Abstract
Genetic diversity is critical to crop breeding and improvement, and dissection of the genomic variation underlying agronomic traits can both assist breeding and give insight into basic biological mechanisms. Although recent genome analyses in plants reveal many structural variants (SVs), most current studies of crop genetic variation are dominated by single-nucleotide polymorphisms (SNPs). The extent of the impact of SVs on global trait variation, as well as their utility in genome-wide selection, is not yet understood. In this study, we built an SV data set based on whole-genome resequencing of diverse sorghum lines (n = 363), validated the correlation of photoperiod sensitivity and variety type, and identified SV hotspots underlying the divergent evolution of cellulosic and sweet sorghum. In addition, we showed the complementary contribution of SVs for heritability of traits related to sorghum adaptation. Importantly, inclusion of SV polymorphisms in association studies revealed genotype–phenotype associations not observed with SNPs alone. Three-way genome-wide association studies (GWAS) based on whole-genome SNP, SV, and integrated SNP + SV data sets showed substantial associations between SVs and sorghum traits. The addition of SVs to GWAS substantially increased heritability estimates for some traits, indicating their important contribution to functional allelic variation at the genome level. Our discovery of the widespread impacts of SVs on heritable gene expression variation could render a plausible mechanism for their disproportionate impact on phenotypic variation. This study expands our knowledge of SVs and emphasizes the extensive impacts of SVs on sorghum.
High-throughput sequencing technologies have sped up the process of discovery for natural genetic variation. However, as a consequence of limited read length and variant calling algorithms, single-nucleotide polymorphisms (SNPs) and small indels are disproportionately overrepresented within characterized sequence variation (Audano et al. 2019). Nevertheless, a growing number of research projects indicate that structural variations (SVs), including large (>30-bp) deletions (DELs), insertions (INSs), duplications (DUPs), inversions (INVs), and translocations (TRAs) (Feuk et al. 2006), greatly contribute to crop phenotypic diversity and selection for physiological and morphological phenotypes (Alonge et al. 2020; Li et al. 2020). Two major SV classes have been proposed to explain how SVs are formed and how they impact phenotypes. The first involves genome rearrangement, such as INVs and TRAs; the second includes large DELs, INSs, and DUPs, collectively referred to as copy number variations (CNVs) (Scherer et al. 2007; Alkan et al. 2011). Because SVs are diverse and influence gene sequence and expression via a myriad of mechanisms, it has been challenging to assess the impact of SVs systematically and comprehensively. In addition, current sequencing and detection technologies leave the bulk of SVs poorly resolved, so they are often not included in studies of genome-wide variation.
Because of their low cost, mature and reliable technology, and proven high accuracy reads, second or “next-generation” short-read sequencing technologies are still the main technology for most studies. Long-read sequencing techniques, such as Pacific Biosciences (PacBio) HiFi and ultralong Oxford Nanopore Technologies (ONT), are both more expensive and more demanding of DNA quantity and quality. Numerous tools have been developed to detect SVs using paired-end short reads over the past decade. There are primarily three strategies used in popular algorithms for SV calling based on short-read sequencing: read-pair technologies, read-depth methods, and split-read approaches (Alkan et al. 2011). However, there is currently no individual algorithm that is able to successfully identify all types of SVs across the entire range of sizes, as strategies display a diversity of strengths and weaknesses in their ability to detect various types of SVs. Utilization of multiple algorithms based on different strategies for SV detection has been proven a viable way to overcome this issue. Zarate et al. (2020) found that reaching a consensus among multiple short-read SV callers can lead to improved precision without significantly compromising sensitivity in human genome. Alonge et al. (2020) deployed three independent tools to call SVs from the short-read alignments of 847 tomato accessions and successfully identified the diverse modern and domesticated samples that maximize SV diversity. In this study, we developed an ensemble pipeline for SV calling based on five independent algorithms involving different SV detection strategies: Sentieon (Kendig et al. 2019), which uses split-read strategy to call SVs and was also used for SNP calling in this study; DELLY (Rausch et al. 2012), which uses paired-end, split-read, and read-depth strategies to sensitively and accurately delineate SVs; Smoove (https://github.com/brentp/smoove), which is an improved version of lumpy and integrates the paired-end and split-read strategies; Manta (Chen et al. 2016), which combines paired and split-read evidence during SV discovery; and CNVnator (Abyzov et al. 2011), which uses read-depth methods.
The standard assumptions of genome-wide association studies (GWAS) include the concept that each SNP used in the study will capture heritable variation via “tagging” any other SNPs, or SVs, in the genome within the range of local linkage disequilibrium (LD) (Kruglyak 2008). For this reason, it has been widely assumed that causative SVs will be detected in GWAS via being “tagged” by adjacent SNPs in LD. Recent evidence has shed doubt on this assumption in plants, owing to the limited LD of many SVs with surrounding SNPs in soybean (Fliege et al. 2022) and maize (Yang et al. 2019). For this reason, many of the effects of SVs on crop phenotypes may still be unknown.
Sorghum (Sorghum bicolor (L.) Moench) is a versatile crop with wide adaptability and broad applications. It has been selectively bred into different varieties for different end uses such as grain sorghum for human consumption; forage sorghum, which is primarily for feeding livestock; and sweet sorghum, which can be used as a food sweetener or for biofuel and chemical production. These types have been created by selective breeding following sorghum domestication in northern Africa ∼10,000 yr ago and its subsequent spread to a variety of areas across Africa, India, the Middle East, and east Asia (Lobell et al. 2008; Morris et al. 2013a). Broad distribution of SVs in sorghum and correlation with local adaptation has been reported (Songsomboon et al. 2021), and each specific sorghum type is characterized by particular morphological and physiological features. A better understanding of the genetic pathways and mechanisms that underpin these features is essential for accelerating future sorghum breeding and improvement. Here, we aimed to build an SV data set based on whole-genome short-read resequencing of 363 sorghum lines from the global Bioenergy Association Panel (BAP) (Brenton et al. 2016) using a fusion workflow, to investigate the impacts of SVs on sorghum genetics, and to find new knowledge of allelic variation that can be used in crop improvement.
Results
Identification of genome-wide variations in the BAP
To explore the genetics of SVs within sorghum germplasm, we used the Illumina short-read whole-genome resequencing data from 363 global sorghum accessions in the BAP (Supplemental Table S1; Brenton et al. 2016; https://terraref.org/). This panel was developed and characterized as a set of racially, geographically, and phenotypically diverse lines aiming to cover a significant portion of the genetic variation within sorghum (Hu et al. 2019). The panel has been classified into three broad types: cellulosic, grain, and sweet (Brenton et al. 2016). The mean sequencing depth is ∼29×, and the mean breadth of the coverage is ∼91%. Sorghum BTx623 (v3.1.1) from Phytozome (https://phytozome.jgi.doe.gov/) was used as the reference genome in SNP and SV calling. To enhance the accuracy and sensitivity of SV detection, five inference software packages—Sentieon (v202010.01) (Kendig et al. 2019), DELLY (v0.8.1) (Rausch et al. 2012), Smoove (https://github.com/brentp/smoove), Manta (v1.6.0) (Chen et al. 2016), and CNVnator (v0.3.3) (Abyzov et al. 2011)—involving different SV detection strategies were applied to the data. We conducted a simulation study to estimate the recall and precision in SV calling using various thresholds, considering SVs supported by one to five callers (Supplemental Fig. S1; Supplemental Results). Based on the simulation result, only SVs supported by at least two callers were reported by our fusion workflow, and the two calls must agree on the type and the strand of SV. A total of 7,162,000 filtered SNPs and 622,236 high-confidence SVs were identified on 10 chromosomes, including 158,614 DELs, 18,028 DUPs, 216 INSs, 142,219 INVs, and 303,159 TRAs (Supplemental Fig. S2A).
To validate the quality of the identified SVs, three new chromosome-scale de novo assemblies (Supplemental Fig. S2B–D; Supplemental Table S2) and two public whole-genome sequence assemblies available at Phytozome (https://phytozome-next.jgi.doe.gov/) for five BAP accessions (PI 329545, PI 337680, PI 651495, Rio [S. bicolor Rio v2.1], and RTx430 [S. bicolor RTx430 v2.1]) were aligned to the standard reference genome (BTx623 v3.1.1) and SVs called by assembly comparison. SVs identified from whole-genome alignment information were then compared with the SVs detected by the fusion workflow using Illumina data. Overall, we observed a high percentage of overlapping fusion workflow calls with assembly comparison for both DEL/INS and DUP and traceable breakpoints of TRA and INV. We concluded that our fusion pipeline is sufficiently sensitive and accurate for SV detection (see Supplemental Results; Supplemental Fig. S3).
