Abstract
Comprehensive understanding prognostic relevance of distinct tumor microenvironment (TME) remained elusive in colon cancer. In this study, we performed in silico analysis of the stromal components of primary colon cancer, with a focus on the markers of cancer-associated fibroblasts (CAF) and tumor-associated endothelia (TAE), as well as immunological infiltrates like tumor-associated myeloid cells (TAMC) and cytotoxic T lymphocytes (CTL). The relevant CAF-associated genes (CAFG)(representing R index = 0.9 or beyond with SPARC) were selected based on stroma specificity (cancer stroma/epithelia, cS/E = 10 or beyond) and expression amounts, which were largely exhibited negative prognostic impacts. CAFG were partially shared with TAE-associated genes (TAEG)(PLAT, ANXA1, and PTRF) and TAMC-associated genes (TAMCG)(NNMT), but not with CTL-associated genes (CTLG). Intriguingly, CAFG were prognostically subclassified in order of fibrosis (representing COL5A2, COL5A1, and COL12A1) followed by exclusive TAEG and TAMCG. Prognosis was independently stratified by CD8A, a CTL marker, in the context of low expression of the strongest negative prognostic CAFG, COL8A1. CTLG were comprehensively identified as IFNG, B2M, and TLR4, in the group of low S/E, representing good prognosis. Our current in silico analysis of the micro-dissected stromal gene signatures with prognostic relevance clarified comprehensive understanding of clinical features of the TME and provides deep insights of the landscape.
Introduction
Cancer is widely recognized as a genetic disease, arising from a combination of inherited (germline mutations) and acquired (somatic mutations) genetic aberrations. These anomalies include gene amplifications [1–3], gene losses [4–6], and gene mutations [7–11], which cumulatively lead to the transformation of cells. Interestingly, despite the accumulation of these genetic alterations, a mathematical model has suggested a relatively low selective advantage (0.004) during tumor progression [12]. Consistent with these findings, single driver gene, for example c-MYC expression did not show prognostic relevance at all in human colorectal cancer (CRC) [13, 14], although its deletion rescued tumorigenesis of APC deficiency [15]. These finding suggested that single driver gene expression alone can not phenotypically affect cancer metastasis.
On the other hand, minimal functional driver gene heterogeneity of mutations was recently confirmed among the metastasis of the individual cancer patients [16, 17], and hence tumor progression is rather associated not with genetic aberrations, plus with epigenetic [18] and/or tumor microenvironment (TME) abnormality [19]. In our current study, we therefore focused on the TME affecting patient prognosis of CRC. The TME are mainly composed of cancer-associated fibroblasts (CAF), tumor-associated endothelia (TAE), cytotoxic T lymphocytes (CTL), and tumor-associated myeloid cells (TAMC) and their derivatives or subpopulations.
The public databases of stromal expression after microdissection of the 13 CRC tumors (GSE35602) [20] was herein used for in silico landscape of stromal components of colon cancer together with comprehensive prognostic relevance was assessed from the bulky tissues in the 232 colon cancer patients (GSE17538) [21] to clarify their comprehensive prognostic roles of the TME. Objectives of in silico analysis of CRC tumors were to obtain results from the same assessment, and public database can be accessed and obtained by other researchers.
Material and methods
Expression profiles of the microdissection tissues of the 13 CRC tumors (GSE35602)
The public databases of stromal (cancer str) expression after microdissection of the 13 CRC tumors (GSE35602) were used in the microarray (Human Genome, Whole 4x44K, Agilent Inc, Santa Clara, CA) harboring 45 015 genes [20], where cancer str/epi expression ratio (cS/E ratio) was calculated. Signal intensities were adjusted by single GAPDH probe (A_23_P13899).
Prognostic analysis from the bulk tissues of the 232 colon cancer tumors (GSE17538)
The public database (GSE17538) was used for prognostic analysis of 232 colon tumor tissues in the microarray base (Human Genome U133 Plus 2.0 Array, Thermo Fisher Scientific Inc, Waltham, MA) [21]. Signal intensities were adjusted by single GAPDH probe (212581_x_at). This database included clinicopathological factors (age, sex, T factor, N factor, M factor, and prognosis representing death). Area under curve (AUC) was calculated for prediction of death according to the individual probes, and the best optimized cut-off value was determined.
Statistical analysis
Follow up data were evaluated in terms of overall survival. The follow-up time was calculated from the date of surgery to death or end-point. Overall survival was estimated by the Kaplan-Meier method, and compared using the log-rank test. Variables suggesting potential prognostic factors on univariate analyses were subjected to a multivariate analysis using a Cox proportional-hazards model. A P-value <0.05 was considered to indicate statistical significance. All statistical analyses were conducted using the SAS software package (JMP Pro16, SAS Institute, Cary, NC).
Ethics statement
Ethical approval has not been obtained because this is an in silico study using only the Public Database.
Results
CAF-associated genes (CAFG) and other TME-associated genes in cancer str of the CRC tumors by in silico analysis
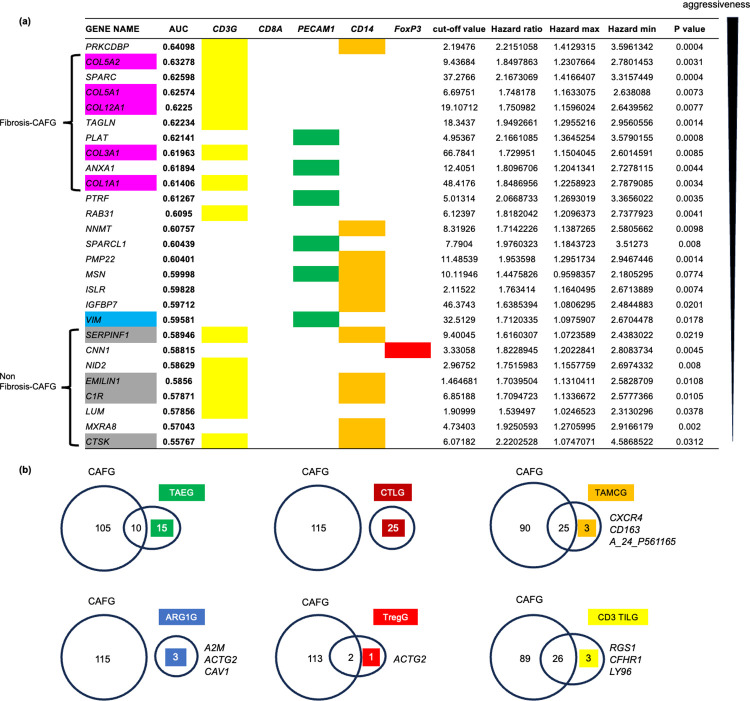
The TME is composed of CAF and TAE representing tumor angiogenesis as well as immunological infiltrates such as myeloid cells, T cells, and B cells or their subpopulations. We initially identified the most relevant CAF-associated genes (CAFG, 256 probes in S1 Table) after selection by stroma specific (cS/E = 10 or beyond) and their strong association (R index = 0.9 or beyond) with well-known stromal marker representing fibroblasts [22], SPARC expression in cancer str of the CRC tumors after microdissection in the array-based public database (GSE35602) [20]. Intriguingly, they included well-established CAF markers including VIM, ACTA2 (aSMA), PDGFRB, and FAP [23] (black color in S1 Table), suggesting that SPARC expression may represent CAF activation in the TME.
