Summary
Objective.
Administrative codes to identify people with rare epilepsies in electronic health records are limited. The current study evaluated the use of keyword search as an alternative method for rare epilepsy cohort creation using electronic health records data.
Methods.
Data included clinical notes from encounters with ICD-9 codes for seizures, epilepsy, and/or convulsions during 2010–2014 across six healthcare systems in New York City. We identified cases with rare epilepsies by searching clinical notes for keywords associated with 33 rare epilepsies. We validated cases via manual chart review. We compared performance of keyword search to manual chart review using positive predictive value (PPV), sensitivity, and F-score. We selected the optimal combinations of keywords with the highest F-scores.
Results.
Data included clinical notes from 77,924 cases with ICD-9 codes for seizures, epilepsy, and/or convulsions. The all-keyword search method identified 6,095 candidates, and manual chart review confirmed that 2,068 (34%) had a rare epilepsy. The optimal keyword combination search method identified 1,862 cases with a rare epilepsy, and this method performed as follows: PPV median = 0.64 (interquartile range, IQR = 0.50–0.81, range = 0.20–1.00), sensitivity median = 0.93 (IQR = 0.76–1.00, range = 0.10–1.00), and F-score median = 0.71 (IQR = 0.63–0.85, range = 0.18–1.00). Using the optimal keyword combination method, we identified four cohorts of rare epilepsies with over 100 individuals, including infantile spasms, Lennox-Gastaut syndrome, Rett syndrome, and tuberous sclerosis complex. We identified over 50 individuals with two rare epilepsies that do not have specific ICD-10 codes for cohort creation (epilepsy with myoclonic atonic seizures, Sturge Weber syndrome).
Significance.
Keyword search is an effective method for cohort creation. These findings can improve identification and surveillance of individuals with rare epilepsies and promote their referral to specialty clinics, clinical research, and support groups.
Keywords: natural language processing, cohort creation, genetic epilepsy, clinical data, automated, observational
INTRODUCTION
Individuals with rare epilepsies are a medically complex and vulnerable population.1 Current knowledge of many rare epilepsies is limited to case reports and case series, in part due to the challenge of finding cases for review. There are opportunities to use electronic health record (EHR) data for cohort creation; however, most rare epilepsies have no specific administrative billing codes. For example, there are no ICD codes specific for Aicardi syndrome, epilepsy with myoclonic-atonic seizures (EMAS) and early infantile developmental and epileptic encephalopathy. Without these codes, researchers depend on manual chart review to identify cases, which is time intensive. In addition, ICD codes are used primarily for billing purposes and can have variable success in epilepsy cohort creation.2 More sophisticated tools are needed to extract information from comprehensive clinical records.
One solution is to use natural language processing (NLP), the computational analysis of text, to search EHR data. An advantage of NLP is that it can analyze narrative clinical text written by clinicians aiming to document and communicate diagnostic patient information in the EHR. NLP has been used previously in epilepsy cohort creation, including in studies evaluating risk factors for sudden unexpected death in epilepsy,3 candidates for epilepsy surgery,4 possible first-time febrile seizures,5 non-epileptic seizures,6 and other epilepsy characteristics.7
The chief disadvantage of NLP is that it can require several complex steps. NLP algorithms often require substantial preprocessing, which can involve cleaning text, segmenting words/sentences, and tagging parts of speech. Further analysis is often done to improve specificity with negation detection (e.g., “no history of seizures”) and name entity recognition (“cousin with Lennox-Gastaut syndrome”). This may be followed by further classification with machine learning.4 This level of complexity limits dissemination.
An alternative is to use “regular expressions”, an NLP building block, that perform text search without the full machinery of more complex algorithms.5 They can be easily shared with other researchers—for example, they are simple enough to send in the body of an email. An example of a regular expression that searches text for seizure descriptors is “(whole|full) body[a-z]{0,80} (shaking|conv[ul]{1,2}s)”. This example matches multiple combinations of words (e.g., whole body shaking, whole body convulsive, full body convulsions), accounts for spelling errors, and matches word pairs separated by 80 characters (e.g., patient had a whole body seizure with stiffening of arms and legs followed by rhythmic shaking).3
The purpose of this study is to develop regular expressions that search clinical text and thereby create cohorts of individuals with rare epilepsies. We identified a broad list of keywords associated with rare epilepsies8 and validated the highest-performing word combinations. We provide regular expressions for others studying rare epilepsies.
METHODS
Data source.
We used the New York City (NYC)-Clinical Data Research Network (CDRN), which includes data from six large academic medical centers in NYC: Weill Cornell Medicine, Columbia Irving University Medical Center (CUMC), Mount Sinai Health System (MSHS), Continuum Hospitals (now merged with Mount Sinai), Montefiore Medical Center, and New York University Langone Medical Center. The NYC-CDRN contains inpatient and outpatient EHR data from 12 million individuals from diverse racial and ethnic backgrounds (41% white, 15% black, and 44% other; 8% Hispanic).9 Each medical center queried the NYC-CDRN for all physician text notes of pediatric and adult patients (any person and any age) with medical encounters for seizures, epilepsy, and/or convulsions (ICD-9 345.x epilepsy or ICD-9 780.39 convulsion) from 2010 to 2014. Clinical data included records from each of the six urban academic hospitals and the affiliated physicians/hospitals within their hospital systems. Data from public city hospitals in NYC were included under the academic hospital system umbrella (e.g., Harlem Hospital – CUMC, Bellevue Hospital – NYU, Metropolitan Hospital – MSHS). Physicians documenting rare epilepsy terms in narrative text were primarily pediatric and adult neurologists. The study was approved by the IRBs at Weill Cornell Medical College, Columbia Vagelos College of Physicians and Surgeons, Mount Sinai Medical System, New York University Medical Center, and Montefiore Medical Center, and facilitated by a central IRB protocol (Biomedical Research Alliance of New York). Data were analyzed using R software.
