ABSTRACT
Infectious bursal disease virus (IBDV) causes immunosuppression and high mortality in young chickens. Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) are important regulators during viral infection. However, detailed the regulatory mechanisms of lncRNA-miRNA-mRNA have not yet been described in IBDV infection. Here, we analysed the role of lncRNA53557/gga-miR-3530-5p/STAT1 axis in very virulent IBDV (vvIBDV) infection. Evidently upregulated expression of lncRNA53557 was observed in bursa of Fabricius and DT40 cells. Meanwhile, overexpression of lncRNA53557 promoted STAT1 expression and inhibited vvIBDV replication and vice versa, indicating that the upregulation of lncRNA53557 was part of the host antiviral defence. The subcellular fractionation assay confirmed that lncRNA53557 can be localized in the cytoplasm. Further, dual-luciferase reporter, RNA pulldown, FISH and RT-qPCR assays revealed that lncRNA53557 were directly bound to gga-miR-3530-5p and had a negative regulatory relationship between them. Subsequent mechanistic analysis showed that lncRNA53557 acted as a competing endogenous RNA (ceRNA) of gga-miR-3530-5p to relieve the repressive effect of gga-miR-3530-5p on its target STAT1, as well as Mx1, OASL, and ISG15, thereby suppressing vvIBDV replication. The study reveals that a network of enriched lncRNAs and lncRNA-associated ceRNA is involved in the regulation of IBDV infection, offering new insight into the mechanisms underlying IBDV-host interaction.
KEYWORDS: Infectious bursal disease virus, bursa of fabricius, lncRNA53557, gga-miR-3530-5p, STAT1, interferon-stimulated genes
Introduction
Infectious bursal disease (IBD), caused by infectious bursal disease virus (IBDV), is an acute, highly contagious disease in chickens aged 3–6 weeks and is thought to be a major infectious disease threatening the poultry industry [1,2]. IBDV is a non-enveloped virus with a double-stranded RNA consisting of two segments: A and B [3–5]. Segment A (3.2 kb) comprises two overlapping open reading frames (ORFs), which encode the viral nonstructural proteins VP4 (28 kDa) and VP5 (17 kDa), and structural proteins VP2 (28 kDa) and VP3 (32 kDa), which are the major components of the virus [3,6]. Segment B (2.8 kb) contains a single ORF and encodes VP1 (90 kDa), an RNA-dependent RNA polymerase [7]. Two serotypes (I and – II) of IBDV have been recognized, and serotype I, which includes very virulent (vv), variant, classically virulent, and attenuated, exhibit varying degrees of pathogenicity and mortality in chickens [8]. IBDV predominantly targets and destroys B lymphocyte precursors in the bursa of Fabricius (BF), depleting the number of B lymphocytes and seriously damaging the bursal tissue [9]. This leads to high mortality, severe immunosuppression, and increased susceptibility of chickens to other pathogens [10,11]. Therefore, exploring the underlying regulatory mechanisms is essential to prevent and control IBDV infection.
The Encyclopedia of DNA Elements indicate that 80% of the genome in an organism is functional and that this majority is transcribed into various types of RNAs, including microRNAs (miRNAs), circular RNAs (circRNAs), and long non-coding RNAs (lncRNAs) [12]. Transcripts longer than 200 nucleotides in length without protein-coding potential are defined as lncRNAs [13,14]. They have been observed in a large diversity of species, including animals [15], plants [16], and even viruses [17]. Moreover, lncRNAs are poorly conserved between species and are generally considered to be transcriptional noise due to their low expression [18,19]. However, lncRNAs have been considered to perform diverse functions by regulating the expression of genes at various levels [20], and playing significant roles in a wide variety of important biological processes [21,22]. Recently, accumulating evidence has confirmed that lncRNAs play a crucial role in viral infection by regulating the expression of antiviral related key genes via the interferon (IFN)/JAK-STAT signalling pathway [23–25]. For example, IFN-α-induced lncRNA-CMPK2 promotes HCV replication by negatively regulating the expression of IFN-stimulation genes (ISGs), IFIT3, ISG15, and IFITM1 [26]. Importantly, our previous study found that loc107051710 promoted the expression of IFN-α and IFN-β by regulating IRF8, thereby suppressing IBDV replication [27], suggesting that lncRNA also plays a crucial role in IBDV infection.
MicroRNAs (miRNAs) are endogenous small non-coding RNAs of approximately 20–25 nt that mainly regulate the degradation or translation inhibition of target genes by binding to the 3′-untranslated regions (UTRs) [28,29]. They exert a wide range of functions associated with inflammation [30], cancer [31], and viral infections [32] by regulating the expression of various host genes at the post-transcriptional level. Moreover, accumulating evidence indicates that miRNAs play an important role in IBDV infection. For example, some host miRNAs can inhibit the translation and replication of IBDV through directly targeting the genomes of the virus, such as gga-miR-454-3p, gga-miR-21-5p and gga-miR-130b-3p [33]. Moreover, host miRNAs also can exhibit the capacity to either enhance or inhibit IBDV replication via regulating the genes involved in the IFN signalling pathway, such as gga-miR-142-5p, gga-miR-9-3p, gga-miR-155-5p and gga-miR-27-3p [33]. These findings illustrated that miRNAs are also involved in the host response to IBDV infection.
One such hypothesis for assigning lncRNA function that is gaining notable attention is the competitive endogenous RNA (ceRNA) hypothesis. The ceRNA hypothesis posits that specific RNAs can serve as miRNA sponges by competitively binding with miRNA response elements (MREs) to impair miRNA activity, thereby upregulating miRNA target gene expression [34]. In particular, lncRNAs are reported in an increasing number of studies as acting as functional ceRNAs. More importantly, studies have suggested that ceRNAs participate in the regulation of various diseases. For example, the lncRNA CHRF can bind miR-489 to regulate cardiac hypertrophy [35]. A number of studies have indicated that a single lncRNA or miRNA can effectively affect IBDV replication by regulating key gene expression in the IFN-I or JAK-STAT signalling pathways [27,33,36,37]. However, it is not yet clear whether lncRNAs are involved in the pathogenesis of IBDV infection, serving as ceRNAs. Here, the probable host lncRNAs and miRNAs involved in vvIBDV infection in BF were investigated and the lncRNA-miRNA-mRNA ceRNA regulatory networks were constructed. The results may shed new light on the regulation of noncoding RNA in vvIBDV infection.
Methods
Animal and treatment
All procedures used in the experiment were approved by the Institutional Animal Care and Use Committee of Northeast Agricultural University (2016NEFU–315, 13 April 2017). Three-week-old SPF White Leghorn chickens were raised in a negative-pressure pathogen-free condition, and the grouping and treatment of all animals was performed as previously described [36]. Briefly, chicken (n = 3) in the IBDV treatment group were infected with 103 ELD50/0.2 mL of vvIBDV strain LJ-5 via eye-nose drops, while chickens (n = 3) in the control group were treated with PBS. The two groups were housed independently. After 3 days, three chickens in the two groups were randomly selected and euthanized, and the bursal tissues were collected and immediately stored in liquid nitrogen.
RNA sequencing
RNA sequencing analysis was performed using Gene Denovo Biotechnology Co. (Guangzhou, China), and detailed sequencing procedures were performed as described previously [36]. Differentially expressed (DE) lncRNAs with |fold change (FC)| > 1.5 and p < 0.01 as well as miRNAs and mRNAs with |FC| > 2 and p value < 0.01 were screened. Furthermore, KEGG databases were used for functional enrichment analysis, and KEGG pathways were determined, with p < 0.05.
Construction of lncRNA-miRNA-mRNA ceRNA network
The lncRNA-miRNA-mRNA ceRNA network was constructed based on the DE lncRNAs, miRNAs, and mRNAs, using Cytoscape v3.6.0 [38].
Cells culture and transfection
DT40 cells, a chicken precursor B cell line, were provided by the Harbin Institute of Veterinary Medicine (Harbin, China), and were cultured in complete RPMI 1640 medium (Thermo Fisher Scientific, USA) supplemented with 10% foetal bovine serum, 2% chicken serum, 50 μM β-mercaptoethanol, 1% tryptose phosphate broth, 1% L-glutamine, and 100 U/mL penicillin G at 37 °C under 5% CO2.
The three specific siRNAs for loc107052552, loc107053874, loc107053557 loc107055286, and loc101749705, STAT1, and gga-miR-3530-5p mimic, and gga-miR-3530-5p inhibitor were designed and synthesized by GenePharma (Shanghai, China). DT40 cells (5 × 105 cells/mL) were seeded into 12-well plates and transfected with 50 nM specific siRNAs and 30 nM gga-miR-3530-5p mimic or inhibitor in Opti-MEM medium using the Lipofectamine RNAi MAX Reagent (3.0 μg/mL, Invitrogen, USA), according to the manufacturer’s protocol. The cells were transfected with negative control (NC) and inhibitor NC as controls. The sequences of NC, specific siRNAs, inhibitor NC, mimics, inhibitor, and the overexpression sequences of lncRNA53557 are summarized in Tables S1–S3. The overexpression sequences of lncRNA53557 were synthesized by Comate Bioscience (Jilin, China) and cloned into the HindIII-EcoRI site of the overexpression vector pcDNA3.1(+) to generate pcDNA3.1-lncRNA53557. In this study, DT40 cells were transfected with 1 μg of pcDNA3.1-lncRNA53557 vector using Lipofectamine 3000 reagent (3.0 μg/mL, Invitrogen, USA) in Opti-MEM medium, and the empty pcDNA3.1 vector with no lncRNA53557 sequence was transfected as a negative control.
Cell viability assay
DT40 cells were transfection into different RNA or plasmid intervention conditions. After transfection, 5000 cells/well were seeded in 96-well plates. Then, the cell viability was measured by a Cell Counting Kit-8 (CCK-8, Sigma) as described previously [39]. Briefly, CCK-8 was added and incubated at 37°C for 2 h, and the absorbance of each well was read at 450 nm with a microplate reader (BioTeke Corporation, China). Cell viability was calculated using the formula: Cell viability = (ODexperimental group/ODcontrol group) × 100%.
