Abstract
The formation of resting cysts commonly found in unicellular eukaryotes is a complex and highly regulated survival strategy against environmental stress that involves drastic physiological and biochemical changes. Although most studies have focused on the morphology and structure of cysts, little is known about the molecular mechanisms that control this process. Recent studies indicate that DNA N6-adenine methylation (6mA) could be dynamically changing in response to external stimuli; however, its potential role in the regulation of cyst formation remains unknown. We used the ciliate Pseudocohnilembus persalinus, which can be easily induced to form cysts to investigate the dynamic pattern of 6mA in trophonts and cysts. Single-molecule real-time (SMRT) sequencing reveals high levels of 6mA in trophonts that decrease in cysts, along with a conversion of symmetric 6mA to asymmetric 6mA. Further analysis shows that 6mA, a mark of active transcription, is involved in altering the expression of encystment-related genes through changes in 6mA levels and 6mA symmetric-to-asymmetric conversion. Most importantly, we show that reducing 6mA levels by knocking down the DNA 6mA methyltransferase PpAMT1 accelerates cyst formation. Taken together, we characterize the genome-wide 6mA landscape in P. persalinus and provide insights into the role of 6mA in gene regulation under environmental stress in eukaryotes. We propose that 6mA acts as a mark of active transcription to regulate the encystment process along with symmetric-to-asymmetric conversion, providing important information for understanding the molecular response to environmental cues from the perspective of 6mA modification.
A microbial cyst is the resting or dormant stage of a microorganism with the ability to sense environmental alterations and to respond appropriately to new situations. Various environmental conditions have been reported to induce the formation of cryptobiotic cysts, such as desiccation, starvation, pH change, sudden changes in temperature, excess of metabolites, crowding, and even tidal cycling (Fenchel 1990; Gutiérrez et al. 1990, 2001; Olendzenski 1999). Starvation, however, seems to be the most universal trigger of encystment (Gutiérrez et al. 1990; Olendzenski 1999). Encystment involves progressive and drastic physiological and biochemical changes, including a reduction in cell volume, condensation of chromatin, and cessation of most activities such as feeding and locomotion (Gutiérrez et al. 1990, 2001; Kaur et al. 2019). Consequently, the process of encystation uses a highly complex array of gene expression alterations to produce a more resistant, differentiated state that enables the organism to maintain viability during variations in the habitat (Gutiérrez et al. 2000, 2001; Fouque et al. 2012). The return to favorable environmental conditions triggers excystment and leads to the emergence of a vegetative, swimming cell (Olendzenski 1999; Gutiérrez et al. 2001; Kaur et al. 2019).
Unicellular eukaryotes such as ciliates serve as good model systems in various biological studies including epigenetics, cell biology, and genomics (Nowacki et al. 2008; Cheng et al. 2019; Liu et al. 2021; Zhao et al. 2021; Zheng et al. 2021; Tian et al. 2022; Wei et al. 2022; Jin et al. 2023; Solberg et al. 2023; Zhang et al. 2023). A characteristic feature of ciliates is their nuclear dimorphism, hosting two functionally distinct nuclei within a single cell: the diploid micronucleus (MIC) and the highly polyploid macronucleus (MAC) (Karrer 2012). Additionally, ciliates are ideal organisms for studying cyst formation, owing to their general ease of culturing and genetic manipulability but also the characteristics of their cysts such as a relatively rapid encystation process and the high morphological variation of cysts (Gutiérrez et al. 1998, 2001; Kaur et al. 2019; Matsuoka 2021; Li et al. 2022). Early studies mainly relied on microscopic and submicroscopic methods, which have accumulated a great deal of morphological and ultrastructural data and revealed various processes involved in encystation, including cyst wall secretion, absorption of ciliary structures, and condensation of the macronuclear chromatin (Repak 1968; Matsusaka et al. 1989; Fenchel 1990; Olendzenski 1999; Leadbeater and Karpov 2000; Watoh et al. 2005; Grisvard et al. 2008; Verni and Rosati 2011; Whang et al. 2011; Fouque et al. 2012, 2015; Gong et al. 2018). More recently, transcriptomic and proteomic analyses identified multiple pathways involved in ciliate encystment, including the mitogen-activated protein kinase (MAPK) pathway, the protein kinase A (PKA) pathway, the cAMP pathway, and the calcium signaling pathway, which regulate encystment-related genes involved in DNA reorganization, autophagy, lipid metabolism, and stress response (Sogame et al. 2011; Jiang et al. 2019; Pan et al. 2020, 2021; Matsuoka 2021). Despite these efforts, the underlying molecular mechanism that enables a subset of genes to be quickly and selectively up- or down-regulated remains poorly understood.
One of the most important ways of altering gene expression is DNA methylation. 5-Methylcytosine (5mC), the most common DNA modification in eukaryotic genomes, has been shown to be a major regulator of genes related to stress response in plants. For example, an increase of 5mC in rice has been shown to contribute to drought adaptation (Zheng et al. 2017), whereas the loss of 5mC can activate the expression of genes that respond to phosphate starvation stress (Kumar et al. 2022). 5mC was also reported to change during cyst formation in the ciliate Colpoda inflata, where it is found in macronuclear DNA and undergoes demethylation during encystation (Palacios et al. 1994). Moreover, treatment with the nucleoside analog 5-azacytidine (5-azaC), a potent inhibitor of DNA methyltransferase (MTase) (Razin and Cedar 1991), was shown to increase the encystment ratio (Palacios et al. 1994). Nonetheless, it remains unclear whether 5mC is physiologically relevant in ciliates, as no homologs of 5mC MTases have been identified so far (Wang et al. 2017b,c; Singh et al. 2018). In contrast, a rediscovered epigenetic mark in eukaryotes, N6-methyadenine (6mA), is highly abundant and universally present in ciliates (Wang et al. 2017b; Beh et al. 2019; Cheng et al. 2019; Wang et al. 2019; Pan et al. 2023). 6mA has been shown to respond to external stress in an expanding list of eukaryotes: It is involved in mitochondrial stress adaptation in the worm Caenorhabditis elegans (Ma et al. 2019), the expression of neuronal genes in the fly Drosophila melanogaster (Yao et al. 2018), salt and temperature tolerance in the rice Oryza sativa (Zhang et al. 2018), hypoxia response in tumor cells (Hsu et al. 2022), and the regulation of stress responsivity of neuronal genes in the mouse Mus musculus (Yao et al. 2017). In the classical model ciliate Tetrahymena thermophila, 6mA also shows changes during starvation, suggesting an evolutionarily conserved response to environmental cues in multi- and unicellular eukaryotes (Sheng et al. 2021). In light of all these clues, it is therefore reasonable to hypothesize that 6mA may regulate cyst formation.
Pseudocohnilembus persalinus is a pathogenic marine ciliate (Xiong et al. 2015; Wei et al. 2018; Liu et al. 2019; Zhang et al. 2020) and has been extensively studied for morphology, behavior, pathology, encystation, and excystation (Repak 1968; Matsusaka et al. 1989; Leadbeater and Karpov 2000; Grisvard et al. 2008; Verni and Rosati 2011; Fouque et al. 2015; Gong et al. 2018). Exponentially growing trophonts have a wide pyriform shape and are relatively sluggish (Fenchel 1990; Liu et al. 2019). When starved, trophonts transform into a slender and more elongated form and significantly increase their swimming speed (Olendzenski 1999). Prolonged starvation induces encystation, forming cysts with a smooth surface that tend to clump together (Olendzenski 1999). Our previous work showed that an abundance of trophonts can be rapidly obtained by feeding with bacteria, and cyst formation can be easily induced by starvation, and thus, a simple and rapid cryopreservation protocol was established (Liu et al. 2019). Moreover, an initial P. persalinus genome database (PPGD; hereafter called PPGD 2015), assembled based on Illumina sequencing data and containing macronuclear genomic and transcriptomic data, has been reported (Xiong et al. 2015). Therefore, P. persalinus is an ideal model for investigating the molecular mechanisms of encystment.
In this study, we present an in-depth analysis of 6mA in P. persalinus that includes genome-wide maps of 6mA at single-nucleotide resolution in two different physiological states (trophont and cyst). Further analysis uncovered drastic alterations in 6mA distribution patterns between trophonts and cysts, suggesting a potential role of 6mA in the regulation of gene expression during cyst formation.
Results
6mA is a dynamic DNA modification in the MAC of P. persalinus
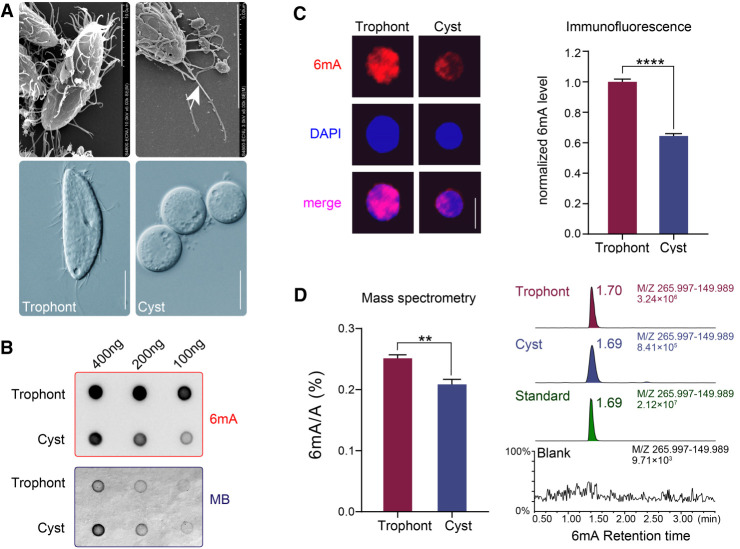
Vegetatively growing P. persalinus cells (trophonts) could be induced into cysts after ∼1 wk of starvation (Fig. 1A). To determine whether 6mA exists in P. persalinus, we first performed dot blot analysis with a 6mA-specific antibody. Strong 6mA signals were detected in DNA isolated from both trophonts and cysts, comparatively weaker in the latter, indicating the presence of 6mA modification in P. persalinus DNA (Fig. 1B). We then performed immunofluorescence (IF) staining, detecting 6mA in the MAC of both trophonts and cysts (Fig. 1C), consistent with previous findings that 6mA is a MAC-specific modification in ciliates (Gorovsky et al. 1973; Wang et al. 2017b). However, observing the MIC in P. persalinus is difficult, a challenge also noted in other Scuticociliatia ciliates (Song 2000). 6mA levels in cysts were significantly lower than in trophonts (∼60% of trophonts), which was confirmed by antibody-free mass spectrometry (MS) analysis (∼83% of trophonts) (Fig. 1D). These observations suggest that the 6mA modification might be dynamic in P. persalinus during the encystment–excystment cycle and prompted us to investigate the role of 6mA in this process.
Figure 1.
6mA levels are reduced in P. persalinus cysts. (A) Photomicrographs of trophonts and cysts of P. persalinus. (Top) Scan electron microscopic images. (Bottom) Phase-contrast images. Note the magnification difference in the top right image. The arrow indicates the caudal cilium. Scale bars, 10 µm. (B) 6mA levels in genomic DNA of trophonts and cysts were detected by dot blot using an anti-6mA antibody (top). Methylene blue hydrate staining was performed to determine the amount of loaded DNA (bottom). (C) Immunofluorescence staining of 6mA (left) and statistical analysis of 6mA signal intensities (right) in trophonts and cysts. Cell images were randomly selected (n = 100) and processed by ZEN 3.0. (****) P < 0.0001 (Student's t-test). Scale bars, 10 µm. (D) Mass spectrometry analysis of 6mA in genomic DNA of trophonts and cysts. The 6mA ratio (6mA/A, %) was defined as the abundance of methylated adenine divided by total adenine. (**) P < 0.01 (Student's t-test). Chromatograms showing the 6mA mass signal in standard, blank (water), and genomic DNA of trophonts and cysts (right).
