Abstract
A retroviral Env molecule consists of a surface glycoprotein (SU) complexed with a transmembrane protein (TM). In turn, these complexes are grouped into oligomers on the surfaces of the cell and of the virion. In the case of murine leukemia viruses (MuLVs), the SU moieties are polymorphic, with SU proteins of different viral isolates directed towards different cell surface receptors. During maturation of the released virus particle, the 16 C-terminal residues of TM (the R peptide or p2E) are removed from the protein by the viral protease; this cleavage is believed to activate the membrane-fusing potential of MuLV Env. We have tested the possibility that different MuLV Env proteins in the same cell can interact with each other, both physically and functionally, in mixed oligomers. We found that coexpressed Env molecules can be precipitated out of cell lysates by antiserum which reacts with only one of them. Furthermore, they can evidently cooperate with each other: if one Env species lacks the R peptide, then it can apparently induce fusion if the SU protein of the other Env species encounters its cognate receptor on the surface of another cell. This functional interaction between different Env molecules has a number of implications with respect to the mechanism of induction of membrane fusion, for the genetic analysis of Env function, and for the design of targeted retroviral vectors for gene therapy.
The entry of an enveloped virus into a mammalian cell occurs in two distinct steps: first, the binding of the virus particle to a cell surface receptor, and second, the fusion of the viral membrane with a cellular membrane.
In retroviruses, entry is mediated by the Env protein complex. This consists of SU, a large glycoprotein displayed on the surface of the particle, and TM, a smaller, membrane-spanning protein. SU and TM are formed by cleavage of a single precursor polyprotein and remain together in a complex after the cleavage event. A number of lines of evidence indicate that the SU component is responsible for the receptor-binding step, while membrane fusion is performed by TM (8, 15). Recent studies demonstrate some striking structural parallels between TM and HA2, the protein required for membrane fusion in influenza virus (6, 11, 31).
In turn, the Env complexes have been shown to exist as oligomers, each containing three or four SU-TM units, both in the cell and in the virion (10, 17, 30; reviewed in reference 9). This physical association raises the possibility that different Env complexes might be able to interact functionally in hetero-oligomers. The present experiments were designed to test this possibility; the results strongly indicate that, in a cell expressing two different molecular species of murine leukemia virus (MuLV) Env, one species is able to perform the initial membrane-binding step, enabling the other to induce membrane fusion. This apparent cooperation between different Env molecules has a number of interesting implications.
In the MuLV Env complex, a 16-residue peptide, termed the R peptide or p2E, is removed from the C-terminal, cytoplasmic domain of TM after the virion is assembled and released from the cell (12, 14, 29). Cells expressing this truncated form of Env, but not the full-length form, induce syncytium formation when they are cocultivated with cells displaying the cognate cell surface receptor (23, 24). It seems likely that the membrane fusion observed in these cocultivation experiments reproduces, in many respects, the fusion process by which Env mediates the entry of a virus particle into a cell. The results suggest that the function of the R peptide is to prevent the MuLV Env complex from becoming fusogenic until the virus has been released from the cell (23, 24, 32). Similar phenomena have also been described in type D retroviruses (3, 4, 28), and somewhat analogous observations have been made in studies of lentiviruses (16, 21, 26, 27, 33).