Analysis of the coding potential
Codon substitution frequencies (CSF) analysis (PhyloCSF) from the University of California Santa Cruz (UCSC; https://genome.ucsc.edu/) was utilized to analyze the coding potential of lncRNA53557. ACTB served as coding RNA control, and metastasis associated lung adenocarcinoma transcript 1 (MALAT1) and X inactive specific transcript (XIST) served as noncoding RNA controls. Moreover, the Coding Potential Calculator (CPC; http://cpc.cbi.pku.edu.cn/) and the Coding-Potential Assessment Tool (CPAT; http://rna-cpat.sourceforge.net/) were also used for potential noncoding analysis [40,41].
Subcellular fractionation
The PARIS™ Kit (Thermo Fisher Scientific, USA) was used to isolate the nucleus and cytoplasm of DT40 cells following the manufacturer’s protocol. Uninfected and IBDV-infected DT40 cells (~1 × 107 cells) were collected and washed twice with cold DEPC-treated PBS. Cold cell fractionation buffer (400 μL) was added to the tube to resuspend the cells and then placed on ice for 5 min, followed by immediate centrifugation at 15,000 × g for 5 min. The supernatant was transferred to a new tube as the cytoplasm extract, and the remainder was considered as the nuclear fraction after washing once with cold DEPC-treated PBS.
Real-time quantitative PCR (RT-qPCR) analysis
Total RNA from the BF tissues and DT40 cells (control and IBDV-infected groups) was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The purity and concentration of RNA samples were determined using Nanodrop 2000 (Thermo Fisher Scientific). Reverse transcription of mRNA and lncRNA was performed as previously described [27]; whereas that of miRNA was performed using miRcute miRNA First-strand cDNA Synthesis Kit (Tiangen, China). Further, RT-qPCR was performed as previously described [27]. Primer sequences are presented in Table S4. The relative mRNA levels of these genes were calculated using the 2−ΔΔCt method, and β-actin or U6 was used as an internal control.
Additionally, the viral loads of IBDV in infected cells were detected by RT-qPCR. The primers were designed to target the VP2 gene of IBDV using Oligo 6 software (forward: 5′-GCT ACA ATG GGT TGA TGT CTG-3′; reverse: 5′-ACG GTC CCT CTC ACT CAG TAT-3′). A standard curve was generated using a ten-fold serially diluted recombinant plasmid containing the IBDV-VP2 gene from 2 to 8 log10 copies, and viral loads of IBDV were quantified according to the standard curve.
Western blotting analysis
Cells were lysed and total protein was extracted using RIPA (Beyotime, China) and then denatured by heating. Total proteins were separated using 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes (Millipore, MA, USA). The membranes were blocked with 5% (w/v) skim milk at 25 °C for 2 h and incubated with diluted the following primary antibodies at 4 °C overnight: mouse anti-β-actin monoclonal (TaKaRa, China), rabbit anti-STAT1 (ProteinTech, China), rabbit anti-phosphorylated-STAT1 (Tyr701; Affinity, China), and mouse monoclonal anti-IBDV-VP2 (produced and preserved in our laboratory). Subsequently, after being washed thrice with PBST, the membranes were incubated with HRP-conjugated goat anti-rabbit IgG (H+L) (Thermo Fisher Scientific) or goat anti-mouse IgG (H+L) antibody (Thermo Fisher Scientific) for 1 h. Finally, the signal was detected using an ECL chemiluminescent system (Cheml Scope 5300, China) and related abundance was analysed and normalized to the level of β-actin by Image Lab Software.
Double luciferase reporter assay
The sequences of STAT1 3′-UTR and lncRNA53557 containing the gga-miR-3530-5p binding sites and their corresponding mutated sequences were designed, synthesized, and individually inserted into the pMIR-REPORT plasmid (Thermo Fisher Scientific), and termed STAT1-WT, STAT1-Mut, lncRNA53557-WT, and lncRNA53557-Mut, respectively. These sequences are listed in Table 1. All the above plasmids and phRL-TK plasmid were co-transfected with gga-miR-3530-5p mimics to DT40 cells using the Lipofectamine 3000 reagent. After 24 h of transfection, luciferase activity was evaluated using the Dual-Luciferase Reporter Assay Kit (Promega, USA). The activity of firefly luciferase was normalized to that of Renilla luciferase for each group.
Table 1.
Wild-type (WT) and mutant-type (mut) primers of STAT1 3’UTR and lncRNA53557.
| Names | Primer sequences (5’-3’) |
|---|---|
| Wild-type (WT) and mutant-type (Mut) primers of STAT1 3’UTR | |
| WT Forward | AGCTTGTATCTCGGATGGTGGATCCAGCAGAGATTGACACAGTAATGTGTTCAGAGCT |
| WT Reverse | CTGAACACATTACTGTGTCAATCTCTGCTGGATCCACCATCCGAGATACA |
| Mut Forward | AGCTTGTATCTCGGATGGTGGATCCATTAGCTATTGACACAGTAATGTGTTCAGAGCT |
| Mut Reverse | CTGAACACATTACTGTGTCAATAGCTAATGGATCCACCATCCGAGATACA |
| Wild-type (WT) and mutant-type (Mut) primers of lncRNA53557 | |
| WT Forward | AGCTTGGAAATATTCTGTGTCTGCTCAGAGCAGAGCCTCCAGCCTGAGTCTCCCAGAGCT |
| WT Reverse | CTGGGAGACTCAGGCTGGAGGCTCTGCTCTGAGCAGACACAGAATATTTCCA |
| Mut Forward | AGCTTGGAAATATTCTGTGTCTGCTCAGATTATCGCCTCCAGCCTGAGTCTCCCAGAGCT |
| Mut Reverse | CTGGGAGACTCAGGCTGGAGGCGATAATCTGAGCAGACACAGAATATTTCCA |
Note: Four sequences inserted in the pMIR-REPORT plasmids. The red colour indicates wild-type bases (for the target location), and green indicates the mutated bases.
RNA fluorescence in situ hybridization (FISH) analysis
FISH assay was performed to observe the location of lncRNA53557 and gga-miR-3530-5p in the BF. CY-3-labelled probes specific to lncRNA53557 and FAM-labelled locked nucleic acid gga-miR-3530-5p probes were designed and synthesized by GenePharma (Shanghai, China). The sequences of lncRNA53557 and gga-miR-3530-5p probes for FISH were as follows: lncRNA53557 (CY3-TCTTATTTATGCCCCTCGTAGTCCGTTCCTTCCTC-CY3) and gga-miR-3530-5p (FAM-CCACAATGGTGCGAGCAGAGC-FAM). The bursal tissues were fixed with FISH fixative at 4 °C for 24 h, and then were embedded in paraffin wax as previously described [39]. Hybridization was performed overnight with lncRNA53557 and gga-miR-3530-5p probes (GenePharma, China) according to the manufacturer’s instructions. Images were acquired using an inverted fluorescence microscope (Nikon, Tokyo, Japan).
RNA pulldown assay
The lncRNA53557 probe (the same as those used in RNA-FISH) and NC (3’-GTGTAACACGTCTATACGCCCA-5’) probe modified with 3,’ 5’ biotins. RNA pulldown assay was performed by RNA pulldown kit (Gene Pharma, China). LncRNA53557 and miRNAs (gga-miR-3530-5p and gga-miR-1808) bound to the pulldown beads were measured by RT-qPCR.
Statistical analysis
Graph generation and data analyses were performed using GraphPad Prism version 7.0. Student’s t-test or one-way analysis of variance and Tukey’s post-hoc pairwise test were used to perform statistical analysis. Data are presented as the mean ± standard deviations, and differences were considered significant at p < 0.05.
Results
Construction and analysis of ceRNA interaction network
To clarify the potential regulatory roles of DE lncRNAs and miRNAs in IBDV-infected BF, expression profiles of mRNAs, lncRNAs, and miRNAs were analysed by RNA sequencing; the results showed that 411 mRNAs, 272 lncRNAs, and 77 miRNAs were differentially expressed between the two groups, which has been determined in our previous study [36]. Subsequently, 10 differentially expressed lncRNAs and 10 miRNAs were randomly selected and verified using RT-qPCR (Figure 1(a,b)). The verification results were similar to the sequencing results, confirming the reliability of the RNA sequencing.
Figure 1.
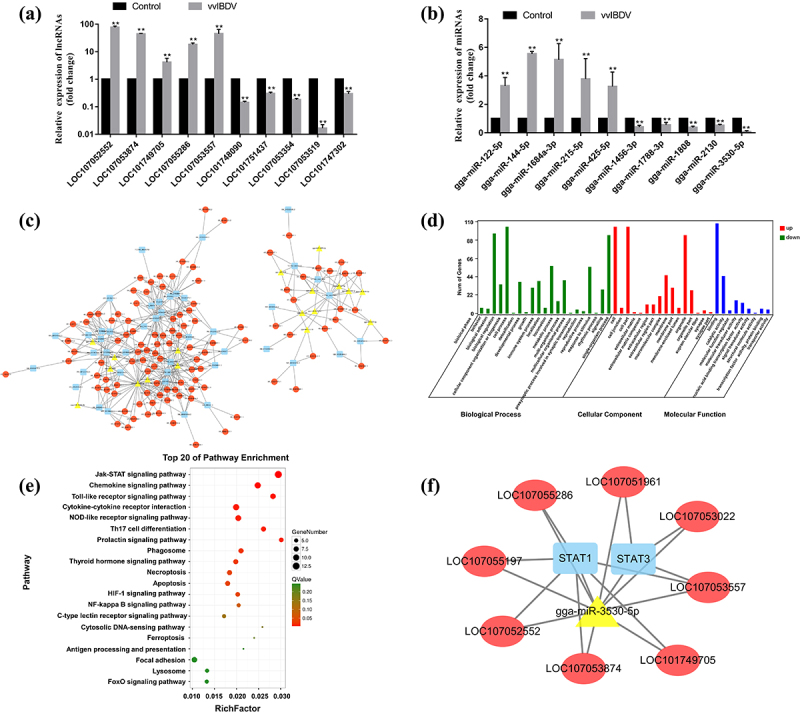
Effects of vvIBDV infection on differentially expressed lncRnas and miRnas, and the construction of ceRNA regulatory networks. (a and b) validation of the differentially expressed lncRnas and miRnas using RT-qPCR. (c) The lncRNA-miRNA-mRNA regulatory network was constructed and generated by cystoscope software. The red ellipses, yellow triangles, and blue rectangles represent lncRnas, miRnas, and mRnas, respectively. (d) Significantly enriched GO pathways with p values of < 0.05. (e) Significantly enriched KEGG pathways with p values of < 0.05. (f) The lncRNA-miRNA-mRNA (JAK-STAT signalling pathway) regulatory network.
