Abstract
Woodchuck hepatitis virus (WHV) enhancer II (EnII) is located upstream of the major pregenomic RNA promoter and is thought to play an important role in the insertional activation of the N-myc2 gene during WHV hepatocarcinogenesis. WHV EnII is recognized by at least three host transcription factors: HNF-1, HNF-4, and Oct-1. Here, the roles of these EnII-binding factors in viral transcription and replication have been further examined. In HepG2 cells transiently transfected with a chloramphenicol acetyltransferase (CAT) gene whose expression is dependent upon EnII, mutations in either the HNF-1 or the HNF-4 site strongly reduced CAT activity, while ablation of the Oct-1 site decreased CAT expression only twofold. Mutations in more than one site completely abolished reporter expression. These same mutations were also tested in an overlength WHV genome for their impact on viral replication and gene expression. In transfected HepG2 cells, lesions in the HNF-1 site inactivated pregenomic RNA expression and viral reverse transcription, with only minimal effects on the expression of other viral mRNAs. By contrast, Oct-1 site lesions had no effect on either viral RNA synthesis or DNA replication, and HNF-4 site lesions produced a modest reduction of pregenomic RNA but had no impact on viral DNA synthesis. Testing of the mutants in susceptible woodchucks revealed that, as expected, viruses with lesions in the HNF-1 site were nearly noninfectious, while mutants with lesions at the Oct-1 site were fully replication competent. HNF-4 site mutants were replication competent but may display reduced levels of replication in the intact animal host. We conclude that (i) EnII is primarily devoted to the regulation of pregenomic RNA in WHV, (ii) HNF-1 is essential for EnII function in vivo, and (iii) HNF-4 plays a demonstrable but adjunctive role in EnII function.
Hepatitis B viruses (HBVs; hepadnaviruses) are small, enveloped DNA viruses that produce persistent hepatic infections and are strongly associated with the development of hepatocellular carcinoma (5). The viral genome is a partially duplex, relaxed circular species of approximately 3 kb that replicates within cells via reverse transcription. Upon entry into cells, the viral genome is converted into covalently closed circular DNA that serves as the template for the synthesis of subgenomic RNA and pregenomic RNA (pgRNA) by host RNA polymerase (19). Studies on transcription of human HBV have shown that this step is controlled by regulatory elements that include four promoters and two enhancers (1, 11, 13, 15, 16, 24, 26, 27). The preS, S, and X promoters drive the transcription of subgenomic RNAs encoding the envelope and X proteins. The C promoter controls the production of the pgRNA that encodes the core protein and polymerase and that also serves as the template for viral reverse transcription.
Two enhancers, enhancer I (EnI) and EnII, located upstream of the HBV X and C promoters, respectively, have been shown to be capable of upregulating homologous and heterologous promoters in transient transfection (15, 21, 29). EnI has been proposed to be widely involved in the regulation of HBV transcription (18). However, despite the shared genomic organization and replication cycle of mammalian hepadnaviruses, EnI activity has not been found in other viruses of the family (2, 22). EnII is known to enhance the C promoter in transient assays, suggesting that it might regulate the transcription of pgRNA (18, 28). The expression of pgRNA is highly liver specific, a fact that correlates well with the known liver specificity of EnII activity (28). Thus, it is attractive to speculate that EnII, together with the C promoter, may account for the liver-specific expression of pgRNA that determines (at least in part) the hepatotropism of hepadnaviruses (6, 14, 28).
Woodchuck hepatitis virus (WHV) is a mammalian hepadnavirus that is closely related to HBV and is strikingly oncogenic in its natural host (12, 20). Analyses of WHV-induced woodchuck hepatocellular carcinomas have demonstrated that activation of the proto-oncogene N-myc2 by viral DNA insertion is commonly involved in carcinogenesis (4, 7, 25). Our studies of viral DNA sequences in N-myc2 activation suggested that the region corresponding to HBV EnII played a major role in the activation (22). Accordingly, we and others have mapped and characterized EnII in WHV (3, 23). The activity of WHV EnII maps to an 88-nucleotide DNA fragment (nucleotides 1772 to 1859) upstream from the transcription initiation site of pgRNA. Biochemical and genetic studies have identified three host transcription factors that recognize elements within this region: the liver-enriched factors HNF-1 and HNF-4 and the ubiquitous factor Oct-1 (23). Deletion analyses suggested that HNF-1 and HNF-4 are the main contributors to the EnII activity in transient assays and together account for its strong liver specificity.