We then surveyed the distribution of genes and variants. Annotated genes are primarily located toward the telomeres, and most of the identified SNPs are distributed in the gene-sparse regions flanking the centromeres (Fig. 1A). In contrast, detected SVs were mainly situated in the gene-rich regions (Fig. 1A,B). Frequent TRAs and INVs were observed from the breakpoints in gene-rich regions (Fig. 1B). Even though the density of called SVs is higher in gene-rich regions, only 0.2% of these SVs affected exons directly.
Figure 1.
Distribution of genome-wide variations in the sorghum Bioenergy Association Panel (BAP). (A) Distribution of gene density and copy number variant (CNV) type structural variants (SVs), including deletions (DELs), duplications (DUPs), and insertions (INSs). From the outermost layer to the innermost layer of the Circos (Krzywinski et al. 2009) plot are chromosomes (a), DEL density (b), DUP density (c), INS density (d), single-nucleotide polymorphism (SNP) density (e), and gene density (f). Annotated genes were primarily located flanking centromeres as expected. Most of the identified SNPs were distributed in the gene-sparse regions. CNV-type SVs showed a different distribution pattern than did SNPs and were mainly situated in the gene-rich regions. The densities of the genes and CNV-type SVs were calculated in 500-kb windows. (B) Distribution of gene density and rearrangement (REA)-type SVs, including inversions (INVs) and translocations (TRAs). From the outermost layer to the innermost layer of the Circos plot are chromosomes (a), gene density (f), INV density (g), and TRA density (h). The core of the Circos plot is a spanning diagram of the identified TRAs. The links show the two breakpoints located in different chromosome positions for each TRA. Each link is colored by the chromosome color of the start position of the corresponding TRA. As with CNV-type variations, identified INVs and TRAs were distributed mainly in gene-enriched zones. Frequent rearrangement flows were observed between chromosomes. The densities of the genes and REA-type variants were calculated in 500-kb windows. The link diagram was evenly thinned (1/256) from the total TRAs.
Because of the limitations of the SV detection algorithms based on short reads, the length of the INV and TRA cannot be precisely inferred from the positions of the two breakpoints of an SV. We further examined the length distribution of DELs, DUPs, and INSs, which showed that most SVs were relatively small, but a substantial minority are large: 30–250 bp, 30.3%; 250–500 bp, 13.1%; 500 bp–1 kb, 13.9%; 1 kb–2 kb, 9%; and >2 kb, 33.6%. Two size bands of enrichment were observed at ∼75 bp and 250 bp for DEL (Supplemental Fig. S4). There were also obvious peaks at ∼150 bp and 60 bp for DUP and INS, respectively. These may reflect specific, abundant mobile elements. Sequence composition survey of the SVs indicated that the two most abundant transposable element sequence signatures were Gypsy and EnSpm (Supplemental Fig. S5). These well-known LTR transposable elements play significant roles in plant genome structure and evolution.
The “domestication syndrome” in sorghum: photoperiod sensitivity and variety type
To explore the population structure of the BAP based on SVs, we first investigated the distribution of SVs across the BAP. Structural changes identified in sweet sorghum and typical grain sorghum were fewer than those observed in cellulosic sorghum (Fig. 2A; Supplemental Fig. S6). Photoperiod sensitivity is a key trait that must be modified to reconcile environmental cues, reproductive cycles, and planting/harvest during crop domestication and radiation from center of origin. Modification of photoperiod sensitivity is accompanied by the occurrence of other domestication traits, considered collectively the “domestication syndrome” (Allaby et al. 2008; Liu et al. 2015; Song et al. 2017; Lu et al. 2020). Genotype data from a total of 339 sorghum lines with variety type and photoperiod information were used for population structural analyses. Principal component analysis (PCA) based on SNP, SV, or combined SNP + SV data sets showed a similar population structure pattern (Fig. 2B; Supplemental Fig. S7). We examined the first two principal components in a region deviating from the main population (PC1 > 50 and PC2 <−50) in the SV PCA results and found, as expected, that the photoperiod-sensitivity feature is strongly linked with cellulosic sorghum whereas the derived sweet sorghum has photoperiod insensitive characteristics (Fig. 2B). Sorghum, unusually, has bidirectional gene flow between wild/weedy relatives and cultivated sorghum lines in sympatric and allopatric species (Mace et al. 2013). Exceptions to the population clusters may reflect gene flow. Grain and sweet sorghum are not well differentiated, although SVs show somewhat better separation than SNPs for these variety types. This finding prompted us to explore the relationship between photoperiod sensitivity and sorghum variety types via haplotype network analysis. As shown in Figure 2C, the edges connecting cellulosic sorghum varieties appear to correspond to those for photoperiod sensitivity, whereas the edges for sweet sorghum correspond to those for photoperiod insensitivity in minimum spanning trees derived from both SNP and SV data sets.
Figure 2.
Structural variation (SV) distributions in different sorghum variety types and population structural analyses. (A) Violin and boxplot for SVs count distributions in cellulosic, grain, and sweet sorghum groups. DEL, DUP, INS, INV, and TRA count distributions were calculated separately in cellulosic (green, left), grain (yellow, center), and sweet (red, right) sorghum groups. Compared with the other two sorghum variety types, cellulosic sorghum contained the most called SVs, indicating that sweet sorghum may be closer to grain sorghum than cellulosic sorghum in SV content as the reference BTx623 is a typical grain sorghum. (B) Principal component analysis (PCA) based on SNP (left) and SVs (right). Photoperiod sensitivity—Photoperiod insensitive (circle), Photoperiod sensitive (triangle), and unknown (square)—and sorghum variety type information—cellulosic (green), grain (yellow), and sweet (red)—were differentiated by PCA based on SNPs and SVs. In SV PCA, the corner in the upper antidiagonal with the translucent green background shows the zones with PC1 > 50; the corner in the lower antidiagonal with the translucent red background shows the area with PC2 < −50. The percentages in both colored corners represent the proportions of different sorghums with the corresponding attributes. (C) Minimum spanning trees. Minimum spanning trees were shown based on both SNPs (top) and SVs (bottom). In the first column, sorghum variety type information is coded: cellulosic (green), grain (yellow), and sweet (red). In the second column, photoperiod-sensitivity information is coded: photoperiod insensitive (sky blue) and photoperiod sensitive (purple). In the third column, distribution of the selected representative cellulosic (green) and representative sweet (red) is shown from the PCA analysis. In general, sweet sorghum spreading branches matched those of photoperiod-sensitive sorghum lines, whereas cellulosic sorghum spreading branches matched those of photoperiod-insensitive sorghum lines and variety type.
