make pGAL4, pSGGALVP was digested with BglII and BamHI and then self-ligated. To construct pSRαRexM64, a 500-bp fragment generated by the digestion of pSRαRex with NcoI/AvrII and a fragment encoding the C-terminal region of Rex derived from pSRαTAgRexM64 by digestion with NcoI/EcoRI were ligated with pSRα296 that had been linearized with AvrII/EcoRI. Plasmid pSRαRexM90 was constructed in the same way, except that pSRαTAgRexM90 was used as the source of the fragment encoding the C-terminal region of Rex. In this way, pSRαRexM64 carried a protein containing amino acid substitutions of Asp and Leu for Tyr-64 and Trp-65, respectively, and pSRαRexM90 carried a protein containing single Gly substitutions for Leu-90, Ser-91, Leu-92, and Asp-93.
pSRαRex, pSRαTAgRexM64, pSRαTAgRexM90, pSRαTAgRexM6490, pCDM-β-gal, pGAL-Rex, pRex-VP, pGAL-RexM64, pRexM64-VP, pGAL-RexM90, pRexM90-VP, and pTU50RXE were described previously (19, 21).
Cell culture and transfection.
HeLa cells were maintained in RPMI medium supplemented with 10% fetal calf serum in a 5% CO2 atmosphere at 37°C. Plasmid DNA was transfected with DOTAP (Boehringer Mannheim Co. Ltd.) according to the manufacturer’s instructions. In order to normalize for variations in transfection efficiency and nonspecific effects of various treatments, 0.1 μg of pCDM-β-gal was included in all samples and the total amount of DNA was kept constant by adding pSRα296.
In vivo assay of protein-protein interactions.
The two-hybrid system was used to analyze protein-protein interactions in mammalian cells (12). HeLa cells were cotransfected with 0.2 μg of the plasmid that expresses the GAL fusion protein, 0.2 μg of the plasmid that expresses the VP16 fusion protein, 0.6 μg of pG5BCAT as a reporter, and 0.1 μg of pCDM-β-gal. Twenty-four hours after transfection, the cells were harvested, the amount of chloramphenicol acetyltransferase (CAT) and the activity of β-galactosidase were quantified with a CAT enzyme-linked immunosorbent assay (ELISA) kit (Boehringer) and standard colorimetric methods, respectively, and the ratio of CAT to β-galactosidase was calculated.
Env ELISA.
HeLa cells were transfected with various amounts of pSRαRex or with a pSRαRex mutant along with 0.5 μg of pTU50RXRE as a reporter plasmid; the latter expresses the HIV type 1 Env protein in the presence of a functional Rex protein. The Env protein was mutated to remove the transmembrane domain and was secreted into the medium (21). Forty-eight hours after transfection, the culture medium of each sample was transferred to a microcentrifugation tube and centrifuged at a low speed. The cells remaining on the bottom of a six-well plate were lysed with the lysis buffer contained in the CAT ELISA kit. One-fifth of the supernatant was added to a 96-well plate which had been coated with anti-Env monoclonal antibody 0.5 β, and the plate was incubated at 37°C for 1 h. The plate was washed five times with the washing buffer contained in the CAT ELISA kit. Human anti-HIV antisera diluted 100-fold were added to the wells of the plate, and the plate was incubated. Finally, peroxidase-conjugated anti-human immunoglobulin G antibody was added. The cell lysates were used to measure β-galactosidase activity, and the ratio of Env to β-galactosidase was calculated.
Immunofluorescence.
HeLa cells were transfected with pSRαRex, pSRαRexM64, of pSRαRexM90 in the presence or absence of pSRαhCRM1. Twenty-four h after transfection, the cells were fixed and incubated with rabbit anti-Rex C terminus antibody, and the immunocomplexes were stained with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G antibody as previously described (19).
Western blotting.
HeLa cells were transfected with 0.2 μg of each plasmid DNA. Forty-eight hours later, the cells were lysed, and the extracted proteins were separated on 10% polyacrylamide gels. The proteins were then transferred to a nitrocellulose filter and incubated with rabbit anti-Rex C terminus-antibody followed by alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G antibody (19).
RESULTS
Promotion of nuclear export of the Rex protein by hCRM1.