Subsequently, a putative lncRNA-miRNA-mRNA regulatory network was constructed, in which 45 DE mRNAs, 108 DE lncRNAs, 18 DE miRNAs (Figure 1(c)). Gene Ontology (GO) analysis of differentially expressed genes (DEGs) in the network showed that cell process, biological regulation, and single-organism process were significantly enriched (Figure 1(d)). The KEGG analysis results showed that DEG in the network were predominantly enriched in the JAK-STAT signalling pathway (Figure 1(e)). DEGs in the JAK-STAT signalling pathway were then selected to construct a ceRNA integration network, and 19 pairs of interaction relationships containing eight lncRNAs, two mRNAs (STAT1 and STAT3), and one miRNA (gga-miR-3530-5p) were identified (Figure 1(f)). Additionally, in the lncRNA-miRNA-mRNA network, STAT1 (fold change = 9.34) and STAT3 (fold change = 2.89) could be regulated by six and three lncRNAs, respectively. Previous studies have indicated that STAT1 plays a vital role in innate antiviral immunity by inducing the expression of interferon-stimulated genes (ISGs) [42,43]. Thus, STAT1 was selected for further study.
Knockdown of STAT1 inhibited ISGs expression and promoted vvIBDV replication
The relative expression level of STAT1 was determined by RT-qPCR and Western Blotting. Evaluating the expression of STAT1 showed that both the mRNA (p < 0.01) and protein levels of STAT1 (p < 0.01) were upregulated in vvIBDV-infected BF (Figure 2(a)). To further investigate the effect of the IBDV LJ-5 strain on the expression level of STAT1 in immune cells, the DT40 cell line (a chicken precursor B cell line) was selected for in vitro study. DT40 cells were infected with 102 ELD50/200 μL vvIBDV for 48 h, and the results suggested that compared to the control group, both the mRNA (p < 0.01) and protein levels of STAT1 (p < 0.01) were significantly increased in vvIBDV-infected DT40 cells (Figure 2(b)). The result in vitro was consistent with the in vivo study.
Figure 2.
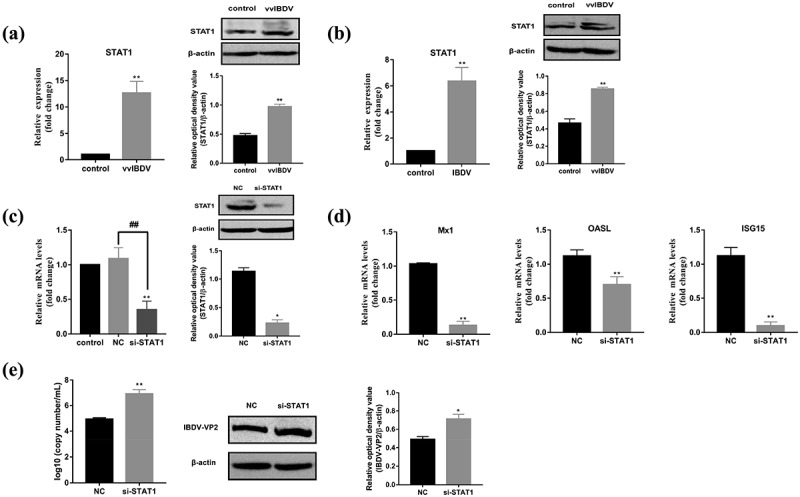
Knockdown of STAT1 inhibited ISGs expression and enhanced vvIBDV replication. (a) Relative mRNA and protein levels of STAT1 was analysed by RT-qPCR and Western Blotting in BF. (b) Relative mRNA and protein levels of STAT1 was analysed in DT40 cells with vvIBDV infection for 48 h. (c) siRNA-based knockdown of STAT1 was analysed by RT-qPCR and Western Blotting in DT40 cells. (d) Relative mRNA levels of Mx1, OASL, and ISG15 were analysed by RT-qPCR in STAT1 knocked-down DT40 cells infected with vvIBDV. (e) Expression of vvIBDV-VP2 gene and protein were measured by RT-qPCR and Western Blotting in STAT1 knocked-down DT40 cells infected with vvIBDV. Data are shown as the mean ± SD for three independent experiments. *p < 0.05, **p < 0.01.
To identify how STAT1 functions in vvIBDV infection, we generated STAT1-specific siRNA to silence STAT1 expression in vitro, and the results of RT-qPCR and Western Blotting revealed that si-STAT1 has a high inhibitory efficiency (Figure 2(c)). Subsequently, the effect of silencing STAT1 on the mRNA levels of Mx1, OASL, and ISG15 and the replication of vvIBDV were evaluated, and the results indicated that the siRNA-mediated knockdown of STAT1 resulted in a decrease in M×1(p < 0.01), OASL (p < 0.01), and ISG15 (p < 0.01) mRNA levels, compared to the NC group (Figure 2(d)); however, vvIBDV replication was significantly increased (Figure 2(e)).
Identification of lncRNA53557 as a candidate lncRNA
According to the lncRNA-miRNA-mRNA regulatory network, loc107052552 (GenBank accession number: XR_001465108.2), loc107053874 (GenBank accession number: XR_001467622.2), loc107053557 (GenBank accession number: XR_001466920.2), loc107055286 (GenBank accession number: XR_001470337.2), and loc101749705 (GenBank accession number: XR_001467871.2) regulated STAT1. Three specific siRNAs for the above lncRNAs were transfected into DT40 cells, and the knockdown efficiency was detected by RT-qPCR. Because of their high inhibitory efficiency (Figure 3(a)), si-loc107052552–1, si-loc107053874–2, si-loc107053557–3, si-loc107055286–1, and si-loc101749705–3 were chosen for the following experiments. Specific siRNAs were transfected into DT40 cells and the expression of STAT1 was detected by RT-qPCR and Western Blotting. Notably, the interference of lncRNAs described above suppressed the expression of STAT1 compared with the control or NC groups (p < 0.05 or p < 0.01), especially loc107053557 (Figure 3(b)), indicating that loc107053557 (lncRNA53557) had a strong regulatory effect on STAT1. Therefore, this uncharacterized lncRNA53557 and its role in gga-miR-3530-5p and STAT1 was studied.
Figure 3.
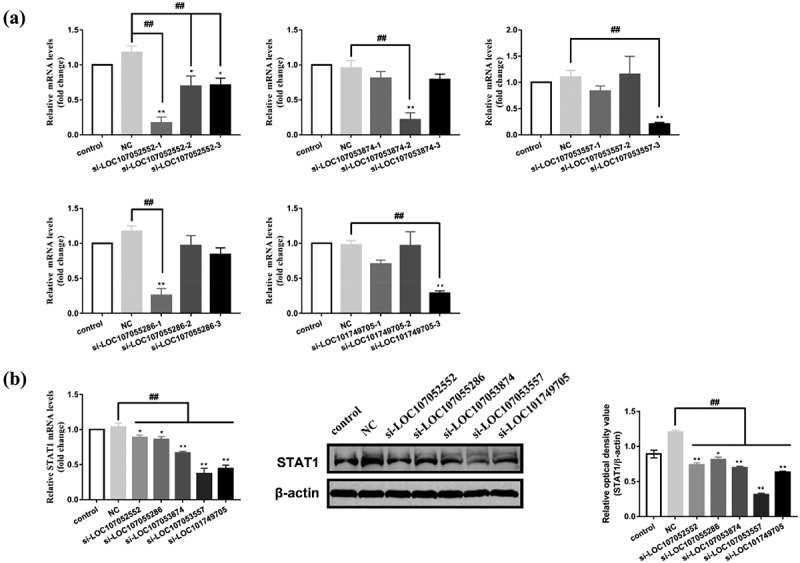
Screening and identification of the lncRnas. (a) The knockdown efficiency of loc107052552, loc107053874, loc107053557, loc107055286, and loc101749705 in DT40 cells treated with specific siRnas were analysed by RT-qPCR. (b) Effect of lncRnas (loc107052552, loc107053874, loc107053557, loc107055286, and loc101749705) on the mRNA and protein levels of STAT1 in DT40 cells was determined by RT-qPCR or Western blotting. Data are shown as the mean ± SD for three independent experiments. *p < 0.05, **p < 0.01 represents a significant difference compared to control group. ##P < 0.01 represents a significant difference compared to the negative control (NC) group.
Characterization of lncRNA53557
LncRNA53557 is located in chromosomal locus NC_052536.1. Online coding ability analysis by PhyloCSF demonstrated low lncRNA53557 coding potential (Figure 4(a)); meanwhile, Coding Potential Calculator (CPC) and coding potential Assessment Tool (CPAT) result also corroborated this lower coding potential (Figure 4(b,c)). Furthermore, the subcellular fractionation assay showed that lncRNA53557 was localized both in the cytoplasm and nucleus (Figure 4(d)), indicating that lncRNA53557 might function in the cytoplasm and nucleus.
Figure 4.
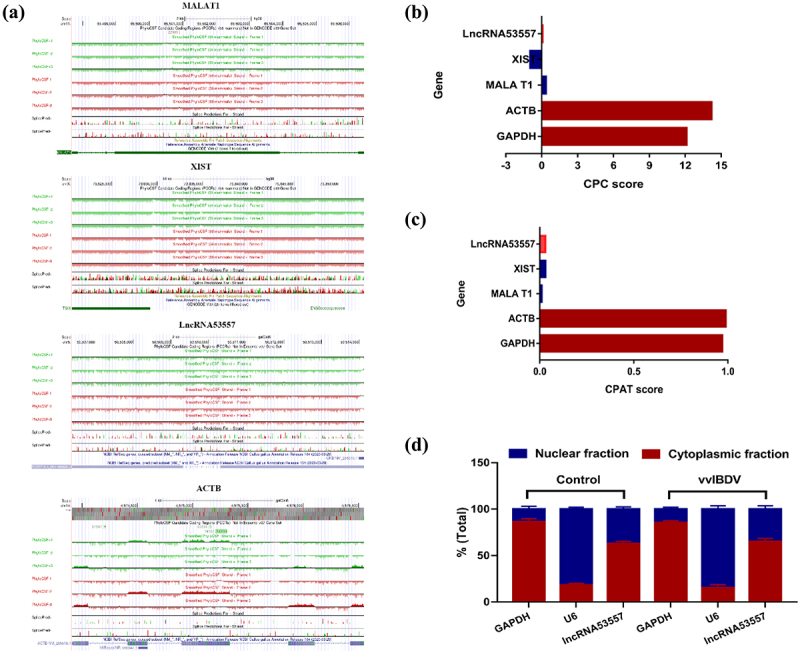
The characteristics of lncRNA53557 in DT40 cells. (a) The coding potential of lncRNA53557 was analysed by the PhyloCSF analysis. ACTB served as coding RNA control, and MALAT1 and XIST served as noncoding RNA controls. The regions with a score less than 0 (below the baseline) were predicted to be noncoding, while regions with a score greater than 0 (above the baseline) were predicted to be coding. (b and c) the coding potential of lncRNA53557 was analysed by CPC and CPAT. ACTB and GAPDH served as coding RNA controls, and MALAT1 and XIST served as noncoding RNA controls. (d) Subcellular fractionation experiment to analysed the localization of lncRNA53557 in DT40 cells. GAPDH and U6 were used as cytoplasmic and intranuclear reference, respectively. Data are shown as the mean ± SD for six independent experiments.