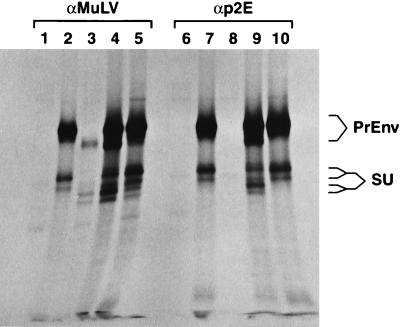
We initially looked for evidence of physical association between two types of MuLV Env molecules which were coexpressed in the same cells. The two molecules were full-length Moloney (Mo)-MuLV Env and p2E− 10A1 Env; thus, they differed both in their SU domains (and therefore in their receptor specificities [25]) and at the C termini of their TM proteins. We precipitated the Env proteins from these cells with anti-p2E antiserum, which should react with the full-length Mo-MuLV Env but not with the p2E− 10A1 Env. Control precipitations were performed with anti-MuLV antiserum. The p15ETM protein was not detectable in these experiments, presumably because insufficient radioactivity was incorporated into it (data not shown); therefore, we investigated the possibility that Env complexes, containing PrEnv and/or SU, were precipitated by the anti-MuLV and anti-p2E antisera. The results of this experiment are shown in Fig. 1. As can be seen, these proteins are present in the precipitates. Further, the Mo-MuLV PrEnv and SU proteins are easily distinguished from the corresponding 10A1 proteins by their lower mobilities in our sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (Fig. 1, lanes 2 vs. 3 and 7 vs. 8) (each of the SU proteins appears to form a doublet in these experiments; this may represent heterogeneity in the glycosylation of the proteins). In lysates of cells expressing both env genes, the p2E− 10A1 proteins (as well as the full-length Mo-MuLV proteins) were precipitated by anti-p2E antiserum (Fig. 1, lane 9). This was not due to nonspecific reactivity between the antibodies and the 10A1 Env proteins, since these Env proteins were not precipitated when expressed alone (Fig. 1, lane 8), even when lysates containing these proteins (Fig. 1, lanes 3 and 5) were mixed with lysates containing full-length Mo-MuLV Env protein (Fig. 1, lane 10) before the immunoprecipitation. The coprecipitation of 10A1 Env proteins by anti-p2E antiserum when these proteins were present in cells together with full-length Mo-MuLV Env proteins (Fig. 1, lane 9) provides strong evidence for the existence of hetero-oligomers of Env proteins in these cells.
FIG. 1.
Coprecipitation of p2E− 10A1 PrEnv and SU with full-length Mo-MuLV PrEnv and SU by anti-p2E antiserum. HeLa cells in six-well plates were infected with a vaccinia virus encoding T7 polymerase and then transfected with 4 μg of plasmids containing a full-length Mo-MuLV env gene and/or a p2E− 10A1 env gene under the control of the T7 promoter, as described previously (32). (Details of the construction of these plasmids are available upon request.) Twelve hours after transfection, the cells were starved for methionine and cysteine for 45 min, labeled for 1 h with 200 μCi of [35S]methionine and [35S]cysteine per ml (Amersham cell-labeling mix, Arlington Heights, Ill.), and chased for 2.5 h with unlabeled medium. They were then lysed as described (32), and the lysates were analyzed by radioimmunoprecipitation (32) with goat anti-MuLV antiserum (32) (lanes 1 to 5) or rabbit anti-p2E antiserum (a kind gift from John Elder) (12, 29) (lanes 6 to 10). Cultures were transfected with plasmid vector lacking env sequences (lanes 1 and 6), the Mo-MuLV env gene (lanes 2 and 7), the p2E− 10A1 env gene (lanes 3 and 8), or a mixture of the Mo-MuLV and p2E− 10A1 env gene plasmids (lanes 4 and 9). Lanes 5 and 10 show the results obtained by mixing the lysates of cells transfected with the Mo-MuLV and p2E− 10A1 env genes and then performing the radioimmunoprecipitations on the mixture. The precipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on an 8% polyacrylamide gel.
We also tested the possibility that different Env molecules can interact with each other functionally, as well as physically, when they are coexpressed in the same cells. As noted above, p2E− Mo-MuLV Env protein can induce syncytium formation when cells expressing it are cocultivated with cells displaying the ecotropic MuLV receptor (i.e., the cell surface receptor used by Mo-MuLV for entry into the cell) but not upon cocultivation with cells whose ecotropic receptor is blocked (23, 24). In the present experiments, we constructed cells expressing both p2E− Mo-MuLV Env and full-length, wild-type 10A1 Env (10A1 and Mo-MuLV infect cells via different cell surface receptors [25]). We then tested the abilities of these cells, containing two types of MuLV Env, to induce fusion with NIH 3T3 cells which were chronically infected with Mo-MuLV and hence did not display the ecotropic receptor. Such fusion would presumably require the functional cooperation of the two Env molecules, since the p2E− Mo-MuLV Env is unable to fuse cells lacking an available ecotropic receptor, while the wild-type 10A1 protein, possessing the R peptide at its C terminus, is not fusogenic while it is on the cell surface.