Selecting keywords.
We used a published list of keywords associated with rare epilepsies.8 These terms were generated using results from national surveys, manual review of online resources, manual review of medical vocabularies, correspondence with rare epilepsy-related national community service networks or advocacy working groups, and independent clinician review.
Inclusion criteria.
Cases with at least one keyword associated with a rare epilepsy in clinical notes were included in the study. We electronically searched the text of clinical notes and counted notes containing each keyword. We removed proposed keywords if they had zero matches since they did not help with identifying cases. We also removed words if they were unlikely to be meaningfully significant (i.e., high number of matches in clinical notes or used to describe multiple diagnoses). Investigators added additional words to create a more refined list.
Preparing text for manual review.
We created Hypertext Markup Language (HTML) files for each reviewer to facilitate rapid manual chart review (Figure 1). For each patient, text from the entire clinical note was in each HTML file. We extracted snippets of text within 70 characters before and after keywords from clinical notes and presented in the HTML files. Some words were highlighted in blue for better visualization during manual review, including keywords related to rare epilepsies, seizure related terms (seiz, epilep, infantile spasms, myoclon, convuls, spell, episode, status epilepticus, tonic, clonic, GTC, grand mal, regression), and note headings (assessment, impression, plan, index, findings).
Figure 1.

Excerpt of HTML file created to facilitate rapid manual chart review.
Manual review.
Two people (NB, co-author; and DH, contributor) independently reviewed HTML files to determine if individuals had a rare epilepsy (Yes, No). When clinical notes documented the diagnosis (e.g., “4yo girl with tuberous sclerosis”), it was considered a rare epilepsy diagnosis (Yes). When a reviewer was uncertain, it was considered a No. Disagreements were resolved by a third reviewer (ZG, co-author). HTML file creation failed for a small subset of cases (371 of 6,095) due to an error copying a block of text over a section of subject IDs in our HTML file case list, and therefore these were not included in the first round of chart review. KB (co-author) and ZG performed a second round of chart review for these individuals. Disagreements were resolved by consensus review with KB and ZG.
We considered cases possible Lennox-Gastaut syndrome (LGS) when clinical notes documented slow spike and waves on electroencephalogram (EEG) and/or multiple seizure types. These cases were re-reviewed for strict diagnostic criteria and a diagnosis of LGS was confirmed when individuals had (1) multiple types of seizures characteristic of LGS (e.g., tonic, atonic, generalized tonic clonic, absence), (2) developmental delay or intellectual disability, and (3) slow spike and waves ≤ 2.5 Hz on the EEG report.
Performance of search tool: The all-keyword method.
Individuals were considered by the NLP search tool as having a rare epilepsy if they had any of the associated rare epilepsy keywords. To evaluate performance, we compared this finding to the finding achieved using manual chart review, which is considered the gold standard method. We counted the number of true positive (TP) and false positive (FP) cases for each rare epilepsy, and measured the positive predictive value (PPV), PPV = TP / (TP + FP).
Performance of search tool: The optimal combination method.
For each rare epilepsy, we evaluated every combination of keywords and selected the highest-performing keyword combinations as measured by the F-score, a measure of accuracy. For example, a rare epilepsy with three keywords (A, B, and C) had seven possible combinations: A or B or C; A or B; A or C; B or C; A; B; C. We measured the number of TPs and FPs for each combination. We estimated false negative values (eFN) by comparing the number of individuals missed using each combination vs. all keyword method (eFN = TPall – TPcombo). Performance was evaluated with PPV, sensitivity, and F-score, compared to the gold standard of manual chart review. Equations were: PPV = TP / (TP + FP), sensitivity = TP / (TP + eFN), and F-score = 2 × (PPV × sensitivity) / (PPV + sensitivity).
Final regular expressions.
The optimal combination method used keywords selected for the highest F-score measure. However, the best statistical combination can be inferior to clinical judgement, particularly when sample sizes are small. Therefore, we adopted a final, more comprehensive keyword list that includes the optimal statistical combination of keywords plus additional clinically important words. We created regular expressions using this list of keywords. When combinations of keywords had equal performance, we selected the combination with the most complete list of keywords to maximize sensitivity. For each rare epilepsy, we also note if a specific ICD-10 code is available for case ascertainment.
RESULTS
Data Sample.
Data included clinical text from 77,924 cases with administrative codes for seizures, epilepsy, and/or convulsions.
All-Keyword Method
Selecting keywords.
Following a search of clinical notes for the presence of 898 terms associated with rare epilepsies (Table S1), including 834 terms from a published list8 and 64 words added by authors, some terms (e.g., “Rett Disease”) were not found in clinical notes and removed. Some terms were nonspecific and removed. For example, “aphasia” and “regression” for epileptic encephalopathy with spike-and-wave activation in sleep (EE-SWAS) and/or electrical status epilepticus in sleep (ESES) and the abbreviation “AS” was nonspecific for Aicardi syndrome because it erroneously identified the word “as”. The term “Fragile X” frequently matched patients undergoing screening tests for developmental delay and therefore had a high number of erroneous matches; we removed it. After removing these terms, our final list included 226 terms associated with 33 rare epilepsies, and we used these for further analysis (Table S2). We identified 6,095 individuals with at least one of the 226 rare epilepsy keywords in clinical notes.
