Abstract
The Epstein-Barr virus (EBV)-encoded BARF0 open reading frame gene products are consistently expressed in EBV-positive Burkitt’s lymphoma (BL) cell lines, nasopharyngeal carcinoma cell lines, and lymphoblastoid cell lines (LCLs). Here we show that the BARF0 sequence includes an HLA A*0201-restricted cytotoxic T-lymphocyte (CTL) epitope. By using theoretically predicted HLA A2 binding motifs and peptide-loaded antigen presentation-deficient T2 cells, polyclonal BARF0-specific CD8+ CTLs were isolated from four different healthy EBV-seropositive donors but not from two seronegative donors. These CTL lines recognized the peptide epitope LLWAARPRL, which was found to be conserved in 33 of 34 virus strains originating from Caucasian, African, and Asian individuals. The BARF0-specific CTL lines could lyse EBV-negative BL cells stably transfected with the BARF0 gene but did not kill HLA A2-matched EBV-positive BL cells and LCLs in a standard 51Cr release assay. Reverse transcriptase PCR analysis demonstrated that these EBV-positive cell lines expressed significantly lower levels of BARF0 mRNA than transfected cells. This data indicated that the BARF0 epitope could be endogenously processed; however, antigen levels in the target cell were a limiting factor for the effective interaction between BARF0-expressing cells and CTLs. The limited expression of BARF0 antigen in EBV-infected BL cells and LCLs might contribute to the escape of immune recognition from virus-specific CTLs present in the host.
Epstein-Barr virus (EBV) is a human gammaherpesvirus which infects at least 90% of the world’s population. EBV is the etiological agent of infectious mononucleosis and is associated with a variety of lymphoid and epithelial cancers, including Burkitt’s lymphoma (BL), nasopharyngeal carcinoma (NPC), and Hodgkin’s disease. After primary infection, EBV establishes a lifelong persistent infection which is thought to be controlled primarily by EBV-specific cytotoxic T lymphocytes (CTLs) (reviewed in reference 27). The oncogenic potential of EBV is reflected in its ability to efficiently transform human B cells in vitro. The resulting latently infected lymphoblastoid cell lines (LCLs) express a restricted set of EBV genes, including those for six nuclear antigens (EBNA-1 to -6), three latent membrane proteins (LMP-1, LMP-2A, and LMP-2B) (reviewed in reference 15), and the novel gene products of the BARF0 open reading frame (ORF) (5). Three types of latency have been described for lymphoid cell lines and infected tissues (reviewed in reference 26). The pattern of latent EBV gene expression present in latently infected LCLs and some BL cell lines is termed type III and is characterized by the expression of the full array of latent proteins. In contrast, some BL cell lines as well as BL biopsies express only EBNA-1 and BARF0, a pattern which is called latency type I. Type II latency represents an intermediate phenotype. These patterns of latent expression are also found during primary B-lymphocyte infection in vivo and in EBV-associated lymphoproliferative disease, emphasizing that latent gene expression is likely to be important in the process of EBV-mediated cell transformation, virus persistence, and lymphomagenesis.
EBV is detected in the malignant epithelial cells of NPC, which are thought to be derived from clonal expansion of a single EBV-infected progenitor cell, thereby implying an etiological role for this virus in these carcinomas (25). It was the cDNA analysis of an NPC sample which subsequently led to the recent discovery of two C-terminally overlapping proteins encoded by the BARF0 ORF of EBV. BARF0 RNA and protein not only were expressed in EBV-infected B lymphocytes and epithelial cells, but, most importantly, they were also detected in protein extracts of NPC and BL biopsies and in a latency type I BL cell line (5, 6, 9, 10). Thus, the BARF0 proteins, EBNA-1, and the LMPs are the only viral proteins consistently expressed in NPC and BL. As EBNA-1 is not recognized by HLA class I-restricted EBV-specific CTLs (11, 19), the BARF0-encoded proteins have become prime candidates for CTL-mediated recognition and killing of NPC and BL tumors. There is an increasing body of evidence that latent EBV infection is controlled primarily by HLA class I-restricted memory CTL responses. These can be reactivated in vitro by stimulating lymphocytes from EBV-seropositive donors with HLA-matched target cells presenting HLA class I- and II-restricted epitopes on the cell surface (reviewed in reference 12). Recently, we published a novel protocol for the identification of HLA-restricted CTL epitopes which led to the discovery of epitopes in LMP-1 (13). The protocol takes advantage of the peptide transporter (TAP)-negative T2 cell line, which is characterized by low HLA class I cell surface expression and impaired endogenous peptide presentation (30). HLA expression on T2 cells can be stabilized by exogenously delivered synthetic peptides, and these peptide-coated T2 cells can then be used to specifically stimulate memory CTLs. Using this approach, we have isolated CTLs specific for the BARF0 antigen from four different healthy EBV-seropositive individuals, and these BARF0-specific CTLs could recognize endogenously processed BARF0 protein expressed in a transfected EBV-negative BL cell line.
MATERIALS AND METHODS
Cell lines.
The TAP-negative BxT hybrid cell line 1.74xCEM (referred to as T2) (30) was used for peptide stabilization assays. The EBV-positive BL cell lines Eli (29) and MutuI c59 (7) express latency type I EBV genes, and the BL cell line MutuIII c62 (7) expresses type III latency genes. BJAB cells originate from an EBV-negative B lymphoma (17). BL30 (18) and DG75 (2) are EBV-negative BL cell lines expressing a nonactivated, BL group I-like phenotype. LCLs were established by exogenous transformation of peripheral B cells (23). All cells were propagated biweekly in RPMI 1640 medium containing 2 mM glutamine, 60 μg of benzylpenicillin per ml, 100 μg of streptomycin per ml, and 10% fetal calf serum (growth medium) at 37°C in a 5% CO2 atmosphere.
Synthesis of peptides.
Peptides with unblocked C and N termini were synthesized by the Merrifield solid phase method (Chiron Mimotopes, Melbourne, Australia) on a 1-mg scale, dissolved in 50 μl of dimethyl sulfoxide followed by 600 μl of water, and finally diluted to 200 μg/ml in serum-free RPMI 1640 medium.
HLA stabilization assay.