Overexpression of lncRNA53557 upregulated STAT1 expression and weakened vvIBDV replication
The relative expression level of lncRNA53557 was determined by RT-qPCR. The results revealed that lncRNA53557 (p < 0.01) was significantly upregulated in vvIBDV-infected BF compared with the control group (Figure 1(a)). Subsequently, the normal DT40 cells were treated with vvIBDV and the expression of lncRNA5357 was measured at various time points. The results showed that vvIBDV infection induced a robust increase in lncRNA53557 in DT40 cells at 48 h (p < 0.01, Figure 5(a)). To explore the biological function of lncRNA53557 in DT40 cells, the overexpression vector of pcDNA3.1-lncRNA53557 were constructed and transfected into DT40 cells. After transfection for 48 h, the mRNA levels of lncRNA53557 was measured by RT-qPCR, and the result demonstrated that the expression of lncRNA53557 was significantly increased (p < 0.01, Figure 5(b)). Meanwhile, the effect of transfection on cytotoxicity was evaluated by the CCK-8 kit. The result showed that no significant difference in cell activity between-group (Figure 5(c)). Subsequently, both lncRNA53557-overexpressed group cells and the PcDNA3.1 group cells were infected with 102 ELD50/200 μL vvIBDV for 48 h, and cell lysates were harvested to evaluate the expression of STAT1 using RT-qPCR and Western Blotting. The results demonstrated that the overexpression of lncRNA53557 markedly increased both the mRNA and protein levels of STAT1 and phospho-STAT1 (p < 0.01, Figure 5(d)) compared to the PcDNA3.1 group.
Figure 5.
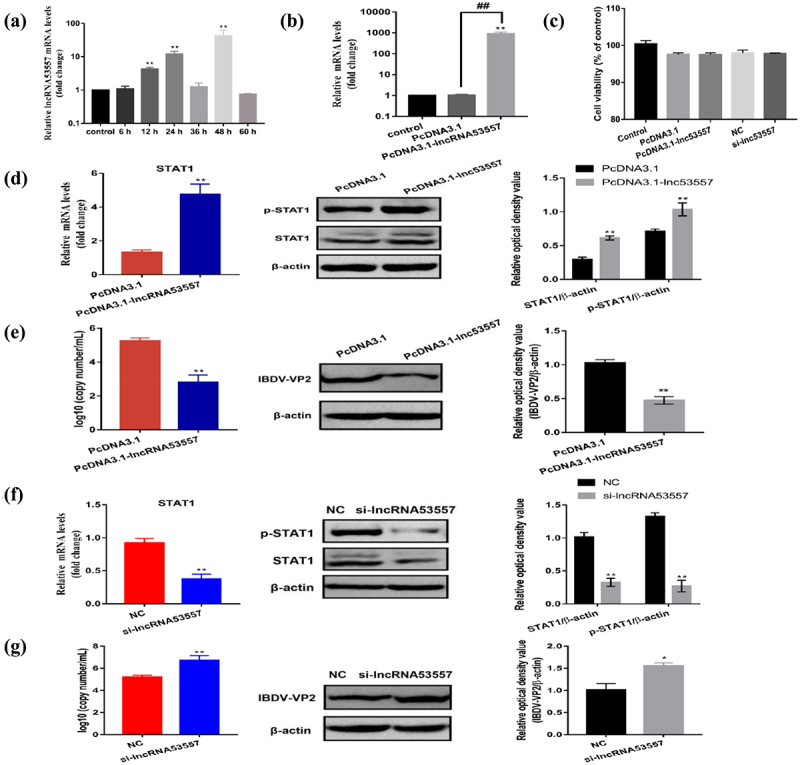
LncRNA53557 upregulated STAT1 expression and inhibited vvIBDV replication. (a) Relative expression levels of lncRNA5357 at various time points was measured in IBDV-infected DT40 cells by RT-qPCR. (b) Verification of the overexpression of the lncRNA53557 by RT-qPCR. (c) The effect of transfection on cell viability. (d) Relative mRNA and protein expression of STAT1 were determined by RT-qPCR and Western Blotting in lncRNA5357 overexpressed DT40 cells infected with vvIBDV. (e) Expression of vvIBDV-VP2 gene and protein were determined by RT-qPCR and Western Blotting in lncRNA5357 overexpressed DT40 cells infected with vvIBDV. (f) Relative mRNA and protein expression of STAT1 were determined by RT-qPCR and Western Blotting in lncRNA5357 knocked-down DT40 cells infected with vvIBDV. (g) Expression of vvIBDV-VP2 gene and protein were determined by RT-qPCR and Western Blotting in lncRNA5357 knocked-down DT40 cells infected with vvIBDV. Data are shown as the mean ± SD for three independent experiments. *p < 0.05, **p < 0.01.
To evaluate the influence of lncRNA53557 on IBDV proliferation, pcDNA3.1-lncRNA53557-transduced DT40 cells were treated and measured vvIBDV replication by RT-qPCR and Western Blotting. Results implied that the overexpression of lncRNA53557 was accompanied by a decrease in vvIBDV replication compared to the PcDNA3.1 group (p < 0.01, Figure 5(e)), indicating that lncRNA53557 plays a critical role in antiviral responses.
Knockdown of lncRNA53557 inhibited STAT1 expression and promoted vvIBDV replication
To further validate the functionality of lncRNA53557 on both STAT1 expression and vvIBDV replication, siRNA against lncRNA53557 was transfected into DT40 cells to silence lncRNA53557 ectopically. Both lncRNA53557 knocked-down cells and the NC group cells were infected with vvIBDV (102 ELD50/200 μL), and cell lysates were harvested to evaluate the expression of STAT1 using RT-qPCR and Western Blotting. The results showed that downregulation of lncRNA53557 considerably decreased both the mRNA and protein levels of STAT1, and phospho-STAT1 (p < 0.01, Figure 5(f)) compared to the NC group. Furthermore, the knockdown of lncRNA53557 significantly enhanced vvIBDV replication in DT40 cells compared to the NC group (p < 0.01, Figure 5(g)).
LncRNA53557 were a direct target of gga-miR-3530-5p
The function of lncRNA is closely related to its subcellular location [44]. Based to the results showed that lncRNA53557 can be located in the cytoplasm and influenced STAT1 expression in the same trend, we speculated that STAT1 might be the target molecule of lncRNA53557. Numerous cytoplasmic lncRNAs have been reported to perform as competing endogenous RNAs (ceRNAs) by binding common miRNAs [45].
TargetScan, an miRNA target prediction tool, was used to analysed the target relationship between lncRNA53557, STAT1 and gga-miR-3530-5p. The results suggested that both lncRNA53557 and STAT1 had the potential to bind gga-miR-3530-5p (Figure 6(a) and 8(a)); that is, they might be connected by the same miRNA response element (gga-miR-3530-5p) in cytoplasm. Additionally, we evaluated the expression of gga-miR-3530-5p in vvIBDV-infected BF (Figure 1(b)) and DT40 cells (Figure 6(b)). Data indicated that infection of vvIBDV induced lower expression of gga-miR-3530-5p in vivo and in vitro.
Figure 6.
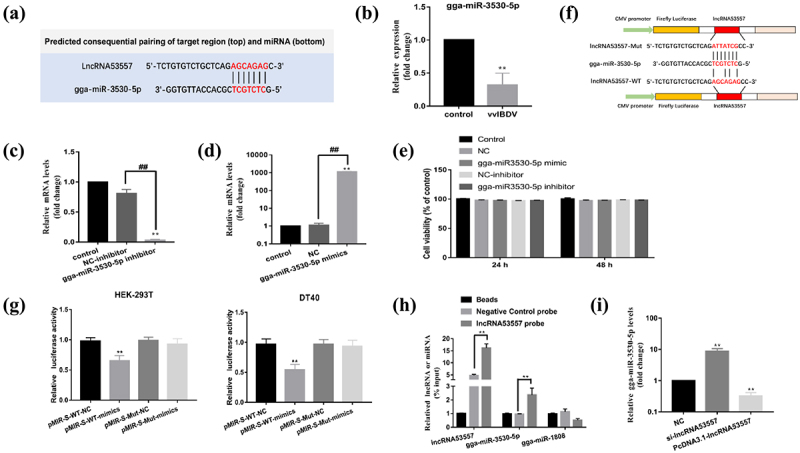
LncRNA53557 was interacted with gga-miR-3530-5p. (a) The binding sites of lncRNA53557 and gga-miR-3530-5p (marked in red). (b) Relative mRNA level of STAT1 was analysed by RT-qPCR in DT40 cells with vvIBDV infection for 48 h. (c and d) the knockdown (c) or overexpression (d) of gga-miR-3530-5p in DT40 cells were analysed by RT-qPCR. (e) The effect of transfection on cell viability. (f) Schematic illustration of lncRNA53557-WT and lncRNA53557-mut luciferase reporter vectors. (g) Luciferase activities were detected in HEK-293T or DT40 cells co-transfected with gga-miR-3530-5p and luciferase reporters containing nothing, lncRNA53557, or mutant transcript. Data are presented as the relative ratio of firefly luciferase activity to renilla luciferase activity. (h) DT40 cell lysates were incubated with biotin-labelled lncRNA53557; after pulldown, lncRNA53557, gga-miR-3530-5p, and gga-miR-1808 were extracted and assessed by RT-qPCR. (i) The effect of lncRNA53557 on the expression of gga-miR-3530-5p were determined by RT-qPCR in lncRNA53557 knocked-down or overexpressed DT40 cells. Data are shown as the mean ± SD for three independent experiments. *p < 0.05, ** or ##P < 0.01.