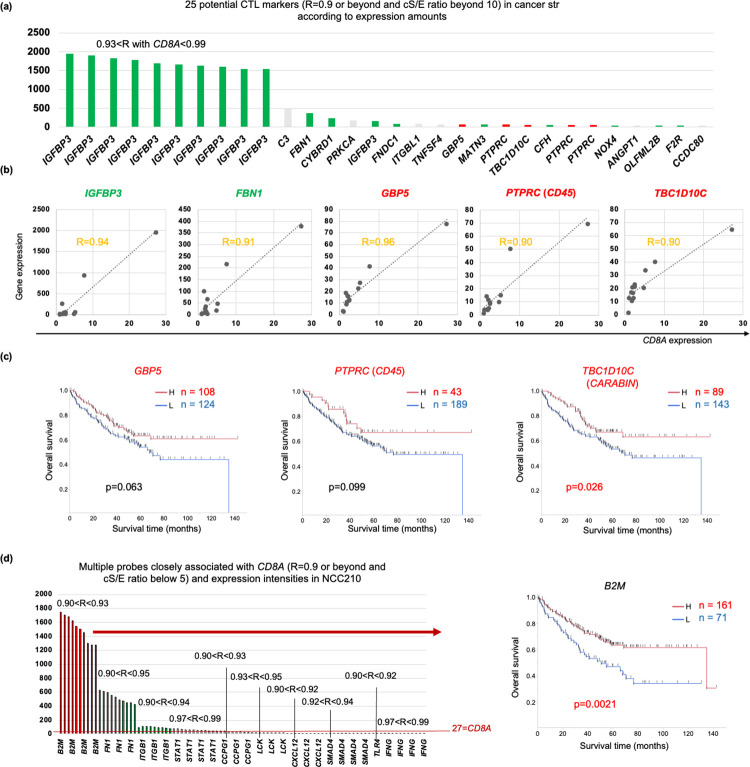
The 4 CAF markers have been all demonstrated to be pan-CAF ones, where single cell RNA sequencing (scRNA) clarified that they were expressed in all 4 CAF subpopulations (vascular CAF, matrix CAF, cycling CAF, and developmental CAF) [24]. The 161 CAFG probes with the highest expression amounts (signal intensity/GAPDHx100 = 40 or beyond in NCC210, S1 Table, red letters) included 115 SYMBOL genes (including SPARC), among which the top 76 genes according to expression amounts were finally selected for prognostic analysis using the public database of 232 colon cancer patients (GSE17538) [21]. Among the 76 genes, 62 genes showed negative prognostic impact (statistically significant, p<0.05), and the 27 genes were commonly shared with any TME markers-associated genes (selected by cS/E ratio = 10 or beyond and high expression amounts = 40 or beyond similarly with CAFG) (Fig 1A).
Fig 1. Relationship between CAFG and other TME-associated genes in cancer str of the CRC tumors (GSE35602).
(a) The 27 CAFG that also highly (R = 0.9 or beyond) correlate with TME markers. They are listed in order of the best AUC values determined individually by prognostic analysis. In the contest of CD3G, fibrosis CAFG ranked as the most aggressive negative prognostic factors in contrast to non-fibrosis CAFG. (b) Common and unique relations to CAFG in the individual TME-associated genes.
The individual TME markers and their associated genes were selected from S2 Table for PECAM1 (CD34) representing TAE (ex: PTRF, VIM, COL4A2, ANXA1) [25], S3 Table for CD8A representing CTL (ex: IGFBP3, C3, FBN1, CYBRD1), S4 Table for CD14 representing TAMC (ex: DCN, IGFBP7, NNMT, DKK3) [22], S5 Table for CD3G representing tumor infiltrating T lymphocytes (CD3 TIL)(ex: DCN, SPARC, LUM, COL1A1), S8 Table for ARG1 representing functional myeloid derived suppressor cell (fMDSC)(ex: A2M, ACTG2, CAV1), S9 Table for CD33 representing immature myeloid derived suppressor cell (iMDSC)(ex: IGFBP7, VIM, ACTA2, LAMA4), S10 Table for FoxP3 representing regulatory T cell (Treg)(ex: ACTG2, CNN1, MEG3), S11 Table for MS4A1 representing tumor infiltrating B lymphocytes (B-TIL)(ex: NOX1) [26], and S12 Table for S100A9 representing tumor-associated neutrophil (TAN)(ex: DEFB1) [25].
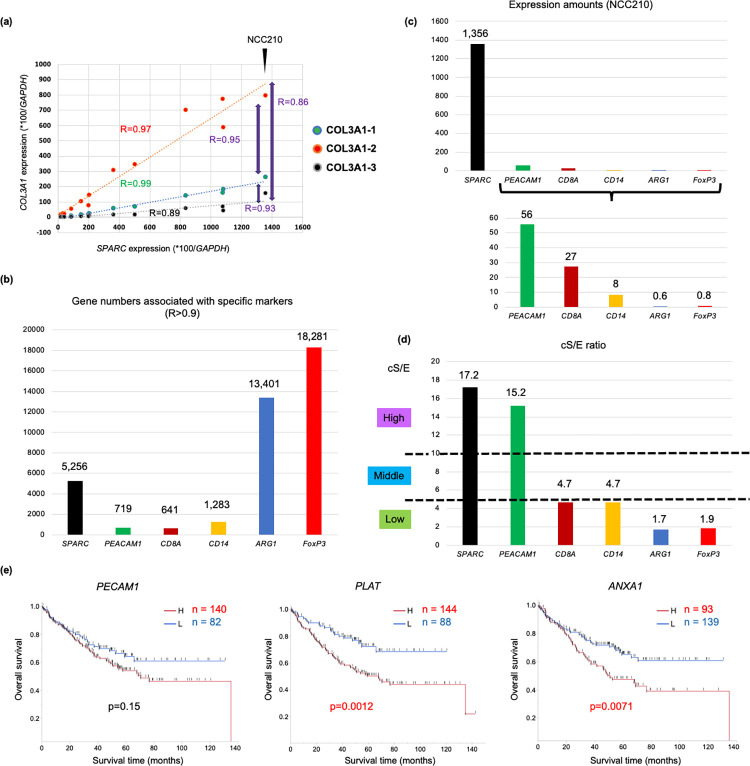
In the microarray-based database, for example, one of the CAFG, COL3A1 expression was closely associated with SPARC (0.89<R<0.99) among the 3 different sets (10 separate kinds of sequences/set) of microarray probes and R indexes between the 3 probe sets were ranged between 0.86 and 0.95 each other (Fig 2A), suggesting that association indicating R = 0.9 may represent same molecular relevance. We therefore thought that gene numbers of the specific TME marker-associated genes (R index = 0.9 or beyond) may represent their molecular signature impacts representing the similar stromal molecular phenotypes (Fig 2B).
Fig 2. TME markers and their associated group genes in cancer str of the CRC tumors (GSE35602).
(a) The 3 separate probe sets of the same gene (COL3A1) showed reproducible R-index near 0.90 with SPARC, and even between the 2 different probe sets among the 3 probes in cancer str of the CRC tumors. (b) The numbers of genes that closely (R>0.9) correlate with representative TME markers in cancer str of the CRC tumors. (c) Expression amounts of each gene in NCC210 case in cancer str of the CRC tumors. (d) cS/E of the representative TME markers. cS/E was classified into the 3 groups as High (cS/E = 10 or beyond), Middle (cS/E = 5 or beyond and below 10), and Low group (cS/E below 5). (e) High PECAM1 expression showed poorer prognosis than its low expression in colon cancer using the best cut-off value, however there was no statistical difference (p = 0.15). On the other hand, CAFG overlapped with TAEG (PLAT and ANXA1) showed significant difference regarding prognosis in colon cancer (p = 0.0012 and p = 0.0071, respectively).
CAFG are partially shared with TAE-associated genes (TAEG) representing tumor angiogenesis in colon cancer
As shown in Fig 2B, SPARC expression putatively representing CAF activation was closely associated with 5 256 genes (R = 0.9 or beyond) (S1 Table), while PECAM1 (CD34) expression represented as TAE activation was associated with 719 genes (S2 Table). Moreover, SPARC expression amount was much higher than that of PECAM1 as well as other representative TME markers (in NCC210) (Fig 2C). Thus, CAF biology should play a central role in TME activation among the stromal components of the CRC tumors.
As PECAM1 showed high cS/E ratio of 15.2 like CAFG, tumor angiogenesis was highly specific to cancer str of the CRC tumors (Fig 2D). We therefore explored genes closely associated with PECAM1 with R = 0.9 or beyond and with cS/E = 10 or beyond (same condition with CAFG) as TAEG according to high expression amounts with signal intensity (40 or beyond, purple color in S2 Table), which identified 36 gene probes (25 SYMBOL).