Manual review.
Among 6,095 patients who had a rare epilepsy keyword in the clinical notes, 5,724 underwent first review, and 371 underwent the second review due to HTML file creation error (Figure 2). The final cohort of 6,095 individuals included 2,068 with a rare epilepsy and 4,027 without a rare epilepsy.
Figure 2.
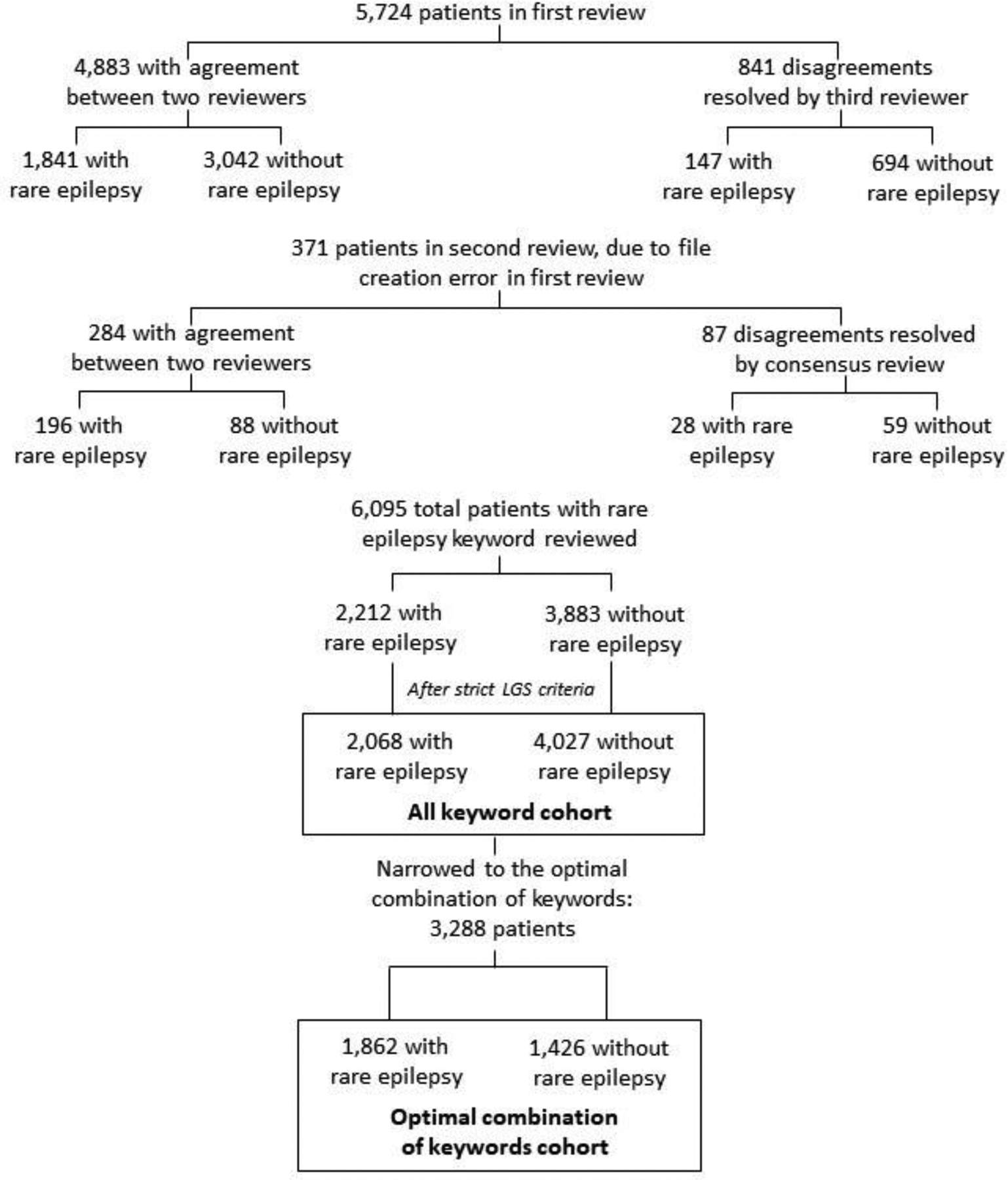
Identifying rare epilepsy cases using keyword search in electronic health record data, followed by manual chart review to validate cases.
Performance of search tool.
We reported the number of TPs, FPs, and PPVs for each rare epilepsy using the all-keyword method (Table 1). These terms identified relatively sizable cohorts of TPs for several rare epilepsies, including 10 rare epilepsies with ≥ 50 individuals: Angelman syndrome, epilepsy with myoclonic atonic seizures (EMAS), Dravet syndrome, infantile spasms, EE-SWAS and/or ESES, LGS, neuronal ceroid lipofuscinosis, Rett syndrome, Sturge-Weber syndrome, and tuberous sclerosis complex. However, six rare epilepsies had five or fewer individuals (Alpers disease, Fragile X syndrome, myoclonic epilepsy with ragged red fibers, PCDH19, ring chromosome 14, and SCN8A). Three rare epilepsies (SLC13a5, SYNGAP, Unverricht-Lundborg disease) had no cases; we removed them from further analysis. We also removed Fragile X syndrome from further analysis because we did not identify cases as expected using the search tool (only identified one case). The all-keyword method had PPVs with median = 0.31, IQR = 0.14 – 0.44, and range = 0.03 – 0.81 (Table 1).
TABLE 1.