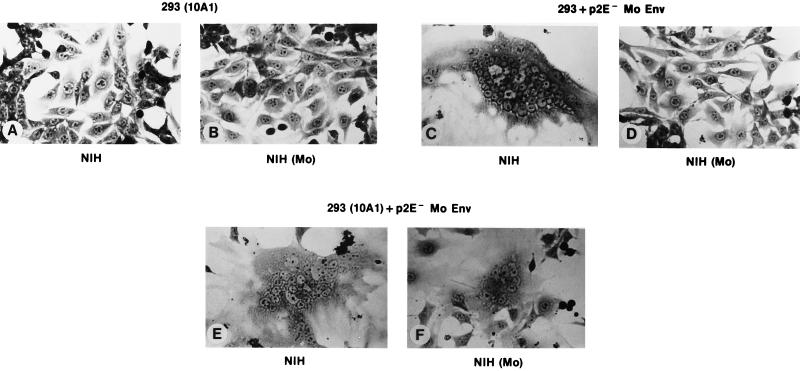
The results of these tests were as follows. When cells infected with wild-type 10A1 MuLV and transiently transfected with a plasmid encoding p2E− Mo-MuLV Env were cocultivated with Mo-MuLV-infected NIH 3T3 cells, a number of syncytia were observed. This level of fusion, although limited, was quite reproducible, and these cultures could easily be distinguished from controls, even by an observer unfamiliar with the identities of the cultures. An example of the syncytia seen here is shown in Fig. 2F. The controls (Fig. 2A to D) showed that, as expected, neither of the two Env molecules could induce detectable fusion in the chronically infected cells when expressed alone (Fig. 2B and D), although the p2E− Mo-MuLV Env induced widespread fusion in uninfected NIH 3T3 cells (Fig. 2C).
FIG. 2.
Cooperation of p2E− Mo-MuLV and wild-type 10A1 Env in syncytium formation. 293 cells expressing simian virus 40 large T antigen were infected with 10A1 MuLV, and the virus was allowed to spread through the culture. These cells, as well as uninfected 293 cells, were seeded at 1 × 105 cells per 60-mm-diameter dish. The following day, they were transfected with pCDEnv encoding p2E− Mo-MuLV Env (24) by the calcium phosphate technique. Two days later, the cells were overlaid with 8 × 105 NIH 3T3 cells which were chronically infected with wild-type Mo-MuLV or with 8 × 105 uninfected NIH 3T3 cells. The plates were fixed and stained as described previously (24) after 30 h of cocultivation. (A) 10A1-infected, untransfected 293 cells with NIH 3T3 cells. (B) 10A1-infected, untransfected 293 cells with Mo-MuLV-infected NIH 3T3 cells. (C) Uninfected 293 cells, transfected with pCDEnv encoding p2E− Mo-MuLV, with NIH 3T3 cells. (D) Uninfected 293 cells, transfected as described for panel C, with Mo-MuLV-infected NIH 3T3 cells. (E) 10A1-infected 293 cells, transfected as described for panel C, with NIH 3T3 cells. (F) 10A1-infected 293 cells, transfected as described for panel C, with Mo-MuLV-infected NIH 3T3 cells.