Intriguingly, the 25 genes identified as TAEG were overlapped with 10 CAFG (Fig 1B), among which 7 genes were negative prognostic factors (p<0.05) and PLAT, ANXA1, and PTRF (CAVIN1) were the strongest regarding AUC (Fig 1A). On the other hand, PECAM1 itself was not statistically significant for prognosis (p = 0.15) (Fig 2E). These findings suggested that tumor angiogenesis may be greatly affected by CAF biology, putatively because vascular CAF is the most dominant CAF subpopulation [24].
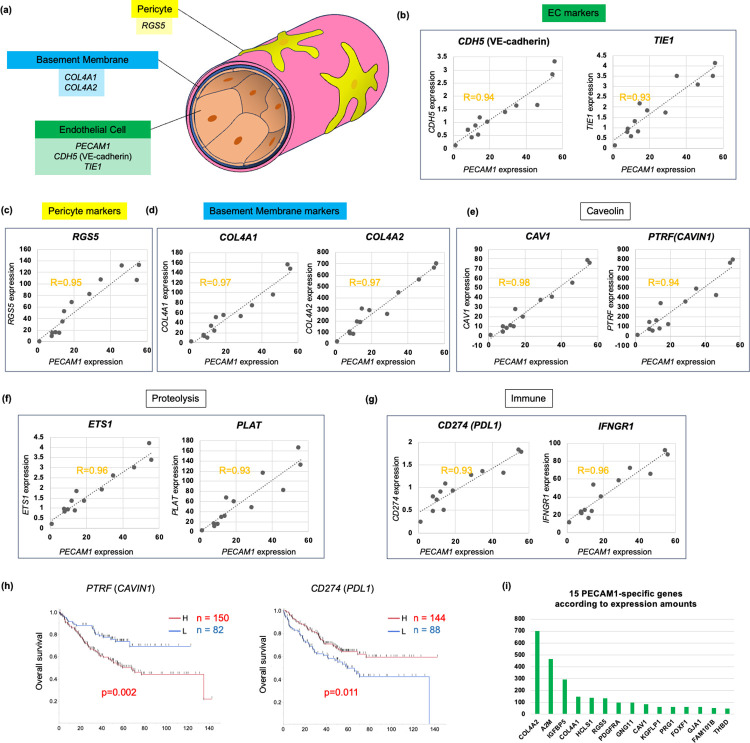
Blood vessels are composed of endothelia, pericytes, and surrounding basement membrane (BM) including type IV collagen [27] (Fig 3A), where they are marked by PECAM1/CDH5 (VE-cadherin)/TIE1, RGS5, and COL4A1/COL4A2 in the scRNA analysis [24], respectively. PECAM1 expression was closely associated with other well-established TAE markers such as CDH5 (R = 0.94) and TIE1 (R = 0.93) in cancer str of the CRC tumors (Fig 3B). Pericyte-specific marker (RGS5) expression is also associated with PECAM1 (R = 0.95) (Fig 3C), and BM markers (COL4A1/COL4A2) were tightly correlated in expression with PECAM1 (R = 0.97), either (Fig 3D). These findings suggested that PECAM1 may represent mature vascular structure in cancer str.
Fig 3. TAE-associated genes (TAEG) in cancer str of the CRC tumors (GSE35602).
(a) Blood vessels are composed of endothelial cells (EC), pericytes, and the surrounding extracellular matrix of basement membrane. (b-d) In cancer str of the CRC tumors, PECAM1 expression was significantly correlated with the expression of other TAE markers, such as CDH5 and TIE1. (c) PECAM1 expression was also closely associated with the Pericyte marker RGS5 in cancer str of the CRC tumors. (d) PECAM1 expression was closely associated with both COL4A1 and COL4A2, the basement membrane components. (e) CAV1 was a gene the most closely correlated with PECAM1 expression, as was PTRF (CAVIN1), a component of caveolin in cancer str of the CRC tumors. (f) PECAM1 expression was closely correlated with ETS1/PLAT in cancer str of the CRC tumors, which may be involved in unique proteolysis. (g) PECAM1 expression was closely correlated with CD274 (PDL1) / IFNGR1 in cancer str of the CRC tumors, which may be involved in tumor immunity. (h) Kaplan-Meier curves for TAEG such as PTRF (CAVIN1) and CD274 (PDL1) in colon cancer. (i) List of 15 genes of TAEG which were not overlapped with CAFG according to expression amounts of NCC210.
Among the 719 TAEG (Fig 2B), on the other hand, PECAM1 was the most strongly associated with CAV1 (0.97<R<0.99) in cancer str of the CRC tumors (S6 Table according to R index), suggesting that CAV1 (12<cS/E<17) plays a critical role in tumor angiogenesis in the TME as previously shown [28, 29]. Multiple different probes for CAV1 were enriched (10/20) as the top priority genes, which are accompanied by its close association with PTRF (CAVIN1), a component of caveola, ascribed to CAFG [30] (Fig 3E). These data indicated significant caveola contribution to tumor angiogenesis in CRC.
PECAM1 expression was also closely associated with ETS1/PLAT (Fig 3F) and CD274 (PDL1)/IFNGR1 (Fig 3G), indicating close involvement of tumor angiogenesis with unique proteolysis [31] and tumor immunity [32]. Nevertheless, their prognostic relevance was conditional; PTRF showed a potent negative prognostic factor putatively reflecting CAFG molecular features, whereas PDL1 was rather a positive prognostic factor in the same database (GSE17538) (Fig 3H). Finally, the 15 unique TAEG in Fig 1B (green color) are shown according to expression amounts (Fig 3I). Intriguingly, PDGFRA, a matrix-CAF marker [24] was included among the TAEG.
CAFG was prognostically independent of CTL-associated genes (CTLG) in colon cancer
The tumor infiltrates marked by CD8A and CD14 were then explored as markers representing CTL and TAMC, because they were demonstrated to be their specific markers by scRNA analysis in CRC, respectively [22]. CD8A and CD14 expressions were closely associated with 641 and 1 283 genes (R = 0.9 or beyond), respectively (Fig 2B), while CD8A expression amount was higher than CD14 expression in this microarray database (NCC210) (Fig 2C). They were both specific to cancer str (showing cS/E = 4.7), but the values were much lower than those of SPARC and PECAM1 (Fig 2D).
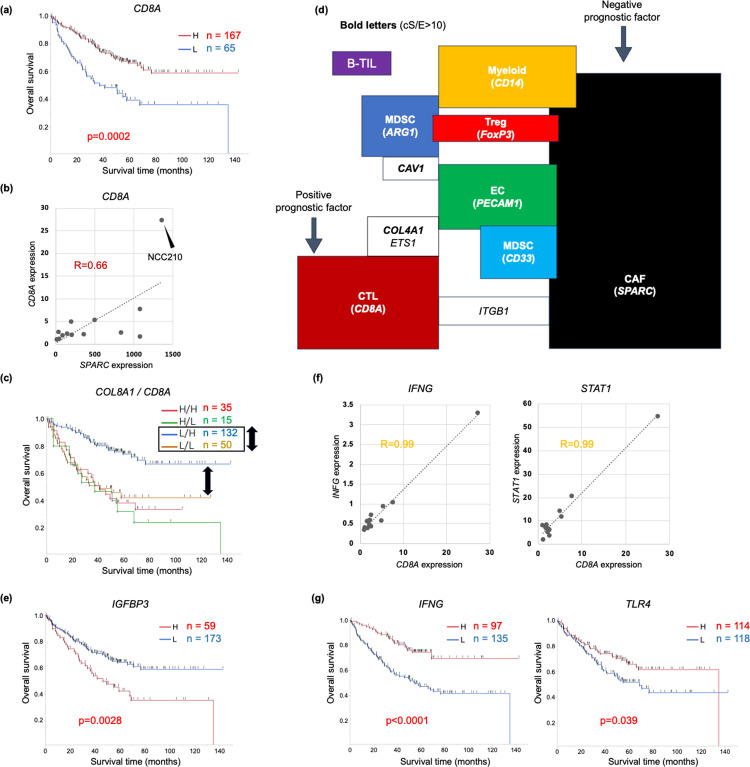
As expectedly, high CD8A expression showed significantly better prognosis than low CD8A expression in colon cancer (Fig 4A), as CD8 CTL was demonstrated to suppress tumorigenesis immunologically [33]. On the other hand, CD8A expression was not associated with a CAFG, SPARC except NCC210 in cancer str of the CRC tumors (R = 0.66, Fig 4B). This finding suggested that CAFG and CD8A may exhibit their independent contribution to prognosis, because most CAFG (75 among the 76 top CAF markers) were negative prognostic factors differently from CD8A. As expectedly, CD8A in combination with the strongest negative prognostic CAFG, COL8A1 expression exhibited additional stratification of prognosis, especially in cases with low COL8A1 expression (Fig 4C, both side black arrows). Interestingly, CD8A did not show such stratification in case of high COL8A1 expression, suggesting that CTL effect can not overcome that of CAF for prognosis.