Figure 8.
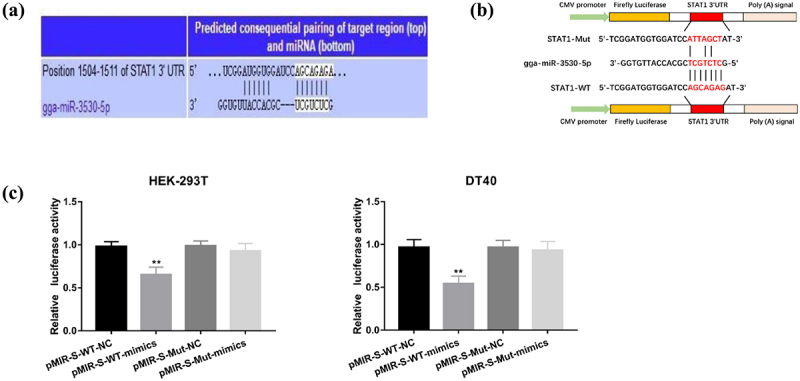
gga-miR-3530-5p directly targets STAT1 3’UTR. (a) Target relationship between gga-miR-3530-5p and STAT1 was predicted by TargetScan. (b) Schematic illustration of STAT1-WT and STAT1-mut luciferase reporter vectors. (c) Luciferase activities were detected in HEK-293T or DT40 cells co-transfected with gga-miR-3530-5p and luciferase reporters containing nothing, STAT1, or mutant transcript. Relative firefly luciferase expression was normalized to renilla luciferase. Data are shown as the mean ± SD for three independent experiments. *p < 0.05, ** or ##P < 0.01.
To investigate the interactions between lncRNA53557 and gga-miR-3530-5p, a series of experiments were performed. First, gga-miR-3530-5p mimics and inhibitor were generated and verified by RT-qPCR, and the effect of transfection on cytotoxicity was evaluated by CCK-8 kit. After transfection for 24 h, gga-miR-3530-5p level was significantly decreased in the inhibitor group, whereas it was significantly increased in the mimics group, compared with the control group (Figure 6(c,d)). Moreover, no significant difference in cell activity between-group (Figure 6(e)). Considering that lncRNA could serve as miRNA sponges in the cytoplasm, the binding between lncRNA53557 and gga-miR-3530-5p were validated in the study. The lncRNA53557-WT and lncRNA53557-Wut (without gga-miR-3530-5p binding sites) were subcloned into the luciferase reporter vector pMIR-REPORT (Figure 6(f)). The results indicated that the luciferase activity of the lncRNA53557-WT group was significantly decreased in the gga-miR-3530-5p mimic group (p < 0.01) compared with the NC group in DT40 or HEK-295T cells; however, there was no difference among gga-miR-3530-5p mimics and NC group in the luciferase activity of lncRNA53557-Mut group (Figure 6(g)). Second, the direct association between lncRNA53557 and gga-miR-3530-5p was investigated in this study. A biotin-labelled lncRNA53557 probe and a NC probe were generated, and DT40 cell lysates were incubated with the probes overnight. The specific binding was confirmed by affinity pulldown of endogenic gga-miR-3530-5p and gga-miR-1808 (used as a control). Then, gga-miR-3530-5p and gga-miR-1808 were then extracted and assessed by RT-qPCR. Meanwhile, the lncRNA53557 level was detected by RT-qPCR to evaluate the efficacy of lncRNA53557 pulldown (Figure 6(h)). The result indicated that endogenic gga-miR-3530-5p, other than gga-miR-1808, could be pulled down by biotin-labelled lncRNA53557. These results suggested that a direct interaction might exist between lncRNA53557 and gga-miR-3530-5p. Furthermore, knockdown of lncRNA53557 negatively regulated gga-miR-3530-5p expression (p < 0.01), while the overexpression of lncRNA53557 significantly suppressed gga-miR-3530-5p expression (p < 0.01, Figure 6(i)) compared to the NC group. Simultaneously, the expression of lncRNA53557 and gga-miR-3530-5p was also confirmed by FISH, and the results showed that after vvIBDV infection, lncRNA53557 (red) was significantly upregulated, whereas gga-miR-3530-5p (green) was downregulated, which was in line with the RT-qPCR results (Figure 7).
Figure 7.
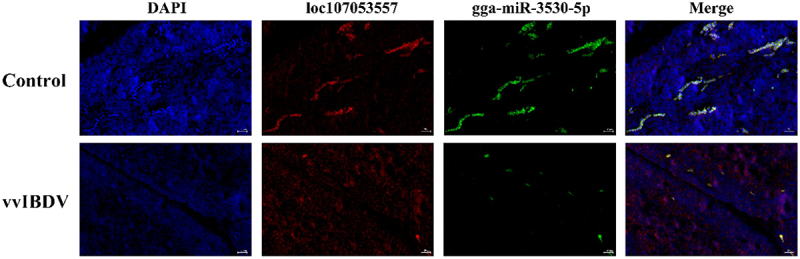
The expression of lncRNA53557 (red) and gga-miR-3530-5p (green) were evaluated by fluorescence in situ hybridization (FISH).
gga-miR-3530-5p inhibited STAT1 expression and promoted vvIBDV replication by targeting STAT1
To further validate the binding between STAT1 and gga-miR-3530-5p, STAT1 (S)-wild type (WT) and STAT1 (S)-mutated type (Mut) were cloned into the pMIR-REPORT plasmids and then co-transfected with gga-miR-3530-5p mimics or NC into DT-40 or HEK-293T cells (Figure 8(b)). The results showed that the activity of the luciferase reporter vector carrying the STAT1 3’UTR-WT sequence was significantly decreased in the gga-miR-3530-5p mimic group (p < 0.01); however, these effects disappeared in mutated binding sites of STAT1 (Figure 8(c)).
Next, the biological function of gga-miR-3530-5p in DT40 cells was explored in the study. NC, gga-miR-3530-5p mimics, NC inhibitor and gga-miR-3530-5p inhibitor groups cells were infected with 102 ELD50/200 μL vvIBDV for 48 h, and cell lysates were harvested to evaluate the expression of STAT1 using RT-qPCR and Western Blotting. The results indicated that compared with the NC group, gga-miR-3530-5p mimics significantly decreased mRNA and protein levels of STAT1, and phospho-STAT1 (p < 0.01, Figure 9(a)). Whereas gga-miR-3530-5p inhibitor markedly increased both the mRNA and protein levels of STAT1, and phospho-STAT1 (p < 0.01, Figure 9(b)) compared to NC inhibitor group. In addition, the effect of gga-miR-3530-5p on vvIBDV replication was measured, and the results demonstrated that gga-miR-3530-5p mimics significantly increased vvIBDV replication in DT40 cells compared to NC (p < 0.01, Figure 9(c)), whereas the gga-miR-3530-5p inhibitor decreased vvIBDV replication (p < 0.01, Figure 9(d)).
Figure 9.
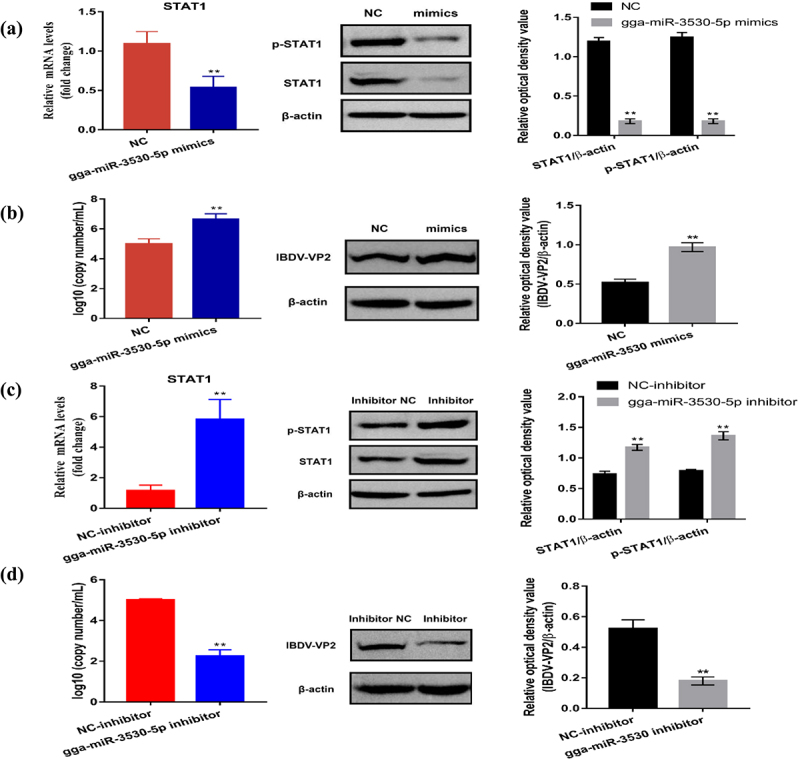
gga-miR-3530-5p inhibited STAT1 expression and promoted vvIBDV replication. (a) Relative mRNA and protein expression of STAT1 were determined by RT-qPCR and Western Blotting in gga-miR-3530-5p overexpressed DT40 cells infected with vvIBDV. (b) Expression of vvIBDV-VP2 gene and protein were determined by RT-qPCR and Western Blotting in gga-miR-3530-5p overexpressed DT40 cells infected with vvIBDV. (c) Relative mRNA and protein expression of STAT1 were determined by RT-qPCR and Western Blotting in lncRNA5357 knocked-down DT40 cells infected with vvIBDV. (d) Expression of vvIBDV-VP2 gene and protein were determined by RT-qPCR and Western Blotting in lncRNA5357 knocked-down DT40 cells infected with vvIBDV. Data are shown as the mean ± SD for three independent experiments. *p < 0.05, ** or ## p < 0.01.