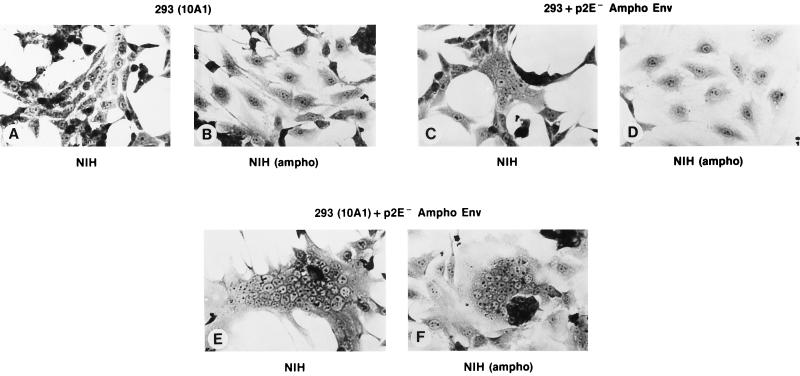
It seemed possible that the fusions seen in Fig. 2F were somehow peculiar to the combination of wild-type 10A1 and p2E− Mo-MuLV Env molecules. To determine whether other combinations could also induce fusion upon coexpression, we replaced the p2E− Mo-MuLV Env with p2E− amphotropic MuLV Env and tested the abilities of the cells containing both Env molecules to induce fusion in NIH 3T3 cells chronically infected with amphotropic MuLV. As shown in Fig. 3, results exactly parallel to those shown in Fig. 2 were obtained.
FIG. 3.
Cooperation of p2E− amphotropic MuLV and wild-type 10A1 Env in syncytium formation. The experiment was identical to that whose results are shown in Fig. 2, except that cells were transfected with pCDEnv encoding p2E− amphotropic MuLV Env (obtained from a molecular clone of 4070A MuLV [22a]) rather than p2E− Mo-MuLV Env, and cocultivations were with NIH 3T3 cells which were chronically infected with amphotropic MuLV rather than with Mo-MuLV. Plates were fixed after 24 h of cocultivation.
The results shown in Fig. 2 (and Fig. 3) demonstrate that a cell containing two different Env molecules can induce fusion with a target cell which neither of the Env molecules is capable of fusing with on its own. Presumably, the fusion we observed here occurs when the wild-type 10A1 Env protein in a hetero-oligomer binds to one of the 10A1 receptors on the NIH 3T3 cells, and the p2E− Mo-MuLV Env protein in the hetero-oligomer then induces membrane fusion.
The cooperation between different Env molecules in mixed oligomers has several significant implications. First, it may provide useful information regarding the mechanism by which MuLV Env molecules induce membrane fusion. In influenza virus, it is known that exposure of the hemagglutinin (HA) complex to the low pH of the endosome triggers a major rearrangement of the HA molecule into the active fusogenic conformation by exposing the fusion peptide of HA2 (5, 19). While some have suggested that low pH is also required for the induction of fusogenicity in Mo-MuLV Env (1, 20, 22), the ability of the p2E− form of this molecule to cause syncytium formation at neutral pH (23, 24) would seem to argue against this hypothesis. In turn, if low pH is not involved in the activation of Mo-MuLV Env, then the most plausible alternative trigger for the exposure of the fusion peptide in TM is probably the contact between SU and the cell surface receptor. Our results thus suggest that, in a mixed oligomer, contact of one SU molecule with its cognate receptor induces the activation of fusogenicity in one or more other Env molecules in the oligomer. We do not know whether the exposure of a single fusion peptide of a p2E− TM protein in the oligomer is sufficient for membrane fusion, but it is also conceivable that all of the fusion peptides in an oligomer, including those in TM proteins which retain the R peptide, are exposed by a conformational change following contact of a single SU protein in the oligomer with its receptor. Interestingly, elegant studies on influenza HA by Boulay et al. (2) have shown that activation of mixed HA oligomers by low pH is a concerted, highly cooperative event.
These results also open the way for a better definition of the elements in Env required for receptor binding and membrane fusion. In essence, we have described complementation between two Env molecules, in which one carries out the receptor-binding step and the other induces membrane fusion. This complementation could be exploited in the genetic analysis of Env function, in which mutant Env molecules could be tested for their abilities to function in receptor binding in the presence of a second Env molecule capable of mediating membrane fusion. (Conversely, of course, mutants could be tested for their abilities to carry out membrane fusion together with another, receptor-binding Env molecule.)