Most analyses of HBV and WHV enhancer activity have been carried out by using transient assays with cell lines transfected with artificial reporter constructs. The role(s) of the host transcription factors that regulate viral En elements in viral replication in vivo have not been fully defined in the mammalian viruses, although efforts in this direction have been made for duck HBV (9). In this study, we undertook in vitro and in vivo analyses of the role of WHV EnII in the viral life cycle. First, we introduced mutations in WHV EnII that abolish the binding of HNF-1, HNF-4, or Oct-1 to the element and assessed the effects of these mutations, singly and in combination, on EnII activity, as assayed on a chloramphenicol acetyltransferase (CAT) reporter driven by a minimal promoter linked to EnII. We then constructed 1.5-mer WHV genomes bearing these mutations and tested their abilities to transcribe and replicate viral DNA in cultured cells. Finally, selected mutant genomes were injected intrahepatically into woodchucks to investigate their replication competence in vivo. We show here that EnII primarily governs the production of pgRNA in vivo and that HNF-1 is required for this activity.
MATERIALS AND METHODS
Plasmids.
All of the WHV EnII mutants we used are based on pBluescript II KS+ (Stratagene). The fragment from nucleotide 1697 to nucleotide 1919 (the nucleotide sequence and numbering are according to Kodama et al. [8]) encompassing EnII was prepared by PCR on the template WHV monomer (pWHVmono), in which 5′-end primer WBam1697-1711 (5′-GACTGGATCCCTCCGGTCCGTGTTG-3′) and 3′-end primer WBam1919-1905 (5′-GACTGGATCCTCGCATGCATTTATG-3′) were used. The PCR product was gel purified and cloned into the BamHI site in pBSKS+. The resulting plasmid, pW1697-1919WT, served as a template for PCR to introduce mutations into the EnII region.
To mutate transcriptional factor binding sites, PCR was performed in two rounds. In the first round, two independent PCRs were carried out with two pairs of primers. The primers were designed such that the 3′-end primer of one pair has the complementary sequence of the 5′-end primer of another pair, and desired mutations were introduced into these two primers. To mutate the HNF-4 binding site (designated the A site), 5′-end primer WBam1697-1711 and 3′-end primer IIA/AS (5′-GATCTTTTATATAAGGAGTCCAcAGaTCCTTACTTGG-3′ [nucleotides 1833 to 1797; nucleotides in lowercase represent mutations] were used in one PCR, while 5′-end primer IIA/S (5′-CATGCCAAGTAAGGAtCTgTGGACTCCTTATATAAAA-3′ [nucleotides 1793 to 1829]) was paired with 3′-end primer WBam1919-1905. To mutate the HNF-1 binding site (designated the B site), primer WBam1697-1711 was paired with primer IIB/AS (5′-GCCCTCCTCCCATTTcGTgAggAgcTGATCTTT-3′ [nucleotides 1859 to 1827]) and primer IIB/S (5′-ATAAAAGATCAgcTccTcACgAAATGGGAGGAG-3′ [nucleotides 1824 to 1856]) was paired with primer WBam1919-1905. To mutate the Oct-1 binding site (designated the C site), primers WBam1697-1711 and IIC/AS (5′-GCATGCCAAGTTGACGgTTgGCgTGCCAGGAGACAAAG-3′ [nucleotides 1797 to 1760]) were paired, while primers IIC/S (5′-GAACTTTGTCTCCTGGCAcGCcAAcCGTCAACTTGGC-3′ [nucleotides 1757 to 1793]) and WBam1919-1905 were paired. The products of the first-round PCR were pooled and used as template DNA in second-round PCRs in which primers WBam1697-1711 and WBam1919-1905 were used. The final PCR products were then gel purified, digested with BamHI, and cloned into pBSKS+ cut with BamHI, resulting in plasmids pW1697-1919IIA(−), pW1697-1919IIB(−), and pW1697-1919IIC(−).
To mutate both the HNF-4 and HNF-1 sites, pW1697-1919IIB(−) was used as the template in the first-round PCRs, in which primer WBam1697-1711 paired with primer IIA/AS and primer IIA/S paired with primer WBam1991-1905 were used. To mutate both the HNF-4 and Oct-1 sites, pW1697-1919IIC(−) was used as the template in PCRs in which primer WBam1697-1711 paired with primer IIA/AS and primer IIA/S paired with primer WBam1991-1905 were used. To mutate both the HNF-1 and Oct-1 sites, pW1697-1919IIB was used as the template in PCRs in which primer WBam1697-1711 paired with primer IIC/AS and primer IIC/S paired with primer WBam1919-1905 were used. The PCR products were pooled and used as templates in second-round PCRs in which primer WBam1697-1711 was paired with primer WBam1919-1905. The final PCR products were cloned into pBSKS+, resulting in plasmids pW1697-1919IIAB(−), pW1697-1919IIAC(−), and pW1697-1919IIBC(−).