High-quality genome assembly and annotation of the P. persalinus genome
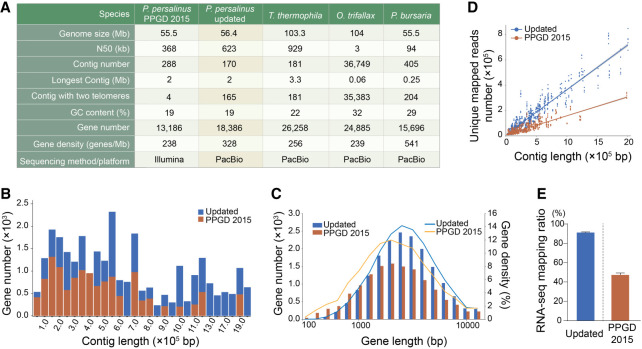
The published P. persalinus genome assembly (PPGD 2015) provided important information about the genome of P. persalinus and gave new insights into the pathogenesis of scuticociliates; however, it is segmented into broken contigs that contain many unknown regions and only limited gene annotations (Xiong et al. 2015; Wei et al. 2018). We therefore used SMRT sequencing to improve the genome assembly for downstream 6mA analysis (Supplemental Fig. S1A), obtaining a 56.4 Mb genome assembly consisting of 170 contigs, including 165 telomere-to-telomere contigs and five contigs with telomeric repeats at one end (Fig. 2A). The genome size and GC content of this updated assembly are nearly identical to PPGD 2015 (Fig. 2A; Supplemental Fig. S2A; Xiong et al. 2015), but the N50 length was increased about twofold from 368 kb to 623 kb, which was second only to T. thermophila (929 kb) (Fig. 2A; Sheng et al. 2020). Notably, a large fraction of contigs with intra- and inter-scaffold gaps present in the PPGD 2015 were assembled into longer supercontigs in our updated assembly (Supplemental Fig. S2B).
Figure 2.
Updated genome assembly of P. persalinus. (A) Statistical details of the new P. persalinus MAC genome assembly and comparison to the PPGD 2015 assembly (Xiong et al. 2015) and three other sequenced ciliate genomes (Lindblad et al. 2019; Sheng et al. 2020; Pan et al. 2023), including Oxytricha trifallax and Paramecium bursaria. (B) Distribution of genes in contigs in the updated genome assembly and PPGD 2015. The x-axis represents the contigs ordered by length; the y-axis, the number of genes. (C) Length distribution of genes in the updated genome assembly and PPGD 2015. The x-axis represents the length of genes; the y-axis, the number/density of genes. Lines represent the gene density; columns, the gene number. (D) RNA-seq mapping variation between the updated genome assembly and PPGD 2015. The x-axis represents the length of contigs; the y-axis, the number of unique reads mapped to the corresponding contigs. (E) RNA-seq mapping ratio of the two genome assemblies. RNA-seq data of trophonts and cyst were mapped to the updated genome assembly or PPGD 2015, respectively.
We then improved the genome annotation by using the LoReAn2 pipeline and incorporating protein MS data (Supplemental Fig. S1B). In total, we obtained 18,386 protein-coding genes and 380 noncoding RNA genes, which is a significant improvement over the PPGD 2015 annotation (13,186 protein-coding genes and zero noncoding RNA genes) (Fig. 2A). The average gene number per contig (108 vs. 46) (Fig. 2B) and the median gene length (2987 bp vs. 2584 bp) (Fig. 2C), as well as the exon number (117,897 vs. 72,961) and the average exon length (328 bp vs. 299 bp) (Supplemental Fig. S2C), were all higher than in the PPGD 2015 annotation. The improvement of the genome assembly and gene annotation was notably manifested by the higher mapping ratio of RNA sequencing (RNA-seq) reads (91.16% and 91.01% for trophont and cyst vs. 45.88% and 48.55% in PPGD 2015) (Fig. 2D,E). To further facilitate downstream analysis, we used our RNA-seq data to predict untranslated regions (UTRs) and annotated a total of 17,273 5′ UTRs and 17,235 3′ UTRs with an average length of 319 bp and 340 bp, respectively (Supplemental Fig. S2D). This largely optimized genome assembly and annotation laid the foundation for investigating the mechanism of 6mA regulation in encystment.
Encystment alters the genome-wide pattern of 6mA
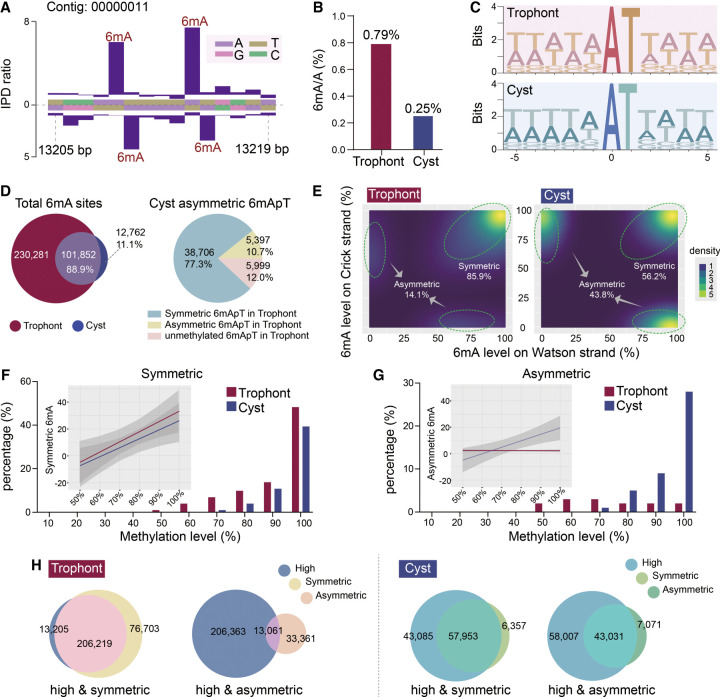
Using our improved de novo map of the P. persalinus genome, we next performed analyses of 6mA sites to determine the distribution of this modification across the genome. 6mA was detected with high confidence (coverage > 25× and Qv > 30 for 100× coverage) on 332,133 adenines in trophonts and 114,614 in cysts (FDR < 0.01), all with the typical kinetic signature (Fig. 3A), corresponding to 0.79% of total adenines in trophonts and 0.25% in cysts (Fig. 3B; Supplemental Fig. S3). This result strengthened our previous observation of global 6mA reduction in cysts revealed by dot blot, IF, and MS analyses (Fig. 1B–D). The slight differences between SMRT-seq and MS arise from two factors: (1) SMRT-seq mitigates the risk of overestimating 6mA levels by filtering out bacterial reads from dam−/dcm− Escherichia coli, which serves as the food source for P. persalinus and contains ∼0.02%–0.07% 6mA/A (O'Brown et al. 2019), and (2) SMRT-seq uses stringent cut-offs for identifying high-confidence 6mA sites, effectively disregarding those with low methylation or inconsistent methylation patterns. In comparison to other ciliates, the 6mA ratio in trophonts (0.79%) was higher than those previously reported in the vegetative cells of T. thermophila (0.54%) (Wang et al. 2017a; Wang et al. 2019) and Oxytricha trifallax (0.38%) (Beh et al. 2019) but lower than Paramecium bursaria (1.29%) (Pan et al. 2023) and Paramecium tetraurelia (1.06%) (Hardy et al. 2021), using the same cut-off value.
Figure 3.
Comparison of 6mA features in trophonts and cysts. (A) Representative inter-pulse duration (IPD) ratios of SMRT sequencing data. Columns indicate the IPD peaks representing the detected 6mA signals on both Watson and Crick strands based on the IPD ratio. (B) 6mA ratios in trophont and cyst genomes. The 6mA ratio was defined as the number of methylated adenine sites divided by the number of total adenine sites. (C) Sequence logos of 6mA sites (at position zero) in trophonts and cysts. (D) Venn diagram of shared 6mA sites in trophonts and cysts (left) and 6mA pattern conversion of asymmetric 6mApT in cysts (right). Three sources of asymmetric 6mApT are shown. (E) Density plot of 6mA distribution, according to methylation levels on Watson (x-axes) or Crick strands (y-axes) in trophonts (left) and cysts (right). Percentages of symmetric and asymmetric 6mA are shown. (F,G) Comparison of 6mA methylation levels in trophonts and cysts for symmetric 6mA (F) and asymmetric 6mA (G). Methylation levels were ranked from low to high and divided into 10 quantiles. Lines represent the percentage of 6mA; columns, the number of 6mA sites. Percentages represents the number of 6mA sites in each quantile divided by the total number of 6mA sites. Linear regression trendlines and 95% confidence intervals are shown as insets. (H) Area-proportional Venn diagram depicting highly methylated 6mA (80%–100%) and symmetric/asymmetric 6mA sites in trophonts (left) and cysts (right).
Despite the global reduction in cysts, the majority of 6mA is located on the 5′-ApT-3′ consensus sequence in both trophonts (99.16%) and cysts (99.82%), similar to other reported ciliates, namely, T. thermophila (82.38% in AT motif), P. bursaria (99.34%), P. tetraurelia (99.63%), and O. trifallax (95.98%) (Fig. 3C; Wang et al. 2017a; Beh et al. 2019; Wang et al. 2019; Hardy et al. 2021; Pan et al. 2023). As expected, we found 6mA sites to be closely linked between cysts and trophonts (Fig. 3D), with 88.9% of 6mA-containing ApT (6mApT) sites in cysts overlapping with those in trophonts (Fig. 3D). However, closer examination revealed that 85.9% of 6mApT sites in trophonts were symmetrically methylated (6mA on both Watson and Crick strands) (Fig. 3E), which is in strong contrast to a much higher proportion of asymmetric 6mA in cysts (43.8%) (Fig. 3E). To further explore this shift in 6mA patterns, we traced the source of individual asymmetric 6mA sites in cysts and found that 77.3% of these sites underwent a shift from the corresponding symmetric 6mApT site in trophonts (Fig. 3D), depicting a symmetric-to-asymmetric 6mA conversion during encystment.
We next divided 6mA sites into 10 quantiles according to their methylation level. In trophonts, 66.07% of 6mA sites had high methylation levels (80%–100%), 33.88% were intermediate (20%–80%), and 0.05% were low (10%–20%) (Supplemental Fig. S4A). Compared with trophonts, 6mA sites in cysts showed a significant increase in the high methylation category (from 66.07% to 88.16%) and a corresponding decrease in intermediate (from 33.88% to 11.83%) (Supplemental Fig. S4A). In trophonts, symmetrically methylated 6mA sites were enriched for high methylation levels (Fig. 3F), with a strong positive association (R2adj = 0.59) and a significant overlap (62.09% of total 6mA; representation factor: 0.96) (Fig. 3H). In cysts, however, highly methylated 6mA sites not only contained symmetric 6mA (R2adj = 0.57; 50.65% of total 6mA; representation factor: 0.92) but also contained asymmetric 6mA (R2adj = 0.65; 37.61% of total 6mA; representation factor: 0.92) (Fig. 3F–H). Taken together, these results reveal a clear shift from highly methylated symmetric 6mA in trophonts to highly methylated asymmetric 6mA in cysts.