Finally, the cooperation between Env molecules can potentially be exploited in the design of targeted retroviral vectors for gene therapy. It would be highly desirable to design modified Env molecules with the ability to bind to new cell surface receptors, since such modified Env molecules might target the virus particle to specific types of cells within the body. However, attempts to replace the natural receptor-binding region of SU with other domains capable of binding to other cell surface ligands have met with very limited success to date. One major reason for the difficulty of targeting may be the delicate interrelationship and interaction between SU and TM in an Env molecule: perhaps in a modified, targeting Env complex, SU can indeed bind a novel receptor, but TM fails to induce membrane fusion. The cooperation between Env molecules might significantly alleviate this problem. Thus, vectors with a targeting Env might retain more infectivity if they also contain a second Env to perform membrane fusion. In fact, successful targeting of vectors has been described with particles which included a wild-type Env in addition to the modified, targeting Env (7, 13, 18). We would propose that the wild-type Env performs the membrane fusion function in these mosaic virions.
After this work was submitted for publication, a paper describing somewhat similar experiments appeared (32a). This paper (32a) was a careful, detailed analysis of functional interactions between defective SU proteins of Mo-MuLV and defective TM proteins of Mo-MuLV. The findings we have presented here are entirely consistent with those reported in this paper (32a) and extend them by providing evidence of functional interaction between Env molecules directed toward different cell surface receptors.
Acknowledgments
Research was sponsored in part by the National Cancer Institute, DHHS under contract with ABL, and NIH grant CA18611.
REFERENCES
- 1.Andersen K B, Nexo B A. Entry of murine retrovirus into mouse fibroblasts. Virology. 1983;125:85–98. doi: 10.1016/0042-6822(83)90065-x. [DOI] [PubMed] [Google Scholar]
- 2.Boulay F, Doms R W, Webster R G, Helenius A. Posttranslational oligomerization and cooperative acid activation of mixed influenza hemagglutinin trimers. J Cell Biol. 1988;106:629–639. doi: 10.1083/jcb.106.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brody B A, Rhee S S, Hunter E. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J Virol. 1994;68:4620–4627. doi: 10.1128/jvi.68.7.4620-4627.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brody B A, Rhee S S, Sommerfelt M A, Hunter E. A viral protease-mediated cleavage of the transmembrane glycoprotein of Mason-Pfizer monkey virus can be suppressed by mutations within the matrix protein. Proc Natl Acad Sci USA. 1992;89:3443–3447. doi: 10.1073/pnas.89.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 6.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 7.Chu T-H T, Dornburg R. Retroviral vector particles displaying the antigen-binding site of an antibody enable cell-type-specific gene transfer. J Virol. 1995;69:2659–2663. doi: 10.1128/jvi.69.4.2659-2663.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickson C, Eisenman R, Fan H, Hunter E, Teich N. Protein biosynthesis and assembly. In: Weiss R, Teich N, Varmus H, Coffin J, editors. Molecular biology of tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1984. pp. 513–648. [Google Scholar]
- 9.Einfeld D. Maturation and assembly of retroviral glycoproteins. In: Krausslich H-G, editor. Morphogenesis and maturation of retroviruses. New York, N.Y: Springer-Verlag; 1996. pp. 133–176. [DOI] [PubMed] [Google Scholar]
- 10.Einfeld D, Hunter E. Oligomeric structure of a prototype retrovirus glycoprotein. Proc Natl Acad Sci USA. 1988;85:8688–8692. doi: 10.1073/pnas.85.22.8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 angstrom resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 12.Green N, Shinnick T M, Witte O, Ponticelli A, Sutcliffe J G, Lerner R A. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han X, Kasahara N, Kan Y W. Ligand-directed retroviral targeting of human breast cancer cells. Proc Natl Acad Sci USA. 1995;92:9747–9751. doi: 10.1073/pnas.92.21.9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson L E, Sowder R, Copeland T D, Smythers G, Oroszlan S. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J Virol. 1984;52:492–500. doi: 10.1128/jvi.52.2.492-500.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 16.Johnston P B, Dubay J W, Hunter E. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J Virol. 1993;67:3077–3086. doi: 10.1128/jvi.67.6.3077-3086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamps C A, Lin Y-C, Wong P K Y. Oligomerization and transport of the envelope protein of Moloney murine leukemia virus-TB and of ts1, a neurovirulent temperature-sensitive mutant of MoMuLV-TB. Virology. 1991;184:687–694. doi: 10.1016/0042-6822(91)90438-h. [DOI] [PubMed] [Google Scholar]