To mutate the triple binding sites, pW1697-1919IIBC(−) was amplified in PCRs in which primer WBam1697-1711 paired with primer IIA/AS and primer IIA/S paired with primer WBam1919-1905 were used. The second-round PCR was performed by using the first-round PCR products as the template and WBam1697-1711 and WBam1919-1905 as the primers. The final PCR product was cloned into a pBSKS+ vector, giving rise to plasmid pW1697-1919IIABC(−).
Plasmids EnIIWTE1bCAT, EnIIA(−)E1bCAT, EnIIB(−)E1bCAT, EnIIC(−)E1bCAT, EnIIAB(−)E1bCAT, EnIIAC(−)E1bCAT, EnIIBC(−)E1bCAT, and EnIIABC(−)E1bCAT were constructed by cloning the insert fragments of pW1697-1919WT, pW1697-1919IIA(−), pW1697-1919IIB(−), pW1697-1919IIC(−), pW1697-1919IIAB(−), pW1697-1919IIAC(−), pW1697-1919IIBC(−) and pW1697-1919IIABC(−) into the XhoI and XbaI sites in E1bCAT, which has been described elsewhere (23).
The RsrII-NsiI fragments (nucleotides 1701 to 1914) of pW1697-1919 mutants were cut out and cloned into pWHVmono cut with RsrII and NsiI. The monomer mutants, as well as the wild type (WT), were then cut with ApaI and EcoRI. The ApaI-EcoRI fragments (nucleotides 891 to 3320) were gel purified and cloned into the ApaI and EcoRI sites in pBSKS+. The pApaI-EcoRI constructs were then digested with EcoRI and SmaI and used as vectors to receive the EcoRI-BglII (nucleotides 1 to 2531) fragments generated from the monomer mutants (the termini from BglII digestion having been blunted by treatment with Klenow polymerase). The resulting 1.5-mer WHV genomes contain two copies of the region from nucleotide 891 to nucleotide 2531.
The nucleotide sequences of the fragments generated by PCR were confirmed by conventional dideoxy sequencing.
EMSA.
The fragments used as probes for electrophoretic mobility shift assay (EMSA) were generated by PCRs in which pW1697-1919WT, -IIA(−), -IIB(−), -IIC(−), -IIAB(−), IIAC(−), -IIBC(−), and -IIABC(−) were amplified with Taq polymerase and primers with the sequences 5′-CTTTGTCTCCTGGC-3′ (nucleotides 1760 to 1773) and 5′-GCCCTCCTCCCATT-3′ (nucleotides 1859 to 1846). The fragments (spanning nucleotides 1760 to 1859) were then labeled with T4 polynucleotide kinase and [32P]ATP and incubated with nuclear extracts from HepG2 cells as previously described (23).
Transfection of HepG2 cells and CAT assay.
HepG2 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% bovine serum and penicillin-streptomycin. Transfection was performed by the calcium phosphate coprecipitation method. At 16 h after transfection, cells were washed twice with phosphate-buffered saline without calcium and magnesium and incubated for another 24 h. The CAT assay was carried out as previously described (23). All assays were performed in duplicate, and each experiment was replicated at least three times.
Nucleic acid analysis.
Total RNA was isolated from transfected HepG2 cells with RNAzol (TEL-TEST), electrophoresed through a standard 1% agarose–2.2 M formaldehyde gel, and transferred to a Hybond N membrane (Amersham). Nucleic acids in cytoplasmic nucleocapsids were prepared as previously described (10, 17) and analyzed by electrophoresis, transfer into Hybond N+ (Amersham) in 0.4 N NaOH, and hybridization with WHV genomic DNA. To prepare DNA from woodchucks, liver biopsy samples were incubated in 10 mM Tris-HCl (pH 8.0)–10 mM NaCl–10 mM EDTA–1% sodium dodecyl sulfate–120-μg/ml proteinase K at 4°C overnight, phenol extracted three times, chloroform extracted once, and ethanol precipitated. DNA was digested with PvuII and subjected to Southern blotting.
In situ transfection of woodchucks.
The experimental woodchucks were born and raised in laboratory animal facilities and, prior to inoculation, were negative for serologic markers of infection (woodchuck hepatitis surface antigen [WHsAg], anti-woodchuck hepatitis core antigen antibody [anti-WHc], and anti-WHsAg antibody). DNA of the 1.5-mer WHV genome was prepared by using the Qiagen plasmid kit. For transfection in vivo, laparotomies were performed under general anesthesia and 50 μg of DNA (suspended in 0.5 ml of phosphate-buffered saline was injected at multiple sites into the parenchyma of the left lateral lobe of the liver. Groups of three animals were transfected with the mutant EnIIA(−), EnIIB(−), EnIIC(−), or EnIIABC(−) or the WT 1.5-mer WHV genome.
Serological assay.