6mA is involved in gene regulation as a transcriptional activator
Similar to the case in other ciliates (Wang et al. 2017b; Beh et al. 2019; Wang et al. 2019; Pan et al. 2023), 6mA could only be detected with high confidence on protein-coding genes transcribed by RNA polymerase II (Pol II), but not on genes transcribed by Pol I or Pol III (Supplemental Table S1), as determined by both SMRT-seq analysis and a separate GATC-qPCR experiment (Supplemental Fig. S4B). We found 6mA to be enriched on the 5′ end of the gene body (18,386 well-annotated genes) (Supplemental Fig. S4C), within 0.7 kb from the transcription start sites (TSSs) in both trophonts and cysts (Supplemental Fig. S4D). Moreover, 6mA showed a periodic distribution in linker DNA (∼50 bp) with a frequency of ∼200 bp and was strongly excluded from nucleosomal DNA (Supplemental Fig. S4D). This distribution pattern resembles that of 6mA in other ciliates (T. thermophila, P. bursaria, P. tetraurelia, and O. trifallax) (Wang et al. 2017a, 2019; Cheng et al. 2019; Hardy et al. 2021; Pan et al. 2023) and the algae Chlamydomonas reinhardtii (Fu et al. 2015) and may indicate a common 6mA distribution pattern among lower eukaryotes. We further investigated the location of symmetric methylation sites in trophonts that were retained or converted to asymmetric sites in cysts and found that both categories of methylation sites were enriched at the 5′ end of genes, which is consistent with the distribution pattern of total 6mA (Supplemental Fig. S4E). Further analysis revealed a global reduction in the degree of nucleosome positioning in cysts relative to trophic cells, in which cysts displayed more dispersed nucleosomes (Supplemental Fig. S4F). In both trophonts and cysts, 6mA sites were progressively enriched in linker DNA as their methylation levels increased, with the sharpest distribution upon methylation saturation (Supplemental Fig. S4G). However, 6mA distributions at the 30%–60% methylation levels were more diffuse in cysts than in trophonts (Supplemental Fig. S4G). These results revealed a correlation between 6mA and nucleosome positioning in two different morphological stages of P. persalinus.
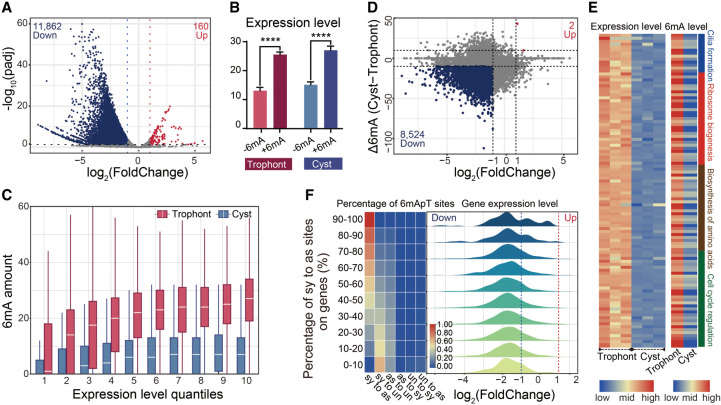
To determine if differences in 6mA patterns may explain why models of genetically identical P. persalinus display enormous gene expression changes during encystment, we used spike-in RNA-seq to determine which genes are differentially expressed between the two physiological states. As previously reported, minimal metabolic activity was observed in cysts, and numerous genes associated with biosynthesis, energy metabolism, cell growth, and division were shut down during the formation of resting cysts (Gutiérrez et al. 1990; Palacios et al. 1994; Jiang et al. 2019; Pan et al. 2019, 2020). In accordance with this observation, we noted a global reduction in gene expression. Out of 17,373 annotated genes, the vast majority (11,862) were down-regulated, and only 160 were up-regulated, in cysts compared with trophonts (Fig. 4A). To investigate the association between 6mA and gene expression, we classified genes by the presence or absence of 6mA and found that genes with 6mA have significantly higher expression levels than do genes without (Fig. 4B). We further divided genes into 10 quantiles based on their expression levels and found that the methylation level of 6mA was positively correlated with gene expression in both trophonts (P < 2.20 × 10−16, R = 0.35) and cysts (P < 2.20 × 10−16, R = 0.22) (Fig. 4C), supporting the notion that 6mA is a mark of active transcription (Fu et al. 2015; Mondo et al. 2017; Liang et al. 2018; Pan et al. 2023).
Figure 4.
The active transcriptional mark 6mA correlates with gene expression alterations. (A) Global transcriptome analysis in trophonts and cysts based on spike-in RNA-seq data. Differentially expressed genes (DEGs) are highlighted (log2(FoldChange) ≥ 1 or ≤−1, P ≤ 0.05; down-regulated in blue and up-regulated in red). (B) Statistical analysis of expression levels (FPKM) of genes with 6mA (+6mA) or without 6mA (−6mA) was performed for trophonts and cysts, respectively. (****) P < 0.0001 (Student's t-test). (C) 6mA amount in genes with different expression levels in trophonts and cysts. Genes are ranked from low to high by their expression levels (FPKM) and divided into 10 quantiles. (D) Plots of log2(FoldChange) and Δ6mA. Genes with up-regulated expression (log2(FoldChange) ≥ 1) and up-regulated 6mA (Δ6mA ≥ 10) are highlighted in red. Genes with down-regulated expression (log2(FoldChange) ≤ −1) and down-regulated 6mA (Δ6mA ≤ −10) are highlighted in blue. Δ6mA was defined as the 6mA site number in cysts minus 6mA in trophonts. (E) Correlation between expression (left) and 6mA (right) levels of genes associated with encystment. (F) All possible 6mApT conversions from trophont to cyst were counted in each gene: (sy to as) symmetric to asymmetric, (sy to un) symmetric to unmethylated, (as to sy) asymmetric to symmetric, (as to un) asymmetric to unmethylated, (un to sy) unmethylated to symmetric, and (un and as) unmethylated to asymmetric. Genes were divided into 10 quantiles by their proportion of symmetric to asymmetric conversion in all 6mApT conversions, and the proportion of each 6mApT conversion is shown in a heatmap (left). (Right) Log2(FoldChange) distribution for each group of genes.
This finding prompted us to further investigate the potential correlation between the global decrease in 6mA levels and the global decrease in gene expression during encystment. We focused on genes that showed alterations in both transcription and 6mA levels (log2(FoldChange) ≥ 1 or ≤−1, Δ6mA ≥ 10 or ≤−10) and identified a large number of 6mA-associated differentially expressed genes (DEGs). Among these 6mA-associated DEGs, 8524 showed concurrent down-regulation in both 6mA levels and gene expression, whereas only two displayed up-regulation in both (Fig. 4D). GO analysis of 6mA-associated DEGs indicated an enrichment in crucial signal transduction pathways, including those mediated by TP53, G protein-coupled receptors, and processes related to cell cycle regulation and cellular respiration metabolism (Supplemental Fig. S5A). Enriched terms were also associated with cellular structures, such as the P-body, mitochondria, mitotic spindle pole, and ciliary plasm, and enzyme activities, specifically protein–glycine ligase and acid–amino-acid ligase (Supplemental Fig. S5A). Notably, we also found genes associated with stress-response pathways to correlate with 6mA alterations, suggesting a direct involvement in cyst formation. For example, the down-regulation of genes involved in cilia formation, ribosome biogenesis, amino-acid biosynthesis, and cell cycle regulation were associated with a decrease in 6mA levels (Fig. 4E). Of note, these genes participate in stress-response pathways to reduce metabolic rates, arrest the cell cycle, and accelerate cilia shedding to form cysts (Jiang et al. 2019; Matsuoka 2021). We conclude that 6mA is associated with DEGs that may allow the organism to adapt to the drastic changes in morphology, physiology, and biochemical processes that accompany the encystment process.
Moreover, we found that 89% of asymmetric 6mApT sites on 6mA-associated DEGs were converted from symmetric 6mApT during encystment (Supplemental Fig. S5B) and that this conversion was more significant than that in the total pool of asymmetric 6mApTs in cysts (77.3%) (Fig. 3D). We therefore examined gene expression changes following the conversion of 6mA and found that a decrease in cyst 6mA was associated with a corresponding down-regulation in gene expression (Fig. 4F). Furthermore, a higher conversion rate (higher percentage of symmetric 6mApT sites converted to unmethylated) was linked to a greater likelihood of down-regulation in their corresponding genes (Fig. 4F). Hence, the conversion between symmetric and asymmetric 6mA may be a means of rapid gene regulation in response to environmental changes during cyst formation.
PpAMT1-KD reduces 6mA and accelerates cyst formation
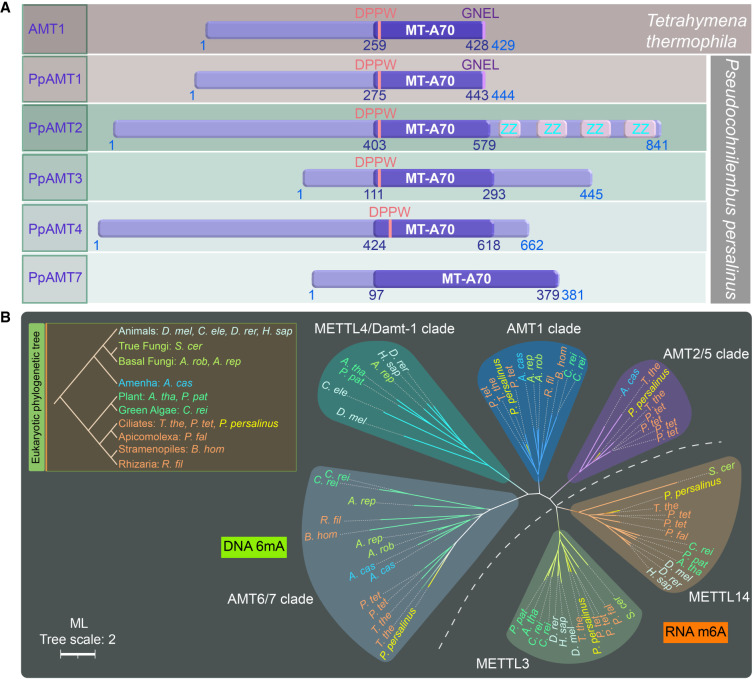
Given the fact that 6mA levels in cysts are significantly lower than in trophonts, we hypothesized that if 6mA levels were reduced, cyst formation would be accelerated. We therefore aimed to find the specific enzyme responsible for 6mA methylation in P. persalinus. Our previous study identified seven genes encoding for 6mA MTases containing the MT-A70 domain in T. thermophila (Wang et al. 2019). Likewise, we found five proteins from the MT-A70 family predicted to be putative MTases in P. persalinus, which we thereafter called P. persalinus DNA adenine MTases (PpAMTs) (Fig. 5A). Phylogenetic analysis of MTase orthologs showed that PpAMTs were well clustered together on each ortholog branch (Fig. 5B): PpAMT1, -3, and -4 in corresponding clades; PpAMT2 in the AMT2/5 clade; and PpAMT7 in the AMT6/7 clade (Fig. 5B; Wang et al. 2019). Sequence alignment and domain structure analysis revealed that PpAMT1–PpAMT4, but not PpAMT7, contain a 6mA MTase signature DPPW motif ([DNSH]PP[YFW]) required for substrate recognition and catalytic activity (Fig. 5A; Iyer et al. 2016).
Figure 5.
Phylogenetic analysis and domain structure comparison of PpAMT proteins. (A) Domain structures of PpAMT proteins. MT-A70 domains of PpAMT1–PpAMT4 were predicted by CD-Search, whereas that of PpAMT7 was inferred from sequence alignment with PpAMT1–PpAMT4. The structure of AMT1 (MT-A70 family methyltransferase in T. thermophila) is also shown for comparison. The MT-A70 domain, DPPW motif, and the conserved GNEL motif in AMT1 and PpAMT1 are highlighted. (B) Phylogenetic analysis of MT-A70 proteins. The DNA 6mA (clades AMT2/5, AMT1, METTL4/DAMT1, and AMT6/7) and RNA m6A (clades METTL3 and METTL14) methyltransferase candidates are separated by a dotted line. Species are marked by different colors based on their phylogenetic position in the eukaryotic tree. PpAMT1–PpAMT4 and PpAMT7 of P. persalinus are shown in yellow. The scale bar corresponds to two expected amino-acid substitution per site. For details, see Supplemental Table S7 (species full name and NCBI GenBank number).