Performance of all-keyword search method compared to manual chart review
| Rare Epilepsy | TP | FP | Total | PPV |
|---|---|---|---|---|
| Aicardi syndrome | 41 | 249 | 290 | 0.14 |
| Alpers disease | 5 | 11 | 16 | 0.31 |
| Angelman syndrome | 52 | 76 | 128 | 0.41 |
| CDKL5 | 28 | 49 | 77 | 0.36 |
| Dravet syndrome | 89 | 115 | 204 | 0.44 |
| Dup15q syndrome | 13 | 11 | 24 | 0.54 |
| Early infantile developmental and epileptic encephalopathy | 25 | 80 | 105 | 0.24 |
| EE-SWAS/ESES | 68 | 454 | 522 | 0.13 |
| EMAS | 107 | 442 | 549 | 0.19 |
| Epilepsy in infancy with migrating focal seizures | 6 | 6 | 12 | 0.50 |
| Fragile X syndrome | 1 | 13 | 14 | - |
| Glut1 deficiency | 14 | 67 | 81 | 0.17 |
| Holoprosencephaly | 10 | 280 | 290 | 0.03 |
| Hypothalamic hamartoma with seizures | 40 | 43 | 83 | 0.48 |
| Infantile spasms | 434 | 700 | 1134 | 0.38 |
| KCNQ2 related epilepsy | 19 | 45 | 64 | 0.30 |
| Lennox-Gastaut syndrome | 511 | 265 | 776 | 0.66 |
| Myoclonic epilepsy with ragged red fibers | 1 | 23 | 24 | 0.04 |
| Neuronal ceroid lipofuscinosis | 56 | 205 | 261 | 0.21 |
| PCDH19 | 3 | 13 | 16 | 0.19 |
| Phelan-McDermid syndrome | 8 | 17 | 25 | 0.32 |
| Prader Willi syndrome | 15 | 102 | 117 | 0.13 |
| Rasmussen syndrome | 25 | 31 | 56 | 0.45 |
| Rett syndrome | 180 | 349 | 529 | 0.34 |
| Ring Chromosome 14 | 2 | 64 | 66 | 0.03 |
| Ring Chromosome 20 | 6 | 131 | 137 | 0.04 |
| SCN2A | 9 | 12 | 21 | 0.43 |
| SCN8A | 1 | 13 | 14 | 0.07 |
| SLC13a5 | 0 | 0 | 0 | - |
| Sturge-Weber syndrome | 99 | 66 | 165 | 0.60 |
| SYNGAP | 0 | 1 | 1 | - |
| Tuberous sclerosis complex | 200 | 48 | 248 | 0.81 |
| Unverricht-Lundborg Disease | 0 | 46 | 46 | - |
| Total | 2,068 | 4,027 | 6,095 | - |
Abbreviations: true positive (TP), false positive (FP), positive predictive value (PPV), inter-quartile range (IQR), epileptic encephalopathy with spike-and-wave activation in sleep (EE-SWAS) and/or electrical status epilepticus in sleep (ESES), epilepsy with myoclonic atonic seizures (EMAS).
Optimal Combination of Keywords Method
Selecting keywords.
After narrowing our keyword list to the optimal statistical combination of words, our list included 102 terms associated with 29 rare epilepsies (Table 2). We identified 3,288 individuals with at least one of these 102 rare epilepsy keywords in clinical notes.
TABLE 2.
Recommended keyword list includes the optimal statistical combination of keywords plus additional clinically important words in italics
| Rare Epilepsy | Specific ICD-10 code available | Recommended keyword list |
|---|---|---|
| Aicardi syndrome | No | Aicardi’s syndrome, Aicardi, Aicardi’s, retinal lacunae |
| Alpers disease | Yes | Alpers Disease, Alpers syndrome, Alpers-Huttenlocher, Alpers-Huttenlocher syndrome |
| Angelman syndrome | Yes | Angelman’s Syndrome, 15q11, Angelman syndrome |
| CDKL5 | Yes | x-linked infantile spasm, CDKL5 |
| Dravet syndrome | Yes | GABRD, severe myoclonic epilepsy of infancy, SMEB, Dravet |
| Dup15q syndrome | No | idic 15, dup15q syndrome, dup15q, 15q11 duplication, 15q11 microduplication, duplication of 15q, duplication 15q, 15q11.2 |
| Early infantile developmental and epileptic encephalopathy | No | Ohtahara, early infantile developmental epileptic encephalopathy, EIDEE, early infantile epileptic encephalopathy, EIEE |
| EE-SWAS/ESES | Yes | electrographic status epilepticus in sleep, electrical status epilepticus of sleep, eses index, continuous slow spike and wave of sleep, spike-index, acquired epilep, continuous spike-wave in sleep, electrographic status epilepticus of sleep, epileptic aphasia, ESES with language regression, LK syndrome, LKS, epileptic encephalopathy with spike-and-wave activation in sleep, epileptic encephalopathy with spike and wave activation in sleep, EE-SWAS, EESWAS |
| EMAS | No | Doose, myoclonic-astatic epilepsy, myoclonic atonic epilepsy, Doose syndrome, myoclonic astatic, EMAS, epilepsy with myoclonic atonic seizures |
| Epilepsy in infancy with migrating focal seizures | No | KCNT1, malignant migrating partial seizures in infancy, migrating partial epilepsy of infancy, MMPSI, migrating partial, epilepsy in infancy with migrating focal seizures, EIMFS |
| Glut1 deficiency | Yes | dystonia 9, glucose transporter type 1 deficiency, glut-1 deficiency syndrome, SLC2A1, SLC2A1 mutation, glut1, glut 1 |
| Holoprosencephaly | Yes | semilobar, holoprosencephaly |
| Hypothalamic hamartoma with seizures | No | hypothalamic hamartoma, gelastic epilepsy, gelastic seizures |