LncRNA53557 acts as a molecular sponge for gga-miR-3530-5p to regulate the STAT1 expression
To explore whether lncRNA53557 regulated STAT1 expression via binding gga-miR-3530-5p, luciferase plasmid pMIR-REPORT, pMIR-STAT1-WT, or pMIR-STAT1-Mut was transfected into cells. A dual luciferase reporter assay showed that overexpression of lncRNA53557 enhanced luciferase activity of pMIR-STAT1-WT. In the rescue experiment, gga-miR-3530-5p mimics abrogated high luciferase activities in lncRNA53557 overexpressed cells (Figure 10(a)).
Figure 10.
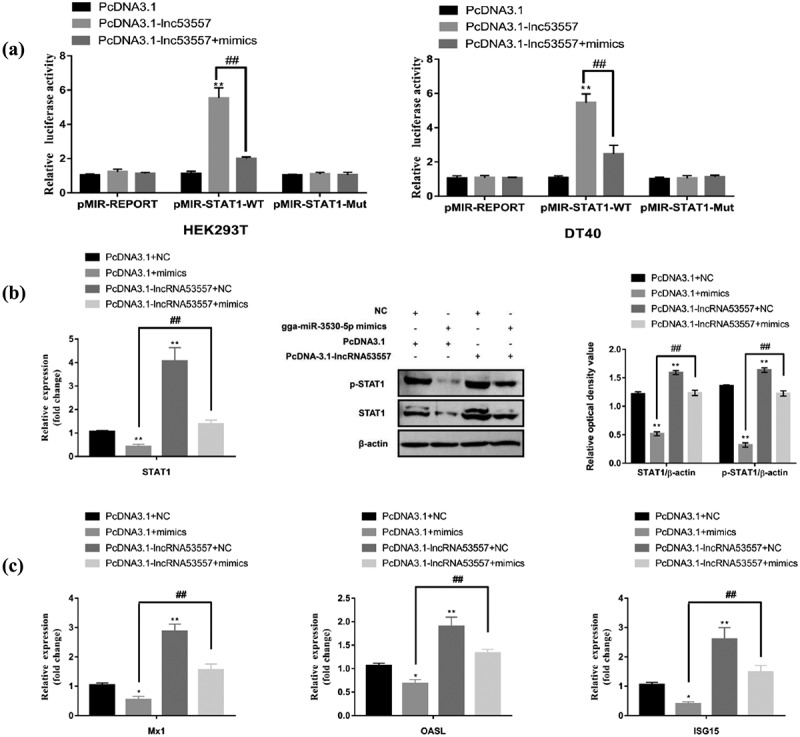
LncRNA53557 serves as a miRNA sponge of gga-miR-3530-5p to regulate STAT1 expression. (a) Luciferase activities were evaluated in HEK-293T or DT40 cells transfected with luciferase reporters containing STAT1 3’ UTR, mut or nothing. Data are presented as the relative ratio of firefly to renilla luciferase activity. (b) Relative mRNA and protein levels of STAT1 were measured by RT-qPCR and Western Blotting in DT40 cells co-transfected with pcDNA3.1 and NC or gga-miR-3530-5p mimics, or co-transfected with pcDNA3.1-lncRNA53557 and NC or gga-miR-3530-5p mimics. (c) Relative mRNA levels of Mx1, OASL, and ISG15 was measured by RT-qPCR and Western Blotting in DT40 cells co-transfected with pcDNA3.1 and NC or gga-miR-3530-5p mimics, or co-transfected with pcDNA3.1-lncRNA53557 and NC or gga-miR-3530-5p mimics. Data are shown as the mean ± SD for three independent experiments. *p < 0.05, **p < 0.01.
Subsequently, we detected such a relationship through RT-qPCR and Western Blotting assays. The pcDNA3.1 empty vector or pcDNA3.1-lncRNA53557 with NC or gga-miR-3530-5p mimics were co-transfected into DT40 cells. After transfection for 48 h, the cells listed above were infected with 102 ELD50/200 μL vvIBDV for 48 h, and cell lysates were harvested to evaluate the expression of STAT1 and ISGs. The results demonstrated that the co-transfection of pcDNA3.1 and gga-miR-3530-5p mimics significantly decreased the mRNA levels of STAT1 (p < 0.05), M×1(p < 0.05), OASL (p < 0.05), and ISG15 (p < 0.05), and the protein levels of STAT1 (p < 0.01) and phospho-STAT1 (p < 0.01) compared to co-transfection of pcDNA3.1 and NC groups (Figure 10(b,c), compare lane 1 to lane 2), whereas co-transfection of pcDNA3.1-lncRNA53557 and NC increased the above (compare lane 1 to lane 3). Interestingly, the co-transfection of pcDNA3.1-lncRNA53557 and gga-miR-3530-5p mimics (the rescue group) reversed low both the mRNA (p < 0.01) and protein (p < 0.01) expression levels of STAT1 and ISGs (compare lanes 2 to 4) compared to co-transfection of pcDNA3.1 and mimics groups. These data convincingly demonstrate that lncRNA53557 was positively associated with STAT1 via competitive binding of gga-miR-3530-5p in DT40 cells, which was called the lncRNA53557/gga-miR-3530-5p/STAT1 axis.
LncRna53557/gga-miR-3530-5p association regulated vvIBDV replication
We further explore whether lncRNA53557 regulated vvIBDV replication via modulating gga-miR-3530-5p. Results indicated that compared to co-transfection of pcDNA3.1 and NC groups, the co-transfection of pcDNA3.1 and gga-miR-3530-5p mimics significantly increased both the mRNA (p < 0.01) and protein (p < 0.01) levels of vvIBDV in the DT40 cells (Figure 11, compare lane 1 to lane 2), whereas co-transfection of pcDNA3.1-lncRNA53557 and NC decreased the above (compare lane 1 to lane 3). Moreover, co-transfection of pcDNA3.1-lncRNA53557 and gga-miR-3530-5p mimics reversed high both the mRNA (p < 0.01) and protein (p < 0.01) levels of vvIBDV (compare lanes 2 to 4) compared to co-transfection of pcDNA3.1 and mimics groups. Accordingly, the results revealed that lncRNA53557 was able to reverse the effect of gga-miR-3530-5p on vvIBDV replication through lncRNA53557/gga-miR-3530-5p interaction.
Figure 11.
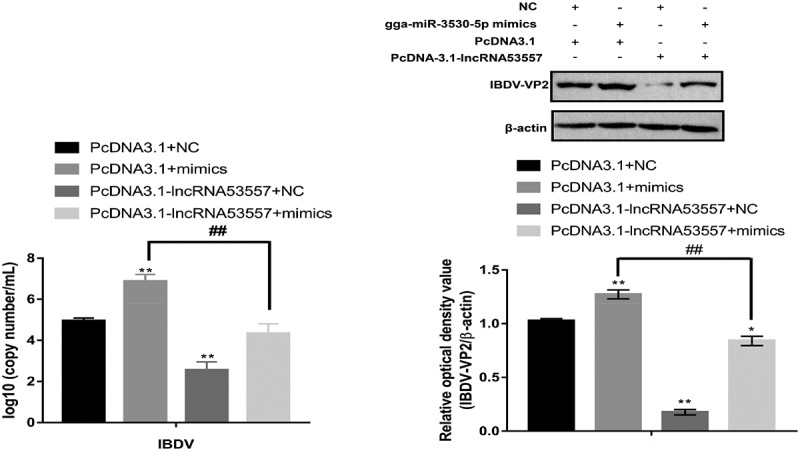
LncRNA53557/gga-miR-3530-5p association regulated vvIBDV expression. Expression of vvIBDV-VP2 gene and protein were determined by RT-qPCR and Western Blotting in DT40 cells co-transfected with pcDNA3.1 and NC or gga-miR-3530-5p mimics, or co-transfected with pcDNA3.1-lncRNA53557 and NC or gga-miR-3530-5p mimics.
Discussion
IBDV infection causes severe damage to the BF in chickens, leading to susceptibility of chickens to other diseases. Recently, lncR NAs and miRNAs have been extensively expressed in IBDV-infected tissues and cells [27,33,36] and determined to play key roles as regulators in various cellular processes. Increasing data has revealed that lncRNAs and miRNAs participate in the regulation of host – pathogen interactions through complex mechanisms. In the present study, the RNA-seq was first used to reveal that vvIBDV-induced the differential expression of lncRNAs and miRNAs in the bursa of Fabricius, and a putative lncRNA-miRNA-mRNA crosstalk network was constructed. Secondly, STAT1 was obviously upregulated in IBDV-infected samples and inhibited vvIBDV replication. Finally, we identified that the lncRNA53557/gga-miR-3530-5p/STAT1 axis is involved in vvIBDV replication (Figure 12).
Figure 12.
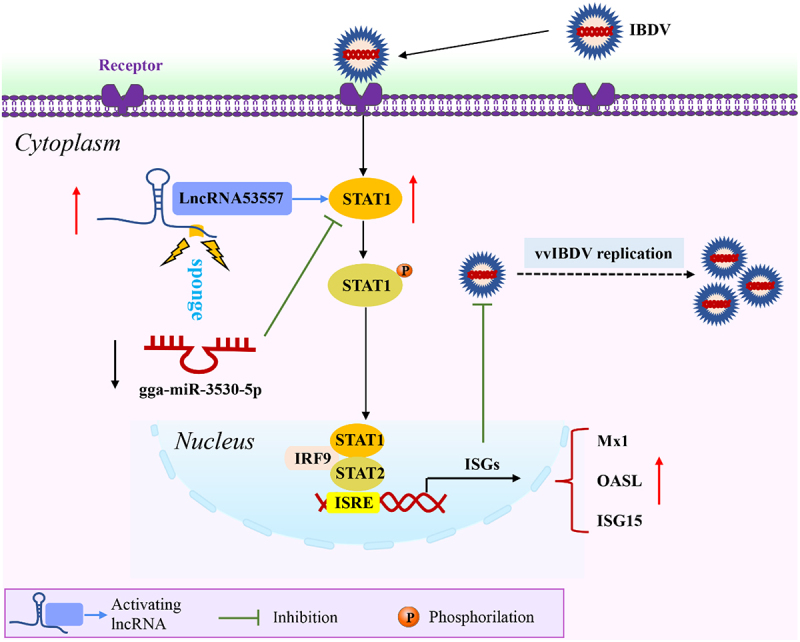
A schematic model of the lncRna53557/gga-miR-3530-5p/STAT1 axis in vvIBDV infection.