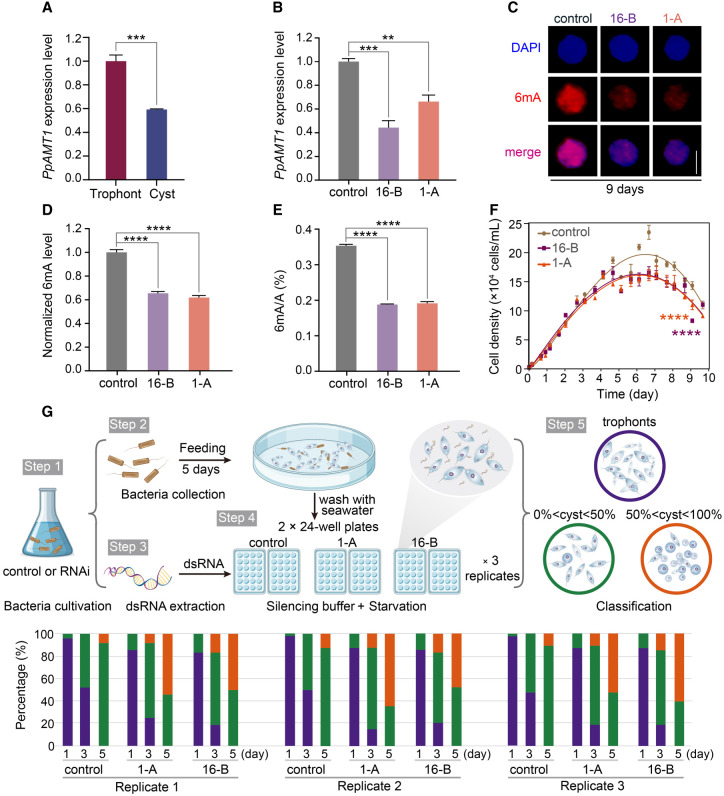
We previously reported that the knockout of the 6mA methyltransferase 1 (AMT1) in T. thermophila resulted in a significant reduction in 6mA levels and impaired cell fitness (Wang et al. 2019). Of note, PpAMT1 clusters within the AMT1 clade and contains a ciliate-specific GNEL motif at the C terminus (Fig. 5A; Wang et al. 2019; Pan et al. 2023). Most importantly, PpAMT1 expression significantly decreased during encystation, shown by both RT-qPCR and RNA-seq analyses (Fig. 6A; Supplemental Table S2). We therefore asked whether 6mA levels could be regulated by PpAMT1 in P. persalinus. We generated two independent RNAi plasmids (1-A and 16-B) targeting PpAMT1 sequences flanking the DPPW motif. Silencing was performed by feeding cells with E. coli containing RNAi plasmids that produce dsRNA. The dsRNA yields in three E. coli systems (two RNAi plasmids and one control plasmid) showed that our RNAi expression system had a high expression efficiency (Supplemental Fig. S6A). We then compared the silencing efficiency after 9 d of RNAi treatment by detecting the expression of PpAMT1 mRNAs using RT-qPCR, which revealed a strong effect after 9 d of silencing for both RNAi plasmids (Fig. 6B). These data show that the RNAi gene knockdown (KD) system can be readily applied in P. persalinus, which will constitute a powerful tool for studying the molecular mechanism of 6mA regulation.
Figure 6.
PpAMT1 knockdown (KD) reduces 6mA levels and accelerates encystation. (A) Expression levels of PpAMT1 in trophonts and cysts, calculated by normalized quantitative RT-PCR data. (***) P < 0.001 (Student's t-test). (B) Expression levels of PpAMT1 in control and PpAMT1-KD cells fed for 9 d. Expression levels were calculated by normalized quantitative RT-PCR data. (**) P < 0.01, (***) P < 0.001 (Student's t-test). (C) Immunofluorescence staining of 6mA in control and PpAMT1-KD cells fed for 9 d. Scale bar, 10 µm. (D) Statistical analysis of 6mA immunofluorescence images in C. (****) P < 0.0001 (Student's t-test). (E) Mass spectrometry analysis of 6mA levels in control and PpAMT1-KD cells fed for 9 d. 6mA ratio (6mA/A, %) was defined as the abundance of methylated adenine divided by the total number of adenine. (****) P < 0.0001 (Student's t-test). (F) Growth curves of control and PpAMT1-KD cells. Cell densities were counted using a counting chamber at indicated time points. (****) P < 0.0001 (two-way ANOVA test). (G) Assessment of cyst formation in control and PpAMT1-KD cells. (Top) Workflow of cell feeding, starvation, and RNAi treatment. (Bottom) Statistical results of encystment progression. Cells were fed with E. coli HT115 for 5 d, adding fresh bacteria on a daily basis. After washing with fresh seawater, cells were transferred to two 24-well plates and treated with silencing buffer containing dsRNA daily. The proportion of cysts in individual wells was counted at the indicated time points (n = 48 wells for each replicate) (Supplemental Table S3). The experiment was repeated three times under the same conditions.
6mA levels were assessed using IF and MS after 9 d of continuous RNAi. IF staining showed greatly reduced 6mA levels in the two PpAMT1-KD cultures (1-A and 16-B) compared with the control (Fig. 6C,D). This was further corroborated by the MS analysis, showing that global DNA 6mA levels in two biological replicates of PpAMT1-KD were reduced to ∼53%–54% (Fig. 6E). Of note, we observed no significant changes in RNA m6A levels by MS analysis in PpAMT1-KD cells (Supplemental Fig. S6B), consistent with our previous finding that DNA is the specific substrate for AMT1 clade MTases (Wang et al. 2019). The PpAMT1-KD cultures displayed a slow-growing phenotype that progressively worsened as RNAi continued, resembling the phenotype observed in T. thermophila after AMT1 knockout (Fig. 6F; Wang et al. 2019). It was previously reported that the 6mA MTase is a multisubunit MTase complex consisting of MTA1 (AMT1), MTA9 (AMT7), p1, and p2, each indispensable for catalyzing 6mA (Beh et al. 2019). To investigate whether this may also be the case in P. persalinus, we performed RNAi knockdown of PpAMT7, the P. persalinus homolog of MTA9 (AMT7), which also resulted in reduced levels of 6mA (Supplemental Fig. S6C,D). Consistent with its contribution, the expression level of PpAMT7 was reduced during encystment (Supplemental Table S2). These findings suggest that PpAMT1 and PpAMT7 may also act in a multisubunit MTase complex in P. persalinus.
To confirm the involvement of 6mA in cyst formation, we monitored the progression of encystment during RNAi treatment. The RNAi-treated cells were examined daily and the percentage of trophonts and cysts recorded. By day three of silencing, only ∼45% of the control wells had begun to form cysts (0% < cyst percentage < 50% in individual wells), whereas this number increased to ∼70% of the PpAMT1-KD wells, some of which had already reached a greater proportion of cysts (∼10% of wells with cyst percentage > 50% in individual wells), indicating that silencing of PpAMT1 induced premature encystation (Fig. 6G; Supplemental Fig. S6E; Supplemental Table S3). As the silencing continued, the differences between the RNAi treatments became more apparent: By day five, 50% of the PpAMT1-KD wells were induced into cysts with a greater proportion (cyst percentage > 50% in individual wells), whereas only ∼10% of the control wells had reached the same proportion (Fig. 6G; Supplemental Fig. S6E; Supplemental Table S3). Taken together, encystation was accelerated in PpAMT1-KD cells compared with control cells under the same starvation conditions, consistent with the highly reduced levels of PpAMT1 and 6mA in cysts (Fig. 6G; Supplemental Fig. S6E). These results suggest that 6mA in P. persalinus depends on the AMT1-clade MTase PpAMT1 and may be involved in regulating the encystment–excystment cycle.
Discussion
Encystment is an epigenetically regulated differentiation process that generates a resistant, differentiated resting cyst in response to stress (Gutiérrez et al. 1990; Pan et al. 2019, 2020, 2021). In this study, we investigated the distribution of 6mA in trophonts and cysts by generating a novel P. persalinus genome assembly with single-nucleotide resolution of 6mA sites. We found that 6mA is a dynamic modification with the following characteristics in the genomes of two different physiological states: (1) 6mA is enriched in complementary ApT dinucleotide motifs, distributed mainly as symmetric methylation sites in trophonts, which are then converted to asymmetric methylation sites in cysts; (2) 6mA is enriched at the 5′ end of the gene body downstream from TSSs and anti-correlates with nucleosomes in both trophonts and cysts; and (3) 6mA acts as an active transcriptional mark, is associated with RNA Pol II transcription, and is positively correlated with gene expression changes during cyst formation.
Comprehensive phylogenomic analyses revealed one potential 6mA MTase in P. persalinus, which belongs to the AMT1-clade: the conserved DNA 6mA MTase PpAMT1 (Fu et al. 2015; Wang et al. 2017a, 2019). By adapting and using RNAi to investigate its function, we found the level of 6mA to be significantly reduced in PpAMT1-KD cells, and that these cells displayed slower growth than the wild-type. These phenotypes resemble the AMT1-KO phenotype in Tetrahymena (Wang et al. 2019). We also observed a shortened encystment time of trophonts upon the knockdown of PpAMT1 and found that PpAMT1-dependent 6mA plays an important role in regulating genes involved in the encystment–excystment cycle, suggesting that the function of the AMT1 clade is conserved in ciliates. Moreover, the ability to perform efficient RNAi in P. persalinus to silence genes of interest, as we have shown using RNAi against PpAMT1, will pave the way for future studies to unravel the mechanism of the encystment–excystment cycle and gene regulation. Moving forward, we hope to develop more genetic manipulation techniques in P. persalinus, such as the application of a gene gun to knockout, rescue, or overexpress target genes.
We also found that the reduction of 6mA in cysts may be attributed to a cooperative effect of passive and active demethylation. In the former, we investigated whether encysting cells undergo DNA replication by performing BrdU labeling of trophonts and starved cells, the latter of which represent cells undergoing encystment. We found that DNA replication occurs during periods of starvation (Supplemental Fig. S7A,B), suggesting that symmetric 6mA sites in trophonts may be converted into asymmetric sites in cysts as a result of passive dilution (77.3% of asymmetric 6mApT sites in cysts are converted from the corresponding symmetric sites in trophonts) (Fig. 3D). To investigate the potential relationship between 6mA and DNA replication, we focused on symmetric sites in trophonts that were retained or converted to asymmetric sites in cysts and evaluated the frequency of symmetric and asymmetric 6mApT sites surrounding these methylation sites. Although the retained symmetric sites did not show a preference for clustering with symmetric or asymmetric sites, converted sites tended to cluster with other asymmetric sites (Supplemental Fig. S7C). This result indicates that passive dilution of 6mA is related to the DNA replication process, converting a substantial proportion of symmetric sites to asymmetric sites. Corroborating this finding, we found a significant reduction in the expression of the 6mA MTase PpAMT1 in cysts, which may contribute to the loss of methylation maintenance during encystment. However, reduced 6mA levels may also be caused by other factors, such as changes to the chromatin structure or DNA-binding proteins, which may contribute to the reduction of 6mA levels observed during the initial stage of encystment. Although passive demethylation may contribute to a dilution of 6mA, it is not sufficient to explain the global reduction in 6mA levels (332,133 in trophonts to 114,614 in cysts) (Fig. 3D). We therefore examined the outcomes of 6mA sites in trophonts during encystment in more detail and found 53% of symmetric 6mA sites in trophonts to convert to an unmethylated state in cysts (Supplemental Fig. S7D), suggesting active demethylation. The removal of 6mA from specific genomic regions or genes that are involved in the encystment process may facilitate encystation, which we speculate is achieved through the action of demethylases. We identified six putative demethylases through homologous sequence alignment with the AlkB family (Supplemental Table S2). However, the identity of demethylases has not been verified in ciliates, and the regulation of demethylase expression and the specific demethylation mechanisms remains to be elucidated. Moreover, the exact mechanism by which 6mA levels are regulated during encystment, including the potential contributions of both active and passive demethylation, needs further investigation. Taken together, our findings highlight the importance of unravelling the complex network of factors involved in DNA methylation maintenance and regulation of the encystment process.