| Infantile Spasms | Yes | jacknife, hypsarrhythmia, hyps, infantile spasm |
| KCNQ2 related epilepsy | No | fifth day fits, familial neonatal seizures, early infantile epileptic encephalopathy, benign neonatal epilepsy, benign familial neonatal seizures, KCNQ2 |
| Lennox-Gastaut syndrome | Yes | lennox syndrome, lennox gastaut, slow spike-wave, slow spike and wave |
| Myoclonic epilepsy with ragged red fibers | Yes | MERRF syndrome, MERRF |
| Neuronal ceroid lipofuscinosis | Yes | Batten’s disease, batten disease, LINCL, NCL |
| PCDH19 | No | EFMR, PCDH19 |
| Phelan-McDermid syndrome | No | Phelan-McDermid Syndrome, 22q13 deletion, Phelan-McDermid, 22q13 |
| Prader Willi syndrome | Yes | Prader Willi Syndrome, Prader Willi |
| Rasmussen syndrome | No | Rasmussen’s encephalitis, Rasmussen’s syndrome, Rasmussen |
| Rett syndrome | Yes | Rett’s disease, Rett’s, Rett syndrome |
| Ring Chromosome 14 | No | ring 14, ring chromosome 14 |
| Ring Chromosome 20 | No | ring chromosome 20 syndrome, ring 20, ring chromosome 20 |
| SCN2A | No | SCN2A, SCN2A mutations |
| SCN8A | No | SCN8A |
| Sturge-Weber syndrome | No | Sturge Weber Syndrome, Sturge-Weber Syndrome, Sturge Weber, Sturge-Weber |
| Tuberous sclerosis complex | Yes | TSC, tuberous sclerosis complex, TSC1, TSC2, multifocal micronodular pneumocyte hyperplasia, radial migration lines, tuberous sclerosis |
Abbreviations: epileptic encephalopathy with spike-and-wave activation in sleep (EE-SWAS) and/or electrical status epilepticus in sleep (ESES), epilepsy with myoclonic atonic seizures (EMAS).
Manual review.
The cohort of 3,288 individuals included 1,862 with a rare epilepsy and 1,426 without a rare epilepsy.
Performance of search tool.
The optimal combination of keywords method had acceptable performance estimates as measured by TP, FP, eFN, PPV, sensitivity, and F-score (Table 3). We observed PPVs with median = 0.64, IQR = 0.50–0.81, and range = 0.20–1.00; sensitivities with median = 0.93, IQR = 0.76–1.00, and range = 0.10–1.00; and F-scores with median = 0.71, IQR = 0.63–0.85, and range = 0.18–1.00.
TABLE 3.
Performance of keyword search using the optimal combination of words compared to manual chart review
| Rare Epilepsy | TP | FP | Total | eFN | PPV | Sens | F-score |
|---|---|---|---|---|---|---|---|
| Aicardi syndrome | 31 | 17 | 48 | 10 | 0.65 | 0.76 | 0.70 |
| Alpers disease | 5 | 3 | 8 | 0 | 0.63 | 1.00 | 0.77 |
| Angelman syndrome | 34 | 21 | 55 | 18 | 0.62 | 0.65 | 0.64 |
| CDKL5 | 24 | 33 | 57 | 4 | 0.42 | 0.86 | 0.56 |
| Dravet syndrome | 85 | 47 | 132 | 4 | 0.64 | 0.96 | 0.77 |
| Dup15q syndrome | 13 | 10 | 23 | 0 | 0.57 | 1.00 | 0.72 |
| Early infantile developmental and epileptic encephalopathy | 22 | 5 | 27 | 3 | 0.81 | 0.88 | 0.85 |
| EE-SWAS/ESES | 50 | 125 | 175 | 18 | 0.29 | 0.74 | 0.41 |
| EMAS | 73 | 24 | 97 | 34 | 0.75 | 0.68 | 0.72 |
| Epilepsy in infancy with migrating focal seizures | 5 | 1 | 6 | 1 | 0.83 | 0.83 | 0.83 |
| Glut1 deficiency | 9 | 10 | 19 | 5 | 0.47 | 0.64 | 0.55 |
| Holoprosencephaly | 1 | 0 | 1 | 9 | 1.00 | 0.10 | 0.18 |
| Hypothalamic hamartoma with seizures | 39 | 39 | 78 | 1 | 0.50 | 0.98 | 0.66 |
| Infantile spasms | 434 | 688 | 1122 | 0 | 0.39 | 1.00 | 0.56 |
| KCNQ2 related epilepsy | 18 | 14 | 32 | 1 | 0.56 | 0.95 | 0.71 |
| Lennox-Gastaut syndrome | 477 | 159 | 636 | 34 | 0.75 | 0.93 | 0.83 |
| Myoclonic epilepsy with ragged red fibers | 1 | 0 | 1 | 0 | 1.00 | 1.00 | 1.00 |
| Neuronal ceroid lipofuscinosis | 44 | 2 | 46 | 12 | 0.96 | 0.79 | 0.86 |
| PCDH19 | 2 | 1 | 3 | 1 | 0.67 | 0.67 | 0.67 |
| Phelan-McDermid syndrome | 8 | 0 | 8 | 0 | 1.00 | 1.00 | 1.00 |
| Prader Willi syndrome | 9 | 36 | 45 | 6 | 0.20 | 0.60 | 0.30 |
| Rasmussen syndrome | 19 | 12 | 31 | 6 | 0.61 | 0.76 | 0.68 |
| Rett syndrome | 144 | 82 | 226 | 36 | 0.64 | 0.80 | 0.71 |
| Ring Chromosome 14 | 2 | 0 | 2 | 0 | 1.00 | 1.00 | 1.00 |
| Ring Chromosome 20 | 6 | 7 | 13 | 0 | 0.46 | 1.00 | 0.63 |
| SCN2A | 9 | 11 | 20 | 0 | 0.45 | 1.00 | 0.62 |
| SCN8A | 1 | 0 | 1 | 0 | 1.00 | 1.00 | 1.00 |
| Sturge-Weber syndrome | 97 | 31 | 128 | 2 | 0.76 | 0.98 | 0.85 |
| Tuberous sclerosis complex | 200 | 48 | 248 | 0 | 0.81 | 1.00 | 0.89 |
| Total | 1,862 | 1,426 | 3,288 | 205 | - | - | - |
Abbreviations: true positive (TP), false positive (FP), estimated false negative (eFN), positive predictive value (PPV), sensitivity (sens), inter-quartile range (IQR), epileptic encephalopathy with spike-and-wave activation in sleep (EE-SWAS) and/or electrical status epilepticus in sleep (ESES), epilepsy with myoclonic atonic seizures (EMAS).