Fig 4. Prognostic relevance of CTL marker, CD8A, in combination with CAFG.
(a) High CD8A expression showed significantly better prognosis than low CD8A expression in colon cancer. (b) CD8A expression did not show close association with SPARC, a representative CAFG except NCC210 in cancer str of the CRC tumors. (c) CD8A stratified prognosis in cases with low COL8A1 (black double arrows), the strongest negative prognostic factor among the CAFG, while it did not stratify in those with high COL8A1 in colon cancer. (d) Shema representing correlation of the individual TME-associated genes in cancer str of the CRC tumors. (e) Among the CTLG (cS/E = 10 or beyond, and expression amounts = 40 or beyond), IGFBP3, showing the highest amounts (see Fig 5A), was a strong negative prognostic factor, unlike CD8A in colon cancer. (f) CD8A expression was closely associated with IFNG, and STAT1, in cancer str of the CRC tumors. (g) High expressions of IFNG and TLR4 showed significantly better prognosis than their low expression in colon cancer.
CTLG were initially selected similarly as 37 gene probes (25 SYMBOL genes) with cS/E ratio = 10 or beyond and expression amounts = 40 or beyond (purple colors in S3 Table), where the 25 CTLG were not overlapped with any CAFG (red brown box in Fig 1B). These findings suggested that CTL activity is independent of CAF activation (Fig 4D). On the other hand, intriguingly, ITGB1 (cS/E ratio = 4.0) association with CD8A was shared with CAFG (Fig 4D), and ITGB1 was of negative prognostic relevance (p = 0.037), suggesting that ITGB1 may link CAF activation to CTL induction. Thus, CTL mobilization may be mediated by CAFG-associated ITGB1 induction of T lymphocytes. On the other hand, COL4A1/ETS1 association with CD8A was shared with TAEG as shown in Fig 4D, indicating that CAF activation may mediate angiogenesis with tissue destruction to CTL mobilization.
Among the top37 gene probes according to expression amounts (S3 Table), multiple probe sets of IGFBP3 were enriched as top genes (11/37) of the CTLG (S3 Table, and Fig 5A). Although IGFBP3 expression was strongly associated with CD8A expression (0.93<R<0.99) in cancer str of the CRC tumors (representative in Fig 5B), IGFBP3 knockdown (KD) unexpectedly increased CTL mobilization accompanied by attenuated tumorigenesis [34]. These findings suggested that IGFBP3 expression is not the cause of CTL mobilization during tumorigenesis, and may rather reflect host responses to CTL accumulation.
Fig 5. CTL-associated genes (CTLG) in cancer str of the CRC tumors (GSE35602).
(a) CTLG was closely associated with a CTL marker, CD8A (R = 0.9 or beyond), in cancer str of the CRC tumors, and the top 25 gene probes of CTLG with cS/E = 10 or beyond are shown according to expression amounts of NCC210. (b) Representative genes showed correlation to CD8A in cancer str of the CRC tumors. Green letters represent negative prognostic factors, while red letters represent positive prognostic factors in colon cancer (GSE17358). (c) Survival curves of the positive prognostic CTLG with cS/E = 10 or beyond in colon cancer (GSE17358). (d) Representative genes of CTLG with cS/E below 5 according to expression amounts of NCC210 (left panel), and red bars (B2M, TLR4, and IFNG) represent positive prognostic factors in colon cancer (GSE17358). High B2M expression showed significantly better prognosis than its low expression in colon cancer (right panel).
Consistent with the report [34], in primary colon cancer, high IGFBP3 expression showed significantly poorer prognosis than low IGFBP3 expression (p = 0.0028) totally differently from CD8A (Fig 4E). This may be partially explained by different cS/E between the 2 genes (the former was high cS/E, while the latter was low cS/E). Similarly negative prognostic effects were also confirmed for the CTLG (with high cS/E) such as FBN1 (p = 0.0014), CYBRD1 (p = 0.0004), FNDC1 (p = 0.013), MATN3 (p = 0.01), CFH (p = 0.022), NOX4 (p = 0.022), OLFML2B (p = 0.011), and F2R (p = 0.012)(green bars indicating significant negative prognostic factors in Fig 5A). These findings suggested that CTLG with high cS/E have promoting role during tumor progression, and did not reflect the antitumor function of CTL.
CTLG affecting good prognosis in colon cancer
CD8A expression was the most strongly associated with IFNG expression (0.97<R<0.99, 2.1<cS/E<2.5, S7 Table according to R index and Fig 4F), suggesting that IFNG could be an excellent indicator of CTL activity as previously shown [35]. As in S7 Table, multiple 10 different probes for IFNG were enriched as the top association with CD8A as well as 11 probes for STAT1 (R>0.97), a representative IFN-stimulated gene (ISG). S3 Table actually included other IFN pathway genes (TLR3,4,7/IFI44L/IFIT3/BST2/CXCL10) [36–38], suggesting that CTL activity may represent IFN pathway activation. From a prognostic point of view, however IFNG expression was the most likely to represent CTL activity (p<0.0001) followed by TLR4 (p = 0.039) than STAT1 (p = 0.09) (Fig 4G).
We further explored CTLG associated with good prognosis like CD8A and IFNG, because such genes may alternatively represent CTL activity in vivo. Among the CTLG with cS/E = 10 or beyond and high expression (40 or beyond) (Fig 5A), GBP5, PTPRC (CD45), and TBC1D10C (CARABIN) were identified (red bars), among which prognostic difference of only TBC1D10C was statistically significant (p = 0.026) (Fig 5C). TBC1D10C knockout mice showed accumulated CTL like IGFBP3 [39]. We then explored group of genes with low cS/E for CTL activation indicators (red letters in S3 Table), because CD8A is ascribed to the group (Fig 2D).
Among the low cS/E genes, CD8A expression was closely (R = 0.9 or beyond) associated with B2M, followed by FN1, ITGB1, STAT1, CCPG1, LCK, CXCL12, SMAD4, and IFNG as the multiple different probes (S3 Table), among which expression intensity of B2M was uniquely the highest (Fig 5D, left panel). Intriguingly, high B2M expression showed significantly better prognosis than low B2M expression in colon cancer (p = 0.0021, Fig 5D, right panel) similarly with CD8A, IFNG and TLR4 (Fig 4A and 4G).
The CD8 CTL is a subpopulation of tumor infiltrating T lymphocytes (TIL), which were commonly marked by CD3 (CD3G was used in our study, because it was the most abundant in the microarray). CD3 TIL-associated genes (CD3 TILG) were overlapped with 26 CAFG that included COL family (S5 Table), the most potent negative prognostic factors among the CAFG (designated as fibrosis-CAFG in Fig 1A). CD3 TILG were alternately overlapped with CAFG related to TAMCG definitely from fibrosis-CAFG (non-fibrosis-CAFG in Fig 1A). These findings suggested that CD3 TIL may have heterogenous subpopulations that are prognostically distinct with or without COL family association. Thus, we did not perform further subpopulation analysis of CD3G.