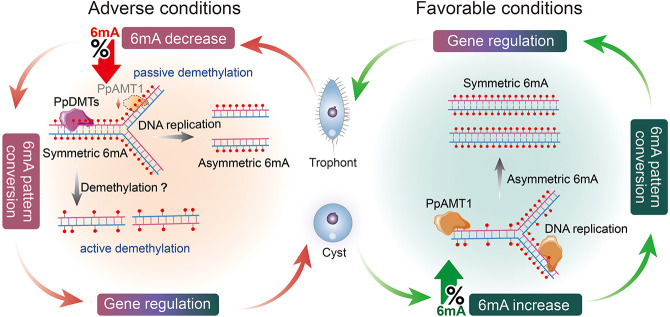
Based on our findings, we propose an encystment–excystment cycle model involving 6mA-regulated gene expression (Fig. 7): Once trophonts are induced by an exogenous trigger (e.g., starvation), passive demethylation of 6mA during DNA replication and/or active demethylation by an unknown demethyltransferase PpDMT is likely to occur. Additionally, the activity of the PpAMT1 MTase is also reduced. These changes result in a decrease in 6mA levels and conversion of symmetric 6mA to asymmetric and/or unmethylated 6mA (Fig. 7), causing subsequent gene expression alterations that promote cyst formation. Conversely, once cysts are induced by favorable conditions (e.g., nutrition), their DNA is rapidly methylated, resulting in an increase of 6mA and conversion of asymmetric 6mA to symmetric 6mA, which we speculate is catalyzed by the 6mA MTase PpAMT1 (Fig. 7). 6mA sites associated with excystment-related genes would be converted from asymmetric to symmetric in order to activate gene expression, which leads to alterations in the intracellular structure of cysts to rapidly recover the mobile cellular state (Fig. 7). The utilization of 6mA as a regulator of gene expression allows the cell to respond to unpredictable environments and may therefore constitute an essential adaptation strategy. This mechanism seems very plausible as it shortens the time required to regulate gene expression in response to environmental changes while simultaneously reducing excessive energy consumption by de novo methylation or complete demethylation. These results and speculation provide important information for uncovering the mechanisms underlying cyst formation and understanding how ciliates use the important epigenetic mark 6mA to respond to environmental changes.
Figure 7.
Proposed model of 6mA regulation in the encystment–excystment cycle. Encystment (left) is induced under unfavorable conditions such as starvation. During this process, 6mA levels are decreased and symmetric 6mA sites are converted to asymmetric and/or unmethylated sites, which results in gene expression alteration and promotes cyst formation. The global 6mA change is likely a combined effort of passive demethylation coupled with DNA replication and/or active demethylation by an unidentified demethylase (PpDMT), along with the reduced activity of the PpAMT1 methyltransferase. Excystment (right) occurs under favorable conditions. 6mA levels are increased and asymmetric 6mA sites are converted to symmetric sites, catalyzed by PpAMT1. The rapid conversion of 6mA sites facilitates the expression of excystment-related genes and helps cysts to recover their mobile state rapidly.
Methods
Cell culture and encystment induction
The scuticociliatid ciliate P. persalinus was collected from surface seawater in Qingdao, northern China (36°03′43′′ N; 120°19′12′′ E) at a water temperature of 24°C and a salinity of 30‰. The species was identified through its morphology (Liu et al. 2019) and through phylogenetic analysis based on the SSU rDNA sequence (Supplemental Fig. S8). To reduce the risk of contamination by bacterial 6mA, the dam−/dcm− E. coli strain (HST04, TaKaRa 9129) with DNA adenine and cytosine MTase deletions was used as a food source for P. persalinus (O'Brown et al. 2019). P. persalinus cells were cultured in sterilized sea water at 25°C in a stationary incubator (Liu et al. 2019). Encystment was induced by starving P. persalinus cells (3 d after the initiation of culture, ∼ × 105 cells/mL) in sterilized seawater for 1 wk. Living cells and cysts were observed using bright field and differential interference contrast microscopy at 100×–1000× magnification. Measurements were made under 1000× magnification with a calibrated ocular micrometer.
IF staining and imaging
IF staining of trophonts and cysts followed previously described procedures (Gao et al. 2013; Wang et al. 2017a, 2019; Zhao et al. 2017; Liu et al. 2019, 2021). Cells were fixed with 2% paraformaldehyde and permeabilized by the addition of 0.4% Triton X-100. For the BrdU staining, trophonts and starved cells were labeled with 0.4 mM BrdU for 2 h and treated with DNase (1:100, Invitrogen AM1907) for 15 min at room temperature. The primary antibodies used in this study were α-6mA (1:2000, Synaptic Systems 202003) and α-BrdU (1:1000, Rockland 600-401-C29), and the secondary antibody was goat antirabbit IgG (H + L) (1:4000, Invitrogen A-21428). Digital images were acquired using a ZEISS imager.M2 microscope with an Axiocam 506 color camera. The 6mA signal intensity of 100 cells for each sample was quantified using the image processing mode of ZEN 3.0 microscopy software (Zeiss).
Preparation of DNA and RNA samples
Genomic DNA was extracted from about 5 × 105 cells using phenol/chloroform/isoamyl alcohol following the manufacturer's instructions (Thermo Fisher Scientific 15593031). For cysts, a cell scraper was used to collect cells. Cysts were snap-frozen in liquid nitrogen, and grinding was used to disrupt the cyst structure and release the cellular components for extraction. The collected trophonts and cysts were filtered with a 40-µm cell strainer before extraction to remove big clusters of bacteria. Separating MAC and MIC in P. persalinus is difficult, making it hard to accurately determine the copy number of the MAC. Nonetheless, given its close relationship with other model ciliates like Tetrahymena and Paramecium (Xiong et al. 2015), it is plausible to assume that the MAC of P. persalinus also has a high ploidy level, far exceeding the diploid MIC. Considering that 6mA is a MAC-exclusive modification, the impact of MIC contamination on MAC is likely negligible.
Total RNA was extracted from about 3 × 105 cells using TRIzol (Invitrogen 15596026) (Rio et al. 2010). To assess the global transcriptional level changes between cysts and trophonts, we conducted spike-in RNA-seq as follows: cysts and trophonts were mixed with T. thermophila cells (CU428) at a ratio of 10:1 based on cell number, followed by RNA extraction. The quality and concentration of the DNA and RNA samples were analyzed by agarose gel electrophoresis, a NanoDrop One/OneC microvolume spectrophotometer, and a Qubit 4 fluorometer.
Cloning and plasmid construction for RNAi
Total RNA was extracted using TRIzol as described above. cDNA was synthesized with the RevertAid first strand cDNA synthesis kit (Thermo Fisher Scientific K1622). The oligo(dT) primer was used to reduce bacterial contamination in the samples because bacterial mRNAs lack polyadenine tails (Lakey et al. 2002). Two RNAi plasmids containing sequences of different lengths, namely, 1-A (298 bp) and 16-B (409 bp), were constructed to specifically target the conserved DPPW motif of PpAMT1. Sequences of PpAMT1 and PpAMT7 (Supplemental Table S4) used for silencing the target gene were amplified from cDNA, with the primers listed in Supplemental Table S5. The purified fragment was ligated into the L4440 plasmid that contains two inverted T7 promoters using ClonExpress ultra one step cloning kit (Vazyme Biotech C115-02) and then transformed into DH5α competent cells (Tsingke Biotechnology TSC-C14). Positive clones were selected for plasmid extraction. Subsequently, the RNAi plasmid was transformed into HT115 competent cells (Shanghai Weidi Biotechnology EC2010). Positive clones were cultured for the RNAi experiment.
Purification of expressed double-stranded RNA
The E. coli HT115 RNAi system was grown in a 37°C shaking incubator under the selection of ampicillin. The production of dsRNA was induced by the addition of 0.4 mM IPTG (Sigma-Aldrich I5502) to 5 mL cultures when they reached OD600 = 0.5 and further incubated for 4 h. Total RNA was extracted using TRIzol as described above. The extracted RNA was resuspended in 20 μL of DEPC-treated water and the quality assessed by 2% agarose gel electrophoresis.
RNAi-mediated knockdown
Fresh control and RNAi feeding E. coli were cultured and induced by IPTG to express dsRNA of the target gene as described above. After induction, bacteria were washed and resuspended in 0.5 mL sterile filtered seawater and then fed to trophonts that had been previously starved for 12–24 h. Induction and feeding with bacteria were repeated daily for 6–9 d. The negative control used for RNAi experiments was an empty vector.
Statistical analysis of the cyst formation ability
To monitor the ability of cyst formation after continuous RNAi for 5 d, PpAMT1-KD (1-A and 16-B) and control (empty vector) cells were collected and washed once with fresh sterile seawater. The samples were equally divided into 24-well plates (500 μL/well, 48 wells for one replicate), and placed in a 25°C incubator for static culture. Each well was supplemented with silencing buffer (3 mg/mL dsRNA and 50 μM spermidine) every day, and the proportion of cysts in individual wells was calculated. Cells in individual wells were categorized in three states: trophonts, 0% < cyst < 50%, and 50% < cyst < 100%. For detailed statistics, see Supplemental Table S3. The proportion of cysts in individual wells was assigned a score for significant differences analysis, zero for trophonts, one for 0% < cyst < 50%, and two for 50% < cyst < 100%.
Dot blot
Dot blot was performed as previously described (Wang et al. 2017a). Briefly, genomic DNA was treated with RNase A (Thermo Fisher Scientific R4642) to prevent RNA m6A contamination, and DNA was denatured at 95°C before spotting on an Amersham Hybond-N+ membrane (GE Healthcare RPN303B). The membrane was baked at 80°C and DNA cross-linked to the membrane with ultraviolet light (2000 mJ/cm2) before incubation with an α-6mA antibody (1:2000, Synaptic Systems 202003) for 1 h at room temperature. After three washes with PBST, the membrane was incubated with a 1:4000 dilution of an HRP-conjugated antirabbit IgG secondary antibody for 1 h at room temperature. Methylene blue hydrate (Molecular Research Center MB119) staining was performed to determine the amount of input DNA.
UHPLC–QQQ–MS/MS analysis
UHPLC–QQQ–MS/MS analysis followed previously described procedures (Wang et al. 2019). Genomic DNA was enzymatically digested into single nucleosides using a mixture of enzymes, including DNase I (1 U, New England Biolabs M0303L), FastAP (1 U, thermosensitive alkaline phosphatase, Thermo Fisher Scientific EF0651), and phosphodiesterase I (0.005 U, Sigma-Aldrich P4506), for 12 h at 37°C. Total RNA was enzymatically digested into single nucleosides using 1 μL nuclease P1 (100 U, New England Biolabs M0660S) and 5 μL 100 mM NH4OAc for 12 h at 37°C. Following digestion, enzymes were removed by ultrafiltration, and the digested samples were analyzed by ultra-high-performance liquid chromatography tandem MS on an Acquity BEH C18 column (100 mm × 2.1 mm, 1.7 μm, Waters), using a Xevo TQ-XS triple-quadrupole mass spectrometer (Waters). The mass spectrometer was set to multiple reaction monitoring (MRM) in positive electrospray mode. The selective MRM transitions were detected under m/z 266/150 for 6mA (DNA), m/z 252/136 for A (DNA), m/z 282/150 for m6A (RNA), and m/z 268/136 for A (RNA). The ratio of 6mA/A (DNA) or m6A/A (RNA) was quantified by the calibration curves of nucleoside standards running in parallel.