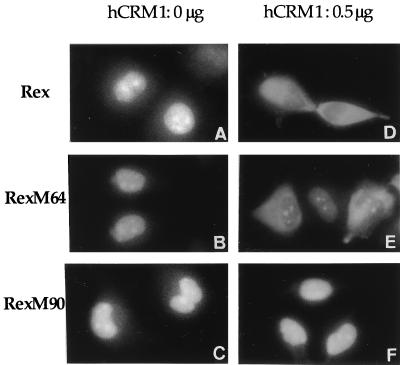
In order to study whether hCRM1 is involved in the nuclear export of the Rex protein, the effect of overexpression of hCRM1 on the localization of the Rex protein was examined by immunofluorescence microscopy. We analyzed not only wild-type Rex but also RexM64, which has a mutation in the multimerization domain, and RexM90, which has a mutation in the activation domain (19). All of these Rex-related proteins were found to be predominantly located in the nucleus and were especially concentrated in the nucleolus, confirming previous reports (33). However, when hCRM1 was overexpressed by transfection, Rex and RexM64 tended to be localized in the cytoplasm, whereas the location of RexM90 was not affected (Fig. 1). Quantitative evaluation confirmed the above observations, but we noticed that hCRM1 enhanced the cytoplasmic localization of wild-type Rex more efficiently than RexM64 (Table 1). This finding may be attributable to the slightly reduced affinity of RexM64 compared to wild-type Rex for hCRM1 (see Fig. 3) or the adverse effect of the mutation in the multimerization domain. These results indicate that the intact activation domain of Rex is required for hCRM1 to localize Rex to the cytoplasm. The results are also consistent with previous reports showing that hCRM1 exports various proteins possessing an NES sequence (11, 13, 22, 28, 29, 35, 39).
FIG. 1.
Subcellular localization of wild-type and mutant Rex proteins. At 24 h after transfection, cells transfected with 0.1 μg of pSRαRex (A and D), pSRαRexM64 (B and E), or pSRαRexM90 (C and F) in combination with 0.5 μg of pSRαhCRM1 (D, E, and F) or 0.5 μg of pSRα296 instead of pSRαhCRM1 in order to adjust the total amounts of the plasmids (A, B, and C) were subjected to immunofluorescence microscopy. Magnification, ×1,720.
TABLE 1.
Subcellular localization of Rex, RexM64, and RexM90 with or without overexpression of hCRM1a
| Protein | hCRM1 (μg) | % Localization in:
|
|
|---|---|---|---|
| Nucleus and nucleolus | Cytoplasm | ||
| Rex | 0 | 100 | 0 |
| 0.3 | 33 | 67 | |
| 0.5 | 15 | 85 | |
| RexM64 | 0 | 100 | 0 |
| 0.3 | 41 | 59 | |
| 0.5 | 37 | 63 | |
| RexM90 | 0 | 100 | 0 |
| 0.3 | 100 | 0 | |
| 0.5 | 100 | 0 | |
Cells stained as described in the legend to Fig. 1. were counted, and the percentages of stained nucleus and nucleolus or cytoplasm were calculated. At least 100 cells were counted. These assays were independently done twice, and essentially the same results were obtained.
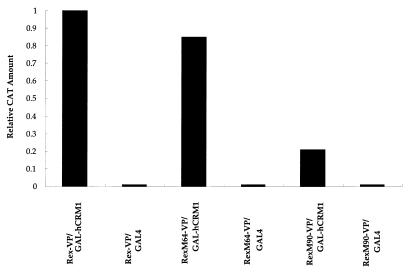
FIG. 3.
Interaction of hCRM1 with Rex, RexM64, and RexM90 in the nuclei of mammalian cells. A mammalian version of the two-hybrid assay with transient expression by transfection into HeLa cells was performed. The hCRM1 protein was expressed as a GAL4 fusion, and the other proteins were expressed as VP16 fusions. pCDM-β-gal (0.1 μg) was included in all samples as an internal control. At 24 h after transfection, the cells were harvested, and the amount of CAT and the activity of β-galactosidase were quantified. The amount of CAT and the activity of β-galactosidase obtained after transfection of Rex-VP and GAL-hCRM1 were 370 pg and 2.5 × 10−3 U, respectively, and the ratio (CAT/β-galactosidase) was arbitrarily set at 1. GAL4 represents the plasmid expressing only the GAL4 region.
Abrogation of the DN effect of TAgRex by hCRM1.