TAMC-associated genes (TAMCG) were partially overlapped with CAFG
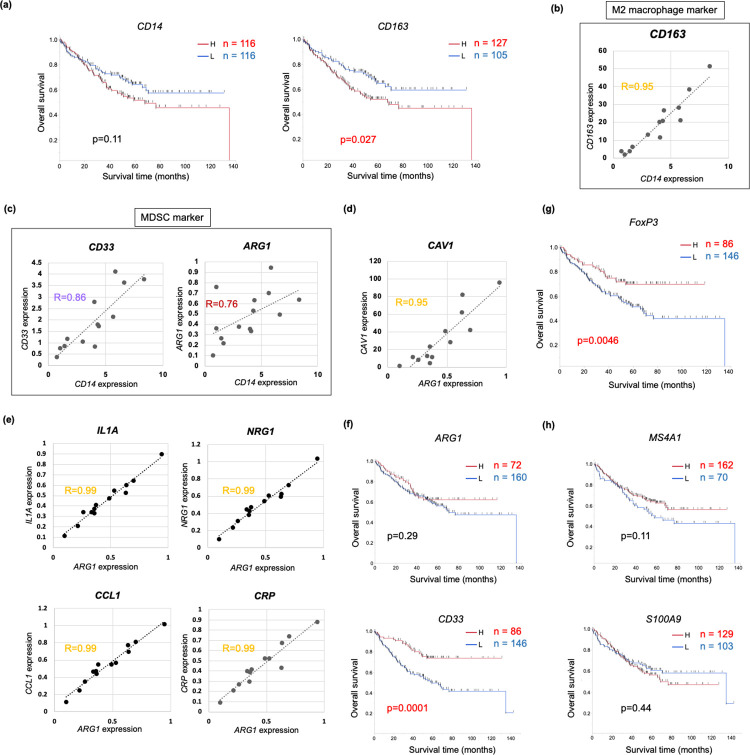
In this study, we used CD14 as a myeloid cell marker according to the previous report [24]. High CD14 expression showed poorer prognosis than low CD14 expression in colon cancer as expectedly, because TAMC was demonstrated to be conditionally involved in cancer promotion [40]. However, the prognostic difference was not statistically significance (p = 0.11), suggesting that TAMC contribution to prognosis may be weaker like TAE than CAF and/or CTL (Fig 6A).
Fig 6. Tumor-associated myeloid cells (TAMC) and other TME markers in cancer str of the CRC tumors (GSE35602).
(a) Survival curves for the TAMC marker CD14 and the M2 macrophage marker CD163 in colon cancer. (b) CD14 and CD163 expressions were closely associated in cancer str of the CRC tumors. (c) CD33, an iMDSC marker, and ARG1, a fMDSC marker, correlation with CD14 are shown in cancer str of the CRC tumors. (d) CAV1 expression was the most closely associated with ARG1 in cancer str of the CRC tumors. (e) ARG1 expression was tightly (R = 0.99) associated with expressions of inflammatory cytokines such as IL1A and NRG1 including unique TAMC markers (CCL1 and CRP) in cancer str of the CRC tumors. (f) Survival curves are shown for ARG1 (upper panel) and CD33 (lower panel) in colon cancer. (g) Survival curve is shown for FoxP3 expression in colon cancer. (h) Survival curves are shown for MS4A1, a B TIL marker (upper panel) and S100A9, a TAN marker (lower panel) in colon cancer.
TAMCG were identified as 32 gene probes (27 SYMBOL genes) (S4 Table), and intriguingly, the 27 TAMCG were overlapped with 25 CAFG, excluding CXCR4 and CD163. CD163 is an alternate well-established M2 macrophage marker, and its significance of prognostic stratification was confirmed differently from CD14 (p = 0.027, Fig 6A). This finding may represent functional aspects of CD163 [41, 42] in contrast to CD14 [43], although their expression was closely associated with each other in the TME (R = 0.95, Fig 6B). Intriguingly, CD163 expression was much higher than CD14.
Recent scRNA analysis revealed that CXCR4 was expressed in tumor infiltrates including myeloid cells as well as T-cells and B-cells in the TME [26], suggesting that CD14 TAMC uniquely may be accompanied by lymphocytes marked by CXCR4. Consistent with this hypothesis, non-fibrosis CAFG were commonly characterized by association with CD3 TIL and TAMC markers (Fig 1A). Prognostic relevance was not overlapped between CD3 TIL alone, TAMC alone, and both-associated CAFG (Fig 1A), suggesting that CAFG may represent subpopulations with differential immune infiltrates.
ARG1 and CD33 putatively representing differential myeloid derived suppressor cells (MDSC) subpopulations unexpectedly exhibited good prognosis in colon cancer
Among the myeloid cells, MDSC inhibit tumor immunity by being mobilized from myeloid to peripheral tissues, and they can be marked by CD33 or arginase1 (ARG1), while all the cells marked by them did not strictly represent MDSC [40]. CD33 expression represented immature myeloid cells recruited from bone marrow, and showed more closely associated with CD14 (R = 0.86), than ARG1 (R = 0.76) did (Fig 6C). Moreover, expressions of both MDSC markers, especially ARG1, were much lower than CD14 (Fig 2C). These findings suggested that ARG1 macrophage may be small subpopulation among the TAMC.
ARG1 has been demonstrated to be a fMDSC marker, because nutritional use of L-arginin, and L-arginin depletion in the TME reduces nutrition of other immunological cells such as CTL [44]. ARG1-associated genes (ARG1G) putatively representing fMDSC were not overlapped with any CAFG totally differently from CD14 (Fig 1B). On the other hand, CAV1 expression was enriched as top priority (S8 Table), and was closely associated with ARG1 expression (Fig 6D). As a result, ARG1G were shared with TAEG representing tumor angiogenesis (Fig 4D). This finding is consistent with the hypothesis that angiogenesis is critical for mobilization of ARG1 macrophage from myeloid tissues into the tumor stroma.
ARG1 expression was the most strongly associated with IL1A, NRG1, CCL1, and CRP in cancer str of the CRC tumors (Fig 6E) among the numerous associated genes (13 401 genes, Fig 2B). CCL1 could be an aggressive TAMC marker as recently shown [45], and fluorescent double immunostaining revealed that CCL1 with myeloid markers of CD204 were colocalized in human cancer tissues [45]. Nevertheless, unexpectedly ARG1 expression did not exhibit negative prognostic relevance in colon cancer (Fig 6F, top panel).
CD33, an alternate MDSC marker, had molecular characteristics (CD33-associated genes = 1 753, expression amounts in NCC210 = 3.76, and cS/E = 5.2 as shown in S9 Table), which were rather close to CD14 than ARG1, and CD33-associated genes were shared with the most abundantly expressed CAFG, IGFBP7. On the other hand, CD33-associated genes expression was uniquely associated with angiogenic CAFG such as VIM, PLAT, and MSN, suggesting that CAFG-induced angiogenesis may be involved in mobilization of CD33 macrophage. CD33 was not significant negative prognostic factors, either, and rather a good prognostic factor (Fig 6F, lower panel), suggesting that MDSC mobilization may be insufficient to immunologically suppress tumors in the TME of clinical tumors.
T cell subpopulations (Treg), B-TIL, and TAN and prognosis in colon cancer
FoxP3 expression representing Treg was very low like ARG1 expression representing fMDSC (Fig 2C) and showed low cS/E = 1.9 (Fig 2D). Both markers had a large number of the related genes (18 281 and 13 401 genes) in contrast to other TME markers (Fig 2B), and were overlapped with very few CAFG with high cS/E (S8 and S10 Tables, and Fig 1B). ARG1G were partially shared with Treg-associated genes (TregG) marked by FoxP3, and interestingly, many common genes between them were ascribed to low cS/E group (green box, S8 Table and S1A Fig), which included both ATF4 and H3F3A as representatives. Both gene expressions were closely associated with ARG1 expression (S1B Fig) and FoxP3 expression (S1C Fig), respectively, in cancer str of the CRC tumors.