MNase-seq sample preparation and analysis
Approximately 5 × 106 cells were incubated in lysis buffer (0.25 M sucrose, 10 mM MgCl2, 10 mM Tris-HCl at pH 7.4, 1 mM dithiothreitol [DTT], 0.2% NP-40, 0.1 mg/mL protease inhibitor, and 1 mM PMSF) and homogenized with a douncer for 5 min until all cells were confirmed lysed through microscopic examination. The nuclei were washed once with 1 mL lysis buffer without NP-40 and once with cold MNase buffer (50 mM Tris-HCl at pH 8.0, 5 mM CaCl2, 1 mM DTT, 0.2% NP-40). The chromatin was digested with 800 U MNase for 15 min at 25°C on a rotor. Digested DNA was collected by phenol-chloroform extraction for sequencing. After mapping the reads, only mono-nucleosome-size fragments (120 bp–260 bp) were used for the analysis. Nucleosome dyads were defined as previously reported to calculate the nucleosome distribution around TSSs (Wang et al. 2017a). The degree of nucleosome positioning was calculated as previously described and defined as “the number of fragment centers within ±20 bp of the called nucleosome dyad, relative to the number of all fragment centers within the 147 bp called nucleosome footprint” (Xiong et al. 2016; Wang et al. 2019).
Quantitative reverse transcription PCR
Total RNA after DNase I treatment (New England Biolabs M0303L) was reverse-transcribed using the RevertAid first strand cDNA synthesis kit (Thermo Fisher Scientific K1621). RT-PCR was performed using the 2× Phanta max master mix (Vazyme Biotech P525). Quantitative reverse transcription PCR (qRT-PCR) was performed in a 96-well plate (Applied Biosystems) with 12.5 μL SYBR green PCR mix (Invitrogen), 5 μL primers (forward and reverse 1 μmol/L), and 7.5 μL cDNA, in a total volume of 25 μL. Parameters for qPCR were as followed: 2 sec at 50°C; 10 sec at 95°C; 40 cycles of 2 sec at 95°C, 1 sec at 50°C, and 1 sec at 60°C; followed by a dissociation step. The constitutively expressed gene g10245 (a gene with stable expression between trophonts and cysts in RNA-seq data) (Supplemental Table S4) was used as an internal control and for normalization. This gene is a homolog of Bax inhibitor-1 (BI-1). The linear standard curve was generated by plotting the cycle threshold (Ct) values of the standard versus the log of the cDNA template concentration (Supplemental Table S6). For primers, see Supplemental Table S5.
DpnI/DpnII digestion and quantitative PCR analysis
DpnI/DpnII digestion and quantitative PCR (qPCR) analysis followed previously described procedures (Wang et al. 2017a). Approximately 2 μg of purified genomic DNA was treated with 40 U DpnI (New England Biolabs R0176V) overnight or with 40 U DpnII (New England Biolabs R0543L) overnight at 37°C. Samples were heat-inactivated for 20 min at 80°C (DpnI) or 65°C (DpnII) after digestion. A total of 4 ng digested and nondigested DNA was subjected to qPCR analysis using the PowerUp SYBR green master mix (Thermo Fisher Scientific A25741). Primers flanking selected GATC sites were used to validate the methylation status at specific positions (Supplemental Table S5). Primers matching the coding DNA sequence (CDS) of the constitutively expressed gene g10245 were used as an internal control. Methylation status was assessed by the normalized Ct differences (ΔCt) between digested and undigested samples.
Phylogenetic analysis of PpAMTs and PpDMTs
The AMT1–AMT7 amino-acid sequences of T. thermophila were queried against the P. persalinus proteins to identify putative MTases by PSI-BLAST v2.9.0+ (Altschul et al. 1997). The AlkB, ALKBH1, ALKBH4, and ALKBH5 amino-acid sequences were queried against the P. persalinus proteins to identify the putative demethyltransferases using the same method. The resulting sequences of putative MTases were used to perform alignments of MT-A70 candidates from Wang et al. (2019). Retrieved hits were collapsed to remove redundant sequences using CD-HIT (Li and Godzik 2006), and the sequences were aligned by MUSCLE (Edgar 2004; https://www.ebi.ac.uk/Tools/msa/muscle/). The phylogenetic tree was constructed using the approximate maximum-likelihood method implemented on the CIPRES Science Gateway v.3.3 (Stamatakis et al. 2008; Miller et al. 2010; http://www.phylo.org/subsections/portal). For all protein sequences used for the phylogenetic analysis, see in Supplemental Table S7. The NCBI conserved domain search (Marchler-Bauer and Bryant 2004; https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) was used to predict the conserved domain of proteins. IBS v1.0 (Liu et al. 2015) was used to plot the sketch.
Genome assembly
Genome assembly followed previously reported methods (Sheng et al. 2020). SMRT and Illumina sequencing libraries of P. persalinus were constructed according to manufacturer-recommended protocols and sequenced by Novogene. Previously published Illumina sequencing and RNA-seq data of P. persalinus were retrieved from the PPGD 2015 (Wei et al. 2018; http://ciliates.ihb.ac.cn/database/home/#pp). Jellyfish version 1.1.11 (Marçais and Kingsford 2011) was used to produce frequency distributions of 17-, 21-, 25-, and 31-mers for all Illumina reads, and genome scope version 1 (Vurture et al. 2017) was used to estimate the genome size. Bacterial contamination in cysts was removed by BLAST to the NCBI bacteria nucleic acid database before genome assembly. SMRT sequencing reads of trophonts (257×) and cysts (137×), with an average subread length of 15.4 kb and 8.6 kb, were assembled using Canu into preliminary assemblies consisting of 1077 and 1518 contigs (Koren et al. 2017). Subsequently, repetitive contigs and contigs with low mapped reads were removed by personal custom Perl scripts (Supplemental Code). Telomeres were identified by searching contigs for exact matches to a 12-mer encompassing two telomeric repeats (GGGGTTGGGGTT or CCCCAACCCCAA), and the remaining 170 contigs were separated into two groups: 114 contigs with telomeres on both sides and 56 contigs with telomeres on only one side or no telomeres on either side. Among these 56 contigs, 46 contigs were completed by BLAST to the SMRT subreads. PCR primers (Supplemental Table S5) were designed for another 10 contigs to amplify the uncompleted sequences and were sequenced by Sanger sequencing, and five contigs were completed. Finally, PE150 reads from Illumina sequencing were imported to Pilon to correct the draft genome (Walker et al. 2014). The assembly workflow is shown in Supplemental Figure S1A.
Gene prediction and annotation
To identify protein-coding genes, de novo gene prediction, homology-based, and transcriptome-based methods were combined using the LoReAn2 annotation pipeline (Cook et al. 2019). In detail, the transcriptomic data were aligned to the genome using the Program to Assemble Spliced Alignments (PASA) (Haas et al. 2003) and the Genomic Mapping and Alignment Program (GMAP) (Wu and Watanabe 2005). For protein alignment, the analysis and annotation tool (AAT) (Huang et al. 1997) was used to align protein sequences to the genome. For RNA-seq alignment, genome-guided transcripts were reconstructed by Trinity (Grabherr et al. 2011). For ab initio gene prediction, the Evidence Modeler (EVM) (Haas et al. 2008) was used to combine the outputs of SNAP (Korf 2004), AUGUSTUS (Stanke and Morgenstern 2005), and GeneMark-ET (Lomsadze et al. 2014) to generate the combined annotation model. PASA was used to update the gene models by identifying UTRs to generate an initial annotation. The UTR information was further examined and supplemented by customized Perl scripts (Supplemental Code) using RNA-seq data. The final genome annotation was obtained by further refining the initial annotation using protein MS data. All predicted proteins were automatically annotated using eggNOG-mapper v2 (Cantalapiedra et al. 2021) against eggNOG 5.0 (Huerta-Cepas et al. 2017) using DIAMOND v2.0.4 (Buchfink et al. 2015) and the SWISSPROT database. The annotation workflow is shown in Supplemental Figure S1B.
SMRT sequencing data analysis
Our updated P. persalinus genome was used as a reference for read mapping. 6mA was identified using the base modification and motif analysis protocol with default parameters in the SMRT link v5.10 (Pacific Biosciences). All data were normalized to 100× while using strict cut-offs (Qv >30 and coverage >25) to filter out unauthentic modifications. As previously defined (Wang et al. 2017a), 6mA was divided into different groups according to their methylation levels (0%–100%) or motifs (symmetric/asymmetric/non-ApT). All 6mA calculations of sites and percentage were implemented by customized Perl scripts (Supplemental Code), and diagrams were plotted by GraphPad Prism 7.0 (Mitteer et al. 2018). The 6mA Circos plot was generated using the R package circlize v0.4.4 (Gu et al. 2014). The 6mA ratio was defined as the number of methylated adenine sites divided by total adenine sites (6mA/A). The density plot of 6mA methylation levels on Watson and/or Crick strands was generated by the R package ggplot2 (Wickham 2009). To analyze the distribution of 6mA, we scaled the gene body length to unit length and extended one unit to each side for locus statistical analysis by customized Perl scripts (bin size = 0.05) (Supplemental Code). The number of 6mA sites was counted from 500 bp upstream of to 2000 bp downstream from TSSs to explore patterns of 6mA distribution. Sequences from 5 bp upstream of to 5 bp downstream from the methylated adenines were selected, and conserved motifs were identified by WebLogo3 (Crooks et al. 2004). RNA polymerase I (Pol I) and III (Pol III) transcribed genes were detected in the P. persalinus genome using RNAmmer v1.2 (Lagesen et al. 2007), tRNAscan-SE v2.0.5 (Chan and Lowe 2019), and Rfam v11.0 (Kalvari et al. 2020) as previously described (Wang et al. 2017a).
Spike-in RNA-seq and data analysis
Three replicates each of trophont and cyst RNA, mixed with T. thermophila RNA, were sequenced at Novogene. Trimming of sequencing adapters and filtering of low-quality reads were performed using Trimmomatic (Bolger et al. 2014). Remaining reads were mapped to the P. persalinus genome and the T. thermophila genome using HISAT2 (Kim et al. 2019). StringTie 1.3.4 (Pertea et al. 2015) was used for assembling potential transcripts as a reference, and featureCounts (Liao et al. 2013) was implemented for counting reads to genomic features with the assembled transcripts as the reference. Effective expression levels (FPKM > 1) were calculated based on RNA-seq coverage using DESeq2 (Love et al. 2014). DEGs were identified by DESeq2 (log2(FoldChange) > 1 or <−1, P < 0.05) (Love et al. 2014). Gene Ontology (GO) enrichment analysis of DEGs was performed using the using TBtools (Chen et al. 2023). The number of reads in each sample were used to calculate size factors (normalized to 109) and perform normalized differential expression analysis using DESeq2 with the parameters (sizeFactor(data) < − c()) instead of default parameters.
Statistical analysis
Significance was calculated using a one-sided Student's t-test and ordinary two-way ANOVA multiple comparison tests. Asterisks indicate statistical significance in comparison with controls: (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, and (****) P < 0.0001. Plotted results were based on the average of at least three biological replicates. Plots were generated using GraphPad prism 7.0 (GraphPad Software) (Mitteer et al. 2018).