Identification of SVs underlying the divergent evolution of cellulosic and sweet sorghum
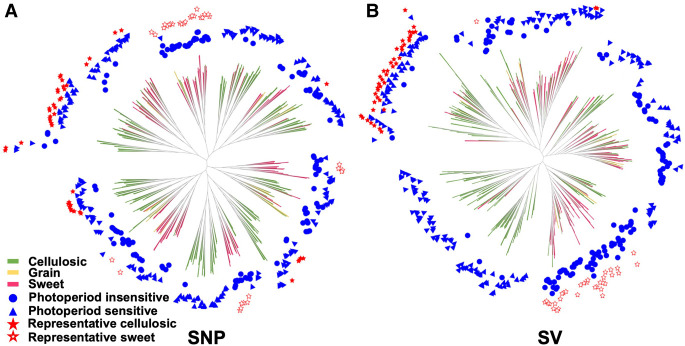
Structural sequence divergence, initiating from hotspots along chromosomes and subsequently expanding through the accumulation of minor genomic variants, has been found to be an important driver of divergent evolution (Song et al. 2002). For the purpose of investigating the location of structural genetic differences that may underlie the divergent evolution of cellulosic and sweet sorghum, we curated 43 cellulosic (PC1 > 50) and 33 sweet (PC2 < −50) sorghum lines from the BAP with consistent genetic clustering based on the SV PCA results (Fig. 2B; Supplemental Tables S3, S4). Genetic-relatedness analyses based on both the SNP and SV data sets were performed. The maximum likelihood tree based on the SV data set shows as expected that the selected cellulosic (solid red pentagram) and sweet (hollow red pentagram) sorghums were each grouped into one cluster (Fig. 3A,B). These results indicate that the curated sorghum lines potentially underwent strong variety-specific selections during sorghum domestication and breeding. To investigate the fixation index of the SVs between selected cellulosic and sweet sorghum groups, FST for each site was estimated between the cellulosic group and the sweet group in the BAP based on whole-genome SNPs. Before establishing the selection threshold, we examined the FST distribution in our study and found that it captured the top 1% of the SNP FST distribution when FST ≥ 0.15. Hence, we considered FST ≥ 0.15 a robust threshold for our selection analysis. There were 1637 SNPs shown to be highly differentiated between the cellulosic and sweet subpopulations with FST ≥ 0.15 (from 0.15 to 0.36). SVs between the curated 43 cellulosic and 33 sweet sorghum lines were then compared with the loci of the 1637 highly differentiated SNPs. Comparison showed that 76% (1250/1637) of the highly differentiated SNPs were adjacent to at least one SV (range from one to 45 SVs) within 10 kb (Supplemental Table S5). This result indicates that the SVs identified between the curated 43 cellulosic and 33 sweet sorghum lines likely underwent strong selection while accompanied by the closely linked SNP loci, and the 43 cellulosic and 33 sweet sorghums selected based on the SV PCA results were representative lines that underwent differential selection during the divergent improvement of cellulosic (tropical landraces) and sweet sorghum subpopulations.
Figure 3.
Phylogenetic trees of 339 sorghum lines in the BAP. Phylogenetic trees were conducted using SNPs (A) and SVs (B) as characters. Sorghum variety type and photoperiod sensitivity were marked as different colors and shapes: cellulosic (green line), grain (yellow line), sweet (red line), photoperiod insensitive (blue solid circle), photoperiod sensitive (blue solid triangle), selected representative cellulosic accessions (red solid pentagram), and selected representative sweet accessions (red hollow pentagram). The maximum likelihood phylogenetic tree based on the SV data set shows a clearer classification of phylogeny, sorghum variety type, and photoperiod sensitivity than the maximum likelihood phylogenetic tree based on SNPs, with selected cellulosic and sweet sorghums being almost monophyletic based on SV data.
To find potential hypervariable regions across the groups, we then examined the SV detection frequency in these two groups, consisting of the 43 curated cellulosic and 33 curated sweet sorghum lines respectively, across 1-Mb windows. Common SVs that were present in both the cellulosic and sweet sorghum groups were excluded to reduce the background noise. Genomic regions with obvious variable SV frequency between the representative cellulosic and sweet sorghum groups were observed (Fig. 4A). The heatmap of SV detection frequency manifested that 186 out of 688 SV frequency windows, including 73 continuous genomic regions, showed significant differences (adjusted P-value < 0.01 and average SV difference between two groups was ≥20) in frequency between representative cellulosic and sweet sorghums (Fig. 4B; Supplemental Table S6). Some hotspots of SV frequency we detected have been reported in previous publications; 56–57 Mb on Chromosome 1 and 61–62 Mb on Chromosome 2 have been identified as hotspots for controlling protein, starch, and amylose content (Ayalew et al. 2022). In addition, 52.23–61.18 Mb on Chromosome 1, 2.52–11.43 Mb on Chromosome 2, and 1.32–3.95 Mb on Chromosome 3 were also hotspots for source-sink-related traits (Chiluwal et al. 2022). Boatwright et al. (2022) identified 18 genomic regions under selection across six generic sorghum subpopulations underlying the evolutionary divergence during domestication. Six out of 10 selection regions with prior QTL information were covered by our identified SV hotspots, whereas only one out of eight selection regions without prior QTL information was covered by our identified SV hotspots.
Figure 4.
Typical SVs in the divergent evolution of cellulosic and sweet sorghum. (A) Circos (Krzywinski et al. 2009) plot for the SV frequency differences between the selected representative cellulosic sorghum group and the sweet sorghum group: (a) chromosomes, (b) SV frequency of cellulosic group, and (c) SV frequency of sweet group. SV frequencies were calculated in 1-Mb sliding windows in each group. Hypervariable genomic regions were observed between representative cellulosic and sweet sorghum groups. (B) Heatmap of SV frequency for selected representative cellulosic and sweet sorghum lines. SV frequencies were detected individually and chromosome by chromosome in 1-Mb sliding windows. The vertical axis stands for the stacked heatmaps for each sorghum line per chromosome. A cyan bar shows the range of the stacked heatmaps for cellulosic sorghum lines in each chromosome. A magenta bar shows the range of the stacked heatmaps for sweet sorghum lines in each chromosome. The x-axis stands for the physical distance for every chromosome. High SV detection frequencies were observed toward the telomeres in each chromosome for both the cellulosic group and sweet group. One hundred eighty-six out of 688 SVs frequency windows were tested as significant difference windows between representative cellulosic and sweet sorghum accessions. A red dash box indicates the hotspots previously reported covered by the 186 significant difference windows.
SVs reveal extensive contributions to heritability
Decades of studies have provided evidence that, despite their rarity compared with SNPs, SVs account for a substantial fraction of characterized molecular genetic variation with phenotypic consequences (Freeman et al. 2006). To examine the likely impact of the identified SVs on gene function, we evaluated the predicted functional effects of the variants in our SV and SNP data sets. As shown in Supplemental Figure S8, SVs were more likely to have large impacts on gene function, for example gene duplication, exon loss, codon frame shift, and transcript ablation, whereas SNPs generally were predicted to have lower impacts. The annotation of the predicted impacts of SNPs and SVs on sorghum gene function suggested that SVs could have a significant impact on functional genetic variation in sorghum.
We then investigated the potential contributions of our SV set to the inheritance of 29 quantitative traits and one binary trait (Supplemental Table S7; Brenton et al. 2016, 2020). The overall proportion of variance explained by the additive effect of genomic variants (narrow-sense heritability) was estimated by using mixed model analysis for each trait for whole-genome SNP variation only, as well as a combined set of SNPs and SVs, that is, SNP + SV. The estimated heritability ranged from 2%–57% (median, 20%) when we considered only SNP variation. However, the estimated heritability increased substantially, by 16%–99% (median, 26.5%) for all but one trait (2015_ADF, which stands for acid detergent fiber content in 2015), when taking both SNP and SVs into account. The additive effect of SNP + SV was particularly marked for the trait of photoperiod sensitivity and for the sorghum variety type itself when used as a phenotype (Fig. 5A). Compared with the heritability contributed by SNP data alone, the reduced heritability of 2015_ADF for SNP + SV (from 57% to 33%) likely resulted from the opposite additive effects contributed by SNP and SV data sets separately. Overall, CNV-type variations consistently produced higher heritability estimates than SNPs for nearly all traits and explained 6.2% more of the phenotypic variance than REA-type variations (Fig. 5B). These findings show that, although SNPs are generally able to capture the bulk of the heritable genetic effects on phenotype, SVs accounted for a substantial proportion of the missing heritability in SNP-based analysis for most traits.
Figure 5.