We identified B-TIL-associated genes as only 46 genes as close association with B-TIL marker, MS4S1 (S11 Table), all of which did not show high or middle cS/E. In our prognostic analysis, MS4S1 expression did not show negative prognostic relevance (Fig 6H, upper panel). TAN-associated genes were also identified as only 1 gene (DEFB1) as its close association with S100A9 (S12 Table). S100A9 expression showed negative prognostic trend, however the difference was not statistically different (Fig 6H, lower panel).
Discussion
Although contribution of the differential TME components to patient prognosis remains elusive, our current study proposed that CAF activation represented by COL8A1 in addition to CTL activation reflected by CD8A are critical determinants of prognosis in colon cancers. Endothelial marker, PECAM1, and myeloid marker, CD14, did not show such strong negative prognostic relevance, but their associated genes are partially shared with CAFG that were of prognostic importance (Fig 1A), and the shared TAEG and TAMCG may be involved in tumor aggressiveness controlled by CAFG.
scRNA analysis recently demonstrated that CAF subpopulations were composed of vascular CAF, matrix CAF, cycling CAF, and developmental CAF, and the most major component (~60%) was vascular CAF [24]. This finding suggested that CAF plays an important role in tumor angiogenesis, and the common genes between CAFG and TAEG included PLAT followed by ANXA1 and PTRF according to their prognostic importance (Fig 1A). Angiogenesis has been demonstrated to be promoted recently by PLAT [46] and classically by ANXA1 [47]. These findings suggested that angiogenesis itself is not a potent prognostic relevance, but overlapped features of CAFG and TAEG represent aggressiveness of CRC.
The TAEG well represented tumor angiogenesis. The established TAE markers (PECAM1, CDH5, and TIE1) were correlated each other, accompanied by close association with pericyte marker (RGS5) and BM markers (COL4A1/COL4A2). The TAEG included genes involved in unique proteolysis (ETS1 and PLAT) and immunity (IFNGR1 and PDL1) as well as caveolin formation (Fig 3E–3G). Caveolin has been well known to be required for angiogenesis [28, 29], while unique proteolysis by ETS1 was recently demonstrated to be involved in transcriptional regulation of tissue destructive genes [31] that might include PLAT. Interestingly, degradation of BM including type IV COL is rate-limiting step for cancer intravasation into blood in metastasis [48], and PLAT as well as PLAU may be the initial members of the protease cascade [49].
In our current study, CAFG always showed negative prognostic relevance, among which COL family genes were uniquely enriched as the most aggressive phenotypes (designated as fibrosis CAFG in this study). Previous reports suggested that CAF induce collagen fibers and fibrosis of the tissue, which hardens the ECM and contributes to malignancy, suggesting that Fibrosis-CAFG is a poor prognostic factor via the COL family [50]. Such fibrosis CAFG comprising of COL5A2, COL5A1, COL12A1, COL3A1, and COL1A1 have recently demonstrated that they have unique molecular mechanism to CAF activation, respectively [51–53]. Moreover, fibrosis CAFG may include SPARC and TAGLN in addition to COL family genes themselves (Fig 1A), because they have been demonstrated to be involved in COL family expression induction [54, 55].
The fibrosis CAFG were shared with CD3 TILG but not with TAMCG, indicating that cancer prognosis may be greatly affected by definite components of TME. From a prognostic point of view, association with CD3 TILG alone, TAMCG alone and both may have differential CAFG molecular features, hence exhibiting different tumor immunity. In our data, CXCR4 was included among the TAMCG, and scRNA assay recently clarified that CXCR4 is expressed in lymphocytes as well as myeloid cells [26], suggesting that CD14 TAMC uniquely may be accompanied by lymphocytes marked by CXCR4. Consistent with this hypothesis, non-fibrosis CAFG were commonly characterized by association with CD3 TIL and TAMC markers (Fig 1A).
Among the CTLG, positive prognostic factors similarly with CD8A were rather few, where we identified such genes as B2M (the highest expression amounts of 40 or beyond) followed by TLR4 and IFNG (all of which belonged to low cS/E group, Fig 5D). IFNG is a well-established CTL activation indicator [35], and TLR4 was demonstrated to be involved in IFN pathway activation, mediated by IRF3 and IRF7 [38]. Intriguingly IFNGR1 was included among the TAEG, indicating that CTL effects may be the most greatly demanded at angiogenic sites (Fig 3G).
B2M truncating mutations were recently discovered in melanoma, resulting in loss of surface expression of major histocompatibility complex (MHC Class I) [56], and loss of such MHC Class I-mediated antigen presentation frequently recognized in MMR-deficient colon cancer rendered these tumors resistant to CTL-mediated tumor immunity [57]. Intriguingly, γδ T cells are proved to be effectors of immunotherapy in cancers with HLA Class I defects [58]. Our data showed that B2M expression was strongly associated with CD8A expression in the multiple probes (0.9<R<0.93, Fig 5D) not in the tumor cells, but in cancer str of the CRC tumors. The association in our study was not necessarily accompanied by MHC Class I antigen expression, suggesting that B2M expression may be response against CTL in stromal cells. As tumor of host sensing of IFNG was redundant and tumors were controlled without direct T cell cytotoxicity, multiple cell type targeted by IFNG should be controlled for tumor equilibrium [59]. Thus, IFNG expression was more potent than CD8A expression itself as a prognostic factor.
In this study, we explored ARG1G marked by ARG1 and TregG marked by FoxP3. They are considered to be subpopulations of TAMC and CD3 TIL, respectively and the marker genes showed very small amounts of expression as compared of CD14 and CD3G expression (Fig 2C) with low cS/E ratio (Fig 2D), however such trace expressions had many related genes (Fig 2B). ACTG2 was commonly associated with both markers (Fig 1B), although there have been no reports describing relations between both markers and ACTG2. On the other hand, many common genes associated with both ARG1 and FoxP3 were identified in gene groups with low cS/E expression ratio (S1A Fig), among which ATF4 is of particular interest, because MDSC function was recently demonstrated to be regulated by ATF4 [60].
MDSC was demonstrated to promote cross-tolerance in cancer by expanding Treg [61], and immune tolerance to tumors is often associated with accumulation of MDSC and an increase in the number of Treg [62]. Consistent with this, both ARG1G and TregG included common genes (S1A Fig). Moreover, Treg marked by FoxP3 showed potent positive prognostic factor in colon cancer (p = 0.0046, Fig 6G), which recapitulated the previous report [63]. These findings suggested that Treg mobilization may be insufficient to immunologically suppress tumors in the TME of clinical tumors like MDSC, either.
Our findings clarified that that both CAFG and CTLG, but other components of the TME were dependent on either factor. Among them, CTLG may be a good marker to predict Immune checkpoint Inhibitors efficacy, while CAFG remains elusive to control, as previous reports have shown that CRC patients with CAF infiltration have a poor prognosis [64, 65]. Among CAFG, the COL family is a particularly poor prognostic factor, suggesting that it could be used as a prognostic marker.
Many common genes were identified in both CAFG and CD3 TILG, indicating a heterogeneous genetic subpopulation (Fig 1A). In previous reports, these common genes are associated with poor prognosis in colorectal cancer. For example, COL1A1 has been reported to be linked with immune infiltrating cells [66], and RAB31 is expressed in CAFs, contributing to the malignant potential of colorectal cancer through the secretion of HGF in the tumor stroma [67].
In the present analysis, the results show that CTL is more strongly related to prognosis than TAE. This is consistent with a previous report that showed a clinical prognostic benefit of immune checkpoint inhibitors over anti-VEGF antibodies [68, 69]. In addition, although no treatment to control CAF has been realized, there is a report showing that control of secretome of CAF has anti-tumor effects [70], which may contribute to the development of novel therapies in the future.