Serum samples were collected serially, beginning 4 weeks postransfection, at ca. 2-week intervals for a total of 7 months. The sera were tested for the presence of WHsAg and anti-WHc by using enzyme-linked immunosorbent assays (1a). At 13 weeks after transfection, the animals were anesthetized and percutaneous needle biopsies of the liver were performed under ultrasound guidance.
RESULTS AND DISCUSSION
Effect of EnII mutations in transient transfection assays.
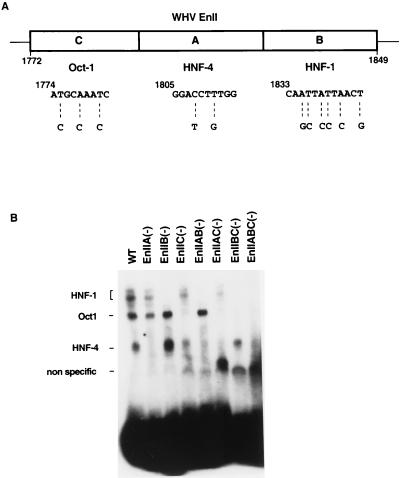
In previous studies (23), we mapped WHV EnII to an 88-bp fragment (nucleotides 1772 to 1859) and demonstrated the binding of transcription factors Oct-1, HNF-1, and HNF-4 to subregions designated IIC, IIB, and IIA, respectively (Fig. 1A). In preliminary deletion analyses using CAT reporter genes driven by EnII and a minimal TATA box, we showed that the absence of the binding site for Oct-1 reduced EnII activity only twofold, while more extensive deletions involving the A and B sites resulted in more dramatic losses of activity (23).
FIG. 1.
Mutations abolishing binding of transcription factors to WHV EnII. (A) schematic representation of the EnII region. The mutated nucleotides in the binding sites of Oct-1, HNF-4, and HNF-1 are shown below the diagram. (B) Mutations block binding of the factors. The indicated WT and mutant labeled EnII fragments were incubated with HepG2 nuclear extract and then examined by EMSA as described in Materials and Methods. IIA(−), HNF-4 binding site mutated; IIB(−), HNF-1 binding site mutated; IIC(−), Oct-1 binding site mutated.
To assess more precisely the effects of these transcription factors on the function of WHV EnII, we constructed point mutations in EnII which selectively destroyed the binding sites for Oct-1, HNF-1, and HNF-4. (The mutations generated were chosen so as not to alter the coding sequence of the overlapping X gene, so that these lesions could later be studied in the context of the intact genome [Fig. 1A].) The mutations were built back into EnII either singly, pairwise, or as the triple mutant; the resulting mutant EnII fragments were tested for the ability to bind to host nuclear factors by EMSA. WT or mutant EnII fragments were end labeled with [32P]ATP and incubated with HepG2 nuclear extract; complexes were then fractionated by nondenaturing acrylamide gel electrophoresis. Figure 1B shows the result of this analysis. As previously demonstrated (23), three complexes involving HNF-1, Oct-1, and HNF-4 were formed on the WT EnII fragment (lane WT). The mutations in the HNF-4, HNF-1, and Oct-1 binding sites selectively blocked the binding of the corresponding factors [lanes EnIIA(−), EnIIB(−), and EnIIC(−)]. Similarly, the expected combinations of shifted complexes were disrupted by the double and triple mutations [lanes EnIIAB(−), EnIIAC(−), EnIIBC(−), and EnIIABC(−)]. These results confirmed that the mutations introduced into each site inactivated the binding of the corresponding factor to EnII DNA.
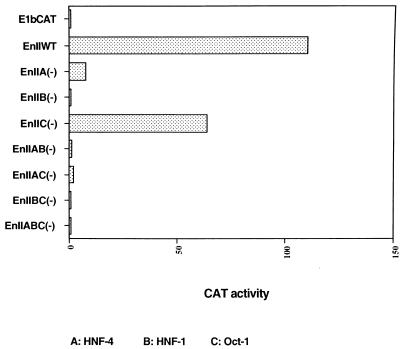
To test the effects of the mutations on EnII activity, we cloned these same mutant EnII DNA segments upstream of an E1b TATA box driving the CAT gene and transfected the constructs into HepG2 cells; the parental E1b TATA-CAT vector was transfected in parallel. The human growth hormone gene (driven by the thymidine kinase promoter of herpes simplex virus) was used as an internal control for transfection efficiency. After 48 h, cells were assayed for CAT activity. The values in Fig. 2 represent the mean results of three independent experiments. As previously described (23), WT EnII strongly enhanced the minimal E1B promoter when linked to it in cis (Fig. 2). Knockout of the HNF-4 binding site in EnII strongly decreased CAT activity (14-fold), while the mutations in the HNF-1 binding site resulted in almost complete inactivation of EnII function. In contrast, an only twofold drop in CAT activity was observed in the transfection with the construct bearing mutations in the Oct-1 binding site (Fig. 2). All combination (double or triple) mutants rendered EnII unable to enhance the minimal promoter activity more than twofold (Fig. 2).