The optimal combination method used fewer search words than the all-keyword method, and therefore, identified a smaller number of cases (1,862 vs. 2,068). Likewise, across rare epilepsies, we observed slightly fewer TPs using the optimal combination vs. all-keyword method (median = 19 vs. 25). However, the optimal keyword method resulted in higher PPVs for nearly all rare epilepsies (28 of 29).
Final Regular Expressions
We provide a keyword search list, which includes the optimal statistical combination of keywords and additional clinically important words (Table 2), as well as the corresponding regular expressions for each rare epilepsy (Table S3).
DISCUSSION
We demonstrated that a simple keyword search using regular expressions is an effective method for creating rare epilepsy cohorts. We reported the performance of two methods. The first used a more complete list of keywords associated with rare epilepsies to maximize sensitivity and therefore is best for very rare conditions. Using this method, some conditions showed high PPVs, including for Sturge-Weber syndrome, tuberous sclerosis complex, and LGS. The second method used the optimal statistical combination of keywords to maximize overall performance. This approach is best for large, multi-institutional clinical data where both sensitivity and specificity are important. Using this approach, we observed adequate performance with sensitivities ≥ 0.80 and PPVs ≥ 0.60 for most rare epilepsies. Both methods identified relatively large cohorts of rare epilepsies, including over 100 individuals with EMAS, Rett syndrome, infantile spasms, and LGS.
NLP algorithms often use negation detection (e.g., “no history of seizures”) and name entity recognition (“cousin with Lennox-Gastaut syndrome”) to improve specificity. We were surprised this was not required to achieve adequate specificity using regular expressions. We suspect this is because of the infrequent occurrence of rare epilepsies. More common conditions are documented in the differential even though some individuals may not have the diagnosis (e.g., “the differential includes heart failure”). More common conditions are also documented in screening tests (e.g., “sent Fragile X testing for development delay”). It would be unusual for a rare genetic diagnosis to be documented in these ways. Instead, documentation would be more general, for example, “suspected genetic etiology”. In addition, keyword search can have false positive matches when text refers to family members with the diagnosis, and this would also occur infrequently in rare conditions. This could explain why we saw the best performance for more rare conditions (e.g., dup15q syndrome) and high erroneous matches for more commonly tested conditions (e.g., Fragile X syndrome).
The use of NLP is a time-efficient method of cohort creation. We used regular expressions to rapidly search narrative text of clinical notes from over 77,000 individuals. This would not have been feasible with manual chart review alone. A combination of NLP to screen data followed by manual chart review validation is a way to maintain time-efficiency while also maximizing accuracy.
We expected to observe larger cohort sizes compared to previous studies relying on more time-intensive manual chart review. As expected, some of our cohorts were larger than previous studies (e.g., 511 individuals with LGS and 68 with EE-SWAS/ESES).10–17 Other cohorts were similar to previous studies (e.g., 107 individuals with EMAS and 25 with Rasmussen syndrome).18–21 For a few rare epilepsies, we were unable to identify individuals in clinical data (i.e., SLC13a5, SYNGAP, Unverricht-Lundborg disease).
Now is the opportune time for development of NLP tools for rare epilepsy cohort creation, since new treatment options are becoming available.22,23 For example, cerliponase alfa was recently approved as an enzyme replacement for neuronal ceroid lipofuscinosis type 2 and initial results show potential for slowing progression of disease.24 Fenfluramine was also recently approved for treatment of seizures in Dravet syndrome and LGS.25, 26 Additional preclinical and clinical trials are ongoing.22 NLP tools can be helpful by identifying individuals for referral to clinical trials and creating cohorts for comparative effectiveness research. The methods we present can be used to better understand the burden of rare epilepsy diseases and inform public health interventions.