To identify potential HLA A2 binding sites within BARF0, the computer program HLA Peptide Binding Predictions was employed as described elsewhere (http://bimas.dcrt.nih.gov/molbio/hla_bind) (24). Peptides containing these predicted motifs were then used in a standard HLA stabilization assay with T2 cells as described recently (3, 13). Briefly, T2 cells (5 × 105) were incubated with 100 μl of each of the peptides (50 to 100 μg/ml) for 14 to 16 h at 26°C, followed by incubation at 37°C for 2 to 3 h. HLA A2 expression on the peptide-coated T2 cells was then measured on a FACSscan (Becton Dickinson, San Jose, Calif.) with an HLA A2-specific monoclonal antibody (MAb) (MA2.1; American Type Culture Collection) and an anti-mouse immunoglobulin G fluorescein isothiocyanate-conjugated antibody (Silenus, Victoria, Australia).
Establishment of polyclonal BARF0-specific CTLs and PHA blasts.
Peripheral blood mononuclear cells (PBMC) from whole blood of HLA A*0201-positive human donors were separated on Ficoll-Paque, and polyclonal BARF0-specific CTL lines were generated from the PBMC as described recently (13). Briefly, 2 × 106 PBMC were cocultivated with 4 × 104 gamma-irradiated (8,000 rads) T2 cells sensitized with peptide in growth medium for 7 days. The cultures were then restimulated with peptide-sensitized, gamma-irradiated T2 cells once per week, fed twice per week with growth medium containing recombinant interleukin-2 (rIL-2) (10 U/ml), and used as polyclonal effectors in a standard 51Cr release assay after 17 days. For long-term cultivation (up to 7 weeks) in T-cell medium, rIL-2 (15 U/ml) and 30% MLA-144 cell supernatant (TIB-201; American Type Culture Collection) were added. T-cell cultures were routinely phenotyped with a FACSscan and murine MAbs (all from Becton Dickinson) detecting the antigens CD4 (clones SK3 and SK4; anti-human Leu-3a,b–fluorescein isothiocyanate), CD8 (clone SK2; anti-human Leu-2b–phycoerythrin), and CD3 (clone SK7; anti-human Leu-4–peridinin–chlorophyll–protein A).
Phytohemagglutinin (PHA) blasts were prepared by stimulating 5 × 105 PBMC with PHA (20 μg/ml) (CSL, Melbourne, Australia) in 2 ml of growth medium. After 3 days, growth medium containing MLA-144 supernatant (30%) and rIL-2 (15 U/ml) was added. The PHA blasts were maintained for up to 6 weeks with biweekly replacement of rIL-2 and MLA supernatant without the PHA.
Cytotoxicity assay.
CTL lines were tested in duplicate for cytotoxicity in a standard 51Cr release assay (23). Briefly, 106 target cells were incubated with 50 μl of 51Cr (1 mCi/ml of sodium chromate [250 to 500 mCi/mg of Cr]; Amersham, Sydney, Australia) in a total of 200 μl and, if applicable, with synthetic peptide (40 to 60 μg/ml) for 90 min at 37°C. Cells were washed twice in growth medium, thereby removing excess unbound peptide, and resuspended at 105 per ml. Alternatively, in the peptide titration experiment, peptides were added directly to the resuspended 51Cr-labelled targets and incubated for 1 h at 37°C, and they remained present throughout the assay. Target cells (100 μl) were combined with effector cells (100 μl) at different effector/target (E/T) ratios in 96-well round-bottomed microtiter plates and, after centrifugation, incubated for 5 h at 37°C. After a further centrifugation, 30 μl of supernatant per well was harvested and dried onto 96-well solid scintillation microtiter plates before the radioactivity was counted in a Topcount Microplate scintillation counter (Packard Instrument Company, Meridan, Conn.). In some experiments, target cells were preincubated with an anti-HLA class I MAb (W6/32, ATCC HB-95; final dilution of 1/10 of the ascites fluid) for 1 h at 37°C before CTL addition.
Generation of BARF0 cell transfectants.
The BARF0 nucleotide sequence derived from an NPC tumor xenograft (C15) (5) was cloned into the end-filled NotI restriction site within the simian virus 40 transcriptional cassette of the expression vector EBO-pLPP. This vector contains the EBNA-1/oriP replicon of EBV for episomal replication and the hygromycin resistance gene for eucaryotic selection (20). Exponentially growing DG75 cells (5 × 106) were washed in growth medium and transfected in growth medium with 12 μg of DNA of BARF0 expression vector or EBO-pLPP vector by using a BioRad Gene Pulser (960 μF, 240 V, 0.4-cm-gap electrode, room temperature, 350-μl assay volume). The cells were then resuspended in 5 ml of growth medium and after 2 days were selected with 600 μg of hygromycin B (Boehringer Mannheim, Castle Hill, Australia) per ml. Polyclonal transfectants grew out after 2 to 3 weeks and were stably maintained in hygromycin B-containing growth medium.
RT-PCR analysis.
Total RNA was prepared from exponentially growing cells by using Total RNA Isolation Reagent (Advanced Biotechnologies, Surrey, United Kingdom). The RNA was then treated with DNase I, which we have recently shown to be important for the complete removal of episome-based DNA (16). Conditions for the reverse transcriptase PCR (RT-PCR) assay were recently outlined in detail (16). Briefly, the total RNA was reverse transcribed in the presence of oligo(dT) primers, deoxynucleoside triphosphates, and SuperScript-II enzyme according to the protocol of the manufacturer (GibcoBRL, Melbourne, Australia) in a 20-μl assay mixture. For RT-negative controls, the SuperScript enzyme was omitted. For amplification of a 227-bp cDNA of BARF0 (position 160586 to 160812 of the B95.8 sequence [1]), primers BARF-LLW5′ (5′-TGTCCAGCGCTCTGGTCG) and BARF-2 (5′-CCACGGCAACCCTTCCAC) were used. To semiquantify BARF0 expression, β2-microglobulin (β2-M) was also amplified with primers β2-M5′ (5′-CCCCCACTGAAAAAGATGAG) and β2-M3′ (5′-TCACTCAATCCAAATGCGGC), generating a 131-bp cDNA fragment. This control was used because the β2-M primer pair flanks a large intron and there are no pseudogenes of β2-M leading to false-positive signals or competion effects (21). PCR amplification (20-μl mixture) was performed in a 9600 GeneAmp PCR instrument (Perkin-Elmer, Norwalk, Conn.) with 2.5 U of Taq DNA polymerase (Promega, Madison, Wis.), 2 μl of RT sample, 200 μM deoxynucleoside triphosphates, 0.5 μM each primer, and 23 cycles for β2-M or 30 cycles for BARF0 amplification, with the following cycle protocol: 5 min of denaturation at 95°C; 23 or 30 cycles of 1 min at 95°C, 30 s at 60°C, and 1 min at 72°C; and 5 min of extension at 72°C. The amplified cDNAs were separated by electrophoresis on 2.5% agarose gels containing ethidium bromide in TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0). The gel was photographed under UV light with Polaroid T-55 film, and the relative amount of each DNA band was quantified by using a Computing Densitometer 300B system (Molecular Dynamics, Sunnyvale, Calif.).