FIG. 2.
Effects of EnII mutations on CAT reporter gene activity in transient transfection assays. EnII mutant and WT DNA segments were cloned into an E1b TATA-CAT vector. The results of CAT assay were normalized to the level of CAT activity of E1b TATA-CAT (arbitrarily set at 1.0).
These data are in good accord with our earlier deletion analyses (23) and confirm and extend those results. Because of the central position of the HNF-4 binding site in EnII (Fig. 1A), our earlier deletion studies were unable to assess the impact of a solo mutation in this element. The present findings reveal that HNF-4 plays an important role in the regulation of EnII function, at least in transient reporter assays using heterologous promoters.
HNF-1 is required for the production of pgRNA in HepG2 cells.
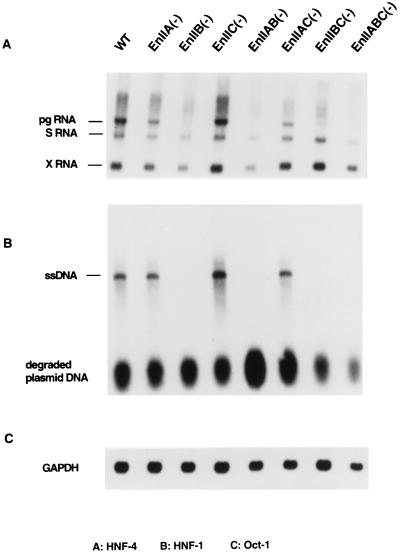
To investigate the effects of these mutations on viral replication in cultured cells, we recombined the mutations into overlength (1.5-mer) viral genomes and transfected them into HepG2 cells, which are known to be permissive for WHV replication (14); WT WHV DNA transfected in parallel served as a positive control. Three days after transfection, total cellular RNA was examined by Northern blot hybridization with WHV DNA. The results of three independent experiments showed that viral genomes bearing lesions in any of the three individual sites had only modest effects (twofold or less) on the production of the S mRNA transcripts (Fig. 3A). In sharp contrast, some of the lesions had pronounced effects on the production of pgRNA. Most dramatic was the impact of mutations in the HNF-1 site: these lesions nearly totally ablated detectable pgRNA synthesis [Fig. 3A, lanes EnIIB(−), EnIIAB(−), EnIIBC(−), and EnIIABC(−)]. Mutations in the Oct-1 site had no impact on pgRNA [or on the subgenomic transcripts; lane EnIIC(−)]. HNF-4 site mutations had a subtler phenotype: they reduced the levels of pgRNA to levels that were roughly equimolar with the S mRNA [lanes ENIIA(−) and EnIIAC(−)]. This phenotype, while modest, was clearly different from that of WT WHV, in which pgRNA was clearly present in a two- to fourfold excess over S mRNA [Fig. 3A, lanes WT and EnIIC(−)].
FIG. 3.
Effects of the mutations on virus expression and replication in cultured cells. (A) Northern blot analysis of total RNA from HepG2 cells transfected with WT or mutant 1.5-mer viral genomes, as indicated above the gels; following transfer, the filters were probed with 32P-labeled WHV DNA. (B) Analysis of DNA in viral nucleocapsids from transfected cells by Southern blot hybridization with a WHV DNA probe. Each lane corresponds to the same transfection as in the RNA samples in panel A. ssDNA, single-stranded DNA. Degraded plasmid DNA is shown as a loading control. (C) Control for RNA loading, 32P-labeled glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe hybridized to the Northern blot of panel A.
These results indicate that EnII has only modest effects on the regulation of subgenomic mRNA transcription in the intact viral genome. This suggests that the C promoter is the major target of its action, a result that agrees well with recent studies of Fourel et al. (3). HNF-1 plays a decisive role in the activation of the C promoter. The small impact of HNF4 site lesions on the C promoter contrasts markedly with the effect of such lesions on activation of the E1b promoter in the CAT reporter context (Fig. 2). The reasons for this difference are not known but could reflect differences in TFIID binding to the different TATA boxes or other differences between the promoters or the rest of the transcription units.