This study has limitations. The selection criteria, such as the stroma/epithelia ratio or R-index of other CAFG, may introduce bias. SPARC was used as a criterion for CAFG selection because SPARC has been identified as an important fibroblast marker in previous reports [22]. It is possible that the selection of CAFG may differ slightly if other markers are used as criteria. Future research should explore alternative criteria for a more comprehensive understanding.
In conclusion, our current in silico analysis of the micro-dissected stromal molecular signatures with prognostic relevance elucidated comprehensive interrelations among the TME components and provides deep insights of the beautiful molecular landscape of stromal biology.
Supporting information
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
GSE35602: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35602 GSE17538: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17538.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Kinzler K.W., et al., Amplification units containing human N-myc and c-myc genes. Proc Natl Acad Sci U S A, 1986. 83(4): p. 1031–5. doi: 10.1073/pnas.83.4.1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong A.J., et al., Gene amplification of c-myc and N-myc in small cell carcinoma of the lung. Science, 1986. 233(4762): p. 461–4. doi: 10.1126/science.3014659 [DOI] [PubMed] [Google Scholar]
- 3.Zehnbauer B.A., et al., Characterization of N-myc amplification units in human neuroblastoma cells. Mol Cell Biol, 1988. 8(2): p. 522–30. doi: 10.1128/mcb.8.2.522-530.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fearon E.R., et al., Loss of genes on the short arm of chromosome 11 in bladder cancer. Nature, 1985. 318(6044): p. 377–80. doi: 10.1038/318377a0 [DOI] [PubMed] [Google Scholar]
- 5.Fearon E.R., Hamilton S.R., and Vogelstein B., Clonal analysis of human colorectal tumors. Science, 1987. 238(4824): p. 193–7. doi: 10.1126/science.2889267 [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B., et al., Genetic alterations during colorectal-tumor development. N Engl J Med, 1988. 319(9): p. 525–32. doi: 10.1056/NEJM198809013190901 [DOI] [PubMed] [Google Scholar]
- 7.Baker S.J., et al., Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science, 1989. 244(4901): p. 217–21. doi: 10.1126/science.2649981 [DOI] [PubMed] [Google Scholar]
- 8.Bos J.L., et al., Prevalence of ras gene mutations in human colorectal cancers. Nature, 1987. 327(6120): p. 293–7. doi: 10.1038/327293a0 [DOI] [PubMed] [Google Scholar]
- 9.Markowitz S., et al., Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science, 1995. 268(5215): p. 1336–8. doi: 10.1126/science.7761852 [DOI] [PubMed] [Google Scholar]
- 10.Powell S.M., et al., APC mutations occur early during colorectal tumorigenesis. Nature, 1992. 359(6392): p. 235–7. doi: 10.1038/359235a0 [DOI] [PubMed] [Google Scholar]
- 11.Howe J.R., The impact of DNA testing on management of patients with colorectal cancer. Ann Gastroenterol Surg, 2022. 6(1): p. 17–28. doi: 10.1002/ags3.12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bozic I., et al., Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci U S A, 2010. 107(43): p. 18545–50. doi: 10.1073/pnas.1010978107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satoh K., et al., Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc Natl Acad Sci U S A, 2017. 114(37): p. E7697–e7706. doi: 10.1073/pnas.1710366114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokota K., et al., WiNTRLINC1/ASCL2/c-Myc Axis Characteristics of Colon Cancer with Differentiated Histology at Young Onset and Essential for Cell Viability. Ann Surg Oncol, 2019. 26(13): p. 4826–4834. doi: 10.1245/s10434-019-07780-3 [DOI] [PubMed] [Google Scholar]
- 15.Sansom O.J., et al., Myc deletion rescues Apc deficiency in the small intestine. Nature, 2007. 446(7136): p. 676–9. doi: 10.1038/nature05674 [DOI] [PubMed] [Google Scholar]
- 16.Makohon-Moore A.P., et al., Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet, 2017. 49(3): p. 358–366. doi: 10.1038/ng.3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiter J.G., et al., Minimal functional driver gene heterogeneity among untreated metastases. Science, 2018. 361(6406): p. 1033–1037. doi: 10.1126/science.aat7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamashita K., et al., Epigenetic biomarkers of promoter DNA methylation in the new era of cancer treatment. Cancer Sci, 2018. 109(12): p. 3695–3706. doi: 10.1111/cas.13812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto Y., et al., The heterogeneity of cancer-associated fibroblast subpopulations: Their origins, biomarkers, and roles in the tumor microenvironment. Cancer Sci, 2023. 114(1): p. 16–24. doi: 10.1111/cas.15609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishida N., et al., Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res, 2012. 18(11): p. 3054–70. doi: 10.1158/1078-0432.CCR-11-1078 [DOI] [PubMed] [Google Scholar]
- 21.Smith J.J., et al., Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology, 2010. 138(3): p. 958–68. doi: 10.1053/j.gastro.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., et al., Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet, 2017. 49(5): p. 708–718. doi: 10.1038/ng.3818 [DOI] [PubMed] [Google Scholar]
- 23.Sahai E., et al., A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer, 2020. 20(3): p. 174–186. doi: 10.1038/s41568-019-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartoschek M., et al., Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat Commun, 2018. 9(1): p. 5150. doi: 10.1038/s41467-018-07582-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo Q., et al., Apatinib remodels the immunosuppressive tumor ecosystem of gastric cancer enhancing anti-PD-1 immunotherapy. Cell Rep, 2023. 42(5): p. 112437. doi: 10.1016/j.celrep.2023.112437 [DOI] [PubMed] [Google Scholar]
- 26.Hornburg M., et al., Single-cell dissection of cellular components and interactions shaping the tumor immune phenotypes in ovarian cancer. Cancer Cell, 2021. 39(7): p. 928–944.e6. doi: 10.1016/j.ccell.2021.04.004 [DOI] [PubMed] [Google Scholar]
- 27.De Palma M., Biziato D., and Petrova T.V., Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer, 2017. 17(8): p. 457–474. doi: 10.1038/nrc.2017.51 [DOI] [PubMed] [Google Scholar]
- 28.Woodman S.E., et al., Caveolin-1 knockout mice show an impaired angiogenic response to exogenous stimuli. Am J Pathol, 2003. 162(6): p. 2059–68. doi: 10.1016/S0002-9440(10)64337-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tahir S.A., et al., Tumor cell-secreted caveolin-1 has proangiogenic activities in prostate cancer. Cancer Res, 2008. 68(3): p. 731–9. doi: 10.1158/0008-5472.CAN-07-2668 [DOI] [PubMed] [Google Scholar]
- 30.Nassar Z.D., et al., Caveola-forming proteins caveolin-1 and PTRF in prostate cancer. Nat Rev Urol, 2013. 10(9): p. 529–36. doi: 10.1038/nrurol.2013.168 [DOI] [PubMed] [Google Scholar]
- 31.Yan M., et al., ETS1 governs pathological tissue-remodeling programs in disease-associated fibroblasts. Nat Immunol, 2022. 23(9): p. 1330–1341. doi: 10.1038/s41590-022-01285-0 [DOI] [PubMed] [Google Scholar]
- 32.Alburquerque-Bejar J.J., et al., MYC activation impairs cell-intrinsic IFNγ signaling and confers resistance to anti-PD1/PD-L1 therapy in lung cancer. Cell Rep Med, 2023. 4(4): p. 101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Titu L.V., Monson J.R., and Greenman J., The role of CD8(+) T cells in immune responses to colorectal cancer. Cancer Immunol Immunother, 2002. 51(5): p. 235–47. doi: 10.1007/s00262-002-0276-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scully T., et al., Enhancement of mammary tumour growth by IGFBP-3 involves impaired T cell accumulation. Endocr Relat Cancer, 2018. 25(2): p. 111–122. doi: 10.1530/ERC-17-0384 [DOI] [PubMed] [Google Scholar]
- 35.Yang J., et al., Application of the ELISPOT assay to the characterization of CD8(+) responses to Epstein-Barr virus antigens. Blood, 2000. 95(1): p. 241–8. [PubMed] [Google Scholar]