Sequencing of the BARF0 epitopes derived from different EBV isolates.
Genomic DNA was prepared from LCLs and BL cell lines by a salt extraction method (22). NPC DNA was prepared from biopsies by using a QIAamp tissue kit (Qiagen, Clifton Hill, Australia). One microgram of DNA was subjected to 30 cycles of DNA PCR amplification with the BARF0 primers BARF-LLW5′ and BARF-2 and the cycle conditions outlined in “RT-PCR analysis” above. The BARF0 DNA fragment was separated on a 2.5% TAE-agarose gel and purified by using the QUIEX II gel extraction kit (Qiagen). Approximately 150 ng of each purified BARF0 fragment was sequenced by using primer BARF-LLW5′, the PRISM Reader DyeDeoxy terminator cycle sequencing kit, and an ABI 377 DNA sequencing system (Applied Biosystems, Foster City, Calif.).
RESULTS
Generation of HLA A2-restricted BARF0-specific polyclonal CTL lines.
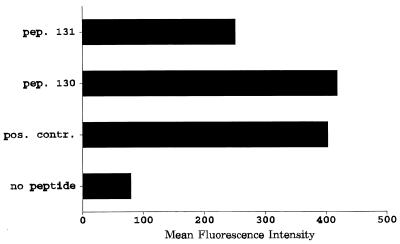
In order to identify potential HLA A2-restricted CTL epitopes within the BARF0 ORF, the 174-amino-acid (aa) sequence of BARF0 (nucleotide residues 160470 to 160994 of the B95.8 strain [1]) was analyzed for HLA A*0201 peptide binding motifs by the method described recently by members of our laboratory (13). The two sequences RLLLSLQQV and LLWAARPRL were identified as potential HLA A2 binding sites. To determine whether these sequences would bind to HLA A2, two overlapping 20-aa peptides (peptides 130 and 131) which corresponded to the BARF0 protein sequence and included the predicted sequences were synthesized and tested for HLA A2 binding efficiency by using T2 cells and an anti-HLA A2 MAb. Both peptides 130 (PPRARDRALLWAARPRLLLS) and 131 (WAARPRLLLSLQQVPEPRLA) (predicted sequences are underlined) significantly increased the expression of HLA A2 on T2 cells compared to untreated cells (Fig. 1). Peptide 130 stabilized HLA A2 expression to a similar strength as the control peptide YLLEMLWRL, which we have recently shown to be an HLA A2-restricted CTL epitope within LMP-1 (13).
FIG. 1.
HLA A2 stabilization analysis with T2 cells. T2 cells were either untreated (no peptide) or incubated with BARF0 peptide (pep.) 130 (PPRARDRALLWAARPRLLLS) or 131 (WAARPRLLLSLQQVPEPRLA). For a positive control (pos. contr.), peptide YLLEMLWRL from LMP-1 was used. HLA A2 expression on these cells was then analyzed by using a FACSscan and an HLA A2-specific MAb.
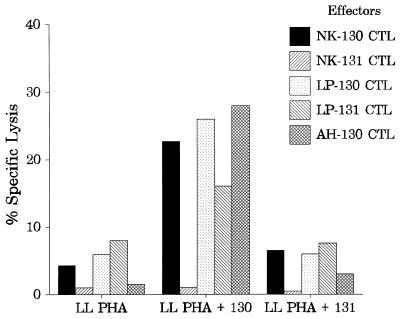
To determine whether those BARF0 peptide sequences included CTL epitopes, PBMC from HLA A*0201 EBV-seropositive, healthy donors were stimulated with T2 cells coated with peptide 130 or 131. After 17 days of culture, these polyclonal effector cells were used in a standard 51Cr release assay against peptide-sensitized PHA blasts from an HLA A2-matched donor (LL). Representative data from polyclonal CTL cultures derived from three different donors (NK, LP, and AH) are illustrated in Fig. 2. All of the cultures stimulated by T2 cells coated with peptide 130 lysed peptide 130-coated PHA blasts but not PHA blasts coated with peptide 131 or untreated targets. In contrast, CTL cultures stimulated with T2 cells coated with peptide 131 did not recognize peptide 131-coated PHA blasts. This data indicated that peptide 130 but not peptide 131 included a CTL epitope. Surprisingly, CTLs from donor LP stimulated with T2 cells coated with peptide 131 showed some recognition of peptide 130-loaded PHA blasts. This recognition may have been due to the partially overlapping regions of peptide 131 and 130 sharing parts of the LLWAARPRL epitope.
FIG. 2.
Specificities of polyclonal BARF0-specific CTL lines. CTL lines derived from PBMC from three seropositive donors (NK, LP, and AH) were tested after 17 days of culture in a standard 51Cr release assay against HLA A2-matched PHA blasts (from donor LL) which were either untreated or coated with BARF0 peptide 130 (PPRARDRALLWAARPRLLLS) or 131 (WAARPRLLLSLQQVPEPRLA). Data from one representative experiment of three is presented. An E/T ratio of approximately 10:1 was used in the assay. The effectors were designated according to their donor origin and sensitizing peptide, e.g., NK-130 CTL indicates BARF0-specific CTLs from donor NK stimulated by peptide 130.
In parallel, the potential of another HLA class I-restricted CTL epitope was investigated by using an HLA B*3501 binding peptide in the BARF0 sequence. Although the predicted peptide specifically increased HLA B35 stabilization on T2 cells, stimulation of PBMC from an HLA B35-positive donor with T2-coated peptide did not generate a significant CTL response (data not shown). This underlined the specificity of the HLA A2-restricted BARF0 CTL epitope.
Definition of the minimal BARF0 CTL epitope.