Patient registries and learning healthcare systems (LHS) are other potential data sources for studying rare epilepsies. A strength of patient registries is that they include a relatively large number of individuals from geographically diverse areas over a long period of time. Some rare epilepsies already have large registries, including CDKL5, ring 14, Aicardi syndrome, Sturge-Weber syndrome, tuberous sclerosis complex, and PCDH19. However, there are potential sources of bias using patient registries. They rely on patient reported outcomes which may be influenced by recall bias. There may be selection bias if more severely affected individuals have families more active in family support groups, research, and registries. An LHS is another excellent data source for clinical research.27 Data can be higher quality since physicians document a uniform template of clinical information at the point of care. The main challenge of an LHS is that it requires buy in from clinicians to document clinical information and needs to be integrated into clinical workflow.
A limitation of this study is that regular expressions become outdated when terminology changes, and may not generalize to geographic regions that use different terminology. Validation studies are needed to evaluate performance of regular expressions in other datasets. A second limitation is that our keyword search only captures cases that have diagnostic terms documented in EHRs. More work is needed to identify potential undiagnosed cases in EHRs, i.e., patients who need a referral to neurology or genetics for diagnostic workup. Lastly, there is a need for new ICD codes for rare epilepsies to standardize diagnostic coding and facilitate clinical research.
Conclusion.
Keyword search using regular expressions is a time-efficient and effective method to identify individuals with rare epilepsies using clinical data. Keyword search performs better for more rare conditions. Anyone with access to clinical notes from their own institution, or using a publicly available clinical research data warehouse, can use these keywords or regular expressions to create rare epilepsy cohorts and improve epidemiology, surveillance, clinical care, and research for rare epilepsy. From a public health standpoint, these studies are important to understand rare disease burden, support people affected by rare epilepsies (e.g., facilitate referral to the Rare Epilepsy Network),28 and educate the public. Additional work is needed to integrate methods into clinical workflow to facilitate referrals to disease specialists, clinical trials, and advocacy groups.
Supplementary Material
KEY POINTS BOX.
Keyword search is an effective method to create relatively large rare epilepsy cohorts.
We report the highest-performing keyword search combinations that identify individuals with rare epilepsies in electronic health records.
We observed adequate performance for keyword search combinations with sensitivities ≥ 0.80 and PPVs ≥ 0.60 for most rare epilepsies.
Methods can be used for improving identification of rare epilepsies and referral to specialists, clinical research, and support groups.
Acknowledgments.
This publication was supported by the Cooperative Agreement Number, U01DP006089, funded by the Centers for Disease Control and Prevention of the U.S. Department of Health and Human Services (HHS) as a financial assistance award totaling $899,766 with 100% percent funded by CDC/HHS.
Thank you to Dr. Dale Hesdorffer for guidance and support with this project.
Footnotes
Disclaimer: The findings and conclusions of this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Disclosures of conflicts of interest. Patricia McGoldrick has served as a speaker for Eisai, Greenwich and Sunovion and on advisory boards to UCB, Neuropace and Supernus. Steven Wolf has served as speaker or consultant for Greenwich Pharma, Eisai Pharma, Neuropace, Lundbeck, UCB, Biomarin, Aquestive, and Zogenix, Neurilis. Tristan Sands has served as a consultant for Biomarin. Zachary Grinspan receives funding for his research from Weill Cornell Medicine, the Pediatric Epilepsy Research Foundation, Clara Inspired, and the Orphan Disease Center. He has performed consulting work for Alpha Insights and Bio-Pharm Solutions (South Korea). The remaining authors have no conflicts of interest.
Ethical Publication Statement. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
REFERENCES
- 1.Ho NT, Kroner B, Grinspan Z, Fureman B, Farrell K, Zhang J, et al. Comorbidities of Rare Epilepsies: Results from the Rare Epilepsy Network J Pediatr. 2018. Dec;203:249–258 e245. [DOI] [PubMed] [Google Scholar]
- 2.Pan S, Wu A, Weiner M, Z MG. Development and Evaluation of Computable Phenotypes in Pediatric Epilepsy:3 Cases J Child Neurol. 2021. Oct;36:990–997. [DOI] [PubMed] [Google Scholar]
- 3.Barbour K, Hesdorffer DC, Tian N, Yozawitz EG, McGoldrick PE, Wolf S, et al. Automated detection of sudden unexpected death in epilepsy risk factors in electronic medical records using natural language processing Epilepsia. 2019. Jun;60:1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen KB, Glass B, Greiner HM, Holland-Bouley K, Standridge S, Arya R, et al. Methodological Issues in Predicting Pediatric Epilepsy Surgery Candidates Through Natural Language Processing and Machine Learning Biomed Inform Insights. 2016;8:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimia AA, Capraro AJ, Hummel D, Johnston P, Harper MB. Utility of lumbar puncture for first simple febrile seizure among children 6 to 18 months of age Pediatrics. 2009. Jan;123:6–12. [DOI] [PubMed] [Google Scholar]
- 6.Hamid H, Fodeh SJ, Lizama AG, Czlapinski R, Pugh MJ, LaFrance WC Jr., et al. Validating a natural language processing tool to exclude psychogenic nonepileptic seizures in electronic medical record-based epilepsy research Epilepsy Behav. 2013. Dec;29:578–580. [DOI] [PubMed] [Google Scholar]
- 7.Cui L, Bozorgi A, Lhatoo SD, Zhang GQ, Sahoo SS. EpiDEA: extracting structured epilepsy and seizure information from patient discharge summaries for cohort identification AMIA Annu Symp Proc. 2012;2012:1191–1200. [PMC free article] [PubMed] [Google Scholar]
- 8.Grinspan ZM, Tian N, Yozawitz EG, McGoldrick PE, Wolf SM, McDonough TL, et al. Common terms for rare epilepsies: Synonyms, associated terms, and links to structured vocabularies Epilepsia Open. 2018. Mar;3:91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.PCORI. New York City Clinical Data Research Network (NYC-CDRN). Available at: https://www.pcori.org/research-results/2015/new-york-city-clinical-data-research-network-nyc-cdrn.