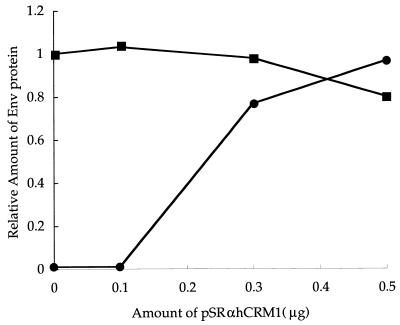
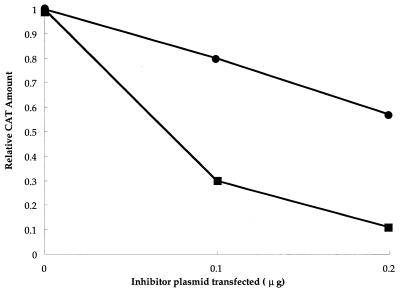
In previous studies, we showed that TAgRex inhibits Rex function by sequestering cellular cofactors and that the intact activation domain of TAgRex is responsible for this inhibition (19, 21). This system may be used to identify cellular cofactors that are involved in the functioning of Rex through its activation domain, since overexpression of these cofactors would be expected to abrogate the DN effect of TAgRex (19). To explore the role of hCRM1 in Rex function, we used this approach with some modifications, including the use of pTU50RXRE as a reporter plasmid and TAgRexM64 as an inhibitor. These modifications allowed quantification of the amount of Env protein produced as a result of the action of Rex and straightforward interpretation, since TAgRexM64 does not form hetero-oligomers with Rex (21). We reproduced our previous results (19, 21), showing that TAgRexM64 interferes with Rex function efficiently: transfection of 0.1, 0.2, and 0.3 μg of pSRαTAgRexM64 reduced the production of Env protein to 37, <1, and <1%, respectively, that normally observed in the absence of TAgRexM64 (data not shown). Next, we examined the effect of the overexpression of hCRM1 on Rex function in the presence of TAgRexM64 (Fig. 2). Rex-dependent production of Env protein was restored by the overexpression of hCRM1 in a dose-dependent manner. Notably, the transfection of 0.5 μg of pSRαhCRM1 achieved full restoration of Env production. On the other hand, the overexpression of hCRM1 in the absence of TAgRexM64 did not affect Rex function significantly, probably because the amount of intrinsic hCRM1 was sufficient to support Rex function. These results suggest that hCRM1 is a major cofactor involved in Rex function.
FIG. 2.
Overexpression of hCRM1 restores Rex activity inhibited by TAgRexM64. HeLa cells were transfected with 0.05 μg of pSRαRex, 0.5 μg of pTU50RXRE as a reporter plasmid, and various amounts of pSRαhCRM1 with (•) or without (▪) 0.2 μg of pSRαTAgRexM64. pCDM-β-gal (0.1 μg) was included in all samples as an internal control. At 48 h after transfection, the cells were harvested, the amount of Env and the activity of β-galactosidase were quantified, and the ratios of Env to β-galactosidase were calculated. The ratio of the amounts of Env and β-galactosidase obtained in the sample in which TAgRexM64 and pSRαhCRM1 were omitted was arbitrarily set at 1.
Interaction of hCRM1 with Rex proteins.
To obtain evidence for an interaction between Rex and hCRM1, two-hybrid analysis was performed with HeLa cells. In the two-hybrid experiments, we prepared cell lysates 24 h after transfection to prevent the overaccumulation of RexM90 in the nucleus. This step we hoped would reduce the deleterious effects of RexM90 overexpression on cell functions, which are due to the export defect of RexM90, which lacks an intact NES. Indeed, earlier preparation of the samples ensured more reproducible results than did preparation 48 h after transfection (data not shown) (2, 19). As shown in Fig. 3, wild-type Rex and RexM64 interacted similarly with hCRM1. In contrast, RexM90 had a greatly reduced affinity for hCRM1, suggesting that the intact activation domain of Rex is required for efficient interaction with hCRM1. Since it was difficult to produce a large amount of hCRM1 protein in Escherichia coli because of its instability, we could not analyze the direct interaction of Rex and hCRM1 in an in vitro protein-protein interaction assay.
Rex mutants fail to multimerize.