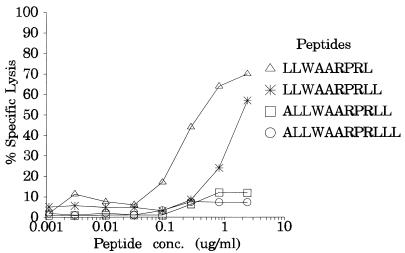
To determine the minimal length of the CTL epitope present within peptide 130, overlapping peptides (9 to 12 aa long), which included the LLWAARPRL sequence, were synthesized. These peptides were applied to PHA blasts and analyzed in a 51Cr release assay with an autologous polyclonal CTL line raised against peptide 130 from donor NK (referred to as NK-130). At standard peptide concentrations (>40 μg/ml), all of the BARF0 peptides containing the LLWAARPRL epitope rendered effective killing of the targets (data not shown). In contrast, at limiting peptide concentrations, both the 9-mer LLWA ARPRL and the 10-mer LLWAARPRLL were efficiently recognized by the NK-130 CTL line, while the 12-mer ALLWAARPRLLL and the 11-mer ALLWAARPRLL elicited very low levels of CTL-mediated cell lysis. However, the peptide titration showed that the 9-mer LLWAARPRL was the most active peptide (Fig. 3).
FIG. 3.
Titration analysis for the minimal BARF0 epitope. 51Cr-labelled HLA A2-positive PHA blasts from donor NK were coated with different concentrations of overlapping 9-, 10-, 11-, or 12-aa-long peptide (all of which had the LLWAARPRL sequence) and assayed for recognition by the autologous polyclonal NK-130 CTL line (E/T ratio, approximately 15:1). Representative results from one of two experiments are shown. Conc., Concentration.
EBV-seropositive, but not -seronegative, donors display a strong BARF0-specific CTL response.
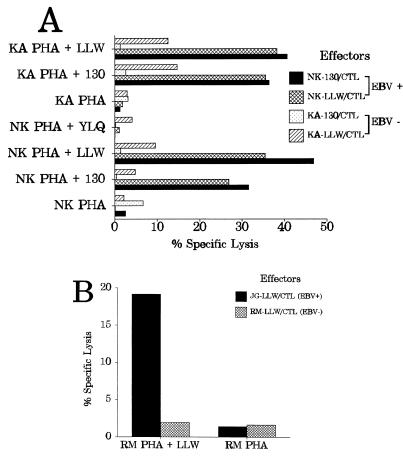
To find out whether a BARF0-specific CTL response was present in seronegative individuals, PBMC of an EBV-negative but HLA A*0201-positive donor (KA) were stimulated with T2 cells sensitized with BARF0 peptide 130 or LLWAARPRL. For positive controls, PBMC of the EBV-positive donor NK were processed in parallel. After 17 days of culture, these polyclonal effector cells raised against peptide 130 or LLWAARPRL (referred to as KA-130, KA-LLW, NK-130, or NK-LLW) were analyzed against peptide-coated PHA blasts from donor NK in a standard 51Cr release assay. As illustrated in Fig. 4A, the CTL lines NK-130 and NK-LLW, both activated from the EBV-positive donor NK, specifically lysed the autologous NK PHA blasts sensitized with peptide 130 or LLWAARPRL but not NK PHA blasts coated with an HLA A2-restricted control peptide from LMP-1 (YLQQNWWTL) (13) or untreated targets. In contrast, the two CTL cultures (KA-130 and KA-LLW) originating from the seronegative donor KA did not significantly recognize the HLA A2-matched NK PHA blasts coated with either the BARF0 or the LMP-1 peptides. To confirm that both donors indeed had the same HLA A2 subtype, the reciprocal experiment was performed with KA PHA blasts as targets (Fig. 4A). Both BARF0-specific CTL lines of the seropositive donor (NK-130 and NK-LLW) strongly reacted with the BARF0 peptide (130 and LLWAARPRL)-coated KA PHA blasts, in contrast to the case for the KA-130 and KA-LLW CTL cultures originating from the seronegative donor. Although the KA-LLW CTL culture showed some killing of the BARF0 peptide-coated targets, it was significantly (approximately threefold) lower than that of the NK CTL lines.
FIG. 4.
EBV-seropositive but not -seronegative individuals display a strong BARF0-specific CTL response. Polyclonal CTL lines (effectors) derived from PBMC from HLA A2, EBV-seropositive (EBV+) donors NK and JG or from seronegative (EBV−) donors KA and RM were tested in a standard 51Cr release assay against HLA A2 PHA blasts as targets. For effector designations, see the legend to Fig. 2. An E/T ratio of either 16:1 (for NK-130/CTL and KA-130/CTL) or 13:1 (for NK-LLW/CTL, KA-LLW/CTL, JG-LLW/CTL, and RM-LLW/CTL) was used. (A) The PHA blasts (from donors NK and KA) were either untreated or coated with BARF0 peptide 130 (PPRARDRALLWAARPRLLLS) or LLWAARPRL (LLW). For negative controls, the HLA A2-restricted peptide YLQQNWWTL (YLQ) from LMP-1 (13) was used. The effector cells were tested after 17 days of culture. (B) PHA blasts from donor RM were either untreated or coated with BARF0 peptide LLWAARPRL (LLW). The effector cells were tested after 18 days of culture.
To confirm these results, a second EBV-seronegative HLA A*0201 donor (RM) was tested in parallel with a fourth seropositive donor (JG). The PBMC of both donors were stimulated with T2 cells sensitized with BARF0 peptide LLWAARPRL and analyzed in a standard 51Cr release assay after 18 days of culture (Fig. 4B). Clearly, the JG-LLW CTL line (from the seropositive donor) specifically lysed the LLWAARPRL peptide-coated HLA-matched RM PHA blasts. In contrast, the RM-LLW CTL culture, originating from the seronegative donor, did not recognize the BARF0 peptide-coated autologous targets.
In addition, a limiting-dillution assay with PBMC from donor KA or NK and T2 cells sensitized with BARF0 peptide 130 or LLWAARPRL was set up and analyzed after 11 days of culture as described recently (13). With autologous PHA blasts either coated with the BARF0 peptide targets or not coated, no BARF0-specific CTLs could be detected in cultures raised from the seronegative donor KA, in contrast to significant CTL-induced lysis from the seropositive donor NK. The precursor frequency of these BARF0-specific CTL clones was calculated as 1:534,956 to 1:538,750 with a 95% confidence limit. Although this frequency was very low, it was significant and in a range similar to that observed with the minimal CTL epitope YLQQNWWTL of LMP-1 (13) when tested in parallel with peptide-coated T2 cells against PBMC of donor NK (data not shown).