- 18.Kasahara N, Dozy A M, Kan Y W. Tissue-specific targeting of retroviral vectors through ligand-receptor interactions. Science. 1994;266:1373–1376. doi: 10.1126/science.7973726. [DOI] [PubMed] [Google Scholar]
- 19.Marsh M, Helenius A. Virus entry into animal cells. Adv Virus Res. 1989;36:107–151. doi: 10.1016/S0065-3527(08)60583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClure M O, Sommerfelt M A, Marsh M, Weiss R A. The pH independence of mammalian retrovirus infection. J Gen Virol. 1990;71:767–773. doi: 10.1099/0022-1317-71-4-767. [DOI] [PubMed] [Google Scholar]
- 21.Mulligan M J, Yamshchikov G V, Ritter G D, Jr, Gao F, Jin M J, Nail C D, Spies C P, Hahn B H, Compans R W. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J Virol. 1992;66:3971–3975. doi: 10.1128/jvi.66.6.3971-3975.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nussbaum O, Roop A, Anderson W F. Sequences determining the pH dependence of viral entry are distinct from the host range-determining region of the murine ecotropic and amphotropic retrovirus envelope proteins. J Virol. 1993;67:7402–7405. doi: 10.1128/jvi.67.12.7402-7405.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Ott D, Rein A. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70su. J Virol. 1992;66:4632–4638. doi: 10.1128/jvi.66.8.4632-4638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragheb J A, Anderson W F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rein A, Schultz A. Different recombinant murine leukemia viruses use different cell surface receptors. Virology. 1984;136:144–152. doi: 10.1016/0042-6822(84)90255-1. [DOI] [PubMed] [Google Scholar]
- 26.Rice N R, Henderson L E, Sowder R C, Copeland T D, Oroszlan S, Edwards J F. Synthesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J Virol. 1990;64:3770–3778. doi: 10.1128/jvi.64.8.3770-3778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritter G D, Jr, Mulligan M J, Lydy S L, Compans R W. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology. 1993;197:255–264. doi: 10.1006/viro.1993.1586. [DOI] [PubMed] [Google Scholar]
- 28.Sommerfelt M A, Petteway S R, Jr, Dreyer G B, Hunter E. Effect of retroviral proteinase inhibitors on Mason-Pfizer monkey virus maturation and transmembrane glycoprotein cleavage. J Virol. 1992;66:4220–4227. doi: 10.1128/jvi.66.7.4220-4227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutcliffe J G, Shinnick T M, Green N, Liu F T, Niman H L, Lerner R A. Chemical synthesis of a polypeptide predicted from nucleotide sequence allows detection of a new retroviral gene product. Nature. 1980;287:801–805. doi: 10.1038/287801a0. [DOI] [PubMed] [Google Scholar]
- 30.Tucker S P, Srinivas R V, Compans R W. Molecular domains involved in oligomerization of the Friend murine leukemia virus envelope glycoprotein. Virology. 1991;185:710–720. doi: 10.1016/0042-6822(91)90542-j. [DOI] [PubMed] [Google Scholar]
- 31.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 32.Yang C, Compans R W. Analysis of the cell fusion activities of chimeric simian immunodeficiency virus-murine leukemia virus envelope proteins: inhibitory effects of the R peptide. J Virol. 1996;70:248–254. doi: 10.1128/jvi.70.1.248-254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Zhao Y, Lee S, Anderson W F. Functional interactions between monomers of the retroviral envelope protein complex. J Virol. 1997;71:6967–6972. doi: 10.1128/jvi.71.9.6967-6972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zingler K, Littman D R. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases Env incorporation into particles and fusogenicity and infectivity. J Virol. 1993;67:2824–2831. doi: 10.1128/jvi.67.5.2824-2831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]