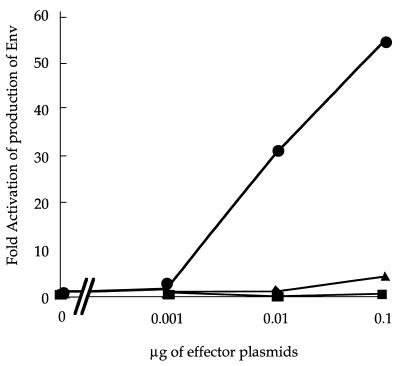
Although the above results are consistent with reports describing hCRM1 as an NES receptor (11, 13, 22, 28, 29, 35, 39), the residual ability of RexM90 (approximately 20% that of wild-type Rex) to interact with hCRM1 was unexpected. Thus, we examined whether RexM90 retained the residual activity to produce Env protein compared to wild-type Rex and RexM64, since quantitative analysis was possible with pTU50RXRE as a reporter. As shown in Fig. 4, RexM90 showed very low activity (approximately 3% that of wild-type Rex at most) and RexM64 had virtually no activity at all.
FIG. 4.
Activities of Rex, RexM64, and RexM90. Cells were transfected with 0.5 μg of pTU50RXRE and increasing quantities of pSRαRex (•), pSRαRexM64 (▪), or pSRαRexM90 (▴). The culture medium was harvested at 48 h posttransfection, the amount of Env protein produced was assayed, and the cell lysates were used for the quantification of β-galactosidase expression levels. The Env/β-galactosidase ratio for each sample was divided by that for the sample without Rex and expressed as fold activation.
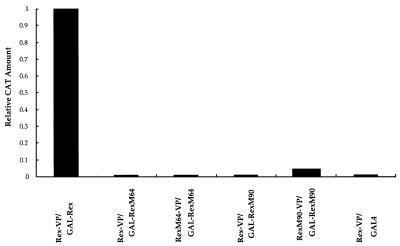
The low activity of RexM90 appeared to be inconsistent with its ability to interact with hCRM1. In order to explore the cause of the low activity of RexM90, we reanalyzed the capacity of Rex-related proteins to multimerize. As depicted in Fig. 5, only the combination of GAL-Rex with Rex-VP showed significant multimerization. No other combination, including Rex-RexM64, RexM64-RexM64, Rex-RexM90, and RexM90-RexM90, resulted in the formation of hetero- or homo-oligomers. It was ascertained by Western blotting that all of the fusion proteins were correctly synthesized and were present in a stable form in the transfected cells (Fig. 6). These results are in accord with the results reported by Bogerd and Greene (2). Thus, the multimerization defect of RexM90 may account for its very weak capacity to induce Env production.
FIG. 5.
Analysis of in vivo multimerization of the wild-type and mutant Rex proteins. HeLa cells were cotransfected with the expression plasmids for GAL and VP16 fusion proteins as indicated. The cells were harvested 24 h after transfection, and the amount of CAT and the activity of β-galactosidase were quantified. The amount of CAT and the activity of β-galactosidase resulting from the coexpression of GAL-Rex and Rex-VP were 270 pg and 4.8 × 10−3 U, respectively, and the ratio was assigned a value of 1.
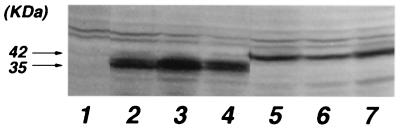
FIG. 6.
Western blot analysis of GAL-Rex and Rex-VP fusion protein expression in HeLa cells. The results for the control (lane 1) and for RexM90-VP (lane 2), RexM64-VP (lane 3), Rex-VP (lane 4), GAL-RexM90 (lane 5), GAL-RexM64 (lane 6), and GAL-Rex (lane 7) are shown.
Restoration of Rex multimerization by the overexpression of hCRM1.
The above results raise the possibility that a cofactor may be involved in the multimerization of the Rex protein. If so, the activity of RexM64, which has a mutation in the multimerization domain, may be complemented by the overexpression of hCRM1. We tested this possibility by cotransfecting pSRαhCRM1 along with a wild-type Rex- or mutant Rex-expressing plasmid. As shown in Table 2, the overexpression of hCRM1 restored the activity of RexM64 up to one third that of wild-type Rex in a dose-dependent manner, whereas it had a little effect on RexM90. These results are consistent with the observation that RexM64 can associate with hCRM1 more efficiently than RexM90 (Fig. 3).
TABLE 2.