To confirm the HLA A2 restriction of the epitope LLWAARPRL, an anti-class I antibody inhibition assay was performed with HLA A2-matched PHA blasts and the polyclonal CTL lines NK-130 and NK-LLW. Preincubation of LLWAARPRL-coated PHA blasts from donor LL with an anti-HLA class I MAb significantly reduced the specific lysis of these targets. Moreover, fluorescence-activated cell sorter analysis showed that both the NK-130 and NK-LLW CTL lines were primarily CD8 positive, with more than 75% of cells expressing both CD3 and CD8 and only 3 to 5% CD16- and CD56-positive LAK cells (data not shown). Taken together, this data identified the minimal BARF0 epitope as the 9-mer LLWAARPRL, which could reactivate an HLA A2-restricted CTL response in EBV-seropositive donors.
Characterization of the BARF0 epitope in EBV-infected BL cells, LCLs, and NPC biopsies.
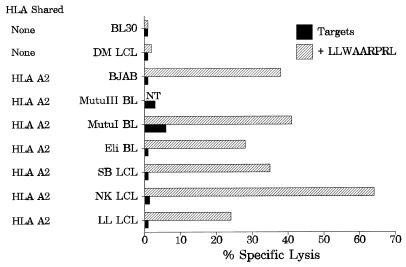
We and others have recently shown expression of the BARF0 ORF in a broad spectrum of EBV-infected cells (5, 6, 9, 10). This raised the question of whether EBV-positive cells could also endogenously process the LLWAARPRL epitope. A panel of HLA A2-matched BL cells expressing latency type I EBV genes (Eli and MutuI), a BL cell line of type III latency (MutuIII), and LCLs (SB, NK, and LL) were tested as targets in a standard 51Cr release assay with the BARF0-specific CTL line NK-130. Surprisingly, none of these targets appeared to be recognized and lysed (Fig. 5). This lack of CTL recognition of these targets was not due to a general impairment in their capacity to endogenously process antigen, since CTLs directed against another HLA A2-restricted epitope of EBV (from LMP-1) effectively lysed the BL and LCL targets (reference 13 and data not shown). Furthermore, exogenous addition of peptide LLWAARPRL to these target cells led to significant CTL-mediated lysis, whereas peptide-coated HLA A2-negative target cells (DM LCL and BL30) were not recognized (Fig. 5). Thus, the lack of CTL recognition of the EBV-positive cells was not due to impaired HLA A2 expression of the target cells.
FIG. 5.
EBV-positive BL cells and LCLs poorly present the BARF0 epitope. LCLs were generated by infection of B lymphocytes with EBV isolate B95.8 (LL LCL and SBLCL) or QIMR-Wil (NK LCL and DM LCL). The BL BL30 and the B-cell lymphoma BJAB are EBV-negative cell lines, whereas MutuI BL and Eli BL are EBV-positive BLs expressing viral latency I genes. MutuIII is an EBV-positive BL demonstrating a latency type III pattern. All of these cells were used as targets which were either untreated or incubated with peptide LLWAARPRL and analyzed in a standard 51Cr release assay against the polyclonal CTL line NK-130 (E/T ration, 10:1). Data from one representative experiment of three is shown. The HLA match between targets and effectors is indicated. NT, not tested.
Another possible explanation for the lack of CTL recognition of these EBV-positive cell lines could relate to strain variations in the BARF0 epitope sequence expressed in these target cells. Therefore, the DNA region of BARF0 encoding the LLWAARPRL peptide was sequenced, but no sequence variation in this region was detected in these target cells. Moreover, a sequence analysis of a panel of 34 different virus isolates from three separate geographical areas showed a nearly perfect (except for one sample) conservation of the LLWAARPRL epitope (Table 1).
TABLE 1.
Sequence analysis of the HLA A2-restricted BARF0 CTL epitopes present in EBV isolates from different geographical regions
| Reference strain or no. of isolates | Epitope sequencea | Virus origin | HLA A2 frequency (%)b |
|---|---|---|---|
| B95.8 | CTC CTC TGG GCC GCC CGG CCT CGC CTT | ||
| L L W A A R P R L | |||
| 13 | --- --- --- --- --- --- --- --- --- | Caucasianc | 46 |
| - - - - - - - - - | |||
| 1 | -G- --- --- --- --- --- --- --- --- | Caucasianc | |
| R - - - - - - - - | |||
| 16 | --- --- --- --- --- --- --- --- --- | Chinesed | 54 |
| - - - - - - - - - | |||
| 4 | --- --- --- --- --- --- --- --- --- | Africane | 30 |
| - - - - - - - - - |
The DNA and amino acid (boldface) sequences are compared to those of reference strain B95.8 (residues 160644 to 160670) (1). −, identity.
From reference 4.
IARC290B and EBV-QIMR strains derived from LCLs of healthy Australian individuals.
NPC isolates from Hong Kong.
BL isolates IARC/BL36, Eli, Mutu, and Ag876.
BARF0-transfected BL cells were killed by CTLs.