To examine the consequences of these lesions for viral genomic replication, cytoplasmic WHV nucleocapsids were prepared from HepG2 cells transfected with the mutant and WT genomes; their nucleic acids were then extracted and subjected to Southern blot analysis. As shown in Fig. 3B, the only replication intermediates we were able to detect were single-stranded DNA species (lane WT). (We do not know the reason for the absence of duplex WHV DNA in HepG2 cells; perhaps it is due to the failure to complete minus strand synthesis or to delayed kinetics of plus strand initiation or elongation.) As expected, replication was normal in the Oct-1 site mutant [lane EnIIC(−)] and was completely absent in the transfections of viral genomes with mutations in the HNF-1 site [lanes EnIIB(−), EnIIAB(−), EnIIBC(−), and EnIIABC(−)]. Interestingly, viral DNA synthesis was not affected by the mutations in the HNF-4 binding site [lanes EnIIA(−), and EnIIAC(−)], implying that the modest reduction of pgRNA levels observed in Fig. 3A had no important consequence for the production of core or polymerase proteins or of functional templates for reverse transcription. We presume that the concentrations of these products generated by the mutant genome were all above the rate-limiting concentrations for reverse transcription, at least in transfected HepG2 cells (but see below).
Mutations in the HNF-1 and HNF-4 binding sites affect viral replication in woodchucks.
HepG2 is a human hepatoma cell line and therefore is derived from a heterologous host for WHV replication. In addition, hepatoma cell lines (and hepatocyte cultures generally) are known to lack many phenotypic features of fully differentiated hepatocytes in vivo. Both of these considerations raise the possibility that the regulation of viral replication in HepG2 cells is different from that observed in the livers of intact host animals. We therefore investigated the impact of some of the preceding EnII mutations on viral replication in woodchucks. Given the limited availability of susceptible woodchucks, we selected the single mutations in the HNF-1, HNF-4, and Oct-1 sites and the triple mutation, which inactivates all three sites. Each of the mutant DNAs was transfected into three individual woodchucks by intrahepatic inoculation. Starting at 4 weeks posttransfection, serum samples were collected at 2-week intervals for 7 months and assayed for WHsAg antigenemia and anti-WHc by enzyme immunoassay (Table 1).
TABLE 1.
WHV serology and hepatic nucleic acid analysis of woodchucks following in situ transfection of the liver with WT and EnII mutant WHV DNAs
| No. | Mutation (construct) | Incubation period (wk)a | WHV serology
|
Hepatic WHV DNAb
|
Comments | ||
|---|---|---|---|---|---|---|---|
| WHsAg | Anti-WHc antibody | PCR | Southern blot | ||||
| 988 | None (WT) | 10 | + | + | + | +++ | |
| 3278 | 12 | + | + | + | +++ | ||
| 4279 | 14 | + | + | NTc | NT | Died 20 wks following in situ hepatic transfection from ruptured aortic aneurysm | |
| 981 | HNF-4 [EnIIA(−)] | − | − | + | + | Died 13 wks following in situ hepatic transfection from abdominal hemorrhage caused by liver biopsy | |
| 1896 | − | − | − | − | |||
| 4424 | 16 | + | + | + | + | ||
| 989 | HNF-1 [EnIIB(−)] | − | − | NT | NT | Euthanized 32 wks following in situ hepatic transfection because of severe otitis media | |
| 1890 | 24 | − | + | NT | NT | ||
| 4436 | − | − | NT | NT | |||
| 980 | Oct-1 [EnIIC(−)] | 10 | + | + | + | +++ | |
| 1891 | 27 | − | + | NT | NT | ||
| 4469 | 10 | − | + | − | − | ||
| 3658 | Triple EnII mutation [EnIIA(−), EnIIB(−), EnIIC(−)] | − | − | NT | NT | ||
| 3263 | − | − | NT | NT | |||
| 4448 | − | − | − | − | Anti-WHc antibody detected at wks 10 and 12 following in situ hepatic transfection but at no other time | ||
Time between in situ hepatic transfection and detection of first serologic marker of WHV infection.
Ultrasound-directed needle biopsies of the liver were obtained 13 weeks following in situ hepatic transfection.
NT, not tested.
As expected, all three woodchucks transfected with WT DNA developed WHsAg antigenemia and anti-WHc and had clear evidence of intrahepatic viral replication (see below). All woodchucks receiving mutants in which the HNF-1 binding site was inactivated [IIB(−) and IIABC(−)] were seronegative for WHsAg for the entire testing period, and all but one were negative for anti-WHc as well. It is of note that one animal (no. 1890) displayed the extremely late anti-WHc development, possibly indicative of a very low-level infection, although subsequent analysis of this animal’s liver DNA by PCR revealed no evidence of WHV sequences (data not shown). Among animals transfected with genomes lacking the Oct-1 site [IIC(−)], all developed anti-WHc and one had readily detectable circulating WHsAg. Interestingly, among the woodchucks injected with viral DNA containing mutations in the HNF-4 site [mutant IIA(−)], two were never surface antigenemic and the third was not positive for WHsAg until 4 months postinoculation; this animal also displayed a prolonged delay in the development of anti-WHc, which did not become detectable until 2 months after the appearance of WHsAg (Table 1).