Restoration of the activity of RexM64 by overexpression of hCRM1a
| Protein | Env/β-galactosidase ratio with the following amt (μg) of pSRαhCRM1:
|
|||
|---|---|---|---|---|
| 0 | 0.1 | 0.3 | 0.5 | |
| Rex | 1 | 0.94 | 0.72 | 0.60 |
| RexM64 | <0.01 | 0.19 | 0.33 | 0.32 |
| RexM90 | 0.01 | 0.04 | 0.06 | 0.06 |
HeLa cells were transfected with 0.05 μg of pSRαRex, pSRαRexM64, or pSRαRexM90, 0.5 μg of pTU50RXRE as a reporter plasmid, and various amounts of pSRαhCRM1. The ratio of the amounts of Env and β-galactosidase obtained with transfection of pSRαRex without pSRαCRM1 was arbitrarily set at 1. pSRαRex increased the Env/β-galactosidase ratio to 44 times that in the sample without pSRαRex.
We examined the ability of wild-type and mutant Rex proteins to multimerize under conditions of hCRM1 overexpression (Table 3). The overexpression of hCRM1 allowed RexM64 to multimerize, albeit at a level that was still inefficient compared to that of wild-type Rex. In contrast, it did not affect the ability of RexM90 to multimerize. The simultaneous restoration of RexM64 activity and multimerization by overexpression of hCRM1 supports the hypothesis that hCRM1 is involved in the multimerization of the Rex protein and that this process is a prerequisite for its biological activity.
TABLE 3.
Restoration of the multimerization of RexM64 by overexpression of hCRM1a
| Fusion proteins | CAT/β-galactosidase ratio with the following amt (μg) of pSRαhCRM1:
|
|||
|---|---|---|---|---|
| 0 | 0.1 | 0.3 | 0.5 | |
| GAL-Rex–Rex-VP | 1 | 1.92 | 1.74 | 1.35 |
| GAL-RexM64–RexM64-VP | <0.01 | 0.09 | 0.20 | 0.20 |
| GAL-RexM90–RexM90-VP | 0.07 | 0.08 | 0.05 | 0.05 |
HeLa cells were cotransfected with pSRαhCRM1 and plasmids expressing various GAL and VP16 fusion proteins. The CAT/β-galactosidase ratio resulting from the cotransfection of pGAL-Rex and pRex-VP without pSRαhCRM1 was assigned a value of 1.
Effect of a DN mutant and hCRM1 on the multimerization of wild-type Rex.
If hCRM1 is involved in the multimerization not only of mutant Rex but also of wild-type Rex, it is conceivable that TAgRexM64 may abrogate the multimerization of wild-type Rex in spite of the inability of RexM64 and Rex to form hetero-oligomers (Fig. 5). We tested this possibility by cotransfecting pSRαTAgRexM64 in the two-hybrid assay. As a negative control, we used TAgRexM6490, which lacks both intact multimerization and intact activation domains (19). Figure 7 shows that the multimerization of Rex was severely inhibited by the coexpression of TAgRexM64, in contrast to the marginal effect of TAgRexM6490. These results suggest that overexpression of the intact activation domain may be required for the inhibition of multimerization, implying an involvement of hCRM1 in multimerization.
FIG. 7.
Inhibitory effect of TAgRexM64 and TAgRexM6490 on Rex-Rex multimerization. HeLa cell cultures were transfected with pGAL-Rex and pRex-VP in combination with various amounts of pSRαTAgRexM64 (▪) or with pSRαTAgRexM6490 (•). At 24 h posttransfection, the cells were harvested, and the amount of CAT and the activity of β-galactosidase were quantified. The amount of CAT and the activity of β-galactosidase obtained with transfection of pRex-VP and pGAL-Rex without any TAgRex expression plasmid were 230 pg and 4.5 × 10−3 U, respectively, and the ratio was arbitrarily set at 1.
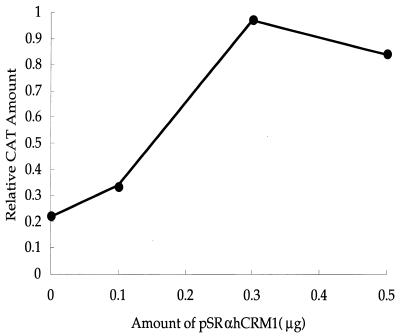
Next, we examined whether or not the overexpression of hCRM1 suppresses the inhibitory effect of TAgRexM64 on multimerization. As shown in Fig. 8, the extent of multimerization of Rex gradually increased with the amount of pSRαhCRM1 cotransfected into cells. These results suggest that hCRM1 may be required for the multimerization of wild-type Rex protein.
FIG. 8.