SV contributes substantially to heritability. (A) Heritability estimates are improved by the addition of SVs. Narrow-sense heritability was estimated for 29 quantitative traits and one binary trait. The upper diagonal colored by melon is the area in which the heritability of SNP + SV is greater than the heritability of SNP only (h2SNP + SV > h2SNP). The lower diagonal colored by spring green is the area in which the heritability of SNP + SV is less than the heritability of SNP only (h2SNP + SV < h2SNP). The diagonal line illustrates where heritability estimates with and without SVs are the same (h2SNP + SV = h2SNP). Thirty traits were dotted by different colors in the plot. The embedded upper triangular dot plot shows the magnification of the shaded area. All of traits, except for 2015_ADF, were observed in the upper h2SNP + SV > h2SNP area, which indicates the predicted total heritability increase for most traits when taking both SNP and SVs into account compared with taking SNPs only into consideration. This was particularly marked for two traits: photoperiod sensitivity (pointed by blue arrow) and sorghum variety type (pointed by red arrow). (B) A bar plot for estimation of heritability contribution from SNP, copy number variations (CNVs), and REA-type variation. Narrow-sense heritability was estimated for 29 quantitative traits and one binary trait (Supplemental Table S7): (2014_ADF) acid detergent fiber content in 2014; (2014_Days_to_harvest) days to harvest in 2014; (2014_Dry_Weight) dry weight of biomass in 2014; (2014_Lignin) lignin content in 2014; (2014_NDF) neutral detergent fiber in 2014; (2014_NFC) nonfibrous carbohydrates content in 2014; (2014_WSC) water-soluble carbohydrates content in 2014; (2015_ADF) acid detergent fiber content in 2015; (2015_Days_to_harvest) days to harvest in 2015; (2015_Dry_Weight) dry weight of biomass in 2015; (2015_Lignin) lignin content in 2015; (2015_NDF) neutral detergent fiber in 2015; (2015_NFC) nonfibrous carbohydrates content in 2015; (2015_WSC) water-soluble carbohydrates content in 2015; (ADF) average acid detergent fiber content of 2014 and 2015; (Anthesis_date) date of anthesis; (Brix_maturity) brix content in maturity stage; (Brix_milk) brix content in milk stage; (Days_to_harvest) average days to harvest of 2014 and 2015; (Dry_tons_per_acre) dry tons per acre; (Dry_Weight) dry weight of biomass; (Lignin) lignin content; (NDF) average neutral detergent fiber content of 2014 and 2015; (NFC) average nonfibrous carbohydrates content of 2014 and 2015; (Pericarp_pigmentation) pericarp pigmentation; (Photoperiod_sensitivity) photoperiod sensitivity; (Sorghum_type) sorghum variety type (sweet, grain, and cellulosic); (Stalk_height) stalk height; (Total_fresh_weight) total fresh weight; and (WSC) average water-soluble carbohydrates content of 2014 and 2015. Blue bars, orange bars, and gray bars with green frame indicate the heritability contributions from SNPs, CNV-type variations, and REA-type variations, respectively. For most of the traits, CNV-type variations explained more variance than REA-type variations.
SV data allow detection of new GWAS associations
To further investigate the causative genomic loci associated with the increased heritability gained by adding SVs to the polymorphism data set, we performed GWAS based on the whole-genome SNP, SV, and combined SNP and SV data sets. First, we investigated associations with a sorghum seed pericarp pigmentation trait, “Pericarp_pigmentation,” a well-studied trait whose global variation is owing largely to the Y locus, which encodes a MYB transcription factor Yellow seed1 (Y1), although the causative variants in this gene have not been definitely identified (Ibraheem et al. 2010; Morris et al. 2013b; Rhodes et al. 2014). GWAS based on SV found three significant association signals for seed pericarp pigmentation, including an SV underlying the Y1 gene (Sobic.001G397900) as expected (Fig. 6A), whereas SNP and SNP + SV analyses did not detect association at this locus (Fig. 6B,C). The SV (1.5 kb downstream from Y1) underlying the Y1 locus was called as a TRA from Chromosome 1 to Chromosome 4 (Fig. 6D). The breakpoint on Chromosome 4 was also detected by SV-based GWAS. Another substantial SV association signal was detected on Chromosome 8. The polymorphism associated with this locus is a 2.6-kb DEL/INS located 3.2 kb upstream of TIM22-2 (Sobic.008G111800), a mitochondrial import inner membrane translocase and a homolog of a protein involved in seed development in Arabidopsis (Zhang et al. 2023b). Further haplotype analyses of the TRA allele underlying the Y1 locus on Chromosome 1 and the 2.6-kb DEL/INS on Chromosome 8 validated their significant correlation with phenotypic variance in “Pericarp_pigmentation” (Supplemental Figs. S9A–D, S10A–D; Supplemental Tables S8 and S9; see Supplemental Results for details). Our GWAS results for seed pericarp pigmentation based on SVs thus not only found a significant SV association for the well-studied Y1 locus, which was not detected in SNP GWAS, but also identified a potential TRA involved in the genesis of this locus and a compelling new candidate gene for the control of seed pericarp pigmentation.
Figure 6.
Manhattan plots of genome-wide association study (GWAS) results for “Pericarp Pigmentation.” (A) GWAS result for the pericarp pigmentation trait based on SVs alone. Three significant signals were detected using a compressed mixed linear model (CMLM) including a signal underlying the well-known pericarp pigmentation–related Y1 gene. The corresponding signal underlying the Y1 was a TRA variation between Chromosome 1 and Chromosome 4. The signal at the other breakpoint on Chromosome 4 of the TRA underlying Y1 was also detected (solid red inverted triangle). The signal on Chromosome 8 was a 2.6-kb DEL/INS located near TIM22-2 (Sobic.008G111800). (B,C) GWAS results for the “Pericarp_pigmentation” based on SNPs alone (B) and SNPs + SVs (C). The red dotted lines in the Manhattan plots show the Bonferroni-corrected threshold of α = 0.05. The red numbers near the red dotted lines were the corresponding values of the Bonferroni-corrected threshold of α = 0.05 based on different data sets; no loci reached the corrected significance threshold. (D) A diagram for the TRA underlying Y1. The corresponding signal underlying Y1 was a TRA with a ∼10.9-kb span including a coding gene (Sobic.004G350800) located on Chromosome 4 in the reference genome.
To further confirm the enhanced detectable heritability conferred by SVs in GWAS, we surveyed the number of significant variations detected in GWAS based on each of the SV, SNP, and SNP + SV data sets for an additional 29 morphological and physiological traits (Supplemental Table S7; Brenton et al. 2016, 2020). We detected the largest number of GWAS associations using the combined SNP + SV data set, including 234 SV hits and 43 SNP hits. This was substantially larger than the number of signals (212 hits) detected in SV-alone GWAS. By far the fewest signals were detected in SNP-only GWAS (50 SNP hits). The number of significantly associated loci in SNP-only GWAS was by far the lowest for all traits except days to harvest. SV or SNP + SV found the largest number of significant association signals for all traits (Supplemental Table S10). SNP hits in SNP + SV GWAS were also observed in SNP-based GWAS for most traits (except for “Total_fresh_weight,” 3/5), with SNP-only GWAS finding more associated SNPs for several traits (likely as a result of an altered multiple-testing correction). Interestingly, however, SVs between SNP + SV and SV-based GWAS results had only 32.6% of loci in common (median across traits, 19.1%) (Fig. 7; Supplemental Table S10). This finding indicates that association analysis based on SNPs and SVs separately, as well as the integrated SNP + SV data set, can each yield distinct and potentially important associations.
Figure 7.
Number of significant genotype–phenotype associations detected in GWAS. Number of significant associations for 28 traits (there were 29 traits in total being analyzed, but there was no significant signal detected for “Brix_milk”) (Supplemental Table S10) detected in GWAS based on the SV (red columns, the first column per three column set), SNP (blue columns, the second column per three column set), and SNP + SV (the third stacked column per three column set, including both SVs [SV#, green] and SNPs [SNP#, orange]) data sets. (2014_ADF) Acid detergent fiber content in 2014, (2014_Days_to_harvest) days to harvest in 2014, (2014_Dry_Weight) dry weight of biomass in 2014, (2014_Lignin) lignin content in 2014, (2014_NDF) neutral detergent fiber in 2014, (2014_NFC) nonfibrous carbohydrates content in 2014, (2014_WSC) water-soluble carbohydrates content in 2014, (2015_ADF) acid detergent fiber content in 2015, (2015_Days_to_harvest) days to harvest in 2015, (2015_Dry_Weight) dry weight of biomass in 2015, (2015_Lignin) lignin content in 2015, (2015_NDF) neutral detergent fiber in 2015, (2015_NFC) nonfibrous carbohydrates content in 2015, (2015_WSC) water-soluble carbohydrates content in 2015, (ADF) average acid detergent fiber content of 2014 and 2015, (Anthesis_date) date of anthesis, (Brix_maturity) brix content in maturity stage, (Days_to_harvest) average days to harvest of 2014 and 2015, (Dry_tons_per_acre) dry tons per acre, (Dry_Weight) dry weight of biomass, (Lignin) lingnin content, (NDF) average neutral detergent fiber content of 2014 and 2015, (NFC) average nonfibrous carbohydrates content of 2014 and 2015, (Pericarp_pigmentation) pericarp pigmentation, (Photoperiod_sensitivity) photoperiod sensitivity, (Sorghum_type) sorghum variety type (sweet, grain, and cellulosic), (Stalk_height) stalk height, (Total_fresh_weight) total fresh weight, and (WSC) average water-soluble carbohydrates content of 2014 and 2015. Data labels on the top of each tripartite column set indicate the percentage of SVs (the value before the colon) and SNPs (the value behind the colon) detected in SNP + SV GWAS that were also detected in SV GWAS or SNP GWAS. (NA) There was no signal detected in SNP + SV GWAS. The number of identified signals in SNP GWAS was always the lowest compared with other data sets for all phenotypes. The detected SNP signals in SNP + SV GWAS mostly overlapped with the results of SNP GWAS. However, SVs detected in SV and SNP-SV GWAS were far from identical. This indicates that association analysis based on all three of the SNP, SV, and integrated SNP + SV data sets is necessary to dissect genetic mechanisms thoroughly.