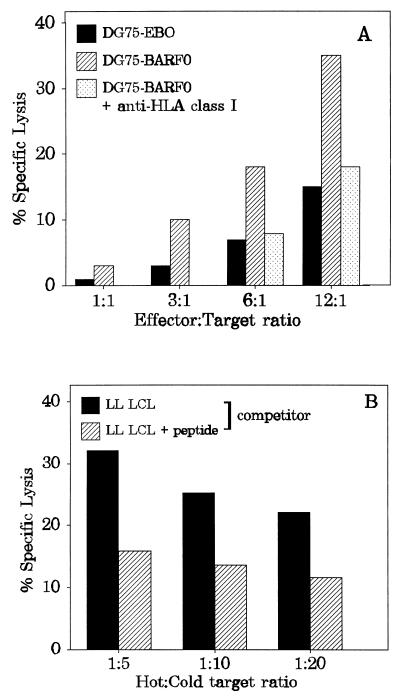
The level of expression of the LLWAARPRL determinant in BL cells and LCLs provides an alternative explanation for the lack of recognition by CTLs. In order to test this hypothesis, a BL cell line was stably transfected with a vector overexpressing the BARF0 ORF. The BARF0 nucleotide sequence, derived from an NPC tumor xenograft, was cloned into the expression vector EBO-pLPP. The BARF0 expression vector or EBO-pLPP alone was introduced into the EBV-negative, HLA A2-positive BL cell line DG75 by electroporation, and stable cell transfectants were selected. With the NK-130 CTL line as effector cells at different E/T ratios in a standard 51Cr release assay, the BARF0-expressing cells were recognized and specifically killed (Fig. 6A). Preincubation of the BARF0-expressing targets with an antibody specific for the nonpolymorphic determinants of HLA class I significantly reduced the specific lysis. Moreover, the CTL lysis of BARF0-expressing DG75 cells was diminished in a cold target inhibition assay by peptide 130-coated, unlabelled HLA A2-positive LCLs (from donor LL) (Fig. 6B). With different ratios of competitors (cold) to BARF0-expressing targets (hot), LL LCLs coated with peptide 130, but not untreated LL LCLs, reduced the specific lysis of the BARF0-expressing targets to control levels. These results indicated that the LLWAARPRL epitope could be endogenously processed and efficiently presented in transfected cells overexpressing the viral BARF0 gene.
FIG. 6.
BARF0-transfected BL cells present the LLWAARPRL epitope. (A) The BL cell line DG75 (EBV negative, HLA A2 positive) was stably transfected with either a control vector (DG75-EBO) or a BARF0 expression vector (DG75-BARF0). These targets were analyzed in a standard 51Cr release assay with the CTL line NK-130 at different E/T ratios. DG75-BARF0 cells were additionally preincubated with an anti-HLA class I-specific antibody before the addition of effector cells. (B) 51Cr-labelled DG75-BARF0 cells were subjected to a cold target inhibition assay with either untreated or peptide 130-coated LL LCLs (competitor) at different hot/cold target ratios. CTL lysis was induced by the effector NK-130 CTL bulk culture at an E/T ratio of 10:1.
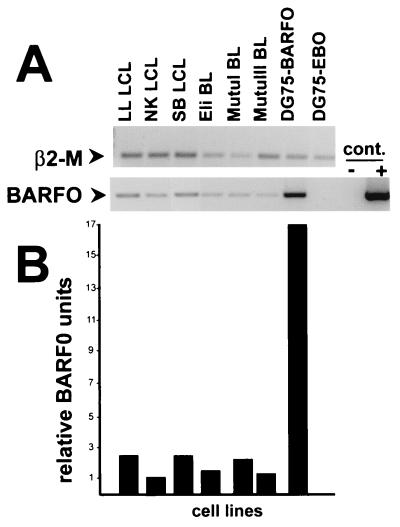
The level of antigen density on target cells is a critical parameter for CTL lysis measured in a standard 51Cr release assay. The significant lysis of the BARF0-transfected DG75 cells compared to the undetectable lysis of EBV-positive cells suggested that differences in the levels of BARF0 expression in these targets may explain the poor CTL recognition of LCLs and BLs. Therefore, the BARF0 expression level was analyzed by using RT-PCR. A protocol for reproducible amplification of the cDNA fragments of BARF0 encoding the LLWAARPRL epitope region and of the constitutively expressed β2-M was established with nonsaturated PCR cycle numbers. As demonstrated in Fig. 7A, no BARF0 cDNA could be detected in the control vector transfectants or in the RT-negative control samples. In contrast, the RT-PCR samples showed BARF0 cDNA which was similar in size to the PCR product amplified from BARF0 DNA. However, EBV-positive BL cells as well as LCLs demonstrated lower levels of BARF0 cDNA than the BARF0 transfectants. Quantitation of the BARF0 signal (normalized to β2-M) demonstrated that there was 7- to 17-fold more BARF0 cDNA present in the DG75-BARF0 cells than in the EBV-positive BL cells or LCLs (Fig. 7B). In summary, these results suggested that the level of antigen expression was critical for the efficient presentation of the LLWAARPRL epitope.
FIG. 7.
RT-PCR of BARF0 expression. (A) Total RNA was reverse transcribed from three EBV-positive LCLs (LL, NK, and SB), two BL cell lines of type I viral latency (Eli and MutuI), a BL cell line of type III viral latency (MutuIII), the EBV-negative DG75 cells expressing the BARF0 gene (DG75-BARF0), or the vector control (DG75-EBO). For RT-negative controls, the RT enzyme was omitted (cont.−). Both a 131-bp fragment of β2-M and a 227-bp fragment of BARF0 were PCR amplified from the first-strand cDNAs and the BARF0 gene DNA (cont.+). These were separated and visualized by electrophoresis on an ethidium bromide-containing agarose gel. Data from one representative experiment of three is shown. (B) The RT-PCR fragments obtained from each of the cell lines were quantified, and the BARF0 signals were standardized against β2-M.
DISCUSSION
The latent persistent infection of EBV is controlled primarily by the HLA class I-restricted memory CTL response, and many EBV-associated neoplasms express viral antigens that can act as targets for CTL-mediated killing of these tumor cells. There is increasing evidence that most of the EBV-associated malignancies escape this potent virus-specific CTL response by restricting viral gene expression (reviewed in references 12 and 27). EBV-positive BL biopsies or NPCs express EBNA-1 and BARF0 or the combination of EBNA-1, LMP-1, LMP-2A, LMP-2B, and BARF0, respectively (5, 15). As EBNA-1 does not include HLA class I-restricted CTL epitopes (11, 19) and only a few HLA class I-restricted CTL epitopes are known to be included in LMP-1 (13), the novel BARF0 protein might be an important source of CTL epitopes for immunotherapeutic treatment of BL and NPC.