Overexpression of hCRM1 restores Rex-Rex multimerization inhibited by TAgRexM64. HeLa cell cultures were transfected with pGAL-Rex, pRex-VP, and 0.2 μg of pSRαTAgRexM64 in combination with various amounts of pSRαhCRM1. The amount of CAT and the activity of β-galactosidase obtained with transfection of pRex-VP and pGAL-Rex without pSRαTAgRexM64 were 380 pg and 6.8 × 10−3 U, respectively, and the ratio was arbitrarily set at 1.
DISCUSSION
In this paper we describe two roles for hCRM1 in Rex function. First, hCRM1 facilitates the export of the Rex protein from the nucleus to the cytoplasm, since the overexpression of hCRM1 localized the Rex protein to the cytoplasm. This effect required the intact activation domain of the Rex protein, since RexM90, which has a mutation in the activation domain, remained in the nucleus. Also, the ability of Rex-related proteins to localize to the cytoplasm was dependent on their association with hCRM1. Furthermore, the overexpression of hCRM1 fully restored the function of Rex inhibited trans dominantly by TAgRexM64, suggesting that hCRM1 is a major cofactor for Rex. Since Rev and Rex have been shown to share the same route for the transport of their cognate mRNAs (19), these results are consistent with hCRM1 being an NES receptor for the export of shuttling proteins, including Rev (11, 13, 22, 29, 35).
Our previous report suggested that the translation initiation factor eIF-5A abrogates the inhibitory effect of TAgRex on Rex function in Cos7 cells (19). In recent experiments, however, we found that the physiological condition of Cos7 cells greatly influences the effect of eIF-5A, and we could not reproduce the effect in HeLa cells. Thus, more extensive study will be required to discover the role of eIF-5A in Rex function.
The second role for hCRM1 in the multimerization of the Rex protein was shown by three experimental lines of evidence. First, RexM90 could not multimerize, suggesting a role for the activation domain in multimerization. Second, the activity of RexM64, which has a mutation in the multimerization domain, was partially restored by the overexpression of hCRM1, and this effect coincided with the partial restoration of multimerization. Third, TAgRexM64 abrogated the ability of the wild-type Rex protein to oligomerize, a defect that could be corrected by furnishing exogenous hCRM1.
We used a two-hybrid assay to measure the ability to oligomerize. The use of this assay to study Rev protein function has been criticized on the basis of the abnormally high accumulation in the nucleus and nucleolus of activation domain mutant proteins, which impair cellular functions and lead to reduced CAT production (37). However, we always cotransfected a plasmid expressing β-galactosidase as a standard for normalization. Accordingly, the nonspecific impairment of cellular function by RexM90 could not account for the reduction in CAT production. In addition, the fact that RexM14, which has a mutation in the activation domain, did not dominantly inhibit wild-type Rex function in terms of RXRE-dependent gene expression (5), a situation that differs from the DN phenotype of the Rev activation domain mutants, suggested that a normal cellular environment existed during the overexpression of the Rex activation domain mutant.
Oligomerization has been proposed to position individual activation domains to create a domain sufficient for interaction with the cellular cofactor (2, 23), namely, hCRM1. In this study, we demonstrated that high concentrations of hCRM1 instead complement the poor multimerization ability of Rex mutants and that even the wild-type Rex protein requires a sufficient amount of hCRM1 to multimerize. Thus, the intrinsic capability of Rex proteins to multimerize may be necessary but not sufficient for multimerization in vivo. Moreover, these results suggest that the multimerization of Rex proteins is not a prerequisite for their interaction with hCRM1. For these reasons, we propose that a Rex protein initially interacts with an hCRM1 molecule, leading to the multimerization of Rex proteins in a step that is favored by the intrinsic capacity of Rex proteins to oligomerize. This process in turn would favor the formation of a more favorable structure for interaction with supplementary hCRM1 molecules. We could not demonstrate homo-oligomerization of hCRM1 because a domain of VP16 inactivated this function of hCRM1. However, Ran has been reported to multimerize (9), suggesting the formation of a large trimeric complex including hCRM1, Ran, and Rex. Alternatively, the binding of hCRM1 may induce conformational changes in Rex proteins, leading to more efficient multimerization. Although we did not address the role of RNA in this study, RNA could conceivably take part in the multimerization process, as suggested previously (23, 26).
ACKNOWLEDGMENTS
We thank Y. Okuda for technical assistance.