In virus-infected cells, lncRNAs and miRNAs encoded by host genes form a regulatory network in the interaction between host and virus. Consequently, the interaction between lncRNAs and miRNAs has provoked great research interest. As many lncRNAs contain MREs, there is increasing evidence to suggest that lncRNAs can adsorb target miRNAs through their own MREs and suppress the targeting mRNAs degradation mediated by miRNAs [46–48]. For example, it has been reported that lncRNA IFITM4P, as a ceRNA, involved in the innate immunity against viral infection through the lncRNA IFITM4P-miR-24-3p-IFITM1/2/3 regulatory network [49]. LncRNA-NRAV, a negative regulator of antiviral response, serves as a ceRNA that promoted respiratory syncytial virus replication through targeting miR-509-3p/Rab5c axis [47]. These studies suggested that lncRNAs plays an important role in viral infection as a miRNA sponge.
Herein, the constructed putative lncRNA-miRNA-mRNA network showed that these aberrantly expressed genes were predominantly enriched in the Jak-STAT signalling pathway. Notably, STAT1 was significantly differentially expressed in the Jak-STAT signalling pathway after IBDV infection. STAT1 is a member of the STAT family and is activated by type I IFN (IFN-I) [50–52]. When activated, STAT1 can induce the transcription of ISGs through a series of signal transduction steps, which includes PKR, ISG15, IFITM3, Mx, and OAS (L), playing critical roles in antiviral responses [53–55]. For example, the STAT1 pathway is crucial for clearing the initial viral load of the dengue virus [56]. However, inhibiting STAT1 phosphorylation antagonizes the IFN antiviral signalling in HRTV and PEDV infection [57,58], and attenuates the immune response to SARS-CoV-2 in moDCs [59]. Recent study revealed STAT1 was also differentially expressed in the IBDV-infected BF and DF-1 cell line [27,60], indicating an important role for STAT1 in IBDV infection. In the present study, knockdown of STAT1 significantly decreased the mRNA levels of Mx1, OASL, and ISG15, and increased IBDV replication, further confirming that STAT1 plays a critical role in anti-IBDV infection.
In the study, it was confirmed that lncRNA53557 strongly regulated STAT1; thus, we focused on lncRNA53557 for functional study. Accumulating researches showed that lncRNAs participate in multiple cellular functions via complicated mechanisms, and its function has been closely linked to the localization in subcellular components. The localization of lncRNA53557 in subcellular components was assessed, and the result showed that both the cytoplasm and nucleus were the component where lncRNA53557 localized, suggested a role of lncRNA53557 in both transcriptional and post-transcriptional regulation. Moreover, functional experiments were carried out in vitro, and results indicated that the overexpression of lncRNA53557 significantly increased the expression of STAT1 and decreased vvIBDV replication; however, opposite results were obtained in the knockdown of lncRNA53557 group, suggesting that lncRNA53557 acts as a positive transcriptional regulator of STAT1.
LncRNAs have been reported to play an active role in the cytoplasm, where they participate in post-transcriptional regulatory processes as miRNA sponges [61]. Based on the results that lncRNA53557 can be located in the cytoplasm, showed the same trend with STAT1, and the lncRNA53557-gga-miR-3530-5p-STAT1 interaction network, we tentatively put forward the idea that lncRNA53557 and STAT1 might share the same miRNA. In the light of bioinformatics analysis, lncRNA53557 was predicted to bind to gga-miR-3530-5p’s seed region. Subsequently, dual luciferase reporter, RNA pulldown, FISH and RT-qPCR assays confirmed that lncRNA53557 were directly bound to gga-miR-3530-5p and had a negative regulatory relationship between them.
There is increasing evidence that miRNAs can coordinate host defence against viral infections by directly targeting viral genomes or triggering the innate immune response [62–64]. Indeed, the role of chicken miRNA in IBDV infection has been extensively studied, and nine miRNAs (gga-miR-21-5p, gga-miR-155, gga-miR-130b-3p, gga-miR-27b-3p, gga-miR-454-3p, gga-miR-2127-3p, gga-miR-16-5p, gga-miR-9-3p, and gga-miR-142-5p) have been found to be involved in IBDV replication [33], among which gga-miR-21-5p, gga-miR-155, gga-miR-130b-3p, gga-miR-27b-3p, and gga-miR-454-3p exert antiviral effects on IBDV infection by targeting the negative regulators or viral genomes. However, gga-miR-2127-3p, gga-miR-16-5p, gga-miR-9-3p, and gga-miR-142-5p promote IBDV replication by targeting antiviral genes of IFN-I-associated pathways. In this study, the results of bioinformatic analysis and dual luciferase reporter assays showed that gga-miR-3530-5p can bind to the 3’UTR region of STAT1. Moreover, gga-miR-3530-5p mimics significantly decreased the expression of STAT1and increased vvIBDV replication. Opposite results were obtained for the gga-miR-3530-5p inhibitor group. These data indicate that gga-miR-3530-5p can affect the replication of vvIBDV by targeting STAT1, which is a critical negative regulator of STAT1.
Subsequently, dual luciferase reporter was performed to verify that STAT1 could be positively influenced by lncRNA53557, and similarly, the influence could be abolished by gga-miR-3530-5p. Moreover, rescue experiments further confirmed that overexpression of lncRNA53557 reversed the inhibitory effect of gga-miR-3530-5p on STAT1. Furthermore, overexpression of gga-miR-3530-5p promoted vvIBDV replication, whereas overexpression of lncRNA53557 achieved the opposite result. These results indicated that lncRNA53557 can act as a sponge for gga-miR-3530-5p to regulate the STAT1 expression and affect vvIBDV replication.
This is the first study to thoroughly investigate the expression, regulation, and function of lncRNA53557, which is also the first study to investigate the relationship between gga-miR-3530-5p and STAT1. These findings may shed light on host anti-vvIBDV infection. However, this study has several limitations. First, the mechanism underlying the interaction of host ncRNAs and viruses is intricate, and the present study only demonstrated that lncRNA53557 can act as a ceRNA to inhibit vvIBDV replication by targeting STAT1 in vitro. Therefore, before lncRNA53557 can be considered as an ideal therapeutic target in vvIBDV infection, further in vivo investigation of lncRNA53557 is needed. In addition, lncRNA53557 can also be localized in the nucleus, suggesting that lncRNA53557 is able to regulate STAT1 in multiple ways. However, whether lncRNA53557 can regulate STAT1 in transcriptional level will further investigation. Second, the present study confirmed that lncRNA53557 binds gga-miR-3530-5p. However, whether other chicken miRNAs were bound to lncRNA53557 and affected IBDV replication was not predicted by bioinformatics analyses and requires further study. Third, it is not clear whether chicken lncRNA53557 is specific to IBDV infection. Therefore, a deeper understanding of the antiviral potential of lncRNA53557 in vvIBDV infection requires further exploration.
Conclusion
LncRNA53557 was upregulated in vvIBDV-infected bursal tissues and DT40 cells, and functions as a positive transcriptional regulator of STAT1. Moreover, lncRNA53557 could exert a crucial regulatory role by blocking the activity of gga-miR-3530-5p in the lncRNA53557/gga-miR-3530-5p/STAT1 axis during vvIBDV infection, thus inhibiting vvIBDV replication, suggesting that the upregulation of lncRNA53557 in vvIBDV infection was part of the host antiviral defence. The results may facilitate improvement in exploring a potential noncoding RNA target for diagnosis and treatment of vvIBDV infection.
Supplementary Material
Acknowledgements
We thank Novel Bioinformatics Ltd., Co for the support of bioinformatics analysis and we also thank GENE DENOVO for the support of uploading the sequencing data.
Funding Statement
This work is supported by the National Key Research and Development Program of China [Grant number 2022YFD1800304] and the Natural Science Foundation of Shandong Province [Grant number ZR2022QC116].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
XWH and XNW designed the experiments. XWH, ZFS, and LW performed the experiment, XWH, YPJ, WC, and XYQ analysed the data. XWH, YJL, HZ, and LJT drafted and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Animal experiments were carried out in accordance with the recommendations in the institutional and national guidelines for animal care and use. The protocol was approved by the Committee on the Ethics of Animal Experiments of Northeast Agricultural University, Harbin, China (2016NEFU–315, 13 April 2017).
Data Availability statement
The raw data presented in this study can be found in the NCBI short reads archive and accession number is PRJNA635782. For information linking please refer to: https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA635782.
Supplemental data
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2024.2333237.