- 36.Kaplan G., et al., The expression of a gamma interferon-induced protein (IP-10) in delayed immune responses in human skin. J Exp Med, 1987. 166(4): p. 1098–108. doi: 10.1084/jem.166.4.1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blasius A.L., et al., Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol, 2006. 177(5): p. 3260–5. doi: 10.4049/jimmunol.177.5.3260 [DOI] [PubMed] [Google Scholar]
- 38.Ryu S., et al., The matricellular protein SPARC induces inflammatory interferon-response in macrophages during aging. Immunity, 2022. 55(9): p. 1609–1626.e7. doi: 10.1016/j.immuni.2022.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen A.O., et al., Tbc1d10c is a selective, constitutive suppressor of the CD8 T-cell anti-tumor response. Oncoimmunology, 2022. 11(1): p. 2141011. doi: 10.1080/2162402X.2022.2141011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bronte V., et al., Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun, 2016. 7: p. 12150. doi: 10.1038/ncomms12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kristiansen M., et al., Identification of the haemoglobin scavenger receptor. Nature, 2001. 409(6817): p. 198–201. doi: 10.1038/35051594 [DOI] [PubMed] [Google Scholar]
- 42.Kim W.K., et al., CD163 identifies perivascular macrophages in normal and viral encephalitic brains and potential precursors to perivascular macrophages in blood. Am J Pathol, 2006. 168(3): p. 822–34. doi: 10.2353/ajpath.2006.050215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bochkov V.N., et al., Protective role of phospholipid oxidation products in endotoxin-induced tissue damage. Nature, 2002. 419(6902): p. 77–81. doi: 10.1038/nature01023 [DOI] [PubMed] [Google Scholar]
- 44.Bronte V., et al., L-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol, 2003. 24(6): p. 302–6. doi: 10.1016/s1471-4906(03)00132-7 [DOI] [PubMed] [Google Scholar]
- 45.Fujikawa M., et al., Chemokine (C-C Motif) Ligand 1 Derived from Tumor-Associated Macrophages Contributes to Esophageal Squamous Cell Carcinoma Progression via CCR8-Mediated Akt/Proline-Rich Akt Substrate of 40 kDa/Mammalian Target of Rapamycin Pathway. Am J Pathol, 2021. 191(4): p. 686–703. doi: 10.1016/j.ajpath.2021.01.004 [DOI] [PubMed] [Google Scholar]
- 46.Noh K., et al., The hidden role of paxillin: localization to nucleus promotes tumor angiogenesis. Oncogene, 2021. 40(2): p. 384–395. doi: 10.1038/s41388-020-01517-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi M. and Schnitzer J.E., Impaired tumor growth, metastasis, angiogenesis and wound healing in annexin A1-null mice. Proc Natl Acad Sci U S A, 2009. 106(42): p. 17886–91. doi: 10.1073/pnas.0901324106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liotta L.A., et al., Metastatic potential correlates with enzymatic degradation of basement membrane collagen. Nature, 1980. 284(5751): p. 67–8. doi: 10.1038/284067a0 [DOI] [PubMed] [Google Scholar]
- 49.Hosen S.M.Z., et al., Metastatic phenotype and immunosuppressive tumour microenvironment in pancreatic ductal adenocarcinoma: Key role of the urokinase plasminogen activator (PLAU). Front Immunol, 2022. 13: p. 1060957. doi: 10.3389/fimmu.2022.1060957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piersma B., Hayward M.K., and Weaver V.M., Fibrosis and cancer: A strained relationship. Biochim Biophys Acta Rev Cancer, 2020. 1873(2): p. 188356. doi: 10.1016/j.bbcan.2020.188356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baghdadi M.B., et al., Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche. Nature, 2018. 557(7707): p. 714–718. doi: 10.1038/s41586-018-0144-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papanicolaou M., et al., Temporal profiling of the breast tumour microenvironment reveals collagen XII as a driver of metastasis. Nat Commun, 2022. 13(1): p. 4587. doi: 10.1038/s41467-022-32255-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi J.H., et al., Single-cell transcriptome profiling of the stepwise progression of head and neck cancer. Nat Commun, 2023. 14(1): p. 1055. doi: 10.1038/s41467-023-36691-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popova A.P., et al., Autocrine production of TGF-beta1 promotes myofibroblastic differentiation of neonatal lung mesenchymal stem cells. Am J Physiol Lung Cell Mol Physiol, 2010. 298(6): p. L735–43. doi: 10.1152/ajplung.00347.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei H.Y., et al., SPARC modulates expression of extracellular matrix genes in human trabecular meshwork cells. Acta Ophthalmol, 2012. 90(2): p. e138–43. doi: 10.1111/j.1755-3768.2011.02283.x [DOI] [PubMed] [Google Scholar]
- 56.Zaretsky J.M., et al., Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med, 2016. 375(9): p. 819–29. doi: 10.1056/NEJMoa1604958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sade-Feldman M., et al., Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun, 2017. 8(1): p. 1136. doi: 10.1038/s41467-017-01062-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Vries N.L., et al., γδ T cells are effectors of immunotherapy in cancers with HLA class I defects. Nature, 2023. 613(7945): p. 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walsh M.J., et al., IFNγ is a central node of cancer immune equilibrium. Cell Rep, 2023. 42(3): p. 112219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Halaby M.J., et al., GCN2 drives macrophage and MDSC function and immunosuppression in the tumor microenvironment. Sci Immunol, 2019. 4(42). doi: 10.1126/sciimmunol.aax8189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serafini P., et al., Myeloid-derived suppressor cells promote cross-tolerance in B-cell lymphoma by expanding regulatory T cells. Cancer Res, 2008. 68(13): p. 5439–49. doi: 10.1158/0008-5472.CAN-07-6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan P.Y., et al., Immune stimulatory receptor CD40 is required for T-cell suppression and T regulatory cell activation mediated by myeloid-derived suppressor cells in cancer. Cancer Res, 2010. 70(1): p. 99–108. doi: 10.1158/0008-5472.CAN-09-1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salama P., et al., Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol, 2009. 27(2): p. 186–92. doi: 10.1200/JCO.2008.18.7229 [DOI] [PubMed] [Google Scholar]
- 64.Dienstmann R., et al., Relative contribution of clinicopathological variables, genomic markers, transcriptomic subtyping and microenvironment features for outcome prediction in stage II/III colorectal cancer. Ann Oncol, 2019. 30(10): p. 1622–1629. doi: 10.1093/annonc/mdz287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guinney J., et al., The consensus molecular subtypes of colorectal cancer. Nat Med, 2015. 21(11): p. 1350–6. doi: 10.1038/nm.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeng X., et al., Expression, prognostic value and potential immunotherapeutic target of COL1A1 in colon cancer. Cell Mol Biol (Noisy-le-grand), 2023. 69(15): p. 120–125. doi: 10.14715/cmb/2023.69.15.21 [DOI] [PubMed] [Google Scholar]
- 67.Yang T., et al., Increased RAB31 Expression in Cancer-Associated Fibroblasts Promotes Colon Cancer Progression Through HGF-MET Signaling. Front Oncol, 2020. 10: p. 1747. doi: 10.3389/fonc.2020.01747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saltz L.B., et al., Bevacizumab in Combination With Oxaliplatin-Based Chemotherapy As First-Line Therapy in Metastatic Colorectal Cancer: A Randomized Phase III Study. J Clin Oncol, 2023. 41(21): p. 3663–3669. doi: 10.1200/JCO.22.02760 [DOI] [PubMed] [Google Scholar]
- 69.André T., et al., Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N Engl J Med, 2020. 383(23): p. 2207–2218. doi: 10.1056/NEJMoa2017699 [DOI] [PubMed] [Google Scholar]
- 70.Kumar V., et al., Cancer-Associated Fibroblasts Neutralize the Anti-tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors. Cancer Cell, 2017. 32(5): p. 654–668.e5. doi: 10.1016/j.ccell.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]