This investigation was supported by grants from the Ministry of Education, Science and Culture, Japan, and the Ministry of Health and Welfare, Japan.
REFERENCES
- 1.Ballaun C, Farrington G K, Dobrovnik M, Rusche J, Hauber J, Böhnlein E. Functional analysis of human T-cell leukemia virus type 1 Rex response element: direct RNA binding of Rex protein correlates with in vivo activity. J Virol. 1991;65:4408–4413. doi: 10.1128/jvi.65.8.4408-4413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogerd H, Greene W C. Dominant negative mutants of human T-cell leukemia virus type 1 Rex and human immunodeficiency virus type 1 Rev fail to multimerize in vivo. J Virol. 1993;67:2496–2502. doi: 10.1128/jvi.67.5.2496-2502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogerd H P, Huckaby G L, Ahmed Y F, Hanly S M, Greene W C. The type I human T-cell leukemia virus (HTLV-I) Rex trans-activator binds directly to the HTLV-I Rex and the type 1 human immunodeficiency virus Rev RNA response elements. Proc Natl Acad Sci USA. 1991;88:5704–5708. doi: 10.1073/pnas.88.13.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogerd H P, Fridell R A, Benson R E, Jian H, Cullen B R. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Böhnlein S, Pirker F P, Hofer L, Zimmermann K, Bachmayer H, Böhnlein E, Hauber J. Transdominant repressors for human T-cell leukemia virus type 1 Rex and human immunodeficiency virus type 1 Rev function. J Virol. 1991;65:81–88. doi: 10.1128/jvi.65.1.81-88.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cochrane A W, Perkins A, Rosen C A. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J Virol. 1990;64:881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen B R. Regulation of human immunodeficiency virus replication. Annu Rev Microbiol. 1991;45:219–250. doi: 10.1146/annurev.mi.45.100191.001251. [DOI] [PubMed] [Google Scholar]
- 8.Cullen B R, Malim M H. The HIV-1 Rev protein: prototype of a novel class of eukaryotic post-transcriptional regulators. Trends Biochem Sci. 1991;16:346–350. doi: 10.1016/0968-0004(91)90141-h. [DOI] [PubMed] [Google Scholar]
- 9.Dingwall C, Kandel-Lewis S, Seraphin B. A family of Ran binding proteins that includes nucleoporins. Proc Natl Acad Sci USA. 1995;92:7525–7529. doi: 10.1073/pnas.92.16.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a Rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;19:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 12.Fujii M, Tsuchiya H, Chuhjo T, Akizawa T, Seiki M. Interaction of HTLV-I Tax1 with p67SRF causes the aberrant induction of cellular immediate early genes through CArG boxes. Genes Dev. 1992;6:2066–2076. doi: 10.1101/gad.6.11.2066. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;20:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 14.Grassman R, Berchtold S, Aepinus C, Ballaun C, Boehnlein E, Fleckenstein B. In vitro binding of human T-cell leukemia virus Rex proteins to the Rex response element of viral transcripts. J Virol. 1991;65:3721–3727. doi: 10.1128/jvi.65.7.3721-3727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanly S M, Rimsky L T, Malim M H, Kim J J, Hauber J, DucDodon M, Le S-Y, Maizel J V, Cullen B R, Greene W C. Comparative analysis of the HTLV-I Rex and HIV-1 Rev trans-regulatory proteins and their RNA response elements. Genes Dev. 1989;3:1534–1544. doi: 10.1101/gad.3.10.1534. [DOI] [PubMed] [Google Scholar]
- 16.Hidaka M, Inoue J, Yoshida M, Seiki M. Post-transcriptional regulator (Rex) of HTLV-I initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 1988;7:519–523. doi: 10.1002/j.1460-2075.1988.tb02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hope T J, Klein N P, Elder M E, Parslow T G. trans-Dominant inhibition of human immunodeficiency virus type 1 Rev occurs through formation of inactive protein complexes. J Virol. 1992;66:1849–1855. doi: 10.1128/jvi.66.4.1849-1855.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue J-I, Yoshida M, Seiki M. Transcriptional (p40×) and post-transcriptional (p27×-III) regulators are required for the expression and replication of human T-cell leukemia virus type I genes. Proc Natl Acad Sci USA. 1987;84:3653–3657. doi: 10.1073/pnas.84.11.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katahira J, Ishizaki T, Sakai H, Adachi A, Yamamoto K, Shida H. Effects of translation initiation factor eIF-5A on the functioning of human T-cell leukemia virus type 1 Rex and human immunodeficiency virus Rev inhibited trans dominantly by a Rex mutant deficient in RNA binding. J Virol. 