Data access
The SMRT sequencing, MNase-seq, and RNA-seq data of P. persalinus trophonts and cysts generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA989345. The genome, genome annotation files, and the associated custom Perl scripts generated in this study are available at GitHub (https://github.com/yefei521/pper-6mA.git) and as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Dr. Dahua Chen (Yunnan University) for the convenience provided in performing the relevant experiments, Dr. Wenxin Zhang (Yunnan University) for assistance with the UHPLC–QQQ–MS/MS experiments, Dr. Yalan Sheng (Hong Kong Baptist University) and Dr. Xiao Chen (Shandong University) for their kind suggestions in data analysis, Dr. Xinpeng Fan (East China Normal University) for gifting the P. persalinus images, three anonymous reviewers for their comments, and Dr. Weibo Song (Ocean University of China, OUC) for his suggestions during the manuscript drafting. We appreciate the computing resources provided by the Institute of Evolution and Marine Biodiversity in OUC, the Center for High Performance Computing and System Simulation at Laoshan Laboratory, and the Marine Big Data Center of Institute for Advanced Ocean Study of OUC. This work was supported by the Natural Science Foundation of China (32125006, 32070437), the Science and Technology Innovation Project of Laoshan Laboratory (no. LSKJ202203203), the Postdoctoral Fellowship Program of the China Postdoctoral Science Foundation (CPSF) under Grant Number GZC20232503), and the World Premier International Research Center Initiative (WPI), MEXT, Japan. Y.L. was supported by the China Scholarship Council (NSCIS no. 202106330059) for exchange PhD study at the University of Bern, Switzerland.
Author contributions: S.G., Y.L., and J.N. conceived the study; Y.L. and J.N. performed the experiments; J.N. and F.Y. performed the data analysis; T.S. and M.N. reviewed and edited the manuscript; B.L. provided the P. persalinus images; C.W. participated in cell culture and RNA extraction; and Y.L., J.N., F.Y., and S.G. wrote and revised the manuscript. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.278796.123.
Competing interest statement
The authors declare no competing interests.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh LY, Debelouchina GT, Clay DM, Thompson RE, Lindblad KA, Hutton ER, Bracht JR, Sebra RP, Muir TW, Landweber LF. 2019. Identification of a DNA N6-adenine methyltransferase complex and its impact on chromatin organization. Cell 177: 1781–1796.e25. 10.1016/j.cell.2019.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12: 59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol 38: 5825–5829. 10.1093/molbev/msab293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. In Gene prediction: methods and protocols (ed. Kollmar M), Vol. 1962, pp. 1–14. Humana, New York. 10.1007/978-1-4939-9173-0_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Wu Y, Li J, Wang X, Zeng Z, Xu J, Liu Y, Feng J, Chen H, He Y, et al. 2023. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining. Mol Plant 16: 1733–1742. 10.1016/j.molp.2023.09.010 [DOI] [PubMed] [Google Scholar]
- Cheng T, Wang Y, Huang J, Chen X, Zhao X, Gao S, Song W. 2019. Our recent progress in epigenetic research using the model ciliate, Tetrahymena thermophila. Mar Life Sci Technol 1: 4–14. 10.1007/s42995-019-00015-0 [DOI] [Google Scholar]
- Cook DE, Valle-Inclan JE, Pajoro A, Rovenich H, Thomma B, Faino L. 2019. Long-read annotation: automated eukaryotic genome annotation based on long-read cDNA sequencing. Plant Physiol 179: 38–54. 10.1104/pp.18.00848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5: 113. 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenchel T. 1990. Adaptive significance of polymorphic life cycles in protozoa: responses to starvation and refeeding in two species of marine ciliates. J Exp Mar Biol Ecol 136: 159–177. 10.1016/0022-0981(90)90159-A [DOI] [Google Scholar]
- Fouque E, Trouilhé MC, Thomas V, Hartemann P, Rodier MH, Héchard Y. 2012. Cellular, biochemical, and molecular changes during encystment of free-living amoebae. Eukaryot Cell 11: 382–387. 10.1128/EC.05301-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouque E, Yefimova M, Trouilhé MC, Quellard N, Fernandez B, Rodier MH, Thomas V, Humeau P, Héchard Y. 2015. Morphological study of the encystment and excystment of Vermamoeba vermiformis revealed original traits. J Eukaryot Microbiol 62: 327–337. 10.1111/jeu.12185 [DOI] [PubMed] [Google Scholar]
- Fu Y, Luo GZ, Chen K, Deng X, Yu M, Han D, Hao Z, Liu J, Lu X, Doré LC,et al. 2015. N6-Methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161: 879–892. 10.1016/j.cell.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Xiong J, Zhang C, Berquist BR, Yang R, Zhao M, Molascon AJ, Kwiatkowski SY, Yuan D, Qin Z, et al. 2013. Impaired replication elongation in Tetrahymena mutants deficient in histone H3 Lys 27 monomethylation. Genes Dev 27: 1662–1679. 10.1101/gad.218966.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z-W, Fan X-P, Ma R, Ni B. 2018. Ultrastructure of vegetative cells and resting cysts, and live observations of the encystation and excystation processes in Diophrys oligothrix Borror, 1965 (Protista, Ciliophora). J Morphol 279: 1397–1407. 10.1002/jmor.20851 [DOI] [PubMed] [Google Scholar]
- Gorovsky MA, Hattman S, Pleger GL. 1973. N6-Methyl adenine in the nuclear DNA of a eucaryote, Tetrahymena pyriformis. J Cell Biol 56: 697–701. 10.1083/jcb.56.3.697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisvard J, Lemullois M, Morin L, Baroin-Tourancheau A. 2008. Differentially expressed genes during the encystment–excystment cycle of the ciliate Sterkiella histriomuscorum. Eur J Protistol 44: 278–286. 10.1016/j.ejop.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, Brors B. 2014. circlize implements and enhances circular visualization in R. Bioinformatics 30: 2811–2812. 10.1093/bioinformatics/btu393 [DOI] [PubMed] [Google Scholar]
- Gutiérrez JC, Martin-Gonzalez A, Matsusaka T. 1990. Towards a generalized model of encystment (cryptobiosis) in ciliates: a review and a hypothesis. BioSystems 24: 17–24. 10.1016/0303-2647(90)90025-V [DOI] [PubMed] [Google Scholar]
- Gutiérrez JC, Martin-González A, Callejas S. 1998. Nuclear changes, macronuclear chromatin reorganization and DNA modifications during ciliate encystment. Eur J Protistol 34: 97–103. 10.1016/S0932-4739(98)80018-7 [DOI] [Google Scholar]
- Gutierrez JC, Callejas S, Borniquel S, Martin-Gonzalez A. 2000. DNA methylation in ciliates: implications in differentiation processes. Int Microbiol 3: 139–146. [PubMed] [Google Scholar]
- Gutierrez JC, Callejas S, Borniquel S, Benítez L, Martin-Gonzalez A. 2001. Ciliate cryptobiosis: a microbial strategy against environmental starvation. Int Microbiol 4: 151–157. 10.1007/s10123-001-0030-3 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31: 5654–5666. 10.1093/nar/gkg770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol 9: R7. 10.1186/gb-2008-9-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy A, Matelot M, Touzeau A, Klopp C, Lopez-Roques C, Duharcourt S, Defrance M. 2021. DNAModAnnot: a R toolbox for DNA modification filtering and annotation. Bioinformatics 37: 2738–2740. 10.1093/bioinformatics/btab032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K-W, Lai JC-Y, Chang J-S, Peng P-H, Huang C-H, Lee D-Y, Tsai Y-C, Chung C-J, Chang H, Chang C-H, et al. 2022. METTL4-mediated nuclear N6-deoxyadenosine methylation promotes metastasis through activating multiple metastasis-inducing targets. Genome Biol 23: 249. 10.1186/s13059-022-02819-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Adams MD, Zhou H, Kerlavage AR. 1997. A tool for analyzing and annotating genomic sequences. Genomics 46: 37–45. 10.1006/geno.1997.4984 [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34: 2115–2122. 10.1093/molbev/msx148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Zhang D, Aravind L. 2016. Adenine methylation in eukaryotes: apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays 38: 27–40. 10.1002/bies.201500104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Wei W, Yan G, Shi T, Miao W. 2019. Transcriptome analysis reveals the molecular mechanism of resting cyst formation in Colpoda aspera. J Eukaryot Microbiol 66: 212–220. 10.1111/jeu.12643 [DOI] [PubMed] [Google Scholar]
- Jin D, Li C, Chen X, Byerly A, Stover NA, Zhang T, Shao C, Wang Y. 2023. Comparative genome analysis of three euplotid protists provides insights into the evolution of nanochromosomes in unicellular eukaryotic organisms. Mar Life Sci Technol 5: 300–315. 10.1007/s42995-023-00175-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Nawrocki EP, Ontiveros-Palacios N, Argasinska J, Lamkiewicz K, Marz M, Griffiths-Jones S, Toffano-Nioche C, Gautheret D, Weinberg Z, et al. 2020. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res 49: D192–D200. 10.1093/nar/gkaa1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer KM. 2012. Nuclear dualism. In Methods in cell biology (ed. Collins K), Vol. 109, pp. 29–52. Academic Press, San Francisco. [DOI] [PubMed] [Google Scholar]
- Kaur H, Iqbal S, Inga E, Yawe D. 2019. Encystment and excystment in ciliated protists: multidimensional approach. Curr Sci 117: 198–203. 10.18520/cs/v117/i2/198-203 [DOI] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinform 5: 59. 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Seem K, Kumar S, Vinod KK, Chinnusamy V, Mohapatra T. 2022. Pup1 QTL regulates gene expression through epigenetic modification of DNA under phosphate starvation stress in rice. Front Plant Sci 13: 871890. 10.3389/fpls.2022.871890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35: 3100–3108. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakey DL, Zhang Y, Talaat AM, Samten B, DesJardin LE, Eisenach KD, Johnston SA, Barnes PF. 2002. Priming reverse transcription with oligo(dT) does not yield representative samples of Mycobacterium tuberculosis cDNA. Microbiology 148: 2567–2572. 10.1099/00221287-148-8-2567 [DOI] [PubMed] [Google Scholar]
- Leadbeater BS, Karpov SA. 2000. Cyst formation in a freshwater strain of the choanoflagellate Desmarella moniliformis kent. J Eukaryot Microbiol 47: 433–439. 10.1111/j.1550-7408.2000.tb00071.x [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- Li Y, Wang Y, Zhang S, Maurer-Alcalá XX, Yan Y. 2022. How ciliated protists survive by cysts: some key points during encystment and excystment. Front Microbiol 13: 785502. 10.3389/fmicb.2022.785502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Shen L, Cui X, Bao S, Geng Y, Yu G, Liang F, Xie S, Lu T, Gu X, et al. 2018. DNA N6-adenine methylation in Arabidopsis thaliana. Dev Cell 45: 406–416.e3. 10.1016/j.devcel.2018.03.012 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2013. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lindblad KA, Pathmanathan JS, Moreira S, Bracht JR, Sebra RP, Hutton ER, Landweber LF. 2019. Capture of complete ciliate chromosomes in single sequencing reads reveals widespread chromosome isoforms. BMC Genomics 20: 1037. 10.1186/s12864-019-6189-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y, Zhao Y, et al. 2015. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31: 3359–3361. 10.1093/bioinformatics/btv362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Nan B, Duan L, Cheng T, Bourland WA, Liu M, Zhao Y. 2019. A simple and rapid cryopreservation technique for ciliates: a long-term storage procedure used for marine scuticociliates. J Eukaryot Microbiol 66: 836–848. 10.1111/jeu.12730 [DOI] [PubMed] [Google Scholar]
- Liu Y, Nan B, Niu J, Kapler GM, Gao S. 2021. An optimized and versatile counter-flow centrifugal elutriation workflow to obtain synchronized eukaryotic cells. Front Cell Dev Biol 9: 664418. 10.3389/fcell.2021.664418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A, Burns PD, Borodovsky M. 2014. Integration of mapped RNA-seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res 42: e119–e119. 10.1093/nar/gku557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Niu R, Huang T, Shao LW, Peng Y, Ding W, Wang Y, Jia G, He C, Li CY, et al. 2019. N6-Methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat Cell Biol 21: 319–327. 10.1038/s41556-018-0238-5 [DOI] [PubMed] [Google Scholar]