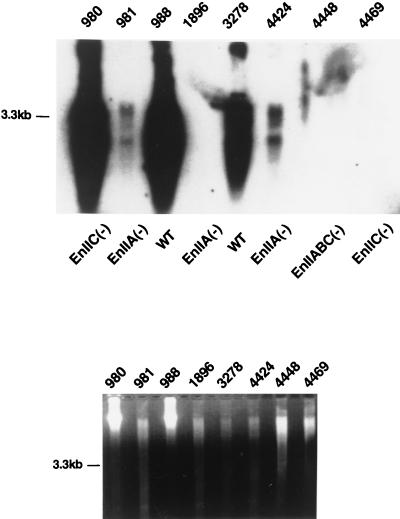
To further evaluate viral replication, we performed liver biopsies on selected woodchucks. Particular attention was paid to the three recipients of the HNF-4 site mutant [IIA(−)], since this mutant had appeared to grow well in HepG2 cells (Fig. 3B) but less so in the animal. DNA was extracted from biopsy samples and examined by PCR (Table 1). Two of the three HNF-4 site mutant recipient animals had viral DNA in their liver by this test, including one that had not displayed antigenemia. To confirm that this represented true viral replication and not simply residual inoculated plasmid DNA, we performed Southern blot analysis on DNA from the biopsy samples. DNA was digested with PvuII, transferred onto a nylon membrane, and hybridized with WHV DNA. As shown in Fig. 4 and summarized in Table 1, of the three woodchucks transfected with the IIA(−) mutant, the two which gave positive PCR signals displayed the characteristic pattern of viral replicative intermediates by Southern blotting (Fig. 4, lanes 981 and 4424). However, in keeping with the serologic results, the signal strength in these mutants was strongly reduced compare to that observed in WT infections (compare lanes 981 and 4424 with the WT in lane 3278, which were loaded with comparable amounts of genomic DNA [as revealed by ethidium bromide staining in the lower panel]). Taken together, these results show that while the IIA(−) mutant is indeed replication competent in vivo, it appears to be less active than its WT parent, in contrast to its behavior in HepG2 cells. However, we caution that this conclusion is based on a small number of inoculees and must be considered provisional.
FIG. 4.
Replication of mutant viral genomes in woodchuck liver. DNA from liver biopsies was digested with PvuII and analyzed by Southern blot hybridization with a WHV DNA probe. Animal numbers and corresponding mutants injected are presented above and below the gel, respectively (upper panel). The lower panel shows ethidium bromide-stained DNA as a loading control.
In summary, our results indicate that WHV EnII plays a crucial role in the regulation of pgRNA but not subgenomic RNA and that binding of HNF-1 to the element is indispensable for this activity. HNF-4 appears to play a demonstrable but secondary role in EnII function.
ACKNOWLEDGMENTS
We thank the National Institutes of Health and the Howard Hughes Medical Institute for support.
REFERENCES
- 1.Cattaneo R, Will H, Schaller H. Hepatitis B virus transcription in the infected liver. EMBO J. 1984;3:2191–2196. doi: 10.1002/j.1460-2075.1984.tb02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Cote P, Ronecker C, Cass K, Schodel F, Tennant B, deNoronha F, Gerin J. New enzyme immunoassays for the serologic detection of woodchuck hepatitis virus infection. Viral Immunol. 1993;6:161–169. doi: 10.1089/vim.1993.6.161. [DOI] [PubMed] [Google Scholar]
- 2.Di Q, Summers J, Burch J, Mason W. Major differences between WHV and HBV in the regulation of transcription. Virology. 1997;229:25–35. doi: 10.1006/viro.1996.8422. [DOI] [PubMed] [Google Scholar]
- 3.Fourel G, Ringeisen F, Flajolet M, Tronche F, Pontoglio M, Tiollais P, Buendia M-A. The HNF1/HNF4-dependent We2 element of woodchuck hepatitis virus controls viral replication and can activate the N-myc2 promoter. J Virol. 1996;70:8571–8583. doi: 10.1128/jvi.70.12.8571-8583.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fourel G, Trépo C, Bougueleret L, Henglein B, Ponzetto A, Tiollais P, Buendia M A. Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumours. Nature. 1990;347:294–298. doi: 10.1038/347294a0. [DOI] [PubMed] [Google Scholar]
- 5.Ganem D. Hepadnaviridae: the viruses and their replication. In: Fields B, et al., editors. Virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1995. pp. 2703–2737. [Google Scholar]