1995;69:3125–3133. doi: 10.1128/jvi.69.5.3125-3133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim F J, Beeche A A, Hunter J J, Chin D J, Hope T J. Characterization of the nuclear export signal of human T-cell lymphotropic virus type 1 Rex reveals that nuclear export is mediated by position-variable hydrophobic interactions. Mol Cell Biol. 1996;16:5147–5155. doi: 10.1128/mcb.16.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiyokawa T, Umemoto T, Watanabe Y, Matsushita S, Shida H. Two distinct pathways for intronless mRNA expression: one related, the other unrelated to human immunodeficiency virus Rev and human T cell leukemia virus type I Rex functions. Biol Signals. 1997;6:134–142. doi: 10.1159/000109119. [DOI] [PubMed] [Google Scholar]
- 22.Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem. 1997;21:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- 23.Madore S J, Tiley L S, Malim M H, Cullen B R. Sequence requirements for Rev multimerization in vivo. Virology. 1994;202:186–194. doi: 10.1006/viro.1994.1334. [DOI] [PubMed] [Google Scholar]
- 24.Malim M H, Böhnlein S, Hauber J, Cullen B R. Functional dissection of the HIV-1 Rev trans-activator—deriviation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 25.Malim M H, Tiley L S, McCarn D F, Rusche J R, Hauber J, Cullen B R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 26.Malim M H, Cullen B R. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 27.Malim M H, McCarn D F, Tiley L S, Cullen B R. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neville M, Stutz F, Lee L, Davis L, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 29.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 30.Roth J, Dobbelstein M. Export of hepatitis B virus RNA on a Rev-like pathway inhibition by the regenerating liver inhibitory factor IkappaB alpha. J Virol. 1997;71:8933–8939. doi: 10.1128/jvi.71.11.8933-8939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seiki M, Inoue J-I, Hidaka M, Yoshida M. Two cis-acting elements responsible for posttranscriptional trans-regulation of gene expression of human T-cell leukemia virus type I. Proc Natl Acad Sci USA. 1988;85:7124–7128. doi: 10.1073/pnas.85.19.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siomi H, Shida H, Nam S H, Nosaka T, Maki M, Hatanaka M. Sequence requirements for nucleolar localization of human T-cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell. 1988;55:197–209. doi: 10.1016/0092-8674(88)90043-8. [DOI] [PubMed] [Google Scholar]
- 34.Solomin L, Felber B K, Pavlakis G N. Different sites of interaction for Rev, Tev, and Rex proteins within the Rev responsive element of human immunodeficiency virus type 1. J Virol. 1990;64:6010–6017. doi: 10.1128/jvi.64.12.6010-6017.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 36.Stauber R, Gaitanaris G A, Pavlakis G N. Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology. 1995;213:439–449. doi: 10.1006/viro.1995.0016. [DOI] [PubMed] [Google Scholar]
- 37.Szilvay A M, Brokstad K A, Boe S O, Haukenes G, Kalland K H. Oligomerization of HIV-1 Rev mutants in the cytoplasm and during nuclear import. Virology. 1997;235:73–81. doi: 10.1006/viro.1997.8671. [DOI] [PubMed] [Google Scholar]
- 38.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 40.Venkatesh L K, Chinnadurai G. Mutants in a conserved region near the carboxy-terminus of HIV-1 Rev identify functionally important residues and exhibit a dominant negative phenotype. Virology. 1990;178:327–330. doi: 10.1016/0042-6822(90)90414-m. [DOI] [PubMed] [Google Scholar]
- 41.Weichselbraun I, Farrington G K, Rusche J R, Böhnlein E, Hauber J. Definition of the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type 1 Rex protein activation domain by functional exchange. J Virol. 1992;66:2583–2587. doi: 10.1128/jvi.66.4.2583-2587.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weichselbraun I, Borger J, Dobrovnik M, Bogerd H, Grassmann R, Greene W C, Hauber J, Böhnlein E. Dominant-negative mutants are clustered in a domain of the human T-cell leukemia virus type 1 Rex protein: implications for trans dominance. J Virol. 1992;66:4540–4545. doi: 10.1128/jvi.66.7.4540-4545.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zapp M L, Green M R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature (London) 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]