- 10.Asadi-Pooya AA, Bazrafshan M, Farazdaghi M. Long-term medical and social outcomes of patients with Lennox-Gastaut syndrome Epilepsy Res. 2021. Nov 13;178:106813. [DOI] [PubMed] [Google Scholar]
- 11.Calvo A, Buompadre MC, Gallo A, Gutierrez R, Valenzuela GR, Caraballo R. Electroclinical pattern in the transition from West to Lennox-Gastaut syndrome Epilepsy Res. 2020. Nov;167:106446. [DOI] [PubMed] [Google Scholar]
- 12.Goldsmith IL, Zupanc ML, Buchhalter JR. Long-term seizure outcome in 74 patients with Lennox-Gastaut syndrome: effects of incorporating MRI head imaging in defining the cryptogenic subgroup Epilepsia. 2000. Apr;41:395–399. [DOI] [PubMed] [Google Scholar]
- 13.Herranz JL, Casas-Fernandez C, Campistol J, Campos-Castello J, Rufo-Campos M, Torres-Falcon A, et al. [Lennox-Gastaut syndrome in Spain: a descriptive retrospective epidemiological study] Rev Neurol. 2010. Jun 16;50:711–717. [PubMed] [Google Scholar]
- 14.Kang JW, Eom S, Hong W, Kwon HE, Park S, Ko A, et al. Long-term Outcome of Resective Epilepsy Surgery in Patients With Lennox-Gastaut Syndrome Pediatrics. 2018. Oct;142. [DOI] [PubMed] [Google Scholar]
- 15.Caraballo RH, Cejas N, Chamorro N, Kaltenmeier MC, Fortini S, Soprano AM. Landau-Kleffner syndrome: a study of 29 patients Seizure. 2014. Feb;23:98–104. [DOI] [PubMed] [Google Scholar]
- 16.Cockerell I, Bolling G, Nakken KO. Landau-Kleffner syndrome in Norway: long-term prognosis and experiences with the health services and educational systems Epilepsy Behav. 2011. Jun;21:153–159. [DOI] [PubMed] [Google Scholar]
- 17.Riccio CA, Vidrine SM, Cohen MJ, Acosta-Cotte D, Park Y. Neurocognitive and behavioral profiles of children with Landau-Kleffner syndrome Appl Neuropsychol Child. 2017. Oct-Dec;6:345–354. [DOI] [PubMed] [Google Scholar]
- 18.Nickels K, Kossoff EH, Eschbach K, Joshi C. Epilepsy with myoclonic-atonic seizures (Doose syndrome): Clarification of diagnosis and treatment options through a large retrospective multicenter cohort Epilepsia. 2021. Jan;62:120–127. [DOI] [PubMed] [Google Scholar]
- 19.Tang S, Addis L, Smith A, Topp SD, Pendziwiat M, Mei D, et al. Phenotypic and genetic spectrum of epilepsy with myoclonic atonic seizures Epilepsia. 2020. May;61:995–1007. [DOI] [PubMed] [Google Scholar]
- 20.Caraballo RH, Fortini S, Cersosimo R, Monges S, Pasteris MC, Gomez M, et al. Rasmussen syndrome: an Argentinean experience in 32 patients Seizure. 2013. Jun;22:360–367. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman CE, Ochi A, Snead OC 3rd, Widjaja E, Hawkins C, Tisdal M, et al. Rasmussen’s encephalitis: advances in management and patient outcomes Childs Nerv Syst. 2016. Apr;32:629–640. [DOI] [PubMed] [Google Scholar]
- 22.Auvin S, Avbersek A, Bast T, Chiron C, Guerrini R, Kaminski RM, et al. Drug Development for Rare Paediatric Epilepsies: Current State and Future Directions Drugs. 2019. Dec;79:1917–1935. [DOI] [PubMed] [Google Scholar]
- 23.Orsini A, Valetto A, Bertini V, Esposito M, Carli N, Minassian BA, et al. The best evidence for progressive myoclonic epilepsy: A pathway to precision therapy Seizure. 2019. Oct;71:247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulz A, Ajayi T, Specchio N, de Los Reyes E, Gissen P, Ballon D, et al. Study of Intraventricular Cerliponase Alfa for CLN2 Disease N Engl J Med. 2018. May 17;378:1898–1907. [DOI] [PubMed] [Google Scholar]
- 25.Lagae L, Sullivan J, Knupp K, Laux L, Polster T, Nikanorova M, et al. Fenfluramine hydrochloride for the treatment of seizures in Dravet syndrome: a randomised, double-blind, placebo-controlled trial Lancet. 2019. Dec 21;394:2243–2254. [DOI] [PubMed] [Google Scholar]
- 26.Specchio N, Pietrafusa N, Doccini V, Trivisano M, Darra F, Ragona F, et al. Efficacy and safety of Fenfluramine hydrochloride for the treatment of seizures in Dravet syndrome: A real-world study Epilepsia. 2020. Nov;61:2405–2414. [DOI] [PubMed] [Google Scholar]
- 27.Grinspan ZM, Patel AD, Shellhaas RA, Berg AT, Axeen ET, Bolton J, et al. Design and implementation of electronic health record common data elements for pediatric epilepsy: Foundations for a learning health care system Epilepsia. 2021. Jan;62:198–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.REN. Rare Epilepsy Network. Available at: https://www.rareepilepsynetwork.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