Considering that sorghum variety type is associated with photoperiod sensitivity (Figs. 2B,C, 3), we further investigated the genetic mechanisms that may underlie their divergent evolution by using three GWAS analyses based on the SNP, SV, and SNP + SV data sets for six photoperiod-sensitivity-related traits, and 23 traits related to the differentiated sorghum variety types (Supplemental Table S7). There were 171 significantly trait-associated SVs detected in SV GWAS; 33 SNPs were detected in SNP GWAS; and 182 variants including 152 SVs and 30 SNPs were detected in GWAS based on SNP + SV data set, of which just 21 SVs were common with those from SV GWAS, whereas all significant SNPs found were in common with those detected using SNP-based GWAS (Supplemental Table S10). In total, 238 polymorphisms, containing 228 SVs and 10 SNPs, were identified as significantly associated with the sorghum differentiated variety type–related traits, whereas 97 variants, including 74 SVs and 23 SNPs, were found to be significantly associated with photoperiod-sensitivity traits. There were 65 polymorphisms, including 54 SVs and 11 SNPs, that were associated with at least two different traits. Among these variations, we identified a potentially pleiotropic SV associated with multiple traits, sv_529156_Chr09_59249767, a 1.3-kb DEL from 59,249,767 bp to 59,252,667 bp on Chromosome 9, located 11.3 kb upstream of a CCT domain–containing gene, Sobic.009G259100. Not only is this locus significantly associated with days to harvest (in both 2014 and 2015) and stalk height, but it is also linked with multiple variety type–related traits: “Dry_tons_per_acre,” “Dry_Weight,” and “Total_fresh_weight.”
Candidate genes within 20 kb of each breakpoint were then investigated for each significant polymorphism. We found 242 candidate genes, such as dof21, SNAC1, and TEOSINTE BRANCHED 1 (tb1), close to SVs associated with sorghum variety type–related features and 69 candidate genes, including likely orthologs of the Arabidopsis genes FL and FAR-RED ELONGATED HYPOCOTYL 3 (FHY3) adjacent to SVs associated with photoperiod-sensitivity traits (Supplemental Table S11). We noted that certain genes were annotated as potentially involved in agronomic variety traits but were also associated with the photoperiod-sensitivity traits, whereas some known photoperiod-related genes were adjacent to SVs associated with usage-related traits. This finding illustrates the relationship between the sorghum usage or variety type and photoperiod sensitivity; for example, modern grain or sweet sorghum varieties will be expected to flower at different latitudes and times than forage or biomass sorghum. Based on analysis of all traits, we selected 13 candidate loci that were correlated with both photoperiod sensitivity and sorghum variety usage type (Supplemental Table S12).
SVs have widespread impacts on gene expression
By modifying the sequence or location of cis-regulatory elements, splicing of a gene, copy number, or regulatory RNA molecules, SVs can readily alter the expression pattern of genes (Li et al. 2012; Alaei-Mahabadi et al. 2016; Chiang et al. 2017; Alonge et al. 2020). To explore the impact of SVs on gene expression, we performed RNA sequencing (RNA-seq) on four sorghum inbred lines included in the BAP: BTx623, which is a typical grain sorghum and also used as the standard reference genome in our study; RTx430, a grain sorghum inbred with a repeat-rich genome (Deschamps et al. 2018); and Tracy and Ramada, which are typical sweet sorghum lines. Gene expression profiles were generated for both leaf and stem, at three stages: preflowering, flowering, and milk. Because of the limitation of associating other types of SV with specific genes, only CNV-type variations (DEL, DUP, INS) were taken into consideration for this analysis. Hypergeometric testing was used for enrichment analysis of differentially expressed genes (DEGs) in SV-associated genes. The P-values were adjusted using the Bonferroni correction. More DEGs were associated with SVs than not. The percentage of SV-associated DEGs, as a percentage of all genes, was notably higher than that of the non-SV-associated DEGs across all tissues and developmental stages, and the DEGs were significantly enriched in the SV-associated genes (Fig. 8A; Supplemental Fig. S11A; Supplemental Table S13). SVs with higher predicted impact on the sorghum genome were associated with more DEGs than the SVs with lower predicted impact (Fig. 8B; Supplemental Fig. S11B); however, the percentage of the DEGs associated with the lower predicted impact SVs was still higher than the percentage of non-SV-associated DEGs, with an average of 9.31% versus 3.21% in leaves and 9.57% versus 4.96% in stems across different accessions and development stages (Supplemental Table S13). Some previously reported genes of phenotypic interest were found among the identified SV-associated DEG set, such as the Dry gene, which is an important gene controlling the stem pithy/juicy trait (Zhang et al. 2018); SUT5, which encodes a sucrose transporter (Cooper et al. 2019); Heading Date 1 (Liu et al. 2015); lipid-transfer protein 1 (Pelèse-Siebenbourg et al. 1994); gs, which is a glutamine synthetase gene that affects growth and development in sorghum (Urriola and Rathore 2015); and Ae1, which is associated with grain quality in sorghum (Figueiredo et al. 2010). These findings suggest that SVs are strongly associated with heritable differential gene expression across varieties, giving a plausible mechanism by which SVs may have a disproportionate impact on phenotypic variation.
Figure 8.
SVs have a widespread impact on gene expression. (A) SVs have an impact on gene expression in sorghum leaf across all developmental stages. The differentially expressed gene (DEG) analysis was performed by comparison of expression profiles in RTx430, Tracy, and Ramada with the expression profile in Tx623 (as control) in leaf tissue at three development stages. The blue and pink bars represent the percentages of SV-associated and non-SV-associated DEGs, respectively. The P-values on the top of SV-associated DEG bars, which were adjusted using Bonferroni correction, indicate the hypergeometric testing results for enrichment of DEGs in SV-associated genes. DEGs were significantly enriched in SV-associated genes, with SV-associated DEGs increased 1.1%∼4.3% compared with non-SV-associated DEGs in different sorghum lines. Only the results in leaf tissue were showed here. Similar results were also observed in stem tissue (see Supplemental Fig. S11A,B). (B) SV-associated DEG count changed according to different impact predictions. Different classes of variant effects were predicted by SnpEff (v5.0) (Cingolani et al. 2012). The vertical axis showed the SV-associated DEG count. The blue, pink, and green bars represent the DEG counts associated by high impact SVs (impact_HIGH), moderate impact SVs (impact_MODERATE), and low impact SVs (impact_LOW), respectively, in leaf tissue of different sorghum lines in three developmental stages (preflower, flower, and milk). The P-values show the significance levels between groups (see Methods). Differential DEG counts between “impact_HIGH” and “impact_MODERATE” were all statistically significant. The significant level of DEG counts between “impact_MODERATE” and “impact_LOW” varied depending on lines and stages. In general, higher impact SVs associated more DEGs.