Indeed, this report presents, for the first time, evidence that the EBV-encoded BARF0 protein contains a CTL epitope and that EBV-positive individuals have an HLA A*0201-restricted CTL response to the 9-mer epitope LLWAARPRL. Starting from theoretically predicted HLA class I binding motifs, peptides were synthesized and exogenously loaded onto antigen presentation-deficient T2 cells. The BARF0 peptide-coated T2 cells strongly activated in vitro polyclonal CTL lines from four healthy EBV-seropositive donors but not those from two seronegative individuals. The BARF0-specific CTL lines recognized the endogenously presented LLWAARPRL epitope in transfected BL cells, indicating that this BARF0 CTL epitope is of biological relevance. The use of peptide-sensitized T2 cells is a very powerful approach to reactivate memory CTL responses; however, this method is also known to induce primary immune responses, as reported recently (32). Indeed, prolonged cultivation and repeated stimulation of the PBMC from the seronegative donor KA with peptide-sensitized T2 cells led to CTL cultures which killed BARF0 peptide-coated PHA blast targets (data not shown). So far, we do not know if this indicates the induction of a genuine primary immune response against the LLWAARPRL epitope or if this was due to a cross-reactivity in the in vitro cell cultures of the seronegative donor. However, the significant differences between the seropositive and seronegative donors in the time kinetics and recognition strengths of the stimulated BARF0 CTLs suggested that BARF0-specific CTLs were reactivated in EBV-positive donors from a memory response.
The HLA A2-restricted BARF0 epitope sequence was found to be conserved in 33 of 34 virus strains originating from Caucasian, African, and Asian individuals. Moreover, the HLA A2 allele has a high frequency in virtually all human populations (Table 1). This epitope therefore has the potential to be used in a CTL-based vaccine designed to control EBV-associated tumors such as Hodgkin’s disease and NPC. In contrast, this potential is limited in the case of BLs, as these tumors cannot endogenously process antigens due to downregulated HLA class I and TAP expression (28). In addition, BARF0-specific CTLs were unable to lyse EBV-positive BL cell lines, in contrast to peptide LLWAARPRL-loaded target cells and BARF0-transfected BL cells, all of which were killed. The encouraging observation that the BARF0-transfected cells could endogenously process the LLWAARPRL epitope implies that it was not an intrinsic BARF0 structure (as seen in the case of EBNA-1, in which antigen processing is prevented by internal repeats [19]) which inhibited HLA class I-restricted BARF0-specific CTL lysis of EBV-positive cells. Rather, this suggested that the amount of BARF0 protein present in the target cell might be important. Indeed, the RT-PCR data demonstrated lower levels of BARF0 mRNA expression in EBV-infected cells than in the BARF0 transfectants. Two recent papers reported the failure of HLA class I-restricted CTLs raised against EBNA antigens to lyse autologous EBV-infected cells (8, 31). Although these LCLs presented enough antigen to stimulate the outgrowth of CTL lines in vitro, both groups demonstrated that either superinfection with recombinant vaccinia virus encoding the appropriate target gene or incubation with the peptide epitope was necessary to detect lysis of the LCLs in a standard 51Cr release assay. As this assay is insensitive if fewer than 10 to 20% of the target cells express the antigen, the authors postulated that the amount of antigen presented on the EBV-positive cells may be heterogenous within the population of LCLs or BL cells. Since antigen density on the target cell seems to be a critical factor for the avidity of interaction between target cells and effector CTLs, it will be necessary to explore possible ways to increase BARF0 gene expression in EBV-positive BL cells and LCLs. It is tempting to speculate that the suboptimal expression of BARF0 antigen in these EBV-infected cells might contribute to the escape of immune recognition from virus-specific CTLs present in the host.
On the other hand, very high levels of BARF0 RNA and protein expression have been observed in NPC (5, 6, 9), suggesting that BARF0-specific CTLs might be of immunotheraputic value for the treatment of this malignancy. Moreover, NPC cells not only express TAP and HLA proteins, both of which are limiting factors in antigen presentation, but also display normal endogenous processing function and are efficiently recognized by virus-specific CTLs in vitro (14). It is important to mention here that our limiting-dilution assay data indicated that the CTL response to the BARF0 epitope constituted a minor component of the virus-specific CTL response in EBV-seropositive individuals. Nevertheless, the continuous presence of this CTL memory response implies constant exposure to endogenously processed BARF0 antigen. This could be occurring during the persistent stage of EBV infection, when the EBV latency proteins are expressed and presented to the immune system. Additionally, as the levels of BARF0 protein were shown to increase after induction of EBV viral replication (5), the low but continuous lytic cycle of EBV in healthy asymptomatic individuals could aid in the presentation of BARF0 antigen. If the high level of BARF0 expression in NPC cells renders them susceptible to lysis by BARF0-specific CTLs, it may be possible to amplify this component of the virus-specific CTL response by vaccination with the relevant peptide or to adoptively transfer BARF0-specific CTLs to control this common malignancy.
ACKNOWLEDGMENTS
We are grateful for the support of the members of the EBV unit at the Queensland Institute of Medical Research, particularly for technical and intellectual help provided by L. Morrison, S. R. Burrows, J. Whitson, and D. J. Moss, and we are indebted to the generosity of the blood donors.
This work was supported by grants from the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund (QCF), and the Mayne Bequest, University of Queensland, Australia. R.K. is supported by an R.D. Wright fellowship from NHMRC.