- 6.Guo W, Chen M, Yen T S B, Ou J H. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol Cell Biol. 1993;13:443–448. doi: 10.1128/mcb.13.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen L J, Tennant B C, Seeger C, Ganem D. Differential activation of myc gene family members in hepatic carcinogenesis by closely related hepatitis B viruses. Mol Cell Biol. 1993;13:659–667. doi: 10.1128/mcb.13.1.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kodama K, Ogasawara N, Yoshikawa H, Murakami S. Nucleotide sequence of a cloned woodchuck hepatitis virus genome: evolutional relationship between hepadnaviruses. J Virol. 1985;56:978–986. doi: 10.1128/jvi.56.3.978-986.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Mason W S, Burch J B E. Identification of factor-binding sites in the duck hepatitis B virus enhancer and in vivo effects of enhancer mutations. J Virol. 1994;68:2286–2296. doi: 10.1128/jvi.68.4.2286-2296.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeb D D, Hirsch R C, Ganem D. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses: implications for other reverse transcribing elements. EMBO J. 1991;10:3533–3540. doi: 10.1002/j.1460-2075.1991.tb04917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Cabrera M, Letovsky J, Hu K Q, Siddiqui A. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1990;87:5069–5073. doi: 10.1073/pnas.87.13.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popper H, Roth L, Purcell R H, Tennant B C, Gerin J L. Hepatocarcinogenicity of the woodchuck hepatitis virus. Proc Natl Acad Sci USA. 1987;84:866–870. doi: 10.1073/pnas.84.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rall L B, Standring D N, Laub O, Rutter W J. Transcription of hepatitis B virus by RNA polymerase II. Mol Cell Biol. 1983;3:1766–1773. doi: 10.1128/mcb.3.10.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeger C, Baldwin B, Tennant B C. Expression of infectious woodchuck hepatitis virus in murine and avian fibroblasts. J Virol. 1989;63:4665–4669. doi: 10.1128/jvi.63.11.4665-4669.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaul Y, Rutter W J, Laub O. A human hepatitis B viral enhancer element. EMBO J. 1985;4:427–430. doi: 10.1002/j.1460-2075.1985.tb03646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siddiqui A, Jameel S, Mapoles J. Expression of the hepatitis B virus X gene in mammalian cells. Proc Natl Acad Sci USA. 1987;84:2513–2517. doi: 10.1073/pnas.84.8.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staprans S, Loeb D D, Gamen D. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J Virol. 1991;65:1255–1262. doi: 10.1128/jvi.65.3.1255-1262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su H, Yee J K. Regulation of hepatitis B virus gene expression by its two enhancers. Proc Natl Acad Sci USA. 1992;89:2708–2712. doi: 10.1073/pnas.89.7.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 20.Summers J, Smolec J M, Synder R. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc Natl Acad Sci USA. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treinin M, Laub O. Identification of a promoter element located upstream from the hepatitis B virus X gene. Mol Cell Biol. 1987;7:545–548. doi: 10.1128/mcb.7.1.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueda K, Wei Y, Ganem D. Activation of N-myc2 gene expression by cis-acting elements of oncogenic hepadnaviral genomes: key role of enhancer II. Virology. 1996;217:413–417. doi: 10.1006/viro.1996.0133. [DOI] [PubMed] [Google Scholar]
- 23.Ueda K, Wei Y, Ganem D. Cellular factors controlling the activity of woodchuck hepatitis virus enhancer II. J Virol. 1996;70:4714–4723. doi: 10.1128/jvi.70.7.4714-4723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Chen P, Wu X, Sun A-L, Wang H, Zhu Y-A, Li Z-P. A new enhancer element, ENII, identified in the X gene of hepatitis B virus. J Virol. 1990;64:3977–3981. doi: 10.1128/jvi.64.8.3977-3981.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei Y, Fourel G, Ponzetto A, Silvestro M, Tiollais P, Buendia M-A. Hepadnavirus integration: mechanisms of activation of the N-myc2 retrotransposon in woodchuck liver tumors. J Virol. 1992;66:5265–5276. doi: 10.1128/jvi.66.9.5265-5276.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Will H, Reiser W, Weimer T, Pfaff E, Büscher M, Sprengel R, Cattaneo R, Schaller H. Replication strategy of human hepatitis B virus. J Virol. 1987;61:904–911. doi: 10.1128/jvi.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yee J K. A liver-specific enhancer in the core promoter region of human hepatitis B virus. Science. 1989;246:658–661. doi: 10.1126/science.2554495. [DOI] [PubMed] [Google Scholar]
- 28.Yuh C H, Ting L P. Differentiated liver cell specificity of the second enhancer of hepatitis B virus. J Virol. 1993;67:142–149. doi: 10.1128/jvi.67.1.142-149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuh C H, Ting L P. The genome of hepatitis B virus contains a second enhancer: cooperation of two elements within this enhancer is required for its function. J Virol. 1990;64:4281–4287. doi: 10.1128/jvi.64.9.4281-4287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]