Discussion
Recent studies have revealed an abundance of large-scale genomic variants in many plant species, but the effects of SV on global variation of quantitative traits are not yet established. Here we built an SV data set based on Illumina whole-genome data for 363 sorghum lines. The apparent discrepancy between detected SNP and SV distribution in the genome (Fig. 1A,B) may illustrate the different mechanisms of creation and mutation of SVs and SNPs. We examined in detail the representative 43 cellulosic and 33 sweet sorghum lines from the BAP. Structural genetic differences underlying the divergent evolution of the representative cellulosic and sweet sorghum lines helped us show the extent of the role played by SVs in sorghum variety type differentiation, and provide potential targets for sorghum breeding and engineering. GWAS based on whole-genome SV revealed novel genetic associations and new candidate genes for sorghum seed pericarp pigmentation, which were not detected in previous GWAS or our SNP-alone analysis. Strong and extensive correlations between SVs and sorghum phenotypes were observed in subsequent association analysis for 29 additional traits. For most of these traits, heritability was improved by the addition of SVs to the extensive set of SNPs, in some cases substantially so, and in many cases, associations were detected that were not seen in SNP data alone. RNA-seq analysis of four sorghum lines in two tissues and three developmental stages showed impacts of SVs on gene expression in the sorghum genome. These findings show that the SV data set we built is a powerful addition to GWAS analysis in sorghum, providing insights into key loci underlying sorghum adaptation and improvement, mechanisms of variation in gene expression, and improved methodologies to maximize discovery of causative genetic alleles.
Limitations in sensitivity and specificity are perhaps the main reason why SV analysis has not yet been more widely used in crop genetics. There are three strategies partly or completely applied to SV calling in current popular algorithms for short-read sequencing data sets: read-pair technologies, read-depth methods, and split-read approaches, all of which are based on aligning sequencing reads to a reference genome and detecting discordances underlying the SVs (Alkan et al. 2011). Depending on the type of variants or the features of the underlying sequence at the SV locus, each algorithm has different strengths and disadvantages in terms of SV detection. The weaknesses can be overcome to some extent by extracting the consensus of multiple algorithms based on different strategies in an ensemble approach, as applied here (Zarate et al. 2020). The common limitation of the short-read reference-based SV callers is that they are heavily biased against INSs relative to the reference, because inserted sequences do not appear in the reference genome (The 1000 Genomes Project Consortium 2010; Mills et al. 2011). Long-read sequencing technologies and assembly-based methods are therefore necessary to provide complete coverage of SVs, particularly insertional polymorphisms, and to fully understand the sequence underlying the different allelic forms of SV. We show here that the ensemble approach, although necessarily incomplete, is nonetheless a powerful addition to understanding causative genetic variation; pangenome construction using long-read technologies will further validate our results and help complete the SV data sets in the future.
As a comparator for the short-read based methods, we used whole-genome alignment of assemblies based on long-read technologies. The MUMmer system, as well as the genome sequence aligner NUCmer included within it, has been widely used for alignment at genome scale (Marçais et al. 2018). Many approaches for variant calling by assembly comparison use the MUMmer system for the genome-scale alignment step. In this study, we used MUM&Co (v3.7) (O'Donnell and Fischer 2020) to call the SVs from five genome assemblies against BTx623 to provide a ground truth in order to evaluate the SV calling approach we deployed. A substantial number of SVs were identified by the mate-pair-based fusion pipeline that did not have a clear match with any of the SVs called by MUM&Co. To cross-reference the accuracy and validity of MUM&Co, we compared the SVs data sets called by MUM&Co to those identified by Assemblytics (Nattestad and Schatz 2016), which is also derived from the MUMmer system. The interpretation of complex SVs posed challenges for these evolving whole-genome comparison methods. We found substantial discrepancies even between the SVs data sets called by MUM&Co and by Assemblytics for the five genomes we compared to the BTx623 reference. The SVs called by Assemblytics also heavily depend on the “unique sequence anchor” and “maximum variant size” parameters, whereas MUM&Co can produce very large artifactual SVs, again making the maximum size threshold a critical parameter. Even when they use the same widely accepted aligner, the inconsistency and parameter sensitivity of whole-genome comparison methods limit their utility, especially for larger variants. We therefore conclude that short-read methods remain a valid and cost-effective approach for SV detection, with no decisive disadvantages when using a single reference approach.
SV GWAS and heritability
Importantly, by adding SV data to GWAS analysis, we found additional significant association peaks. In other words, SNPs alone do not identify all the detectable LD blocks in association with the target traits. This violates the basic assumptions of GWAS (Lipka et al. 2015), because genome-wide SNP data should provide multiple polymorphisms within the range of LD for each causative locus, even if the causative locus is an SV not detected by SNP genotyping. However, recent studies have shown that SVs causing important trait variation in crops (e.g., soybean protein and oil content) (Fliege et al. 2022) are not always in strong LD with surrounding SNPs, because of transposon excision, illegitimate recombination, and other mechanisms independent of the Mendelian assumptions underlying LD calculations. Notably, by including SVs in our GWAS, we were not only able to identify more loci in significant association with traits but also substantially increased the measured narrow-sense heritability for some traits, in one case approaching the maximum value of one. Previous studies in other species have also shown the power of SVs to identify missing heritability (Jeffares et al. 2017; Alonge et al. 2020). The capacity of SVs to capture missing heritability could be attributed, at least in part, to their frequent direct impact on gene expression. Chiang et al. (2017) performed the eQTLs mapping using joint analysis of SVs, SNVs, and indels in humans and observed a notable abundance of SV-associated gene expression. Our findings confirm in sorghum that missing heritability may be at least partially owing to SVs that are not in strong LD with any local SNPs.
Potential for SV-driven breeding of sorghum
Sorghum is a good resource for bioenergy production, and production of lipids is of increasing interest to remedy the world-wide energy crisis (Sandesh and Ujwal 2021). To verify the possibility of sorghum as a feedstock for oil production by SV-driven breeding, we identified 331 orthologs characterized as involving oil synthesis in Arabidopsis (Supplemental Table S14). We predicted the potential functional effects of SVs on the oil-related genes. Ninety-six percent (323/331) and 99% (328/331) of the oil gene orthologs were associated with CNV-type SVs and rearrangement-type SVs, respectively (Supplemental Tables S15, S16). Almost half of the orthologs (48%, 159/331) are predicted to be highly impacted by CNV type SVs (Supplemental Fig. S12). We found also that DEGs are strongly associated with SVs, even in populations outside the BAP (Supplemental Figs. S13–S15; Supplemental Table S17; for details, see Supplemental Results) These results suggest that bioenergy traits, including oil traits, could be enhanced via breeding endeavors and that specific targeting of SVs via marker-assisted selection could allow modification of gene expression levels in many cases.
Altogether, our study highlights the complementary contribution of the underexplored SVs in heritability of important traits, reveals their widespread impacts on gene expression, and shows their crucial role in shaping population genetic diversity as well as trait determination. The findings in our study have significant implications for crop breeding and improvement, underscoring the indispensable role of SVs in future studies.
Methods
Resequencing data set and phenotypes
The Illumina short-read sequence data set and phenotypes of the sorghum lines used in this study were collected by the TERRA-REF project (https://terraref.org) (Brenton et al. 2016); 339 sorghum lines with population information were considered for population genetic analysis. Information for each line is included in Supplemental Table S1.
Plant tissue and sequencing
Leaves from the seedlings of sorghum were sampled in the greenhouse. At least 10 g of leaf tissue for each sorghum accession was sent to the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign. Raw HIFI sequence data in BAM format were generated by the PacBio Sequel IIe platform.
Variant calling
SNPs were called using the Sentieon (version 202010.01) (Kendig et al. 2019) DNA-seq pipeline. Ensemble variant calling using five independent tools based on different algorithms was used to call SVs. For details, see the Supplemental Methods and Code.
De novo assembly and comparison
BAM files were converted to FASTQ files by SAMtools (Li et al. 2009). Reads <1 kb were identified and filtered by SeqKit tools (Shen et al. 2016). Genome de novo assembly was performed by hifiasm (Cheng et al. 2021). Genome assembly quality was evaluated by quast (Gurevich et al. 2013) and BUSCO (Simão et al. 2015). MUM&Co (v3.7) (O'Donnell and Fischer 2020) was used to evaluate SVs based on assembly comparison.
The heatmap of SV detection frequency
The heatmap of SV detection frequency was built individually in 1-Mbp sliding windows for the representative 43 cellulosic and 33 sweet sorghum lines to identify regions with elevated genetic differentiation. To reduce the noise from the background, SVs that were present in both cellulosic sorghum lines and sweet sorghum lines were excluded individually. The P-values for the difference tests were adjusted using Bonferroni correction; significance hypervariable regions were defined as adjusted P-value < 0.01; and average SV difference between two groups was ≥20.