- Marçais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764–770. 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32: W327–W331. 10.1093/nar/gkh454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T. 2021. Early signaling pathways mediating dormant cyst formation in terrestrial unicellular eukaryote Colpoda. FEMS Microbiol 368: fnab019. 10.1093/femsle/fnab019 [DOI] [PubMed] [Google Scholar]
- Matsusaka T, Noguchi O, Yonezawa F. 1989. Cortical morphogenesis during encystment in a ciliate, Euplotes encysticus Yonezawa, 1985. Eur J Protistol 24: 133–137. 10.1016/S0932-4739(89)80041-0 [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), pp. 1–8. IEEE, New Orleans. 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Mitteer DR, Greer BD, Fisher WW, Cohrs VL. 2018. Teaching behavior technicians to create publication-quality, single-case design graphs in graphpad prism 7. J Appl Behav Anal 51: 998–1010. 10.1002/jaba.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondo SJ, Dannebaum RO, Kuo RC, Louie KB, Bewick AJ, LaButti K, Haridas S, Kuo A, Salamov A, Ahrendt SR, et al. 2017. Widespread adenine N6-methylation of active genes in fungi. Nat Genet 49: 964–968. 10.1038/ng.3859 [DOI] [PubMed] [Google Scholar]
- Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. 2008. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature 451: 153–158. 10.1038/nature06452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brown ZK, Boulias K, Wang J, Wang SY, O'Brown NM, Hao Z, Shibuya H, Fady PE, Shi Y, He C, et al. 2019. Sources of artifact in measurements of 6mA and 4mC abundance in eukaryotic genomic DNA. BMC Genomics 20: 445. 10.1186/s12864-019-5754-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olendzenski LC. 1999. Growth, fine structure and cyst formation of a microbial mat ciliate: Pseudocohnilembus pusillus (Ciliophora, Scuticociliatida). J Eukaryot Microbiol 46: 132–141. 10.1111/j.1550-7408.1999.tb04596.x [DOI] [Google Scholar]
- Palacios G, Martin-Gonzalez A, Gutierrez JC. 1994. Macronuclear DNA demethylation is involved in the encystment process of the ciliate Colpoda inflata. Cell Biol Int 18: 223–228. 10.1006/cbir.1994.1067 [DOI] [PubMed] [Google Scholar]
- Pan N, Niu T, Bhatti MZ, Zhang H, Fan X, Ni B, Chen J. 2019. Novel insights into molecular mechanisms of Pseudourostyla cristata encystment using comparative transcriptomics. Sci Rep 9: 19109. 10.1038/s41598-019-55608-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Bhatti MZ, Zhang H, Ni B, Fan X, Chen J. 2020. The encystment-related microRNAs and its regulation molecular mechanism in Pseudourostyla cristata revealed by high throughput small RNA sequencing. Int J Mol Sci 21: 2309. 10.3390/ijms21072309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Bhatti MZ, Zhang W, Ni B, Fan X, Chen J. 2021. Transcriptome analysis reveals the encystment-related lncRNA expression profile and coexpressed mRNAs in Pseudourostyla cristata. Sci Rep 11: 8274. 10.1038/s41598-021-87680-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Ye F, Wei F, Warren A, Wang Y, Gao S. 2023. Role of N6-adenine DNA methylation in alternative splicing and endosymbiosis in the unicellular eukaryote Paramecium bursaria. iScience 26: 106676. 10.1016/j.isci.2023.106676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A, Cedar H. 1991. DNA methylation and gene expression. Microbiol Rev 55: 451–458. 10.1128/mr.55.3.451-458.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repak AJ. 1968. Encystment and excystment of the heterotrichous ciliate Blepharisma stoltei Isquith. J Protozool 15: 407–412. 10.1111/j.1550-7408.1968.tb02148.x [DOI] [Google Scholar]
- Rio DC, Ares M Jr, Hannon GJ, Nilsen TW. 2010. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harbor Protoc 2010: pdb.prot5439. 10.1101/pdb.prot5439 [DOI] [PubMed] [Google Scholar]
- Sheng Y, Duan L, Cheng T, Qiao Y, Stover NA, Gao S. 2020. The completed macronuclear genome of a model ciliate Tetrahymena thermophila and its application in genome scrambling and copy number analyses. Sci China Life Sci 63: 1534–1542. 10.1007/s11427-020-1689-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Pan B, Wei F, Wang Y, Gao S. 2021. Case study of the response of N6-methyladenine DNA modification to environmental stressors in the unicellular eukaryote Tetrahymena thermophila. mSphere 6: e0120820. 10.1128/mSphere.01208-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Vancura A, Woycicki RK, Hogan DJ, Hendrick AG, Nowacki M. 2018. Determination of the presence of 5-methylcytosine in Paramecium tetraurelia. PLoS One 13: e0209707. 10.1371/journal.pone.0209707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogame Y, Kinoshita E, Matsuoka T. 2011. Ca2+-dependent in vivo protein phosphorylation and encystment induction in the ciliated protozoan Colpoda cucullus. Eur J Protistol 47: 208–213. 10.1016/j.ejop.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Solberg T, Mason V, Wang C, Nowacki M. 2023. Developmental mRNA clearance by PIWI-bound endo-siRNAs in Paramecium. Cell Rep 42: 112213. 10.1016/j.celrep.2023.112213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W. 2000. Morphological and taxonomical studies on some marine scuticociliates from China sea, with description of two new species, Philasterides armatalis sp.n, and Cyclidium varibonneti sp.n. (Protozoa: Ciliophora: Scuticociliatida). Acta Protozool 39: 295–322. [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57: 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33: W465–W467. 10.1093/nar/gki458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Cai X, Liu Y, Liucong M, Howard-Till R. 2022. A practical reference for studying meiosis in the model ciliate Tetrahymena thermophila. Mar Life Sci Technol 4: 595–608. 10.1007/s42995-022-00149-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verni F, Rosati G. 2011. Resting cysts: a survival strategy in Protozoa Ciliophora. Ital J Zool 78: 134–145. 10.1080/11250003.2011.560579 [DOI] [Google Scholar]
- Vurture GW, Sedlazeck FJ, Nattestad M, Underwood CJ, Fang H, Gurtowski J, Schatz MC. 2017. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33: 2202–2204. 10.1093/bioinformatics/btx153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen X, Sheng Y, Liu Y, Gao S. 2017a. N6-Adenine DNA methylation is associated with the linker DNA of H2A.Z-containing well-positioned nucleosomes in Pol II-transcribed genes in Tetrahymena. Nucleic Acids Res 45: 11594–11606. 10.1093/nar/gkx883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng Y, Liu Y, Pan B, Huang J, Warren A, Gao S. 2017b. N6-Methyladenine DNA modification in the unicellular eukaryotic organism Tetrahymena thermophila. Eur J Protistol 58: 94–102. 10.1016/j.ejop.2016.12.003 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Sheng Y, Huang J, Chen X, Al-Rasheid KAS, Gao S. 2017c. A comparative study of genome organization and epigenetic mechanisms in model ciliates, with an emphasis on Tetrahymena, Paramecium and Oxytricha. Eur J Protistol 61: 376–387. 10.1016/j.ejop.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Wang Y, Sheng Y, Liu Y, Zhang W, Cheng T, Duan L, Pan B, Qiao Y, Liu Y, Gao S. 2019. A distinct class of eukaryotic MT-A70 methyltransferases maintain symmetric DNA N6-adenine methylation at the ApT dinucleotides as an epigenetic mark associated with transcription. Nucleic Acids Res 47: 11771–11789. 10.1093/nar/gkz1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watoh T, Sekida S, Yamamoto K, Kida A, Matsuoka T. 2005. Morphological study on the encystment of the ciliated protozoan Colpoda cucullus. J Protozool Res 15: 20–28. [Google Scholar]
- Wei W, Chen K, Miao W, Yang W, Xiong J. 2018. Pseudocohnilembus persalinus genome database: the first genome database of facultative scuticociliatosis pathogens. BMC Genomics 19: 676. 10.1186/s12864-018-5046-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Pan B, Diao J, Wang Y, Sheng Y, Gao S. 2022. The micronuclear histone H3 clipping in the unicellular eukaryote Tetrahymena thermophila. Mar Life Sci Technol 4: 584–594. 10.1007/s42995-022-00151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whang I, Kang HS, Lee J. 2011. Morphological and molecular characterization of Pseudocohnilembus longisetus Thompson, 1965 from farmed black rockfish Sebastes schlegelii in Korea. Vet Parasitol 179: 227–233. 10.1016/j.vetpar.2011.02.009 [DOI] [PubMed] [Google Scholar]
- Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. [Google Scholar]
- Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875. 10.1093/bioinformatics/bti310 [DOI] [PubMed] [Google Scholar]
- Xiong J, Wang G, Cheng J, Tian M, Pan X, Warren A, Jiang C, Yuan D, Miao W. 2015. Genome of the facultative scuticociliatosis pathogen Pseudocohnilembus persalinus provides insight into its virulence through horizontal gene transfer. Sci Rep 5: 15470. 10.1038/srep15470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Gao S, Dui W, Yang W, Chen X, Taverna SD, Pearlman RE, Ashlock W, Miao W, Liu Y. 2016. Dissecting relative contributions of cis- and trans-determinants to nucleosome distribution by comparing Tetrahymena macronuclear and micronuclear chromatin. Nucleic Acids Res 44: 10091–10105. 10.1093/nar/gkw684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B, Cheng Y, Wang Z, Li Y, Chen L, Huang L, Zhang W, Chen D, Wu H, Tang B, et al. 2017. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat Commun 8: 1122. 10.1038/s41467-017-01195-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B, Li Y, Wang Z, Chen L, Poidevin M, Zhang C, Lin L, Wang F, Bao H, Jiao B, et al. 2018. Active N6-methyladenine demethylation by DMAD regulates gene expression by coordinating with polycomb protein in neurons. Mol Cell 71: 848–857.e6. 10.1016/j.molcel.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Liang Z, Cui X, Ji C, Li Y, Zhang P, Liu J, Riaz A, Yao P, Liu M, et al. 2018. N6-Methyladenine DNA methylation in Japonica and Indica rice genomes and its association with gene expression, plant development, and stress responses. Mol Plant 11: 1492–1508. 10.1016/j.molp.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Zhang J, Chen K, Jiang C, Yang W, Gu S, Wang G, Lu Y, Miao W, Xiong J. 2020. Bacteria-derived hemolysis-related genes widely exist in scuticociliates. Microorganisms 8: 1838. 10.3390/microorganisms8111838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhao Y, Zheng W, Nan B, Fu J, Qiao Y, Zufall RA, Gao F, Yan Y. 2023. Genome-wide identification of ATP-binding cassette transporter B subfamily, focusing on its structure, evolution and rearrangement in ciliates. Open Biol 13: 230111. 10.1098/rsob.230111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wang Y, Wang Y, Liu Y, Gao S. 2017. Histone methyltransferase TXR1 is required for both H3 and H3.3 lysine 27 methylation in the well-known ciliated protist Tetrahymena thermophila. Sci China Life Sci 60: 264–270. 10.1007/s11427-016-0183-1 [DOI] [PubMed] [Google Scholar]
- Zhao L, Gao F, Gao S, Liang Y, Long H, Lv Z, Su Y, Ye N, Zhang L, Zhao C, et al. 2021. Biodiversity-based development and evolution: the emerging research systems in model and non-model organisms. Sci China Life Sci 64: 1236–1280. 10.1007/s11427-020-1915-y [DOI] [PubMed] [Google Scholar]
- Zheng X, Chen L, Xia H, Wei H, Lou Q, Li M, Li T, Luo L. 2017. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant's adaptation to drought condition. Sci Rep 7: 39843. 10.1038/srep39843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Wang C, Lynch M, Gao S. 2021. The compact macronuclear genome of the ciliate Halteria grandinella: a transcriptome-like genome with 23,000 nanochromosomes. mBio 12: e01964-20. 10.1128/mBio.01964-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.