References
- [1].Sharma JM, Kim IJ, Rautenschlein S, et al. Infectious bursal disease virus of chickens: pathogenesis and immunosuppression. Dev Comp Immunol. 2000;24(2–3):223–20. doi: 10.1016/s0145-305x(99)00074-9 [DOI] [PubMed] [Google Scholar]
- [2].Pitcovski J, Gutter B, Gallili G, et al. Development and large-scale use of recombinant VP2 vaccine for the prevention of infectious bursal disease of chickens. Vaccine. 2003;21(32):4736–4743. doi: 10.1016/s0264-410x(03)00525-5 [DOI] [PubMed] [Google Scholar]
- [3].Luque D, Rivas G, Alfonso C, et al. Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc Natl Acad Sci U S A. 2009;106(7):2148–2152. doi: 10.1073/pnas.0808498106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mahgoub HA, Bailey M, Kaiser P.. An overview of infectious bursal disease. Arch Virol. 2012;157(11):2047–2057. doi: 10.1007/s00705-012-1377-9 [DOI] [PubMed] [Google Scholar]
- [5].Vukea PR, Willows-Munro S, Horner RF, et al. Phylogenetic analysis of the polyprotein coding region of an infectious South African bursal disease virus (IBDV) strain. Infect Genet Evol. 2014;21:279–286. doi: 10.1016/j.meegid.2013.11.017 [DOI] [PubMed] [Google Scholar]
- [6].Azad AA, Barrett SA, Fahey KJ. The characterization and molecular cloning of the double-stranded RNA genome of an Australian strain of infectious bursal disease virus. Virology. 1985;143(1):35–44. doi: 10.1016/0042-6822(85)90094-7 [DOI] [PubMed] [Google Scholar]
- [7].von Einem UI, Gorbalenya AE, Schirrmeier H, et al. VP1 of infectious bursal disease virus is an RNA-dependent RNA polymerase. J Gen Virol. 2004;85(Pt 8):2221–2229. doi: 10.1099/vir.0.19772-0 [DOI] [PubMed] [Google Scholar]
- [8].McFerran JB, McNulty MS, McKillop ER, et al. Isolation and serological studies with infectious bursal disease viruses from fowl, turkeys and ducks: demonstration of a second serotype. Avian Pathol. 1980;9(3):395–404. doi: 10.1080/03079458008418423 [DOI] [PubMed] [Google Scholar]
- [9].Ruby T, Whittaker C, Withers DR, et al. Transcriptional profiling reveals a possible role for the timing of the inflammatory response in determining susceptibility to a viral infection. J Virol. 2006;80(18):9207–9216. doi: 10.1128/jvi.00929-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Alkie TN, Rautenschlein S. Infectious bursal disease virus in poultry: current status and future prospects. Vet Med. 2016;7:9–18. doi: 10.2147/vmrr.S68905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu M, Vakharia VN. VP1 protein of infectious bursal disease virus modulates the virulence in vivo. Virology. 2004;330(1):62–73. doi: 10.1016/j.virol.2004.09.009 [DOI] [PubMed] [Google Scholar]
- [12].An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42(5):792–804. doi: 10.1016/j.immuni.2015.05.004 [DOI] [PubMed] [Google Scholar]
- [14].Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35(9):408–419. doi: 10.1016/j.it.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brown CJ, Hendrich BD, Rupert JL, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m [DOI] [PubMed] [Google Scholar]
- [16].Swiezewski S, Liu F, Magusin A, et al. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462(7274):799–802. doi: 10.1038/nature08618 [DOI] [PubMed] [Google Scholar]
- [17].Reeves MB, Davies AA, BP M, et al. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science. 2007;316(5829):1345–1348. doi: 10.1126/science.1142984 [DOI] [PubMed] [Google Scholar]
- [18].Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22(1):1–5. doi: 10.1016/j.tig.2005.10.003 [DOI] [PubMed] [Google Scholar]
- [19].Wang J, Zhang J, Zheng H, et al. Mouse transcriptome: neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004;431(7010):1. d oi: [PubMed] [Google Scholar]
- [20].Akhade VS, Pal D, Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv Exp Med Biol. 2017;1008:47–74. doi: 10.1007/978-981-10-5203-3_2 [DOI] [PubMed] [Google Scholar]
- [21].Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22(4):366–376. doi: 10.1016/j.semcdb.2011.01.001 [DOI] [PubMed] [Google Scholar]
- [22].Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- [23].Ouyang J, Zhu X, Chen Y, et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014;16(5):616–626. doi: 10.1016/j.chom.2014.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nishitsuji H, Ujino S, Yoshio S, et al. Long noncoding RNA #32 contributes to antiviral responses by controlling interferon-stimulated gene expression. Proc Natl Acad Sci U S A. 2016;113(37):10388–10393. doi: 10.1073/pnas.1525022113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Carnero E, Barriocanal M, Segura V, et al. Type I interferon regulates the expression of long non-coding RNAs. Front Immunol. 2014;5:548. doi: 10.3389/fimmu.2014.00548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kambara H, Niazi F, Kostadinova L, et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 2014;42(16):10668–10680. doi: 10.1093/nar/gku713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Huang X, Xu Y, Lin Q, et al. Determination of antiviral action of long non-coding RNA loc107051710 during infectious bursal disease virus infection due to enhancement of interferon production. Virulence. 2020;11(1):68–79. doi: 10.1080/21505594.2019.1707957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lee Y, Ahn C, Han J, et al. The nuclear RNase III drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- [29].Ohtani K, Dimmeler S. Control of cardiovascular differentiation by microRnas. Basic Res Cardiol. 2011;106(1):5–11. doi: 10.1007/s00395-010-0139-7 [DOI] [PubMed] [Google Scholar]
- [30].O’Connell RM, Taganov KD, Boldin MP, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104(5):1604–1609. doi: 10.1073/pnas.0610731104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by Adenosines, indicates that thousands of human genes are MicroRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- [32].Trobaugh DW, Klimstra WB. MicroRNA Regulation of RNA virus replication and pathogenesis. Trends Mol Med. 2017;23(1):80–93. doi: 10.1016/j.molmed.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li J, Zheng SJ. Role of MicroRNAs in host defense against infectious bursal disease virus (IBDV) infection: a hidden front line. Viruses. 2020;12(5):543. doi: 10.3390/v12050543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang K, Liu F, Zhou LY, et al. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014;114(9):1377–1388. doi: 10.1161/circresaha.114.302476 [DOI] [PubMed] [Google Scholar]
- [36].Huang X, Zhang J, Liu Z, et al. Genome-wide analysis of differentially expressed mRnas, lncRnas, and circRnas in chicken bursae of Fabricius during infection with very virulent infectious bursal disease virus. BMC Genomics. 2020;21(1):724. doi: 10.1186/s12864-020-07129-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Huang X, Li Y, Wang X, et al. Genome-wide identification of chicken bursae of Fabricius miRnas in response to very virulent infectious bursal disease virus. Arch Virol. 2022;167(9):1855–1864. doi: 10.1007/s00705-022-05496-6 [DOI] [PubMed] [Google Scholar]
- [38].Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Huang X, Liu W, Zhang J, et al. Very virulent infectious bursal disease virus-induced immune injury is involved in inflammation, apoptosis, and inflammatory cytokines imbalance in the bursa of fabricius. Dev Comp Immunol. 2021;114:103839. doi: 10.1016/j.dci.2020.103839 [DOI] [PubMed] [Google Scholar]
- [40].Wang L, Park HJ, Dasari S, et al. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41(6):e74. doi: 10.1093/nar/gkt006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kang YJ, Yang DC, Kong L, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45(W1):W12–w6. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Stark GR, Darnell JE Jr.. The JAK-STAT pathway at twenty. Immunity. 2012;36(4):503–514. doi: 10.1016/j.immuni.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Schindler C, Plumlee C. Inteferons pen the JAK-STAT pathway. Semin Cell Dev Biol. 2008;19(4):311–318. doi: 10.1016/j.semcdb.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41(9):761–772. doi: 10.1016/j.tibs.2016.07.003 [DOI] [PubMed] [Google Scholar]
- [45].Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cheng Q, Wang L. LncRNA XIST serves as a ceRNA to regulate the expression of ASF1A, BRWD1M, and PFKFB2 in kidney transplant acute kidney injury via sponging hsa-miR-212-3p and hsa-miR-122-5p. Cell Cycle. 2020;19(3):290–299. doi: 10.1080/15384101.2019.1707454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li J, Li M, Wang X, et al. Long noncoding RNA NRAV promotes respiratory syncytial virus replication by targeting the MicroRNA miR-509-3p/Rab5c axis to regulate vesicle transportation. J Virol. 2020;94(10). doi: 10.1128/jvi.00113-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi: 10.1016/j.ccr.2014.03.010 [DOI] [PubMed] [Google Scholar]
- [49].Xiao M, Chen Y, Wang S, et al. Long noncoding RNA IFITM4P regulates Host antiviral responses by acting as a competing endogenous RNA. J Virol. 2021;95(21):e0027721. doi: 10.1128/jvi.00277-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hertzog PJ, Williams BR. Fine tuning type I interferon responses. Cytokine Growth Factor Rev. 2013;24(3):217–225. doi: 10.1016/j.cytogfr.2013.04.002 [DOI] [PubMed] [Google Scholar]
- [51].Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36–49. doi: 10.1038/nri3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32(1):513–545. doi: 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(Pt 1):1–47. doi: 10.1099/vir.0.83391-0 [DOI] [PubMed] [Google Scholar]
- [54].Verhelst J, Hulpiau P, Saelens X. Mx proteins: antiviral gatekeepers that restrain the uninvited. Microbiol Mol Biol Rev. 2013;77(4):551–566. doi: 10.1128/mmbr.00024-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ghosh A, Shao L, Sampath P, et al. Oligoadenylate-synthetase-family protein OASL inhibits activity of the DNA sensor cGAS during DNA virus infection to limit interferon production. Immunity. 2019;50(1):51–63.e5. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Shresta S, Sharar KL, Prigozhin DM, et al. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J Immunol. 2005;175(6):3946–3954. doi: 10.4049/jimmunol.175.6.3946 [DOI] [PubMed] [Google Scholar]
- [57].Feng K, Deng F, Hu Z, et al. Heartland virus antagonizes type I and III interferon antiviral signaling by inhibiting phosphorylation and nuclear translocation of STAT2 and STAT1. J Biol Chem. 2019;294(24):9503–9517. doi: 10.1074/jbc.RA118.006563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Guo L, Luo X, Li R, et al. Porcine epidemic diarrhea virus infection inhibits interferon signaling by targeted degradation of STAT1. J Virol. 2016;90(18):8281–8292. doi: 10.1128/jvi.01091-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yang D, Chu H, Hou Y, et al. Attenuated interferon and proinflammatory response in SARS-CoV-2-Infected Human Dendritic Cells Is Associated with Viral Antagonism of STAT1 Phosphorylation. J Infect Dis. 2020;222(5):734–745. d oi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Mohd Isa F, Al-Haj N A, Mat Isa N, et al. Differential expression of immune-related genes in the bursa of Fabricius of two inbred chicken lines following infection with very virulent infectious bursal disease virus. Comp Immunol Microbiol Infect Dis. 2020;68:101399. doi: 10.1016/j.cimid.2019.101399 [DOI] [PubMed] [Google Scholar]
- [61].Wang L, Cho KB, Li Y, et al. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 2019;20(22). doi: 10.3390/ijms20225758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Thounaojam MC, Kundu K, Kaushik DK, et al. MicroRNA 155 regulates Japanese encephalitis virus-induced inflammatory response by targeting src homology 2-containing inositol phosphatase 1. J Virol. 2014;88(9):4798–4810. doi: 10.1128/jvi.02979-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Guo XK, Zhang Q, Gao L, et al. Increasing expression of microRNA 181 inhibits porcine reproductive and respiratory syndrome virus replication and has implications for controlling virus infection. J Virol. 2013;87(2):1159–1171. doi: 10.1128/jvi.02386-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ren Z, Ambros VR. Caenorhabditis elegans microRnas of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc Natl Acad Sci U S A. 2015;112(18):E2366–75. doi: 10.1073/pnas.1422858112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data presented in this study can be found in the NCBI short reads archive and accession number is PRJNA635782. For information linking please refer to: https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA635782.