Heritability estimation
LDAK (v5.1) (Zhang et al. 2021) was used to estimate the trait heritability explained by the SNP and SV polymorphisms.
Population genetics analysis
SNPRelate (Zheng et al. 2012) was used for data handling and format conversion. SVs were converted to present–absent binary representation before conducting PCA. FST was calculated by using VCFtools (v0.1.16) (Danecek et al. 2011). PCA was performed using the R function prcomp() (R Core Team 2022). A minimum spanning tree was created using the R package Poppr (Kamvar et al. 2014). SNPhylo (Lee et al. 2014) was used to create maximum likelihood phylogenetic trees.
GWAS
GWAS was performed by GAPIT3 using the compressed mixed linear model (CMLM) model (Zhang et al. 2010; Wang and Zhang 2021). For details on SV association methods, see the Supplemental Methods.
Haplotype analyses
The R package geneHapR (Zhang et al. 2023a) was used to perform analyses.
RNA-seq analysis
Tissues samples for RNA were collected from plants grown in the field at the Energy Farm at the University of Illinois at Urbana-Champaign in 2018. RNA-seq data were analyzed using the DESeq2 package (Love et al. 2014), and plots were drawn by ggplot2 (Wickham 2016).
Data access
The raw sequencing data for the 363 TERRA-REF lines are available at Data Commons (https://datacommons.cyverse.org/browse/iplant/home/shared/terraref/genomics/raw_data/bap/resequencing). The raw gene expression data are available at JGI Genome Portal (https://genome.jgi.doe.gov/portal/SorbicEProfiling_31_FD/SorbicEProfiling_31_FD.info.html and https://genome.jgi.doe.gov/portal/SorbicEProfiling_30_FD/SorbicEProfiling_30_FD.info.html). The SV and SNP data sets used in this study are available as Supplemental Material.
Supplementary Material
Acknowledgments
We thank Dr. Amy Marshall-Colon for her assistance with the data storage and computation resources, Drs. Todd Mockler and Jeremy Schmutz for assistance with data access, and Drs. John Vogel and Peggy Lemaux for prepublication access to the RTx430 genome information. This work was funded by the DOE Center for Advanced Bioenergy and Bioproducts Innovation (U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under award no. DE-SC0018420). Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of the U.S. Department of Energy. The work (proposal: 10.46936/10.25585/60001277) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under contract no. DE-AC02-05CH11231.
Author contributions: Z.Z. helped design the study, performed the analysis, and wrote the manuscript. J.P.G.V. offered R script recommendations. B.Z. contributed to the protocols of RNA extraction and purification and quantification and samples collection. K.K.O.W. offered advice for genome de novo assembly. H.M.P. provided suggestions for Python scripts. S.P.M. contributed to sample collection and RNA sample submission to JGI. G.P.M. assisted with sorghum genetics and phenotype information and provided the phenotypic data sets of BAP. C.D., K.W.B., and N.S. produced and coordinated DNA and RNA sequence data. M.E.H. obtained funding, designed the study, assisted with the analysis, and edited the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.278396.123.
Competing interest statement
The authors declare no competing interests.
References
- The 1000 Genomes Project Consortium. 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov A, Urban AE, Snyder M, Gerstein M. 2011. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21: 974–984. 10.1101/gr.114876.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaei-Mahabadi B, Bhadury J, Karlsson JW, Nilsson JA, Larsson E. 2016. Global analysis of somatic structural genomic alterations and their impact on gene expression in diverse human cancers. Proc Natl Acad Sci 113: 13768–13773. 10.1073/pnas.1606220113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Coe BP, Eichler EE. 2011. Genome structural variation discovery and genotyping. Nat Rev Genet 12: 363–376. 10.1038/nrg2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allaby RG, Fuller DQ, Brown TA. 2008. The genetic expectations of a protracted model for the origins of domesticated crops. Proc Natl Acad Sci 105: 13982–13986. 10.1073/pnas.0803780105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, Wang X, Benoit M, Soyk S, Pereira L, Zhang L, Suresh H, Ramakrishnan S, Maumus F, Ciren D, et al. 2020. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182: 145–161.e23. 10.1016/j.cell.2020.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AE, Dougherty ML, Nelson BJ, Shah A, Dutcher SK, et al. 2019. Characterizing the major structural variant alleles of the human genome. Cell 176: 663–675.e19. 10.1016/j.cell.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew H, Peiris S, Chiluwal A, Kumar R, Tiwari M, Ostmeyer T, Bean S, Jagadish SVK. 2022. Stable sorghum grain quality QTL were identified using SC35 × RTx430 mapping population. Plant Genome 15: e20227. 10.1002/tpg2.20227 [DOI] [PubMed] [Google Scholar]
- Boatwright JL, Sapkota S, Jin H, Schnable JC, Brenton Z, Boyles R, Kresovich S. 2022. Sorghum association panel whole-genome sequencing establishes cornerstone resource for dissecting genomic diversity. Plant J 111: 888–904. 10.1111/tpj.15853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenton ZW, Cooper EA, Myers MT, Boyles RE, Shakoor N, Zielinski KJ, Rauh BL, Bridges WC, Morris GP, Kresovich S. 2016. A genomic resource for the development, improvement, and exploitation of sorghum for bioenergy. Genetics 204: 21–33. 10.1534/genetics.115.183947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenton ZW, Juengst BT, Cooper EA, Myers MT, Jordan KE, Dale SM, Glaubitz JC, Wang X, Boyles RE, Connolly EL, et al. 2020. Species-specific duplication event associated with elevated levels of nonstructural carbohydrates in Sorghum bicolor. G3 (Bethesda) 10: 1511–1520. 10.1534/g3.119.400921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32: 1220–1222. 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, Zhang H, Li H. 2021. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods 18: 170–175. 10.1038/s41592-020-01056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Scott AJ, Davis JR, Tsang EK, Li X, Kim Y, Hadzic T, Damani FN, Ganel L, Consortium GT, et al. 2017. The impact of structural variation on human gene expression. Nat Genet 49: 692–699. 10.1038/ng.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiluwal A, Perumal R, Poudel HP, Muleta K, Ostmeyer T, Fedenia L, Pokharel M, Bean SR, Sebela D, Bheemanahalli R, et al. 2022. Genetic control of source-sink relationships in grain sorghum. Planta 255: 40. 10.1007/s00425-022-03822-5 [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper EA, Brenton ZW, Flinn BS, Jenkins J, Shu S, Flowers D, Luo F, Wang Y, Xia P, Barry K, et al. 2019. A new reference genome for Sorghum bicolor reveals high levels of sequence similarity between sweet and grain genotypes: implications for the genetics of sugar metabolism. BMC Genomics 20: 420. 10.1186/s12864-019-5734-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps S, Zhang Y, Llaca V, Ye L, Sanyal A, King M, May G, Lin H. 2018. A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nat Commun 9: 4844. 10.1038/s41467-018-07271-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. 2006. Structural variation in the human genome. Nat Rev Genet 7: 85–97. 10.1038/nrg1767 [DOI] [PubMed] [Google Scholar]
- Figueiredo LF, Sine B, Chantereau J, Mestres C, Fliedel G, Rami JF, Glaszmann JC, Deu M, Courtois B. 2010. Variability of grain quality in sorghum: association with polymorphism in Sh2, Bt2, SssI, Ae1, Wx and O2. Theor Appl Genet 121: 1171–1185. 10.1007/s00122-010-1380-z [DOI] [PubMed] [Google Scholar]
- Fliege CE, Ward RA, Vogel P, Nguyen H, Quach T, Guo M, Viana JPG, Dos Santos LB, Specht JE, Clemente TE, et al. 2022. Fine mapping and cloning of the major seed protein quantitative trait loci on soybean chromosome 20. Plant J 110: 114–128. 10.1111/tpj.15658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME, et al. 2006. Copy number variation: new insights in genome diversity. Genome Res 16: 949–961. 10.1101/gr.3677206 [DOI] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Olatoye MO, Marla S, Morris GP. 2019. An integrated genotyping-by-sequencing polymorphism map for over 10,000 Sorghum genotypes. Plant Genome 12: 1–15. 10.3835/plantgenome2018.06.004 [DOI] [PubMed] [Google Scholar]