REFERENCES
- 1.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 2.Ben Bassat H, Goldblum N, Mitrani S, Goldblum T, Yoffey J M, Cohen M M, Bentwich Z, Ramot B, Klein E, Klein G. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt like” malignant lymphoma (line D.G.-75) Int J Cancer. 1977;19:27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- 3.Burrows J M, Khanna R, Sculley T B, Alpers M P, Moss D J, Burrows S R. Identification of a naturally occurring recombinant Epstein-Barr virus isolate from New Guinea that encodes both type 1 and type 2 nuclear antigen sequences. J Virol. 1996;70:4829–4833. doi: 10.1128/jvi.70.7.4829-4833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Central Data Analysis Committee. The data book of the 11th International Histocompatibility Workshop. Oxford, United Kingdom: Oxford University Press; 1991. pp. 807–814. [Google Scholar]
- 5.Fries K L, Sculley T B, Webster Cyriaque J, Rajadurai P, Sadler R H, Raab Traub N. Identification of a novel protein encoded by the BamHI A region of the Epstein-Barr virus. J Virol. 1997;71:2765–2771. doi: 10.1128/jvi.71.4.2765-2771.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilligan K J, Rajadurai P, Lin J C, Busson P, Abdel Hamid M, Prasad U, Tursz T, Raab Traub N. Expression of the Epstein-Barr virus BamHI A fragment in nasopharyngeal carcinoma: evidence for a viral protein expressed in vivo. J Virol. 1991;65:6252–6259. doi: 10.1128/jvi.65.11.6252-6259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregory C D, Rowe M, Rickinson A B. Different Epstein-Barr virus-B cell interactions in phenotypically distinct clones of a Burkitt’s lymphoma cell line. J Gen Virol. 1990;71:1481–1495. doi: 10.1099/0022-1317-71-7-1481. [DOI] [PubMed] [Google Scholar]
- 8.Hill A B, Lee S P, Haurum J S, Murray N, Yao Q Y, Rowe M, Signoret N, Rickinson A B, McMichael A J. Class I major histocompatibility complex-restricted cytotoxic T lymphocytes specific for Epstein-Barr virus (EBV)-transformed B lymphoblastoid cell lines against which they were raised. J Exp Med. 1995;181:2221–2228. doi: 10.1084/jem.181.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitt M M, Allday M J, Hara T, Karran L, Jones M D, Busson P, Tursz T, Ernberg I, Griffin B E. EBV gene expression in an NPC-related tumour. EMBO J. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karran L, Gao Y, Smith P R, Griffin B E. Expression of a family of complementary-strand transcripts in Epstein-Barr virus-infected cells. Proc Natl Acad Sci USA. 1992;89:8058–8062. doi: 10.1073/pnas.89.17.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khanna R, Burrows S R, Kurilla M G, Jacob C A, Misko I S, Sculley T B, Kieff E, Moss D J. Localization of Epstein-Barr virus cytotoxic T-cell epitopes using recombinant vaccinia: implications for vaccine development. J Exp Med. 1992;176:169–176. doi: 10.1084/jem.176.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khanna R, Burrows S R, Moss D J. Immune regulation in Epstein-Barr virus-associated diseases. Microbiol Rev. 1995;59:387–405. doi: 10.1128/mr.59.3.387-405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khanna R, Burrows S R, Nicholls J, Poulsen L M. Identification of cytotoxic T cell epitopes within Epstein-Barr virus (EBV) oncogene latent membrane protein 1 (LMP1): evidence for HLA A2 supertype-restricted immune recognition of EBV-infected cells by LMP1-specific cytotoxic T lymphocytes. Eur J Immunol. 1998;28:451–458. doi: 10.1002/(SICI)1521-4141(199802)28:02<451::AID-IMMU451>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 14.Khanna R, Busson P, Burrows S R, Raffoux C, Moss D J, Nicholls J M, Cooper L. Molecular characterization of antigen-processing function in nasopharyngeal carcinoma (NPC): evidence for efficient presentation of Epstein-Barr virus cytotoxic T-cell epitopes by NPC cells. Cancer Res. 1998;58:310–314. [PubMed] [Google Scholar]
- 15.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 16.Kienzle N, Young D, Zehntner S, Bushell G, Sculley T B. DNaseI treatment is a prerequisite for the amplification of cDNA from episomal-based genes. BioTechniques. 1996;20:612–616. doi: 10.2144/19962004612. [DOI] [PubMed] [Google Scholar]
- 17.Klein G, Lindahl T, Jondal M, Leibold W, Memzes J, Nilsson K, Sundstrom C. Continuous lymphoid cell lines with characteristics of B cells (bone-marrow-derived), lacking the Epstein-Barr virus genome and derived from three human lymphomas. Proc Natl Acad Sci USA. 1974;71:3283–3286. doi: 10.1073/pnas.71.8.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenoir G M, Vuillaume M, Bonnardel C. The use of lymphomatous and lymphoblastoid cell lines in the study of Burkitt’s lymphoma. IARC Sci Publ. 1985;60:309–318. [PubMed] [Google Scholar]
- 19.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald Mullen P M, Klein G, Kurilla M G, Masucci M G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 20.Margolskee R F, Kavathas P, Berg P. Epstein-Barr virus shuttle vector for stable episomal replication of cDNA expression libraries in human cells. Mol Cell Biol. 1988;8:2837–2847. doi: 10.1128/mcb.8.7.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthes T, Werner Favre C, Tang H, Zhang X, Kindler V, Zubler R H. Cytokine mRNA expression during an in vitro response of human B lymphocytes: kinetics of B cell tumor necrosis factor alpha, interleukin (IL)6, IL-10, and transforming growth factor beta 1 mRNAs. J Exp Med. 1993;178:521–528. doi: 10.1084/jem.178.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller S A, Dykes D D, Polesky H F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moss D J, Misko I S, Burrows S R, Burman K, McCarthy R, Sculley T B. Cytotoxic T-cell clones discriminate between A- and B-type Epstein-Barr virus transformants. Nature. 1988;331:719–721. doi: 10.1038/331719a0. [DOI] [PubMed] [Google Scholar]
- 24.Parker K C, Bednarek M A, Coligan J E. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 25.Raab Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 26.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 27.Rickinson A B, Moss D J. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu Rev Immunol. 1997;15:405–431. doi: 10.1146/annurev.immunol.15.1.405. [DOI] [PubMed] [Google Scholar]
- 28.Rowe M, Khanna R, Jacob C A, Argaet V, Kelly A, Powis S, Belich M, Croom Carter D, Lee S, Burrows S R, Moss D J, Rickinson A B. Restoration of endogenous antigen processing in Burkitt’s lymphoma cells by Epstein-Barr virus latent membrane protein-1: coordinate up-regulation of peptide transporters and HLA-class I antigen expression. Eur J Immunol. 1995;25:1374–1384. doi: 10.1002/eji.1830250536. [DOI] [PubMed] [Google Scholar]
- 29.Rowe M, Rooney C M, Rickinson A B, Lenoir G M, Rupani H, Moss D J, Stein H, Epstein M A. Distinctions between endemic and sporadic forms of Epstein-Barr virus-positive Burkitt’s lymphoma. Int J Cancer. 1985;35:435–441. doi: 10.1002/ijc.2910350404. [DOI] [PubMed] [Google Scholar]
- 30.Salter R D, Cresswell P. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y, Smith K D, Kurilla M G, Lutz C T. Cytotoxic CD8+ T cells recognize EBV antigen but poorly kill autologous EBV-infected B lymphoblasts: immunodominance is elicited by a peptide epitope that is presented at low levels in vitro. J Immunol. 1997;159:1844–1852. [PubMed] [Google Scholar]
- 32.van der Burg S H, Klein M R, van de Velde C J, Kast W M, Miedema F, Melief C J. Induction of a primary human cytotoxic T-lymphocyte response against a novel conserved epitope in a functional sequence of HIV-1 reverse transcriptase. AIDS. 1995;9:121–127. [PubMed] [Google Scholar]