- Ibraheem F, Gaffoor I, Chopra S. 2010. Flavonoid phytoalexin-dependent resistance to anthracnose leaf blight requires a functional yellow seed1 in Sorghum bicolor. Genetics 184: 915–926. 10.1534/genetics.109.111831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, Balloux F, Dessimoz C, Bähler J, Sedlazeck FJ. 2017. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun 8: 14061. 10.1038/ncomms14061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamvar ZN, Tabima JF, Grünwald NJ. 2014. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2: e281. 10.7717/peerj.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendig KI, Baheti S, Bockol MA, Drucker TM, Hart SN, Heldenbrand JR, Hernaez M, Hudson ME, Kalmbach MT, Klee EW, et al. 2019. Sentieon DNASeq variant calling workflow demonstrates strong computational performance and accuracy. Front Genet 10: 736. 10.3389/fgene.2019.00736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L. 2008. The road to genome-wide association studies. Nat Rev Genet 9: 314–318. 10.1038/nrg2316 [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T-H, Guo H, Wang X, Kim C, Paterson AH. 2014. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15: 162. 10.1186/1471-2164-15-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Varala K, Moose SP, Hudson ME. 2012. The inheritance pattern of 24 nt siRNA clusters in Arabidopsis hybrids is influenced by proximity to transposable elements. PLoS One 7: e47043. 10.1371/journal.pone.0047043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xiang X, Huang Y, Zhou Y, An D, Dong J, Zhao C, Liu H, Li Y, Wang Q, et al. 2020. Long-read sequencing reveals genomic structural variations that underlie creation of quality protein maize. Nat Commun 11: 17. 10.1038/s41467-019-14023-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka AE, Kandianis CB, Hudson ME, Yu J, Drnevich J, Bradbury PJ, Gore MA. 2015. From association to prediction: statistical methods for the dissection and selection of complex traits in plants. Curr Opin Plant Biol 24: 110–118. 10.1016/j.pbi.2015.02.010 [DOI] [PubMed] [Google Scholar]
- Liu H, Liu H, Zhou L, Zhang Z, Zhang X, Wang M, Li H, Lin Z. 2015. Parallel domestication of the Heading Date 1 gene in cereals. Mol Biol Evol 32: 2726–2737. 10.1093/molbev/msv148 [DOI] [PubMed] [Google Scholar]
- Lobell DB, Burke MB, Tebaldi C, Mastrandrea MD, Falcon WP, Naylor RL. 2008. Prioritizing climate change adaptation needs for food security in 2030. Science 319: 607–610. 10.1126/science.1152339 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Dong L, Fang C, Liu S, Kong L, Cheng Q, Chen L, Su T, Nan H, Zhang D, et al. 2020. Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat Genet 52: 428–436. 10.1038/s41588-020-0604-7 [DOI] [PubMed] [Google Scholar]
- Mace ES, Tai S, Gilding EK, Li Y, Prentis PJ, Bian L, Campbell BC, Hu W, Innes DJ, Han X, et al. 2013. Whole-genome sequencing reveals untapped genetic potential in Africa's indigenous cereal crop sorghum. Nat Commun 4: 2320. 10.1038/ncomms3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol 14: e1005944. 10.1371/journal.pcbi.1005944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. 2011. Mapping copy number variation by population-scale genome sequencing. Nature 470: 59–65. 10.1038/nature09708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, Riera-Lizarazu O, Brown PJ, Acharya CB, Mitchell SE, et al. 2013a. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci 110: 453–458. 10.1073/pnas.1215985110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GP, Rhodes DH, Brenton Z, Ramu P, Thayil VM, Deshpande S, Hash CT, Acharya C, Mitchell SE, Buckler ES, et al. 2013b. Dissecting genome-wide association signals for loss-of-function phenotypes in sorghum flavonoid pigmentation traits. G3 (Bethesda) 3: 2085–2094. 10.1534/g3.113.008417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattestad M, Schatz MC. 2016. Assemblytics: a web analytics tool for the detection of variants from an assembly. Bioinformatics 32: 3021–3023. 10.1093/bioinformatics/btw369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell S, Fischer G. 2020. MUM&co: accurate detection of all SV types through whole-genome alignment. Bioinformatics 36: 3242–3243. 10.1093/bioinformatics/btaa115 [DOI] [PubMed] [Google Scholar]
- Pelèse-Siebenbourg F, Caelles C, Kader J-C, Delseny M, Puigdomènech P. 1994. A pair of genes coding for lipid-transfer proteins in Sorghum vulgare. Gene 148: 305–308. 10.1016/0378-1119(94)90703-X [DOI] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28: i333–i339. 10.1093/bioinformatics/bts378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2022. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rhodes DH, Hoffmann L Jr, Rooney WL, Ramu P, Morris GP, Kresovich S. 2014. Genome-wide association study of grain polyphenol concentrations in global sorghum [Sorghum bicolor (L.) Moench] germplasm. J Agric Food Chem 62: 10916–10927. 10.1021/jf503651t [DOI] [PubMed] [Google Scholar]
- Sandesh K, Ujwal P. 2021. Trends and perspectives of liquid biofuel: process and industrial viability. Energy Convers Manag X 10: 100075. 10.1016/j.ecmx.2020.100075 [DOI] [Google Scholar]
- Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, Hurles ME, Feuk L. 2007. Challenges and standards in integrating surveys of structural variation. Nat Genet 39(Suppl 7): S7–S15. 10.1038/ng2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Le S, Li Y, Hu F. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One 11: e0163962. 10.1371/journal.pone.0163962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Song R, Llaca V, Messing J. 2002. Mosaic organization of orthologous sequences in grass genomes. Genome Res 12: 1549–1555. 10.1101/gr.268302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Zhang T, Stelly DM, Chen ZJ. 2017. Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons. Genome Biol 18: 99. 10.1186/s13059-017-1229-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songsomboon K, Brenton Z, Heuser J, Kresovich S, Shakoor N, Mockler T, Cooper EA. 2021. Genomic patterns of structural variation among diverse genotypes of Sorghum bicolor and a potential role for deletions in local adaptation. G3 (Bethesda) 11: jkab154. 10.1093/g3journal/jkab154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urriola J, Rathore KS. 2015. Overexpression of a glutamine synthetase gene affects growth and development in sorghum. Transgenic Res 24: 397–407. 10.1007/s11248-014-9852-6 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang Z. 2021. GAPIT version 3: boosting power and accuracy for genomic association and prediction. Genomics Proteomics Bioinformatics 19: 629–640. 10.1016/j.gpb.2021.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. https://ggplot2.tidyverse.org. [Google Scholar]
- Yang N, Liu J, Gao Q, Gui S, Chen L, Yang L, Huang J, Deng T, Luo J, He L, et al. 2019. Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat Genet 51: 1052–1059. 10.1038/s41588-019-0427-6 [DOI] [PubMed] [Google Scholar]
- Zarate S, Carroll A, Mahmoud M, Krasheninina O, Jun G, Salerno WJ, Schatz MC, Boerwinkle E, Gibbs RA, Sedlazeck FJ. 2020. Parliament2: accurate structural variant calling at scale. GigaScience 9: giaa145. 10.1093/gigascience/giaa145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, et al. 2010. Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42: 355–360. 10.1038/ng.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LM, Leng CY, Luo H, Wu XY, Liu ZQ, Zhang YM, Zhang H, Xia Y, Shang L, Liu CM, et al. 2018. Sweet Sorghum originated through selection of dry, a plant-specific NAC transcription factor gene. Plant Cell 30: 2286–2307. 10.1105/tpc.18.00313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Privé F, Vilhjálmsson B, Speed D. 2021. Improved genetic prediction of complex traits from individual-level data or summary statistics. Nat Commun 12: 4192. 10.1038/s41467-021-24485-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Jia G, Diao X. 2023a. geneHapR: an R package for gene haplotypic statistics and visualization. BMC Bioinformatics 24: 199. 10.1186/s12859-023-05318-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hu Y, Wang Z, Lin X, Li Z, Ren Y, Zhao J. 2023b. The translocase of the inner mitochondrial membrane 22-2 (AtTIM22-2) is required for mitochondrial membrane functions during Arabidopsis seed development. J Exp Bot 74: 4427–4448. 10.1093/jxb/erad141 [DOI] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28: 3326–3328. 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.